ABSTRACT
The immune escape mutations of SARS-CoV-2 variants emerged frequently, posing a new challenge to weaken the protective efficacy of current vaccines. Thus, the development of novel SARS-CoV-2 vaccines is of great significance for future epidemic prevention and control. We herein reported constructing the attenuated Mycobacterium smegmatis (M. smegmatis) as a bacterial surface display system to carry the spike (S) and nucleocapsid (N) of SARS-CoV-2. To mimic the native localization on the surface of viral particles, the S or N antigen was fused with truncated PE_PGRS33 protein, which is a transportation component onto the cell wall of Mycobacterium tuberculosis (M.tb). The sub-cellular fraction analysis demonstrated that S or N protein was exactly expressed onto the surface (cell wall) of the recombinant M. smegmatis. After the immunization of the M. smegmatis-based COVID-19 vaccine candidate in mice, S or N antigen-specific T cell immune responses were effectively elicited, and the subsets of central memory CD4+ T cells and CD8+ T cells were significantly induced. Further analysis showed that there were some potential cross-reactive CTL epitopes between SARS-CoV-2 and M.smegmatis. Overall, our data provided insights that M. smegmatis-based bacterial surface display system could be a suitable vector for developing T cell-based vaccines against SARS-CoV-2 and other infectious diseases.
KEYWORDS: Mycobacterium smegmatis, bacterial surface display system, SARS-CoV-2, vaccine, T cell immunity
Introduction
The pandemic of Coronavirus Disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), remains a severe threat to global public health. The establishment of herd immunity by mass vaccination is thought of as the most cost-effective intervention to eventually stop this pandemic,1 and thus it is of great significance to develop novel vaccines for the prevention and control of SARS-CoV-2 transmission. Currently, many COVID-19 vaccines have been urgently approved by the WHO for human use (https://www.who.int/teams/regulation-prequalification/eul/covid-19). However, a rapid mutation accumulation of SARS-CoV-2 variants is emerging frequently, including B.1.1.7 strain in the UK, B.1.351 in Africa, P.1 in Brazil, B.1.617 strain in India, and B.1.1.529 strain in South Africa, resulting in immune escape from existing neutralizing antibodies.2 Different from antibodies response, T-cell immune response to the SARS-CoV-2 variants is relatively preserved in most individuals.3,4 Thus, to prevent these variations, vaccines that induce a specific T-cell immune response against SARS-CoV-2 are needed.
Mycobacterium bovis Bacillus Calmette-Guérin (BCG) is well known as a live attenuated vaccine against Mycobacterium tuberculosis (M.tb). Recent studies indicated that in response to BCG vaccination, the subsets of CCR6+ CD4+ T cells, central memory (Tcm) and effector memory (Tem) CD4+ T cells and CD8+ T cells were significantly induced, but the subsets of naïve T cells and regulatory T cells were significantly diminished,5,6 suggesting that BCG immunization could effectively modulate the T cell-based immunity. Moreover, recombinant BCG expressing various antigens could enhance T cell immune response to protect against various diseases, including measles virus,7 rodent malaria,8 cancer,9 etc. Importantly, the recombinant BCG expressing nucleocapsid antigen from SARS-CoV-2 could elicit antigen-specific adaptive and trained immunity.10
Mycobacterium smegmatis (M. smegmatis) is a mycobacterium with genetic relationships with BCG and other Mycobacterium species, but it is more easy to be cultivated and genetically manipulated in the laboratory environment. Specifically, M. smegmatis can propagate 10 times faster than BCG.11 Importantly, M. smegmatis-based HIV vaccine could elicit specific T-cell immune responses.12–14 In addition, unlike the BCG to survive in the host cells by inhibiting phagocytosis maturation, M. smegmatis can be quickly killed by the protease in the phagosome, and thus it is usually harmless to healthy people.15 However, M. smegmatis might cause diseases in immunocompromised people. To further improve its safety, we modified an attenuated M. smegmatis strain ZWY2 as a gene delivery vector. The ZWY2, in which the virulence gene esx-3 operon is deleted, had a good safety even in the immunodeficiency mice.16 In addition, the ZWY2 is a drug-susceptible strain and thus can be well controlled by multiple antibiotics. Therefore, ZWY2 is more suitable as a potential vector for vaccine development. To our knowledge, there is no report on the immunogenicity of M. smegmatis-based SARS-CoV-2 vaccine yet.
SARS-CoV-2 consists of four major structural proteins, including the spike glycoprotein (S), nucleoprotein (N), envelope protein (E), and membrane protein (M). The S protein contains S1 and S2 subunits. Of these, the receptor-binding domain (RBD) of the S1 subunit binds to the human angiotensin-converting enzyme 2 (hACE2) receptor to help the virus enter the host, and S2 mediates the subsequent membrane fusion.17 Full-length S or RBD protein plays a major role in developing the COVID-19 vaccine to generate neutralizing antibodies.18,19 However, neutralizing antibodies alone might be not sufficient to control SARS-CoV-2 mutants, and thus T cell immune responses, which usually tolerate more mutants, should be induced for the next generation of SARS-CoV-2 vaccines. Compared with S protein, N protein is relatively conservative and possesses more T cell epitopes. SARS-CoV-2 vaccines based on the N antigen induced strong T cell protective immune response in mice and macaques in recent studies.20,21 Herein, we proposed to construct an attenuated M. smegmatis as a bacterial surface display system to carry the S and N of SARS-CoV-2 and explored the feasibility of M. smegmatis-based vector for SARS-CoV-2 vaccine candidate.
Materials and methods
Bacterial strains and medium
Escherichia coli strain T1 was grown in Luria-Bertani medium at 37°C with 100 μg/mL ampicillin (Meilunbio) when necessary. M. smegmatis Ms_0615 knockout attenuated strain ZWY222 was cultured at 37°C in Middlebrook 7H9 medium (BD Biosciences) supplemented with 10% of oleic acid, albumin, dextrose complex (OADC, BD Biosciences), 0.2% glycerol (Macklin), and 0.05% Tween 80 (Amresco) or Middlebrook 7H11 agar (BD Biosciences) supplemented with 10% OADC and 0.2% glycerol, and 150 μg/mL Hygromycin B (Roche) when necessary.
DNA manipulation reagents
PCR reactions were performed using KOD One PCR Master Mix (Toyobo). Restriction enzymes were purchased from Takara. Seamless cloning was performed using pEASY-Basic Seamless Cloning and Assembly Kit (Transgen).
Assembly of recombinant vectors
Plasmid pI1818 was constructed for surface display in BCG and ZWY2 under the control of the strong mycobacterium hsp60 promoter followed by an N-terminal truncated Rv1818’, a transportation protein anchored in the cell wall of the bacteria. A linker (GGGS)4 was added between the transportation protein and the N terminus of the inserted antigen to avoid possible interference with protein folding. The pI1818 is an integrative vector that can replicate in E.coli and integrate into the attachment site of the mycobacterium genome through L5 mycobacterium phage integrase. The vector pI1818 was digested with KpnI and EcoRI and then inserted with the hsp60-Rv1818c’-linker fragment and the full-length codon-optimized spike gene (Sopt) or nucleocapsid gene (Nopt) of SARS-CoV-2 through seamless cloning. The influenza hemagglutinin (HA) epitope tag was fused with the C-terminal of Sopt and Nopt by PCR for protein detection (Figure 1). Primers were designed using CE Design V1.04 (Vazyme) and synthesized in Sangon (Shanghai, China).
Figure 1.
