Summary
Response to hypoxia is a highly regulated process but little is known about single cell responses to hypoxic conditions. Using fluorescent reporters of hypoxia response factor-1α (HIF-1α) activity in various cancer cell lines, and patient derived cancer cells, we show that hypoxic responses in individual cancer cells can be highly dynamic and variable. These responses fall into three classes, including oscillatory activity. We identify a molecular mechanism that can account for all three response classes, implicating reactive oxygen species-dependent chaperone-mediated autophagy of HIF-1α in a subset of cells. Furthermore, we show that oscillatory response is modulated by the abundance of extracellular lactate in a quorum sensing-like mechanism. We show that oscillatory HIF-1α activity can help rescue hypoxia- mediated inhibition of cell division, and causes broad suppression of genes downregulated in cancers, and activation of genes upregulated in many cancers, suggesting a mechanism for aggressive growth in a subset of hypoxic tumor cells.
eTOC Blurb
This study shows emergence of oscillations in HIF-1a transcriptional activity in dense, hypoxic cancer cell populations. Lactate, a byproduct of HIF-1a induced glycolysis, acts as a quorum sensing signal driving HIF-1a oscillations allowing cells to escape cell cycle arrest. HIF-1a oscillations decrease expression of tumor suppressor genes in many cancers.
Graphical Abstract
INTRODUCTION
Specific response to lack of oxygen (hypoxia) is a critical correlate and regulator of various physiological processes1–3. These responses are mediated by stabilization of a subunit of hypoxia inducible factor, HIF-1α, ubiquitously expressed in diverse tissues4. HIF-1α interacts with hypoxia regulatory elements (HRE) transcriptionally controlling expression of multiple hypoxia-responsive genes, and regulates a wide range of cellular functions1,5. In particular, HIF-1α plays a crucial role in the onset of anaerobic glycolysis, regulating the expression of pyruvate dehydrogenase kinases (PDKs) isoforms. PDKs phosphorylate and inhibit pyruvate dehydrogenase diverting pyruvate away from acetyl-CoA towards lactate synthesis, decreasing respiration and permitting acute metabolic adaptation to hypoxia6. Paradoxically, cells can also maintain reduced, but active oxidative phosphorylation in hypoxia7. HIF-1α can enhance mitochondrial respiration efficiency, e.g., by controlling the expression of the subunits of the cytochrome c oxidase type 4 (COX4)8,9 and potentially other regulators. Currently, the paradox of up-regulation of both glycolytic and respiratory states by the same transcriptional factor remains unresolved.
This paradox might be resolved if individual cells in a hypoxic population could display diverse single cell responses. Phenotypic plasticity permits adoption of distinct cell states even for cells with the same genetic background, exposed to the same environmental cues10. E.g., such states could reflect phenotypic differentiation into the Tip and Stalk fates of endothelial cells within a growing angiogenic sprout or the epithelial-to-mesenchymal transition in tumor progression11,12. These states might be defined by dynamic activity profiles of the underlying networks, rather than static responses11,13,14. However, little is known about the single cell level dynamics of HIF-1α mediated regulation.
In this study, using live cell fluorescent HIF-1α probes in cancer cells, we describe findings that may help resolve the paradox of seemingly contradicting roles of HIF-1α in the control of distinct metabolic cell states. We show that subpopulations of hypoxic cells display distinct temporal dynamics of HIF-1α abundance and transcriptional activity, including oscillations. We find that these oscillations are associated with continued oxidative phosphorylation and respiration in the hypoxic environment, promoted by lactate conversion to pyruvate, whereas cells in the glycolytic state display steady increases in HIF-1α levels. Moreover, we demonstrate that gene expression specific to the oscillatory HIF-1α response partially rescues anti-proliferative effect of hypoxia. Notably, we also show that oscillatory hypoxia can promote expression of oncogenes and downregulate tumor suppressors across multiple cancers. We demonstrate that lactate can act as a mediator of intercellular communication enabling a quorum sensing-like response promoting fluctuation in HIF-1α transcriptional activity through pH- and ROS-sensitive chaperone-mediated autophagy. These findings suggest that different, potentially contradictory metabolic and phenotypic states, can indeed co-exist and be controlled by differential dynamics of HIF-1α activity simultaneously present in hypoxic cell populations.
RESULTS
HIF-1α activity and abundance display three types of lactate-dependent dynamic responses to hypoxia
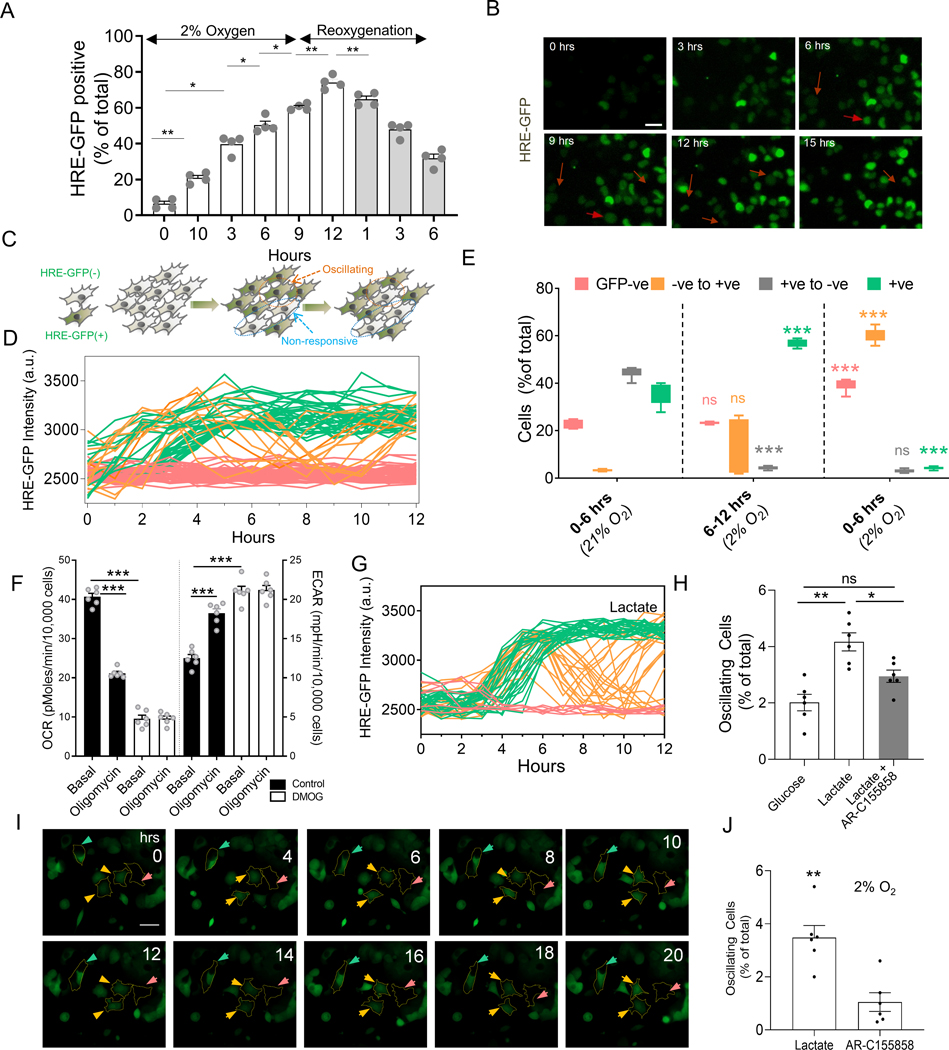
Cell responses to hypoxia measured at a specific point of time can show considerable variability15–17. Using HRE-GFP transfected cells18, we found that in human cell lines, a fraction of cells showed no HIF-1α stabilization for hours after switching from normoxia (21% O2) to hypoxia (2% O2) (Fig 1A, S1A–D). The lack of uniform response is not due to an inherent inability of cells to upregulate HIF-1α, since after treatment with 5 mM Dimethyloxalylglycine (DMOG), which stabilizes HIF-1α by inhibiting prolyl-4-hydroxylase (PHD), nearly all cells express elevated HIF-1α (Fig S1A–D). These findings could reflect static variability in cellular responses, or might represent a ‘snapshot’ of a much more dynamic response to hypoxia. To assess whether responses to hypoxic conditions also have variable dynamics, we studied single cell HIF-1α-mediated transcriptional activity in cells perfused with hypoxic gaseous mixture, while monitoring a transcriptionally encoded biosensor. The biosensor, coupling 5 copies of HRE to expression of an unstable variant of GFP 18, was packaged in a lentiviral vector, and expressed in HEK293 (HRE-HEK293) and HeLa (HRE-HeLa) cells. Cells were selected for their reversibility in HRE-GFP expression in hypoxia followed by reoxygenation, and clonally expanded. Cells showed dose dependent increase in expression of HRE-GFP with decreasing atmospheric O2 tension (Fig S1E). We then observed these cells using time lapse microscopy in glass-bottomed plates either directly, or with microfluidic PDMS caps allowing continuous perfusion of gas (2% oxygen) in the medium19. Microfluidics-mediated oxygen accessibility, assessed with Tris (4,7-diphenyl-1,10-phenanthroline) Ruthenium dichloride (Ru(dpp)3Cl2) quenching20, could be actively controlled on time scales much faster than the expected protein expression dynamics (Fig S1F). Although the population-wide HRE-mediated response to hypoxia displayed a steady increase followed by a steady decline after re-oxygenation (Fig 1A), analysis of individual hypoxic cells with microscopy revealed three different types of dynamic response at the single cell level. Most cells underwent a strong and stable increase in HRE mediated GFP expression (HRE-GFPhi cells) over the first few hours of hypoxic conditions (Fig 1B–C). A smaller fraction of cells displayed no observable change in GFP throughout the course of the observation (HRE-GFPlo cells). Unexpectedly, the remaining cells underwent oscillatory fluctuations between high and low levels of GFP expression over the same time course (Fig 1D–E). Consistent with the average response dynamics, transitions to the HRE-GFPhi state were rapidly reversible after switching back to normoxic conditions (Fig 1E). However, despite reoxygenation, a small percentage of cells continued to change their state from GFPlo to GFPhi (Fig 1E). Overall, these observations suggest that cells can display different co-occurring dynamics in HIF-1α-dependent transcription even under sustained hypoxic inputs.
Fig 1. Single cell responses to hypoxia display complex and varied dynamic profiles.
(A) Time course analysis of HRE-GFP transfected HEK293 cells exposed to hypoxia (2% O2) for 12 hours, followed by reoxygenation for 6 hours; n > 500 cells from 4 experiments (exp); (B) Representative time course images of HRE-GFP in HEK293 cells in sustained hypoxia show a subset of cells exhibit reduction in HRE-GFP levels (arrowheads); Scale bar = 25μm. (C) Schematic showing putative dynamic diversity of HRE responses, with a subset of cells switching from HRE-GFPhi to HRE-GFPlo state. (D) Representative HRE-GFP intensity traces in HEK293 cells in 12 hours of hypoxia, showing cells that remain in HRE-GFPlo state (red), those that monotonically increase and maintain their HRE-GFP levels (green), and those that exhibit oscillations between HRE-GFPhi to HRE-GFPlo (orange); (E) Live cell imaging analysis of HRE-GFP transitions in sustained hypoxia (12 hours) followed by reoxygenation (6 hours); Shown is percentage of HEK293 cells in 6 hours segments belonging to subclasses in D; statistical significance shown for each subclass (colored) vs previous 6h time segment; n = 7 exp with more than 500 cells/exp. (F) Live metabolic analysis by Seahorse of HeLa cells shows oxygen consumption rate (OCR) decreases following 5 mM DMOG treatment vs control, and rate of acid efflux (ECAR) levels increase with DMOG treatment similar to oligomycin treatment; n = 6 exp; (G) Representative traces of HRE-GFP intensity in HEK293 cells in low density for 18 hours in hypoxia with 10 mM lactate (colors are defined as in (D)). (H) Fractions of HEK293 cells displaying HRE-GFP oscillation during 12 hours of hypoxia in 25 mM glucose, 10 mM lactate (without or with 100 nM AR-C155858, an inhibitor against MCT-1 transporter); n = 5 exp; (I) Imaging of HRE-GFP patient-derived breast cancer cells in the presence of 10mM lactate; colored arrows point to bounded cells representative of stably high (green), stably low (pink), and oscillating (orange) response; Phase-contrast images shown in Fig S1L; (J) Percentage of oscillating patient-derived cells with lactate, without or with AR-155858; n = 6 exp (>300 cells/exp). Error bars: s.e.m.; *: p < 0.05, **: p < 0.01, ***: p < 0.001.
Sustained hypoxia is thought to result in HIF-1α-mediated glycolysis6,21,22. Indeed, stabilization of HIF-1α by DMOG decreases the oxygen consumption rate (OCR) and simultaneously increases the extracellular acidification rate (ECAR) in HeLa cells (Fig 1F). Increased glycolysis enhances lactate production, accounting for high lactate concentrations in tumors 23,24. We hypothesized that lactate, the end product of anaerobic glycolysis, might be responsible for the observed dynamic variation in HIF-1α activity. We evaluated intracellular lactate using Laconic biosensor 25, calibrated in different doses of extracellular lactate (Fig S1G–H). We observed a marked lactate dose-dependent increase in the fraction of cells showing oscillatory HIF-1α responses up to 10mM lactate (comparable to the levels generated by hypoxic cells (Fig S1I), and reported for various tumors26,27). This lactate concentration was used in subsequent experiments. Notably, addition of lactate increased the proportion of HRE-GFPhi cells, while also increasing the oscillatory subpopulation in 2% hypoxia (Fig S1J). We observed similar oscillatory responses in A375 melanoma cells, and MCF7A breast cancer cells, also in lactate dependent fashion (Fig S1K). Addition of 100 nM AR-C518858, an inhibitor of monocarboxylate transporter (MCT-1), which shunts lactate into cells28, reversed this lactate effect, suggesting a direct role for lactate in regulating oscillatory HIF-1α activity (Fig 1G–H). To determine whether oscillatory responses also occur in primary tumor cells, we isolated breast cancer epithelial cells from an ER/PR+HER2− breast cancer patient biopsy, transduced them with a HRE-GFP-expressing lentivirus and observed the cells in 10 mM lactate and 2% O2. We again observed HIF-1α fluctuations (Fig 1I–J, S1L), mitigated by MCT-1 inhibitor (Fig 1J). We then set out to explore the mechanisms of these dynamic hypoxic responses, focusing on how lactate can help revert HIF-1α stabilization in a sub-population of hypoxic cancer cells.
