Abstract
The extent to which newly developed blood-based biomarkers could reduce screening costs in secondary prevention trials of Alzheimer’s disease is mostly unexplored. We collected plasma amyloid-β42/40, apolipoprotein E ε4 status and amyloid PET at baseline in 181 cognitively unimpaired participants [the age of 72.9 (5.3) years; 61.9% female; education of 11.9 (3.4) years] from the Swedish BioFINDER-1 study. We tested whether a model predicting amyloid PET status from plasma amyloid-β42/40, apolipoprotein E status and age (combined) reduced cost of recruiting amyloid PET + cognitively unimpaired participants into a theoretical trial. We found that the percentage of cognitively unimpaired participants with an amyloid PET + scan rose from 29% in an unscreened population to 64% [(49, 79); P < 0.0001] when using the biomarker model to screen for high risk for amyloid PET + status. In simulations, plasma screening also resulted in a 54% reduction of the total number of amyloid PET scans required and reduced total recruitment costs by 43% [(31, 56), P < 0.001] compared to no pre-screening when assuming a 16× PET-to-plasma cost ratio. Total savings remained significant when the PET-to-plasma cost ratio was assumed to be 8× or 4×. This suggests that a simple plasma biomarker model could lower recruitment costs in Alzheimer’s trials requiring amyloid PET positivity for inclusion.
Keywords: Alzheimer’s disease, clinical trials, plasma biomarkers, amyloid, PET
Cullen et al. demonstrated that combining plasma amyloid-β42/40 with APOE status and age could be used to effectively screen cognitively unimpaired individuals in the Swedish BioFINDER study for amyloid PET abnormality. These results add to the evidence that this panel could greatly improve recruitment for clinical trials.
Graphical Abstract
Graphical abstract.
Introduction
Alzheimer’s disease is the most common form of dementia and is expected to increase in prevalence during the coming decades due to a growing elderly population.1 A disease-modifying compound that lowers brain amyloid β (Aβ) load in mild Alzheimer’s disease recently gained regulatory approval,2 and other candidate treatments also look promising.3 However, clinical trials have been largely unsuccessful when targeting patients at later stages of Alzheimer’s disease. These failures have led to a shifting focus towards patients at earlier disease stages where cognitive symptoms are minor and Alzheimer’s disease pathology may be present but not widespread.4,5
In the future, the target population of Alzheimer’s disease therapies is likely to include elderly individuals who are at risk for Alzheimer’s disease but do not exhibit any cognitive impairment.6 Such secondary prevention trials in cognitively unimpaired (CU) individuals may still require confirmation of Aβ pathology before the start of treatment to ensure target engagement, although recent trials in the symptomatic phase of the disease suggest that tau pathology could also be useful for selecting an appropriate trial population.3 The gold standard of detecting Aβ pathology is through PET scanning or cerebrospinal fluid collection, but these modalities are likely to remain prohibitively expensive as a front-line screening tool for secondary prevention (both in trials and in clinical practice) since the prevalence of Aβ pathology is estimated to be around 25% in enriched Alzheimer’s disease study cohorts and as low as 15% in the general population (depending on age).7
Therefore, there is a need for pre-screening tools which can effectively and affordably identify CU individuals at high risk for Aβ pathology before a more invasive and expensive measurement is obtained for confirmation. Effective pre-screening biomarkers could lower overall recruitment costs by reducing the number of negative (i.e. normal) Aβ PET scans which result in exclusion from trials. Recently established plasma biomarkers of Alzheimer’s disease show great promise in filling this role as inexpensive predictors of abnormal Aβ pathology as measured by PET.5 In particular, plasma Aβ42/Aβ40—considered to directly reflect Aβ pathology—has been shown to provide significant prognostic value for Alzheimer’s disease-related outcomes in a CU population.8 Apolipoprotein E (APOE) is also the strongest genetic risk predictor for the risk of Alzheimer’s disease in healthy elderly individuals and may be included in prognostic models which are utilized for early screening of the disease.9,10
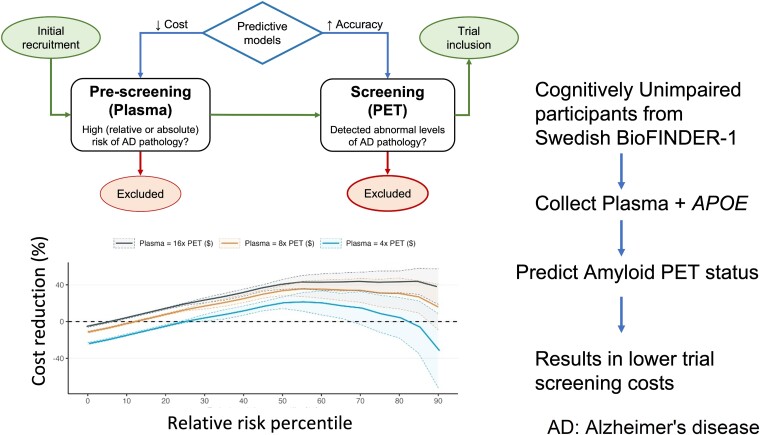
Although it is clear that these biomarkers are individually related to Aβ PET risk in a CU population, more studies are needed to fully understand how a panel of non-invasive biomarkers can be realistically operationalized for trial screening while still acknowledging the eventual necessity of Aβ PET collection. One recent paper demonstrated the efficiency of combining plasma Aβ42/Aβ40, APOE status and age to predict Aβ PET status in a mixed group of individuals with and without cognitive impairment.11 In this study, we expand on this approach to predict the risk of abnormal Aβ PET scans using this same plasma biomarker panel but focusing on a CU population and exploring a novel cost-benefit perspective throughout a variety of clinical trial designs. Validation of Aβ screening panels in CU populations is of high importance to facilitate its establishment in clinical care and secondary prevention trials. We hypothesized that this biomarker panel could reduce the cost of recruiting Aβ PET + CU participants in a simulated clinical trial. We also placed our risk prediction model into the context of a larger framework of trial recruitment in Alzheimer’s disease involving separate ‘pre-screening’ and ‘screening’ phases (see Fig. 1).
