Abstract
We have previously shown that the activity of the Escherichia coli rRNA promoter rrnB P1 in vitro depends on the concentration of the initiating nucleotide, ATP, and can respond to changes in ATP pools in vivo. We have proposed that this nucleoside triphosphate (NTP) sensing might contribute to regulation of rRNA transcription. To test this model, we have measured the ATP requirements for transcription from 11 different rrnB P1 core promoter mutants in vitro and compared them with the regulatory responses of the same promoters in vivo. The seven rrnB P1 variants that required much lower ATP concentrations than the wild-type promoter for efficient transcription in vitro were defective for response to growth rate changes in vivo (growth rate-dependent regulation). In contrast, the four variants requiring high ATP concentrations in vitro (like the wild-type promoter) were regulated with the growth rate in vivo. We also observed a correlation between NTP sensing in vitro and the response of the promoters in vivo to deletion of the fis gene (an example of homeostatic control), although this relationship was not as tight as for growth rate-dependent regulation. We conclude that the kinetic features responsible for the high ATP concentration dependence of the rrnB P1 promoter in vitro are responsible, at least in part, for the promoter's regulation in vivo, consistent with the model in which rrnB P1 promoter activity can be regulated by changes in NTP pools in vivo (or by hypothetical factors that work at the same kinetic steps that make the promoter sensitive to NTPs).
In rapidly dividing Escherichia coli, transcription from the seven rRNA operons accounts for over half of all transcription in the cell (16, 26, 27, 37). rRNA operons are transcribed by two tandem promoters, rrn P1 and rrn P2, with P1 being the predominant promoter at medium to fast growth rates. Several features of rrn P1 promoters contribute to their unusual strength. All seven P1 core promoters contain exact matches to the consensus −10 hexamer (TATAAT) and close matches to the consensus −35 hexamer (TTGACA). All of the P1 promoters also derive much of their strength from UP elements, A+T-rich sequences (located from approximately −40 to −60 with respect to the transcription start site) that interact with the α subunit C-terminal domain of RNA polymerase (RNAP) and activate transcription ∼20- to 50-fold (28, 32, 52). In addition, the transcription factor FIS binds to three to five sites upstream of the UP element in each operon and activates transcription approximately fivefold (32, 53).
While the rrn P1 promoters can be exceptionally strong, they are also subject to regulatory controls that ensure that energy is not wasted synthesizing excess translation machinery under less favorable growth conditions (16, 26, 27, 37). There are multiple ways in which these control systems have been assayed: responses of the promoters to amino acid starvation (stringent control), to different steady-state growth rates (growth rate-dependent control), and to conditions that elicit a homeostatic response (see below).
Multiple molecular mechanisms likely underlie these regulatory responses (27). For example, guanosine 5′-diphosphate, 3′-diphosphate (ppGpp) is responsible for inhibiting rRNA transcription during the stringent response (12) but other regulatory responses utilize different effectors. ppGpp is not essential for growth rate-dependent or homeostatic regulation, since rrn P1 promoter activity increases with growth rate and responds to at least one type of homeostatic response in strains devoid of ppGpp (5, 8, 22; see also reference 49).
rrn P1 promoters require much higher concentrations of the initiating NTP (ATP or GTP) for maximal transcription in vitro than most other promoters (7, 21), and rrnB P1 and rrnD P1 promoter activities correlate with the levels of their initiating NTPs (ATP and GTP, respectively) in strains with altered NTP pools (21; D. A. Schneider and R. L. Gourse, unpublished data). These results indicate that rRNA transcription can respond directly to variations in ATP and GTP concentrations in vivo. We have proposed that NTP levels could serve as indicators of the translational capacity of the cell and that regulation by changing NTP concentrations, referred to as NTP sensing, might be responsible for the increase in rrn P1 promoter activity with growth rate. Furthermore, NTP sensing might also contribute to the regulation of other promoters involved in the synthesis of the translational machinery (49, 61). However, since no methods are available with which to measure the concentration of free NTPs in growing cells, and since measurements of total NTP pools as a function of growth rate vary with the extraction procedures used (21, 48; Schneider and Gourse, unpublished data), it has been difficult to assess the role of NTP sensing in growth rate-dependent regulation.
Homeostatic regulation (feedback control) of rRNA core promoter activity keeps total rRNA synthesis relatively constant under conditions that might be expected to perturb it. Genetic alterations that would lead to under- or overproduction of ribosomes result in compensating changes in rRNA core promoter activity. For example, rrn gene dose increases (by addition of rrn operons on multicopy plasmids) result in corresponding decreases in rrn P1 activity (29, 33) and rrn gene dose decreases (by deletion of several rrn operons) increase rrn P1 promoter activity (3, 15). Likewise, decreases in upstream activation of rrn P1 promoters (by mutation of fis or rpoA) or decreases in rRNA elongation (by mutation of genes for Nus factors) increase rrn P1 promoter activity (50, 52, 53, 56). Consistent with the homeostatic regulation model, rrn P1 promoter activity is stimulated by the protein synthesis inhibitor chloramphenicol or spectinomycin in a futile attempt to compensate for the resulting reduction in translational capacity (57; Schneider and Gourse, unpublished data).
The molecular effector(s) responsible for homeostatic control is unclear and could, in principle, be different in different experimental situations. rrn P1 promoter activity decreases with an increase in translationally competent ribosomes (but not with an increase in translationally defective ribosomes), suggesting that the feedback signal is either generated or consumed during protein synthesis (14, 33, 62). Since translation is a major consumer of ATP and GTP, we have proposed that homeostatic control, like growth rate-dependent regulation, might be mediated by changes in the concentration of available ATP and GTP (21). Consistent with this hypothesis, treatment with protein synthesis inhibitors not only increases rRNA transcription (see above) but also increases ATP pools (Schneider and Gourse, unpublished data).
In this study, we explored whether two rrn P1 regulatory responses are consistent with the predictions of the NTP sensing model. That is, if NTP sensing were responsible for the changes in rrn P1 promoter activity observed with increasing growth rates or deletion of the fis gene, then it would be expected that mutant rrn P1 promoters with altered responses to the NTP concentration in vitro should display altered regulation in vivo. We show that the ATP concentration dependences in vitro of 11 previously identified rrnB P1 promoter variants (17, 20, 35) correlate with their responses to growth rate and to deletion of fis. These results are consistent with a simple model in which growth rate-dependent regulation and, perhaps, feedback control are mediated directly, at least in part, by changing concentrations of the initiating nucleotide in vivo (or by a changing parameter that works at the same kinetic steps that make the promoter sensitive to NTPs).
MATERIALS AND METHODS
Promoter mutants, plasmids, strains, and bacteriophage.
