Abstract
The uptake and stable maintenance of extracellular DNA, genetic transformation, is universally recognized as a major force in microbial evolution. We show here that extracellular DNA, both homospecific and heterospecific, can also serve as the sole source of carbon and energy supporting microbial growth. Mutants unable to consume DNA suffer a significant loss of fitness during stationary-phase competition. In Escherichia coli, the use of DNA as a nutrient depends on homologs of proteins involved in natural genetic competence and transformation in Haemophilus influenzae and Neisseria gonorrhoeae. Homologs of these E. coli genes are present in many members of the γ subclass of Proteobacteria, suggesting that the mechanisms for consumption of DNA may have been widely conserved during evolution.
Horizontal gene transfer in bacteria can occur between organisms of the same or different species via one of three mechanisms: conjugation, transduction, or transformation (7). The last mechanism relies on the cell being able to take up and stably maintain extracellular DNA. Many bacteria are “naturally competent” for genetic transformation and, at least under some environmental conditions, can take up and integrate extracellular DNA. The mechanisms of DNA uptake in several naturally competent gram-negative and gram-positive bacteria have been extensively studied and reviewed (8, 9, 12, 14, 27, 29, 31). The process of transformation in naturally competent bacteria involves several steps. First, double-stranded DNA (dsDNA) is bound to the surface of the cell and enters a compartment where it becomes resistant to exogenous nuclease. Next, one strand of the DNA enters the cytoplasm while the other strand is degraded (9). Some bacteria, such as Haemophilus influenzae and Neisseria gonorrhoeae, preferentially take up homospecific DNA. Specificity of DNA uptake in these organisms is determined by the presence of “uptake signal sequences,” which are overrepresented conserved sequences found throughout the genome (28). Finally, after a recombination event, the new DNA is integrated into the chromosome.
Natural competence and transformation have not been observed to occur in many bacterial species, including Escherichia coli. It has been proposed that natural competence, in addition to playing a role in genetic recombination, might serve to allow the use of extracellular DNA for a nutritional purpose (22, 24, 29, 30). That is, the uptake of DNA into the cell may have two non-mutually exclusive functions: to provide DNA for genetic transformation and to provide nutrients. While studying mechanisms of survival of E. coli during long-term starvation, we identified a transposon insertion mutant that demonstrated an inability to survive when competing with its wild-type parent during extended stationary-phase incubation. The mutated gene showed high homology with a putative competence gene in H. influenzae. Since for some naturally transformable bacteria, genetic competence is induced during starvation, and since no such natural induction of competence under standard laboratory conditions has yet to be described for E. coli, we chose to address whether the competitive disadvantage of this mutant was due to an inability of that strain to compete for a nutrient resource, namely, extracellular DNA.
MATERIALS AND METHODS
Transposon insertion mutagenesis and initial screen for stationary phase-specific competition-defective mutants.
All experiments were performed using strains derived from E. coli K-12 strain ZK126 (W3110 ΔlacU169 tna-2) (33). ZK1142 is a nalidixic acid-resistant (Nalr) derivative of ZK126. Transposon insertion mutagenesis of ZK126 using λNK1324 was performed as previously described (16), resulting in a pool of mutant cells carrying a mini-Tn10d-Camr insert conferring resistance to chloramphenicol. Using this transposon, a screen for “stationary-phase-specific competition-defective” mutants was performed: mutant candidates were identified after coculture at 37°C of individual transposon insertion mutant strains with wild-type ZK1142 (Nalr) cells in 200 μl of Luria-Bertani (LB) broth in 96-well microtiter plates. Both the Nalr allele (16) and the presence of the chloramphenicol resistance (Camr) marker (S. Finkel and R. Kolter, unpublished data) are neutral in the absence of drug selection. Transposon insertions which resulted in the loss of mutant cells, as determined by detecting Nalr cells and few or no Camr cells, after 5 days of competition were then rescreened in 5-ml batch cultures (see below). Mutant candidates were reconstructed by bacteriophage P1 transduction (19) and rescreened. The ZK126 hofQ::Tn10d-Camr mutant was kindly provided by L. Pratt and R. Kolter (unpublished data).
