Abstract
In this study we cloned CTA1, the gene encoding peroxisomal catalase, from the methylotrophic yeast Candida boidinii and studied targeting of the gene product, Cta1p, into peroxisomes by using green fluorescent protein (GFP) fusion proteins. A strain from which CTA1 was deleted (cta1Δ strain) showed marked growth inhibition when it was grown on the peroxisome-inducing carbon sources methanol, oleate, and d-alanine, indicating that peroxisomal catalase plays an important nonspecific role in peroxisomal metabolism. Cta1p carries a peroxisomal targeting signal type 1 (PTS1) motif, -NKF, in its carboxyl terminus. Using GFP fusion proteins, we found that (i) Cta1p is transported to peroxisomes via its PTS1 motif, -NKF; (ii) peroxisomal localization is necessary for Cta1p to function physiologically; and (iii) Cta1p is bimodally distributed between the cytosol and peroxisomes in methanol-grown cells but is localized exclusively in peroxisomes in oleate- and d-alanine-grown cells. In contrast, the fusion protein GFP-AKL (GFP fused to another typical PTS1 sequence, -AKL), in the context of CbPmp20 and d-amino acid oxidase, was found to localize exclusively in peroxisomes. A yeast two-hybrid system analysis suggested that the low transport efficiency of the -NKF sequence is due to a level of interaction between the -NKF sequence and the PTS1 receptor that is lower than the level of interaction with the AKL sequence. Furthermore, GFP-Cta1pΔnkf coexpressed with Cta1p was successfully localized in peroxisomes, suggesting that the oligomer was formed prior to peroxisome import and that it is not necessary for all four subunits to possess a PTS motif. Since the main physiological function of catalase is degradation of H2O2, suboptimal efficiency of catalase import may confer an evolutionary advantage. We suggest that the PTS1 sequence, which is found in peroxisomal catalases, has evolved in such a way as to give a higher priority for peroxisomal transport to peroxisomal enzymes other than to catalases (e.g., oxidases), which require a higher level of peroxisomal transport efficiency.
Catalase, which degrades H2O2 to oxygen and H2O, is present along with various types of H2O2-generating oxidases in the matrix of the peroxisome, an organelle found in virtually all eukaryotic cells. Because of this, catalase has been used for a long time as a marker enzyme for peroxisomes (6, 24, 52). A lack of catalase in peroxisomes is thought to result in accumulation of toxic H2O2 and/or other reactive oxygen species derived from H2O2 (7, 18, 44, 53, 60), and the physiological importance of catalase activity is shown by the existence of a genetic disease, known as acatalasemia (52). In addition, a defect in catalase import into the peroxisome has been reported to lead to a severe neurological disorder (44). The peroxiredoxine Pmp20 family is another peroxisomal antioxidant system capable of degrading H2O2 that has recently been identified in both mammalian and yeast cells (20, 57). The physiological significance of catalase is reevaluated in this report and compared with the function of members of the Pmp20 family of proteins.
Peroxisomal proteins are encoded by nuclear genes, are synthesized on free polyribosomes, and, following translation, are imported into the peroxisome (24). The targeting of proteins to peroxisomes is mediated by cis-acting peroxisomal targeting signals (PTSs) and their corresponding receptors. PTSs are necessary and sufficient for peroxisomal targeting. At least three types of PTSs are known, including a C-terminal tripeptide, PTS1 (16); an NH2-terminal peptide, PTS2 (12, 13, 50); and mPTS, which is specific for an integral peroxisomal membrane protein (8, 27, 28). The PTS1 receptor, Pex5p, and the PTS2 receptor, Pex7p, have been shown to bind and recruit PTS1 and PTS2 proteins to peroxisomes, respectively (46).
Another feature of peroxisomal protein import is the presence of some proteins that can be imported into peroxisomes in a folded oligomeric state. Such oligomeric transport has been demonstrated with artificial PTS1-tagged chloramphenicol acetyltransferase in yeast and human cells (29) and also with some native PTS1 and PTS2 proteins, including Saccharomyces cerevisiae thiolase (13), S. cerevisiae malate dehydrogenase 3 (9), human alanine/glyoxylate amino transferase 1 (26), and plant isocitrate lyase (25). In these studies, a reporter protein from which PTS had been deleted was shown to be transported into peroxisomes only when the same reporter protein harboring a PTS was coexpressed in the same cell. This finding was supported by the results of microinjection studies of human cell lines in which colloidal gold particles (diameter, 9 nm) that were coated with a human serum albumin-PTS1 conjugate were shown to be imported into the peroxisome (54). In contrast, two major methanol-induced peroxisomal proteins, dihydroxyacetone synthase and alcohol oxidase (AOD), seemed to differ in that these two proteins fold within peroxisomes after they are imported into these organelles (40, 56).
There is no direct evidence that oligomeric transport of catalase occurs, but there is indirect evidence which suggests that oligomeric transport of catalase could occur. For example, in cells of patients with Zellweger syndrome, in which catalase and other peroxisomal matrix proteins are synthesized on ribosomes normally but are not transported across the peroxisomal membrane (52), catalase assembles into catalytically active tetramers in the cytosol (55). These cells can be divided into distinct complementation groups so that fusion of cells from different groups results in the appearance of catalase-containing peroxisomes (4, 45). However, studies performed with monoclonal antibodies specific for tetrameric or dimeric-monomeric catalase subunits showed that in contrast to what happens in rodent liver, human skin fibroblasts assemble cytosolic tetrameric catalase in the cytosol (within 1 h of synthesis), and this catalase is then targeted to peroxisomes for disassembly and import (30).
We have used the methylotrophic yeast Candida boidinii extensively as a model organism to study peroxisomal metabolism and protein import. C. boidinii can grow on three metabolically distinct peroxisome-inducing carbon sources, methanol, fatty acids, and d-alanine (15, 38). When this organism is used in combination with a gene manipulation system (37, 41), the metabolic significance of a specific protein can be evaluated by examining the growth defect with cells grown on a peroxisome-inducing carbon source. For example, in a previous study we showed that the function of CbPmp20 is specific for methanol metabolism (20). In the present study we sought to evaluate the physiological contributions of peroxisomal catalase in various types of peroxisome metabolism. In addition, the following two aspects of peroxisomal import of catalase were studied: the efficiency of transport; and oligomeric transport in which green fluorescent protein (GFP) was used, which enabled us to visualize the localization of GFP-Cta1p fusion protein in living cells. We were able to show that the efficiency of catalase import into peroxisomes depends on the growth conditions (i.e., the carbon source). Furthermore, our findings suggest that the variations in PTSs in different proteins are related to the metabolic significance of each protein. In this report we show that not only peroxisomal metabolism but also the efficiency of peroxisomal protein transport can change depending on the environmental conditions.
MATERIALS AND METHODS
Microorganisms and growth conditions.
C. boidinii TK62 (ura3) (37) was used as the host for transformation. C. boidinii GC (39) was used as the wild-type strain. Synthetic MI medium was used as the basal medium on which C. boidinii was cultured (42). One or more of the following compounds was used as the carbon source in each experiment: 1% (wt/vol) glucose, 1% (vol/vol) methanol, 0.6% (wt/vol) d-alanine, or 0.5% (vol/vol) oleic acid. Tween 80 was added to the oleate medium at a concentration of 0.05% (vol/vol). The initial pH of the medium was adjusted to 6.0. Complex YP medium containing 2% Bacto Peptone (Difco Laboratories, Detroit, Mich.) and 1% yeast extract (Difco) was also used as the basal medium in some experiments. YPD medium contained 2% glucose and YPMGy medium contained 0.5% methanol and 0.5% glycerol as the carbon source(s). The C. boidinii strains were incubated under aerobic conditions at 28°C with reciprocal shaking, and growth of the yeast was monitored by measuring the optical density at 610 nm.
Escherichia coli DH5α (43) was routinely used for plasmid propagation.
Cloning and sequencing of CTA1 from C. boidinii S2.