Figure 1.
A schematic workflow of clinical trial recruitment in secondary prevention trials. This figure shows a schematic workflow of clinical trial recruitment in secondary prevention trials of Alzheimer’s disease. It demonstrates how both pre-screening and screening phases of trial recruitment can benefit from clinical prediction models. It also shows how the priority of pre-screening is to have a low cost model, while the priority of screening is to have high accuracy biomarkers which can truly detect individuals who have abnormal Alzheimer’s disease pathology.
Materials and methods
Study participants
Participants from the Swedish BioFINDER-1 (Biomarkers for Identifying Neurodegenerative Disorders Early and Reliably; clinical trial no. NCT01208675, www.biofinder.se) cohort were included in the present analysis. The CU group consisted of (i) cognitively normal (CN), healthy control participants with no objective evidence of cognitive impairment at baseline and (ii) participants with subjective cognitive decline (SCD) who were referred to the memory clinic for investigation but deemed to not have any cognitive impairment after undergoing an extensive neuropsychological battery.12,13 The inclusion criteria were the following: (i) age ≥60 years for CN, 60–80 years for SCD; (ii) absence of objective cognitive impairment as assessed by a physician with a special interest in cognitive disorders; (iii) mini-mental state examination score of at least 28 for CN participants and at least 24 for SCD participants at screening visit; (iv) fluent in Swedish and (v) does not fulfil the criteria for mild cognitive impairment or dementia according to DSM-5. Exclusion criteria included (i) a significant unstable systemic illness that makes it difficult to participate in the study and (ii) current significant alcohol or substance misuse.
The study was approved by the institutional review board at Lund University and written informed consent was received from all participants. All data were collected between July 2008 and June 2019. Only participants with complete data for all variables were included in the present study.
Biomarker and genetic measurements
Plasma Aβ42/Aβ40 was measured at baseline at the Bateman laboratory at Washington University using a method which has been previously described.14 Briefly, plasma samples were spiked with 15N-Aβ40 and 15N-Aβ42 for use as analytical reference standards. Aβ42 and Aβ40 isoforms were immunoprecipitated using a monoclonal anti-Aβ mid-domain antibody (HJ5.1, anti-Aβ13-28). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) and analysis of MS data were then performed.14 Average intra-assay coefficient of variation was 0.72% and the average inter-assay coefficient of variation was 3.46%. There were no failed samples.
To facilitate easier interpretation of results, plasma Aβ42/Aβ40 levels were negated (i.e. multiplied by −1) so that higher values indicate worsening Aβ pathology. Aβ pathology in the brain was measured at baseline using 18F-flutemetamol PET conducted on a Philips Gemini TF 16 scanner. A global neocortical composite standardized uptake ratio (SUVR) was calculated for each individual using cerebellar cortex as the reference region. A pathologically abnormal (‘positive’) Aβ PET scan was defined as SUVR > 0.742 as defined previously through mixture modelling.15 Further, APOE genotype was measured at baseline and, due to a low number of APOE ε4 homozygotes in the sample, was treated as a binary variable indicating presence of at least one ε4 allele or not.
Statistical analysis
A logistic regression model was first fit with Aβ PET status [normal (‘PET-’) versus abnormal (‘PET+’)] as an outcome and with plasma Aβ42/Aβ40, APOE status and age as predictors. We tested whether using this fitted model to choose which CU participants should undergo amyloid PET scanning could reduce the cost to recruit 500 Aβ PET + CU participants into a theoretical clinical trial compared to obtaining Aβ PET scans on everyone. A value of 500 participants was selected to reflect reasonable sample sizes for recruitment in clinical trials in this population.16
We did this by comparing the percentage of Aβ PET + participants in the entire study population (representing a scenario without pre-screening) with the expected percentage of Aβ PET + participants found when using the biomarker model for pre-screening across a range of risk thresholds from 0 to 90%. This analysis was done first using a relative risk approach where an individual’s risk value was calculated relative to the entire study population, and also using an absolute risk approach where risk was represented as the absolute probability of being Aβ PET+ . In other words, relative risk is interpreted as an individual’s risk relative to others in the study population (e.g. 40% relative risk means the individual is in the 40th percentile in the population) while the absolute risk is interpreted as an individual’s risk relative in absolute terms (e.g. 40% absolute risk means the individual has a 40% probability of being Aβ PET positive). Subsequently, we calculated the total cost of recruitment at each risk threshold across a range of PET-to-plasma cost ratios (4×, 8×, 16×).
While the primary goal was to validate a pre-specified and well-established panel, a data-driven approach to variable selection was also undertaken where non-significant and non-trending variables were removed from the screening model.
All analysis was done in the R programming language (v4.0.3) and all statistical tests were two-sided with P < 0.05. All confidence intervals (CIs) and P-values were derived from 1000 trials of bootstrap sampling to extrapolate what model performance would be on new data. The performance of the logistic regression model was additionally validated using 5-fold cross-validation.
Results
Study participants
A total of 52 of 180 (28.9%) participants had abnormal Aβ PET scans at baseline and 80 of 180 (44.4%) participants had SCD. The distribution of Aβ PET positivity in the SCD group (32 of 80; 40%) compared to the CN group (20 of 100; 20%) was significantly different (P = 0.005). The average age was 73.0 ± 5.3 years, with 111 (61.7%) female participants and an average educational attainment of 11.9 ± 3.3 years. The cohort is further described in Table 1.
Table 1.
