Abstract
We have developed a series of powerful and versatile conditional-replication, integration, and modular (CRIM) plasmids. CRIM plasmids can be replicated at medium or high copy numbers in different hosts for making gene (or mutant) libraries. They can be integrated in single copies into the chromosomes of Escherichia coli and related bacteria to study gene function under normal physiological conditions. They can be excised from the chromosome, e.g., to verify that phenotypes are caused by their presence. Furthermore, they can be retrieved singly or en masse for subsequent molecular analyses. CRIM plasmids are integrated into the chromosome by site-specific recombination at one of five different phage attachment sites. Integrants are selected as antibiotic-resistant transformations. Since CRIM plasmids encode different forms of resistance, several can be used together in the same cell for stable expression of complex metabolic or regulatory pathways from diverse sources. Following integration, integrants are stably maintained in the absence of antibiotic selection. Each CRIM plasmid has a polylinker or one of several promoters for ectopic expression of the inserted DNA. Their modular design allows easy construction of new variants with different combinations of features. We also report a series of easily curable, low-copy-number helper plasmids encoding all the requisite Int proteins alone or with the respective Xis protein. These helper plasmids facilitate integration, excision (“curing”), or retrieval of the CRIM plasmids.
Multicopy plasmids have greatly facilitated gene structure-function studies. However, the use of such plasmids can lead to high-copy-number artifacts, especially in physiological studies. Thus, several methods have been developed for recombining genes on bacterial chromosomes in order to study their functions in single copies. Such methods are frequently used to construct novel Escherichia coli strains that stably express foreign genes for use in both basic research and biotechnology (5, 18, 27). However, the development of strains encoding complex metabolic or regulatory pathways poses special problems that often require manipulating many genes and expressing them individually at different levels or under separate regulatory controls. To address these concerns, we have developed a series of plasmid-host systems for the introduction of multiple genes into the same cell in single copies. Our approach is based on genome targeting systems that utilize plasmids carrying a conditional-replication origin and a phage attachment (attP) site (17). We refer to our plasmids as CRIM (conditional-replication, integration, and modular) plasmids. CRIM plasmids can be integrated into or retrieved from their bacterial attachment (attB) site by supplying phage integrase (Int) without or with excisionase (Xis) in trans.
Advantages of our CRIM plasmid-host systems include the use of alternative attP and attB sites (for phages λ, HK022, φ80, P21, and P22) and different selectable markers (for chloramphenicol, gentamicin, kanamycin, spectinomycin and streptomycin, tetracycline, and trimethoprim resistance) in conjunction with a polylinker or promoter (ParaB, PrhaB, PrhaS, Ptac, Psyn1, and Psyn4) for ectopic expression of the cloned gene(s). These CRIM plasmids have the γ replication origin of R6K, which requires the trans-acting Π protein (encoded by pir) for replication. So, they replicate at a medium (15 per cell) or high (250 per cell) plasmid copy number in pir+ or pir-116 (high-copy-number mutant) E. coli hosts (28), respectively. Int helper plasmids are used for integration of CRIM plasmids into the corresponding chromosomal attB sites of normal (non-pir) hosts, which are nonpermissive for CRIM plasmid replication. Xis/Int helper plasmids are used for excision (“curing”) of the respective CRIM plasmids from the chromosome, e.g., to verify that phenotypes are due to their presence. Xis/Int helper plasmids are also used for retrieval (cloning) of CRIM plasmids from the chromosome, e.g., to recover a particular CRIM plasmid after screening of CRIM plasmid or mutant libraries.
Since integration and retrieval involve phage-site-specific recombination events, the original and recovered plasmids are identical. CRIM plasmids can therefore be used for the construction of gene (or mutant) libraries that can be directly integrated into bacterial chromosomes in single copies for screening or selection purposes. Afterwards, CRIM plasmids can be retrieved from individual cells or en masse. The recovered plasmids can then be propagated as plasmids for molecular analysis or integrated directly into the chromosomes of other hosts for subsequent processing without further in vitro manipulation steps. We previously found similar oriRγ attλ plasmids to be extremely useful in mutagenesis studies, especially when it was important that the mutated gene be free of plasmid copy number effects (16). We also found them to be useful in studying genes from diverse bacteria, including gram-negative and -positive cells (14, 15, 25, 34). Our versatile CRIM plasmid-host systems should be widely useful in gene structure-function studies. Here we describe our basic set of CRIM plasmids, the requisite helper plasmids, and how to use them.
MATERIALS AND METHODS
Media and culture conditions.
Luria-Bertani (LB) broth (without glucose), tryptone-yeast extract agar (pH 7.0), and glucose M63 agar were used as complex and minimal media (38). SOB and SOC were prepared as described elsewhere (30). To maintain plasmids, antibiotics (from Sigma, St. Louis, Mo.) were added as follows: ampicillin at 100 μg/ml, chloramphenicol at 25 μg/ml, gentamicin at 15 μg/ml, kanamycin at 50 μg/ml, spectinomycin and streptomycin (together) at 100 μg/ml, trimethoprim at 300 μg/ml, or tetracycline at 12.5 μg/ml. Single-copy integrants were selected using chloramphenicol at 6 μg/ml, gentamicin at 5 μg/ml, kanamycin at 10 μg/ml, spectinomycin and streptomycin (together) at 35 μg/ml, trimethoprim at 25 μg/ml, or tetracycline at 8 μg/ml. We found that proper medium pH is especially important for selection of gentamicin-resistant (Gmr) integrants. Complex media (tryptone-yeast extract agar) were used for selection of all forms of resistance except trimethoprim resistance, for which minimal media were used. Following primary selection, integrants were routinely maintained in the absence of antibiotics.
Bacteria.
All strains are derivatives of E. coli K-12. Normal (self-replicating) plasmids were propagated in DH5α (12), BW5328, or BW25141. Strains from this laboratory are fully described Table 1. The DE(araBAD)567 and DE(rhaBAD)568 mutations correspond to the ΔaraBADAH33 and ΔrhaBADLD78 alleles (14), respectively. The adjacent rrnB3 ΔlacZ4787 mutations were previously called rrnBT14 ΔlacZWJ16 (14). The ΔendA9 allele corresponds to the ΔendA8::tetAR mutation (from BT333 [9]) after Flp-mediated excision of tetAR with pCP20 (9). The recD1014 mutation originated from V355 (from G. C. Walker [33]). The attP22(EcoB) allele refers to the attP22 site of E. coli B, which had been introduced into BW25368 (proA::Tn10) by using P1kc grown on NC3 (E. coli B/r hsdR, also called BW9688 [39]) by selecting proline-independent transductants to make BW25676. Our standard wild-type E. coli K-12 strain is BD792 (36), which is a direct F− descendant of W1485 (2). The rph-1 allele refers to the rph frameshift mutation (19), which is also present in E. coli BD792 (data not shown). Strain BD792, like both its parent, W1485, and wild-type E. coli K-12 EMG2 (20), carries the rpoS396(Am) allele (data not shown). Several strains were therefore made rpoS+. This was done in two steps. A strain was first made tetracycline resistant (Tcr) and kanamycin resistant (Kmr) by using P1kc grown on ZK1001 (cysC95::Tn10 rpoS::kan; from R. Kolter). A resultant Cys− transductant was then made cysteine independent and kanamycin sensitive by using P1kc grown on MG1655. E. coli K-12 strains BW25113, BW25141, BW25142, and BW25695 are descendants of BD792 derivatives that were made rpoS+.
