Abstract
The role of tabanids as potential transmitters of animal trypanosomiasis (AAT) has not yet been established in Cameroon. The objectives of this study were: (i) to trap and determine the species richness and abundance of tabanids, (ii) to identify circulating trypansomes in cattle and tabanids in a tsetse free area. A three year (2015 to 2017) tabanid survey in six regions of Cameroon was conducted. In Galim village, which is in a tsetse free area, both tabanids and cattle blood samples were screened by PCR for the presence of trypanosome DNA. Tabanids were diverse in Littoral (13 species) and in Adamawa (13 species), but were abundant in the Far North region (36.37 to 145.58 tabanids per trap per day (t/t/d)). In Galim, the tabanid trypanosomal DNA presence was 24.4% (95% CI: 11.25–37.53), while the bovine trypanosomal DNA presence was 4.8% (95% CI: 1.68–11.20). In this village, the Trypanosoma spp. identified in tabanids were T. theileri, T. vivax and T. evansi, while those in cattle were T. theileri and T. vivax. The control of tabanids is required to stop the mechanical spread of AAT in tsetse free areas.
Keywords: Animal trypanosomiasis, Distribution, Prevalence, Tabanids, Transmission
Graphical abstract
1. Introduction
Tabanids (Diptera, Tabanidae) are biting dipterous flies with about 4434 species and > 144 genera (Sevidzem et al., 2021). In Cameroon, tabanids have been identified in all the agro-ecozones where 92 species have been confirmed (Lendzele et al., 2017; Sevidzem et al., 2021). In the Far North region of Cameroon, tabanids are found in marshy, woodland, grassland where they have been observed biting cattle, horses, donkeys, humans, and poultry (Mamoudou et al., 2016). This broad spectrum of tabanid hosts and their ability to obtain blood meals from many different vertebrate species depicts the high risk for the potential transmission of zoonotic disease causing agents such as Loa loa (Kouam and Kamgno, 2017), Trypanosoma spp. (Baldacchino et al., 2014), Bacillus anthracis (Baldacchino et al., 2014) in regions where they are abundant (Getahun et al., 2020).
The primary consequences of biting flies such as tabanids include energy loss, reduction in food intake, stress and blood losses, resulting in the reduction of meat, milk and manure production as well as draught power which is epitizing in a consequent loss of productivity with significant economic impact (Baldacchino et al., 2014). Tabanids are often interrupted while taking a bloodmeal leading to repeated attempts which could result in mechanical transmission of dangerous diseases of medical and veterinary importance (Baldacchino et al., 2014). Mechanical transmission is defined as the transfer of a disease causing agent from an infected host to a susceptible counterpart without any development or transformation of the infectious agent inside the vector. One of the important parasitic diseases in sub-Saharan Africa (SSA) mechanically transmitted by tabanids is AAT (Mounioko et al., 2018). In Cameroon, AAT causes high mortality in cattle herds in the Adamawa plateau than accidents such as vehicle knocks, fighting, and falling from hills (Abah, 2020). Although AAT is cyclically transmitted (cyclical or biological transmission is defined as the transfer of a disease causing agent from an infected host to a susceptible counterpart with the development of the infectious agent inside the vector) by glossines (Glossina spp.), mechanical transmission could be conducted by biting flies of the genera-Hippobosca, Stomoxys, Haematopota, Tabanus, Pangonia, Chrysops (Getahun et al., 2020) where tsetse flies are absent (Wells, 1972).
AAT or ‘nagana’ is a hemo-parasitic disease caused by Trypanosoma spp. The pathogenic trypanosomes such T. congolense, T. vivax and T. brucei brucei have been reported to be circulating in animals in Cameroon (Mamoudou et al., 2015). >90% of cattle in Cameroon are at risk of trypanosomiasis (Paguem et al., 2019). For the past decades, animal trypanosomiasis control has been conducted in the major cattle rearing Northern regions by the Special Mission for Tsetse Fly Eradication (MSEG) and most often by farmers themselves. Because of the frequent use of trypanocides by farmers that often purchase them from black markets, the risk of development of trypanocide resistance by the local strains of animal trypanosomes was suspected in Cameroon. In 2005, Mamoudou et al. (2008) reported the resistance of tryanocides against trypanosomes in cattle in the Adamawa Plateau. Fifteen years later, the resistance profiles of trypanosomes to trypanocides in domestic animals was established in Yoko (Center region) (Mewamba et al., 2020). Despite the recent reports on trypanocides resistances, they are still used for the treatment of trypanosomiasis cases in Cameroon.