Oscillatory HIF-1α activity dynamics is prevalent in cells respiring in hypoxia
We hypothesized that effects of an increasing lactate dose would be related to the metabolic effects of this molecule. Consistent with previous reports29, microarray analysis showed higher expression of genes related to oxidative phosphorylation (Oxphos), and lower expression of genes related to glycolysis in HRE-GFPlo than HRE-GFPhi cells sorted for GFP levels after 24 hours of hypoxia 17 (Fig S2A). To explore this phenomenon further at the single cell level, we assessed the mitochondrial membrane potential (expecting distinct values in oxphos and glycolysis) by monitoring the relative intensity of TMRM sensor in HeLa cells. Controls demonstrated increased TMRM levels (i.e. membrane potential hyperpolarization) following ATP synthase inhibition (Fig S2B–C). In bulk population, TMRM was elevated in sustained hypoxia, along with HRE-GFP levels (Fig S2D). At the individual cell level, however, dynamic patterns of both signals emerged (Fig S2E). In most cells, both HRE-GFP and TMRM stably increased in hypoxia (Fig S2F–G (case i)). In a smaller subset, TMRM either did not change, or increased after delay (Fig S2F–G), and it was in these cells that HRE-GFP underwent oscillations as long as TMRM remained relatively low (Fig S2F–G (case ii)). A sizable fraction of cells also displayed hyperpolarized mitochondrial potential even prior to hypoxia onset, consistent with the Warburg effect (Fig S2F–H). In these cells, HRE-GFP increased with the onset of hypoxia and remained stably high (Fig S2G (case iii)). Overall, these dynamic patterns indicate that maintenance of consistently lower mitochondrial membrane potential correlates with oscillating HRE-GFP levels.
To test whether transition to Oxphos is accompanied by increased reactive oxygen species (ROS) levels, we used DCF-DA biosensor in HeLa cells cultured in pyruvate for 24 hours to induce mitochondrial respiration. DCF-DA accumulation was markedly increased compared to cells cultured in glucose, which was reversed by shutting down Oxphos by rotenone, indicating that Oxphos in cancer cells results in intracellular ROS accumulation (Fig 2A). Adding peroxide to the medium increased the percentage of cells with HRE-GFP oscillations, which was reversed by anti-oxidant L-N-acetylcysteine (LNAC), suggesting that ROS accumulation promotes the oscillation phenotype (Fig 2B). To explore whether intracellular ROS promote HRE-driven oscillations, we sorted cells based on intensity levels of a fluorogenic indicator of oxidative stress, CellROX, and observed HRE-GFP dynamics. The subpopulation with high initial ROS levels had a higher percentage of oscillating cells than that with low initial ROS levels, showing that Oxphos-induced ROS accumulation can promote HIF-1α oscillations (Fig 2C).
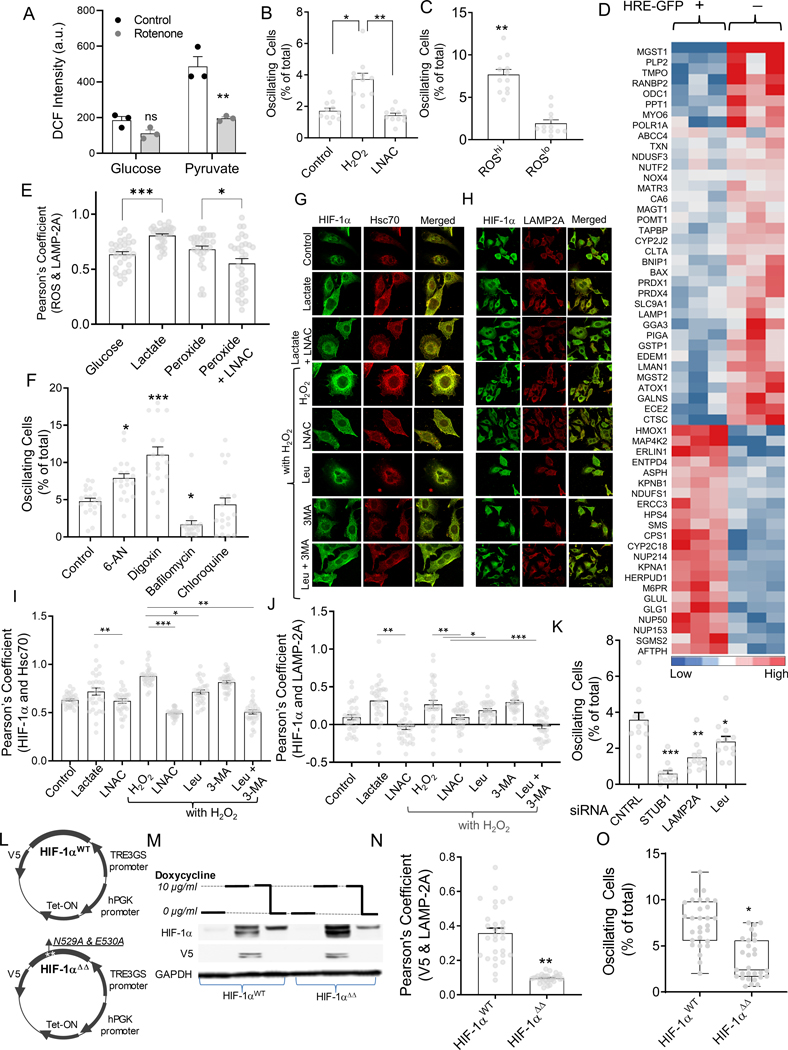
Fig 2. ROS-induced CMA controls HIF-1α dynamics in hypoxic cells.
(A-D) ROS increases oscillations in HIF-1α activity; (A) Average DCF-DA intensities in HeLa cells cultured with 25 mM glucose or pyruvate, before and after treatment with Rotenone for 30 minutes; n = 3 exp; (B) Fraction of HRE-GFP cells displaying oscillation in the presence of 5 μM H2O2 with or without 1 mM LNAC; n = 10 exp; (C) Fraction of oscillatory cells in subpopulations sorted for high CellROX and low CellROX, imaged after 6 hours subsequent to sorting; n =12 exp. (D) Microarray analysis of gene transcripts related to redox, and chaperone-mediated autophagy (CMA) pathways in HypoxCR-HEK293 cells 17 cultured in hypoxia for 12 hours, and sorted for GFPhi and GFPlo levels. (E-J) ROS induced CMA mediates HIF-1α oscillations; (E) Pearson’s coefficient of correlation between ROS levels and LAMP-2A levels in individual HeLa cells shown on the right; n > 28 cells; (F) Percentage of oscillating cells in the presence of activators (50 mM 6-AN, and 200 nM Digoxin), and inhibitors of lysosomal degradation (10nM Bafilomycin, and 50 μM Chloroquin). (G-H) Representative images showing HIF-1α, along with lysosomal Hsc70 13D3 (G), and LAMP-2A (H) in HeLa cells cultured for 12 hours in 1% O2, 10 mM lactate, lactate with LNAC, as well as 25 μm H2O2 without or with LNAC, or CMA inhibitor Leupeptin (50 μM); (I-J) Pearson’s coefficient for colocalization of lysosomal Hsc70, or LAMP-2A and HIF-1α; n = 30 cells; (K) Live cell imaging analysis of GFP oscillations in HIF-1α-GFP transfected HeLa cells cultured in hypoxia (control), or with Leupeptin (50 μM), or after siRNA mediated knockdown of STUB1 and LAMP-2A; n = 12 cells; (L) Schematic showing plasmid maps for HIF-1αWT and HIF-1αΔΔ with mutation in the KFERQ-like motif in the HIF-1α encoding sequence. (M) Immunoblot showing abundance of V5 tagged-HIF-1α, and total HIF-1α in HIF-1αWT and HIF-1αΔΔ cells maintained in 5% O2 and treated with doxycline for 8 hours and then withdrawn for 2 hours; GAPDH shown as loading control; (N) Pearson’s coefficient of correlation between HIF-1α and LAMP-2A in HeLa cells stably expressing HIF-1αWT and HIF-1αΔΔ; n > 25 cells; (O) Live cell imaging analysis of GFP oscillations in HeLa cells transduced with wild-type HIF-1αWT and mutated HIF-1αΔΔ; n > 20 samples. Error bars: s.e.m.; *: p < 0.05, **: p < 0.01, ***: p < 0.001.
Oscillation in HIF-1α activity is controlled by ROS-dependent chaperone-mediated autophagy
Microarray analysis indicated that cells with lower HRE-GFP expression during sustained hypoxia have increased expression of genes associated with ROS metabolism and chaperone-mediated autophagy (CMA) (Fig 2D). CMA is a mechanism of lysosomal degradation reported to be associated with increased ROS levels30–32. Since proteins targeted for CMA-mediated lysosomal degradation include HIF-1α33,34, we asked whether CMA is activated under the conditions that lead to HIF-1α oscillations. When we simultaneously measured DCF and LAMP-2A levels in HeLa cells35, we observed that an increase in ROS was indeed correlated with an enhanced lysosome buildup under different culture conditions, but particularly in the presence of lactate and peroxide, an effect partially reversed by LNAC (Fig 2E, S3A).
We next explored whether CMA is indeed involved in regulation of dynamic HIF-1α fluctuations. Inhibiting lysosomal degradation with bafilomycin (a specific vacuolar-type H+ ATPase inhibitor) led to modest increase in HIF-1α levels (Fig S3B). Promoting lysosomal degradation with 6-aminonicotinamide (6-AN) or digoxin increased the percentage of oscillating cells (Fig 2F). Inhibition of lysosomal protein degradation in hypoxic HeLa cells subjected to H2O2 resulted in a decreased colocalization of HIF-1α with lysosomal Hsc70 as well as LAMP-2A (Fig 2G–J). Both lactate and H2O2 reduced the nuclear localization of HIF-1α (Fig S3C). On the other hand, addition of 10mM 3-MethylAdenine (3-MA, an inhibitor of macroautophagy) did not further decrease HIF-1α colocalization with lysosomal Hsc70 and LAMP-2A (Fig 2G–J), suggesting that CMA rather than macroautophagy is primarily responsible for HIF-1α degradation in oxidative stress. Finally, we observed that adding lactate to hypoxic HeLa cells increased colocalization of HIF-1α with lysosomal Hsc70, and LAMP-2A, an effect reversed by LNAC, suggesting that lactate causes lysosomal degradation of HIF-1α by increasing oxidative stress (Fig 2G–J).
We next sought to provide genetic evidence for the role of CMA in controlling HIF-1α response dynamics. HIF-1α is targeted for lysosomal degradation by CMA via K63 linked ubiquitination mediated by STUB1, a ubiquitin ligase and co-chaperone36. Silencing of STUB1, as well as LAMP-2A significantly reduced percentage of oscillating cells (Fig 2K, S3D). Because targeting the protein to the lysosomes by Hsc70 is dependent on the KFERQ motif in HIF-1α 36,37, we investigated the responses of cells expressing HIF-1α with a mutation disrupting this motif (N529EFKL533 mutated to A529AFKL533) (Fig 2L). Both the mutated version (HIF-1αΔΔ) and the wild type control (HIF-1αWT) were expressed under inducible control of the tetracycline dependent EF-1 promoter (Fig S3E), allowing direct control of HIF-1α levels (Fig 2M). As expected, we found that HIF-1α accumulation in the lysosomes (after bafilomycin treatment), was significantly reduced in the HIF-1αΔΔ cells vs. HIF-1αWT cells. (Fig 2N, S3F). Immunoblotting also showed reduced abundance of HIF-1α (both endogenous and encoded by the HIF-1αWT vector) after treatment with lactate, reversed by leupeptin, a CMA inhibitor (Fig S3G). In contrast, in HIF-1αΔΔ cells, addition of lactate or leupeptin did not affect HIF-1α abundance, supporting the role of CMA in HIF-1α degradation (Fig S3G). Oscillations were reduced in cells expressing HIF-1αΔΔ, even in the presence of lactate (Fig 2O). In summary, these data, relying on both genetic and pharmacological perturbations strongly suggested the critical role of CMA in oscillatory HIF-1α dynamics.
Lactate regulates ROS dependent CMA-mediated control of HIF-1α activity and abundance
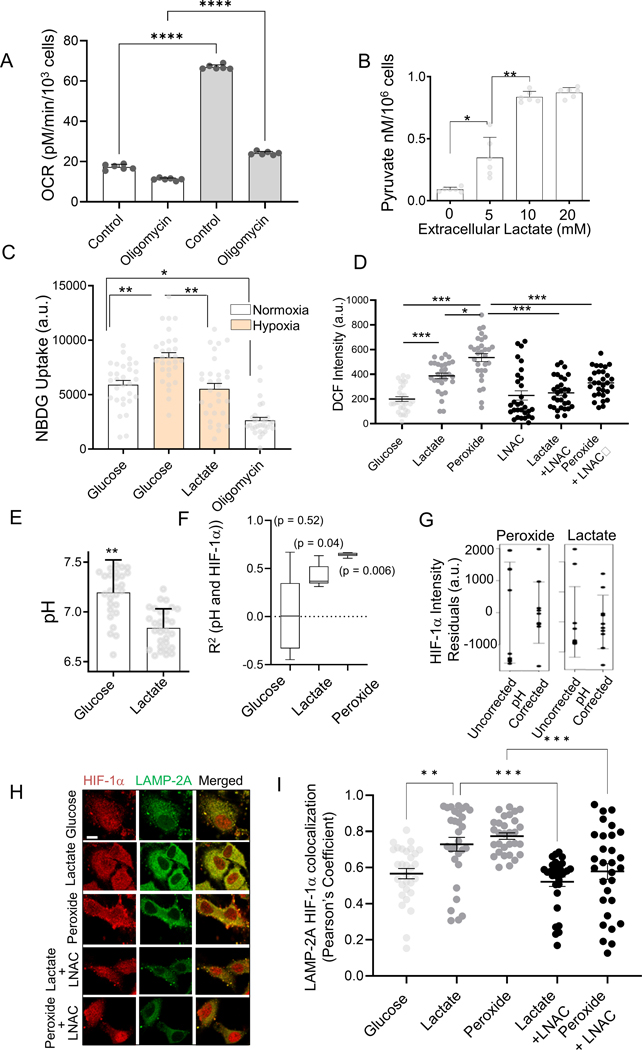
Accumulating extracellular lactate can be converted into intracellular pyruvate, which can further lead to intracellular acidification due to concomitant reduction of NAD+ to NADH, and generation of H+ 38. We therefore tested whether extracellular lactate can promote HIF-1α oscillations via CMA in a ROS and pH dependent fashion. First, we confirmed an increased oxygen consumption rate of HeLa cells in lactate (Fig 3A), consistent with previous reports 39,40. Inhibition of Oxphos by oligomycin for 30 minutes reduced OCR for cells in lactate, but not in glucose (Fig 3A). Oligomycin treatment also resulted in massive death of cells in lactate-containing medium within 3 hours, indicating their critical dependence on Oxphos. This result is consistent with putative extracellular lactate conversion into intracellular pyruvate. Pyruvate levels indeed showed dose dependence on lactate (Fig 3B). Whereas hypoxia increased glucose uptake in cells, addition of lactate decreased it (Fig 3C). We further confirmed that intracellular ROS levels were increased in the presence of 10 mM lactate, an effect that was reversed by the antioxidant LNAC (Fig 3D).
