Abstract
A growing body of evidence indicates that RNA plays a critical role in orchestrating DNA double-strand break repair (DSBR). Recently, we showed that homologous nascent RNA can be used as a template for error-free repair of double-strand breaks (DSBs) in the transcribed genome and to restore the missing sequence at the break site via the transcription-coupled classical nonhomologous end-joining (TC-NHEJ) pathway. TC-NHEJ is a complex multistep process in which a reverse transcriptase (RT) is essential for synthesizing the DNA strand from template RNA. However, the identity of the RT involved in the TC-NHEJ pathway remained unknown. Here, we report that DNA polymerase eta (Pol η), known to possess RT activity, plays a critical role in TC-NHEJ. We found that Pol η forms a multiprotein complex with RNAP II and other TC-NHEJ factors, while also associating with nascent RNA. Moreover, purified Pol η, along with DSBR proteins PNKP, XRCC4, and Ligase IV can fully repair RNA templated 3′-phosphate-containing gapped DNA substrate. In addition, we demonstrate here that Pol η deficiency leads to accumulation of R-loops and persistent strand breaks in the transcribed genes. Finally, we determined that, in Pol η depleted but not in control cells, TC-NHEJ-mediated repair was severely abrogated when a reporter plasmid containing a DSB with several nucleotide deletion within the E. coli lacZ gene was introduced for repair in lacZ-expressing mammalian cells. Thus, our data strongly suggest that RT activity of Pol η is required in error-free DSBR.
Keywords: double strand break repair, transcription-coupled NHEJ, RNA-templated repair, RNA-templated DNA polymerase activity, DNA polymerase eta
Abbreviations: ATXN3, Ataxin-3; BER, base excision repair; ChIP, chromatin immunoprecipitation; C-NHEJ, classical nonhomologous end-joining; co-IP, coimmunoprecipitation; DMEM, Dulbecco’s modified Eagle’s medium; DMSO, dimethyl sulfoxide; DNA-PKcs, DNA-dependent protein kinase, catalytic subunit; dNTP, deoxynucleoside triphosphate; DSB, double-strand break; DSBR, double-strand break repair; gRNA, guide RNA; HR, homologous recombination; IC, immunocomplex; iPSC, induced pluripotent stem cell; LA-qPCR, long amplicon quantitative PCR; Lig IV, Ligase IV; NE, nuclear extract; NSC, neural stem cell; PNKP, polynucleotide kinase 3′-phosphatase; Pol η, DNA polymerase η; RT, reverse transcriptase; SB, strand break; TC-NHEJ, transcription-coupled classical nonhomologous end-joining; TLS, translesion DNA synthesis; XP-V, xeroderma pigmentosum variant
Mammalian cells are continuously exposed to various endogenous and exogenous genotoxic agents (1) that constitute an inevitable challenge for maintaining genome integrity (2). Among various DNA lesions incurred by these genotoxic agents, DNA double-strand breaks (DSBs) leading to loss of nucleotides, particularly in the transcribed region, could be lethally toxic to the cells and lead to multiple pathologies including cancer, aging, and neurodegenerative diseases (3, 4, 5, 6). DSBs are repaired either by homologous recombination (HR) in the S/G2 phase (7, 8) or via the classical nonhomologous end-joining (C-NHEJ) pathway (9), which is operative in all stages of the cell cycle and is most active in G2/M phase (10). Over the last few years, the field of double-strand break repair (DSBR) has been revolutionized, particularly with the discovery of the critical role of RNA in such repair pathways (11, 12, 13, 14, 15). Recent studies provide compelling evidence for RNA-templated HR in yeast and human cells (11, 16, 17, 18, 19). Rad52-mediated inverse strand exchange between homologous dsDNA and RNA has been established as a potential mechanism for RNA-templated HR in yeast (20). On the other hand, our own studies provide evidence of nascent RNA-templated error-free DSBR of the transcribed genome via the transcription-coupled NHEJ (TC-NHEJ) pathway in mammalian cells (6, 21). Hence, a polymerase with reverse transcriptase (RT) activity is essential for generating the nascent DNA strand using an RNA template in mammalian cells.
Various reverse transcriptases have been implicated in diverse DSBR pathways in higher eukaryotes and mammalian cells. Among these RTs, a majority are involved in a process called “Translesion DNA Synthesis” (TLS), a mechanism that cells have adopted to bypass various DNA lesions, such as pyrimidine dimers, oxidized DNA bases, and abasic sites, during replication fork movement (22, 23, 24, 25). The role of translesion DNA polymerase ζ was shown in utilization of RNA as a donor in RNA–DNA recombination during HR in yeast (26). Furthermore, another TLS polymerase, DNA polymerase θ (Pol θ), implicated in microhomology-mediated end joining (27, 28, 29, 30, 31), was also shown to possess RT activity. RT activity of Pol θ plays a critical role in microhomology-mediated end joining–mediated RNA-templated error-prone DSBR in mammalian cells (32, 33). Complementary DNA sequencing confirmed nucleotide misincorporation and insertion–deletions (indels) in Pol θ–mediated RNA-dependent DNA synthesis, indicating low-fidelity RT activity of Pol θ (32). However, the RT polymerase for the RNA-templated TC-NHEJ pathway in mammalian cells has not been identified and characterized.
Among the various TLS polymerases, DNA polymerase η (Pol η) plays a unique, highly error-free role in replicative bypass of UV-induced pyrimidine dimers and 8-oxoguanine lesions (34, 35) in mammalian cells (36). Consequently, mutations in Pol η are responsible for the variant form of xeroderma pigmentosum (XP-V) (37) that ultimately leads to high incidence of sunlight-induced skin cancer (38, 39, 40). Apart from the canonical TLS function, growing evidence also suggests a TLS-independent cellular function of Pol η (41). This evidence includes the function of Pol η in D-loop extension during HR (42, 43), in synthesis of non-B DNA structures like G4 DNA (44), and in telomere DNA synthesis involving a process called “Alternative Lengthening of Telomeres” via an HR-dependent pathway (45). Interestingly, human Pol η plays a role in RNA synthesis (reviewed in (41)) and has been shown to utilize RNA as one of the two strands during primer extension, mainly by inserting deoxynucleoside monophosphate opposite unmodified templates or DNA lesions (46). In addition, a recent study showed that purified human Pol η adds rNTPs to DNA primers at physiological rNTP concentrations and in the presence of competing deoxynucleoside triphosphates (dNTPs) (47). Most notably, HEK293T cell extracts could add dNTPs to DNA primers hybridized to RNA, but this RT activity was abrogated in Pol η–depleted cells (47). Also, this RT activity in mammalian cells was found to be specific for Pol η among the canonical Y-family TLS polymerases, such as Pol ι and Pol κ (47), indicating some uncharacterized physiological role of this reverse transcriptase function of Pol η. A recent study demonstrated that Pol η could be targeted to etoposide-induced DSBs and that depletion of Pol η in cells caused increased sensitivity to etoposide (48). Furthermore, this depletion of Pol η also caused a defect in C-NHEJ repair, suggesting the crucial role of Pol η in such repair. Notably, in budding yeast, the role of Pol η was implicated in transcription elongation (49).
Based on these established instances of RT activity in TLS polymerases and their role in various DSBR pathways, we reasoned that Pol η might be involved in TC-NHEJ-mediated repair of DSBs. Our in vitro and in vivo biochemical and cell biology studies indeed identify Pol η as a candidate RT-polymerase for TC-NHEJ and here, we provide evidence for its involvement in RNA-templated error-free DSBR in mammalian cells.
Results
Pol η associates with RNAP II and key C-NHEJ proteins forming a multiprotein complex
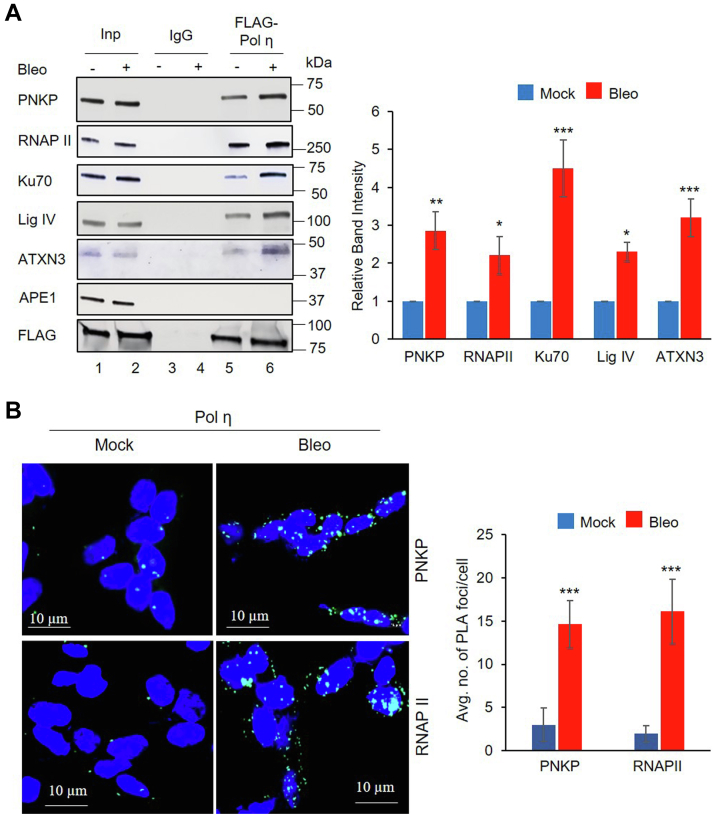
We reported earlier that repair pathway–specific, preformed, multiprotein complexes exist in mammalian cells under physiological conditions (6, 21, 50). Hence, we performed coimmunoprecipitation (co-IP) from nuclear extracts (NEs) of HEK293 cells to characterize the Pol η immunocomplex (IC), in order to gain functional insight into the potential role of Pol η in RNA-templated TC-NHEJ. Owing to apparent unavailability of an IP-grade Pol η antibody (Ab) from commercial vendors, we generated an HEK293 stable cell line where an N-terminal FLAG sequence was introduced in-frame with the endogenous PolH gene (expressing Pol η) using CRISPR technology. We then performed co-IP with anti-FLAG Ab to characterize the Pol η IC from cells after Bleomycin (Bleo; a DSB inducing radiomimetic agent) treatment. It was found that Pol η does associate with the key C-NHEJ proteins, including Polynucleotide kinase 3′-phosphatase (PNKP) (Fig. 1A; lane 5; PNKP, Ku70, Ligase IV [Lig IV]) along with RNAP II. Interestingly, we detected Ataxin-3 (ATXN3) in the Pol η IC, which is consistent with our previous report of ATXN3’s role in TC-NHEJ-mediated DSBR (6). In addition, we observed that the association of most of these proteins with Pol η was enhanced upon DSB induction (3- to-5-fold, Fig. 1A, lane 6 versus 5). The absence of APE1 (involved in OGG1/NTH1-mediated base excision repair (BER) (51, 52) in the Pol η IC demonstrated DSBR-specific association of these proteins. To confirm this association in mammalian cells, we generated HEK293 cells stably expressing Pol η containing an in-frame C-terminal HA-tag and performed co-IP using anti-HA Ab. We detected PNKP, Ku70, Lig IV, and XRCC4 along with RNAP II in complex with Pol η, and the association was enhanced significantly upon Bleo treatment (8- to-12.5-fold, Fig. S1A, lane 6 versus 5), confirming our results with FLAG-Pol η IP. To further corroborate the presence of Pol η in the TC-NHEJ complex, we performed reverse co-IPs with anti-RNAP II and anti-Lig IV Ab and indeed found the presence of Pol η along with other C-NHEJ factors (Fig. S1, B and C, lane 5). The association of Pol η and other C-NHEJ proteins were enhanced significantly post Bleo treatment as we observed earlier (2- to 4-fold, Fig. S1, B and C; lane 6 versus 5). These data indicate a stable association of Pol η with RNAP II and TC-NHEJ components under physiological conditions.
Figure 1.
Pol η associates with RNAP II and transcription-coupled classical nonhomologous end-joining proteins.A, left panel, nuclear extracts (NEs) (benzonase-treated) from FLAG-POLH HEK293 cells (either mock or Bleo treated) were immunoprecipitated (IP’d) with anti-FLAG Ab (lanes 5 and 6) or mouse IgG (lanes 3 and 4) and tested for the presence of associated proteins with specific Abs as indicated to the left of each row. One percent of NE was used as inputs (Inp) (lanes 1 and 2). IP experiment was repeated at least three times from separate batches of cells, and one representative figure is shown. Right panel, the quantitation of the IP’d bands after normalization with FLAG-Pol η band intensity. Error bars represent ±SD of the mean. The increase in IP band intensity following Bleo treatment was significant at the level of ∗∗∗p < 0.005, ∗∗p < 0.01, or ∗p < 0.05. B, left panel, detection of FLAG-Pol η′s (anti-mouse FLAG Ab) association with PNKP and RNAP II (both anti-rabbit Abs) in mock- or Bleo-treated FLAG-POLH HEK293 cells by proximity ligation assay. Nuclei were counterstained with DAPI (blue). Right panel, average number of proximity ligation assay foci per cell was calculated from 25 randomly selected cells in each sample. Error bars represent ±SD of the mean. The data were significant at ∗∗∗p < 0.005 between mock and Bleo treatment (n = 3). The scale bar represents 10 μm.
Next, we performed in situ proximity ligation assay in mock- or Bleo-treated FLAG-Pol η expressing HEK293 cells (described earlier) to investigate in-cell association of Pol η with PNKP (a key DNA end processing enzyme with 3′-phosphatase and 5′-kinase activities (53, 54) and involved in TC-NHEJ (21)) and RNAP II. Mock-treated cells had an average of 1 to 3 ± 2 foci/cell, whereas Bleo-treated cells had 9 to 14 ± 3 foci/cell for various interactions (Fig. 1B), indicating a significant increase in the association of Pol η with PNKP and RNAP II upon DSB induction. Collectively, these data suggest formation of a DSBR-specific multiprotein complex involving Pol η, RNAP II, and C-NHEJ factors in mammalian cells.
