Abstract
Halophilic archaea, such as Halobacterium salinarum and Natronobacterium pharaonis, alter their swimming behavior by phototaxis responses to changes in light intensity and color using visual pigment-like sensory rhodopsins (SRs). In N. pharaonis, SRII (NpSRII) mediates photorepellent responses through its transducer protein, NpHtrII. Here we report the expression of fusions of NpSRII and NpHtrII and fusion hybrids with eubacterial cytoplasmic domains and analyze their function in vivo in haloarchaea and in eubacteria. A fusion in which the C terminus of NpSRII is connected by a short flexible linker to NpHtrII is active in phototaxis signaling for H. salinarum, showing that the fusion does not inhibit functional receptor-transducer interactions. We replaced the cytoplasmic portions of this fusion protein with the cytoplasmic domains of Tar and Tsr, chemotaxis transducers from enteric eubacteria. Purification of the fusion protein from H. salinarum and Tar fusion chimera from Escherichia coli membranes shows that the proteins are not cleaved and exhibit absorption spectra characteristic of wild-type membranes. Their photochemical reaction cycles in H. salinarum and E. coli membranes, respectively, are similar to those of native NpSRII in N. pharaonis. These fusion chimeras mediate retinal-dependent phototaxis responses by Escherichia coli, establishing that the nine-helix membrane portion of the receptor-transducer complex is a modular functional unit able to signal in heterologous membranes. This result confirms a current model for SR-Htr signal transduction in which the Htr transducers are proposed to interact physically and functionally with their cognate sensory rhodopsins via helix-helix contacts between their transmembrane segments.
Halobacteria exhibit phototaxis responses to changes in light intensity and color using the seven-transmembrane retinylidene photoreceptors sensory rhodopsins I and II (SRI and SRII) (8). Light-activated SRI and SRII transmit signals to their cognate transducers, HtrI and HtrII, respectively. The Htr proteins contain two transmembrane helices and cytoplasmic methyl-accepting and His-kinase-activating domains (21, 38) homologous to those of chemotaxis transducers of eubacteria, such as the Escherichia coli and Salmonella enterica serovar Typhimurium Tsr and Tar, chemotaxis transducers for serine and aspartate, respectively (20, 29). Sensory rhodopsin II from Natronobacterium pharaonis (NpSRII) is very similar in spectroscopic and functional properties to the repellent receptor SRII in Halobacterium salinarum, and it has been found to be more stable in response to variation in external conditions such as pH and ionic strength (23). The NpSRII protein mediates a repellent response to blue-green light (maximum λ, 497 nm) when it is coexpressed with its transducer, NpHtrII, in H. salinarum (16). Also, when expressed in E. coli, NpSRII is capable of binding all-trans retinal to form a blue-green-light-absorbing pigment (9, 24).
The SR and Htr proteins appear to be subunits of a stable molecular complex (14, 19), and transducer chimeras show that the specificity of interaction between SRI and HtrI and between SRII and HtrII is encoded in the transmembrane portion of the transducers (39). Based on this finding and studies with mutants, a model for SR-Htr signal transduction has been proposed in which the Htr transducers physically and functionally interact with their cognate sensory rhodopsins via helix-helix contacts within the membrane (27). As a test of this proposal we reasoned that the seven helices of the receptor fused to the two transmembrane helices of the transducer should be sufficient to form a photosignal-transducing module which could couple functionally to the cytoplasmic domain of eubacterial chemotaxis transducers, which have been shown to contain exchangeable cytoplasmic domains with eubacterial homologs (2, 15, 35). We therefore undertook to express both the NpSRII receptor and its transducer, NpHtrII, in a form able to mediate phototaxis signaling in E. coli.
MATERIALS AND METHODS
Bacterial strains and plasmids.
E. coli strains were grown in Luria-Bertani medium at 30 or 37°C, and halobacterial strains were grown in complex medium (CM) (pH 6.0) medium at 37°C (28).
The wild-type proteins and the fusion constructs of NpSRII and NpHtrII were expressed under the fdx or bop promoter by transformation of the H. salinarum strain Pho81Wr− (31) with pKJ305 derivatives (11). The NpSRII and NpSRII-NpHtrII-StTar or -EcTsr fusion chimeras were expressed under the plac1 promoter of pMS107 (30) in E. coli strains RP437 (wild type for chemotaxis) and UT5600 (5). E. coli DH5α was used for cloning the cytoplasmic portion of StTar from the plasmid pMS107 (provided by Jeffry Stock, Princeton University) and the cytoplasmic portion of EcTsr from the plasmid JC3 (provided by John S. Parkinson, University of Utah).
Construction of plasmids encoding the fusion and fusion chimera proteins.
Restriction enzymes and T4 ligase were from Promega (Madison, Wis.), and Pfu DNA polymerase was from Stratagene (La Jolla, Calif.). Oligonucleotides were purchased from Fisher-Genosys (The Woodlands, Tex.). Mevinolin was a gift from A. W. Alberts (Merck Sharp & Dohme, Whitehouse Station, N.J.), and ampicillin was from Sigma (St. Louis, Mo.).
