Abstract
Background
To assess whether artificial intelligence (AI)-based decision support allows more reproducible and standardized assessment of treatment response on MRI in neuro-oncology as compared to manual 2-dimensional measurements of tumor burden using the Response Assessment in Neuro-Oncology (RANO) criteria.
Methods
A series of 30 patients (15 lower-grade gliomas, 15 glioblastoma) with availability of consecutive MRI scans was selected. The time to progression (TTP) on MRI was separately evaluated for each patient by 15 investigators over two rounds. In the first round the TTP was evaluated based on the RANO criteria, whereas in the second round the TTP was evaluated by incorporating additional information from AI-enhanced MRI sequences depicting the longitudinal changes in tumor volumes. The agreement of the TTP measurements between investigators was evaluated using concordance correlation coefficients (CCC) with confidence intervals (CI) and P-values obtained using bootstrap resampling.
Results
The CCC of TTP-measurements between investigators was 0.77 (95% CI = 0.69,0.88) with RANO alone and increased to 0.91 (95% CI = 0.82,0.95) with AI-based decision support (P = .005). This effect was significantly greater (P = .008) for patients with lower-grade gliomas (CCC = 0.70 [95% CI = 0.56,0.85] without vs. 0.90 [95% CI = 0.76,0.95] with AI-based decision support) as compared to glioblastoma (CCC = 0.83 [95% CI = 0.75,0.92] without vs. 0.86 [95% CI = 0.78,0.93] with AI-based decision support). Investigators with less years of experience judged the AI-based decision as more helpful (P = .02).
Conclusions
AI-based decision support has the potential to yield more reproducible and standardized assessment of treatment response in neuro-oncology as compared to manual 2-dimensional measurements of tumor burden, particularly in patients with lower-grade gliomas. A fully-functional version of this AI-based processing pipeline is provided as open-source (https://github.com/NeuroAI-HD/HD-GLIO-XNAT).
Keywords: Artificial intelligence (AI)-based decision support, RANO, tumor response assessment, tumor volumetry
Key Points.
Artificial intelligence (AI)-based decision support improved the concordance of TTP ratings over Response Assessment in Neuro-Oncology (RANO) alone.
AI-based decision support was more useful for lower-grade gliomas as compared to glioblastoma.
Less experienced investigators judged the AI-based decision support as more helpful.
Importance of the Study.
The Response Assessment in Neuro-Oncology (RANO) criteria are widely adopted in neuro-oncology, however the prescribed manual measurements of tumor burden on MRI may be challenging and potentially limit the reproducibility of the RANO criteria for reliable assessment of treatment response. There has been long-standing interest in using volumetric assessment of tumor burden with previous studies indicating that volumetric measurements may be more reliable and accurate as compared to 2-dimensional measurements of tumor diameters in arbitrarily chosen slices. The present study demonstrates that artificial intelligence (AI)-based decision support has the potential to yield more reproducible and standardized assessment of treatment response in neuro-oncology as compared to manual 2-dimensional measurements of tumor burden. Particularly the evaluation of lower-grade gliomas where reliable assessment of the TTP may be challenging due to the slow-growing nature of these tumors may benefit from AI-based decision support. A fully functional version of this AI-based processing pipeline is provided as open-source (https://github.com/NeuroAI-HD/HD-GLIO-XNAT).
Magnetic resonance imaging (MRI) is used extensively in cancer research during drug development, including clinical trials, as well as for the routine management of cancer patients.1 It is particularly valuable for brain tumors, which are located in one of the most vulnerable and hard-to-reach regions of the human body. However, the assessment of imaging data by radiologists still relies primarily on qualitative (subjective) visual interpretation, which may increase the burden of time and expenditure on clinical trials, and which may also hamper the validity of imaging biomarkers used in clinical trials and clinical practice for assessing treatment response. The criteria for assessing treatment response and efficacy in neuro-oncology are essentially based on longitudinal measurements of the largest diameters of contrast-enhancing target lesions on imaging as formalized by the Response Assessment in Neuro-Oncology (RANO) criteria.2,3 The RANO criteria are widely adopted in neuro-oncology clinical trials to yield a standardized and reproducible assessment of treatment response. Underlying the use of RANO is the assumption that the two-dimensional measurement of a contrast-enhancing lesion’s largest diameter on MRI is a surrogate marker of the overall tumor burden. However, this assumption is not always accurate, since brain tumors frequently display very complex shapes and anisotropic growth, influenced in part by the surrounding anatomic boundaries, host tissue–tumor interface, or treatment related effects (eg, areas of necrosis and surgical cavities). Consequently, reproducible assessment of tumor burden and treatment response and/or disease progression between different radiologists using RANO criteria may be challenging and thus potentially limiting its value for clinical decision making. Reproducible assessment is further complicated by the assessment of nonenhancing T2/FLAIR lesions as an additional criterion besides contrast-enhancing lesions for evaluating treatment response and/or disease progression.