The plasmids and strains used in this study are listed in Table 1. Wild-type and variant rrnB P1 promoters (−66 to +9 with respect to the transcription start site) were generated by PCR from plasmids containing the wild-type rrnB P1 promoter or previously identified promoter variants (17, 20, 35). The variants are described in Results, and for consistency, the nomenclature used is the same as that employed previously (17, 20, 35). DNA fragments were generated with an EcoRI site at the upstream junction with the promoter sequence and with a HindIII site at the downstream junction, ligated into the transcription vector pRLG770 (53), and then cloned into bacteriophage λ to form promoter-lacZ fusions in strain VH1000 (VH1000 = MG1655 pyrE+ lacI lacZ; courtesy of V. J. Hernandez, State University of New York, Buffalo) as previously described (50). Transductions of fis::kan-767 mutations were performed with phage P1vir (44), using RJ1617 as the donor strain (34).
TABLE 1.
Promoters, plasmids, and lysogens used in this study
| Promotera | Plasmidb | Promoter-lacZ fusions
|
|
|---|---|---|---|
| VH1000 | fis::kan | ||
| Wild-type rrnB P1 | pRLG6555 | RLG6558 | RLG6556 |
| rrnB P1 variants | |||
| CGC-5-7ATA | pRLG6120 | RLG5651 | RLG6573 |
| C-5A | pRLG6128 | RLG6133 | RLG6563 |
| T-33A/Ains-22 | pRLG6122 | RLG6134 | RLG6564 |
| C-4T/A-3G | pRLG6127 | RLG6135 | RLG6565 |
| T-33A | pRLG6121 | RLG6136 | RLG6566 |
| C-1T | pRLG6123 | RLG6137 | RLG6567 |
| Tins-23 | pRLG6124 | RLG6138 | RLG6568 |
| C-17T | pRLG6125 | RLG6139 | RLG6569 |
| C-19T | pRLG6126 | RLG6140 | RLG6570 |
| G-25T | pRLG6130 | RLG6141 | RLG6571 |
| C-4T | pRLG6131 | RLG6142 | RLG6572 |
| Controls | |||
| λPR | RLG6641 | RLG6646 | |
| PhisG | RLG4418 | RLG6642 | |
| No promoter | pRLG770 | RLG4999 | RLG6576 |
The promoters tested were wild-type rrnB P1 (endpoints −66 to +9), rrnB P1 variants (−66 to +9) with the indicated mutations, λPR (−40 to +20), and PhisG (−60 to +16), and a no-promoter control (M13 polylinker) was included. λPR and PhisG promoter-lacZ fusions were constructed previously (2, 5).
Plasmids used for in vitro transcription contained the indicated promoters inserted into pRLG770 (see Materials and Methods).
ATP dependence assay.
E. coli RNAP (Eς70) was a generous gift from R. Landick and was purified as previously described (11). Multiple-round transcription reactions were performed essentially as previously described (5, 6), using solution conditions in which the rate constants affected by the initiating NTP are rate limiting. Transcription was started by addition of 8 nM RNAP (∼50% active) to 0.5 nM supercoiled plasmid in a mixture of 150 mM NaCl, 40 mM Tris-Cl (pH 8.0), 10 mM MgCl2, 1 mM dithiothreitol, 0.1 μg of bovine serum albumin per μl, 200 μM CTP, 200 μM GTP, 10 μM UTP, 2 μCi of [α-32P]UTP, and various concentrations of ATP (5 μM to 2 mM) in 10-μl reactions at 30°C. Transcription was terminated 20 min after RNAP addition with an equal volume of formamide loading buffer. The reaction mixtures were electrophoresed on 5% polyacrylamide–7 M urea gels, and the dried gels were visualized and quantified by phosphorimaging (ImageQuant Software; Molecular Dynamics). Fits to data points were made using Sigmaplot (Jandel Scientific).
We measured the ATP dependence of the C-4T/A-3G promoter using a concentration of CTP and GTP (20 μM) lower than those used for the other promoters (see above and Results). Control experiments examining the ATP dependences of the wild-type promoter and several variants at 20 μM CTP and GTP showed that the [ATP]1/2max values of the variant promoters relative to that of the wild type were the same at both high and low CTP and GTP concentrations (data not shown).
Promoter activities at different growth rates.
Cells were grown in Luria broth (LB) or in M9 minimal medium (44) containing 0.4% glucose or glycerol, with or without 0.8% Casamino Acids (Difco) plus tryptophan (40 μg/ml). Liquid cultures were inoculated to an A600 of 0.025 from fresh colonies. Cultures were grown at 30°C for about four generations to an A600 of 0.35, harvested, and sonicated, and β-galactosidase activity was measured (44). Cells can tolerate the extremely active rrnB P1 promoters fused to lacZ using system I (50), unlike the case for some other lacZ fusion systems (58). However, the background activity in system I fusions is relatively high. Background activity (60 to 120 Miller units, depending on the growth rate and genotype) was estimated from the β-galactosidase activities of a lysogen containing an M13 polylinker-lacZ fusion instead of a promoter-lacZ fusion. Appropriate background activities were subtracted from all of the reported values, although it is possible that the insertion of the promoter into the cloning site, in itself, eliminates the background. In any case, the conclusions were not qualitatively different with or without the subtraction of background activities.
Promoter activities in strains lacking the fis gene.
β-Galactosidase activities were measured as described above for lysogens containing either a wild-type fis gene or the fis::kan-767 insertion-deletion (34). Cells were grown as described above in LB. We noted that the activities of non-FIS-regulated promoter-lacZ fusions (e.g., λPR and PhisG) increased 2.6-fold in fis::kan strains for unknown reasons, as observed previously (53). Therefore, only an increase in promoter activity significantly greater than 2.6-fold (such as that observed for the wild-type rrnB P1 promoter lacking FIS sites) was considered a specific response to the absence of fis.
RESULTS
Choice of mutant rrnB P1 promoters.
The rrnB P1 sequence endpoints required for regulation of transcription initiation by growth rate in vivo are limited to the core promoter (approximately −41 to +1 with respect to the transcription start site) (8). Furthermore, our previous mutagenesis studies suggested that the sequence requirements for growth rate-dependent regulation involve some of the sequence features that are conserved among rrn P1 core promoters (17, 20, 35). We chose 11 previously identified rrnB P1 core promoter mutants (17, 20, 35) for analysis of NTP sensing in vitro (Fig. 1). These included a 1-bp substitution mutant (T-33A) with a consensus −35 hexamer, a 1-bp insertion mutant (Tins-23) with an increase in the length of the spacer between the −10 and −35 hexamers to the Eς70 consensus (17 bp), and a double mutant that combines a 17-bp spacer with a perfect −35 hexamer (T-33A/Ains-22), thus creating a consensus core promoter. Three additional mutants with substitutions in positions within the spacer were examined (C-17T, C-19T, and G-25T). To investigate the region between the −10 hexamer and the transcription start site, termed the discriminator because its high G+C-content is characteristic of rRNA and most tRNA promoters (59), we examined a triple mutant and two single substitutions that diminished this region's G+C content (CGC-5–7ATA, C-5A, and C-4T) and a double mutant that altered the discriminator sequence but preserved the G+C content (C-4T/A-3G). Lastly, we examined a mutant with a substitution at position −1 (C-1T); this position, which is conserved as a C in all seven rrn P1 promoters, was previously implicated in regulation by NTPs (21).
