Abstract
The oxidative cleavage of heme to release iron is a mechanism by which some bacterial pathogens can utilize heme as an iron source. The pigA gene of Pseudomonas aeruginosa is shown to encode a heme oxygenase protein, which was identified in the genome sequence by its significant homology (37%) with HemO of Neisseria meningitidis. When the gene encoding the neisserial heme oxygenase, hemO, was replaced with pigA, we demonstrated that pigA could functionally replace hemO and allow for heme utilization by neisseriae. Furthermore, when pigA was disrupted by cassette mutagenesis in P. aeruginosa, heme utilization was defective in iron-poor media supplemented with heme. This defect could be restored both by the addition of exogenous FeSO4, indicating that the mutant did not have a defect in iron metabolism, and by in trans complementation with pigA from a plasmid with an inducible promoter. The PigA protein was purified by ion-exchange chromotography. The UV-visible spectrum of PigA reconstituted with heme showed characteristics previously reported for other bacterial and mammalian heme oxygenases. The heme-PigA complex could be converted to ferric biliverdin in the presence of ascorbate, demonstrating the need for an exogenous reductant. Acidification and high-performance liquid chromatography analysis of the ascorbate reduction products identified a major product of biliverdin IX-β. This differs from the previously characterized heme oxygenases in which biliverdin IX-α is the typical product. We conclude that PigA is a heme oxygenase and may represent a class of these enzymes with novel regiospecificity.
Many pathogenic microorganisms possess specific heme uptake systems that use heme-iron for metabolic needs. The common finding in bacterial heme assimilation is that the intact heme molecule is internalized into the cell (32). While the transport pathways of heme have been well described, little was known about the fate of heme in bacteria until recently. The discovery of heme oxygenases, enzymes that degrade heme to biliverdin, iron, and carbon monoxide, in corynebacteriae and neisseriae established heme degradation as one of the pathways of heme processing in the bacterial cytoplasm (21, 34, 39, 40). However, it is not clear whether the finding of heme oxygenases in these microorganisms represents an unusual mechanism shared by only a few organisms or a common mechanism likely to be present in other pathogens. The list of microorganisms that are able to use heme as a source of iron is expanding constantly, while the mechanisms by which these bacteria utilize iron from heme are still poorly understood.
One heme-assimilating bacteria is the soil bacterium Pseudomonas aeruginosa (15). This microorganism is carried by approximately 10% of the healthy human population, but when it is found in certain types of patients, such as burn victims and those inflicted with cystic fibrosis, it presents a formidable challenge to treatment and eradication. Although pseudomonads produce and secrete extremely efficient siderophores, they are also able to utilize heme and heme-containing proteins as sole sources of iron (16, 18, 31). P. aeruginosa cells produce at least two fur-regulated systems of heme transport across the outer membrane, Has and Phu (16). These systems are similar to heme assimilation pathways in other gram-negative bacteria, such as Serratia marcescens and Yersinia enterocolitica (11, 26, 35).
In this communication, we characterize the product of the pigA gene of P. aeruginosa. We show that this gene can complement for Neisseria meningitidis heme oxygenase in vivo. PigA mutants are unable to use heme as a source of iron, which together with biochemical data showing the ability of PigA to catalyze heme degradation in vitro indicate that the PigA protein is a heme oxygenase. Quite unexpectedly, we found that biliverdin produced by the PigA-catalyzed reaction is structurally different from biliverdins produced by other, previously characterized heme oxygenases. These data provide another example of heme degradation by heme oxygenase in distantly related bacteria, suggesting that this is a common mechanism of iron acquisition in bacteria.
MATERIALS AND METHODS
Bacteria, media, and antibiotics.
Strains and plasmids used in this study are listed in Table 1. Luria agar (Difco) was routinely used for the growth and maintenance of Escherichia coli strains. Minimal A (MinA) agar was routinely used for the growth and maintenance of P. aeruginosa strains. MinA agar was prepared as described by Kagami et al. (10). Gonococcal broth (GCB) agar (Difco) was routinely used for the growth and maintenance of neisseriae. All neisseriae were incubated at 36.5°C under 5% CO2 in the atmosphere. GCB agar plates lacking iron supplements but containing 50 μM the iron chelator desferoxamine mesylate (Desferal; Ciba Geigy) were used for heme utilization plate assays for neisseriae. Succinate minimal medium was used for all growth curves of P. aeruginosa and was supplemented with various concentrations of hemin (ferriprotoporphyrin IX-Cl; Aldrich) prepared fresh in 100% dimethyl sulfoxide (DMSO) or FeSO4 · 7H2O (Sigma) in the presence or absence of dipyridyl. Succinate medium was prepared as described by Meyer and Abdallah (13). Antibiotics were used at the following concentrations for the maintenance of E. coli strains: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; gentamicin, 20 μg/ml. Antibiotics were used at the following concentrations for the maintenance of P. aeruginosa strains: carbenicillin, 200 μg/ml; gentamicin, 50 μg/ml. Isopropyl-1-thiol-β-d-galactopyranoside (IPTG; Fisher) was used at a final concentration of 1 mM for E. coli and N. meningitidis and 20 mM for P. aeruginosa.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and characteristic(s)a | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | Routine cloning vector | Stratagene |
