Abstract
Simple and rapid multiresidue trace detection of organophosphate pesticides (OPs) is extremely important for various reasons, including food safety, environmental monitoring, and national health. Here, a catalytic hairpin self-assembly (CHA)-based competitive fluorescent immunosensor was developed to detect OPs in agricultural products, involving enabled dual signal amplification followed by a CHA reaction. The developed method could detect 0.01–50 ng/mL triazophos, parathion, and chlorpyrifos, with limits of detection (LODs) of 0.012, 0.0057, and 0.0074 ng/mL, respectively. The spiked recoveries of samples measured using this assay ranged from 82.8% to 110.6%, with CV values ranging between 5.5% and 18.5%. This finding suggests that the CHA-based competitive fluorescent immunosensor is a reliable and accurate method for detecting OPs in agricultural products. The results correlated well with those obtained from the liquid chromatography-tandem mass spectrometry (LC–MS/MS) method, indicating that the CHA-based biosensor is able to accurately detect OPs and can be used as a reliable alternative to the LC‒MS/MS method. Additionally, the CHA-based biosensor is simpler and faster than LC‒MS/MS, which makes it a more practical and cost-effective option for the detection of OPs. In summary, the CHA-based competitive fluorescent immunosensor can be considered a promising approach for trace analysis and multiresidue determination of pesticides, which can open up new horizons in the fields of food safety, environmental monitoring, and national health.
Keywords: Catalyzed hairpin self-assembly, Fluorescent immunosensors, Multiresidue detection, Organophosphate pesticides, Gold nanoparticles
1. Introduction
Organophosphate pesticides (OPs) are one of the most widely used classes of pesticides and account for more than one-third of the total global pesticide use (Costa, 2006). Due to their broad-spectrum activity and relatively low cost, OPs are used in a wide range of agricultural, horticultural, and forestry applications. They are also used to control pests in public health and urban settings. Poorly justified or overuse of pesticides can have a range of negative impacts on agricultural products, livestock (including poultry), and the environment (including the air, water and soil) (Md Meftaul, Venkateswarlu, Dharmarajan, Annamalai, & Megharaj, 2020; Racke, 1992). Furthermore, residual pesticides can accumulate in the food chain and pose a severe threat to human health and environmental quality (Fang et al., 2015; Kaushal, Khatri, & Arya, 2021). Triazophos, parathion, and chlorpyrifos are considered to be highly hazardous pesticides (https://pubchem.ncbi.nlm.nih.gov), of which parathion is on the list of carcinogens. It is considered a potential human carcinogen by the International Agency for Research on Cancer (IARC) and the US Environmental Protection Agency (EPA). Due to their toxicities, these pesticides are currently banned or restricted in many countries, including the EU, the US, and China (Khairy, Ayoub, & Banks, 2018; Ore, Adeola, Bayode, Adedipe, & Nomngongo, 2023). In many developing countries, the use of highly hazardous pesticides, including parathion, is still a major problem. Despite being banned or restricted in many countries, these pesticides are still illegally used in daily agricultural production to meet production needs. This is often due to a lack of enforcement of regulations and a lack of education and resources for farmers to use safer and more sustainable pest management practices. Sprayed pesticides are often formulated with a mixture of multiple pesticides, which is known as a pesticide cocktail. The use of these pesticide cocktails can lead to the presence of multiple pesticides in food products, water and air. This can have serious consequences for human health, the environment and agricultural production, as it can increase the risk of exposure to multiple toxic compounds simultaneously. To address this problem, it is important to have methods that can detect multiple pesticides simultaneously (Parra-Arroyo et al., 2022). Therefore, a low-cost, sensitive and rapid strategy for determining OPs needs to be developed.
GC‒MS (Chunrui, Liping, & Yali, 2021; Acosta-Dacal, Rial-Berriel, Díaz-Díaz, Suárez, & Luzardo, 2020) and LC‒MS/MS (Kecojević, Đekić, Lazović, Mrkajić, Baošić, & Lolić, 2021; Silva, De Menezes, De Castro, Nobre, Milhome, & Do Nascimento, 2019) are currently considered the gold standards for the confirmatory detection of OP residues in agricultural products. These are highly sensitive and specific analytical methods that can detect and quantitate OP residues in agricultural products at very low concentrations. However, these methods can be time-consuming and expensive and require a high level of expertise to operate.
Technological progress and the development of the discipline have facilitated the emergence of other rapid detection techniques for bioanalysis, such as enzyme inhibition assays, surface plasmon resonance, enzyme-linked immunosorbent assay (ELISA), and fluorescence methods (Guo et al., 2018; Korram et al., 2020; Yao, Liu, Liu, Wei, & Liu, 2019; Yue, Chen, Zhang, Yin, & Dong, 2022). These methods offer a range of advantages and can be used in conjunction with or as an alternative to GC‒MS and LC‒MS/MS for the detection of OPs in agricultural products. While these methods have shown promise as rapid and sensitive methods for the detection of OPs in agricultural products, the stability and reproducibility of these methods still require further exploration. Therefore, developing rapid, simple, and sensitive methods for the simultaneous detection of multiple pesticide residues is necessary to meet the demands of food safety, environmental monitoring, and national health. Biosensors and immunoassays are analytical methods that utilize biological molecules, such as enzymes, antibodies, and nucleic acids, to detect and quantify specific compounds (Tsagkaris, Pulkrabova, & Hajslova, 2021, Cui et al., 2018). These techniques are widely used in the analysis of pesticide residues and environmental monitoring because they are simple, rapid, sensitive, and specific. They have the potential to be used in a variety of settings, including food safety, clinical diagnostics, and environmental monitoring. A biobarcode immunoassay is a type of assay that utilizes nucleotide biobarcodes, which are short DNA or RNA sequences, to detect and quantify specific analytes. The biobarcodes are immobilized on the surface of a carrier, such as a bead or a microarray, through chemical bonding. Quantitative detection of the analyte is achieved through specific antigen-antibody interactions (direct or indirect) between the biobarcode-analyte complex and a detection antibody and signal amplification of the biobarcode (Nam & J.-M., 2003). Biobarcode immunoassay has been increasingly applied in the detection of a wide range of biomolecules, such as proteins, nucleic acids, and small-molecule compounds, such as pesticides and veterinary drugs. For instance, Du et al. (2016) compared two different assay models: a traditional sandwich assay and a competitive assay to detect triazophos. They found that both assay models were able to detect triazophos with high sensitivity and specificity, but the competitive assay had a higher dynamic range and a lower limit of detection than the sandwich assay. The study also found that the competitive assay was more resistant to interference from other OPs. Moreover, Zhang et al. (2020) developed a multiplexed biobarcode immunoassay for the detection of three OP pesticides: triazophos, chlorpyrifos, and parathion. They used oligonucleotide chains as biobarcodes, which were designed to specifically bind to the OPs. To increase the sensitivity and specificity of the assay, they selected three fluorescent substances with high fluorescence intensity and no apparent cross-reactivity as markers of the oligonucleotide chains: 6-FAM, Cy3, and Texas Red. They found that the multiplexed assay was able to detect all three OPs in a single reaction, with a low limit of detection and no cross-reactivity with other compounds. They also showed that the assay was able to detect OPs in real-world samples such as soil and water with high accuracy and precision. Using these, Chen et al. (2021) developed a multiresidue method for detecting OPs using a bimetallic Au@Pt nanozyme. The study used a bimetallic Au@Pt nanozyme as a substrate, which was able to catalyze the hydrolysis of OPs, generating a detectable signal. They then used a biobarcode immunoassay to capture the OPs, which were then hydrolyzed by the nanozyme, and the generated signal was detected by a fluorescence reader. The development of DNA nanotechnology and nucleic acid signal amplification techniques has greatly expanded the capabilities of biobarcode immunoassays. Thermal cycling amplification techniques, such as polymerase chain reaction (PCR), and isothermal amplification techniques, such as hybridization chain reaction (HCR), loop-mediated isothermal amplification (LAMP), and rolling circle amplification (RCA), have been developed to amplify nucleic acid signals in a highly specific and sensitive manner (Liang, Wu, Chen, Liu, Aguilar, & Xu, 2020; Sukphattanaudomchoke et al., 2020; M. Zhang et al., 2018). Catalytic hairpin self-assembly (CHA) is a nucleic acid isothermal amplification technique that was proposed by Yin, Choi, Calvert, and Pierce in 2008. CHA is based on the hybridization chain reaction (HCR) and uses the principle of strand displacement and hairpin formation to amplify nucleic acid sequences. One of the key differences between CHA and HCR is that in CHA, the target chain can be replaced to promote the next round of the reaction cycle, which allows for a cyclic catalytic reaction. CHA has several advantages over other nucleic acid amplification techniques, including a high target initiation chain, high signal amplification efficiency and low background signal. It can also be combined with various detection techniques, such as fluorescence and electrochemical detection techniques, thus enabling the possible detection of large molecules, such as pathogenic bacteria and proteins (Luo, Li, Dai, Lu, He, & Wang, 2020; S. Wang, Zhang, Chen, & Cai, 2019), as well as various small molecules, such as biotoxins and environmental pollutants (Yuan et al., 2021; Zhang, Li, Pan, & Han, 2020).