Cohort characteristics
| Level | Value | |
|---|---|---|
| n | 180 | |
| Diagnosis [n (%)] | CN | 100 (55.6) |
| SCD | 80 (44.4) | |
| Age [mean (sd)] | 73.01 (5.29) | |
| Sex [n (%)] | Male | 69 (38.3) |
| Female | 111 (61.7) | |
| Education [mean (sd)] | 11.90 (3.32) | |
| APOE ε4 alleles [n (%)] | 0 | 119 (66.1) |
| 1 | 51 (28.3) | |
| 2 | 10 (5.5) | |
| Aβ PET, SUVR [mean (sd)] | 0.73 (0.15) | |
| Aβ PET, status [n (%)] | Normal | 128 (71.1) |
| Abnormal | 52 (28.9) | |
| PACC, baseline [mean (sd)] | −0.13 (3.63) | |
| Plasma Aβ42/Aβ40 [mean (sd)] | 0.13 (0.01) | |
| Plasma P-tau217 [mean (sd)] | 0.17 (0.14) | |
| Plasma NfL [mean (sd)] | 21.33 (9.70) | |
| Plasma GFAP [mean (sd)] | 1247.75 (621.75) |
This table describes the cohort used for analysis. All continuous values are reported as mean and standard deviation while all categorical values are reported as count and percentage.
CN, cognitively normal; GFAP, glial fibrillary acidic protein; PACC, preclinical Alzheimer cognitive composite; SCD, subjective cognitive decline.
Prediction of amyloid PET status
The logistic regression model with plasma Aβ42/Aβ40, APOE status and age as predictors showed high performance in predicting Aβ PET status [area under the curve (AUC) = 0.87, 95% CI = (0.82, 0.92)] with bootstrap sampling. Moreover, evaluating model performance in a predictive context using 5-fold cross-validation resulted in an out-of-sample AUC of 0.86 [95% CI (0.85, 0.88)]. In this combined model, there was a significant effect for plasma Aβ42/Aβ40 [odds ratio (OR) = 5.98 (3.27, 12.32), P < 0.0001] and APOE status [OR = 3.67 (1.64, 8.46), P = 0.002], but not age [OR = 1.11 (0.73, 1.69), P = 0.62]. The effect of each variable on Aβ PET status is visualized in Fig. 2A, the distribution of predicted Aβ PET risk separated by true Aβ PET status is visualized in Fig. 2B, and a receiver operating characteristics (ROC) curve is shown in Fig. 2C.
Figure 2.
Ability to estimate Aβ PET status from plasma biomarkers. A logistic regression model was fit to predict Aβ PET status from age, plasma Aβ42/Aβ40 and APOE ε4 status. The effect of each variable in the model is shown for all individuals in subplot A (note that the upper line in the far-left subplot shows the effect of positive APOE status), the distribution of predicted amyloid PET risk (i.e. the kernel density estimate based on the probability density function) derived from the logistic regression model is visualized in subplot B, and a ROC curve showing results of the fitted model is visualized in subplot C. Note that plasma Aβ42/Aβ40 values have been negated to improve interpretability so that higher levels are associated with worsening risk for Aβ PET positivity.
Restricting the logistic regression analysis to CN participants led to only a slight drop in the performance of the combined model for predicting amyloid PET status [AUC = 0.84 (0.75, 0.92)], although the effect of plasma Aβ42/Aβ40 was still significant [OR = 4.21 (1.93, 11.02), P = 0.0011] and the effect of APOE status trended towards significant [OR = 1.55 (0.92, 2.64), P = 0.098].
Relative risk approach to pre-screening
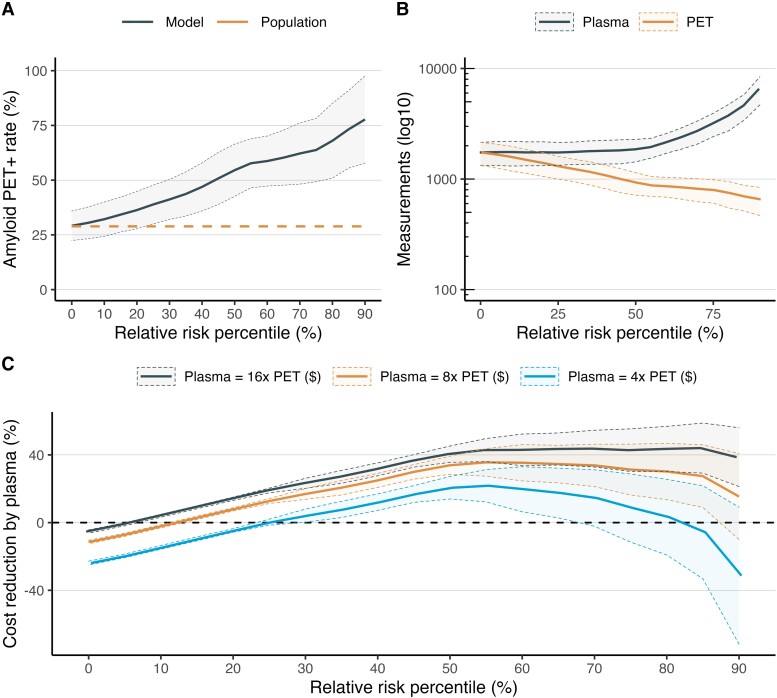
When using a relative risk approach to pre-screening where predicted Aβ PET risk was normalized based on percentiles derived from the entire population, the expected Aβ PET + rate increased from 28.9% with no pre-screening (i.e. the overall rate in the study population) to 38.7% [CI (29.8, 47.7); P < 0.0001 versus no pre-screening] with a 25th percentile risk cutoff (i.e. the 75% of individuals with highest estimated Aβ PET risk relative to others in the pre-screened population are invited for PET scanning), increased to 54.6% [CI (43.0, 66.2); P < 0.0001] with a 50th percentile risk cutoff, and increased to 63.7% [CI (48.8, 78.6); P < 0.0001] with a 75th percentile risk cutoff. These results are displayed in Fig. 3A.
Figure 3.
Pre-screening with a relative risk threshold approach. This figure shows the effect of different thresholds for relative risk (i.e. predicted probability from the logistic regression model normalized across the study population) on trial recruitment. Panel A shows the Aβ PET + rate as a function of relative risk (i.e. the lower threshold, so 0% means everyone gets a PET scan). Panel B shows the trade-off between the total number of tests (on log10 scale) needed in the pre-screening phase (i.e. plasma) versus the screening phase (i.e. PET) for a trial with 500 PET + CU participants. Panel C shows the total cost savings by pre-screening across different cost ratios. For example, a trial using the relative risk threshold of 25% would have an Aβ PET + rate of 38.6% and a threshold of 75% would have an Aβ PET + rate of 64.2%. The expected Aβ PET + rate with no pre-screening is 28.6%.