A 0.8-kb DNA fragment of the peroxisomal catalase gene from a related Candida strain (32) was used as the probe for cloning of the CbCTA1 gene. This fragment was obtained by PCR by using primers CTAN1 and CTAR1 (Table 1) and Candida tropicalis pK233 genomic DNA as the template. The PCR-amplified fragment was gel purified, 32P labeled by the method of Feinberg and Vogelstein (10), and used for cloning experiments. A ca. 5.0-kb EcoRV-digested genomic DNA pool was gel purified and ligated into the EcoRV site of pBluescript II KS+ (Stratagene, San Diego, Calif.). E. coli transformants were transferred onto a Biodyne nylon membrane (Pall Bio Support, New York, N.Y.). After lysis of the bacteria, the liberated DNA was bound to the nylon membrane, and the blots were then used for colony hybridization under high-stringency hybridization conditions, using Church-Gilbert buffer (1% bovine serum albumin, 1 mM EDTA, 0.25 M NaCl, 0.25 M Na3PO4 [pH 7.2], 7% sodium dodecyl sulfate) (5). Hybridization was performed at 65°C overnight, and then the membranes were washed six times in 0.2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at the same temperature. Clones that showed strong positive signals were found to harbor a reactive 5.0-kb EcoRV fragment. Nested deletion mutants were derived as previously described (58), and the complete CTA1 gene was sequenced in both directions by using a 7-deaza sequencing kit (Thermo Sequence fluorescent labeled primer cycle sequencing kit) from Amersham Pharmacia Biotech (Little Chalfont, Buckinghamshire, England) and a model DSQ-2000L DNA sequencer (Shimadzu Co. Ltd., Kyoto, Japan).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′-3′) |
|---|---|
| CTAN1 | CATGGATCCGACGTTTGTGCCGCAAAG |
| CTAR1 | CATGGATCCCAATCTGTGTCTGTGAGT |
| CBCAT-N | ATAAGAATGCGGCCGCAATATGTCTAATCCTCCAACTTAT |
| CBCAT-R1 | ATAAGAATGCGGCCGCTTAAAATTTATTCTTAGAAGCACC |
| CBCAT-R2 | ATAAGAATGCGGCCGATTACTTAGAAGCACCTCTTGGAC |
| CTA1PstI3′ | AACTGCAGTTAAAATTTATTCTTAGAAGCACC |
| AKLPstI3′ | AACTGCAGTTATAATTTAGCCTTAGAAGCACCTCTTGGAC |
| DNKFPstI | AACTGCAGTTACTTAGAAGCACCTCTTGGAC |
| 5′Sa1ICTA1 | ACGCGTCGACTGTCTAATCCTCCAACTTATAC |
| CTA1GFP | GTTGTATAAGTTGGAGGATTAGAACCTCCGGACTTGTATAGTTCATC |
| GFPCTA1 | GATGAACTATACAAGTCCGGAGGTTCTAATCCTCCAACTTATACAAC |
| TH-Pex5-N | CTCGAGAAATGTCATTCATAGGTGGAGGT |
| TH-Pex5-C | TCTAGATTAAATCCCGTTAGATGTGCG |
| TH-GFP-N | ACGCGTCGACTGGGTAAAGGAGAAGAACTTTT |
| GFP-ATG | ACGCGTCGACATGGGTAAAGGAGAAGAACTTT |
| GFP-AKL | AACTGCAGTTATAATTTAGCACCTCCGGA |
| GFP-NKF | AACTGCAGTTAAAATTTATTACCTCCGGACTTGTATAGTTC |
| GFP-Stop | AACTGCAGTTAACCTCCGGACTTGTATAG |
Construction of the disruption cassette and one-step disruption of CTA1.
pCTA1, carrying 5.0-kb EcoRV fragment harboring the C. boidinii CTA1 gene, was digested with SpeI and HindIII, and the 2.5-kb SpeI-HindIII fragment containing the 5′ flanking region and truncated C-terminal coding region of C. boidinii CTA1 was cloned into the multiple cloning site of pBluescript II SK+ (Stratagene), which yielded pCTA1SH. This plasmid was digested with StyI to delete a 1.4-kb fragment which included the 5′ half of the C. boidinii CTA1 gene. The remaining fragment was gel purified, blunt ended with T4 DNA polymerase, and ligated to the XhoI-SacI fragment of pSPR (C. boidinii URA3 gene having a repeated sequence in the 5′ and 3′ flanking regions) (41). The ligation reaction generated the C. boidinii CTA1 disruption vector pCTA1SPR. After propagation of pCTA1SPR in E. coli, the inserted DNA fragment was isolated following SacI-XhoI digestion, and this fragment was used to transform C. boidinii TK62 to uracil prototrophy. The disruption of CTA1 was confirmed by genomic Southern analysis of BglII-HindIII-digested DNA from a Ura+ transformant, using the 0.4-kb StyI-HincII fragment from pCTA1 as a probe. The strain in which CTA1 was disrupted (cta1Δ strain) which was obtained with pCTA1SPR was reverted to uracil auxotrophy by using a previously described procedure (41), which yielded the cta1Δ ura3 strain.
Construction of Cta1p and Cta1pΔnkf overexpression vectors.
NotI sites were added to both ends of the C. boidinii CTA1 coding sequence by PCR performed with primers CBCAT-N and CBCAT-R1 (Table 1), and C. boidinii CTA1 DNA was used as the template in the reaction. The amplified fragment was purified from an agarose gel, digested with NotI, and introduced into the C. boidinii expression vector pNoteI (35). The plasmid construct was designated pNotCta1p-NKF. pNotCta1pΔnkf, an expression vector of Cta1pΔnkf lacking the coding sequence of the C-terminal three amino acids of Cta1p, was constructed in the same way by using primers CBCAT-N and CBCAT-R2 (Table 1). The correct orientation of the inserted fragment was confirmed by physical mapping. Plasmid constructs were linearized with BamHI and introduced into the cta1Δ ura3 strain by the modified lithium acetate method (36).
Construction of GFP fusion proteins and their expression vectors.
A PCR technique was used to construct GFP fusion proteins and their expression vectors. To construct pGFP-Cta1p-NKF, two rounds of PCR were performed by using primers GFP-ATG and GFPCTA1 (Table 1) with pGFP-C1 (Clontech, Palo Alto, Calif.) as the template and primers CTA1GFP and CTA1PstI3′ (Table 1) with C. boidinii CTA1 DNA as the template. Next, the two amplified fragments were gel purified and used as PCR templates with primers GFP-ATG and CTA1PstI3′ (Table 1). The amplified fragment was gel purified, digested with SalI and PstI, and introduced into the C. boidinii expression vector pACT1 (42). pACT1 harbored the C. boidinii ACT1 promoter and terminator sequences with a unique SalI-PstI site to insert coding sequences for expression (42). pGFP-Cta1pΔnkf and pGFP-Cta1p-AKL were constructed in exactly the same manner, except that primer DNKFPstI was used instead of CTA1PstI3′ and AKLPstI3′, respectively (Table 1). pGFP-NKF was constructed by cloning the PCR product obtained by using primers GFP-ATG and GFP-NKF (Table 1) with pGFP-C1 as the template into the SalI-PstI site of pACT1. The plasmid constructs were linearized with XbaI and introduced into C. boidinii strain TK62 and the cta1Δ ura3 strain by the modified lithium acetate method (36). Strains GFP-AKL/wt and GFP-Stop/wt (42) were used as the wild-type strains for comparison of peroxisome morphology.
Protein methods and antibody preparations.
Size exclusion chromatography was performed by using HiLoad 16/60 Superdex pg (Amersham Pharmacia Biotech) with 0.1 M potassium phosphate buffer (pH 7.5) at a flow rate of 1.0 ml/min at 4°C. The molecular mass standards used were glutamate dehydrogenase (290 kDa), lactate dehydrogenase (142 kDa), enolase (67 kDa), adenylate kinase (32 kDa), and cytochrome c (12.4 kDa).
Standard 9% polyacrylamide Laemmli gels (22) with a pH 9.2 separating gel were used. Immunoblotting was performed by the method of Towbin et al. (49), using an ECL detection kit (Amersham, Arlington Heights, Ill.). IVA7 monoclonal anti-Pmp47 antibody and anti-Aod1p antibody were kindly provided by J. M. Goodman (University of Texas Southwestern Medical Center, Dallas). Anti-GFP antibody was kindly provided by M. Fransen (Katholieke Universiteit, Louvain, Belgium).
Enzyme assays.
The levels of catalase and cytochrome c oxidase activities were determined as described previously (1, 48). Protein quantification was performed by the method of Bradford (3) with a protein assay kit (Bio-Rad Laboratories, Hercules, Calif.), using bovine serum albumin as the standard.
H2O2 concentration and methanol concentration.
Fluorescent measurement was used for horseradish peroxidase-catalyzed oxidation of homocanillic acid to determinine the concentration of H2O2 (17, 59). Because thiols are substrates for horseradish peroxidase (19), sulfhydryl groups were alkylated with N-ethylmaleimide prior to the determination of H2O2 concentration. Calibration curves were generated with known amounts of H2O2. Methanol concentrations were determined by gas-liquid chromatography with a Porapak Q column (inside diameter, 0.55 cm; length, 2 m) by using a Shimadzu GC 7A as previously described (47).
Organelle fractionation.
C. boidinii cells were grown on YPMGy medium overnight, converted to spheroplasts with Zymolyase 100T (Seikagaku Co., Tokyo, Japan), and osmotically lysed by the method of Goodman et al. (14), as described previously (33). Unlysed cells, nuclei, and other cell debris were removed from the lysate by centrifugation at 1,000 × g and 4°C for 10 min. The supernatant was centrifuged at 20,000 × g and 4°C for 20 min to obtain a crude pellet containing mainly peroxisomes and mitochondria.
To prepare a continuous Nycodenz (Sigma Chemical Co., St. Louis, Mo.) gradient solution, a 10.6-ml stepwise gradient (1.3 ml of 60% [wt/vol] Nycodenz, 2 ml of 50% [wt/vol] Nycodenz, 4 ml of 35% [wt/vol] Nycodenz, and 3.3 ml of 28% [wt/vol] Nycodenz) was frozen once in liquid nitrogen and then thawed. Next, the organelle suspension was layered on top of the 10.6-ml gradient, and samples were centrifuged at 100,000 × g for 2 h at 4°C in a VTi 65.1 vertical rotor (Beckman Instruments, Inc., Palo Alto, Calif.). The gradients were fractionated from the bottom.