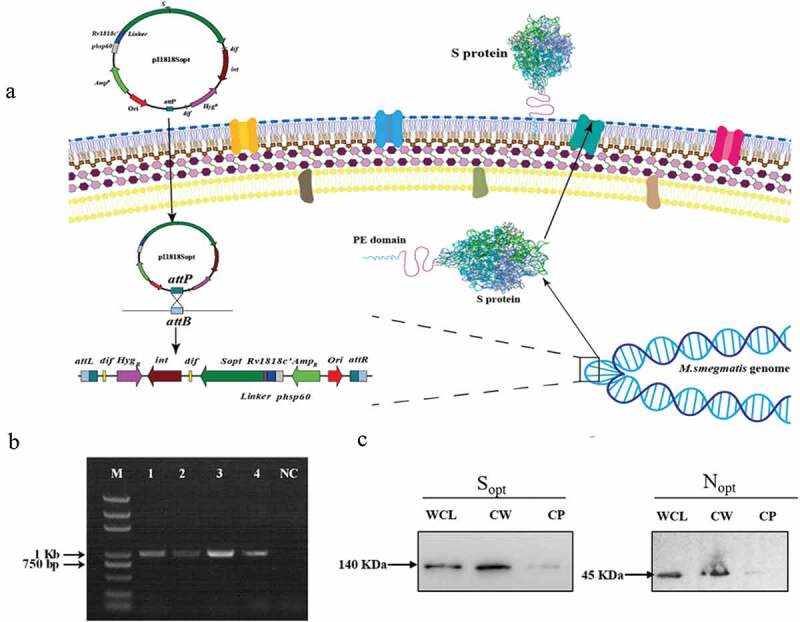
Construction of ZWY2-based recombinant M.smegmatis strains displaying the S and N on the surface. (a) Schematic illustration of the integration process of recombinant pI1818 plasmid into the M. smegmatis chromosome, and then the fused S protein with the transportation protein of the truncated PE_PGRS33 was located onto the cell surface. The attB is the Mycobacteriophage L5 integrase(int) recognition site located near the tRNAGly in the mycobacterium’s genome. The attP is the attachment site on Mycobacteriophage’s genome. After the integration, the fused protein was expressed and translocated to the cell wall through the PE domain of the truncated PE_PGRS33 protein. (b) Identification of ZWY2-based recombinant mycobacterial strains using FX and pF1 primers (Table S1). The FX is located in the gene HygR in the integrative plasmid, and the pF1 is located upstream of the attB site in the M. smegmatis genome. Lane1: DNA marker; Lane 2: ZWY2-S; Lane 3: ZWY2-S-HA; Lane 4: ZWY2-N; Lane 5: ZWY2-N-HA; NC: negative control (ZWY2). (c) Western blotting analysis of the subcellular fractions of ZWY2-S-HA and ZWY2-N-HA. WCL: whole cell lysate; CW: cell wall; CP: cell plasma; NC: negative control (the cell wall of ZWY2).
Construction of recombinant mycobacterial strains
Competent cells of ZWY2 for electroporation were prepared as previously described.23 The four plasmids pI1818Sopt, pI1818Nopt, pI1818Sopt-HA, and pI1818Nopt-HA were electroporated into ZWY2 respectively to obtain the corresponding recombinant strains ZWY2-S, ZWY2-N, ZWY2-S-HA, and ZWY2-N-HA. Recombinant mycobacterial clones were selected on 7H11 agar containing 150 μg/mL of Hygromycin B. Single colony was picked up for PCR identification to make sure the corresponding plasmids had been integrated into the genome of ZWY2 successfully. All primers are described in Supplementary Table S1.
Subcellular fractions isolation and Western Blotting
ZWY2-S-HA and ZWY2-N-HA strains were cultured in the 37°C shaker for 2 to 3 days until the optical density at 600 nm (OD600) approximately reached 1.0. Cells from 1 L culture were harvested and washed with 0.16 M of NaCl solution. After weighing the corresponding wet cells, 1 mL lysis buffer (0.05 M potassium phosphate, 0.022% (v/v) β-mercaptoethanol, pH 6.5) was added for each gram of bacteria and supplement with lysozyme (Roche) at a final concentration of 2.4 mg/mL. Subsequently, the cells were incubated at 37°C for 2 h and sonicated 4 times for 15 minutes each time in an ice bath ultrasonic cell disruptor JY92-IIN (Scientz). The lysates were centrifugal at a low speed of 1,000 × g for 10 minutes to remove the unruptured cells. Then centrifugation was repeated twice for 1 h at 27,000 × g, the precipitate is the cell wall, and the supernatant is the cell plasm.24 The isolation of subcellular fractions was done at 4°C. The expression level of the S and N antigens in the different mycobacterial subcellular fractions lysates were assessed by western blotting using a mouse anti-HA monoclonal antibody (MAb) (Abcam), Goat anti-Mouse IgG HRP (Abclonal), and a chemiluminescence HRP substrate (Millipore).
Cytokine determination from infected antigen-presenting cells
Raw264.7 cells (from macrophage of a male adult mouse) and DC2.4 cells (mouse bone marrow-derived dendritic cells) were cultured in the complete RPMI 1640 medium with 10% fetal bovine serum (FBS), penicillin-streptomycin (100 units/mL and 100 μg/ml), at 37°C, 5% CO2. The 1 × 106 cells of Raw264.7 or DC2.4 were infected with ZWY2-S or ZWY2-N (10 M.O.I.; multiplicity of infection) in RPMI 1640 medium only complemented with 10% FBS for 4 hours at 37°C. The culture medium treated was assigned as a negative control. After washing with phosphate-buffered saline (PBS), cells were maintained with fresh complete RPMI 1640 medium for 24 h. Total RNA was extracted using TRIzol reagent (Vazyme) from recombinant M. smegmatis-infected Raw264.7 and DC2.4 cells. After synthesizing the cDNA from 1 μg total RNA by using an HifairTM III 1st Strand cDNA Synthesis SuperMix kit (Yeasen), the quantitative real-time PCR was performed by a QuantStudio 7 Flex quantitative real-time PCR (qPCR) detection system (Applied Biosystems) using the PerfectStart SYBR Green qPCR supermix (Transgen) to determine the fold change in expression level for each transcript with the 2−ΔΔCt method. The sequence information of primers used in this study, including the IL-1β, IL-6, TNF-α, IFN-β, β-actin, was described in Supplementary Table S1.
Flow cytometry assay for antigen-presenting cells mature
ZWY2-S or ZWY2-N infected Raw264.7 cells and DC2.4 cells were collected and blocked with CD16/32 antibody (Biolegend) for 30 min on ice. After being washed with PBS, the cells were stained with anti-mouse CD86-BV421, anti-mouse CD80-APC-750, anti-mouse CD40-APC, and anti-mouse major histocompatibility complex (MHC) II-APC-700 (Biolegend) for 30 min on ice. The stained Raw264.7 cells and DC2.4 cells were resuspended in fluorescence-activated cell sorting (FACS) buffer and analyzed by CytExpert (Beckman). The data were analyzed with FlowJo software.
Mice and immunizations
To evaluate the immunogenicity of these M. smegmatis-based vaccines, six-week-old female pathogen-free BALB/c mice were obtained from the Laboratory Animal Center, Sun Yat-sen University. All animal studies were conducted following guidelines for animal use and care established by the Laboratory Animal Ethics Committee, Sun Yat-sen University (Assurance number: SYSU-IACUC-2021–000258). ZWY2 and recombinant ZWY2-S and ZWY2-N strains were cultured in 7H9 medium until OD600 approximately reached 0.85 while we estimated the CFU of bacteria is 5 × 108. Forty mice were randomly allocated into four groups. Group 1 and Group 2 respectively were injected with 200 μL PBS and 108 CFU ZWY2 as the sham group and negative group. Group 3 and Group 4 were respectively vaccinated with ZWY2-S and ZWY2-N in 200 μL of PBS via the subcutaneous route. Immunization was carried out at 0, 3, and 6 weeks. In the 6th week, the mice were euthanasia and the splenocytes were subjected to enzyme-linked immunosorbent assays (ELISpot), and intracellular cytokine staining (ICS). In the 9th week, the splenocytes were subjected to ICS and T cell phenotype analysis, and the mouse serum was analyzed by enzyme-linked immunosorbent assays (ELISA).
