Abstract
Flagella purified from Salmonella enterica serovar Typhimurium contain FliG, FliM, and FliN, cytoplasmic proteins that are important in torque generation and switching, and FliF, a transmembrane structural protein. The motor portion of the flagellum (the basal body complex) has a cytoplasmic C ring and a transmembrane M ring. Incubation of purified basal bodies at pH 4.5 removed FliM and FliN but not FliG or FliF. These basal bodies lacked C rings but had intact M rings, suggesting that FliM and FliN are part of the C ring but not a detectable part of the M ring. Incubation of basal bodies at pH 2.5 removed FliG, FliM, and FliN but not FliF. These basal bodies lacked the C ring, and the cytoplasmic face of the M ring was altered, suggesting that FliG makes up at least part of the cytoplasmic face of the M ring. Further insights into FliG were obtained from cells expressing a fusion protein of FliF and FliG. Flagella from these mutants still rotated but cells were not chemotactic. One mutant is a full-length fusion of FliF and FliG; the second mutant has a deletion lacking the last 56 residues of FliF and the first 94 residues of FliG. In the former, C rings appeared complete, but a portion of the M ring was shifted to higher radius. The C-ring–M-ring interaction appeared to be altered. In basal bodies with the fusion-deletion protein, the C ring was smaller in diameter, and one of its domains occupied space vacated by missing portions of FliF and FliG.
The engine of the bacterial flagellum is a proton-powered, rotary motor (for a review of the field, see reference 14). Five proteins (FliG, FliM, FliN, MotA, and MotB) are thought to be directly involved in the generation of torque and the reversal of the direction of motor rotation. FliG is thought to be primarily involved in torque generation, while FliM and FliN are thought to be more involved in switching the direction of rotation (12). The inclusion of FliN as an integral component in torque generation or switching has been questioned (17, 21). MotA and MotB are transmembrane proteins that form a complex, which conducts protons from the periplasm to the cytoplasm (2, 18). The MotA-MotB complex is thought to be anchored to the peptidoglycan layer and hence to be part of the stator (3). In freeze fracture images of the flagellar motor, the presence of ca. 10 membrane particles (called studs) encircling the flagellar shaft depends on the expression of both MotA and MotB, suggesting that the studs correspond to the MotA-MotB complex (10).
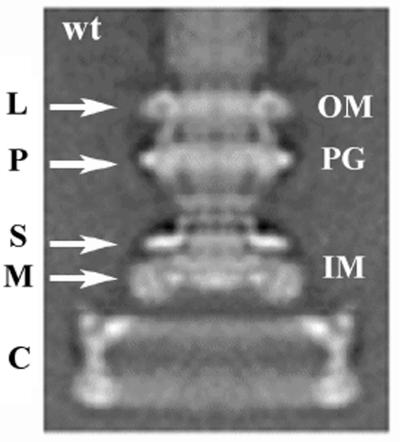
Flagella purified from cells retain three of the five proteins required for torque generation, namely, FliG, FliM, and FliN (6). In electron micrographs, the basal body (the cell-proximal portion of the flagellum) consists of a set of coaxial rings named for their locations relative to the partitions of the bacterial cell (Fig. 1). The L ring lies in the outer lipopolysaccharide layer, the P ring lies in the periplasmic space and is believed to be associated with the peptidoglycan layer, the S ring is a supramembrane feature (i.e., just outside the cytoplasmic membrane of the cell), the M ring crosses the cytoplasmic membrane, and the C ring lies in the cytoplasm. FliF is a transmembrane protein that is responsible for both the S ring and the M ring. Using antibody labeling and electron microscopy, Francis et al. (5) demonstrated that FliG is associated with the cytoplasmic face of the M ring to form the extended (or thicker) M ring. (Hereafter, when we refer to the M ring we include the extension due to FliG). The proteins FliM and FliN were identified by Francis et al. (6) as components of the C ring. Although these authors presented a structure of the basal body with the C ring, they were not able to determine whether parts of FliM or FliN also contribute to the M ring or whether parts of FliG contribute to the C ring.
FIG. 1.
An averaged image of a basal body from a wild-type strain (SJW880). The L, P, S, M, and C rings are labeled. The label “wt” is used for wild type in all of the figures. The positions of the outer membrane (OM), peptidoglycan (PG), and inner membrane (IM) are marked.
In addition, Francis et al. carried out studies of two mutant strains that make FliF-FliG fusion proteins (5). The mutants appeared as spontaneous deletions that eliminated the stop codon in fliF and shifted the adjacent fliG gene to a location that is in frame with respect to fliF. The proteins made from the mutants are single polypeptides containing both FliF and FliG sequences. In one of the two FliF-FliG fusion mutants, the fusion protein contains essentially the entirety of both FliF and FliG with only 5 amino acids deleted from the C terminus of FliF accompanied by the insertion of an isoleucine at the junction of FliF and FliG. This is called the full-length fusion protein. Flagellar motors having the full-length protein are vigorously motile. In the second mutant, there is a large deletion in which 56 residues from the C-terminal part of FliF and 94 residues from the N-terminal part of FliG are absent. This is known as the fusion-deletion protein. The deleted portion of FliG contains the residues (i.e., residues 1 to 40) required for binding of FliG to FliF (11). Therefore, were it not for the covalent linkage of FliG to FliF, FliG would be lost from the basal bodies. Flagellar motors having the fusion-deletion protein rotate, but such cells have a less vigorous and more variable motility. The presence of the fusion-deletion protein provided an opportunity to determine the location of the terminal sequences of the FliF and FliG proteins.
In order to define more accurately the locations of the component proteins, we undertook a structural study of basal bodies purified from wild-type cells and cells making the fusion and fusion-deletion proteins. The basal body preparations were then treated to remove some of the component proteins: for example, incubation at pH 4.5 removed FliM and FliN and hence the C ring, but not FliG; CsCl density gradient centrifugation also produced the same result, whereas incubation at pH 2.5 also removed FliG. We compared images of differentially extracted basal bodies in order to map the locations of these proteins.