Fluorescence microscopy.
Cells expressing GFP fusion protein were placed on a microscope slide and examined with an Axioplan 2 fluorescence microscope (Carl Zeiss, Oberkochen, Germany) equipped with a Plan-NEOFLUAR 100×/1 · 30 (oil) objective and Nomarski attachments and set at the fluorescein isothiocyanate channel. Images were acquired with a charge-coupled device camera (Carl Zeiss ZVS-47DE) and a CG7 frame grabber (Scion Corp., Frederick, Md.).
Two-hybrid procedures and assays.
A PCR technique was employed to construct vectors expressing fusion proteins with the GAL4 DNA binding domain or the transcriptional activation domain. pBD-Cta1p-NKF was constructed by cloning the PCR product by using primers 5′SalICTA1 and CTA1PstI3′ (Table 1) with pCTA1 as the template and ligating it to the SalI-PstI site of the pBD-GAL4 Cam phagemid vector (Stratagene). pBD-Cta1pΔnkf was constructed in exactly the same way, except that primer DNKFPstI was used instead of CTA1PstI3′ (Table 1). pBD-GFP-NKF was constructed by cloning the PCR product by using primers TH-GFP-N and GFP-NKF (Table 1) with pGFP-C1 as the template and ligating it to the SalI-PstI site of the pBD-GAL4 Cam phagemid vector. pBD-GFP-AKL and pBD-GFP-Stop were constructed in exactly the same way, except that primer GFP-NKF was replaced by primers GFP-AKL and GFP-Stop, respectively (Table 1). pAD-CbPex5p was constructed by cloning the PCR product by using primers TH-Pex5-N and TH-Pex5-C (Table 1) with C. boidinii genomic DNA as the template and ligating it into the XhoI-XbaI site of the pAD-GAL4 Cam phagemid vector (Stratagene). Cotransformation of two-hybrid vectors into strain YRG-2 (Stratagene) was performed by using the protocols of the manufacturer. Double transformants were selected on SD medium lacking tryptophan and leucine.
Yeast cells were harvested, washed with distilled water, and lysed by freezing in liquid nitrogen. Cell debris was removed by centrifugation, and the protein concentration of the supernatant was determined. β-Galactosidase activity was measured photometrically at 420 nm by using the substrate o-nitrophenyl-β-d-galactopyranoside, as described previously (31). Two transformants harboring each of the plasmids were tested in duplicate, and the β-galactosidase activities presented below are averages based on three determinations (the standard deviations were less than 15%). One unit was defined as the amount of protein which resulted in hydrolysis of 1 μmol of o-nitrophenyl-β-d-galactopyranoside per min (31).
Nucleotide sequence accession number.
The nucleotide sequence of the CTA1 gene determined in this study has been deposited in the DDBJ/GenBank/EMBL databases under accession number AB064338.
RESULTS
Cloning and disruption of C. boidinii CTA1 gene coding for peroxisomal catalase.
The 5.0-kb EcoRV fragment cloned from the C. boidinii genomic library harbored an open reading frame (length, 504 amino acids) with high levels of similarity to peroxisomal catalases from other sources (e.g., 76, 65, 52, and 48% amino acid sequence similarities with peroxisomal catalases from Hansenula polymorpha, C. tropicalis, S. cerevisiae, and humans, respectively). This open reading frame was designated the C. boidinii CTA1 gene. The molecular mass of the protein was calculated to be 57,092 Da, which is in close agreement with the molecular mass of purified C. boidinii catalase as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (51). Cta1p had conserved amino acid residues which are thought to be of considerable importance for construction and functioning of the catalase active site (His63, Ser102, and Asn136) and for interaction with heme (Val62, Thr103, Phe141, Pro323, Arg341, and Tyr345). The extreme C-terminal sequence of Cta1p is -NKF, which is a PTS1 motif.
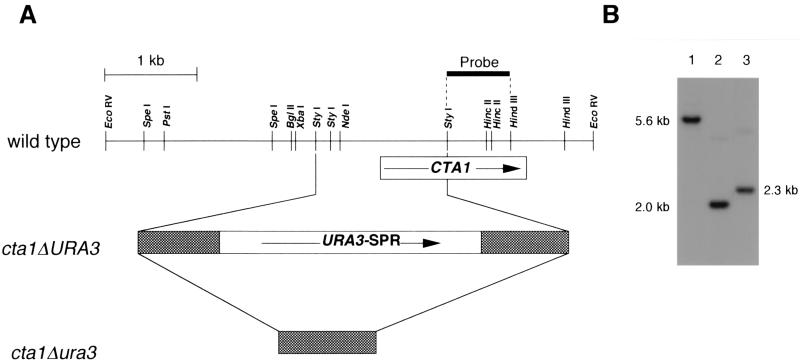
A CTA1 disruption vector, pCTA1SPR, was constructed and introduced into C. boidinii TK62 (Fig. 1A). Gene disruption and subsequent excision of the URA3 sequence were confirmed by Southern analysis with BglII-HindIII-digested genomic DNA from the transformant, in which the 0.4-kb StyI-HincII fragment from pCTA1 was used as a probe (Fig. 1B). The 2.3-kb hybridizing band in the host strain (Fig. 1B, lane 3) shifted to 5.6 kb in the cta1Δ strain (Fig. 1B. lane 1), and this 5.6-kb band shifted to 2.0 kb upon regeneration of uracil auxotrophy (Fig. 1B, lane 2). The levels of catalase enzyme activity in the C. boidinii wild-type strain were maximally enhanced in methanol-grown cells (2,530 U/mg of protein), followed by oleate-grown cells (1,010 U/mg of protein) and d-alanine-grown cells (117 U/mg of protein), while enzymatic activity was lowest in glucose-grown cells (10.2 U/mg of protein). It has been shown previously that catalase activity is regulated at the mRNA level, most likely via transcription, as in S. cerevisiae (42). In contrast, methanol-induced cells of the cta1Δ strain exhibited a detectable loss of catalase activity (data not shown). (Using our assay conditions, we could not detect cytosolic catalase or cytochrome peroxidase activity and estimated that more than 98% of the H2O2-degrading activity could be attributed to Cta1p.) These results and results described below indicate that the cloned gene encodes peroxisomal catalase of C. boidinii.
FIG. 1.
One-step disruption of the CTA1 gene in the C. boidinii genome. (A) Physical map of the cloned fragment and disruption strategy. The arrows indicate the directions of the coding sequences. The shaded boxes at the two ends of URA3 represent repeated sequences for homologous recombination to remove the URA3 gene after gene disruption (41). (B) Genomic Southern analysis of BglII-HindIII-digested total DNAs (3 μg each) of the cta1Δ strain (lane 1), host strain TK62 (lane 2), and the cta1Δ ura3 strain (lane 3) probed with the 32P-labeled 0.8-kb StyI-HindIII fragment, including the 3′ flanking region of CTA1.
Growth defect of the cta1Δ strain in the presence of peroxisome-inducing carbon sources.
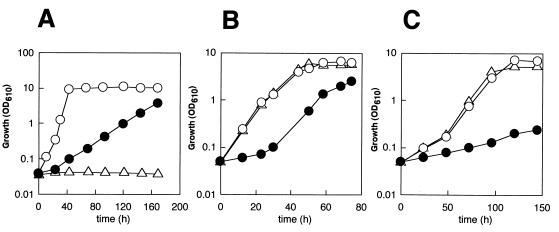
To evaluate the physiological significance of Cta1p in peroxisome metabolism, growth of the cta1Δ strain on various peroxisome-inducing carbon sources was examined and was compared with growth of the wild-type strain and the pmp20Δ strain. CbPmp20 is a recently identified peroxisomal antioxidant enzyme, glutathione peroxidase, which also degrades H2O2 and is specifically induced in methanol medium (20, 42). As shown in Fig. 2, growth of the cta1Δ strain was clearly inhibited in the presence of all of the peroxisome-inducing carbon sources tested (methanol, oleic acid, and d-alanine). On the other hand, although the pmp20Δ strain was not able to grow at all on methanol, growth on oleate or d-alanine was not inhibited. When the cta1Δ strain was grown in methanol medium, H2O2 gradually accumulated in the medium for up to 110 h, reaching a concentration of 0.67 mM. In addition, the cta1Δ strain was able to grow on glucose medium as well as the wild-type strain (data not shown). On the basis of these results, we propose that the main physiological role of Cta1p is to degrade H2O2 generated by peroxisomal oxidases (i.e., alcohol oxidase, acyl coenzyme A oxidase, and d-amino acid oxidase).
FIG. 2.
Growth of the C. boidinii cta1Δ strain on peroxisome-inducible carbon sources, including methanol (A), oleate (B), and d-alanine (C). Symbols: ○, wild-type strain; ●, cta1Δ strain; ▵, pmp20Δ strain. OD610, optical density at 610 nm.
C-terminal -NKF sequence is necessary for peroxisomal transport of Cta1p and its physiological function.