Here, we combined a competitive immunoassay with a CHA reaction to achieve multiresidue detection of three OPs (triazophos, parathion, and chlorpyrifos) through double amplification of the fluorescent detection signal of different fluorescent substances modified on hairpin-structured DNA strands (Scheme 1). The surface coating of three pesticide antigens in 96-well microtitre plates forms an immunocompetitive system by competing with pesticide molecules to bind antibodies modified by three AuNP probes. The unbound free material is removed, the biobarcode modified on the AuNPs is released, and a hairpin structure is then added. Driven by free energy, the released biobarcode acts as the chain initiator and forms a double-stranded structure after complementary pairing with the corresponding hairpin structure, thereby displacing the target chain, which can continue to circulate in a catalytic hybridization reaction to achieve double-signal amplification. The double-stranded structure is labeled with a fluorescent substance, and quantitative multiresidue detection of OPs is based on the measured fluorescence signal.
Scheme 1.
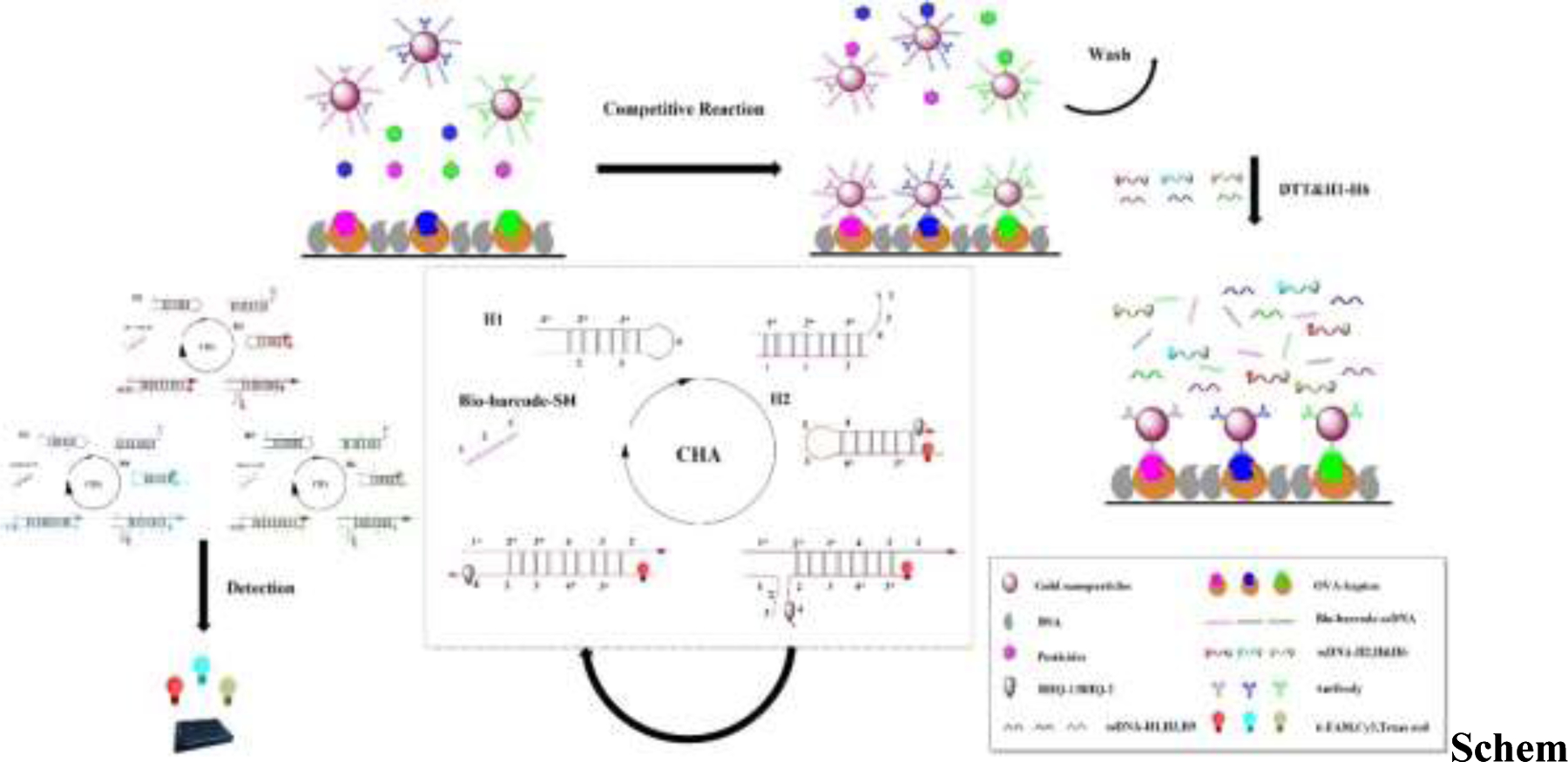
Schematic of the competitive fluorescent immunosensors for OP detection.
2. Experimental
2.1. Materials and reagents
OVA-haptens and mAbs against parathion, chlorpyrifos and triazophos were generously donated by Beijing Kwinbon Biotechnology Co., Ltd. (Beijing, China) and the Institute of Pesticide and Environmental Toxicology (Zhejiang University, Zhejiang, China). Polyethylene glycol 20000 (PEG 20000) and Tris-EDTA (TE) buffer (pH 7.4) were purchased from SolarBio (Beijing, China). Tris(2-carboxyethyl)phosphine (TCEP), dithiothreitol (DTT), chloroauric acid (HAuCl4‧3H2O), trisodium citrate (sodium citrate), and bovine serum albumin (BSA) were purchased from Sigma–Aldrich (St. Louis, MO, USA). Triazophos (analytical standard, 98%) was provided by Dr. Ehrenstorfer GmbH (Augsburg, Germany). Concentrated hydrochloric acid and nitric acid were purchased from the Beijing Chemical Reagent Factory (Beijing, China). Methanol, HPLC-grade acetonitrile (ACN), formic acid, and ammonium acetate were purchased from Thermo Fisher Scientific (MA, USA). Sodium chloride (NaCl), magnesium chloride (MgCl2), potassium chloride (KCl), potassium carbonate (K2CO3), disodium hydrogen phosphate (Na2HPO4), potassium dihydrogen phosphate (KH2PO4), sodium bicarbonate (NaHCO3), anhydrous magnesium sulfate (MgSO4), sodium carbonate (NaCO3), and Tween-20 were purchased from Beijing Chemical Reagent Co., Ltd. (Beijing, China). N-propylethylenediamine (PSA) and octadecyltrimethoxysilane (C18) were purchased from Bonna-Agela Technologies (Tianjin, China).