Fig 3. HIF-1α dynamics is regulated by lactate through control of intracellular ROS and pH.
(A) Oxygen consumption rate (OCR) in HeLa cells with 10 mM glucose, or 10 mM lactate + 5 mM glucose, or oligomycin; n = 6 exp; (B) Intracellular pyruvate levels in HeLa cells to extracellular lactate after 12 hours; n = 6 exp; (C) NBDG uptake in HEK293 cells in normoxia or hypoxia, with10 mM lactate in 12 hours; n = 30 cells; (D) Average DCF intensity in HeLa cells cultured in medium containing a combination of glucose, lactate, H2O2, and LNAC after 12 hours; n > 25 cells. (E-G) Lactate mediated acidosis contributes to variation in HRE-GFP levels; (E) Intracellular pH (pHi, measured by Snarf-1 dye) of HeLa cells in the presence of glucose, lactate, H2O2, and LNAC; n = 30 cells; (F) Correlation of pHi and HIF-1α-GFP levels in HeLa cells cultured in glucose, lactate, or H2O2; (G) Spread in HIF-1α-GFP intensities attributed to pH-mediated degradation; Total noise is the observed GFP intensity values minus the mean; pH corrected noise is the residual deviation remaining after regressing to the pH; error bars denote one standard deviation on each side; n = 10 samples. (H) Representative immunostained confocal images showing colocalization of LAMP-2A and HIF-1α-GFP in HeLa cells treated with 10nM Bafilomycin, and cultured for 12 hours in the presence of glucose, lactate, and H2O2 without, or in the presence of LNAC; green: LAMP-2A, red: HIF-1α-GFP; Scale bar = 2 μm; Pearson’s coefficient of spatial correlation shown in (I); Concentrations where applicable: 25 mM glucose; or 2 mM glucose added to 10 mM lactate, 25 μM H2O2, 10mM LNAC; n = 30 cells. Error bars: s.e.m.; *: p < 0.05, **: p < 0.01, ***: p < 0.001.
We next explored if lactate conversion to pyruvate can induce intracellular acidosis, as has been reported previously 41. We evaluated the intracellular pH (pHi) of cells using the ratiometric Snarf-1 substrate (pKα = 7.2) (Fig S3H–I), and found that pHi was significantly lower in cells cultured in lactate vs glucose (Fig 3E). Co-immunostaining with HIF-1α revealed that under elevated lactate and peroxide conditions, HIF-1α levels were correlated with the pHi (Fig 3F). We then calculated the residuals in HIF-1α abundance (deviation from the mean) both from raw observations, and after normalizing HIF-1α levels using pHi as a linear predictor (Fig 3G). We indeed observed a decrease in variation in HIF-1α levels after accounting for pHi variation, for cells cultured in lactate and peroxide, indicating that pHi partially explained the variation in HIF-1α levels in the same cells (Fig 3G). Overall, these results suggested elevated pHi can indeed lead to an increase in HIF-1α abundance.
Finally, we explored whether lactate regulated oscillatory HIF-1α activity in a CMA dependent manner. Lysosomes isolated from bafilomycin-treated HEK293 cells showed increased HIF-1α accumulation when cultured in lactate or H2O2 vs. glucose, while LNAC reversed the effect (Fig S3J). Similarly, HIF-1α and LAMP-2A displayed significantly higher co-localization in cells cultured in lactate vs. glucose. This effect was also reversed by LNAC suggesting that lactate increased CMA-mediated degradation of HIF-1α in a ROS dependent manner (Fig 3H–I). Response to lactate was similar to the response to H2O2, further supporting a link to ROS production (Fig 3H–I). These results suggested that extracellular lactate can enhance HRE dynamics in hypoxia by promoting intracellular acidification and ROS generation, and the resulting CMA upregulation.
Mathematical modeling predicts all three types of hypoxia-induced HIF-1α dynamics
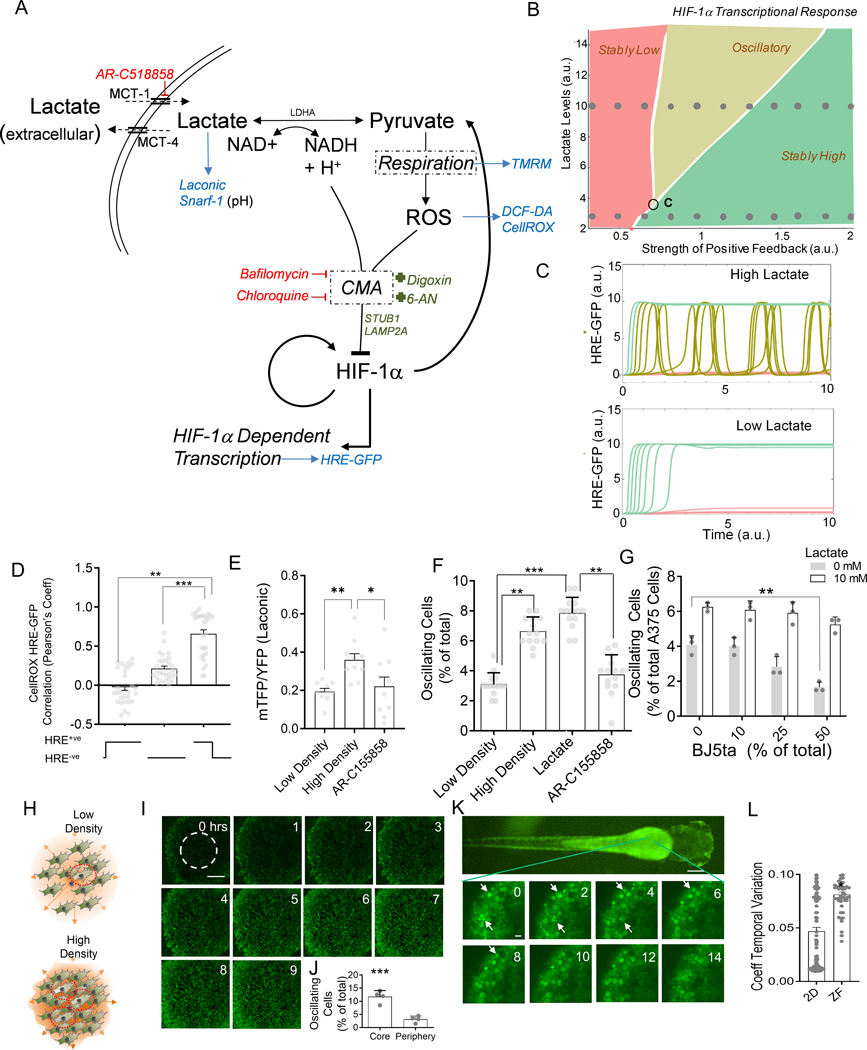
Our findings suggest that, in hypoxic cells, HIF-1α activity can undergo complex dynamic changes in abundance and activity, which fall into three general classes: no response, steady upregulation and oscillatory fluctuations. To explore if a single mechanism can underlie these diverse responses, we developed a simple mathematical model combining two critical regulatory mechanisms controlling HIF-1α. One is the presence of positive feedbacks in HIF-1α accumulation, many of which have been previously identified 41–44. Positive feedback can help explain the concurrent presence of stably non-responsive and stably responsive cell subpopulations, but not the oscillatory behavior. The other regulatory mechanism constitutes a negative feedback identified in the experiments described above, whereby in a subset of cells, HIF-1α-induced expression of LDH leads to accumulation of pyruvate that can in turn lead to an increase in ROS and lowering of pH – triggering CMA-mediated HIF-1α degradation and continued respiration (Fig 4A). Very generally, a combination of a positive and negative feedback regulation is expected to lead to a range of dynamic responses, including sustained oscillations 45. We found that all three experimentally observed dynamic characteristics of HIF-1α transcriptional responses were predicted by a model combining a positive and negative feedback loops, provided that the strength of the positive feedback exceeds a critical value (Fig 4B). Variability of cells within a population can ensure that all types of dynamical responses can occur simultaneously, with cells ‘exploring’ distinct regions of the corresponding phase diagram. Furthermore, the model predicted that the fraction of the cells undergoing oscillatory response would rise with the increasing strength of the positive feedback or at higher lactate levels (Fig 4B–C), with the latter prediction providing a test for the model. As noted above, we indeed observed that elevated lactate concentrations led to an increase in the fraction of cells undergoing oscillations (Fig 1H–I). The relatively small fraction of cells undergoing oscillations under the experimental conditions used here also suggested that the parameters characterizing the cell population were close to the critical response regime, at which the oscillations begin to emerge (point ‘C’ in Fig 4B). The model further predicted that other components of the negative feedback loop would undergo oscillations in concert with HIF-1α, including ROS. To validate the prediction, we tracked HRE-GFP HeLa cells co-stained with CellROX Deep Red (Fig 4D), and found that CellROX and HRE-GFP levels were time-wise correlated in oscillating cells with CellROX intensity rising and falling along with HRE-GFP levels over time (Fig 4D, Fig S4A–B). Although this model was not meant to be fully quantitative, it nevertheless provided a semi-quantitative basis for the proposed mechanism of different co-occurring dynamic single cell responses of HIF-1α abundance and kinetics in hypoxic cells.
Fig 4. Modeling CMA mediated HIF-1α degradation accounts for diverse dynamics in HIF-1α activity mediated by cell density.
(A) Hypothesized molecular circuit controlling the dynamics of HIF-1α responses; CMA: chaperone mediated autophagy; Also listed are the main perturbations used in the study: Red: inhibitors, Green: activators, Blue: reporters. (B) Bifurcation diagram of the mathematical model corresponding to (A), see Methods, demonstrating existence of different dynamic response modes; ‘C’ denotes the critical point of the phase diagram corresponding to the onset of oscillatory responses; (C) Model predictions for dynamic HRE-GFP intensity profiles in hypoxic cells in the presence of low and high levels of lactate; Traces correspond to the sets of parameter values indicated by two rows of circular dots in the bifurcation diagram in (B). (D) Pearson’s coefficient showing correlation of HRE-GFP and CellROX levels; n = 30 cells; (E-G) The effect of cell density on variation in HRE-GFP expression; (E) Laconic FRET ratio in A375 cells; n = 12 cells; (F) HRE-GFP transitions in HEK293 cells cultured at low density, at high density with and without 10 mM lactate, or MCT-1 transporter inhibitor; n > 12 exp; (G) Percentage of HRE-GFP oscillations in A375 in co- culture with BJ5ta fibroblasts at different relative percentages, also with lactate; n = 3 exp; (H) Schematic showing that high density in solid tumors can result in lactate buildup in the core, causing increased oscillations in HIF-1α activity; (I-J) Confocal time lapsed images of HRE-GFP HEK293 spheroids in CoCl2 and frequency of GFP switches in the core (area shown within dotted line); Scale bar = 100 μm; n = 4 spheroids. (K) Confocal images showing dynamics of HRE-GFP HEK293 cells in a zebrafish embryo, imaged 1 day after microinjection into the yolk sac; white arrows show cells with changes in GFP expression; (L) Coefficient of temporal variation (standard deviation normalized by mean) over 14 hours of observation for detectable cells in 2D cultures, and zebrafish; n > 40 locations. Error bars: s.e.m.; *: p < 0.05, **: p < 0.01, ***: p < 0.001.
Lactate can act as a cell density dependent signal modulating the dynamics of HIF-1α response
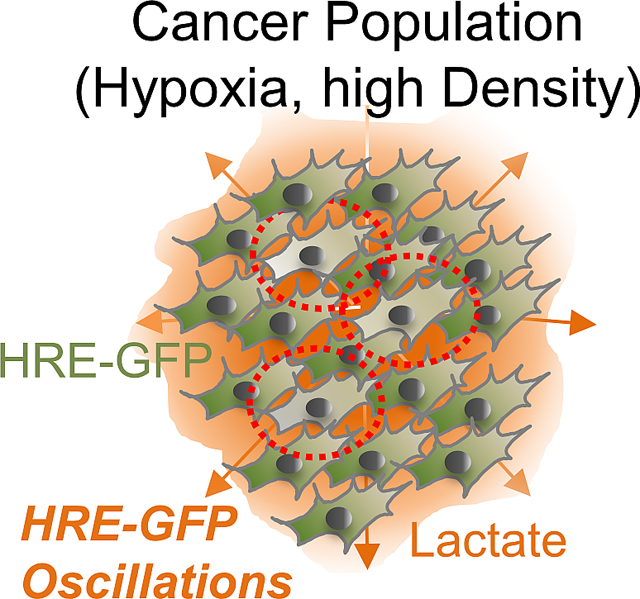
Lactate secreted by glycolytic cells accumulates to high local levels in hypoxic tumors, modulated by the local cell density. To experimentally examine cell density dependence of dynamic HIF-1α responses, we cultured HRE-HEK293 cells at low and high cell density conditions, limiting cell growth area and diffusion of secreted factors using a microfabricated stencil (Fig S4C–D). Cells cultured at higher densities showed a more rapid (2-fold change) increase in GFP levels, with a modestly higher variability (Fig S4E–F). Dense cultures in hypoxia showed higher lactate accumulation, reversed by inhibition of the lactate transporter (Fig 4E). At higher cell density, the fraction of oscillating cells increased significantly (Fig 4F). This effect could not be further enhanced by adding exogenous lactate, but was reversed by AR-C155858 (Fig 4F), supporting a critical role for lactate accumulation in this density-dependent outcome. The density dependence of a phenotypic feature (e.g., the probability to undergo oscillatory responses) on accumulation of a cell-secreted signal is a common way to ensure collective responses, as e.g., in bacterial quorum sensing, or eukaryotic community effects46,47, which similarly result from cell density-dependent build-up of secreted signals. To further explore the density dependence of HIF-1α activity in the context of a tumor model – where cancer cell density can be indirectly modulated by less glycolytic stromal cells – we co-cultured HRE-GFP transfected A375 human melanoma cells with BJ5ta human skin fibroblast cells, varying their relative fractions in confluency. Percentage of A375 cells displaying HIF-1α oscillations declined with relative fractional increase in BJ5ta cells, in a way that was reversed by adding exogenous lactate (Fig 4G). To test this effect in 3D, we created spheroids of HRE-GFP HEK293 cells and observed them in the presence of a chemical hypoxia inducer, CoCl2 (Fig 4H–I). We indeed found a higher fraction of oscillatory responses in HRE-GFP levels in the spheroids, with the percentage of oscillating cells substantially higher at the spheroid cores vs. their peripheries (Fig 4J), suggesting that local accumulation of lactate can indeed increase in denser areas of a 3D cell colony, creating spatially heterogeneous population responses. To observe this phenomenon in vivo, we used a zebrafish xenograft model, which allows for dynamic imaging of live cells in vivo. We microinjected HRE-HEK293 cells into a developing zebrafish embryo yolk sac, 48 hours after hatching, and observed the cells 24 hours thereafter under confocal microscope. Live observation for more than 12 hours revealed that oscillations were indeed significantly enhanced in this in vivo xenograft model (Fig 4K, L). Overall, these results collectively suggested that high cell density, in different culture conditions and in vivo, can increase the probability of oscillatory responses to hypoxia, in a lactate dependent, quorum sensing-like fashion.