MATERIALS AND METHODS
Bacterial strains.
The Salmonella enterica serovar Typhimurium strains used for these experiments are shown in Table 1. SJW134 is wild type except for the lack of an expressed flagellin gene, which makes it nonmotile. SJW880 has the polyhook phenotype and as a result is nonmotile. Both strains are otherwise wild type in motor function. The basal bodies prepared from SJW134 and SJW880 are identical except for a mutation in the fliK gene, which affects hook length. SJW3063-1 is a spontaneous pseudorevertant of a nonchemotactic (cheY) mutant. In this mutant, there is a small deletion that places fliF and fliG in the same reading frame. The mutant bacterium produces an essentially full-length fusion of the proteins FliF and FliG (FliF-FliG) (5). SJW3063-1 has a filament and is motile, though not chemotactic. MYR1101 is the same strain as SJW3063-1 but has had the filament gene knocked out by transposon insertion. It is, of course, nonmotile. SJW2334 also makes a FliF-FliG fusion protein but with a 150-amino-acid deletion, of which 56 are the C-terminal residues of FliF and 94 are the N-terminal residues of FliG. SJW2334 is weakly motile. SJW3821 is the same strain as SJW2334 but has had the filament gene knocked out by transposon insertion. The filament-free, full-length FliF-FliG mutant MYR1101 was given to us by R. Macnab (Yale University). The SJW3281 fusion-deletion strain was provided to us by S. Yamaguchi (Meiji University, Tokyo, Japan).
TABLE 1.
Strains used
| Strain | Parent strain/gene(s) altered | FliF, FliGa | Filament present | Motility |
|---|---|---|---|---|
| SJW134 | WTa | WT | No | None |
| SJW880 | SJW134/fliK | WT | No | None |
| SJW3063-1 | SJW1103/fliF and fliG | Full-length fusion | Yes | Vigorous |
| MYR1101 | SJW3063-1/fliC | Full-length fusion | No | None |
| SJW2334 | SJW806/fliF and fliG | Fusion with 150-aac deletion | Yes | Variable |
| SJW3821b | SJW2334/fliC | Fusion with 150-aa deletion | No | None |
WT, wild type.
Two separate but identical isolates both were purified. Basal bodies were the same, but only SJW3821 is presented here.
aa, amino acids.
Extraction and purification of flagellar motors.
The preparation of flagellar motor complexes from these strains followed procedures previously described (6) with some modification. Cells were grown as previously described in 8-liter cultures. At late log stage (8 − 10 h), the cells were pelleted at 4,000 × g for 20 min. The pellets were resuspended in 500 ml of an ice-cold solution of 0.5 M sucrose–50 mM Tris-HCl at pH 8.0. The preparation was divided in two and kept on ice. We added 8 ml of 0.5 M EDTA at pH 8.0 to each of the two aliquots to bring their final concentrations of EDTA to ca. 8 mM. The solutions were stirred for 30 min, after which 15 ml of lysozyme at 2 mg/ml was added, and the incubation continued for 30 to 60 min on ice with stirring. We added 0.5 M Mg2 SO4 to a final concentration of 20 mM to activate the endogenous DNase. The solutions were then incubated while stirring the mixtures on ice for 30 min. Unlysed cells were removed by centrifugation at 4,000 × g for 20 min. The supernatant was adjusted to pH 11 by dropwise addition of 5 M NaOH. The solution was centrifuged for 70 min at 90,000 × g. The pellets were resuspended in 30 ml of buffer containing 100 mM KCl, 10% (wt/vol) sucrose, 0.1% (vol/vol) Triton X-100, and 10 mM Tris-HCl (pH 8.0). The resuspended pellets were centrifuged at 4,000 × g for 10 min to remove debris. The supernatants were then centrifuged at 120,000 × g for 60 min, and the pellets resuspended in 100 to 500 μl of 0.1% (vol/vol) Triton X-100, 5 mM EDTA, and 10 mM Tris-HCl pH 8.0 (TET) buffer. The basal body preparation was then further purified by density gradient centrifugation in 50% (vol/vol) Percoll (Bio-Rad, Hercules, Calif.) in TET buffer. The Percoll beads were removed by gel filtration using an S-1000 column (Bio-Rad) as described earlier (9). The complexes are stable on ice for several weeks.
Aliquots of preparations from the fusion-deletion mutant, and the wild-type strains were purified by CsCl density gradient centrifugation as described earlier (1). Aliquots of the wild-type preparations were extracted at pH 4.5 or 2.5 as described previously (6). An aliquot was added to a 50 mM glycine buffer containing 0.1% (vol/vol) Triton X-100 adjusted to pH 2.5 or 4.5. The solutions were then centrifuged at 200,000 × g, and the pellets were resuspended and centrifuged again in the low-pH buffer. The extracted basal bodies were then resuspended in TET buffer and subjected to an additional round of centrifugation and resuspension in TET buffer.
Frozen hydrated samples were prepared in a cold room by placing 4 μl of a basal body preparation on a copper grid covered by a holey carbon film. The carbon films were made according to the procedure of Fukami and Adachi (8). Samples in which the C rings were removed from the basal bodies tended to adhere to uncharged carbon films. These samples were applied to glow-discharged holey carbon films. After 30 to 60 s, excess fluid was blotted away by using Whatman no. 40 filter paper (Whatman, Inc., Clifton, N.J.); the grid was plunged into liquid nitrogen-cooled ethane and stored under liquid nitrogen. Frozen grids were transferred into a CM12 electron microscope (Philips Electronic Instruments Co., Mahwah, N.J.) equipped with a model 651 anticontaminator and a model 626 cold stage (Gatan, Inc., Pleasanton, Calif.). Images were recorded at ×60,000 on SO-163 film (Eastman Kodak Co., Rochester, N.Y.) with a dose of 5 to 10 e/Å2 at ∼1 μm under focus. Images were digitized at 3.3 Å/pixel on an Eikonix (Bedford, Mass.) densitometer.