Elisa
ELISA assay was performed to detect the total IgG antibodies in the mice serum as we previously reported.25 96-well ELISA plates were coated with 0.1 μg full-length S or N protein of SARS-CoV-2 (Sino Biological Inc.) at 4°C overnight and blocked with PBST (PBS supplemented with 0.05% Tween-20) supplemented with 5% skimmed milk powder at 37°C for 1 h. After washing with PBST, the mice serum samples diluted at 1:20 were added per well and incubate at 37°C for 2 h, and then the HRP-labeled goat anti-mouse IgG (Abcam) was added and incubated for 1 h. Finally, 3,3,’5,5’-tetramethylbenzidine (TMB) substrate was added and reacted for 10 min, and then determined at 450 nm after the termination of the reaction with 1 M H2SO4.
IFN-γ ELISpot assays
The gamma interferon (IFN-γ) ELISpot assays were performed with freshly isolated mouse splenic lymphocytes as we previously reported.26 In brief, a sterile PVDF 96-well plate (Millipore) was coated with mouse IFN-γ coating antibody (U-CyTech) at 4°C overnight. After washing with PBS, the plate was blocked with R10 medium for 2 h at 37°C. Mouse splenic lymphocytes isolated by using a density gradient medium (Dakewe Biotech) were seeded in the plate at 1 × 106 cells per well. S or N peptide pools were 15 amino acids in length and overlapped by 11 amino acids covering the full-length S or N peptide pools respectively, and were synthesized by GenScript company. The vaccine groups were specifically stimulated with total S peptide or N peptide pools at 4 μg/ml per peptide. Dimethyl sulfoxide (DMSO) and concanavalin A (ConA) were used as mock and positive stimulation respectively. Subsequently, the plate was incubated with biotinylated detection antibody (U-CyTech) at 4°C overnight, and then alkaline phosphatase-conjugated streptavidin (U-CyTech) and NBT/BCIP reagent (Pierce) were added for color reaction. Finally, the spots were analyzed with an ELISpot reader (Bioreader4000, BIOSYS, Germany).
Intracellular cytokine staining (ICS) and T cell phenotype analysis
The ICS assays were performed as we previously reported.27 In brief, fresh mouse splenic lymphocytes were seeded in a 96-well cell culture plate at 2 × 106 cells per well and stimulated with S1 peptide pool (peptides 1–134), S2 peptide pool (peptides 135–253) together covering the full-length S protein] or N peptide pools at a final concentration of 4 μg/ml per peptide for 2 h, as well as corresponding mock (DMSO) and positive control [Phorbol myristate acetate (PMA) and Ionomycin] group were constructed. Protein transportation was blocked by Brefeldin A (BFA) and then further incubated for 16 h. Cells were collected and stained with anti-mouse CD3e-FITC (BD Biosciences), anti-mouse CD4-BB700 (BD Biosciences), anti-mouse CD8a-PE Cy7 (BD Biosciences), anti-mouse CD107a-BV510 (Biolegend) for 30 min at room temperature in the dark. After stained, cells were permeabilized/fixated (BD Cytofix/CytopermTM) for 20 min at 4°C in the dark, and then 1× Perm/WashTM buffer (BD Biosciences) was added for washing. Then, cells were stained with anti-mouse IFN-γ-Alexa Fluor 647 (BD Biosciences), anti-mouse TNF-α-PE (BD Biosciences), and anti-mouse IL-2-BV421 (BD Biosciences) for 1 h at 4°C in the dark. For T cell phenotype analysis, mock (DMSO), S or N peptides-stimulated mouse splenocytes were stained with anti-mouse CD3e-FITC, anti-mouse CD4-BB700, anti-mouse CD8a-PE Cy7, anti-mouse CD44-KO525 (BD Biosciences), and anti-mouse CD62 L-BV780 (BD Biosciences). Flow cytometry was performed by Beckman CytExpert (Beckman) and data was subsequently analyzed with CytExpert software. To avoid the experimental background, the data were calculated by subtracting the corresponding DMSO-simulated value from the peptide-simulated value.
Identification of the cross-reactive cytotoxic T lymphocyte epitopes between SARS-CoV-2 and M.Smegmatis
To identify the cross-reactive cytotoxic T lymphocyte (CTL) epitopes, SARS-CoV-2 S protein sequence (Genebank: YP_009724390.1), N protein sequence (Genebank: YP_009724390.1), and 6427 protein sequences of M.smegmatis were obtained from the National Center for Biotechnology Information (NCBI). Subsequently, we performed sequence alignment of SARS-CoV-2 structure protein and M.smegmatis homologs in which a series of conserved sequence fragments have been found. We then submitted the conserved sequence to the NetMHCpan-4.0 EL 4.1 algorithm and selected a reference panel of 27 HLA alleles.28,29 The epitope was determined based on the Percentile Rank, which was lower than 2% (0.5% is the specified threshold for strong binders and 0.5%–2% is the specified threshold for weak binders).
Data analysis
Flow cytometric data were analyzed using CytExpert software or Flowjo software. Statistical analyses and graphs were performed with GraphPad Prism 8 software (GraphPad Software, Inc). One-way ANOVA tests and post-hoc tests (Turkey HSD) were performed to compare the immune responses between different groups, and a two-tailed p-value less than 0.05 was considered statistically significant (*: P < .05; **: P < .01; ***: P < .001).
Results
Construction of recombinant M.Smegmatis ZWY2 strains displaying the S and N antigens on the surface
The pI1818 plasmid backbone was used to obtain the recombinant constructs carrying the codon-optimized S gene or N gene of SARS-CoV-2. To display them on the surface of ZWY2 bacteria, the S or N antigen was fused with the transportation protein of the truncated PE_PGRS33 (Figure 1a), which is encoded by Rv1818’ gene from M. tuberculosis. All the plasmids contained the hsp60 promoter to drive the expression of these fusion genes. Then, these plasmids were transformed into ZWY2, and the ZWY2-based recombinant M. smegmatis strains (ZWY2-S, ZWY2-S-HA, ZWY2-N, ZWY2-N-HA) were screened and identified by PCR assay (Figure 1b). To validate the expression and location of the targeted antigens, we isolated the subcellular fractions of the recombinant M. smegmatis strains and got the whole cell lysate, cell wall, and cell plasma respectively. These fractions were then detected by Western blotting. As expected, our results showed that these recombinant bacteria could appropriately express the S or N antigens onto the cell wall of ZWY2 bacteria (Figure 1c).
The recombinant M. smegmatis-based vaccine induced the activation and maturation of antigen-presenting cells
To determine whether recombinant M. smegmatis can stimulate the maturation of antigen-presenting cells (APCs), Raw264.7 and DC2.4 cells were infected with ZWY2-S or ZWY2-N, and then the activation and maturation marker including CD40, CD80, CD86, MHC II on the cell surface were assessed. Results showed that the proportion of Raw264.7 and DC2.4 cells expressing CD40, CD80, CD86, MHC II was greatly increased after being infected with recombinant M. smegmatis (Figure 2a). Meanwhile, a higher mRNA level of IFN-β and some inflammatory factors, such as IL-6, TNF-α, and IL-1β, were observed in Raw264.7 cells and DC2.4 cells (Figure 2b). These findings demonstrated that the recombinant M. smegmatis could stimulate the expression of co-stimulatory molecules on the surface of macrophages and dendritic cells in vitro, and thus promote the maturation of APCs and stimulate the secretion of inflammatory cytokines.
Figure 2.