HIF-1α oscillations promote differential gene expression and proliferation in hypoxia
We next asked whether oscillations in HIF-1α abundance and activity in hypoxia can specify gene expression and phenotypic responses that qualitatively differ from those observed in cells displaying steadily low or high HIF-1α activity. There is much evidence that the responses seen when signaling inputs oscillate can be quite distinct from those triggered by sustained inputs48,49. Since oscillations were observed only in a small cell sub-population, we enriched the oscillating responses by entraining HeLa cells to oscillatory hypoxic inputs, mimicking the frequency of the naturally observed HIF-1α oscillations (6 hours of hypoxia followed by 3 hours of reoxygenation for a total of 24 hours) (Fig 5A). HIF-1α abundance increased, as expected, after 6 hours of hypoxia, and rapidly decreased upon reoxygenation, so that there was no detectable HIF-1α after 1 hour of normoxia, confirming oscillatory changes in HIF-1α abundance in a large number of cells (Fig 5B). We then explored gene expression using RNA sequencing. A total of 1702 genes were significantly differentially regulated between the three conditions (false discovery rate < 0.1). Hierarchical clustering revealed distinct gene expression profiles (Fig 5C). For the majority of these genes, we observed that their expression levels in oscillating cells were intermediate between cells subjected to steady hypoxia and normoxia, indicating cell response to an integrated (averaged, and thus intermediate) oscillatory hypoxic input. However, crucially, we also found groups of genes whose expression profiles were qualitatively distinct from (e.g., higher or lower than) those triggered by both steady hypoxic or steady normoxic conditions (oscillation-responsive genes), indicating that oscillatory HIF-1α activity may indeed differentially regulate gene expression vs. either persistently high or low levels (Fig S5, 5C).
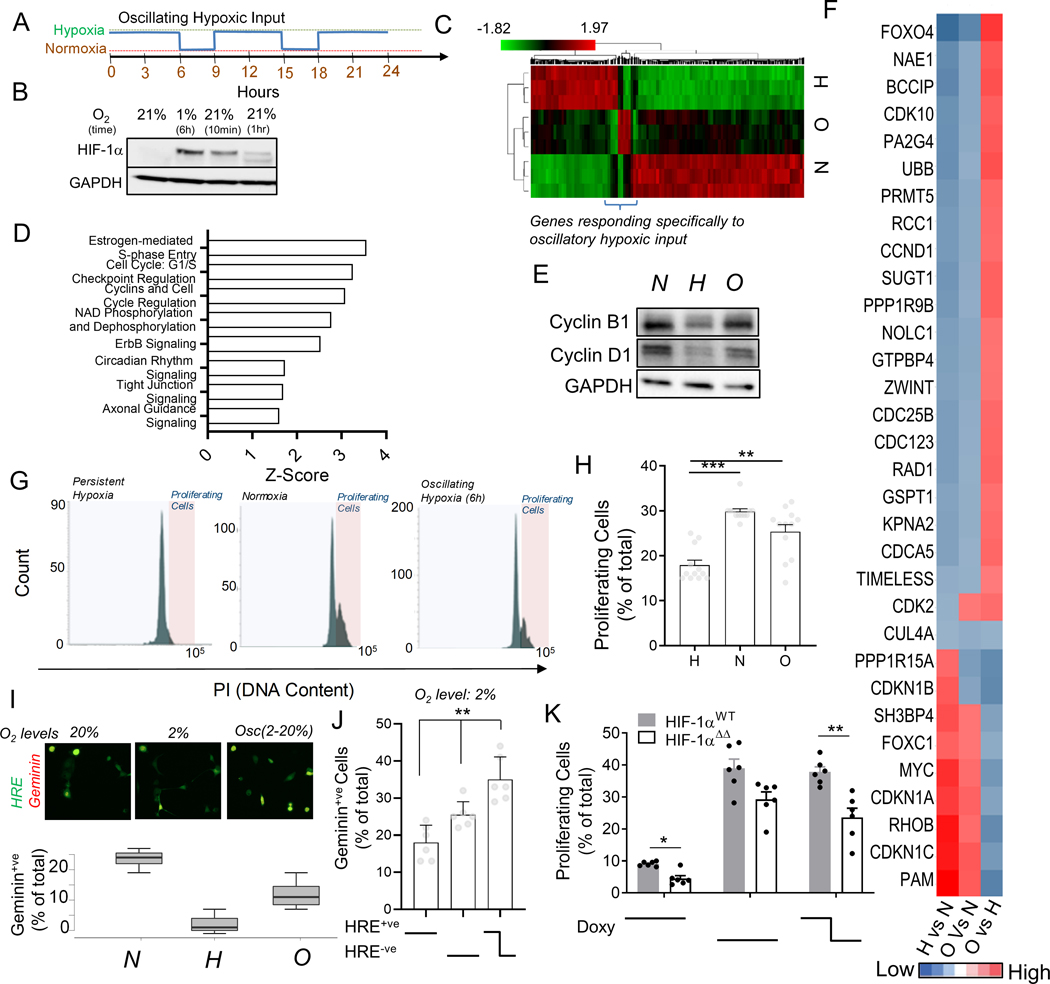
Fig 5. Oscillating hypoxic response can lead to differential gene regulation and allow proliferation under hypoxia.
(A) Experimental protocol to entrain intracellular hypoxic response by imposing oscillations of extracellular O2. (B) Immunoblot showing HIF-1α abundance in HeLa cells after 1% hypoxia for 6 hours, and after re-oxygenation for 10 min, and 1 hour, GAPDH is loading control; (C) RNA-Seq based hierarchical clustering of gene expression in HeLa cells kept in normoxia, and stable or oscillating hypoxia for > 24 h; Genes differentially regulated by oscillatory hypoxia shown in bracket. (See Fig S5). (D) Key Ingenuity pathways upregulated in gene-set differentially regulated by oscillatory hypoxia (referred to in B) vs persistent hypoxia. (E-J) Oscillations in HIF-1α abundance allows cells to continue to proliferate in hypoxia; (E) Immunoblot showing abundance of Cyclin B1 and Cyclin D1 in cells in normoxia, and stable or oscillating hypoxia; GAPDH is loading control. (F) RNA-Seq-based analysis of pairwise fold-changes between cells in above conditions for cell-cycle related genes. (G) Propidium iodide levels in HEK293 cells after conditioning with stably high, low or oscillatory input of 2% oxygen for 18 h; Quantification of gated cycling cells shown in (H); n = 12 exp. (I) Percentage of HypoxCR-GFP transfected HeLa cells positive for Geminin-mCherry after 12 h each of environmental normoxia, hypoxia, and oscillating hypoxia; above panel shows a representative location. (J) Percentage of HypoxCR-GFP transfected HEK293 cells positive for Geminin-mCherry within HRE-GFP+ve, HRE-GFP-ve subpopulations, and cells that spontaneously oscillate in persistent hypoxia; n = 6 exp. (K) Percentage of proliferating cells in HeLa cells stably transfected with Tet-inducible wild-type HIF-1α (HIF-1αWT) and HIF-1α with mutation in KFERQ-like lysosomal targeting motif (HIF-1αΔΔ), with stable or oscillating doxycycline concentrations; n = 6 exp. Error bars: s.e.m.; *: p < 0.05, **: p < 0.01, ***: p < 0.001.
The oscillation-responsive genes encoded various key transcriptional and signaling molecules, controllers of cell cycle, protein stability and intracellular trafficking, indicating that key cellular phenotypes can indeed be differentially controlled in cells with oscillatory hypoxic responses (Fig S5). Gene ontology enrichment analysis further showed that, in comparison to genes upregulated by persistent hypoxia, the oscillation-responsive genes showed a significantly higher enrichment in regulators of cell cycle, NAD metabolism, metastasis, and circadian rhythm (Fig 5D). Recent works, including ours, have highlighted the interaction between hypoxia and circadian rhythm50–52. To elucidate the functional significance of HIF-1α oscillations, we therefore focused on cell cycle regulation. The canonical response to hypoxia results in accumulation of HIF-1α, leading to inhibition of cell cycle53–56. Paradoxically, cell population studies have also revealed that some cancer cells can continue to respire, as well as divide in hypoxia17,54,57. Key genes related to glycolysis and Oxphos were differentially regulated in oscillatory hypoxia vs persistent hypoxia (Fig S6A). We hypothesized that this paradox can be resolved if a sub-population of cells displays HIF-1α dynamics that are distinct from the bulk response – as observed in this study. Abundance of Cyclin B1 and Cyclin D1, two essential regulators of cell cycle, were increased in cells subjected to oscillatory vs persistent hypoxia (Fig 5E). Key cell cycle activating genes are differentially upregulated in cells undergoing oscillatory hypoxic responses, including CDK2, CDC25B, CDK10, CDC25B, CDC123, CCND1, NAE1 and others, whereas many inhibitors for cell cycle were downregulated, including the CDKN1 isoforms (Fig 5F). Propidium Iodide accumulation in DNA after 24 hours of cell culture revealed that, whereas the steady hypoxia inhibited cell cycle, this effect was largely rescued in the oscillatory hypoxia (Fig 5G–H). Using our published HypoxCR biosensor reporting both on the HIF-1α activity and the cell cycle dependent Geminin expression (which is absent in G1, accumulates in S2, G2 and M phases, disappearing in metaphase-anaphase transition17, we confirmed that cells subjected to oscillatory hypoxia begin to re-enter cell cycle (Fig 5I). Notably, using HypoxCR biosensor, we confirmed that cells undergoing spontaneous HRE-GFP oscillations in stable hypoxia preferentially enter cell cycle (Fig 5J). Finally, to test whether oscillating HIF-1α can directly enable escape from cell cycle arrest, we studied HeLa cells stably transfected with plasmids expressing HIF-1αWT or HIF-1αΔΔ under Tet-on promoter control.. After 24 hours of an oscillatory doxycycline exposure (with concentration varying between high (5 mM) and low (0 mM)) with frequency matching natural oscillation (Fig 5A), we examined the percentage of cycling cells by Propidium iodide staining. Oscillatory doxcycyline exposure promoted cell cycle in hypoxia, confirming that oscillations in HIF-1α expression can allow cells to escape hypoxia induced cell cycle arrest (Fig 5K, S6B). We also found that the percentage of cycling cells was higher in HIF-1αWT compared to HIF-1αΔΔ populations, for both steady high and oscillating doxycycline levels, suggesting that inhibition of CMA reduces escape of cell cycle arrest (Fig 5K). In combination, these data strongly argue that CMA-mediated oscillations in HIF-1α abundance in a cell subpopulation can allow these cells to continue to enter cell cycle at high rates even in a hypoxic environment.
Expression of Genes Differentially Regulated by Oscillatory Hypoxia is Directionally Enriched in Human Cancers
If genes exhibiting HIF-1α oscillation-specific responses can enable or enhance cancer-related phenotypic responses, their differential expression may be enriched in clinical tumor samples. We thus examined expression profiles of these genes across diverse cancers, available through the human cancer genome atlas program (TCGA). We grouped oscillation responsive genes into two classes: those that increased their expression in stable hypoxia, but downregulated in oscillatory hypoxia (Oscdown genes), and those that decreased in expression in stable hypoxia, but increased in oscillatory hypoxia (Oscup genes). Using GEPIA 58, which collates TCGA transcriptomic data, we explored if Oscdown and Oscup genes are enriched in the tumor samples vs. the corresponding non-cancerous tissue samples. We found that Oscdown genes showed significantly lower expression in many tumor types (Fig 6A–C), with a particularly high statistical power for cancers of the breast, bladder, prostate, lung, skin, liver and head-and-neck, and the only significant exception being pancreatic adenocarcinoma and lymphoma. Furthermore, the pattern was reversed for Oscup genes, which, as a group, were significantly up-regulated across several cancers, particularly those of the pancreas, stomach, liver, and head-and-neck, with no cancer displaying significant down-regulation (Fig 6D–F). To note, the head-and-neck (HNSC) and liver (LIHC) cancers displayed both significant up-regulation of the Oscup genes and down-regulation of Oscdown genes vs. non-cancerous controls. These results broadly held when we calculated the tumor vs normal expression of Oscdown genes (Fig 6G), and Oscup genes (Fig 6H) compared to the expression changes for all genes to remove systemic bias in transcriptomic analysis.