FIG. 1.
DNA sequences in the core promoter region of wild-type and mutant rrnB P1 promoters. Numbers above the wild-type sequence refer to promoter positions with respect to the transcription initiation site. The −10 and −35 hexamers are underlined and in bold in the wild-type promoter and underlined in mutant promoters. Mutations are indicated in bold uppercase letters and underlined. The rrnB P1 promoters analyzed extended from −66 to +9.
In order to maximize basal promoter activity and thereby improve measurement accuracy, both in vivo and in vitro, we examined the 11 mutations in the context of promoters containing the UP element. The presence of an UP element has little or no effect on the regulation of wild-type rrnB P1 by growth rate, feedback, or NTP sensing (8, 21, 50, 52; M. M. Barker, T. Gaal, W. Ross, and R. L. Gourse, unpublished data).
Several rrnB P1 variants display altered NTP sensing in vitro.
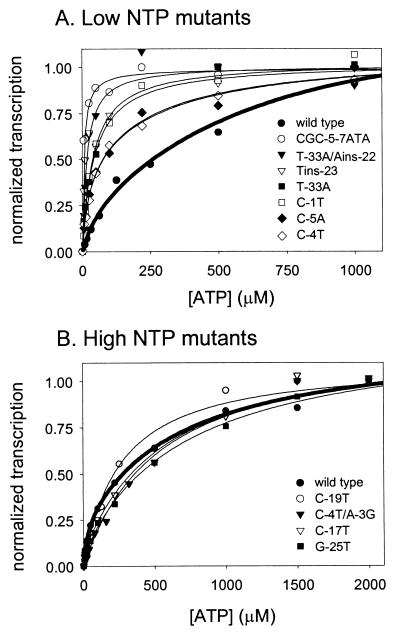
To determine whether the variant promoters require different initiating NTP concentrations than the wild-type rrnB P1 promoter, we used an in vitro transcription assay in which the concentration of ATP was varied from 5 to 2,000 μM while the concentrations of CTP, GTP, and UTP were kept constant (Fig. 2). As shown previously (21), the wild-type rrnB P1 promoter requires a relatively high ATP concentration for half-maximal transcription (under the solution conditions employed, [ATP]1/2max was ∼250 μM). Seven mutant promoters (CGC-5–7ATA, T-33A/Ains-22, Tins-23, T-33A, C-1T, C-5A, and C-4T) required at least threefold less ATP than wild-type rrnB P1 for half-maximal transcription ([ATP]1/2max, <10 to 81 μM) (Fig. 2A). We refer to the seven promoters with low initiating NTP concentration requirements as low-NTP mutants. Four variants (C-19T, C-4T/A-3G, C-17T, and G-25T) required ATP concentrations similar to or greater than that required by wild-type rrnB P1 ([ATP]1/2max, ∼205 to 430 μM ATP) (Fig. 2B). We refer to the promoters requiring high concentrations of the initiating NTP in vitro (like wild-type rrnB P1) as high-NTP mutants. For a summary of the [ATP]1/2max values of the wild-type and mutant promoters and the other regulatory properties of the promoters (see below), see Fig. 4.
FIG. 2.
Effect of ATP concentration on in vitro transcription of wild-type and mutant rrnB P1 promoters. Transcription was normalized to the highest level for each promoter. The wild-type regression fit is in bold. (A) Mutants that required at least threefold lower ATP concentrations for half-maximal transcription than wild-type rrnB P1. (B) Mutants that required ATP concentrations similar to or higher than that required by the wild type. Note the different x-axis scale than in panel A.
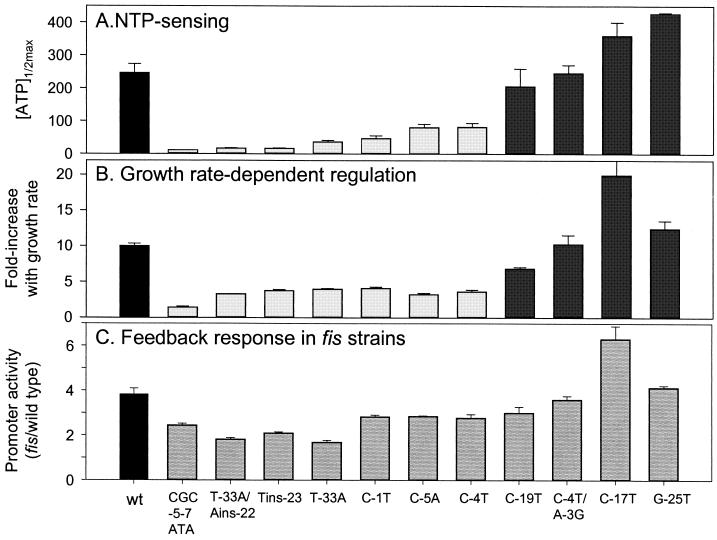
FIG. 4.
Summary of regulation of wild-type (wt) and mutant rrnB P1 promoters. Promoter mutants are listed in the same order for each panel, from the promoter with the lowest [ATP]1/2max to that with the highest. The bars for the wild-type promoter are black, those for the seven low-NTP, growth rate regulation-defective mutants are light gray, and those for the four high-NTP, growth rate regulated mutants are dark gray. The bars for the promoter mutants in panel C are not separated into classes because distinctions are somewhat ambiguous in this assay. (A) The mean [ATP]1/2max values were determined from at least two independent experiments. Variation was less than 20%, except for C-19T (27%). (B) Average fold increase with growth rate was calculated for the wild type and mutants (except C-17T) by dividing the β-galactosidase activity in LB (μ = ∼1.4) by the activity in M9 glycerol (μ = ∼0.33). The averages and standard deviations (less than 14%) were calculated for three independent experiments. The activity of the C-17T promoter is too close to the background in M9 glycerol for accurate assessment, so its fold increase was estimated by dividing the activity in LB by the activity in M9 glucose (μ = 0.58). C-17T increased approximately twofold more than the wild-type promoter in this growth rate range (standard deviation = 26%). Therefore, its fold increase with growth rate may be an underestimate. (C) Feedback derepression in fis mutant strains. fis/wild-type promoter activity ratio is reported as in Table 2.
We emphasize the relative (and not absolute) [ATP]1/2max values of the different promoters, since the absolute [ATP]1/2max values vary dramatically with the temperature, identity, and concentration of anions and cations in the reaction mixture and the superhelicity of the DNA template (21) (data not shown). At relatively high salt concentrations (e.g., 170 mM NaCl) and/or with nonsupercoiled templates, millimolar ATP concentrations (i.e., in the range of the total ATP concentrations present in cells) are required for maximal activity of the wild-type rrnB P1 promoter in vitro (21) (data not shown). Most importantly, rrnB P1 activity responds to both increases and decreases in the total ATP concentration in vivo (21; Schneider and Gourse, unpublished data). Therefore, the free ATP concentration in cells is likely not saturating for initiation at this promoter.
