Abstract
Background
Improved treatment of glioblastoma (GBM) needs to address tumor invasion, a hallmark of the disease that remains poorly understood. In this study, we profiled GBM invasion through integrative analysis of histological and single-cell RNA sequencing (scRNA-seq) data from 10 patients.
Methods
Human histology samples, patient-derived xenograft mouse histology samples, and scRNA-seq data were collected from 10 GBM patients. Tumor invasion was characterized and quantified at the phenotypic level using hematoxylin and eosin and Ki-67 histology stains. Crystallin alpha B (CRYAB) and CD44 were identified as regulators of tumor invasion from scRNA-seq transcriptomic data and validated in vitro, in vivo, and in a mouse GBM resection model.
Results
At the cellular level, we found that invasive GBM are less dense and proliferative than their non-invasive counterparts. At the molecular level, we identified unique transcriptomic features that significantly contribute to GBM invasion. Specifically, we found that CRYAB significantly contributes to postoperative recurrence and is highly co-expressed with CD44 in invasive GBM samples.
Conclusions
Collectively, our analysis identifies differentially expressed features between invasive and nodular GBM, and describes a novel relationship between CRYAB and CD44 that contributes to tumor invasiveness, establishing a cellular and molecular landscape of GBM invasion.
Keywords: CRYAB, glioblastoma, invasion, postoperative recurrence, single-cell RNA sequencing
Key Points.
Invasive GBM are less dense and proliferative than noninvasive counterparts.
GBM invasion is associated with unique transcriptomic features.
CRYAB drives GBM invasion and contributes to postoperative recurrence.
Importance of the Study.
Glioblastoma (GBM) is the most common primary brain cancer and is highly invasive, which greatly complicates the development of effective treatments. In this study, we profiled GBM invasion through integrative analysis of histological and single-cell RNA sequencing data from 10 patients. At the cellular level, we found that invasive GBM are less dense and proliferative than their noninvasive counterparts. At the molecular level, we described unique transcriptomic features that significantly contribute to GBM invasion. Specifically, we identified a novel relationship between crystallin alpha B and CD44 that drives GBM invasion and contributes to postoperative recurrence. Taken together, this work establishes a cellular and molecular landscape of GBM invasion that can guide the development of more effective therapies.
Glioblastoma (GBM) is the most common and aggressive primary brain cancer.1,2 Current treatment options at diagnosis are multimodal and typically include surgical resection, radiation, and chemotherapy. Despite extensive efforts, GBM patients have poor prognosis with a median survival rate of less than 15 months and a 5-year survival rate of 2%.1,2
Two major characteristics of GBM have significantly hindered the development of effective therapies. First, GBM is highly invasive by nature.3 As a result, complete surgical resection of the tumor is almost impossible, and tumor recurrence is inevitable.4 Second, GBM, and specifically cancer stem cells (CSCs) within the tumor, are remarkably heterogeneous, which complicates the development of therapeutics strategies since the composition of the tumor greatly affects its response to targeted therapies.5 CSCs are known to recapitulate the heterogeneity of the parental tumor and drive tumor invasion.6,7 Several markers, such as CD1338 and L1CAM,9 are highly expressed in CSCs; however, no markers have been identified to exclusively mark CSCs or differentiate them by their functional role in the tumor (ie, invasion). Therefore, improved treatment of GBM needs to address the biology of GBM invasion and the heterogeneity of CSCs, specifically how they contribute to tumor invasion.
In this study, we investigated the biology of GBM invasion through multi-platform profiling. We first collected and analyzed human histology and patient-derived xenograft (PDX) mouse histology samples from 10 GBM patients and classified them into 2 cohorts, invasive and nodular, based on their histological features. Next, we performed single-cell RNA sequencing (scRNA-seq) analysis of those samples and validated their classification at the transcriptional level. Further analysis showed that GBM invasion is negatively associated with tumor density and proliferation, and the CD44-expressing subpopulation within CSCs, but not others, contributes significantly to tumor invasion. Lastly, we determined crystallin alpha B (CRYAB) contributes to GBM invasion and postoperative recurrence, and is highly co-expressed with CD44, suggesting that the interaction between CRYAB and CD44 may be a promising molecular target for GBM invasion intervention.