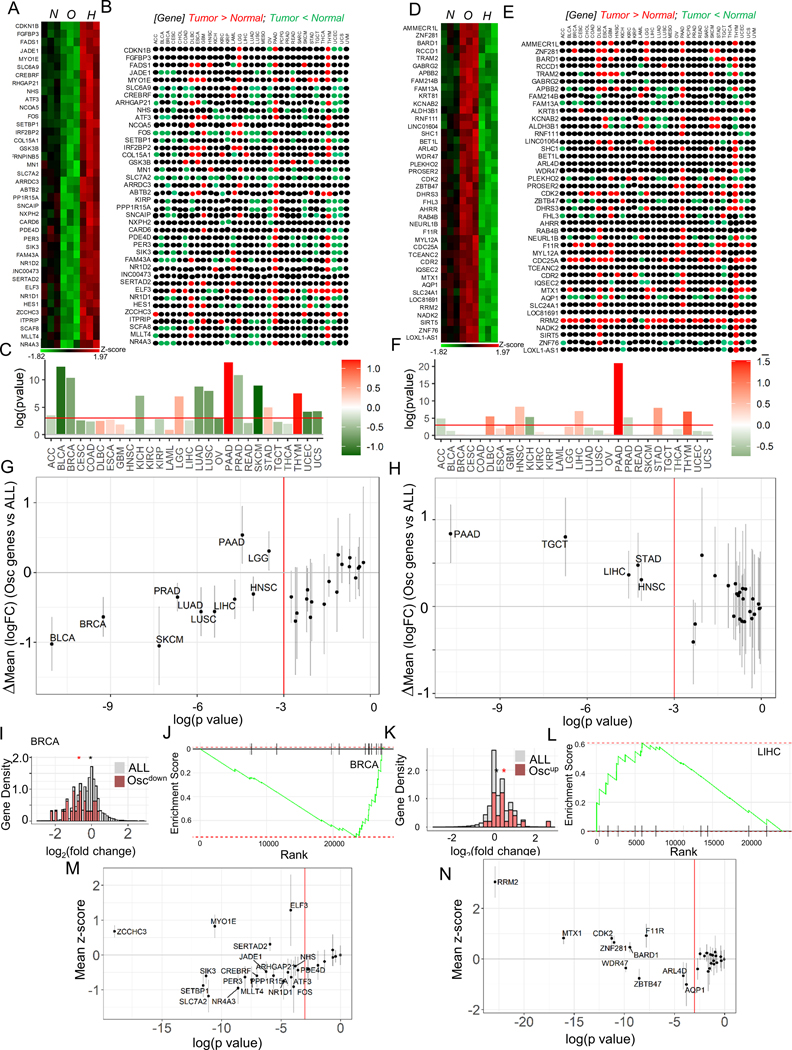
Fig 6. Oscillating hypoxic response dynamics regulates gene expression congruent to many human cancers.
(A-C) Genes downregulated by oscillating hypoxia show a correlatively downregulated signature in many human cancer types. (A) Heatmap showing genes upregulated by hypoxia, but downregulated by oscillatory hypoxia (Oscdown genes), and (B) their relative expression in cancers vs normal tissue in the TCGA tumor database; colors refer to a TCGA study where the given gene is expressed significantly higher (red), or significantly lower (green) in tumor compared to normal tissue sample, with significance for the whole set of Oscdown genes in (C). (D-F) Genes upregulated by oscillating hypoxia also show a correlatively upregulated signature in many human cancer types: (D) Heatmap showing genes downregulated in hypoxia, but upregulated in oscillatory hypoxia (Oscup genes), and (E) direction of their expression in TCGA tumors vs normal samples; color code same as B. (F) Significance of the whole set of Oscup genes shown in tumor vs normal; In C, and F, bars show overall fold change of the geneset, colorbar represents the mean log2fold change between cancer and normal samples. (G-H) Genes differentially regulated by oscillatory hypoxia are significantly downregulated (G), or upregulated (H) when compared against the directional expression of all other genes in those TCGA tumors vs control samples; In both G, and H, each dot represents a tumor comparison with normal tissue; x-axis refer to log(pvalue) of tumor vs normal comparison; bars show 95% confidence interval for mean vs all genes difference; vertical red line marks threshold of pvalue = 0.05. (I-L) Representative examples of Oscdown and Oscup gene sets in breast cancer BRCA, and liver hepatocellular carcinoma LIHC: (I) Histogram showing density of Oscdown genes (green), or all genes (pink); Genes are normalized by total gene numbers; BRCA tumor samples = 1085, normal breast samples = 291. (J) GSEA enrichment analysis in Oscdown gene expression in BRCA vs normal samples; all genes are ranked in order of decreasing tumor/normal fold changes on x-axis, with bars representing Oscdown genes. (K) Histogram showing relative distribution of fold change of Oscdown genes (green), or all genes (pink); Genes are normalized by total gene numbers; LIHC tumor samples = 369, normal breast samples = 160. In I, and K, asterisk represents the mean log2fold change between cancer and normal samples for the corresponding gene set (all or Oscdown /Oscup). (L) GSEA enrichment analysis in Oscup gene expression in LIHC cancers. (M) Oscdown genes are significantly downregulated in many TCGA cancer types when compared to all the other genes; bars show a 95% confidence interval for the difference. (N) Many Oscup genes are significantly upregulated in many TCGA cancer types compared to all the other genes; bars show a 95% confidence interval.
The relative downregulation of Oscdown genes was also evident when fold-change expression was analyzed more in detail for individual genes rather than gene groups (Fig 6I, S7A), and when we performed gene set enrichment analysis (GSEA) (Fig 6J, S7B). Conversely, we found significant and consistent upregulation of Oscup genes across multiple cancers (Fig 6H, K–L, S7C–D). We next analyzed whether individual Oscdown and Oscup genes might show consistent changes of expression in multiple cancers. We found that although the changes of expression of several genes were statistically significant when analyzed vs. all genes across all cancers, the statistical power (as judged by the mean Z-score value being greater than 1 or smaller than −1) was particularly high for up-regulation of an Oscup gene and putative oncogene, RRM259, and down-regulation of an Oscdown gene, a putative tumor suppressor SLC7A260,61. These results suggested that although that Oscdown and Oscup genes may have collective roles as suppressors or promoters of oncogenesis in diverse cancers, individual genes in these sets can be particularly consequential for pan-cancer oncogenesis. Together these results suggested a trend for significant up-regulation of Oscup genes and downregulation of Oscdown genes across multiple cancers, suggesting that they may have potential role in tumorigenesis, or inhibition of tumor suppressors in many cancer types.
Discussion
Our results suggest that HIF-1α expression and activity can display complex dynamic responses to hypoxia. The diversity of these dynamic responses, seen at the single cell level within a hypoxic cell population, is associated with the diversity of metabolic states, so that the cells that continue to respire in hypoxia undergo increased HIF-1α degradation through elevated ROS and, consequently, CMA. Competition between hypoxia-mediated canonical stabilization of HIF-1α and its CMA-mediated destabilization can result in an oscillatory response observed in this subset of cells. The number of cells in this ‘oscillatory’ subset can substantially increase in response to endogenously produced or exogenously added lactate. Lactate can be converted within cells to pyruvate leading to cellular acidosis, ROS production and CMA, promoting HIF-1α oscillations. Such oscillations could emerge, and be strengthened by other mechanisms driving HIF-1α destabilization, including changes in PHD activity62. This process allows a cancer cell population to undergo a cell density-dependent response, in which accumulating lactate produced by an increasing number of hypoxic cells leads to an elevated proportion of oscillating cells, in a manner analogous to quorum sensing or community effects. Thus, the relative number of cells displaying HIF-1α oscillations can increase with increasing tumor mass, potentially leading to adaptive responses distinct from those associated with the canonical, steady and persistent HIF-1α activation. Other existent heterogeneities within tumor, including differences in expression of LDHA and MCT1 genes63, encoding for pyruvate to lactate conversion, and lactate transport respectively, may further complicate the individual responses of cancer cells in regulating HIF-1α dependent behaviors.
What is the functional role of HIF-1α oscillations? Oscillatory vs. steady activities of signaling proteins and transcription factors may trigger expression of distinct gene subsets, thus achieving differential phenotypic outcomes, as seen in other biological systems with oscillatory dynamics48,49.We enriched the oscillatory cells by entraining them to environmental hypoxia, which resulted in a similar oscillation in HIF-1α abundance, though it could potentially affect other oxygen dependent, but HIF-1α independent responses64. Examining gene expression, we found that large subset of HIF-1α-responsive genes are expressed in cells with oscillatory hypoxic responses at levels between sustained hypoxia and normoxia. However, crucially, many genes also displayed divergent expression patterns from those triggered by sustained hypoxia, suggesting a specific response to oscillatory hypoxia. Many of these genes, when expressed, may support/reinforce the oscillatory state itself, representing a special kind of positive feedback, including TXNIP, a gene coding for a key controller of ROS levels, and H6PD, which could support CMA-mediated HIF-1α degradation. Perhaps, more importantly, we found that the expression of an isoform of pyruvate dehydrogenase kinase (PDK4) was downregulated in cells with oscillatory hypoxic response, suggesting that these cells would maintain Oxphos. PDK4 expression is reported to be directly induced by hypoxia 65, and its downregulation reduces lactate production66. Furthermore, PDK isoforms are key controllers of the switch between glycolytic and respiring states, with the high activity of the enzyme associated with directing carbon flux from the Oxphos state to glycolysis67. Our observation That PDK4 is downregulated specifically in oscillatory cells provides a putative resolution to the paradox of HIF-1α regulation of both glycolysis and respiration-related genes. Indeed, our results suggest that this divergent regulation can occur in different subsets of cells with dynamically distinct HIF-1α regulation (with the glycolysis induced by steady and hypoxic respiration by oscillatory HIF-1α activity).
Another key observation stemming from the differential gene expression analysis is the key relevance of oscillatory HIF-1α activity to tumor progression. Consistent with upregulation of cell cycle promoting genes and downregulation of negative cell cycle regulators, we find that HIF-1α oscillations can help cells escape from HIF-1α induced cell cycle arrest. As a result, within the context of increasing hypoxia and high lactate concentrations, many cancer cells may resume cell division contributing to tumor growth. Furthermore, we find that components of many key signaling pathways (e.g., ATF3, GLI-3, Calcineurin A), cytoskeleton components (e.g. Col15A), cell adhesion molecules (e.g. F11R), which are strongly implicated in carcinogenesis and tumor progression, are also differentially controlled in cells with the oscillatory response. These also include RhoA, and Rho activating protein ArhGAP21, essential for tumor metastasis. Furthermore, we previously found that respiring hypoxic cells are particularly tumorigenic in the animal models 17. These results are of potential clinical importance, since we found that these genes were either consistently downregulated (for Oscdown genes) or upregulated (for Oscup genes) across many cancers vs. normal tissue controls. This finding held when contrasted with expression of all other genes, confirming that these gene expression patterns were highly unlikely to be obtained by chance.
Our findings also may help introduce a new context for reexamining prior surprising observations related to the interplay between hypoxia and tumor progression. For instance, intermittent hypoxia can potentially entrain quasi-periodic HIF-1α activity. Interestingly, prior reports associated this phenomenon with increased cancer progression in mouse models of melanoma68 and breast cancer 69. Unstable hypoxia in tumor mass, therefore, may be a more common phenomenon than is appreciated. More generally, it will be potentially informative to examine whether Oscdown and Oscup genes identified here may constitute a useful diagnostic and prognostic clinical signature. Additionally, it will be of interest to explore whether our analysis can explain why cells with dysregulated intracellular pH, associated in our analysis with oscillatory HIF-1α responses, display more aggressive, invasive behavior and promote metastatic progression 70.
The analysis presented here also highlights the increasingly prominent role of lactate as a signaling molecule. More specifically, our findings suggest that elevated lactate may carry two important facets of information about the cell microenvironment: the hypoxic input and cell density. Both these environmental ‘variables’ can lead to higher lactate levels. Increasing lactate concentration can then, in a feedback loop, lead to an increased fraction of hypoxic cells undergoing hypoxic respiration and oscillatory deviations from the steady HIF-1α levels, decreasing the rate of lactate production. Notably, this phenomenon, confined to hypoxic cells, is distinct from the so-called metabolic symbiosis observed in various tumors, where lactate is shuttled between hypoxic glycolytic cells and more normoxic respiring cells adjacent to blood vessels 23. Signaling rather than purely metabolic role of lactate has also been implicated in polarization of tumor associated macrophages71. In combination, these results suggest that lactate can mediate diverse cell-cell communication processes, both homotypic and heterotypic, underlying various aspects of tumor biology.
Overall, our findings argue that the analysis of dynamic single cell responses can be critical for better understanding of tissue-level responses to dramatic changes in cell micro-environment, such as hypoxia. More specifically, cell sub-populations can display variability not only in the amplitude of response, but in the dynamic response characteristics, which might help diversify the effects of master regulators, such as HIF-1α, and permit complex phenotypes resulting from interactions between cells with differentiated responses. In particular, engaging in oscillatory HIF-1α-mediated hypoxic responses can generate a particularly aggressive cell sub-population, which resumes cell cycle and rewires diverse signaling networks, leading to increased tumorigenesis across multiple cancers. These findings can thus contribute to further understanding of fundamental processes controlling hypoxic responses and inform clinical strategies of cancer treatment.
STAR Methods
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled to the best of capabilities by the lead contact, Kshitiz (kshitiz@uchc.edu)
Materials Availability
New cell lines, and plasmids will be made available by lead contact on request.
Data and Code Availability
RNAseq data have been deposited at Mendeley, and are publicly available. DOI: 10.17632/m8ctygyjtm.1
ODE model is available at DOI: 10.5281/zenodo.7300158
All other data reported in this paper will be shared by the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture and maintenance.
HEK293, HeLa, A375, and MCF7A cells were purchased from ATCC, and cultured in DMEM supplemented with 10% FBS, 1% penicillin/streptomycin, and maintained at 37°C at 5% CO2. HRE-GFP cell lines were selected using 5 μg/ml puromycin, sorted using FACS (BD Biosciences), and maintained in 1 μg/ml puromycin. HyopxCR-GFP Geminin-mCherry cell lines were selected with 5μg/ml puromycin selection. These cells were subsequently maintained in 1μg/ml puromycin. For all the cell lines, we selected GFPhi cells after 18 hours of hypoxia using FACS, and then resorted GFPlo cells after 24 hours of normoxia to ensure that only the cells that can exhibit dynamics in GFP levels driven by hypoxia are used. In low cell densities, cells were cultured at 50000 cells/cm2, while for high density, cells were cultured at 500,000 cells/cm2.
Patient derived cancer cells.
Patient derived breast cancer cells were obtained from a biopsy from the UConn Health Biorepository derived from a 64 years old woman with invasive ductal carcinoma (ER/PR+HER2−). The solid tumor had an overall tumor grade II of III, and had a non-identified lymphovascular invasion.
Zebrafish xenograft and imaging.
Zebrafish were raised and maintained at 28.5°C using standard methods, and according to protocols approved by Yale University Institutional Animal Care and Use Committee (# 2017–11473). AB was used as the wild type genetic background. HRE-GFP expressing HEK293T cells were suspended in 5 μl of PBS and injected in the perivitelline space of 2 days post fertilization (dpf) zebrafish AB embryos as described previously72,73. Imaging were performed after 36 hour post injection.
METHOD DETAILS
Oxygen Reperfusion and Dynamic Imaging.