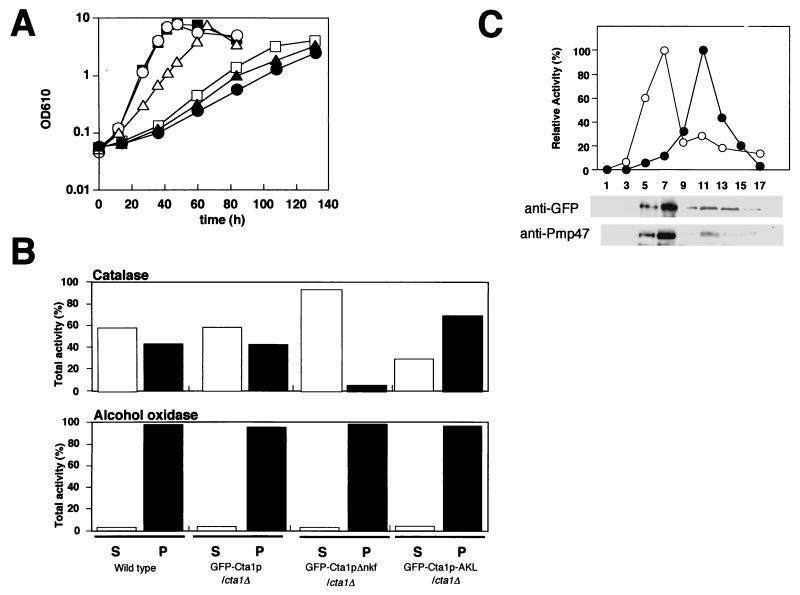
Next, we asked whether peroxisomal localization of Cta1p is necessary for its physiological function. When Cta1p was expressed in cta1Δ cells, the Cta1p/cta1Δ strain was able to grow in methanol medium at a level comparable to the level of the wild-type strain. In contrast, Cta1pΔnkf, in which the PTS1 motif sequence -NKF is not present, could not complement the growth defect of the cta1Δ strain (Fig. 3A). Since Cta1pΔnkf was present in an enzymatically active form and its enzyme activity was much higher than the activity of the wild-type strain due to overexpression under the control of the AOD1 promoter (data not shown), the loss of the physiological function of Cta1pΔnkf may have been due to the deficiency of peroxisomal transport. To confirm this, we expressed two GFP-Cta1p fusion proteins, GFP-Cta1p and GFP-Cta1pΔnkf, in cta1Δ cells under the control of the actin promoter (the C. boidinii GFP-Cta1p/cta1Δ strain and the GFP-Cta1pΔnkf/cta1Δ strain, respectively). These GFP-tagged proteins showed catalase activity in vivo, and GFP-Cta1p could complement the growth defect of the cta1Δ strain on methanol medium (Fig. 3A), indicating that these fusion proteins were physiologically functional. The partial complementation may have been due to the low level of enzymatic activity of the C. boidinii GFP-Cta1p/cta1Δ strain (1/70 of the wild-type level), whose expression was driven by the actin promoter. The localization of GFP-Cta1p and GFP-Cta1pΔnkf was analyzed by subjecting each strain to differential centrifugation, which separated the intracellular components into a cytosolic supernatant and an organelle pellet fraction consisting mainly of peroxisomes and mitochondria (Fig. 3B). About 40% of the catalase activity was detected in the organelle pellet fraction in the GFP-Cta1p/cta1Δ cells, while more than 90% of the catalase activity in the GFP-Cta1pΔnkf/cta1Δ strain was found in the cytosolic supernatant fraction (Fig. 3B). These results indicate that transport of GFP-Cta1p fusion protein into the peroxisome was necessary to complement the growth defect of the cta1Δ strain.
FIG. 3.
Carboxyl-terminal -NKF of Cta1p is necessary for peroxisomal transport. (A) Growth on methanol medium of cta1Δ strains expressing Cta1p and GFP-tagged Cta1ps. Symbols: ○, wild-type strain; ▪, Cta1p/cta1Δ strain; □, Cta1pΔnkf/cta1Δ strain; ▵, GFP-Cta1p/cta1Δ strain; ▴, GFP-Cta1pΔnkf/cta1Δ strain; ●, cta1Δ URA3 strain. OD610, optical density at 610 nm. (B) Differential centrifugation of organelle pellet and supernatant fractions from the wild type, the GFP-Cta1p/cta1Δ strain, the GFP-Cta1pΔnkf/cta1Δ strain, and the GFP-Cta1p-AKL/cta1Δ strain. Methanol-induced cells were lysed by osmotic shock, and cell debris was removed by centrifugation at 500 × g. The resulting supernatant was separated by centrifugation at 20,000 × g into a pelletable fraction (P), including peroxisomes and mitochondria, and supernatant (S) and resuspended in the same volume of buffer. Catalase and AOD activities of each fraction were then assayed as described in Materials and Methods. (C) The organelle pellet fraction of the GFP-Cta1p/cta1Δ strain after centrifugation at 20,000 × g was further fractionated by Nycodenz equilibrium density gradient centrifugation. A relative activity of 100% for catalase (○) corresponded to 453 U/ml for the GFP-Cta1p/cta1Δ strain, and a relative activity of 100% for cytochrome c oxidase (●) corresponded to 0.156 U/ml for this strain. Peroxisomal localization of GFP-Cta1p was confirmed by Western blotting by using anti-GFP antibody with the GFP-Cta1p/cta1Δ strain.
Efficiency of catalase import into the peroxisome depends on the growth carbon source.
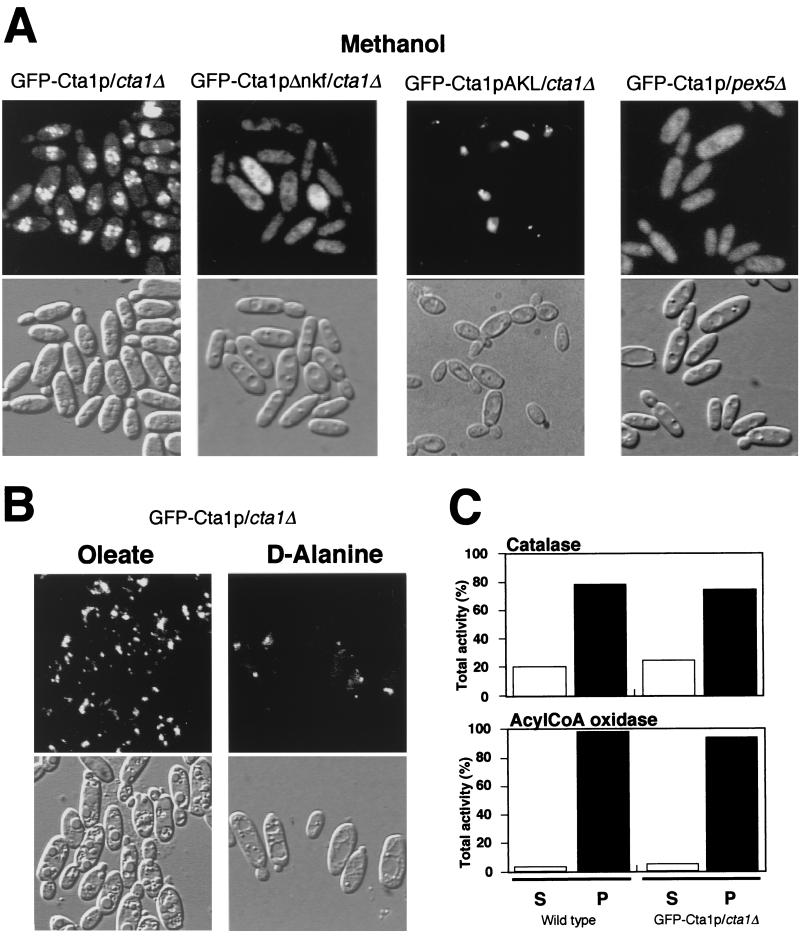
To further confirm the subcellular localization of GFP-Cta1p fusion proteins in C. boidinii, methanol-grown cells expressing GFP-Cta1p or GFP-Cta1pΔnkf were observed by fluorescent microscopy. While GFP fluorescence was distributed throughout the cytosol in the GFP-Cta1pΔnkf/cta1Δ cells, the GFP-Cta1p/cta1Δ cells had a bimodal distribution of GFP fluorescence in peroxisomes and the cytosol. These results were in accordance with the results of the biochemical experiments described above (Fig. 3B). Since more than 90% of peroxisomal AOD activity was detected in the organelle pellet fraction in both strains and a similar low pelletability of catalase in the methanol-induced wild-type strain was observed (Fig. 3B), the lower pelletability of catalase activity than of AOD activity in the GFP-Cta1p/cta1Δ strain was not due to rupture of peroxisomes or fusion of GFP. The pelletable catalase activity of GFP-Cta1p comigrated with the peroxisomal marker Pmp47 protein during Nycodenz equilibrium density gradient centrifugation (Fig. 3C). On the basis of these results, we concluded that catalase is present both in peroxisomes and in the cytosol in methanol-grown cells of C. boidinii. In the pex5Δ strain, most catalase activity was present in the cytosolic fraction, and GFP-Cta1p had a cytosolic distribution (33) (Fig. 4A). This indicated that the imported portion of Cta1p was transported to peroxisomes via the PTS1 pathway in C. boidinii.