Batch culture competition assays.
E. coli wild-type (ZK1142 Nalr) and mutant (Camr) strains were each grown overnight in LB broth (reaching a density of ∼5 × 109 CFU/ml.) Cultures were then inoculated 1:1,000,000 (vol/vol) into fresh LB broth, either in coculture or alone. Cell titers were determined by serial dilution on LB agar plates supplemented with nalidixic acid (20 μg/ml) or chloramphenicol (30 μg/ml) as appropriate. The limit of detection of this titration method is <100 CFU/ml.
DNA sequencing of transposon insertion sites.
The DNA sequence of the region flanking the transposon insertion was obtained using an arbitrary PCR-based technique (4). The primers specific to the mini-Tn10d-Camr element were primer 1L (CTGCCTCCCAGAGCCTG) and primer OUT 1L (CAGGCTCTCCCCGTGGAGG).
Preparation of conditioned medium.
Filter-sterilized conditioned medium was prepared as follows. LB cultures (50 ml) were inoculated 1:1,000 (vol/vol) with cells from a fresh overnight culture of ZK126 and incubated for 5 days in 250-ml Erlenmeyer flasks at 37°C with vigorous aeration. After 5 days, cells were pelleted and the supernatant was removed and filtered through a 0.2-μm NYL filter unit (Nalgene). It was essential that filters be rinsed with at least 100 ml of sterile distilled water prior to use to ensure removal of trace contaminants on the filter which are metabolizable by E. coli (data not shown). Supernatants treated with DNase I were incubated at 37°C with 10 μg of DNase I (Sigma Chemical Co., St. Louis, Mo.) per ml for 20 min prior to inoculation. Cultures were then inoculated, and titers were determined after overnight incubation.
Preparation of minimal medium supplemented with purified DNA.
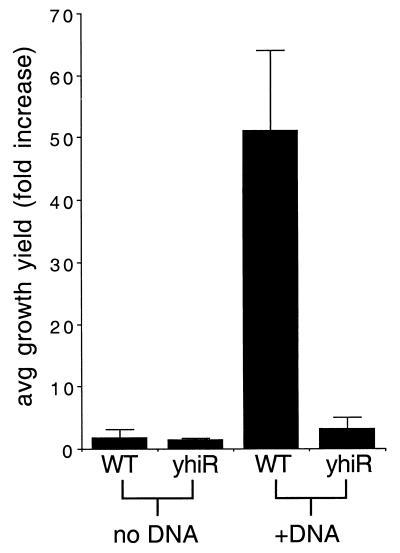
M63 minimal medium (1×) was prepared as described (19) and supplemented with 1 mM MgSO4 and 1 μg of vitamin B1 per ml. E. coli chromosomal DNA was prepared as described previously (2). Isolated DNA was sonicated to an average length of 300 to 500 bp and extensively extracted with phenol, phenol-chloroform, chloroform, and ethyl ether. DNA was then precipitated and reprecipitated with ethanol and resuspended in sterile distilled water immediately before use. It was essential to use freshly precipitated DNA, most likely because DNA stored for long periods of time contained easily metabolized nucleotides or other breakdown products from the dsDNA. For the experiments in Fig. 3, chromosomal DNA was added at a concentration of ∼6 μg/ml. Cultures were inoculated and titers were determined as described above.
FIG. 3.
Growth of E. coli utilizing DNA as the sole carbon source. Cells were grown in M63 minimal medium in the presence or absence of purified E. coli chromosomal DNA as the sole carbon source. Results are averages of three experiments. WT, wild type.
DNA sequence analysis.
DNA sequence similarity searches using the basic BLAST, tBLASTn (1), and Microbial Genomes BLAST algorithms and open reading frame searches using the ORF Finder program were performed at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/). Protein sequence alignments using the ClustalW 1.8, MAP, and PIMA algorithms were performed at the Baylor College of Medicine Molecular Biology website (http://www.hgsc.bcm.tmc.edu/SearchLauncher/).
RESULTS
Identification of the yhiR mutant.