Depletion of Pol η leads to DNA damage response in mammalian cells
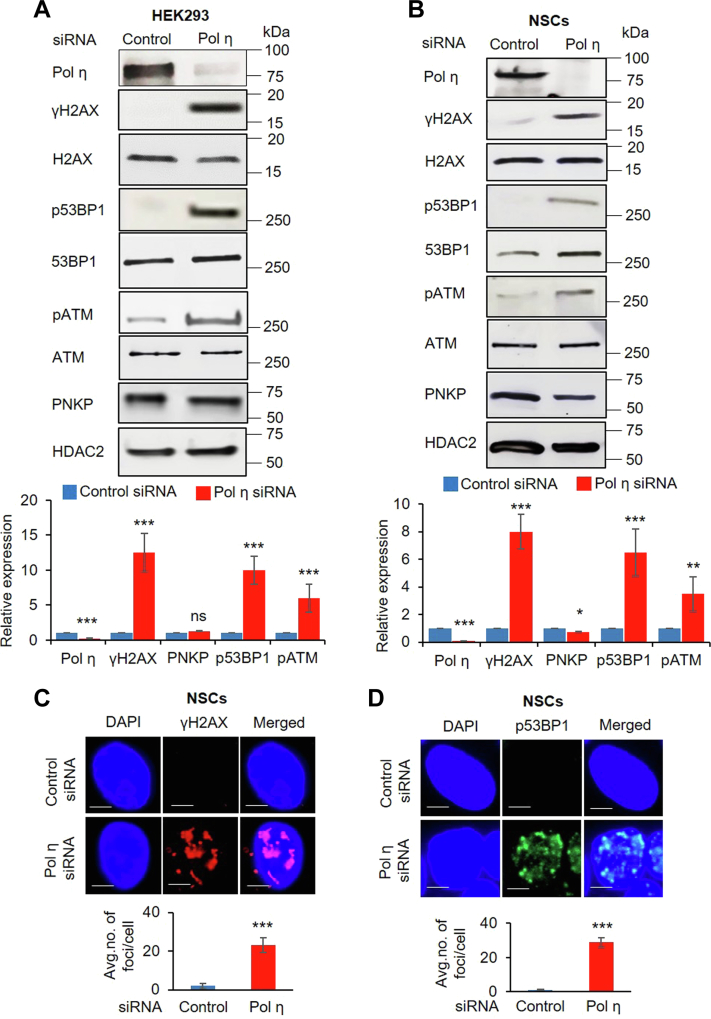
To further assess the role of Pol η in DSBR, we validated individual siRNAs from multiple vendors to determine the efficiency of Pol η depletion as well as the specificity of siRNAs by complementation with ectopic expression of Pol η post siRNA transfection. We indeed observed significant depletion of Pol η (70–80%); however, its level was restored following ectopic expression (Fig. S2, A and B, lane 3 versus lane 2). We then depleted Pol η in HEK293 cells using these validated siRNAs and found widespread induction of phosphorylated variant of the histone H2A protein family (γH2AX) and phospho-ATM (pATM), both hallmarks of DSB accumulation in these cells (Fig. 2A, upper and lower panels). Moreover, we observed induction of phosho-p53 binding protein 1 (p53BP1) in the NE of Pol η-depleted HEK293 cells. Phosphorylation of 53BP1, in response to genotoxic stress, is known to promote C-NHEJ-mediated DSBR (6, 55, 56). Thus, persistent induction of p53BP1 indicated DSB accumulation under Pol η–deficient condition (Fig. 2A). Moreover, we observed phosphorylation of DNA-dependent protein kinase, catalytic subunit (DNA-PKcs) (a C-NHEJ marker (9)) in Pol η–depleted cells. However, the extent of phosphorylation for all critical C-NHEJ factors decreased significantly upon ectopic expression of Pol η, which suggested C-NHEJ-mediated DSBR progression and Pol η′s specific role therein (Fig. S2, A and B). We would like to mention here that replication stress due to TLS deficiency under Pol η–deficient conditions leads to activation of ataxia telangiectasia and Rad3-related (ATR) kinase (57, 58). Thus, γH2AX, pATM, and p53BP1-mediated DNA damage response as observed here is due to Pol η′s DSBR-specific role. We conducted a similar assay as well as a few more key experiments in human induced pluripotent stem cell (iPSC)-derived neural stem cells (NSCs). These cells are among the best characterized mammalian stem cells with robust DNA damage response signaling (59). We indeed observed similar results in the NEs of NSCs following siRNA-mediated depletion of Pol η (Fig. 2B, upper and lower panels). Immunofluorescence analyses further provided evidence of widespread formation of γH2AX and p53BP1 foci in NSCs upon Pol η depletion (Fig. 2, C and D). These results reiterate that Pol η plays a crucial role in C-NHEJ-mediated DSBR.
Figure 2.
Depletion of Pol η leads to DNA damage response.A, upper panel, Western blots showing the levels of proteins (as indicated in the left of each row) in the nuclear extract of Pol η–depleted HEK293 cells. HDAC2, used as nuclear loading control. The lower panel shows the quantitation of Pol η depletion and concurrent increase in γH2AX, p53BP1, and pATM levels (normalized with the corresponding unmodified H2AX, 53BP1, and ATM levels) with expression in control siRNA-treated samples arbitrarily considered as unity (n = 3, ∗∗∗p < 0.005, ns = nonsignificant, p > 0.05). B, similar study involving neural stem cells (NSCs) (∗p < 0.05, ∗∗p < 0.01). In both A and B, error bars represent ±SD of the mean. C, upper pane, γH2AX foci formation in Pol η–depleted neural stem cells. Nuclei were counterstained with DAPI (blue) (the scale bar represents 5 μm). Lower panel, the average number of γH2AX foci per cell is shown in the bar diagram, calculated from ten randomly selected cells/sample. Error bars represent ±SD of the mean. The increase in foci/cell in Pol η–depleted cells was significant at the level of ∗∗∗p < 0.005. D, similar study in neural stem cells to show p53BP1 foci formation upon Pol η depletion (the scale bar represents 5 μm).
Pol η associates with the nascent RNA transcripts and Pol η deficiency leads to R-loop accumulation
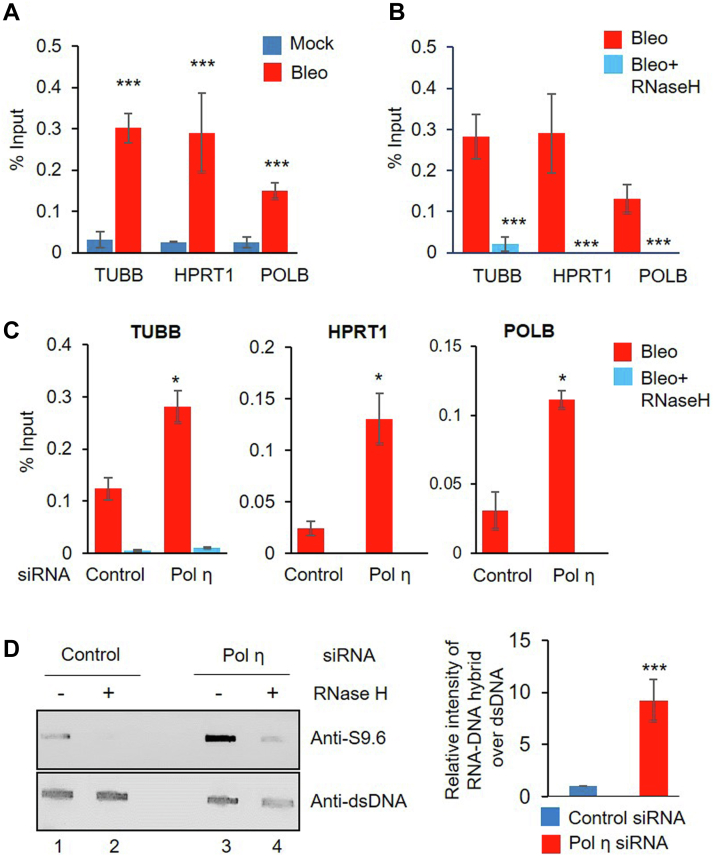
We previously showed that several C-NHEJ proteins associate with nascent RNA following DSB induction in mammalian cells, in order to promote RNA-templated DSBR (6, 21). Given that Pol η is a component of the TC-NHEJ complex and was shown to possess RT activity in mammalian cells, we examined whether nascent RNA associates with Pol η. Therefore, we performed RNA–chromatin immunoprecipitation (ChIP) using anti-FLAG Ab from either mock- or Bleo-treated HEK293 cells expressing endogenous FLAG-Pol η, followed by reverse-transcriptase quantitative PCR (RT-qPCR) with intron-specific primers for three randomly selected transcribing genes (TUBB, HPRT1, and POLB). Indeed, we detected a significant increase in the association of nascent RNA with Pol η post Bleo treatment (Fig. 3A). Control reactions without reverse transcriptase (-RT) ruled out the possibility of genomic DNA contamination in the samples (Fig. S3A). In order to serve as a template, RNA should pair with DNA in an RNA–DNA hybrid (R-loop), and we previously confirmed such hybrid formation at DSB sites (6, 21). Hence, to confirm that Pol η indeed uses nascent RNA as a template, we treated FLAG-Pol η expressing cells with Bleo followed by RNase H treatment, which specifically degrades RNA in an RNA–DNA hybrid. We then performed RNA-ChIP assays using anti-FLAG Ab. We observed a significant decrease in the association of nascent RNA with Pol η after RNase H treatment (Fig. 3B) for all the genes tested, suggesting an association of Pol η with RNA–DNA hybrid at DSB sites. A no RT control reaction ruled out the possibility of genomic DNA contamination (Fig. S3B). However, the association of Pol η with DNA was not affected by RNase H treatment, as analyzed by DNA-ChIP performed under similar conditions (Fig. S3C), thus suggesting RNase H mediated specific reduction of Pol η′s association with RNA–DNA hybrids only. Notably, we detected an increase in the accumulation of RNA–DNA hybrids at DSB sites in Pol η–depleted cells (Fig. S3D) post Bleo treatment (Fig. 3C), as shown by RNA-ChIP assay using anti-PNKP Ab. RNase H treatment significantly decreased the association of nascent RNA with PNKP, suggesting R-loop accumulation occurs at DSB sites as a direct consequence of Pol η deficiency. Control reactions without RT ruled out the possibility of genomic DNA contamination in the samples (Fig. S3E). To further confirm that Pol η depletion leads to R-loop accumulation, genomic DNA (including RNA–DNA hybrids) isolated from Pol η–depleted HEK293 cells (Fig. S3F) was subjected to slot-blot analysis using S9.6 RNA–DNA hybrid–specific Ab. We observed a significant increase in the R-loop accumulation in Pol η–depleted cells compared with control cells (Fig. 3D, lane 3 versus 1, upper panel). Notably, the R-loop pool was significantly decreased in RNase H treatment (Fig. 3D, lanes 2 and 4, upper panel), but no effect on the dsDNA pool was detected (Fig. 3D, lower panel), demonstrating the specificity of the R-loop signal. Collectively, these results strongly indicate that Pol η specifically interacts with nascent RNA at DSB sites and provide support for its potential role in RNA-templated DSBR.
Figure 3.
Association of Pol η with nascent RNA and R-loops.A, FLAG-POLH HEK293 cells were mock or Bleo treated and their nuclear extracts subjected to RNA-ChIP using the anti-FLAG Ab. Real-time PCR was carried out for TUBB, HPRT1, and POLB genes using intron-specific primers. Data were presented as % input; error bars show ±SD of the mean (n = 3, ∗∗∗p < 0.0001). B, Bleo-treated FLAG-POLH HEK293 cells were incubated with RNase H before RNA-ChIP using FLAG Ab to detect RNA–DNA hybrids, and real-time quantitative PCRs were done using intron-specific primers as described above. Data were presented as % input where error bars show ±SD of the mean (n = 3, ∗∗∗p < 0.0001). C, control or Pol η siRNA-transfected HEK293 cells were Bleo treated and incubated with RNase H before RNA-ChIP using PNKP Ab to detect RNA–DNA hybrids, and real-time quantitative PCRs were done using intron-specific primers as described above. Data were presented as % input where error bars show ±SD of the mean (n = 3, ∗p < 0.05). D, left panel, HEK293 cells were transfected with control or Pol η siRNA. Genomic DNA was isolated and digested with HindIII, EcoR1, XbaI, and BsrGI followed by mock or RNase H treatment. Five hundred nanograms from each of the digested DNA was blotted onto nylon membrane. The DNA was cross-linked by UV (Stratalinker) and incubated with a S9.6 antibody to detect RNA–DNA hybrid and a dsDNA-specific Ab as loading control. Right panel, bar diagram shows quantification of R-loop-specific signal normalized to dsDNA-specific signals. Error bars show ±SD of the mean (n = 3, ∗∗∗p < 0.005).