The behavior of the C-4T/A-3G variant was somewhat complicated. Unlike that of the wild-type promoter, this mutant's transcription start site appeared to switch as the relative concentrations of ATP, GTP, and CTP were varied (data not shown). As a result, promoter activity was dependent on the ATP, GTP, and CTP concentrations, depending on their relative concentrations in vitro. Since the start site switch occurred when the ATP concentration was much lower than the concentrations of the other NTPs, and since this condition is unlikely to occur in vivo, we did not explore the properties of the start site switch in more detail. The ATP dependence of C-4T/A-3G was measured at low GTP and CTP concentrations in order to keep the transcription start site the same as for the wild-type promoter (see Materials and Methods). Under these conditions, the C-4T/A-3G mutant required high levels of ATP ([ATP]1/2max, ∼300 μM), like wild-type rrnB P1.
Low-NTP promoter mutants are defective for growth rate-dependent regulation.
The NTP sensing model predicts that an rrnB P1 variant requiring much less ATP than the wild type would be impaired for growth rate regulation, since the free ATP concentration present in cells would always be higher than that required for maximal transcription (21). Conversely, the model predicts that an rrnB P1 variant sensitive to changes in the concentration of free ATP present in cells would be fully subject to growth rate regulation.
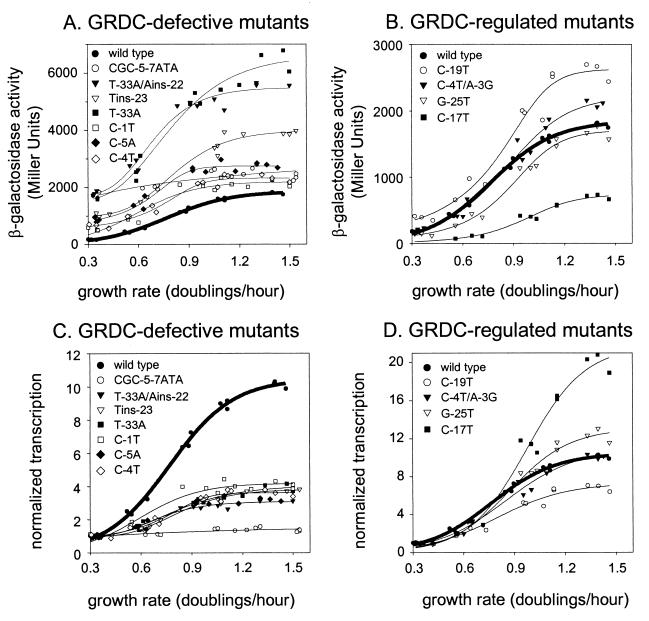
In order to test whether there is a correlation between the promoter sequences required for NTP sensing and growth rate-dependent regulation, single-copy lacZ fusions to the wild-type and mutant promoters were constructed on the chromosome and their activities were measured as a function of the growth rate (Fig. 3A and B). Since there are differences in the mutant promoters' intrinsic strengths, it is difficult to visualize the effects of the mutations on regulation without normalization for promoter activity. Figure 3C and D illustrate the growth rate dependence of the wild-type and mutant promoters after their activities were scaled to the same value at the lowest growth rate.
FIG. 3.
Growth rate-dependent control (GRDC) of wild-type and mutant rrnB P1 promoters. Each panel includes data points from three independent experiments for each promoter. The wild-type regression fit is in bold. Note that all of the panels have different ordinate scales. The top panels display the actual β-galactosidase activities versus the growth rate (doublings per hour). The bottom panels display the promoter activities normalized at the lowest growth rate in order to facilitate visualization of defects in regulation (see text). (A and C) Mutants that are defective for growth rate-dependent regulation. (B and D) Mutants with activities that increase with growth rate similarly to that of the wild type.
The responses of the rrnB P1 variants to changes in growth rate fell into two major classes. Whereas the wild-type promoter's activity increased about 10-fold over growth rates ranging from 0.3 to 1.5 doublings per h, seven mutants (CGC-5–7ATA, T-33A/Ains-22, Tins-23, T-33A, C-1T, C-5A, and C-4T) increased only 1.4- to 4-fold with increasing growth rates, much less than wild-type rrnB P1. In contrast, four mutants (C-19T, C-4T/A-3G, C-17T, and G-25T) increased 7- to 20-fold with increasing growth rates, to extents similar to or greater than that of the wild-type promoter.
The average fold increases from the lowest to the highest growth rate for the wild-type and variant promoters are summarized in Fig. 4B. Although this type of representation tends to exaggerate small differences in the primary data (e.g., the differences in NTP sensing of the four high-NTP mutants appears to be much more significant in Fig. 4A than in Fig. 2B), it is clear from comparison of Fig. 4A and B that the seven mutants most defective for growth rate-dependent regulation are the same mutants most defective for NTP sensing. In contrast, the four mutants that are growth rate regulated most like the wild-type promoter are the ones most sensitive to the same range of ATP concentrations as the wild type. Thus, although there may be some minor differences in the rank order of the extents of growth rate-dependent regulation and NTP sensing (see Discussion), we concluded that the cis-acting sequences in rrnB P1 needed for NTP sensing correlate well with the sequences required for growth rate-dependent regulation.
Low-NTP promoter mutants are defective for feedback derepression in fis::kan strains.
We next examined the relationship between a promoter's NTP concentration requirement and its response in one of the several assays that have been used to estimate feedback (homeostatic) control. In strains with the fis gene deleted, rRNA core promoter activity increases to partially compensate for the loss of activation (50, 53). If the mechanism responsible for this feedback regulation is NTP sensing, then the promoter sequences important for a response to the loss of FIS-dependent activation should correlate with those required for NTP sensing. Therefore, we compared the activities of the wild-type and mutant rrnB P1 promoters in wild-type strains or strains with fis deleted.
As observed previously (50, 53), for reasons that are unclear (see Materials and Methods), the activities of all promoter-lacZ fusions increased in fis::kan mutants. However, as also observed previously, the activity of the wild-type rrnB P1 promoter lacking FIS sites increased in a fis::kan strain 1.5-fold more than the activities of control promoters (3.8-fold versus 2.6-fold; Table 2). In general, the mutants that were defective for NTP sensing in vitro responded to the loss of fis more like the control promoters than like wild-type rrnB P1; i.e., the promoters requiring the highest ATP concentrations (C-17T, G-25T, and C-4T/A-3G) increased the most in response to the loss of fis, the promoters requiring the lowest ATP concentrations (CGC-5–7ATA, T-33A/Ains-22, Tins-23, and T-33A) increased the least in response to the loss of fis, and the promoters with intermediate ATP concentration requirements (C-1T, C-5A, C-4T, and C-19T) had intermediate responses to the loss of fis. However, because of differences in rank order and the relatively modest regulatory effect observed in this assay, we have not grouped the promoters into the same classes as for NTP sensing in vitro and for growth rate-dependent control in vivo (Table 2; Fig. 4C). Nevertheless, we conclude that the promoters' response to the loss of the fis gene correlates qualitatively with their response to changing NTP concentrations. Potential explanations for the less than perfect correlation between NTP sensing in vitro and the response to the loss of fis in vivo are discussed further below.