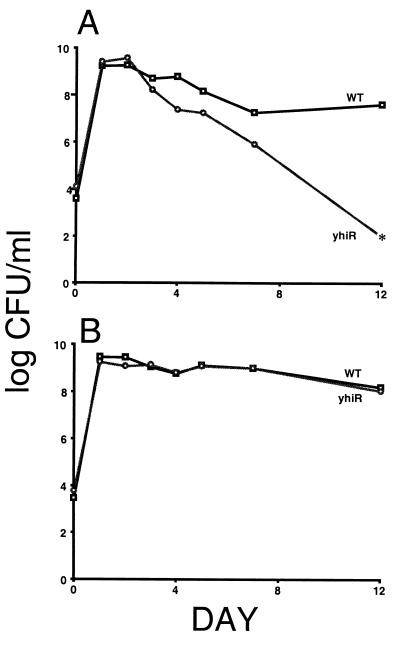
We performed a genetic screen for stationary-phase-specific competition-defective mutants; the analysis of one of those mutants is presented here. The transposon insertion mutant exhibits no fitness loss during the exponential phase of growth or upon entry into stationary phase when cocultured with the wild-type parent. However, after 2 days of coincubation in stationary phase, the mutant is outcompeted by wild-type cells and is completely lost from the culture after 10 to 12 days (Fig. 1A). Yet, when cultured separately, the mutant and the wild-type strains show identical patterns of exponential-phase growth and survival during long-term stationary phase incubation (Fig. 1B).
FIG. 1.
Survival patterns of yhiR and the wild-type (WT) parental strain in the presence or absence of competition. LB cultures were incubated for 12 days. (A) Cells grown in coculture; (B) cells grown separately. The asterisk indicates no detectable counts (limit of detection of <100 CFU/ml).
Analysis of the DNA sequence flanking the insertion mutation showed that the transposon inserted into yhiR, a gene of previously unassigned function, located at 78.5 min on the E. coli chromosome. Sequence comparisons identified this gene as a homolog of the comJ (orfJ) gene of H. influenzae; the predicted amino acid sequences of ComJ and YhiR are ∼66% identical and ∼85% similar. comJ, located at chromosomal nucleotide position 463327, was first identified as a gene adjacent to the H. influenzae competence locus, a cluster of genes that mediate natural competence of this organism (32). Though direct evidence that comJ plays a role in competence is lacking, a deletion mutation removing part of comJ and an adjacent gene causes a transformation-deficient phenotype in H. influenzae (5). Since the yhiR gene appears to be present in a single gene operon and transduction of the yhiR::Tn10d-Camr mutation into fresh wild-type strains confers the competition-defective phenotype, we can assume that the effect of the transposon insertion is due directly to the loss of YhiR activity.
Conditioned medium experiments indicate the use of DNA as a nutrient.
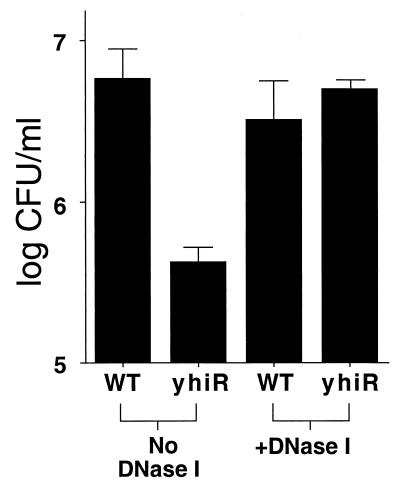
During long-term incubation in a rich medium (LB broth), viable cell counts of E. coli ZK126 reach a plateau of ∼5 × 109 CFU per ml at the end of exponential phase, drop to ∼5 × 107 CFU/ml after ∼3 days (the death phase), and level off at that density for many weeks (10, 33). We hypothesized that during the death phase, DNA might be released from dead cells into the medium and serve as a nutrient for the minority of cells that are still alive. (It has already been shown that amino acids are released for catabolic use, presumably from dead cells, during this time [34].) To determine whether DNA was available as a nutrient source and whether the yhiR mutant is defective in the utilization of released DNA, we prepared conditioned media by filter sterilization of 5-day-old cultures of wild-type E. coli. When inoculated at ∼5 × 103 CFU/ml, the wild-type cultures reach a density of ∼8 × 106 CFU/ml after overnight incubation (Fig. 2). In contrast, yhiR mutant cells reach a density of only ∼6 × 105 CFU/ml, less than 1/10 that of the wild type. Yet if the conditioned medium is pretreated with DNase I for 20 min and then inoculated with either wild-type or yhiR mutant cells, the growth yields are identical to that of the wild type growing without added DNase I. These results suggest that DNA is present in the conditioned medium and is metabolized by wild-type E. coli but not by the yhiR mutant.
