Abstract
Pholiota microspora (“nameko” in Japanese) is one of the most common edible mushrooms, especially in Japan, where sawdust-based cultivation is the most dominant method accounting for 99% of the production. The current strains for sawdust cultivation in Japan are considered to have been derived from a single wild strain collected from Fukushima, Japan, implying that commercial nameko mushrooms are derived from a severe genetic bottleneck. We tested this single founder hypothesis by developing 14 microsatellite markers for P. microspora to evaluate the genetic diversity of 50 cultivars and 73 wild strains isolated from across Japan. Microsatellite analysis demonstrated that sawdust-cultivated strains from Japan were significantly less genetically diverse than the wild strains, and the former displayed a significant bottleneck signature. Analyzing the genetic relationships among all genotypes also revealed that the sawdust-cultivated samples clustered into one monophyletic subgroup. Moreover, the sawdust-cultivated samples in Japan were more closely related than full-sibs. These results were consistent with the single founder hypothesis that suggests that all commercial nameko mushrooms produced in Japan are descendants of a single ancestor. Therefore, we conclude that cultivated P. microspora originated from a single domestication event that substantially reduced the diversity of commercial nameko mushrooms in Japan.
Keywords: domestication event, genetic diversity, log-cultivated strains, microsatellite markers, sawdust-cultivated strains, wild strains
1. Introduction
Pholiota microspora (Berk.) Sacc. (synonym P. nameko) (“nameko” in Japanese) is one of the most popular edible mushrooms, especially in Japan. This species is a type of white-rot fungus that grows on dead or rotten trees in cool-temperate deciduous forests of East Asia, ranging from the Himalayas and China to Japan (Berkeley, 1850; Neda, 2008; Adgujarum, Watanabe, & Parajuli, 2014; GBIF.org, 2022). According to records, the nameko mushroom was first artificially log-cultivated in 1921 at the Tohoku region of northeastern Japan (Yoshimi, 1979; Pegler, 2003), where this mushroom was traditionally consumed. Modern sawdust-based cultivation of nameko mushroom was established in 1930 by Hikosaburo Morimoto (US patent No. 1,833,089), the pioneer of sawdust-based cultivation of edible mushrooms (Hamada & Hagimoto, 1962; Yoshimi, 1979). Then, commercial production of selected strains in Japan expanded with the development of wooden tray cultivation during the 1960s up to the 1970s. Since the 1980s, the year-round production of mushrooms was possible using sawdust cultivation with air-conditioning, making it the dominant method accounting for 99.7% of its production in Japan (Forestry Agency, 2019). Subsequently, the cultivation of nameko mushroom was introduced to the southern part of the Liaoning Province, China, in the middle of the 1970s (Meng et al., 2019). Since then, China has become its largest producer. Finally, in 2012, China and Japan produced over 700,000 and 20,000 tons of nameko mushroom, respectively (Zhang, Chen, Huang, Gao, & Qu, 2015), causing the current spread of its cultivation to other parts of Asia, North America, and Europe.
Therefore, it has been reported that the cultivation of edible mushrooms has a relatively short history compared to crop plants and farm animals. For example, the cultivation of the shiitake mushroom (Lentinula edodes), one of the oldest cultivated mushrooms, likely occurred about 800 years ago in China (Chang & Miles, 1987). However, the nameko mushroom has been cultivated for approximately one hundred years. Although the first domesticated strains of nameko mushroom were isolated from wild fruit bodies sampled in 1929 in Yamagata Prefecture, Japan (Morimoto, 1930), these strains were proposed to have disappeared during the chaotic times after World War II. There are few records on the domestication process of P. microspora. However, it is likely that the current spawn strains used for sawdust cultivation are derived from a single wild strain, F27 (Kanno, Matsuki, Shigihara, Kimura, & Suyama, 2016), that was collected from Kawairi, Ichinoki, Yamato-machi, Fukushima Prefecture, Japan on Oct 16, 1962 (Nakamoto, Ito, Shoji, & Otake, 1967). Therefore, the vast majority of commercial nameko mushrooms are presumably derived from this severe bottleneck event, resulting in a very low level of genetic variation. However, a comparison of the genetic diversity of cultivars and wild strains has not been conducted.
Hence, a proper understanding of the evolutionary history of domesticated species is required to be able to use their genetic resources and facilitate further breeding. Such understanding requires knowledge of the genetic diversity and population structure of natural populations of domesticated organisms. Obatake, Matsumoto, Mimura and Fukumasa-Nakai (2002) investigated the genetic relationship among 36 wild isolates of P. microspora sampled from across Japan using RAPD and PCR-RFLP markers. They discovered no geographic basis in the population structure, and estimates of genetic diversity were uncertain. Thus, the level of genetic diversity in wild P. microspora has remained uncertain because of the low reproducibility of RAPD profiles (Jones, Edwards, & Castaglione, 1997) and the weak polymorphism of RFLP markers. Besides, although STS markers based on RAPD and ISSR analysis were developed to discriminate P. microspora strains (Sasaki et al., 2007), they were uninformative in assessing the genetic diversity. Therefore, we developed microsatellite markers for P. microspora to evaluate the genetic diversity of wild strains isolated from across Japan, including cultivars, and we further assessed the putative origin of these cultivars.
Microsatellite markers are highly polymorphic and reproducible, easy-to-score, and are assumed to be neutral and co-dominant. Thus, they have been invaluable in many fields of biology, including population genetics, forensic science, and food science. Another approach, genome-wide SNP genotyping, offers high-density information and has been more available in recent years. However, microsatellite analysis is better suited for individual-unit operations, requires a very small amount of sample DNA, and can analyze damaged DNA samples, such as those from processed food products. Thus, microsatellite markers have been developed for some economically important mushrooms (e.g., Agaricus bisporus and L. edodes) to facilitate genetic and breeding studies (e.g., Foulongne-Oriol, Spataro, & Savoie, 2009; Xiao, Liu, Dai, Fu, & Bian, 2010), even though microsatellites occur less frequently in the genomes of fungal species, and they often exhibit lower levels of polymorphism than those in angiosperms and mammals (Dutech et al., 2007). In the last decade, rapid developments in genome sequencing have accelerated the discovery of microsatellites in various organisms, but especially in fungal species, due to their relatively small-sized genomes. In fact, the draft genome sequences of P. microspora, as well as 89 other mushrooms in China, were published simultaneously (Li et al., 2018). This genome sequence assembly of a nameko mushroom in China can be used to develop microsatellite markers in this species, as well as provide insight into the domestication history of P. microspora.
In this study, we aimed to (1) develop reliable microsatellite markers for P. microspora; (2) evaluate the genetic diversities of cultivars mainly from Japan and China, including wild strains isolated from across Japan; and (3) evaluate the consistency of the domestication history of nameko mushroom with the single founder hypothesis stating that commercial nameko mushrooms descended from a single wild strain collected from Japan. Finally, we considered the importance of assessing the unexplored genetic resource of this recently domesticated organism and its role in enhancing breeding and cultivation.
2. Materials and methods
2.1. The fungal material
First, 124 P. microspora samples comprising 51 cultivars mainly from Japan and China, in addition to 73 wild strains collected from across Japan, were analyzed using microsatellite genotyping (see detailed in Table 1). Of the 51 cultivars, 18 strains were isolated from spawn for sawdust cultivation in Japan, including not only most of the current strains in circulation but also past-historical strains, such as the putative founder strain F27. However, of the other 33 cultivars, seven strains were from spawn for log-bed cultivation in Japan, one strain was from spawn for sawdust cultivation in China, one strain was from spawn for log-bed cultivation in China, and the remaining 22 and two strains were commercial mushrooms produced in Japan and China, respectively. Alternatively, 73 wild strains were collected between 1993 and 2020 from 13 prefectures, covering much of the distribution range of P. microspora in Japan. Of the 124 samples, 82 samples were extracted for genomic DNA using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. Then, DNA was extracted from the remaining 32 samples for subsequent experiments, as reported in previous studies (Sasaki et al., 2007; Sasaki et al., 2013; Suyama & Matsuki, 2015; Kanno et al., 2016).