The DNA sequences corresponding to the three pesticides tested herein are listed in Table S1 in reference to Chen et al. (2020) and Tang et al. (2018). All oligonucleotides were manufactured by Shanghai Sangon Biotechnology Co. Ltd. (Shanghai, China).
2.2. Functionalization of AuNP probes
The preparation of AuNPs was reported in our previous study (Zhang et al., 2018). The characterization of AuNPs and their particle size distribution are shown in Fig. S1. In brief, the three sulfhydryl-modified ssDNAs were centrifuged at 4,000×g for 1 min, dissolved, mixed with TE buffer solution and TCEP solution to a final concentration of 50 µM, and shaken for more than 1 h for DNA activation. The bare AuNPs were adjusted to pH 8.5–9.0 by adding 0.2 mol/L K2CO3 solutions and left to stand for 15 min. Six microlitres (7.57 µg/mL) parathion mAb, 8 µL (10.2 µg/mL) chlorpyrifos mAb, and 14.4 µL (1.4 µg/mL) triazophos mAb were added to the three tubes and incubated for 1 h to form mAb-AuNP complexes. After activating the three sulfhydryl ssDNA strands, the mixture was added and reacted overnight at 4°C. The following day, PEG 20000 was added to the AuNPs at a final concentration of 0.5%, and the mixture was aged for more than 12 h by adding PBS at 4°C. After aging, BSA was added to the mixture to a final concentration of 1%, blocked at 37°C for 1 h, and then centrifuged at 4°C and 12,000×g. The supernatant was discarded, and the unmodified antibody and biobarcode DNA sulfhydryl chain were removed from the surface of the AuNPs. Next, 400 µL of probe resuspension was added and homogenized by slowly blowing and pipetting it more than 10 times. After thorough mixing, the solution was stored at 4°C.
2.3. Preparation of hairpin probes
Denaturation–renaturation pretreatment of hairpin probes is needed. All hairpin structures (H1–H6) were centrifuged at 4,000×g for 1 min. Stock solutions were prepared using Milli-Q ultrapure water, followed by denaturation at 95°C for 5 min before use and then slowly cooled to room temperature to fold into stem–loop structures and stored at 4°C. The stock solutions were diluted to a working concentration of 0.5 µM using 40 mmol/L Tris-HCl (pH 8.5, 8 mmol/L MgCl2, and 100 mmol/L NaCl).
2.4. Gel electrophoresis
Agarose gel electrophoresis was performed to verify whether the hairpin structure and its corresponding target initiation strand underwent CHA. After gel solidification, the sample was mixed with 10× DNA loading buffer at a ratio of 1:9, loaded into each well, and electrophoresed at 120 mV in 1×TAE buffer (40 mM Tris-acetate, 1 mM EDTA, pH 8.0) for 40 min. Bio-Rad ChemDoc XRS was used to image the gels.
2.5. Competitive biobarcode immunoassay based on CHA
Triazophos-OVA-hapten, parathion-OVA-hapten, and chlorpyrifos-OVA-hapten were diluted 8000-, 4000-, and 8000-fold, respectively, with carbonate buffer solution and shaken thoroughly. Next, 100 µL of each dilution was added to the wells of a 96-MicroWell™ black plate and incubated overnight. The following day, the coating solution was poured out, and the plate was washed three times with washing buffer (0.01 mol/L phosphate-buffered saline-Tween 20, PBST) and patted dry. Excess protein binding sites were blocked with 1% BSA for 1 h at 37°C in a constant humidity chamber. After blocking, the solution was poured from the plate, and the plate was washed three times with PBST and patted dry. Fifty microliters of the three pesticide AuNPs (10 × diluted) and an equal amount of pesticide diluent or the sample solution to be tested were added to each well of the patted dry plate. After gentle shaking for 2 min, plates were incubated at 37°C for 1 h. When the competition reaction was complete, the previous steps were repeated to clean the well again and patted dry. Subsequently, 100 µL of 12.5 µM DTT solution was added to each well to dissociate the SH-DNA bound to the AuNPs, followed by the addition of a 1:3 ratio (5 µL:15 µL) of 2 µM hairpin structure. The mixture was then lightly shaken for 2 min and incubated at 37°C for 90 min. After the reaction, fluorescence was detected at 489/521 nm, 532/568 nm, and 592/622 nm emission/absorption wavelengths by using an Infinite M200 PRO microplate reader (TECAN, Männedorf, Switzerland).
2.6. Sample pretreatment
To confirm the accuracy and applicability of the competitive fluorescent immunosensors, apple, cabbage, cucumber, and rice samples were procured from Beijing local markets and confirmed to be free from triazophos, parathion, and chlorpyrifos by LC–MS/MS. The pretreatment method uses safe, efficient, fast, and simple QuEChERS (Perestrelo et al., 2019).
The samples (10 g) were weighed in 50 mL centrifuge tubes, and triazophos, parathion, and chlorpyrifos standard solutions were added at final concentrations of 10, 50, and 100 µg/kg. The mixture was mixed well and allowed to stand for more than 4 h. Subsequently, 10 mL acetonitrile was added. After mixing and shaking, 1 g NaCl and 4 g anhydrous MgSO4 were added, vortexed for 5 min, and centrifuged at 5,000×g for 5 min at 4°C. The supernatant was extracted with 100 mg PSA and 100 mg C18 for 5 min and centrifuged at 5,000×g for 5 min at 4°C. The supernatant was collected, and part of it was used for LC‒MS/MS analysis. The other part was blown dry with nitrogen, redissolved in PBS (5% methanol) and detected using a fluorescent immunosensor based on CHA.
2.7. LC‒MS/MS conditions
The column used in the experiments was a 3.5 µM, 100 × 2.1 mm XBndge™ C18 column (Waters, Ireland). The detailed parameters were as follows: injection volume: 2.0 µL; mobile phase A: containing 5 mM ammonium acetate and 0.1% formic acid in distilled water; mobile phase B: acetonitrile; elution program: flow rate: 0.3 mL/min, elution program: (B): 0 min: 10%; 3.5 min: 10%; 4.5 min: 10%; 4.6 min: 80%; 8 min: 10%. The mass spectrometry system used multiple reaction monitoring modes: interface ion source: ESI; scan mode: positive ion; voltage: 3000 V; atomization gas flow rate: 3 L/min; heating gas flow rate: 10 L/min; drying gas flow rate: 10 L/min; and interface temperature: 350°C. The mass spectrometry parameters for the analysis of the three pesticide compounds are shown in Table S2.