| BL21(DE3) | gal ompT, lysogen of DE3 carrying T7 polymerase gene | 29 |
| S17-1 (IR747) | Carries RP4 tra genes and λ pir gene | 25 |
| IR1638 | IR747 harboring pWMZ1637 | This study |
| P. aeruginosa | ||
| IA614 | PAO1 derivative, siderophore deficient | 1 |
| IR1648 | IA614 derivative, chromosomal knockout of pigA, Genr | This study |
| IR4309 | IR1648, harbors pMMB66HE | This study |
| IR4310 | IR1648, harbors pMMB66HE | This study |
| IR4311 | IR1648, harbors pMCR2050(pigA) | This study |
| N. meningitidis | ||
| IR1072 | WT laboratory strain, serogroup C | 40 |
| 2855 | IR1072 derivative, chromosomal knockout of hemO | 40 |
| 4093 | 2855 derivative, chromosomal insertion of pigA | This study |
| Plasmids | ||
| pCR 2.1 Topo II | PCR cloning vector, Ampr Kanr | Invitrogen |
| pBluescript II SK(+) | ColE1 cloning phagemid, Ampr | Stratagene |
| pRK2013 | Kmr, ColE1 with Tra+ Mob+ | 6 |
| pET21a | T7 expression vector, Ampr | Novagene |
| pGMΩ | 0.9-kb gentamicin cassette cloned into pHP45Ω, Ampr Genr | 23 |
| pGP704 | pir-dependent suicide vector, Ampr | 14 |
| pCVD442 | pir-dependent suicide vector, Sucs, Ampr | 5 |
| pMMB66HE | Broad host range, tacP expression vector, Ampr | 8 |
| pWMZ1591a | 3-kb erm-lacIOP cassette cloned 5′ of hemO | 24 |
| pWMZ1605 | PCR product encoding PigA on pCR 2.1 Topo II | This study |
| pWMZ1606 | BamHI fragment containing pigA in pBluescript II SK(+) | This study |
| pWMZ1613 | HindIII-SacI fragment from pWMZ1606 in pUC18 | This study |
| pWMZ1621 | Gentamicin cassette cloned into KpnI site of pWMZ1613 | This study |
| pWMZ1629 | EcoRI fragment of pWMZ1621 cloned into pGP704 | This study |
| pWMZ1637 | XbaI-SmaI fragment from pWMZ1621 cloned into pCVD442 | This study |
| pWMZ1614 | 0.8-kb EcoRI-HindIII PCR product of hmbR in pCR 2.1 Topo II | This study |
| pWMZ1616 | 4-kb EcoRI-KpnI from pWMZ1591 and hmbR from pWMZ1614 in pBlueScript II SK(+) | This study |
| pWMZ1619 | pCR 2.1 Topo II minus EcoRI restriction site | This study |
| pWMZ1635 | 5-kb KpnI-HindIII fragment of pWMZ1616 into pWMZ1619 | This study |
| pWMZ1639a | EcoRI pigA fragment of pWMZ1605 in pWMZ1635 | This study |
| pWMZ1655 | PCR product encoding PigA-His tagged in pCR 2.1 Topo II | This study |
| pWMZ1656 | NdeI-XhoI fragment from pWMZ1655 cloned into pET21a | This study |
| pWMZ1665 | PCR product encoding native PigA in PCR 2.1 Topo II | This study |
| pWMZ1666 | NdeI-XhoI fragment from pWMZ1666 cloned into pET21a | This study |
| pMCR2050 | EcoRI fragment carrying pigA cloned into pMMB66HE | This study |
WT, wild type; Amp, ampicillin; Kan, kanamycin; Gen, gentamicin; Suc, sucrose.
Construction of strains and plasmids.
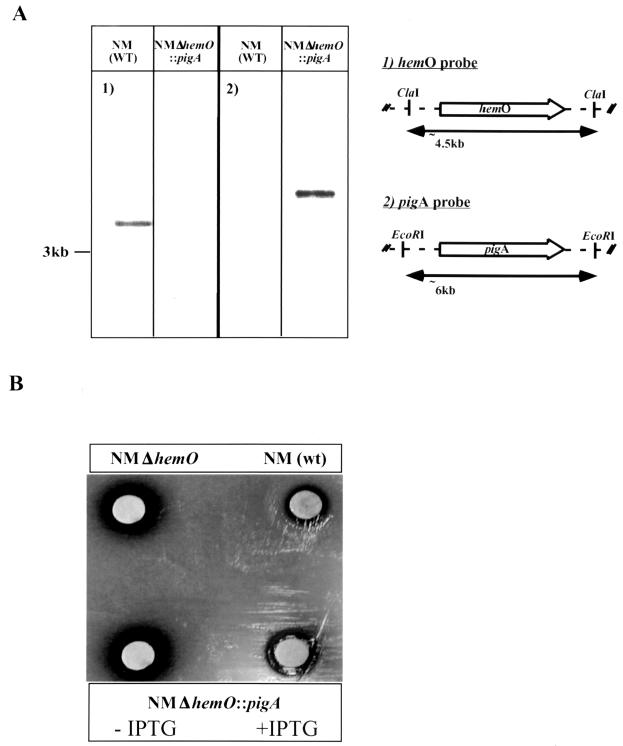
Plasmid pWMZ1635 carrying an ermC-lacIOP cassette (23) inserted upstream of the putative ribosome binding site of a cloned hemO gene was previously described (40). The hemO sequence was removed by EcoRI digestion and replaced with the pigA gene sequence from pWMZ1605 to generate plasmid pWMZ1639a. This construct now carried an IPTG-inducible pigA gene. pWMZ1639a was transformed into IR2855 (wild-type N. meningitidis), and replacement of the hemO sequence with pigA was confirmed by Southern blot analysis of chromosomal DNA from the mutant by using both a pigA probe and a hemO probe.
The structural gene for pigA was disrupted by cassette mutagenesis in P. aeruginosa. A 600-bp PCR fragment encoding PigA was generated with the following primers containing engineered BamHI restriction sites: 5′CGATCCCTTTTCTGGTCAATGGCAGTCGGT and 3′GGATCCCCCCTCAGGCGAAGGTACGTCCA. The BamHI fragment was cloned into the BamHI site of pBluescript II SK(+). A HindIII-SacI fragment containing pigA was then subcloned into pUC18. A gentamicin cassette of 850 bp was inserted into a central KpnI site within the pigA sequence by blunt-end ligation to disrupt the pigA gene (23). An EcoRI fragment was cloned into pGP704, which was then used to clone an XbaI-SmaI restriction fragment containing the disrupted pigA gene into pCVD442, a vector conferring sucrose sensitivity and ampicillin resistance (5, 14). This plasmid, pWMZ1637, was transformed into S17.1 (IR747), an E. coli strain carrying a chromosomal copy of the pir gene, for recombination into P. aeruginosa (25). Upon selection and screening for Carbs, Sucr, and Genr ex-conjugates, IR1648, a P. aeruginosa pigA knockout, was isolated. The disruption was confirmed by Southern blot hybridization using a pigA probe and a gentamicin cassette probe (data not shown).