In the light of that, there has been long-standing interest in using volumetric assessment of tumor burden3,4 with previous studies indicating that volumetric measurements may be more reliable and accurate as compared to 2-dimensional measurements of tumor diameters in arbitrarily chosen slices (Figure 1).5,6 In the present study we investigated the clinical utility of artificial intelligence (AI)-based decision support with automated volumetric quantification of tumor burden on MRI in neuro-oncology and evaluated whether it enables more reproducible and standardized assessment of treatment response as compared to manual 2-dimensional measurements of tumor burden using the RANO criteria.
Fig. 1.
Use of automated AI-based volumetric quantification of tumor burden to overcome the interrater variability of RANO measurements of tumor diameters towards a more standardized & reproducible assessment of treatment efficacy in neuro-oncology.
Methods
Study Design and Participants
This study was institutional review board-approved and informed consent was waived (S-784/2018). For the present study, a nonconsecutive series of n = 30 adult brain tumor patients (including n = 15 glioblastoma WHO °IV and n = 15 lower-grade gliomas, the latter encompassing n = 2 IDH-mutant astrocytoma WHO °II, n = 3 IDH-mutant astrocytoma WHO °III, n = 8 IDH-mutant 1p/19q codeleted oligodendroglioma WHO °II and n = 2 IDH-mutant 1p/19q codeleted oligodendroglioma WHO °III) previously treated at Heidelberg University Hospital. The selection of patients that were included for the present study was performed on consensus by three local investigators from Heidelberg University Hospital (P.V., W.W., and M.B.) aiming at representing different clinical scenarios from different disease stages in neuro-oncology. The MRI exams were acquired during the period of 09/2009 and 02/2019 with a standardized imaging protocol7 and included 3D T1-weighted images before (T1-w) and after contrast agent administration (cT1-w) as well as axial 2D FLAIR and T2-weighted (T2-w) images as well as diffusion-weighted MRI with apparent diffusion coefficient (ADC) maps. To increase the diversity of the dataset, longitudinal MRI scans were selected from the primary treatment setting in 12 cases (with the postradiation MRI scan used as the first imaging timepoint; except in patients that did not receive radiation therapy the postsurgery MRI scan was used as the first imaging timepoint) and the recurrent treatment setting in 18 cases (with the MRI scan prior to change of therapy as the first imaging timepoint). The last imaging timepoint was the MRI scan showing definitive tumor progression with subsequent change of treatment. Specifically, a median of 5 consecutive MRI scans (IQR, 5–10) was selected for each patient with a median of interval of 3.1 months (IQR, 2.5–3.6 months) between the scans. The interval between scans was significantly longer for lower-grade gliomas as compared to glioblastoma (p < .0001) with 3.5 months (IQR, 3.0–5.6 months) for lower-grade gliomas and 2.8 months (IQR, 1.5–3.1 months) for glioblastoma. Supplementary Table 1 contains information on patient and treatment characteristics.
The 15 participating investigators were neuroradiologists (namely P.V., R.Y.H., F.B., J.E.P., Y.W.P., S.S.A., R.J., M.S., W.B.P., M.B.) or neuro-oncologists (namely N.G., M.J.v.d.B, M.W., P.Y.W., W.W.) and the majority (11/15, 73%) are active members of the RANO working group and/or the brain tumor group (BTG) from the European Organization for Research and Treatment of Cancer (EORTC). The investigators represented 11 institutions from 5 countries (Germany: n = 4, USA: n = 4, Netherlands: n = 3, South Korea: n = 3, Switzerland: n = 1) and all of them are authors of this article. None of the investigators used AI-based decision support for assessment of treatment response in neuro-oncology prior to this study. Prior to the start of the study, all investigators reached consensus on the number and composition of patients to be included (ie, n = 30 patients including both lower-grade gliomas and glioblastomas from both primary and recurrent treatment settings). This consensus decision enabled each of the 15 participating investigators would manage to interpret all cases in a reasonable timeframe while still including patients from a broad range of clinical scenarios.
Image Interpretation
In the first assessment round of the study investigators were provided with the consecutive MRI scans (including T1-w, cT1-w, FLAIR, T2-w, DWI, and ADC sequences for each timepoint) as well as relevant clinical information (integrated diagnosis according to the WHO 2016 classification of CNS tumors, current tumor-specific treatment, and in case of recurrent tumors the number of recurrences) from each patient. The investigators received no information regarding at what timepoint when treatment was changed during the period covered by the available MRI scans.