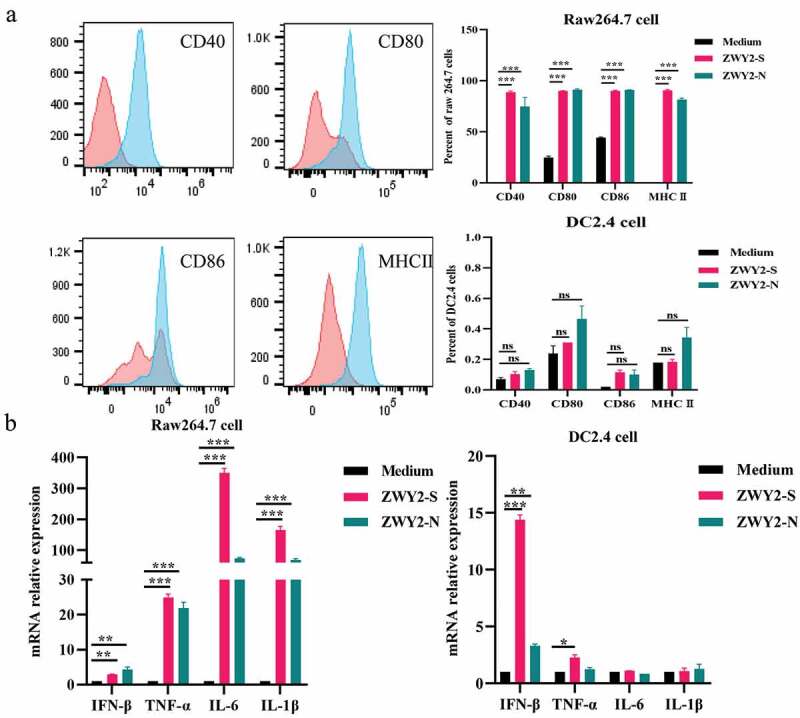
Recombinant M. Smegmatis-based vaccine could induce the activation and maturation of antigen-presenting cells. (a) the expression level of co-stimulatory molecules on Raw264.7 and DC 2.4 cells after being infected with ZWY2-S or ZWY2-N bacteria. The red peak represents the medium control group, and the blue one represents the recombinant M. smegmatis treated group. (b) the mRNA levels of IFN-β, TNF-α, IL-6, and IL-1β in Raw264.7 and DC2.4 cells after being infected with ZWY2-S or ZWY2-N. These data were expressed as the mean ± SEM from two individual trials (*: P < .05; **: P < .01; ***: P < .001).
The recombinant ZWY2-S and ZWY2-N elicited strong T-cellular immune responses in mice
To evaluate the immunogenicity of ZWY2-S and ZWY2-N in vivo, forty mice were randomly allocated into four groups. Among them, Group 1 and Group 2 were injected with PBS and ZWY2 respectively as control. Group 3 and Group 4 were immunized with ZWY2-S or ZWY2-N respectively. The immunization procedure was conducted at 0, 3, and 6 weeks, and then the T-cellular immune response was detected (Figure 3a, Table S2). The results demonstrated that ZWY2-S or ZWY2-N could elicit the increased IFN-γ-secreting cells in response to S or N peptide pool by ELIspot assay at week 6 (Figure 3b), indicating that this M. smegmatis-based COVID-19 vaccine can induce an antigen-specific T-cellular immune response in mice. To further identify which domain of S antigen might mainly contribute to this immunogenicity, the total S peptides were divided into two peptide pools, S1 (peptides 1–134) and S2 (peptides 135–253) for the subsequent stimulation. Our data showed that a higher frequency of CD107a+ and IL-2+ CD4+ T cells was produced in response to S2 peptides stimulation in the ZWY2-S group than that of the ZWY2 or PBS group, and a higher frequency of IL-2+ CD4+ T cells, IFN-γ+ CD8+ T cells, and TNF-α+ CD8+ T cells were observed in the ZWY2-N group than that of ZWY2 group. Interestingly, ZWY2-N also showed a higher frequency of CD8+ T cells expressing CD107a, indicating that the ZWY2-N vaccine could elicit an N-specific CTL response (Figure 3c,d). At week 9, there was a higher frequency of TNF-α+ CD4+ T cell and TNF-α+ CD8+ T cell simulated with S2 peptide pools in the ZWY2-S group. Moreover, a significantly increased frequency of IL2-secreting CD4+ and CD8+ T cells in response to the stimulation of the N peptide pool was observed in the ZWY2-N group (Figure 3e). Besides the T cell immune response, an increased antibody level was also observed by ZWY2-S and ZWY2-N than that of PBS group, though the OD450 value of ELISA-based antibodies stayed at low levels (Figure S1). Collectively, these data implied that ZWY2-S and ZWY2-N could elicit the S or N antigen-specific T cell-biased cellular immune responses.
Figure 3.
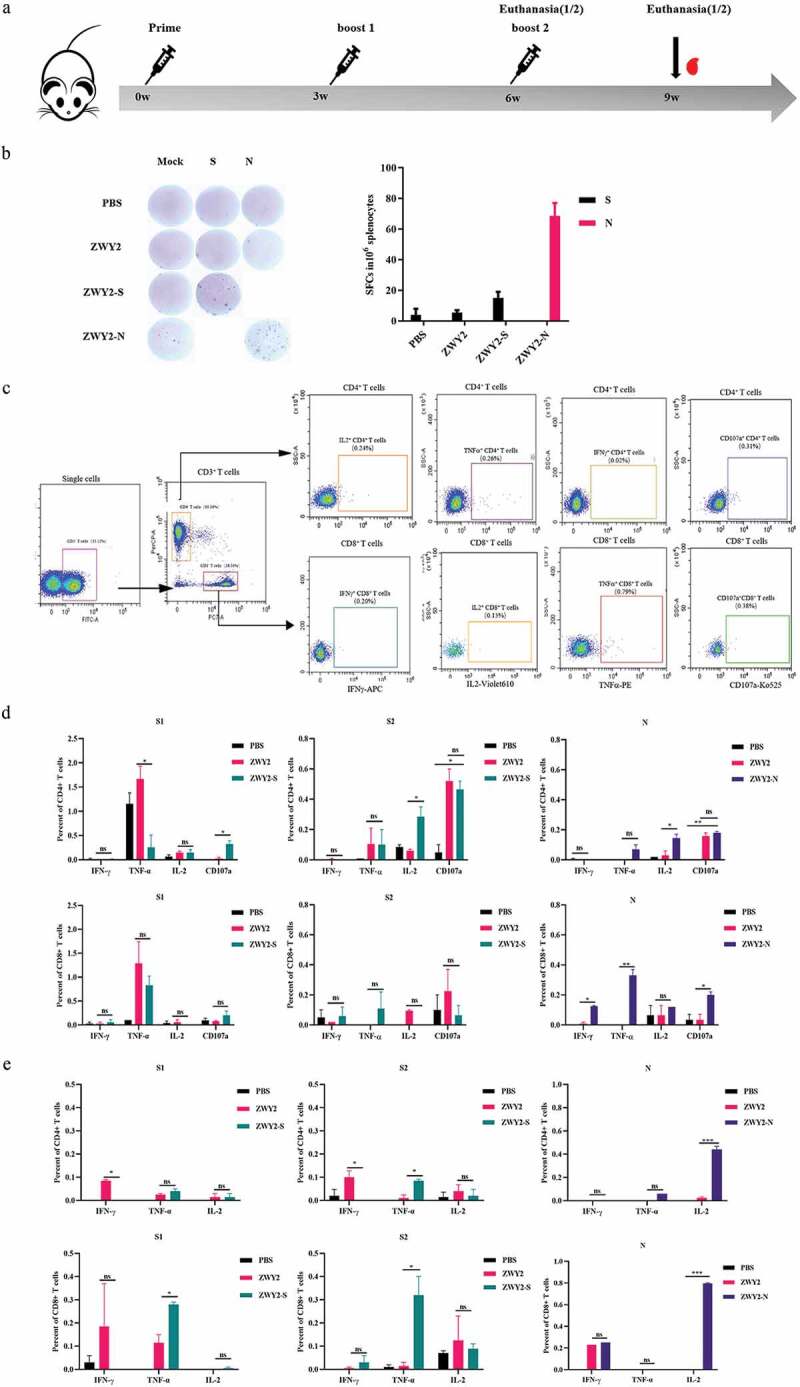
SARS-CoV-2 antigen-specific T-cellular immune responses were elicited by the recombinant ZWY2-S and ZWY2-N in mice. (a) Schedule for vaccination and the immunogenicity detecting procedure in mice. Six-week-old BALB/c mice were vaccinated with PBS, ZWY2, ZWY2-S, and ZWY2-N using the indicated dosage through a subcutaneous route. The symbols of red blood drops indicate the time points for collecting the serum samples. At 6 weeks and 9 weeks after the initial vaccination, five mice in each group were sacrificed, and the splenocytes were harvested and subjected to the following immunological assays. (b) at week 6, SARS-CoV-2 S or N antigen-specific T cell immune responses were detected using IFN-γ ELISpot assays following stimulation with the corresponding peptide pools. SFCs: spot-forming cells. (c) Gating strategy to analyze the frequency of CD4+ T and CD8+ T cells secreting the IFN-γ, TNF-α, IL-2 cytokines, and expressing CD107a marker by ICS assay. Column graphs depict the frequency of cytokine-positive CD4+ T cells and CD8+ T cells, which is calculated by subtracting the mock-stimulated sample at week 6 (d) and week 9 (e). Two independent experiments for animal immunization were repeated. The frequency of antigen-specific cytokine-positive T cell ratio These data were expressed as the mean ± SEM (*: P < .05; **: P < .01; ***: P < .001).
