Abstract
Human immunodeficiency virus 1 (HIV-1) is a fast-evolving, genetically diverse virus. The HIV-1 evolution rate is also significantly influenced by the frequency of HIV-1 spread in a population. Transmission via homosexual contact has become the predominant transmission route, leading to an increase in the HIV-1 epidemic in Hebei province, China. In this study, we report three novel HIV-1 CRF01_AE/CRF07_BC recombinant forms isolated from three men who have sex with men (MSM) in the cities of Shijiazhuang (20747) and Langfang (20809 and 20820). Phylogenetic analysis based on HIV-1 near-full-length genome (NFLG) sequences indicated that the three novel recombinant forms formed a distinct monophyletic branch that was separate from all known HIV-1 subtypes and circulating recombinant forms (CRFs). Breakpoint analysis showed that the three NFLGs displayed different recombinant patterns. NFLGs 20747 and 20809 had a recombinant pattern with subtype CRF01_AE gene fragments inserted into a CRF07_BC backbone, spanning from the gag to env gene regions, whereas NFLG 20820 had a recombinant pattern with subtype CRF07_BC gene fragments inserted into a CRF01_AE backbone. Subregion phylogenetic analysis confirmed that these three NFLGs comprised CRF01_AE and CRF07_BC. Our findings confirm the emergence of novel recombinant forms and highlight the need for continuous monitoring of the diversity of HIV-1 among sexually active populations, especially MSM, to better control the HIV-1 epidemic.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00705-023-05738-1.
Keywords: HIV-1, CRF01_AE, CRF07_BC, MSM, Hebei
Since the first acquired immune deficiency syndrome (AIDS) case caused by human immunodeficiency virus (HIV) was detected in the USA in the early 1980s, HIV has infected 77.5 million people worldwide and claimed 34.7 million lives over the past four decades [1]. Among the HIV types, HIV-1 is the most dominant contributor to the global AIDS pandemic [2]. HIV-1 is a fast-evolving, genetically diverse virus that has diversified into several groups and subtypes as a result of various factors, including an error-prone polymerase, a high frequency of recombination, and selection resulting from the host immune response [3–5]. Substitution rates vary substantially both within an individual HIV-1 RNA genome and among the main subtypes and recombinants, ranging 3.5-fold from 1.34 × 10˗3 to 4.72 × 10˗3 substitutions site˗1 year˗1 in the env region and 2.3-fold from 0.95 × 10˗3 to 2.18 × 10˗3 substitutions site˗1 year˗1 in the pol region [6]. The rate of HIV-1 evolution is also significantly influenced by the frequency of HIV-1 spread in a population [7]. The diversity of HIV-1 has become a major obstacle to HIV vaccine development [7, 8], antiretroviral treatment, diagnostic testing, and viral load assays [9].
In addition to sequence variation due to nucleotide substitutions, recombination between subtypes has resulted in the generation of circulating recombinant forms (CRFs) and unique recombinant forms (URFs) [10]. Since 1990, global infections caused by HIV-1 recombinant strains have rapidly increased from 9.3% (1990–1999) to 22.8% (2010–2015), and East Asia, including China, has reported the highest HIV-1 inter-subtype recombination rate in the world, at 80.5% [2]. To date, 132 different CRFs have been published worldwide (https://www.hiv.lanl.gov/content/sequence/HIV/CRFs/crfs.comp), and these CRFs have accounted for 16.7% of all HIV-1 infections reported globally [11]. In China, the number of people living with HIV has increased from 64,170 in 2016 to 818,360 in 2020 [12]. Different from the most prevalent strains in other areas worldwide (which include simple subtype strains such as A, B, or C) [13], CRF01_AE and CRF07_BC are the predominant subtypes in China, with CRF01_AE progressing faster than CRF07_BC. The co-circulation of these two CRFs provides the opportunity for dual infections. The epidemic pattern of HIV in Hebei is consistent with that nationally in China [14]. In particular, in some cities of Hebei, several CRFs and URFs have been identified continually, such as CRF123_0107 [15] and CRF01_AE/CRF07_BC [16, 17].