Pol η–mediated RNA-templated DNA polymerase (reverse transcriptase) activity in mammalian cells
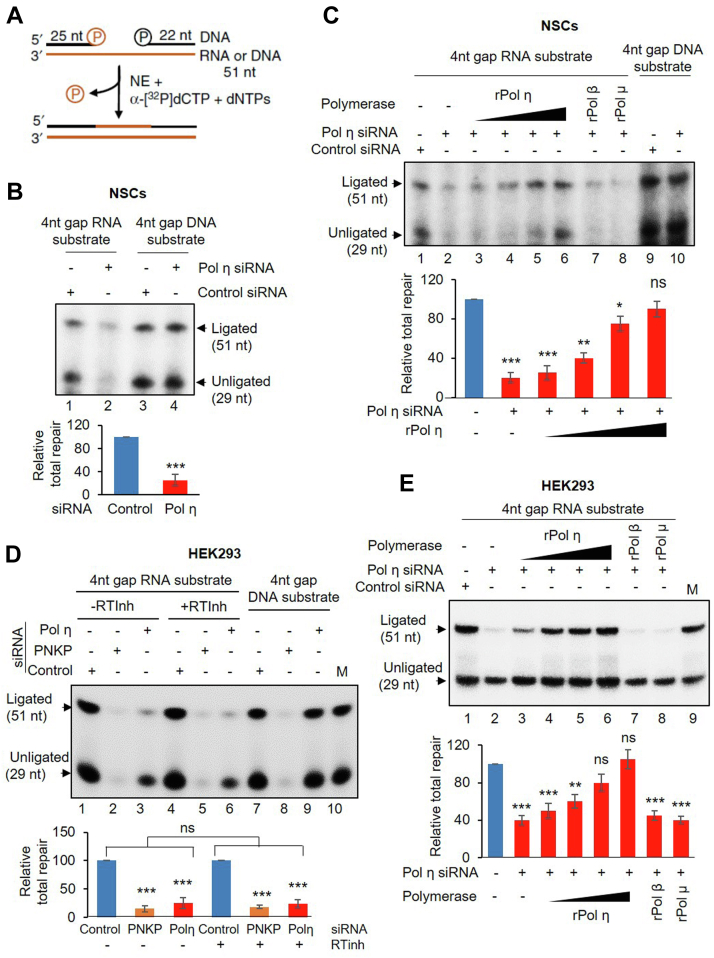
To provide evidence of RNA-templated repair in mammalian cells, NEs from NSCs were incubated with an RNA oligo (as template) annealed to two different complementary DNA oligos to generate a 4-nt gap as shown schematically in Figure 4A. We then measured α-32P-dCMP incorporation to assess repair synthesis, as in our previous study (21). Pol η–depleted NEs (Fig. 2B) from NSCs showed significantly less repair synthesis compared with control NEs (Fig. 4B, lane 2 versus 1), indicating that Pol η is the relevant RT responsible for this RNA-templated repair synthesis. Our previous results showed that loss of PNKP function impairs this repair synthesis due to deficiency of 3′-P end processing (21). To exclude such possibility, we examined the PNKP level in NEs from control versus Pol η siRNA-treated cells and found the level to be comparable (Fig. 2B). Finally, we examined the 3′-phosphatase activity of PNKP in these NEs and found that there is no significant difference in PNKP activity (Fig. S4A, lane 3 versus 2, lane 6 versus 5), indicating that the decreased repair synthesis is specifically due to depleted levels of Pol η. To further confirm the specific role of Pol η in this RNA-templated repair synthesis, we purified N-terminal His-tagged full-length recombinant Pol η (rPol η, Fig. S4B, left panel, lane 1) and supplemented the Pol η–depleted NE with increasing amounts of rPol η, then followed RNA-templated repair synthesis. We observed a dose-dependent, complete restoration of the repair synthesis activity in the Pol η–depleted NEs, indicating the specific role of Pol η in RNA-templated DNA polymerase activity (Fig. 4C, lanes 3–6 versus 2). To further establish specificity for Pol η RT activity, we added two purified recombinant DNA polymerases individually, one involved in DNA BER/single-strand break repair (DNA polymerase β; rPol β; Fig. S4B, left panel, lane 2) and another involved in DSBR via C-NHEJ (DNA polymerase μ; rPol μ; Fig. S4B, left panel, lane 3). However, these two recombinant polymerases failed to complement the RNA-templated repair synthesis even when added at the highest dose used for rPol η (Fig. 4C, lanes 7 and 8, respectively). Moreover, Pol η depletion had no effect on the DNA template–directed DNA polymerase activity in the NEs (Fig. 4B, lane 4 versus 3; Fig. 4C, lane 10 versus 9). Hence, our data suggest that Pol η is a key reverse transcriptase in human cells.
Figure 4.
Pol η–mediated RT activity in mammalian cells.A, schematic representation of the RNA-templated total repair assay with 4-nt gapped oligos. B, upper panel, twenty picomoles of annealed RNA-templated substrate (shown schematically in A) was used to assess total repair activity in the nuclear extracts (NEs) (500 ng) from control or Pol η siRNA-transfected neural stem cells (NSCs) (lanes 1 and 2). Similar assays were performed with NEs from control or Pol η siRNA-transfected NSCs in DNA-templated (lanes 3 and 4; as control) substrates (5 pmol). The upper arrow indicates the 51-mer repaired product, and the lower arrow, unligated product (after nt incorporation, 29 nt). Lower panel, quantitation of the total repaired product (n = 3, ∗∗∗p < 0.005) expressed as the combined band intensities of the unligated (29 nt long) and ligated (51 nt long) products in control (arbitrarily set as 100, lane 1) versus Pol η–depleted nuclear extract. C, similar assay in NSCs where complementation of Pol η–depleted NE by recombinant Pol η (100, 200, 400, 500 nmol) (lanes 3–6), Pol β (500 nmol, lane 7), and Pol μ (500 nmol, lane 8) is shown. rPol η complementation shows gradual restoration of the total repair activity (n = 3, ∗∗∗p < 0.005, ∗∗p < 0.01, ∗p < 0.05, and ns = nonsignificant, p > 0.05, between control and Pol η complemented NE). D, a similar RNA-templated total repair assay carried out in mock (-RTinh)/Line1-specific RT inhibitor-treated (+RTinh) NEs from Control (lanes 1 and 4) or PNKP (lanes 2 and 5) or Pol η (lanes 3 and 6)-depleted NE from HEK293 cells. The lower panel shows quantitation of the relative total repair activity calculated as described in B. Error bars represent ±SD of the mean (n = 3, difference in total repair activity in PNKP or Pol η–depleted versus control NE was significant (∗∗∗p < 0.005), whereas difference between –RTinh and +RTinh NEs remained insignificant (ns = nonsignificant, p > 0.05) between similar samples. 5′ End-labeled 29-nt (lower) and 51-nt (upper) (lane 10, M) oligos were used as markers. E, similar assay was conducted where complementation of Pol η–depleted NE by recombinant Pol η (100, 200, 400, 500 nmol) (lanes 3–6), Pol β (500 nmol, lane 7), and Pol μ (500 nmol, lane 8) are shown. Lower panel, quantitation of the relative total repair activity (n = 3, ∗∗∗p < 0.005, ∗∗p < 0.01, and ns = nonsignificant, p > 0.05 between control versus recombinant protein complemented NE) shows gradual restoration of the total repair activity with rPol η.
To further investigate RT activity mediated by Pol η in mammalian cells, we conducted the following experiments in HEK293 derived control versus Pol η–depleted NEs (Fig. 4D). Long Interspersed Elements (LINE1) is a well-known reverse transcriptase in mammalian cells and has been reported to play a role in DSBR through its RT activity (13). We have previously shown a modest expression of LINE1 retrotransposon in HEK293 cells compared with a robust expression in HCT116 colon cancer cell line (21). Hence, to investigate a possible role of LINE1 in RNA-templated repair assay, we treated Pol η–depleted HEK293 cells with mock or LINE1 inhibitor (denoted as RTinh) and could not detect any significant difference in RT activity between mock and RTinh-treated control versus Pol η–depleted NEs (Fig. 4D, lane 4 versus 1 and lane 6 versus 3). Thus, this RNA-templated repair activity was independent of LINE1. However, we observed significant abrogation of repair synthesis in Pol η–depleted NEs from HEK293 cells (Fig. 4D, lane 3 versus 1 and lane 6 versus 4). We used PNKP depletion as a positive control, since PNKP-depleted extracts are unable to process 3′-P ends and, therefore, such repair synthesis is abrogated (Fig. 4D, lane 2 versus 1) (21). We observed no significant difference in PNKP’s 3′-phosphatase activity (Fig. S4C, lane 3 versus 2) or its level (Fig. 2A) in control versus Pol η–depleted NEs, ultimately indicating the abrogated repair synthesis is due to specific depletion of Pol η activity. Next, we performed “rescue of repair synthesis” by supplementing rPol η and observed dose-dependent complementation of the RT activity (Fig. 4E, lanes 3–6 versus 2). Interestingly, rPol β and rPol μ failed to restore repair synthesis individually (Fig. 4E, lanes 7 and 8, respectively), indicating Pol η to be the relevant RT polymerase in mammalian cells. Finally, we depleted Pol μ in HEK293 cells using specific siRNA (Fig. S4D) and performed RNA-templated repair synthesis in parallel with Pol η–depleted NEs. We observed an insignificant abrogation of RNA-templated repair in Pol μ–depleted NEs as compared with Pol η–depleted NEs (Fig. S4E, lanes 3 and 2 versus 1).
The in vitro RNA-templated repair assay, as described earlier, relied on the incorporation of radioactive dCMP with an equal amount of RNA-templated gapped substrate used in all assay reaction mixtures. So, to further confirm Pol η–mediated RNA-templated repair, we used an alternative assay system where the 3′-phosphate-containing 25-nt oligo was 5′-end-labeled with radioactive phosphate and annealed with the complementary oligos (as described in the Experimental procedures section) to produce the 4-nt gapped RNA-templated substrate. Repair was then followed via nonradiolabeled dNTP incorporation (schematically shown in Fig. S5A). In this assay, 29-nt and 51-nt products represented gap-filled unligated and fully repaired ligated products, respectively. Supplementation of purified Pol η restored the RNA-templated repair in Pol η–depleted NEs from HEK293 cells in a dose-dependent manner (Fig. S5B, lanes 4–5 versus lane 3); however, purified Pol β and Pol μ failed to restore the RNA-templated repair (Fig. S5B, lanes 6 and 7, respectively). We then performed a similar “rescue of repair synthesis” in XP-V human skin fibroblast cells that are devoid of Pol η (60, 61). We utilized a 3′-OH end containing oligo (5′-end-labeled with radioactive phosphate as described above) to assess the Pol η–mediated RNA-templated polymerase activity as a proof of principle. NEs from XP-V cells show deficient RNA-templated repair compared with control human fibroblasts, and such deficiency can be rescued in a dose-dependent manner by purified Pol η (Fig. S5C, lanes 4–5 versus lane 3) but not by purified Pol β or Pol μ (Fig. S5C, lanes 6 and 7, respectively). These results provide evidence of Pol η–mediated RNA-templated repair in mammalian cell-free NEs.
In vitro reconstitution of Pol η–mediated RNA-templated DNA polymerase activity
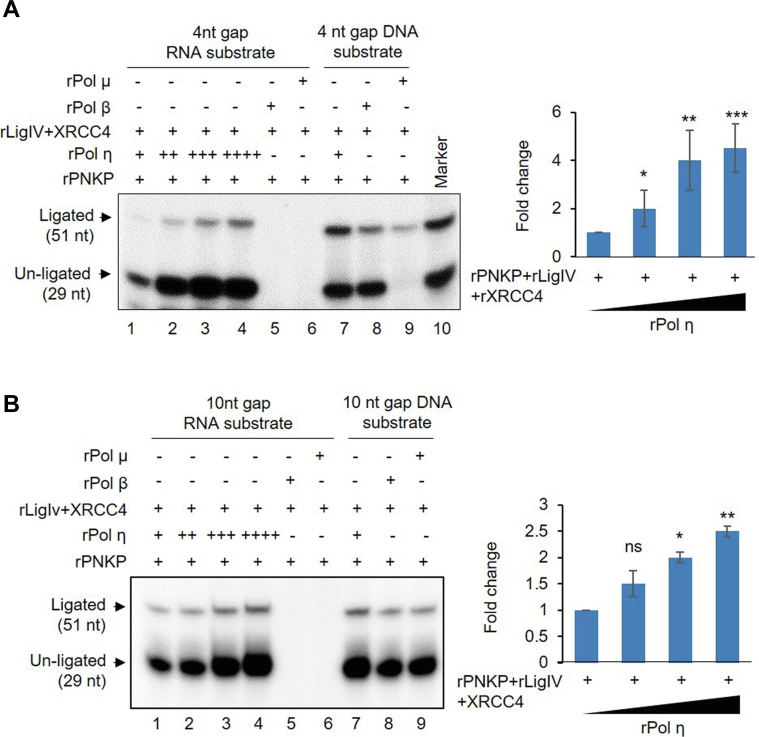
Since mammalian cells show Pol η–mediated RT activity, we reconstituted this repair synthesis in vitro using recombinant proteins. DNA strand breaks (SBs) generated due to endogenous or exogenous sources rarely harbor canonical 3′-OH and 5′-P; as in most cases, the ends undergo chemical modifications. 3′-P is one of the major blocked DNA ends, and we therefore used it as a model system in our assays to mimic in vivo conditions. Because the RNA-templated oligo substrate containing a 4-nt gap contains 3′-P, PNKP-mediated 3′-P end processing is necessary. Followed by end processing, gap filling requires a polymerase that will reverse transcribe the sequence information from the RNA template. Finally, a DNA ligase is necessary to seal the nick and complete the repair process. Thus, using these minimum components to reconstitute RNA-templated repair in vitro, we incubated the oligo substrate with increasing amounts of rPNKP, rPol η, and rLig IV-rXRCC4 (Fig. S4B, left panel, lanes 5 and 1; Fig. S4B, right panel). We indeed observed an increase in the formation of the 51-nt ligated product (the complete RNA-templated repair product) with increasing amounts of component recombinant proteins (Fig. 5A, lanes 1–4). However, when we replaced rPol η with rPol β or rPol μ, individually, we did not observe any such repair synthesis product (Fig. 5A, lanes 5 and 6), indicating that only rPol η can incorporate the deoxynucleotides using the sequence information on an RNA template. However, we observed formation of the 51-nt complete repair product on a DNA template with the addition of rPol η, rPol β, or rPol μ (individually) in the reaction mixture (Fig. 5A, lanes 7–9), indicating the individual polymerases being otherwise active on a DNA template.
Figure 5.
In vitro reconstitution of RNA-templated DNA repair activity by Pol η.A, left panel, assessment of the RNA-templated total repair activity on 4-nt gapped oligo substrate (Fig. 4A) with recombinant proteins: PNKP (25 fmol), Pol η (100, 200, 400, 500 nmol; lanes 1–4), Pol β (500 nmol, lane 5), Pol μ (500 nmol, lane 6), XRCC4 (100 nmol), and Lig IV (250 nmol). Lanes 7 to 9: Similar assays in DNA-templated substrate (lane 7 with Pol η, lane 8 with Pol β, lane 9 with pol μ, all 100 nmol). Right panel, quantitation of the relative total repair activity with rPol η (n = 3, ∗∗∗p < 0.005, ∗∗p < 0.01, ∗p < 0.05) expressed as fold change over the combined band intensities of the unligated (29 nt long) and ligated (51 nt long) products where repair with lowest concentration of Pol η arbitrarily set as unity. 5′ End-labeled 29-nt and 51-nt (lane 10) oligos were used as markers. B, left and right panels, similar RNA-templated repair assay with 10-nt gapped annealed oligos (ns, nonsignificant, p > 0.05).