3. Results and discussion
3.1. Feasibility analysis
Agarose gel electrophoresis is a simple and reliable method to confirm the integrity and structure of the DNA strand, and thus, it is an important step in verifying that the CHA reaction can be initiated and the results are reliable. As shown in Fig. S2, when no target strand DNA was present, only slight amplification was observed in lanes 4, 9, and 14. These lanes are labeled H2, H3, H4, H5, and H6. When target strand DNA (A), DNA (B) and DNA (C) were added, the H1-H6 strands were complementary to the target strand, which caused the hairpin structure to open and undergo a catalytic self-assembly reaction. This reaction led to DNA amplification. This means that the experiment was successful and the feasibility of the methodology has been achieved.
3.2. Optimization of antibody addition in the AuNPs
The protocol used for optimizing triazophos-mAb-conjugated AuNPs was based on our previously reported study (Wang et al., 2022), and the optimal antibody concentration for the triazophos pesticide was determined to be 20.16 mg/mL. AuNPs were prepared by adding varying amounts of antibodies (4, 6, 8, 10, and 12 µL) against parathion and chlorpyrifos to colloidal gold. The method used for preparing the AuNPs was the fluorometric method, and fluorescence was measured. The results are shown in Fig. 1. The optimal amounts of antibodies to add for the preparation of parathion and chlorpyrifos AuNP probes were 6 µL (45.42 mg/mL) and 8 µL (81.6 mg/mL), respectively.
Fig. 1.
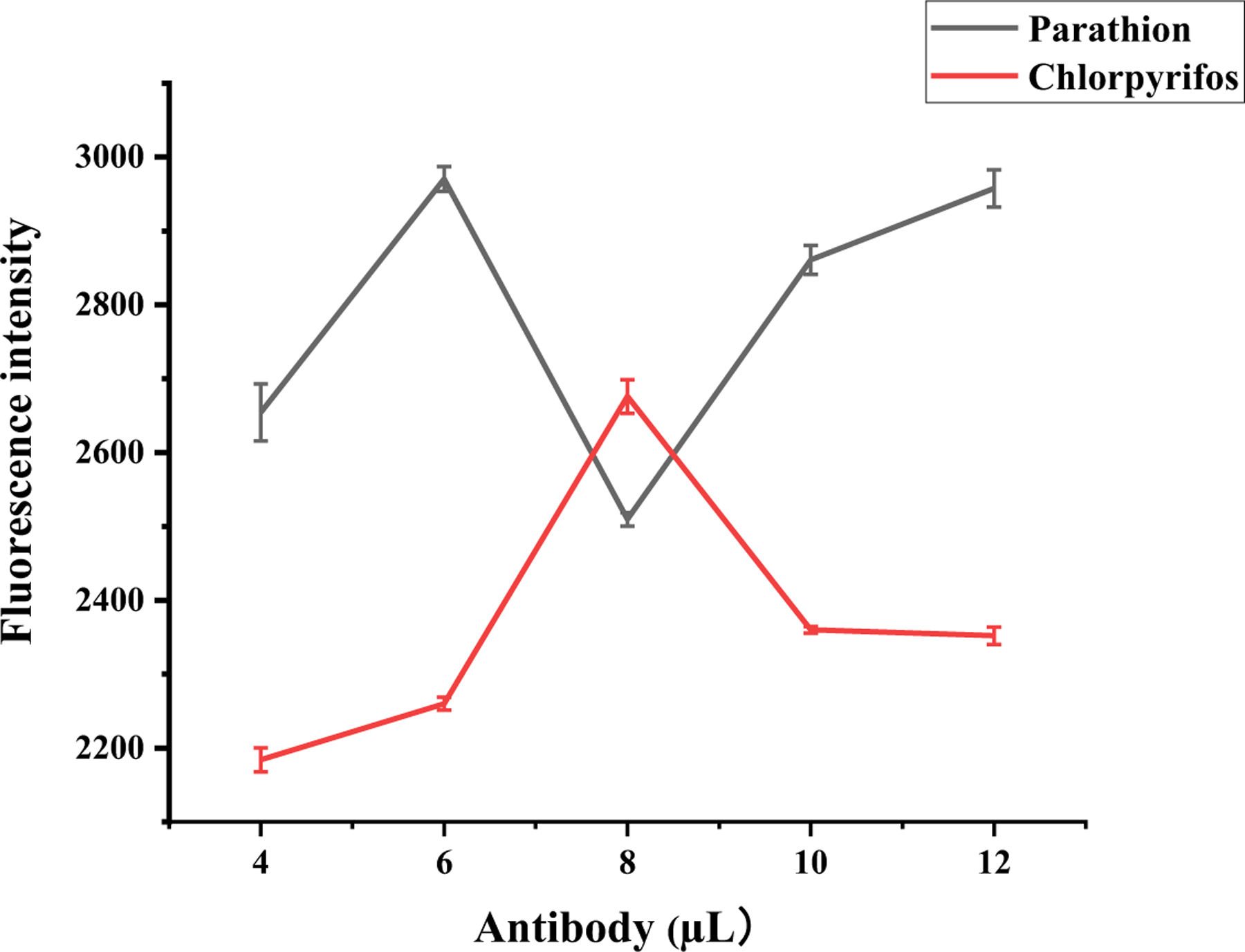
Optimization of antibody addition for parathion and chlorpyrifos AuNP probes.
3.3. Cross-reactivity of hairpin probe fluorescence
In this experiment, the cross-reactivity of the fluorescence of the hairpin probe was verified. As shown in Fig. 2, all three fluorescent substances were able to be excited to the maximum extent, and no cross-reactions were observed. This suggests that the hairpin probe is specific for the intended target and does not bind to other fluorescent molecules, leading to false positive results. Therefore, three different fluorescent groups—6-FAM, Cy3, and Texas Red—were selected because they have high fluorescence intensity at specific excitation/emission wavelengths. 6-FAM is excited at 489 nm and emits at 521 nm, Cy3 is excited at 532 nm and emits at 568 nm, and Texas Red is excited at 592 nm and emits at 622 nm. These different excitation/emission wavelengths allow multiplexing, meaning that we can detect multiple targets at the same time using different probes that emit at different wavelengths. These bands are not affected by interference and can be modified onto the hairpin structure DNA strand. This allows for the multiresidue fluorescence determination of OPs.
Fig. 2.