In the present work, three novel HIV-1 second-generation recombinant forms derived from CRF01_AE and CRF07_BC were detected and characterized using near-full-length genome (NFLG) sequence analysis. These URFs were isolated from three MSM individuals. As shown in Table 1, HIV-1 infection in these MSM was detected via voluntary counseling and testing (VCT). Subject 20747, who resided in the city of Shijiazhuang, was an 18-year-old unmarried student with a high school education level. Subjects 20809 and 20820, who resided in the city of Langfang, were 33-year-old and 38-year-old married farmers, respectively, with a primary school education level. Their initial CD4 cell counts were more than 400 cells/mm3. They were diagnosed with HIV-1 infection based on a western blot test in 2020. The capital of Hebei province, Shijiazhuang, is the area most severely affected by HIV-1. An outbreak of HIV-1 occurred in Langfang from 1993–1995 that resulted from blood transmission. In recent years, sexual contact among MSM has become the dominant transmission route that has led to an increase in the HIV-1 epidemic in these two cities, and the dominant HIV-1 subtype was CRF01_AE, followed by CRF07_BC and subtype B [14, 18]. The current study was approved by the local ethics committee of Hebei Provincial Centers for Disease Control and Prevention. All participants signed written informed consent statements before sample collection.
Table 1.
Demographic characteristics of the three participants infected with HIV-1
| Sample ID | Age (years) | Gender | Marital status | City | Infectious route | Occupation | Educational level | Sample source | Initial CD4 count (cells/mm 3) |
|---|---|---|---|---|---|---|---|---|---|
| 20747 | 18 | Male | Unmarried | Shijiazhuang | MSM | Student | High school | VCT | 665 |
| 20809 | 33 | Male | Married | Langfang | MSM | Farmer | Primary school | VCT | 479 |
| 20820 | 38 | Male | Married | Langfang | MSM | Farmer | Primary school | VCT | 467 |
MSM, men who have sex with men; VCT, voluntary counseling and testing
Viral RNA was extracted from 200 µl of the plasma samples using MagNA Pure 2.0 and Original Reagent (Roche Diagnostic Ltd., Rotkreuz, Switzerland). HIV-1 NFLG amplification, sequencing, and sequence assembly were performed as described previously [13]. Multiple sequence comparisons were made using ClustalW and edited manually using Bio Edit 7.0 software. Standard reference sequences for HIV-1 subtypes were downloaded from the HIV databases (http://www.hiv.lanl.gov/content/index), including all full-length CRFs_01C and CRFs_0107 sequences. A phylogenetic tree and subregion trees were constructed by the neighbor-joining (N-J) method in MEGA 6.0, using the Kimura two-parameter model with 1000 bootstrap replications. Recombination breakpoints were identified using the online resources jpHMM (http://jpHMM.gobics.de/submission_hiv.html), RIP 3.0 (https://hiv.lanl.gov/contens/sequence/RIP/RIP.html), and Simplot 3.5.1. The mosaic recombinant structure was elucidated using the Recombinant HIV-1 Drawing Tool (www.hiv.lanl.gov/content/sequence/DRAWCRF/recom_mapper.html).