The epidemiological landscape of AAT in SSA is very complex due to the occurrence of several climatic conditions that favours the development of tsetse and the presence of diverse hosts and biting flies capable of transmitting trypanosomes mechanically. The successful elimination of AAT requires the possession of relevant and robust data on the presence and distribution of the disease and its vectors. Through the Pan-African Tsetse and Trypanosomiasis Eradication Campaign (PATTEC) collaboration, Cameroon will develop a national atlas that will form part of the continental atlas aiming to eradicate tsetse flies and AAT from the continent. In the Far North and the Adamawa plateau where major tsetse eradication campaigns were conducted in the past, some tsetse free areas with ongoing AAT transmission are present. Such tsetse free zones harbour huge tabanid populations and have been suggested to be responsible for the transmission of AAT-causing trypanosomes such as T. vivax (Fongho et al., 2017). Mechanical transmission of trypanosomes by tabanids have already been reported in some African countries such as Nigeria (Odeniran et al., 2020), Kenya (Getahun et al., 2020) and Gabon (Mounioko et al., 2018). Based on the potential risk of tabanids in spreading AAT beyond the tsetse belt of Cameroon especially those harbouring the major livestock production regions, there is a need to determine the abundance of tabanids across the tsetse free belt in Cameroon. There is also need to show the potential transmission risk of AAT by mechanical vectors especially tabanids in tsetse free areas. The present study aims at: (i) determing the abundance of tabanids in the different regions of Cameroon, and (ii) to screen for the presence of Trypanosoma spp. in tabanids caught in tsetse free pastoral area of Galim.
2. Materials and methods
2.1. Description of trapping sites
The study was conducted in Cameroon which is located in central Africa (Fig. 1a). The study sites in the different regions of the country were found in the five main agro-ecological zones (AEZs) (Sudan savanna, Guinee savanna, Mosaic forest, Highland plateau and Rain forest) (Fig. 1b). The characteristics of the different AEZs have already been fully described (Sevidzem et al., 2021).
Fig. 1.
location of Cameroon (a) and the key agro-ecological zones (b). Different trap types used in the study (i:Vavoua, ii: MVT, iii: Biconical, iv: Nzi) (c). Tabanid trapping sites in the tsetse free pastoral area of Galim in Ngaoundere of the Adamawa plateau (d).
2.2. Entomological study
A three year (2015 to 2017) tabanid trapping was conducted in sites of the six regions (Far North, North, Adamawa, East, Littoral and North West) of Cameroon using different trap types, including Nzi, Vavoua, Biconical, and the modified Vavoua (MVT) (n = 144 traps) (Fig. 1c). The Nzi, Vavoua and Biconical traps have already been reported to capture tabanids in Cameroon (Hiol et al., 2019). The MVT, a modified version of the Vavoua trap, designed by Sevidzem SL and tested against the Nzi trap in the tsetse free livestock breeding area of Ngaoundere in Cameroon to manage the populations of stable flies in rangelands, was found to capture more stable flies than the Nzi trap and could be used as a trap for biting and non-biting muscids (Sevidzem et al., 2019). Due to variations in topography, limited number of traps, and personnel for monitoring, the number of traps and trap types varied between sites. The geolocalisation of trap-points was conducted using a GPS handset (GPS eTrex®; Garmin (Europe) Ltd., Southampton, UK). The study regions in the five main AEZs of Cameroon, the study sites (n = 28), characteristics of each trap points, trap types, trap numbers, and trapping period are summarised in Table 1. The study was conducted within 108 days using 144 traps, resulting in trapping effort of 15, 552 traps days.
Table 1.
Summary table for the description of trapping sites, trap types, trap numbers and trapping period in the main AEZs of Cameroon.
| SN | AEZ | Region | Sites | Characteristics of sites | Trap type/number | N° of Traps | Trapping period |
|---|---|---|---|---|---|---|---|
| I | Sudan savanna | Far North | Kalang, Kainide, Diddel Tanne, Doulam and Yanga | Marshy land around livestock drinking and grazing spots, Open grass Savanna and Gallery forest | Nzi | 20 | Traps were set at 8:00 am and emptied at 6:00 pm, for three consecutive days per month |
| North | Mbele, Zone 26 and Zone 27 | Game Reserve, River banks and livestock grazing spots | Vavoua (n = 19), Biconical (n = 10, Nzi (n = 10) | 39 | Traps set at 8:00 am and emptied at 6:00 pm, for three consecutive days per month | ||
| II | Rainforest | Littoral | Abattoir, Palm oil plantation, and Game Reserve | Open forest, cattle grazing spots, humid and closed forest with tall canopy trees | Vavoua (n = 6), Biconical (n = 6), Nzi (n = 6) | 18 | Traps were set at 6:00 am and emptied at 6:00 pm, for three consecutive days per month |
| III | Guinee savanna | Adamawa | Velambai, Mbidjoro, and Vina du Sud Soukourwo |
Lake Djalingo with open grass Savanna representing cattle grazing spots, forest- Savanna mosaic, River vina banks, and gallery forest | Vavoua (n = 24), Biconical, Nzi (n = 20), MVT (n = 12) | 56 | Traps were set at 7:00 am and emptied at 6:00 pm, for three consecutive days per month |
| IV | Mosaic forest | East | SODEPA Ranch Bertoua sites: Minali, Oudou, Camp Général, and Gabong | Gallery forests, cattle corrals and river banks representing animal drinking spots | Nzi (n = 3), Vavoua (n = 3) | 6 | Traps were emptied after 24 h of exposition for three consecutive days per month |
| V | Highland plateau | North West | Bali top quarters, Saphery, Babah, Njinki, Tchaboutchou, Munam and Ntchuobo | Open grass savanna-forest mosaic representing cattle grazing spots | Biconical | 5 | Traps were set at 7:00 am and emptied at 6:00 pm, for three consecutive days per month |
SN: Site number; AEZ: Agro-ecological zone; SODEPA: livestock production and exploitation corporation.