Computer image analysis.
All images of the hook-basal body complexes were processed by using the Brandeis package (15) and the single particle routines in SPIDER (7). All images were normalized by setting the average to zero and the standard deviation to a constant. This gave all images approximately the same statistical properties, and thus all images contributed equally to the averages. No correction for the contrast transfer function (CTF) was applied since the first node of the CTF occurred at ∼1/17 Å for these images recorded at ∼1 μm under focus. The resolution determined by spectral signal-to-noise calculation from the combined L and P rings from the samples used in this study was 22 Å (data not shown). We used the L and P rings as our resolution criterion since they should be the most constant portion of the structure across the various samples.
Averaging images.
Images of basal bodies were extracted from large scanned fields by using a generous mask. The basal bodies were rotated into an approximately upright position by eye. This upright position was later determined to be within ∼5° of vertical. The images were then masked from their surroundings so that all images had the same dimensions. Separate averages were produced based on alignment of the L and P rings, the M and S rings, and the C rings, and these separate averages were spliced into a single image (16). The reason for averaging parts separately is that the images of the basal bodies are not quite superposable so that alignment and averaging of the parts produced sharper features than alignment of the whole structure. For example, when basal bodies were aligned by using the L and P rings, the averages of the M and S rings or of the C rings were noticeably worse than when these rings were used for alignment. The individually masked rings were aligned against a “best” particle. Subsequent rounds of alignment and averaging used the mirror-symmetrized intermediate average as a reference image. The cycles of alignment and averaging were stopped when the alignment parameters ceased to change significantly. A summary of the numbers of images analyzed and averaged for each specimen is included in Table 2.
TABLE 2.
Data collected and averaged
| Specimen | No. of images analyzed | No. of images included in average |
|---|---|---|
| SJW134, pH 2.5 treated | 84 | 63 |
| SJW3063-1, pH 2.5 treated | 60 | 53 |
| SJW134 with C ring | 74 | 50 |
| SJW134 with CsCl purified | 103 | 82 |
| SJW134, pH 4.5 treated | 131 | 90 |
| SJW880 with C ring | 109 | 90 |
| SJW880 without C ring | 200 | 200 |
| SJW3281 with C ring | 78 | 63 |
| SJW3281 without C ring | 65 | 50 |
| SJW3281 CsCl purified | 61 | 40 |
| MYR1101 with C ring | 28 | 14 |
| MYR1101 without C ring | 145 | 62 |
Computing difference images.
The differences were calculated between pairs of mirror-symmetrized averaged images. The most difficult aspect of calculating differences among data from different data sets was scaling the averages to one another so that the differences observed represent true structural differences. The best approach was to use histogram fitting with the function CE FIT in the SPIDER package on the actual windowed images to be compared, using the wild-type without a C ring as the reference when M rings were compared. The result is a fit based on the following parameters: x′ = Ax + B, where x and x′ represent the densities in the input and output images, respectively, and A and B are the scaling constants determined from the histograms. The corresponding variance map was multiplied by A2 prior to determining the significance levels. The statistical significance was determined by using the program DIFFTTEST (20).
RESULTS
In order to locate the features that had been altered by the removal of particular proteins, we compared images of the corresponding basal bodies. To obtain a representative image of a particular preparation, we selected and then averaged images of dozens of individual basal bodies. In computing the average, we also computed a variance map, in which regions of structural variation appear as bright features. Positive (bright) features arise from variations among the images in a preparation. Such variations can result from noise, from differences in orientation (e.g., tilt out of the plane of view), from variations in the degree of defocus used to obtain the electron micrographs, and from true variations in the structure, position, and numbers of the component parts. To look for features that were altered in different preparations, we subtracted the corresponding averaged images, thereby generating difference maps. Positive (bright) or negative (dark) features might indicate changes in the structures. To assess the significance of the differences, we computed significance maps in which bright and dark features corresponded to statistically significant positive and negative differences, respectively (20). Variations in the background surrounding the basal bodies were generally not significant because the backgrounds tended to be the same. Thus, when averages taken from the corresponding background areas were subtracted, the difference was no more than expected by chance given the noise. On the other hand, if one set of images had a feature not present in the other, then there was a significant change in one set of images relative to the other. Differences corresponding to this region of true structural variation were larger than expected by chance. Hence, statistical maps helped us sort out which differences correspond to structural changes and which arise just by chance due to noise in the images.
Averaged images of basal bodies, the cell-proximal portion of the flagellum.
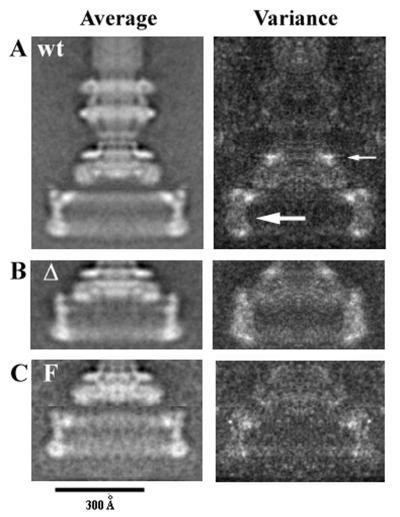
The averaged images and variance images of basal bodies with C rings from the wild-type and the gene-fusion strains are shown in Fig. 2. Both variation in the diameters of the C rings (19) and a slight tilting of the structure in some of the images lead to increases in the C-ring portions of the variance maps (see arrows in Fig. 2A). The source of variations in the S ring is unknown.
FIG. 2.
