Abstract
Background:
The reversible phosphorylation of proteins regulates many key functions in eukaryotic cells. Phosphorylation is catalyzed by protein kinases, with the majority of phosphorylation occurring on side chains of serine and threonine residues. The phosphomonoesters generated by protein kinases are hydrolyzed by protein phosphatases. In the absence of a phosphatase the half-time for the hydrolysis of alkyl phosphate dianions at 25° C is over 1 trillion years; knon ~2 x 10−20 sec−1. Therefore, ser/thr phosphatases are critical for processes controlled by reversible phosphorylation.
Methods:
This review is based on a search of the literature in available databases. We compare the catalytic mechanism of PPP-family phosphatases (PPPases) and the interactions of inhibitors that target these enzymes.
Results:
PPPases are metal-dependent hydrolases that enhance the rate of hydrolysis ([kcat/kM]/knon ) by a factor of ~1021, placing them among the most powerful known catalysts on earth. Biochemical and structural studies indicate the remarkable catalytic proficiencies of PPPases are achieved by 10 conserved amino acids, DXH(X)~26DXXDR(X)~20-26NH(X)~50H(X)~25-45R(X)~30-40H. Six act as metal-coordinating residues. Four position and orient the substrate phosphate. Together, two metal ions and the 10 catalytic residues position the phosphoryl group and an activated bridging water/hydroxide nucleophile for inline attack upon the substrate phosphorous atom. The PPPases are conserved among species, and many structurally diverse natural toxins co-evolved to target these enzymes.
Conclusion.
Although the catalytic site is conserved, opportunities for the development of selective inhibitors of this important group of metalloenzymes exist.
Keywords: Phosphatase, inhibitor, okadaic acid, fostriecin, cantharidin, tautomycin, microcystin crystal structures
1. INTRODUCTION
Reversible phosphorylation of structural and regulatory proteins plays a crucial role in the regulation of many cellular functions in eukaryotes. Protein phosphorylation occurs principally upon serine, threonine, and tyrosine residues and is mediated by members of an extremely large group of ser/thr- and tyr-protein kinases. Protein kinases catalyze the transfer of a gamma phosphate from adenosine triphosphate (ATP) to a side chain hydroxyl. There are >500 genes in humans known to encode protein kinases [1], with the number of potential gene products being much greater due to alternative splicing [2,3]. Phosphorylation affects an extremely large and diverse panoply of cellular processes including: cell growth and proliferation, gene transcription, transcript processing, translation, protein-protein interactions, subcellular localization, programmed cell death, turnover of cellular proteins and organelles, metabolism, muscle contraction, and cytoskeleton assembly and function [4-11]. Most kinases target substrates in a sequence specific manner [12-14].
The removal of phosphate is accomplished by one or more of the >150 ser/thr-, tyr-, or dual-specificity phosphatases, which catalyze the hydrolysis of seryl, threonyl, and/or tyrosyl phosphomonoesters [8,15-18]. The biological activity of a protein, its location within a cell, its stability, and interactions with other macromolecules can all be affected by its phosphorylation state, which is governed dynamically via coordinated regulation of the kinases and phosphatases that utilize it as a substrate.
Most tyrosine-specific and dual-specificity protein phosphatases share a common catalytic mechanism in which a covalent thiophosphate intermediate forms with the side chain of a catalytic cysteine (one family instead utilizes an aspartyl phosphate intermediate) [17,19-21]. In contrast, serine/threonine-specific protein phosphatases are metaloenzymes that utilize metal-ligated water/hydroxide as a nucleophile in the phosphomonoester hydrolysis reaction. Modern nomenclature divides the ser/thr phosphatases into two genetically and structurally distinct lineages, the PPP family and the PPM family. This review will focus on the biochemistry and structural biology of the PPP family phosphatases (PPPases) in mammals, discussing the common catalytic mechanism, interactions with natural toxins and small molecule inhibitors, and prospects for developing more selective inhibitors.
1.1. Serine/Threonine Protein Phosphatases:
Deciphering the PPP-family literature can be bewildering, especially to a novice to the field. Therefore, it should be useful to start with a short discussion of the evolution of the phosphatase nomenclature. Historically, both protein kinases and protein phosphatases were first named by the substrates they acted upon. The kinase that phosphorylates phosphorylase was named phosphorylase kinase, and phosphorylase phosphatase seemed to be an appropriate name for the phosphatase that catalyzed dephosphorylation [22]. However, later it became apparent that, in vitro, more than one phosphatase could catalyze the same reaction, and many phosphatases demonstrate little substrate specificity. To establish order in the field, Ingebritsen and Cohen in the 1980s proposed a system of nomenclature that is currently obsolete, but the names are still pertinent because their “traditional” nomenclature lives on and is still in common usage within the field [23].
The traditional nomenclature was based upon the notion that the ser/thr phosphatase activities identified and characterized in the literature up to that time were the result of only four principal enzymes with phosphatase activity: type 1 (PP1), type 2A (PP2A), type 2B (PP2B), and type 2C (PP2C). This classification was based upon biochemical characteristics, sensitivity to endogenous inhibitory proteins, and the limited substrate specificity that could be demonstrated in vitro. In the 1990’s we learned that PP1, PP2A, and PP2B are highly conserved across taxa and are closely related to each other, sharing ~40% sequence identity within the ~250 residue conserved core domain. Genomic analysis would also reveal separate genes encoding isoforms of PP1 (α, β, γ), PP2A (α, β) and PP2B (α, β, γ). However, the lack of substrate discriminatory power raised doubts about the ability of these enzymes to meaningfully participate in complex signaling pathways, as highly specific interactions between components are necessary to minimize interference and unregulated cross-talk between different pathways, which is needed to ensure high-fidelity transmission. Therefore, for years many erroneously viewed ser/thr protein phosphatases as simple “house keeping” enzymes.
Advances in DNA sequencing technologies allowed the entire genome of many organisms to be sequenced in their entirety, and today we know that PP1, PP2A and PP2B and their isoforms are actually catalytic subunits that are shared by many different holoenzymes. The current nomenclature is based on amino acid similarity and conservation of catalytic mechanisms. This classification system divides phospho-ser/thr phosphatases into two major families (PPP and PPM). PPP has an extra P because the PP designation was given to protein proteases. In humans there are also eight CTD aspartate-based phosphatases that have a different catalytic mechanism and will not be addressed further in this review.
PP1, PP2A, and PP2B share a common catalytic mechanism and represent the “founding’ members of the PPP-family. Novel mammalian PPP phosphatases include PP4, PP5, PP6, and the two isoforms of PP7/PPEF: PPPEF1 and PPPEF2 [8,16,24,25]. The primary amino acid sequence of PP2C shares no sequence similarity to the PPP family and was placed as a member of the separate and distinct PPM-family [16]. The reclassification is borne out by the crystal structures of PPP and PPM phosphatases produced from the 1990s onward, which reveal that PPP-family members show great structural similarity to one another and possess a fold distinct from the PPM family.
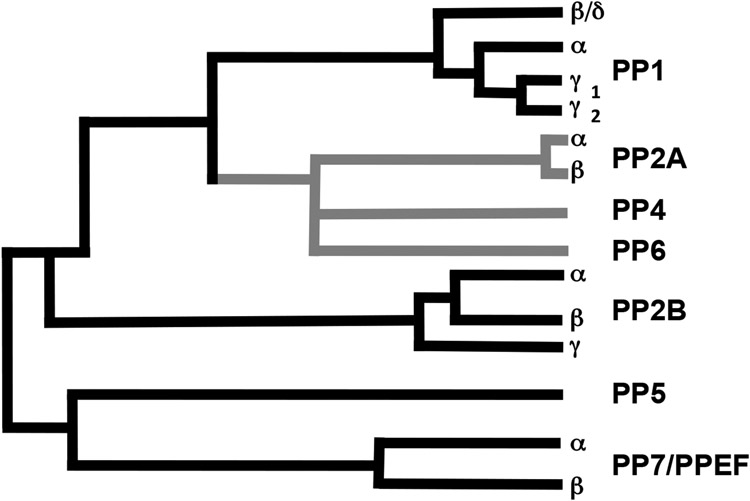
Based upon sequence comparisons, the mammalian PPP phosphatases and their isoforms can be further placed into four basic groups or subfamilies [16,18,26]: PP1, PP2A-like (which includes PP4 and PP6), PP2B, and PP5-like (which includes PP7/PPEF; Figure 1).
Figure 1. Mammalian branch of the PPP-family.
Examples of each of the four subfamilies are found in yeast as well as mammals, strongly suggesting the entrenchment of all four subfamilies across the entirety of the Eukarya. Similar to PP1 and PP2A, PP4 and PP6 serve as the catalytic subunits of PPP-holoenzymes. Therefore, the name of the subunit with the catalytic site was designated with the letter C (e.g. PP1 is now PP1C/PPP1C with the gene often designated in italic lettering. A confusing point in the literature is that, when it was realized that PP2A acted as a common catalytic subunit shared among many PPP2-holenzymes, PP2A was designated as PP2AC or PP2Ac to indicate which subunit of a holoenzyme was being discussed. However, in the current system PP2A/PP2AC is now PPP2CA or PPP2CB, with A and B replacing the α β designation for the PP2AC isoforms. PP2B officially became PPP3C, but in the literature it is often referred to as PP2BC or calcineurin, which was the first name given to this protein. Nomenclature issues aside, all PPP-family enzymes display a great deal of sequence similarity within the central conserved core domain (associated with the catalytic activity) but their N- and C- termini are divergent. This is particularly striking for PPP3C, PPP5C, and PPP7C, which have large N- or C- terminal extensions forming accessory domains that regulate catalytic activity and/or protein-protein interactions.
Although many of the PPP phosphatases display little substrate specificity in vitro, it is now apparent that diversity of function and discriminatory power in the PPP phosphatases is primarily due to the existence of numerous distinct accessory domains having targeting and/or regulatory functions, which act to adapt the catalytic subunits to particular physiological roles by controlling sub-cellular localization, catalytic activity, and/or substrate specificity [8,24,27,28]. In some cases (e.g. PPP5C and PPP7/EF), the catalytic domain has been fused directly to such an accessory domain [25,29,30]. More typically, however, the accessory domains are provided by separate polypetide subunits that form multimeric holoenzymes [24,27,28,31-34]. In this way, a small number of evolutionally conserved catalytic subunits are able to assemble into a diverse array of functionally specific holoenzymes. This stands in stark contrast to the serine/threonine protein kinases, where functional diversity and specificity of interaction appear to have evolved via gene duplication and subsequent divergence that allowed the florescence of a vast multiplicity of different catalytic isoforms [1,35-37].
Although it is beyond the scope of this review to address the assembly all PPP-family holoenzymes, we will provide a few examples for each family to address holoenzyme differences and to introduce the nomenclature of the regulatory and scaffold proteins.
1.1.1. PPP1C:
PPP1 type phosphatases are involved in regulation of actions as diverse as glycogen metabolism, synaptic plasticity, cell cycle progression, RNA splicing, and smooth muscle contraction [7,8,27,28,38]. Isoforms of the PP1 catalytic subunit are expressed from three genes in humans: PPP1CA, PPP1CB and PPP1CC that encode PP1α, PP1β/δ and two PP1γ isoforms, respectively. Transcripts from PPP1CC undergo tissue-specific alternative splicing to produce either the ubiquitously expressed PPP1Cγ1or the testis-specific PPP1Cγ2 variant [39-41]. Overall, the PPP1C isoforms are very closely related (>90% identity) the main differences being near the N- and C-termini. Unlike their protein kinase counterparts, PPP1 catalytic subunits exhibit little substrate selectivity and lack any obvious consensus sequence for phospho-site recognition. In the cellular context, protein phosphatase 1 enzymes exist as multi-subunit holoenzyme complexes composed of a highly conserved catalytic subunit in complex with one or two variable regulatory subunits. A very large number of regulatory subunits have been identified so far [27,33,34], including proteins involved in subcellular targeting of the holenzyme complex, modifying substrate specificity, or inhibiting catalytic function. Mixing and matching catalytic subunits with such a large variety of accessory proteins allows for an incredibly diverse array of functionally distinct PP1 holoenzymes to be assembled from a rather meager set of catalytic subunit gene products.
1.1.2. PP2A/PPP2CA/PPP2CB:
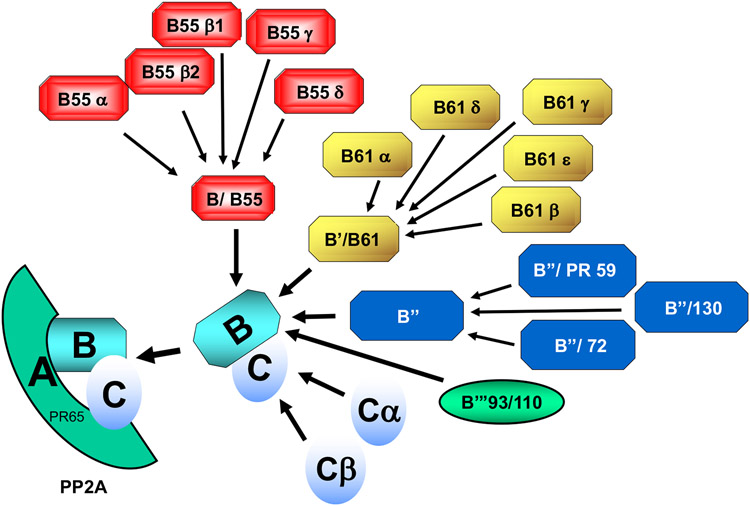
Like PP1, PP2A type catalytic subunits lack an obvious consensus sequence and can dephosphorylate a variety of substrates in vitro and in vivo. Two isoforms, α and β, are expressed in human cells from two genes; PPP2CA and PPP2CB, respectively. PP2A enzymes function in vivo principally as heterocomplexes, each consisting of a PP2A catalytic subunit (C subunit), one of two A-type subunits (α or β; encoded by PPP2R1A and PPP2R1B) and one of ~18 or more regulatory B-type subunits from four distinct families (B, B’, B”, and B’”) expressed from ~14 genes [24,28,32,42] (Figure 2). The A subunit acts as a molecular scaffold allowing close association of the catalytic subunit with a B subunit, and specific roles for most of the many possible holoenzyme complexes have yet to be clearly elucidated. However, PPP2AC containing complexes participate in many signal transduction pathways involved in regulation of the cell cycle, cell size, migration, metabolism, and apoptosis [32,42-47].
Figure 2.
Myriad of PP2A/PPP2-holoenzymes generated from different combinations of catalytic and regulatory subunits. PP2A-holoenzymes are composed of a catalytic subunit (C) a scaffold protein (A/PR65) and any one of several targeting/regulatory proteins (B). There are two isoforms of the A and two isoforms of the C (α, β) subunits. The B-regulatory/targeting subunits are derived from one of four structurally unrelated families (B/B55; B’/B61; B”; B’”/93/110). Each of the different B families contains several isoforms, some of which are expressed in a tissue or developmentally specific manner. Given the existence of two isoforms of A, two isoforms of C and at least 18 different B subunits, >75 different trimeric forms of PPP2 can be generated with the known proteins. Table 1 provides a cross reference for the old and new (assigned based on the gene) names for the A, B and C subunits
1.1.3. PP2B/calcineurin/PPP3C:
The PP2B type phosphatases (also referred to as calcineurin) play a major role in Ca2+-dependent signaling pathways in eukaryotes. Three isoforms of the catalytic subunit are expressed from three genes (PPP3CA, PPP3CB, PPP3CC) in humans. In contrast to PPP1 and PPP2, PP2B demonstrates relatively narrow substrate specificity, efficiently dephosphorylating inhibitor 1 and the RII regulatory subunit of cyclic AMP dependent protein kinase (PKA). PP2B/PPP3C holoenzymes exist as heterodimers comprised of a 58-64 kDa A subunit contains the catalytic and calmodulin-binding domains and a 19 kDa Ca2+-binding regulatory subunit (calcineurin B). The phosphatase activity of PP2B/PPP3C is regulated by two structurally similar calcium binding proteins (calmodulin and calcineurin B). Under conditions of low intracellular Ca2+ concentration (<10−7 M) calcineurin B is bound to PP2B and the A-C dimer is inactive, with the calmodulin binding domain situated in the active site cleft blocking substrate access. At high intracellular calcium concentration, Ca2+ binding to calmodulin causes conformational changes enabling binding to PP2B resulting in abrogation of autoinhibition. For reviews of PP2B/calcinurin activity see Rusnak and Mertz [48], Musson and Smit [49], and Nygren and Scott [50].
