Summary
The public-domain International Tree-Ring Data Bank (ITRDB) is an under-utilized dataset to improve existing estimates of global tree longevity. We used the longest continuous ring-width series of existing ITRDB collections as an index of maximum tree age for that species and site. Using a total of 3,689 collections, we obtained longevity estimates for 237 unique tree species, 157 conifers and 80 angiosperms, distributed all over the world. More than half of the species (167) were represented by no more than 10 collections, and a similar number of species (144) reached longevity greater than 300 years. Maximum tree ages exceeded 1,000 years for several species (22), all of them conifers, whereas angiosperm longevity peaked around 500 years. Given the current emphasis on identifying human-induced impacts on global systems, detailed analyses of ITRDB holdings provide one of the most reliable sources of information for tree longevity as an ecological trait.
Subject areas: Biological database, Plant Biology, Methodology in biological sciences
Graphical abstract

Highlights
-
•
The International Tree-Ring Data Bank (ITRDB) can improve tree longevity estimates
-
•
Maximum tree lifespans were obtained for 157 conifers and 80 angiosperms
-
•
22 conifers exceeded 1,000 years of age, whereas angiosperms peaked around 500 years
-
•
Based on the data, about 20 sites were enough to obtain stable longevity estimates
Biological database; Plant Biology; Methodology in biological sciences
Introduction
Tree longevity is an essential ecological trait for understanding forest vegetation dynamics,1 climatic impacts on woody species,2 and terrestrial carbon cycling.3 Although there is no research program specifically designed to investigate tree longevity, all research efforts aimed at predicting the fate of terrestrial ecosystems depend, more or less explicitly, on understanding and quantifying demographic patterns and traits, which include maximum tree lifespans. The emphasis currently being placed on modeling the future response of forest stands to climatic changes (especially atmospheric warming) and disturbance events (from droughts to extreme storms and wildfires) has prompted researchers to investigate resilience and resistance of woody species.4,5 For individual trees, mortality is complementary to longevity,6 hence baseline information on maximum tree lifespan provides an index whose variability in time and space can reveal environmental and human impacts on forest species.7
For our purposes, tree age is defined as stem (or trunk) age, which is the cumulative duration of secondary growth because pith formation at a specified height from the ground.8 Using this definition, tree longevity can be determined for wood-forming species, either clonal or non-clonal, by means of dendrochronological methods, radiocarbon dating, or a combination of both.9 Among existing big-data resources, the International Tree-Ring Data Bank (ITRDB10) has accumulated, over the past half century, thousands of tree-ring measurements from studies dealing with woody species in a number of scientific fields, from hydroclimatology to ecology, from archaeology to volcanology, etc.11 Dendrochronological work, especially when focused on reconstructing climate variability and ecological disturbance, has been traditionally focused on the oldest individuals of a species, but existing tree-ring data have rarely been analyzed in terms of potential maximum lifespans. For instance, Zhao et al.12 reviewed in detail and quantified ITRDB holdings in terms of species representation, spatial distribution, and potential improvements for macroecological research purposes, yet they did not address the issues connected with maximum tree lifespans.
Accessing and analyzing ITRDB data is not a simple task, even though some recent efforts have made them more readily accessible over the internet13. Our objective was therefore to investigate maximum tree lifespans using the information contained in the ITRDB, which is publicly available but not yet searchable for demographic traits. We present in this short communication the results of our analysis as a contribution to baseline data used for modeling and simulation purposes, and as a starting point for future in-depth analyses of tree eco-physiology and evolutionary ecology.
Materials and methods
Ring-width data were obtained from the public-domain ITRDB repository in mid-March 2022. To enhance replication, we did not introduce any additional information besides what was available on the ITRDB ftp server (ftp.ncdc.noaa.gov/pub/data/paleo/treering). The conventional four-letter code used in ITRDB collections, which are based on the first two letters of the scientific (Latin) genus and species names (binomial nomenclature), were compared with their original meanings14 as an initial check for potential coding errors.
The maximum length of all samples included in a collection was used to estimate tree longevity. To evaluate how reliable this index was, we compared it with a more refined estimate of longevity that was based on grouping ring-width series by tree code. This analysis was performed on a subset of the ITRDB collections, including a total of 519 sites from Canada, Africa, and from the Updates subdirectory, and that were diverse enough to be representative of the entire ITRDB dataset. The maximum number of tree rings in a continuous sample exactly matched the tree-based estimate in most cases, with differences only in 64 collections, and with only two of these differences being greater than 100 years (Figure S1). As expected, the index we used was a conservative measure of tree longevity, because it was always less than the value estimated by grouping measurements for individual trees (Figure S1).
Additional checks were performed on the species name to avoid duplicates, incorrect entries, and collections where only the genus was given. A final comparison was made between the maximum sample length of a collection and the difference between the overall first and last year, which is included in the standard metadata information for each collection. When this difference exceeded the maximum series length by more than 100 years, we analyzed the collection using the COFECHA software.15,16
Summary statistics were calculated for all species as well as by separating angiosperms (Magnoliophyta) from conifers (Pinophyta). For comparison with previous studies,2 the extra-tropics were not the regions outside ±23.5° latitude, but were instead all areas above 30°N or below 30°S. We also examined the relationship between maximum tree age and number of ITRDB collections. To quantify the minimum number of sites that should generate reliable estimates of tree longevity, a resampling analysis was applied to the most represented species in the database. Data processing was performed using the R numerical environment,17 the SAS software,18 and MATLAB.19
Results
Geographical and taxonomic distribution
Overall a total of 3689 out of 5444 collections could be analyzed, which is a considerably larger number than previous summaries of ITRDB holdings (e.g., 2624 in2). The excluded files were affected either by non-standard data organization, end-of-line and end-of-record issues that could not be resolved, or both. Longevity estimates were obtained for 237 unique tree species (Table S1), 157 conifers (3043 collections) and 80 angiosperms (646 collections). Many species (75) were represented by only one collection, more than half of the species (168) were represented by no more than 10 collections, and a handful of species (7) were represented by more than 100 collections, with Douglas fir (Pseudotsuga menziesii) being the species with the highest number (311). The most recent ITRDB collection that included a species’ maximum longevity was made in 2019, and the oldest one in 1978.