Averaged images (left) and variance maps (right) of basal bodies with a C ring. (A) Basal bodies from strain SJW880. (B) Basal bodies from strain SJW2381, the FliF-FliG fusion-deletion mutant. (C) Basal bodies from strain MYR1101, the full-length FliF-FliG fusion mutant. The markings in the upper left-hand corner of the average images are used in all figures: “wt” for wild type, “Δ” for the fusion-deletion mutant, and “F” for the full-length fusion mutant. Any additional treatment will be indicated in the upper right-hand corner of the image, e.g., 4.5 to indicate treatment with pH 4.5 buffer. The density in each pixel in an average map represents the average of the optical densities of the corresponding pixels in the individual images. Along with the average density at each pixel, we computed a variance of the densities that are averaged at that pixel. Peaks in the variance map indicate positions at which there is the most variation among the contributing densities. The averages and variances are displayed in the identical pixels in the two maps. In the variance map in panel A, there is what appears to be a ghost of the C ring (large arrow) and also one of the S ring (small arrow). The former is due to variations in the size of the C ring and/or to the tilt of the C ring out of the plane of view. The source of the variation in the S ring is not known.
Changes associated with the loss of FliM and FliN.
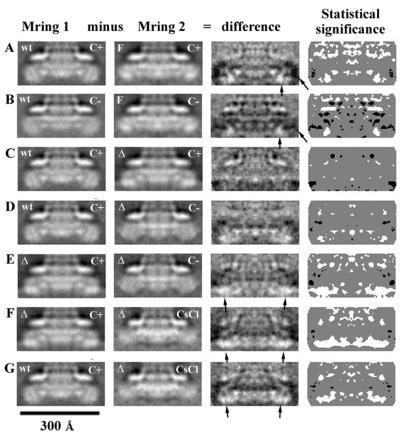
Treatment of the basal body preparations by either incubation at pH 4.5 or by CsCl density gradient centrifugation results in the loss of FliM and FliN (6). In the electron microscope, all basal bodies in these preparations lack the C ring (Fig. 3B and C). In untreated samples, ca. 30% of the basal bodies have no C ring (Fig. 3A). Using difference images, we compared the M rings of basal bodies with C rings to those lacking C rings, whether the C rings had been removed by incubation at pH 4.5 or by subjection to CsCl density gradient centrifugation or simply were not present in the untreated samples (Fig. 4A to D). The differences in densities comprising the M ring are very close to the background level in Fig. 4A to D. When there are true differences in the M ring, there are strong features in the difference map, as is the case in Fig. 4E (see below). Thus, loss of the C ring does not appreciably alter the M ring.
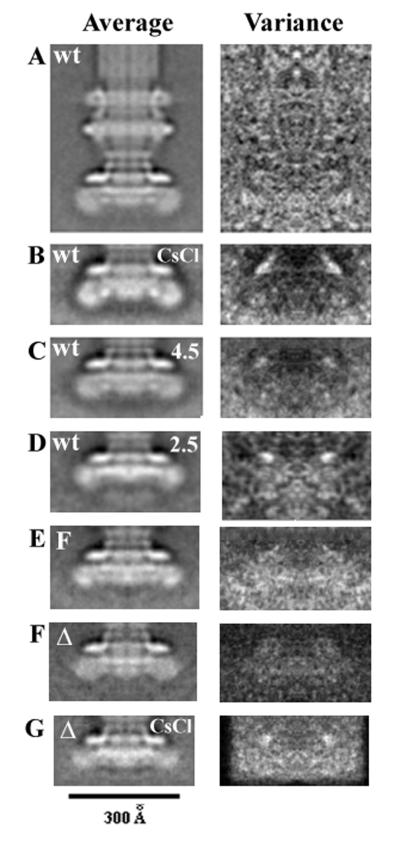
FIG. 3.
Averaged images (left) and variance maps of basal bodies lacking the C ring. (A) Basal bodies that lack C ring in preparations from wild-type strain (SJW880) in which most have a C ring. (B) Basal bodies (from SJW134) that have had the C ring removed by CsCl density gradient centrifugation. (C) Basal bodies (from SJW134) that have had the C ring removed by incubation at pH 4.5. (D) Basal bodies (from SJW134) that have had the C ring and FliG removed by incubation at pH 2.5. (E) Basal bodies that lack C rings from the full-length fusion mutant MYR1101. (F) Basal bodies that lack C rings from the fusion-deletion mutant SJW3821. (G) Basal bodies (from the fusion-deletion mutant SJW3821) that have had the C ring removed by CsCl density gradient centrifugation.
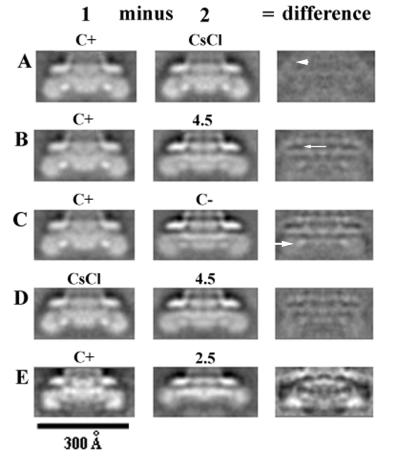
FIG. 4.
Difference images of M rings from wild-type basal bodies. The average in the middle column is subtracted from the average in the left column. The difference is shown in the right-side column. The label “C+” indicates that the C ring was present (but boxed out). The label “C−” indicates that the image is an average of M rings of basal bodies that lack C rings but that are found in preparations in which most basal bodies had C rings. The labels “CsCl,” “4.5,” and “2.5” refer to treatments used to remove the C ring and FliG. (A) Basal bodies with C rings (Fig. 2A) minus basal bodies in which C rings were removed by CsCl density gradient centrifugation (Fig. 3B). (B) Basal bodies with C rings (Fig. 2A) minus basal bodies in which C rings were removed by incubation at pH 4.5 (Fig. 3C). (C) Basal bodies with C rings (Fig. 2A) minus untreated basal bodies that lacked C rings (Fig. 3A). (D) Basal bodies that had the C ring removed by CsCl density gradient centrifugation (Fig. 3B) minus basal bodies in which C rings had been removed by incubation at pH 4.5 (Fig. 3C). (E) Basal bodies with C rings (Fig. 2A) minus basal bodies in which C rings and FliG had been removed by incubation at pH 2.5 (Fig. 3D). The differences are all shown with the same gray scale so that the large differences in panel E are much bigger than those found in panels A through D, as expected. Regions of the image that are black correspond to negative regions. Regions that are light gray (such as seen in the region outside the particle) are approximately zero. The small arrows indicate the negative difference peaks that occur in the region containing the S ring. The larger arrows indicate differences in the region containing the M ring. These differences (except in panel E) are about equal to those found outside the basal body, such as the region marked with the arrowhead, and are therefore not significant. The difference shown in panel E is significant.