Glass bottomed 24-well plates (LiveAssay, and MatTek) were capped with polydimethylsiloxane (PDMS) caps that fit tightly and contained fluidic channels connected to an external gas perfusion driven either by a controller, or directly connected to a 2% O2 tank (5% CO2, remaining N2). Tubing connected the channel opening in the PDMS cap and perfused the gaseous mixtures directly into the medium by gentle bubbling. Pressure of gas was maintained at < 1 psi for continuous bubbling, without adversely affecting the cultured cells in the wells. Large volumes of medium was maintained in the wells to minimize any effect of gaseous bubbling, and to maintain a stable perfused gaseous milieu. In a control well, Tris(4,7-diphenyl-1,10-phenanthroline) Ruthenium dichloride (Ru(dpp)3Cl2) dye (Sigma-Aldrich, 36309–88-3) provided continuous indicator of oxygen levels.
Immunoblot.
Cultured cells were lysed using ice-cold RIPA buffer with pre-mixed protease inhibitor (Thermo Scientific, 78430), and total protein concentration was normalized using BCA Assay kit (Thermo Scientific, 23227). After appropriate dilution with sample buffer, heated to 70°C for 10 minutes, and cooled to room temperature, proteins were separated on a 4–20% w/v SDS PAGE gel (BioRad). Resolved gels were transferred to a nitrocellulose membrane (BioRad). Membrane was blocked with 3% BSA in TBST (10 mM Tris, pH 8.0, 1% Tween 20), and incubated in anti-LAMP-2A (Abcam, ab18528) or anti-HIF-1α antibodies (BD Biosciences, 610959, or ThermoFisher 16H4L13) with 1:100 dilution of the stock at 4°C overnight. Membranes were washed for 1 hour in TBST, blocked, and incubated with HRP-conjugated secondary antibodies (Pierce, anti-mouse 31160, and anti-rabbit 31188) diluted 1:5000 in blocking buffer overnight at 4°C. Membranes were washed for 2 hours in TBST and analyzed using BioRad gel imaging system, ChemiDoc XRS+ with ECL Western Blotting Substrate (Pierce, 32106). Primary antibodies used were anti-LAMP-2A (abcam, ab18528), anti-Cyclin D1 (Santa Cruz, sc-20044), anti-cyclin B1 (Santa Cruz, sc-752), and anti-GAPDH (Abcam, G8795). For analysis of lysosomal proteins, cells were dislodged using .05% Trypsin-EDTA (Life Technologies), and gently lysed. Lysosomes were isolated using the Lysosomal enrichment kit (Thermo Fisher, #89839). Lysosomes were lysed using RIPA buffer (Thermo Scientific) with addition of 1% protease inhibitor cocktail (Sigma-Aldrich, S7830).
qRT-PCR.
RNA was isolated using RNeasy microkit (QIAGEN, 74004) and resuspended in 10 ml DI H2O. cDNA was synthesized from 1 mg mRNA using Superscript IV VILO Master Mix (Thermofisher), and DNA was removed after transcription with ezDNase. qRT-PCR was performed using Primers (IDT) with Taqman Gene Expression Assays (Thermofisher), and efficiency of siRNA mediated knockdown was analyzed. Primers used were: LAMP2 (Hs00174474-m1), STUB1 (Hs01071598-g1), and GAPDH (Hs99999905-m1). Percent knockdown was calculated using comparative Ct method (ΔΔCt). GAPDH was used as an internal control, and the normalized expression value of the siRNA treatment sample was compared with the negative scrambled control to derive the (ΔΔCt, which was used to calculate the percent knockdown.
pH and ROS levels dynamic monitoring.
pH was monitored using carboxy Snarf-1 substrates (Thermo Fisher, S22801), by incubating cells in their experimental conditions with the dye at 10 μM for 45–60 minutes. For standards, cells were incubated in the dye along with 20 μM Nigericin (Invitrogen, N-1495) in precalibrated PBS buffers prepared at different pH. Cells were imaged using a Leica Laser Scanning Confocal microscope at 580 nm, and 640 nm post excitation at 514 nm using a White Laser. pH of experimental cells was calculated against the ratio of emission at 580nm and 640 nm for standard conditions. ROS was measured using either DCF-DA (Invitrogen, D339) labeling of cells at 50 μM for 15 minutes, or by CellROX deep-red (Invitrogen, C10422) at 10 μM for 30 minutes. Cells were imaged using Leica Widefield Epifluorescence microscope with LED based excitation at 620nm, and emission collected at 665nm. HIF-1α abundance was also imaged by co-immunostaining along with Snarf-1 labeling. Deviation from the mean was calculated for HIF-1α levels and residual deviations were calculated after regressing to pH (residuals that remain when predicting the HIF-1α using pH as a linear predictor). The shrinkage of the spread for the pH corrected values shows that part of the spread in HIF-1α levels is explained by pH (Fig 3F–G).
Flow cytometry.
Cells were detached from the substrate using 0.05% trypsin-EDTA, quenched with excess medium and prepared for flow cytometry with the relevant controls. Flow cytometry was performed with FACSDiva (BD Biosciences), and analyses were performed with FACSDiva 6.0 (BD Biosciences) or MATLAB 2015. Statistical analyses were performed and graphs were plotted with Prism (GraphPad).
Epifluorescence and confocal microscopy.
Both HRE-HEK293 and HIF-1α-GFP (for certain colocalization experiments) transfected HEK293 cells were cultured in glass bottomed dishes (LiveAssay) coated with 5 μg/ml fibronectin for time-lapse microscopy. Live cell epifluorescence microscopy was performed with a Zeiss Axiovert 2000 with 10x, 20x, and 40x apochromat objective lenses with a numerical aperture (NA) of 1.4. A Lambda LS Xenon arc lamp (Sutter Instrument) was used for excitation, and acquisition was performed with a Cascade 512BII camera (Roper Scientific Inc.). Additionally, two-colored imaging (CellROX/ HRE-GFP, and TMRM/HRE-GFP) was performed with Leica Widefield Fluorescence Microscope with 20x apochromat objective lenses with a numerical aperture of 1.4. Excitation was provided by a white LED with appropriate filter sets. Images were acquired and processed with SlideBook 4.2.0 (Intelligent Imaging Innovations Inc.), and Leica AFX software (Leica Microscopy Inc.). Images were processed using ImageJ and custom tracking code programmed in MATLAB. Control of oxygen levels during live cell microscopy was achieved using a custom poly demethoxy silane (PDMS) cap over the glass bottomed dish connected to a custom created gas mixer combining 5% CO2, 95% N2 with air to create the desired concentration oxygen concentration. Oxygen levels were measured using an oxygen-quenching dye [Tris(4,7-diphenyl-1,10-phenanthroline) ruthenium dichloride (Ru(dpp)3Cl2)] in a separate adjoining chamber for most experiments.
Confocal microscopy was performed on Zeiss LSM510 Meta CLSM microscope, and Leica Laser Scanning SP8 microscope. With Zeiss LSM510, a blue diode (405nm), a green argon laser (488 nm), and a red HeNe laser (568 nm) were used and imaged processed with Zeiss Meta software 3.5. For Leica SP8, we used a EL6000 white light with continuous wavelength 470~670 nm and detected using a combination of PMTs and Hybrid Detectors. Objectives used were APO 63x Oil objective (1.4 NA), and APO 40x Oil (1.3 NA).
FRET Analysis of pyronic and laconic transfected cells.
A375 cells cultured on glass-bottomed multiwell chambers (LiveAssay) were transiently transfected with the probes using Fugene 6 (Roche) based transfection reagent 24 hours before the experiment. Perfusion with 2% oxygen was started 8 hours prior to observation. Drugs, or metabolites were added 3 hours prior to imaging. We imaged the sample using Leica confocal microscope, and fluorophores were excited using a Whitelight laser at 462nm, and measured for mTFP emission in a HyDD based adjustable bandpass filter between 475–495nm, and for Venus between 520–540nm. FRET ratios were calculated using ImageJ.
Measuring mitochondrial membrane potential levels.
To measure mitochondrial membrane potential levels in cells, cells were cultured in glass-bottomed dishes (LiveAssay) and pre-incubated with 25 nM TMRM (Invitrogen, T668) for 15 minutes before microscopy. Cells were washed 2x times with medium to remove excess dye and then re-cultured in 5 nM TMRM to allow for possible exchange of potential in cells and extracellular medium. Samples were thereafter mounted on the microscope for live cell imaging.
Immunocytochemistry and immunohistology.
For immunocytochemistry, samples were washed with ice-cold PBS, fixed with 4% paraformaldehyde (Sigma-Aldrich, F8775) for 20 min at room temperature, permeabilized with 0.05% Triton X-100 (Sigma-Aldrich, 9002931) for 15 min, washed twice with PBS, and blocked with 10% goat serum and 1% AlbuMAX (Gibco, 11020021) for 1 hour. Samples wereincubated with primary antibody for 1 hour followed by three washes and incubation with Alexa Fluor–conjugated secondary antibody (Molecular Probes) for 1 hour. Primary antibodies used were anti- LAMP-2A (abcam, ab18528), anti-V5 (Abcam, ab27671), anti- HIF-1α (BD, 610958, and Abcam, ab2185), anti-Hsc70 (Abcam, ab2788). Samples were washed, mounted to microscopy slides with a drop of SlowFade (Invitrogen, S26938), and sealed with medical adhesive (Hollister, 7730).
Plasmids.
HRE-GFP plasmid contained 5 copies of 35 bp fragment from the hypoxia responsive element (HRE) of the human VEGF gene driven by a human cytomegalovirus minimal promoter and tagged to d2EGFP with a half-life of 2 hours (Addgene). The insert was cloned in gateway compatible lentiviral pLenti X1 backbone with puromycin selection. HypoxCR plasmid, and HEK293T cells stably transfected with the plasmid were used as described previously12. KFERQ mutant of HIF-1α ( HIF-1αΔΔ) was generated using QuikChange Lightning Site Directed Mutagenesis Kit (Agilent). We mutated amino acids Asn and Glu to Alanine in pcDNA3 HIF-1α wt-V5 using 5′-CAAAAAGTTTTTCTACCAATTCCAACTTGAATGCAGCGACCATATCACTATCCACATAAAAACAATATT −3′ and 5′- AATATTGTTTTTATGTGGATAGTGATATGGTCGCTGCATTCAAGTTGGAATTGGTAGAAAAACTTTTTG −3′ primers to generate KRERQ mutant HIF-1α-V5 (HIF-1αΔΔ). The mutation was verified by DNA Sanger sequencing. Both HIF-1αWT and HIF-1αΔΔ were PCR amplified (from pcDNA3 HIF-1αWT- wt- V5 using CCCTCGTAAAGAATTCATGGAGGGCGCCGG and GAGGTGGTCTGGATCCCTACGTAGAATCGAGACCGAGGAGAG and cloned into linearized (BamHI and EcoRI) pLVX-TetOne-Puro (Takara Bio).
Zebrafish xenograft imaging:
Imaging were performed after 36 hour post injection. Zebrafish embryos imaged by confocal microscopy were raised in 0.003% PTU starting after the gastrulation stage to prevent pigmentation. Embryos imaged live by confocal microscopy were anesthetized in 0.1% tricaine and mounted in 1% low melt agarose. Fluorescent images were captured using a Nikon TiE spinning disk confocal microscope using a 20x objective.
Pyruvate Titration.
Pyruvate levels were measured after specific treatments (e.g., culture in different dosage of lactate, and 5 mM glucose for 6 hours) using Biovision pyruvate assay kit (Biovision, K609). Each measurement was based on nearly 2 million cells from a 6 cm culture dish. Cells were rapidly lysed on ice, and pyruvate levels measured after substrate hybridization in a Perkins Elmer plate reader at 570 nm absorbance. Quantification was done using a standard curve from known concentrations of pyruvate.
Seahorse XF respiration assay.
Detailed methods of cellular respiration are mentioned earlier74. Briefly, Seahorse Bioscience XF-96 instrument (Agilent) was used to measure OCR or oxygen consumption rate and ECAR or extracellular acidification rate (change in pH). Each experiment was performed at least 3 times and 6 replicates were used in each run. Basal rates were measured before injection of inhibitors of the mitochondrial electron transport chain (ETC) or glycolysis. Oligomycin (4μM) was used to inhibit mitochondrial ATP synthase, Rotenone (2 μM) to inhibit Complex 1, Antimycin A (2μM) to inhibit complex 3 of ETC, FCCP (500 nM) to uncouple mitochondria and Iodoacetate (100μM) to inhibit glycolysis (glyceraldehyde-3-phosphate dehydrogenase). The data was normalized to DNA content using Picogreen DNA assay kit (ThermoFisher Scientific) following manufacturer’s instructions.
Microarray Analysis.
Microarray analysis was performed on data from Le et al.17 using Galaxy Open Source Platform, and MATLAB. The gene ontologies used were GO:0006979/response to oxidative stress, and GO:0007040/lysosome organization and biogenesis. KEGG database was used to map gene expression data to specific categories: Peroxisome (04146), Lysosome (04142), Oxidative Phosphorylation (00190), Glycolysis (00010), beta-oxidation and acyl-CoA synthesis (M00086). The Genelist categories used from Gene Ontology were: GO response to oxidative stress, GO lysosome organization and biogenesis.
RNA Sequencing and Analysis.
Cultured cells were manually treated with hypoxia, normoxia, or oscillating hypoxia as indicated. Cells were immediately treated with RNAProtect (Qiagen, 76526) to stabilize mRNA. Dissociated cells were collected and RNA isolated using Qiagen RNeasy mini kit (Qiagen, 74106). RNA Q/A and Sequencing were performed at Yale Center for Genomic Analysis using Illumina HISEQ Rapid Sequencing. Transcriptomic reads were mapped to UCSC hg38 reference genome using STAR alignment, and quantified and annotated the reads using an expectation-maximization algorithm (Partek E/M model). Normalized expression values of genes were expressed as RPKM (reads per kilobase per million reads) values using Partek E/M quantification and normalization.
ODE based Mathematical Model.
We modeled the HIF-1α dynamics based on interactions within a simple dynamical system with a relatively fast positive and delayed negative feedback (see details below) 45,75. The model was simulated with SimBiology in MATLAB 2014b (MathWorks). All simulations were integrated by ode15s stiff solver. We modeled the effect of lactate addition by modifying the endogenous, HIF-1α-dependent lactate production rate. Details of all model equations along with the parameter values can be found as equations and the parameter table below. The model reflects the positive and negative feedback interactions described in the text, with the HIF-1α- dependent regulation assumed to be saturable and non-linear. GFP represents the explicit signal from HRE-GFP construct measured experimentally.