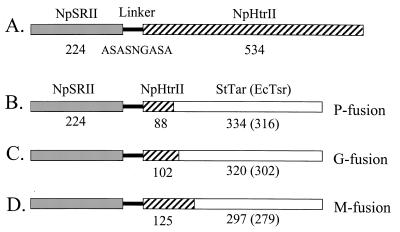
Recombinant PCR was used to introduce 27 nucleotides encoding a 9-amino-acid residue linker (Ala-Ser-Ala-Ser-Asn-Gly-Ala-Ser-Ala; 5′-GCGTCGGCGTCGAACGGCGCGTCGGCG-3′) (10) between the C-terminal residue of NpSRII and the N-terminal residue of NpHtrII (Fig. 1). The linker replaced the C-terminal 15 residues of NpSRII, which we have found to be dispensable for normal signaling function in H. salinarum.
FIG. 1.
Construction of the fusion proteins. Numbers indicate numbers of residues in the indicated segment. P, G, and M fusions (named for the final residue in the NpHtrII portion at the junctions) are defined by the different junctions between the haloarchaeal and eubacterial protein domains.
Three different junctions were used for the chimera constructs (P, G, and M fusion) (Fig. 1) for E. coli expression based on studies of dimerization (6, 32) and the conservation in the region linking the second transmembrane helix and the methylation domain (in bacterial chemotaxis this region is named after the domain present in histidine kinases, adenylyl cyclases, methyl-accepting proteins, and phosphatase [HAMP domain] [1]; we will refer to it as the stimulus relay domain [8] to avoid confusion with the nine-residue linker joining NpSRII and NpHtrII in this study). Nucleotides encoding the N-terminal portion of NpHtrII and C-terminal portion of S. enterica serovar Typhimurium Tar (StTar) or E. coli Tsr (EcTsr) were fused by overlapping primers (top forward and bottom backward primers) in which the first 20 bases derive from N. pharaonis htrII and bases 21 to 40 are from tar or tsr. The first round of PCR was performed at 31 cycles of 95°C for 1 min, 55°C 1 for min, and 72°C for 2 min, with a primer encoding the N-terminal portion of NpSRII (M fusion, 5′-GATATACAT-ATGGTGGGACTTACGACCTC-3′) and a backward overlapping primer (M fusion of StTar, 5′-TCAATCAGCGAGCGTTGCAT-ATGGTCGAAGGCCGCATAGA-3′), along with a forward overlapping primer (5′-TCTATGCGGCCTTCGACCAT-ATGCAACGCTCGCTGATTGA-3′) and primer encoding the C- terminal portion of the eubacterial transducer (5′-GGCGGAGGCGATTTCGCCC-3′). The conditions for the second round of recombinant PCR (7, 36) consisted of 31 cycles of 95°C for 1 min, 50°C for 1 min, and 72°C for 5 min with two first-round PCR products. The recombinant PCR product was purified and digested with NdeI (CATATG) and inserted into the E. coli expression vector under the control of isopropyl-β-d-thiogalactopyranoside (IPTG) induction.
All of the cloned PCR products were sequenced to confirm that no mutations were introduced during the PCR amplification.
Motion analysis.
The swimming behavior of cells was monitored by a computerized cell-tracking system (Motion Analysis Systems, Santa Rosa, Calif.). Early stationary phase cultures of halobacterial cells were diluted 1:13 in fresh CM (pH 6.0), incubated for 0.5 to 2 h at 37°C with agitation, and analyzed as described previously (26).
Transformed E. coli cells were grown to an optical density at 600 nm of 0.4 in Luria-Bertani medium supplemented with 50 μg of ampicillin/ml at 30°C on a rotating incubator followed by the addition of 1 mM IPTG and 1 to 2 μM all-trans retinal. At 2 h postinduction in the dark, cells carrying the fusion proteins in a wild-type background (RP437) were diluted 1:10 in chemotaxis motility buffer (20 mM potassium phosphate, 0.1 mM K-EDTA [pH 7.0]) and analyzed for their swimming responses to phototaxis stimuli using computerized motion analysis as described previously (13).
Phototaxis stimuli were delivered through a Nikon 100W He/Xe short arc lamp. Each stimulus was a pulse of 100 ms or 2 to 4 s of 500-nm light (500 ± 5 nm; 1.8 × 106 ergs/cm2) delivered to the cell in an infrared monitoring light (>750 nm). Data were collected and processed by a SUN SPARC-IPC workstation (SUN Microsystems, Mountain View, Calif.).
Membrane vesicle preparation.
The halobacterial cells containing wild-type and fusion proteins of NpHtrII and NpSRII were grown in 200 ml of CM in a 250-ml flask with mevinolin (1 μg/ml) at 37°C on a gyratory shaker at 240 rpm for 5 to 7 days in the presence of the light. Membrane vesicles were prepared by sonication as described previously (4).
Transformed E. coli RP437 and UT5600 cells were grown at 30°C. Cells were induced by adding 1 mM IPTG and 5 μM all-trans retinal. After an induction period of 3.5 h the cells were harvested, resuspended in 50 mM Tris containing 10% glycerol, and stored at −20°C. Cells were lysed by sonication at 4°C in the presence of 2 mM phenylmethylsulfonylfluoride, 1 μM leupeptin, 1 μM pepstatin, 2 to 5 mM 1,10-phenanthroline, 300 μM p-hydroxymecuribenzoate, and 2 to 5 mM EDTA additives at their final respective concentrations in 50 mM Tris buffer, pH 8.0.