Changes in the M ring associated with treatment at pH 2.5.
Incubation of basal body preparations at pH 2.5 results in the loss of FliG, as well as FliM and FliN (5, 6). Images of the preparations reveal a loss of the C ring and a thinning of the M ring (Fig. 3D). Difference images between pH 2.5-treated basal bodies and untreated basal bodies reveal a loss of matter on the cytoplasmic face of the M ring (Fig. 4E).
Changes in the M ring of basal bodies containing the full-length FliF-FliG fusion protein.
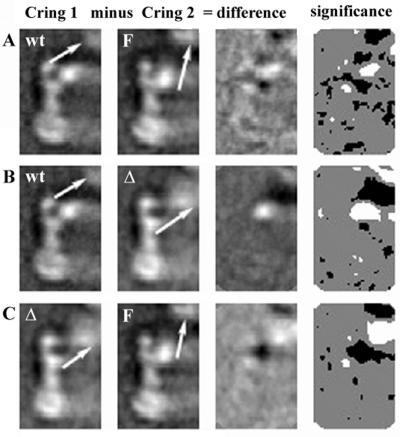
Most of the basal bodies with the full-length fusion protein lacked C rings, suggesting that the mutation destabilizes the C-ring–M-ring interactions. The averages of the M and C rings are shown in Fig. 2C. An averaged image of M ring lacking the C ring is shown in Fig. 3E. No difference was observed between the M rings of basal bodies with the C ring and those lacking it (data not shown). Figure 5A and B are difference images between basal bodies from the full-length fusion strain and those from wild type. As is the case in Fig. 4E, the maps show differences in the M ring, as well as unexplained differences in the S ring. The differences in the M ring suggest a redistribution of matter in the cytoplasmic part of the M ring, the region thought to contain FliG. There is a shift of matter from an inner part of the M ring to an outer part of the M ring, as evidenced by the presence of a positive peak in the difference maps at an inner portion of the M ring and a negative peak at the outer edges of the M ring (Fig. 5A and B). This is what one would expect to see in the difference image if there were a net movement of density from one place to another.
FIG. 5.
Difference images of M rings from wild-type basal bodies and basal bodies from the FliF-FliG mutants strains. Columns 1 and 2 contain MS ring averages. Column 2 is subtracted from column 1 to generate the difference image shown in column 3. Column 4 contains a statistical significance map. White represents positive differences exceeding the 99% significance level as determined by using a Student's t test. Black represents negative differences exceeding the 99% significance level. Gray represents differences that are not significant at the 99% level. (A) Basal bodies that have C rings (from wild-type strain SJW880 in Fig. 2A) minus basal bodies that have the C ring (from the full-length fusion mutant in Fig. 2C). (B) Basal bodies that lack C rings (from the wild-type strain) minus basal bodies that have the C ring (from the full-length mutant in Fig. 2C). In panels A and B, arrows indicate the positions of positive peaks and negative peaks, which suggest a shift of matter from an outer to an inner radius. (C) Basal bodies that have C rings (from wild-type strain SJW880 in Fig. 2A) minus basal bodies that have the C ring (from the fusion-deletion mutant in Fig. 2B). (D) Basal bodies with C ring (from wild-type strain SJW880 in Fig. 2A) minus basal bodies lacking C ring (from the fusion-deletion mutant in Fig. 3F). In place of basal bodies with the C ring, we could have substituted wild-type basal bodies lacking the C ring (Fig. 3A) with the same result, since the M rings of both are identical. (E) Basal bodies with a C ring (from the fusion-deletion mutant) minus basal bodies that lack the C ring (from the fusion-deletion mutant). (F) Basal bodies with a C ring (from the fusion-deletion mutant) minus basal bodies (from the fusion-deletion mutant) that have had the C rings removed by CsCl density gradient centrifugation (Fig. 3G). (G) Basal bodies that have C rings (from wild-type strain SJW880 in Fig. 2A) minus basal bodies (from the fusion-deletion mutant) that have had C rings removed by CsCl density gradient centrifugation (Fig. 3G). In panels D to F, arrows indicate the position of positive peaks arising from the loss of material in the M ring.
Changes in the M rings of basal bodies containing the FliF-FliG fusion-deletion protein.
We expected that differences due to the deletion would be confined to the M ring because FliF and FliG are thought to make up the M ring (5, 6). Instead there is a large rearrangement of matter involving part or parts of the C ring. Figure 5C shows the difference images between the M rings with wild-type proteins and the M ring with the fusion-deletion protein when the C ring is present. There are again changes in the S ring but essentially no change in the M ring (see the statistical significance map in Fig. 5C). But when the C ring is absent, the M ring with the fusion-deletion protein does indeed appear different. The difference images in the absence of the C ring show positive peaks that indicate material is missing from the cytoplasmic face of the M ring (Fig. 5D). Using basal bodies with the FliF-FliG fusion-deletion protein, we compared those lacking a C ring to those having a C ring (Fig. 5E) and again found a change in the cytoplasmic portion of the M ring. These same differences are more clearly seen when the same comparisons are made with basal bodies purified by CsCl density gradient centrifugation from the fusion-deletion mutant, as shown in Fig. 5F and G. These differences are similar to those shown in Fig. 4E in the comparison of the wild-type basal body with the C ring to the pH 2.5-treated basal body, which lacks the C ring and FliG.