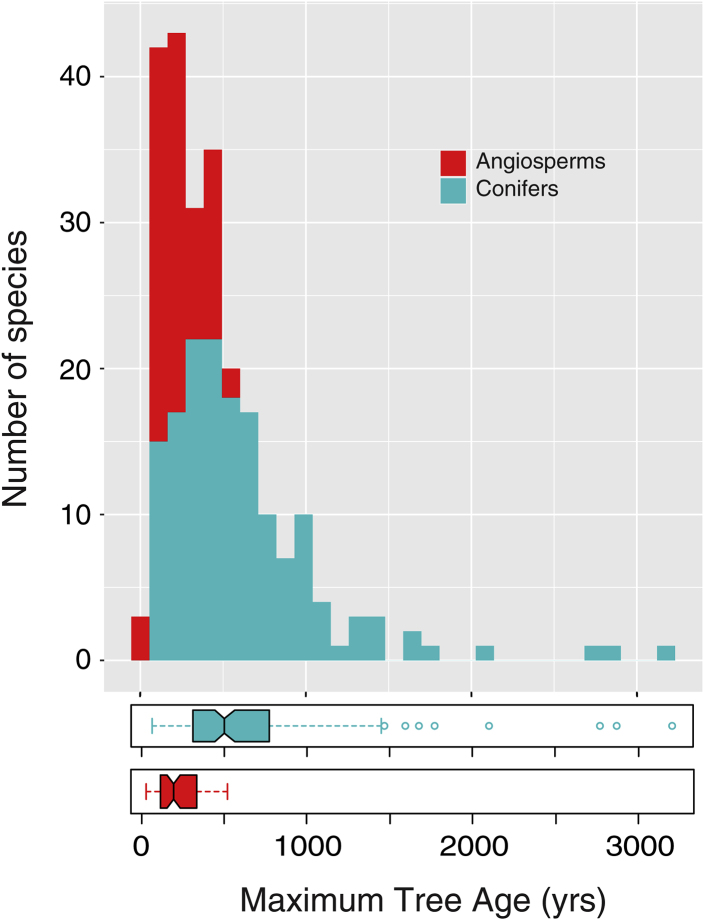
Collections that included maximum tree ages were distributed all over the world, but with greater density in the mid- and high-latitudes (Figure 1) because tropical trees often form indistinct growth layers, which are difficult to recognize and measure.20 Areas with latitude above 30° N or below 30° S included 194 species, of which 65 were angiosperms (9 in the southern hemisphere) and 129 were conifers (19 in the southern hemisphere). The majority of species (144) reached longevity greater than 300 years, and maximum tree ages exceeded 1000 years for several species (22), all of them conifers, whereas angiosperm longevity peaked around 500 years (Figure 2 and Table 1). This very large taxonomic difference in realized longevity is well known, albeit its causes are still being investigated.8,21,22 Based on stochastic modeling of the theoretical relationship between average mortality rate and age structure in old-growth forests, maximum tree ages of a few centuries in angiosperms and of a few millennia in conifers are consistent with differences in their average mortality rate.23
Figure 1.
Map of 237 ITRDB collections (solid dots) that provided the maximum estimated tree age by species (80 angiosperms: red; 157 conifers: green)
Sites cover most of the globe, from the Arctic (69.5°N) to the sub-Antarctic (54.9°S, Campbell Island), but with higher density in the extra-tropics (i.e., areas with latitude above 30°N or below 30°S), which included 194 species.
Figure 2.
Distribution of tree longevity estimates, showing differences between angiosperms (red bars and boxplot) and conifers (green bars and boxplot)
Table 1.
Summary of maximum tree ages estimated from ITRDB collections
| Taxa | Species (#) | Sites (#) | Min-Max (yrs) | Mean (yrs) | St.Err.Mean (yrs) | St.Dev. (yrs) | Median (yrs) | IQRa (yrs) |
|---|---|---|---|---|---|---|---|---|
| Angiosperms | 80 | 646 | 28–518 | 229 | 15 | 132 | 194 | 119–331 |
| Conifers | 157 | 3033 | 64–3205 | 616 | 40 | 495 | 501 | 310–766 |
IQR: Inter-Quartile Range (first-third quartile).
Tree longevity and species replication
Future changes in maximum tree lifespans should be relatively small for species already represented by several sites in the ITRDB. To clarify this issue, we investigated further how replication influences estimated tree longevity. The slope of a curve fit to the scatterplot of maximum tree ages against number of ITRDB collections was positive (Figure 3), also reflecting the dendrochronological preference for sampling sites with long-lived species. In fact, few collections (<50) were sufficient to identify conifer species with exceptional longevity. Overall, a larger percentage of angiosperms appeared capable of reaching the maximum lifespan of Magnoliophyta (a few centuries) compared to Pinophyta (Figure 3), because only a handful of species attained the conifer maximum lifespan (a few millennia; see Figure 2).
Figure 3.
Nonlinear fit [y = ln(x)] to the scatterplot of estimated tree longevity (Maximum tree age) against the number of collections for each species
The 80 angiosperm species were represented by 1–76 collections and the 157 conifers by 1–311 collections.
The most represented species in the ITRDB was Pseudotsuga menziesii (the rightmost green dot in Figure 3), with 311 collections whose maximum tree ages ranged from 51 to 1,001 years with a bell-shaped distribution (Figure 4A). Resampling of the 311 maximum tree ages 5,000 times without replacement for sample sizes j = 1, 2 , …, 310 indicated a rapid, nonlinear increase in estimated longevity with number of collections (Figure 4B). The median maximum age reached a plateau at about 20 collections, when it was already greater than 850 years, i.e. within 20% of the observed maximum (1,001 years). Additional collections improved the estimate marginally, so for example the median of 5,000 maximum tree ages for j = 63 collections was 903 years. Such an assessment is however species-specific, and depends on the distribution of maximum tree ages. If that distribution is more concentrated around the mean, fewer chronologies are needed for reaching a stable approximation of maximum lifespan.