All MRI scans were provided in DICOM format, stripped of patient information (with allocated patient identifiers being subject_01 to subject_30), and delivered to investigators. Investigators used their personal workstations with RadiAnt (Medixant, Poland) or OsiriX Lite (Pixmeo, Switzerland) as DICOM viewer. The investigators were asked to assess the timepoint of tumor progression on MRI in each patient by applying the 2D RANO concept with bi-perpendicular measurements of tumor burden as a general guide as outlined in the RANO criteria for high-grade3 and lower-grade gliomas4 and complete all reads at a typical clinical pace within a 2-month timeframe. The investigators received no feedback following submission of their readings. An illustrative case depicting the MRI sequences from consecutive timepoints during the first round of the assessment is shown in Supplementary Figure 1.
The second assessment round of the study started after a 1-month wash out period. Patient identifiers within the image and clinical data were reordered to impede the re-use of the TTP assessment from the first round. The investigators received all the information from the first assessment round (MRI scans as well as relevant clinical information) as well as additional MRI sequences for each MRI scan generated using a previously developed and validated in-house AI-based processing pipeline, including deep-learning based skull stripping (https://github.com/NeuroAI-HD/HD-BET) and deep-learning based tumor segmentation (https://github.com/NeuroAI-HD/HD-GLIO) as core components.6,8 To allow unbiased evaluation of the performance we did not perform any manual adjustments to the output of the AI-based processing pipeline (eg, editing of tumor segmentation masks). Thereby for each MRI scan three additional MRI sequences were provided: (1–2) cT1-w and FLAIR sequences with color-coded overlays that indicate the contrast-enhancing and T2/FLAIR-hyperintense tumor identified by the AI-based processing pipeline, and (3) a DICOM sequence depicting a graph with the absolute and relative change in these tumor volumes over time (plotting contrast-enhancing and T2/FLAIR-hyperintense tumor volumes from the current and all previous MRI scans). Identical to the first assessment round the investigators were asked to assess the timepoint of tumor progression on MRI in each patient by incorporating this additional information and complete all reads at a typical clinical pace within a 2-month timeframe. As a general guideline, investigators were told that a 25% increase in the bi-perpendicular diameter would correspond to a 40% increase in the tumor volume (assuming spherical configuration of the tumor),6 however final judgment (strict adherence to this volumetric threshold vs. subjective interpretation of the growth curve) was done at the discretion of the investigators. An illustrative case depicting the additional AI-enhanced MRI sequences from consecutive timepoints provided during the second round of the assessment are shown in Supplementary Figure 2. Moreover, investigators were asked to record whether the additional information from the AI-based decision support was perceived as helpful or not (specified as “1” or “0”) for each of the assessed patients. The investigators received no feedback following submission of their readings. Subsequently, a questionnaire was circulated among the investigators to collect information about the years of their experience with neuro-oncology imaging.
Statistical Analysis
All statistical analyses were performed with R version 4.1.2 (R Foundation for Statistical Computing, Vienna, Austria). The agreement as well as disagreement in the individual readings for the timepoint of tumor progression between first and second round of the assessment (overall 450 pairs ie, from 30 patients * 15 investigators) were analyzed with descriptive metrics (absolute and relative agreement); differences were assessed with a 2-sample test for equality of proportions.
The time to progression (TTP) for each reading was calculated from the date of baseline MRI until the timepoint of tumor progression specified by the readings from each of the investigators. For those cases where the investigator did not judge tumor progression until the last MRI timepoint, an interval of 3 months (equivalent to one follow-up interval) was added as a workaround to the TTP measurement prior calculating the concordance correlation coefficient (CCC) and the standard deviation (SD) of the TTP measurements. This procedure allowed that none of the readings needed to be excluded when calculating these metrics while preserving statistical validity. The agreement of the TTP measurements between the investigators (separately for the first and second round of the assessment) was evaluated for the whole patient cohort as well as for the glioblastoma and lower-grade glioma subgroups using the CCC.9 The reported 95% confidence intervals (CI) were calculated using bootstrapping (with n = 1000 iterations) with the bias-adjusted and accelerated bootstrap method. Empirical P-values were computed from the bootstrap distribution to assess differences between the CCC from the first and second round for the whole patient cohort as well as for the glioblastoma and lower-grade glioma subgroups. The SD of the TTP measurements from all investigators was computed for each patient (separately for the first and second round of the assessment) and was used as an additional metric beyond the CCC to evaluate agreement between investigators on a per-patient level. The reported 95% CI were calculated using bootstrapping (with n = 1000 iterations) with the bias-adjusted and accelerated bootstrap method. A Pearson correlation test was used to evaluate the association (1) between the percentage of investigators judging AI-based decision support as helpful for assessing the TTP in patients and the standard deviation of the TTP measurements in the second round of the assessment, as well as (2) between the percentage of patients where investigators judged AI-based decision as helpful for assessing the TTP and the experience of the investigators with neuro-oncology imaging. P-values <.05 were considered significant.