TABLE 1.
Bacterial strains
| Straina | Genotypeb | Pedigreec | Reference(s) and/or derivationd |
|---|---|---|---|
| BW37 | IN(rrnD-rrnE)I tna bglR::ISe trpR ilv rpsL rph-1 | W3110 via BW33 | 38, 40 |
| BW5328 | Δ(lacIZYA argF)U169 rph-1 rpoS396(Am) recA1 robA1 creC510 hsdR514 | BD792 via BW5206 | 37 |
| BW23473 | Δ(lacIZYA argF)U169 rph-1 rpoS396(Am) robA1 creC510 hsdR514 ΔendA9 uidA(ΔMluI)::pir(wt) recA1 | BD792 via BW23438 | 15, 16 |
| BW23838 | lacIqrrnB3 ΔlacZ4787 ΔphoBR580 ΔcreABCD154 hsdR514 Δ(pta ackA hisQ hisP)TA3516phn(EcoB) DE(araBAD)567 DE(rhaBAD)568 rph-1 rpoS396(Am) uidA(ΔMluI)::pir(wt) ΔendA9 recD1014 recA1) | BD792 via BW23832 | Srl+ with P1kc on BW8078; 14 |
| BW24249 | lacIqrrnB3 ΔlacZ4787 ΔphoBR580 ΔcreABCD154 hsdR514 Δ(pta ackA hisQ hisP)TA3516phn(EcoB) DE(araBAD)567 DE(rhaBAD)568 rpoS396(Am) rph-1ΔendA9 galU95 uidA(ΔMluI)::pir(wt) recA1 | BD792 via BW24217 | Srl+ with P1kc on BW8078 |
| BW24304 | lacIqrrnB3 ΔlacZ4787 ΔphoBR580 ΔcreABCD154 hsdR514 Δ(pta ackA hisQ hisP)TA3516phn(EcoB) DE(araBAD)567 DE(rhaBAD)568 rph-1 rpoS396(Am) ΔendA9 galU95 uidA(ΔMluI)::pir-116 recA1 | BD792 via BW24296 | Srl+ with P1kc on BW8078 |
| BW24320 | lacIqrrnB3 ΔlacZ4787 ΔphoBR580 ΔcreABCD154 DE(araBAD)567 DE(rhaBAD)568 rph-1 | BD792 via BW24310 | Cys+ with P1kc on MG1655; 24 |
| BW25113 | lacIqrrnB3 ΔlacZ4787 hsdR514 DE(araBAD)567 DE(rhaBAD)568 rph-1 | BD792 via BW25083 | Pro+ with P1kc on BW24321; 10, 24 |
| BW25141 | lacIqrrnB3 ΔlacZ4787 hsdR514 DE(araBAD)567 DE(rhaBAD)568 ΔphoBR580 rph-1 galU95 ΔendA9 uidA(ΔMluI)::pir(wt) recA1 | BD792 via BW25140 | Srl+ with P1kc on BW8078; 10, 24 |
| BW25142 | lacIqrrnB3 ΔlacZ4787 hsdR514 DE(araBAD)567 DE(rhaBAD)568 ΔphoBR580 rph-1 galU95 ΔendA9 uidA(ΔMluI)::pir-116 recA1 | BD792 via BW25137 | Srl+ with P1kc on BW8078 |
| BW25695 | lacIqrrnB3 ΔlacZ4787 hsdR514 attP22(EcoB) DE(araBAD)567 DE(rhaBAD)568 rph-1 | BD792 via BW25367 | Pro+ with P1kc on BW25676 |
All strains are λ− and F−.
All known mutations are indicated.
The ancestral strain from another lab and its immediate ancestor in our lab are given.
Additional information is given in Materials and Methods.
IS, insertion sequence.
CRIM (oriRγ) plasmids were propagated at a medium copy number in BW23473, BW24249, or BW25141 or at a high copy number in BW23474, BW24304, or BW25142. As standard wild-type hosts, we used BW25113 and BW25695 [like BW25113, except with attP22(EcoB)]. CRIM plasmids were retrieved from integrants of BW24320 by using helper plasmid transformants of BW23473 (pir+ recA) or BW25141 (pir+ recA) when we used P1kc transduction (P1-Int-Xis [PIX] cloning [16]) or helper plasmid transformants of BW23838 (pir+ recD) when we used transformation (see below). Strain BW37 was used as the recipient for selection of Ilv+ transductants when we determined the Ilv+ transducing titers of P1kc lysates.
Molecular biology methods.
PCR fragments for cloning were generated by using Vent (New England Biolabs, Beverly, Mass.) or Pfu DNA polymerase (Stratagene, La Jolla, Calif.) and oligonucleotide primers (from IDT Inc., Coralville, Iowa). Other enzymes were from New England Biolabs or Promega (Madison, Wis.). Qiagen (Hilden, Germany) products were used for the isolation of plasmid DNA, extraction of DNA fragments from agarose gels, and purification of PCR fragments.
Plasmids.