1.1.4. PP4/PPP4C:
PPP4C has diverse functions in the cell, including spliceosome assembly, regulation of microtubule growth and nucleation, growth and maturation of the centrosome during cell division, cell migration, insulin signaling and regulation of RNA polymerase III [51-55]. The PPP4 catalytic subunit has only one isoform that is expressed from a single gene (PPP4C). PPP4C is highly homologous to PPP2CA/PPP2CB, sharing ~65% identity at the level of their primary amino acid sequences. Like PPP1 and PPP2, functional PPP4 enzymes in vivo consist of multimeric holoenzyme complexes formed from a catalytic subunit combined with a variety of regulatory subunits. There are five genes encoding the principal PPP4 regulatory subunits: PPP4R1, PPP4R2, PPP4R3A, PPP4R3B, and PPP4R4 [51,56-58]. The roles of individual PPP4-holoenzymes are poorly understood.
1.1.5. PP5/PPP5C:
PPP5C is involved in regulation of many stress- and hormone-induced signaling cascades and can act as a negative regulator of apoptosis. PPP5C forms part of the hsp90 chaperone machinery, regulating the phosphorylation states of chaperone complexes and is involved in heat shock protein 90 (HSP90) client maturation [59-67]. PPP5 is expressed from a single gene (PPP5C in humans) and contains a catalytic domain sharing 42-43% sequence identity with PPP1C/PPP2CA/PPP2CB /PPP3C. However, PPP5C has a unique N-terminal domain that both regulates catalytic activity and mediates interactions with binding partners. Structural studies indicate that the N-terminal domain of PPP5C is connected to the catalytic domain by a flexible linker/tether sequence that allows a tripartite tetratricopeptide-repeat (TPR) motif within the N-terminal domain to adopt a conformation that occludes the active site via the formation of stabilizing interactions with the catalytic domain and an adjacent C-terminal J-helix [68,69]. Rather than forming stable heterocomplexes with myriad regulatory subunits, PPP5's TPR-domains are involved in the binding of PPP5C to “TPR-docking” sites on proteins such as heat shock protein 90, the best characterized interaction partner to date. When PPP5 is displaced from its binding partners, the N-terminal/C-terminal interaction is inhibitory. When in a complex with other proteins, a binding-induced conformational change opens the active site to substrates, thereby “activating” PPP5C. Removal of the autoinhibitory N-terminal domain by proteolysis results in a bare catalytic domain with greatly increased activity [70,71]. PPP5C is also activated by the addition of micellar concentrations of unsaturated fatty acids (e.g. arachidonic acid, palmitate) or micromolar concentrations of fatty acid CoA esters, which may act by ablating the auto-inhibitory function of the N-terminal domain [70,72,73].
1.1.6. PP6/PPP6C:
PPP6C also has diverse functions in the cell, including the regulation of mitotic progression, meiosis II exit, NF-kB signaling, autophagy, and T cell development. The disruption of PPP6 expression has also been linked as a driver mutation in the development of melanoma [74-80]. Like PPP4C, there is only one isoform of the catalytic subunit (PPP6C) that is highly similar to PPP2CA/B, sharing >50% identity. Like PPP2, PPP6 functions as a trimeric holoenzyme that consist of PPP6C, one of three regulatory proteins (PPP6R1, PPP6R2, PPP6R3), and one of three ankrin repeat domain containing scafolding proteins (ANR28, ANR44 and ANR52) [81,82]. The combinatorial assembly results in nine distinct PPP6C holoenzymes, and the functions of the individual holoenzymes are poorly characterized.
1.1.7. PP7/PPEF1/2:
PP7 type phosphatase, like PP2B/PPP3C, is regulated by calcium [8,25]. In humans there are two highly homologous isoforms (PP7C/EF1 and PPEF2) expressed from two genes (PPP7C/PPPEF1 and PPPEF2, respectively). PPP7 expression is restricted to primary sensory neurons, and it has unique N- and C-terminal regions. The N-terminal domain contains a calmodulin binding site, while the extended C-terminal domain contains five EF-hand motifs and has been implicated in activation of catalytic activity by calcium [25,83]. Human PPP7 shares homology with Drosophila retinal degeneration C Gene product (rdgC). However, to date, the physiological roles for PPP7C have remained elusive.
1.2. Architecture of the Phosphatase Domain:
Several excellent reviews discuss the structural biology of PP1, PP2A, and PP2B holoenzyme complexes and their function/assembly [48,49,84-86]. However, the details of subunit interactions within PPPase holoenzyme complexes is beyond the scope of this review, which will focus upon the catalytic mechanism and structures of the conserved phosphatase domain with a view to inform efforts to develop type-specific inhibitors of these enzymes.
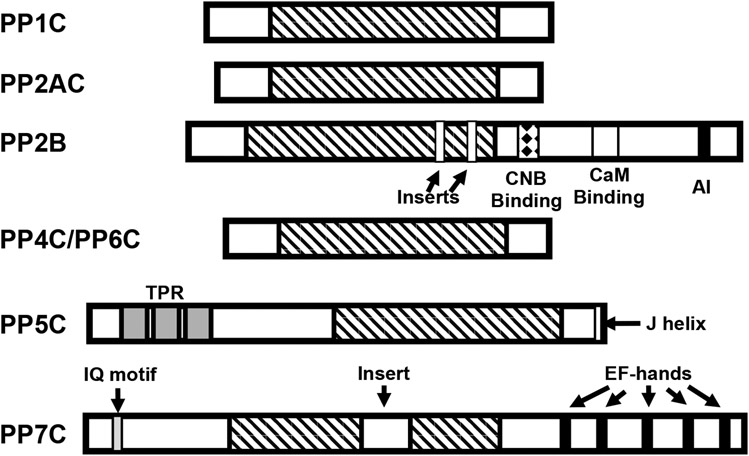
PPP family phosphatases share a conserved catalytic core domain of ~250 residues (Figure 3). Catalytic subunits of PPP1C, PPP2C, PPP4C, and PPP6C are of roughly similar size and display high homology within this core domain, differing from one another mainly within short non-conserved N- and C-terminal sequences. Homology is particularly high (~60-65% identity) between PPP2C, PPP4C, and PPP6C. PP2B/PPP3C departs from this trend by having a long non-conserved C-terminal extension containing regulatory domains (e.g. a calmodulin binding domain) as well as two small stretches of non-conserved sequence within the otherwise conserved core that are inserted into the β7-β8 and β11-β12 loops, respectively, which are positioned far from both the catalytic center and the regulatory B subunit binding interface. PPP5C differs from other members of the family in that it possesses a large N-terminal regulatory/targeting domain, and a unique C-terminal alpha helix. PPP7C isoforms have a ~40 residue non-conserved insert in the catalytic core and also diverge in the C-terminal region, which contains a calcium-binding regulatory domain with EF hand motifs. To date, crystal structures have been reported for PPP5C and the catalytic subunits PPP1CA (PP1α), PPP1CC (PP1γ1), PPP2CA (PP2Aα), PPP3CA (PP2Bα/calcineurin) and PPP3CB (PP2Bβ/calcineurin). As the catalytic domain (residues 169-499) of PPP5C has yielded many of the highest resolution structures from this family [68,87,88], the overall architecture of the phosphatase fold and the basic catalytic mechanism will be discussed here primarily with reference to PPP5C.
Figure 3. Homology and domain organization of PPP-family phosphatases.
PPPase share a highly conserved catalytic core domain of ~250 amino acids (indicated by diagnal stripes). PPP1, PPP2, PPP4, and PPP6 catalytic subunits are highly homologous enzymes, differing primarily in their C- and N-terminal regions. PP2B catalytic subunits (calcineurin A) differs in that it has a large C-terminal extension containing a regulatory subunit (calcineurin B; CNB) binding site (indicated by a diamond filled box), a Ca2+-calmodulin (CaM) binding domain (indicated by a white box), a short autoinhibitory (AI) domain in its C-terminal region and two small divergent regions (inserts) in the catalytic core. PPP5C possesses three tetratricopeptide repeats (TPRs - indicated by grey boxes) in its N-terminal region and a short regulatory helix (J helix; indicated by a white box) at its C-terminus. PPP7 contains a CaM binding IQ motif (gray box) in its N-terminal region, Ca2+ binding EF-hand motifs (indicated by black boxes) in its C-terminal region and a 43-amino acid insert in the catalytic core domain (indicated by an open box).
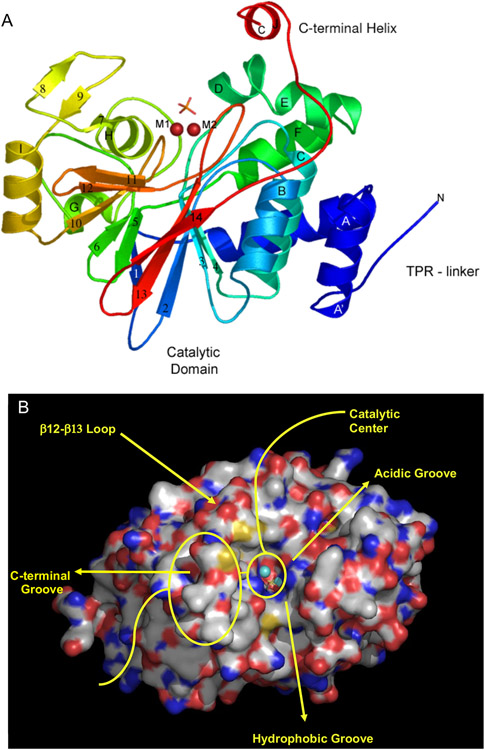
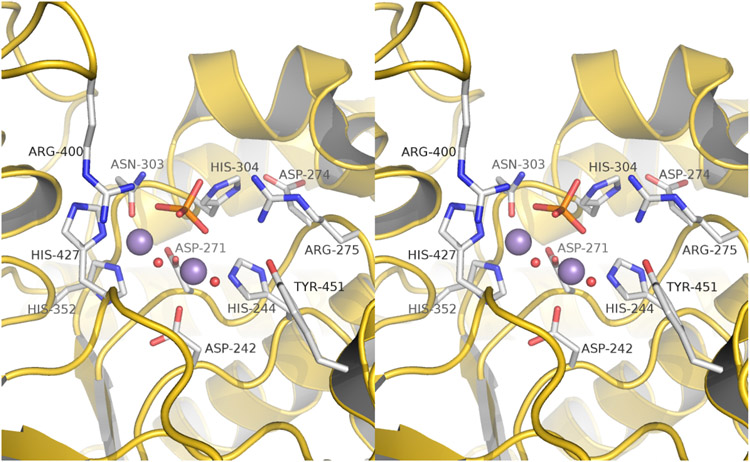
The PPP phosphatase fold, as exemplified by the PPP5- catalytic domain [68,69,87,88], has a compact α/β fold comprised of 11 α-helices and 14 β-strands (Figure 4). Eleven of 14 strands (β1–β6 and β10–β14) form two β-sheets that are arranged in a β-sandwich structure in the center of the molecule. One side of this β-sandwich is occupied by three short helices (αG–αI) and a three-stranded β-sheet (β7–β9) that nearly forms a separate subdomain. On the other side of this β-sandwich there is an α-helical bundle that includes six helices (αA–αF) plus the A′-helix, which is immediately C-terminal to a non-conserved TPR linker sequence. The first helix is designated A′ because, in spite of very low sequence homology within the PPP family in this region, significant structural similarity still exists. PPP5C also possesses a non-conserved C-terminal regulatory subdomain composed of the J-helix tethered by a short linker extending from the end of the β14-strand.
Figure 4.
Structure of the PPP-phosphatase fold as represented by PPP5C. The catalytic domain of serine/threonine protein phosphatase in complex with orthophosphate (PDB code: 1S95) provides an example of the overall structure of the phosphatase domain. A) A cartoon representation of the PP5c structure with is shown in rainbow colors starting with blue at the N-terminus and ending with red at the C-terminus. Divalent metal ion cofactors, M1 and M2 (red spheres) and a phosphate ion indicate the active site location. Alpha helices are labeled A' to J and beta strands are labeled 1 to 14. PPP5C specific features, such as the C-terminal J helix and the TPR domain linker, are also labeled. B) Surface representation of the structure of PPP5C derived from PDB code 1S95. Bound orthophosphate is displayed as sticks, and the catalytic metal ions are depicted as spheres.
1.3. Catalytic Mechanism:
For ease of discussion, in this section the catalytic mechanism is discussed using the amino acid sequence numbering from PPP5C. Much work has established that alkyl phosphate dianions, which are found in phosphorylated serine or threonine residues, are markedly stable. The half-time for hydrolysis of such groups at 25°C is estimated to be over 1 trillion years; knon ~2 x 10−20 sec−1 [89]. Substrate turnover rates (kcat) for PPPases typically range from ~1 to ~100 sec−1, implying a profound ~1021-fold rate enhancement. These enzymes are therefore among the most powerful known catalysts, with catalytic proficiencies ([kcat/kM]/knon) of ~1025–1026 M−1.[89-92] rate enhancements require not only increasing the nucleophilicity of water but also an exquisite alignment of the attacking nucleophile with bound substrate as well as profound stabilization of the transition state.
The catalytic center is dominated by two closely apposed metal ions that are tightly bound to the enzyme through interactions with multiple active site residues. Most of the existing crystal structures of PPPases have been derived from catalytically active recombinant proteins that were heterologously expressed in the presence of Mn2+. X-ray fluorescence of recombinant PPP5C crystals revealed the presence of Fe, Zn, and Mn [68], proton-induced x-ray emission analysis showed Fe and Mn were present in recombinant PPP1CA crystals [93], and ICP atomic emission spectroscopy of recombinant PPP2CA expressed in insect cells identified bound metal as Mn [94]. However, metal analyses of native PPPase enzymes purified from mammalian tissues/cells have consistently shown a combination of Fe and Zn bound to the catalytic subunits [95-98]. Electron paramagnetic resonance spectroscopy studies strongly suggest an Fe2+/Zn2+ binuclear site in native PP2B/PPP3C [96] and, by extension, this is likely to also be the case for the rest of the family. Even though a Fe/Zn bimetal site seems to exist natively, enzyme activity appears to be tolerant of substitution at these sites by manganese.