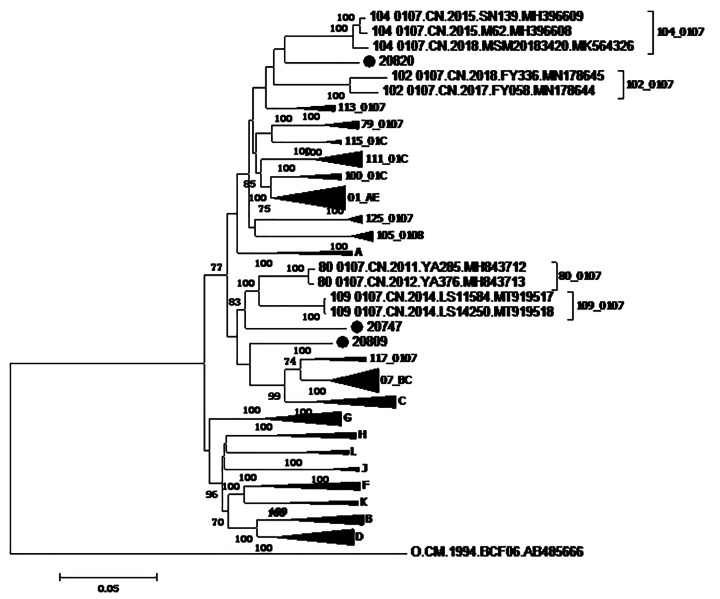
The N-J tree (Fig. 1) based on HIV-1 NFLG sequences indicated that the three NFLG sequences of interest form a distinct monophyletic branch, separate from all known HIV-1 subtypes and CRFs. According to the results obtained for the recombinant breakpoints (Supplementary Figs. S1 and S2) and the subregion trees (Supplementary Fig. S3), we deduced that the NFLGs of subjects 20747, 20809, and 20820 represented three novel recombinant forms composed of subtypes CRF01_AE and CRF07_BC. Supplementary Figure S4 shows the mosaic maps of these three NFLGs, generated using the online Recombinant HIV-1 Drawing Tool (https://www.hiv.lanl.gov./content/sequence/DRAW_CRF/recom_mapper.html). Breakpoint analysis (Supplementary Figs. S1–S4) revealed that the three NFLGs had different recombination sites. NFLGs 20747 and 20809 had a recombinant pattern with subtype CRF01_AE gene fragments inserted into a CRF07_BC backbone, spanning from the gag gene to the env gene, whereas, NFLG 20820 had a recombinant pattern with subtype CRF07_BC gene fragments inserted into a CRF01_AE backbone. Four breakpoints within NFLG 20747 were distributed in the gag, pol, and env gene regions, six breakpoints within NFLG 20809 were distributed in the pol, vif, vpr, and env gene regions, and seven breakpoints within NFLG 20820 were distributed in the pol, vif, vpr, and gag gene regions. In this study, no breakpoints were found in the nef, tat, and vpu gene regions.
Fig. 1.
Phylogenetic analysis based on HIV-1 NFLG sequences. A neighbor-joining tree was constructed using MEGA 6.0 with 1000 bootstrap replicates. The standard reference sequences of HIV-1 subtypes were downloaded from the HIV Database (http://www.hiv.lanl.gov/content/index). Bootstrap values ≥ 70% are indicated on the tree. The scale length indicates 5% nucleotide sequence divergence. Black dots indicate sequences from this study
Supplementary Fig. S1 Intersubtype recombinant analysis. Similarity distance analyses of the NFLGs 20747 (A), 20809 (B), and 20820 (C) were carried out using RIP (version 3.0; Siepel AC, Halpern AL, Macken C, Korber BT, http://hiv-web.lanl.gov). Color images are available online.
Supplementary Fig. S2 Bootscan analysis of the NFLGs 20747 (A), 20809 (B), and 20820 (C). The reference sequences of CRF01_AE, CRF07_BC, and subtype L were obtained from HIV databases. Color images are available online.
Supplementary Fig. S3 Subregion phylogenetic trees of the NFLGs 20747 (A), 20809 (B), and 20820 (C). The neighbor-joining trees were constructed using MEGA 6.0 with 1000 bootstrap replicates. Black dots indicate sequences from this study. Bootstrap values ≥ 70% are shown at the corresponding nodes. The scale bar represents 5% genetic distance.
Supplementary Fig. S4 Mosaic maps of the NFLGs 20747 (A), 20809 (B), and 20820 (C). Mosaic structures were mapped using the Recombinant HIV-1 Drawing Tool (http://www.hiv.lanl.gov/content/sequence/DRAW_CRF/recom_ mapper. html).