Table 1. Collections of Pholiota microspora used in this study.
| ID | MLG a | Type | Organ | Date b | Collector c | Location / Source |
| 218 | G14 | Wild strain | Hypha | 1995/10/01 | SG | Etanbetsu, Asahikawa, Hokkaido, Japan |
| 242 | G03 | Wild strain | Hypha | 1998/10/15 | KB | Kamui-yama, Okushiri, Hokkaido, Japan |
| 243 | G38 | Wild strain | Hypha | 1998/10/15 | KB | Kamui-yama, Okushiri, Hokkaido, Japan |
| 171 | G30 | Wild strain | Hypha | 1996/10/24 | AK | Nisimeya-mura, Aomori, Japan |
| 172 | G12 | Wild strain | Hypha | 1996/10/24 | AK | Ammonnotaki, Nisimeya-mura, Aomori, Japan |
| 173 | G39 | Wild strain | Hypha | 1996/10/24 | AK | Ammonnotaki, Nisimeya-mura, Aomori, Japan |
| 174 | G34 | Wild strain | Hypha | 1996/10/24 | AK | Ammonnotaki, Nisimeya-mura, Aomori, Japan |
| 192 | G25 | Wild strain | Hypha | 1996/11/06 | KB | Yagen-onsen, Ohata-machi, Aomori, Japan |
| K32 | KG62 | Wild strain | DNA extract | 2014 | KX | 26-99 strain, Hirosaki, Aomori, Japan |
| K07 | KG45 | Wild strain | DNA extract | ― | KX | 62-2 strain, Mt. Hakkoda, Aomori, Japan |
| 216 | G04 | Wild strain | Hypha | 1997/10/21 | KB | Hayasaka-kogen, Morioka, Iwate, Japan |
| 217 | G06 | Wild strain | Hypha | 1997/10/21 | KB | Hayasaka-kogen, Morioka, Iwate, Japan |
| 121 | G33 | Wild strain | Hypha | 1994/11/01 | AK | Yuri-gun, Akita, Japan |
| K27 | KG06 | Wild strain | DNA extract | 2006 | KX | 19-25 strain, Towada, Akita, Japan |
| K28 | KG60 | Wild strain | DNA extract | 2009 | KX | 21-48 strain, Mt. Funagata, Miyagi, Japan |
| K29 | KG07 | Wild strain | DNA extract | 2014 | KX | 26-59 strain, Miyagi, Japan |
| K30 | KG07 | Wild strain | DNA extract | 2014 | KX | 26-60 strain, Miyagi, Japan |
| K31 | KG07 | Wild strain | DNA extract | 2014 | KX | 26-61 strain, Miyagi, Japan |
| 111 | G46 | Wild strain | Hypha | 1993/10/30 | AK | Gassan, Yamagata, Japan |
| 112 | G26 | Wild strain | Hypha | 1993/10/30 | AK | Gassan, Yamagata, Japan |
| 248 | G49 | Wild strain | Hypha | 1999/10/29 | KB | Nishikawa-machi, Yamagata, Japan |
| 255 | G17 | Wild strain | Hypha | 1999/10/29 | AK | Nishikawa-machi, Yamagata, Japan |
| 257 | G29 | Wild strain | Hypha | 1999/10/29 | AK | Nishikawa-machi, Yamagata, Japan |
| 262 | G50 | Wild strain | Hypha | 1999/10/29 | AK | Nishikawa-machi, Yamagata, Japan |
| H01 | GH1 | Wild strain | Fruit body | 2020/10/25 | AH | Nukumidaira, Oguni-machi, Yamagata, Japan |
| H02 | GH2 | Wild strain | Fruit body | 2020/10/25 | AH | Nukumidaira, Oguni-machi, Yamagata, Japan |
| 202 | G27 | Wild strain | Hypha | 1997/10/29 | AK | Tengendai, Yonezawa, Yamagata, Japan |
| K04 | KG32 | Wild strain | DNA extract | 2013/07/19 | KX | Iide, Yamagata, Japan |
| K33 | KG23 | Wild strain | DNA extract | 2015/10/22 | KX | 27-111 strain, Yamagata, Japan |
| K34 | KG36 | Wild strain | DNA extract | 2015/10/22 | KX | 27-122 strain, Yamagata, Japan |
| K35 | KG56 | Wild strain | DNA extract | 2015/10/26 | KX | 27-140 strain, Mogami-gun, Yamagata, Japan |
| K36 | KG52 | Wild strain | DNA extract | 2015/10/26 | KX | 27-141 strain, Mogami-gun, Yamagata, Japan |
| K37 | KG08 | Wild strain | DNA extract | 2015/10/26 | KX | 27-144 strain, Mogami-gun, Yamagata, Japan |
| 222 | G22 | Wild strain | Hypha | 1998/11/11 | AK | Mt. Kinpoku-san, Sado, Niigata, Japan |
| 224 | G48 | Wild strain | Hypha | 1998/11/11 | AK | Mt. Kinpoku-san, Sado, Niigata, Japan |
| 226 | G10 | Wild strain | Hypha | 1998/11/11 | AK | Mt. Kinpoku-san, Sado, Niigata, Japan |
| 290 | G40 | Wild strain | Hypha | 2000/10/17 | TK | Akiyamago, Tsunan-machi, Niigata, Japan |
| 291 | G42 | Wild strain | Hypha | 2000/10/17 | TK | Akiyamago, Tsunan-machi, Niigata, Japan |
| 114 | G51 | Wild strain | Hypha | 1993/10/30 | AK | Mori-toge, Koriyama, Fukushima, Japan |
| 012 | G23 | Wild strain | Hypha | 1982/10/21 | MW | Yaheishiro, Nishiaizu, Fukushima, Japan |
| 013 | G13 | Wild strain | Hypha | 1982/10/21 | MW | Yaheishiro, Nishiaizu, Fukushima, Japan |
| 022 | G18 | Wild strain | Hypha | 1982/10/21 | MW | Yaheishiro, Nishiaizu, Fukushima, Japan |
| 015 | G19 | Wild strain | Hypha | 1982/10/21 | MW | Yaheishiro, Nishiaizu, Fukushima, Japan |
| 020 | G19 | Wild strain | Hypha | 1982/10/21 | MW | Yaheishiro, Nishiaizu, Fukushima, Japan |
| 021 | G19 | Wild strain | Hypha | 1982/10/21 | MW | Yaheishiro, Nishiaizu, Fukushima, Japan |
| 017 | G41 | Wild strain | Hypha | 1982/10/21 | MW | Yaheishiro, Nishiaizu, Fukushima, Japan |
| 018 | G37 | Wild strain | Hypha | 1982/10/21 | MW | Yaheishiro, Nishiaizu, Fukushima, Japan |
| 294 | G20 | Wild strain | Hypha | 2000/11/09 | AK | Hinoemata-mura, Fukushima, Japan |
| 237 | G43 | Wild strain | Hypha | 1998/10/08 | AK | Furaisawa, Kaneyama-machi, Fukushima, Japan |
| 270 | G44 | Wild strain | Hypha | 1999/11/10 | AK | Tadami-machi, Fukushima, Japan |
| 199 | G47 | Wild strain | Hypha | 1997/10/19 | AK | Tenei-mura, Iwase-gun, Fukushima, Japan |
| 188 | G31 | Wild strain | Hypha | 1995/11/08 | KM | Gokayama, Taira-mura, Toyama, Japan |
| 116 | G28 | Wild strain | Hypha | 1993/10/28 | KM | Izumi-mura, Onogun, Fukui, Japan |
| 117 | G28 | Wild strain | Hypha | 1993/10/28 | KM | Izumi-mura, Onogun, Fukui, Japan |
| 118 | G28 | Wild strain | Hypha | 1993/10/28 | KM | Izumi-mura, Onogun, Fukui, Japan |
| 119 | G28 | Wild strain | Hypha | 1993/10/28 | KM | Izumi-mura, Onogun, Fukui, Japan |
| 115 | G36 | Wild strain | Hypha | 1993/10/28 | KM | Izumi-mura, Onogun, Fukui, Japan |
| 283 | G02 | Wild strain | Hypha | 2000/10/18 | KB | Akiyamago, Sakae-mura, Nagano, Japan |
| 284 | G02 | Wild strain | Hypha | 2000/10/18 | SO | Akiyamago, Sakae-mura, Nagano, Japan |
| 287 | G05 | Wild strain | Hypha | 2000/10/18 | KM | Akiyamago, Sakae-mura, Nagano, Japan |
| 288 | G01 | Wild strain | Hypha | 2000/10/18 | KB | Akiyamago, Sakae-mura, Nagano, Japan |
| 106 | G09 | Wild strain | Hypha | 1993/10/17 | AK | Otari-mura, Nagano, Japan |
| 107 | G16 | Wild strain | Hypha | 1993/10/28 | AK | Otari-mura, Nagano, Japan |
| 108 | G32 | Wild strain | Hypha | 1993/10/28 | AK | Otari-mura, Nagano, Japan |
| 109 | G32 | Wild strain | Hypha | 1993/10/28 | AK | Otari-mura, Nagano, Japan |
| 105 | G45 | Wild strain | Hypha | 1993/10/28 | AK | Otari-mura, Nagano, Japan |
| 189 | G21 | Wild strain | Hypha | 1996/10/24 | KB | Daisen, Tottori, Japan |
| 190 | G08 | Wild strain | Hypha | 1996/10/24 | KB | Daisen, Tottori, Japan |
| 204 | G11 | Wild strain | Hypha | 1997/10/30 | AK | Ishizuchi-yama, Kochi, Japan |
| 205 | G00 | Wild strain | Hypha | 1997/10/30 | AK | Ishizuchi-yama, Kochi, Japan |