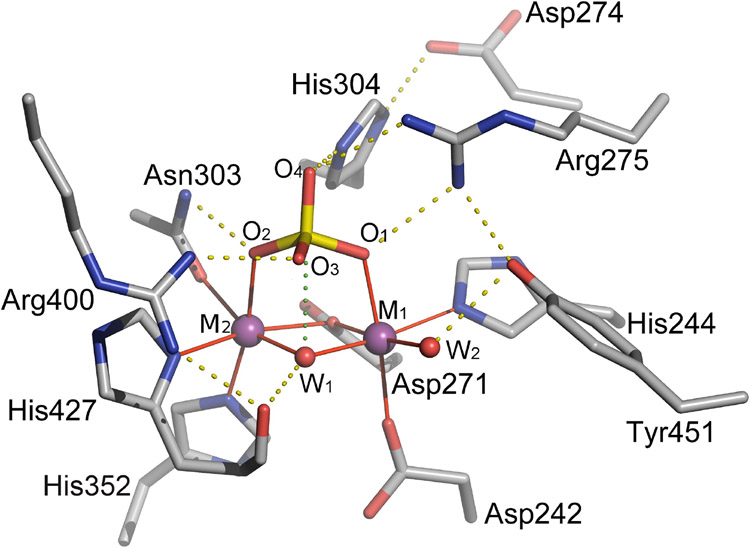
The bimetal site is located at the base of a shallow depression on the PPPase surface formed by the residues of four loops; β4-αD, αG-αH, β10-β11, and β12-β13 (Figure 4). A structural comparison of eukaryotic PPPases reveals an active site motif consisting of 10 conserved amino acid residues; Asp242, His244, Asp271, Asp274, Arg275, Asn303, His304, His352, Arg400, and His427 (PPP5C numbering; Figure 5).
Figure 5. A detailed representation of contacts within the active site.
Hydrogen bonds are shown as dotted yellow lines and coordination bonds to metal ions are shown as solid red lines. The dotted green line between the hydroxide (W1) ion and the phosphorus atom (P) represents a close contact suggestive of the near-attack configuration in metal ionmediated hydrolysis of phosphoprotein substrates. Adapted from Swingle et al 2004 [68].
As both enzyme kinetics [99] and kinetic isotope effect studies [100] of phosphomonoester hydrolysis by PP2B indicate that the substrate dianion is the catalytically active species, the bound phosphate (primarily dianionic at the crystallization pH) offers a reasonable structural analog of the substrate phosphoryl moiety. The structure of the PPP5C catalytic domain with bound phosphate (PDB code 1S95) thus gives a model of substrate-enzyme interactions in a near attack configuration. Six of the 10 conserved residues (Asp242, His244, Asp271, Asn303, His352, and His427) are metal-coordinating and four (Arg275, Asn303, His304, and Arg400) are observed to be directly involved in substrate binding via strong hydrogen bonds to the phosphoryl oxygens [68] (Figure 6). Mutation of either equivalent arginine in PPP1C or PP2B/PPP3C adversely affects substrate binding and catalysis and mutation of Asp274 or His304 equivalent residues severely impacts catalytic activity [90,101]. In the substrate analog complex, His304 forms a short, strong hydrogen bond (2.6 Å) with O4 of phosphate (equivalent to the substrate leaving group oxygen) and is well positioned to act as a general acid in the hydrolysis reaction while Asp274 hydrogen bonds with the His304 imidazole and is thought to increase the imidazole pKa, stabilizing the histidine side chain in a cationic state that is capable of protonating the leaving group [68]. Alternatively, even without a general acid function, positively charged His304, along with Arg275 and Arg400, could contribute to electrostatic stabilization of the transition state by neutralization of the negative charge developing on the leaving group oxygen.
Figure 6. Catalytic site of PPP5C.
A stereo representation of the active site of the catalytic domain of PPP5C. Pertinent active site residues are labeled and shown in stick representation with carbon atoms colored white, oxygen atoms colored red, nitrogen atoms colored blue and phosphorus atoms colored orange. The active-site metal ions are shown as purple spheres and metal ligated waters are shown as small red spheres. Adapted from Swingle et al 2004 [68].
In early work on the enzymology of PP2B, Martin et al [102] proposed three possible types of catalytic mechanisms. The first required nucleophilic attack by an active site residue to form a phosphoenzyme intermediate. This type of mechanism occurs in some families of protein phosphatases (e.g. protein tyrosine phosphatases) but attempts to detect evidence of such intermediates in PPPases [99,102,103] have failed, thereby eliminating this mechanism from consideration. Another possibility considered was the unimolecular breakdown of monoanionic substrate ligated to active site metal ions. However, subsequent kinetic isotope effect data indicated that PPPases utilize dianions, rendering this mechanism unlikely [100]. Finally we are left with the most viable type of mechanism considered, that of nucleophilic attack by a metal-coordinated water (or hydroxide) upon the tetrahedral ground state phosphorus [68]. Water-derived species generally display nucleophilicities in the following order: H3O+ << metal-ligated water < unligated water << metal-ligated hydroxide < unligated hydroxide [104,105]. Coordination of water to a metal ion greatly reduces its pKa, thus the catalytic effect here derives from the generation of local concentrations of the nucleophile far in excess of what would otherwise be possible at a given pH rather than direct activation of the nuclophile.
Returning to the substrate analog complex (1S95), we see that phosphate is coordinated to the active site metal ions in a bidentate fashion (Figure 6).
This may contribute to rate enhancement through stabilization of negative charges building up on the phosphoryl oxygens in the transition state and may also make the phosphorus more electrophilic by polarizing the P-O bonds in the ground state. Also coordinating the metals are two waters/hydroxides that comprise the most likely candidates for the catalytic nucleophile. One of these is terminally ligated while the other sits at a bridging position between the two metals. While bridging two Lewis-acidic metal centers may reduce the nucleophilicity of a bound hydroxide below that of the corresponding terminally-ligated species, the pKa of water at the bridging position would be expected to be much lower than at the terminal position. Thus, at the approximately neutral pH optima of PPPases, the bridging position is likely to produce a much greater population of reactive hydroxide nucleophiles. This suggests the bridging hydroxide is the more likely catalytic nucleophile. Geometric considerations also argue for this [68]. The bridging hydroxide in 1S95 is very close to the phosphate phosphorus (3.0 Å, less than the van der Waals contact distance) and is directly in line (i.e. a nearly 180° angle) with phosphorus and O4 (equivalent to the leaving group oxygen). Additionally, there is a short hydrogen bond between the bridging hydroxide and the backbone carbonyl of His427 with a ~109° angle between the carbonyl O, hydroxide O, and P. This appears to be an example of orbital steering [106], with the metals and His427 acting to fix the orientation of a lone pair of electrons from the hydroxide toward the substrate phosphorus atom. The likely reaction mechanism, derived from available data, is show schematically in Figure 7.
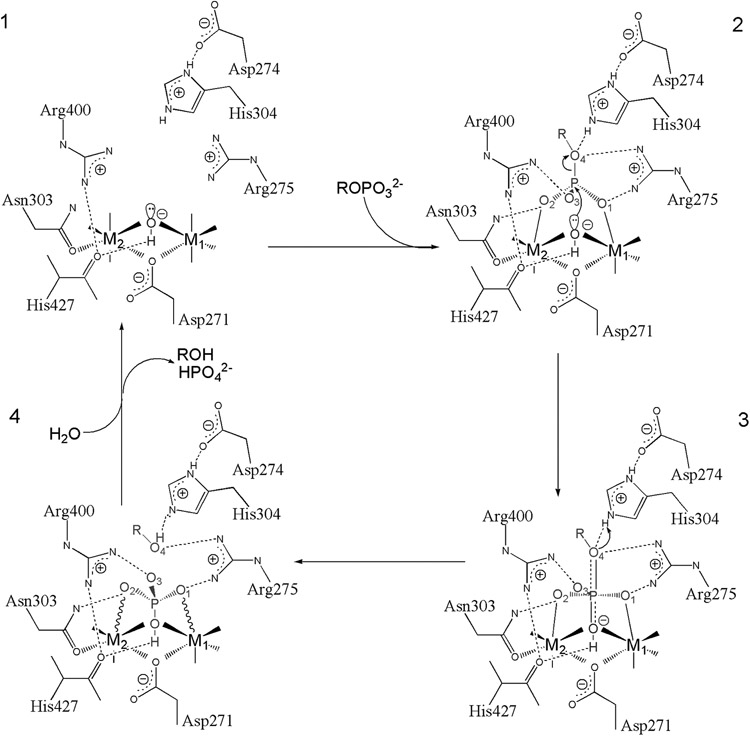
Figure 7. PPPase Reaction mechanism.
A Schematic representation of phosphomonoester hydrolysis mediated by metal ions and active site residues is presented. PP5 residue numbering is used. Solid lines to the metal ions denote metal-ligand bonds, and solid or dashed wedges indicate metal-ligand bonds directed above or below the plane of the page, respectively. Wavy lines to the metal ions indicate strained contacts with poor coordination geometry. Dotted lines indicate hydrogen bonds, and the nearly dissociated axial bonds in the transition state are shown by half-dotted double-lines. Adapted from Swingle et al. the Journal of Biological Chemistry 2004 [68].
The bridging hydroxide [1] is precisely positioned for in-line nucleophilic attack upon the phosphorus of incoming substrate by the active site metals and carbonyl oxygen of His427. Kinetic isotope effects measured for pNPP hydrolysis by PP2B [100] and the structurally related bacteriophage lambda phosphatase [107] indicate a highly dissociative, metaphosphate-like transition state (as is also the case for the uncatalyzed reaction), with advanced bond dissociation to the leaving group and little bond formation to the attacking nucleophile. Since the attacking nucleophile and the leaving group oxygen are held in place by multiple strong interactions with the enzyme, neither are likely to move relative to one another during the evolution of the transition state. As the reaction progresses from step [2] to [3], the phosphorus moves along the reaction coordinate toward the bridging hydroxide, while the angles between the equatorial P-O bonds increase, forming a planar metaphosphate-like dissociative transition state [3] with the attacking nucleophile and leaving group oxygen sitting at axial positions with low bond order to P. In the transition state the equatorial P-O bonds have little double-bond character and are highly polarized. These polarized bonds are stabilized through interactions with Arg275, Asn303, and Arg400 as well as the active site metal ions. The negative charge developing on the leaving group oxygen as that P-O bond dissociates is stabilized through interactions with Arg275 and His304. Strong electrostatic effects produced by the dinuclear metal center underlie a major part of its catalytic role. By stabilizing negative charges, the metals not only promote deprotonation of the bridging water but can also make the phosphoryl moiety more susceptible to attack by stabilizing the polarized P-O bonds that develop during the reaction. In a similar way, Arg275 and Arg400 are not only necessary for binding and orienting the substrate, but also provide stabilizing interactions for the transition state.
As the reaction continues past step [3] toward step [4], the phosphoryl group continues its inversion about phosphorus. Bond order to the attacking nucleophile increases and bond dissociation to the leaving group nears completion. At some point during bond dissociation, the proton affinity of the leaving group oxygen increases enough to accept a proton from His304, which acts as a general acid. At completion [4], the incoming hydroxide nucleophile has become a hydroxyl group of the phosphate dianion product bound to the dinuclear metal center in an unstable tridentate mode and is subsequently expelled while the bridging coordinating position is repopulated from bulk solvent. The high solvent accessibility of the shallow active site combined with the relatively low turnover (kcat) of phosphoprotein/phosphopeptide substrates by PPPases [89] allows enough time and opportunity during the catalytic cycle for regeneration of the active ionization state (i.e. bridging hydroxide nucleophile and protonated His304 general acid) to take place through simple exchange of protons with intracellular bulk solvent and/or buffer molecules (e.g. organic acids, phosphates, and free amino acids) since proton exchange reactions can proceed at rates far greater than typical PPPase kcat's [108].
1.4. Inhibitors of PPP family phosphatases:
Going back in history ~25 years, the dogma of the time was that protein kinases were not suitable targets for drug development. Fortunately, not everyone followed the dogma, and today after an extensive effort there are > 20 drugs targeting protein kinases (i.e. gefitinib, dabrafenib, STI-571/Gleevec, ceritinib) that are used in a clinical setting to treat human disease, most for the medical management of human cancers. Currently, there are many that argue the PPPases are not valid drug targets. However, like the view of PPPases as simple house keeping enzymes, this opinion is slowly falling out of favor. Still. developing PPPase inhibitors into drugs will be challenging because as discussed above the PPP-family phosphatases share a catalytic mechanism, and many family members retain a high degree of structural similarity in the regions surrounding the active site. In addition, as will be discussed in more detail below, high affinity non-selective inhibitors of PPP-catalytic activity are highly toxic to most, if not all, human cells.
1.4.1. Immunophilin ligands:
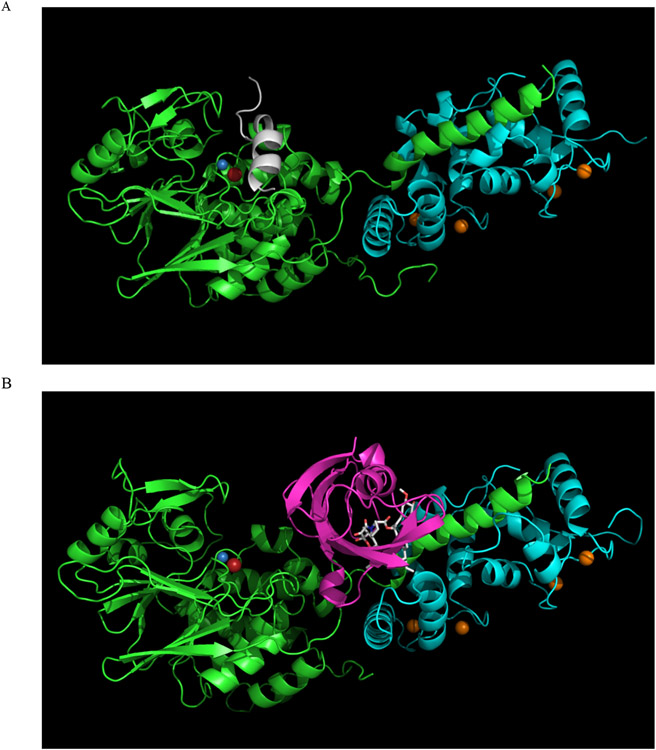
Proof that PPP-family phosphatases can be targeted for drug development already exist, in the respect that the cyclic undecapeptide cyclosporin A and the macrolide tacrolimus (FK506) have already been developed into highly effective immunosuppressant drugs. Both are potent and selective inhibitors of PP2B/PPP3C holoenzymes in vivo [109]. However, their mode of action is not typical in that binding to an accessory protein (an immunophilin) is first necessary in order to make the drug competent for phosphatase inhibition. Immunophilins are highly conserved chaperone proteins typically having peptidylprolyl isomerase activity [110-112]. The two main families are the cyclosporin A binding cyclophilins (Cyp) and the FK506 binding proteins (FKBP), which bind macrolides such as rapamycin and tacrolimus (FK506). The binary complex of CypA with cyclosporin A or FKBP-12 with tacrolimus is able to dock to and inhibit the PP2B holoenzyme (catalytic and regulatory subunits; Figure 8). Inhibition of PP2B by these immunophilin/immunosuppressant complexes is extremely selective due to extensive interactions between the inhibitor complex and a composite surface formed by the non-conserved C-terminal helix of the PP2B catalytic subunit and the regulatory B subunit, which is not shared by other PPP-ases [48,49,112-116].
Figure 8. Inhibition of PP2B by immunophilin/immunosuppressant complex.