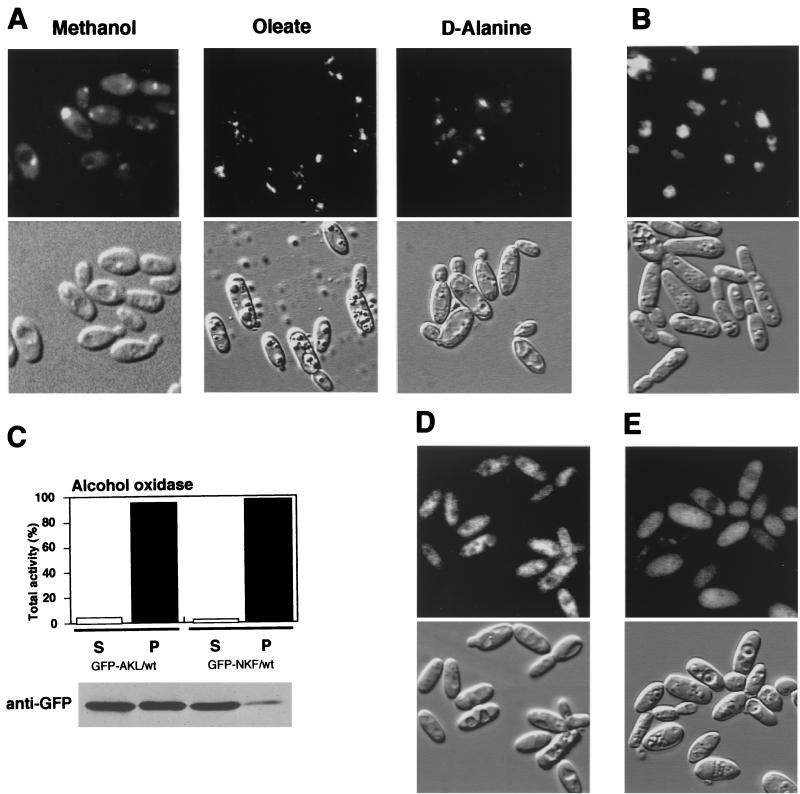
FIG. 4.
Import efficiency of peroxisomal catalase. (A) Fluorescence and Nomarski images of the GFP-Cta1p/cta1Δ strain, the GFP-Cta1pΔnkf/cta1Δ strain, the GFP-Cta1p-AKL/cta1Δ strain, and the GFP-Cta1p/pex5Δ strain in methanol medium. (B) Fluorescence and Nomarski images of the GFP-Cta1p/cta1Δ strain in oleate and d-alanine media. (C) Enzyme activities after differential centrifugation of the wild-type and GFP-Cta1p/cta1Δ strains grown on oleate medium that resulted in cytosolic supernatant (S) and an organelle pellet fraction (P). AcylCoA, acyl coenzyme A.
Another significant observation was that the bimodal distribution of GFP-Cta1p fluorescence did not occur in cells grown on oleate or d-alanine (Fig. 4B). Oleate- or d-alanine-grown cells had a clear punctate pattern of fluorescence representing peroxisomal localization, and these cells did not exhibit cytosolic fluorescence like that observed in methanol-grown cells. Since the actin promoter used gave rise to constant gene expression and the expressed protein level was also constant as determined by Western blot analysis (data not shown), the change in the localization was not likely to be the result of a change in the level of expression of the proteins. Oleate-grown cells of the GFP-Cta1p/cta1Δ strain and of the wild-type strain were also subjected to subcellular fractionation (Fig. 4C). In both strains, more than 95% of the acyl coenzyme A oxidase activity and 75 to 80% of the catalase activity were detected in the organelle pellet fraction. These biochemical and fluorescence analyses showed that the efficiency of catalase import is high in oleate- and d-alanine-grown cells and low in methanol-grown cells.
Peroxisomal transport efficiency of PTS1 sequences and their interaction with CbPex5p.
Next, we asked whether the -NKF sequence is sufficient for peroxisomal transport in C. boidinii. This question was addressed by fusing the -NKF sequence to the C terminus of GFP and expressing it in the wild-type strain. While GFP-NKF was observed to be imported into peroxisomes in oleate- and d-alanine-grown cells (Fig. 5A), much of the GFP-NKF fluorescence was observed in the cytosol, as well as in peroxisomes, of methanol-grown cells (Fig. 5A). Another PTS1 sequence, -AKL, which is present in Pmp20 and the d-amino acid oxidase of C. boidinii, delivers GFP to peroxisomes efficiently in methanol-grown cells (Fig. 5B) (42). Subcellular fractionation experiments performed with anti-GFP antibody also revealed that the amount of GFP fusion protein in the organelle pelletable fraction was larger for GFP-AKL than for GFP-NKF (Fig. 5C). In contrast, both the GFP without the PTS1 sequence (Fig. 5D) and the GFP-AKL expressed in the pex5Δ strain (Fig. 5E) showed cytosolic fluorescence. These experiments revealed that the efficiency of peroxisomal transport for GFP-NKF is lower than that for GFP-AKL.
FIG. 5.
Import efficiency of PTS motifs. (A) Fluorescence and Nomarski images of the GFP-NKF/wt strain in methanol, oleate, and d-alanine media. (B) Fluorescence and Nomarski images of the GFP-AKL/wt strain in methanol medium. (C) Enzyme activity after differential centrifugation of the GFP-AKL/wt and GFP-NKF/wt strains grown on methanol medium that resulted in cytosolic supernatant (S) and an organelle pellet fraction (P). The AOD activity of each fraction was assayed as described in Materials and Methods. The GFP-PTS1 proteins were detected by Western blotting using anti-GFP antibody. (D) Fluorescence and Nomarski images of the GFP-Stop/wt strain in methanol medium. (E) Fluorescence and Nomarski images of the GFP-AKL/pex5Δ strain in methanol medium.
Next, the interactions of the two PTS1 sequences with the PTS1 receptor CbPex5p were analyzed by using a yeast two-hybrid system, in which β-galactosidase activity was previously shown to reflect the binding of various PTS1 sequences to Pex5p proteins (23). Significant interactions with CbPex5p were detected for Cta1p (39.7 U/mg of protein), GFP-NKF (31.2 U/mg of protein), and GFP-AKL (68.5 U/mg of protein) (Table 2). On the other hand, Cta1pΔnkf, GFP-Stop, and other control constructs exhibited background levels of interaction. These experiments confirmed that Cta1p interacted with CbPex5p through its carboxyl-terminal -NKF sequence and that the level of interaction of GFP-NKF with CbPex5p is about half that of GFP-AKL.
TABLE 2.
Interaction between GFP-PTS1s and CbPex5p in the two-hybrid systema
| Binding domain | Activation domain | Growth | β-Galactosidase activity (U/mg of protein) |
|---|---|---|---|
| Cta1p | Pex5p | + | 39.7 ± 2.2 |
| Cta1p | None | − | 4.22 ± 1.01 |
| Cta1pΔnkf | Pex5p | − | 5.81 ± 0.39 |
| Cta1pΔnkf | None | − | 5.71 ± 0.47 |
| GFP-NKF | Pex5p | + | 31.2 ± 2.6 |
| GFP-NKF | None | − | 6.12 ± 1.08 |
| GFP-AKL | Pex5p | ++ | 68.5 ± 1.5 |
| GFP-AKL | None | − | 4.33 ± 0.87 |
| GFP-Stop | Pex5p | − | 5.25 ± 0.65 |
| GFP-Stop | None | − | 6.65 ± 0.26 |
| None | Pex5p | − | 3.87 ± 1.02 |
S. cerevisiae YRG-2 cells were transformed with plasmids as indicated and were grown on glucose media lacking histidine (but containing 20 mM 3-amino-1,2,4-triazole). ++, single colonies after 3 days of incubation at 30°C; +, single colonies after 5 days of incubation at 30°C; −, no growth. β-Galactosidase activities were measured photometrically at 420 nm by using the substrate o-nitrophenyl-β-d-galactopyranoside. The β-galactosidase activities shown are averages based on triplicate measurements for three independent transformants harboring each set of plasmids.
Finally, we expressed a GFP-Cta1p fusion protein having an optimal PTS1 -AKL sequence under the control of the actin promoter (C. boidinii GFP-Cta1p-AKL/cta1Δ strain). The results of a biochemical fractionation experiment (Fig. 3B) and fluorescence analysis (Fig. 4A) confirmed that GFP-Cta1p-AKL was transported to peroxisomes more efficiently than GFP-Cta1p having the original PTS1 sequence, -NKF. These experiments showed that the carboxyl-terminal PTS1 sequence primarily determines the efficiency of peroxisomal transport of catalase.
Oligomeric transport of GFP-Cta1pΔnkf and oligomer formation with native Cta1p.
We monitored import of the oligomeric form of catalase into peroxisomes by using GFP-tagged Cta1pΔnkf in living cells. When GFP-Cta1pΔnkf was introduced into the C. boidinii wild-type strain expressing full-length catalase (GFP-Cta1pΔnkf/wt strain), GFP-Cta1pΔnkf exhibited fluorescence similar to that in the GFP-Cta1p/cta1Δ strain, showing that GFP-Cta1pΔnkf was transported into peroxisomes even though it lacked the PTS1 sequence (Fig. 6A). A subcellular fractionation experiment performed with GFP-Cta1pΔnkf/wt cells confirmed that the GFP-Cta1pΔnkf protein was present in the peroxisomal fraction (Fig. 6B). In contrast, the GFP-Cta1pΔnkf/cta1Δ strain showed cytosolic GFP fluorescence (Fig. 4A).