FIG. 2.
Growth yields for overnight cultures of wild-type (WT) or yhiR cells incubated in conditioned medium. Culture medium was prepared from 5-day-old LB cultures without or with DNase I treatment prior to inoculation. Results are the averages of four experiments.
Use of defined minimal media supplemented with DNA.
While it is clear that prior to DNase I treatment the conditioned medium contains a nutrient that only the wild-type cells can utilize, we chose to directly address the question of whether this nutrient was DNA. Minimal medium which contained only inorganic salts, vitamin B1, and purified E. coli chromosomal DNA as the sole source of carbon and energy was prepared. This medium was inoculated with wild-type or yhiR mutant cells at ∼5 × 103 CFU/ml and incubated for 4 days. As shown in Fig. 3, the growth yields of wild-type cells are more than 50-fold greater than those of the yhiR mutant; wild-type cells reach final densities of 2 × 105 to 5 × 105 CFU/ml. This directly demonstrates that E. coli has the ability to take up and utilize exogenous DNA as a carbon source. Similar results were obtained using heterologous DNAs, including sonicated salmon sperm DNA or synthetic double-stranded oligonucleotides (data not shown). However, when minimal medium containing added DNA is first pretreated with DNase I, both wild-type and mutant cells grow similarly, reaching final cell densities of ∼2 × 109 CFU/ml (data not shown). No growth of either wild-type or mutant strains when cells were incubated in M63 minimal medium plus DNase I alone, with no added DNA or other source of carbon, was observed (data not shown). A similar result is observed when wild-type and yhiR mutant strains are grown in minimal medium with 100 mM concentrations of deoxynucleoside triphosphates as the sole carbon source; both strains grow to the same density (data not shown). Together, these results indicate that yhiR mutants cannot catabolize dsDNA but can consume DNA breakdown products. This defect in dsDNA consumption causes the yhiR mutant to have a competitive disadvantage during coculture with its wild-type parent.
Identification of other com gene homologs in E. coli.
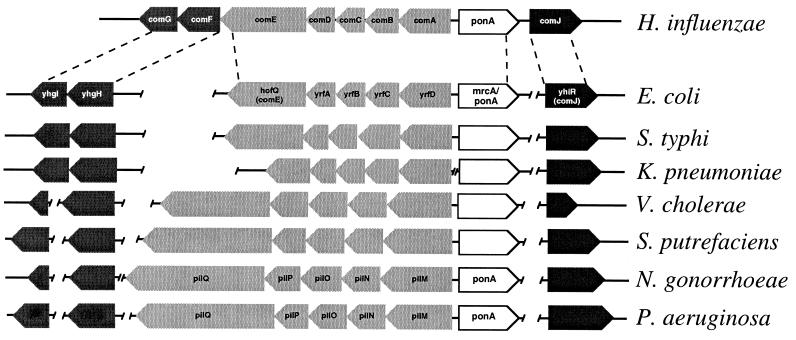
The experiments presented here demonstrate that E. coli can grow using DNA as a source of carbon and energy and that this ability depends on a homolog of a gene in the competence locus of another organism, H. influenzae. This prompted us to determine whether other H. influenzae competence gene homologs are present in the E. coli genome. In fact, E. coli has homologs to eight H. influenzae genes believed to be involved in competence and transformation (Table 1). The H. influenzae com (11) locus includes seven genes (comABCDEFG) in a putative operon plus comJ, which is transcribed divergently and separated from the cluster by the ponA gene, encoding penicillin-binding protein 1a (Fig. 4). The eight E. coli com gene homologs show various degrees of amino acid sequence similarity to the H. influenzae com cluster gene products, ranging from 12 to 74% identity (Table 1). In E. coli, these genes are found in three loci: five genes (yrfDCBA and hofQ) in an apparent operon located at 75.8 min, two genes (yhgHI) in a putative bicistronic operon at 76.4 min, and yhiR at 78.5 min transcribed monocistronically.