E. coli membranes were sedimented at 100,000 × g for 1 h at 4°C, and the pellet was resuspended in buffer containing 2 M NaCl and again pelleted by centrifugation at 100,000 × g for 30 min at 4°C. Yields of 10 to 12 mg of total membrane protein per g of frozen cell paste were typical. The salt wash enriched ∼2-fold the integral membrane proteins by removal of peripheral membrane proteins. The fusion protein was extracted from the washed membranes with gentle shaking in extraction buffer (1% octyl glucoside, 300 mM NaCl, 10% glycerol, 50 mM Tris buffer [pH 8.0] containing 1 mM phenylmethylsulfonyl fluoride and 1 μM [each] pepstatin and leupeptin) for 4 to 6 h at 4°C, and the solubilized fraction was collected as supernatant after centrifugation at 20,000 × g. The yield of solubilized membrane proteins was ∼70%. Protein assays for membrane preparations were performed with the membrane protein estimation kit from Bio-Rad (Hercules, Calif.). The amount of fusion protein was assessed by using flash photolysis signals at 500 nm calibrated with NpSRII in 1% octyl glucoside, 300 mM NaCl, and 50 mM morpholinoethanesulfonic acid (MES) buffer, pH 6.8.
Purification of His-tagged fusion protein.
The NpSRII-NpHtrII fusion protein containing six histidine residues at the C terminus was purified by using Ni-nitrilotriacetic acid (NTA)–agarose beads. The halobacterial membrane was solubilized in 1% octyl glucoside with 300 mM NaCl, 5 mM imidazole, and 50 mM MES, pH 6.0, and incubated with beads at 4°C for 16 h. The protein-bound beads were washed with 5 mM imidazole and eluted by 300 mM imidazole, 1% octyl glucoside, and 50 mM Tris buffer, pH 7.5. E. coli fusion chimeras containing nine histidine residues at the C terminus were purified by Ni2+-NTA resins. The fusion chimeras were solubilized from the E. coli membrane with extraction buffer at 4°C for 8 to 10 h. Protein-bound beads were incubated with 5 mM imidazole in extraction buffer to wash out proteins bound nonspecifically. Finally, the protein was eluted by using 100 mM imidazole in extraction buffer. Purified proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using anti-His-tag antibody.
Photocycle measurements.
Flash-induced absorption changes were measured with a laboratory-constructed cross-beam flash spectrophotometer (26), with a frequency-doubled Nd-YAG laser (532 nm, 6 ns, 40 mJ) providing the actinic flash. Thirty-two or sixty-four transients were recorded and averaged at a constant temperature (18°C) with a Nicolet (Madison, Wis.) Integra 20. The amplitudes and half-life values were calculated by fitting of single or double exponential curve fitting programs from Igor Pro version 3.1 (WaveMetrics, Lake Oswego, Ore.).
RESULTS AND DISCUSSION
Expression of fusion proteins and fusion chimera proteins.
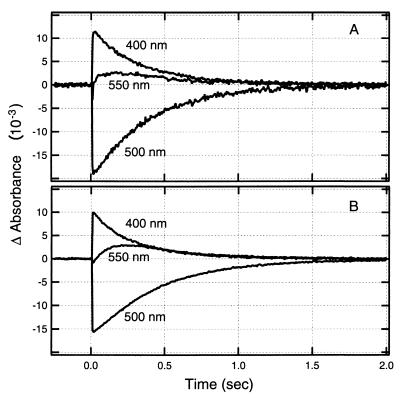
The NpSRII-NpHtrII fusion and NpSRII-NpHtrII-StTar fusion chimera proteins were expressed in H. salinarum and E. coli, respectively. As an initial assessment of photochemical activity and expression level, we analyzed H. salinarum and E. coli envelope membranes for their transient absorption changes following a laser flash at 532 nm (Fig. 2). No flash-induced absorbance changes in the millisecond-to-second time window were detected in membranes from either organism when NpSRII was not present (data not shown). In both membranes, reactions characteristic of NpSRII in its native membrane (3, 34) were observed, namely, an absorbance transient at 400 nm with a decay half-life of <100 ms, indicative of formation and decay of a deprotonated Schiff base form (M) of the pigment, and a slower-decaying transient at 550 nm characteristic of the final intermediate (O), which decays back into the dark state of the pigment as seen at 500 nm. The O decay of NpSRII-NpHtrII in the H. salinarum membrane is the slowest step in the photocycle (362-ms half-life) (Fig. 2). Due to the greater light scattering of the E. coli membranes, assessment of NpSRII-NpHtrII-StTar required addition of 1% octyl glucoside, after which clear photocycle signals were obtained showing an O decay of 365 ms (Fig. 2). When assayed under identical conditions (4 M NaCl and 1% octyl glucoside), the O decays of NpSRII-NpHtrII and NpSRII-NpHtrII-StTar were nearly identical at 694 and 640 ms, respectively. The NpSRII protein alone when expressed in Pho81Wr− showed photochemistry similar to that of the fusion protein (data not shown). We observed no large effect of NpHtrII on NpSRII photochemistry. The lack of a significant effect of the transducer on NpSRII photocycling differs from the case of HsHtrI and HsSRI, in which the transducer protein HtrI increases light-induced production of the photocycle intermediate S373 (an SRI signaling conformation) and modulates the rate of return of S373 to the prestimulus state, rendering this return pH independent (26). Similarly, only a small effect of HsHtrII on HsSRII was observed previously for the corresponding H. salinarum proteins (22). The photocycling rate of NpSRII is therefore not greatly affected by its being fused to the NpHtrII protein or by its being expressed in the E. coli membrane, despite its very different lipid composition from that of H. salinarum.