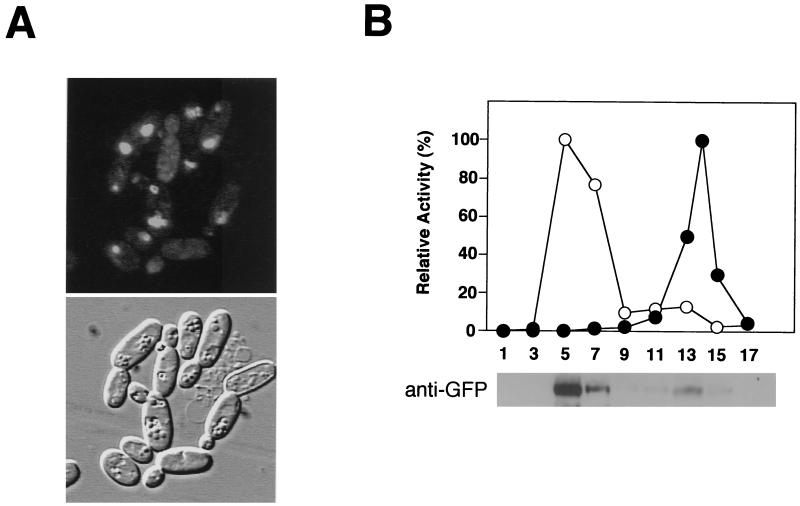
FIG. 6.
-NKF-truncated Cta1p is transported into peroxisomes in wild-type cells. (A) Fluorescence and Nomarski images of the GFP-Cta1pΔnkf/wt strain. (B) Methanol-induced cells were lysed by osmotic shock, and cell debris was removed by centrifugation at 500 × g. The resulting supernatant was separated by centrifugation at 20,000 × g into a pelletable fraction, including peroxisomes and mitochondria, and supernatant. The organelle pellet fraction after centrifugation at 20,000 × g was further fractionated by Nycodenz equilibrium density gradient centrifugation. A relative activity of 100% for catalase (○) corresponded to 1,970 U/ml for the GFP-Cta1pΔnkf/wt strain, and a relative activity of 100% for cytochrome c oxidase (●) corresponded to 0.208 U/ml for the GFP-Cta1pΔnkf/wt strain. Peroxisomal localization of GFP-Cta1pΔnkf was confirmed by Western blotting using anti-GFP antibody with the GFP-Cta1pΔnkf/wt strain.
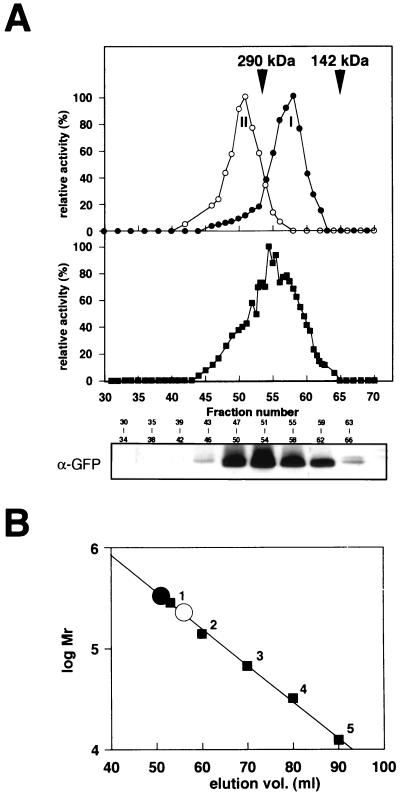
Next, we asked whether the GFP-Cta1pΔnkf protein could form an active heterooligomer with a native Cta1p subunit in peroxisomes, and gel permeation chromatography was performed with the peroxisomal fractions from the different strains. In the wild-type peroxisomal fraction, the peak of catalase activity was detected around an elution volume of 56 ml (Fig. 7A), corresponding to ca. 230 kDa, which coincided with the molecular mass of a tetrameric form of Cta1p (Fig. 7B) (peak I). On the other hand, the peroxisomal fraction from the GFP-Cta1p/cta1Δ strain had a peak of catalase activity around an elution volume of 51 ml (Fig. 7A) (peak II), which represented a higher molecular mass than that of the 290-kDa marker protein (around ca. 350 kDa), which is consistent with the molecular mass of a tetrameric form of the GFP-Cta1p protein (Fig. 7). When the peroxisomal fraction from the GFP-Cta1pΔnkf/wt strain was used, several catalase activity peaks were detected between peaks I and II, and Western blot analysis with anti-GFP antibody showed that the fusion protein was indeed present in these fractions (Fig. 7A). These results strongly suggested that the GFP-Cta1pΔnkf protein could form active hybrid oligomers with the Cta1p subunit and that the GFP-Cta1pΔnkf protein was transported to peroxisomes after oligomer formation with Cta1p.
FIG. 7.
Formation of hybrid oligomers of Cta1p and GFP-Cta1pΔnkf in peroxisomes of the GFP-Cta1pΔnkf/wt strain. (A) Size exclusion chromatography with Cta1p and GFP-Cta1pΔnkf. Cell extracts of methanol-induced cells of the wild-type strain (●) and the GFP-Cta1pΔnkf/cta1Δ strain (○) and a peroxisomal fraction (▪) (Fig. 6B, fraction 5) of the GFP-Cta1pΔnkf/wt strain were loaded onto a HiLoad 16/60 Superdex pg column that had been equilibrated with a buffer containing 100 mM potassium phosphate buffer (pH 7.5). The catalase activity of the column effluent was assayed. The arrows indicate the positions of proteins used as molecular mass standards (see Materials and Methods). A relative activity of 100% for catalase corresponded to 312 U/ml for the cell extract from the wild-type strain, to 214 U/ml for the cell extract from the GFP-Cta1pΔnkf/cta1Δ strain, and to 106 U/ml for the cell extract from the GFP-Cta1pΔnkf/wt strain. The GFP-Cta1pΔnkf protein was also detected by Western blotting using anti-GFP antibody. (B) Estimation of the sizes of the native tetramer (○) and GFP-Cta1pΔnkf tetramer (●) on the Hi Load 16/60 Superdex pg column. The calibration line was determined with the following standards: glutamate dehydrogenase (290 kDa) (1), lactate dehydrogenase (142 kDa) (2), enolase (67 kDa) (3), adenylate kinase (32 kDa) (4), and cytochrome c (12.4 kDa) (5). Each calibration point is the result of at least two independent measurements in the same buffer containing 100 mM potassium phosphate (pH 7.5).
DISCUSSION
In this study, we monitored transport of peroxisomal catalase in the methylotrophic yeast C. boidinii, and on the basis of the results obtained, we were able to elucidate its metabolic significance.
The CTA1 gene encoding peroxisomal catalase was cloned from the C. boidinii genome. The fact that growth of the cta1Δ strain was inhibited with all of the peroxisome-inducible carbon sources used indicates that catalase plays a general metabolic role in scavenging H2O2 produced by peroxisomal oxidase. This result was distinct from the results obtained for another peroxisomal antioxidant protein, CbPmp20 (20), as gene knockout studies established that this protein plays a role only in methanol medium. The affinity of Cta1p for H2O2 (Km for H2O2, 25 mM) is lower than that of CbPmp20 (Km for H2O2, 2.86 mM). However, since the cta1Δ strain accumulated H2O2 during incubation in methanol medium, peroxisomal catalase apparently plays a major role in the degradation of H2O2 (20).
The transport of Cta1p was monitored by using GFP fusion proteins as fluorescent and immunogenic tags. Although in some previous studies workers have used GFP-PTS1 proteins to monitor peroxisomal transport in vivo, our results demonstrate that GFP fluorescence images can be used to evaluate the efficiency of peroxisomal import. The transport efficiency of peroxisomal proteins has also been analyzed in biochemical fractionation experiments by procedures that require careful manipulation and skill of the investigators to compensate for the fragile nature of peroxisomes. Our GFP fluorescence analysis procedure, although qualitative, can readily be used to analyze peroxisomal and cytosolic localization of GFP fusion proteins in vivo, and the results are consistent with those of conventional biochemical fractionation experiments. Digital image analyses should provide more quantitative information regarding transport efficiency.
One line of evidence has suggested that peroxisomal catalase enters peroxisomes after oligomer formation, but this has not been confirmed directly. In the present study we were able to confirm by fluorescent microscopy in a relatively simple and straightforward manner that catalase transport to peroxisomes occurs in an oligomeric form.
The efficiency of peroxisomal transport in relation to various PTS1 signals was studied previously with S. cerevisiae in a homologous context (9). Subsequently, randomly synthesized PTS1 sequences were extensively studied to determine their ability to target the reporter protein to peroxisomes and their ability to bind to Pex5p in human and S. cerevisiae cells (23). These previous studies revealed that the functional PTS1 sequences are species specific, based on the binding activity to the Pex5p of the organism. Indeed, the interaction of the PTS1 sequence from Cta1p, -NKF, with CbPex5p was about half that of the Pmp20 and d-amino acid oxidase sequence, -AKL, in the yeast two-hybrid system. This finding further emphasizes the point that the binding activity of the PTS1 sequence directly affects the efficiency of peroxisomal transport.