2.3. Tabanid collection in tsetse free pastoral area of Galim
The tabanid trapping sites in the tsetse free pastoral zone of Galim were in an experimental cattle herd (this herd was put in place by German researchers of the programme onchocercoses to monitor the population dynamics of Onchocerca ochengi worms and vectors (Simulium spp.)). This village is located approximately 25 km from the town of Ngaoundere and falls within latitude 07° 11, 887′ north and longitude 013° 34, 919′ east. It is a major cattle rearing village that is adjacent to the Institute of Agricultural Research for Development (IRAD) Wakwa, the Wakwa zootechnical station, the Ngaoundere modern abattoir, and two ranches (Amao and Dawa) are found here. This village falls within the tsetse free belt of Cameroon (Abah, 2020). During the trapping period, the experimental herd (Fig. 1d) consisted of 42 Zebu Goudali heads of cattle that originated from this village and have never received trypanocide treatment since birth. Other animals found roaming around this herd were domestic animals (goats, sheep, horses, dogs) and wildlife (monkeys). A 14 days consecutive tabanid collection in November and December 2017 was conducted using one Nzi and one Vavoua set in upland and lowland areas that represented cattle grazing spots and cattle exposition sites to study biting flies bionomics (Fig. 1d). Traps were emptied every 24 h throughout the study period.
2.4. Fly identification
All specimens were conserved in ethanol (Votýpka et al., 2019) and identified using a stereo microscope (Carl Zeiss™ STEMI 2000-C). The identification of tabanid flies was conducted following published morphological identification keys (Surcouf and Ricardo, 1909; Oldroyd, 1954; Oldroyd, 1957).
2.5. Blood collection from cattle in Galim
Cattle sampling was conducted in November 2017 during fly collection. Blood was collected through the jugular vein of 42 cattle, using a vacutainer tube containing ethylene diamine tetra-acetate (EDTA) as anticoagulant. To prepare the buffy coat, micro-capillary tubes were filled with whole blood and centrifuged at 12,000 rpm for 5 min using a micro-haematocrit centrifuge (Hawksley, UK). The capillary tubes were cut with a diamond micro-haematocrit tube cutter, few millimetres below the buffy coat. Buffy coat samples were stored at −20 °C prior to DNA extraction.
2.6. Fly preparation
Trapped flies were individually placed in 2 ml eppendorf tubes containing 70% alcohol and stored at −20 °C prior to DNA extraction. The flies were individually transferred into sterile 2 ml eppendorf tubes with phosphate buffered saline (PBS) and a steel bead (5 mm) and then centrifuged at full speed for 10 min.
2.7. PCR amplification and gel electrophoresis of amplicons
Genomic DNA was extracted from the buffy coat and whole flies using the Wizard Genomic DNA Purification Kit (Promega, Germany) according to the manufacturer's instructions, and then stored at −20 °C. Generic primers were used in a nested PCR targeting kinetoplastid ITS-1 region of the trypanosome ribosomal DNA as described by Ngomtcho et al. (2017). Briefly, the first 25 μl volume reaction contained 2 μM of each outer primers (ITS1-Out forward and ITS1-Out Reverse) (Table 2) with product size between 180 and 640 bp, 0.2 mM dNTP mix, 0.5 U DreamTaq DNA polymerase (Thermo Scientific, Dreieich, Germany), 1× DreamTaq buffer, and 1 μl of extracted DNA isolates. Water and genomic DNA of T. grayi were added as negative and positive controls respectively. PCR amplification was carried out as follows: initial denaturation step at 95 °C for 60 s, followed by 30 amplification cycles with 94 °C for 60 s, 52 °C for 60 s, 72 °C for 30 s, and final extension 72 °C for 5 min. After that, the second PCR reaction was carried out with 1 μl of first 1:80 diluted PCR template under the same cycling conditions as described above, except an annealing temperature of 54 °C, and using the inner primer pairs (ITS1-Inner Forward and ITS1-Inner Reverse) (Table 2). Amplified products were subjected to electrophoresis on 2% agarose gels. The confirmation of the different Trypanosoma species was conducted by making reference to amplicons band sizes (bp) as from Ngomtcho et al. (2017) in supplementary Table 1.
Table 2.