Inhibition of PP2B by immunophilin/immunosuppressant complex. A. The structure of a PP2B holoenzyme is shown in a cartoon representation. The PP2B catalytic subunit is green, the regulatory B subunit is cyan, and the autoinhibitory domain is white. Metal ions are shown as spheres: Fe is red, Zn is blue, and Ca is orange. B. The structure of a PP2B holoenzyme bound to the inhibitory FKBP12:tacrolimus complex is shown in a cartoon representation. The PP2B catalytic subunit is green, the regulatory B subunit is cyan, and FKBP12 is magenta. Metal ions are shown as spheres: Fe is red, Zn is blue, and Ca is orange. Tacrolimus (FK506) is shown as sticks
1.4.2. Natural compounds targeting PPPases.
PPPases are evolutionarily-primitive enzymes that emerged with the development of Archaea. During evolution many structurally diverse natural compounds targeting these enzymes evolved, likely as a means of chemical defense to prevent predation. The first natural toxin shown to act as a PPPase inhibitor [117] was okadaic acid, a complex polyether produced by marine dinoflagellates (e.g Prorocentrum sp. and Dinophysis sp.). Microcystin-LR, a cyclic heptapeptide produced by fresh water cyanobacteria (Microcystis sp.) and nodularin, a structurally related compound produced by marine cyanobacteria (i.e. Nodularia sp.), were shown to inhibit the same phosphatases (PPP1C and PPP2) a few years later [118,119]. Analysis of extracts from red algae and marine sponges identified thyrsiferyl 23-acetate, calyculins and dragmacidins [120-123]. Extracts from Streptomyces sp.) yielded fostriecin, phostriecin, sultriecin [124-126], cytostatins [127], phospholine [128], leustroducsins [129], phoslactomycins [130], tautomycin [131] and tautomycetin [132]. Extracts of ~1500 species of blister beetles contain cantharidin [133]. Although structurally diverse, (Figure 9), the aforementioned compounds all act by inhibiting a subset of the “toxin sensitive PPP-family members (PPP1C, PPP2CA/B, PPP4C, PPP5C and PPP6C) and the binding affinity and inhibitory activity of several differ substantially among the sensitive enzymes. To date, inhibitor-PPPase co-crystal structures have been reported for okadaic acid, microcystin-LR, nodularin, tautomycin, calyculin A, cyclosporine A, cantharidin, and analogues of cantharidin. Considerable insight into means of targeting PPPases can be gained from examining the structures that currently exist.
Figure 9. Natural compounds with inhibitory activity against PPP-family phosphatases.
1.4.3. Cyanobacterial toxins:
Microcystins are a family of toxic cyclic heptapeptides produced by certain cyanobacteria, with toxicity comparable to organophosphate nerve agents [134]. Microcystin was first identified as a highly toxic compound produced by Microcystis aeruginosa called fast death factor [135,136]. In vivo, microcystins primarily target the liver, because transport into hepatocytes occurs actively, via specific organic anion transporting polypeptides (OATPs) [137]. Microcystin-LR (MCLR) was first reported to potently inhibit PPP1C and PPP2CA/B [118]. However, later it became clear that it also inhibits PPP4, PPP5, and PPP6, acting on all with sub- to low-nM IC50 [138-140]. Now there are over 50 chemical variants of microcystin that act as potent, but rather non-selective inhibitors of these PPPases. PP2B is much less sensitive, having an IC50 of ~200 nM [141], and PPP7 is very insensitive, being unaffected by 100 nM MCLR [25].
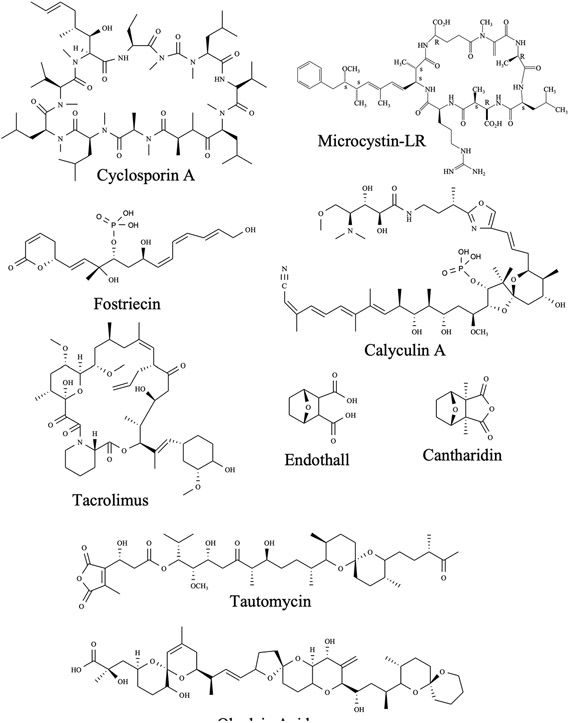
The crystal structure of the PPP1Cα:MCLR complex was among the first structures of a PPPase catalytic domain to be reported [142]. A view of MCLR in its binding site is presented in Figure 10 (Top; PDB code 1FJM). MCLR interacts with three distinct regions on PPP1Cα. First, it interacts both directly and indirectly with the highly conserved areas in and around the active site. Microcystin interacts with the active site metal ions by forming hydrogen bonds between two coordinated waters and the cyclic peptide glutamate side chain carboxyl and adjacent backbone carbonyl. It also interacts with Tyr134 and the absolutely conserved catalytic Arg96 via interactions with the D-erythro-β-methylaspartate (Masp) moeity of the ligand. These interactions help position the cyclic peptide backbone directly over the catalytic center, blocking substrate access. Second, the Adda side chain of MCLR interacts with residues in the hydrophobic groove of PPP1Cα, some of which are not conserved across the PPP family. The close packing of the Adda moiety suggests a significant contribution to binding affinity arises from desolvation/hydrophobic interactions. Third, the β12-β13 loop adjacent to the active site engages in important interactions with MCLR side chains. Tyr272 (conserved across the family) of PPP1Cα participates in a hydrophobic interaction with the cyclic peptide leucine side chain while the thiolate of Cys273, conserved only in the MCLR-sensitive PPPases (PPP1C, PPP2C, PPP4C, PPP5C, and PPP6C), is seen to react with the α,β-unsaturated carbonyl of methyldehydroalanine (Mdha) to form a covalent adduct. Comparison with later structures of PPP1Cα or PPP1Cγ1 also show that the β12-β13 loop shifts slightly in the MCLR complex to avoid a steric clash with the conserved Tyr272, suggesting that the flexibility of this loop may be important for high affinity binding [143].
Figure 10. Microcystin-LR bound to PP1C and PP2A.
The structure of the PPP2C:MCLR complex has also been reported [94] (Figure 10; bottom; PDB code 2IE3). Intermolecular interactions in this complex are, overall, quite similar to what is observed for PPP1Cα:MCLR. The bound conformation of MCLR is virtually identical in both cases and is positioned similarly within the active site and hydrophobic groove. A major difference is observed in the interactions with the β12-β13 loop, which has a small sequence segment that is not conserved with PPP1C. In the PPP2C:MCLR complex, the β12-β13 loop adopts a different conformation and Cys269, present only in PPP2C, PPP4C, and PPP6C, forms the covalent adduct with the electrophilic methyldehydroalanine residue of the inhibitor. Other differences include interactions with non-conserved residues in the hydrophobic groove.
1.4.5. Okadaic acid:
Okadaic acid (OA) and its close structural analogs, the dinophysistoxins (DTXs), are complex polyether fatty acids produced by several phytoplanktonic species and subject to bioaccumulation in filter feeders and their predators [144,145]. OA was first isolated from the black sponge Halichondria okadai as one of the principal causative agents of diarrhetic shellfish poisoning (DSP). DSP is characterized by incapacitating diarrhea and vomiting. The symptoms are usually self-limiting without medical intervention and subside in 1-2 days. OA is membrane permeable, and the discovery that OA acts as a potent inhibitor of PPP1C and PPP2C catalytic subunits, with considerable selectivity for PPP2C [117,146], greatly advanced our understanding of the roles played by OA sensitive PPPases. OA rapidly become a workhorse inhibitor for the study of PPP family protein phosphatases and their roles in cellular processes. Although, widely reported as a specific or highly selective inhibitor of PP2A, IC50s vs PPP2AC, PPP4C, and PPP6C catalytic subunits are all sub-nanomolar (~0.1 nM). PPP1C is less sensitive (15-50 nM IC50) and PPP5C has an intermediate sensitivity (~1 nM IC50) [138-140,147]. PP2B and PPP7 are far less sensitive than even PPP1C, having IC50s above 1 μM [25,146].
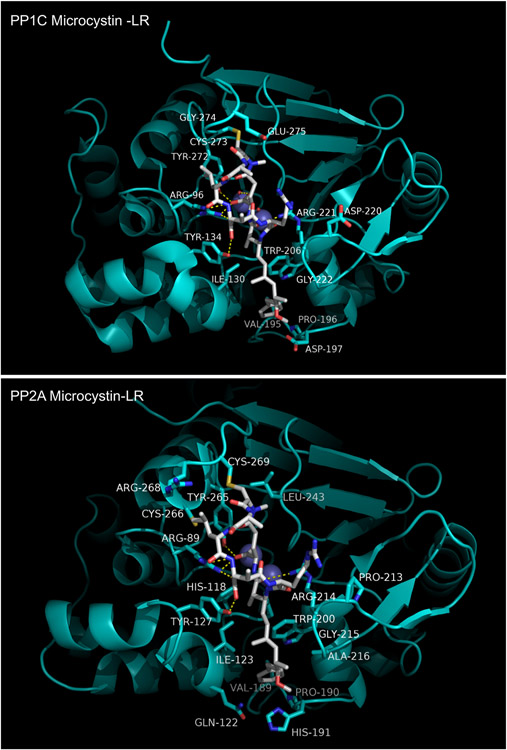
The crystal structures of OA in complex with PPP1Cγ and a PPP2 heterodimer have been reported [94,143]. The PPP1Cγ:OA structure is shown in Figure 11. From a comparison with the MCLR complex shown in Figure 10 it is readily apparent that the binding sites for the two inhibitors coincide quite closely. Indeed, rather than binding in an extended linear conformation, OA adopts a psuedo-macrocyclic shape, containing two of the inhibitor's three spiroketal moieties, that is stabilized by an intra-molecular hydrogen bond between its C1 carboxylic acid moiety and the C24 hydroxyl. The remaining spiroketal group and a short linking segment form a hydrophobic tail projecting out from the psuedo-macrocycle. OA makes contacts with three absolutely conserved active site residues that are involved in substrate phosphoryl group binding: Arg96, His125, and Arg221. Like MCLR, OA also interacts with residues in and near the hydrophobic groove, principally Ile130, Tyr134, and Trp206. Finally, several residues in the β12-β13 loop make close contacts with the inhibitor, most notably Tyr272, a conserved residue known from mutagenesis studies to be important for OA binding [148].
Figure 11. Okadaic acid bound to PP1C and PP2A.
The PPP2C:OA complex is shown in Figure 11. The bound conformation of OA is nearly identical to that seen in the PPP1Cγ:OA complex. Many interactions are shared with the PPP1Cγ:OA complex. Conserved active site residues Arg89, His118, and Arg214 (Arg96, His125, and Arg221 in PPP1Cγ) make similar contacts with OA in both core complexes. Similarly, Tyr265 and Cys266 in the β12-β13 loop and Ile123, Tyr127, and Trp200 in the hydrophobic groove are seen to recapitulate the interactions of their counterparts in PPP1Cγ. However, there are non-conserved sequences in the β12-β13 loop and hydrophobic groove that give rise to differential sets of interactions with the OA pseudomacrocyle and the hydrophobic tail, respectively. In the β12-β13 loop, PPP1Cγ has glutamate and phenylalanine side chains contacting hydrophobic groups in a spiroketal moeity within the pseudomacrocycle while in PPP2CA the interacting residues are arginine and cysteine. The polypeptide sequence immediately C-terminal to the conserved (F/W)SA(P/S)NY motif in the β12-β13 loop abuts the active site and exhibits significant divergence across the PPP family. Thus, divergent sequences in the β12-β13 loop form very different interacting surfaces for this portion of the inhibitor. Similarly, in the hydrophobic groove, non-conserved residues produce significantly different binding pockets for the OA hydrophobic tail. Gln122 and His191, along with Ile123 and Trp200 (conserved in PPP1C) help form a hydrophobic cage in the PPP2C:OA complex (but absent from PPP1C) that has much better shape complementarity to the inhibitor (compare in Figure 11).
1.4.6. Tautomycin:
Tautomycin (TM) is a cytotoxic natural product polyketide produced by the soil bacterium Streptomyces verticillatus, having partial structural similarity to okadaic acid [149]. It is a sub-nanomolar inhibitor of PPP1C, PPP2CA/PPP2CB, and PPP4C, a somewhat less potent inhibitor of PPPC5 (IC50 ~10 nM), and a very weak ( IC50 > 10 μM) inhibitor of PP2B/PPP3C [139,149,150].
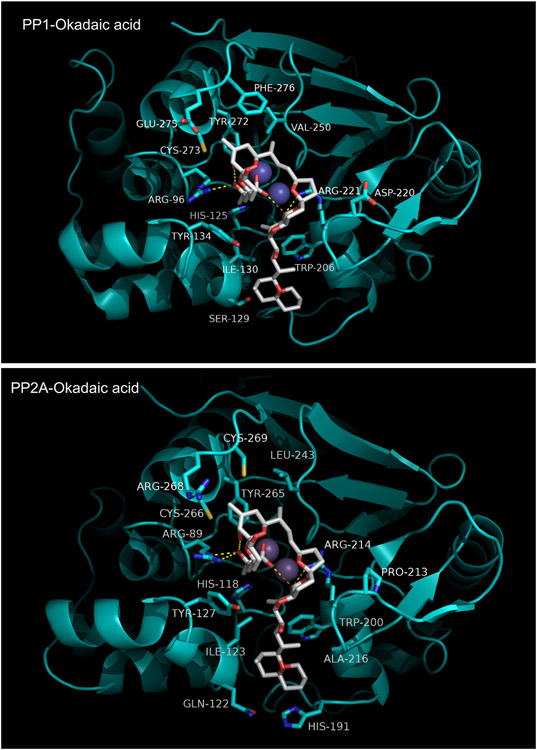
The crystal structure of the PPP1Cα:TM complex was reported by Kelker et al in 2008 [151]. TM is found to bind in its diacid form and is shown in Figure 12. Tautomycin can be conceptually divided into two segments: a hydrophobic fragment containing a spiroketal group, which binds in the hydrophobic groove of PPP1α, and a hydrophillic fragment containing the diacid, as well as other polar groups, which interacts with PPP1α in and around the active site. The hydrophillic segment forms six hydrogen bonds with four residues in or near the catalytic site: Arg96, Arg221, Val250, and Tyr272. The hydrogen bond with Tyr272 is a feature shared with OA:PPPase complexes and mutation of this residue increases the IC50 of TM by 500-1500-fold [148]. The hydrophobic segment of TM is anchored in the hydrophobic groove through interactions with Cys127, Ser129, Ile133, Tyr134, Trp206, Gly222, and Val223. The reported selectivity of TM for PPP1C vs PPP2CA is quite modest and structural comparisons do not reveal any obvious reasons for selectivity. On the other hand, the markedly lower affinity of TM for PP2B may be due, in part, to substitution of bulky, hydrophobic leucine for a cysteine residue (Cys273 in PPP1C: conserved in all PPPases except PP2B and PPP7) in the β12-β13 loop. This substitution likely produces a steric clash with the diacid moiety of TM.
Figure 12. Tautomycin bound to PPP1C.