In practical terms, recruiting 500 Aβ PET + CU individuals without plasma pre-screening would therefore require an estimated 1749 Aβ PET scans to fulfil recruitment. Meanwhile, using plasma biomarkers for pre-screening with a relative risk approach would require only 1312 PET scans (∼25% reduction) with a 25th percentile risk cutoff, 926 PET scans (∼47% reduction) with a 50th percentile risk cutoff and 796 PET scans (∼54% reduction) with a 75th percentile risk cutoff. However while no plasma measurements would be required without plasma pre-screening, 1750 plasma measurements would be required with a 25th percentile risk cutoff, 1850 plasma measurements with a 50th percentile risk cutoff and 3184 plasma measurements with a 75th percentile risk cutoff. These results are displayed in Fig. 3B.
When assuming that Aβ PET scanning would be four-times (4×) as expensive as plasma biomarker measurement, there were no significant cost savings by employing pre-screening with the plasma panel and a 25th percentile risk cutoff [Δcost = −8.7% (−12.1, 29.5), P = 0.18], or a 75th percentile risk cutoff [Δcost = −0.27% CI (−1.3, + 5.6), P = 0.05], but there was a significant cost saving with a 50th percentile risk cutoff [Δcost = −20.8% CI (−26.9, −14.7), P < 0.001]. However, plasma pre-screening always resulted in significant cost savings when PET scanning was assumed to be at least 8 × as expensive as plasma measurement [Δcost = −31.3% (−46.4, −16.2), P = 0.002 with a 25th percentile risk cutoff; Δcost = −33.9% (−38.9, −28.9), P < 0.001 with a 50th percentile risk cutoff; Δcost = −12.8% (−14.3, −11.2), P < 0.001 with a 75th percentile risk cutoff]. When Aβ PET scanning was assumed to be 16 × as expensive as plasma biomarker measurement, the savings were strongly significant for plasma [Δcost = −42.7% (−55.3, −30.2), P < 0.001 with a 25th percentile risk cutoff; Δcost = −40.6% (−45.3, −35.9), P < 0.001 with a 50th percentile risk cutoff; Δcost = −18.9% (−20.4, −17.5), P < 0.001 with a 75th percentile risk cutoff]. These results are displayed graphically in Fig. 3C.
Absolute risk approach to pre-screening
With an absolute risk approach to pre-screening where an unnormalized predicted probability of Aβ PET positivity was used, the expected Aβ PET positivity rate increased from 28.9% with no plasma pre-screening (i.e. the overall rate in the study population) to 58.3% [CI (47.5, 69.2); P < 0.0001 versus no pre-screening] with a 25% risk cutoff (i.e. individuals with at least a 25% predicted probability of being Aβ PET + would be invited for PET scanning), increased to 63.8% [CI (50.5, 77.1); P < 0.0001] with a 50% risk cutoff, and increased to 80.2% [CI (58.9, 100); P < 0.0001] with a 75% risk cutoff. These results are displayed in Fig. 4A.
Figure 4.
Pre-screening with an absolute risk threshold approach. This figure shows the effect of different thresholds for absolute risk (i.e. raw predicted probability from the logistic regression model) on trial recruitment. Panel A shows the Aβ PET + rate as a function of the absolute risk (i.e. the lower threshold, so a 0% risk cutoff means everyone gets invited for a PET scan). Panel B shows the trade-off between the total number of tests (on log10 scale) needed in the pre-screening phase (i.e. plasma) versus the screening phase (i.e. PET) for a trial with 500 PET + CU participants. Panel C shows the total cost savings by pre-screening across different cost ratios. For example, a trial with absolute risk threshold of 25% would have an Aβ PET + rate of 58.0% and a threshold of 75% would have an Aβ PET + rate of 80.3%. The expected Aβ PET + rate with no pre-screening is 28.6%.
In practical terms, recruiting 500 Aβ PET + CU individuals without plasma pre-screening would require an estimated 1749 Aβ PET scans to fulfil recruitment as reported above. Meanwhile, using plasma biomarkers for pre-screening with an absolute risk approach would require only 865 PET scans with a 25% risk cutoff, 792 PET scans with a 50% risk cutoff and 637 PET scans with a 75% risk cutoff. And while no plasma measurements would be required without plasma pre-screening, 1153 plasma measurements would be required with a 25% risk cutoff, 1585 plasma measurements with a 50% risk cutoff, and 2548 plasma measurements with a 75% risk cutoff. These results are displayed in Fig. 4B.
Additionally, when assuming that Aβ PET scanning would be four-times (4×) as expensive as plasma biomarker measurement, there were significant cost savings by employing pre-screening with the plasma panel and a 25% risk cutoff [Δcost = −33.5% CI (−44.2, −22.4), P < 0.001 compared to no pre-screening], with a 50% risk cutoff [Δcost = −31.7% CI (−46.9, 16.5), P < 0.001] and with a 75% risk cutoff [Δcost = −26.6% CI (−50.9, −2.3), P = 0.028]. The same results were found when Aβ PET scanning was assumed to be 8 × and 16 × more expensive than plasma measurement. These results are displayed graphically in Fig. 4C.
Sensitivity analysis of model predictors
The primary aim of our analysis was to expand on the use of a combination of plasma Aβ42/Aβ40, APOE status and age according to previously published work.11 However, as shown above, age was not a significant predictor in our fitted model, so we conducted a sensitivity analysis using plasma Aβ42/Aβ40 and APOE status as predictors, but without age.
For pre-screening, the results found using the relative risk approach were almost identical to those found when also including age (see Supplementary Fig. 1). In terms of performance of the logistic regression model, the model with only plasma Aβ42/Aβ40 and APOE status performed similarly to the full model [AUC = 0.868, 95% CI (0.817, 0.920)]. Fitting a logistic regression model with only APOE status and age led to a significantly worse model performance than the model with plasma Aβ42/Aβ40 [AUC = 0.734, 95% CI (0.654, 0.814)].