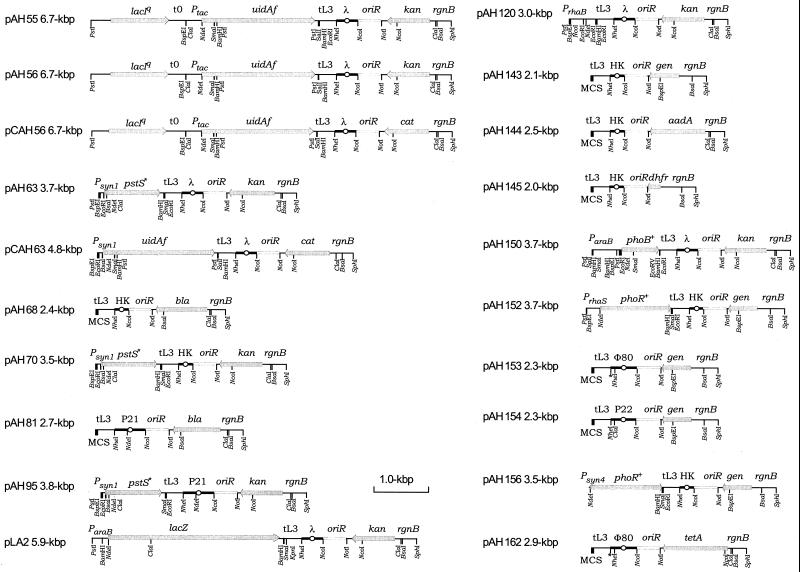
CRIM (Fig. 1) and CRIM helper plasmids (Fig. 2; Table 2) were assembled by standard techniques. pLA2 was constructed by L. Avramova and B. L. Wanner (unpublished). Various fragments were subcloned directly or cloned as PCR-generated fragments containing restriction site extensions (Table 3). The bla, cat, kan, tet, and oriRγ segments were from pANTSγ, pCANTSγ, pKANTSγ, and pTANTSγ (32) (from M. Koob); the attλ (with a destroyed NdeI site) and lacIq-Ptac segments were from pCANTSγΔNdeI and pCANTSγlacIqSLP (26) (both from A. S. Lynch); the lacZ cassette in pAH125 was from pCS3 (29); the promoterless uidAf (the uidA2 fusion in pSK49Δ::uidA2) cassette was from pWM3 (29); the ParaB fragment in pAH150 was from pAH31 (14); and PrhaB in pAH120 was from pLD78 (14). The lacZ gene in pLA2 was constructed in a series of steps that involved introducing an NdeI site overlapping its Met start codon and eliminating a native NdeI site; the lacZ gene was generated using pOD (31) as the template, so it has a mutated lacO2 region; and the ParaB fragment in pLA2 was generated as a PCR fragment (Table 3). Our initial CRIM plasmid pAH55 is a derivative of pKANTSγ in which we introduced a mutated attλ segment (lacking its native NdeI site), lacIq, and uidAf segments in sequential steps. The CRIM helper plasmids were assembled using pINT-ts (17) as the backbone. PCR fragments were generated by using as templates pHH7013 and pHH7009 (from J. C. Hu) for Psyn1 and Psyn4; a derivative of pLD78 (14) called pLD81 (from L. Daniels) for PrhaS; p34E-Tp (11) (from D. E. Woods) for dhfr; pBBR1MCS-5 (22) (from M. Kovach) for gen; λ (from R. Somerville) for xisλ; pBAD33 (13) (from L.-M. Guzman) and pBS1attP6-1 (6) (from A. Campbell) for attP21, xisP21, and intP21; pEY109 (42) (from E. Yagil) for attHK022, xisHK022, and intHK022; pJL10 and pJL110 (23) (from A. Landy) for attφ80, xisφ80, intφ80, and attP22; and P22 (from S. Maloy) for xisP22 and intP22.
FIG. 1.
Structures of CRIM plasmid series. Gene designations include aadA (aminoglycoside adenyltransferase for spectinomycin and streptomycin resistance), bla (β-lactamase for ampicillin resistance), cat (chloramphenicol acetyltransferase), dhfr (dihydrofolate reductase for trimethoprim resistance), gen (gentamicin-3-acetyltransferase for gentamicin resistance), kan (aminoglycoside 3′-phosphotransferase for kanamycin resistance), pstS∗ (a mutant pstS, Pi-specific binding protein), tetA (tetracycline resistance), and uidAf (the uidA2 fusion in pSK49Δ::uidA2 [16]). The multiple cloning site (MCS) is from pUC18 (44). Unique sites within the MCS of pAH68 include, from left to right, SphI, PstI, SalI, XbaI, BamHI, SmaI, KpnI, SacI, and EcoRI. All sites are illustrated for the enzymes shown. Sites destroyed during construction are marked with an asterisk. Modules are flanked by SphI, EcoRI (BamHI or NdeI), NheI, NcoI, NotI, and ClaI (and/or BsaI) sites. Plasmids with aadA, bla, or gen facilitate certain constructions as they have a unique NcoI site. Due to the manner in which these plasmids were constructed, the ParaB region of pAH150, but not of pLA2, encodes an N-terminal portion of AraC as a fusion protein. As a consequence, ParaB is expressed at a normal level in pLA2 but at a much reduced level in pAH150 (see the text). All attP sites were designed taking into account information on important DNA binding sites and structure (23, 35, 43). Accordingly, the attP21 sequence encodes the C terminus of icd and the attP22 sequence includes sequences for the thrW tRNA gene (6). Unexpectedly, we found that CRIM plasmids carrying attP22s or attφ80s (Table 3) failed to integrate or gave very few integrants, respectively, suggesting that additional att sequences are required (data not shown). Plasmids carrying a longer attφ80 site (such as pAH153 and pAH162) integrated efficiently, while those carrying the longer attP22 site (such as pAH154) integrated less frequently than others (see the text). Primers routinely used to verify cloned inserts by PCR or DNA sequencing include rgnB-f (TTGTCGGTGAACGCTCTCCT), ParaB-f (CACATTGATTATTTGCACGG), PrhaB-f (CGTTCATCTTTCCCTGGT), and tL3-r (AGGATGCGTCATCGCCATTA). Priming sites rgnB-f and tL3 are common to all CRIM plasmids and are useful for sequencing inserts in all CRIM plasmids, except those containing lacIq, in which only tL3-r remains useful.
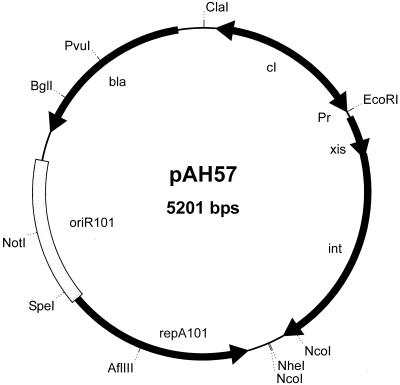
FIG. 2.
CRIM helper plasmid. All helper plasmids express int alone or with xis from λpR under cI857 control, which is also borne by these plasmids, and are temperature sensitive for plasmid replication. As described in Materials and Methods, all are derivatives of pINT-ts, whose complete DNA sequence we determined in this study. With one exception, the xis-int plasmids express these genes in the same orientation as that found naturally. The construction of pAH129 resulted in placing xisφ80 upstream of intφ80 to create an xis-intφ80 operon (data not shown). We also deliberately destroyed the native BamHI site in intλ during the construction of pAH57 by introducing a silent mutation with the xisλ-5′ primer (Table 2). Pr, phage λ promoter.
TABLE 2.
CRIM helper plasmids
| Plasmida | Function(s) |
|---|---|
| pINT-ts | Intλ |
| pAH57 | Xis and Intλ |
| pAH69 | IntHK022 |
| pAH83 | Xis and IntHK022 |
| pAH121 | IntP21 |
| pAH122 | Xis and IntP21 |
| pAH123 | Intφ80 |
| pAH129 | Xis and Intφ80 |
| pAH130 | IntP22 |
| pAH131 | Xis and IntP22 |
The structure of an example is shown in Fig. 2.
TABLE 3.