Results
The CCC of TTP measurements between investigators was 0.77 (95% CI = 0.69–0.88) in the second round of the assessment without AI-based decision support and increased to 0.91 (95% CI = 0.82–0.95) with AI-based decision support (p = .005) (Figure 2). This effect was more pronounced for patients with lower-grade gliomas, where the CCC was 0.70 (95% CI = 0.56–0.85) without AI-based decision support, as compared to 0.90 (95% CI = 0.76–0.95) with AI-based decision support (p = .008). In contrast, for patients with glioblastoma the CCC was 0.83 (95% CI = 0.75–0.92) without AI-based decision support, as compared to 0.86 (95% CI = 0.78–0.93) with AI-based decision support (p = .016). Similarly, the median SD for the TTP measurements between the investigators was 6.1 months (95% CI = 4.3–9.6 months) without AI-based decision support and decreased to 4.8 months (95% CI = 3.7–6.2 months) with AI-based decision support (p = .004) (Figure 3). Thereby a greater decrease in the SD when using additional AI-based decision support was observed for patients with lower-grade gliomas (−1.7 months [95% CI: −4.2 to −1.1 months]) as compared to glioblastoma (−0.1 months [95% CI: −0.5 to 0.0 months]) (p < .001). Illustrative cases from two representative cases which demonstrate improved agreement in the TTP among investigators when using additional AI-based decision support are shown in Figures 4 and 5 and Supplementary Figure 3.
Fig. 2.
Concordance correlation coefficients (CCC) of tumor response assessment between investigators in the first round of the study without AI-based decision support and the second round of the study with AI-based decision support. The central line of the boxplot denotes the median and the edges of the boxplot denote the first and the third quartile of the bootstrap distribution of the CCC. The lines extending from the boxes (whiskers) indicating variability outside the upper and lower quartiles. The outliers are denoted by black dots at the end of the whisker lines.
Fig. 3.
Standard deviation (SD) of tumor response assessment between investigators in the first round of the study without AI-based decision support and the second round with AI-based decision support. The difference in the SD between the first and the second round is shown in blue.
Fig. 4.
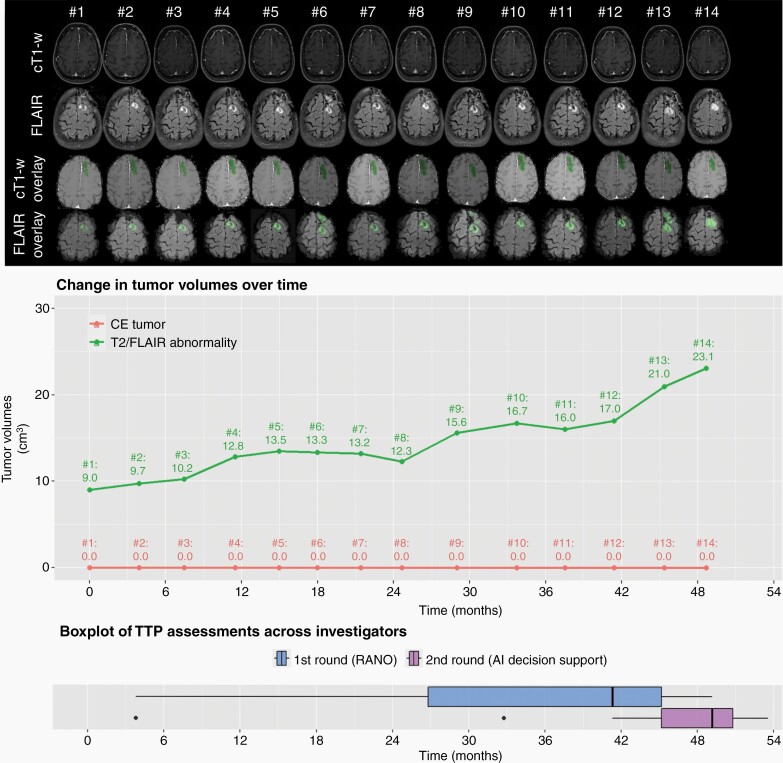
Illustrative case (patient #17, oligodendroglioma WHO°III) depicting the change in tumor burden over time on cT1-w and FLAIR sequences (1st and 2nd row). The cT1-w overlay and FLAIR overlay sequences (3rd and 4th row) as well as the corresponding tumor volume plot were provided in the second round of the assessment and visualize the contrast-enhancing tumor volumes and T2-w/FLAIR abnormality volumes which were automatically generated by the AI-based decision support for each timepoint. The last row visualizes the time to progression (TTP) measurements from the 15 investigators based on RANO alone (first round; blue colored boxplot) vs. additional AI-based decision support (second round; purple colored boxplot). The boxplots demonstrate higher agreement of the TTP measurements from the 15 investigators with additional AI-based decision support.