TABLE 2.
Feedback derepression of wild-type rrnB P1 and mutant rrnB P1 variants in fis::kan strains
| Promoter | β-Galactosidase activity (Miller units)
|
fis::kan/wild-type ratiob | |
|---|---|---|---|
| fis::kana | Wild typea | ||
| Wild-type rrnB P1 | 6,480 ± 480 | 1,690 ± 160 | 3.8 ± 0.3 |
| rrnB P1 variants | |||
| CGC-5-7ATA | 6,550 ± 110 | 2,690 ± 140 | 2.4 ± 0.1 |
| T-33A/Ains-22 | 9,530 ± 340 | 5,280 ± 280 | 1.8 ± 0.1 |
| Tins-23 | 8,900 ± 110 | 4,270 ± 90 | 2.1 ± 0.1 |
| T-33A | 10,600 ± 370 | 6,320 ± 200 | 1.7 ± 0.1 |
| C-1T | 6,210 ± 120 | 2,210 ± 120 | 2.8 ± 0.1 |
| C 5A | 7,580 ± 470 | 2,680 ± 210 | 2.8 ± 0.1 |
| C-4T | 6,350 ± 310 | 2,290 ± 250 | 2.8 ± 0.2 |
| C-19T | 7,690 ± 270 | 2,570 ± 150 | 3.0 ± 0.3 |
| C-4T/A-3G | 7,150 ± 340 | 2,000 ± 190 | 3.6 ± 0.2 |
| C-17T | 3,830 ± 78 | 610 ± 46 | 6.3 ± 0.6 |
| G-25T | 6,430 ± 480 | 1,560 ± 130 | 4.1 ± 0.1 |
| Controls | |||
| λPR | 5,160 ± 130 | 2,010 ± 50 | 2.6 ± 0.1 |
| PhisG | 3,360 ± 50 | 1,300 ± 40 | 2.6 ± 0.1 |
The mean number of Miller units was calculated from two or more independent experiments. Variation was less than 11%.
The fis::kan/wild-type ratio was calculated separately for each experiment. These ratios were then used to calculate the mean and variation shown.
DISCUSSION
Regulation by NTPs in vitro correlates with growth rate-dependent regulation and feedback derepression in fis::kan strains in vivo.
By analysis of the regulation of rrnB P1 promoter variants, we have shown that the concentration of the initiating NTP required in vitro for efficient transcription of a particular promoter correlates well with its susceptibility to growth rate-dependent regulation and more qualitatively with feedback control in vivo. As a result, our primary conclusion is that the kinetic features responsible for the dependence of rrn P1 promoters on high initiating NTP concentrations in vitro make these promoters sensitive, at least in part, to changes in growth rate and loss of fis in vivo. We noted that different mutations likely alter the topology and salt concentration dependences of each promoter to somewhat different extents. Thus, it is all the more remarkable that the ATP concentration dependences of the different promoters under any single condition in vitro correlate as well as they do with regulation in vivo.
Models of rrn P1 regulation by growth rate and feedback.
There are two general models to explain the mechanism of rRNA regulation that are consistent with the observed correlations. The simplest model is that rrn P1 promoters monitor the levels of free initiating NTP pools to coordinate rRNA synthesis rates with protein synthesis rates. This model is attractive because it explains how rRNA synthesis can be regulated, at least in part, by the overall biosynthetic energy capacity of the cell as a function of growth rate, and it also suggests the identity of one of the feedback signals generated by transient under- or overproduction of ribosomes.
We cannot exclude a more complicated alternative model in which one or more unidentified regulatory signals are elicited in response to growth rate changes and loss of the fis gene and these signals work on the same kinetic steps that make the promoter dependent on high concentrations of the initiating NTP for maximal transcription. We have used mutants with altered purine or pyrimidine metabolism to demonstrate that rrn P1 promoter activity changes when total NTP pools are altered (21; Schneider and Gourse, unpublished data). Although these studies suggest that NTP concentrations are not saturating for rrn P1 promoters in vivo, we cannot rule out the possibility that free NTP pools do not change in vivo as a function of growth rate or in fis::kan strains. In any case, because free NTP pools are likely subsaturating, hypothetical factors that affect the same kinetic steps that make the promoter sensitive to NTPs could theoretically alter the kinetics of initiation in vivo.
In lieu of an assay capable of measuring free NTP pools, our finding that there is a good correlation between NTP requirements in vitro and regulation of rRNA transcription in vivo supports the NTP sensing model. We note that it is well established that NTP concentration changes can affect the transcription of pyrimidine biosynthetic operons (although by using mechanisms different from that proposed for the regulation of rRNA transcription) (13, 30, 39, 43).
Kinetic basis for regulation by NTPs.
During the multistep process of transcription initiation (51), RNAP first binds the promoter to form a short-lived closed complex. The closed complex can then isomerize through at least one intermediate to form an open complex in which the DNA in the −10 hexamer and start site region is locally unwound. rrnB P1 and other similarly regulated rRNA and tRNA promoters form open complexes with half-lives of a few seconds or minutes under solution conditions in which most other promoters form open complexes with half-lives of several hours (5, 25, 31, 38, 40, 41, 49).
The unusually short-lived open complex formed by rrn P1 promoters makes them susceptible to regulatory molecules that alter open-complex occupancy. For example, ppGpp shortens the half-life of all RNAP-promoter open complexes in vitro, independently of whether or not the promoter is regulated by ppGpp in vivo (5). However, ppGpp only inhibits transcription from those promoters, like rrnB P1, that form intrinsically short-lived complexes (i.e., where the open-complex half-life is rate limiting for transcription initiation), explaining its specificity in vivo (5). We propose that the effect of initiating NTP concentration on rRNA promoters can be explained in a similar way: although all promoters bind NTPs to initiate transcription, the free NTP concentrations present in vivo are limiting for those promoters whose open complexes are exceptionally short-lived (21).
We have characterized the kinetic properties of the rrnB P1 promoter mutants described here (M. M. Barker, T. Gaal, W. Ross, and R. L. Gourse, unpublished data). The seven low-NTP promoters form complexes 4-fold to more than 30-fold longer-lived than wild-type rrnB P1. In contrast, the high-NTP mutants form open complexes with short lifetimes, similar to the wild-type promoter. Therefore, we propose that for a promoter to be susceptible to regulation by the initiating NTP concentration, the rate of open-complex collapse must be competitive with the time required for initiation. In theory, there could be promoters with short half-lives and with very fast forward rate constants. Therefore, we speculate that NTP sensing might have a second requirement: the forward rate constants cannot be so fast that the open complex accumulates even in the absence of the initiating NTP.