Previous studies focusing exclusively on the transport efficiency of PTS1 signals have not taken into account the metabolic aspects of the native PTS1 proteins. By using C. boidinii, three independent peroxisomal metabolic processes could be controlled by the carbon sources used, and we were able to assess the transport efficiency of Cta1p in relation to peroxisomal metabolism (i.e., the content and amount of peroxisomal proteins). Oleate- or d-alanine-grown cells showed normal localization of both GFP-Cta1p and Cta1p exclusively in peroxisomes. In contrast, these same proteins were distributed bimodally between the cytosol and peroxisomes in methanol-grown cells. Methanol-induced peroxisomes contain large amounts of PTS1 proteins, AOD, and dihydroxyacetone synthase, which account for nearly 80% of the total soluble protein. Oleate- or d-alanine-induced peroxisomes do not contain such large amounts of peroxisomal proteins. Hence, in methanol-induced cells, the majority of CbPex5p and other peroxin molecules may be used for transport of two major peroxisomal proteins, and as a result, the transport efficiency of Cta1p is low due to its weak interaction with CbPex5p. Furthermore, cytosolic catalase activity is very low or not detectable in C. boidinii cells, and due to its bimodal distribution Cta1p in methanol-grown cells may help scavenge cytosolic H2O2 which has leaked from peroxisomes. In fact, expression of Cta1pΔnkf slightly stimulated the growth of the cta1Δ strain on methanol (Fig. 3A). Thus, the bimodal distribution of H2O2-scavenging peroxisomal catalase does not confer a physiological disadvantage on the cells. Clearly, this is different from the situation in which there is aberrant localization of H2O2-generating oxidases in the cytosol which should result in major cellular damage. In addition, even under the same conditions in methanol-induced cells, CbPmp20 was efficiently transported into peroxisomes, where it could carry out its antioxidant function, probably because the loss of CbPmp20 from methanol-induced peroxisomes resulted in immediate cell death (20). PTS1 motif sequences which do not function with the minimum three amino acids have been reported for peroxisomal catalyzes from humans and S. cerevisiae (-KANL and -SSNSKF, respectively) (21, 34), in which four or six carboxyl-terminal amino acid sequences are necessary for peroxisomal transport. Residues upstream of the tripeptide influenced the strength of the interaction, and the molecular basis of the interaction was recently revealed by the three-dimensional structure of human Pex5p (11). The catalase PTS1 sequences may have evolved in such a way that peroxisomal enzymes other than catalases (i.e., enzymes requiring exclusive localization to peroxisomes) have priority for peroxisomal transport. Differences in PTS1 motif sequences may reflect the priority of peroxisomal transport.
The efficiency of peroxisomal transport can be considered an important determinant in peroxisomal disorders. The loss of catalase import into peroxisomes has been reported to cause severe peroxisomal dysfunction in human skin fibroblast cell lines in which the import of other PTS1 and PTS2 signal-containing proteins was normal (44). Since peroxisomal functions in a mutant cell line could not be restored by normal catalase (catalase-KANL protein) but could be restored by catalase fused to the efficient PTS1 sequence (catalase-SKL protein) (44), we speculated that catalase import deficiency in the cell line is due to retarded efficiency of general peroxisomal transport caused by mild peroxine mutation (e.g., Pex5p). Indeed, Bottger et al. showed that in an S. cerevisiae pex5 mutant catalase was completely mislocalized to the cytosol while other PTS1 proteins were unaffected (2). In these cases, catalase transport, whose priority is low, may be the first to be affected. Although peroxisomal transport efficiency has not been fully considered due to technical difficulties, GFP technology as described in this paper should provide a new approach for assessing the transport efficiencies of various peroxisomal proteins, and the resulting information may help establish effective gene therapy for peroxisomal disorders.
ACKNOWLEDGMENTS
We are very grateful to Joel M. Goodman (University of Texas Southwestern Medical Center, Dallas) for his generous gift of valuable reagents and to M. Fransen (Katholieke Universiteit, Louvain, Belgium) for his generous gift of the anti-GFP antibody.
This research was supported by a grant-in-aid for scientific research from the Ministry of Education, Science, Sports, and Culture of Japan.
REFERENCES
- 1.Bergmeyer H U. Zur Messung von Katalase Aktivitaten. Biochem Z. 1955;327:255–258. [PubMed] [Google Scholar]
- 2.Bottger G, Barnett P, Klein A T J, Kragt A, Tabak H F, Distel B. Saccharomyces cerevisiae PTS1 receptor Pex5p interacts with the SH3 domain of the peroxisomal membrane protein Pex13p in an unconventional, non-PXXP-related manner. Mol Biol Cell. 2000;11:3963–3976. doi: 10.1091/mbc.11.11.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Brul A, Wiemer E A, Westerveld A, Strijland A, Wanders R J, Schram A W, Heymans H S, Schutgens R B, van den Bosch H, Tager J M. Kinetics of the assembly of peroxisomes after fusion of complementary cell lines from patients with the cerebro-hepato-renal (Zellweger) syndrome and related disorders. Biochem Biophys Res Commun. 1988;152:1083–1089. doi: 10.1016/s0006-291x(88)80395-4. [DOI] [PubMed] [Google Scholar]
- 5.Church G M, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Duve C, Baudhuin P. Peroxisomes (microbodies and related particles) Physiol Rev. 1966;46:323–357. doi: 10.1152/physrev.1966.46.2.323. [DOI] [PubMed] [Google Scholar]
- 7.Didion T, Roggenkamp R. Targeting signal of the peroxisomal catalase in the methylotrophic yeast Hansenula polymorpha. FEBS Lett. 1992;303:113–116. doi: 10.1016/0014-5793(92)80500-g. [DOI] [PubMed] [Google Scholar]
- 8.Dyer J M, McNew J A, Goodman J M. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol. 1996;133:269–280. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elgersma Y, Vos A, van den Berg M, van Roermund C W T, van der Sluijs P, Distel B, Tabak H F. Analysis of the carboxyl-terminal peroxisomal targeting signal 1 in a homologous context in Saccharomyces cerevisiae. J Biol Chem. 1996;271:26375–26382. doi: 10.1074/jbc.271.42.26375. [DOI] [PubMed] [Google Scholar]
- 10.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 11.Gatto G J J, Geisbrecht B V, Gould S J, Berg J M. Peroxisomal targeting signal-1 recognition by the TPR domains of human PEX5. Nat Struct Biol. 2000;7:1091–1095. doi: 10.1038/81930. [DOI] [PubMed] [Google Scholar]
- 12.Gietl C, Faber K N, van der Klei I J, Veenhuis M. Mutational analysis of the N-terminal topogenic signal of watermelon glyoxysomal malate dehydrogenase using the heterologous host Hansenula polymorpha. Proc Natl Acad Sci USA. 1994;91:3151–3155. doi: 10.1073/pnas.91.8.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glover J R, Andrews D W, Rachubinski R A. Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc Natl Acad Sci USA. 1994;91:10541–10545. doi: 10.1073/pnas.91.22.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodman J M, Scott C W, Donahue P N, Atherton J P. Alcohol oxidase assembles post-translationally into the peroxisome of Candida boidinii. J Biol Chem. 1984;259:8485–8493. [PubMed] [Google Scholar]
- 15.Goodman J M, Trapp S B, Hwang H, Veenhuis M. Peroxisomes induced in Candida boidinii by methanol, oleic acid and d-alanine vary in metabolic function but share common integral membrane proteins. J Cell Sci. 1990;97:193–204. doi: 10.1242/jcs.97.1.193. [DOI] [PubMed] [Google Scholar]
- 16.Gould S J, Keller G A, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guibault G G, Brignac P J, Jr, Juneau M. New substrates for the fluorometric determination of oxidative enzymes. Anal Chem. 1968;40:1256–1263. doi: 10.1021/ac60264a027. [DOI] [PubMed] [Google Scholar]
- 18.Hansen H, Roggenkamp R. Functional complementation of catalase-defective peroxisomes in a methylotrophic yeast by import of the catalase A from Saccharomyces cerevisiae. Eur J Biochem. 1989;184:173–179. doi: 10.1111/j.1432-1033.1989.tb15004.x. [DOI] [PubMed] [Google Scholar]
- 19.Harman L S, Carver D K, Schreiber J, Manson R P. One- and two-electron oxidation of reduced glutathione by peroxidases. J Biol Chem. 1986;261:1642–1648. [PubMed] [Google Scholar]
- 20.Horiguchi H, Yurimoto H, Kato N, Sakai Y. Antioxidant system within yeast peroxisome: biochemical and physiological characterization of CbPmp20 in the methylotrophic yeast Candida boidinii. J Biol Chem. 2001;276:14279–14288. doi: 10.1074/jbc.M011661200. [DOI] [PubMed] [Google Scholar]