Fig. 5.
Illustrative case (patient #18, astrocytoma WHO°III) depicting the change in tumor burden over time on cT1-w and FLAIR sequences (1st and 2nd row). The cT1-w overlay and FLAIR overlay sequences (3rd and 4th row) as well as the corresponding tumor volume plot were provided in the second round of the assessment and visualize the contrast-enhancing tumor volumes and T2-w/FLAIR abnormality volumes which were automatically generated by the AI-based decision support for each timepoint. The last row visualizes the time to progression (TTP) measurements from the 15 investigators based on RANO alone (first round; blue colored boxplot) vs. additional AI-based decision support (second round; purple colored boxplot). The boxplots demonstrate higher agreement of the TTP measurements from the 15 investigators with additional AI-based decision support.
Comparison of all available pairs of TTP assessments from the first and second round of the assessment (450 pairs ie, 30 patients × 15 raters) showed that the assessment performed with RANO alone was kept unchanged with additional AI-based decision support in 251/450 instances (56%) and were changed for the remaining 199/450 instances (44%) (Supplementary Table 2). Thereby, the probability of changing the TTP assessment with additional AI-based decision support was higher for the subset of patients with lower-grade gliomas (114/225 [51%]) as compared to glioblastoma (85/225 assessments [38%], p = .008). The AI-based decision support did not systematically shift the judgment of tumor progression towards an earlier or later timepoint, instead the probability between shifting towards an earlier timepoint (105/450 instances [23%]) as compared towards shifting to a later timepoint (94/450 instances [21%]) with additional AI-based decision support was balanced (p = .42).
The percentage of patients where individual investigators judged AI-based decision as helpful (median, 57% [IQR, 47–63%]) was negatively correlated with the experience of the investigators with neuro-oncology imaging (median of 19 years [IQR, 12–24 years]; Pearson correlation coefficient = −0.52; p = .02) that is, investigators with less years of experience judged the AI-based decision support as more helpful (Figure 6A). Moreover, the percentage of investigators who judged the information provided through AI-based decision support as helpful for assessing the TTP in individual patients (median, 64% [IQR: 45–79%]) was negatively correlated with the SD of the TTP measurements in the second of the assessment (Pearson correlation coefficient = −0.34; p = .03) that is, the more investigators who judged the AI-based decision support to be helpful for a given patient, the better the agreement on TTP measurements for that patient (Figure 6B).
Fig. 6.
(A) Correlation between the percentage of investigators judging AI-based decision support as helpful for assessing the TTP in individual patients and the standard deviation of the corresponding TTP measurements in round 2 (RANO + AI). (B) Correlation between the percentage of patients where investigators judged AI-based decision as helpful for assessing the TTP and the corresponding experience of the investigators with neuro-oncology imaging.
A fully functional version of AI-based processing pipeline that was used in the present study (illustrative case shown in Supplementary Figure 2) is provided through https://github.com/NeuroAI-HD/HD-GLIO-XNAT as open-source and allows seamless manufacturer neutral integration into existing radiological infrastructures through the XNAT framework as a Container Service Plugin10 (Supplementary Figure 4).
Discussion
The importance and meaningful clinical use of AI algorithms for automated quantification of tumor burden in neuro-oncology is reflected in the growing body of literature showing that accurate automated delineation of the various tumor sub-compartments can offer the basis for generating quantitative and reproducible imaging endpoints in neuro-oncology.5,6,11–13 Specifically, AI algorithms for automated volumetric segmentation of tumor burden proved to be highly accurate with spatial overlap agreement between the predicted and the expert ground truth tumor annotation of more than 90% for the segmentation of contrast-enhancing tumor, as well as nonenhancing T2/FLAIR signal abnormality,5,6,11 even when applying the AI algorithm to unseen data from a multicenter phase II/III trial.6 The findings from the present study now provide additional evidence regarding the clinical value of AI-based decision support towards establishing high-quality imaging endpoints in neuro-oncology. Specifically, we demonstrate within the setting of an international multi-reader study with 15 investigators that automated AI-based volumetric quantification of tumor burden allows to improve the reproducibility and agreement of tumor response assessment measurements as compared to standard RANO criteria within a simulated clinical setting. We demonstrate that particularly lower-grade gliomas where reliable assessment of the tumor progression may be challenging due to their slow growing nature of these tumors may benefit from AI-based decision support with a potentially clinically meaningful and relevant decrease in the SD (by a median of 1.7 months) of the TTP measurements between the investigators. In contrast, when investigators used the AI-based decision support in patients with glioblastoma, there was comparatively less impact on the reproducibility of tumor response assessment. Potentially, this may reflect that tumor growth dynamics are comparatively more robust to discern when assessing tumors with a faster growth trajectory, thereby limiting the impact of AI-based decision support.