| 209 | G00 | Wild strain | Hypha | 1997/10/30 | AK | Ishizuchi-yama, Kochi, Japan |
| 210 | G00 | Wild strain | Hypha | 1997/10/30 | AK | Ishizuchi-yama, Kochi, Japan |
| 211 | G00 | Wild strain | Hypha | 1997/10/30 | AK | Ishizuchi-yama, Kochi, Japan |
| F27 | CG1 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | Putative original strain for indoor sawdust cultivation |
| Tohoku-N104 | CG1 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | Tohoku N104 strain, Kinokkusu Co., Miyagi, Japan |
| KX-N006 | CG2 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | KX-N006 strain, Kinokkusu Co., Miyagi, Japan― |
| KX-N007 | CG3 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | KX-N007 strain, Kinokkusu Co., Miyagi, Japan |
| KX-N008 | CG3 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | KX-N008 strain, Kinokkusu Co., Miyagi, Japan |
| KX-N009 | CG4 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | KX-N009 strain, Kinokkusu Co., Miyagi, Japan |
| KX-N010 | CG2 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | KX-N010 strain, Kinokkusu Co., Miyagi, Japan |
| Tohoku-N123 | CG5 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | Tohoku N123 strain, Kinokkusu Co., Miyagi, Japan |
| Tohoku-N127 | CG6 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | Tohoku N127 strain, Kinokkusu Co., Miyagi, Japan |
| 007 | CG1 | Cultivated strain for sawdust cultivation | Hypha | ― | FPFRC | Inoculum for wooden tray cultivation, Japan |
| 008 | CG6 | Cultivated strain for sawdust cultivation | Hypha | ― | FPFRC | Inoculum for wooden tray cultivation, Japan |
| N103 | KG02 | Cultivated strain for log cultivation | DNA extract | ― | KX | N103 strain, Kinokkusu Co., Miyagi, Japan |
| KX-N109 | KG58 | Cultivated strain for log cultivation | DNA extract | ― | KX | KX-N109 strain, Kinokkusu Co., Miyagi, Japan |
| Mori-13 | CG6 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | Mori-13 strain, Mori Sangyo Co. Ltd, Gunma, Japan |
| N245 | CG7 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | N245 strain, Kawamura-shokuyokin-kenkyujo Co., Yamagata, Japan |
| N258 | CG5 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | N258 strain, Kawamura-shokuyokin-kenkyujo Co., Yamagata, Japan |
| Hokken-N108 | CG5 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | N108 strain, Hokken Co. Ltd, Tochigi, Japan |
| Hokken-N160 | CG6 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | N160 strain, Hokken Co. Ltd, Tochigi, Japan |
| Fukushima-N1 | CG8 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | Fukushima N1 strain, FPFRC, Fukushima |
| Fukushima-N2 | KG42 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | Fukushima N2 strain, FPFRC, Fukushima |
| KM-86 | KG02 | Cultivated strain for log cultivation | DNA extract | 2012 | KX | KM-86 strain, Kagawashiitake Co., Miyagi, Japan |
| KM-88 | KG09 | Cultivated strain for log cultivation | DNA extract | 2012 | KX | KM-88 strain, Kagawashiitake Co., Miyagi, Japan |
| K10 | KG02 | Cultivated strain for log cultivation | DNA extract | 2013/07/19 | KX | 5-126 strain, Miyagi, Japan |
| N133 | CG6 | Cultivated strain for sawdust cultivation | DNA extract | ― | KX | N133 strain, Sakegawa, Yamagata, Japan |
| K25 | KG02 | Cultivated strain for log cultivation | DNA extract | ― | KX | Middle-maturing strain, Sakata-Kawamura, Japan |
| K26 | KG02 | Cultivated strain for log cultivation | DNA extract | ― | KX | Middle-maturing strain, Amarume-Kawamura, Japan |
| CP06 | CG3 | Commercial product | Fruit body | 2019/11 | AH | Donguri farm Coop., Miyagi, Japan |
| K17 | CG5 | Commercial product | DNA extract | 2021/07/06 | KX | Fresh-nameko, SugawaraReizou Co., Ltd., Yamagata, Japan |
| K18 | CG5 | Commercial product | DNA extract | 2014/05/08 | KX | Okki-nameko, SugawaraReizou Co., Ltd., Yamagata, Japan |
| K19 | CG5 | Commercial product | DNA extract | 2014/05/08 | KX | Chokai-nameko, SugawaraReizou Co., Ltd., Yamagata, Japan |
| K40 | CG5 | Commercial product | Fruit body | 2021/08/04 | AH | Chokai-nameko, SugawaraReizou Co., Ltd., Yamagata, Japan |
| K41 | CG3 | Commercial product | Fruit body | 2021/07/11 | AH | JA-Oishii-mogami Coop., Yamagata, Japan |
| CP09 | CG3 | Commercial product | Fruit body | 2019/11 | AH | Oak farm Coop., Yamagata, Japan |
| CP01 | CG4 | Commercial product | Fruit body | 2019/11 | AH | JA-Fukushima-mirai Coop., Fukushima, Japan |
| K24 | CG5 | Commercial product | DNA extract | 2014 | KX | Suzuki farm Co., Ltd., Fukushima, Japan |
| CP03 | CG5 | Commercial product | Fruit body | 2019/11 | AH | Suzuki farm Co., Ltd., Fukushima, Japan |
| CP04 | CG5 | Commercial product | Fruit body | 2019/11 | AH | Suzuki farm Co., Ltd., Fukushima, Japan |
| CP05 | CG5 | Commercial product | Fruit body | 2019/11 | AH | Suzuki farm Co., Ltd., Fukushima, Japan |
| CP07 | CG3 | Commercial product | Fruit body | 2019/11 | AH | An individual farmer, Koriyama, Fukushima, Japan |
| CP08 | CG3 | Commercial product | Fruit body | 2019/11 | AH | Happy farm Co., Ltd., Fukushima, Japan |
| CP02 | CG3 | Commercial product | Fruit body | 2019/11 | AH | JA-Tokamachi Coop., Niigata, Japan |
| K43 | CG3 | Commercial product | Fruit body | 2021/12/12 | AH | JA- Tanofuji Coop., Gunma, Japan |
| K44 | CG3 | Commercial product | Fruit body | 2021/07/11 | AH | JA-Nagano Coop., Nagano, Japan |
| K46 | CG3 | Commercial product | Fruit body | 2021/12/18 | TT | Nameko farm Hida Coop., Gifu, Japan |
| K22 | CG3 | Commercial product | DNA extract | 2010/11/29 | KX | Akaike farm Coop., Shizuoka, Japan |
| K45 | CG3 | Commercial product | Fruit body | 2021/12/16 | AH | Kamiyukawa kinoko-seisan Coop., Nara, Japan |
| K42 | CG3 | Commercial product | Fruit body | 2021/12/16 | AH | Oyamacho-nokyo Coop., Oita, Japan |
| CP10 | CG5 | Commercial product | Fruit body | 2021/07 | SK | Marukome Co., Ltd., a processed food made in China |
| K23 | NA | Commercial product | DNA extract | 2017/11/27 | KX | Marukome Co., Ltd., a processed food made in China |
| K38 | KG11 | Cultivated strain for sawdust cultivation | DNA extract | 2013 | KX | 25-1 strain, China |
| No137 | KG02 | Cultivated strain for log cultivation | DNA extract | ― | KX | No137 strain, China |
a MLG: Multi-locus genotype based on 14 microsatellite markers. b Date: sampling date for wild strain or purchase date for commercial product. c Collectors: AH, Akira Hirao; AK. Atsushi Kumata; FPFRC, Fukushima Prefecture Forest Research Center; KB, Katsuhiko Babasaki; KX, Kinokkusu Co.;KM; Kazuhiko Masuno; SG, Seiki Gisusi; SK, Shingo Kaneko; SO, Shinjiro Oya; TK, Takaaki Kishimoto; TT, Toshihito Takagi; MW, Masaaki Watanabe.