The promoter sequence determinants for regulation are complex.
The sequence determinants for regulation are multipartite and involve several of the sequence and structural characteristics common to rrn P1 promoters, including nonconsensus −35 hexamers, 16-bp spacers, G+C-rich discriminators, and a C at position −1 (see also references 5, 19, 31, 36, 37, 47, 49, and 63). Other promoter positions also could be important for regulation, since our screens were not exhaustive, and promoter context is likely to play an important role. In addition, transcription initiation occurs farther from the −10 hexamer in rrn P1 promoters than in most promoters and it is possible that atypical positioning of the start site plays a role in regulation.
As part of an extensive survey of a large number of promoter mutants, we previously characterized the growth rate-dependent regulation (but not the feedback regulation or NTP concentration requirements) of the promoter mutants described here (8, 17, 35). For 3 of the 11 mutants characterized here (C-5A, C-17T, and G-25T), the degree of growth rate-dependent regulation differs somewhat from that reported previously. For technical reasons, we believe that the results reported here are more accurate.
Multiple mechanisms contribute to regulation of rRNA transcription.
Some of the same kinetic features that make rRNA promoters sensitive to changing NTP concentrations also make rRNA promoters sensitive to other potential regulators (4–6). Although strains lacking ppGpp or FIS retain relatively normal growth rate-dependent regulation of rRNA transcription, these regulators could work in conjunction with the effects of NTPs. Moreover, since the level of negative supercoiling and the concentrations of monovalent and divalent cations also affect the rrnB P1 open-complex lifetime in vitro (25, 41; M.M.B. and R.L.G., unpublished data), systematic changes in superhelicity or osmolarity in vivo (if they occur) have the potential to work in conjunction with NTPs and ppGpp during changes in growth rate or in fis deletion strains. However, unlike the case with NTPs, which are consumed by protein synthesis and thus could serve as indicators of the ribosome level, it is unclear why or how supercoiling and osmolarity should change under these conditions.
We think it unlikely that another recently proposed regulatory mechanism, changes in the free RNAP concentration (42), is a major contributor to the control of rRNA transcription in vivo. rrn P1 promoters require lower concentrations of RNAP for transcription in vitro than most other promoters (4), and rRNA transcription does not respond in vivo to changes in the RNAP concentration that affect transcription from mRNA promoters (4, 9, 46).
Close examination of Fig. 4 reveals that the rank order and extent of the mutant promoters' responses to NTP concentration changes in vitro, to growth rate changes, and/or to deletion of fis in vivo are not always exactly the same. These disparities could result simply from compounding of errors associated with comparison of ratios or from the inability of in vitro assays to exactly duplicate conditions in cells. Most notably, while the most NTP-responsive mutants responded most to the loss of fis and the least NTP-responsive mutants responded least to the loss of fis, the relatively narrow window of the regulatory response in this assay makes quantitative assessment difficult. We also emphasize that the changes in rrnB P1 promoter activity associated with changes in growth rate or loss of the fis gene could be mediated only in part by variations in NTPs, and quantitative discrepancies in the correlations between the in vitro NTP-sensing behavior of the promoters and their in vivo regulatory responses could reflect the participation of other components in the system.
Squires and coworkers have suggested that feedback elicited by an increased or decreased rRNA gene dose might be mediated by a mechanism distinct from that responsible for growth rate-dependent regulation and NTP sensing (60). They studied four rrnB P1 variants from our collection (T-33A, Ains-22, C-1T/C-15G, and C-1T) (17) and found an incomplete correlation between the cis-acting sequences required for growth rate-dependent regulation and those required for a response to gene dose changes. Given the complexity intrinsic to rrn P1 promoters and their regulation, it seems entirely reasonable that different molecular mechanisms might operate under different conditions eliciting a feedback response. For example, part of the compensating increase in rRNA transcription that occurs in fis deletion strains probably is an increase in rrn P2 promoter activity (42a).
Many regulators of rRNA transcription influence the levels or activities of other contributors. For example, deletions of the fis or hns gene (H-NS is a histone-like protein that appears to negatively regulate rrnB P1 under some conditions [1, 55]) can lead to changes in DNA supercoiling (45, 54). FIS not only recruits RNAP to rrn P1 promoters (10) but also reduces the ATP concentration required for rrn P1 transcription (6). Induction of ppGpp synthesis reduces ATP and GTP pools (23, 24), and the ATP/ADP ratio may regulate supercoiling levels in vivo (18). Thus, the output from all of the regulatory mechanisms affecting rRNA transcription may be more than the simple sum of the inputs.
ACKNOWLEDGMENTS
We thank Tamas Gaal, Wilma Ross, David Schneider, and other members of our laboratory for insightful discussions and/or comments on the manuscript. We also thank Bob Landick for the generous gift of RNAP and Christine Hirvonen for cloning the λ lysogen containing the M13 polylinker-lacZ fusion.
This work was supported by NIH grant GM37048 to R.L.G. and a fellowship from Pfizer Biotechnology to M.M.B.