The principal benefits of AI-based decision support may be useful not only in a routine clinical scenario, but especially in the context of clinical trials, where the assessment of treatment efficacy on MRI is – besides overall survival – a key endpoint for the approval of new treatment concepts. Therefore, blinded central assessment of treatment efficacy by independent radiologists is frequently requested by regulatory authorities14 to mitigate over- or under-estimation of the true effect of treatments (ie, systematic bias) when only relying on the local RANO readings where investigators are not blinded to the patients’ treatment assignments and clinical information.15 Moreover, central RANO reading by expert radiologists is labor and time intensive and thus increases the burden of time and expenditure on clinical trials. Consequently, AI algorithms for automated volumetric delineation of tumor burden and tumor response assessment may assist investigators during central reading of the imaging data to yield high-quality imaging endpoints in neuro-oncology. As part of this study, we provide a fully functional version of the AI-based processing pipeline as open-source, enabling seamless manufacturer neutral integration into existing radiological infrastructures through the XNAT framework10 (Supplementary Figure 4) and thus may hold great promise for enhancing future research efforts in the field of neuro-oncology.
Our study also demonstrates that the information provided through the AI-based decision support is perceived as more helpful by comparatively less experienced investigators. Moreover, perceiving AI-based decision support as helpful by a greater number of investigators for determining the TTP in a specific patient, directly translated into a better agreement in the TTP measurements for this patient. Both findings taken together, highlight (1) that the confidence and validity of tumor response assessment readings could be augmented through AI-based decision support especially for less experienced investigators, and (2) that investigators were able to readily identify appropriate cases where AI-based decision support is helpful and thereby leading to a more reproducible assessment of treatment efficacy in neuro-oncology.
Our study has some limitations. First, we acknowledge the retrospective nature of the study and the selection of a nonconsecutive patient series. However, we aimed to simulate a realistic clinical scenario including patients with different tumor subtypes from both primary and recurrent treatment situations and a broad range of treatment scenarios. Although further validation in a prospective clinical scenario is needed to better establish the value of AI-based decision support and to specifically assess whether more reliable surrogate endpoints can be obtained from MRI, it may be challenging to adopt a rigorous prospective multi-reader design with/without AI-based decision support as performed in the present study.
Second, the AI-based processing pipeline applied in the present study makes use of our previously trained and validated artificial neural networks for automated skull-stripping8 and automated tumor segmentation which has been developed using >3000 MRI examinations from >1400 brain tumor patients.6 However, the potential underrepresentation of atypical or particularly challenging in these data used for training the artificial neural networks may affect the performance in a real-world clinical scenario and potentially lead to false-positive or false-negative detection of tumor burden. Consequently, this may have negatively affecting the perceived usefulness of the AI-based decision support among the investigators in the present study. Although data sharing initiatives with public deposition of annotated cases (eg, through collaborative efforts such as the Cancer Genome Imaging Archive [TCIA] or the Brain Tumor Segmentation Challenge [BraTS] for gliomas11,16) is a crucial first step to address this limitation, medical data privacy regulations often pose a significant challenge towards establishing a centralized data repository.17 Recent technical developments in the field of AI, specifically federated learning which allows multiple healthcare institutions to share their data to train an AI model while still guaranteeing medical data privacy, aim to address this challenge.17–20
Third, the differentiation of T2/FLAIR hyperintensities as well as contrast-enhancing lesions during the follow-up into treatment or tumor-related changes, may still be a challenge in the field of neuro-oncology, particularly with treatment concepts that incorporate immunotherapies or anti-angiogenic drugs.21,22 Consequently, the future incorporation of advanced MRI modalities such as diffusion or perfusion-weighted imaging23,24 or metabolic imaging with radiolabeled molecules from positron emission tomography (PET)25 will be important to overcome limitations of structural MRI and may allow to further optimize the clinical value of the AI-based decision support applied in the present study.