A pigA-complementing plasmid was generated by cloning the EcoRI fragment from pWMZ1605 carrying pigA into the EcoRI site of pMMB66HE. The resultant construct, pMCR2050, carries pigA under an IPTG-inducible promoter.
pWMZ1656 and pWMZ1666 were generated by cloning pigA carrying a carboxy-terminal, six-histidine tag or native pigA, respectively. The genes were cloned by PCR amplification using Pfx DNA polymerase (Promega) and the following primers containing the listed restriction sites: NdeI, 5′CGCGCAT ATGGATACCCTGGCCCCTGAATCC, and XhoI, 3′CGCGCTCGAGGGCGAAGGTACGCTCCAGCAG. Poly(A) overhangs were generated by the addition of 1 μM dATP and 1 μl of Taq polymerase to the completed PCR mixture and incubation at 72°C for 15 min. The 600-bp PCR products were cloned into the pCR 2.1-TOPO II vector (Invitrogen) to generate pWMZ1655 and pWMZ1665. The resultant PCR fragments containing native and His-tagged versions of pigA were subcloned into pET21a expression vector by NdeI-XhoI digestion and religation, generating pWMZ1656 and pWMZ1666.
Chromosomal DNA preparations.
A 100-μl aliquot of an overnight Luria broth culture was pelleted by centrifugation for 1 min at 6,000 × g. The cell pellet was resuspended in 50 mM Tris-EDTA, pH 8.0, and 50 μl of lysozyme (10 mg/ml). The cells were allowed to incubate at room temperature for 10 min followed by the addition of 25 μl of proteinase K (100 mg/ml) and 5 μl of 10% sodium dodecyl sulfate (SDS) and incubation at 65°C for 1 h. To each lysate, 200 μl of phenol-chloroform-alcohol was added, and it was vortexed and then centrifuged at 12,000 × g for 5 min. The supernatant was transferred to a new tube, and the DNA was precipitated with a 1:10 (vol/vol) volume of 4 M LiCl and 2.5 volumes of 100% ethanol. The DNA pellet was resuspended in 400 μl of Tris-EDTA and 2 μl of RNase (10 mg/ml) and incubated for 1 h at 37°C. The DNA was again precipitated and resuspended in 100 μl of Tris-EDTA and stored at −20°C.
PCR, DNA sequencing, and Southern blot hybridization.
Standard procedures for plasmid DNA preparation and restriction analysis were used as described by Sambrook et al. (20). A 600-bp fragment containing pigA was amplified using primers 5′CGATCCCTTTTCTGGTCAATGGCAGTCGGT and 3′GGATCCCCCCTCAGGCGAAGGTACGTCCA with the following reaction cycle: 94°C, 30 s; 50°C, 1 min; 72°C, 1 min (28 cycles). The PCR product was sequenced by dye terminator cycle sequencing on an ABI model 377 automated sequencer with M13 forward and M13 reverse primers to obtain the nucleotide sequences of both strands. Southern blot analysis procedure was performed as described by Sambrook et al. (20). The pigA-specific probe and gentamicin-specific probe were generated by digoxigenin nonradioactive DNA labeling (Genius system; Boehringer GmbH, Mannheim, Germany) during PCR with the pigA primers listed above.
Heme utilization assays (for N. meningitidis) and growth studies (for P. aeruginosa).
To determine the ability of the N. meningitidis Δ hemO::pigA mutant to utilize heme, filter disc assays were performed as described by Zhu et al. (39). Growth studies and heme utilization of the Pseudomonas strains were carried out by seeding 1 ml of succinate medium with a single colony for each strain. A 100-μl aliquot of the cell suspensions was used to inoculate 100 μl of succinate media present in a sterile, round-bottom, microtiter plate well. The plate was incubated with vigorous shaking at 38°C for 18 h in a Biotek 808 ELx incubator and plate reader. When specified, various concentrations of a freshly prepared hemin-DMSO solution were added to the culture wells. Iron-restricted media were achieved with the addition of 0.5 mM dipyridyl. Iron-rich media were prepared by the addition of various concentrations of FeSO4 · 7H2O. For the complementation assays, IPTG was added to a final concentration of 20 mM in each well, regardless of hemin concentration.
Expression, purification, and characterization of PigA heme oxygenase.
The overexpression of PigA from vector pET21a and purification were done as described previously (39). The heme-PigA heme oxygenase [heme-PigA(HO)] complex was prepared as described previously (33). The millimolar extinction coefficient (ɛ405) for the heme-PigA(HO) complex was determined as previously described (7). The absorbance of a purified heme-PigA(HO) sample at 405 nm was measured. An excess of dithionite was added, and the spectrum of the reduced ferrous pyridine hemochrome was then recorded. The concentration was calculated from the absorbance maxima at 418.5, 526, and 555 nm using millimolar extinction coefficient values of 170, 17.5, and 34.4, respectively. The reaction of the heme-PigA(HO) complex in presence of NADPH reductase was carried out as described previously (34, 39). Following completion of the reaction, the product was extracted for high-performance liquid chromatography (HPLC) analysis as described below.
The ascorbic acid-dependent conversion of heme to biliverdin was also monitored. Ascorbic acid at a final concentration of 5 mM was added directly to the heme-PigA(HO) complex (10 μM) in 20 mM Tris-HCl buffer (pH 7.5). The spectral changes between 300 and 750 nm were recorded over a 20-min time period. The products of the reaction were extracted and subjected to HPLC analysis as described below.