Generic primers used for the PCR amplification.
| Primers | 5′- 3′ sequence | Sequence size | Reference |
|---|---|---|---|
| ITS1-OutR | CTT TGC TGC GTT CTT | ||
| ITS1-OutF | TGC AAT TAT TGG TCG CGC | Adams et al., 2006 | |
| ITS1-InF | TAG AGG AAG CAA AAG | (640–180) bp | |
| ITS1-InR | AAG CCA AGT CAT CCA TCG |
2.8. Determination of abundance
The abundance of trapped tabanids was defined as their apparent density (ADT), stated as the number of tabanids per trap per day (t/t/d) and calculated using the formula from Sevidzem et al. (2021) as follows:
Where:
ADT: Apparent density,
NTC: Number of tabanids captured,
NT: Number of traps,
NTD: Number of trapping days.
3. Results
3.1. Species richness and abundance of tabanids
This study recorded 25, 280 tabanid specimens, with 25 identified species regrouped under 5 genera (Tabanus (13 species), Chrysops (5 species), Haematopota (4 species), Ancala (2 species) and Atylotus (1 species)) (Table 3). Regarding the number of species, tabanids were diverse in the Adamawa Plateau (n = 13 species) and the Littoral (n = 13 species). The abundant tabanid species was A. agrestis (35.55 to 136.64 t/t/d) that was caught in sites of the Far North region. Tabanids were abundant in the Far North region (36.37 to 145.58 t/t/d) (Table 3).
Table 3.
The ADT of tabanids captured in the different regions of Cameroon.
| Far North | North | Adamawa | East | Littoral | North West | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus/Species | S1 | S2 | S3 | S4 | S5 | S6 | S7 | S8 | S9 | S10 | S11 | S12 | S13 | S14 | S15 | S16 | S17 | S18 | S19 | S20 | S21 | S22 | S23 | S24 | S25 |
| Tabanus (13 spp.) | |||||||||||||||||||||||||
| T. taeniola | 0 | 0.82 | 9.25 | 9.25 | 6.49 | 0.44 | 0.45 | 0.40 | 0.35 | 0.50 | 0.76 | 0.36 | 0.22 | 0 | 0.08 | 48.63 | 12.02 | 3.09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. par | 0 | 0 | 0.04 | 0.04 | 0.23 | 0.15 | 0.27 | 0.23 | 0.26 | 0.2 | 0.27 | 0.06 | 0.03 | 0 | 0 | 38.63 | 7.92 | 3.48 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. socius | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.08 | 0.06 | 0 | 0 | 3.23 | 0.27 | 0.26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. latipes | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.73 | 0.24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. argenteus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.28 | 0.09 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. biggutatus | 0 | 0 | 0 | 0 | 0 | 0.21 | 0.37 | 0.26 | 0 | 0 | 0 | 0.03 | 0.08 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. gratus | 0 | 0 | 0 | 0 | 0 | 1.10 | 1.67 | 1.15 | 0.01 | 0 | 0.09 | 0.06 | 0.11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. fasciatus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.02 | 0.03 | 0.03 | 0.03 | 0.06 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. secedens | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. ruficrus | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. ricardae | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.05 | 0.07 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| T. sufis | 0 | 0 | 0 | 0 | 0 | 0.21 | 0.31 | 0.26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Tabanus sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.39 | 0.45 | 0 | 0.01 | 0.01 | 0.10 | 0.01 | 1.01 | 0.10 | 0.04 |
| Chrysops (5 spp.) | |||||||||||||||||||||||||
| C. silacea | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.65 | 0.12 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. funebris | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.19 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. dimidiata | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.56 | 0.19 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. longicornis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.53 | 0.06 | 0.35 | 0 | 0 | 0 | 0 | 3.06 | 0.12 | 0.12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| C. distinctipennis | 0 | 0 | 0 | 0 | 0 | 0.6 | 0.56 | 0.65 | 0.31 | 0.23 | 0.51 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Haematopota(4 spp.) | |||||||||||||||||||||||||
| H. decora | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.06 | 0.02 | 0.12 | 0 | 0 | 0 | 0 | 0.79 | 0.17 | 0.05 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H. negripennis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.03 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| H. pluvialis | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Haematopota sp. | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.48 | 0.08 | 0.02 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ancala (2 spp.) | |||||||||||||||||||||||||
| A. fasciata | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.15 | 0.03 | 0.19 | 0 | 0 | 0.53 | 0.17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Ancala sp. | 0 | 0 | 0.04 | 0.04 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Atylotus (1 sp.) | |||||||||||||||||||||||||
| A. agrestis | 136.64 | 35.55 | 136.25 | 70.44 | 0 | 0 | 0 | 0 | 0.02 | 0 | 0.01 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total | 136.64 | 36.37 | 145.58 | 79.77 | 6.72 | 2.71 | 3.63 | 2.95 | 1.54 | 1.06 | 2.35 | 0.65 | 0.91 | 0.09 | 0.17 | 99.96 | 21.95 | 7.07 | 0.01 | 0.01 | 0.10 | 0.01 | 1.01 | 0.10 | 0.04 |
S1 to S25: study sites where:
S1: Kainide, S2: Kalang, S3: Diddel Tanne, S4:Yanga, S5: Doulam, S6: Mbele, S7: Zone 26, S8: Zone 27, S9: Mbidjoro, S10: Velambai, S11: Galim, S12: Campe Général, S13: Oudou, S14: Gabong, S15: Minali, S16: Abattoir, S17: Palm oil plantation, S18: Game Reserve, S19: Bali Top Quarters, S20: Saphery, S21: Babah, S22: Njinki, S23: Tchaboutchou, S24: Munam, S25: Ntchuobo.