The NFLGs 20747, 20809, and 20820 were divided into five, seven, and eight subregions, respectively. According to their positions based on HXB2 numbering, the mosaic recombinant structures of the three NFLG sequences were described as follows: NFLG 20747: ICRF07_BC (HXB2, nt 790–1044), IICRF01_AE (HXB2, nt 1045–1169), IIICRF07_BC (HXB2, nt 1170–2690), IVCRF01_AE (HXB2, nt 2691–6316), and VCRF07_BC (HXB2, nt 6317–9411); NFLG 20809: ICRF07_BC (HXB2, nt 790–3455), IICRF01_AE (HXB2, nt 3456–4529), IIICRF07_BC (HXB2, nt 4530–4711), IVCRF01_AE (HXB2, nt 4712–5189), VCRF07_BC (HXB2, nt 5190–5754), VICRF01_AE (HXB2, nt 5755–8370), and VIICRF07_BC (HXB2, nt 8371–9411); NFLG 20820: ICRF07_BC (HXB2, nt 790–1023), IICRF01_AE (HXB2, nt 1024–1721), IIICRF07_BC (HXB2, nt 1722–2858), IVCRF01_AE (HXB2, nt 2859–3706), VCRF07_BC (HXB2, nt 3707–4731), VICRF01_AE (HXB2, nt 4732–5459), VIICRF07_BC (HXB2, nt 5460–5787), and VIIICRF01_AE (HXB2, nt 5788–9411). The parental genetic origins of all segments within NFLGs 20747 (Supplementary Fig. S3A), 20809 (Supplementary Fig. S3B), and 20820 (Supplementary Fig. S3C) were confirmed using subregion phylogenetic analysis. Supplementary Figure S3 shows that each gene segment was closely clustered with the respective reference sequence of CRF01_AE or CRF07_BC. These findings confirmed that NFLGs 20747, 20809, and 20820 represent three novel CRF01_AE/CRF07_BC recombinant forms.
In conclusion, we report three novel HIV-1 CRF01_AE/CRF07_BC recombinant forms isolated from three MSM individuals in the cities of Shijiazhuang (20747) and Langfang (20809 and 20820) in Hebei province, China. In recent years, CRF07_BC and CRF01_AE have become the most frequent subtypes in China. The co-circulation and dual infections of subtypes CRF01_AE and CRF07_BC in the sexually active population, especially among MSM, will undoubtedly provide opportunities for the generation of recombinant strains. Since the first CRF, composed of CRF07_BC and CRF01_AE (CRF79_0107), was identified in China in 2017, nine CRFs_0107 have been confirmed worldwide (https://www.hiv.lanl.gov/content/sequence/HIV/CRFs/crfs.comp). Of the nine CRFs_0107, eight were isolated from the MSM population, and one was isolated from the heterosexual population. It is predicted that novel recombinant forms derived from CRF07_BC and CRF01_AE will continue to be identified in the future, which will lead to the occurrence of new CRF_0107 variants. Our findings suggest that we should continuously monitor the diversity of HIV-1 among sexually active populations, especially among MSM, to better control the HIV-1 epidemic.
Electronic Supplementary Material
Below is the link to the electronic supplementary material
Acknowledgments
The authors thank all staff involved in this study. We also thank Liwen Bianji (Edanz) (https://www.liwenbianji.cn) for editing the language of a draft of this manuscript.
Author Contribution
XL: manuscript writing, conception of the study project, and data analysis. QL: experiment design, manuscript correction. NA: sample information collection. YZ: manuscript correction. ML: laboratory procedures, experimental operation. XZ: experimental operation, sequence assembly, and data analysis. YW: CD4 cell counts. YL: sample information collection. XT: sequence assembly, data analysis
Funding
This study was supported by the Key Science and Technology Planning Project of Hebei province (192777107D).
Data Availability
The three NFLG sequences reported in this study have been submitted to the GenBank database under accession numbers OP390172–OP390174.
Declarations
Conflict of Interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
The study was approved by the local ethics committee at Hebei Provincial Centers for Disease Control and Prevention. All of the experimental methods were implemented in accordance with the approved regulations and guidelines, and the experimental protocols were approved by the institutional review boards of Hebei Provincial Center for Disease Control and Prevention. The protocol number was IRB(S)2020-002.