FIG. 2.
Flash-induced absorption difference transients. Absorption transients of the halobacterial (A) and E. coli (B) membranes were measured at 400, 500, and 550 nm at 5-ms time resolution. Membranes containing the halobacterial NpSRII-NpHtrII fusion protein were resuspended at a concentration of 0.12 mg of protein/ml in a solution containing 4 M NaCl and 25 mM Tris, pH 6.8, and membranes containing the E. coli NpSRII-NpHtrII-StTar fusion were resuspended at a concentration of 0.11 mg of protein/ml in a solution containing 1% octyl-glucoside, 100 mM NaCl, and 100 mM MES, pH 6.8. The half-life values for O-rise and decay are, respectively, 29 and 362 ms for the NpSRII-NpHtrII fusion and 50 and 365 ms for the NpSRII-NpHtrII-StTar fusion.
From the flash photolysis data we can calculate the level of expression of photoactive fusion proteins. Assuming that the extinction coefficient and flash yield from the NpSRII part of the fusion are the same as those in H. salinarum membranes, we calculate 1.6 × 104 molecules/cell for the fusion in H. salinarum and 1.4 × 104 molecules/cell for the fusion chimera in E. coli. These values are consistent with the purification yields of the NpSRII-NpHtrII fusion in H. salinarum, 1.0 mg/liter of culture, and of the NpSRII-NpHtrII-StTar fusion in E. coli, of 0.8 to 1.0 mg/liter of culture.
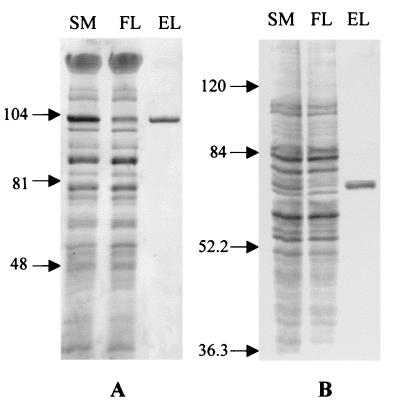
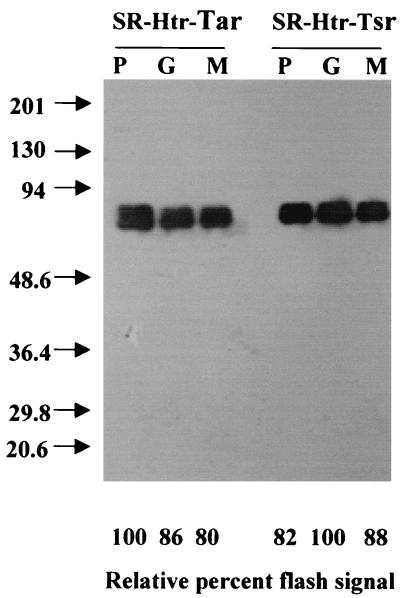
Polyhistidine tags were engineered on the C termini of the NpSRII-NpHtrII fusion and NpSRII-NpHtrII-StTar fusion chimera proteins, and Ni2+-affinity chromatography was applied to purify these octyl-glucoside-extracted membrane proteins from the host cells. The 110-kDa NpSRII-NpHtrII fusion protein from H. salinarum and the 72-kDa NpSRII-NpHtrII-StTar fusion protein from E. coli were present in imidazole-eluted material and migrated as single Coomassie-stained bands on SDS-PAGE (Fig. 3) and as single bands on immunoblots (Fig. 4). There was no indication of degradation to smaller-size His-tagged products in either case and in particular no indication of cleavage of the linker that would release NpHtrII or NpHtrII-EcTsr molecules. The similar flash yields (Fig. 4) indicate uniform expression of protein in the various fusion chimeras.
FIG. 3.
Expression of the His-tagged fusion proteins in H. salinarum (A) and E. coli (B) analyzed by SDS–8% PAGE. SM, solubilized membrane; FL, flowthrough after sample was bound to the resin; EL, eluate with imidazole. A comparable amount of protein (10 μg) eluted by imidazole was loaded in each case and was taken from the same samples that were used for the absorption spectra measurement of Fig. 5. An equal number and 2.5× the number of cell equivalents in the samples were loaded in the eluate lanes of panels A and B, respectively.
FIG. 4.
Immunoblot analysis of the His-tagged fusion proteins in E. coli. An identical amount of membrane protein in each lane (4 μg) was separated by SDS–12% PAGE. The immunoblot used anti-His-tag antibody. Relative percent flash yields were calculated from the maximum laser flash-induced depletion values at 500 nm. P, G, and M fusions are defined by the fusion junction position as shown in Fig. 1.