3.2. Tabanid species identified in Galim
From the study in the tsetse free zone of Galim, 46 tabanids regrouped under two genera were identified as Chrysops (deer flies), Tabanus (horse flies) and Haematopota (clegs). The apparent density of the tabanids caught was C. longicornis (1.2 t/t/d), H. decora (0.6 t/t/d), T. taeniola (0.5 t/t/d), and T. gratus (0.3 t/t/d).
3.3. Trypanosoma species identified in tabanids in Galim
In this study, 41 tabanids were tested for the presence of trypanosomes. The tabanid trypanosomal infection rate was 24.4% (95% CI: 11.25–37.53). Three Trypanosoma spp. notably T. theileri, T. vivax, and T. evansi were identified in tabanids. Chrysops longicornis was the only tabanid that harboured all the three trypanosomes and the others only harboured T. theileri (Table 4). Interestingly, a co-infection with T. theileri and T. evansi occurred in C. longicornis (Fig. 2).
Table 4.
Trypanosoma spp. identified in tabanids from tsetse free Galim.
| N(%) of tabanids infected with Trypanosoma spp. |
|||||
|---|---|---|---|---|---|
| Species | Number trapped | Number tested | T. theileri | T. vivax | T. evansi |
 C. longicornis |
21 | 21 | 2 (9.5) | 1 (4.8) | 1 (4.8) |
 H. decora |
10 | 9 | 5 (55.6) | 0 | 0 |
 T. taeniola |
9 | 7 | 1 (14.3) | 0 | 0 |
 T. gratus |
6 | 4 | 1 (25) | 0 | 0 |
Fig. 2.
Gel electrophoresis results of tabanid samples. Each lane numbers indicates are samples from different tabanid species. The red rectangle on the two bands of sample 3 shows a co-infection of C. longicornis with T. theileri and T. evansi. C + ve = control positive (T. grayi), C-ve = control negative. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.4. Trypanosomes identified in cattle sampled in Galim
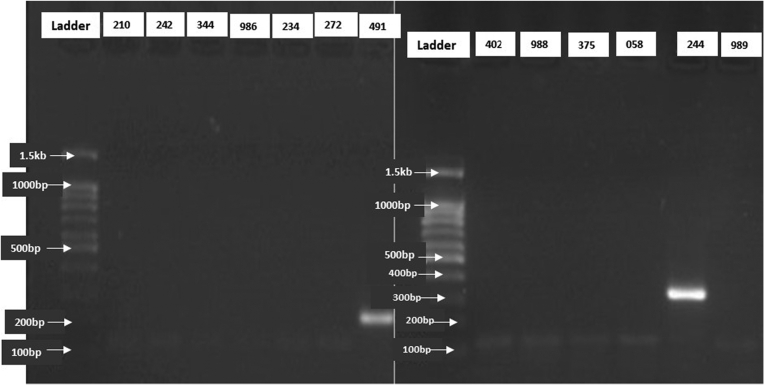
Only two animal samples were positive out of the 42 tested with overall molecular prevalence of animal trypanosomiasis of 4.8% (95% CI: 1.68–11.20). One of the positive sample was for Trypanosoma theileri (animal 244) and the other was for T. vivax (animal 491) (Fig. 3).
Fig. 3.
Gel electrophoresis showing T. vivax (animal 491) and T. theileri (animal 244). The numbers in each lane represents the code for each bovine sample. The same control positive (T. grayi) used here was that used in fig. 4.