DSBs can lead to loss of several nucleotides that need to be faithfully restored for error-free repair to maintain normal physiological functions. Hence, we tested the ability of rPol η to incorporate up to 10 nt on an RNA-templated oligo substrate using a 10-nt gapped oligo. We observed that rPol η can incorporate 10 nt on an RNA template to produce the 51-nt ligated product in a dose-dependent manner when utilized in combination with rPNKP and rLig IV-rXRCC4 (Fig. 5B, lanes 1–4). Consistent with our previous results, rPol β or rPol μ failed to reconstitute this repair synthesis on an RNA template (Fig. 5B, lanes 5 and 6); however, these DNA polymerases can fill a 10-nt gap on a DNA template (Fig. 5B, lanes 7–9). Furthermore, we performed in vitro reconstitution experiments with a 5′-end-labeled 25-nt oligo containing 4-nt and 10-nt gapped substrate as described earlier (Fig. S5, D and E). The results were consistent, and we did observe Pol η–mediated efficient repair synthesis up to 10 nt. Collectively, our results further demonstrate that Pol η is the major reverse transcriptase involved in RNA-templated DSBR.
Pol η preferentially associates with the transcribed genes and is involved in their preferential repair
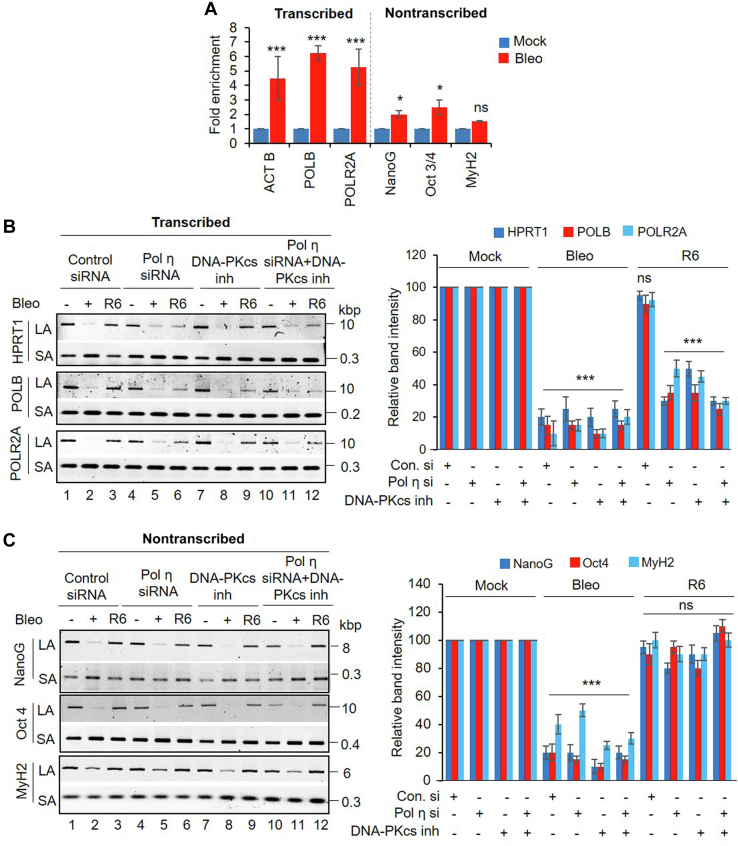
Because Pol η associates with RNAP II in a multiprotein complex and is involved in the RNA-templated repair of DSBs, we anticipated its role in the preferential repair of the transcribed genome. As a first evidence of a transcribed genome-specific role in DSBR, we performed DNA-ChIP using anti-FLAG Ab for immunoprecipitation of Pol η from an HEK293 cell line carrying N-terminally FLAG-tagged Pol η, following mock or Bleo treatment. We indeed observed significantly enhanced association of Pol η with the transcribed genes (ACTB, POLB, POLR2A) as compared with nontranscribed genes (NanoG, Oct3/4, and MyH2; 21), post Bleo treatment in HEK293 cells (Fig. 6A). To assess the role of Pol η in transcribed genome-specific repair, we analyzed Bleo-induced SB accumulation in transcribed (HPRT1, POLB, and POLR2A) versus nontranscribed (NanoG, Oct4, MyH2) genes using long amplicon quantitative PCR (LA-qPCR) (6, 21, 50, 62, 63) from the genomic DNA isolated from the control versus Pol η–depleted cells at 6 h post Bleo treatment. We selected 6 h recovery based on our previous report where we observed significant repair at this time point in control cells post Bleo treatment (21). Despite comparable SB levels in both transcribed and nontranscribed genes after Bleo treatment, depletion of Pol η significantly abrogated the repair of DNA SBs in the transcribed (Fig. 6B; lane 6 versus lane 3) but not in the nontranscribed genes (Fig. 6C; lane 6 versus lane 3). However, both transcribed and nontranscribed genes show repair of the DNA SBs equally well in control siRNA-transfected cells (Fig. 6, B and C; lane 3 versus lane 1). To further confirm that DSBR proceeds through the C-NHEJ pathway, we additionally inhibited DNA-PKcs (a critical C-NHEJ protein involved in the early steps (9)) using NU7441 (KU-57788), followed by Bleo treatment to induce SBs, and then assessed repair by LA-qPCR. We indeed observed abrogated SB repair, specifically in the transcribed genes, in cells where DNA-PKcs was inactivated (Fig. 6, B and C, lane 9 versus lane 3), indicating deficiency of the C-NHEJ pathway that led to impediment of DSBR. This result further confirms our recent report of transcribed region-specific C-NHEJ repair in nonreplicating cells (21), as inactivation of a dedicated C-NHEJ protein, DNA-PKcs, leads to abrogation of DSBR, specifically in the transcribed genes. Interestingly, this transcribed genome-specific repair deficiency was further evident in cells where Pol η was depleted prior to DNA-PKcs inactivation (Fig. 6, B and C, lane 12 versus 9; Fig. S6A). These results provide strong evidence that Pol η plays a key role in TC-NHEJ-mediated repair of DSBs in the transcribed genome. To define the repair kinetics more specifically under the Pol η–deficient condition, we depleted Pol η (Fig. S6B) and treated the cells with Bleo. We then allowed the cells to undergo repair and followed the repair kinetics at various time points (4, 8, 12 h post Bleo treatment). While control cells mostly repaired the SBs within 8 to 12 h post DSB induction, Pol η–depleted cells were inefficient in repairing the DSBs as evident from the enhanced levels of γH2AX and pATM even at 12 h post Bleo treatment (Fig. S6, B and C). In addition, we observed unresolved p53BP1 in the Pol η–depleted cells, further indicating C-NHEJ deficiency in these cells (Fig. S6, B and C). An LA-qPCR-based assay corroborated these results, where Pol η depletion showed transcribed genome-specific abrogation of SB repair (Fig. S7A, lanes 8–10 versus lanes 3–5) but not in nontranscribed genome (Fig. S7B, lanes 8–10 versus lanes 3–5). However, the control cells showed efficient SB repair at 4 to 8 h post Bleo treatment, in both transcribed and nontranscribed genes (Fig. S7, C and D).
Figure 6.
Pol η associates with and preferentially repairs transcribed genome.A, FLAG-POLH HEK293 cells were mock or Bleo treated, and ChIP was performed with FLAG Ab. Binding to the exonic regions of transcribed (β-Actin, POLB, and POLR2A) versus nontranscribed (NanoG, Oct3/4, and MyH2) genes was quantified by quantitative PCR (qPCR) from immunoprecipitated DNA. The data are represented as fold enrichment of % input over IgG with mock-treated samples considered as unity. Error bars represent ±SD of the mean (n = 3). ∗∗∗p < 0.005, ∗p < 0.05 represent statistical significance between mock and Bleo treatment for each gene (ns, nonsignificant, p > 0.05). B, left panel, representative gel images of long amplicon qPCR assay showing each long amplicon (10–12 kb) and a short amplicon (∼200–300 bp) of the transcribed (HPRT1, POLB, and POLR2A) genes from the DNA isolated from control siRNA (lanes 1–3) or Pol η siRNA (lanes 4–6) or DNA-PKcs inhibitor (lanes 7–9) or Pol η siRNA in conjunction with DNA-PKcs inhibitor–treated (lanes 10–12) HEK293 cells either mock or Bleo treated or at 6 h recovery (R6) following Bleo treatment. Right panel, the bar diagram represents the normalized (with short PCR amplicon) relative band intensity with the mock-treated sample in each case arbitrarily set as 100 (n = 3, persistence of strand break accumulation after 6 h recovery remains significant except control siRNA transfected samples, ∗∗∗p < 0.005; ns = nonsignificant, p > 0.05). C, left and right panels, similar long amplicon qPCR assay with nontranscribed (NanoG, Oct4, and MyH2) genes. n = 3, persistence of strand break accumulation after 6 h recovery remains insignificant (ns = nonsignificant, p > 0.05), indicating repair in the nontranscribed genes even in the absence of Pol η and/or presence of DNA-PKcs inhibitor. Error bars represent ±SD of the mean in both (B) and (C).
To provide further evidence for abrogation of Bleo-induced SB repair in Pol η–deficient cells, we assessed such repair in control versus XP-V fibroblasts at 6 h post Bleo treatment. We indeed observed abrogated SB repair in XP-V cells (Fig. S7E, lane 6 versus 3), which corroborates our earlier data showing impeded RNA-templated repair in NEs of XP-V cells and provides additional evidence of the critical role of Pol η in TC-NHEJ-mediated DSBR in human cells.
Plasmid-based in-cell DSBR assay shows Pol η–mediated RNA-templated error-free TC-NHEJ
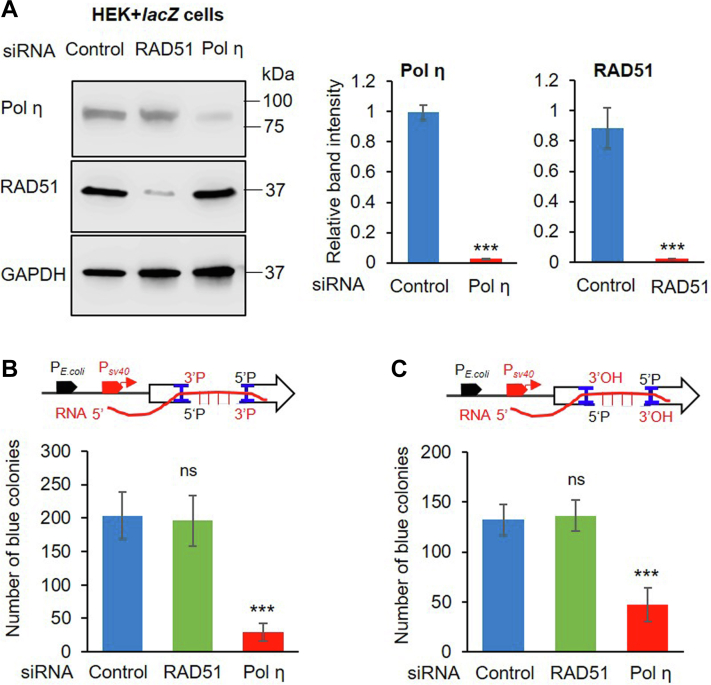
We have previously shown error-free TC-NHEJ in mammalian cells using a plasmid-based in-cell repair assay (6, 21). This assay utilizes a bacterial plasmid containing a DSB with 3′-P ends (the substrate for PNKP) within an Escherichia coli lacZ gene, leading to deletion of several nucleotides and consequent inactivation of lacZ (Fig. S8A and Experimental procedures section). The linearized plasmid was transfected for repair into HEK293 cells stably expressing E. coli lacZ under the CMV promoter (HEK293-pcmv-lacZ). The plasmid is under the control of both mammalian and bacterial promoters and is transcription competent; however, the linearized plasmid only generates RNA up to the DSB point (21). Thus, the lacZ sequence on the plasmid can only be faithfully restored if lacZ RNA from stable cells provide the template, as shown previously (6, 21). To test the specific role of Pol η in error-free DSBR using the stably expressing lacZ cells (HEK293-pcmv-lacZ), we depleted Pol η by siRNA (Fig. 7A) and assessed the in-cell repair of the DSB-containing plasmid. To achieve this, we first transfected HEK293-pcmv-lacZ cells with Pol η–specific siRNA followed by transfection with the DSB-containing plasmid. We previously showed that in-cell repair takes place via the TC-NHEJ, but not the HR pathway, as RAD51 depletion had no significant effect on repair activity (21). Hence, we used RAD51 depletion (Fig. 7A) as our control to exclude the possibility of HR-mediated DSBR. A recent report provided evidence in favor of RAD52-mediated RNA-templated HR via an inverse strand exchange mechanism (20). Thus, we further assessed the effect of its depletion on RNA-templated repair. After 16 h of repair, the plasmids recovered from HEK293-pcmv-lacZ cells were introduced into rec−lacZ− E. coli and blue colonies were scored as a measure of error-free repair of the lacZ sequence on the repaired plasmid. We found a significant decrease in blue colony numbers (29 ± 12) in Pol η–depleted cells compared with control siRNA-transfected cells (203 ± 34) (Fig. 7B). RAD51 depletion had no significant effect on the error-free repair (196 ± 37), consistent with our previous findings (21). Similarly, a significant decrease in blue colonies was observed when a linearized plasmid containing a 3′-OH group was transfected into Pol η–depleted HEK293-pcmv-lacZ cells for repair, and then recovered plasmid was introduced into rec−lacZ− E. coli for screening, as compared with control, or RAD51 siRNA-transfected cells (Fig. 7C). Interestingly, we did not observe any significant effect of RAD52 depletion (Fig. S8B) on error-free repair of either 3′-OH or 3′-P end containing DSBs compared with the control group (Fig. S8C). The result is corroborative with our earlier data that showed RAD52 is not part of the PNKP-mediated TC-NHEJ complex (21). To further confirm that repair under these conditions takes place via the C-NHEJ pathway, we performed plasmid-based in-cell repair assay following specific inhibition of DNA-PKcs. We indeed observed a significant decrease in blue colonies when linearized plasmid containing either 3′-OH or 3′-P ends was transfected individually into DNA-PKcs-inhibited cells, indicating abrogated repair under these conditions (Fig. S8D) and thus further confirming that the repair of this plasmid specifically proceeded via TC-NHEJ pathway. Collectively, the in-cell plasmid-based repair assay in combination with our in vitro biochemical experiments strongly suggest Pol η as the candidate RT for error-free TC-NHEJ.
Figure 7.