Oligonucleotide primers used for plasmid constructions
| Region | Template | Primer, sequencea |
|---|---|---|
| aadA | pWM5 | aadA-5′, GCAATCGATACGGATGAAGGCACGAACC; |
| aadA-3′, GCAGCGGCCGCTCGGCTTGAACGAATTGTTAG | ||
| dhfrIIb | p34E-Tp | dhfrIIb-5′, GCAGGCGCCACGAACCCAGTTGACATAAGC; |
| dhfrIIb-3′, GCAGCGGCCGCTTAGGCCACACGTTCAAGT | ||
| gen | pBBR1MCS-5 | gen-5′, GCAGGCGCCCACGAACCCAGTTGACATA; |
| gen-3′, GCAGCGGCCGCCTTGAACGAATTGTTAGG | ||
| ParaB | pBAD33 | ParaB-5′, AACTGCAGCGCCATTCAGAGAAGAAACCAA; |
| ParaB-3′, GCAGTCGACCATATGAATTCCTCCATCCAAAAAAACGGGTATGGAGA | ||
| PrhaS | pAH151 | PrhaS-5′, GCATCCGGAAATTCGCGACCTTCTCG; |
| PrhaS-3′, GCATCTAGAGCATATGGGCCTCCTGATGTCGTC | ||
| Psyn1-pstS∗ | pLD81 | Psyn1-5′, GCATCCGGAACTAGTGTCTTCAAGAATTCTAGG; |
| pstS-3′, CGGGATCCACGCGTTTACAAAGTC | ||
| xisλ | λ | xisλ-5′, CCGGAATTCTTGCGTGTAATTGCGGAGAC; |
| xisλ-3′, GGAAGATCTCCTTCGAAGGAAAGACCTGATGC | ||
| attPHK022 | pEY109 | attPHK022#1, GCAGCTAGCTAATGCTCTGTCTCAGGTC; |
| attPHK022#2, GCACCATGGGACAAAATTGAAATCG | ||
| intHK022 | pEY109 | intHK022-5′, GCACCATGGTAAGTAGGTCATTATTAGTC; |
| intHK022-3′, GCAGAATTCGTAGCCTTTTGAAGAGG | ||
| xis-intHK022 | pEY109 | xisHK022-5′, GCAGAATTCTGCGGAGACTTTGC; |
| intHK022-3′, see above | ||
| attPP21 | pBS1attP6-1 | attPP21#1, GCACCATGGAATGACCGACCGATA; |
| attPP21#2, GCAGCTAGCATAAGGCCTCGCAA | ||
| intP21 | pBS1attP6-1 | intP21-5′, GCAGAATTCAGAACCGCAACTCCCAA; |
| intP21-3′, GCACCATGGATAACGGGCGTATAACA | ||
| xis-intP21 | pBS1attP6-1 | xisP21-5′, GCAGAATTCGCAGCTAAGAGGAGGAC; |
| intP21-3′, see above | ||
| attPP22s | pJL110 | attPP22#1, GCAGCTAGCCGTTGTTACCGATCAAT; |
| attPP22#2, GCACCATGGAAGGCACGAATAATACG | ||
| attPP22 | P22 | attPP22#3, GCAGCTAGCCATTATGAAAATCAGCGGATTCGGA; |
| attPP22#4, CATGCCATGGAATCACCTGACTGAACATGCTCGAC | ||
| intP22 | P22 | intP22-5′, GCAGAATTCCACCACACGACAAGCCT; |
| intP22-3′, GCACCATGGACTCCTATTATCGGCAC | ||
| xis-intP22 | P22 | xisP22-5′, GCAGAATTCCTACGACATGCCTAACG; intP22-3′, see above |
| attPφ80s | pJL10 | attPφ80#1, AACCGGTGAATCACGAC; |
| attPφ80#2, GCACCATGGACATCCTTGAAAGCCTG | ||
| attPφ80 | pJL10 | attPφ80#3, GCATCTAGAGGTGAATCACGACAAAGCGTATC; |
| attPφ80#4, CATGCCATGGGCCCGTGCGAATCAGAAATAAT | ||
| intφ80 | pJL10 | intφ80-5′, GCAGAATTCGCTAGCAACCCTTATCCAGCA; |
| intφ80-3′, GCACCATGGCAATCAAAGAGTGGGAG | ||
| xisφ80 | pJL10 | xisφ80-5′, GCAGAATTCGGCAACTGGAGAGAGCTAT; |
| xisφ80-3′, GCAGCTAGCAAGAGATCATCGGAGAG |
Restriction sites are italicized. Bases complementary to the template are underlined.
CRIM plasmid integration.
Cells carrying a CRIM helper plasmid were grown in 5 ml of SOB cultures with ampicillin at 30°C to an optical density of 600 nm of ca. 0.6 and then made electrocompetent. Following electroporation, cells were suspended in SOC without ampicillin, incubated at 37°C for 1 h and at 42°C for 30 min, and then spread onto selective agar and incubated at 37°C. Colonies were purified once nonselectively and then tested for antibiotic resistance for stable integration and loss of the helper plasmid and by PCR for copy number (see below).
CRIM plasmid excision.
Cells were transformed with the respective Xis/Int CRIM helper plasmid and then spread on ampicillin agar media at 30°C. Colonies were purified once or twice nonselectively on plates that were incubated for 1 h at 42°C and overnight at 37°C. They were then tested for antibiotic sensitivities and by PCR for loss of the integrated plasmid.
CRIM plasmid retrieval.
P1kc lysates were made on integrants by using standard procedures. Recipient cells carrying the corresponding Xis/Int CRIM helper plasmid were grown in LB agar with ampicillin at 30°C to early stationary phase and then infected with a P1kc lysate. Following phage absorption, centrifugation, and resuspension as described elsewhere (38), the infected cells were incubated for 1 h at 37°C, 30 min at 42°C, and an additional hour at 37°C and then spread onto selective media (without ampicillin) for the CRIM plasmid and incubated at 37°C. To recover plasmids by transformation, chromosomal DNAs were isolated from integrants and subjected to shearing by sonication or DNase I digestion in the presence of divalent manganese (1). Recipient cells carrying a helper plasmid were grown in 5-ml SOB cultures with ampicillin at 30°C to an optical density at 600 nm of ca. 0.6 and then made electrocompetent. Following electroporation, cells were suspended in SOC without ampicillin, incubated at 37°C for 1 h and at 42°C for 30 min, and then spread onto selective agar and incubated at 37°C.
PCR verification of integrant copy number.
Isolated colonies were picked up with a plastic tip and suspended in 20 μl of water. Five microliters of the cell suspension, 10 pmol of each primer (P1 to P4 together), and 0.5 U of Taq DNA polymerase (New England Biolabs) were combined in 1× PCR buffer–2.5 mM MgCl2 with deoxynucleoside triphosphates in a final volume of 25 μl. PCR was carried out for 25 cycles with denaturing for 1 min at 94°C, annealing for 1 min (Table 4), and extension for 1 min at 72°C.
TABLE 4.