The recombinant ZWY2-S and ZWY2-N induced the formation of central memory T-cell subsets
It is critical to induce the formation of antigen-specific memory T cell subsets for rapid viral clearance and long-term immune protection by an effective vaccine candidate. Of note, recent data showed that SARS-CoV-2-specific memory T cell responses were elicited in COVID-19 convalescent patients and persist for long-term,30 and thus memory T cells might play important role in a potential SARS-CoV-2 vaccine. Therefore, we further characterize the phenotype of memory T cell subsets by assessing the expression of CD62 L and CD44 using the corresponding monoclonal antibodies (Figure 4a). Although there was no obvious statistical difference in the effector memory T subset (Tem, CD44high CD62 L-) when compared with the sham group, there was an increased percentage of the central memory T subset (Tcm, CD44high CD62 L+) in the vaccinated group (Figure 4b). The percentage of CD4+ and CD8+ Tcm subset was higher than that of Tem subset in the vaccinated group, implying that the recombinant M.smegmatis-based COVID-19 vaccines could induce the differentiation and formation of Tcm subsets, which is important to sustain a long-term protective T cell immunity.
Figure 4.
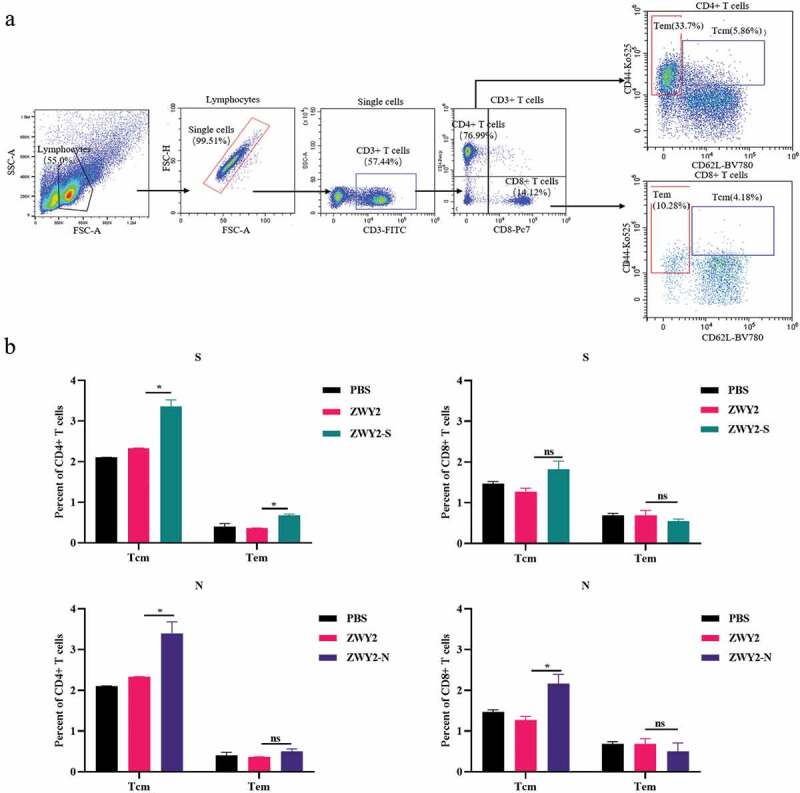
The central memory T subsets were induced by recombinant ZWY2-S and ZWY2-N in mice. (a) Gating strategy for analyzing the percentage of different T cell subsets, including Tcm (Cd44high CD62 L+), Tem (Cd44high CD62 L-) of CD4+ T cells, and CD8+ T cells. (b) Column graphs of the different cell subsets percentage of CD4+ T cells and CD8+ T cells. Two independent experiments for animal immunization were repeated. These data were expressed as the mean ± SEM (*: P < .05; **: P < .01; ***: P < .001).
Identification of the potential cross-reactive CTL epitopes between SARS-CoV-2 and M. smegmatis
Considering that BCG vaccination might provide an extent of cross-protection against SARS-CoV-2 infection and mortality, our data also implied that the mice vaccinated with ZWY2 vector bacteria could elicit an extent of SARS-CoV-2-specific T cell immune responses, suggesting that there might be some cross-reactive epitopes between the SARS-CoV-2 and ZWY2 bacteria. To further clarify this hypothesis, we analyzed the potential cross-reactive CTL epitopes between SARS-CoV-2 and M. smegmatis. As predicted in Table 1, there were 14 cross-reactive epitopes between S protein and M.smegmatis, and 2 cross-reactive epitopes between N protein and M.smegmatis.
Table 1.
The cross-epitope between SARS-CoV-2 and M. smegmatis.
