ABSTRACT
Monitoring mammalian macroautophagic/autophagic flux is necessary in most autophagy studies but has generally been difficult to do. Here, we discuss our recent report of a HaloTag-based processing method that offers a straightforward readout for autophagic flux. We found that the self-labeling protein HaloTag becomes resistant to proteolysis when labeled with its ligand. Fusing HaloTag to an autophagy protein such as LC3 results in a reporter that is completely degraded when delivered into lysosomes but, when pulse-labeled with HaloTag ligand, releases free HaloTagligand when processed by lysosomal enzymes. The quantifiable amount of free HaloTagligand, observed by immunoblotting or in-gel fluorescence detection, reflects autophagic flux. Besides being compatible with fluorescence microscopy and flow cytometry applications, this quantitative assay can be readily adapted to monitor most autophagy pathways or the autophagic degradation of a protein of interest.
KEYWORDS: Autophagic activity, autophagic flux, autophagy, HaloTag, lysosomal degradation, lysosome, processing assay, protein degradation, protein turnover, pulse-labeling
Main
Mammalian autophagy research would benefit from a more straightforward method of monitoring autophagic flux in mammalian cells. Conventional assays are generally difficult to carry out correctly, giving ambiguous data otherwise. An excellent alternative would be a mammalian version of the yeast GFP-Atg8 processing assay, with which autophagic flux is measured from the amount of free GFP generated from the processing of GFP-Atg8 in the yeast vacuole. This assay requires no lysosomal inhibitors and gives a result that is produced entirely dependent on autophagic activity. However, it cannot be implemented in mammalian cells due to the unsuitability of typical fluorescent protein tags: GFP is quickly degraded in mammalian lysosomes and RFP accumulates in lysosomes under basal conditions, which obscures detection of free RFP generated after autophagy induction.
In a recent study, we show that the reporter processing assay can be implemented in mammalian cells with HaloTag (Halo), a protein tag that becomes resistant to proteolysis when labeled with its ligand [1]. We demonstrated this property with Halo-LC3. The protein levels of unlabeled Halo-LC3 were reduced by starvation-induced autophagy, as observed by immunoblotting. Pulse-labeling Halo-LC3 by briefly exposing the reporter to its ligand followed by starvation induces a reduction in Haloligand-LC3 protein levels as well as the production of a free Haloligand band, which can also be observed by in-gel fluorescence imaging when a fluorescent ligand is used (Figure 1A,B). This processing depends on the lysosomal enzymatic activity and the amount of free Haloligand corresponds closely to the duration of autophagy activity. Hence, mammalian autophagic flux can be monitored by labeled Halo reporter processing.
Figure 1.
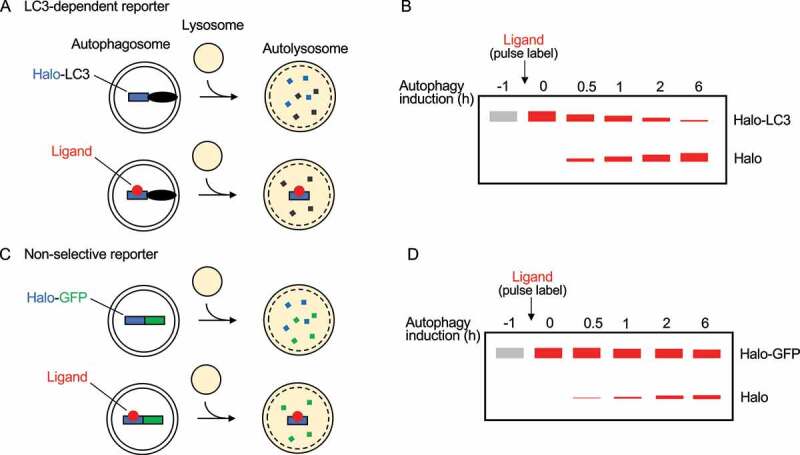
The HaloTag processing assay. (A) Halo-LC3 is completely degraded in autolysosomes but pulse-labeled Haloligand-LC3 releases free Haloligand when processed by lysosomal enzymes. (B) Haloligand-LC3 processing is observed by SDS-PAGE methods, namely immunoblotting and in-gel fluorescence imaging. Along with a decrease in Haloligand-LC3 protein levels, the amount of free Haloligand produced after autophagy induction increases over time, indicating that it is a clear indicator of autophagic flux. (C) As described in (A), pulse-labeling the bulk non-selective reporter Halo-GFP is required for free Haloligand to persist in autolysosomes after the reporter has been processed in autolysosomes. (D) As described in (B), Haloligand-GFP processing is detected by SDS-PAGE methods. The decrease in Haloligand-GFP protein levels over time is not as obvious as Halo-LC3 because Haloligand-GFP is a cytosolic reporter that is randomly incorporated into phagophores. However, bulk nonselective autophagy can still be readily detected by the production of free Haloligand.