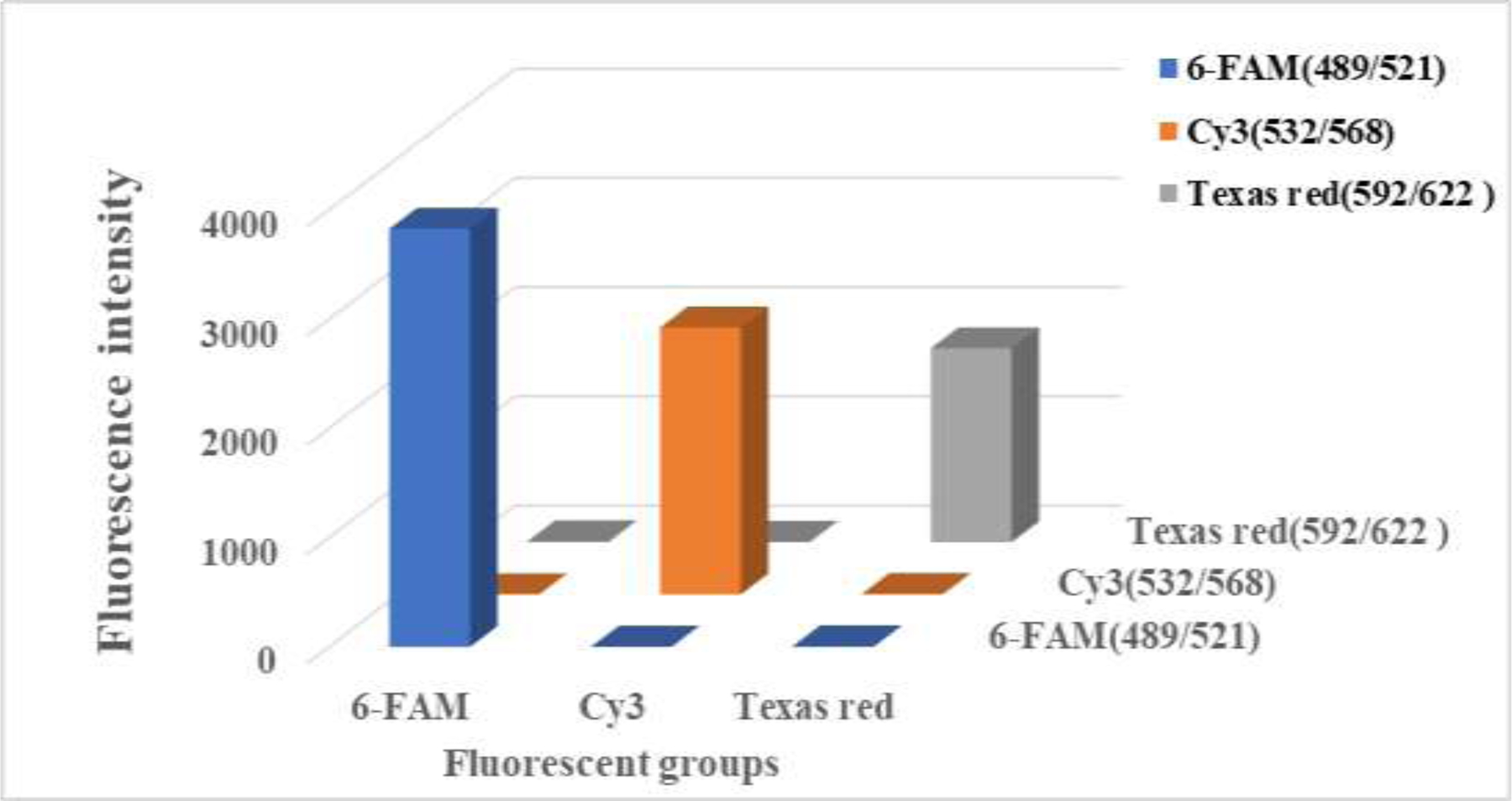
Fluorescence intensity and cross-reactivity of the three fluorescent groups.
3.4. Optimization of competition detection reaction parameters
The working concentrations of the OVA haptens and antibodies are crucial for the performance of the immunocompetitive assay. Hence, the working concentrations of these two parameters in relation to the OPs triazophos, parathion, and chlorpyrifos were optimized.
With 0.05 mol/L carbonate buffered saline, the three OVA-haptens were diluted 4000-, 8000-, and 16 000-fold, respectively, while the three AuNPs were diluted 10-, 20-, and 40-fold with PBS (0.01 mol/L). The optimal dilutions of OVA-hapten and AuNPs for triazophos were 8000- and 10-fold, for parathion were 4000- and 10-fold, and for chlorpyrifos were 8000- and 10-fold, respectively (Table S3).
3.5. Specificity of three pesticides in a competitive immune system
The specific recognition of OVA haptens, antibodies, and hairpin structures is crucial for proper responses to the three pesticides (triazophos, parathion, and chlorpyrifos) in an immunocompetent hybridization reaction system. This recognition allows for appropriate responses to the pesticide. Therefore, to determine the specificity of a competitive fluorescent immunosensor for the detection of OPs based on CHA, pesticides with similar structures to triazophos, parathion, and chlorpyrifos were chosen as test compounds. This is to evaluate the sensitivity and specificity of the sensor for these specific OPs. (Tables S4, S5, S6). The cross-reactivity (CR) values were calculated using equation (1). Here, the IC50 value (half-maximal inhibitory concentration) was calculated for triazophos, parathion, and chlorpyrifos. The IC50 values for each of these pesticides were 0.55 ng/mL, 4.79 ng/mL, and 1.45 µg/mL, respectively. None of the tested analogs were recognized by the competitive fluorescent immunosensor. This is indicated by the CR (cross-reactivity) value being less than 0.1%. This result indicates that the immunosensor has a high degree of specificity for detecting OPs and does not produce false positive results when exposed to analogs.
| (1) |
In addition, the OPs and their corresponding OVA haptens, antibodies, and hairpin structures were grouped in experiments to investigate the fluorescence cross-reactivity of their specific recognition and verify the feasibility of the multiresidue assay. The OVA-haptens of three pesticides were diluted 8000-, 4000-, and 8000-fold, and the three AuNPs were each diluted 10-fold; all of these were added at a concentration of 5 ng/mL pesticide for the reaction. Experimental groups 1–8, 9–16, and 17–24 contained eight combinations of triazophos, parathion, and chlorpyrifos in the immunocompetitive system—single/mixed OVA-haptens, single/mixed AuNPs, and single/mixed hairpin structures, respectively—and each group underwent five parallel studies.
The experimental fluorescence detection results are shown in Fig. 3. The fluorescence values for the three pesticides across the eight experiments were consistent, and the fluorescence values of the multiresidue immunoassay for all three pesticides were slightly higher than those of the single-residue immunoassay. Slight cross-reactivity may have occurred, but the cross-reactivity rate was low and within acceptable limits, demonstrating the feasibility of the immunocompetitive and catalytic hairpin reactions.
Fig. 3.
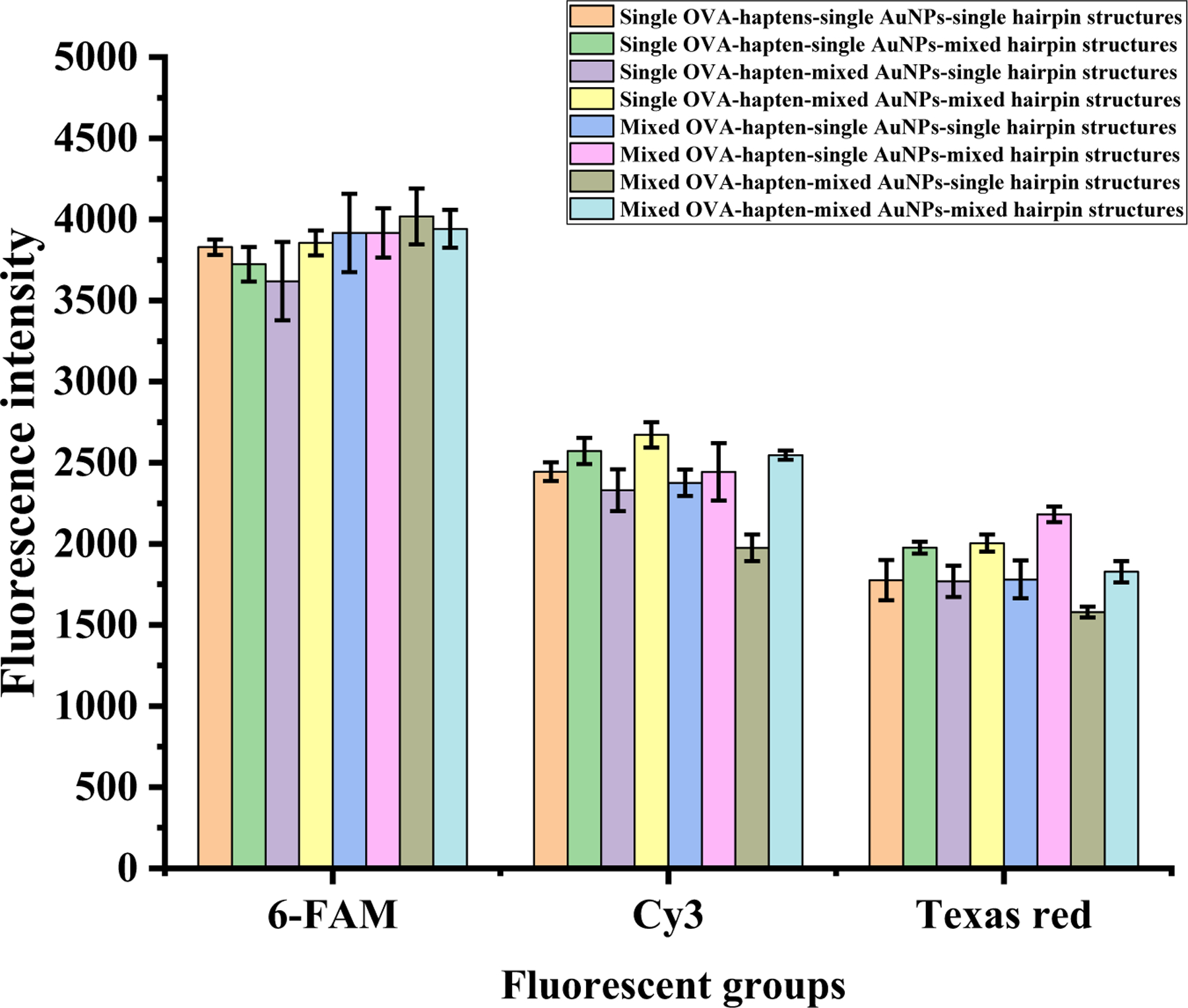
Fluorescence detection for three pesticide specificities.