The reaction of the heme-PigA(HO) complex with H2O2 was monitored as previously described (39) except that 10 equivalents of 10 mM H2O2 (10 μl) in Tris-HCl (pH 7.5) were added to the heme-PigA(HO) complex (10 μM) in the same buffer.
HPLC analysis of the heme-PigA(HO) reaction products was performed as described previously (39).
RESULTS
Homologues of neisserial heme oxygenase in gram-negative bacteria.
It has been previously demonstrated that the hemO gene of N. meningitidis encodes a protein capable of catalytic degradation of heme to iron, CO, and α-biliverdin (39). Analysis of several bacterial genomes for open reading frames similar to that of the HemO protein did not reveal many homologues: only Pseudomonas spp. contained potential heme oxygenases (Fig. 1). An iron-inducible protein of unknown function in P. aeruginosa, PigA, was 37% identical in its amino acid sequence to the HemO protein. Further, the genome of another pseudomonad, Pseudomonas putida, contained two open reading frames with a high homology to HemO and PigA proteins HemO-PP and PigA-PP, respectively (Fig. 1). The two putative P. putida heme oxygenases shared 41% identical amino acid residues. Interestingly, HemO-PP was more homologous to the neisserial heme oxygenase (51% identity) than to the P. aeruginosa PigA (44% identity). Conversely, two Pseudomonas PigA homologues shared 56% identity at the amino acid level. All four heme oxygenases shown in Fig. 1 possess a highly conserved histidine residue that serves as the proximal ligand to the heme in all heme oxygenases characterized thus far (2, 22, 22a, 27, 39).
FIG. 1.
An amino acid sequence alignment of the N. meningitidis heme oxygenase (HemO-MC) and putative P. aeruginosa and P. putida heme oxygenases (HemO-PP, PigA-PP, and PigA-PA). ∗, identical residues; dot, similar residues. The alignment was performed using the ClustalW 1.8 program on the Baylor College of Medicine Search Launcher.
Replacement of neisserial hemO with P. aeruginosa pigA.
To determine if the pigA gene of P. aeruginosa encodes a protein with similar function, pigA was cloned under an IPTG-regulatable promoter between flanking regions of hemO. The resultant plasmid construct was homologously recombined into the chromosome of N. meningitidis to replace hemO, and the replacement was confirmed by Southern blot analysis (Fig. 2A). The replacement strain has a complete pigA gene and no longer contains the hemO gene sequence. Heme utilization of the pigA replacement mutant of neisseriae was assayed on GCB-Desferal plates. A 50-μg aliquot of a fresh hemin-DMSO solution was applied to a sterile filter disk, and the plates were incubated for 24 h under 5% CO2 in the presence and absence of IPTG. As shown in Fig. 2B, wild-type N. meningitidis can utilize heme on these plates whereas a hemO deletion mutant cannot. Additionally, the pigA replacement mutant can utilize heme on these plates only in the presence of IPTG, indicating that the expression of pigA is under the control of the lac promoter as intended (see Materials and Methods).
FIG. 2.
Chromosomal complementation of the N. meningitidis hemO deletion mutant with pigA gene from P. aeruginosa. (A) Southern blot analysis was performed on chromosomal DNA from the wild type and the mutant; hybridization and detection were achieved with either a hemO probe (panel 1) or a pigA probe (panel 2). (B) For each construct, complementation is shown by growth around a heme disk on GCB-Desferal media in the presence of IPTG.
Characterization of P. aeruginosa pigA knockout mutant.
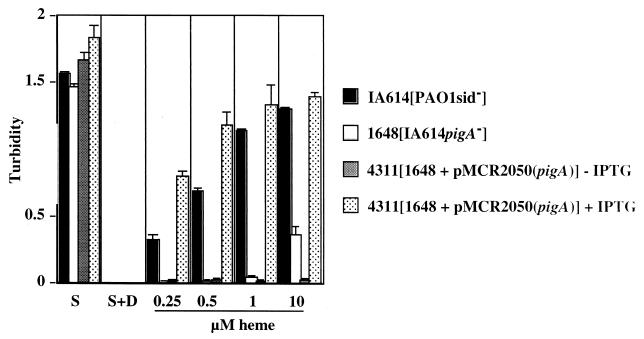
To study the role of PigA in heme catabolism, the pigA gene was disrupted by the insertion of a gentamicin antibiotic cassette in the siderophore-deficient P. aeruginosa IA614 strain. The insertion was confirmed by Southern blot analysis using probes specific to pigA and the cassette (data not shown). Heme utilization was assayed by determining growth of the mutants in iron-restricted succinate medium in the absence and presence of heme (free iron was chelated with 0.5 mM dipyridyl). Both strains grew well in normal succinate medium and did not grow in the succinate medium supplemented with dipyridyl (Fig. 3). The IA614 cells were capable of growth above an optical density at 600 nm (OD600) of 1.0 with heme supplementation, whereas the mutant reached an OD600 of only 0.2. The growth observed with IA614 appears to correlate directly with the increasing concentrations of heme. To determine that the defect in the pigA mutant affected heme utilization and not iron metabolism or sensitivity of the mutant to dipyridyl, the cells were grown in the dipyridyl-succinate media made iron rich by the addition of FeSO4 · 7H2O. Both IA614 and the pigA mutant demonstrated growth near an OD600 of 1.0 (data not shown).
FIG. 3.
Growth analysis of P. aeruginosa PAO1 versus PAO1 pigA insertion mutant under various culture conditions. Aliquots of 106 cells were incubated with aeration at 38°C in succinate minimal medium in the presence of absence of 0.5 mM dipyridyl. Where indicated, cells were grown in succinate-dipyridyl medium supplemented with increasing concentrations of heme for 18 h. Cultures were grown in the presence or absence of 20 mM IPTG. Strain IA614, PAO1sid−; strain IR1648, PAO1sid− pigA::Gen; strain 4311, PAO1sid− pigA::Gen + pMCR2050 (pigA). S, succinate medium; S+D, succinate medium with dipyridyl only.
Complementation of pigA knockout mutant.