TABLE 1.
Degree of similarity and location of E. coli and H. influenzae competence genes
|
H. influenzae
|
E. coli
|
% Identity (similarity) | ||
|---|---|---|---|---|
| Gene | Position (nt) | Gene | Position (min) | |
| comA | 460543 | yrfD | 75.8 | 19.8 (33.2) |
| comB | 459748 | yrfC | 75.8 | 21.6 (59.2) |
| comC | 459245 | yrfB | 75.8 | 24.2 (49.7) |
| comD | 458727 | yrfA | 75.8 | 11.8 (44.2) |
| comE | 458304 | hofQ | 75.8 | 36.9 (68.9) |
| comF | 456958 | yhgH | 76.4 | 39.5 (69.5) |
| comG/orfG | 456193 | yhgI | 76.4 | 73.7 (91.9) |
| comJ/orfJ | 463327 | yhiR | 78.5 | 65.8 (85.1) |
FIG. 4.
Genetic maps of the H. influenzae com locus genes and their homologs. Dashed lines indicate the limits of gene clusters between H. influenzae and E. coli. Arrows indicate direction of transcription, and arrow length is proportional to gene length (with the exception of ponA/mrcA homologs, which are not to scale). Genes that are not contiguous are indicated by breaks between genes or clusters. Annotated sequence information from the completed genomes of H. influenzae and E. coli was obtained from The Institute for Genomic Research (www.tigr.org) and the University of Wisconsin (www.genetics.wisc.edu) websites, respectively. Except for the published pil locus sequences (6, 18, 25), unannotated sequence data were obtained as follows: for S. enterica serovar Typhi, www.sanger.ac.uk/projects/S.typhi; for V. cholerae and S. putrefaciens, www.tigr.org; for N. gonorrhoeae, dna1.chem.ou.edu/gono.html; for P. aeruginosa, www.pseudomonas.com; for K. pneumoniae, genome.wustl.edu/gsc/Projects/bacterial/klebsiella/klebsiella.shtml.
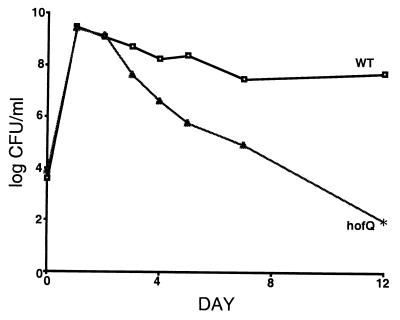
The identification in E. coli of homologs of the H. influenzae competence apparatus encouraged us to determine if another com gene homolog is essential for growth on DNA as the sole carbon source. An insertional mutant with a mutation in the E. coli hofQ gene, a homolog of H. influenzae comE, was tested for its ability to compete with its wild-type parent and to utilize DNA as a sole carbon source. hofQ is located in a different operon from yhiR, over 2.5 min away. Under all conditions tested it behaved identically to the yhiR mutant, showing a stationary-phase competition defect during coculture with the wild type (Fig. 5) and an inability to utilize extracellular DNA as the sole source of carbon or energy (data not shown). Importantly, both yhiR and hofQ mutants can be artificially induced to competence, by treatment with calcium chloride or by electroporation (3, 13, 15), as efficiently as the wild type (data not shown). This indicates that the mechanism of DNA uptake when DNA is used as a nutrient is distinct from the uptake mechanism used during induction of artificial competence.
FIG. 5.
Survival patterns of the hofQ mutant and the wild-type parental strain (WT) during competition in stationary phase. LB cultures were incubated for 12 days.