Materials and Methods
Patient-derived GBM samples and primary cell culture
Patient-derived GBM samples were obtained as previously described.10,11 All GBM samples were provided by Yale Neuropathology Service. Fresh patient-derived GBM cells were isolated and cultured from GBM patient surgical specimens with approval from the Institutional Review Board at Yale-New Haven Hospital. Extensively rinsed tumor specimens were finely minced and placed in DMEM/F-12 medium (Gibco, #11330032) with 25 unit/ml Papain (Worthington Biochemical Corp, #LS003124). A series of mechanical dissociations was used to obtain a single-cell suspension. Resuspended cells were cultured in complete Neurobasal medium, which is serum-free Neurobasal medium (Gibco, #21103049) supplemented with B27 (0.5X, Gibco, #17504044), fibroblast growth factor (20 ng/ml, Peprotech), and epidermal growth factor (20 ng/ml, Peprotech). Brain tumor-derived neurospheres were evident as early as 1 week after plating. Cells were maintained in complete neurobasal medium and passaged every 3–4 days.
PDX GBM Mouse Model
This study was performed under a protocol approved by the Institutional Animal Care and Use Committee of Yale University. Female athymic nude mice (BALB/c nu/nu, 6 weeks old) were purchased from Charles River Laboratories and maintained at the Yale Animal Resource Center. To establish intracranial patient-derived GBM mouse xenografts models, mice were anesthetized via intraperitoneal injection of ketamine and xylazine. Forty-thousand cells in 5 μl of phosphate-buffered saline (PBS) were injected into the right striatum 2 mm lateral and 0.5 mm posterior to the bregma and 3 mm below the dura using a stereotactic apparatus with an UltraMicroPump (World Precision Instruments, #UMP3).
Histology Staining
Brains were harvested and fixed in 4% formaldehyde for 48 h, stored at 4°C in 70% ethanol, and sectioned at a thickness of 20 µm. Hematoxylin and eosin (H&E), Ki-67, and SOX2 stainings were performed on sections for histological analysis. Slides were scanned using an Aperio whole slide scanner (Leica) at 40× resolution.
Histopathology Quantification
A machine learning-based approach was used to quantitatively analyze histology samples as previously described.12 We used this algorithm to quantify H&E, Ki-67, and SOX2 stainings. Our method consisted of three main steps: (1) selecting 5 representative windows from 2 to 3 independent sections of each tumor xenograft sample, (2) training an algorithm to identify tumor and normal nuclei, and (3) segmenting the input images to quantify the area occupied by tumor nuclei and apply a color mask to visualize nuclei for each sample. Tumor boundary was defined as less than 90% of cell nuclei compared to that of the tumor core. Tumor invasion (%) was calculated as % cells outside tumor boundary/% cells within tumor boundary × 100. Using this approach, this algorithm was able to identify a tumor as invasive or nodular with high confidence (P < .01).
scRNA-seq Collection and Analysis
The scRNA-seq data was collected as previously described.10 Briefly, patient-derived GBM cells were collected from 10 GBM patients at the Yale-New Haven Hospital after approval from the Yale University Institutional Review Board. Fresh tumor specimens were minced by a series of mechanical dissociations until a single-cell suspension was obtained. Single cells from patient-derived GBM neurospheres were sequenced by using 75 bp pair-end reads on a HiSeq2500 instrument (Illumina) in HighOutput Mode V4. Raw reads were preprocessed for cell barcodes and unique molecular identifiers (UMIs), and then aligned to the human genome(hg19) using STAR v2.5.2b as described in Dropseq method.13 A digital expression matrix was generated for the cells with over 10 000 reads per cell. In total, sequencing data from 26 018 single cells from 10 GBM patients were collected.
The Seurat package (V3.1.5) in R (V3.6.2) was used to analyze the digital expression matrices generated from the GBM samples.14 Cells that met quality control conditions (unique feature counts between 200 and 2 500 features and <5% mitochondrial counts) were included for downstream analysis. Seven thousand seven hundred fifty-seven cells encoding 25 729 genes passed quality control. Dimensionality reduction using principal component analysis was performed with 12 statistically significant principal components using the top 1000 highly variable genes from all samples. Principal components were chosen heuristically based on percentage of variance determined from ElbowPlot(). We then projected single cells onto a two-dimensional map using uniform manifold approximation and projection (UMAP). Genes that were differentially expressed in each cluster were identified using a Wilcoxon Rank Sum test from FindMarkers(), which returns the name, average log fold-change, and adjusted P for significant genes in each cluster.