4. Discussion
This current study builds on from the previous publication (Sevidzem et al., 2021) by looking at the trypanosome presence in this sample set of flies as well as some cattle. Tabanids are present in all the six regions of Cameroon and this could be attributed to the diverse climatic conditions and the presence of high density and diversity of vertebrate blood meal hosts as already indicated by Sevidzem et al. (2021). The species number and abundance were not the same with sites where a high species number was encountered in Adamawa/Littoral (13 species) while a high abundance was recorded in the Far North region. A high observed daily landing rate (ODLR) of tabanids on experimental cattle in the Guinee savanna of the Adamawa plateau indicated high risk for livestock health in this area (Lendzele et al., 2019). Adamawa is a major cattle rearing region of Cameroon (Mamoudou and Sevidzem, 2017) with high cattle density and forest reserves such as the Faro and Kontcha that harbours high densities of wild animals that are blood meal hosts for tabanids. Also, the sites in the Littoral region were located around the Game reserve where wild animals and trypanotolerant cattle breed (N'Dama) were present and served as bloodmeal for tabanids. According to Baldacchino et al. (2014), host density and favourable climatic conditions favours the development and survival of tabanids and was the case of Adamawa, Far North, and Littoral regions. >90% of cattle in the major cattle rearing regions (Far North, North, Adamawa, North West) (Mamoudou and Sevidzem, 2017) of Cameroon are exposed to AAT infections, reason why it is considered as one of the important diseases affecting cattle health in the country (Mamoudou et al., 2008). Because it continues to affect livestock and slow the development of the livestock industry of Cameroon, the country's MSEG is actively involved in activities to control the disease and its vectors.
The distributon map of tsetse in Cameroon shows that there exist tsetse free areas in the Northern regions (Abah, 2020). The AAT data of Cameroon in 2020 shows that the disease occurs beyond the tsetse belt (Abah, 2020). A near-worldwide Trypanosoma species distribution map by Magez et al. (2021) revealed the presence of only T. congolense, T. vivax, T. brucei brucei, and T. brucei gambiense in Cameroon, but previous molecular studies reveals the circulation of T. evansi, T. grayi, T. theileri, T. simiae in animals and tsetse (Nimpaye et al., 2011; Ngomtcho et al., 2017). AAT-causing T. vivax have been reported in cattle in the tsetse free (tabanid infested) area of the Far North region and it was suggested to be transmitted by tabanids that were observed around domestic animals (Mamoudou et al., 2016; Fongho et al., 2017; Fongho et al., 2019). The molecular entomological data obtained from the tsetse free zone of Galim showed that the DNA of T. theileri, T. vivax, T. evansi were identified in tabanids (C. longicornis, H. decora, T. taeniola and T. gratus) caught around cattle that were diagnosed positive for the DNA of T. vivax and T. theileri. This interesting finding clearly shows that T. vivax and T. theileri DNA circulates in cattle and is possibly transmitted mechanically by tabanids in this tsetse free area. Infact, T. vivax and T. theileri has been reported in cattle of Ngaoundere (Paguem et al., 2019). Furthermore, cattle from ranches in the tsetse free Ngaoundere were detected with T. vivax (Ebanga et al., 2020). Moreover, the detection of T. theileri has already been realised in field collected tabanids of Senegal (Keita et al., 2020). In the tsetse free area of kenya, T. vivax and Trypanozoon were detected in Tabanus (Getahun et al., 2020).
The current study isolated high trypanosomal DNA in tabanids than cattle. This high prevalence of trypanosomes in tabanids could be related to the fact that in the study area, other vertebrate hosts were present such as monkeys, horses, goats, dogs, sheep that could be more preferred for feeding by tabanids, hence high risk of contaminative transmission from the diverse hosts. It is important to add that the movement of animals between tsetse infested and non-infested zones could have contributed in contamination of animals with trypanosomes as already reported in Adamawa (Mamoudou et al., 2008), but the present study herd was sedentary and animals never went on transhumance so there was no possibility to move into tsetse infested zones that were > 100 km from Galim. Also, animals in the study herd were born in Galim so there was no possibility for them to have been infected elsewhere. Furthermore, the high prevalence of trypanosome DNA in tabanids could very likely have been due to either the lingering of DNA even after trypanosomes have been through the digestive tract/ not persisted and the potential for DNA contamination between flies in the same trap. However, future studies in Cameroon will include: (i) identifying trypanosome DNA in non-biting flies collected by traps, (ii) identifying blood meal source of tabanids and (iii) to collect blood from all domestic animals to compare infection rates. Moreover, the detection of AAT-causing trypanosomes in tabanids have already been reported in some neighbouring countries of Cameroon such as Nigeria (Odeniran et al., 2020) and Gabon (Mounioko et al., 2018).The importance of mechanical vectors in the epizootiology of AAT has already been established by Wells (1972). It is therefore important to include tabanids in the fly control programme of Cameroon.
5. Conclusion
A high tabanid species richness was recorded in the Adamawa and Littoral regions while a high abundance occured in the Far North region. Trypanosoma vivax and T. theileri were detected in both bovine and tabanid samples collected from the tsetse free Galim village. There is need to control tabanids in areas where AAT transmission is ongoing in the absence of tsetse flies. Future studies will include the identification of blood meal sources of tabanids as well as to collect and screen animal and tabanid samples from all the ten regions of Cameroon to identify trypanosomes.
Authors' contributions
Conceived and designed the experiments: SSL. Performed the experiments: SSL. Performed entomological survey: SSL, CN, SA, KAB. Analysed the data: SSL, AAK. Wrote and revised the paper: SSL, JFM, KAB. All authors read and approved the final manuscript.