As demonstrated above, iron-restricted minimal medium supplemented with heme was unable to support growth of the pigA knockout mutant. To demonstrate that the heme utilization defect is due to the loss of PigA, the pigA gene was cloned onto an IPTG-inducible plasmid, pMMB66HE, and conjugated into the mutant. The growth of the complemented pigA mutant was tested in the presence of heme and the inducer (Fig. 3). In comparison with the controls, pMMB66HEpigA (pMCR2050) was able to restore heme utilization capability (growth in iron-poor media supplemented with heme) to near-wild-type levels only in the presence of the inducer. These data suggest that the product of the pigA gene is essential for the ability of P. aeruginosa to use heme as a source of iron. The high level of amino acid identity and the ability of the pigA gene to complement the heme oxygenase mutant of N. meningitidis indicate strongly that the pigA gene product may be a heme oxygenase.
Characterization of the putative P. aeruginosa heme oxygenase.
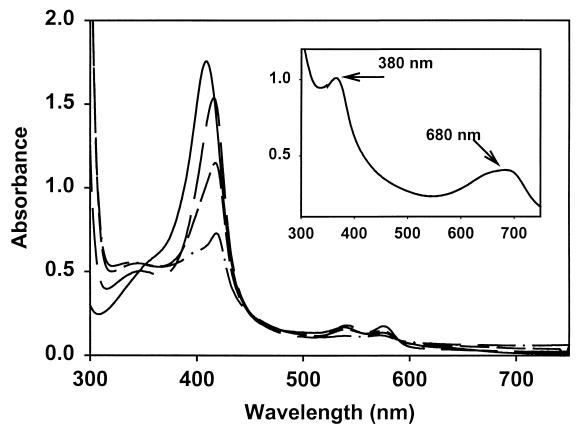
The wild-type PigA(HO) was expressed as a soluble catalytically active protein, evidenced from the accumulation of biliverdin within the cells. Purification of PigA(HO) by ion-exchange and gel-filtration chromatography yielded a protein band with a molecular mass of 23 kDa by SDS-polyacrylamide gel electrophoresis (PAGE) (Fig. 4, inset). The yield of purified protein from a liter of cells was in the range of 20 to 30 mg. Reconstitution of the protein with heme yielded a 1:1 complex with a Soret maximum at 406 nm (Fig. 4). Reduction of the heme with dithionite yielded a reduced ferrous-deoxy complex with a Soret maximum at 434 nm; addition of CO to the reduced complex resulted in a shift of the Soret band to 419 nm, with the appearance of α/β-bands at 537 and 567 nm (Fig. 4). The spectral properties of the heme-PigA(HO) complex are similar to those reported for the bacterial and mammalian heme oxygenases in which the proximal ligand to the heme is a histidine (2, 33–36, 38). With both the mammalian and bacterial enzymes, it has been shown that a water molecule occupies the sixth ligand position. With increasing pH, a shift in the Soret and the appearance of α/β bands on transition from a six-carbon high-spin (6CHS) to six-carbon low-spin (6CLS) system indicates that the sixth ligand in the P. aeruginosa HO is also a water molecule. The transition from a high-spin to low-spin system results from the deprotonation of an H2O molecule to a hydroxide molecule with pKa of 8.0 (data not shown). This is in agreement with that previously observed for both C. diphtheriae HmuO (2) and human HO (28). The millimolar extinction coefficient of the heme-PigA(HO) complex was calculated to be 129 mM−1.
FIG. 4.
Absorption spectra of the heme-PigA(HO) complexes and SDS-PAGE gel of the purified PigA(HO) protein (inset). The spectra are ferric (—), ferrous deoxy (—), and ferrous CO bound (—) heme-PigA(HO). (Inset) SDS-PAGE gel of the purified PigA(HO). Lane 1, markers; lane 2, purified native PigA(HO).
Catalytic activity of heme-PigA(HO) complex.
The heme-PigA(HO) complex was converted to ferric-biliverdin in the presence of ascorbate as an exogenous reductant. The reaction was monitored by UV-visible spectroscopy, and the products were determined by HPLC analysis following extraction and methylation. The ferric heme-PigA(HO) complex was quantitatively converted to ferric-biliverdin (Fig. 5, inset). Initial formation of the ferrous-dioxygen complex was observed with a shift in the Soret from 406 to 410 nm and the appearance of α/β bands at 570 and 540 nm, respectively. Over a period of 30 min, the reaction proceeded with a decrease in the Soret and a shift back to 404 nm and the disappearance of the α/β bands on further conversion of the ferrous-dioxygen complex to ferric-biliverdin. Acidification of the product resulted in a spectrum with maxima at 380 nm and 680 nm, indicative of iron-free biliverdin (Fig. 5, inset).
FIG. 5.
Conversion of heme-PigA(HO) to biliverdin in the presence of ascorbate. Spectra were taken at 10-min intervals after the addition of ascorbate (5 mM) to the ferric heme-HemO complex. (Inset) The UV/visible spectra of a final biliverdin product after acidification.
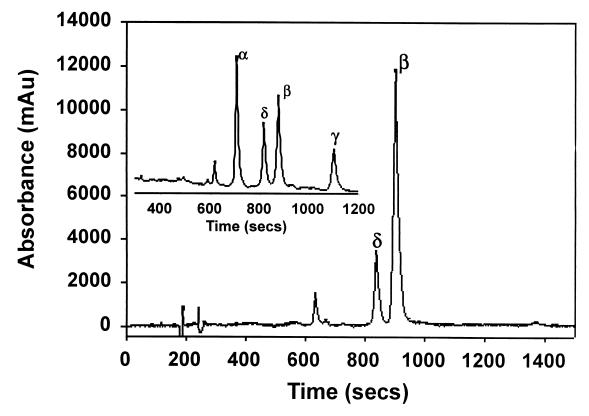
HPLC analysis of the products yielded a major and a minor peak with retention times identical to those of biliverdin IX-β and -δ, respectively (Fig. 6). The reaction products eluted with biliverdin IX-β and -δ on coinjection with the known standards (data not shown). The biliverdin standards for comparison are shown in Fig. 6 (inset).