DISCUSSION
We have presented direct evidence supporting a model (9, 22, 24, 30) in which the DNA uptake function of a competence system can be used for nutrient acquisition rather than (or in addition to) obtaining and processing DNA for genetic transformation. While the potential evolutionary fitness advantages conferred by the acquisition of a beneficial gene by horizontal transfer are obvious (23), also as significant might be the potential advantage of taking up extracellular DNA solely for nutritional purposes, particularly when that DNA is heterologous and less likely to recombine onto the chromosome. Whereas there is a chance to lose an essential function or acquire a deleterious allele when taking in extracellular DNA as genetic material, using DNA solely as a nutrient source might pose little threat to the cell. Since so many organisms have developed mechanisms for natural competence and genetic transformation, it seems that over evolutionary time, the benefits of maintaining a system for horizontal genetic transfer outweigh the costs. However, having an additional, and not mutually exclusive, system for nutrient uptake could also provide great benefit. It has been noted that because in naturally transformable bacteria, such as B. subtilis, Streptococcus pneumoniae, H. influenzae, and N. gonorrhoeae, only a single strand enters the cytoplasm during transformation (with the other strand being degraded), this mechanism lacks efficiency as a nutrient acquisition system (9). That is, if the mechanism for DNA uptake, when the DNA is being used as a source of nutrients rather than genetic information, is the same for “noncompetent” bacteria as for naturally transformable species, then these bacteria would only be able to take up “half” of the DNA as food. While this may be more of an issue for gram-positive organisms, which do not have an outer membrane, we feel that this may be less of a problem for gram-negative organisms, particularly because current models of natural transformation for these bacteria suggest that DNA degradation of the single strand which does not enter the cytoplasm may take place in the periplasm (9), thus allowing the retention and possible transport of the resulting nucleotides into the cytoplasm for catabolic use. It has also been proposed that bacteria may produce an extracellular nuclease which digests dsDNA for catabolic purposes. However, we do not believe that our data support this model in E. coli, since we would expect the products of such a nuclease to help support the growth of not only wild-type strains but also the yhiR or hofQ mutants when grown in coculture with the wild-type.
It is interesting that the organization of these genes has been conserved in many organisms. For example, the relative position of the E. coli ponA homolog, mrcA, adjacent to the comABCDE homologs, is similar to that of H. influenzae ponA (Fig. 4). In fact, this organization, with the ponA homolog transcribed divergently from a five-gene cluster containing homologs of competence genes, is conserved in a wide variety of gram-negative bacteria, including Salmonella enterica serovar Typhi, Vibrio cholerae, Shewanella putrefaciens, N. gonorrhoeae, Klebsiella pneumoniae, and Pseudomonas aeruginosa (Fig. 4). What is particularly striking about these homologs is that the genetic organization of the cluster is highly conserved but levels of sequence conservation and gene sizes are quite variable. This suggests that these genes were present in a common ancestor and that this genetic organization has been conserved over evolutionary time, but the functions encoded by these genes may have diverged. As observed by A. Pugsley, homologs of comE are part of a superfamily with roles in cellular processes involving the movement of macromolecules across membranes (21), including pilus biogenesis, protein secretion, competence, and twitching motility; this prompted D. Dubnau, in a review of mechanisms of DNA uptake in bacteria, to term them PSTC proteins (9). For example, members of this family play roles in competence in H. influenzae and N. gonorrhoeae, type IV pilus biogenesis in N. gonorrhoeae and P. aeruginosa, and twitching motility in P. aeruginosa (6, 18, 21, 25). It is interesting that P. aeruginosa is not known to be naturally competent.
Several investigators have noted that the E. coli yrfDBCA/hofQ genes are homologs of genes involved in the production of type IV pili in pseudomonads (18, 25, 26). In an effort to address the lack of expression of type IV pili in K-12 E. coli, Sauvonnet and coworkers (26) recently measured mRNA levels expressed from the putative hofQ-containing operon (they refer to the five-gene cluster as hofMNOPQ). No significant expression was observed in cells grown in LB medium during exponential or early stationary phase, as determined by lacZ fusions or reverse transcription-PCR techniques. However, the fact that we observe a phenotype under conditions of competition in stationary-phase LB cultures and of outgrowth in minimal media supplemented with DNA suggests that these genes are, in fact, expressed. These differences may be due to several factors, including the time points during stationary phase when cells were harvested and the possibility that extracellular DNA may act as an inducer of yrfD/hofM operon gene expression.