3.6. Sensitivity of OP detection
Under optimized conditions, standard solutions of three OPs were diluted to concentration gradients from 0.01 to 1000 ng/mL. A multiresidue immunoassay method was established for the determination of OPs. The standard curve was plotted with the log value of pesticide concentration (LogC) as the horizontal coordinate and the inhibition rate (see Equation 1 for the calculation method) as the vertical coordinate. The IC10, the concentration of pesticides causing 10% inhibition of the maximum fluorescence, was used to express the detection sensitivity of the method (Fig. S3).
Table 1 indicates that the multiresidue immunoassay method has good linearity. The calibration curve for triazophos was y = 10.334x + 29.773, with an LOD (IC10) of 0.012 ng/mL; that for parathion was y = 12.527x + 38.145, with an LOD of 0.0057 ng/mL; and that for chlorpyrifos was y = 11.694x + 34.94, with an LOD of 0.0074 ng/mL. These values were calculated using equation (2):
| (2) |
where “I” represents the rate of inhibition, “Fmax” is the fluorescence intensity without pesticides, “Fmin” is the fluorescence intensity of the blank control well, and “Fx” is the fluorescence intensity at pesticide concentration x.
Table 1.
Results from three pesticide standard curves for OPs.
| Pesticides | Linear equation | Correlation coefficient (R2) | Linear range (μg/L) | IC10 (ng/mL) |
|---|---|---|---|---|
| Triazophos | y = 10.334x + 29.773 | 0.974 | 0.01–50 | 0.012 |
| Parathion | y = 12.527x + 38.145 | 0.975 | 0.01–50 | 0.0057 |
| Chlorpyrifos | y = 11.694x + 34.94 | 0.966 | 0.01–50 | 0.0074 |
3.7. Precision and accuracy of the method
To further determine the precision and accuracy of the established immunoassay method for OP multiresidues, this study used four representative matrices to simulate contaminated fruit, vegetable, and grain samples to determine the concentrations of triazophos, parathion and chlorpyrifos in the actual samples. The results of the method established in this work were contrasted with those of LC–MS/MS.
Table S7 shows the comparison results. The recoveries of the spiked samples measured using this method ranged from 82.8% to 110.6%, with CVs from 5.5% to 18.5% at three spiked concentrations of 10, 50, and 100 µg/kg. The recoveries of the samples measured using the instrument confirmation method ranged from 81.6% to 110.4%, with CVs from 1.3% to 15.7%. This indicates that our immunocompetitive method has good applicability and relevance in terms of the precision and accuracy of detection in agricultural products, such as apples, cucumbers, cabbages, and rice.
3.8. Comparison with other methods
With the development of science and technology, optical detection has become the most powerful analytical tool in various fields. It has been widely used for the sensing detection of OPs. As one of the most studied and relatively mature rapid detection techniques, acetylcholinesterase inhibition, an enzyme inhibition-mediated optical biosensor, responds to light signals through hydrolysis of the substrate. Currently, many materials are used as fluorescent tracers, including fluorescent dyes, semiconductor nanomaterials, metal nanomaterials, and carbon nanomaterials, exhibiting excellent optical properties. Compared with other fluorescent immunoassay methods for detecting OPs (Table 2), this method is the same as FPIA in that it uses antigen antibodies with very high affinity and specific binding ability for OPs. Although it is not as novel and portable as FIBSHM and FCDSP, the method uses an enzyme marker that is simple and inexpensive. The rapid and sensitive detection of triazophos pesticide is accomplished on a 96-well microtiter plate without enzymes, and the catalytic reaction can be performed at a constant temperature. The combination with the CHA method further improves the detection limit and sensitivity, with high specificity and suitability for detecting various agricultural products within a wide linear range, which can meet the sensitivity requirements of increasingly stringent standards. In addition, for pesticide multiresidue immunoassay technology, the specificity of pesticide antigen-antibodies can be used to develop their synthesis and preparation, which has the potential to achieve more small molecule multiresidue immunoassay detection.
Table 2.
Comparison with previously reported fluorescent methods aimed at detecting OPs.
| Method | Pesticides | Spiked samples | Recovery (%) | RSD (%) | Fluorescence materials | LOD (ng/mL) | Ref |
|---|---|---|---|---|---|---|---|
| FIBSHHMs | fenitrothion, chlorpyrifos-methyl, fenthion, carbaryl and metolcarb | Cucumber, lettuce, apple, cabbage | 82.6–106.3 | 4.7–14.2 | SHHMs | 0.012–0.1 | Wang, Mu, Shangguan, Liu, Pu, & Yin, 2014 |
| BFIA | methyl parathion, chlorpyrifos, and trichlorfon | pear, carrot, kiwifruit, and banana | 73.1–119.3 | 1.0–13.0 | quantum dots | 0.21–0.44 | Jiang, He, Gong, Gao, & Xu, 2019 |
| FPIA | triazophos | water, brown rice, cabbage and apple | 72.1–104.4 | 3.1–17.0 | fluorescent tracers (THBu-AMF & THBu-EDF) | 0.29 | Ying, Rui, Boroduleva, Eremin, & Guo, 2016 |
| FCDSP | Glyphosate | rice, corn, cabbage, apple, milk, tap water, and river water | 90.8–122.4 | 1.2–3.4 | rQDs@SiO2 @gQDs probes | 0.476–0.561 | Wei et al., 2021 |
| CHA- FI | Triazophos, Parathion, Chlorpyrifos | apple, cucumber, cabbage, and rice | 82.8–110.6 | 5.5–18.5 | fluorescent groups—6-F AM, Cy3, and Texas Red | 0.0057–0.012 | This method |
FIBSHHMs: fluorescent immunoassay based on silica–hydrogel hybrid microbeads
BFIA: biomimetic fluorescence immunoassay
FPIA: fluorescence polarization immunoassay
FCDSP: fluorescent-colorimetric dual-signal platform
CHA- FI: CHA-based fluorescent immunoassay
4. Conclusion
In summary, we proposed CHA-based fluorescent immunosensor multiresidue assays to detect OPs and validated the applicability and accuracy of the method in analyzing practical samples. We have demonstrated the coupling of the dual signal amplification of the immunoassay method with a nucleic acid amplification technique, expanding a new avenue in the trace analysis and detection of pesticide small-molecule multiresidues. Different fluorescent substances were modified on the DNA strands of three hairpin structures, and fluorescence signals were detected. The development of competitive fluorescent immunosensors based on catalytic hairpin self-assembly (CHA) offers great potential for the simultaneous detection of multiple organophosphate pesticides (OPs) in agricultural products. However, further research and development are needed to address the challenges and limitations of these methods and to ensure their reliability and accuracy.