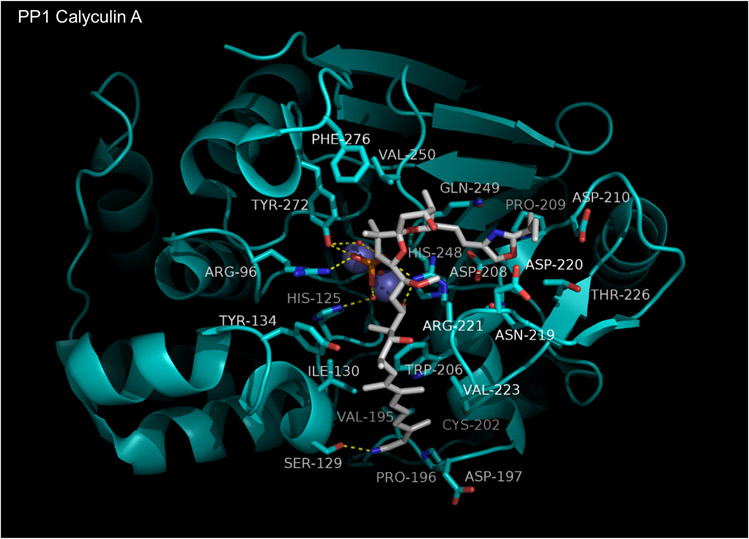
1.4.7. Calyculin A:
Calyculin A (CalA), first isolated from the sponge Discodermia calyx, is a cytotoxic agent consisting of a dipeptide segment plus a phosphorylated polyketide moiety [120,121]. It is a potent, sub-nM to low nM, inhibitor of PP1, PPP2C, PPP4C, PPP5C, and PPP6C [138-140,150], with PP2B and PPP7C having much lower affinity (IC50 > 1μM). Structure-activity relationship (SAR) studies of CalA derivatives have demonstrated the importance of the phosphate, the C13 hydroxyl, and the polyketide tail, for phosphatase inhibition. The dipeptide segment, however, seems to make only a minor contribution [152].
The PPP1Cγ:CalA complex structure (Figure 13) was solved by Kita et al in 2002 [153]. CalA is observed to make contacts with groups in the active site, the hydrophobic groove, and the acidic groove. The CalA phosphate participates in many polar contacts with the enzyme, interacting indirectly with the active site metals and His125 via hydrogen bonds to structural waters and directly with Arg96, Arg222, and Tyr272 side chains via hydrogen bonds. the hydrophobic tail of CalA (N1–C11) makes numerous non-polar contacts with residues in the hydrophobic groove, including the side chains of Tyr134, Trp206, Ile130, Val223, Cys202, and Asp197. Ser129 and the backbone carbonyl of Arg221 form hydrogen bonds with the C1 nitrile and C11 hydroxyl, respectively. The oxazole moiety makes weak hydrophobic contacts in the acidic groove with Asn219, Asp220, Thr226, Gln249, Asp208, and Asp210. Polar interactions with the enzyme were not observed in the acidic groove. Part of CalA (C33 to C37) was not modeled due to poor/uninterpretable density and is presumed to contribute little to binding affinity.
Figure 13. Calyculin A bound to PPP1C.
1.4.8. Fostriecin:
Fostriecin is a phosphate monoester isolated from the fermentation beer of Streptomyces pulveraceus. The antitumor activity of fostriecin has been evaluated extensively [124,154-156] and marked cytotoxicity against many cancer cell lines as well as potent antitumor activity in animals has been demonstrated. Its potential use as an antitumor agent in humans has been evaluated in Phase 1 clinical trials. However, trials were discontinued before the maximal tolerated dose was established due to the loss of momentum for further development following corporate mergers of the pharmaceutical companies developing fostriecin, which lead to production limitations. There were also concerns raised about storage stability [154]. Fostriecin acts as a potent (~1-3 nM IC50) inhibitor of PP2A/PPP2CA [157-159] and PPP4C [138], a weak inhibitor of PPP1C and PPP5C (~50 μM IC50) [158, 159] and at 100 μM has no apparent effect on PP2B/PPP3C [138,157-159]. Assessments of fostriecin inhibition of PPP6C and PPP7C have not been reported. Due to its high degree of homology with PPP2CA and PPP4C, PPP6C is expected to be highly sensitive fostriecin. Thus fostriecin appears to be a specific inhibitor of the PPP2/PPP4/PPP6 subfamily, with > 10,000 fold selectivity for PPP2/PPP4/PPP6 vs other PPPases. No structures of fostriecin in complex with PPPases have been reported. However, mass spectrometry analysis of fostriecin:PPP2CA covalent complexes have established that the electrophilic α,β-unsaturated δ-lactone moiety of fostriecin undergoes nucleophilic attack by Cys269 (conserved in PPP2, PPP4, and PPP6) in the β12-β13 loop to form a covalent adduct[160]. Mutagenesis studies and SAR of fostriecin derivatives indicate that the reaction between Cys269 and the lactone is one of the principal determinants of fostriecin's potency and selectivity [158,161]. Mutation of PPP1C or PPP5C to introduce an appropriately placed cysteine dramatically increases fostriecin affinity. In addition derivatives of fostriecin that contain a hydrolyzed lactone experience a 105 -fold loss in activity [159]. This provides a high degree of confidence in the positioning of the lactone ring in the active site. However, the position of the hydrophobic tail is not clear at this time.
1.4.9. Cantharidin and the 7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid scaffold:
Cantharidin (2,3-dimethyl-7-oxabicyclo[2.2.l]heptane-2.3-dicarboxylic acid anhydride) is the active constituent in a traditional Chinese medicine. It is naturally occurring in the bodies of ~1500 species of blister beetles, which use it as a chemical defense mechanism [162-164]. Medical use of cantharidin also has a history in Europe where it was first isolated in the early 1800’s [165]. Although occasionally utilized as a topical vesicant for the removal of warts [166], cantharidin was considered as obsolete and too toxic for internal use by the early 1900’s. Cantharidin acts is a semi-selective inhibitor of PPPases and is known to strongly inhibit the catalytic activity of PPP1C, PPP2CA, PPP5C and PPP6C (IC50 ~ 0.2–8 μM), while having much lower affinity for PP2BC/PPP3C, PPP4C, or PPP7/PPPEF (IC50 >> 20 μM) [25,87,133,139,167-169]. Unlike the previously discussed inhibitors, which are all complex natural products that are difficult to synthesize/derivatize, cantharidin class inhibitors, containing a core 7-oxabicyclo[2.2.1]heptane-2,3-dicarboxylic acid scaffold, are synthetically accessible and production of derivatives is relatively straightforward.
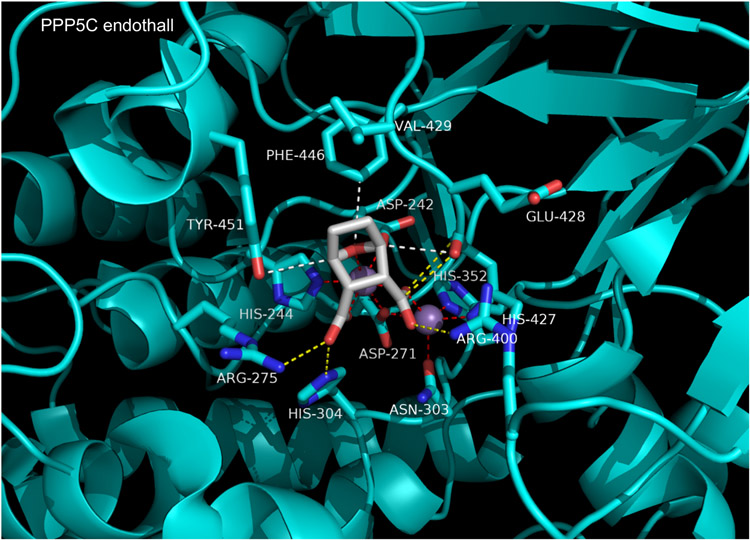
Structures of cantharidin and related derivatives (e.g. the aquatic herbicide, endothall) in complex with PP5C have been reported by Bertini et al [88]. Some members of this class are synthesized or isolated as anhydrides, while others are diacids. However, only the diacid forms are observed bound in co-crystal structures. Forms with anhydride rings, when incubated in aqueous buffer with enzyme, appear to be readily hydrolyzed to the ring opened diacid. The observed binding modes for inhibitors in this class are all highly similar. As an example, in Figure 14 we show the complex of PPP5C with endothall (PDB 3H61). Endothall is small, with a low molecular weight (186 g/mol) and high ligand efficiency. It binds tightly to PPP5C right at the catalytic center, directly coordinating the metal ions with both carboxylic acids and the bridging O7. One metal ion is pentacoordinate with trigonal bipyramidal geometry and the other maitains an octahedral coordination sphere. The 2- and 3-carboxy groups of endothall also form hydrogen bonds with conserved PPP5C catalytic residues Arg275, Arg400, and His304. A few somewhat unusual interactions are also on display. An anti-parallel dipole-dipole stacking interaction occurs between the 2-carboxyl of endothall and the backbone carbonyl of His427. Three weak CH---O hydrogen bonds help stabilized the position of the oxabicyclo ring: one between the Tyr451 hydroxyl and the C1 hydrogen, one between the C4 hydrogen an the backbone carbonyl of His427, and one between an aromatic ring hydrogen of Phe446 and the bridging oxygen (O7). Also observed are hydrophobic contacts between the inhibitor and side chains of Val429, Phe446, and Tyr451. Almost all of the inhibitor contacts described above are with residues absolutely conserved across the PPP family. The exceptions are Val429 and Phe446, which might account for some of the observed IC50 differences. Indeed, substituting tryptophan for Phe446 in PPP5C greatly increases the IC50 for cantharidin and PPP4C, which has a tryptophan at this site, is relatively insensitive [87]. Molecular modeling of endothall bound to PPP4 suggests that the much larger tryptophan side chain may produce a steric clash with the inhibitor that perturbs the high-affinity binding mode it is free to adopt in the PPP5C complex.
Figure 14. Endothall bound to PPP5C.
1.5. Future development of selective inhibitors:
Based on the interactions discussed above, sequence divergence in the β12-β13 loop, hydrophobic groove, and acidic groove appear to provide opportunities for developing non-conserved contacts in derivatives of existing inhibitors or novel scaffolds. The small size and simple methods needed for the synthesis of nor-cantharidin derivatives makes it an attractive scaffold. However, merely “decorating” the cantharidin core scaffold with small groups may not be adequate. A fragment linking strategy may prove more fruitful. Methods to improve the storage stability of fostriecin may be all that is needed to renew interest in this compound with previously demonstrated marked antitumor activity. Because we now have structural information about how complex natural product inhibitors interact with targeted PPPases, fragments of existing large natural inhibitors that are toxic in their entirely may also provide novel leads for the development of highly specific inhibitors. This may facilitate deriving synthetically accessible fragments from okadaic acid, tautomycin, calyculin A, and microcystin (maybe directly, maybe indirectly via similarity searches on interesting substructures) that could be linked up to a core scaffold.
It may also prove useful to conduct virtual screening and biophysical screening (SPR, NMR, MS etc.) to identify compounds with improved selectivity. As the structures of PPP1 and PPP2 holoenzymes are reported, it will likely be fruitful to explore the development of compounds that interact at the interface where the substrate, catalytic and regulator subunits converge. Such compounds will likely demonstrate greatly reduced systemic toxicity and thus have better target specific utility.
CONCLUSION:
Although PPP-family phosphatases share a common catalytic mechanism, insights obtained from the examination of PPPase-inhibitor co-crystal complexes indicate that the development of more selective or even specific inhibitors should be possible.
Table 1.
Nomenclature associated with PPP2-phosphatases
| Subunit | Gene | Isoform | Other Name(s) |
|---|---|---|---|
| Scaffold (A) | PPP2R1A | α | PP2AA α |
| Scaffold (A) | PPP2R1B | β | PP2AAβ |
| Catalytic (C) | PPP2CA | α | PP2Acα |
| Catalytic (C) | PPP2CB | β | PP2Acβ |
| Regulatory (B) | PPP2R2A | α | B55α, PR55α, PP2ABα |
| Regulatory (B) | PPP2R2B | β | B55β, PR54β, PP2ABβ |
| Regulatory (B) | PPP2R2C | γ | B55γ, PR55γ, PP2ABγ |
| Regulatory (B) | PPP2R2D | δ | B55δ, PR55δ, PP2ABδ |
| Regulatory (B’) | PPP2R5A | α | B56α, PR56/61α, PP2AB’α |
| Regulatory (B’) | PPP2R5B | β | B56β, PR56β/PP2AB’β |
| Regulatory (B’) | PPP2R5C | γ,1,2,3 | B56γ, PR56/61γ, PP2AB’γ |
| Regulatory (B’) | PPP2R5D | δ | B56δ, PR56/61δ, PP2AB’δ |
| Regulatory (B’) | PPP2R5E | ε | B56ε, PR56/61ε, PP2AB’ε |
| Regulatory (B’’) | PPP2R3A | α | PR130, B”α1 |
| Regulatory (B’’) | PPP2R3A | α | PR72, B”α2 |
| Regulatory (B’’) | PPP2R3B | β | PR70, PR48,B”β |
| Regulatory (B’’) | PPP2R3C | γ | G5PR, G4-1 |
| Regulatory (B’’) | PPP2R3D | δ | PR59, B”δ |
| Regulatory (B’’’) | PPP2R6A | STRN | Striatin, PR110, |
| Regulatory (B’’’) | PPP2R6B | STRN3, SG2NA | |
| Regulatory (B’’’) | PPP2R6C | STRN4 | |
| Regulatory (B’’’) | PPP2R4 | PTPA, PR53 |
ACKNOWLEDGEMENTS
This work was supported by in part by grants from the National Institutes of Health (NCI Grant CA 60750 and NS071533 to REH) and funds provided by the University of South Alabama Cancer Center Research Fund.
LIST OF ABBREVIATIONS
- PPPase
PPP-family serine/threonine protein phosphatase
- OA
okadaic acid
- MCLR
microcystin-LR
- CalA
calyculin A
- TM
tautomycin
Biography

Footnotes
CONFLICT OF INTEREST The authors have nothing to disclose.