| SARS-CoV-2 | Mycolicibacterium smegmatis protein | Predicted epitope in SARS-CoV-2 protein | Allele | Rank | |
|---|---|---|---|---|---|
| SARS-CoV-2 N | ASSRSSSRSR | >WP_014877465.1 DNA translocase FtsK | 183SSRSSSRSR191 | HLA-A *31:01 | 0.13 |
| HLA-A *30:01 | 0.08 | ||||
| HLA-A *68:01 | 1.4 | ||||
| HLA-A *33:01 | 0.78 | ||||
| HLA-A *03:01 | 1.2 | ||||
| 182ASSRSSSRSR191 | HLA-A *31:01 | 0.59 | |||
| HLA-A *30:01 | 0.86 | ||||
| HLA-A *11:01 | 1.2 | ||||
| SARS-CoV-2 S | RDIADTTDAV | RDIADTTDIT (>WP_003896768.1 ectoine/hydroxyectoine ABC transporter ATP-binding protein EhuA) | 568DIADTTDAV576 | HLA-A *68:02 | 0.09 |
| HLA-A *26:01 | 0.68 | ||||
| HLA-B × 51:01 | 2 | ||||
| 567RDIADTTDAV576 | HLA-A *02:03 | 1.6 | |||
| HLA-A *02:06 | 1.6 | ||||
| ISVTTEILPV | IYVTAEILPV (>WP_011726763.1 MFS transporter) | 721SVTTEILPV729 | HLA-A *02:06 | 0.24 | |
| HLA-A *68:02 | 0.43 | ||||
| HLA-A *02:03 | 0.64 | ||||
| HLA-A *02:01 | 0.74 | ||||
| SVTTEILPVS | NPTTEILPVS (>WP_011728271.1 hydrogenase nickel incorporation protein HypB) | 721SVTTEILPV729 | HLA-A *02:06 | 0.24 | |
| HLA-A *68:02 | 0.43 | ||||
| HLA-A *02:03 | 0.64 | ||||
| HLA-A *02:01 | 0.74 | ||||
| TTEILPVSMT | TTEILPVSAR (>WP_011728271.1 hydrogenase nickel incorporation protein HypB) | 723TTEILPVSM731 | HLA-A *01:01 | 0.5 | |
| HLA-B × 35:01 | 1.1 | ||||
| HLA-A *68:02 | 1.3 | ||||
| HLA-A *26:01 | 1.5 | ||||
| HLA-B × 53:01 | 1.8 | ||||
| 724TEILPVSMT732 | HLA-B × 40:01 | 1.3 | |||
| HLA-B × 44:03 | 1.4 | ||||
| HLA-B × 44:02 | 1.8 | ||||
| 723TTEILPVSMT732 | HLA-A *68:02 | 1.9 | |||
| VTTEILPVSM | PTTEILPVSA (>WP_011728271.1 hydrogenase nickel incorporation protein HypB) | 722VTTEILPVSM731 | HLA-A *68:02 | 1.2 | |
| HLA-B × 57:01 | 1.3 | ||||
| HLA-B × 58:01 | 1.4 | ||||
| EMIAQYTSAL | EMVAQYTPAL (>WP_011729237.1 FAD-binding protein) | 869MIAQYTSAL877 | HLA-B × 08:01 | 0.31 | |
| HLA-A *68:02 | 0.34 | ||||
| HLA-A *02:03 | 0.4 | ||||
| HLA-B × 07:02 | 0.43 | ||||
| HLA-A *26:01 | 0.51 | ||||
| HLA-B × 35:01 | 0.63 | ||||
| HLA-A *02:06 | 0.68 | ||||
| HLA-A *02:01 | 0.91 | ||||
| HLA-A *32:01 | 0.98 | ||||
| HLA-B × 15:01 | 1.4 | ||||
| 868EMIAQYTSAL877 | HLA-A *26:01 | 0.89 | |||
| HLA-A *68:02 | 0.91 | ||||
| 868EMIAQYTSA876 | HLA-A *68:02 | 1.1 | |||
| HLA-B × 08:01 | 1.3 | ||||
| HLA-A *02:03 | 1.6 | ||||
| AAEIRASANL | ASAIRASANL (>WP_011728932.1 acyl-CoA dehydrogenase family protein) | 1016AEIRASANL1024 | HLA-B × 40:01 | 0.01 | |
| HLA-B × 44:03 | 0.1 | ||||
| HLA-B × 44:02 | 0.11 | ||||
| 1015AAEIRASANL1024 | HLA-B × 40:01 | 0.21 | |||
| HLA-B × 44:03 | 0.68 | ||||
| HLA-B × 44:02 | 0.65 | ||||
| RLNEVAKNLN | KLLEVAKNLN (>WP_003892935.1 propanediol/glycerol family dehydratase large subunit) | 1185RLNEVAKNL1193 | HLA-A *02:03 | 0.06 | |
| HLA-A *32:01 | 0.1 | ||||
| HLA-A *02:01 | 0.16 | ||||
| HLA-A *02:06 | 0.51 | ||||
| HLA-B × 15:01 | 1.1 | ||||
| HLA-A *30:01 | 1.4 | ||||
| HLA-A *30:02 | 1.6 | ||||
| HLA-B × 08:01 | 1.9 | ||||
| HLA-B × 07:02 | 1.9 |
Note: The underline fonts represent different amino acids.
Collectively, these data indicate that this M. smegmatis-based SARS-CoV-2 vaccine candidate effectively induced a potential SARS-CoV-2-specific cellular immunity, and thus could be a suitable vector for developing T cell-based vaccines against SARS-CoV-2.
Discussion
With the continued emergence of the SARS-CoV-2 variants, the protective efficacy of neutralizing antibodies elicited by current vaccines has been greatly compromised. Increasing data has demonstrated that antigen-specific CTL responses also play a vital role in controlling SARS-CoV-2 infection and replication.31 Previous studies found that a poor T cell response was associated with the progression of severe COVID-19 patients, and a high level of SARS-CoV-2-specific CD4+ and CD8+ T cells were present in convalescent individuals.32–34 Moreover, the latest work demonstrated that antigen-specific CTL potentially recognized different SARS-CoV-2 variants.35 Therefore, more attention should be paid to T-cell-based immunity in the development of the next generation of COVID-19 vaccines.
A rational gene delivery system plays a critical role in developing an effective vaccine. A variety of strategies, including inactivated vaccine, subunit protein vaccine, viral vector vaccine, and mRNA vaccine, and, have been developed against SARS-CoV-2 infection.36–39 Herein, for the first time, we developed a live attenuated M. smegmatis ZWY2 as a bacterial surface display system for the COVID-19 vaccine candidate. M. smegmatis, ZWY2, is a safe and rapid-growing mycobacterium. Compared with viral vector vaccines and mRNA vaccines, M. smegmatis-based vaccines have many advantages, such as a high capacity carrying antigen genes, ease of production to a high yield, thermal stability, and good safety. In this study, we demonstrated that recombinant ZWY2-S and ZWY2-N could successfully display S and N proteins onto bacterial surfaces, and effectively elicited the S and N antigen-specific immune responses. In addition, the recombinant M. smegmatis-based vaccine could induce the differentiation and maintenance of memory T cells in the immunized mice. Consistent with previous data on BCG vector,40 our work showed that the subsets of central memory CD4+ T cells and CD8+ T cells were effectively induced by recombinant M. smegmatis-based COVID-19 vaccine. Mechanistically, M. smegmatis effectively induce the differentiation and maturation of dendritic cells (DCs) by the upregulation of MHC I and costimulatory molecules and thus elicit the T cell-biased immune responses.41 Recent epidemiologic data also suggested that BCG vaccination might alleviate the COVID-19 disease progression and mortality, which may be related to trained immunity and cross immunity.42–45 Interestingly, our study also revealed the existence of some potential cross-reactive CTL epitopes between SARS-CoV-2 and M.smegmatis, which is consistent with the previous report that SARS-CoV-2 shared some T-cell and B-cell epitopes with BCG strain.45 These findings implied that M. smegmatis might be an attractive bacterial vector for T cell-based SARS-CoV-2 vaccine.
There are some limitations to this study. For example, we used the full-length Spike protein to construct the recombinant ZWY2-S vaccine in this study. It’s well known that the S protein is about 140kDa, and thus might not be good enough to display on the surface of the recombinant bacteria. This may account for the low immunogenicity of the ZWY2-S construct in this study. Given these issues, a recombinant ZWY2 carrying the RBD domain should be needed to improve its immunogenicity. In addition, the protective efficacy was not evaluated in an animal infection model. Nevertheless, our data showed that the M. smegmatis-based COVID-19 vaccine effectively induced T cell immune responses, which warranted a further study on the development of a universe SARS-CoV-2 vaccine. Of note, it has been shown that priming the recombinant M. smegmatis vaccine could elicit a memory CD8+ T-lymphocyte response,46 implying that it could be a potential priming vector in the sequential vaccination strategy.