2.2. Microsatellite identification, primer design, and genotyping
Microsatellites were identified based on a draft genome assembly of P. microspora (GenBank/EMBL/DDBJ accession number QFFD00000000: Li et al., 2018) using QDD (ver. 3.1.2) software (Meglécz et al., 2014). Primers were designed using the Primer3 algorithm (Rozen & Skaletsky, 1999) with the following parameters: amplicon sizes of 90-350 bp, GC contents of 30%-60% with an optimum at 50%, primer melting temperatures (Ta) of 56 °C-62 °C with an optimum at 60 °C, and primer lengths of 18-28 bp.
PCR amplification was performed in a total volume of 6 μL, containing 1 μL of template DNA, 3 μL of Multiplex PCR Master Mix (QIAGEN), and 0.2 μM of each primer. Each forward primer was labeled with either FAM, VIC, PET, or NED fluorescent dye. We also prepared unlabeled forward primers and mixed them with the fluorescent ones, as recommended by Suyama (2012). Five reverse primers were appended with the “PIG-tail” sequence (GTTTCTT) at the 5' end to promote non-templated A addition and facilitate subsequent genotyping (Brownstein, Carpten, & Smith, 1996) (see Table 2 for details). The amplification parameters were initial denaturation at 95 °C for 15 min followed by 45 cycles of 95 °C for 10 s, 58 °C for 90 s, and 72 °C for 1 min; and a final elongation step of 60 °C for 30 min. Allele-specific fragment sizes of PCR amplicons were determined using GeneMapper ver. 4.0 (Applied Biosystems) and GeneMaker ver. 1.6 (SoftGenetics, State College, PA, USA). To confirm the power and accuracy of the microsatellite markers, we plotted a genotype accumulation curve with 1000 iterations, then calculated the pairwise linkage disequilibrium between loci, and finally determined the multi-locus genotypes (MLGs) with the genetic distance threshold set at 5%, using the “Poppr” package (Kamvar, Tabima, & Grünwald, 2014) in the R 4.1.2 software environment (R Core Team, 2021).
Table 2. Characteristics of 14 microsatellite markers developed for Pholiota microspora.
| Locus | Primer sequences (5'-3') | Repeat motif | Ta (°C) | Allele size range (bp) | Fluorescent label | Primer ratioa | Multiplex combination | A |
| Phmi01 | F-AAAGCTTATTTCACGCCAGG R-CGGCGTTAGTCGTTACATTG |
(ATC)10 | 59 | 85-94 | FAM | 1:7 | A | 4 |
| Phmi02 | F-TGTGCCCTTTCTCCTTGTCT R-TCCGAAACGCGATTTCTTAC** |
(AG)10 | 60 | 99-107 | VIC | 1:7 | A | 5 |
| Phmi03 | F-AGCTGGCGATCCCTGAAC R-GCCTCACCACCAGCCATC* |
(AGC)9 | 61 | 106-115 | PET | 1:3 | A | 4 |
| Phmi05 | F-GGGTTATCTCGATAATTTAACGGC R-GAGTCCATAAGTTTATCATCTAGCAAA |
(AG)9 | 61 | 102-110 | NED | 1:7 | A | 4 |
| Phmi07 | F-TACCTCAGCCACACTGGCA R-TGCCTTCAGTTCCGGAATAC** |
(AC)9 | 61 | 120-159 | FAM | 1:3 | B | 11 |
| Phmi08 | F-TTAACGGGTTTCTAGACCGC R-AAATCAATCTCAGAGCGCC |
(AC)9 | 59 | 135-147 | PET | 1:3 | B | 5 |
| Phmi09 | F-ACATGCGCATCATCGAAGT R-GCGCTGAGCTACAGTGAGACT* |
(AT)9 | 60 | 146-154 | VIC | 1:7 | B | 5 |
| Phmi10 | F-TAACACCACAGTTTGCATGG R-GACTTATGGTGCTGGCCC |
(AAC)9 | 58 | 133-145 | NED | 1:5 | B | 4 |
| Phmi13 | F-GGTTCCATGTCCGTGTTCTT R-GGGCTACCTTGCCTTCATCT |
(ACC)9 | 60 | 151-175 | PET | 1:3 | A | 8 |
| Phmi14 | F-GATGCCAGCCATTAGACGAT R-TGTATCCGCTTTCCTCTTGC** |
(AGC)10 | 60 | 142-183 | VIC | 1:7 | A | 8 |
| Phmi17 | F-CCGAGTGAACATATCCAGCA R-TTGTCTTCCTCCCTTCCTGA |
(AC)12 | 60 | 213-238 | FAM | 1:3 | A | 16 |
| Phmi20 | F-CGCTTAGAGGCACCAATACG R-GCTGAACGTGGACACTCTGA |
(AT)9 | 61 | 282-292 | FAM | 1:3 | B | 7 |
| Phmi23 | F-ACAAGACCCAACGTACAGGG R-ACAGGTGAAAGGGTCATGGA |
(AGG)9 | 60 | 286-313 | NED | 1:7 | B | 5 |
| Phmi24 | F-GCAATCTCATGAGCCTTTCA R-GAGATCCTGAAGGTTGTGACG |
(AT)9 | 59 | 309-321 | FAM | 1:3 | A | 7 |
Ta, annealing temperature; A, number of alleles across the 123 samples analyzed.
a Ratio of fluorescent and unlabeled forward primers for multiplex PCR. See text for details.
*, modified with a “PIG-tail” (GTTTCTT) at the 5' end (Brownstein et al., 1996). **, modified with an adjusted “PIG-tail” (GTTTCT) at the 5' end.
2.3. Genetic data analysis
We used “hierfstat” (Goudet, 2005) and “adegenet” (Jombart, 2008) packages in R to calculate the percentages of polymorphic loci (P), number of alleles (A), and allele richness (AR), including the observed and expected heterozygosity (Ho and He, respectively) among genotypes within each of the following group of strains: wild strains, sawdust-cultivated strains from Japan, and the other cultivated strains from Japan and China. During the grouping of strains, Fukushima-N2 for sawdust cultivation was exceptionally assigned to “the other cultivated strains” because although the Fukushima-N2 strain was not a descendant of a single founder, it was established from a hybrid between the wild strain and Tohoku-N127 strain (PVP Office of Japan, 2022). Thus, to test the evidence accounting for recent bottleneck events in each group of strains, we used the Wilcox signed-rank test of heterozygosity excess (Cornuet & Luikart, 1996) under the two-phase model with 95% single-step mutations, 5% multi-step mutations, and a variance of 12%, as implemented in the BOTTLENECK program (v. 1.2.02) (Piry, Luikart, & Cornuet, 1999).