The following are the supplementary data related to this article.
Trypanosoma species specific amplicons band sizes.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
We thank CARPA, IRAD Wakwa and MSEG for providing Nzi, Biconical and Vavoua traps. A big thanks to the Production Laitière et d'Embouche Bovine (PLEB) de Troua Belel, Cameroun, a non-governmental organisation for sponsoring the development and field testing of the Modified Vavoua Trap (MVT) in Cameroon.
References
- Abah S. Annual Scientific report of the Special Mission for Tsetse Fly Eradication (MSEG) for 2020. 2020. Tsetse control activities in Cameroon since November 2018; p. 12. [Google Scholar]
- Adams E.R., Malele I.I., Msangi A.R., Gibson W.C. Trypanosome identification in wild tsetse populations in Tanzania using generic primers to amplify the ribosomal RNA ITS1 region. Acta Trop. 2006;100:103–109. doi: 10.1016/j.actatropica.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Baldacchino F., Desquesnes M., Mihok S., Foil L.D., Duvallet G., Jittapalapong S. Tabanids: neglected subjects of research, but important vectors of disease agents! Infect. Genet. Evol. 2014;28:596–615. doi: 10.1016/j.meegid.2014.03.029. [DOI] [PubMed] [Google Scholar]
- Ebanga E.J., Akumbu A.J., Oumarou F., Abba S., Mahamat O. Animal trypanosomiasis in cattle of good body condition score in the Adamawa region of Cameroon: Haematological and immunological implications. Int. J. Trop. Dis. 2020;3:041. [Google Scholar]
- Fongho P.S., Njiokou F., Mamoudou A., Ahmadou T.M., Mouhaman A., Garabed R. Bovine trypanosomiasis in tsetse-free pastoral zone of the FarNorth region. Cameroon. J. Vector Borne. Dis. 2017;54:263–269. doi: 10.4103/0972-9062.217618. [DOI] [PubMed] [Google Scholar]
- Fongho S.P., Njiokou F., Garabed R., Mamoudou A., Arabi M., Malam A.T. Etude préliminaire sur les vecteurs mechqniques potentiels des trypanosomes animaux dans la region de l’Extreme – Nord du Cameroun. Rev. Elev. Méd. Vét. Pays Trop. 2019;72:133–136. [Google Scholar]
- Getahun M.N., Villinger J., Bargul1 J.L., Orone A., Ngiela J., Ahuya P.O., Muema J.M., Rajinder K.S., Torto B., Masiga D.K. 2020. Molecular Characterization of Pathogenic African Trypanosomes in Biting Flies and Camels in Surra-endemic Areas Outside the Tsetse Fly Belt in Kenya; pp. 1–37. [DOI] [Google Scholar]
- Hiol V., Sieumeni A.D., Mamoudou A., Sevidzem S.L., Njan-Nloga A.M., Nukenine E.N. Spatio-temporal dynamics of Glossinidae, Tabanidae and Stomoxyidae around the Douala-Edea wildlife reserve in Cameroon. American J. Entomol. 2019;3:36–42. [Google Scholar]
- Keita M.L., Medkour H., Sambou M., Dahmana H., Mediannikov O. Tabanids as possible pathogens vectors in Senegal (West Africa) Parasit. Vectors. 2020;13:500. doi: 10.1186/s13071-020-04375-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouam M.K., Kamgno J. Biological Control of Pest and Vector Insects. Intechopen Science. 2017. The African Chrysops; pp. 286–298. [DOI] [Google Scholar]
- Lendzele S.S., Abdoulmoumini M., Lydie A.Y. Spatial repartition of tabanids (Diptera: Tabanidae) in different ecological zones of North Cameroon. Biodiversity Int. J. 2017;1:64–68. [Google Scholar]
- Lendzele S.S., Eisenbarth A., Zinga K.R.C., Mavoungou J.F., Renz A. Aspects of the bionomics of hematophagous symbovine dipterans in a hyper infested rangeland of Ngaoundere (Adamawa-Cameroon) J. Asia-Pacific Entomol. 2019;22:1019–1030. [Google Scholar]
- Magez S., Pinto Torres J.E., Oh S., Radwanska M. Salivarian trypanosomes have adopted intricate host-pathogen interaction mechanisms that ensure survival in plain sight of the adaptive immune system. Pathogens. 2021;10:679. doi: 10.3390/pathogens10060679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamoudou A., Sevidzem S.L. In: Advances in Medicine and Biology Vol. 105. Berhardt Leon V., editor. Nova Science Publishers, Inc; 2017. Bovine trypanosomiasis prevalence and its major vector density in the major cattle rearing regions of Cameroon (Extreme North, North, Adamawa and North West): Past, Current and Future; p. 203. [Google Scholar]