Changes in the C rings of basal bodies with the FliF-FliG full-length fusion protein.
The C ring in this mutant has the same dimensions as the wild-type C ring, although the yield of C ring containing basal bodies, ∼10% versus ∼70% for the wild type, would suggest the connection between the C ring and basal body is less stable. While the outer part of the C ring appears unaltered, the inner domain of the C ring shows an upward tilt toward the outer edge of the M ring, as if the angle or direction of the connection between C and M ring had been changed. We have indicated the angle of the connection with arrows. The connection in the wild type and the fusion-deletion mutant (Fig. 6, left-hand columns) is at ca. 2 o'clock, whereas that in the full-length fusion is at ca. 12 to 1 o'clock. The movement of density associated with the change in orientation is indicated by the presence of both positive and negative peaks in the difference images.
FIG. 6.
Difference images of C rings from wild-type basal bodies and basal bodies from the FliF-FliG mutants strains. Columns 1 and 2 contain C-ring averages. Column 2 is subtracted from column 1 to generate the difference image shown in column 3. Column 4 contains a statistical significance map. White represents positive differences exceeding the 99% significance level as determined by using a Student's t test. Black represents negative differences exceeding the 99% significance level. Gray represents differences that are not significant at the 99% level. (A) Basal bodies from the wild-type strain (SJW880) versus basal bodies from the full-length fusion mutant (MYR1101). The elongated inner C-ring feature or domain appears rotated, and its connection to the M ring has been shifted radially outward from the flagellar axis. The arrows above and then below this feature in the left and middle columns, respectively, indicate how the angular orientation changes. Note the positive and negative difference peaks in the region of the C ring's inner domains. (B) Basal bodies from the wild-type strain (SJW880) versus basal bodies from the fusion-deletion mutant (SJW2381). The arrows indicate how the inner domain of the C ring is shifted inward and upward in the fusion-deletion mutant. (C) Basal bodies from the fusion-deletion mutant versus basal bodies from the full-length fusion mutant. The arrows indicate the direction of the connection between the inner domain of the C ring and the outer rim of the M ring. The direction of the connection in the wild-type basal body is oriented at ca. 2 o'clock. In the full-length fusion mutant, however, the inner domain of the M ring appears to be tilted upward so that the connection is oriented somewhere between 12 and 1 o'clock.
Changes in the C rings of basal bodies from the fusion-deletion mutant.
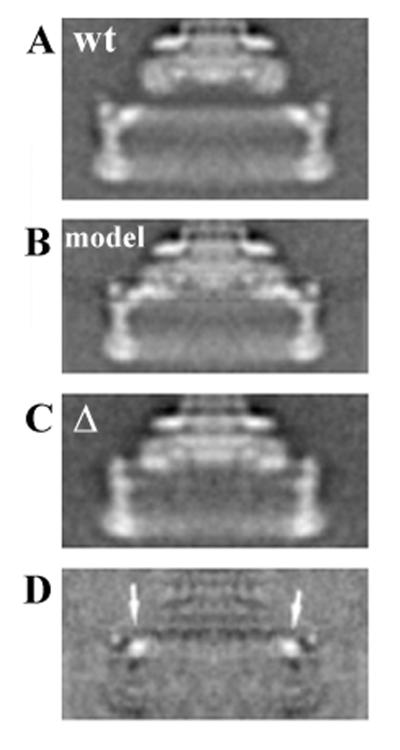
The averaged images of basal bodies having the fusion-deletion protein (Fig. 2B) shows that there has been a ∼6 nm axial shift of the C ring relative to the M ring so that the top of the C ring is now opposite the bottom of the M ring. The C ring is also reduced by 10% in diameter. Previously, we showed that this reduction was the result of having 10% fewer subunits in the ring (19). The innermost domain of the C ring appears to be missing, but the M ring appears to be complete. When the C ring is absent, however, mass is missing from the M ring. Recalling that the loss of the C ring produces no change in the M ring from wild-type strains, we conclude that the inner domain of the C ring occupies a gap in the M ring resulting from the loss of the C-terminal portion of FliF and the N-terminal portion of FliG. In this rearrangement, the inner C-ring domain must be shifted relative to its position in basal bodies from the wild-type strain. To demonstrate this, we altered the image of the C ring from wild-type preparations to mimic the diameter and position of the C ring of the mutant (Fig. 7B). The inner domain of the C ring after the appropriate axial shift and reduction in radius still lies outside the M ring (compare Fig. 7B and C) in a position that is unoccupied in the mutant. The difference image shows a clear peak associated with absence of this domain but no negative peak that should indicate the missing portions of FliF and FliG. This domain therefore appears to be shifted to smaller radius and hence into the M ring, relative to the rest of the C ring. The length of the connection between the outer part of the C ring and what should be the inner domain thus would appear to be stretched by at least 2.2 nm.
FIG. 7.
Evidence that the C ring of the fusion-deletion mutant is distorted relative to the C ring in the wild-type strain. To reveal the changes in the C ring, we reduced the diameter to that of the C ring in the fusion-deletion mutant (∼10%), and we shifted it upward to the position of the C ring in the fusion-deletion mutant. Thus, except for distortions of the C ring, the two images should be the same. (A) Unaltered wild-type SJW880 basal body. (B) Computer-altered image of the basal body from the wild type. (C) Basal body from the fusion-deletion mutant. (D) Panel B minus panel C. The appearance of the positive peak in the difference image shows that the conformation of the C ring must be altered in the mutant relative to the wild type.
DISCUSSION
FliM and FliN make no appreciable contribution to the M ring.