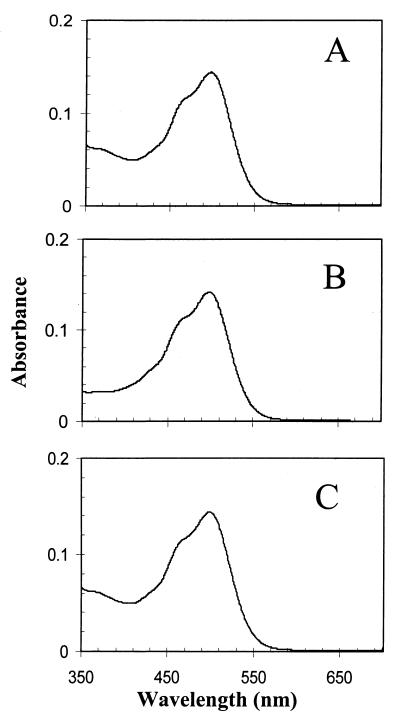
The absorption spectra of the purified fusion proteins from H. salinarum and E. coli are very similar to each other and to that of purified NpSRII (34). Each shows a vibrational fine structure (31) with a main band at 501 nm (H. salinarum fusion) and at 503 nm (E. coli fusion chimera) (Fig. 5). The presence of a vibrational fine structure is a characteristic feature of SRIIs and has not been observed in other archaeal rhodopsins. The absorption spectra of the proteins extracted in nondenaturing detergent and the flash photolysis data from the membrane preparations argue for proper insertion and folding of the NpSRII portion of the fusion proteins in both H. salinarum and E. coli.
FIG. 5.
Absorption spectra of purified His-tagged fusion proteins. (A) NpSRII protein in E. coli membrane. The absorption maximum is at 503 nm. (B) NpSRII-NpHtrII fusion protein. The absorption maximum is at 501 nm. (C) NpSRII-NpHtrII-StTar fusion chimera. The absorption maximum at is 503 nm. Matching molar concentrations of the different proteins were used in the spectral measurements.
Phototaxis responses in H. salinarum.
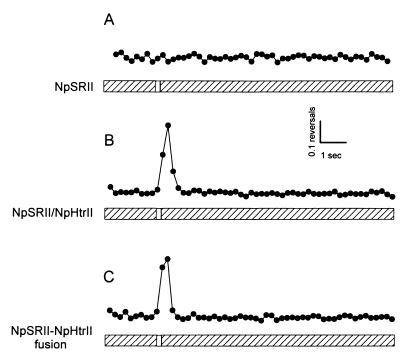
Pho81Wr− cells carrying either NpSRII, NpSRII and NpHtrII, or the NpSRII-NpHtrII fusion were analyzed for their response to blue-green (500 nm) photostimuli. As expected, Pho81Wr− cells expressing NpSRII alone did not show a response; their frequency of swimming reversals, by which they reorient swimming direction when exposed to a repellent stimulus, was unaffected by a pulse of light (Fig. 6A). The cells exhibited a strong repellent response, evident as a high reversal frequency in the population of cells peaking at ∼400 ms after the pulse, when the wild-type N. pharaonis NpHtrII and NpSRII proteins were heterologously coexpressed in H. salinarum (Fig. 6B), confirming functional interaction in vivo between NpSRII, NpHtrII, and the H. salinarum cytoplasmic signal transduction proteins, as reported in a previous study (16). Furthermore, the NpSRII-NpHtrII fusion mediates normal repellent phototaxis responses in H. salinarum (Fig. 6C). Therefore, the flexible linker does not inhibit the functional interaction between the receptor and the transducer.
FIG. 6.
Phototaxis responses for H. salinarum. Two seconds after initiation of data collection, a 500-nm photostimulus was delivered to the cells for 100 ms. Five points per second were collected and used to calculate the reversal frequency. Response data are the averages for 16 repetitive stimuli delivered every 1 min.
Phototaxis responses in E. coli.
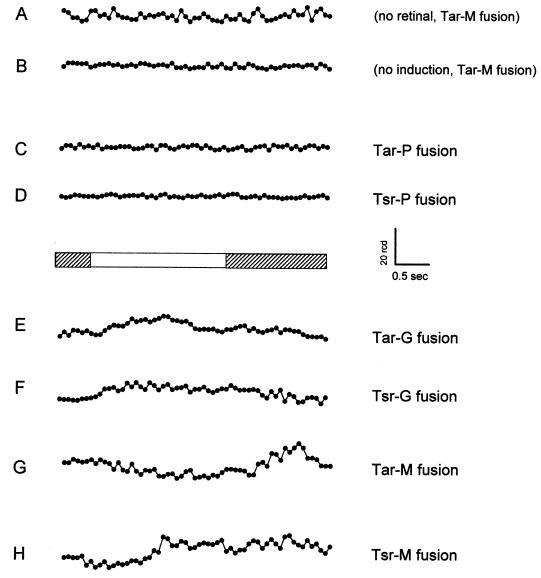
The NpSRII-NpHtrII-StTar fusion chimeras and the NpSRII-NpHtrII-EcTsr fusion chimeras were tested for function in E. coli cells by monitoring their swimming behavior in response to a step-up and step-down to blue-green light. E. coli cells reorient their swimming direction by an erratic subsecond motion called tumbling, and attractant and repellent chemotaxis stimuli, respectively, inhibit and augment tumbling frequency in a population of cells. Tumbling frequency was not affected by light in cells containing the fusion chimeras without the addition of retinal (shown for NpSRII-NpHtrII-StTar in Fig. 7A). Also, no light effect was observed without induction (Fig. 7B), nor was it observed with either of the P fusions (Table 1 and Fig. 7C and D). These four sets of data (Fig. 7A to D) illustrate the variation in the signal/noise ratio that we observed with assays of the cell populations used in these experiments, ranging from the most stable (Fig. 7D) to least stable (Fig. 7A) signals that we obtained.