Taken together, we developed a simple and effective strategy to display the SARS-CoV-2 antigen onto the surface of M. smegmatis as a novel COVID-19 vaccine candidate, and these constructs effectively elicited the SARS-CoV-2 antigen-specific T cell-biased cellular immune responses. In particular, the induction of the subsets of central memory T cells could be expected to contribute to a long-lasting protective T cell immunity. In addition, M. smegmatis-based vector is also promising as a component in prime/boost vaccination regimens for the induction of a balanced humoral and cellular immune response.
Supplementary Material
Acknowledgment
We appreciate all other members of our group for their helpful advice and discussion to improve this project.
Funding Statement
This research was funded by the National Key R&D Program of China (2022YFE0203100), the National Natural Science Foundation of China (82271786, 81971927, 82061138019, 82061128001), the Chinese Academy of Sciences (154144KYSB20190005, YJKYYQ20210026), and the Science and Technology Planning Project of Shenzhen City (20190804095916056, JSGG20200225152008136).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
Project design and supervised by C.S., T.Z.; experiment performed by Z.W. C.F., X.L., Y.L., M.L., Y.Y., Z.H., C.W.; data analyzing by Z.W., C.F.; materials and reagents contributed by C.S., T.Z.; Writing by Z.W., C.S.; all authors have read and agreed to the published version of the manuscript.
Institutional review board statement
Animal experiments were carried out in accordance with the recommendations in the institutional and national guidelines for animal care and use. The protocol was approved by the Committee on the Ethics of Animal Experiments of Sun Yat-sen University, Guangzhou, China (SYSU-IACUC-2021–000258). All procedures were performed under ether anesthesia and made to minimize suffering.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2023.2171233
References
- 1.Randolph HE, Barreiro LB.. Herd Immunity: understanding COVID-19. Immunity. 2020;52:737–12. doi: 10.1016/j.immuni.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdool Karim SS, de Oliveira T.. New SARS-CoV-2 variants – clinical, public health, and vaccine implications. N Engl J Med. 2021;384:1866–68. doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hajnik RL, Plante JA, Liang Y, Alameh MG, Tang J, Bonam SR, Zhong C, Adam A, Scharton D, Rafael GH. Dual spike and nucleocapsid mRNA vaccination confer protection against SARS-CoV-2 Omicron and Delta variants in preclinical models. Sci Transl Med 2022;14:eabq1945. doi: 10.1126/scitranslmed.abq1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naranbhai V, Nathan A, Kaseke C, Berrios C, Khatri A, Choi S, Getz MA, Tano-Menka R, Ofoman O, Gayton A, et al. T cell reactivity to the SARS-CoV-2 Omicron variant is preserved in most but not all individuals. Cell. 2022;185:1041–51. doi: 10.1016/j.cell.2022.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar NP, Padmapriyadarsini C, Rajamanickam A, Bhavani PK, Nancy A, Jayadeepa B, Selvaraj N, Asokan D, Renji RM, Venkataramani V, et al. BCG vaccination induces enhanced frequencies of memory T cells and altered plasma levels of common γc cytokines in elderly individuals. PLoS One. 2021;16(11):e0258743. doi: 10.1371/journal.pone.0258743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singhania A, Dubelko P, Kuan R, Chronister WD, Muskat K, Das J, Phillips EJ, Mallal SA, Seumois G, Vijayanand P, et al. CD4+CCR6+ T cells dominate the BCG-induced transcriptional signature. EBioMedicine. 2021;74:103746. doi: 10.1016/j.ebiom.2021.103746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu YD, Fennelly G, Miller C, Tarara R, Saxe I, Bloom B, McChesney M. Recombinant bacille Calmette-Guérin expressing the measles virus nucleoprotein protects infant rhesus macaques from measles virus pneumonia. J Infect Dis. 1997;176:1445–53. doi: 10.1086/514140. [DOI] [PubMed] [Google Scholar]
- 8.Nurul AA, Norazmi MN. Immunogenicity and in vitro protective efficacy of recombinant Mycobacterium bovis bacille Calmette Guerin (rBCG) expressing the 19 kDa merozoite surface protein-1 (MSP-1(19)) antigen of Plasmodium falciparum. Parasitol Res. 2011;108:887–97. doi: 10.1007/s00436-010-2130-5. [DOI] [PubMed] [Google Scholar]
- 9.Jeong H, Lee SY, Seo H, Kim BJ. Recombinant Mycobacterium smegmatis delivering a fusion protein of human macrophage migration inhibitory factor (MIF) and IL-7 exerts an anticancer effect by inducing an immune response against MIF in a tumor-bearing mouse model. J Immunother Cancer. 2021;9(8):e003180. doi: 10.1136/jitc-2021-003180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soto JA, Díaz FE, Retamal-Díaz A, Gálvez NMS, Melo-González F, Piña-Iturbe A, Ramírez MA, Bohmwald K, González PA, Bueno SM, et al. BCG-based vaccines elicit antigen-specific adaptive and trained immunity against SARS-CoV-2 and Andes orthohantavirus. Vaccines (Basel). 2022;10(5):721. doi: 10.3390/vaccines10050721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shiloh MU, Champion PA. To catch a killer. What can mycobacterial models teach us about Mycobacterium tuberculosis pathogenesis? Curr Opin Microbiol. 2010;13(1):86–92. doi: 10.1016/j.mib.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim BJ, Gong JR, Kim GN, Kim BR, Lee SY, Kook YH, Kim BJ. Recombinant Mycobacterium smegmatis with a pMyong2 vector expressing human immunodeficiency virus Type I Gag can induce enhanced virus-specific immune responses. Sci Rep. 2017;7:44776. doi: 10.1038/srep44776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim BJ, Kim BR, Kook YH, Kim BJ. Development of a live recombinant BCG expressing human immunodeficiency virus Type 1 (HIV-1) gag using a pMyong2 vector system: potential use as a novel HIV-1 vaccine. Front Immunol. 2018;9:643. doi: 10.3389/fimmu.2018.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cayabyab MJ, Hovav AH, Hsu T, Krivulka GR, Lifton MA, Gorgone DA, Fennelly GJ, Haynes BF, Jacobs WR, Letvin NL. Generation of CD8+ T-cell responses by a recombinant nonpathogenic Mycobacterium smegmatis vaccine vector expressing human immunodeficiency virus type 1 Env. J Virol. 2006;80(4):1645–52. doi: 10.1128/JVI.80.4.1645-1652.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuehnel MP, Goethe R, Habermann A, Mueller E, Rohde M, Griffiths G, Valentin-Weigand P. Characterization of the intracellular survival of Mycobacterium avium ssp. paratuberculosis: phagosomal pH and fusogenicity in J774 macrophages compared with other mycobacteria. Cell Microbiol. 2001;3(8):551–66. doi: 10.1046/j.1462-5822.2001.00139.x. [DOI] [PubMed] [Google Scholar]