FIG. 6.
HPLC chromatogram of the product of the heme-pigA(HO) reaction with ascorbate following extraction and methylation. (Inset) An HPLC chromatogram of a mixture of all four biliverdin dimethyl esters as standards is shown. The UV-visible spectra and the HPLC product analysis conclude that oxidative cleavage of heme by PigA(HO) is regiospecific with the biliverdin IX-β and that small amounts of biliverdin IX-δ isomers are formed.
Reaction of heme-PigA(HO) complex with H2O2.
The reaction of the heme-PigA(HO) complex with 10 equivalents of H2O2 resulted in the formation of verdoheme, as previously described for the bacterial and mammalian HO enzymes (30, 34). A decrease in the Soret band at 404 nm and an increase in the visible absorption in the 640- to 680-nm region were observed (data not shown). Addition of pyridine to a final concentration of 20% and extraction into chloroform yielded a spectrum typical of a pyridine-verdohemochrome (data not shown). Hydrolytic conversion of the product to biliverdin and subsequent HPLC analysis verified that the initial H2O2-dependent hydroxylation occurred solely at the β-meso carbon (data not shown).
DISCUSSION
How widespread are heme oxygenases in bacterial genomes?
The identification of a heme oxygenase (HmuO) in corynebacteriae and more recently in neisseriae (HemO) provide evidence that oxidative cleavage of heme is a mechanism by which pathogenic bacteria can acquire iron (21, 32, 39, 40). However, the analysis of 83 completed and uncompleted bacterial genomes for HmuO and HemO homologues identified an unexpectedly small number of orthologs: only Pseudomonas spp. and legionellae contained potential heme oxygenases. Another possible homologue was found in the genome of Deinococcus radiodurans (data not shown). This finding raises the argument that not all microorganisms use heme oxygenases to obtain iron for metabolic needs. Alternatively, the enzymes that degrade heme in these microorganisms might not share significant homology with currently identified heme oxygenases or they may employ a different mechanism of extracting iron from heme. It can be concluded that the currently characterized or identified bacterial heme oxygenases group into two subfamilies, those characterized by HemO-PigA of neisseriae and Pseudomonas and by HmuO of corynebacteriae. When compared, the members of these two subfamilies do not share significant amino acid identity apart from the histidine residue, which functions as the proximal ligand to the heme, and a conserved motif (G-S-X-L-G) in which both a serine and leucine are conserved (Fig. 1). The conserved glycine motif, as evidenced from the crystal structures of both the human HO-1 and the neisserial HemO, introduces a kink in the distal helix directly above the heme, presumably to allow binding of the substrate and release of the product (22, 22a). Similarly, the human and neisserial heme oxygenases show low sequence identity but still retain the same overall fold (22, 40). Thus, although the prevalence of heme oxygenases in bacterial genomes seems very low, this is probably a result of search tool limitations and not the rarity of oxygen-dependent degradation of heme in prokaryotes.
Product of the pigA gene is a heme oxygenase.
The current study was aimed at characterizing the putative heme oxygenase of P. aeruginosa. Four independent results lend strong support to the claim that the product of the pigA gene is involved in heme catabolism: (i) a high degree of amino acid homology (37% identity) with a known heme oxygenase, HemO, (ii) the ability to complement the heme utilization defect of a hemO mutant of N. meningitidis, (iii) the inability of a P. aeruginosa pigA mutant to use heme as a source of iron, and (iv) the complementation of the growth defect of a pigA mutant by functional copy of pigA or by inorganic iron supplementation of growth media. A previous study by Ochsner and Vasil established that the pigA gene is negatively regulated by Fur and iron, which is consistent with the role of PigA in iron assimilation (17).
The biochemical characterization of the protein provides further evidence that PigA possesses heme oxygenase activity and can catalyze the final step in the utilization of heme by P. aeruginosa. The UV-visible spectra of the P. aeruginosa heme-HO complexes are similar to those reported for the bacterial and mammalian enzymes, in which the proximal ligand to the heme is a histidine, with a water bound in the sixth ligand position (3, 27, 28, 33, 34, 39). In the presence of ascorbate, the heme-PigA(HO) complex is quantitatively converted to ferric-biliverdin which remains associated with the protein. It has been reported for the C. diphtheriae heme oxygenase that both the ascorbate-driven and NADPH cytochrome P450 reductase-driven reactions yield biliverdin as the product (2, 21). However, it must be noted that in the ascorbate-driven reaction, free biliverdin was observed only following a 2-h incubation (2). Indeed, the product of the reaction catalyzed by the human HO-1 in the presence of ascorbate is ferric-biliverdin, which yields free biliverdin only after the addition of iron chelators (12). In contrast to the C. diphtheriae and N. meningitidis enzymes, P. aeruginosa PigA(HO) on reconstitution with the human NADPH cytochrome P450 reductase yielded very little ferric-biliverdin under the same conditions as used previously for the bacterial heme oxygenases (data not shown).
PigA(HO)-catalyzed reaction yields novel biliverdin isomer pattern.
HPLC analysis of the product from the ascorbate dependent reaction yielded biliverdin IX-β as the primary product with a minor peak due to the δ-isomer (Fig. 6). The appearance of the δ-isomer most likely arises from the 180° rotation around the α/γ axis previously observed in nuclear magnetic resonance studies of the human HO-1 protein (9). In contrast to the isozymes with α-selectivity, the rotation around the α/γ-axis does not alter the regiospecificity, whereas in the P. aeruginosa heme oxygenase, the β- and δ-meso-carbons would be exchanged. It is not known if the rotation around the α/γ-axis is of relevance in vivo, as the heme may be delivered to heme oxygenase in a concerted manner from an as-yet-unidentified transport protein, which may preclude rotation around the α/γ-axis. It is reasonable to assume that physiological oxidative cleavage of heme by P. aeruginosa occurs at the β-meso-edge (Fig. 7). This is the first report of a heme oxygenase with regiospecificity other than that for the α-meso-carbon. The physiological relevance of the β-selectivity is unknown given that both the C. diphtheriae and N. meningitidis enzymes retain regiospecificity for the α-meso-edge as previously observed for the eukaryotic heme oxygenases. Reaction of the heme-HO complex with H2O2 in the presence of oxygen yields verdoheme as the final product. Hydrolytic conversion of the verdoheme product to biliverdin and subsequent HPLC analysis again identified biliverdin-IX-β as the final product. The ability of H2O2 to substitute for NADPH and oxygen in the initial hydroxylation of heme, which in the presence of oxygen rapidly reacts to verdoheme, is consistent with that previously reported for the human (36, 37) and bacterial (3, 39) enzymes.