Consent for publication
All participants signed written informed consent statements before sample collection. Using a blood sample for further related studies was also permitted.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.UNAIDS (2021) Global HIV & AIDS Statistics—Fact Sheet. Available online: https://www.unaids.org/en/resources/fact-sheet(accessed on 19 August 2021)
- 2.Hemelaar J, Elangovan R, Yun J, et al. Global and Regional Molecular Epidemiology of HIV-1, 1990–2015: A Systematic Review, Global Survey, and Trend Analysis. Lancet Infect Dis. 2019;19(2):143–155. doi: 10.1016/S1473-3099(18)30647-9. [DOI] [PubMed] [Google Scholar]
- 3.Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18(3):182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Leitner T. The puzzle of HIV neutral and selective evolution. Mol Biol Evol. 2018;35(6):1355–1358. doi: 10.1093/molbev/msy089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alizon S, Fraser C. Within-host and between-host evolutionary rates across the HIV-1 genome. Retrovirology. 2013;10:49. doi: 10.1186/1742-4690-10-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arshan N, Mira D, Ethan R-S, et al. Large Evolutionary Rate Heterogeneity among and within HIV-1 Subtypes and CRFs. Viruses. 2021;13(9):1689. doi: 10.3390/v13091689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdur R, Kang L, Yi F, et al. HIV-1 genetic diversity a challenge for AIDS vaccine development: a retrospective bibliometric analysis. Hum Vaccin Immunother. 2022;18(1):2014733. doi: 10.1080/21645515.2021.2014733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rashid A, Li K, Feng Y, et al. HIV-1 genetic diversity a challenge for AIDS vaccine development: a retrospective bibliometric analysis. Hum Vaccin Immunother. 2022;18(1):2014733. doi: 10.1080/21645515.2021.2014733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemelaar J, Loganathan S, Elangovan R, et al. Country level diversity of the HIV-1 pandemic between 1990 and 2015. J Virol. 2020;95(2):e01580–e01520. doi: 10.1128/JVI.01580-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robertson DL, Anderson JP, Bradac JA, et al. HIV-1 nomenclature proposal. Science. 2000;288(5463):55–56. doi: 10.1126/science.288.5463.55d. [DOI] [PubMed] [Google Scholar]
- 11.Hemelaar J, Elangovan R, Yun J, et al. Global and regional molecular epidemiology of HIV-1, 1990–2015: a systematic review, global survey, and trend analysis. Lancet Infect Dis. 2019;19(2):143–155. doi: 10.1016/S1473. [DOI] [PubMed] [Google Scholar]
- 12.Ye J, Chen J, Wang J et al (2022) CRF07_BC is associated with slow HIV disease progression in Chinese patients. Sci Rep. Mar 8;12(1):3773. 10.1038/s41598-022-07518-4 [DOI] [PMC free article] [PubMed]
- 13.Bbosa N, Kaleebu P, Ssemwanga D. HIV subtype diversity worldwide. Curr Opin HIV AIDS. 2019;14(3):153–160. doi: 10.1097/COH.0000000000000534. [DOI] [PubMed] [Google Scholar]
- 14.Lu X, Kang X, Liu Y, et al. HIV-1 molecular epidemiology among newly diagnosed HIV-1 individuals in Hebei, a low HIV prevalence province in China. PLoS ONE. 2017;12(2):e0171481. doi: 10.1371/journal.pone.0171481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xing Y, Wang L, Li Y, et al. Identification of a new HIV-1 intersubtype circulating recombinant form (CRF123_0107) in Hebei province, China. J Infect. 2022;84(3):e36–e39. doi: 10.1016/j.jinf.2022.01.017. [DOI] [PubMed] [Google Scholar]
- 16.Han L, Li H, Wang L et al (2021) Near Full-Length Genomic Characterization of Two Novel HIV-1 Unique Recombinant Forms (CRF01_AE/CRF07_BC) Among Men Who Have Sex with Men in Shijiazhuang City, Hebei Province, China. AIDS Res Hum Retroviruses 37(12): 978–984. 10.1089/AID. 2021.0100 [DOI] [PubMed]
- 17.Yang X, Zhu H, An W, et al. Genetic characterization of a novel HIV–1 CRF01_AE/ CRF07_BC recombinant form found among men who have sex with men in Baoding City, Hebei Province, China. Arch Virol. 2022;167:2395–2402. doi: 10.1007/s00705-022-05563-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang X, Liu X, Li F, et al. Epidemiological surveillance of HIV-1 transmitted drug resistance among newly diagnosed individuals in Shijiazhuang, northern China, 2014–2015. PLoS ONE. 2018;13(6):e0198005. doi: 10.1371/journal.pone.0198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The three NFLG sequences reported in this study have been submitted to the GenBank database under accession numbers OP390172–OP390174.