Figure 4.
Maximum tree ages of Pseudotsuga menziesii collections included in the ITRDB
(A) Histogram showing a bell-shaped distribution of the 311 estimates ranging from 51 to 1001 years.
(B) Lines showing the median and 0.05 quantile of maximum tree ages for 5,000 random samples (taken without replacement) with size j = 1, 2, …, 310 collections.
Because only 10 angiosperms and 35 conifers were represented by more than 20 ITRDB collections, it is therefore possible that additional contributions will increase estimated tree longevity. Considering the very large number (3,689) of ITRDB collections we analyzed, and that our results included 20% more species for the extra-tropics than previously reported (1612), it is also plausible that most changes in tree longevity estimates derived from tree-ring data will be caused by adding new species to the ITRDB holdings. Yet, we note that our overall estimated mean longevity of trees in all extra-tropical biomes was 516 ± 34 years, which is considerably higher than Locosselli et al.2’s estimate of 322 ± 200 years. At the same time, the global average maximum tree lifespan we calculated (229 years for angiosperms and 616 years for conifers; Table 1) was lower than what Liu et al.24 have reported using a variety of sources (334 and 714 years).
Discussion
Dendrochronologists have usually targeted the oldest trees in a stand; thus, the ITRDB public-domain data are bound to offer better estimates of maximum tree age than those available from randomized plots, grid-based inventories, or the most complex, state-of-the-art simulation models. As an example, based on a global analysis of forest inventories and climate data, Besnard et al.25 defined as “old growth” any stand older than 300 years, which is an order of magnitude less than the maximum tree ages we uncovered for some conifer species. Although several large geographic areas were not included in Besnard et al.’s global analysis (“Africa, Indonesia and Australia were either underrepresented or not represented”), regions where data were relatively abundant, such as the US, could include unmanaged forests in remote areas that were not well represented. Even when old-growth forests, rather than longevity per se, are under consideration,26,27 under-estimating tree lifespans results in over-simplifying the long-term processes at work in forest ecosystems. Therefore, the ITRDB ring-width data, as shown in our relatively simple analysis, help clarifying the role of tree longevity as an ecological trait.
Previous analyses of ITRDB data have already pointed out its limited coverage of tropical areas.12 For a number of reasons, including wood anatomy peculiarities, lack of dating control, limited research funding, and required data formatting, ITRDB collections are scarce in the tropical regions of South America, Oceania, Asia, and Africa. Tree ages in these regions usually need to be estimated by means of radiocarbon dating28 or by locating well-dated historical injuries in the wood structure,29 because even tree species that appear to form relatively clear growth increments may in fact not follow an annual ring pattern.30 Although tree-ring based age determinations have given maximum ages of not more than 600 years for broad-leaf trees in tropical lowlands,31 tropical dendrochronology is still an active area of investigation.32
In the tropics and elsewhere, as new collections are constantly being added to the ITRDB, estimates of tree longevity should increase. For example, we performed an in-house evaluation of some species’ maximum tree ages using collections that we developed but have not yet been properly archived in the ITRDB. Chronologies that have been published in connection with research projects in the Sierra Nevada33 and the Great Basin34 of North America provided estimates that in some cases exceeded the current ITRDB ones, but ultimately did not result in large changes. For instance, single-needle pinyon (Pinus monophylla) reached 784 years (ITRDB: 653 years; Table S1), big-cone Douglas-fir (Pseudotsuga macrocarpa) peaked at 683 years (ITRDB: 658 years), and blue oak (Quercus douglasii) topped at 539 years (ITRDB: 496 years). On the other hand, a large difference emerged for Fagus sylvatica (ITRDB: 407 years), which has a tree-ring-based maximum age of 625 years.35
The need for science-based information on maximum tree lifespans, which are based on dendrochronological and/or radiocarbon analyses,8,24 is made yet more urgent by the current over-abundance of popular reports, either in press or on the internet, that exaggerate the age of the oldest trees. Occasionally these unscientific claims are repeated in the most prestigious scientific journals, as shown by recent news that oaks older than a millennium can be found in the UK and in Fennoscandia.36,37 Denmark’s King Oak (Quercus robur) is an example of charismatic megaflora, but the notion that it could be “around 1,900 years of age” is nothing more than myth, because it is not based on either dendrochronological or radiocarbon analyses, and it is more than twice the science-based maximum reported ages of northern hemisphere, mid-latitude angiosperms in general, and of oaks in particular.38 Another peer-reviewed article recently published in a top-tier scientific journal39 has claimed that a deciduous species, Ficus religiosa, can reach up to 1,500 years of age, but such an assertion appears to be based, according to the references included in that article, on popular tradition and religious beliefs.
Unrealistic tree ages, especially for very large stems, have often been obtained by assuming a constant radial growth rate (i.e., a constant ring width). The assumption of a constant ring width from stem pith to bark is biologically untenable, as it has already been explained in detail elsewhere.8,40,41,42 Furthermore, this constant annual growth rate is often calculated using only the outermost wood increments, which are typically smaller than the previous ones. Besides making this incorrect assumption, and despite available evidence that the age:girth ratio for Castanea sativa is quite unpredictable,43 Nunziata et al.44 used a constant ring width estimated from published studies of aboveground trunks, and erroneously applied it to the size of an underground, enormous tree stump – today at least two separate stumps are visible aboveground – rather than to the size of the individual sucker stems. By compounding their mistakes, they were able to proclaim an estimated age of 2000–3000 years for the monumental chestnut (C. sativa) named “Castagno dei Cento Cavalli”.