Discussion
We demonstrated that a blood-based biomarker panel including plasma Aβ42/Aβ40, APOE status and age could be employed in the pre-screening phase of Alzheimer’s disease clinical trial recruitment to reduce future costs associated with Aβ PET screening. A sensitivity analysis also showed that plasma Aβ42/Aβ40 alone without age may even be equally as effective while removing plasma Aβ42/Aβ40 results in clearly worse performance. This suggests that the contribution of plasma biomarkers is clearly beneficial for pre-screening, while the role of age remains unsettled in the context of highly selected cohorts.
In the pre-screening phase, the ideal biomarkers are those which are non-invasive and inexpensive to collect, thereby allowing for measurement in a large number of individuals.17 For secondary prevention trials of Alzheimer’s disease, an important pre-requisite to recruitment is the identification of individuals who are at high risk of having amyloid pathology and are therefore likely to have an abnormal Aβ PET scan.
Our results demonstrated that accessible biomarkers can effectively identify high-risk and low-risk individuals and that pre-screening is cost-effective under the assumption that PET scanning is at least 4 × –8 × as expensive as plasma biomarker measurement. The expectation of PET scanning being 4 × as expensive as plasma biomarker measurement appears quite conservative given the high cost of Aβ PET scanning (considering both costs for equipment, consumables and staff) compared with the high competition for developing plasma biomarkers which may place downward price pressure.18 For example, an Aβ PET scan costs between $5000 and $8000 in clinical practice and can be higher in a research setting, while a blood test costs approximately $500–$1000.18 With the expectation of PET scanning being 16 × more expensive than plasma measurement, total costs of patient recruitment were estimated to be reduced by more than 40%. In reality, the PET-plasma cost ratio will likely vary depending on the situation and reducing the number of PET scans may also be beneficial in non-monetary ways considering the radiation and time consumption associated with PET scanning. To note, this cost-benefit analysis was done in terms of the relative cost between PET and plasma measurement to ensure generalizability of our results across clinics or locations where the absolute costs of such tests may differ greatly (e.g. between Europe and the US).
Our cost-benefit analysis suggests that the optimal pre-screening risk threshold is around 50% in terms of relative risk (i.e. individuals in the top 50% of predicted risk levels relative to those in a similar population) and 20% in terms of absolute risk (i.e. individuals with at least a 20% probability of being Aβ PET+). In the relative risk scenario, around half of the individuals who are pre-screened using plasma would be invited to receive an Aβ PET scan. This threshold increased the likelihood of Aβ PET positivity from 28.9 to 54.6%. Since almost every second scan would still be negative with this approach, these results suggest that simply maximizing positive predictive value or negative predictive value is not necessarily the optimal strategy for biomarker-based screening in a CU population. The size of the clinical trial population is also likely to play a role as larger fixed costs are more readily tolerated with larger trials, but a full exploration of this factor was outside the scope of the current analysis.
Previous efforts in the area of reducing unnecessary Aβ PET scans have considered widely available biomarker modalities such as cognition and MRI.19–21 Other studies have also investigated how Aβ risk assessment using simple cognitive tests and demographics can reduce the economic burden of PET scanning.22,23 However, our study contributes to the existing literature in its analysis of how a novel but readily available plasma biomarker reflecting amyloid pathology can be leveraged together with APOE status and Aβ PET scanning during trial recruitment to reduce overall costs of trial inclusion.24,25 Our results are also novel in the targeting of CU individuals at the earliest stages of Alzheimer’s disease pathological development, where recruitment is likely to be difficult without effective pre-screening tools. Our study built upon a previously suggested combination of plasma Aβ42/Aβ40, APOE status and age, but in a sensitivity analysis, we noted that the cost-benefit results were largely unchanged using a more sparse model with only plasma Aβ42/Aβ40 and APOE status. This may suggest that age is not a necessary component in prediction models in CU populations when efficient biochemical tests are employed. Alternatively, the lack of an effect of age in our study (in contrast to Ref. 11) could be a cohort-specific effect. Besides a more thorough evaluation of model variables, additional novel contributions of our study include an emphasize on cost-benefit calculations across a variety of inclusion thresholds and relevant clinical trial scenarios.
One important previous study did look at a similar panel of plasma Aβ42/Aβ40 and APOE by evaluating its ability to predict and screen for Aβ PET status.26 The result presented in that article was validated nicely by our model since the AUC values were similar between studies (0.84 for their study, 0.87 for our study). The main addition of our study was the use of a more sensitive and well-validated MS-based assay for measuring plasma Aβ42/Aβ40 that is already being implemented for clinical care and clinical trial recruitment.
Practically speaking, we explored two contrasting methods of applying biomarker-based algorithms in a pre-screening context. The first method is to use the relative risk for screened individuals, where the predicted risk for amyloid PET positivity for each individual is compared to all other screened individuals within a batch of biomarker samples. After biomarkers have been analysed, only a certain percentage of individuals in terms of predicted risk for amyloid positivity are carried forward to PET. The advantage of this approach is that it provides a consistent sample size for downstream screening and is likely to be more robust against the variability of assays or demographic compositions between sites. Ensuring equitable inclusion in clinical trials is a major goal given the acknowledgement that biomarker cutoffs are not necessarily applicable across different socio-economic groups.27,28 When using the relative risk approach, however, there may be a delay between plasma analysis and the determination of eligibility for PET scanning for the individual participants.
The second option would be to use the absolute risk of Aβ PET positivity for each screened individual. Here, the statistical model is used to generate a predicted Aβ PET risk score based on each individual’s biomarker data, using previously derived model specifications. Only e.g. those with >75% absolute risk of Aβ positivity are carried forward to PET. This approach would require highly standardized assays and generalizable clinical prediction models. The advantage of this approach is that it allows for greater control of positive predictive value or negative predictive value for inclusion of actually amyloid PET + participants compared to the relative risk strategy. This approach also allows for quicker differentiation of participants into those who should move on to PET scanning versus those who are not eligible but requires careful calibration and monitoring of prediction models.