ODEs:
Parameter Values:
| Parameter Name | Parameter Value | Annotation |
|---|---|---|
| Ksxs | 0.02 | Basal production rate of HIF-1α |
| Ksx | 1 | Max HIF-1α production rate affected by HIF-1α |
| Kpx | 1 | The constant defining the HIF-1α positive feedback |
| Kdx | 0.2 | Basal HIF-1α degradation rate |
| Kdxs | 1 | HIF-1α degradation rate affected by lactate |
| ksy | 0.01 | Lactate production rate |
| kpy | 1 | The regulating for lactate production |
| kdy | 0.01 | Basal lactate degradation rate |
| HIF[0] | 0.1 | The initial HIF-1α concentration |
| Lactate[0] | 0.2 | The initial lactate concentration |
| GFP[0] | 0 | The initial GFP concentration |
| Vg | 1 | The GFP production rate |
| kg | 0.05 | The constant defining GFP production |
| dg | 0.1 | GFP degradation rate |
QUANTIFICATION AND STATISTICAL ANALYSIS
Students t-test was performed unless specified. For grouped data, one way ANOVA was applied. Data are presented as ±SEM, or ±SD. Statistical significance is defined as: p < 0.05 (*), p < 0.01 (**), or p < 0.001 (***). For specific figure panels, statistical analyses are described below.
Statistics for RNAseq analysis.
All gene expression TPM values were analyzed after ANOVA, and filtered for p < 0.01. Gene profile clusters were created using Partek Heirarchical Clustering algorithm with average linkage was used as a metric to determine distance between features. Z-score was determined for each gene analyzed after filtering for false discovery rate < 0.1 (total 1702 genes filtered), and represented in a heat map. Gene sets for cell cycle were analyzed with KEGG gene set hsa04110, and pairwise comparisons were noted when p < 0.01, and relative fold changes expressed as corresponding color values on a heat map.
Gene expression in TCGA tumor and normal tissue types.
We used GEPIA analysis58 to identify gene expression in TCGA tumor subtypes and matched normal tissue subtypes in the TCGA database. The abbreviations used in Fig 5 are standard name assignments of TCGA studies. Number of samples used for the GEPIA differential gene expression analysis are listed as follows:
| S.No | TCGA | Study Name | N (tumor) | N (normal) |
|---|---|---|---|---|
| 1 | ACC | Adrenocortical carcinoma | 77 | 128 |
| 2 | BLCA | Bladder Urothelial Carcinoma | 404 | 28 |
| 3 | BRCA | Breast invasive carcinoma | 1085 | 291 |
| 4 | CESC | Cervical squamous cell carcinoma & endocervical adenocarcinoma | 305 | 13 |
| 5 | CHOL | Cholangiocarcinoma | 36 | 9 |
| 6 | COAD | Colon adenocarcinoma | 275 | 349 |
| 7 | DLBC | Lymphoid Neoplasm Diffuse Large B-cell Lymphoma | 47 | 337 |
| 8 | ESCA | Esophageal carcinoma | 182 | 286 |
| 9 | GBM | Glioblastoma multiforme | 163 | 207 |
| 10 | HNSC | Head and Neck squamous cell carcinoma | 519 | 44 |
| 11 | KICH | Kidney Chromophobe | 66 | 53 |
| 12 | KIRC | Kidney renal clear cell carcinoma | 523 | 100 |
| 13 | KIRP | Kidney renal papillary cell carcinoma | 286 | 60 |
| 14 | LAML | Acute Myeloid Leukemia | 173 | 70 |
| 15 | LGG | Brain Lower Grade Glioma | 518 | 207 |
| 16 | LIHC | Liver hepatocellular carcinoma | 369 | 160 |
| 17 | LUAD | Lung adenocarcinoma | 483 | 347 |
| 18 | LUSC | Lung squamous cell carcinoma | 486 | 338 |
| 19 | MESO | Mesothelioma | 87 | 426 |
| 20 | OV | Ovarian serous cystadenocarcinoma | 88 | 179 |
| 21 | PAAD | Pancreatic adenocarcinoma | 179 | 171 |
| 22 | PCPG | Pheochromocytoma and Paraganglioma | 182 | 3 |
| 23 | PRAD | Prostate adenocarcinoma | 492 | 152 |
| 24 | READ | Rectum adenocarcinoma | 92 | 318 |
| 25 | SARC | Sarcoma | 262 | 2 |
| 26 | SKCM | Skin Cutaneous Melanoma | 461 | 558 |
| 27 | STAD | Stomach adenocarcinoma | 408 | 211 |
| 28 | TGCT | Testicular Germ Cell Tumors | 137 | 165 |
| 29 | THCA | Thyroid carcinoma | 512 | 337 |
| 30 | THYM | Thymoma | 118 | 339 |
| 31 | UCEC | Uterine Corpus Endometrial Carcinoma | 174 | 91 |
| 32 | UCS | Uterine Carcinosarcoma | 57 | 78 |
| 33 | UVM | Uveal Melanoma | 79 | 0 |
Statistical tests of comparison between oscillation-specific genes vs the rest of the genes were conducted using t-tests of difference in means of log2 fold changes in cancer vs normal samples. Enrichment of genes was tested using the Kolmogorov Smirnov tests76 by the Gene Set Enrichment Analysis method77, using the fgsea package78. Error bars were plotted for the t test confidence intervals of the difference of means.
Supplementary Material
KEY RESOURCE TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-LAMP2A | Abcam | ab18528 |
| anti-HIF1a | BD Biosciences | 610959 |
| anti-HIF1a | ThermoFisher | 16H4L13 |
| anti-GAPDH | Abcam | G8795 |
| anti-Cyclin D1 | Santa Cruz | sc-20044 |
| anti-Cyclin B1 | Santa Cruz | sc-752 |
| anti-V5 | Abcam | ab27671 |
| anti-HIF1a | Abcam | ab2185 |
| anti-Hsc70 | Abcam | ab2788 |
| Biological Samples | ||
| Breast Cancer Sample | UConn Biorepository | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trypsin-EDTA | Life Technologies | |
| Protease inhibitor cocktail | Sigma Aldrich | S7830 |
| RIPA Buffer | Thermo Fisher | |
| Snarf-1 | Thermo Fisher | S22801 |
| CellROX deep red | Invitrogen | C10422 |
| DCF-DA | Invitrogen | D339 |
| Nigericin | Invitrogen | N-1495 |
| Tris(4,7-diphenyl-1,10-phenanthroline) Ruthenium dichloride (Ru(dpp)3Cl2) | Sigma-Aldrich | 36309–88-3 |
| Protease inhibitor | Thermo Fisher | 78430 |
| 4–20% SDS PAGE gel | BioRad | 4561096 |
| Nitrocellulose 0.2um | BioRad | 1620112 |
| PFA | Sigma-Aldrich | F8775 |
| Triton X-100 | Sigma-Aldrich | 9002931 |
| AlbuMAX | Gibco | 11020021 |
| SlowFade | Invitrogen | S26938 |
| Medical Adhesive for Sealing | Hollister | 7730 |
| Critical Commercial Assays | ||
| Lysosomal enrichment kit | Thermo Fisher | 89839 |
| RNeasy microkit | Qiagen | 74004 |
| BCA Assay kit | Thermo Fisher | 23227 |
| QuikChange Lightning Site Directed Mutagenesis Kit | Agilent | 210519 |
| Pyruvate Assay Kit | Biovision | K609 |
| Picogreen DNA assay kit | Thermofisher | P11496 |
| RNAProtect Kit | Qiagen | 76526 |
| RNeasy mini kit | Qiagen | 74106 |
| Deposited Data | ||
| RNA Sequencing Raw Data | Mendeley Data | 10.17632/m8ctygyjtm.1 |
| Experimental Models: Cell Lines | ||
| HEK293 | ThermoFisher | R70007 |
| HeLa | ATCC | CCL2 |
| A375 | ATCC | CRL-1619 |
| MCF7 | ATCC | HTB-22 |
| BJ5ta | ATCC | CRL-4001 |
| Experimental Models: Organisms/Strains | ||
| zebrafish (Danio rerio) embryo tumor xenograft assay | Nicoli Lab, Yale University | NA |
| Oligonucleotides | ||
| CCCTCGTAAAGAATTCATGGAGGGCGCCGG | V5 forward | |
| GAGGTGGTCTGGATCCCTACGTAGAATCGAGACCGAGGAGAG | V5 reverse | |
| 5′-CAAAAAGTTTTTCTACCAATTCCAACTTGAATGCAGCGACCATATCACTATCCACATAAAAACAATATT −3′ | HIF1a forward | |
| 5′- AATATTGTTTTTATGTGGATAGTGATATGGTCGCTGCATTCAAGTTGGAATTGGTAGAAAAACTTTTTG −3′ | HIF1a reverse | |
| Recombinant DNA | ||
| pLVX-TetOne-puro | Takara Bio | 631849 |
| CMV-d2eGFP-empty | Addgene | 26164 |
| 5HRE-GFP | Addgene | 46926 |
| HypoxCR | Addgene | 59946 |
| pDONR223_HIF1A_WT | Addgene | 82129 |
| Software and Algorithms | ||
| Partek Heirarchical Clustering Algorithm | Partek Flow Inc. Missourie | |
| GEPIA2 | Zemin Zhang Lab | gepia2.cancer-pku.cn |
| ImageJ | NIH | imagej.nih.gov |
| Zeiss Meta | Zeiss Inc | 3.5 |
| Prism | Graphpad | NA |
| Slidebook | Intelligent Imaging Innovations Inc. | 4.2.0 |
| FACSDIVA | BD Biosciences | 6.0 |
| MATLAB | Mathworks | ver 2015 |
| Model Code: DOI: 10.5281/zenodo.7300158 | ||
Highlights.
HIF-1a transcriptional response can be dynamic in subpopulation of cancer cells
Lactate, a byproduct of HIF-1a induced glycolysis, drives HIF-1a oscillations
Lactate can serve as a quorum sensing signal to convey information about cell density
Emergent HIF-1a oscillations allow tumor to escape HIF-1a induced cell cycle arrest
Acknowledgements
We would like to acknowledge the funding obtained from the National Cancer Institutes through the following grants: NCI R37248161 (PI: Kshitiz), NCI U54CA209992 (PI: Andre Levchenko), and NCI R01CA51497 (PI: Chi V Dang). We would also like to thank Dr. Alexander Groisman (University of California, San Diego) for helping us in creating the microfluidic O2 perfusion system.
Footnotes
Declaration of Interests
Authors have no competing interests.
REFERENCES
- 1.Semenza GL (2002). HIF-1 and tumor progression: pathophysiology and therapeutics. Trends Mol Med 8, S62–67. [DOI] [PubMed] [Google Scholar]
- 2.Semenza GL (2003). Targeting HIF-1 for cancer therapy. Nat Rev Cancer 3, 721–732. 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 3.Semenza GL (2010). Vascular responses to hypoxia and ischemia. Arterioscler Thromb Vasc Biol 30, 648–652. 10.1161/ATVBAHA.108.181644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Semenza GL (2010). Oxygen homeostasis. Wiley Interdiscip Rev Syst Biol Med 2, 336–361. 10.1002/wsbm.69. [DOI] [PubMed] [Google Scholar]
- 5.Semenza GL (1998). Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev 8, 588–594. [DOI] [PubMed] [Google Scholar]
- 6.Kim JW, Tchernyshyov I, Semenza GL, and Dang CV (2006). HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab 3, 177–185. 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Frezza C, Zheng L, Tennant DA, Papkovsky DB, Hedley BA, Kalna G, Watson DG, and Gottlieb E. (2011). Metabolic profiling of hypoxic cells revealed a catabolic signature required for cell survival. PLoS One 6, e24411. 10.1371/journal.pone.0024411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV, and Semenza GL (2007). HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122. 10.1016/j.cell.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 9.Jose C, Bellance N, and Rossignol R. (2011). Choosing between glycolysis and oxidative phosphorylation: a tumor’s dilemma? Biochim Biophys Acta 1807, 552–561. 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 10.Suhail Y, Cain MP, Vanaja K, Kurywchak PA, Levchenko A, Kalluri R, and Kshitiz (2019). Systems Biology of Cancer Metastasis. Cell Syst 9, 109–127. 10.1016/j.cels.2019.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noren DP, Chou WH, Lee SH, Qutub AA, Warmflash A, Wagner DS, Popel AS, and Levchenko A. (2016). Endothelial cells decode VEGF-mediated Ca2+ signaling patterns to produce distinct functional responses. Sci Signal 9, ra20. 10.1126/scisignal.aad3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nieto MA (2013). Epithelial plasticity: a common theme in embryonic and cancer cells. Science 342, 1234850. 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 13.Shimojo H, Ohtsuka T, and Kageyama R. (2008). Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64. 10.1016/j.neuron.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Marshall CJ (1995). Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–185. [DOI] [PubMed] [Google Scholar]
- 15.Pucciarelli D, Lengger N, Takacova M, Csaderova L, Bartosova M, Breiteneder H, Pastorekova S, and Hafner C. (2016). Hypoxia increases the heterogeneity of melanoma cell populations and affects the response to vemurafenib. Mol Med Rep 13, 3281–3288. 10.3892/mmr.2016.4888. [DOI] [PubMed] [Google Scholar]
- 16.Erapaneedi R, Belousov VV, Schafers M, and Kiefer F. (2016). A novel family of fluorescent hypoxia sensors reveal strong heterogeneity in tumor hypoxia at the cellular level. EMBO J 35, 102–113. 10.15252/embj.201592775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le A, Stine ZE, Nguyen C, Afzal J, Sun P, Hamaker M, Siegel NM, Gouw AM, Kang BH, Yu SH, et al. (2014). Tumorigenicity of hypoxic respiring cancer cells revealed by a hypoxia-cell cycle dual reporter. Proc Natl Acad Sci U S A 111, 12486–12491. 10.1073/pnas.1402012111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vordermark D, Shibata T, and Brown JM (2001). Green fluorescent protein is a suitable reporter of tumor hypoxia despite an oxygen requirement for chromophore formation. Neoplasia 3, 527–534. 10.1038/sj/neo/7900192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polinkovsky M, Gutierrez E, Levchenko A, and Groisman A. (2009). Fine temporal control of the medium gas content and acidity and on-chip generation of series of oxygen concentrations for cell cultures. Lab Chip 9, 1073–1084. 10.1039/b816191g. [DOI] [PubMed] [Google Scholar]
- 20.Rumsey WL, Vanderkooi JM, and Wilson DF (1988). Imaging of phosphorescence: a novel method for measuring oxygen distribution in perfused tissue. Science 241, 1649–1651. [DOI] [PubMed] [Google Scholar]
- 21.Smith H, Board M, Pellagatti A, Turley H, Boultwood J, and Callaghan R. (2016). The Effects of Severe Hypoxia on Glycolytic Flux and Enzyme Activity in a Model of Solid Tumours. J Cell Biochem. 10.1002/jcb.25488. [DOI] [PubMed] [Google Scholar]