The Halo reporter processing assay can be adapted to monitor most autophagy pathways by fusing Halo to a protein of that pathway or, for bulk nonselective autophagy, to another protein tag like GFP. For example, reticulophagy can be monitored specifically with Halo-GFP-KDEL and mitophagy with pSu9-Halo-GFP (pSu9 is derived from the presequence of N. crassa Fo-ATPase subunit 9). Halo-GFP processing reveals bulk nonselective autophagic flux (Figure 1C,D), which reflects total autophagic flux more closely than the processing of other reporters containing proteins that are preferentially degraded by autophagy, such as LC3/GABARAP-family proteins and autophagic substrates. Moreover, examining Halo-GFP processing allows autophagic flux to be monitored in cells deficient in LC3/GABARAP or LC3 lipidation, which could not be done with conventional LC3-based methods. The Halo reporter processing assay cannot be applied to the non-canonical autophagy pathways involving LC3 conjugation to single-membrane organelles because the reporters would not be delivered into lysosomes. Besides this exception, we expect the Halo reporter processing assay to be useful for all known canonical autophagy pathways, including macroautophagy and microautophagy.
In addition to monitoring autophagic flux with the processing assay, examining Halo-GFP-containing reporters with fluorescence microscopy and flow cytometry may provide more information on autophagic flux. For example, after detecting a block in autophagic flux, researchers can observe a number of autophagosomes and autolysosomes with Halo reporters containing LC3/GABARAP-family proteins or autophagic substrates. As GFP is quenched and degraded in lysosomes, autophagosomes will appear as Haloligand and GFP double-positive puncta, while autolysosomes will be puncta that are Haloligand-positive but GFP-negative. Lower numbers of autophagosomes suggest defective initiation, and an accumulation of autophagosomes suggests fusion defects. Furthermore, researchers who would like an even more quantitative readout to, for example, confirm that processing assay readouts can make use of Halo-GFP-LC3-RFP. This ‘three-in-one’ reporter can be used with flow cytometry, providing quantitative GFP:RFP ratios that reflect autophagic flux at the single-cell level.
The major weaknesses of the Halo processing assay are (1) the need for exogenous expression, (2) the eventual degradation of Haloligand, (3) the SDS-PAGE-based readout complicating large-scale screenings, and (4) potential difficulties in implementing it in animal models. Researchers concerned with excessive expression of Halo reporters can knockin Halo, sort for low-expressing populations with FACS, or reduce viral titer. As for the stability of Haloligand, we observe a linear increase in free Haloligand levels over time for up to 12 h after inducing autophagy with starvation, which indicates that Haloligand in lysosomes is relatively stable for at least 12 h. The Haloligand will be degraded eventually so this assay cannot be used for prolonged autophagic flux assays. Nevertheless, for autophagy studies that are testing only for a change in autophagic flux, the stability of Haloligand should not be an issue. Finally, researchers looking to conduct large-scale screens with the Halo reporters can potentially do so by microscopy-based high-content screening, while those seeking to use the assay with animal models can try labeling with more cost-effective non-fluorescent Halo blockers that resemble the ligand or opt for localized injections of the ligand.
In summary, the Halo reporter processing assay is a strong alternative to conventional assays for mammalian autophagic flux. Plasmids of the reporters used in our study can be found on Addgene (https://www.addgene.org/browse/article/28225176/). We anticipate that the autophagy research community will find this tool useful for monitoring and detecting changes in autophagic flux.
Funding Statement
This work was supported by the Exploratory Research for Advanced Technology (ERATO) research funding program of the Japan Science and Technology Agency (JST) (JPMJER1702 to N.M.), a Grant-in-Aid for Transformative Research Areas (A) (21H05256 to H.Y.) and a Grant-in-Aid for Specially Promoted Research (22H04919 to N.M.) from the Japan Society for the Promotion of Science (JSPS). W.W.Y. was supported by the Japanese Ministry of Education, Culture, Sports, Science and Technology.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Reference
- [1].Yim WW, Yamamoto H, Mizushima N.. A pulse-chasable reporter processing assay for mammalian autophagic flux with HaloTag. Elife. 2022. Aug 8;11:e78923. DOI: 10.7554/eLife.78923. [DOI] [PMC free article] [PubMed] [Google Scholar]