PCR tests for integration of CRIM plasmidsa
| attP phage | Primer P1 sequence | Primer P4 sequence | Temp (°C) | Predicted size(s) of PCR fragment(s) for attB withb:
|
||
|---|---|---|---|---|---|---|
| No integrant with P1 and P4 | Single integrant with P1 and P2, P3 and P4 | Multiple integrant with P1 and P2, P3 and P2, P3 and P4 | ||||
| λ | GGCATCACGGCAATATAC | TCTGGTCTGGTAGCAATG | 63 | 741 | 577, 666 | 577, 502, 666 |
| HK022 | GGAATCAATGCCTGAGTG | GGCATCAACAGCACATTC | 59 | 740 | 289, 824 | 289, 373, 824 |
| φ80 | CTGCTTGTGGTGGTGAAT | TAAGGCAAGACGATCAGG | 63 | 546 | 409, 732 | 409, 595, 732 |
| P21 | ATCGCCTGTATGAACCTG | TAGAACTACCACCTGACC | 57 | 506 | 568, 620 | 568, 682, 620 |
| P22 | AAGTGGATCGGCATTGGT | TTCGTATCGACAGGAAGG | 63 | 735 | 376, 926 | 376, 568, 926 |
| e14attR | CGCTTGAAGATGTGTGGT | GTTACGGTCTTGGCATTG | 57 | 862 | 1,226, 389 | 1,226, 682, 389 |
| P22(EcoB) | AAGTGGATCGGCATTGGT | CGATTGAACCGCAGATTACG | 63 | 609 | 376, 801 | 376, 568, 801 |
Primers P2 (ACTTAACGGCTGACATGG) and P3 (ACGAGTATCGAGATGGCA) are the same for all attP sites. “Temp” is the annealing temperature. The P4 priming sites for attP21 and attP22 lie within prophage elements of wild-type E. coli K-12 (Fig. 2). An attP21 CRIM plasmid integrates to the left (counterclockwise) of e14. The e14attR primers were used to test for integration to the right (clockwise) of e14. E. coli K-12, but not E. coli B, has an uncharacterized prophage in attP22. An attP22 CRIM plasmid integrates adjacent (counterclockwise) to this prophage in E. coli K-12. Primers for attP22(EcoB) were used to test for integration of attP22 CRIM plasmids in E. coli K-12 strains (BW25695 and others) with an “empty” attP22 site from E. coli B.
Sizes are in nucleotides.
DNA sequencing.
The DNA sequences of all CRIM and CRIM helper plasmids were deduced in their entirety by verifying the sequences of all modules used in their constructions. PCR-amplified segments were verified by automated DNA sequencing of both strands after initial cloning. Many were initially cloned into SmaI-digested pSPORT1 (from Gibco BRL, Gaithersburg, Md.) or EcoRI- and NcoI-digested pLITMUS29 (from New England Biolabs). Others were cloned directly into a CRIM plasmid and then sequenced. Several additional regions were also sequenced to permit generation of detailed maps of all CRIM and CRIM helper plasmids.
Nucleotide sequence accession numbers.
GenBank accession numbers for the CRIM plasmids are AY048713 (pAH55), AY048714 (pAH56), AY048716 (pAH63), AY048717 (pAH68), AY048719 (pAH70), AY048720 (pAH81), AY048722 (pAH95), AY048723 (pAH120), AY054372 (pAH125), AY048730 (pAH143), AY048731 (pAH144), AY048732 (pAH145), AY048733 (pAH150), AY048734 (pAH152), AY048735 (pAH153), AY048736 (pAH154), AY048737 (pAH156), AY048738 (pAH162), AY048739 (pCAH56), AY048740 (pCAH63), and AY054373 (pLA2). GenBank accession numbers for the CRIM helper plasmids are AY048715 (pAH57), AY048718 (pAH69), AY048720 (pAH83), AY048724 (pAH121), AY048725 (pAH122), AY048726 (pAH123), AY048727 (pAH129), AY048728 (pAH130), AY048729 (pAH131), and AY048741 (pINT-ts).
RESULTS AND DISCUSSION
General description.
Our basic CRIM plasmids are shown in Fig. 1. Each has four general regions in common: a polylinker or a cloning region consisting of a promoter for ectopic expression with or without a regulatory gene, a phage attachment (attP) site, a conditional-replication origin (oriRγ), and a selectable marker. Several already contain an E. coli gene (lacZ, phoB, phoR, pstS, or uidAf) within the cloning region; however, these genes act solely as replaceable (“stuffer”) fragments in new constructions. In addition, all CRIM plasmids have bacterial (rgnB) and phage λ (t0, tL3) terminators flanking their cloning region to protect other segments from transcriptional readthrough. The CRIM plasmids were designed so that standard cloning methods can be used for making new ones with various combinations of these and other features, as necessary.
CRIM plasmid integration.
CRIM plasmids can be simply integrated into the chromosome by direct transformation of normal (non-pir) hosts carrying a CRIM helper plasmid synthesizing the respective Int (Fig. 2; Table 2). Int synthesis from the helper plasmids is induced at elevated temperatures. Since the helper plasmids are also temperature sensitive for replication (see Materials and Methods), the resulting transformants are nearly always simultaneously cured of the helper plasmid. Upon integration at the respective attB site, all CRIM plasmids lie in the same relative orientation on the E. coli chromosome (Fig. 3). Therefore, even though they have sequences in common (tL3, oriRγ, and rgnB), homologous recombination among them does not lead to instability because essential chromosomal genes lie between these attB sites. Due to the high efficiency of site-specific recombination, these homologies also do not interfere with integrating multiple CRIM plasmids at different attB sites in the same strain.
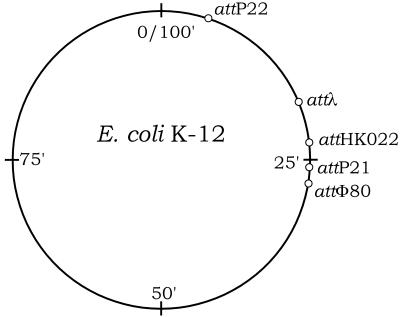
FIG. 3.
Locations of chromosomal attB sites. Wild-type E. coli K-12 contains the prophage element e14 adjacent (clockwise) to attP21 (6). Sites are based on the linkage map (3) and the positions of the appropriate attB core sequences (for attλ, gCTTttTtatActAA; for attHK022, CTTTaggtgaa; for attP21, tGCtGCgcCATAT; for attP22, ATTcgtAATGcGAAG; for attφ80, AACAcTTTcttAAAt; lowercase letters indicate bases that differ from the consensus [6]), in the E. coli K-12 genome sequence (4). E. coli K-12 has two attP22 sites separated by ca. 34 kb (at nucleotides 262125 and 296433 of the genome), which is consistent with their being separated by an uncharacterized phage. Furthermore, by using PCR and P22(EcoB) primers (Table 2), we have shown that the intervening sequences is absent from E. coli B, so it lacks this phage (data not shown).