- 21.Kragler F, Langeder A, Raupachova J, Binder M, Hartig A. Two independent peroxisomal targeting signals in catalase A of Saccharomyces cerevisiae. J Cell Biol. 1993;120:665–673. doi: 10.1083/jcb.120.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 23.Lametschwandtner G, Brocard C, Fransen M, van Veldhoven P, Berger J, Hartig A. The difference in recognition of terminal tripeptides as peroxisomal targeting signal 1 between yeast and human is due to different affinities of their receptor Pex5p to the cognate signal and to residues adjacent to it. J Biol Chem. 1998;273:33635–33643. doi: 10.1074/jbc.273.50.33635. [DOI] [PubMed] [Google Scholar]
- 24.Lazarow P B, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 25.Lee M S, Mullen R T, Trelease R N. Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell. 1997;9:185–197. doi: 10.1105/tpc.9.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieper J M, Oatey P B, Danpure C J. Inhibition of alanine:glyoxylate aminotransferase 1 dimerization is a prerequisite for its peroxisome-to-mitochondrion mistargeting in primary hyperoxaluria type 1. J Cell Biol. 1996;135:939–951. doi: 10.1083/jcb.135.4.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCammon M T, Dowds C A, Orth K, Moomaw C R, Slaughter C A, Goodman J M. Sorting of peroxisomal membrane protein PMP47 from Candida boidinii into peroxisomal membranes of Saccharomyces cerevisiae. J Biol Chem. 1990;265:20098–20105. [PubMed] [Google Scholar]
- 28.McCammon M T, McNew J A, Willy P J, Goodman J M. An internal region of the peroxisomal membrane protein PMP47 is essential for sorting to peroxisomes. J Cell Biol. 1994;124:915–925. doi: 10.1083/jcb.124.6.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNew J A, Goodman J M. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Middelkoop E, Wiemer E A C, Schoenmaker D E T, Strijland A, Tager J M. Topology of catalase assembly in human skin fibroblasts. Biochim Biophys Acta. 1993;1220:15–20. doi: 10.1016/0167-4889(93)90091-3. [DOI] [PubMed] [Google Scholar]
- 31.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 32.Murray W W, Rachubinski R. The nucleotide sequence of complementary DNA and the deduced amino acid sequence of peroxisomal catalase of the yeast Candida tropicalis pK233. Gene. 1987;61:401–413. doi: 10.1016/0378-1119(87)90202-2. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa T, Imanaka T, Morita M, Ishiguro K, Yurimoto H, Yamashita A, Kato N, Sakai Y. Peroxisomal membrane protein Pmp47 is essential in the metabolism of middle-chain fatty acid in yeast peroxisomes and is associated with peroxisome proliferation. J Biol Chem. 2000;275:3455–3461. doi: 10.1074/jbc.275.5.3455. [DOI] [PubMed] [Google Scholar]
- 34.Purdue P E, Lazarow P B. Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J Cell Biol. 1996;134:849–862. doi: 10.1083/jcb.134.4.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai Y, Akiyama M, Kondoh H, Shibano Y, Kato N. High-level secretion of fungal glucoamylase using the Candida boidinii gene expression system. Biochim Biophys Acta. 1996;1308:81–87. doi: 10.1016/0167-4781(96)00075-9. [DOI] [PubMed] [Google Scholar]
- 36.Sakai Y, Goh T K, Tani Y. High-frequency transformation of a methylotrophic yeast, Candida boidinii, with autonomously replicating plasmids which are also functional in Saccharomyces cerevisiae. J Bacteriol. 1993;175:3556–3562. doi: 10.1128/jb.175.11.3556-3562.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakai Y, Kazarimoto T, Tani Y. Transformation system for an asporogenous methylotrophic yeast, Candida boidinii: cloning of the orotidine-5′-phosphate decarboxylase gene (URA3), isolation of uracil auxotrophic mutants, and use of the mutants for integrative transformation. J Bacteriol. 1991;173:7458–7463. doi: 10.1128/jb.173.23.7458-7463.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakai Y, Matsuo H, He K Z, Saiganji A, Yurimoto H, Takabe K, Saiki H, Kato N. Isolation and characterization of mutants of the methylotrophic yeast Candida boidinii S2 that are impaired in growth on peroxisome-inducing carbon sources. Biosci Biotechnol Biochem. 1995;59:869–875. [Google Scholar]
- 39.Sakai Y, Rogi T, Yonehara T, Kato N, Tani Y. High-level ATP production by a genetically-engineered Candida yeast. Bio/Technology. 1994;12:291–293. doi: 10.1038/nbt0394-291. [DOI] [PubMed] [Google Scholar]
- 40.Sakai Y, Saiganji A, Yurimoto H, Takabe K, Sakai H, Kato N. The absence of Pmp47, a putative yeast peroxisomal transporter, causes a defect in transport and folding of a specific matrix enzyme. J Cell Biol. 1996;134:37–51. doi: 10.1083/jcb.134.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai Y, Tani Y. Directed mutagenesis in an asporogenous methylotrophic yeast: cloning, sequencing, and one-step gene disruption of the 3-isopropylmalate dehydrogenase gene (LEU2) of Candida boidinii to derive doubly auxotrophic marker strains. J Bacteriol. 1992;174:5988–5993. doi: 10.1128/jb.174.18.5988-5993.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sakai Y, Yurimoto H, Matsuo H, Kato N. Regulation of peroxisomal proteins and organelle proliferation by multiple carbon sources in the methylotrophic yeast Candida boidinii. Yeast. 1998;14:1175–1187. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1175::AID-YEA319>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 44.Sheikh F G, Pahan K, Khan M, Barbosa E, Singh I. Abnormality in catalase import into peroxisomes leads to severe neurological disorder. Proc Natl Acad Sci USA. 1998;95:2961–2966. doi: 10.1073/pnas.95.6.2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimozawa N, Tsukamoto T, Suzuki Y, Orii T, Shirayoshi Y, Mori T, Fujiki Y. A human gene responsible for Zellweger syndrome that affects peroxisome assembly. Science. 1992;255:1132–1134. doi: 10.1126/science.1546315. [DOI] [PubMed] [Google Scholar]
- 46.Subramani S. Components involved in peroxisome import, biogenesis, proliferation, turnover, and movement. Physiol Rev. 1998;78:171–188. doi: 10.1152/physrev.1998.78.1.171. [DOI] [PubMed] [Google Scholar]
- 47.Tani Y, Sakai Y, Yamada H. Production of formaldehyde by a mutant of methanol yeast, Candida boidinii S2. J Ferment Technol. 1985;63:443–449. [Google Scholar]
- 48.Tolbert N E. Isolation of subcellular organelles of metabolism on isopycnic sucrose gradients. Methods Enzymol. 1974;31:734–746. doi: 10.1016/0076-6879(74)31077-4. [DOI] [PubMed] [Google Scholar]
- 49.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T, Osumi T. Characterization of the signal peptide at the amino terminus of the rat peroxisomal 3-ketoacyl-CoA thiolase precursor. J Biol Chem. 1994;269:6001–6010. [PubMed] [Google Scholar]
- 51.Ueda M, Mozaffar S, Tanaka A. Catalase from Candida boidinii 2201. Methods Enzymol. 1990;188:463–467. doi: 10.1016/0076-6879(90)88074-k. [DOI] [PubMed] [Google Scholar]
- 52.van den Bosch H, Schutgens R B H, Wanders R J A, Tager J M. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–197. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- 53.van der Leij I, van der Berg M, Boot R, Franse M, Distel B, Tabak H F. Isolation of peroxisome assembly mutants from Saccharomyces cerevisiae with different morphologies using a novel positive selection procedure. J Cell Biol. 1992;119:153–162. doi: 10.1083/jcb.119.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walton P A, Hill P E, Subramani S. Import of stably folded proteins into peroxisomes. Mol Biol Cell. 1995;6:675–683. doi: 10.1091/mbc.6.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wanders R J, Strijland A, van Roermund C W, van den Bosch H, Schutgens R B, Tager J M, Schram A W. Catalase in cultured skin fibroblasts from patients with the cerebro-hepato-renal (Zellweger) syndrome: normal maturation in peroxisome-deficient cells. Biochim Biophys Acta. 1987;923:478–482. doi: 10.1016/0304-4165(87)90057-2. [DOI] [PubMed] [Google Scholar]
- 56.Waterham H R, Russell K A, de Vries Y, Cregg J M. Peroxisomal targeting, import, and assembly of alcohol oxidase in Pichia pastoris. J Cell Biol. 1997;139:1419–1431. doi: 10.1083/jcb.139.6.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamashita H, Avraham S, Jiang S, London R, Van Veldhoven P P, Subramani S, Rogers R A, Avraham H. Characterization of human and murine PMP20 peroxisomal proteins that exhibit antioxidant activity in vitro. J Biol Chem. 1999;274:29897–29904. doi: 10.1074/jbc.274.42.29897. [DOI] [PubMed] [Google Scholar]
- 58.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 59.Yim M B, Chae H Z, Rhee S G, Chock P B, Stadtman E R. On the protective mechanism of the thiol-specific antioxidant enzyme against the oxidative damage of biomacromolecules. J Biol Chem. 1994;269:1621–1626. [PubMed] [Google Scholar]
- 60.Zhang J W, Han Y, Lazarow P B. Peroxisome clustering mutants and peroxisome biogenesis mutants of Saccharomyces cerevisiae. J Cell Biol. 1993;123:1133–1147. doi: 10.1083/jcb.123.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]