Supplementary Material
Highlights.
Dual signal amplification was achieved by combining immunoassay and nucleic acid amplification.
The method could occur at a constant temperature without enzyme catalysis.
The detection limit was as low as 0.012, 0.0057, and 0.0074 ng/mL.
Signals were detected through different fluorescent groups modified on DNA strands.
The designed sensor leads to a new direction for trace analysis of multiple pesticide residues.
Acknowledgments
This study was financially supported by the National Natural Science Foundation (32272423), NIEHS Superfund Research Program (No. P42 ES004699), Central Public-interest Scientific Institution Basal Research Fund (No. 1610072021004), Central Public Interest Scientific Institution Basal Research Fund for the Chinese Academy of Agricultural Sciences (No. Y2021PT05) and NIEHS RIVER AWARD (No. R35ES030443).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Credit Author statement
Yuanshang Wang: Writing - Original draft, methodology, investigation. A. M. Abd El-Aty: formal analysis; validation; writing - review & editing. Shanshan Wang: Funding acquisition, supervision. Xueyan Cui: AuNP probe synthesis. Jing Zhao: Investigation. Xingmei Lei: conceptualization, investigation. Lingyuan Xu: TEM analysis. Yongxin She: Visualization, Resources. Fen Jin: Data Curation, Resources. Jong-Bang Eun: Writing - review & editing. Jae-Han Shim: Writing - review & editing. Jing Wang: Funding acquisition, Supervision. Maojun Jin: conceptualization, methodology, funding acquisition, project administration, supervision. Bruce D. Hammock: Writing - Review & Editing, Funding acquisition.
References
- Acosta-Dacal A, Rial-Berriel C, Díaz-Díaz R, Suárez M, & Luzardo OP (2020). Optimization and validation of a QuEChERS-based method for the simultaneous environmental monitoring of 218 pesticide residues in clay loam soil. Science of the Total Environment, 753(3), 142015. 10.1016/j.scitotenv.2020.142015. [DOI] [PubMed] [Google Scholar]
- Chen G, Liu G, Jia H, Cui X, Wang Y, Li D, … Hammock BD (2021). A sensitive bio-barcode immunoassay based on bimetallic Au@Pt nanozyme for detection of organophosphate pesticides in various agro-products. Food Chemistry, 362, 130118. 10.1016/j.foodchem.2021.130118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Sun Y, Huo B, Yuan S, Sun X, Zhang M, … Gao Z (2020). Highly sensitive detection of ochratoxin A based on bio-barcode immunoassay and catalytic hairpin assembly signal amplification. Talanta, 208, 120405. 10.1016/j.talanta.2019.120405. [DOI] [PubMed] [Google Scholar]
- Cui XY, Jin MJ, Du PF, Chen G, Zhang C, Zhang YD, … Wang J (2018). Development of immunoassays for multi-residue detection of small molecule compounds. Food and Agricultural Immunology, 29(1), 638–652. 10.1080/09540105.2018.1428284. [DOI] [Google Scholar]
- Costa LG (2006). Current issues in organophosphate toxicology. Clinica Chimica Acta, 366(1), 1–13. 10.1016/j.cca.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Du PF, Jin MJ, Chen G, Zhang C, Jiang ZJ, Zhang YX, … Wang J (2016). A Competitive Bio-Barcode Amplification Immunoassay for Small Molecules Based on Nanoparticles. Scientific Reports, 6, 8. 10.1038/srep38114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L, Zhang S, Chen Z, Du H, Zhu Q, Dong Z, & Li H (2015). Risk assessment of pesticide residues in dietary intake of celery in China. Regulatory Toxicology and Pharmacology, 73(2), 578–586. 10.1016/j.yrtph.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Guo Y, Liu R, Liu Y, Xiang D, Liu Y, Gui W, … Zhu G (2018). A non-competitive surface plasmon resonance immunosensor for rapid detection of triazophos residue in environmental and agricultural samples. Science of the Total Environment, 613–614, 783–791. 10.1016/j.scitotenv.2017.09.157. [DOI] [PubMed] [Google Scholar]
- Habedank F, Abraham M, Tardel H, Feldhusen F, & Schulz-Bull DE (2017). Determination of organophosphate pesticides in sea and surface water with ultrasound-assisted dispersive liquid-liquid micro-extraction coupled to GC-MS/MS analysis. International Journal of Environmental Analytical Chemistry, 97(9), 819–830. 10.1080/03067319.2017.1361944. [DOI] [Google Scholar]
- Jiang M, He J, Gong J, Gao H, & Xu Z (2019). Development of a quantum dot-labelled biomimetic fluorescence immunoassay for the simultaneous determination of three organophosphorus pesticide residues in agricultural products. Food and Agricultural Immunology, 30(1), 248–261. 10.1080/09540105.2019.1572714. [DOI] [Google Scholar]
- Kaushal J, Khatri M, & Arya SK (2021). A treatise on Organophosphate pesticide pollution: Current strategies and advancements in their environmental degradation and elimination. Ecotoxicology and Environmental Safety, 207, 111483. 10.1016/j.ecoenv.2020.111483. [DOI] [PubMed] [Google Scholar]
- Kecojević I, Đekić S, Lazović M, Mrkajić D, Baošić R, & Lolić A (2021). Evaluation of LC-MS/MS methodology for determination of 179 multi-class pesticides in cabbage and rice by modified QuEChERS extraction. Food Control, 123, 107693. 10.1016/j.foodcont.2020.107693. [DOI] [Google Scholar]
- Khairy M, Ayoub HA, & Banks CE (2018). Non-enzymatic electrochemical platform for parathion pesticide sensing based on nanometer-sized nickel oxide modified screen-printed electrodes. Food Chemistry, 255, 104–111. 10.1016/j.foodchem.2018.02.004. [DOI] [PubMed] [Google Scholar]
- Korram J, Dewangan L, Karbhal I, Nagwanshi R, Vaishanav SK, Ghosh KK, & Satnami ML (2020). CdTe QD-based inhibition and reactivation assay of acetylcholinesterase for the detection of organophosphorus pesticides. RSC Advances, 10(41), 24190–24202. 10.1039/d0ra03055d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang T, Wu X, Chen B, Liu J, Aguilar ZP, & Xu H (2020). The PCR-HCR dual signal amplification strategy for ultrasensitive detection of Escherichia coli O157:H7 in milk. LWT, 130, 109642. 10.1016/j.lwt.2020.109642. [DOI] [Google Scholar]
- Luo F, Li Z, Dai G, Lu Y, He P, & Wang Q (2020). Ultrasensitive biosensing pathogenic bacteria by combining aptamer-induced catalysed hairpin assembly circle amplification with microchip electrophoresis. Sensors and Actuators B: Chemical, 306, 127577. 10.1016/j.snb.2019.127577. [DOI] [Google Scholar]