Furthermore, we constructed a neighbor-joining dendrogram of the genetic relationships among individual genotypes using the Bruvo distance (Bruvo, Michiels, D'Souza, & Schulenburg, 2004) as implemented in the R package, “adegenet” (Jombart, 2008). Then, we visualized the differences in allelic composition among individual genotypes using principal component analysis (PCA), using the “adegenet” package. We also evaluated the pairwise relatedness (rxy) (Queller & Goodnight, 1989) among genotypes within wild strains, sawdust-cultivated strains from Japan, and other cultivated strains, after which the rxy estimate was calculated using the “related” package (Pew, Muir, Wang, & Frasier, 2015) in R (version 3.6.1).All R scripts for data analysis can be obtained from https://github.com/akihirao/nameko_SSR.
3. Results
3.1. Microsatellite markers and identified genotypes
The draft genome of P. microspora yielded a total of 1714 microsatellite regions, with a relative density of 20.5 microsatellites per Mb. The most abundant motif was dinucleotide (55.4%), followed by tri- (35.7%), hexa- (3.4%), tetra- (3.3%), and pentanucleotide (2.1%) motifs. We carried out a preliminary PCR amplification test on 24 microsatellites consisting of 14 di- and 10 trinucleotide microsatellites with a minimum of nine repetitions per unit. Twenty-one out of the 24 candidate markers were amplified, and we selected 14 polymorphic microsatellites with easy-to-score profiles for subsequent analyses (Table 2).
One sample with missing alleles (ID: K23 from China) was excluded from the following analysis. Using the 14 microsatellites across 123 samples yielded 93 alleles, ranging from 4 to 16 alleles per locus with an average of 6.6. Furthermore, we used a 5% genetic distance threshold to identify 74 multi-locus genotypes (MLGs) from 123 samples (Table 1; Supplementary Table S1). Then, we resolved all 74 genotypes, using a genotype accumulation curve that reached a plateau at more than 13 loci (Supplementary Fig. S1). Of the 74 genotypes, 61 were identified from wild-type strains and eight were from sawdust-cultivated P. microspora, mainly in Japan but also in China. Additionally, one genotype was from a hybrid between a wild-type strain and sawdust-cultivated strain in Japan, one genotype was from a sawdust-cultivated strain in China, and the remaining three were from log-cultivated strains. We also observed identical genotypes among the wild strains collected from the same sampling locations, indicating putative clones. Moreover, overlapping genotypes among the cultivated strains existed, some of which were distinct cultivated spawn strains (Table 1). All genotypes from commercial mushroom products in Japan exactly matched the spawn strains used for sawdust cultivation in Japan. Besides, the genotype of a commercial mushroom from China (ID: CP10) also exactly matched that found in cultivated spawn strains and commercial samples in Japan (Supplementary Table S1). Hence, we then used the 74 unique genotypes to assess the genetic diversity of our samples.
3.2. Genetic diversity
Summarized statistics of genetic diversity (Table 3) showed a moderate level of genetic diversity across all the genotypes (He = 0.542). As shown, the cultivars for sawdust cultivation in Japan (A = 1.4, AR = 1.4, He = 0.159) were significantly less genetically diverse than the wild strains (A = 6.6, AR, = 3.3, He = 0.564; all P < 0.05 by Kruskal-Wallis tests). Additionally, the cultivars for sawdust cultivation in Japan had one or two alleles per locus. Both alleles were also found in the wild strains (Supplementary Table S1). In contrast, the bottleneck test detected a bottleneck signal in the sawdust-cultivated samples from Japan (P = 0.03) but not in the wild strains (P = 0.99) and other cultivars (P = 0.19) (Table 3).
Table 3. Genetic diversity in cultivated and wild strains of Pholiota microspora.
| Category | N | G | P | A (SE) | AR5 (SE) | Ho | He | Bottleneck test | |
| Wild strain | 73 | 61 | 1.00 | 6.6 (0.9) | 3.3 (0.3) | 0.352 | 0.564 | 0.999 | |
| Cultivated strain | Sawdust-cultivated strains in Japan | 41 | 8 | 0.36 | 1.4 (0.1) | 1.4 (0.1) | 0.179 | 0.159 | 0.031* |
| The other cultivated strains | 9 | 5 | 086 | 2.8 (0.3) | 2.8 (0.3) | 0.429 | 0.536 | 0.190 | |
| Overall | 123 | 74 | 1.00 | 6.6 (0.9) | 3.2 (0.3) | 0.339 | 0.539 | ― | |
N: number of samples analyzed; G: number of multi-locus genotypes; P: percent of polymorphic loci; A: number of alleles; AR5: allelic richness per five individuals; Ho: observed heterozygosity; He: expected heterozygosity. Bottleneck test: probability of significant heterozygosity excess was calculated using the program BOTTLENECK ver.1.2.02 (Piry et al., 1999). *: P < 0.05.
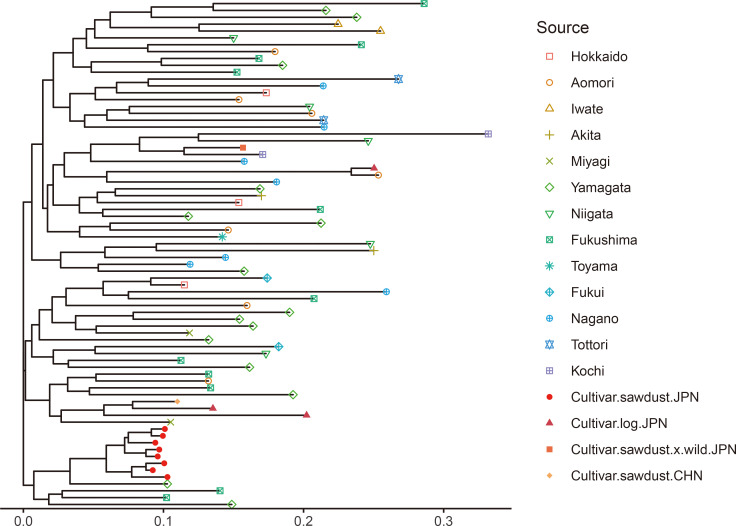
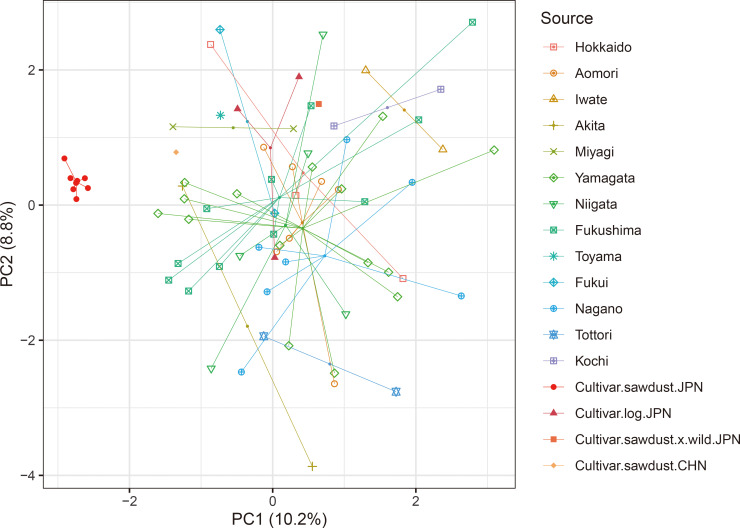
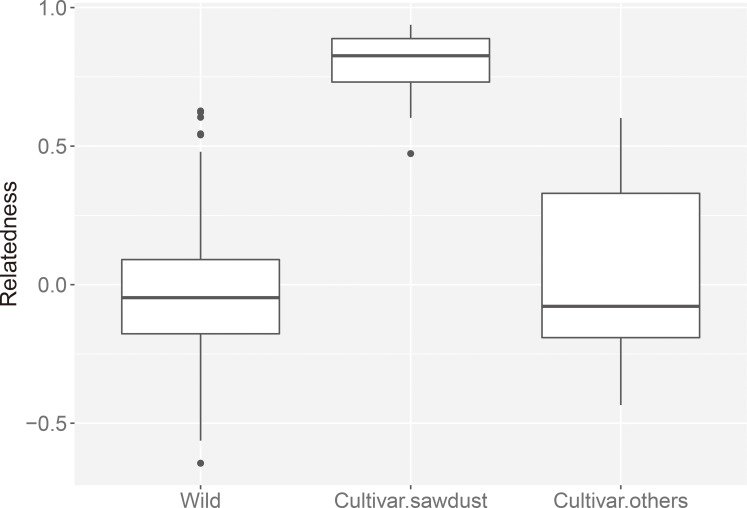
The neighbor-joining dendrogram (Fig. 1) showed sawdust-cultivated samples in Japan clustered in a single subgroup whose branch lengths are shorter than those of other subgroups. The wild strains did not cluster according to geographical regions. Alternatively, the PCA plot (Fig. 2) shows the relationships among the genotypes along the first two PCA axes that explained 10.2% and 8.8% of the total variance, respectively. Therefore, the PCA result agrees with the dendrogram, i.e., that the sawdust-cultivated samples in Japan were tightly clustered, while the wild strains and the other cultivars were scattered and do not group according to source origin. The sawdust-cultivated samples in Japan were also more genetically related (rxy = 0.799 ± 0.112 SD) compared with the wild strains (rxy = −0.040 ± 0.190) and other cultivars (rxy = 0.035 ± 0.368) (P < 0.001 by randomization test; Fig. 3). Besides, all but one relatedness within the sawdust-cultivated samples in Japan had rxy values greater than 0.5 (i.e., indicative of full-sib relationship).