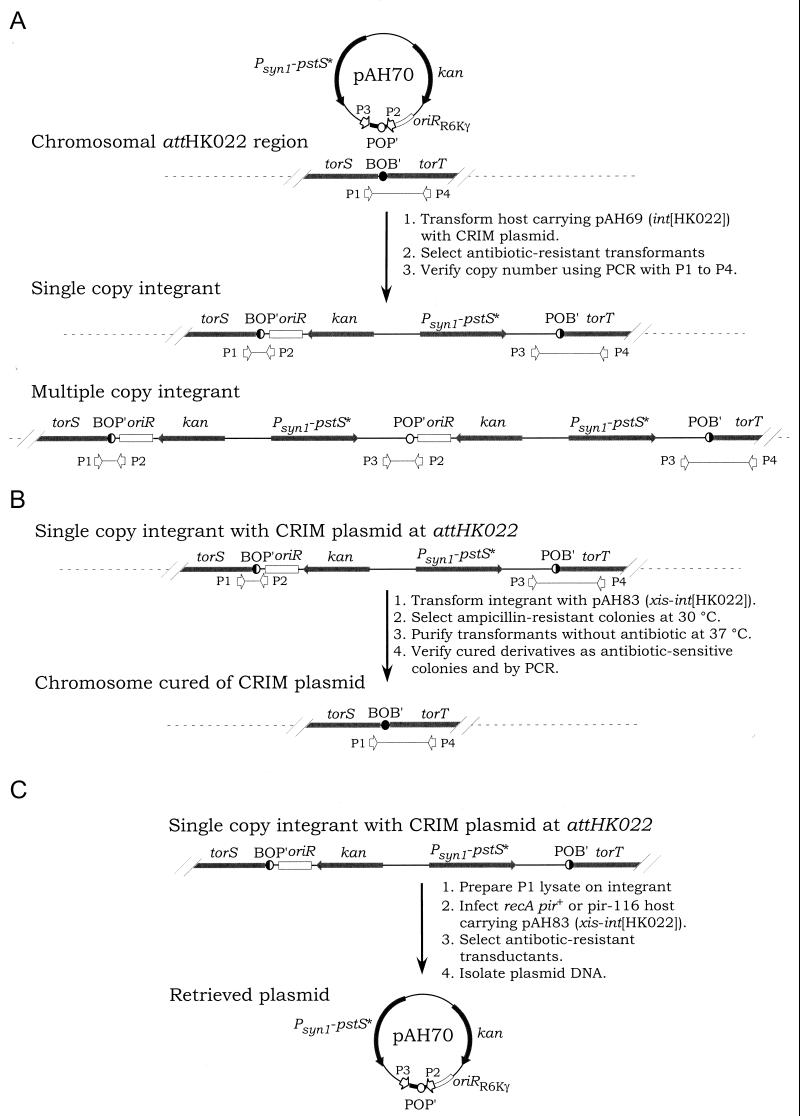
To test for single-copy integration, we routinely use a single PCR with four primers (P1, P2, P3, and P4 in Fig. 4A; see Materials and Methods). Single-copy integrants are revealed as integrants that have lost the fragment corresponding to the respective attB site (the P1 to P4 fragment) and simultaneously gained two new fragments that are characteristic of the attL (BOP′; the P1-to-P2 fragment) and attR (POB′; the P3-to-P4 fragment) junctions. Recombinants with two (or more) CRIM plasmids at the attB site are also easily distinguishable. Such multiple integrants also gain a third fragment characteristic of the attP site of the integrating plasmid (the P2-to-P3 fragment). Recombinants resulting from integration elsewhere on the chromosome yield instead PCR products for both the attB (the P1-to-P4 fragment) and attP (the P2-to-P3 fragment) sites, provided that such integration occurs via homologous recombination or otherwise outside the attP region. Based on these criteria, we have shown that integration occurs primarily at the respective attB site and always requires the corresponding Int. The most common undesirable events are the occurrence of multiple-copy integrants; however, these integrants seldom represent more than a few percentages of the antibiotic-resistant transformants. With one exception, this is true for all CRIM plasmids (data not shown).
FIG. 4.
Integration (A), excision (B), and retrieval (C) of CRIM plasmids from attHK022. POP′ and BOB′ are sites for phage site-specific recombination according to the Campbell model (7, 41). P1, P2, P3, and P4 are priming sites used in PCR tests (see Materials and Methods).
The exception concerns the attP22 CRIM plasmids. These plasmids differ in two ways. First, the attP22 plasmids integrate ca. 100-fold less efficiently than the others. Second, one-half or more of the resulting attP22 plasmid integrants are often incorrect and appear to occur via recombination events that do not involve the attP22 site. Since wild-type E. coli K-12 apparently has an uncharacterized prophage occupying the chromosomal attP22 site (unpublished results), we considered the possibility that this prophage interferes with site-specific integration at this site. However, we obtained similar results with an otherwise isogenic host lacking this prophage, suggesting that an additional factor or sequence is required for efficient attP22 recombination. Nevertheless, attP22 CRIM plasmids have still been quite useful for constructing strains that have multiple CRIM plasmids. In these cases, we have usually integrated attP22 CRIM plasmids before integrating others to prevent the attP22 plasmid from recombining with others via homologous recombination, which can also occur at low frequency. The attP22 CRIM plasmids are therefore less valuable as vectors for library construction or other uses requiring high integration efficiency.
CRIM plasmid excision.
Integrated CRIM plasmids can also be excised very efficiently. CRIM plasmid excision is carried out by using CRIM helper plasmids encoding both Xis and Int (Fig. 4B). We found that all CRIM plasmids were easily eliminated from a specific attP site when using the respective Xis/Int helper plasmid but not when using an Xis/Int helper plasmid for a different attP site. In most cases, 100% of the transformants were cured of the respective CRIM plasmid after a single colony purification step (see Materials and Methods). No aberrant (nonspecific) excision events were detected when we used cells containing multiple CRIM plasmids integrated at different sites (data not shown). We have used excision as a simple way to verify that novel phenotypes result from the presence of particular CRIM plasmids. We have also found excision to be useful in certain strain constructions. For example, when studying complex metabolic or regulatory pathways, it has often been necessary to make strains containing multiple CRIM plasmids in various combinations. In such cases, it has occasionally been more convenient to excise a single CRIM plasmid from a strain containing a combination of different CRIM plasmids in order to introduce an alternative one than to construct an entirely new strain containing most of the same CRIM plasmids by integrating each individually.
CRIM plasmid retrieval.
The ease of retrieving CRIM plasmids from the chromosome is an especially valuable attribute. Because Xis and Int catalyze the excision and circularization of molecules from the corresponding att sites, CRIM plasmids can be retrieved simply by introducing chromosomal DNAs from an integrant into permissive (pir+) hosts that synthesize Xis and Int from a helper plasmid. We have usually done this by using the generalized transducing phage P1kc in a process that we have called PIX cloning (Fig. 4C) (16).
PIX cloning is done using recipients that are pir+ for replication of the CRIM plasmids and recA to avoid homologous-recombination events and carry the appropriate Xis/Int CRIM helper plasmid. We measured PIX cloning efficiencies by determining the number of antibiotic-resistant transductants per infectious phage in standard phage P1 crosses (Table 5). We assayed the transducing titer of the same P1kc lysates by determining the number of Ilv+ transductants. As shown in Table 5, PIX cloning is an extremely efficient process. Efficient retrieval occurs only in the presence of the proper CRIM helper plasmid (data not shown). The recovered plasmids have always been correct, based on restriction enzyme analysis of plasmid DNAs isolated from several representative transductants in numerous such crosses. We have also used PIX cloning to recover plasmids for direct DNA sequence analysis (16; unpublished results). In addition, we have shown that CRIM plasmids can be recovered following transformation of a recD pir+ host carrying the appropriate helper plasmids with chromosomal DNA (Materials and Methods). Accordingly, CRIM plasmids are also retrievable from bacteria that are insensitive to phage P1kc.