Although tree-ring records are science-based data for estimating tree longevity, it is still necessary to point out that the results we have produced on such a fundamental botanical and ecological trait truly represent the minimum boundary for a species. In some cases, tree-ring samples may contain many more rings that are not measured, and are therefore excluded from ITRDB holdings. Furthermore, dendrochronologists may often avoid measuring sections of increment cores or stem sections that are too difficult to crossdate, either because of erratic growth patterns, extremely low growth rates, injuries, branch insertions, rot, or other anatomical imperfections of the wood structure.8 There are also tree longevity records that, albeit relatively famous, are not part of the ITRDB holdings. For example, our results (Table S1) place Pinus longaeva in third place for maximum tree age (2,771 years), after Sequoiadendron giganteum (3,205 years) and Juniperus przewalskii (2,868 years), when in reality P. longaeva (bristlecone pine) can reach a maximum stem age of 4,844 years,45 more than any other species. The unfortunate history associated with the discovery of the “Currey Tree”, also known as Prometheus, the oldest tree on Earth because it was at least 4,900 years of age when it was cut, is well known among dendrochronologists46. Despite the abundant information on maximum tree lifespans that can be obtained from the ITRDB, it is not the only reliable source of tree longevity data (see8,24), especially in tropical regions (see2).
As it happens with other public-domain resources, the ITRDB provides extremely useful data, but it also has limitations, particularly with regard to metadata. In the STAR Methods section, we describe in detail the very limited information that is associated with the actual measurements. Most recently, the web interface for downloading tree-ring data (https://www.ncei.noaa.gov/access/paleo-search/?dataTypeId=18) has been updated to include links to published research and studies that provide additional information on the data themselves. Yet, besides the criticisms and suggestions that have already been made in previous papers,12 it should be noted that ITRDB ring-width data include no information on the size of sampled trees, such as stem diameter or total height. Even scientific users are therefore prone to potential pitfalls when interpreting these data, as it was evident in a recent high-profile publication, where ITRDB measurements were analyzed under the assumption that “the size and height of trees sampled at a given site are usually similar”.47 Without the actual tree records, this type of statement cannot be verified, and it is our personal experience that in many cases it is factually incorrect.
An additional confounding factor is that, even when tree-ring measurements are archived in the ITRDB, researchers may not provide the entire datasets. By doing so, investigators can satisfy funding agency requirements for archiving data while at the same time avoiding to share the most important, i.e. longest-term, information. This issue was noticed in more than one case, but a clear example was provided by the 37 California chronologies coded as CA561-CA597, which all end in 1990-1991 and start in 1879-1880. Since the collections only cover 111-112 years, but were made on species (Abies concolor, A. magnifica, Calocedrus decurrens, Pinus contorta, P. jeffreyi, P. lambertiana, P. ponderosa, Tsuga mertensiana) and in areas (Sierra Nevada of the Western USA) that are known to yield much older trees (see Table S1), it is unlikely that all data were archived. One could argue that perhaps the study was performed in even-aged plantations, or that there were special constraints that forced the investigators to sample young trees or to extract very short increment cores even when the stem was large. As it turns out, one of us (FB) actually participated in some of those field collections as a graduate student, acquiring first-hand knowledge of these stands and of these collections, which were dendroclimatic-oriented and performed in naturally seeded, uneven-aged stands by targeting the largest and oldest looking trees.
When the number of ITRDB collections for a species is large enough, the above mentioned issue should not impact the estimated maximum tree age. However, a potentially large underestimation occurs if data are not fully archived and only a few chronologies are available for a species. Among the collections coded as CA561-CA597 are indeed the only ITRDB holdings for a species, Quercus kelloggii, whose longevity was therefore estimated at 111 years – an unreliably small value. Partial submissions may cause other artifacts, for instance connected to changes in tree longevity over time. Although we did not perform an exhaustive analysis of this problem, one can imagine how the maximum age of tree species included in collections CA561-CA597 could be compared to the longevity of the same species in earlier collections. As reports of the impending doom of ancient trees accumulate,2,48,49 such a comparison could then lead to claims of human-induced reduction in tree longevity even without the presence of a naive observer or one fully vested in promoting an apocalyptic narrative.
The definition of ‘old growth’ stands, which has fundamental implications for conservation efforts and science-based forest management, depends on correctly estimating tree longevity. We emphasize that what ‘old’ means depends both on the tree species, as shown here, and on its realized longevity niche, as we have argued elsewhere.8 Using a fixed threshold, such as 300 years,25 fails to consider ecoclimatic and taxonomic differences, not to mention structural diversity.50 Earlier, detailed analyses of old-growth conditions had already pointed out that old-growth forest ages can range from 50 to 1,150 years,51 making it necessary to design new metrics for evaluating old-growth conditions.52 Although we cannot determine if our overall mean tree longevity estimates differed from other studies2,24 because of recent additions to the ITRDB; different ways to ingest the ITRDB data and/or to estimate longevity; different computer scripts, algorithms, or software; and/or different quality control checks, it remains true that additional submissions of tree-ring data to the ITRDB, and related publications of dendrochronological and radiocarbon-based information on tree longevity, are bound to improve our understanding of tree life histories, forest demographics, old-growth features, and of their complex dependence on multi-scale impacts from natural and human-caused disturbances.
Limitations of the study
The ITRDB, while providing scientific-quality data for investigating tree longevity, cannot be considered the only source of information on maximum tree lifespans, especially because of its limited coverage of tropical regions and species. Data quality also relies almost entirely on the generosity and dedication of individual researchers, and its correct use, preservation, and expansion require additional resources.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited data | ||
| International Tree-Ring DataBank (ITRDB) | World Data Service for Paleoclimatology |
https://www.ncei.noaa.gov/products/paleoclimatology/tree-ring; ftp.ncdc.noaa.gov/pub/data/paleo/treering |
| Analyzed data | This paper (Table S1) | iScience-S-22-04432 |
| Software and algorithms | ||
| R v.4.0.2 | The R Foundation |
http://www.R-project.org; https://cloud.r-project.org/ |
| SAS Studio, Release 3.8 (Enterprise Edition) | SAS Institute Inc., Cary, NC, USA. | https://welcome.oda.sas.com/home |
| MATLAB | The MathWorks Inc. | https://www.mathworks.com |
| COFECHA | Laboratory of Tree-Ring Research, The University of Arizona, Tucson, AZ, USA | https://www.ltrr.arizona.edu/pub/dpl/COFECHA.ZIP |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Franco Biondi (fbiondi@unr.edu).