In conclusion, AI-based decision support has the potential to yield more reproducible and standardized assessment of treatment response in neuro-oncology as compared to manual 2-dimensional measurements of tumor burden. Particularly the evaluation of patients with lower-grade gliomas where reliable assessment of the TTP may be challenging due to their slow growing nature of these tumors may benefit from AI-based decision support. To enhance future research efforts in the field of neuro-oncology imaging, we provide a fully functional version of the AI-based processing pipeline as open-source which can readily be integrated into existing radiological (research) infrastructures.
Supplementary Material
Acknowledgments
None.
Contributor Information
Philipp Vollmuth, Department of Neuroradiology, Heidelberg University Hospital, Heidelberg, Germany.
Martha Foltyn, Department of Neuroradiology, Heidelberg University Hospital, Heidelberg, Germany.
Raymond Y Huang, Department of Radiology, Brigham and Women’s Hospital, Boston, Massachusetts, USA.
Norbert Galldiks, Department of Neurology, Faculty of Medicine, University Hospital Cologne, University of Cologne, Cologne, Germany; Institute of Neuroscience and Medicine (INM-3, -4), Research Center Juelich, Juelich, Germany; Center for Integrated Oncology (CIO), Universities of Aachen, Bonn, Cologne, and Duesseldorf, Germany.
Jens Petersen, Department of Medical Image Computing (MIC), German Cancer Research Center (DKFZ), Heidelberg, Germany.
Fabian Isensee, Department of Medical Image Computing (MIC), German Cancer Research Center (DKFZ), Heidelberg, Germany.
Martin J van den Bent, Brain Tumor Center, Erasmus MC Cancer Institute, Rotterdam, the Netherlands.
Frederik Barkhof, Department of Radiology & Nuclear Medicine, Amsterdam UMC, Vrije Universiteit, Amsterdam, the Netherlands; Institutes of Neurology & Centre for Medical Image Computing, University College London, London, UK.
Ji Eun Park, Department of Radiology and Research Institute of Radiology, Asan Medical Centre, University of Ulsan College of Medicine, Seoul, Republic of Korea.
Yae Won Park, Department of Radiology and Research Institute of Radiological Science and Center for Clinical Imaging Data Science, Yonsei University College of Medicine, Seoul, Republic of Korea.
Sung Soo Ahn, Department of Radiology and Research Institute of Radiological Science and Center for Clinical Imaging Data Science, Yonsei University College of Medicine, Seoul, Republic of Korea.
Gianluca Brugnara, Department of Neuroradiology, Heidelberg University Hospital, Heidelberg, Germany.
Hagen Meredig, Department of Neuroradiology, Heidelberg University Hospital, Heidelberg, Germany.
Rajan Jain, Department of Radiology, New York University School of Medicine, New York, New York, USA.
Marion Smits, Department of Radiology and Nuclear Medicine, Erasmus MC, Rotterdam, the Netherlands.
Whitney B Pope, Department of Radiological Sciences, David Geffen School of Medicine, University of California Los Angeles, Los Angeles, California, USA.
Klaus Maier-Hein, Department of Medical Image Computing (MIC), German Cancer Research Center (DKFZ), Heidelberg, Germany.
Michael Weller, Department of Neurology, University Hospital and University of Zurich, Zurich, Switzerland.
Patrick Y Wen, Center for Neuro-oncology, Dana-Farber Cancer Institute, Boston, Massachusetts, USA.
Wolfgang Wick, Neurology Clinic, Heidelberg University Hospital, Heidelberg, Germany; Clinical Cooperation Unit Neurooncology, German Cancer Consortium (DKTK) within the German Cancer Research Center (DKFZ), Heidelberg, Germany.
Martin Bendszus, Department of Neuroradiology, Heidelberg University Hospital, Heidelberg, Germany.
Funding
This work was supported by Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) through the SFB 1389 - UNITE Glioblastoma, Work Package C02 [Project-ID 404521405] and Priority Programme 2177 “Radiomics: Next Generation of Biomedical Imaging” [Project-ID 428223917].
Conflict of interest statement. None.
Authorship statement. Conceived and designed the research: Philipp Vollmuth, Martha Foltyn, Martin Bendszus; Performed tumor response assessment: Philipp Vollmuth, Raymond Y. Huang, Norbert Galldiks, Martin J. van den Bent, Frederik Barkhof, Ji Eun Park, Yae Won Park, Sung Soo Ahn, Rajan Jain, Marion Smits, Whitney B. Pope, Michael Weller, Patrick Y. Wen, Wolfgang Wick, Martin Bendszus; Performed XNAT implementation: Jens Petersen, Fabian Isensee, Klaus Maier-Hein, Gianluca Brugnara, Hagen Meredig, Philipp Vollmuth; Analyzed and interpreted the data: all authors; Performed statistical analysis: Philipp Vollmuth, Martha Foltyn; Handled funding and supervision: Philipp Vollmuth; Drafted the manuscript: Philipp Vollmuth, Martha Foltyn; Made critical revision of the manuscript for important intellectual content: all authors.