REFERENCES
- 1.Afflerbach H, Schroder O, Wagner R. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol Microbiol. 1998;28:641–653. doi: 10.1046/j.1365-2958.1998.00829.x. [DOI] [PubMed] [Google Scholar]
- 2.Aiyar S E, Gourse R L, Ross W. Upstream A-tracts increase bacterial promoter activity through interactions with the RNA polymerase alpha subunit. Proc Natl Acad Sci USA. 1998;95:14652–14657. doi: 10.1073/pnas.95.25.14652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai T, Condon C, Voulgaris J, Zaporojets D, Shen B, Al-Omar M, Squires C, Squires C L. Construction and initial characterization of Escherichia coli strains with few or no intact chromosomal rRNA operons. J Bacteriol. 1999;181:3803–3809. doi: 10.1128/jb.181.12.3803-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker M M, Gaal T, Gourse R L. Mechanism of regulation of transcription initiation by ppGpp. II. Models for positive control based on properties of RNAP mutants and competition for RNAP. J Mol Biol. 2001;305:689–702. doi: 10.1006/jmbi.2000.4328. [DOI] [PubMed] [Google Scholar]
- 5.Barker M M, Gaal T, Josaitis C A, Gourse R L. Mechanism of regulation of transcription initiation by ppGpp. I. Effects of ppGpp on transcription initiation in vivo and in vitro. J Mol Biol. 2001;305:673–688. doi: 10.1006/jmbi.2000.4327. [DOI] [PubMed] [Google Scholar]
- 6.Bartlett M S, Gaal T, Ross W, Gourse R L. Regulation of rRNA transcription is remarkably robust: FIS compensates for altered nucleoside triphosphate sensing by mutant RNA polymerases at Escherichia coli rrn P1 promoters. J Bacteriol. 2000;182:1969–1977. doi: 10.1128/jb.182.7.1969-1977.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartlett M S, Gaal T, Ross W, Gourse R L. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 8.Bartlett M S, Gourse R L. Growth rate-dependent control of the rrnB P1 core promoter in Escherichia coli. J Bacteriol. 1994;176:5560–5564. doi: 10.1128/jb.176.17.5560-5564.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bedwell D M, Nomura M. Feedback regulation of RNA polymerase subunit synthesis after the conditional overproduction of RNA polymerase in Escherichia coli. Mol Gen Genet. 1986;204:17–23. doi: 10.1007/BF00330181. [DOI] [PubMed] [Google Scholar]
- 10.Bokal A J, Ross W, Gourse R L. The transcriptional activator protein FIS: DNA interactions and cooperative interactions with RNA polymerase at the Escherichia coli rrnB P1 promoter. J Mol Biol. 1995;245:197–207. doi: 10.1006/jmbi.1994.0016. [DOI] [PubMed] [Google Scholar]
- 11.Burgess R R, Jendrisak J J. A procedure for the rapid, large-scale purification of Escherichia coli DNA-dependent RNA polymerase involving Polymin P precipitation and DNA-cellulose chromatography. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 12.Cashel M, Gentry D R, Hernandez V H, Vinella D. The stringent response. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- 13.Cheng Y, Dylla S M, Turnbough C L., Jr A long TA tract in the upp initially transcribed region is required for regulation of upp expression by UTP-dependent reiterative transcription in Escherichia coli. J Bacteriol. 2001;183:221–228. doi: 10.1128/JB.183.1.221-228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cole J R, Olsson C L, Hershey J W, Grunberg-Manago M, Nomura M. Feedback regulation of rRNA synthesis in Escherichia coli. Requirement for initiation factor IF2. J Mol Biol. 1987;198:383–392. doi: 10.1016/0022-2836(87)90288-9. [DOI] [PubMed] [Google Scholar]
- 15.Condon C, Liveris D, Squires C, Schwartz I, Squires C L. rRNA operon multiplicity in Escherichia coli and the physiological implications of rrn inactivation. J Bacteriol. 1995;177:4152–4156. doi: 10.1128/jb.177.14.4152-4156.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condon C, Squires C, Squires C L. Control of rRNA transcription in Escherichia coli. Microbiol Rev. 1995;59:623–645. doi: 10.1128/mr.59.4.623-645.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dickson R R, Gaal T, deBoer H A, deHaseth P L, Gourse R L. Identification of promoter mutants defective in growth rate-dependent regulation of rRNA transcription in Escherichia coli. J Bacteriol. 1989;171:4862–4870. doi: 10.1128/jb.171.9.4862-4870.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drlica K. Control of bacterial DNA supercoiling. Mol Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 19.Figueroa-Bossi N, Guerin M, Rahmouni R, Leng M, Bossi L. The supercoiling sensitivity of a bacterial tRNA promoter parallels its responsiveness to stringent control. EMBO J. 1998;17:2359–2367. doi: 10.1093/emboj/17.8.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gaal T, Barkei J, Dickson R R, deBoer H A, deHaseth P L, Alavi H, Gourse R L. Saturation mutagenesis of an Escherichia coli rRNA promoter and initial characterization of promoter variants. J Bacteriol. 1989;171:4852–4861. doi: 10.1128/jb.171.9.4852-4861.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaal T, Bartlett M S, Ross W, Turnbough C L, Jr, Gourse R L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science. 1997;278:2092–2097. doi: 10.1126/science.278.5346.2092. [DOI] [PubMed] [Google Scholar]
- 22.Gaal T, Gourse R L. Guanosine 3′-diphosphate 5′-diphosphate is not required for growth rate-dependent control of rRNA synthesis in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:5533–5537. doi: 10.1073/pnas.87.14.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallant J, Harada B. The control of ribonucleic acid synthesis in Escherichia coli. 3. The functional relationship between purine ribonucleoside triphosphate pool sizes and the rate of ribonucleic acid accumulation. J Biol Chem. 1969;244:3125–3132. [PubMed] [Google Scholar]
- 24.Gallant J, Irr J, Cashel M. The mechanism of amino acid control of guanylate and adenylate biosynthesis. J Biol Chem. 1971;246:5812–5816. [PubMed] [Google Scholar]
- 25.Gourse R L. Visualization and quantitative analysis of complex formation between E. coli RNA polymerase and an rRNA promoter in vitro. Nucleic Acids Res. 1988;16:9789–9809. doi: 10.1093/nar/16.20.9789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gourse R L, Gaal T, Aiyar S E, Barker M M, Estrem S T, Hirvonen C A, Ross W. Strength and regulation without transcription factors: lessons from bacterial rRNA promoters. Cold Spring Harbor Symp Quant Biol. 1998;63:131–139. doi: 10.1101/sqb.1998.63.131. [DOI] [PubMed] [Google Scholar]
- 27.Gourse R L, Gaal T, Bartlett M S, Appleman J A, Ross W. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu Rev Microbiol. 1996;50:645–677. doi: 10.1146/annurev.micro.50.1.645. [DOI] [PubMed] [Google Scholar]
- 28.Gourse R L, Ross W, Gaal T. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 29.Gourse R L, Takebe Y, Sharrock R A, Nomura M. Feedback regulation of rRNA and tRNA synthesis and accumulation of free ribosomes after conditional expression of rRNA genes. Proc Natl Acad Sci USA. 1985;82:1069–1073. doi: 10.1073/pnas.82.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han X, Turnbough C L., Jr Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription. J Bacteriol. 1998;180:705–713. doi: 10.1128/jb.180.3.705-713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinemann M, Wagner R. Guanosine 3′,5′-bis(diphosphate) (ppGpp)-dependent inhibition of transcription from stringently controlled Escherichia coli promoters can be explained by an altered initiation pathway that traps RNA polymerase. Eur J Biochem. 1997;247:990–999. doi: 10.1111/j.1432-1033.1997.00990.x. [DOI] [PubMed] [Google Scholar]