Bacteria inhabit a wide variety of niches, and within many of these environments extracellular DNA may be available. Estimates of extracellular DNA concentrations in various marine and aquatic environments range from 0.2 to 44 μg/liter (reviewed in reference 17). DNA has been shown to be quite stable when complexed with various clays and soil minerals, binding at concentrations in the microgram to milligram range per gram of material (17). Extracellular DNA concentrations within the mammalian host, including the gastrointestinal tract and the mucosa in normal human lung, are estimated at hundreds of micrograms per milliliter, reaching as much as 4 mg/ml in the lungs of cystic fibrosis patients (20). With an abundance of DNA available in the environment and primarily of heterologous origin, it is unlikely that much of the DNA would be suitable for incorporation into the bacterial chromosome by homologous recombination, hence reducing its potential as a source of genetic diversity. Therefore, it is quite reasonable that E. coli and other organisms would take advantage of this rich nutrient source. Put simply, DNA is “good eating.” It remains to be determined if the system in place for the consumption of DNA arose first and then evolved into a system for genetic transformation, or vice versa. Also remaining to be demonstrated is the possibility that E. coli itself is capable of natural genetic competence and transformation and that we simply have not found the appropriate environmental conditions to observe such a phenomenon in the laboratory. It is possible that once the DNA is taken up, it can undergo two fates: recombination-replication, to yield stable transformation, or digestion, to be used as a nutrient. Since the genes used for this process, regardless of the final fate, appear to be homologs of competence genes, we propose to name the E. coli genes com genes after the designation of the H. influenzae genes. The ability of the com genes to mediate utilization of DNA as a nutrient appears to play a significant role in determining the relative fitness of organisms competing for very limited nutrient resources in natural environments.
ACKNOWLEDGMENTS
We thank members of the Kolter lab and Leah Macfadyen for helpful discussions; Erik Zinser, George O'Toole, Patrick Stragier, Michael Farrell, and Miriam Susskind for comments on the manuscript; and Vyacheslav Palchevskiy for technical assistance.
This work was supported by grants from the NIH and NSF to R.K., a postdoctoral fellowship from the Helen Hay Whitney Foundation to S.E.F., and a grant from the USC/Norris Comprehensive Cancer Center to S.E.F.
REFERENCES
- 1.Altschul S F, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F M, et al., editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987. [Google Scholar]
- 3.Baur B, Hanselmann K, Schlimme W, Jenni B. Genetic transformation in freshwater: Escherichia coli is able to develop natural competence. Appl Environ Microbiol. 1996;62:3673–3678. doi: 10.1128/aem.62.10.3673-3678.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caetano-Annoles G. Amplifying DNA with arbitrary oligonucleotide primers. PCR Methods Appl. 1993;3:85–92. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 5.Dougherty B A, Smith H O. Identification of Haemophilus influenzae Rd transformation genes using cassette mutagenesis. Microbiology. 1999;145:401–409. doi: 10.1099/13500872-145-2-401. [DOI] [PubMed] [Google Scholar]
- 6.Drake S L, Koomey M. The product of the pilQ gene is essential for the biogenesis of type IV pili in Neisseria gonorrhoeae. Mol Microbiol. 1995;18:975–986. doi: 10.1111/j.1365-2958.1995.18050975.x. [DOI] [PubMed] [Google Scholar]
- 7.Dreiseikelmann B. Translocation of DNA across bacterial membranes. Microbiol Rev. 1994;58:293–316. doi: 10.1128/mr.58.3.293-316.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubnau D. Genetic competence in Bacillus subtilis. Microbiol Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dubnau D. DNA uptake in bacteria. Annu Rev Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 10.Finkel S E, Zinser E, Kolter R. Long-term survival and evolution in stationary phase. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 231–238. [Google Scholar]