It was shown previously (5) that basal bodies lacking FliM and FliN did not have C rings. Basal bodies in which the bulk of the C ring was missing could not be decorated with antibodies against FliN or FliM but could be decorated with antibodies against FliG. What was not known from these studies was whether FliM or FliN contributed domains to the M ring and whether FliG indeed was located on the cytoplasmic face of the M ring or whether it gave rise to a portion of the C ring. We show here that difference maps made by subtracting the averaged images of basal bodies lacking FliM and FliN but having FliG from the averaged image obtained from basal bodies with all three proteins reveal no significant differences in the M ring (Fig. 4A to D). The simplest interpretation is that FliM and FliN are components of the C ring, a ring that is missing in basal bodies lacking FliM and FliN. This result is evidence that FliM and FliN do not make up any significant portion of the M ring.
FliG contributes density to the cytoplasmic face of the M ring.
When the preparations of basal bodies are treated with a pH 2.5 buffer, FliG is extracted from the preparations (5). Difference maps reveal loss of material from the cytoplasmic face of the M ring (Fig. 4E). We cannot conclude, however, that all of the missing mass corresponds to FliG since FliE, FliP, and FliR, which have a role in flagellar protein export, are present in our preparations (4) but may also be lost upon extraction.
We hoped that the FliF-FliG fusion protein would enable us to localize FliG. Recall that extraction by incubation at pH 2.5 removes FliG from the wild-type basal body. In the full-length fusion, FliG, which is covalently linked to FliF, is indeed not removed (6). Images of mutant and wild-type flagella incubated at pH 2.5 might be expected to reveal the location of FliG since the loss of other proteins (e.g., export proteins) will not be at issue because both wild-type and mutant preparations were treated the same. The images of the M rings, however, are the same and do not reveal the position of FliG (data not shown). The reason for a lack of visible difference in the M ring, despite the presence of FliG, is likely to be that FliG becomes disordered or denatured. This is a plausible result of treatment with pH 2.5 buffer, which in addition to removing FliG in wild-type preparations also depolymerizes the filament and could disrupt the noncovalent interactions between the FliG and FliF portions in the fusion protein, thereby causing disorder.
The fusion of FliG to FliF causes a conformational change in the M ring and in the C ring.
The full-length fusion mutant provides insight into the location of FliG in the M ring. Comparison of the M ring from the full-length fusion mutant with that of the wild-type strain reveals a shift of an inner feature to an outer position. The feature that is moved is believed to be a part of FliG since this movement occurs in the same region where mass is removed in pH 2.5- treated basal bodies that lack FliG (compare Fig. 4E and 5A and B). The fusion-deletion mutant basal bodies that either naturally lack the C ring or have been purified by CsCl density gradient centrifugation lack mass at the cytoplasmic surface of the M ring, a finding again consistent with the loss of mass observed in pH 2.5-treated basal bodies lacking FliG. These results all suggest that the location of FliG must be on the cytoplasmic surface of the M ring and that FliG is exposed at the outer radius where it can make contact with both the MotA-MotB complex and FliM.
In the fusion-deletion mutant, a domain of the C ring replaces material missing in the M ring.
In the fusion-deletion mutant, the diameter of the C ring is 10% narrower (and has 10% fewer subunits) than the C ring of wild-type strains (19). The M rings in the M ring-C ring complex in basal bodies from the fusion-deletion mutant are indistinguishable in difference maps from wild type, but the inner domain of the C ring in the mutant appears missing (Fig. 5C and 7C). When the C ring is removed, however, the M ring of the fusion-deletion mutant lacks material on its cytoplasmic face (Fig. 5E), a finding similar to that obtained when FliG is removed by incubation at pH 2.5. There are two possibilities to explain this: (i) the “missing” domain in the C ring corresponds to the deleted residues in the fusion-deletion protein and (ii) the “missing” domain in the C ring is now located in the M ring, where it replaced the mass corresponding to the deleted residues. We argue that the latter possibility better explains the observations. First, there is a change in the M ring upon loss of the C ring, when no such change is expected if the first explanation were true. No such change is expected because when the C ring is removed from wild-type basal bodies there is no change in the M ring. If the M rings in the mutant and wild type are essentially identical, as should be the case in the first explanation, then there is no reason to expect that the M ring in the fusion-deletion mutant would be altered by removal of the C ring. The second reason involves the attachment of the C ring to the M ring. If the missing domain of the C ring corresponds to the missing sequences in FliF and FliG, then it is that portion of FliF and FliG that interacts with FliM and FliN. If this portion of FliF and FliG were deleted, we would expect that FliM and FliN would not bind to the M ring and hence that the C ring would be absent. The better interpretation of changes attending removal of the C ring is therefore that the missing domain of the C ring lies in the M ring of the fusion-deletion mutant and that removal of the C ring results in the loss or disordering of this domain so that it is not seen in the averaged image.
We next explored the possibility that changes in only the diameter and the position of the C ring, but not changes in its structure, could account for the features of the basal bodies from the fusion-deletion mutant. To test this possibility, we simulated such a change in position and diameter in the computer. We used the averaged image of the basal body from the wild type. We reduced the diameter of the C ring and shifted it to the position it takes in the mutant (Fig. 7B). The inner domain, however, did not occupy the expected position in the M ring for this possibility to be correct. The difference map (Fig. 7D) has a positive peak that lies in the region that is unoccupied in the fusion-deletion mutant. The inner domain instead must lie at a smaller radius than would occur if the conformations of the subunits in the C ring were unchanged. Thus, there must be distortion of the C ring that accompanies the assembly of the C ring at a smaller radius.
To complete our argument, we made a rough calculation showing that the volume of the shifted inner domain of the C ring is approximately the volume expected for the missing residues of FliF and FliG in the fusion-deletion protein. The shifted domain is ca. 25 Å by 47 Å in two dimensions. We can estimate its size in the third dimension because we know there are 34 such domains packed into a ring that lies at a radius of 165 Å. The third dimension is therefore 330 Å π/34 ≅ 30 Å. The estimated volume of the ellipsoid is then (4π/3)(25 Å · 47 Å · 31 Å/8) ≅ 18,000 Å3. The mass missing in the fusion-deletion protein is ca. 16,000 Da which would correspond to a volume of ca. 16,000 Da/0.75 Da/Å3 ≅ 21,000 Å3, which is close to the estimated volume of the shifted domain.