Pol η–mediated RNA-templated plasmid-based in-cell transcription-coupled classical nonhomologous end-joining assay.A, left panel, Western blots showing the levels of proteins (as indicated in the left of each row) in the whole cell extract of Pol η– or RAD51–depleted pcmv-lacZ HEK293 cells. GAPDH: used as loading control. Right panel, quantitation of Pol η and RAD51 depletion where expression level in control siRNA-treated sample is arbitrarily set as unity (n = 3, ∗∗∗p < 0.0001). B, plasmid-based double-strand break (DSB) repair (3′ phosphate–containing DSB ends) assay in control versus Pol η– or RAD51–depleted HEK293-Pcmv-lacZ stable cells. The number of E. coli blue colonies after transformation of the plasmids recovered from those cells was plotted. The data were the average of at least three independent experiments (n = 3) where p < 0.0001(indicated by ∗∗∗) for Pol η depletion and p > 0.05 (nonsignificant, ns) for RAD51 depletion in comparison with control siRNA-transfected samples. Error bars represent ±SD of the mean. C, similar plasmid-based assay with 3′ OH-containing DSB ends. The data were the average of at least three independent experiments where ∗∗∗p < 0.0001 for Pol η–depleted cells and p > 0.05 (nonsignificant, ns) for RAD51 depleted cells. Upper panels in (B) and (C) show schematic representation of the plasmid-based RNA-templated DSB repair assay.
Discussion
DNA DSBs are generated in both proliferating and nonproliferating postmitotic cells (24). Lack of a sister chromatid and a reduced deoxynucleotide pool impede the error-free HR pathway in nonproliferating cells. Consequently, in these cells, DSBs are primarily repaired by the C-NHEJ pathway, which is usually error-prone (9). However, error-free repair of DSBs in the transcribed sequences is critically important in maintaining normal physiological function of mammalian cells, and we have now shown that DSBs in the transcribed genome can be faithfully repaired in an error-free manner via the RNA-templated TC-NHEJ pathway (6, 21). In the present follow-up study, we report the identification of Pol η as the reverse transcriptase essential for the TC-NHEJ pathway to copy the sequence information from the homologous nascent RNA as the template utilized to fill in gaps in genomic DNA at DSB sites and prevent any loss of sequence information.
While investigating the role of Pol η in TC-NHEJ, we found Pol η to be associated with RNAP II and C-NHEJ factors in a preformed multiprotein complex in mammalian cells under normal physiological conditions. Moreover, association of these proteins with Pol η was significantly enhanced upon DSB induction. Our study further revealed accumulation of DSBs upon depletion of Pol η in mammalian cells, as indicated by enhanced γH2AX (a DSB-specific marker) and p53BP1 (a C-NHEJ marker) accumulation/foci formation and their persistence for 12 h post DSB induction in those cells. This result is consistent with a previous study, which showed that XP-V fibroblast cells, devoid of Pol η, harbor 53BP1 foci even 30 h post DSB induction, in contrast to Pol η–proficient cells (48). The role of Pol η in DSBR was substantiated by our observation of persistently increased pATM levels in Pol η–depleted cells even 12 h post DSB induction. Activated ATM (pATM) coordinates cell cycle progression with damage-response checkpoints and DNA repair to preserve genomic integrity via a well-orchestrated signaling network (64, 65, 66, 67). Considering that ATM phosphorylation is a vital and early transducer of DSB signals via strong activation of the ATM pathway, elevated pATM levels reflect persistence of DSBs and chronic ATM activation when Pol η is depleted in HEK293 as well as NSCs. NSCs primarily stay in a dormant state and can last throughout human lifespan (59). Hence their genomes are prone to damage, including DSBs that can accumulate over a long period of time. A robust error-free repair system, particularly in the transcribed regions, would be critical for the survival and function of these cells. This further prompted us to validate the RT activity of Pol η in the RNA-templated DSBR pathway in NSCs.
Since Pol η is a component of the TC-NHEJ complex and involved in DSBR, we investigated its role in transcribed genome-specific repair. Notably, DNA-ChIP provided evidence for preferential recruitment of Pol η to the transcribed genome following DSB induction, and concomitantly, Pol η–deficient cells show accumulation of DNA SBs in the transcribed genes, whereas the nontranscribed genes are repaired efficiently, indicating a crucial role of Pol η in the maintenance of transcribed genome sequence via TC-NHEJ. Here we confirmed Pol η′s C-NHEJ specific role by providing evidence that inhibition of DNA-PKcs also showed a similar effect on transcribed genome repair and that Pol η inhibition further augments such effect.
In our earlier study (21), we postulated that strand invasion by nascent RNA occurred to form an RNA–DNA hybrid and provided evidence of such RNA–DNA hybrid formation at the DSB sites that promotes RNA-templated DSBR. It has been shown that Pol η can bind to RNA–DNA hybrids with similar affinity as DNA–DNA duplexes (46), which would be essential for its ability to utilize nascent RNA as a repair template. We indeed observed strong association of Pol η with nascent RNA following DSB induction. Our study also showed specific association of Pol η with RNA–DNA hybrid (R-loop) post DSB induction, which is an essential step for Pol η to function as an RT. In Pol η–depleted cells, we observed enhanced accumulation of R-loops and consequent, increased association of C-NHEJ repair proteins with R-loops at DSB sites, compared with control cells. Collectively, these data indicate failure to efficiently repair DSBs in Pol η–deficient cells, which leads to prolonged persistence of DSB markers, γH2AX and p53BP1, and activation of ATM signaling.
Given the role of Pol η in TC-NHEJ, we further tested and observed significant (∼80%) abrogation of RNA-dependent DNA polymerase activity in vitro in Pol η–depleted cells with a 4-nt gapped RNA–DNA hybrid substrate. As expected, this RT activity is restored in a dose-dependent manner by complementation with purified full-length Pol η. We further confirmed such abrogated RT activity in human XP-V cells and subsequent restoration of such activity by supplementation of purified Pol η. These data are consistent with a recent report that showed similar abrogation of RT activity in XP-V cells compared with Pol η–corrected cells as well as in Pol η–depleted HEK293T cells (47). Importantly, the authors of the above study eliminated the possibility of involvement of RT activity of related TLS polymerases, including Pol ι, κ, or ζ, because their individual depletion did not have any effect on RT activity in HEK293T cells. As a follow-up, we tested two other DNA polymerases, polymerase μ (involved in C-NHEJ and reported to incorporate ribonucleotide stretches during NHEJ-mediated repair) (68, 69) and polymerase β (involved in single-strand break repair with a reported unanticipated role in NHEJ) (70) in our assay and found no role of these polymerases in TC-NHEJ.
Our in-cell plasmid-based DSBR assay clearly indicated abrogation of error-free repair of the lacZ plasmid with several nucleotide deletions in Pol η–depleted cells as compared with control cells. Notably, we observed such abrogation of error-free repair in both 3′-P-containing blocked end substrate plasmid (subject to PNKP-mediated end-processing) and 3′-OH-containing substrate plasmid, indicating a generalized role of Pol η in such repair. We also show that this repair proceeds via RNA-templated TC-NHEJ, not the HR pathway, as RAD51 (a dedicated HR protein) and RAD52 (another protein with reported role in RNA-templated HR) depletion had no significant effect on the error-free repair. Since inhibition of DNA-PKcs significantly abrogated error-free repair in these cells, our results provide strong evidence that repair in HEK293-pcmv-lacZ cells proceeded through the TC-NHEJ pathway and Pol η depletion abrogates such error-free repair. Collectively, our data strongly support an RT function of Pol η in TC-NHEJ-mediated error-free DSBR in mammalian cells.
Finally, TC-NHEJ is a highly intricate, multistep process and several other proteins are likely to be involved in subsequent steps. We could envisage the involvement of other proteins in facilitating the process of recruiting and presenting the template RNA to the DSB sites for Pol η to copy the sequence from RNA and an RNA–DNA helicase to resolve the transient R-loop formation, which is necessary for completion of repair. Identification of these proteins remains a priority for the biomedical research community, which may help understanding the repair pathway and the components involved therein, which in turn would be instrumental in development of mechanism-based treatment options for various diseases caused by TC-NHEJ deficiency.
Experimental procedures
Generation of HEK293 stable cells
CRISPR/Cas9-mediated targeted insertion of N-terminal FLAG tag in-frame into the endogenous POLH gene
Incorporation of the FLAG sequence (GAT TAC AAG GAT GAC GAC GAT AAG) was achieved using CRISPR/Cas9 technology, as described (71), to generate an HEK293 clonal cell line (FLAG-POLH HEK293) harboring a genomically encoded N-terminal FLAG-tag within the DNA polymerase η (POLH) gene. Guide RNAs (gRNAs) were designed using the CRISPOR program for gRNA prediction (72) using a 100-bp genomic sequence flanking the translation start site of the POLH gene. Two criteria were followed to choose the gRNAs: (i) the shortest distance between the PAM motif (NGG) and the FLAG insertion site and (ii) the specificity score. The gRNA expression vectors, pGL3-U6-sgRNA-PGK-puromycin (Addgene plasmid # 51133) and pST1374-NLS-FLAG-linker-Cas9 (Addgene plasmid # 44758), were a kind gift from the Eric Wagner lab. The selected gRNAs (Table S1) were synthesized (Integrated DNA Technologies), annealed, and cloned into the pGL3-U6-sgRNA-PGK-puromycin digested with BsaI (New England Biolabs) as per standard protocol (71). The specificity of genome targeting efficiency of gRNAs and the Cas9-mediated genome-editing efficiency was determined by the T7 endonuclease (NEB) assay by amplifying the targeted region using specific primers FP1 and RP1 (Table S1) as described (73). A 200-nucleotide-long single-stranded DNA donor template sequence (Table S1), including 24 bases of the FLAG sequence in the middle surrounding the translation start site, was synthesized (Integrated DNA Technologies). The PAM motif within the donor template was silently mutated to overcome iterative cleavage at the PAM site by the concerted action of transfected gRNA and Cas9. An equal amount of gRNA (350 ng) and Cas9 (350 ng) plasmid was mixed with 10 pmol (∼660 ng) of single-stranded DNA donor template and transfected into HEK293 cells in 24-well plates with Lipofectamine 2000 (Life Technologies) according to the manufacturer's protocol. Selection using antibiotics (10 μg/ml Blasticidin and 1 μg/ml Puromycin) was started 24 h post transfection and completed by 48 h. Then transfected cells were trypsinized and expanded in 6-well plates in complete Dulbecco’s modified Eagle’s medium (DMEM) media, without antibiotics. The clonal cell line expressing FLAG-tagged POLH was determined by Western blotting using an anti-FLAG Ab (Sigma Cat# F1804). The HR of FLAG on all alleles of the target gene was resolved by amplifying the genomic region containing the FLAG-tag in the clonal cell line using specific primers FP2 and RP2 (Table S1) as described (74). Finally, the integration of a FLAG-tag in the N terminus of the POLH gene in genomic DNA was further confirmed by cloning the PCR fragment in pCDNA3 followed by sequencing (UTMB Molecular Genomics Core).
Ectopic expression of Pol η with C-terminal HA tag
Full-length human Pol η (NCBI NM_006502) complementary DNA was amplified from HEK293 cells by RT-PCR using Coding DNA Sequence (CDS)-specific oligos and cloned in a modified pCDNA3 vector using BamHI and XhoI restriction sites with an in-frame C-terminal HA tag. This vector was then used for generating a stable HEK293 cell line (Pol η-HA HEK293) resistant to G418 (Corning Cat# 30-234-CR) following standard procedure (21). Stable cells were selected on 400 μg/ml G418 and maintained at a drug concentration of 50 μg/ml. Ectopic expression of Pol η was confirmed by Western blotting of whole cell extract using anti-HA Ab (Cell Signaling Technology Cat# 3724S). All oligos used for cloning are listed in Table S1.
Cell culture and treatment of cells
Human Embryonic Kidney 293 (HEK293) cells/FLAG-POLH HEK293 cells/Pol η-HA HEK293 cells were grown at 37 °C and 5% CO2 in DMEM:F-12 (1:1, Cellgro) medium containing 10% fetal bovine serum (Bio-Techne), 100 units/ml penicillin, and 100 units/ml streptomycin. Cells were then either mock treated, or treated with Bleomycin (Bleo, 50 μg/ml for 1 h) in reduced serum media (OptiMEM, GIBCO) for co-IP and DNA/RNA-ChIP experiments. Similarly, the cells were mock (dimethyl sulfoxide [DMSO]) treated or treated with DNA-PKcs inhibitor, NU7441 (KU-57788) (Selleckchem.com Cat# S2638) (10 μM final concentration for 2 h) before Bleo treatment to induce DNA SB. All the cell lines (initial source: ATCC) were authenticated by Short Tandem Repeat (STR) analysis in the UTMB Molecular Genomics Core. To maintain the cells free from mycoplasma contamination, we routinely tested our experimental cells using the Mycoalert Mycoplasma Detection Kit (Lonza) according to the manufacturer’s protocol and cells were found to be free from mycoplasma contamination. About 90 to 100% confluent, nongrowing cells were used for making NEs for all co-IP and ChIP experiments.
XP-V (XP30RO with hTERT transfected) cells were grown at 37 °C and 5% CO2 in MEM (GIBCO) medium containing 10% fetal bovine serum (Bio-Techne), 100 units/ml penicillin, and 100 units/ml streptomycin. Control HCA2 human foreskin fibroblasts were immortalized by infecting with lentiviruses expressing hTERT shRNA. Cells were selected and immortalized HCA2-hTERT cells were grown in DMEM supplemented with 10% fetal bovine serum (Invitrogen), 1% penicillin-streptomycin (Cellgro), and 2 mM L-glutamine (Gibco) and maintained at 37 °C with 5% CO2.
To generate NSCs, a normal iPSC line KYOU-DXR0109B (201B7, ATCC) was grown in Cell Matrix basement membrane gel (Gibco) and Pluripotent Stem Cell (PSC) SFM XF/FF media (Gibco) at 37 °C and 5% CO2. Derivation of NSCs was done as described (6, 75, 76). Briefly, Essential 8 media (E8M; GIBCO; required for feeder-free iPSC culture) was replaced with neural induction medium approximately 24 h after passaging iPSCs, which were then maintained in this medium for 7 days. On day 7 of neural induction, NSCs (Passage 0 = P0) were cultured into Geltrex (Gibco)-coated 6-well plates and expanded in StemPRO neural stem cell SFM media. Neural induction efficiency was tested at P3 by immunofluorescence staining with a pluripotent marker (Oct4) and neural lineage stem cell marker (Nestin) as described (75).