Fig. 1. A neighbor-joining dendrogram showing the 74 genotypes of Pholiota microspora. Samples are colored and symbol-coded according to their sources. Cultivar.sawdust.JPN: sawdust-cultivated samples from Japan; Cultivar.log.JPN: log-cultivated samples from Japan; Cultivar.sawdust.x.wild.JPN: a cultivated sample derived from the hybrid between a sawdust-cultivated strain and a wild strain; Cultivar.CHN: cultivated samples from China; The other samples: wild strains.
Fig. 2. Principal component analysis of 74 Pholiota microspora genotypes. Samples are colored and symbol-coded according to their sources. Cultivar.sawdust.JPN: sawdust-cultivated samples from Japan; Cultivar.log.JPN: log-cultivated samples from Japan; Cultivar.sawdust.x.wild.JPN: a cultivated sample derived from the hybrid between a sawdust-cultivated strain and a wild strain; Cultivar.CHN: cultivated samples from China; The other samples: wild strains.
Fig. 3. Genetic relatedness between individual genotypes of Pholiota microspora within the wild samples (Wild), within the sawdust-cultivated samples in Japan (Cultivar.sawdust), or within the other cultivars (Cultivar.others).
4. Discussion
4.1. Domestication footprints of sawdust-cultivated strains in Japan
Sawdust-cultivated strains of P. microspora in Japan (He = 0.16) are significantly less genetically diverse than wild strains (He = 0.56) (Table 3), and the former display a genetic bottleneck signature (Table 3). Analysis of the genetic relationships among all genotypes shows the sawdust-cultivated samples in Japan clustering into a monophyletic subgroup (Figs. 1, 2). Using SNP genotyping at 252 loci, Kanno et al. (2016) showed that among 25 sawdust-cultivated and 3 wild strains of P. microspora in Japan, all 25 cultivated strains are monophyletic (however, the authors did not address their genetic diversity). This study demonstrated that the genetic relatedness among sawdust-cultivated samples in Japan (rxy = 0.799 ± 0.112 SD) exceeds that of full-sib relationships, indicating that the sawdust-cultivated strains in Japan are derived from self-breeding and/or within-family mating. These results agree with the single founder hypothesis stating that commercial nameko mushrooms produced by sawdust cultivation in Japan are the descendants of a single ancestor. Therefore, we conclude that the genetic diversity of commercial nameko mushrooms in Japan is remarkably low because they have originated from a single domestication event.
Strain F27, collected approximately 60 years ago from Fukushima, Japan (Nakamoto et al., 1967), is the putative founder strain of P. microspora cultivars for sawdust cultivation in Japan; this strain was included in the microsatellite genotyping analysis, which shows that all the loci in strain F27 are fixed, i.e., one allele per locus for all the 14 loci analyzed. In contrast, in the sawdust-cultivated samples in Japan, five loci are polymorphic (Pm08, Pm13, Pm14, Pm17, and Pm23) with two types of alleles, and the nine remaining loci are monomorphic (Supplementary Table S2). Thus, strain F27 is not likely to be the original founder because the founder should have five heterozygous loci that are the sources of the five polymorphic loci in the descendant cultivars. In other words, it is possible to determine the genotype of the original founder strain at the time of collection based on the types of alleles found across all the cultivars (see detailed in Supplementary Table S2). We speculate that all loci in strain F27 have been fixed through inbreeding and/or dedikaryotization of the original F27 strain. In growing mycelia (especially in marginal hyphae) of P. microspora, dedikaryotization is a process that has often been observed (Arita, 1979).
It has been reported that the moderate genetic diversity level of wild P. microspora isolates collected from across Japan (He = 0.56) was equivalent to or lower than wild strains of other wood-decay mushroom species. For example, He = 0.57 in Flammulina velutipes (Liu, Feng, Li, Yan, & Yang, 2016); He = 0.52 in Pleurotus ostreatus (Li, Liu, Zhao, & Yang, 2019); and He = 0.73 in L. edodes (Lee, Moon, Ro, Chung, & Ryu, 2020). In this study, compared to these wild mushrooms, the sawdust-cultivated P. microspora strains in Japan were less genetically diverse (He = 0.16). Studies have also shown that the cultivars of the other mushroom species all had multiple domestication origins, i.e., He = 0.36 in F. velutipes (Liu et al., 2016); He = 0.50 in P. ostreatus (Li et al., 2019); and He = 0.76 in L. edodes (Lee et al., 2020). Thus, we propose that the natural populations of P. microspora can serve as a new genetic resource to develop effective strains for sawdust cultivation. Specifically, although the Fukushima-N2 strain as a hybrid of the wild and sawdust-cultivated P. microspora was established at the Fukushima Prefecture Forest Research Center, it was rarely produced in the Fukushima Prefecture, Japan (A. Kumata, personal communication). Hence, we did not find its genotype in the mushroom products analyzed. Our results confirm the result of Obatake et al. (2002), who reported the absence of geographic influence on the population genetic structure of P. microspora across Japan (Figs. 1, 2), suggesting that genetic resource management at the regional scale is not critical for the mushroom breeding strategy. Nevertheless, the genetic signatures of local adaptations to heterogeneous environments may play an important role in breeding for sources of diversity. In the genomics era, the isolation and characterization of functional genetic diversity will play important roles in the breeding and cultivation of P. microspora.
4.2. Genetic characteristics of the other cultivars
The log-bed cultivation of nameko mushrooms accounts for only less than 1% of the total production of the mushroom in Japan (Forestry Agency, 2019). We analyzed eight spawn strains for log-bed cultivation―seven and one from Japan and China, respectively, to identify three multi-locus genotypes (MLGs) existed among them (Table 1; Supplementary Table S1). The three distinct genotypes were scattered on the neighbor-joining dendrogram (Fig. 1) and PCA diagram (Fig. 2), implying multiple origins of domestication. Results also showed that one of the three genotypes (MLG: KG02) was frequently discovered in several spawns from Japan and China, suggesting that one of the preferred spawn strains with a common ancestor was introduced from Japan to China.