Regardless of whether a relative or absolute risk approach is employed, the use of an MS-based plasma assay means that plasma samples would have to be shipped to one (or at most a few) central locations which have the expertise to measure such samples. This is a clear drawback compared to automated platforms or immunoassays where samples can be measured on site. However, to date, there does not appear to be any non-MS-based plasma Aβ assays which meet suitable diagnostic standards for Aβ PET screening.14
The strengths of our study include a well-characterized cohort with longitudinal follow-up over highly relevant timeframes for secondary prevention trials of Alzheimer’s disease.29 Our study also features the latest plasma Aβ42/Aβ40 assay which has been shown to be effective in estimating Alzheimer’s disease-related outcomes on its own and combined.30–36 This assay, along with APOE status and age, combines to make up a well-validated test which has been certified for use in predicting risk for amyloid pathology. The fact that this panel is already commercially available for clinical use and is implemented for clinical trial screening makes it important to validate in independent cohorts. Future studies may further compare this panel with models that also incorporate other promising Alzheimer’s disease biomarkers under development.
In terms of generalizability, the model was evaluated using both bootstrap sampling and cross-validation, which showed good concordance between performance on train and test samples. This indicates that the model is likely to generalize to new data, although performance would depend on assay variability and reliability.
The limitations of our analysis are a relatively small cohort in relation to the heterogeneity likely to be found in a CU elderly population, even if the size of our cohort relative to previous studies is quite large. A larger collection of CU individuals pulled from cohorts unrelated to Alzheimer’s disease could provide important information towards this end in the future.
Additionally, this study included participants who were recruited specifically as part of an Alzheimer’s disease-related study, meaning that the performance of our model may be optimistic compared to a true population screening scenario where specific expertise in plasma biomarkers may not be available. However, when we excluded SCD participants from the analysis to simulate a true population-based screening scenario, the performance of the model dropped only slightly. This indicates that our model was not overly influenced by the inclusion of SCD participants.
We restricted the analyses to a readily available plasma biomarker panel, but the addition of other biomarkers may also be relevant. The optimal biomarker model may likely depend on both the target population and outcome of interest, as previous research has shown that the utility of specific plasma biomarkers varies greatly depending on disease stage.37 Cerebrospinal fluid biomarkers could also theoretically be used to screen for Aβ PET status, as they show much stronger association than plasma biomarkers. However, they are not as accessible or inexpensive as plasma biomarkers, making their utility for Aβ PET screening limited.
Supplementary Material
Acknowledgements
We would like to acknowledge participants and relatives of the Swedish BioFINDER study.
Abbreviations
- Aβ =
Beta-amyloid protein
- APOE =
apolipoprotein E
- AUC =
area under the curve
- BioFINDER =
Biomarkers for Identifying Neurodegenerative Disorders Early and Reliably
- CI =
confidence interval
- CN =
cognitively normal
- CU =
cognitively unimpaired
- MS =
mass spectrometry
- OR =
odds ratio
- SCD =
subjective cognitive impairment
- SUVR =
standardized uptake value ratio
Contributor Information
Nicholas C Cullen, Clinical Memory Research Unit, Department of Clinical Sciences Malmö, Faculty of Medicine, Lund University, 202 13 Lund, Sweden.
Shorena Janelidze, Clinical Memory Research Unit, Department of Clinical Sciences Malmö, Faculty of Medicine, Lund University, 202 13 Lund, Sweden.
Erik Stomrud, Clinical Memory Research Unit, Department of Clinical Sciences Malmö, Faculty of Medicine, Lund University, 202 13 Lund, Sweden; Memory Clinic, Skåne University Hospital, 205 02 Malmö, Sweden.
Randall J Bateman, Department of Neurology, Washington University School of Medicine, St. Louis, MO 63110, USA.
Sebastian Palmqvist, Clinical Memory Research Unit, Department of Clinical Sciences Malmö, Faculty of Medicine, Lund University, 202 13 Lund, Sweden; Memory Clinic, Skåne University Hospital, 205 02 Malmö, Sweden.
Oskar Hansson, Clinical Memory Research Unit, Department of Clinical Sciences Malmö, Faculty of Medicine, Lund University, 202 13 Lund, Sweden; Memory Clinic, Skåne University Hospital, 205 02 Malmö, Sweden.
Niklas Mattsson-Carlgren, Clinical Memory Research Unit, Department of Clinical Sciences Malmö, Faculty of Medicine, Lund University, 202 13 Lund, Sweden; Department of Neurology, Skåne University Hospital, 221 85 Lund, Sweden; Wallenberg Center for Molecular Medicine, Lund University, 221 84 Lund, Sweden.
Supplementary material
Supplementary material is available at Brain Communications online.
Funding
Work at the authors’ research centre was supported by the Swedish Research Council (2016-00906, 2018-02052 and 2021-02219), the Knut and Alice Wallenberg foundation (2017-0383 and WCMM), the Marianne and Marcus Wallenberg foundation (2015.0125), the Strategic Research Area MultiPark (Multidisciplinary Research in Parkinson’s disease) at Lund University, the Swedish Alzheimer Foundation (AF-939932 and AF-940046), the Swedish Brain Foundation (FO2020-0271, FO2021-0293 and FO2020-0275), The Parkinson foundation of Sweden (1280/20), the Skåne University Hospital Foundation (2020-O000028), Regionalt Forskningsstöd (2020-0314 and 2020-0383), the Swedish federal government under the ALF agreement (2018-Projekt0279, 2018-Projekt0226 and 2018-Projekt0054), Bundy Academy, and Konung Gustaf V:s och Drottning Victorias Frimurarestiftelse.