Coimmunoprecipitation
FLAG-POLH/Pol η-HA HEK293/HEK293 cells were mock/Bleo treated and harvested immediately. NEs were prepared as per standard protocols (6, 21, 77) with minor modifications. Briefly, cells were lysed in buffer A (10 mM Tris-HCl [pH 7.9], 340 mM sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM EDTA, 1 mM DTT, 0.1% Nonidet P-40 [NP-40], and 1× protease inhibitor cocktail [Roche]) and centrifuged at 3500g for 15 min. Nuclear pellets were washed with buffer A without NP-40 and then lysed in Buffer B (20 mM HEPES [pH 7.9], 3 mM EDTA, 10% glycerol, 150 mM potassium acetate, 1.5 mM MgCl2, 1 mM DTT, 0.1% NP-40, 1 mM Sodium orthovanadate, 1X protease inhibitors, and 1× phosphatase inhibitors PhosSTOP [Roche]) by homogenization. Supernatants were collected after centrifugation at 15,000g for 30 min, and DNA/RNA in the suspension was digested with 0.15 U/μl benzonase (Novagen) at 37 °C for 1 h. The samples were centrifuged at 20,000g for 30 min, and the supernatants collected as NEs and stored in aliquots at −80 °C. Co-IPs were performed using anti-FLAG affinity gels (Sigma Cat# A2220), anti-HA epitope–tagged agarose (Pierce Cat# 26181), or RNAP II (Santa Cruz Biotechnology Cat# sc 47701) or Lig IV (Santa Cruz Biotechnology Cat# sc 271299) antibodies, followed by three washes with Wash buffer (20 mM Hepes [pH 7.9], 200 mM KCl, 0.5 mM EDTA, 10% glycerol, 0.25% Triton-X-100, and 1× protease and phosphatase inhibitors [Roche]). In the case of HA co-IP, the washes were carried out in 1× Tris-buffered saline with 0.05% Tween 20 (TBST). The immunoprecipitates were eluted in the elution buffer (20 mM Tris-Cl [pH 7.5], 200 mM NaCl, 10% glycerol supplemented with 200 μg/ml FLAG peptides [Sigma Cat# F3290]) or with HA-peptides (Cat# 26184, 1 mg/ml in 1× TBS), respectively, and the eluates were tested for the presence of various interacting proteins using appropriate Abs (PNKP [Bio Bharati Life Science Cat# BB AB0105], RNAP II [pSer2, H5 Ab, Biolegend Cat# 920202], Ku70 [GeneTex Cat# GTX101820], Lig IV [GeneTex Cat# GTX108820], 53BP1 [Santa Cruz Biotechnology Cat# Sc 22760], ATXN3 [Proteintech Cat# 13-505-1 AP], APE1 [in-house Ab (51)], FLAG [Sigma]/HA [Cell Signaling Technology]).
Proximity ligation assay
FLAG-POLH HEK293 cells were cultured onto 8-well chamber slides (Millicell EZ slides, Millipore), fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, and permeabilized with 0.2% Tween 20 in phosphate buffered saline (PBST) for 20 min at room temperature. In situ protein–protein association in close proximity (∼16 nm) was analyzed using a proximity ligation assay (Duolink) kit as described (6, 76). Anti-mouse FLAG Ab [Sigma] was used against anti-rabbit PNKP (BioBharati) and RNAPII (Sigma Cat# PLA0127) Abs. Images were analyzed in an AXIO Observer inverted microscope (Carl Zeiss). Nuclei were counterstained with DAPI.
Immunoblotting
The proteins in the NEs/whole cell extracts were separated onto a Bio-Rad 4 to 20% gradient Bis-Tris gel, then electrotransferred on a nitrocellulose (0.45 μm pore size; GE Healthcare) membrane using 1× Bio-Rad transfer buffer. The membranes were blocked with 5% w/v skimmed milk in TBST buffer (1× Tris-Buffered Saline, 0.1% Tween 20), then immunoblotted with appropriate antibodies (Pol η [Cell Signaling Technology Cat# 13848S], PNKP, γ-H2AX [ser phos 139 residue; Cell Signaling Technology Cat# 9718S], unmodified H2AX [Cell Signaling Technology Cat# 2595S], p53BP1 [S1778, Cell Signaling Technology Cat# 2675S], unmodified 53BP1 [Santa Cruz Biotechnology Cat# sc 517281], pATM [Santa Cruz Biotechnology Cat# sc 47739], unmodified ATM [Santa Cruz Biotechnology Cat# sc 377293], HDAC2 [GeneTex Cat# 109642]). The membranes were extensively washed with 1% TBST followed by incubation with anti-isotype secondary antibody (Cell Signaling Technology) conjugated with horseradish peroxidase in 5% skimmed milk at room temperature. Subsequently, the membranes were further washed three times (10 min each) in 1% TBST, developed, and imaged using Kwikquant image analyzer and image analysis software (ver 5.2) (Kindle Biosciences).
DNA- and RNA-ChIP
ChIP assays were performed as described (6, 21), with some modifications. Briefly, mock- or Bleo-treated FLAG-POLH cells were cross-linked in 1% formaldehyde for 10 min at room temperature and sonicated to an average DNA size of ∼300 bp in 50 mM Tris-HCl (pH 8.0), 10 mM EDTA, and 1% SDS with 1X protease inhibitor cocktail using a Qsonica sonicator. The supernatants were diluted with 15 mM Tris-HCl (pH 8.0), 1.0 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.01% SDS, and 1x protease inhibitors and incubated with FLAG-Ab overnight at 4 °C. Immunocomplexes (ICs) were captured by Magna ChIP Protein-A/G magnetic beads (Millipore) that were then washed sequentially in buffer I (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 1% Triton-X-100, and 0.1% SDS), buffer II (same as buffer I except containing 500 mM NaCl), buffer III (1% NP-40, 1% sodium deoxycholate, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA), and finally with 1× Tris-EDTA (pH 8.0) buffer. The ICs were extracted from the beads with elution buffer (1.0% SDS, and 100 mM NaHCO3) and de-cross-linked for 4 h at 65 °C, and DNA was isolated by phenol–chloroform extraction and ethanol precipitation.
For RNA-ChIP, NEs were prepared before sonication. The cells were incubated in buffer A (5 mM Hepes, pH 7.4, 85 mM KCl, 0.5% NP-40, and 1× protease inhibitor) for 10 min at 4 °C, then washed once with buffer B (buffer A minus NP-40), centrifuged at 2500g for 5 min, and the nuclear pellet was resuspended in sonication buffer (1% [w/v] SDS, 10 mM EDTA, 50 mM Tris-HCl [pH 8]). After sonication, the lysate was diluted in IP buffer (15 mM Tris-HCl [pH 8.0], 1.0 mM EDTA, 150 mM NaCl, 1% Triton X-100, 0.01% SDS, and 1× protease inhibitors) and immuno-pulldown was carried out for 4 h at 4 °C followed by capture of IC in protein A/G PLUS agarose beads overnight, and washed as described for DNA-ChIP. RNase inhibitor (50 U/ml; Roche) was added to buffers A and B, sonication and IP buffers, and 40 U/ml to each wash buffer. De-cross-linking was done for 2 h. RNA isolation was carried out in acidic phenol–chloroform followed by ethanol precipitation with GlycoBlue as a carrier. Genomic DNA was digested, and reverse transcription was performed using a PrimeScript RT Kit with gDNA Eraser (TaKaRa). All the ChIP samples were analyzed by qPCR/PCR using specific primers (Table S1).
RNase H treatment
Cells were washed with PBS, permeabilized in 2% PBST (v/v) for 10 min at room temperature, washed again with PBS, and then incubated with 150 U/ml RNase H (TaKaRa) in 1X buffer (40 mM Tris-HCl [pH 7.7], 4 mM MgCl2, 1 mM DTT, 4% Glycerol) for 20 min at 37 °C. The cells were then harvested and washed twice with PBS, then cross-linked with 1% formaldehyde in PBS for 10 min, and lysates prepared for ChIP assays.
Gene knockdown by siRNA transfection
Depletion of Pol η (Sigma and Santa Cruz Biotechnology siRNAs: SASI Hs02 00352513 and sc 36289, respectively) was carried out in HEK293/NSC cells using siRNAs (80 nM final dose, individually or using a mixture of Sigma and SCBT siRNA preps; transfected twice on consecutive days) and Lipofectamine 2000 (Life Technologies). The control siRNA, used as negative control, was also purchased from Sigma (Mission universal control, SIC001). Nuclear or whole cell extracts were prepared from an aliquot of harvested cells (72 h post transfection) to check for depletion of Pol η by Western analysis with the Ab mentioned earlier. Depletion of Pol μ was carried out with siRNA from Santa Cruz Biotechnology (sc105304) using similar protocol as described, and Western analysis was carried out with GeneTex Ab (GTX116332).
Similar depletion of Pol η, RAD51 (siRNA sequence adapted from (78), custom made from Sigma), or RAD52 (QIAGEN, Cat# GS5893 (79)) was carried out in the HEK293-pcmv-lacZ cell line.
Detection of R-loop by slot blot assay
HEK293 cells were grown in a 6-well plate, and depletion of Pol η was carried out using siRNA as described above. Cell pellets for the slot blot assay were resuspended with 1 ml of PBS supplemented with 0.5% SDS and digested with 0.5 mg of Proteinase K at 37 °C for 6 h. Cell lysates were extracted twice with phenol–chloroform–isoamyl alcohol, and the DNA was precipitated with 1 volume of isopropanol. The DNA pellets were transferred to microcentrifuge tubes and washed with 1 ml of 70% ethanol and dried at room temperature followed by rehydration in water. Nucleic acids were digested with restriction enzyme cocktail (EcoRI, HindIII, BsrGI, and XbaI from NEB) overnight at 37 °C. DNA concentration was measured, and half of the DNA samples were digested with RNase H (Thermo Fisher) at a concentration of 5 units per 10 μg DNA, and incubated at 37 °C overnight. Genomic DNAs from control siRNA and Pol η siRNA-treated cells with or without RNase H digestion were blotted (0.5, 0.25, and 0.125 μg) onto nitrocellulose membrane in duplicate as indicated. DNAs were cross-linked using UV (Stratalinker 1800) with auto cross-link program. The membrane was blocked with 5% nonfat milk in PBST for 1 h. The membrane was probed with RNA–DNA (S9.6) hybrid antibody (Kerafast Cat# ENH001) with a dilution of 1:1000 and anti-dsDNA antibody (Abcam Cat# ab215896) with a dilution of 1:3000 for 3 h followed by secondary antibody (goat anti-mouse HRP conjugated) incubation and detection of DNA by supersignal west pico (Thermo) and DNA–RNA hybrid by ECL select (Cytiva) and imaged with Varsadoc (Bio-Rad).
Long amplicon quantitative PCR
The HEK293 cells were mock or Bleo treated 72 h post transfection of Control or Pol η siRNA and harvested immediately or kept for recovery and harvested at various time points (4, 6, 8, and 12 h) after Bleo treatment. In another experiment, HEK293 cells were mock or Bleo treated 2 h post treatment with DNA-PKcs inhibitor and harvested immediately or kept for recovery and harvested at 6 h after the Bleo treatment. Finally, HEK293 cells were transfected with control or Pol η siRNA, then 72 h post siRNA transfection further mock (DMSO) or DNA-PKcs inhibitor treated for 2 h followed by mock or Bleo treatment and harvested immediately or kept for recovery and harvested at 6 h after Bleo treatment. Control and XP-V cells were also mock or Bleo treated and harvested 6 h post Bleo treatment. Genomic DNA was extracted using the Genomic tip 20/G kit (Qiagen) according to manufacturer’s protocol. This kit ensures minimal DNA oxidation during the isolation steps. The DNA was quantitated by Pico Green (Molecular Probes) in a black-bottomed 96-well plate, and gene-specific LA qPCR assays were performed as described (6, 21, 50, 62, 63) using LongAmp Taq DNA Polymerase (NEB). Three transcribed (HPRT1, POLB, and POLR2A; ∼10, ∼12, and ∼12 kb, respectively) and three nontranscribed genes (NANOG, 8.6 kb; OCT3/4, 10.1 kb; MyH2, 6 kb) were amplified using appropriate oligos (Table S1). The LA-qPCR reaction was set for all the genes under study from the same stock of diluted genomic DNA sample (10–20 ng) to avoid variations in PCR amplification. The final PCR reaction conditions were optimized at 94 °C for 30 s; (94 °C for 30 s, 58–60 °C for 30 s depending on the oligo annealing temperature, 65 °C for 10 min) for 24 cycles; 65 °C for 10 min. Since amplification of a small region is independent of DNA damage, a small DNA fragment (∼200–400 bp) from the corresponding gene(s) was also amplified for normalization of long amplicon. The amplified products were then visualized on gels using ethidium bromide, and the intensity of bands was quantitated with ImageJ software (NIH). The relative extent of DNA damage was determined by calculating the values of the intensity of bands and comparing with the calculated band intensity of a control siRNA/mock-treated sample considered as 100%.
Protein expression and purification
Wildtype recombinant His-tagged PNKP (rPNKP, N-terminal His tagged in pET28a, within NdeI BamHI sites), Pol β (rPol β, N-terminal His tagged in pET30a+, within NdeI BamHI sites), and Pol μ (CDS amplified from HEK293 cells by RT-PCR and cloned into pET22b with C-terminal His tag within NdeI XhoI sites) were purified from E. coli using protocol as described (51, 80, 81). Briefly, the expression vector containing CDS of proteins of interest were transformed into E.coli BL21(DE3) RIPL Codon-plus cells (Agilent). Log-phase culture (A600 = 0.4–0.6) of E. coli was induced with 0.5 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) at 16 °C for 16 h. After centrifugation, the cell pellets were suspended in a lysis buffer (buffer A) containing 25 mM Tris-HCl (pH 7.5), 500 mM NaCl, 10% glycerol, 1 mM ß-mercaptoethanol, 0.25% Tween 20, 5 mM imidazole, 2 mM phenylmethylsulfonyl fluoride (PMSF). After sonication (10–15 pulses at 30 amplitude for 15 s with a 45-s gap after each pulse; QSonica), the lysates were centrifuged at 20,000 rpm and the supernatant was loaded onto HisPur Cobalt Superflow Agarose (Thermo Scientific, Cat# 25228) previously equilibrated with buffer A and incubated for 2 h at 4 °C. After washing with buffer A with gradient increasing concentration of imidazole (10, 20, 30, 40 mM), the His-tagged proteins were eluted with an imidazole gradient (80–500 mM imidazole in buffer containing 25 mM Tris-HCl [pH 7.5], 300 mM NaCl, 10% glycerol, 1 mM ß-mercaptoethanol, 0.25% Tween 20). Most of the proteins eluted at 100 to 200 mM imidazole concentration.