- Md Meftaul I, Venkateswarlu K, Dharmarajan R, Annamalai P, & Megharaj M (2020). Pesticides in the urban environment: A potential threat that knocks at the door. Science of the Total Environment, 711, 134612. 10.1016/j.scitotenv.2019.134612. [DOI] [PubMed] [Google Scholar]
- Nam, & J.-M. (2003). Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science, 301(5641), p.1884–1886. 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- Ore OT, Adeola AO, Bayode AA, Adedipe DT, & Nomngongo PN (2023). Organophosphate pesticide residues in environmental and biological matrices: Occurrence, distribution and potential remedial approaches. Environmental Chemistry and Ecotoxicology, 5, 9–23. 10.1016/j.enceco.2022.10.004. [DOI] [Google Scholar]
- Parra-Arroyo L, González-González RB, Castillo-Zacarías C, Melchor Martínez EM, Sosa-Hernández JE, Bilal M, … Parra-Saldívar R (2022). Highly hazardous pesticides and related pollutants: Toxicological, regulatory, and analytical aspects. Science of the Total Environment, 807, 151879. 10.1016/j.scitotenv.2021.151879. [DOI] [PubMed] [Google Scholar]
- Perestrelo R, Silva P, Porto-Figueira P, Pereira JAM, Silva C, Medina S, & Camara JS (2019). QuEChERS - Fundamentals, relevant improvements, applications and future trends. Analytica Chimica Acta, 1070, 1–28. 10.1016/j.aca.2019.02.036. [DOI] [PubMed] [Google Scholar]
- Racke KD (1992). Degradation of Organophosphorus Insecticides in Environmental Matrices. Organophosphates Chemistry, Fate, and Effects, 15(6),47–78. 10.1016/B978-0-08-091726-9.50018-5. [DOI] [Google Scholar]
- Silva RD, De Menezes MGG, De Castro RC, Nobre CDA, Milhome MAL, & Do Nascimento RF (2019). Efficiency of ESI and APCI ionization sources in LC-MS/MS systems for analysis of 22 pesticide residues in food matrix. Food Chemistry, 297, 7. 10.1016/j.foodchem.2019.06.001. [DOI] [PubMed] [Google Scholar]
- Sukphattanaudomchoke C, Siripattanapipong S, Thita T, Leelayoova S, Piyaraj P, Mungthin M, & Ruang-areerate T (2020). Simplified closed tube loop mediated isothermal amplification (LAMP) assay for visual diagnosis of Leishmania infection. Acta Tropica, 212, 105651. 10.1016/j.actatropica.2020.105651. [DOI] [PubMed] [Google Scholar]
- Tang S, Gu Y, Lu H, Dong H, Zhang K, Dai W, … Zhang X (2018). Highly-sensitive microRNA detection based on bio-bar-code assay and catalytic hairpin assembly two-stage amplification. Analytica Chimica Acta, 1004, 1–9. 10.1016/j.aca.2017.12.004. [DOI] [PubMed] [Google Scholar]
- Tsagkaris AS, Pulkrabova J, & Hajslova J (2021). Optical Screening Methods for Pesticide Residue Detection in Food Matrices: Advances and Emerging Analytical Trends. Foods, 10(1). 10.3390/foods10010088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Zhang F, Chen C, & Cai C (2019). Ultrasensitive graphene quantum dots-based catalytic hairpin assembly amplification resonance light scattering assay for p53 mutant DNA detection. Sensors and Actuators B: Chemical, 291, 42–47. 10.1016/j.snb.2019.04.015. [DOI] [Google Scholar]
- Wang X, Mu Z, Shangguan F, Liu R, Pu Y, & Yin L (2014). Rapid and sensitive suspension array for multiplex detection of organophosphorus pesticides and carbamate pesticides based on silica–hydrogel hybrid microbeads. Journal of Hazardous Materials, 273, 287–292. 10.1016/j.jhazmat.2014.03.006. [DOI] [PubMed] [Google Scholar]
- Wang Y, Abd El-Aty AM, Chen G, Jia H, Cui X, Xu L, … Hammock BD (2022). A competitive immunoassay for detecting triazophos based on fluorescent catalytic hairpin self-assembly. Microchimica Acta, 189(3), 1–12. 10.1007/s00604-022-05325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei D, Wang Y, Zhu N, Xiao J, Li X, Xu T, … Yin D (2021). A Lab-in-a-Syringe Device Integrated with a Smartphone Platform: Colorimetric and Fluorescent Dual-Mode Signals for On-Site Detection of Organophosphorus Pesticides. ACS Applied Materials & Interfaces, 13(41), 48643–48652. 10.1021/acsami.1c13273. [DOI] [PubMed] [Google Scholar]
- Yao T, Liu A, Liu Y, Wei M, Wei W, & Liu S (2019). Ratiometric fluorescence sensor for organophosphorus pesticide detection based on opposite responses of two fluorescence reagents to MnO2 nanosheets. Biosensors and Bioelectronics, 145, 111705. 10.1016/j.bios.2019.111705. [DOI] [PubMed] [Google Scholar]
- Yin P, Choi H, Calvert CR, & Pierce NA (2008). Programming biomolecular self-assembly pathways. Nature, 451(7176), 318–322. 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- Ying L, Rui L, Boroduleva A, Eremin S, & Guo Y (2016). A highly specific and sensitive fluorescence polarization immunoassay for the rapid detection of triazophos residue in agricultural products. Analytical Methods, 8(36). 10.1039/c6ay00908e. [DOI] [Google Scholar]
- Yuan L, Fu Q, Zhou MJ, Ma YQ, Zang LH, Qin YJ, … Zhang FS (2021). Highly sensitive and selective detection of PCB 77 using an aptamer-catalytic hairpin assembly in an aquatic environment. RSC Advances, 11(10), 5506–5511. 10.1039/d0ra10285g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Chen J, Zhang M, Yin Y, & Dong Y (2022). Determination of Organophosphorus Pesticides in Vegetables and Fruit by an Indirect Competitive Enzyme-Linked Immunosorbent Assay (ic-ELISA) and a Lateral-Flow Immunochromatographic (LFIC) Strip Assay. Analytical Letters, 55(11), 1701–1718. 10.1080/00032719.2021.2023170. [DOI] [Google Scholar]
- Zhang C, Du P, Jiang Z, Jin M, Chen G, Cao X, … Wang J (2018). A simple and sensitive competitive bio-barcode immunoassay for triazophos based on multi-modified gold nanoparticles and fluorescent signal amplification. Analytica Chimica Acta, 999, 123–131. 10.1016/j.aca.2017.10.032. [DOI] [PubMed] [Google Scholar]
- Zhang C, Jiang Z, Jin M, Du P, Chen G, Cui X, … Wang J (2020). Fluorescence immunoassay for multiplex detection of organophosphate pesticides in agro-products based on signal amplification of gold nanoparticles and oligonucleotides. Food Chemistry, 126813. 10.1016/j.foodchem.2020.126813. [DOI] [PubMed]
- Zhang M, Huo B, Yuan S, Ning B, Bai J, Peng Y, … Gao Z (2018). Ultrasensitive detection of T-2 toxin in food based on bio-barcode and rolling circle amplification. Analytica Chimica Acta, 1043, 98–106. 10.1016/j.aca.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Zhang SQ, Li KB, Pan YJ, & Han DM (2020). Ultrasensitive detection of ochratoxin A based on biomimetic nanochannel and catalytic hairpin assembly signal amplification. Talanta, 220. 10.1016/j.talanta.2020.121420. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