- 22.De Saedeleer CJ, Copetti T, Porporato PE, Verrax J, Feron O, and Sonveaux P. (2012). Lactate activates HIF-1 in oxidative but not in Warburg-phenotype human tumor cells. PLoS One 7, e46571. 10.1371/journal.pone.0046571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sonveaux P, Vegran F, Schroeder T, Wergin MC, Verrax J, Rabbani ZN, De Saedeleer CJ, Kennedy KM, Diepart C, Jordan BF, et al. (2008). Targeting lactate-fueled respiration selectively kills hypoxic tumor cells in mice. J Clin Invest 118, 3930–3942. 10.1172/JCI36843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doherty JR, and Cleveland JL (2013). Targeting lactate metabolism for cancer therapeutics. J Clin Invest 123, 3685–3692. 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.San Martin A, Ceballo S, Ruminot I, Lerchundi R, Frommer WB, and Barros LF (2013). A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PLoS One 8, e57712. 10.1371/journal.pone.0057712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao B, Hemann MT, and Lauffenburger DA (2014). Intratumor heterogeneity alters most effective drugs in designed combinations. Proc Natl Acad Sci U S A 111, 10773–10778. 10.1073/pnas.1323934111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherz-Shouval R, and Elazar Z. (2011). Regulation of autophagy by ROS: physiology and pathology. Trends Biochem Sci 36, 30–38. 10.1016/j.tibs.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Ovens MJ, Davies AJ, Wilson MC, Murray CM, and Halestrap AP (2010). AR-C155858 is a potent inhibitor of monocarboxylate transporters MCT1 and MCT2 that binds to an intracellular site involving transmembrane helices 7–10. Biochem J 425, 523–530. 10.1042/BJ20091515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbi ME, Hu H, Kshitiz, Ahmed I, Levchenko A, and Semenza GL (2013). Chaperone-mediated autophagy targets hypoxia-inducible factor-1alpha (HIF-1alpha) for lysosomal degradation. J Biol Chem 288, 10703–10714. 10.1074/jbc.M112.414771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hsieh CY, Chen CL, Yang KC, Ma CT, Choi PC, and Lin CF (2015). Detection of reactive oxygen species during the cell cycle under normal culture conditions using a modified fixed-sample staining method. J Immunoassay Immunochem 36, 149–161. 10.1080/15321819.2014.910806. [DOI] [PubMed] [Google Scholar]
- 31.Shacka JJ, Klocke BJ, Shibata M, Uchiyama Y, Datta G, Schmidt RE, and Roth KA (2006). Bafilomycin A1 inhibits chloroquine-induced death of cerebellar granule neurons. Mol Pharmacol 69, 1125–1136. 10.1124/mol.105.018408. [DOI] [PubMed] [Google Scholar]
- 32.Filomeni G, De Zio D, and Cecconi F. (2015). Oxidative stress and autophagy: the clash between damage and metabolic needs. Cell Death Differ 22, 377–388. 10.1038/cdd.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galardo MN, Regueira M, Riera MF, Pellizzari EH, Cigorraga SB, and Meroni SB (2014). Lactate regulates rat male germ cell function through reactive oxygen species. PLoS One 9, e88024. 10.1371/journal.pone.0088024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubbi ME, Gilkes DM, Hu H, Kshitiz, Ahmed, I., and Semenza, G.L. (2014). Cyclin-dependent kinases regulate lysosomal degradation of hypoxia-inducible factor 1alpha to promote cell-cycle progression. Proc Natl Acad Sci U S A 111, E3325–3334. 10.1073/pnas.1412840111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Echigoya Y, Morita S, Itou T, and Sakai T. (2012). Effects of extracellular lactate on production of reactive oxygen species by equine polymorphonuclear leukocytes in vitro. Am J Vet Res 73, 1290–1298. 10.2460/ajvr.73.8.1290. [DOI] [PubMed] [Google Scholar]
- 36.Ferreira JV, Fofo H, Bejarano E, Bento CF, Ramalho JS, Girao H, and Pereira P. (2013). STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy 9, 1349–1366. 10.4161/auto.25190. [DOI] [PubMed] [Google Scholar]
- 37.Ferreira JV, Soares AR, Ramalho JS, Pereira P, and Girao H. (2015). K63 linked ubiquitin chain formation is a signal for HIF1A degradation by Chaperone-Mediated Autophagy. Sci Rep 5, 10210. 10.1038/srep10210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brisson L, Banski P, Sboarina M, Dethier C, Danhier P, Fontenille MJ, Van Hee VF, Vazeille T, Tardy M, Falces J, et al. (2016). Lactate Dehydrogenase B Controls Lysosome Activity and Autophagy in Cancer. Cancer Cell 30, 418–431. 10.1016/j.ccell.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 39.Li J, Wong CL, Vijayasankaran N, Hudson T, and Amanullah A. (2012). Feeding lactate for CHO cell culture processes: impact on culture metabolism and performance. Biotechnol Bioeng 109, 1173–1186. 10.1002/bit.24389. [DOI] [PubMed] [Google Scholar]
- 40.Templeton N, Lewis A, Dorai H, Qian EA, Campbell MP, Smith KD, Lang SE, Betenbaugh MJ, and Young JD (2014). The impact of anti-apoptotic gene Bcl-2 expression on CHO central metabolism. Metab Eng 25, 92–102. 10.1016/j.ymben.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 41.Kelly TJ, Souza AL, Clish CB, and Puigserver P. (2011). A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol 31, 2696–2706. 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang F, Zhang H, Xu N, Huang N, Tian C, Ye A, Hu G, He J, and Zhang Y. (2016). A novel hypoxia-induced miR-147a regulates cell proliferation through a positive feedback loop of stabilizing HIF-1alpha. Cancer Biol Ther, 0. 10.1080/15384047.2016.1195040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fabian Z, Taylor CT, and Nguyen LK (2016). Understanding complexity in the HIF signaling pathway using systems biology and mathematical modeling. J Mol Med (Berl) 94, 377–390. 10.1007/s00109-016-1383-6. [DOI] [PubMed] [Google Scholar]
- 44.Yang F, Zhang H, Mei Y, and Wu M. (2014). Reciprocal regulation of HIF-1alpha and lincRNA-p21 modulates the Warburg effect. Mol Cell 53, 88–100. 10.1016/j.molcel.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 45.Pfeuty B, and Kaneko K. (2009). The combination of positive and negative feedback loops confers exquisite flexibility to biochemical switches. Phys Biol 6, 046013. 10.1088/1478-3975/6/4/046013. [DOI] [PubMed] [Google Scholar]
- 46.Melke P, Sahlin P, Levchenko A, and Jonsson H. (2010). A cell-based model for quorum sensing in heterogeneous bacterial colonies. PLoS Comput Biol 6, e1000819. 10.1371/journal.pcbi.1000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dandekar AA, Chugani S, and Greenberg EP (2012). Bacterial quorum sensing and metabolic incentives to cooperate. Science 338, 264–266. 10.1126/science.1227289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wee KB, Yio WK, Surana U, and Chiam KH (2012). Transcription factor oscillations induce differential gene expressions. Biophys J 102, 2413–2423. 10.1016/j.bpj.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nelson DE, See V, Nelson G, and White MR (2004). Oscillations in transcription factor dynamics: a new way to control gene expression. Biochem Soc Trans 32, 1090–1092. 10.1042/BST0321090. [DOI] [PubMed] [Google Scholar]
- 50.Walton ZE, Patel CH, Brooks RC, Yu Y, Ibrahim-Hashim A, Riddle M, Porcu A, Jiang T, Ecker BL, Tameire F, et al. (2018). Acid Suspends the Circadian Clock in Hypoxia through Inhibition of mTOR. Cell 174, 72–87 e32. 10.1016/j.cell.2018.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Adamovich Y, Ladeuix B, Golik M, Koeners MP, and Asher G. (2017). Rhythmic Oxygen Levels Reset Circadian Clocks through HIF1alpha. Cell Metab 25, 93–101. 10.1016/j.cmet.2016.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Peek CB, Levine DC, Cedernaes J, Taguchi A, Kobayashi Y, Tsai SJ, Bonar NA, McNulty MR, Ramsey KM, and Bass J. (2017). Circadian Clock Interaction with HIF1alpha Mediates Oxygenic Metabolism and Anaerobic Glycolysis in Skeletal Muscle. Cell Metab 25, 86–92. 10.1016/j.cmet.2016.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Goda N, Ryan HE, Khadivi B, McNulty W, Rickert RC, and Johnson RS (2003). Hypoxia-inducible factor 1alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol 23, 359–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hubbi ME, and Semenza GL (2015). Regulation of cell proliferation by hypoxia-inducible factors. Am J Physiol Cell Physiol 309, C775–782. 10.1152/ajpcell.00279.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hubbi ME, Kshitiz, Gilkes DM, Rey S, Wong CC, Luo W, Kim DH, Dang CV, Levchenko A, and Semenza GL (2013). A nontranscriptional role for HIF-1alpha as a direct inhibitor of DNA replication. Sci Signal 6, ra10. 10.1126/scisignal.2003417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, and Dang CV (2001). Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem 276, 7919–7926. 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- 57.Harris AL (2002). Hypoxia--a key regulatory factor in tumour growth. Nat Rev Cancer 2, 38–47. 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 58.Tang Z, Kang B, Li C, Chen T, and Zhang Z. (2019). GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47, W556–W560. 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rasmussen RD, Gajjar MK, Tuckova L, Jensen KE, Maya-Mendoza A, Holst CB, Mollgard K, Rasmussen JS, Brennum J, Bartek J Jr., et al. (2018). Author Correction: BRCA1-regulated RRM2 expression protects glioblastoma cells from endogenous replication stress and promotes tumorigenicity. Nat Commun 9, 5396. 10.1038/s41467-018-07892-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coburn LA, Singh K, Asim M, Barry DP, Allaman MM, Al-Greene NT, Hardbower DM, Polosukhina D, Williams CS, Delgado AG, et al. (2019). Loss of solute carrier family 7 member 2 exacerbates inflammation-associated colon tumorigenesis. Oncogene 38, 1067–1079. 10.1038/s41388-018-0492-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lieu EL, Nguyen T, Rhyne S, and Kim J. (2020). Amino acids in cancer. Exp Mol Med 52, 15–30. 10.1038/s12276-020-0375-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bagnall J, Leedale J, Taylor SE, Spiller DG, White MR, Sharkey KJ, Bearon RN, and See V. (2014). Tight control of hypoxia-inducible factor-alpha transient dynamics is essential for cell survival in hypoxia. J Biol Chem 289, 5549–5564. 10.1074/jbc.M113.500405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Payen VL, Hsu MY, Radecke KS, Wyart E, Vazeille T, Bouzin C, Porporato PE, and Sonveaux P. (2017). Monocarboxylate Transporter MCT1 Promotes Tumor Metastasis Independently of Its Activity as a Lactate Transporter. Cancer Res 77, 5591–5601. 10.1158/0008-5472.CAN-17-0764. [DOI] [PubMed] [Google Scholar]
- 64.Rocha S. (2007). Gene regulation under low oxygen: holding your breath for transcription. Trends Biochem Sci 32, 389–397. 10.1016/j.tibs.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 65.Lee JH, Kim EJ, Kim DK, Lee JM, Park SB, Lee IK, Harris RA, Lee MO, and Choi HS (2012). Hypoxia induces PDK4 gene expression through induction of the orphan nuclear receptor ERRgamma. PLoS One 7, e46324. 10.1371/journal.pone.0046324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu X, Zuo R, Bao Y, Qu X, Sun K, and Ying H. (2017). Down-regulation of PDK4 is Critical for the Switch of Carbohydrate Catabolism during Syncytialization of Human Placental Trophoblasts. Sci Rep 7, 8474. 10.1038/s41598-017-09163-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang S, Hulver MW, McMillan RP, Cline MA, and Gilbert ER (2014). The pivotal role of pyruvate dehydrogenase kinases in metabolic flexibility. Nutr Metab (Lond) 11, 10. 10.1186/1743-7075-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Almendros I, Montserrat JM, Ramirez J, Torres M, Duran-Cantolla J, Navajas D, and Farre R. (2012). Intermittent hypoxia enhances cancer progression in a mouse model of sleep apnoea. Eur Respir J 39, 215–217. 10.1183/09031936.00185110. [DOI] [PubMed] [Google Scholar]
- 69.Chen A, Sceneay J, Godde N, Kinwel T, Ham S, Thompson EW, Humbert PO, and Moller A. (2018). Intermittent hypoxia induces a metastatic phenotype in breast cancer. Oncogene 37, 4214–4225. 10.1038/s41388-018-0259-3. [DOI] [PubMed] [Google Scholar]
- 70.Liu Y, White KA, and Barber DL (2020). Intracellular pH Regulates Cancer and Stem Cell Behaviors: A Protein Dynamics Perspective. Front Oncol 10, 1401. 10.3389/fonc.2020.01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, et al. (2014). Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nicoli S, and Presta M. (2007). The zebrafish/tumor xenograft angiogenesis assay. Nat Protoc 2, 2918–2923. 10.1038/nprot.2007.412. [DOI] [PubMed] [Google Scholar]
- 73.Nicoli S, Ribatti D, Cotelli F, and Presta M. (2007). Mammalian tumor xenografts induce neovascularization in zebrafish embryos. Cancer Res 67, 2927–2931. 10.1158/0008-5472.CAN-06-4268. [DOI] [PubMed] [Google Scholar]
- 74.Afzal J, Liu Y, Du W, Suhail Y, Zong P, Feng J, Ajeti V, Sayyad WA, Nikolaus J, Yankova M, et al. (2022). Cardiac ultrastructure inspired matrix induces advanced metabolic and functional maturation of differentiated human cardiomyocytes. Cell Rep 40, 111146. 10.1016/j.celrep.2022.111146. [DOI] [PubMed] [Google Scholar]
- 75.Novak B, and Tyson JJ (2008). Design principles of biochemical oscillators. Nat Rev Mol Cell Biol 9, 981–991. 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smirnov N. (1948). Table for Estimating the Goodness of Fit of Empirical Distributions. Ann. Math. Statist. 19, 279–281. [Google Scholar]
- 77.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, and Sergushichev A. (2021). Fast gene set enrichment analysis. bioRxiv. 10.1101/060012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNAseq data have been deposited at Mendeley, and are publicly available. DOI: 10.17632/m8ctygyjtm.1
ODE model is available at DOI: 10.5281/zenodo.7300158
All other data reported in this paper will be shared by the lead contact upon request.