FIG. 7.
Phototaxis responses by E. coli containing fusion chimeras. One-half second after initiation of data collection, cells with 1 μM retinal (except row A) were stimulated with 2 s of 500-nm continuous light for recording the attractant or repellent responses of Tar-M fusion (A and B), Tar-P and Tsr-P fusion (C and D), Tar-G and Tsr-G fusion (E and F), and Tar-M and Tsr-M fusion (G and H). Tracking was set at 15 frames per second, and the rcd (degrees/second) was plotted. Response data are the averages for 32 repetitive stimuli delivered every 30 s.
TABLE 1.
Motility responses for fusion chimeras
| Fusion construct | Response with junction
|
||
|---|---|---|---|
| P fusion | G fusion | M fusion | |
| NpSRII-NpHtrII-StTar | None | Weak repellent | Attractant |
| NpSRII-NpHtrII-EcTsr | None | Weak repellent | Repellent |
A similar population to which retinal had been added exhibited suppression of tumbling by 500-nm light and a clear enhancement of tumbling behavior peaking ∼1 s after the light was extinguished (Fig. 7G). Light is therefore acting as an attractant stimulus through the NpSRIItr-NpHtrII-StTar molecule. On the other hand, the NpSRII-NpHtrII-EcTsr protein mediates a repellent response to the same light, evident as an increase in tumbling frequency during the illumination period (Fig. 7F and H). Repellent responses (Fig. 7E) are also seen for the fusion chimera with the Tar cytoplasmic domain when the splice point is changed (G fusion) (Fig. 1 and Table 1). The differences in motility responses among constructions with differing junctions in the signal relay domain (Table 1) point to this region of the transducers as being important in controlling the conformations of their CheW/A kinase-binding domains.
The responses shown in Fig. 7 are small in comparison to those of E. coli bacteria stimulated with photoreleased chemoeffectors (13), which produced ∼10-fold-larger rate-of-change-of-direction (rcd) amplitudes. The rcd as well as the signal/noise ratio are limited by the sluggish motility of the cells, which is caused by the high concentration of retinal that must be added to reconstitute the receptor in E. coli in vivo. A more quantitative assessment of SR-mediated responses in E. coli would be facilitated by use of a strain engineered to produce retinal endogenously, such as has been accomplished recently (37) using plasmid-encoded β-carotene and oxidative cleavage enzymes.
These results demonstrate that the fusion chimeras functionally couple to the CheW/A/Y taxis machinery in E. coli. One interesting question in these experiments was whether the repellent nature of the NpSRII-NpHtrII signal seen in H. salinarum would translate to a repellent signal in E. coli regardless of the origin of the cytoplasmic domain. The answer is that either attractant or repellent signals may be generated depending on the particular cytoplasmic domain selected and the position of the splice point. For example, Tar-G and Tar-M (Table 1) mediate opposite responses (Fig. 7E and G), despite the fact that they differ only in the splice position, with the latter containing 23 more NpHtrII residues and 23 fewer StTar residues than the former.
The irregular nature of the sign of the response may indicate that subtle forces set the bias in the kinase-activating versus kinase-inhibiting conformations of the two cytoplasmic domains that couple to CheW/A. Both Tar (“taxis to aspartate and repellents”) (25) and Tsr (“taxis to serine and repellents”) are dual-function attractant or repellent receptors depending on the nature of the chemoeffector, and therefore an equilibrium mixture of their two conformations must be maintained in order to be able to shift in either direction depending on the ligand. A metastable equilibrium between two conformations that can be shifted by small energy changes in the proteins has been indicated by studies of mutant chemotaxis receptors (18, 33) and by our studies of response inversion in the SRI-HtrI complex in H. salinarum (12). SRI-HtrI is also a dual-function attractant and repellent receptor; the sign of the signal in this case depends on the spectral quality of the light, which determines whether one or two photons are absorbed. Single mutations, resulting in one case in an addition of a single methyl group to residue I61V, I64V, or V71I in HtrI, switches the complex from repellent to attractant signaling in response to orange light activation (12).
The haloarchaeal transducer chimeras demonstrated that the SRI and SRII interaction specificities are encoded in the transmembrane portions of HtrI and HtrII, respectively (39), suggesting that the surfaces of physical interaction between the SR and Htr proteins also reside in the transmembrane region. However, the presence of a receptor interaction domain on the cytoplasmic portion of the transducers, as was suggested based on deletion experiments (14), could not be completely ruled out by the swapping of HtrI and HtrII portions, if such a domain were to be conserved between HtrI and HtrII and not exhibit SR specificity. The results obtained here strongly support the chimeral evidence for interaction between the transmembrane helices, because one would not expect any natural interaction between NpSRII and the Tar or Tsr cytoplasmic domains from enteric eubacteria. Evidently the seven helices of the NpSRII receptor fused to the two transmembrane helices of the transducer constitute a membrane-embedded mobile photosignaling module sufficient in itself to convert light into a conformational signal, which is transmittable to evolutionarily very distant transducer cytoplasmic domains. The function of this module in E. coli provides a potential in vitro assay of the SR-Htr signal relay based on the E. coli chemotaxis system, for which in vitro reactions are well established (17).