- Mamoudou A., Delespaux V., Chepnda V., Hachimou Z., Andrikaye J.P., Zoli A., et al. Assessment of the occurrence of trypanocidal drug resistance in trypanosomes of naturally infected cattle in the Adamaoua region of Cameroon using the standard mouse test and molecular tools. Acta Trop. 2008;106:115–118. doi: 10.1016/j.actatropica.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Mamoudou A., Payne V.K., Sevidzem S.L. Hematocrit alterations and its effects in naturally infected indigenous cattle breeds due to Trypanosoma spp. on the Adamawa plateau- Cameroon. Vet. World. 2015;8:813–818. doi: 10.14202/vetworld.2015.813-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamoudou A., Mbanwei M., Fongho S.P., Sevidzem S.L., Farikou O., Rebecca G., et al. Tabanids (Diptera: Tabanidae) fauna composition in different sites and biotopes of far-north. Cameroon. J. Biol. Nature. 2016;6:146–154. [Google Scholar]
- Mewamba E.M., Oumarou F., Kamga R.M.N., Magang M.E.K., Tume C., Tiofack A.A.Z., et al. Molecular identification of diminazene aceturate-resistant strains of Trypanosoma congolense in naturally infected domestic animals of Yoko in the Centre region of Cameroon. Vet. Parasitol. Regional Stud. Rep. 2020;20 doi: 10.1016/j.vprsr.2020.100405. [DOI] [PubMed] [Google Scholar]
- Mounioko F., Maganga G.D., Mavoungou J.F., Zinga Koumba C.R., Koumba A.A., Sevidzem S.L., et al. Molecular screening of Trypanosoma spp. in Glossina, Stomoxys and Tabanids in the Moukalaba Doudou National Park (south-west, Gabon) World J. Vet. Sci. 2018;6:52–61. [Google Scholar]
- Ngomtcho S.C.H., Weber J., Ngo Bum E., Terlumu G.T., Kelm S., Achukwi M.D. Molecular screening of tsetse flies and cattle reveal different Trypanosoma species including T. grayi and T. theileri in northern Cameroon. Parasit. Vectors. 2017;10:631. doi: 10.1186/s13071-017-2540-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimpaye H., Njiokou F., Njine T., Njitchouang G.R., Cuny G., Herder S., et al. Trypanosoma vivax, T. congolense forest type and T. simiae: prevalence in domestic animals of sleeping sickness foci of Cameroon. Parasit. 2011;18:171–179. doi: 10.1051/parasite/2011182171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odeniran P.O., Onifade A.A., McLeod E.T., Ademola I.O., Alderton S., Welburn S.C. Mathematical modelling and control of African animal trypanosomiasis with interacting populations in West Africa-could biting flies be importantin maintaining the disease endemicity? PLoS One. 2020;15 doi: 10.1371/journal.pone.0242435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd H. The horse-flies of the Ethiopian region (Diptera: Tabanidae). II. Tabanzzs and related genera. Brit. Ilfus. (Nat. Hist.), London. 1954:338. [Google Scholar]
- Oldroyd H. The horse-flies of the Ethiopian region (Diptera: Tabanidae). III. Subfamilies Chrysopinae, Scepsidinae and Pangoniinae and a revised classification. Brit. Mus. (Nat. Hist.), London. 1957:496. [Google Scholar]
- Paguem A., Abanda B., Ndjonka D., Weber J.S., Ngomtcho S.C.H., Manchang K.T. Widespread co-endemicity of Trypanosoma species infecting cattle in the Sudano Sahelian and Guinea Savannah zones of Cameroon. BMC Vet. Res. 2019;15:344. doi: 10.1186/s12917-019-2111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevidzem S.L., Zinga Koumba C.R., Acapovi-Yao G.L., M’batchi B., Mavoungou J.F. Comparative efficacy of Unbaited modified Vavoua and Nzi traps in the capture of S. niger niger M. 1851 and S. calcitrans L. 1758 in Ngaoundere-Cameroon. Arch. Vet. Sci. Med. 2019;2:017–027. [Google Scholar]
- Sevidzem S.L., Koumba A.A., Yao-Acapovi G.L., Mavoungou J.F. A nationwide survey of the tabanid fauna of Cameroon. Parasit. Vectors. 2021;14:392. doi: 10.1186/s13071-021-04894-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surcouf J.M.R., Ricardo G. Masson et Cie, Editeurs; Paris: 1909. Etude monographique des Tabanides d'Afrique (groupe des “Tabanus”) p. 278. [Google Scholar]
- Votýpka J., Brzoňová J., Ježek J., Modrý D. Horse flies (Diptera: Tabanidae) of three west African countries: faunistic update, barcoding analysis and trypanosome occurrence. Acta Trop. 2019;197 doi: 10.1016/j.actatropica.2019.105069. [DOI] [PubMed] [Google Scholar]
- Wells E.A. The importance of mechanical transmission in the epidemiology of nagana: a review. Trop. Anim. Hlth. Prod. 1972;4:74–88. doi: 10.1007/BF02359739. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Trypanosoma species specific amplicons band sizes.