Additionally, two sawdust-cultivated samples from China were successfully genotyped. From the results, while the genotype of a commercial nameko mushroom produced in China (ID: CP10) exactly matched that of some sawdust-cultivated samples from Japan (MLG: CG5), the other genotype of a spawn strain from China (ID: K38) did not uniquely match any sample (Table 1; Supplementary Table S1). These results indicated that although a descendant of the founder strain F27 from Japan partly contributed to the production of nameko mushrooms in China, at least a part of Chinese cultivars underwent different domestication processes. In this study, the Chinese sample of a commercial product was collected from a freeze-dried food product. Thus, its DNA components had suffered considerable damage due to high-temperature pressurization during food processing. Despite the damage to its DNA, the processed food sample was successfully genotyped using the microsatellite marker system. Therefore, microsatellite analysis can be applied to natural samples and food products such as mushrooms. It should be noted that the microsatellite markers for P. microspora were designed from a genome assembled from a nameko mushroom collected from a local fresh market in China (Li et al., 2018). The geographic separation between China and Japan should lead to genetic differentiation between the wild strains of P. microspora from the two regions. Thus, some level of mismatch is expected when applying a specific set of genetic markers on strains from both countries. However, the microsatellite markers designed from the Chinese sample allowed the successful genotyping of all the samples from Japan. Additionally, while developing the markers, we achieved high rates of success in amplifying the primary candidate markers (21/24 = 0.85) as well as the final set of selected markers (14/24 = 0.58). This result was in contrast to the usual process of our microsatellite marker development, where the ratios of the final selected markers to the primary candidate ones were several to dozens in percentages: 18/45 = 0.40 in Gastrodia takeshimensis (Kishikawa et al., 2019); 13/102 = 0.13 in Phraortes elongatus (Nozaki et al., 2021); 15/293 = 0.05 in Pyrola japonica (Shutoh, Izuno, Isagi, Kurosawa, & Kaneko, 2017); and 26/238 = 0.11 in Cypripedium japonicum (Yamashita, Izuno, Isagi, Kurosawa, & Kaneko, 2016). Our results imply that the Chinese sample used for genome assembly is genetically similar to Japanese samples. Specifically, the cultivation of nameko mushroom was introduced from Japan to China in 1970s (Meng et al., 2019). Thus, the Chinese sample used for genome assembly may have the same origin as Japanese cultivars. However, we genotyped only two sawdust-cultivated and one log-cultivated sample from China. Therefore, further investigation on the overall genetic diversity of cultivated nameko mushrooms in China is necessary to reveal its cultivation history outside Japan.
5. Conclusions
Cultivated P. microspora originated from a single domestication event that greatly reduced the genetic diversity of current commercial nameko mushrooms in Japan. Nameko mushroom cultivation has been spreading to China, South-East Asia, North America, and Europe. Clarifying the genetic diversity of nameko mushrooms worldwide will provide important information on the sustainable use of this edible mushroom as well as insight into the evolutionary biology of this recently domesticated organism.
Disclosures
No conflicts of interest among the authors. All the experiments undertaken in this study complied with the current laws of the country where they were performed.
Supplementary Material
Acknowledgments
We are grateful to Katsuhiko Babasaki, Kazuhiko Masuno, Masaaki Watanabe, Seiki Gisusi, Takaaki Kishimoto, and Shinjiro Oya for providing valuable samples, and to Daisuke Sakuma for kindly informing us about Morimoto Mushroom Farm. We thank Yoshito Takagi, Naoko Takagi, Satomi Hirao, and Misuzu Hirao for support with sample collection and Yasuko Hanafusa for advice on the culture media and DNA extraction procedure. We also thank two anonymous referees for suggestions that greatly improved this manuscript.
References
- Adgujarum, M., Watanabe, K., & Parajuli, G. (2014) .A new variety of Pholiota microspora (Agaricales) from Nepal. Biodiversitas Journal of Biological Diversity, 15(1), 101–103. https://doi.org/10.13057/biodiv/d150115 [Google Scholar]
- Arita, I. (1979) .The mechanism of spontaneous dedikaryotization in hyphae of Pholiota nameko. Mycologia, 71(3), 603–611. [Google Scholar]
- Berkeley, M. (1850) .Decades of fungi; decades XXV to XXX. Sikkim Himalaya fungi, collected by Dr. J. D. Hooker. Hooker's Journal of Botany and Kew Garden Miscellany, 2, 76–88. [Google Scholar]
- Brownstein, M. J., Carpten, J. D., & Smith, J. R. (1996) .Modulation of non-templated nucleotide addition by Tag DNA polymerase: primer modifications that facilitate genotyping. BioTechniques, 20, 1004–1010. https://doi.org/https://doi.org/10.2144/96206st01 [DOI] [PubMed] [Google Scholar]
- Bruvo, R., Michiels, N. K., D'Souza, T. G., & Schulenburg, H. (2004) .A simple method for the calculation of microsatellite genotype distances irrespective of ploidy level. Molecular Ecology, 13(7), 2101–2106. https://doi.org/10.1111/j.1365-294X.2004.02209.x [DOI] [PubMed] [Google Scholar]
- Chang, S., & Miles, P. (1987) .Historical record of the early cultivation of Lentinus in China. Mushroom Journal of the Tropics, 7, 31–37. [Google Scholar]
- Cornuet, J. M., & Luikart, G. (1996) .Description and power analysis of two tests fro detecting recent population bottlenecks from allele frequency dat. Genetics, 14, 2001–2014. https://doi.org/https://doi.org/10.1093/genetics/144.4.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutech, C., Enjalbert, J., Fournier, E., Delmotte, F., Barres, B., Carlier, J., Tharreau, D., & Giraud, T. (2007) .Challenges of microsatellite isolation in fungi. Fungal Genetics and Biology, 44(10), 933–949. https://doi.org/10.1016/j.fgb.2007.05.003 [DOI] [PubMed] [Google Scholar]
- Forestry Agency (2019) . [Forest products statistics]. Tokyo: Forestry Agency, Ministry of Agriculture, Forestry and Fisheries (Japan); [accessed 2022 Jun ]. (in Japanese) https://www.maff.go.jp/j/tokei/kouhyou/tokuyo_rinsan/. [Google Scholar]
- Foulongne-Oriol, M., Spataro, C., & Savoie, J.-M. (2009) .Novel microsatellite markers suitable for genetic studies in the white button mushroom Agaricus bisporus. Applied Microbiology and Biotechnology, 84(6), 1125–1135. https://doi.org/10.1007/s00253-009-2030-8 [DOI] [PubMed] [Google Scholar]
- GBIF.org. (2022) .GBIF Occurrence Download https://doi.org/10.15468/dl.j75jex [15 Feburary 2020]. [Google Scholar]
- Goudet, J. (2005) .HIREFSTAT, a package for R to compute and test hierarchical F-statistics. Molecular Ecology Notes, 5, 184–186. https://doi.org/10.1111/j.1471-8286.2004.00828.x [Google Scholar]
- Hamada, M., & Hagimoto, H. (1962) .Kinoko-saibaika Morimoto Hikosaburo shi no syougai to sono-kenkyu ni-tsuite (The life and work of Hikosaburo Morimoto, a mushroom cultivator) (in Japanese). Japanese Journal of Mycology, 3, 145–147. [Google Scholar]
- Jombart, T. (2008) .adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics, 24(11), 1403–1405. https://doi.org/10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- Jones, C., Edwards, K., & Castaglione, S. (1997) .Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Molecular Breeding 3, 381–390. https://doi.org/10.1023/A:1009612517139 [Google Scholar]
- Kamvar, Z. N., Tabima, J. F., & Grünwald, N. J. (2014) .Poppr: an R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ, 2, e281. https://doi.org/10.7717/peerj.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno, S. A., Matsuki, Y., Shigihara, T., Kimura, Y., & Suyama, Y. (2016) .Jisedai-shikensa niyoru SNP maka no tansaku to kinoko hinshu shikibetsu heno riyo (Identification of SNP markers for variety identification in mushroom species using the next generation sequencing platform) (in Japanese). Microbiology and Biotechnology, 24(3), 153–155. [Google Scholar]