Materials availability
This study did not generate new unique materials.
Method details
Data acquisition
In mid-March 2022 we downloaded all available ring-measurement files from the ITRDB ftp server listed in the key resources table. These are measurements in units of .01 mm (the end-of-record is coded as 999) or .001 mm (the end-of-record is coded as -9999) of the ring width for each year in a wood sample. Data format and metadata that are normally included remain those that were first established, back in the 1960s at the Laboratory of Tree-Ring Research of the University of Arizona in Tucson, and are therefore referred to as the “Tucson” format. Files are in ASCII, or text, so that they can be read by any computer operating system.
Metadata are included in the first three lines, or rows, of each file, and are the header records, each 80 characters long, which are then followed by the measurements, each row of them being 72 characters long. The first row of the file (header record #1) includes the “Site ID” (columns 1-6), the “Site Name” (columns 10-61), and the four-letter “Species Code” (columns 62-65). The second row (header record #2) repeats the “Site ID” (columns 1-6) and then lists the “State/Country” (columns 10-22), the “Species” name (columns 23-30), the “Elevation” in m (columns 41-45), and the geographical coordinates (columns 47-57), with both “Latitude” (negative for the southern hemisphere) and “Longitude” (negative to the west of the prime meridian) in degrees and minutes without separation; the line ends with the overall first and last year of the measurements (columns 68-76), with negative values indicating years BCE. The third row (header record #3) repeats the “Site ID” (columns 1-6) and then lists the “Investigators” (columns 10-72) and other optional information. In our analysis we focused on the Site ID, Species Code, Latitude, and Longitude (see Table S1).
Estimation of tree longevity
Tree-ring measurements are listed by calendar decade (hence this data format is also called “decadal”), starting with the fourth row of the file. Each row lists the sample ID (columns 1-6 or 1-8), the decade (columns 9-12), and the annual measurements (columns 13-72, with 6 columns per measurement) as integers. The first and last row of each sample may be shorter if they do not include the full decade; in that case the first decade is replaced by the first year. The sample ID is supposed to include the site code (columns 1-3), the tree code (columns 4-5), and the radius code (column 6), and we used this assumption when we compared the maximum tree lifespan obtained from individual samples or from aggregating samples by tree (see Figure S1).
Irregular coding of sample IDs made it necessary to screen files carefully to correctly associate ring-width measurements with particular trees. Many collections in the ITRDB have more than one character assigned as the radius code (e.g., “NE” and “SW” as cores from the northeastern and southwestern directions). The radius code for some collections is a number (e.g., “1” for the first core and “2” for the second), whereas conventionally sequential letters (“A”, “B”,…) are used to designate radii. Other collections have sample IDs that are all-numeric despite showing one core collected per tree, such that the last character is not a radius number. The ITRDB data we used for the comparison of maximum tree ages with maximum segment length consisted of a total of 519 sites from Canada, Africa, and from the Updates subdirectory of the ITRDB ftp site. No collection from the Updates directory, one from Africa, and about 15% of Canada collections had unconventional sample IDs or other formatting issues that prevented us from confidently associating ring-width series with individual trees. Overall, the subset we were able to analyze covered a large spectrum of environmental and phylogenetic features, thereby being representative of ITRDB collections.
Quality control
There are very few, if any, standard procedures that could be attributed to all ITRDB collections. Data submitted to the ITRDB are annually resolved, and they are supposed to have been accurately dated by visual and/or numerical techniques, hence they are crossdated16, but that is not a requirement for submission, nor it was required for our analysis. Yet, because ITRDB data are annually resolved, there is a lack of tropical records, which often do not have the required dating control or annual resolution. We did not exclude any data a priori, but of the 5444 files we downloaded, a total of 1755 could not be analyzed because of either encoding issues or non-standard formats for the header records, for the measurements, or for both. Among the files we downloaded that could not be read, the end-of-line encoding was often responsible, since it could include both Carriage Return (CR) and Line Feed (LF) characters, or only one of them, but we were able to resolve this issue in several cases. For the files that we were able to read correctly, our approach for identifying maximum tree lifespans did not require information on the stem pith location, which is indeed not included in ITRDB metadata, nor did it require knowing if a wood sample was taken from a live or a dead tree, which is also not part of ITRDB metadata. An in-depth review and evaluation of numerical tree-ring analyses that are linked with pith visibility is provided by 41.
Quality control checks were also performed to compare the metadata we collected from the headers of the data files with the information contained in a summary of all metadata, in Excel format, that was included on the ITRDB ftp site (file “ITRDBmetadata12January2022.xlsx”). We focused on the species name to avoid duplicates, incorrect entries, and collections where only the genus was given. As an example, we considered to be duplicates the following species codes: PINI-PILR, PIHE-PILE, and PIMU-PIMG (see Table S1). A final comparison was made, for each ITRDB collection we analyzed, between the maximum sample length and the difference between the overall first and last year. When this difference exceeded the maximum series length by more than 100 years, we analyzed the collection using the COFECHA software 15,16, and the maximum series length was identified from its output.
Quantification and statistical analysis
Resampling analysis
We used a resampling approach to estimate the number of ITRDB collection that are needed to provide a relatively stable estimate of maximum tree lifespan. The starting point for our analysis was a list of 311 maximum tree ages for the 311 Douglas-fir PSME collections included in our study. The greatest of these maximum ages was 1001 years. If we were to examine a handful, such as k=5, of these 311 PSME collections, our estimate of maximum PSME age would likely be less than 1001 years, because the site with the oldest tree is unlikely to be in a sample of 5 out of 311 collections. The likely maximum PSME age to be arrived at from a random sample of k=5 collections can be estimated by randomly resampling without replacement the 311 PSME maximum ages N times, where N is a very large number, each time drawing a sample of 5 collections, computing the maximum of these 5 numbers, and then examining the empirical probability distribution of the N maxima.