REFERENCES
- [1].Manning G; Whyte DB; Martinez R; Hunter T; Sudarsanam S The Protein Kinase Complement of the Human Genome. Science, 2002, 298, 1912–1934. [DOI] [PubMed] [Google Scholar]
- [2].Milanesi L; Petrillo M; Sepe L; Boccia A; D’Agostino N; Passamano M; Di Nardo S; Tasco G; Casadio R; Paolella G Systematic Analysis of Human Kinase Genes: A Large Number of Genes and Alternative Splicing Events Result in Functional and Structural Diversity. BMC Bioinformatics, 2005, 6, S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Anamika K; Garnier N; Srinivasan N Functional Diversity of Human Protein Kinase Splice Variants Marks Significant Expansion of Human Kinome. BMC Genomics, 2009, 10, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sefton BM Overview of Protein Phosphorylation. Curr. Protoc. Cell Biol. Editor. Board Juan Bonifacino Al, 2001, Chapter 14, Unit 14.1. [DOI] [PubMed] [Google Scholar]
- [5].Brognard J; Hunter T Protein Kinase Signaling Networks in Cancer. Curr. Opin. Genet. Dev, 2011, 21, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Cohen P Protein Kinases--the Major Drug Targets of the Twenty-First Century? Nat. Rev. Drug Discov, 2002, 1, 309–315. [DOI] [PubMed] [Google Scholar]
- [7].Rebelo S; Santos M; Martins F; da Cruz e Silva EF; da Cruz e Silva OAB Protein Phosphatase 1 Is a Key Player in Nuclear Events. Cell. Signal, 2015, 27, 2589–2598. [DOI] [PubMed] [Google Scholar]
- [8].Honkanen RE; Golden T Regulators of Serine/threonine Protein Phosphatases at the Dawn of a Clinical Era? Curr. Med. Chem, 2002, 9, 2055–2075. [DOI] [PubMed] [Google Scholar]
- [9].Cohen P The Origins of Protein Phosphorylation. Nat. Cell Biol, 2002, 4, E127–130. [DOI] [PubMed] [Google Scholar]
- [10].Cohen P The Regulation of Protein Function by Multisite Phosphorylation--a 25 Year Update. Trends Biochem. Sci, 2000, 25, 596–601. [DOI] [PubMed] [Google Scholar]
- [11].Cohen P The Role of Protein Phosphorylation in Human Health and Disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem, 2001, 268, 5001–5010. [DOI] [PubMed] [Google Scholar]
- [12].Ubersax JA; Ferrell JE Mechanisms of Specificity in Protein Phosphorylation. Nat. Rev. Mol. Cell Biol, 2007, 8, 530–541. [DOI] [PubMed] [Google Scholar]
- [13].de Oliveira PSL; Ferraz FAN; Pena DA; Pramio DT; Morais FA; Schechtman D Revisiting Protein Kinase-Substrate Interactions: Toward Therapeutic Development. Sci. Signal, 2016, 9, re3. [DOI] [PubMed] [Google Scholar]
- [14].Kobe B; Kampmann T; Forwood JK; Listwan P; Brinkworth RI Substrate Specificity of Protein Kinases and Computational Prediction of Substrates. Biochim. Biophys. Acta, 2005, 1754, 200–209. [DOI] [PubMed] [Google Scholar]
- [15].Almo SC; Bonanno JB; Sauder JM; Emtage S; Dilorenzo TP; Malashkevich V; Wasserman SR; Swaminathan S; Eswaramoorthy S; Agarwal R; Kumaran D; Madegowda M; Ragumani S; Patskovsky Y; Alvarado J; Ramagopal UA; Faber-Barata J; Chance MR; Sali A; Fiser A; Zhang Z; Lawrence DS; Burley SK Structural Genomics of Protein Phosphatases. J. Struct. Funct. Genomics, 2007, 8, 121–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cohen PT Novel Protein Serine/threonine Phosphatases: Variety Is the Spice of Life. Trends Biochem. Sci, 1997, 22, 245–251. [DOI] [PubMed] [Google Scholar]
- [17].Alonso A; Sasin J; Bottini N; Friedberg I; Friedberg I; Osterman A; Godzik A; Hunter T; Dixon J; Mustelin T Protein Tyrosine Phosphatases in the Human Genome. Cell, 2004, 117, 699–711. [DOI] [PubMed] [Google Scholar]
- [18].Moorhead GBG; De Wever V; Templeton G; Kerk D Evolution of Protein Phosphatases in Plants and Animals. Biochem. J, 2009, 417, 401–409. [DOI] [PubMed] [Google Scholar]
- [19].Rebay I Multiple Functions of the Eya Phosphotyrosine Phosphatase. Mol. Cell. Biol, 2015, 36, 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Barford D; Das AK; Egloff MP The Structure and Mechanism of Protein Phosphatases: Insights into Catalysis and Regulation. Annu. Rev. Biophys. Biomol. Struct, 1998, 27, 133–164. [DOI] [PubMed] [Google Scholar]
- [21].Jung S-K; Jeong DG; Chung SJ; Kim JH; Park BC; Tonks NK; Ryu SE; Kim SJ Crystal Structure of ED-Eya2: Insight into Dual Roles as a Protein Tyrosine Phosphatase and a Transcription Factor. FASEB J., 2010, 24, 560–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Graves DJ; Fischer EH; Krebs EG Specificity Studies on Muscle Phosphorylase Phosphatase. J. Biol. Chem, 1960, 235, 805–809. [PubMed] [Google Scholar]
- [23].Ingebritsen TS; Cohen P The Protein Phosphatases Involved in Cellular Regulation. 1. Classification and Substrate Specificities. Eur. J. Biochem, 1983, 132, 255–261. [DOI] [PubMed] [Google Scholar]
- [24].Shi Y Serine/threonine Phosphatases: Mechanism through Structure. Cell, 2009, 139, 468–484. [DOI] [PubMed] [Google Scholar]
- [25].Huang X; Honkanen RE Molecular Cloning, Expression, and Characterization of a Novel Human Serine/threonine Protein Phosphatase, PP7, That Is Homologous to Drosophila Retinal Degeneration C Gene Product (rdgC). J. Biol. Chem, 1998, 273, 1462–1468. [DOI] [PubMed] [Google Scholar]
- [26].Kennelly PJ Protein Phosphatases--a Phylogenetic Perspective. Chem. Rev, 2001, 101, 2291–2312. [DOI] [PubMed] [Google Scholar]
- [27].Ceulemans H; Bollen M Functional Diversity of Protein Phosphatase-1, a Cellular Economizer and Reset Button. Physiol. Rev, 2004, 84, 1–39. [DOI] [PubMed] [Google Scholar]
- [28].Virshup DM; Shenolikar S From Promiscuity to Precision: Protein Phosphatases Get a Makeover. Mol. Cell, 2009, 33, 537–545. [DOI] [PubMed] [Google Scholar]
- [29].Chen MX; McPartlin AE; Brown L; Chen YH; Barker HM; Cohen PT A Novel Human Protein Serine/threonine Phosphatase, Which Possesses Four Tetratricopeptide Repeat Motifs and Localizes to the Nucleus. EMBO J., 1994, 13, 4278–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kang H; Sayner SL; Gross KL; Russell LC; Chinkers M Identification of Amino Acids in the Tetratricopeptide Repeat and C-Terminal Domains of Protein Phosphatase 5 Involved in Autoinhibition and Lipid Activation. Biochemistry (Mosc.), 2001, 40, 10485–10490. [DOI] [PubMed] [Google Scholar]
- [31].Goldberg Y Protein Phosphatase 2A: Who Shall Regulate the Regulator? Biochem. Pharmacol, 1999, 57, 321–328. [DOI] [PubMed] [Google Scholar]
- [32].Janssens V; Goris J Protein Phosphatase 2A: A Highly Regulated Family of Serine/threonine Phosphatases Implicated in Cell Growth and Signalling. Biochem. J, 2001, 353, 417–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Heroes E; Lesage B; Görnemann J; Beullens M; Van Meervelt L; Bollen M The PP1 Binding Code: A Molecular-Lego Strategy That Governs Specificity. FEBS J., 2013, 280, 584–595. [DOI] [PubMed] [Google Scholar]
- [34].Bollen M; Peti W; Ragusa MJ; Beullens M The Extended PP1 Toolkit: Designed to Create Specificity. Trends Biochem. Sci, 2010, 35, 450–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Caenepeel S; Charydczak G; Sudarsanam S; Hunter T; Manning G The Mouse Kinome: Discovery and Comparative Genomics of All Mouse Protein Kinases. Proc. Natl. Acad. Sci. U. S. A, 2004, 101, 11707–11712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Manning G; Plowman GD; Hunter T; Sudarsanam S Evolution of Protein Kinase Signaling from Yeast to Man. Trends Biochem. Sci, 2002, 27, 514–520. [DOI] [PubMed] [Google Scholar]
- [37].Zulawski M; Schulze G; Braginets R; Hartmann S; Schulze WX The Arabidopsis Kinome: Phylogeny and Evolutionary Insights into Functional Diversification. BMC Genomics, 2014, 15, 548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lesage B; Qian J; Bollen M Spindle Checkpoint Silencing: PP1 Tips the Balance. Curr. Biol. CB, 2011, 21, R898–903. [DOI] [PubMed] [Google Scholar]
- [39].Barker HM; Craig SP; Spurr NK; Cohen PT Sequence of Human Protein Serine/threonine Phosphatase 1 Gamma and Localization of the Gene (PPP1CC) Encoding It to Chromosome Bands 12q24.1-q24.2. Biochim. Biophys. Acta, 1993, 1178, 228–233. [DOI] [PubMed] [Google Scholar]
- [40].Okano K; Heng H; Trevisanato S; Tyers M; Varmuza S Genomic Organization and Functional Analysis of the Murine Protein Phosphatase 1c Gamma (Ppp1cc) Gene. Genomics, 1997, 45, 211–215. [DOI] [PubMed] [Google Scholar]
- [41].Chakrabarti R; Cheng L; Puri P; Soler D; Vijayaraghavan S Protein Phosphatase PP1 Gamma 2 in Sperm Morphogenesis and Epididymal Initiation of Sperm Motility. Asian J. Androl, 2007, 9, 445–452. [DOI] [PubMed] [Google Scholar]
- [42].Lechward K; Awotunde OS; Swiatek W; Muszyńska G Protein Phosphatase 2A: Variety of Forms and Diversity of Functions. Acta Biochim. Pol, 2001, 48, 921–933. [PubMed] [Google Scholar]
- [43].Van Hoof C; Goris J Phosphatases in Apoptosis: To Be or Not to Be, PP2A Is in the Heart of the Question. Biochim. Biophys. Acta BBA - Mol. Cell Res, 2003, 1640, 97–104. [DOI] [PubMed] [Google Scholar]
- [44].Janssens V; Zwaenepoel K; Rossé C; Petit MMR; Goris J; Parker PJ PP2A Binds to the LIM Domains of Lipoma-Preferred Partner through Its PR130/B″ Subunit to Regulate Cell Adhesion and Migration. J. Cell Sci, 2016, 129, 1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Chica N; Rozalén AE; Pérez-Hidalgo L; Rubio A; Novak B; Moreno S Nutritional Control of Cell Size by the Greatwall-Endosulfine-PP2A·B55 Pathway. Curr. Biol. CB, 2016, 26, 319–330. [DOI] [PubMed] [Google Scholar]
- [46].Seshacharyulu P; Pandey P; Datta K; Batra SK Phosphatase: PP2A Structural Importance, Regulation and Its Aberrant Expression in Cancer. Cancer Lett., 2013, 335, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Wang Y; Xia Y; Kuang D; Duan Y; Wang G PP2A Regulates SCF-Induced Cardiac Stem Cell Migration through Interaction with p38 MAPK. Life Sci., 2017, 191, 59–67. [DOI] [PubMed] [Google Scholar]
- [48].Rusnak F; Mertz P Calcineurin: Form and Function. Physiol. Rev, 2000, 80, 1483–1521. [DOI] [PubMed] [Google Scholar]
- [49].Musson REA; Smit NPM Regulatory Mechanisms of Calcineurin Phosphatase Activity. Curr. Med. Chem, 2011, 18, 301–315. [DOI] [PubMed] [Google Scholar]
- [50].Nygren PJ; Scott JD Regulation of the Phosphatase PP2B by Protein-Protein Interactions. Biochem. Soc. Trans, 2016, 44, 1313–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cohen PTW; Philp A; Vázquez-Martin C Protein Phosphatase 4--from Obscurity to Vital Functions. FEBS Lett., 2005, 579, 3278–3286. [DOI] [PubMed] [Google Scholar]
- [52].Voss M; Campbell K; Saranzewa N; Campbell DG; Hastie CJ; Peggie MW; Martin-Granados C; Prescott AR; Cohen PTW Protein Phosphatase 4 Is Phosphorylated and Inactivated by Cdk in Response to Spindle Toxins and Interacts with γ-Tubulin. Cell Cycle Georget. Tex, 2013, 12, 2876–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Martin-Granados C; Philp A; Oxenham SK; Prescott AR; Cohen PTW Depletion of Protein Phosphatase 4 in Human Cells Reveals Essential Roles in Centrosome Maturation, Cell Migration and the Regulation of Rho GTPases. Int. J. Biochem. Cell Biol, 2008, 40, 2315–2332. [DOI] [PubMed] [Google Scholar]
- [54].Xie Y; Jüschke C; Esk C; Hirotsune S; Knoblich JA The Phosphatase PP4c Controls Spindle Orientation to Maintain Proliferative Symmetric Divisions in the Developing Neocortex. Neuron, 2013, 79, 254–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Oler AJ; Cairns BR PP4 Dephosphorylates Maf1 to Couple Multiple Stress Conditions to RNA Polymerase III Repression. EMBO J., 2012, 31, 1440–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hastie CJ; Carnegie GK; Morrice N; Cohen PT A Novel 50 kDa Protein Forms Complexes with Protein Phosphatase 4 and Is Located at Centrosomal Microtubule Organizing Centres. Biochem. J, 2000, 347 Pt 3, 845–855. [PMC free article] [PubMed] [Google Scholar]
- [57].Gingras A-C; Caballero M; Zarske M; Sanchez A; Hazbun TR; Fields S; Sonenberg N; Hafen E; Raught B; Aebersold R A Novel, Evolutionarily Conserved Protein Phosphatase Complex Involved in Cisplatin Sensitivity. Mol. Cell. Proteomics MCP, 2005, 4, 1725–1740. [DOI] [PubMed] [Google Scholar]
- [58].Chen GI; Tisayakorn S; Jorgensen C; D’Ambrosio LM; Goudreault M; Gingras A-C PP4R4/KIAA1622 Forms a Novel Stable Cytosolic Complex with Phosphoprotein Phosphatase 4. J. Biol. Chem, 2008, 283, 29273–29284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Skarra DV; Goudreault M; Choi H; Mullin M; Nesvizhskii AI; Gingras A-C; Honkanen RE Label-Free Quantitative Proteomics and SAINT Analysis Enable Interactome Mapping for the Human Ser/Thr Protein Phosphatase 5. Proteomics, 2011, 11, 1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Schopf FH; Biebl MM; Buchner J The HSP90 Chaperone Machinery. Nat. Rev. Mol. Cell Biol, 2017, 18, 345–360. [DOI] [PubMed] [Google Scholar]