FIG. 7.
Chemical steps in heme degradation as defined by the studies of eukaryotic heme oxygenases and C. diphtheriae HmuO. Me, methyl side chain; V, vinyl side chain; Pr, propionic side chain. All thus far characterized heme oxygenases cleave the porphyrin ring at the α-carbon atom, whereas PigA(HO) cleaves at the β-carbon.
These data taken together clearly identify PigA(HO) as a heme oxygenase enzyme with a novel regiospecificity. This is the first report of a heme oxygenase from any organism that displays selectivity for a meso-carbon other than the α-meso-carbon (Fig. 7). This not only has interesting physiological ramifications but also provides us with an opportunity to study heme oxygenases with two different regiospecificities, and perhaps we can gain a better understanding of the steric and electronic factors controlling the regiospecificity of heme cleavage.
ACKNOWLEDGMENTS
Melanie Ratliff and Wenming Zhu contributed equally to this work.
We thank K. Poole and H. Schweizer for strains. We thank Heather Alexander, Donna Balding-Perkins, and Anthony Richardson for reading the manuscript and for helpful suggestions.
This work is supported by the Public Service grant AI42870 to I.S.
REFERENCES
- 1.Ankenbauer R G, Cox C D. Isolation and characterization of Pseudomonas aeruginosa mutants requiring salicylic acid for pyochelin biosynthesis. J Bacteriol. 1998;170:5364–5367. doi: 10.1128/jb.170.11.5364-5367.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu G C, Katakura K, Zhang X, Yoshida T, Ikeda-Saito M. Heme degradation as catalyzed by a recombinant bacterial heme oxygenase (HmuO) from Corynebacterium diphtheriae. J Biol Chem. 1999;274:21319–21325. doi: 10.1074/jbc.274.30.21319. [DOI] [PubMed] [Google Scholar]
- 3.Chu G C, Tomita T, Sonnichsen F D, Yoshida T, Ikeda-Saito M. The heme complex of HmuO, a bacterial heme degradation enzyme from Corynebacterium diphtheriae. Structure of the catalytic site. J Biol Chem. 1999;274:24490–24496. doi: 10.1074/jbc.274.35.24490. [DOI] [PubMed] [Google Scholar]
- 4.Crusats J, Suzuki A, Mizutani A, Ogoshi T H. Regioselective porphyrin bridge cleavage controlled by electronic effects coupled oxidation of 3-demethyl-3-(trifluoromethyl)mesohemin IX and identification of its four biliverdin isomers. J Org Chem. 1998;63:602–607. doi: 10.1021/jo9714728. [DOI] [PubMed] [Google Scholar]
- 5.Donnenberg M S, Kaper J B. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect Immun. 1991;59:4310–4317. doi: 10.1128/iai.59.12.4310-4317.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Figurski D H, Helinski D R. Replication on an origin-containing derivative of plasmid RK2 dependent on a plasmid function in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuhrhop J H K, Smith M. In: Porphyrins and metalloporphyrins. Smith K M, editor. Amsterdam, The Netherlands: Elsevier; 1975. pp. 804–807. [Google Scholar]
- 8.Fürste J P, Pansegrau W, Frank R, Bloeker H, Scholtz P, Bagdasarian M, Lanka E. Molecular cloning of the plasmid RP4 primase region in a multi-host range tac expresion vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 9.Hernandez G, Wilks A, Paolesse R, Smith K M, Ortiz de Montellano P R, La Mar G N. Proton NMR investigation of substrate-bound heme oxygenase: evidence for electronic and steric contributions to stereoselective heme cleavage. Biochemistry. 1994;33:6631–6641. doi: 10.1021/bi00187a033. [DOI] [PubMed] [Google Scholar]
- 10.Kagami Y, Ratliff M, Surber M, Martinez A, Nunn D N. Type II protein secretion in Pseudomonas aeruginosa: genetic suppression of a conditional mutation in the pilin-like component XcpT by the cytoplasmic component XcpR. Mol Microbiol. 1998;27:221–233. doi: 10.1046/j.1365-2958.1998.00679.x. [DOI] [PubMed] [Google Scholar]
- 11.Letoffe S, Ghigo J M, Wandersman C. Iron acquisition from heme and hemoglobin by a Serratia marcescens extracellular protein. Proc Natl Acad Sci USA. 1994;91:9876–9880. doi: 10.1073/pnas.91.21.9876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Ortiz de Montellano P R. Reaction intermediates and single turnover rate constants for the oxidation of heme by human heme oxygenase-1. J Biol Chem. 2000;275:5297–5307. doi: 10.1074/jbc.275.8.5297. [DOI] [PubMed] [Google Scholar]
- 13.Meyer J M, Abdallah M A. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification, and physicochemical properties. J Gen Microbiol. 1978;107:319–328. [Google Scholar]
- 14.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutants: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noya F, Arias A, Fabiano E. Heme compounds as iron sources for nonpathogenic Rhizobium bacteria. J Bacteriol. 1997;179:3076–3078. doi: 10.1128/jb.179.9.3076-3078.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ochsner U A, Johnson Z, Vasil M L. Genetics and regulation of two distinct haem-uptake systems, phu and has, in Pseudomonas aeruginosa. Microbiology. 2000;146:185–198. doi: 10.1099/00221287-146-1-185. [DOI] [PubMed] [Google Scholar]