TABLE 5.
PIX cloning efficiencies
| attP Phage | CRIM plasmid | No. of Gmr or Kmr transductants per PFUa |
|---|---|---|
| λ | pAH63 | 2.3 × 10−5 |
| HK022 | pAH70 | 1.6 × 10−5 |
| φ80 | pAH153 | 1.9 × 10−4 |
| P21 | pAH95 | 3.9 × 10−5 |
| P22 | pAH154 | 1.8 × 10−7 |
Values are normalized to the I1v+ transducing titers. The same P1kc lysates yielded ca. 10−4 to 10−3 I1v+ transductants per PFU.
Using CRIM plasmids.
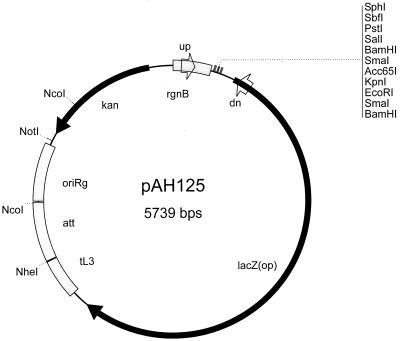
Although CRIM plasmids can be used with most ordinary (non-pir) E. coli strains, we have made standard hosts for their use. These hosts have defined deletions of araBAD, rhaBAD, and lacZ and are lacIq. Hence, they cannot catabolize arabinose or rhamnose and yet encode the regulatory proteins (AraC, RhaR, and RhaS) required for ectopic expression of foreign genes from the corresponding promoters (ParaB, PrhaB, and PrhaS, also called PBAD or araBp, PrhaB, and PrhaS, respectively). These hosts provide tight regulation of these and LacI-controlled promoters. They can also be used with lacZ fusions generated using our standard CRIM lacZ transcriptional fusion vector pAH125 (Fig. 5). Expression levels of ParaB in pLA2, PrhaB in pAH120, and PrhaS in pAH152 were similar to those reported elsewhere (14). We have also shown elsewhere that PrhaB is an especially tightly regulated promoter (14). The synthetic (Psyn1 and Psyn4) promoters provide for low-level unregulated gene expression. These promoters are juxtaposed to a ribosome binding site and AUG start codon that is contained within an NdeI site for convenient construction purposes. With the exception of plasmids carrying attP21, the NdeI site is unique (14).
FIG. 5.
CRIM reporter plasmid for construction of lacZ transcriptional fusions. Primer sites routinely used to sequence inserts are indicated as “up” (TTGTCGGTGAACGCTCTCCT, same as rgnB-f in Fig. 1) and “dn” (down) (AAGTTGGGTAACGCCAGG).
Unexpectedly, we have recently found that ParaB expression is much lower in pAH150 than in pLA2, which shows a normal level of expression (14; L. Avramova and B. L. Wanner, unpublished). Lower expression in pAH150 results from interference by an N-terminal AraC′ fusion protein that is encoded by the ParaB segment in pAH150 but not in pLA2. Nevertheless, both of these ParaB CRIM plasmids have been useful as they both show arabinose-regulated promoter expression. pAH150 has been especially useful for conditional expression of regulatory genes, such as phoB, requiring low-level expression, while pLA2 has been more useful for expression of structural genes requiring high-level expression. Elsewhere, we have recently described E. coli hosts that show homogeneous expression of genes under ParaB control which constitutively synthesize the low-affinity AraE transporter from the chromosome (21).
We have also shown that attP22 and attλ CRIM plasmids integrate into the appropriate attB sites of Salmonella enterica serovar Typhimurium. Others were not tested. Since phages tend to exploit highly conserved and sometimes essential genes (e.g., tRNA genes) as sites for integration (8), several CRIM plasmids can probably integrate into chromosomes of other bacteria, especially in other members of the family Enterobacteriaceae and related families. CRIM plasmids should therefore be useful in many applications involving bacteria other than common laboratory strains.
ACKNOWLEDGMENTS
We thank M. Koob and A. S. Lynch for communicating unpublished results, individuals cited in the text for providing strains and plasmids, Jill Hutchcroft for reading the manuscript, and lab members for helpful discussions. We also thank Cynthia Walchle for technical assistance while B.L.W. was on sabbatical leave with J. J. Mekalanos at Harvard Medical School.
Research was supported by NSF award DMB9108005 to B.L.W., NIH award AI8045 to J. J. Mekalanos, and NIH senior fellowship F33AI10093 to B.L.W.
REFERENCES
- 1.Anderson S. Shotgun DNA sequencing using cloned DNase I-generated fragments. Nucleic Acids Res. 1981;9:3015–3027. doi: 10.1093/nar/9.13.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B J. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2460–2488. [Google Scholar]
- 3.Berlyn M K B. Linkage map of Escherichia coli K-12: edition 10. The traditional map. Microbiol Mol Biol Rev. 1998;62:814–984. doi: 10.1128/mmbr.62.3.814-984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Boyd D, Weiss D S, Chen J C, Beckwith J. Towards single-copy gene expression systems making gene cloning physiologically relevant: lambda InCh, a simple Escherichia coli plasmid-chromosome shuttle system. J Bacteriol. 2000;182:842–847. doi: 10.1128/jb.182.3.842-847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell A, Schneider S J, Song B. Lambdoid phages as elements of bacterial genomes (integrase/phage21/Escherichia coli K-12/icd gene) Genetica. 1992;86:259–267. doi: 10.1007/BF00133724. [DOI] [PubMed] [Google Scholar]
- 7.Campbell A M. Episomes. Adv Genet. 1962;11:101–145. [Google Scholar]
- 8.Campbell A M. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cherepanov P P, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- 10.Datsenko K A, Wanner B L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeShazer D, Woods D E. Broad-host-range cloning and cassette vectors based on the R388 trimethoprim resistance gene. BioTechniques. 1996;20:762–764. doi: 10.2144/96205bm05. [DOI] [PubMed] [Google Scholar]
- 12.Fisher S L, Jiang W, Wanner B L, Walsh C T. Cross-talk between the histidine protein kinase VanS and the response regulator PhoB: characterization and identification of a VanS domain that inhibits activation of PhoB. J Biol Chem. 1995;270:23143–23149. doi: 10.1074/jbc.270.39.23143. [DOI] [PubMed] [Google Scholar]
- 13.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haldimann A, Daniels L L, Wanner B L. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haldimann A, Fisher S L, Daniels L L, Walsh C T, Wanner B L. Transcriptional regulation of the Enterococcus faecium BM4147 vancomycin resistance gene cluster by the VanS-VanR two-component regulatory system in Escherichia coli. J Bacteriol. 1997;179:5903–5913. doi: 10.1128/jb.179.18.5903-5913.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haldimann A, Prahalad M K, Fisher S L, Kim S-K, Walsh C T, Wanner B L. Altered recognition mutants of the response regulator PhoB: a new genetic strategy for studying protein-protein interactions. Proc Natl Acad Sci USA. 1996;93:14361–14366. doi: 10.1073/pnas.93.25.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasan N, Koob M, Szybalski W. Escherichia coli genome targeting, I. Cre-lox-mediated in vitro generation of ori− plasmids and their in vivo chromosomal integration and retrieval. Gene. 1994;150:51–56. doi: 10.1016/0378-1119(94)90856-7. [DOI] [PubMed] [Google Scholar]