H.Z. is a Wallenberg Scholar supported by grants from the Swedish Research Council (#2018-02532), the European Research Council (#681712), Swedish State Support for Clinical Research (#ALFGBG-720931), the Alzheimer Drug Discovery Foundation (ADDF), USA (#201809-2016862), the AD Strategic Fund and the Alzheimer’s Association (#ADSF-21-831376-C, #ADSF-21-831381-C and #ADSF-21-831377-C), the Olav Thon Foundation, the Erling-Persson Family Foundation, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden (#FO2019-0228), the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie grant agreement No 860197 (MIRIADE) and the UK Dementia Research Institute at UCL.
K.B. is supported by the Swedish Research Council (#2017-00915), the Alzheimer Drug Discovery Foundation (ADDF), USA (#RDAPB-201809-2016615), the Swedish Alzheimer Foundation (#AF-742881), Hjärnfonden, Sweden (#FO2017-0243), the Swedish state under the agreement between the Swedish government and the County Councils, the ALF-agreement (#ALFGBG-715986), the European Union Joint Program for Neurodegenerative Disorders (JPND2019-466-236), the National Institute of Health (NIH), USA (grant #1R01AG068398-01) and the Alzheimer’s Association 2021 Zenith Award (ZEN-21-848495).
Doses of 18F-flutemetamol injection were sponsored by GE Healthcare.
The funding sources had no role in the design and conduct of the study; in the collection, analysis, interpretation of the data; or in the preparation, review, or approval of the manuscript. Plasma measures and R.J.B. were supported by an Anonymous Foundation, and the National Institute on Aging grants NIH R56 AG061900 and RF1 AG061900 (R.J. Bateman, PI).
Competing interests
N.C., S.J., E.S., R.J.B., S.P. and N.M.C. have no disclosures. H.Z. has served at scientific advisory boards and/or as a consultant for Abbvie, Alector, Annexon, AZTherapies, CogRx, Denali, Eisai, Nervgen, Pinteon Therapeutics, Red Abbey Labs, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics and Wave, has given lectures in symposia sponsored by Cellectricon, Fujirebio, Alzecure and Biogen and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program. K.B. has served as a consultant, at advisory boards, or at data monitoring committees for Abcam, Axon, Biogen, JOMDD/Shimadzu. Julius Clinical, Lilly, MagQu, Novartis, Prothena, Roche Diagnostics, and Siemens Healthineers and is a co-founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program. S.P. has served on scientific advisory boards and/or given lectures in symposia sponsored by F. Hoffmann-La Roche, Biogen and Geras Solutions. O.H. has acquired research support (for the institution) from AVID Radiopharmaceuticals, Biogen, Eli Lilly, Eisai, GE Healthcare, Pfizer and Roche. In the past 2 years, he has received consultancy/speaker fees from Roche, Genentech, Siemens, Biogen, Alzpath and Cerveau. R.J.B. cofounded C2N Diagnostics. Washington University and R.J.B. have equity ownership interest in C2N Diagnostics and receive income based on technology (stable isotope labelling kinetics and blood plasma assay) licensed by Washington University to C2N Diagnostics. He receives income from C2N Diagnostics for serving on the scientific advisory board. Washington University, with R.J.B. as coinventor, has submitted the US nonprovisional patent application ‘Plasma Based Methods for Determining A-Beta Amyloidosis.’ R.J.B. has received honoraria as a speaker/consultant/advisory board member from Amgen and Roche, and reimbursement of travel expenses from Roche.
Data availability
Anonymized data will be shared by request from a qualified academic investigator for the sole purpose of replicating procedures and results presented in the article if data transfer is in agreement with EU legislation on the general data protection regulation and decisions by the Ethical Review Board of Sweden and Region Skåne, which should be regulated in a material transfer agreement. Code in the R programming language to reproduce the logistic regression modelling and the screening analysis is available in Supplementary Text 1.
References
- 1. Brookmeyer R, Abdalla N, Kawas CH, Corrada MM. Forecasting the prevalence of preclinical and clinical Alzheimer’s disease in the United States. Alzheimer’s Dementia. 2018;14:121–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alexander GC, Knopman DS, Emerson SS, et al. Revisiting FDA approval of aducanumab. New Engl J Med. 2021;385:769–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mintun MA, Lo AC, Evans CD, et al. Donanemab in early Alzheimer’s disease. New Engl J Med. 2021;384:1691–1704. [DOI] [PubMed] [Google Scholar]
- 4. Cummings J. Lessons learned from Alzheimer disease: Clinical trials with negative outcomes. Clin Transl Sci. 2018;11:147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hansson O. Biomarkers for neurodegenerative diseases. Nat Med. 2021;27:954–963. [DOI] [PubMed] [Google Scholar]
- 6. Kryscio RJ. Secondary prevention trials in Alzheimer disease: The challenge of identifying a meaningful end point. JAMA Neurol. 2014;71:947–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jansen WJ, Ossenkoppele R, Knol DL, et al. Prevalence of cerebral amyloid pathology in persons without dementia: A meta-analysis. JAMA. 2015;313:1924–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cullen NC, Leuzy A, Janelidze S, et al. Plasma biomarkers of Alzheimer’s disease improve prediction of cognitive decline in cognitively unimpaired elderly populations. Nat Commun. 2021;12:3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Belloy ME, Napolioni V, Greicius MD. A quarter century of APOE and Alzheimer’s disease: Progress to date and the path forward. Neuron. 2019;101:820–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. O’Donoghue MC, Murphy SE, Zamboni G, Nobre AC, Mackay CE. APOE Genotype and cognition in healthy individuals at risk of Alzheimer’s disease: A review. Cortex. 2018;104:103–123. [DOI] [PubMed] [Google Scholar]