Full-length Pol η (CDS amplified from HEK293 cells by RT-PCR and cloned in pET28a within NdeI BamHI with N-terminal His-tag sequence) was purified by affinity chromatography as described above. However, to purify the protein further, we passed the protein through HiTrap SP-HP (GE Healthcare) cation exchange column (based on its predicted isoelectric point of 8.74) via AKTA pure (GE Healthcare) using a gradient of 100 mM to 1000 mM NaCl. Equilibration and column wash steps were performed as per manufacturer protocol with 5 column volumes and a flow rate of 0.5 ml/min. The Pol η eluted within the 300 to 400 mM salt gradient.
Lig IV CDS was cloned in a modified pCDNA3 vector with in-frame C-terminal HA tag within BamHI and XhoI sites. Similarly, XRCC4 CDS with an N-terminal in-frame FLAG sequence was cloned in the same vector within EcoRI and XhoI sites. These expression vectors were transiently transfected into HEK293 cells individually using lipofectamine 2000 reagent, and proteins were purified from NEs prepared from these cells harvested 72 h post transfection using HA epitope–tagged agarose beads and M2 FLAG affinity gels, respectively (described earlier), and with HA and FLAG-peptide-based elution as per manufacturer protocols.
After elution, all proteins were dialyzed against Buffer C (25 mM Tris-HCl [pH 7.5], 350 mM NaCl, 1 mM DTT, and 50% glycerol).
In vitro repair assays using RNA as template
RNA-templated repair assays were carried out essentially as described (21) with minor modifications. RNA-templated substrate (a 51-mer RNA-oligo with stretches of four DNA nucleotides at the 5′ and 3′ ends to prevent RNase-mediated degradation [Integrated DNA Technologies]), 20 pmol, was annealed to two other complementary DNA oligos containing either 3′-P or 5′-P with a 4-nt or 10-nt gap in the middle. In another case, 3′-P-containing oligo was 5′-end-labeled with γATP and annealed as described above to produce 4- or 10-nt gapped RNA-templated substrate (Table S1). RT activity was assessed in PNKP- or Pol η– or Pol μ–depleted NEs (500 ng) compared with control NE. The reaction mixture (20 μl) contained 1 mM ATP, 50 μM unlabeled dNTPs, and 0.5 pmol [α-32P]dCTPs (the concentration of the corresponding cold dNTP was lowered to 5 μM) in a modified BER buffer containing 40 mM Hepes (pH 7.5), 50 mM KCl, 10 mM MgCl2, 0.1 mM EDTA, 5 mM DTT, 0.1 mM spermidine, 5% glycerol (6, 21) containing 50 U/ml RNase inhibitor (Roche), and the reaction mixture was then incubated for 1 h at 30 °C. For substrates containing 5′-end-labeled oligo, nonradiolabeled dCTP was used in the same concentration as other dNTPs for gap filling. The reaction products were run in 20% urea-PAGE, and the radioactive bands were detected using a Typhoon PhosphorImager (GE). DNA-templated substrates (5 pmol) with 4-nt or 10-nt gaps were used as a positive control, and total repair was assessed in the NEs from control and PNKP/Pol η/Pol μ siRNA-transfected HEK293 cells.
For inhibition of LINE1’s RT activity, HEK293 cells were mock treated or treated with 3′-Azido-3′-deoxythymidine (AZT) and 2′, 3′-dideoxyinosine (ddI) (1 μg/ml each) (13) for 48 h post siRNA transfection, and the NE was used for RNA-templated repair as described above. The reaction buffer also contained AZT and ddI in similar doses for samples with RT inhibition.
For in vitro repair by purified proteins, similar conditions were followed. The protein concentration used were as follows: rPNKP, 25 to 50 fmol; rPol η, 100 to 500 nmol; rPol β, 25 to 50 fmol; rPol μ, 50 to 100 fmol; Lig IV, 250 to 500 nmol; XRCC4, 100 to 200 nmol.
3′-Phosphatase activity of PNKP
The 3′-phosphatase activity of PNKP in the NE (125–250 ng) of control or Pol η–depleted cells was performed as described earlier (6, 51, 63, 82). Five picomoles of the radiolabeled substrate was incubated at 37 °C for 12 min in buffer A (25 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM MgCl2, 1 mM DTT, 10% glycerol, and 0.1 μg/μl acetylated bovine serum albumin). Purified recombinant PNKP (25 fmol) was used as positive control. The radioactive bands were visualized via PhosphorImager (GE Healthcare) and quantitated using ImageQuant software. The data were represented as % product (phosphate) released from the radiolabeled substrate.
In-cell DSBR plasmid assay in lacZ-expressing stable HEK293 cell line
We previously generated a stable WT lacZ-expressing HEK293 cell line and described in detail the construction of a DSB-containing plasmid for in cell repair assay (6, 21). Briefly, a silent mutation in the proline codon (CCC to CCG) was introduced between the BsaBI and BclI sites of the bacterial WT lacZ-containing plasmid pCH110, carrying both bacterial and SV40 promoters, to generate a functionally active variant form of lacZ plasmid pTV123. The necessity of variant plasmid was to distinguish it from the WT lacZ (transcribed in HEK293 stable cell line), and the transcript acted as a template for transferring the missing sequences into the gapped variant plasmid (Fig. S8A). The plasmid pTV123, isolated from E. coli dam−dcm− strain (NEB), was digested separately with the two single-cut enzymes BsaBI and BclI (NEB) bracketing the silent mutation, to produce 3′-OH ends containing DSBs in the plasmid. In another case, two complementary HPLC-purified uracil (U)-containing oligos (10 pmol each, Table S1) with the same silent mutation in the lacZ sequence were annealed and ligated to the BsaBI/BclI-digested vector. To create DSB-containing 3′-P ends, the resulting plasmids were digested with Udg and Fpg (both from NEB) as per manufacturer protocol except 2 mM EDTA was added to the reaction buffer to avoid nonspecific nuclease activity and the linear form of the plasmid was purified from the 0.8% agarose gel. Only the gel-purified linear form was used for transfection into mammalian cells to study error-free repair.
To assess the role of Pol η in RNA-templated DSBR, HEK293 lacZ-expressing stable cells were initially transfected with control or Pol η–specific siRNA and kept for 48 h. The cells (50–60% confluent) were then further transfected with 250 ng (per 60 mm dishes) of the DSB-containing plasmid (3′-OH or 3′-P ends) using Lipofectamine 2000 and kept for 16 h following transfection to allow repair. Plasmids were recovered as per standard protocol (83) and DH5α recA− lacZ− cells (NEB) were transformed with the recovered DNA as described (6, 21). The blue colonies, selected on Ampicillin (50 μg/ml)/X-gal (40 μg/ml) agar plates, were counted, and the presence of WT lacZ was confirmed by sequencing (UTMB Molecular Genomics Core). For control experiments, either RAD51 or RAD52 was depleted and DSB-containing plasmids with 3′-OH or 3′-P ends were used for transfection. In another case, HEK293 lacZ-expressing stable cells were mock (DMSO) treated or treated with DNA-PKcs inhibitor for 2 h before transfection of the DSB containing plasmid.
Statistical analysis
Two-sided unpaired Student’s t test (http://www.ruf.rice.edu/∼bioslabs/tools/stats/ttest.html) was used for analysis of statistical significance between two sets of data. Significance was evaluated at level p > 0.05 (ns), p < 0.05 (∗), p < 0.01 (∗∗), and p < 0.005 or 0.0001 (∗∗∗), as the case may be.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflict of interests involving the contents of this article.
Acknowledgments
This work was supported by National Institutes of Health Grants 2R01 NS073976 to T. H., R01HL145477 (to T. H. and Sanjiv Sur [S. S.], Division of Allergy and Clinical Immunology, Baylor College of Medicine, Houston), R01 NS088645 to M. L. H., DOD grant no. W81XWH-18-1-0743 (to S. S. and T. H.), and University of California Tobacco Related Disease Research Program (TRDRP) grant (26IR-0017 to A. H. S.), and grant (P30 ES 06676) to the NIEHS, National Institutes of Health Center Cell Biology and Molecular Genomics Core of UTMB. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We thank Prof. James E Cleaver for kindly providing us the XP-V (XP30RO) human skin fibroblast cell line. We thank Dr Sankar Mitra for critically reading the manuscript and Dr Katherine Kaus, Research Development Specialist at the University of Texas Medical Branch, for editing this manuscript.
Author contributions
T. H. conceptualization; A. C., N. T., A. I., A. H. S., K. M., J. M., M. L. H., and T. H. methodology; A. C., N. T., A. I., A. H. S., K. M., and J. M. formal analysis; A. C., N. T., A. I., A. H. S., K. M., and J. M. investigation; M. L. H. resources; A. C., T. H., N. T., and A. I. writing – original draft; A. C., N. T., A. I., A. H. S., K. M., J. M., M. L. H., and T. H. writing – review & editing; T. H. supervision.
Reviewed by members of the JBC Editorial Board. Edited by Patrick Sung
Supporting information
References
- 1.Berti M., Vindigni A. Replication stress: getting back on track. Nat. Struct. Mol. Biol. 2016;23:103–109. doi: 10.1038/nsmb.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patra A., Nagy L.D., Zhang Q., Su Y., Müller L., Guengerich F.P., et al. Kinetics, structure, and mechanism of 8-oxo-7,8-dihydro-2’-deoxyguanosine bypass by human DNA polymerase η. J. Biol. Chem. 2014;289:16867–16882. doi: 10.1074/jbc.M114.551820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trenner A., Sartori A.A. Harnessing DNA double-strand break repair for cancer treatment. Front. Oncol. 2019;9:1388. doi: 10.3389/fonc.2019.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu L.-S., Wang D-Q, Cui K, Liu D, Zhu L-Q. Emerging perspectives on DNA double-strand breaks in neurodegenerative diseases. Curr. Neuropharmacol. 2019;17:1146–1157. doi: 10.2174/1570159X17666190726115623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Provasek V.E., Mitra J., Malojirao V.H., Hegde M.L. DNA double-strand breaks as pathogenic lesions in neurological disorders. Int. J. Mol. Sci. 2022;23:4653. doi: 10.3390/ijms23094653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakraborty A., Tapryal N., Venkova T., Mitra J., Vasquez V., Sarker A.H., et al. Deficiency in classical nonhomologous end-joining-mediated repair of transcribed genes is linked to SCA3 pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 2020;117:8154–8165. doi: 10.1073/pnas.1917280117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 8.Heyer W.D., Ehmsen K.T., Liu J. Regulation of homologous recombinationin eukaryotes. Annu. Rev. Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakarougkas A., Jeggo P.A. DNA DSB repair pathway choice: an orchestrated handover mechanism. Br. J. Radiol. 2014;87:20130685. doi: 10.1259/bjr.20130685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen Y., Nandi P., Taylor M.B., Stuckey S., Bhadsavle H.P., Weiss B., Storici F. RNA-driven genetic changes in bacteria and in human cells. Mutat. Res. 2011;717:91–98. doi: 10.1016/j.mrfmmm.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 12.Francia S., Michelini F., Saxena A., Tang D., Hoon M., Anelli V., et al. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onozawa M., Zhang Z., Kim Y.J., Goldberg L., Varga T., Bergsagel P.L., et al. Repair of DNA double-strand breaks by template nucleotide sequence insertions derived from distant regions of the genome. Proc. Natl. Acad. Sci. U. S. A. 2014;111:7729–7734. doi: 10.1073/pnas.1321889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.di Fagagna F.D. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014;24:171–178. doi: 10.1016/j.tcb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Francia S., Cabrini M., Matti V., Oldani A., d'Adda di Fagagna F. DICER, DROSHA and DNA damage response RNAs are necessary for the secondary recruitment of DNA damage response factors. J. Cell Sci. 2016;129:1468–1476. doi: 10.1242/jcs.182188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Storici F., Bebenek K., Kunkel T.A., Gordenin D.A., Resnick M.A. RNA-templated DNA repair. Nature. 2007;447:338–341. doi: 10.1038/nature05720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keskin H., Shen Y., Huang F., Patel M., Yang T., Ashley K., et al. Transcript-RNA-templated DNA recombination and repair. Nature. 2014;515:436–439. doi: 10.1038/nature13682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei L., Nakajima S., Böhm S., Bernstein K.A., Shen Z., Tsang M., et al. DNA damage during the G0/G1 phase triggers RNA-templated, Cockayne syndrome B-dependent homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E3495–E3504. doi: 10.1073/pnas.1507105112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meers C., Keskin H., Storici F. DNA repair by RNA: templated, or not templated, that is the question. DNA Repair (Amst.) 2016;44:17–21. doi: 10.1016/j.dnarep.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazina O.M., Keskin H., Hanamshet K., Storici F., Mazin A.V. Rad52 inverse strand exchange drives RNA-templated DNA double-strand break repair. Mol. Cell. 2017;67:19–29.e3. doi: 10.1016/j.molcel.2017.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakraborty A., Tapryal N., Venkova T., Horikoshi N., Pandita R.K., Sarker A.H., et al. Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat. Commun. 2016;7:13049. doi: 10.1038/ncomms13049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prakash S., Johnson R.E., Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 2005;74:317–353. doi: 10.1146/annurev.biochem.74.082803.133250. [DOI] [PubMed] [Google Scholar]