ACKNOWLEDGMENTS
We thank Shahid Khan for his current motion analysis program of E. coli swimming behavior and Michael Manson, John S. Parkinson, and Jeffry Stock for E. coli and S. enterica serovar Typhimurium plasmids and strains. We also thank Jun Sasaki for early work on htrII-sopII cloning from N. pharaonis DNA and Chii-Shen Yang for comments.
This work was supported by National Institutes of Health grant R01GM27750 to J.L.S.
REFERENCES
- 1.Aravind L, Ponting C P. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol Lett. 1999;176:111–116. doi: 10.1111/j.1574-6968.1999.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 2.Baumgartner J W, Kim C, Brissette R E, Inouye M, Park C, Hazelbauer G L. Transmembrane signalling by a hybrid protein: communication from the domain of chemoreceptor Trg that recognizes sugar-binding proteins to the kinase/phosphatase domain of osmosensor EnvZ. J Bacteriol. 1994;176:1157–1163. doi: 10.1128/jb.176.4.1157-1163.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bivin D B, Stoeckenius W. Photoactive retinal pigments in haloalkaliphilic bacteria. J Gen Microbiol. 1986;132(Pt. 8):2167–2177. doi: 10.1099/00221287-132-8-2167. [DOI] [PubMed] [Google Scholar]
- 4.Bogomolni R A, Spudich J L. Spectroscopic assays for sensory rhodopsin I and II in Halobacterium salinarium cells and enriched membrane preparations. In: Robb F T, Place A R, Sowers K R, Schreier H J, DasSarma S, Fleischmann E M, editors. Archaea: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 63–73. [Google Scholar]
- 5.Elish M E, Pierce J R, Earhart C F. Biochemical analysis of spontaneous fepA mutants of Escherichia coli. J Gen Microbiol. 1988;134(Pt. 5):1355–1364. doi: 10.1099/00221287-134-5-1355. [DOI] [PubMed] [Google Scholar]
- 6.Gardina P J, Manson M D. Attractant signaling by an aspartate chemoreceptor dimer with a single cytoplasmic domain. Science. 1996;274:425–426. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi R. Recombinant PCR. In: Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocol: a guide to methods and applications. San Diego, Calif: Academic Press, Inc.; 1990. pp. 177–183. [Google Scholar]
- 8.Hoff W D, Jung K H, Spudich J L. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 9.Hohenfeld I P, Wegener A A, Engelhard M. Purification of histidine tagged bacteriorhodopsin, pharaonis halorhodopsin and pharaonis sensory rhodopsin II functionally expressed in Escherichia coli. FEBS Lett. 1999;442:198–202. doi: 10.1016/s0014-5793(98)01659-7. [DOI] [PubMed] [Google Scholar]
- 10.Jones P C, Fillingame R H. Genetic fusions of subunit c in the F0 sector of H+-transporting ATP synthase. Functional dimers and trimers and determination of stoichiometry by cross-linking analysis. J Biol Chem. 1998;273:29701–29705. doi: 10.1074/jbc.273.45.29701. [DOI] [PubMed] [Google Scholar]
- 11.Jung K H, Spudich J L. Protonatable residues at the cytoplasmic end of transmembrane helix-2 in the signal transducer HtrI control photochemistry and function of sensory rhodopsin I. Proc Natl Acad Sci USA. 1996;93:6557–6561. doi: 10.1073/pnas.93.13.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung K H, Spudich J L. Suppressor mutation analysis of the sensory rhodopsin I-transducer complex: insights into the color-sensing mechanism. J Bacteriol. 1998;180:2033–2042. doi: 10.1128/jb.180.8.2033-2042.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S, Amoyaw K, Spudich J L, Reid G P, Trentham D R. Bacterial chemoreceptor signaling probed by flash photorelease of a caged serine. Biophys J. 1992;62:67–68. doi: 10.1016/S0006-3495(92)81781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krah M, Marwan W, Oesterhelt D. A cytoplasmic domain is required for the functional interaction of SRI and HtrI in archaeal signal transduction. FEBS Lett. 1994;353:301–304. doi: 10.1016/0014-5793(94)01068-4. [DOI] [PubMed] [Google Scholar]
- 15.Krikos A, Conley M P, Boyd A, Berg H C, Simon M I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luttenberg B, Wolff E K, Engelhard M. Heterologous coexpression of the blue light receptor psRII and its transducer pHtrII from Natronobacterium pharaonis in the Halobacterium salinarium strain Pho81/w restores negative phototaxis. FEBS Lett. 1998;426:117–120. doi: 10.1016/s0014-5793(98)00322-6. [DOI] [PubMed] [Google Scholar]