The median of the N maximum ages was the expected maximum PSME age arrived at with a sample of k=5 collections. Ninety-five percent of the resampled N maximum ages would by definition be older than the 0.05 quantile of the N resampled ages. We plotted the median and 0.05 quantile of maximum ages by resampling N=5000 times (Figure 4B) for sample sizes k=1 to k=310. Of course, only one sample of k=311 is possible, and that would necessarily identify the maximum PSME age as 1001 years. The Matlab function randsample was used for the sampling without replacement, which is appropriate in this context because someone examining k tree-ring measurement sets would deal with unique samples – each of the k would represent a different ITRDB collection.
Acknowledgments
This work was supported by the US National Science Foundation (grant AGS-P2C2-1903561 to F.B. and AGS-P2C2-1903535 to D.M.). The views and conclusions contained in this document are those of the authors and should not be interpreted as representing the opinions or policies of the funding agencies and supporting institutions.
We are grateful to the Contributors of the International Tree-Ring Data Bank, and to the agencies and institutions that have allowed the establishment and maintenance of this exceptional, publicly available resource.
Author contributions
All authors contributed to the study conception and design. Data compilation and analysis were performed by F.B. with contributions by D.M. and G.P. The first draft of the manuscript was written by F.B., and all authors made suggestions to modify it. Answers to comments provided by three reviewers were initially prepared by F.B., who also restructured the manuscript using the STAR Methods format. All authors contributed to, and approved, the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: February 3, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106138.
Supplemental information
Code: Four-letter abbreviation of the scientific (Latin) binomial nomenclature for Genus (first two letters) and Species (last two letters).
Division: botanical distinction between conifers (Pinophyta) and angiosperms (Magnoliophyta).
Order: botanical taxon below the Division.
Family: botanical taxon below the Order.
Genus: botanical taxon below the Family.
Species: botanical taxon below the Genus.
Sites: number of ITRDB collections that were analyzed.
Longevity: maximum tree lifespan estimated from ITRDB data.
Site ID: ITRDB code assigned to the collection that included the Longevity estimate.
Lat. (°): latitude, in decimal degrees, of the ITRDB collection that included the Longevity estimate.
Long. (°): longitude, in decimal degrees, of the ITRDB collection that included the Longevity estimate.
Data and code availability
-
•
This paper analyzes existing, publicly available data. Internet addresses (URLs) for the datasets are listed in the key resources table. Tree longevity data reported in this paper are available as Supplementary Material (Table S1).
-
•
This paper does not report original code. We have provided detailed information for replicating our analysis in the STAR Methods to the best of our knowledge.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Gutsell S.L., Johnson E.A. Accurately ageing trees and examining their height-growth rates: implications for interpreting forest dynamics. J. Ecol. 2002;90:153–166. doi: 10.1046/j.0022-0477.2001.00646.x. [DOI] [Google Scholar]
- 2.Locosselli G.M., Brienen R.J.W., Leite M.d.S., Gloor M., Krottenthaler S., Oliveira A.A.d., Barichivich J., Anhuf D., Ceccantini G., Schöngart J., Buckeridge M. Global tree-ring analysis reveals rapid decrease in tropical tree longevity with temperature. Proc. Natl. Acad. Sci. USA. 2020;117:33358–33364. doi: 10.1073/pnas.2003873117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Körner C. A matter of tree longevity. Science. 2017;355:130–131. doi: 10.1126/science.aal2449. [DOI] [PubMed] [Google Scholar]
- 4.Vitasse Y., Bottero A., Cailleret M., Bigler C., Fonti P., Gessler A., Lévesque M., Rohner B., Weber P., Rigling A., Wohlgemuth T. Contrasting resistance and resilience to extreme drought and late spring frost in five major European tree species. Global Change Biol. 2019;25:3781–3792. doi: 10.1111/gcb.14803. [DOI] [PubMed] [Google Scholar]
- 5.Hessburg P.F., Miller C.L., Parks S.A., Povak N.A., Taylor A.H., Higuera P.E., Prichard S.J., North M.P., Collins B.M., Hurteau M.D., et al. Climate, environment, and disturbance history govern resilience of western North American forests. Front. Ecol. Evol. 2019;7:227–239. doi: 10.3389/fevo.2019.00239. [DOI] [Google Scholar]
- 6.Das A.J., Stephenson N.L., Davis K.P. Why do trees die? Characterizing the drivers of background tree mortality. Ecology. 2016;97:2616–2627. doi: 10.1002/ecy.1497. [DOI] [PubMed] [Google Scholar]
- 7.Xu C., Liu H., Hampe A. Hydraulic adaptability promotes tree life spans under climate dryness. Global Ecol. Biogeogr. 2021;31:51–61. doi: 10.1111/geb.13410. [DOI] [Google Scholar]
- 8.Piovesan G., Biondi F. On tree longevity. New Phytol. 2021;231:1318–1337. doi: 10.1111/nph.17148. [DOI] [PubMed] [Google Scholar]
- 9.Piovesan G., Biondi F., Baliva M., Presutti Saba E., Calcagnile L., Quarta G., D'Elia M., De Vivo G., Schettino A., Di Filippo A. The oldest dated tree of Europe lives in the wild Pollino massif: italus, a strip-bark Heldreich's pine. Ecology. 2018;99:1682–1684. doi: 10.1002/ecy.2231. [DOI] [PubMed] [Google Scholar]