- 11.Fleischmann R D, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Goodgal S H. DNA uptake in Haemophilus transformation. Annu Rev Genet. 1982;16:169–192. doi: 10.1146/annurev.ge.16.120182.001125. [DOI] [PubMed] [Google Scholar]
- 13.Huang R, Reusch R. Genetic competence in Escherichia coli requires poly-beta-hydroxybutyrate/calcium polyphosphate membrane complexes and certain divalent cations. J Bacteriol. 1995;177:486–489. doi: 10.1128/jb.177.2.486-490.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kahn M E, Smith H O. Transformation in Haemophilus: a problem in membrane biology. J Membr Biol. 1984;81:89–103. doi: 10.1007/BF01868974. [DOI] [PubMed] [Google Scholar]
- 15.Kimoto H, Taketo A. Initial stage of DNA-electrotransfer into E. coli cells. J Biochem. 1997;122:237–242. doi: 10.1093/oxfordjournals.jbchem.a021734. [DOI] [PubMed] [Google Scholar]
- 16.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 17.Lorenz M G, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin R, Watson A A, McCaul T F, Mattick J S. Characterization of a five-gene cluster required for the biogenesis of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol. 1995;16:497–508. doi: 10.1111/j.1365-2958.1995.tb02414.x. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 20.Potter J, Spector S, Matthews L, Lemm J. Studies on pulmonary secretions. 3. The nucleic acids in whole pulmonary secretions from patients with cystic fibrosis, bronchiectasis, and laryngectomy. Am Rev Respir Dis. 1963;99:909–916. doi: 10.1164/arrd.1969.99.6.909. [DOI] [PubMed] [Google Scholar]
- 21.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Redfield R. Genes for breakfast: the have-your-cake-and-eat-it-too of bacterial transformation. J Hered. 1993;84:400–404. doi: 10.1093/oxfordjournals.jhered.a111361. [DOI] [PubMed] [Google Scholar]
- 23.Redfield R J. Evolution of bacterial transformation: is sex with dead cells ever better than no sex at all? Genetics. 1988;119:213–221. doi: 10.1093/genetics/119.1.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Redfield R J, Schrag M R, Dean A M. The evolution of bacterial transformation: sex with poor relations. Genetics. 1997;146:27–38. doi: 10.1093/genetics/146.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ropp P A, Nicholas R A P. Cloning and characterization of the ponA gene encoding penicillin-binding protein 1 from Neisseria gonorrhoeae and Neisseria meningitidis. J Bacteriol. 1997;179:2783–2787. doi: 10.1128/jb.179.8.2783-2787.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauvonnet N, Gounon P, Pugsley A P. PpdD type IV pilin of Escherichia coli K-12 can be assembled into pili in Pseudomonas aeruginosa. J Bacteriol. 2000;182:848–854. doi: 10.1128/jb.182.3.848-854.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith H O, Danner D B, Deich R A. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- 28.Smith H O, Tomb J-F, Dougherty B A, Fleischmann R D, Venter J C. Frequency and distribution of DNA uptake signal sequences in the Haemophilus influenzae Rd genome. Science. 1995;269:538–540. doi: 10.1126/science.7542802. [DOI] [PubMed] [Google Scholar]
- 29.Solomon J M, Grossman A D. Who's competent and when: regulation of natural genetic competence in bacteria. Trends Genet. 1996;12:150–155. doi: 10.1016/0168-9525(96)10014-7. [DOI] [PubMed] [Google Scholar]
- 30.Stewart G J, Carlson C A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- 31.Syvanen M, Kado C I. Horizontal gene transfer. London, United Kingdom: Chapman & Hall; 1998. [Google Scholar]
- 32.Tomb J-F, el-Hajj H, Smith H O. Nucleotide sequence of a cluster of genes involved in the transformation of Haemophilus influenzae Rd. Gene. 1991;104:1–10. doi: 10.1016/0378-1119(91)90457-m. [DOI] [PubMed] [Google Scholar]
- 33.Zambrano M M, Siegele D A, Almirón M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 34.Zinser E R, Kolter R. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J Bacteriol. 1999;181:5800–5807. doi: 10.1128/jb.181.18.5800-5807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]