The inner domain appears to be the domain that links the C ring to the M ring because the change in the position of the inner domain alters the diameter of the C ring. In the averaged image in Fig. 1, the C ring seems unattached to the M ring, which of course cannot be true since they are isolated as a complex. There must therefore be a connector from the C ring to the M ring, although it is not visible in our averages. We assume it has insufficient mass to be seen given the fact that we are not determining a true three-dimensional structure from our alignments and are presenting what are in effect cylindrically averaged images. Does this connector arise from a portion of FliG that extends out toward the inner domain of the C ring to make contact with FliM, does it correspond to a portion of FliM that extends inwards to make contact with FliG in the M ring, or does it arise from both FliM and FliG? We have good evidence that FliM and FliN make up no significant part of the M ring. If FliM makes up the connector, it adds little if any mass to the M ring. We do not know whether FliG contributes significantly to the inner domain of the C ring.
We know that FliM and FliN can be removed without loss of FliG. In the averaged images of such preparations, the C ring is completely gone and the M ring is unchanged (Fig. 4A to D). FliG either does not contribute to the C ring or, if it does, that portion is disordered and thus is not seen when the C ring is removed. In principle one might be able to detect the disordered portions of FliG in difference maps or variance maps, since a disordered portion of FliG would increase the average density and the variance relative to the background. Neither is seen, but perhaps the changes are not significant. Thus, the identity of the protein in the inner domain and the connector is unresolved.
How can a motor continue to operate as well as it does in the fusion-deletion mutant? Since the C-terminal domain of FliG is responsible for torque generation (12, 13), whereas FliM is responsible for switching, it may be that the C-terminal torque-generating domain of FliG is largely unchanged by the changes in its N-terminal portion and therefore the motor continues to function. The changes in the other portions of the structure would only affect switching, which is altered in the fusion-deletion mutant. The other possibility is that it is the junction of FliG and FliM at which torque is generated (19) and, although the junction is moved, the relationship and hence the function is retained.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants R01-GM35433 and T32-GM07596 from the National Institute of General Medical Sciences and by funds from the W. M. Keck Foundation.
We thank Noreen Francis for help in preparing samples.
REFERENCES
- 1.Aizawa S I, Dean G E, Jones C J, Macnab R M, Yamaguchi S. Purification and characterization of the flagellar hook-basal body complex of Salmonella typhimurium. J Bacteriol. 1985;161:836–849. doi: 10.1128/jb.161.3.836-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blair D F, Berg H C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 3.Blair D F, Kim D Y, Berg H C. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan F, Ohnishi K, Francis N R, Macnab R M. The FliP and FliR proteins of Salmonella typhimurium, putative components of the type III flagellar export apparatus, are located in the flagellar basal body. Mol Microbiol. 1997;26:1035–1046. doi: 10.1046/j.1365-2958.1997.6412010.x. [DOI] [PubMed] [Google Scholar]
- 5.Francis N R, Irikura V M, Yamaguchi S, DeRosier D J, Macnab R M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization, and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 7.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 8.Fukami A, Adachi K. On an adhering method of thin film specimens to specimen grids. J Electron Microsc. 1964;13:26–27. [Google Scholar]
- 9.Khan I H, Reese T S, Khan S. The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA. 1992;89:5956–5960. doi: 10.1073/pnas.89.13.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan S, Dapice M, Reese T S. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988;202:575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- 11.Kihara M, Miller G U, Macnab R M. Deletion analysis of the flagellar switch protein FliG of Salmonella. J Bacteriol. 2000;182:3022–3028. doi: 10.1128/jb.182.11.3022-3028.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lloyd S A, Tang H, Wang X, Billings S, Blair D F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloyd S A, Whitby F G, Blair D F, Hill C P. Structure of the C-terminal domain of FliG, a component of the rotor in the bacterial flagellar motor. Nature. 1999;400:472–475. doi: 10.1038/22794. [DOI] [PubMed] [Google Scholar]
- 14.Macnab R M. Flagella and motility. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 15.Owen C H, Morgan D G, DeRosier D J. Image analysis of helical objects: the Brandeis helical package. J Struct Biol. 1996;116:167–175. doi: 10.1006/jsbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 16.Sosinsky G E, Francis N R, Stallmeyer M J, DeRosier D J. Substructure of the flagellar basal body of Salmonella typhimurium. J Mol Biol. 1992;223:171–184. doi: 10.1016/0022-2836(92)90724-x. [DOI] [PubMed] [Google Scholar]
- 17.Tang H, Billings S, Wang X, Sharp L, Blair D F. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol. 1995;177:3496–3503. doi: 10.1128/jb.177.12.3496-3503.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang H, Braun T F, Blair D F. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 19.Thomas D R, Morgan D G, DeRosier D J. Rotational symmetry of the C ring and a mechanism for the flagellar rotary motor. Proc Natl Acad Sci USA. 1999;96:10134–10139. doi: 10.1073/pnas.96.18.10134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Trachtenberg S, DeRosier D J. Three-dimensional structure of the frozen-hydrated flagellar filament. The left-handed filament of Salmonella typhimurium. J Mol Biol. 1987;195:581–601. doi: 10.1016/0022-2836(87)90184-7. [DOI] [PubMed] [Google Scholar]
- 21.Vogler A P, Homma M, Irikura V M, Macnab R M. Salmonella typhimurium mutants defective in flagellar filament regrowth and sequence similarity of FliI to F0F1, vacuolar, and archaebacterial ATPase subunits. J Bacteriol. 1991;173:3564–3572. doi: 10.1128/jb.173.11.3564-3572.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]