- [61].Vaughan CK; Mollapour M; Smith JR; Truman A; Hu B; Good VM; Panaretou B; Neckers L; Clarke PA; Workman P; Piper PW; Prodromou C; Pearl LH Hsp90-Dependent Activation of Protein Kinases Is Regulated by Chaperone-Targeted Dephosphorylation of Cdc37. Mol. Cell, 2008, 31, 886–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Oberoi J; Dunn DM; Woodford MR; Mariotti L; Schulman J; Bourboulia D; Mollapour M; Vaughan CK Structural and Functional Basis of Protein Phosphatase 5 Substrate Specificity. Proc. Natl. Acad. Sci. U. S. A, 2016, 113, 9009–9014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Shao J; Hartson SD; Matts RL Evidence That Protein Phosphatase 5 Functions to Negatively Modulate the Maturation of the Hsp90-Dependent Heme-Regulated eIF2alpha Kinase. Biochemistry (Mosc.), 2002, 41, 6770–6779. [DOI] [PubMed] [Google Scholar]
- [64].Golden T; Swingle M; Honkanen RE The Role of Serine/threonine Protein Phosphatase Type 5 (PP5) in the Regulation of Stress-Induced Signaling Networks and Cancer. Cancer Metastasis Rev., 2008, 27, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhou G; Golden T; Aragon IV; Honkanen RE Ser/Thr Protein Phosphatase 5 Inactivates Hypoxia-Induced Activation of an Apoptosis Signal-Regulating Kinase 1/MKK-4/JNK Signaling Cascade. J. Biol. Chem, 2004, 279, 46595–46605. [DOI] [PubMed] [Google Scholar]
- [66].Urban G; Golden T; Aragon IV; Scammell JG; Dean NM; Honkanen RE Identification of an Estrogen-Inducible Phosphatase (PP5) That Converts MCF-7 Human Breast Carcinoma Cells into an Estrogen-Independent Phenotype When Expressed Constitutively. J. Biol. Chem, 2001, 276, 27638–27646. [DOI] [PubMed] [Google Scholar]
- [67].Zhang Y; Leung DYM; Nordeen SK; Goleva E Estrogen Inhibits Glucocorticoid Action via Protein Phosphatase 5 (PP5)-Mediated Glucocorticoid Receptor Dephosphorylation. J. Biol. Chem, 2009, 284, 24542–24552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Swingle MR; Honkanen RE; Ciszak EM Structural Basis for the Catalytic Activity of Human Serine/threonine Protein Phosphatase-5. J. Biol. Chem, 2004, 279, 33992–33999. [DOI] [PubMed] [Google Scholar]
- [69].Yang J; Roe SM; Cliff MJ; Williams MA; Ladbury JE; Cohen PTW; Barford D Molecular Basis for TPR Domain-Mediated Regulation of Protein Phosphatase 5. EMBO J., 2005, 24, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Chen MX; Cohen PT Activation of Protein Phosphatase 5 by Limited Proteolysis or the Binding of Polyunsaturated Fatty Acids to the TPR Domain. FEBS Lett., 1997, 400, 136–140. [DOI] [PubMed] [Google Scholar]
- [71].Sinclair C; Borchers C; Parker C; Tomer K; Charbonneau H; Rossie S The Tetratricopeptide Repeat Domain and a C-Terminal Region Control the Activity of Ser/Thr Protein Phosphatase 5. J. Biol. Chem, 1999, 274, 23666–23672. [DOI] [PubMed] [Google Scholar]
- [72].Skinner J; Sinclair C; Romeo C; Armstrong D; Charbonneau H; Rossie S Purification of a Fatty Acid-Stimulated Protein-Serine/threonine Phosphatase from Bovine Brain and Its Identification as a Homolog of Protein Phosphatase 5. J. Biol. Chem, 1997, 272, 22464–22471. [DOI] [PubMed] [Google Scholar]
- [73].Ramsey AJ; Chinkers M Identification of Potential Physiological Activators of Protein Phosphatase 5. Biochemistry (Mosc.), 2002, 41, 5625–5632. [DOI] [PubMed] [Google Scholar]
- [74].Rusin SF; Schlosser KA; Adamo ME; Kettenbach AN Quantitative Phosphoproteomics Reveals New Roles for the Protein Phosphatase PP6 in Mitotic Cells. Sci. Signal, 2015, 8, rs12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zeng K; Bastos RN; Barr FA; Gruneberg U Protein Phosphatase 6 Regulates Mitotic Spindle Formation by Controlling the T-Loop Phosphorylation State of Aurora A Bound to Its Activator TPX2. J. Cell Biol, 2010, 191, 1315–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hu M-W; Wang Z-B; Teng Y; Jiang Z-Z; Ma X-S; Hou N; Cheng X; Schatten H; Xu X; Yang X; Sun Q-Y Loss of Protein Phosphatase 6 in Oocytes Causes Failure of Meiosis II Exit and Impaired Female Fertility. J. Cell Sci, 2015, 128, 3769–3780. [DOI] [PubMed] [Google Scholar]
- [77].Ziembik MA; Bender TP; Larner JM; Brautigan DL Functions of Protein Phosphatase-6 in NF-κB Signaling and in Lymphocytes. Biochem. Soc. Trans, 2017, 45, 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Wengrod J; Wang D; Weiss S; Zhong H; Osman I; Gardner LB Phosphorylation of eIF2α Triggered by mTORC1 Inhibition and PP6C Activation Is Required for Autophagy and Is Aberrant in PP6C-Mutated Melanoma. Sci. Signal, 2015, 8, ra27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Ye J; Shi H; Shen Y; Peng C; Liu Y; Li C; Deng K; Geng J; Xu T; Zhuang Y; Zheng B; Tao W PP6 Controls T Cell Development and Homeostasis by Negatively Regulating Distal TCR Signaling. J. Immunol. Baltim. Md 1950, 2015, 194, 1654–1664. [DOI] [PubMed] [Google Scholar]
- [80].Hammond D; Zeng K; Espert A; Bastos RN; Baron RD; Gruneberg U; Barr FA Melanoma-Associated Mutations in Protein Phosphatase 6 Cause Chromosome Instability and DNA Damage Owing to Dysregulated Aurora-A. J. Cell Sci, 2013, 126, 3429–3440. [DOI] [PubMed] [Google Scholar]
- [81].Stefansson B; Brautigan DL Protein Phosphatase 6 Subunit with Conserved Sit4-Associated Protein Domain Targets IκBϵ. J. Biol. Chem, 2006, 281, 22624–22634. [DOI] [PubMed] [Google Scholar]
- [82].Stefansson B; Ohama T; Daugherty AE; Brautigan DL Protein Phosphatase 6 Regulatory Subunits Composed of Ankyrin Repeat Domains. Biochemistry (Mosc.), 2008, 47, 1442–1451. [DOI] [PubMed] [Google Scholar]
- [83].Andreeva AV; Kutuzov MA PPEF/PP7 Protein Ser/Thr Phosphatases. Cell. Mol. Life Sci. CMLS, 2009, 66, 3103–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Peti W; Nairn AC; Page R Structural Basis for Protein Phosphatase 1 Regulation and Specificity. FEBS J., 2013, 280, 596–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Verbinnen I; Ferreira M; Bollen M Biogenesis and Activity Regulation of Protein Phosphatase 1. Biochem. Soc. Trans, 2017, 45, 89–99. [DOI] [PubMed] [Google Scholar]
- [86].Xu Y; Xing Y; Chen Y; Chao Y; Lin Z; Fan E; Yu JW; Strack S; Jeffrey PD; Shi Y Structure of the Protein Phosphatase 2A Holoenzyme. Cell, 2006, 127, 1239–1251. [DOI] [PubMed] [Google Scholar]
- [87].Chattopadhyay D; Swingle MR; Salter EA; Wood E; D’Arcy B; Zivanov C; Abney K; Musiyenko A; Rusin SF; Kettenbach A; Yet L; Schroeder CE; Golden JE; Dunham WH; Gingras A-C; Banerjee S; Forbes D; Wierzbicki A; Honkanen RE Crystal Structures and Mutagenesis of PPP-Family Ser/thr Protein Phosphatases Elucidate the Selectivity of Cantharidin and Novel Norcantharidin-Based Inhibitors of PP5C. Biochem. Pharmacol, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bertini I; Calderone V; Fragai M; Luchinat C; Talluri E Structural Basis of Serine/threonine Phosphatase Inhibition by the Archetypal Small Molecules Cantharidin and Norcantharidin. J. Med. Chem, 2009, 52, 4838–4843. [DOI] [PubMed] [Google Scholar]
- [89].Lad C; Williams NH; Wolfenden R The Rate of Hydrolysis of Phosphomonoester Dianions and the Exceptional Catalytic Proficiencies of Protein and Inositol Phosphatases. Proc. Natl. Acad. Sci. U. S. A, 2003, 100, 5607–5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhang J; Zhang Z; Brew K; Lee EY Mutational Analysis of the Catalytic Subunit of Muscle Protein Phosphatase-1. Biochemistry (Mosc.), 1996, 35, 6276–6282. [DOI] [PubMed] [Google Scholar]
- [91].Zhang L; Lee EY Mutational Analysis of Substrate Recognition by Protein Phosphatase 1. Biochemistry (Mosc.), 1997, 36, 8209–8214. [DOI] [PubMed] [Google Scholar]
- [92].Martin BL; Jurado LA; Hengge AC Comparison of the Reaction Progress of Calcineurin with Mn2+ and Mg2+. Biochemistry (Mosc.), 1999, 38, 3386–3392. [DOI] [PubMed] [Google Scholar]
- [93].Egloff MP; Cohen PT; Reinemer P; Barford D Crystal Structure of the Catalytic Subunit of Human Protein Phosphatase 1 and Its Complex with Tungstate. J. Mol. Biol, 1995, 254, 942–959. [DOI] [PubMed] [Google Scholar]
- [94].Xing Y; Xu Y; Chen Y; Jeffrey PD; Chao Y; Lin Z; Li Z; Strack S; Stock JB; Shi Y Structure of Protein Phosphatase 2A Core Enzyme Bound to Tumor-Inducing Toxins. Cell, 2006, 127, 341–353. [DOI] [PubMed] [Google Scholar]
- [95].Heroes E; Rip J; Beullens M; Van Meervelt L; De Gendt S; Bollen M Metals in the Active Site of Native Protein Phosphatase-1. J. Inorg. Biochem, 2015, 149, 1–5. [DOI] [PubMed] [Google Scholar]
- [96].Namgaladze D; Hofer HW; Ullrich V Redox Control of Calcineurin by Targeting the Binuclear Fe(2+)-Zn(2+) Center at the Enzyme Active Site. J. Biol. Chem, 2002, 277, 5962–5969. [DOI] [PubMed] [Google Scholar]
- [97].Nishito Y; Usui H; Shinzawa-Itoh K; Inoue R; Tanabe O; Nagase T; Murakami T; Takeda M Direct Metal Analyses of Mn2+-Dependent and -Independent Protein Phosphatase 2A from Human Erythrocytes Detect Zinc and Iron Only in the Mn2+-Independent One. FEBS Lett., 1999, 447, 29–33. [DOI] [PubMed] [Google Scholar]
- [98].King MM; Huang CY The Calmodulin-Dependent Activation and Deactivation of the Phosphoprotein Phosphatase, Calcineurin, and the Effect of Nucleotides, Pyrophosphate, and Divalent Metal Ions. Identification of Calcineurin as a Zn and Fe Metalloenzyme. J. Biol. Chem, 1984, 259, 8847–8856. [PubMed] [Google Scholar]
- [99].Martin BL; Graves DJ Mechanistic Aspects of the Low-Molecular-Weight Phosphatase Activity of the Calmodulin-Activated Phosphatase, Calcineurin. J. Biol. Chem, 1986, 261, 14545–14550. [PubMed] [Google Scholar]
- [100].Hengge AC; Martin BL Isotope Effect Studies on the Calcineurin Phosphoryl-Transfer Reaction: Transition State Structure and Effect of Calmodulin and Mn2+. Biochemistry (Mosc.), 1997, 36, 10185–10191. [DOI] [PubMed] [Google Scholar]
- [101].Mondragon A; Griffith EC; Sun L; Xiong F; Armstrong C; Liu JO Overexpression and Purification of Human Calcineurin Alpha from Escherichia Coli and Assessment of Catalytic Functions of Residues Surrounding the Binuclear Metal Center. Biochemistry (Mosc.), 1997, 36, 4934–4942. [DOI] [PubMed] [Google Scholar]
- [102].Martin B; Pallen CJ; Wang JH; Graves DJ Use of Fluorinated Tyrosine Phosphates to Probe the Substrate Specificity of the Low Molecular Weight Phosphatase Activity of Calcineurin. J. Biol. Chem, 1985, 260, 14932–14937. [PubMed] [Google Scholar]
- [103].Jackson MD; Denu JM Molecular Reactions of Protein Phosphatases--Insights from Structure and Chemistry. Chem. Rev, 2001, 101, 2313–2340. [DOI] [PubMed] [Google Scholar]
- [104].Bertini I; Luchinat C 2 The Reaction Pathways of Zinc Enzymes and Related Biological Catalysts. [Google Scholar]
- [105].Bertini I; Luchinat C; Rosi M; Sgamellotti A; Tarantelli F pKa of Zinc-Bound Water and Nucleophilicity of Hydroxo-Containing Species. Ab Initio Calculations on Models for Zinc Enzymes. Inorg. Chem, 1990, 29, 1460–1463. [Google Scholar]
- [106].Mesecar AD; Stoddard BL; Koshland DE Orbital Steering in the Catalytic Power of Enzymes: Small Structural Changes with Large Catalytic Consequences. Science, 1997, 277, 202–206. [DOI] [PubMed] [Google Scholar]
- [107].Hoff RH; Mertz P; Rusnak F; Hengge AC The Transition State of the Phosphoryl-Transfer Reaction Catalyzed by the Lambda Ser/Thr Protein Phosphatase. J. Am. Chem. Soc, 1999, 121, 6382–6390. [Google Scholar]
- [108].Rowlett RS; Silverman DN Kinetics of the Protonation of Buffer and Hydration of Carbon Dioxide Catalyzed by Human Carbonic Anhydrase II. J. Am. Chem. Soc, 1982, 104, 6737–6741. [Google Scholar]
- [109].Azzi JR; Sayegh MH; Mallat SG Calcineurin Inhibitors: 40 Years Later, Can’t Live without .. J. Immunol. Baltim. Md 1950, 2013, 191, 5785–5791. [DOI] [PubMed] [Google Scholar]
- [110].Barik S Immunophilins: For the Love of Proteins. Cell. Mol. Life Sci. CMLS, 2006, 63, 2889–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Harikishore A; Yoon HS Immunophilins: Structures, Mechanisms and Ligands. Curr. Mol. Pharmacol, 2015, 9, 37–47. [DOI] [PubMed] [Google Scholar]
- [112].Grigoriu S; Bond R; Cossio P; Chen JA; Ly N; Hummer G; Page R; Cyert MS; Peti W The Molecular Mechanism of Substrate Engagement and Immunosuppressant Inhibition of Calcineurin. PLoS Biol., 2013, 11, e1001492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Kissinger CR; Parge HE; Knighton DR; Lewis CT; Pelletier LA; Tempczyk A; Kalish VJ; Tucker KD; Showalter RE; Moomaw EW Crystal Structures of Human Calcineurin and the Human FKBP12-FK506-Calcineurin Complex. Nature, 1995, 378, 641–644. [DOI] [PubMed] [Google Scholar]
- [114].Griffith JP; Kim JL; Kim EE; Sintchak MD; Thomson JA; Fitzgibbon MJ; Fleming MA; Caron PR; Hsiao K; Navia MA X-Ray Structure of Calcineurin Inhibited by the Immunophilin-Immunosuppressant FKBP12-FK506 Complex. Cell, 1995, 82, 507–522. [DOI] [PubMed] [Google Scholar]
- [115].Ke H; Huai Q Structures of Calcineurin and Its Complexes with Immunophilins-Immunosuppressants. Biochem. Biophys. Res. Commun, 2003, 311, 1095–1102. [DOI] [PubMed] [Google Scholar]