- 17.Ochsner U A, Vasil M L. Gene repression by the ferric uptake regulator in Pseudomonas aeruginosa: cycle selection of iron-regulated genes. Proc Natl Acad Sci USA. 1996;93:4409–4414. doi: 10.1073/pnas.93.9.4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poole K, Heinrichs D E, Neshat S. Cloning and sequence analysis of an EnvCD homologue in Pseudomonas aeruginosa: regulation by iron and possible involvement in the secretion of the siderophore pyoverdine. Mol Microbiol. 1993;10:529–544. doi: 10.1111/j.1365-2958.1993.tb00925.x. [DOI] [PubMed] [Google Scholar]
- 19.Saito S, Itano H A. Verdohemochrome IX alpha: preparation and oxidoreductive cleavage to biliverdin IX alpha. Proc Natl Acad Sci USA. 1982;79:1393–1397. doi: 10.1073/pnas.79.5.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 21.Schmitt M P. Utilization of host iron sources by Corynebacterium diphtheriae: identification of a gene whose product is homologous to eukaryotic heme oxygenases and is required for acquisition of iron from heme and hemoglobin. J Bacteriol. 1997;179:838–845. doi: 10.1128/jb.179.3.838-845.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuller D J, Wilks A, Ortiz de Montellano P R, Poulos T L. Crystal structure of human heme oxygenase-1. Nat Struct Biol. 1999;6:860–867. doi: 10.1038/12319. [DOI] [PubMed] [Google Scholar]
- 22a.Schuller, D. I., W. Zhu, A. Wilks, I. Stojiljkovic, and T. L. Poulos. Crystal structure of heme oxygenase from the Gram-negative pathogen Nesseria meningitidis and a comparison with mammalian heme oxygenase-1. Biochemistry, in press. [DOI] [PubMed]
- 23.Schweizer H P. Small broad-host range gentamycin resistance gene cassettes for site-specific insertion and deletion mutagenesis. BioTechniques. 1993;15:852–853. [PubMed] [Google Scholar]
- 24.Seifert H S. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene. 1997;188:215–220. doi: 10.1016/s0378-1119(96)00810-4. [DOI] [PubMed] [Google Scholar]
- 25.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 26.Stojiljkovic I, Hantke K. Haemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in Gram-negative bacteria. EMBO J. 1992;11:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugishima M, Omata Y, Kakuta T, Sakamoto H, Noguchi M, Fukuyama M K. Crystal structure of rat heme oxygenase-1 in complex with heme. FEBS Lett. 2000;471:61–66. doi: 10.1016/s0014-5793(00)01353-3. [DOI] [PubMed] [Google Scholar]
- 28.Sun J, Wilks A, Ortiz de Montellano P R, Loehr T M. Resonance Raman and EPR spectroscopic studies on heme-heme oxygenase complexes. Biochemistry. 1993;32:14151–14157. doi: 10.1021/bi00214a012. [DOI] [PubMed] [Google Scholar]
- 29.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takahashi S, Wang J, Rousseau D L, Ishikawa K, Yoshida T, Host J R, Ikeda-Saito M. Heme-heme oxygenase complex: structure of the catalytic site and its implication for oxygen activation. J Biol Chem. 1994;269:1010–1014. [PubMed] [Google Scholar]
- 31.Vasil M L, Oschner U A. The response of Pseudomonas aeruginosa to iron: genetics, biochemistry, and virulence. Mol Microbiol. 1999;34:399–413. doi: 10.1046/j.1365-2958.1999.01586.x. [DOI] [PubMed] [Google Scholar]
- 32.Wandersman C, Stojiljkovic I. Bacterial heme sources. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 33.Wilks A, Moenne-Loccoz P. Identification of the proximal ligand His-20 in heme oxygenase (HmuO) from Corynebacterium diphtheriae: oxidative cleavage of the heme macrocycle does not require the proximal histidine. J Biol Chem. 2000;275:11686–11692. doi: 10.1074/jbc.275.16.11686. [DOI] [PubMed] [Google Scholar]
- 34.Wilks A, Schmitt M P. Expression and characterization of a heme oxygenase (HmuO) from Corynebacterium diphtheriae. J Biol Chem. 1998;273:837–841. doi: 10.1074/jbc.273.2.837. [DOI] [PubMed] [Google Scholar]
- 35.Wilks A, Sun J, Loehr T M, Ortiz de Montellano P R. Heme oxygenase His25Ala mutant: replacement of the proximal iron ligand by exogenous bases restores catalytic activity. J Am Chem Soc. 1995;117:2925–2926. [Google Scholar]
- 36.Wilks A, Torpey J, Ortiz de Montellano P R. Heme oxygenase (HO 1). Evidence for electrophilic oxygen addition to the porphyrin ring in the formation of alpha meso-hydroxyheme. J Biol Chem. 1994;269:29553–29556. [PubMed] [Google Scholar]
- 37.Wilks A, Ortiz de Montellano P R. Rat liver heme oxygenase. High level expression of a truncated soluble form and nature of the meso-hydroxylating species. J Biol Chem. 1993;268:22357–22362. [PubMed] [Google Scholar]
- 38.Yoshida T, Kikuchi G. Purification and properties of heme oxygenase from rat liver microsomes. J Biol Chem. 1979;254:4487–4491. [PubMed] [Google Scholar]
- 39.Zhu W, Wilks A, Stojiljkovic I. Degradation of heme in gram-negative bacteria: the product of the hemO gene of Neisseriae is a heme oxygenase. J Bacteriol. 2000;182:6783–6790. doi: 10.1128/jb.182.23.6783-6790.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu W, Hunt D J, Richardson A R, Stojiljkovic I. Use of heme compounds as iron sources by pathogenic Neisseriae requires the product of the hemO gene. J Bacteriol. 2000;182:439–447. doi: 10.1128/jb.182.2.439-447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]