- Kishikawa, K., Suetsugu, K., Kyogoku, D., Ogaki, K., Iga D., Shutoh, K., Isagi, Y., & Kaneko, S. (2019) .Development of microsatellite markers for the completely cleistogamous species Gastrodia takeshimensis (Orchidaceae) that are transferable to its chasmogamous sister G. nipponica. Genes & Genetic Systems, 94(2), 95–98. https://doi.org/10.1266/ggs.18-00057 [DOI] [PubMed] [Google Scholar]
- Lee, H. Y., Moon, S., Ro, H. S., Chung, J. W., & Ryu, H. (2020) .Analysis of genetic diversity and population structure of wild strains and cultivars using genomic SSR markers in Lentinula edodes. Mycobiology, 48(2), 115–121. https://doi.org/10.1080/12298093.2020.1727401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H., Wu, S., Ma, X., Chen, W., Zhang, J., Duan, S., Gao, Y., Kui, L., Huang, W., Wu, P., Shi, R., Li, Y., Wang, Y., Li, J., Guo, X., Luo, X., Li, Q., Xiong, C., Liu, H.,... (2018) .The genome sequences of 90 mushrooms. Scientific reports, 8(1), 9982. https://doi.org/10.1038/s41598-018-28303-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Liu, X.-B., Zhao, Z.-W., & Yang, Z. L. (2019) .Genetic diversity, core collection and breeding history of Pleurotus ostreatus in China. Mycoscience, 60(1), 14–24. https://doi.org/10.1016/j.myc.2018.07.002 [Google Scholar]
- Liu, X. B., Feng, B., Li, J., Yan, C., & Yang, Z. L. (2016) .Genetic diversity and breeding history of Winter Mushroom (Flammulina velutipes) in China uncovered by genomic SSR markers. Gene, 591(1), 227–235. https://doi.org/10.1016/j.gene.2016.07.009 [DOI] [PubMed] [Google Scholar]
- Meglécz, E., Pech, N., Gilles, A., Dubut, V., Hingamp, P., Trilles, A., Grenier, R., & Martin, J. F. (2014) .QDD version 3.1: a user-friendly computer program for microsatellite selection and primer design revisited: experimental validation of variables determining genotyping success rate. Molecular Ecology Resources, 14(6), 1302–1313. https://doi.org/10.1111/1755-0998.12271 [DOI] [PubMed] [Google Scholar]
- Meng, L., Fu, Y., Li, D., Sun, X., Chen, Y., Li, X., Xu, S., Li, X., Li, C., Song, B., & Li, Y. (2019) .Effects of corn stalk cultivation substrate on the growth of the slippery mushroom (Pholiota microspora). RSC Advances, 9(10), 5347–5353. https://doi.org/10.1039/c8ra10627d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, H. (1930) .Shokuyo-take nametake (enokitake) bunashimeji (nameko) hiratake shiitake furansutake jinko-baiyoho (Artificial cultivation methods for editable mushrooms: enokitake, nameko, hiratake, shiitake, and champignon) (in Japanese). Morimoto Yokinen, Kyoto. https://dl.ndl.go.jp/info:ndljp/pid/1035576 [Google Scholar]
- Nakamoto, M., Ito, T., Shoji, A., & Otake, R. (1967) .Nameko no hasseiryou oyobi hasseijiki to keishitsu ni kansuru hikakushiken II. (Screening of wild strains of Pholiota microspora for the yields, timing, and the characters of fruitbody. II)(in Japanese). Bulletein of Fukushima Prefectural Forestry Research Centre, 12, 34–77. https://dl.ndl.go.jp/info:ndljp/pid/9625305 [Google Scholar]
- Neda, H. (2008) .Correct name for “nameko”. Mycoscience, 49(1), 88–91. https://doi.org/10.1007/s10267-007-0391-3 [Google Scholar]
- Nozaki, T., Suetsugu, K., Sato, K., Sato, R., Takagi, T., Funaki, S., Ito, K., Kurita, K., Isagi, Y., & Kaneko, S. (2021) .Development of microsatellite markers for the geographically parthenogenetic stick insect Phraortes elongatus (Insecta: Phasmatodea). Genes & Genetic Systems, 21–00022. https://doi.org/https://doi.org/10.1266/ggs.21-00022 [DOI] [PubMed] [Google Scholar]
- Obatake, Y., Matsumoto, T., Mimura, K., & Fukumasa-Nakai, Y. (2002) .Genetic relationships in the natural population of Pholiota nameko from Japan based on DNA polymorphisms. Mycoscience, 43, 463–469. https://doi.org/10.1007/s102670200067 [Google Scholar]
- Pegler, D. (2003) .Useful fungi of the world: the Shii-take, Shimeji, Enoki-take, and Nameko mushrooms. Mycologist, 17(1), 3–5. https://doi.org/10.1017/S0269915X03001071 [Google Scholar]
- Pew, J., Muir, P. H., Wang, J., & Frasier, T. R. (2015) .related: an R package for analysing pairwise relatedness from codominant molecular markers. Molecular Ecology Resources, 15(3), 557–561. https://doi.org/10.1111/1755-0998.12323 [DOI] [PubMed] [Google Scholar]
- Piry, S., Luikart, G., & Cornuet, J.-M. (1999) .BOTTLENECK: a program for detecting recent effective population size reductions from allele data frequencies. Journal of Heredity, 90(4), 502–503. https://doi.org/https://doi.org/10.1093/jhered/90.4.502 [Google Scholar]
- PVP Office of Japan, (2022) .Plant Variety database of PVP Office MAFF in Japan. Available from http://www.hinshu2.maff.go.jp/en/en_top.html[15 Feburary 2020].
- Queller, D. C., & Goodnight, K. F. (1989) .Estimating relatedness using genetic markers. Evolution, 43(2), 258–275. https://doi.org/https://doi.org/10.1111/j.1558-5646.1989.tb04226.x [DOI] [PubMed] [Google Scholar]
- R Core Team (2021) .R: A language and environment for statistical computing.In R Core Team R Foundation for Statistical Computing . In R Core Team R Foundation for Statistical Computing. http://www.R-project.org/ [Google Scholar]
- Rozen, S., & Skaletsky, H. (1999) .Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics methods and protocols (pp. 365, 386). Springer. https://doi.org/10.1385/1-59259-192-2:365 [DOI] [PubMed] [Google Scholar]
- Sasaki, Y., Itabashi, Y., Shigihara, T., Chiba, N., Nakamura, S., Miyazaki, A., & Kimura, E. (2013) .Examination of molecular tools for discrimination of Pholiota microspora wild strains. Mushroom Science and Biotechnology, 20(4), 199–204. https://doi.org/10.24465/msb.20.4_199 [Google Scholar]
- Sasaki, Y., Shigihara, T., Itabashi, Y., Chiba, N., Nakamura, S., Miyazaki, A., & Kimura, E. (2007) .Development of STS markers for the identification of Pholiota nameko cultivars and wild isolates. Mushroom Science and Biotechnology, 15(4), 177–182. https://doi.org/https://doi.org/10.24465/msb.15.4_177 [Google Scholar]
- Shutoh, K., Izuno, A., Isagi, Y., Kurosawa, T., & Kaneko, S. (2017) .Development of microsatellite markers for partially and putative fully mycoheterotrophic varieties of Pyrola japonica sensu lato (Ericaceae). Genes & Genetic Systems, 16–00048. https://doi.org/https://doi.org/10.1266/ggs.16-00048 [DOI] [PubMed] [Google Scholar]
- Suyama, Y. (2012) .SSR genotyping method (in Japanese). In Tsumura, Y., & Suyama, Y. (Eds.), Molecular Ecology in Forest Ecosystems (pp. 291–323). Bun-ichi Sogo Shuppan. [Google Scholar]
- Suyama, Y., & Matsuki, Y. (2015) .MIG-seq: an effective PCR-based method for genome-wide single-nucleotide polymorphism genotyping using the next-generation sequencing platform. Scientific reports, 5, 16963. https://doi.org/10.1038/srep16963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y., Liu, W., Dai, Y., Fu, C., & Bian, Y. (2010) .Using SSR markers to evaluate the genetic diversity of Lentinula edodes' natural germplasm in China. World Journal of Microbiology and Biotechnology, 26(3), 527–536. https://doi.org/10.1007/s11274-009-0202-4 [Google Scholar]
- Yamashita, Y., Izuno, A., Isagi, Y., Kurosawa, T., & Kaneko, S. (2016) .Isolation and characterization of novel microsatellite loci for the endangered orchid Cypripedium japonicum (Orchidaceae). Applications in Plant Sciences, 4(2), 1500097. https://doi.org/10.3732/apps.1500097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimi, S. (1979) .Mori-no-yousei: kinoko-saibai-no-chichi, Morimoto Hikosabro (A forest fairy: Morimoto Hikosaburo, a pioneer mushroom cultivator) (in Japanese). Kaiseisya, Tokyo. [Google Scholar]
- Zhang, J.-X., Chen, Q., Huang, C.-Y., Gao, W., & Qu, J.-B. (2015) .History, current situation and trend of edible mushroom industry development (in Chinese with English abstruct). Mycosystema, 34(4), 524–540. https://doi.org/10.13346/j.mycosystema.150076 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.