- 10.Grissino-Mayer H.D., Fritts H.C. The International Tree-Ring Data Bank: an enhanced global database serving the global scientific community. Holocene. 1997;7:235–238. doi: 10.1177/095968369700700212. [DOI] [Google Scholar]
- 11.Speer J.H. University of Arizona Press; 2010. Fundamentals of Tree-Ring Research. [Google Scholar]
- 12.Zhao S., Pederson N., D'Orangeville L., HilleRisLambers J., Boose E., Penone C., Bauer B., Jiang Y., Manzanedo R. The International Tree-Ring Data Bank (ITRDB) revisited: data availability and global ecological representativity. J. Biogeogr. 2019;46:355–368. doi: 10.1111/jbi.13488. [DOI] [Google Scholar]
- 13.Zang C. Dendrobox – an interactive exploration tool for the international tree ring Data Bank. Dendrochronologia. 2015;33:31–33. doi: 10.1016/j.dendro.2014.10.002. [DOI] [Google Scholar]
- 14.Grissino-Mayer H.D. An updated list of species used in tree-ring research. Tree-Ring Bull. 1993;53:17–43. [Google Scholar]
- 15.Grissino-Mayer H.D. Evaluating crossdating accuracy: a manual and tutorial for the computer program COFECHA. Tree-Ring Res. 2001;57:205–221. [Google Scholar]
- 16.Holmes R.L. Computer-assisted quality control in tree-ring dating and measurement. Tree-Ring Bull. 1983;43:69–78. [Google Scholar]
- 17.R Core Team R: a language and environment for statistical computing. 2021. http://www.R-project.org
- 18.Delwiche L.D., Slaughter S.J. Sixth Edition. SAS Institute Inc.; 2019. The Little SAS Book: A Primer. [Google Scholar]
- 19.The MathWorks Inc. MATLAB. 2018. https://www.mathworks.com
- 20.Worbes M. How to measure growth dynamics in tropical trees: a review. IAWA J. 1995;16:337–351. doi: 10.1163/22941932-90001424. [DOI] [Google Scholar]
- 21.Munné-Bosch S. Limits to tree growth and longevity. Trends Plant Sci. 2018;23:985–993. doi: 10.1016/j.tplants.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Peñuelas J., Munné-Bosch S. Potentially immortal? New Phytol. 2010;187:564–567. doi: 10.1111/j.1469-8137.2010.03360.x. [DOI] [PubMed] [Google Scholar]
- 23.Cannon C.H., Piovesan G., Munné-Bosch S. Old and ancient trees are life history lottery winners and vital evolutionary resources for long-term adaptive capacity. Nat. Plants. 2022;8:136–145. doi: 10.1038/s41477-021-01088-5. [DOI] [PubMed] [Google Scholar]
- 24.Liu J., Xia S., Zeng D., Liu C., Li Y., Yang W., Yang B., Zhang J., Slik F., Lindenmayer D.B. Age and spatial distribution of the world's oldest trees. Conserv. Biol. 2022;36:e13907. doi: 10.1111/cobi.13907. [DOI] [PubMed] [Google Scholar]
- 25.Besnard S., Koirala S., Santoro M., Weber U., Nelson J., Gütter J., Herault B., Kassi J., N'Guessan A., Neigh C., et al. Mapping global forest age from forest inventories, biomass and climate data. Earth Syst. Sci. Data. 2021;13:4881–4896. doi: 10.5194/essd-13-4881-2021. [DOI] [Google Scholar]
- 26.Spies T.A. Ecological concepts and diversity of old-growth forests. J. For. 2004;102:14–20. doi: 10.1093/jof/102.3.14. [DOI] [Google Scholar]
- 27.O’Brien L., Schuck A., Fraccaroli C., Pötzelsberger E., Winkel G., Lindner M. European Forest Institute; 2021. Protecting Old-Growth Forests in Europe - A Review of Scientific Evidence to Inform Policy Implementation. [DOI] [Google Scholar]
- 28.Vieira S., Trumbore S., Camargo P.B., Selhorst D., Chambers J.Q., Higuchi N., Martinelli L.A. Slow growth rates of Amazonian trees: consequences for carbon cycling. Proc. Natl. Acad. Sci. USA. 2005;102:18502–18507. doi: 10.1073/pnas.0505966102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubau W., De Mil T., Van den Bulcke J., Phillips O.L., Angoboy Ilondea B., Van Acker J., Sullivan M.J.P., Nsenga L., Toirambe B., Couralet C., et al. The persistence of carbon in the African forest understory. Native Plants. 2019;5:133–140. doi: 10.1038/s41477-018-0316-5. [DOI] [PubMed] [Google Scholar]
- 30.Santos G.M., Rodriguez D.R.O., Barreto N.d.O., Assis-Pereira G., Barbosa A.C., Roig F.A., Tomazello-Filho M. Growth assessment of native tree species from the southwestern Brazilian Amazonia by post-ad 1950 14C analysis: implications for tropical dendroclimatology studies and atmospheric 14C reconstructions. Forests. 2021;12:1177. doi: 10.3390/f12091177. [DOI] [Google Scholar]
- 31.Worbes M. One hundred years of tree-ring research in the tropics – a brief history and an outlook to future challenges. Dendrochronologia. 2002;20:217–231. doi: 10.1078/1125-7865-00018. [DOI] [Google Scholar]
- 32.Brienen R.J.W., Schöngart J., Zuidema P.A. In: Tropical Tree Physiology: Adaptations and Responses in a Changing Environment. Goldstein G., Santiago L.S., editors. Springer International Publishing; 2016. Tree rings in the tropics: insights into the ecology and climate sensitivity of tropical trees; pp. 439–461. [DOI] [Google Scholar]
- 33.Meko D.M., Woodhouse C.A., Touchan R. University of Arizona; 2014. Klamath/San Joaquin/Sacramento Hydroclimatic Reconstructions from Tree Rings. [Google Scholar]
- 34.Biondi F. Dendroclimatic reconstruction at km-scale grid points: a case study from the Great Basin of north America. J. Hydrometeorol. 2014;15:891–906. doi: 10.1175/jhm-d-13-0151.1. [DOI] [Google Scholar]