- 18.Huang L C, Wood E A, Cox M M. Convenient and reversible site-specific targeting of exogenous DNA into a bacterial chromosome by use of the FLP recombinase: the FLIRT system. J Bacteriol. 1997;179:6076–6083. doi: 10.1128/jb.179.19.6076-6083.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jensen K F. The Escherichia coli K-12 “wild types” W3110 and MG1655 have an rph frameshift mutation that leads to pyrimidine starvation due to low pyrE expression levels. J Bacteriol. 1993;175:3401–3407. doi: 10.1128/jb.175.11.3401-3407.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaasen I, Falkenberg P, Styrvold O B, Strøm A R. Molecular cloning and physical mapping of the otsBA genes, which encode the osmoregulatory trehalose pathway of Escherichia coli: evidence that transcription is activated by KatF (AppR) J Bacteriol. 1992;174:889–898. doi: 10.1128/jb.174.3.889-898.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khlebnikov, A., K. A. Datsenko, T. Skaug, B. L. Wanner, and J. D. Keasling. Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology, in press. [DOI] [PubMed]
- 22.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 23.Leong J M, Nunes-Düby S, Lesser C F, Youderian P, Susskind M M, Landy A. The φ80 and P22 attachment sites. Primary structure and interaction with Escherichia coli integration host factor. J Biol Chem. 1985;260:4468–4477. [PubMed] [Google Scholar]
- 24.Lessard I A D, Pratt S D, McCafferty D G, Bussiere D E, Hutchins C, Wanner B L, Katz L, Walsh C T. Homologs of the vancomycin resistance d-ala-d-ala dipeptidase VanX in Streptomyces toyocaensis, Escherichia coli, and Synechocystis: attributes of catalytic efficiency, stereoselectivity, and regulation with implications for function. Chem Biol. 1998;5:489–504. doi: 10.1016/s1074-5521(98)90005-9. [DOI] [PubMed] [Google Scholar]
- 25.Lu F, Schumacher M A, Arvidson D N, Haldimann A, Wanner B L, Zalkin H, Brennan R G. Structure-based redesign of corepressor specificity of the Escherichia coli purine repressor by substitution of residue 190. Biochemistry. 1997;37:971–982. doi: 10.1021/bi971942s. [DOI] [PubMed] [Google Scholar]
- 26.Lynch A S, Wang J C. Use of an inducible site-specific recombinase to probe the structure of protein-DNA complexes involved in F plasmid partition in Escherichia coli. J Mol Biol. 1994;236:679–684. doi: 10.1006/jmbi.1994.1179. [DOI] [PubMed] [Google Scholar]
- 27.Martinez-Morales F, Borges A C, Martinez K, Shanmugam K T, Ingram L O. Chromosomal integration of heterologous DNA in Escherichia coli with precise removal of markers and replicons used during construction. J Bacteriol. 1999;181:7143–7148. doi: 10.1128/jb.181.22.7143-7148.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metcalf W W, Jiang W, Wanner B L. Use of the rep technique for allele replacement to construct new Escherichia coli hosts for maintenance of R6Kγ origin plasmids at different copy numbers. Gene. 1994;138:1–7. doi: 10.1016/0378-1119(94)90776-5. [DOI] [PubMed] [Google Scholar]
- 29.Metcalf W W, Wanner B L. Construction of new β-glucuronidase cassettes for making transcriptional fusions and their use with new methods for allele replacement. Gene. 1993;129:17–25. doi: 10.1016/0378-1119(93)90691-u. [DOI] [PubMed] [Google Scholar]
- 30.Miller J H. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 31.Müller J, Oehler S, Müller-Hill B. Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J Mol Biol. 1996;257:21–29. doi: 10.1006/jmbi.1996.0143. [DOI] [PubMed] [Google Scholar]
- 32.Pósfai G, Koob M, Hradecná Z, Hasan N, Filutowicz M, Szybalski W. In vivo excision and amplification of large segments of the Escherichia coli genome. Nucleic Acids Res. 1994;22:2392–2398. doi: 10.1093/nar/22.12.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shevell D E, Abou-Zamzam A M, Demple B, Walker G C. Construction of an Escherichia coli K-12 ada deletion by gene replacement in a recD strain reveals a second methyltransferase that repairs alkylated DNA. J Bacteriol. 1988;170:3294–3296. doi: 10.1128/jb.170.7.3294-3296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva J C, Haldimann A, Prahalad M K, Walsh C T, Wanner B L. In vivo characterization of the type A and B vancomycin-resistant enterococci (VRE) VanRS two-component systems in Escherichia coli: a nonpathogenic model for studying the VRE signal transduction pathways. Proc Natl Acad Sci USA. 1998;95:11951–11956. doi: 10.1073/pnas.95.20.11951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang H, Yang C H, Lee G, Chang F, Wilson H, Del Campillo-Campbell A, Campbell A. Integration specificities of two lambdoid phages (21 and e14) that insert at the same attB site. J Bacteriol. 1997;179:5705–5711. doi: 10.1128/jb.179.18.5705-5711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wanner B L. Overlapping and separate controls on the phosphate regulon in Escherichia coli K-12. J Mol Biol. 1983;166:283–308. doi: 10.1016/s0022-2836(83)80086-2. [DOI] [PubMed] [Google Scholar]
- 37.Wanner B L. Molecular cloning of Mud(bla lacZ) transcriptional and translational fusions. J Bacteriol. 1987;169:2026–2030. doi: 10.1128/jb.169.5.2026-2030.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wanner B L. Gene expression in bacteria using TnphoA and TnphoA′ elements to make and switch phoA gene, lacZ (op), and lacZ (pr) fusions. Methods Mol Genet. 1994;3:291–310. [Google Scholar]
- 39.Wanner B L, Boline J A. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J Bacteriol. 1990;172:1186–1196. doi: 10.1128/jb.172.3.1186-1196.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wanner B L, McSharry R. Phosphate-controlled gene expression in Escherichia coli using Mud1-directed lacZ fusions. J Mol Biol. 1982;158:347–363. doi: 10.1016/0022-2836(82)90202-9. [DOI] [PubMed] [Google Scholar]
- 41.Weisberg R A, Landy A. Site-specific recombination in phage lambda. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 211–250. [Google Scholar]
- 42.Yagil E, Dolev S. Determinants of site-specific recombination in the lambdoid coliphage HK022. An evolutionary change in specificity. J Mol Biol. 1989;207:695–717. doi: 10.1016/0022-2836(89)90238-6. [DOI] [PubMed] [Google Scholar]
- 43.Yagil E, Dorgai L, Weisberg R A. Identifying determinants of recombination specificity: construction and characterization of chimeric bacteriophage integrases. J Mol Biol. 1995;252:163–177. doi: 10.1006/jmbi.1995.0485. [DOI] [PubMed] [Google Scholar]
- 44.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]