- 17.Ninfa E G, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 18.Nishiyama S, Nara T, Homma M, Imae Y, Kawagishi I. Thermosensing properties of mutant aspartate chemoreceptors with methyl-accepting sites replaced singly or multiply by alanine. J Bacteriol. 1997;179:6573–6580. doi: 10.1128/jb.179.21.6573-6580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olson K D, Spudich J L. Removal of the transducer protein from sensory rhodopsin I exposes sites of proton release and uptake during the receptor photocycle. Biophys J. 1993;65:2578–2585. doi: 10.1016/S0006-3495(93)81295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parkinson J S. Signal transduction schemes of bacteria. Cell. 1993;73:857–871. doi: 10.1016/0092-8674(93)90267-t. [DOI] [PubMed] [Google Scholar]
- 21.Rudolph J, Oesterhelt D. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarium. EMBO J. 1995;14:667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki J, Spudich J L. The transducer protein HtrII modulates the lifetimes of sensory rhodopsin II photointermediates. Biophys J. 1998;75:2435–2440. doi: 10.1016/S0006-3495(98)77687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scharf B, Pevec B, Hess B, Engelhard M. Biochemical and photochemical properties of the photophobic receptors from Halobacterium halobium and Natronobacterium pharaonis. Eur J Biochem. 1992;206:359–366. doi: 10.1111/j.1432-1033.1992.tb16935.x. [DOI] [PubMed] [Google Scholar]
- 24.Shimono K, Iwamoto M, Sumi M, Kamo N. Functional expression of pharaonis phoborhodopsin in Escherichia coli. FEBS Lett. 1997;420:54–56. doi: 10.1016/s0014-5793(97)01487-7. [DOI] [PubMed] [Google Scholar]
- 25.Springer M S, Goy M F, Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci USA. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spudich E N, Spudich J L. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J Biol Chem. 1993;268:16095–16097. [PubMed] [Google Scholar]
- 27.Spudich J L. Variations on a molecular switch: transport and sensory signalling by archaeal rhodopsins. Mol Microbiol. 1998;28:1051–1058. doi: 10.1046/j.1365-2958.1998.00859.x. [DOI] [PubMed] [Google Scholar]
- 28.Spudich J L, Spudich E N. Selection and screening methods for halophilic archaeal rhodopsin mutants. In: Robb F T, Place A R, Sowers K R, Schreier H J, DasSarma S, Fleischmann E M, editors. Archaea: a laboratory manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1995. pp. 23–28. [Google Scholar]
- 29.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology, second ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1103–1129. [Google Scholar]
- 30.Surette M G, Stock J B. Role of alpha-helical coiled-coil interactions in receptor dimerization, signaling, and adaptation during bacterial chemotaxis. J Biol Chem. 1996;271:17966–17973. doi: 10.1074/jbc.271.30.17966. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi T, Yan B, Mazur P, Derguini F, Nakanishi K, Spudich J L. Color regulation in the archaebacterial phototaxis receptor phoborhodopsin (sensory rhodopsin II) Biochemistry. 1990;29:8467–8474. doi: 10.1021/bi00488a038. [DOI] [PubMed] [Google Scholar]
- 32.Tatsuno I, Homma M, Oosawa K, Kawagishi I. Signaling by the Escherichia coli aspartate chemoreceptor Tar with a single cytoplasmic domain per dimer. Science. 1996;274:423–425. doi: 10.1126/science.274.5286.423. [DOI] [PubMed] [Google Scholar]
- 33.Taylor B L, Johnson M S. Rewiring a receptor: negative output from positive input. FEBS Lett. 1998;425:377–381. doi: 10.1016/s0014-5793(98)00253-1. [DOI] [PubMed] [Google Scholar]
- 34.Tomioka H, Sasabe H. Isolation of photochemically active archaebacterial photoreceptor, pharaonis phoborhodopsin from Natronobacterium pharaonis. Biochim Biophys Acta. 1995;1234:261–267. doi: 10.1016/0005-2736(94)00292-w. [DOI] [PubMed] [Google Scholar]
- 35.Utsumi R, Brissette R E, Rampersaud A, Forst S A, Oosawa K, Inouye M. Activation of bacterial porin gene expression by a chimeric signal transducer in response to aspartate. Science. 1989;245:1246–1249. doi: 10.1126/science.2476847. [DOI] [PubMed] [Google Scholar]
- 36.Vallette F, Mege E, Reiss A, Adesnik M. Construction of mutant and chimeric genes using the polymerase chain reaction. Nucleic Acids Res. 1989;17:723–733. doi: 10.1093/nar/17.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Lintig J, Vogt K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J Biol Chem. 2000;275:11915–11920. doi: 10.1074/jbc.275.16.11915. [DOI] [PubMed] [Google Scholar]
- 38.Yao V J, Spudich E N, Spudich J L. Identification of distinct domains for signaling and receptor interaction of the sensory rhodopsin I transducer, HtrI. J Bacteriol. 1994;176:6931–6935. doi: 10.1128/jb.176.22.6931-6935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X N, Zhu J, Spudich J L. The specificity of interaction of archaeal transducers with their cognate sensory rhodopsins is determined by their transmembrane helices. Proc Natl Acad Sci USA. 1999;96:857–862. doi: 10.1073/pnas.96.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]