- 16.Sweeney KA, Dao DN, Goldberg MF, Hsu T, Venkataswamy MM, Henao-Tamayo M, Ordway D, Sellers RS, Jain P, Chen B, et al. A recombinant Mycobacterium smegmatis induces potent bactericidal immunity against Mycobacterium tuberculosis. Nat Med. 2011;17(10):1261–68. doi: 10.1038/nm.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, Schiergens TS, Herrler G, Wu N-H, Nitsche A, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–11. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao H, Wang TC, Li XF, Zhang NN, Li L, Zhou C, Deng YQ, Cao TS, Yang G, Li RT, et al. Long-term stability and protection efficacy of the RBD-targeting COVID-19 mRNA vaccine in nonhuman primates. Signal Transduct Target Ther. 2021;6:438. doi: 10.1038/s41392-021-00861-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harris PE, Brasel T, Massey C, Herst CV, Burkholz S, Lloyd P, Blankenberg T, Bey TM, Carback R, Hodge T, Ciotlos S, Wang L, Comer JE, Rubsamen RM. A synthetic peptide CTL vaccine targeting nucleocapsid confers protection from SARS-CoV-2 challenge in Rhesus Macaques. Vaccines (Basel). 2021;9(5):520. doi: 10.3390/vaccines9050520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matchett WE, Joag V, Stolley JM, Shepherd FK, Quarnstrom CF, Mickelson CK, Wijeyesinghe S, Soerens AG, Becker S, Thiede JM, et al. Cutting edge: nucleocapsid vaccine elicits spike-independent SARS-CoV-2 protective immunity. J Immunol. 2021;207(2):376–79. doi: 10.4049/jimmunol.2100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang F, Tan Y, Liu J, Liu T, Wang B, Cao Y, Qu Y, Lithgow T, Tan S, Zhang T. Efficient construction of unmarked recombinant mycobacteria using an improved system. J Microbiol Methods. 2014;103:29–36. doi: 10.1016/j.mimet.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Goude R, Roberts DM, Parish T. Electroporation of mycobacteria. Methods Mol Biol. 2015;1285:117–30. doi: 10.1007/978-1-4939-2450-9_7. [DOI] [PubMed] [Google Scholar]
- 24.Rezwan M, Laneelle M-A, Sander P, Daffe M. Breaking down the wall: fractionation of mycobacteria. J Microbiol Methods. 2007;68(1):32–39. doi: 10.1016/j.mimet.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Li M, Chen J, Liu Y, Zhao J, Li Y, Hu Y, Chen Y-Q, Sun L, Shu Y, Feng F, et al. Rational design of AAVrh10-vectored ACE2 functional domain to broadly block the cell entry of SARS-CoV-2 variants. Antiviral Res. 2022;205:105383. doi: 10.1016/j.antiviral.2022.105383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu T, Ma F, Ma X, Jia W, Pan E, Cheng G, Chen L, Sun C. Regulating innate and adaptive immunity for controlling SIV infection by 25-hydroxycholesterol. Front Immunol. 2018;9:2686. doi: 10.3389/fimmu.2018.02686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun C, Feng L, Zhang Y, Xiao L, Pan W, Li C, Zhang L, Chen L. Circumventing antivector immunity by using adenovirus-infected blood cells for repeated application of adenovirus-vectored vaccines: proof of concept in rhesus macaques. J Virol. 2012;86(20):11031–42. doi: 10.1128/JVI.00783-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M. NetMhcpan-4.0: improved peptide–MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol. 2017;199(9):3360–68. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Yuan Y, Li P, Deng Z, Wen Z, Wang H, Feng F, Zou H, Chen L, Tang S, et al. Comparison of the immunogenicity of HIV-1 CRF07_BC gag antigen with or without a seven amino acid deletion in p6 region. Front Immunol. 2022;13:850719. doi: 10.3389/fimmu.2022.850719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng Y, Mentzer AJ, Liu G, Yao X, Yin Z, Dong D, Dejnirattisai W, Rostron T, Supasa P, Liu C, et al. Broad and strong memory CD4+ and CD8+ T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21(11):1336–45. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.You M, Chen L, Zhang D, Zhao P, Chen Z, Qin E-Q, Gao Y, Davis MM, Yang P. Single-cell epigenomic landscape of peripheral immune cells reveals establishment of trained immunity in individuals convalescing from COVID-19. Nat Cell Biol. 2021;23(6):620–30. doi: 10.1038/s41556-021-00690-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sekine T, Perez-Potti A, Rivera-Ballesteros O, Strålin K, Gorin J-B, Olsson A, Llewellyn-Lacey S, Kamal H, Bogdanovic G, Muschiol S, et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–68. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grifoni A, Weiskopf D, Ramirez SI, Mateus J, Dan JM, Moderbacher CR, Rawlings SA, Sutherland A, Premkumar L, Jadi RS, et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–501. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M, Zeng J, Li R, Wen Z, Cai Y, Wallin J, Shu Y, Du X, Sun C. Rational design of a pan-coronavirus vaccine based on conserved CTL epitopes. Viruses. 2021;13(2):333. doi: 10.3390/v13020333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarke A, Sidney J, Kidd CK, Dan JM, Ramirez SI, Yu ED, Mateus J, da Silva Antunes R, Moore E, Rubiro P, et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep Med. 2021;2(2):100204. doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, et al. An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med. 2020;383(20):1920–31. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voysey M, Costa Clemens SA, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, et al. Single-dose administration and the influence of the timing of the booster dose on immunogenicity and efficacy of ChAdox1 nCov-19 (AZD1222) vaccine: a pooled analysis of four randomised trials. Lancet. 2021;397(10277):881–91. doi: 10.1016/S0140-6736(21)00432-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Logunov DY, Dolzhikova IV, Shcheblyakov DV, Tukhvatulin AI, Zubkova OV, Dzharullaeva AS, Kovyrshina AV, Lubenets NL, Grousova DM, Erokhova AS, et al. Safety and efficacy of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine: an interim analysis of a randomised controlled phase 3 trial in Russia. Lancet. 2021;397(10275):671–81. doi: 10.1016/S0140-6736(21)00234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia S, Duan K, Zhang Y, Zhao D, Zhang H, Xie Z, Li X, Peng C, Zhang Y, Zhang W, et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. JAMA. 2020;324(10):951–60. doi: 10.1001/jama.2020.15543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chege GK, Burgers WA, Stutz H, Meyers AE, Chapman R, Kiravu A, Bunjun R, Shephard EG, Jacobs WR, Rybicki EP, et al. Robust immunity to an auxotrophic Mycobacterium bovis BCG-VLP prime-boost HIV vaccine candidate in a nonhuman primate model. J Virol. 2013;87(9):5151–60. doi: 10.1128/JVI.03178-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheadle EJ, O’Donnell D, Selby PJ, Jackson AM. Closely related mycobacterial strains demonstrate contrasting levels of efficacy as antitumor vaccines and are processed for major histocompatibility complex class I presentation by multiple routes in dendritic cells. Infect Immun. 2005;73(2):784–94. doi: 10.1128/IAI.73.2.784-794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Netea MG, Giamarellos-Bourboulis EJ, Domínguez-Andrés J, Curtis N, van Crevel R, van de Veerdonk FL, Bonten M. Trained immunity: a tool for reducing susceptibility to and the severity of SARS-CoV-2 infection. Cell. 2020;181(5):969–77. doi: 10.1016/j.cell.2020.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rivas MN, Ebinger JE, Wu M, Sun N, Braun J, Sobhani K, Van Eyk JE, Cheng S, Arditi M. BCG vaccination history associates with decreased SARS-CoV-2 seroprevalence across a diverse cohort of health care workers. J Clin Invest. 2021;131(2):131. doi: 10.1172/JCI145157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang B-Z, Shuai H, Gong H-R, Hu J-C, Yan B, Yuen TT, Hu Y-F, Yoon C, Wang X-L, Hou Y, et al. Bacillus Calmette-Guérin–induced trained immunity protects against SARS-CoV-2 challenge in K18-hACE2 mice. JCI Insight. 2022;7(11). doi: 10.1172/jci.insight.157393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Urbán S, Paragi G, Burián K, McLean GR, Virok DP. Identification of similar epitopes between severe acute respiratory syndrome coronavirus-2 and Bacillus Calmette–Guérin: potential for cross-reactive adaptive immunity. Clin Transl Immunol. 2020;9(12):e1227. doi: 10.1002/cti2.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hovav A-H, Cayabyab MJ, Panas MW, Santra S, Greenland J, Geiben R, Haynes BF, Jacobs WR, Letvin NL. Rapid memory CD8+ T-lymphocyte induction through priming with recombinant Mycobacterium smegmatis. J Virol. 2007;81(1):74–83. doi: 10.1128/JVI.01269-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