- 35.Piovesan G., Biondi F., Baliva M., De Vivo G., Marchianò V., Schettino A., Di Filippo A. Lessons from the wild: slow but increasing long-term growth allows for maximum longevity in European beech. Ecology. 2019;100 doi: 10.1002/ecy.2737. [DOI] [PubMed] [Google Scholar]
- 36.Sonne C., Xia C., Lam S.S. Ancient oaks of Europe are archives — protect them. Nature. 2021;594:495. doi: 10.1038/d41586-021-01699-0. [DOI] [PubMed] [Google Scholar]
- 37.Pennisi E. Rare and ancient trees are key to a healthy forest. Science. 2022 doi: 10.1126/science.ada0797. [DOI] [Google Scholar]
- 38.Piovesan G., Baliva M., Calcagnile L., D’Elia M., Dorado-Liñán I., Palli J., Siclari A., Quarta G. Radiocarbon dating of Aspromonte sessile oaks reveals the oldest dated temperate flowering tree in the world. Ecology. 2020;101 doi: 10.1002/ecy.3179. [DOI] [PubMed] [Google Scholar]
- 39.Chakraborty A., Mahajan S., Bisht M.S., Sharma V.K. Genome sequencing and comparative analysis of Ficus benghalensis and Ficus religiosa species reveal evolutionary mechanisms of longevity. iScience. 2022;25:105100–105126. doi: 10.1016/j.isci.2022.105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biondi F. From dendrochronology to allometry. Forests. 2020;11:146. doi: 10.3390/f11020146. [DOI] [Google Scholar]
- 41.Biondi F., Qeadan F. A theory-driven approach to tree-ring standardization: defining the biological trend from expected basal area increment. Tree-Ring Res. 2008;64:81–96. doi: 10.3959/2008-6.1. [DOI] [Google Scholar]
- 42.Biondi F. Comparing tree-ring chronologies and repeated timber inventories as forest monitoring tools. Ecol. Appl. 1999;9:216–227. doi: 10.1890/1051-0761(1999)009[0216:ctrcar]2.0.co;2. [DOI] [Google Scholar]
- 43.Jarman R., Moir A.K., Webb J., Chambers F.M., Russell K. Dendrochronological assessment of British veteran sweet chestnut (Castanea sativa) trees: successful cross-matching, and cross-dating with British and French oak (Quercus) chronologies. Dendrochronologia. 2018;51:10–21. doi: 10.1016/j.dendro.2018.07.001. [DOI] [Google Scholar]
- 44.Nunziata A., Ferlito F., Magri A., Ferrara E., Petriccione M. The Hundred Horses Chestnut: a model system for studying mutation rate during clonal propagation in superior plants. Forestry. 2022;95:ecpac020. doi: 10.1093/forestry/cpac020. [DOI] [Google Scholar]
- 45.Currey D.R. An ancient bristlecone pine stand in eastern Nevada. Ecology. 1965;46:564–566. doi: 10.2307/1934900. [DOI] [Google Scholar]
- 46.Salzer M.W., Baisan C. Second American Dendrochronology Conference; 2013. Dendrochronology of the "Currey Tree". [Google Scholar]
- 47.Au T.F., Maxwell J.T., Robeson S.M., Li J., Siani S.M.O., Novick K.A., Dannenberg M.P., Phillips R.P., Li T., Chen Z., Lenoir J. Younger trees in the upper canopy are more sensitive but also more resilient to drought. Nat. Clim. Change. 2022;12:1168–1174. doi: 10.1038/s41558-022-01528-w. [DOI] [Google Scholar]
- 48.McDowell N.G., Allen C.D., Anderson-Teixeira K., Aukema B.H., Bond-Lamberty B., Chini L., Clark J.S., Dietze M., Grossiord C., Hanbury-Brown A., et al. Pervasive shifts in forest dynamics in a changing world. Science. 2020;368:eaaz9463. doi: 10.1126/science.aaz9463. [DOI] [PubMed] [Google Scholar]
- 49.Lindenmayer D.B., Laurance W.F., Franklin J.F. Global decline in large old trees. Science. 2012;338:1305–1306. doi: 10.1126/science.1231070. [DOI] [PubMed] [Google Scholar]
- 50.Martin M., Valeria O. Old” is not precise enough: airborne laser scanning reveals age-related structural diversity within old-growth forests. Remote Sens. Environ. 2022;278:113098. doi: 10.1016/j.rse.2022.113098. [DOI] [Google Scholar]
- 51.Wirth C., Messier C., Bergeron Y., Frank D., Fankhänel A. In: Old-Growth Forests: Function, Fate and Value. Wirth C., Gleixner G., Heimann M., editors. Springer Berlin Heidelberg; 2009. Old-growth forest definitions: a pragmatic view; pp. 11–33. [DOI] [Google Scholar]
- 52.Di Filippo A., Biondi F., Piovesan G., Ziaco E. Tree ring-based metrics for assessing old-growth forest naturalness. J. Appl. Ecol. 2017;54:737–749. doi: 10.1111/1365-2664.12793. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Code: Four-letter abbreviation of the scientific (Latin) binomial nomenclature for Genus (first two letters) and Species (last two letters).
Division: botanical distinction between conifers (Pinophyta) and angiosperms (Magnoliophyta).
Order: botanical taxon below the Division.
Family: botanical taxon below the Order.
Genus: botanical taxon below the Family.
Species: botanical taxon below the Genus.
Sites: number of ITRDB collections that were analyzed.
Longevity: maximum tree lifespan estimated from ITRDB data.
Site ID: ITRDB code assigned to the collection that included the Longevity estimate.
Lat. (°): latitude, in decimal degrees, of the ITRDB collection that included the Longevity estimate.
Long. (°): longitude, in decimal degrees, of the ITRDB collection that included the Longevity estimate.
Data Availability Statement
-
•
This paper analyzes existing, publicly available data. Internet addresses (URLs) for the datasets are listed in the key resources table. Tree longevity data reported in this paper are available as Supplementary Material (Table S1).
-
•
This paper does not report original code. We have provided detailed information for replicating our analysis in the STAR Methods to the best of our knowledge.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.