- 32.Hirvonen C A, Ross W, Wozniak C E, Marasco E, Anthony J R, Aiyar S E, Newburn V, Gourse R L. Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J Bacteriol. 2001;183:6305–6314. doi: 10.1128/JB.183.21.6305-6314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jinks-Robertson S, Gourse R L, Nomura M. Expression of rRNA and tRNA genes in Escherichia coli: evidence for feedback regulation by products of rRNA operons. Cell. 1983;33:865–876. doi: 10.1016/0092-8674(83)90029-6. [DOI] [PubMed] [Google Scholar]
- 34.Johnson R C, Ball C A, Pfeffer D, Simon M I. Isolation of the gene encoding the Hin recombinational enhancer binding protein. Proc Natl Acad Sci USA. 1988;85:3484–3488. doi: 10.1073/pnas.85.10.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Josaitis C A, Gaal T, Gourse R L. Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc Natl Acad Sci USA. 1995;92:1117–1121. doi: 10.1073/pnas.92.4.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung Y H, Lee Y. Escherichia coli rnpB promoter mutants altered in stringent response. Biochem Biophys Res Commun. 1997;230:582–586. doi: 10.1006/bbrc.1996.6005. [DOI] [PubMed] [Google Scholar]
- 37.Keener J, Nomura M. Regulation of ribosome biosynthesis. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1417–1431. ,. [Google Scholar]
- 38.Kupper H, Contreras R, Khorana H G, Landy A. The tyrosine tRNA promoter. In: Losick R, Chamberlin M, editors. RNA polymerase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1976. pp. 473–484. [Google Scholar]
- 39.Landick R, Turnbough C L, Jr, Yanofsky C. Transcription attenuation. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1263–1286. [Google Scholar]
- 40.Langert W, Meuthen M, Mueller K. Functional characteristics of the rrnD promoters of Escherichia coli. J Biol Chem. 1991;266:21608–21615. [PubMed] [Google Scholar]
- 41.Leirmo S, Gourse R L. Factor-independent activation of Escherichia coli rRNA transcription. I. Kinetic analysis of the roles of the upstream activator region and supercoiling on transcription of the rrnB P1 promoter in vitro. J Mol Biol. 1991;220:555–568. doi: 10.1016/0022-2836(91)90100-k. [DOI] [PubMed] [Google Scholar]
- 42.Liang S, Bipatnath M, Xu Y, Chen S, Dennis P, Ehrenberg M, Bremer H. Activities of constitutive promoters in Escherichia coli. J Mol Biol. 1999;292:19–37. doi: 10.1006/jmbi.1999.3056. [DOI] [PubMed] [Google Scholar]
- 42a.Liebig B, Wagner R. Effects of different growth conditions on the in vivo activity of the tandem Escherichia coli ribosomal RNA promoters P1 and P2. Mol Gen Genet. 1995;249:328–335. doi: 10.1007/BF00290534. [DOI] [PubMed] [Google Scholar]
- 43.Liu J, Turnbough C L., Jr Effects of transcriptional start site sequence and position on nucleotide-sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J Bacteriol. 1994;176:2938–2945. doi: 10.1128/jb.176.10.2938-2945.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 45.Mojica F J, Higgins C F. In vivo supercoiling of plasmid and chromosomal DNA in an Escherichia coli hns mutant. J Bacteriol. 1997;179:3528–3533. doi: 10.1128/jb.179.11.3528-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nomura M, Bedwell D M, Yamagishi M, Cole J R, Kolb J M. RNA polymerase and regulation of RNA synthesis in Escherichia coli: RNA polymerase concentration, stringent control, and ribosome feedback regulation. In: Reznikoff W S, editor. RNA polymerase and the regulation of transcription. New York, N.Y: Elsevier Science Publishing Co., Inc.; 1987. pp. 137–149. [Google Scholar]
- 47.Pemberton I K, Muskhelishvili G, Travers A A, Buckle M. The G+C-rich discriminator region of the tyrT promoter antagonises the formation of stable preinitiation complexes. J Mol Biol. 2000;299:859–864. doi: 10.1006/jmbi.2000.3780. [DOI] [PubMed] [Google Scholar]
- 48.Petersen C, Moller L B. Invariance of the nucleoside triphosphate pools of Escherichia coli with growth rate. J Biol Chem. 2000;275:3931–3935. doi: 10.1074/jbc.275.6.3931. [DOI] [PubMed] [Google Scholar]
- 49.Pokholok D K, Redlak M, Turnbough C L, Jr, Dylla S, Holmes W M. Multiple mechanisms are used for growth rate and stringent control of leuV transcriptional initiation in Escherichia coli. J Bacteriol. 1999;181:5771–5782. doi: 10.1128/jb.181.18.5771-5782.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao L, Ross W, Appleman J A, Gaal T, Leirmo S, Schlax P J, Record M T, Jr, Gourse R L. Factor independent activation of rrnB P1. An “extended” promoter with an upstream element that dramatically increases promoter strength. J Mol Biol. 1994;235:1421–1435. doi: 10.1006/jmbi.1994.1098. [DOI] [PubMed] [Google Scholar]
- 51.Record M T, Jr, Reznikoff W S, Craig M L, McQuade K L, Schlax P J. Escherichia coli RNA polymerase (Eς70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 792–820. [Google Scholar]
- 52.Ross W, Gosink K K, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 53.Ross W, Thompson J F, Newlands J T, Gourse R L. E. coli Fis protein activates ribosomal RNA transcription in vitro and in vivo. EMBO J. 1990;9:3733–3742. doi: 10.1002/j.1460-2075.1990.tb07586.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schneider R, Travers A, Muskhelishvili G. FIS modulates growth phase-dependent topological transitions of DNA in Escherichia coli. Mol Microbiol. 1997;26:519–530. doi: 10.1046/j.1365-2958.1997.5951971.x. [DOI] [PubMed] [Google Scholar]
- 55.Schroder O, Wagner R. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J Mol Biol. 2000;298:737–748. doi: 10.1006/jmbi.2000.3708. [DOI] [PubMed] [Google Scholar]
- 56.Sharrock R A, Gourse R L, Nomura M. Defective antitermination of rRNA transcription and derepression of rRNA and tRNA synthesis in the nusB5 mutant of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:5275–5279. doi: 10.1073/pnas.82.16.5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shen V, Bremer H. Chloramphenicol-induced changes in the synthesis of ribosomal, transfer, and messenger ribonucleic acids in Escherichia coli B/r. J Bacteriol. 1977;130:1098–1108. doi: 10.1128/jb.130.3.1098-1108.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 59.Travers A A. Conserved features of coordinately regulated E. coli promoters. Nucleic Acids Res. 1984;12:2605–2618. doi: 10.1093/nar/12.6.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voulgaris J, Pokholok D, Holmes W M, Squires C, Squires C L. The feedback response of Escherichia coli rRNA synthesis is not identical to the mechanism of growth rate-dependent control. J Bacteriol. 2000;182:536–539. doi: 10.1128/jb.182.2.536-539.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker K A, Atkins C L, Osuna R. Functional determinants of the Escherichia coli fis promoter: roles of −35, −10, and transcription initiation regions in the response to stringent control and growth phase-dependent regulation. J Bacteriol. 1999;181:1269–1280. doi: 10.1128/jb.181.4.1269-1280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yamagishi M, de Boer H A, Nomura M. Feedback regulation of rRNA synthesis. A mutational alteration in the anti-Shine-Dalgarno region of the 16S rRNA gene abolishes regulation. J Mol Biol. 1987;198:547–550. doi: 10.1016/0022-2836(87)90299-3. [DOI] [PubMed] [Google Scholar]
- 63.Zacharias M, Goringer H U, Wagner R. Influence of the GCGC discriminator motif introduced into the ribosomal RNA P2- and tac promoter on growth-rate control and stringent sensitivity. EMBO J. 1989;8:3357–3363. doi: 10.1002/j.1460-2075.1989.tb08498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]