- [116].Huai Q; Kim H-Y; Liu Y; Zhao Y; Mondragon A; Liu JO; Ke H Crystal Structure of Calcineurin-Cyclophilin-Cyclosporin Shows Common but Distinct Recognition of Immunophilin-Drug Complexes. Proc. Natl. Acad. Sci. U. S. A, 2002, 99, 12037–12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Takai A; Bialojan C; Troschka M; Rüegg JC Smooth Muscle Myosin Phosphatase Inhibition and Force Enhancement by Black Sponge Toxin. FEBS Lett., 1987, 217, 81–84. [DOI] [PubMed] [Google Scholar]
- [118].Honkanen RE; Zwiller J; Moore RE; Daily SL; Khatra BS; Dukelow M; Boynton AL Characterization of Microcystin-LR, a Potent Inhibitor of Type 1 and Type 2A Protein Phosphatases. J. Biol. Chem, 1990, 265, 19401–19404. [PubMed] [Google Scholar]
- [119].Honkanen RE; Dukelow M; Zwiller J; Moore RE; Khatra BS; Boynton AL Cyanobacterial Nodularin Is a Potent Inhibitor of Type 1 and Type 2A Protein Phosphatases. Mol. Pharmacol, 1991, 40, 577–583. [PubMed] [Google Scholar]
- [120].Suganuma M; Fujiki H; Furuya-Suguri H; Yoshizawa S; Yasumoto S; Kato Y; Fusetani N; Sugimura T Calyculin A, an Inhibitor of Protein Phosphatases, a Potent Tumor Promoter on CD-1 Mouse Skin. Cancer Res., 1990, 50, 3521–3525. [PubMed] [Google Scholar]
- [121].Ishihara H; Martin BL; Brautigan DL; Karaki H; Ozaki H; Kato Y; Fusetani N; Watabe S; Hashimoto K; Uemura D Calyculin A and Okadaic Acid: Inhibitors of Protein Phosphatase Activity. Biochem. Biophys. Res. Commun, 1989, 159, 871–877. [DOI] [PubMed] [Google Scholar]
- [122].Capon RJ; Rooney F; Murray LM; Collins M; Sim ATR; Rostas JAP; Butler MS; Carroll AR Dragmacidins: New Protein Phosphatase Inhibitors from a Southern Australian Deep-Water Marine Sponge, Spongosorites Sp. J. Nat. Prod, 1998, 61, 660–662. [DOI] [PubMed] [Google Scholar]
- [123].Matsuzawa S; Suzuki T; Suzuki M; Matsuda A; Kawamura T; Mizuno Y; Kikuchi K Thyrsiferyl 23-Acetate Is a Novel Specific Inhibitor of Protein Phosphatase PP2A. FEBS Lett., 1994, 356, 272–274. [DOI] [PubMed] [Google Scholar]
- [124].Mamber SW; Okasinski WG; Pinter CD; Tunac JB Antimycotic Effects of the Novel Antitumor Agents Fostriecin (CI-920), PD 113,270 and PD 113,271. J. Antibiot. (Tokyo), 1986, 39, 1467–1472. [DOI] [PubMed] [Google Scholar]
- [125].Burke CP; Haq N; Boger DL Total Synthesis, Assignment of the Relative and Absolute Stereochemistry, and Structural Reassignment of Phostriecin (Aka Sultriecin). J. Am. Chem. Soc, 2010, 132, 2157–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Ohkuma H; Naruse N; Nishiyama Y; Tsuno T; Hoshino Y; Sawada Y; Konishi M; Oki T Sultriecin, a New Antifungal and Antitumor Antibiotic from Streptomyces Roseiscleroticus. Production, Isolation, Structure and Biological Activity. J. Antibiot. (Tokyo), 1992, 45, 1239–1249. [DOI] [PubMed] [Google Scholar]
- [127].Lawhorn BG; Boga SB; Wolkenberg SE; Colby DA; Gauss C-M; Swingle MR; Amable L; Honkanen RE; Boger DL Total Synthesis and Evaluation of Cytostatin, Its C10-C11 Diastereomers, and Additional Key Analogues: Impact on PP2A Inhibition. J. Am. Chem. Soc, 2006, 128, 16720–16732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ozasa T; Tanaka K; Sasamata M; Kaniwa H; Shimizu M; Matsumoto H; Iwanami M Novel Antitumor Antibiotic Phospholine. 2. Structure Determination. J. Antibiot. (Tokyo), 1989, 42, 1339–1343. [DOI] [PubMed] [Google Scholar]
- [129].Kohama T; Enokita R; Okazaki T; Miyaoka H; Torikata A; Inukai M; Kaneko I; Kagasaki T; Sakaida Y; Satoh A Novel Microbial Metabolites of the Phoslactomycins Family Induce Production of Colony-Stimulating Factors by Bone Marrow Stromal Cells. I. Taxonomy, Fermentation and Biological Properties. J. Antibiot. (Tokyo), 1993, 46, 1503–1511. [DOI] [PubMed] [Google Scholar]
- [130].Fushimi S; Nishikawa S; Shimazu A; Seto H Studies on New Phosphate Ester Antifungal Antibiotics Phoslactomycins. I. Taxonomy, Fermentation, Purification and Biological Activities. J. Antibiot. (Tokyo), 1989, 42, 1019–1025. [DOI] [PubMed] [Google Scholar]
- [131].Hori M; Magae J; Han YG; Hartshorne DJ; Karaki H A Novel Protein Phosphatase Inhibitor, Tautomycin. Effect on Smooth Muscle. FEBS Lett., 1991, 285, 145–148. [DOI] [PubMed] [Google Scholar]
- [132].Cheng XC; Kihara T; Ying X; Uramoto M; Osada H; Kusakabe H; Wang BN; Kobayashi Y; Ko K; Yamaguchi I A New Antibiotic, Tautomycetin. J. Antibiot. (Tokyo), 1989, 42, 141–144. [PubMed] [Google Scholar]
- [133].Honkanen RE Cantharidin, Another Natural Toxin That Inhibits the Activity of Serine/threonine Protein Phosphatases Types 1 and 2A. FEBS Lett., 1993, 330, 283–286. [DOI] [PubMed] [Google Scholar]
- [134].Dawson RM The Toxicology of Microcystins. Toxicon, 1998, 36, 953–962. [DOI] [PubMed] [Google Scholar]
- [135].Bishop CT; Anet EF; Gorham PR Isolation and Identification of the Fast-Death Factor in Microcystis Aeruginosa NRC-1. Can. J. Biochem. Physiol, 1959, 37, 453–471. [PubMed] [Google Scholar]
- [136].Konst H; McKercher PD; Gorham PR; Robertson A; Howell J Symptoms and Pathology Produced by Toxic Microcystis Aeruginosa NRC-1 in Laboratory and Domestic Animals. Can. J. Comp. Med. Vet. Sci, 1965, 29, 221–228. [PMC free article] [PubMed] [Google Scholar]
- [137].Fischer WJ; Altheimer S; Cattori V; Meier PJ; Dietrich DR; Hagenbuch B Organic Anion Transporting Polypeptides Expressed in Liver and Brain Mediate Uptake of Microcystin. Toxicol. Appl. Pharmacol, 2005, 203, 257–263. [DOI] [PubMed] [Google Scholar]
- [138].Hastie CJ; Cohen PT Purification of Protein Phosphatase 4 Catalytic Subunit: Inhibition by the Antitumour Drug Fostriecin and Other Tumour Suppressors and Promoters. FEBS Lett., 1998, 431, 357–361. [DOI] [PubMed] [Google Scholar]
- [139].Swingle M; Ni L; Honkanen RE Small-Molecule Inhibitors of Ser/thr Protein Phosphatases: Specificity, Use and Common Forms of Abuse. Methods Mol. Biol. Clifton NJ, 2007, 365, 23–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Prickett TD; Brautigan DL The alpha4 Regulatory Subunit Exerts Opposing Allosteric Effects on Protein Phosphatases PP6 and PP2A. J. Biol. Chem, 2006, 281, 30503–30511. [DOI] [PubMed] [Google Scholar]
- [141].MacKintosh C; Beattie KA; Klumpp S; Cohen P; Codd GA Cyanobacterial Microcystin-LR Is a Potent and Specific Inhibitor of Protein Phosphatases 1 and 2A from Both Mammals and Higher Plants. FEBS Lett., 1990, 264, 187–192. [DOI] [PubMed] [Google Scholar]
- [142].Goldberg J; Huang HB; Kwon YG; Greengard P; Nairn AC; Kuriyan J Three-Dimensional Structure of the Catalytic Subunit of Protein Serine/threonine Phosphatase-1. Nature, 1995, 376, 745–753. [DOI] [PubMed] [Google Scholar]
- [143].Maynes JT; Bateman KS; Cherney MM; Das AK; Luu HA; Holmes CFB; James MNG Crystal Structure of the Tumor-Promoter Okadaic Acid Bound to Protein Phosphatase-1. J. Biol. Chem, 2001, 276, 44078–44082. [DOI] [PubMed] [Google Scholar]
- [144].Valdiglesias V; Prego-Faraldo MV; Pásaro E; Méndez J; Laffon B Okadaic Acid: More than a Diarrheic Toxin. Mar. Drugs, 2013, 11, 4328–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Prego-Faraldo MV; Valdiglesias V; Méndez J; Eirín-López JM Okadaic Acid Meet and Greet: An Insight into Detection Methods, Response Strategies and Genotoxic Effects in Marine Invertebrates. Mar. Drugs, 2013, 11, 2829–2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Bialojan C; Takai A Inhibitory Effect of a Marine-Sponge Toxin, Okadaic Acid, on Protein Phosphatases. Specificity and Kinetics. Biochem. J, 1988, 256, 283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Ni L; Swingle M; Bourgeois AC; Honkanen RE High Yield Expression of Serine/threonine Protein Phosphatase Type 5, and a Fluorescent Assay Suitable for Use in the Detection of Catalytic Inhibitors. Assay Drug Dev. Technol, 2007, 5, 645–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Zhang L; Zhang Z; Long F; Lee EY Tyrosine-272 Is Involved in the Inhibition of Protein Phosphatase-1 by Multiple Toxins. Biochemistry (Mosc.), 1996, 35, 1606–1611. [DOI] [PubMed] [Google Scholar]
- [149].MacKintosh C; Klumpp S Tautomycin from the Bacterium Streptomyces Verticillatus. Another Potent and Specific Inhibitor of Protein Phosphatases 1 and 2A. FEBS Lett., 1990, 277, 137–140. [DOI] [PubMed] [Google Scholar]
- [150].Honkanen RE; Codispoti BA; Tse K; Boynton AL; Honkanan RE Characterization of Natural Toxins with Inhibitory Activity against Serine/threonine Protein Phosphatases. Toxicon Off. J. Int. Soc. Toxinology, 1994, 32, 339–350. [DOI] [PubMed] [Google Scholar]
- [151].Kelker MS; Page R; Peti W Crystal Structures of Protein Phosphatase-1 Bound to Nodularin-R and Tautomycin: A Novel Scaffold for Structure-Based Drug Design of Serine/threonine Phosphatase Inhibitors. J. Mol. Biol, 2009, 385, 11–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Wakimoto T; Matsunaga S; Takai A; Fusetani N Insight into Binding of Calyculin A to Protein Phosphatase 1: Isolation of Hemicalyculin A and Chemical Transformation of Calyculin A. Chem. Biol, 2002, 9, 309–319. [DOI] [PubMed] [Google Scholar]
- [153].Kita A; Matsunaga S; Takai A; Kataiwa H; Wakimoto T; Fusetani N; Isobe M; Miki K Crystal Structure of the Complex between Calyculin A and the Catalytic Subunit of Protein Phosphatase 1. Struct. Lond. Engl. 1993, 2002, 10, 715–724. [DOI] [PubMed] [Google Scholar]
- [154].Lê LH; Erlichman C; Pillon L; Thiessen JJ; Day A; Wainman N; Eisenhauer EA; Moore MJ Phase I and Pharmacokinetic Study of Fostriecin given as an Intravenous Bolus Daily for Five Consecutive Days. Invest. New Drugs, 2004, 22, 159–167. [DOI] [PubMed] [Google Scholar]
- [155].Jackson RC; Fry DW; Boritzki TJ; Roberts BJ; Hook KE; Leopold WR The Biochemical Pharmacology of CI-920, a Structurally Novel Antibiotic with Antileukemic Activity. Adv. Enzyme Regul, 1985, 23, 193–215. [DOI] [PubMed] [Google Scholar]
- [156].Leopold WR; Shillis JL; Mertus AE; Nelson JM; Roberts BJ; Jackson RC Anticancer Activity of the Structurally Novel Antibiotic Cl-920 and Its Analogues. Cancer Res., 1984, 44, 1928–1932. [PubMed] [Google Scholar]
- [157].Walsh AH; Cheng A; Honkanen RE Fostriecin, an Antitumor Antibiotic with Inhibitory Activity against Serine/threonine Protein Phosphatases Types 1 (PP1) and 2A (PP2A), Is Highly Selective for PP2A. FEBS Lett., 1997, 416, 230–234. [DOI] [PubMed] [Google Scholar]
- [158].Swingle MR; Amable L; Lawhorn BG; Buck SB; Burke CP; Ratti P; Fischer KL; Boger DL; Honkanen RE Structure-Activity Relationship Studies of Fostriecin, Cytostatin, and Key Analogs, with PP1, PP2A, PP5, and( beta12-beta13)-Chimeras (PP1/PP2A and PP5/PP2A), Provide Further Insight into the Inhibitory Actions of Fostriecin Family Inhibitors. J. Pharmacol. Exp. Ther, 2009, 331, 45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Buck SB; Hardouin C; Ichikawa S; Soenen DR; Gauss C-M; Hwang I; Swingle MR; Bonness KM; Honkanen RE; Boger DL Fundamental Role of the Fostriecin Unsaturated Lactone and Implications for Selective Protein Phosphatase Inhibition. J. Am. Chem. Soc, 2003, 125, 15694–15695. [DOI] [PubMed] [Google Scholar]
- [160].Takeuchi T; Takahashi N; Ishi K; Kusayanagi T; Kuramochi K; Sugawara F Antitumor Antibiotic Fostriecin Covalently Binds to Cysteine-269 Residue of Protein Phosphatase 2A Catalytic Subunit in Mammalian Cells. Bioorg. Med. Chem, 2009, 17, 8113–8122. [DOI] [PubMed] [Google Scholar]
- [161].Evans DR; Simon JA The Predicted beta12-beta13 Loop Is Important for Inhibition of PP2Acalpha by the Antitumor Drug Fostriecin. FEBS Lett., 2001, 498, 110–115. [DOI] [PubMed] [Google Scholar]
- [162].Moed L; Shwayder TA; Chang MW Cantharidin Revisited: A Blistering Defense of an Ancient Medicine. Arch. Dermatol, 2001, 137, 1357–1360. [DOI] [PubMed] [Google Scholar]
- [163].Wang GS Medical Uses of Mylabris in Ancient China and Recent Studies. J. Ethnopharmacol, 1989, 26, 147–162. [DOI] [PubMed] [Google Scholar]
- [164].Liao YF; Wang Y; Huang Y; Zha SF; Liu JJ; Wang ZK; Yin YP; Liao YF; Wang Y Isolation and Functional Analysis of McMenA, a Gene Encoding a 1,4-dihydroxy-2-napthoate octaprenyltransferase in Mylabris Cichorii. Arch. Insect Biochem. Physiol, 2015, 89, 127–137. [DOI] [PubMed] [Google Scholar]
- [165].Oaks WW; Ditunno JF; Magnani T; Levy HA; Mills LC Cantharidin Poisoning. Arch. Intern. Med, 1960, 105, 574–582. [DOI] [PubMed] [Google Scholar]
- [166].Al-Dawsari NA; Masterpol KS Cantharidin in Dermatology. Skinmed, 2016, 14, 111–114. [PubMed] [Google Scholar]
- [167].Prickett TD; Brautigan DL The α4 Regulatory Subunit Exerts Opposing Allosteric Effects on Protein Phosphatases PP6 and PP2A. J. Biol. Chem, 2006, 281, 30503–30511. [DOI] [PubMed] [Google Scholar]
- [168].Li YM; Mackintosh C; Casida JE Protein Phosphatase 2A and Its [3H]cantharidin/[3H]endothall Thioanhydride Binding Site. Inhibitor Specificity of Cantharidin and ATP Analogues. Biochem. Pharmacol, 1993, 46, 1435–1443. [DOI] [PubMed] [Google Scholar]
- [169].Swingle MR; Honkanen RE Development and Validation of a Robust and Sensitive Assay for the Discovery of Selective Inhibitors for Serine/threonine Protein Phosphatases PP1α (PPP1C) and PP5 (PPP5C). Assay Drug Dev. Technol, 2014, 12, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]