- 11. West T, Kirmess KM, Meyer MR, et al. A blood-based diagnostic test incorporating plasma Aβ42/40 ratio, ApoE proteotype, and age accurately identifies brain amyloid status: Findings from a multi cohort validity analysis. Mol Neurodegener. 2021;16:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Petrazzuoli F, Vestberg S, Midlöv P, Thulesius H, Stomrud E, Palmqvist S. Brief cognitive tests used in primary care cannot accurately differentiate mild cognitive impairment from subjective cognitive decline. J Alzheimer’s Dis. 2020;75:1191–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mattsson N, Insel PS, Palmqvist S, et al. Increased amyloidogenic APP processing in APOE ɛ4-negative individuals with cerebral β-amyloidosis. Nat Commun. 2016;7:10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Janelidze S, Teunissen CE, Zetterberg H, et al. Head-to-head comparison of 8 plasma amyloid-β 42/40 assays in Alzheimer disease. JAMA Neurol. 2021;78:1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mattsson N, Palmqvist S, Stomrud E, Vogel J, Hansson O. Staging β-amyloid pathology with amyloid positron emission tomography. JAMA Neurol. 2019;76:1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cullen NC, Zetterberg H, Insel PS, et al. Comparing progression biomarkers in clinical trials of early Alzheimer’s disease. Ann Clin Transl Neur. 2020;7:1661–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Visser PJ, Verhey FRJ, Boada M, et al. Development of screening guidelines and clinical criteria for predementia Alzheimer’s disease. Neuroepidemiology. 2008;30:254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Caselli RJ, Woodruff BK. Clinical impact of amyloid positron emission tomography—Is it worth the cost? JAMA Neurol. 2016;73:1396. [DOI] [PubMed] [Google Scholar]
- 19. Sevigny J, Suhy J, Chiao P, et al. Amyloid PET screening for enrichment of early-stage Alzheimer disease clinical trials. Alzheimer Dis Assoc Dis. 2016;30:1–7. [DOI] [PubMed] [Google Scholar]
- 20. Casamitjana A, Petrone P, Tucholka A, et al. MRI-based screening of preclinical Alzheimer’s disease for prevention clinical trials. J Alzheimer’s Dis. 2018;Preprint:1–14. [DOI] [PubMed] [Google Scholar]
- 21. Palmqvist S, Janelidze S, Stomrud E, et al. Performance of fully automated plasma assays as screening tests for Alzheimer disease–related β-amyloid status. JAMA Neurol. 2019;76:1060–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Langford O, Raman R, Sperling RA, et al. Predicting amyloid burden to accelerate recruitment of secondary prevention clinical trials. J Prev Alzheimer’s Dis. 2020;7:213–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Palmqvist S, Insel PS, Zetterberg H, et al. Accurate risk estimation of β-amyloid positivity to identify prodromal Alzheimer’s disease: Cross-validation study of practical algorithms. Alzheimer’s Dementia. 2019;15:194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Veitch DP, Weiner MW, Aisen PS, et al. Understanding disease progression and improving Alzheimer’s disease clinical trials: Recent highlights from the Alzheimer’s disease neuroimaging initiative. Alzheimer’s Dementia. 2019;15:106–152. [DOI] [PubMed] [Google Scholar]
- 25. Anderson RM, Hadjichrysanthou C, Evans S, Wong MM. Why do so many clinical trials of therapies for Alzheimer’s disease fail? Lancet. 2017;390:2327–2329. [DOI] [PubMed] [Google Scholar]
- 26. Keshavan A, Pannee J, Karikari TK, et al. Population-based blood screening for preclinical Alzheimer’s disease in a British birth cohort at age 70. Brain. 2021;144:434–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McDonough IM. Beta-amyloid and cortical thickness reveal racial disparities in preclinical Alzheimer’s disease. Neuroimage Clin. 2017;16:659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Babulal GM, Quiroz YT, Albensi BC, et al. Perspectives on ethnic and racial disparities in Alzheimer’s disease and related dementias: Update and areas of immediate need. Alzheimer’s Dementia. 2019;15:292–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carrillo MC, Brashear HR, Logovinsky V, et al. Can we prevent Alzheimer’s disease? Secondary “prevention” trials in Alzheimer’s disease. Alzheimer’s Dementia. 2013;9:123–131.e1. [DOI] [PubMed] [Google Scholar]
- 30. Jack CR, Wiste HJ, Therneau TM, et al. Associations of amyloid, tau, and neurodegeneration biomarker profiles with rates of memory decline among individuals without dementia. JAMA. 2019;321:2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hampel H, O’Bryant SE, Molinuevo JL, et al. Blood-based biomarkers for Alzheimer disease: Mapping the road to the clinic. Nat Rev Neurol. 2018;14:639–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nakamura A, Kaneko N, Villemagne VL, et al. High performance plasma amyloid-β biomarkers for Alzheimer’s disease. Nature. 2018;554:249. [DOI] [PubMed] [Google Scholar]
- 33. Janelidze S, Mattsson N, Palmqvist S, et al. Plasma P-tau181 in Alzheimer’s disease: Relationship to other biomarkers, differential diagnosis, neuropathology and longitudinal progression to Alzheimer’s dementia. Nat Med. 2020;26:379–386. [DOI] [PubMed] [Google Scholar]
- 34. de Wolf F, Ghanbari M, Licher S, et al. Plasma tau, neurofilament light chain and amyloid-β levels and risk of dementia; a population-based cohort study. Brain. 2020;143:1220–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ovod V, Ramsey KN, Mawuenyega KG, et al. Amyloid β concentrations and stable isotope labeling kinetics of human plasma specific to central nervous system amyloidosis. Alzheimer’s Dementia. 2017;13:841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schindler SE, Bollinger JG, Ovod V, et al. High-precision plasma β-amyloid 42/40 predicts current and future brain amyloidosis. Neurology. 2019;93(17):e1647–e1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mattsson-Carlgren N, Janelidze S, Bateman RJ, et al. Soluble P-tau217 reflects amyloid and tau pathology and mediates the association of amyloid with tau. Embo Mol Med. 2021;13:e14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Anonymized data will be shared by request from a qualified academic investigator for the sole purpose of replicating procedures and results presented in the article if data transfer is in agreement with EU legislation on the general data protection regulation and decisions by the Ethical Review Board of Sweden and Region Skåne, which should be regulated in a material transfer agreement. Code in the R programming language to reproduce the logistic regression modelling and the screening analysis is available in Supplementary Text 1.