References
- 1. O’Connor JP, Aboagye EO, Adams JE, et al. Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol. 2017; 14(3):169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wen PY, Chang SM, Van den Bent MJ, et al. Response assessment in neuro-oncology clinical trials. J Clin Oncol. 2017; 35(21):2439–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010; 28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 4. van den Bent MJ, Wefel JS, Schiff D, et al. Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol. 2011; 12(6):583–593. [DOI] [PubMed] [Google Scholar]
- 5. Chang K, Beers AL, Bai HX, et al. Automatic assessment of glioma burden: a deep learning algorithm for fully automated volumetric and bidimensional measurement. Neuro Oncol. 2019; 21(11):1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kickingereder P, Isensee F, Tursunova I, et al. Automated quantitative tumour response assessment of MRI in neuro-oncology with artificial neural networks: a multicentre, retrospective study. Lancet Oncol. 2019; 20(5):728–740. [DOI] [PubMed] [Google Scholar]
- 7. Ellingson BM, Bendszus M, Boxerman J, et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro Oncol. 2015; 17(9):1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Isensee F, Schell M, Pflueger I, et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum Brain Mapp. 2019; 40(17):4952–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin LA. Concordance correlation coefficient to evaluate reproducibility. Biometrics 1989; 45(1):255–268. [PubMed] [Google Scholar]
- 10. Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007; 5(1):11–34. [DOI] [PubMed] [Google Scholar]
- 11. Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the Brats challenge. arXiv preprint arXiv:1811.02629. 2018. [Google Scholar]
- 12. Peng J, Kim DD, Patel JB, et al. Deep learning-based automatic tumor burden assessment of pediatric high-grade gliomas, medulloblastomas, and other leptomeningeal seeding tumors. Neuro Oncol. 2022; 24(2):289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Baid U, Ghodasara S, Mohan S, et al. The rsna-asnr-miccai brats 2021 benchmark on brain tumor segmentation and radiogenomic classification. arXiv preprint arXiv:2107.02314. 2021. [Google Scholar]
- 14. Food U, Administration D. Guidance for industry: clinical trial endpoints for the approval of cancer drugs and biologics. Fed Regist. 2007; 72:94. [Google Scholar]
- 15. Ford R, Schwartz L, Dancey J, et al. Lessons learned from independent central review. Eur J Cancer. 2009; 45(2):268–274. [DOI] [PubMed] [Google Scholar]
- 16. Bakas S, Akbari H, Sotiras A, et al. Advancing the cancer genome atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017; 4(1):170117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zerka F, Barakat S, Walsh S, et al. Systematic review of privacy-preserving distributed machine learning from federated databases in health care. JCO Clin Cancer Inform. 2020; 4:184–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sheller MJ, Reina GA, Edwards B, Martin J, Bakas S.Multi-institutional deep learning modeling without sharing patient data: a feasibility study on brain tumor segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T, eds. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes 2018. Lecture Notes in Computer Science, vol 11383. Cham: Springer; 2019. 10.1007/978-3-030-11723-8_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rieke N, Hancox J, Li W, et al. The future of digital health with federated learning. arXiv preprint arXiv:2003.08119.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pati S, Baid U, Edwards B, et al. Federated learning enables big data for rare cancer boundary detection. arXiv preprint arXiv:2204.10836.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okada H, Weller M, Huang R, et al. Immunotherapy response assessment in neuro-oncology: a report of the RANO working group. Lancet Oncol. 2015; 16(15):e534–e542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ellingson BM, Wen PY, Cloughesy TF. Modified criteria for radiographic response assessment in glioblastoma clinical trials. Neurotherapeutics. 2017; 14(2):307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kickingereder P, Park JE, Boxerman JL. Advanced physiologic imaging: perfusion – theory and applications. In: Pope WB, ed. Glioma Imaging: Physiologic, Metabolic, and Molecular Approaches. Cham: Springer International Publishing; 2020:61–91. [Google Scholar]
- 24. LaViolette PS. Advanced physiologic imaging: diffusion – theory and applications. In: Pope WB, ed. Glioma Imaging: Physiologic, Metabolic, and Molecular Approaches. Cham: Springer International Publishing; 2020:93–108. [Google Scholar]
- 25. Galldiks N, Lohmann P, Albert NL, Tonn JC, Langen K-J. Current status of PET imaging in neuro-oncology. Neurooncol Adv. 2019; 1(1):vdz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.