- 23.Hoeijmakers J.H. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 24.Ciccia A., Elledge S.J. The DNA damage response: making it safe to play with knives. Mol. Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makridakis N.M., Reichardt J.K.V. Translesion DNA polymerases and cancer. Front. Genet. 2012;3:174. doi: 10.3389/fgene.2012.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meers C., Keskin H., Banyai G., Mazina O., Yang T., Gombolay A.L., et al. Genetic characterization of three distinct mechanisms supporting RNA-driven DNA repair and modification reveals major role of DNA polymerase ζ. Mol. Cell. 2020;79:1037–1050.e5. doi: 10.1016/j.molcel.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateos-Gomez P.A., Gong F., Nair N., Miller K.M., Lazzerini-Denchi E., Sfeir A. Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination. Nature. 2015;518:254–257. doi: 10.1038/nature14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kent T., Chandramouly G., Michael McDevitt S., Ozdemir A.Y., Pomerantz R.T. Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase θ. Nat. Struct. Mol. Biol. 2015;22:230–237. doi: 10.1038/nsmb.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyatt D.W., Feng W., Conlin M.P., Yousefzadeh M.J., Roberts S.A., Mieczkowski P., et al. Essential roles for polymerase θ-mediated end joining in the repair of chromosome breaks. Mol. Cell. 2016;63:662–673. doi: 10.1016/j.molcel.2016.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Black S.J., Ozdemir A.Y., Kashkina E., Kent T., Rusanov T., Ristic D., et al. Molecular basis of microhomology-mediated end-joining by purified full-length Polθ. Nat. Commun. 2019;10:4423. doi: 10.1038/s41467-019-12272-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandramouly G., Liao S., Rusanov T., Borisonnik N., Calbert M.L., Kent T., et al. Poltheta promotes the repair of DNA-protein crosslinks by microhomology-mediated end-joining. Cell Rep. 2021;34:108820. doi: 10.1016/j.celrep.2021.108820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chandramouly G., Zhao J., McDevitt S., Rusanov T., Hoang T., Borisonnik N., et al. Poltheta reverse transcribes RNA and promotes RNA-templated DNA repair. Sci. Adv. 2021;7:eabf1771. doi: 10.1126/sciadv.abf1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen X.S., Pomerantz R.T. DNA polymerase θ: a cancer drug target with reverse transcriptase activity. Genes (Basel) 2021;12:1146. doi: 10.3390/genes12081146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCulloch S.D., Kokoska R.J., Garg P., Burgers P.M., Kunkel T.A. The efficiency and fidelity of 8-oxo-guanine bypass by DNA polymerases δ and η. Nucleic Acids Res. 2009;37:2830–2840. doi: 10.1093/nar/gkp103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodriguez G.P., Song J.B., Crouse G.F., et al. In vivo bypass of 8-oxodG. PLoS Genet. 2013;9:e1003682. doi: 10.1371/journal.pgen.1003682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yoon J.H., Prakash L., Prakash S. Highly error-free role of DNA polymerase eta in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18219–18224. doi: 10.1073/pnas.0910121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T. 2nd Ed. ASM Press; Washington, DC: 2006. DNA Repair and Mutagenesis. [Google Scholar]
- 38.Johnson R.E., Prakash S., Prakash L. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase Poleta. Science. 1999;283:1001–1004. doi: 10.1126/science.283.5404.1001. [DOI] [PubMed] [Google Scholar]
- 39.Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., et al. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 40.Silverstein T.D., Jain R., Johnson R.E., Prakash L., Prakash S., Aggarwal A.K. Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase eta. Structure. 2010;18:1463–1470. doi: 10.1016/j.str.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Acharya N., Manohar K., Peroumal D., Khandagale P., Patel S.K., Sahu S.R., et al. Multifaceted activities of DNA polymerase η: beyond translesion DNA synthesis. Curr. Genet. 2019;65:649–656. doi: 10.1007/s00294-018-0918-5. [DOI] [PubMed] [Google Scholar]
- 42.Kawamoto T., Araki K., Sonoda E., Yamashita Y.M., Harada K., Kikuchi K., et al. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Mol. Cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 43.McIlwraith M.J., Vaisman A., Liu Y., Fanning E., Woodgate R., West S.C. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol. Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 44.Betous R., Rey L., Wang G., Pillaire M-J, Puget N., Selves J.K., et al. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA human cells. Mol. Carcinog. 2009;48:369–378. doi: 10.1002/mc.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Garcia-Exposito L., Bournique E., Bergoglio V., Bose A., Barroso-Gonzalez J., Zhang S., et al. Proteomic profiling reveals a specific role for translesion DNA polymerase eta in the alternative lengthening of telomeres. Cell Rep. 2016;17:1858–1871. doi: 10.1016/j.celrep.2016.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Su Y., Egli M., Guengerich F.P. Human DNA polymerase eta accommodates RNA for strand extension. J. Biol. Chem. 2017;292:18044–18051. doi: 10.1074/jbc.M117.809723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Su Y., Ghodke P.P., Egli M., Li L., Wang Y., Guengerich F.P. Human DNA polymerase η has reverse transcriptase activity in cellular environments. J. Biol. Chem. 2019;294:6073–6081. doi: 10.1074/jbc.RA119.007925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma X., Wang C., Zhou B., Cheng Z., Mao Z., Tang T-S, Guo C. DNA polymerase η promotes nonhomologous end joining upon etoposide exposure dependent on the scaffolding protein Kap1. J. Biol. Chem. 2022;298:101861. doi: 10.1016/j.jbc.2022.101861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gali V.K., Balint E., Serbyn N., Frittmann O., Stutz F., Unk I. Translesion synthesis DNA polymerase eta exhibits a specific RNA extension activity and a transcription-associated function. Sci. Rep. 2017;7:13055. doi: 10.1038/s41598-017-12915-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chakraborty A., Wakamiya M., Venkova-Canova T., Pandita R.K., Aguilera-Aguirre L., Sarker A.H., et al. Neil2-null mice accumulate oxidized DNA bases in the transcriptionally active sequences of the genome and are susceptible to innate inflammation. J. Biol. Chem. 2015;290:24636–24648. doi: 10.1074/jbc.M115.658146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiederhold L., Leppard J.B., Kedar P., Karimi-Busheri F., Rasouli-Nia A., Weinfeld M., et al. AP endonuclease-independent DNA base excision repair in human cells. Mol. Cell. 2004;15:209–220. doi: 10.1016/j.molcel.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 52.Das A., Wiederhold L., Leppard J.B., Kedar P., Prasad R., Wang H., et al. NEIL2-initiated, APE-independent repair of oxidized bases in DNA: evidence for a repair complex in human cells. DNA Repair (Amst.) 2006;5:1439–1448. doi: 10.1016/j.dnarep.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jilani A., Ramotar D., Slack C., Ong C., Yang X.M., Scherer S.W., Lasko D.D. Molecular cloning of the human gene, PNKP, encoding a polynucleotide kinase 3’-phosphatase and evidence for its role in repair of DNA strand breaks caused by oxidative damage. J. Biol. Chem. 1999;274:24176–24186. doi: 10.1074/jbc.274.34.24176. [DOI] [PubMed] [Google Scholar]
- 54.Karimi-Busheri F., Daly G., Robins P., Canas B., Pappin D.J., Sgouros J., et al. Molecular characterization of a human DNA kinase. J. Biol. Chem. 1999;274:24187–24194. doi: 10.1074/jbc.274.34.24187. [DOI] [PubMed] [Google Scholar]
- 55.Jiang Y., Dong Y., Luo Y., Jiang S., Meng F-L, Tan M., et al. AMPK-mediated phosphorylation on 53BP1 promotes c-NHEJ. Cell Rep. 2021;34:108713. doi: 10.1016/j.celrep.2021.108713. [DOI] [PubMed] [Google Scholar]
- 56.Schellenbauer A., Guilly M.-N., Grall R., Le Bars R., Paget V., Kortulewski T., et al. Phospho-Ku70 induced by DNA damage interacts with RNA Pol II and promotes the formation of phospho-53BP1 foci to ensure optimal cNHEJ. Nucleic Acids Res. 2021;49:11728–11745. doi: 10.1093/nar/gkab980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saldivar J.C., Cortez D., Cimprich K.A. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 2017;18:622–636. doi: 10.1038/nrm.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee J.W., Ratnakumar K., Hung K.-F., Rokunohe D., Kawasumi M. Deciphering UV-induced DNA damage responses to prevent and treat skin cancer. Photochem. Photobiol. 2020;96:478–499. doi: 10.1111/php.13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schneider L., Pellegatta S., Favaro R., Pisati F., Roncaglia P., Testa G., et al. DNA damage in mammalian neural stem cells leads to astrocytic differentiation mediated by BMP2 signaling through JAK-STAT. Stem Cell Rep. 2013;1:123–138. doi: 10.1016/j.stemcr.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thakur M., Wernick M., Collins C., Limoli C.L., Crowley E., Cleaver J.E. DNA polymerase eta undergoes alternative splicing, protects against UV sensitivity and apoptosis, and suppresses Mre11-dependent recombination. Genes Chromosomes Cancer. 2001;32:222–235. doi: 10.1002/gcc.1186. [DOI] [PubMed] [Google Scholar]
- 61.de Feraudy S., Limoli C.L., Giedzinski E., Karentz D., Marti T.M., Feeney L., Cleaver J.E., et al. Pol eta is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene. 2007;26:5713–5721. doi: 10.1038/sj.onc.1210385. [DOI] [PubMed] [Google Scholar]
- 62.Santos J.H., Meyer J.N., Mandavilli B.S., Van Houten B. Quantitative PCR-based measurement of nuclear and mitochondrial DNA damage and repair in mammalian cells. Methods Mol. Biol. 2006;314:183–199. doi: 10.1385/1-59259-973-7:183. [DOI] [PubMed] [Google Scholar]
- 63.Gao R., Chakraborty A., Geater C., Pradhan S., Gordon K.L., Snowden J., et al. Mutant huntingtin impairs PNKP and ATXN3, disrupting DNA repair and transcription. Elife. 2019;8:e42988. doi: 10.7554/eLife.42988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kurz E.U., Lees-Miller S.P. DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst.) 2004;3:889–900. doi: 10.1016/j.dnarep.2004.03.029. [DOI] [PubMed] [Google Scholar]
- 65.Matsuoka S., Ballif B.A., Smogorzewska A., McDonald E.R., 3rd, Hurov K.E., Luo J., et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 66.Shiloh Y. ATM: expanding roles as a chief guardian of genome stability. Exp. Cell Res. 2014;329:154–161. doi: 10.1016/j.yexcr.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 67.Shiloh Y., Lederman H.M. Ataxia-telangiectasia (A-T): an emerging dimension of premature ageing. Ageing Res. Rev. 2017;33:76–88. doi: 10.1016/j.arr.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 68.Waters C.A., Strande N.T., Wyatt D.W., Pryor J.M., Ramsden D.A. Nonhomologous end joining: a good solution for bad ends. DNA Repair (Amst.) 2014;17:39–51. doi: 10.1016/j.dnarep.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pryor J.M. Ribonucleotide incorporation enables repair of chromosome breaks by nonhomologous end joining. Science. 2018;361:1126–1129. doi: 10.1126/science.aat2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ray S., Breuer G., DeVeaux M., Zelterman D., Bindra R., Sweasy J.B. DNA polymerase beta participates in DNA end-joining. Nucleic Acids Res. 2018;46:242–255. doi: 10.1093/nar/gkx1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baillat D., Russell W.K., Wagner E.J. CRISPR-Cas9 mediated genetic engineering for the purification of the endogenous integrator complex from mammalian cells. Protein Expr. Purif. 2016;128:101–108. doi: 10.1016/j.pep.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 72.Haeussler M., Schönig K., Eckert H., Eschstruth A., Mianné J., Renaud J-B, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lackner D.H., Al Carré, Guzzardo P.M., Banning C., Mangena R., Henley T., et al. A generic strategy for CRISPR-Cas9-mediated gene tagging. Nat. Commun. 2015;6:10237. doi: 10.1038/ncomms10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang K.-L., Jee D., Stein C.B., Elrod N.D., Henriques T., Mascibroda L.G., et al. Integrator recruits protein phosphatase 2A to prevent pause release and facilitate transcription termination. Mol. Cell. 2020;80:345–358. doi: 10.1016/j.molcel.2020.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vasquez V., Mitra J., Hegde P.M., Pandey A., Sengupta S., Mitra S., et al. Chromatin-bound oxidized α-synuclein causes strand breaks in neuronal genomes in in vitro models of Parkinson’s disease. J. Alzheimers Dis. 2017;60(suppl. 1):S133–S150. doi: 10.3233/JAD-170342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mitra J., Guerrero E.N., Hegde P.M., Liachko N.F., Wang H., Vasquez V., Gao J., et al. Motor neuron disease-associated loss of nuclear TDP-43 is linked to DNA double-strand break repair defects. Proc. Natl. Acad. Sci. U. S. A. 2019;116:4696–4705. doi: 10.1073/pnas.1818415116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dignam J.D., Lebovitz R.M., Roeder R.G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aymard F., Bugler B., Schmidt C.K., Guillou E., Caron P., Briois S., et al. Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 2014;21:366–374. doi: 10.1038/nsmb.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang J., Oh Y.-T., Li Z., Dou J., Tang S., Wang X., et al. RAD52 adjusts repair of single-strand breaks via reducing DNA-damage-promoted XRCC1/LIG3α co-localization. Cell Rep. 2021;34:108625. doi: 10.1016/j.celrep.2020.108625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mandal S.M., Hegde M.L., Chatterjee A., Hegde P.M., Szczesny B., Banerjee D., et al. Role of human DNA glycosylase Nei-like 2 (NEIL2) and single strand break repair protein polynucleotide kinase 3’-phosphatase in maintenance of mitochondrial genome. J. Biol. Chem. 2012;287:2819–2829. doi: 10.1074/jbc.M111.272179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tapryal N., Shahabi S., Chakraborty A., Hosoki K., Wakamiya M., Sarkar G., et al. Intrapulmonary administration of purified NEIL2 abrogates NF-κB-mediated inflammation. J. Biol. Chem. 2021;296:100723. doi: 10.1016/j.jbc.2021.100723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chatterjee A., Saha S., Chakraborty A., Silva-Fernandes A., Mandal S.M., Neves-Carvalho A., et al. The role of the mammalian DNA end-processing enzyme polynucleotide kinase 3′-phosphatase in spinocerebellar ataxia type 3 pathogenesis. PLoS Genet. 2015;11:e1004749. doi: 10.1371/journal.pgen.1004749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patnaik P.K., Kulkarni S.K., Cross G.A. Autonomously replicating single-copy episomes in Trypanosoma brucei show unusual stability. EMBO J. 1993;12:2529–2538. doi: 10.1002/j.1460-2075.1993.tb05908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.