Genes that had been previously reported to positively regulate tumor invasion were used for the invasion heatmap. PDPN,15 CHL1,16 TSPAN7,17 CD44,18 RAP1A,7 LIN7A,7 PROS1,19 ANGPT1,17 SNAI1,18 TAZ,20 SH3KBP1,7 ECM2,17 MYL12A,7 STAT3,21 CSF1,22 CTGF,23 NOTCH1,24 and ACKR325 were the included genes. Figure 1F shows the mean of the normalized expression from these invasion-associated genes.
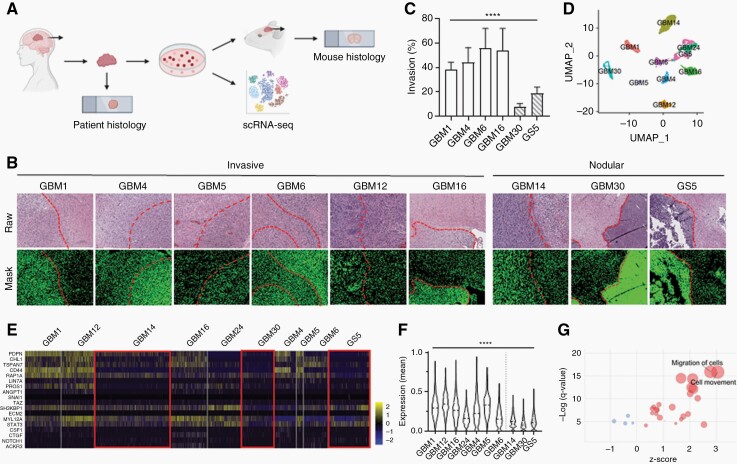
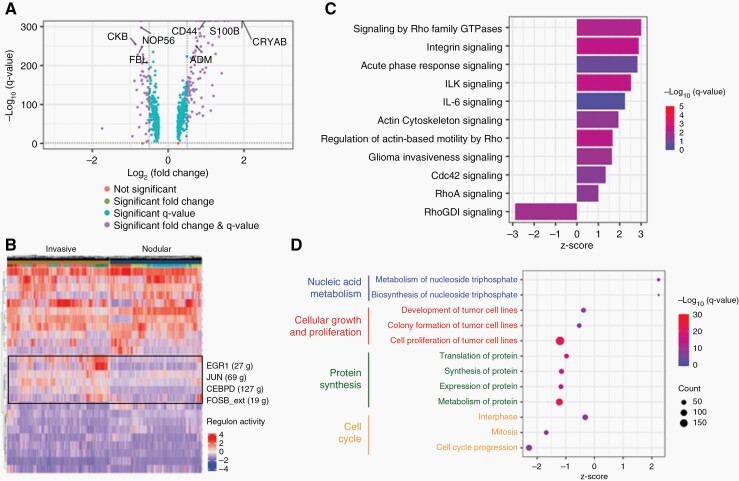
Fig. 1.
Histopathological and scRNA-seq analysis identify two cohorts of GBM samples with distinct phenotypes, invasive and nodular.
(A) Schematic of data collection from human GBM histology samples, PDX mouse GBM histology samples, and scRNA-seq data. (B) Human and PDX mouse GBM H&E samples showing differences in tumor boundaries between invasive and nodular samples. Dotted line: tumor boundary. (C) Quantification of (B) for PDX mouse GBM samples. Migration (%) = positive pixels outside tumor boundary/ positive pixels within tumor boundary. (D) UMAP of scRNA-seq data clustered by most differentially expressed genes. (E) Heatmap of genes related to tumor invasion. Red boxes highlight that nodular GBM samples have lower expression of invasion-related genes. (F) Quantification of (E). y-axis shows the mean of the normalized expression of invasion-related genes in each sample. White: invasive samples. Striped: nodular samples. (G) Dot plot showing that pathways related to cell migration and movement are significantly upregulated in invasive samples. Red: positive z-score. Blue: negative z-score. Size of circle correlates with number of significant genes in pathway.
Ingenuity Pathway Analysis
Differentially expressed genes and their average log fold-change between nodular and invasive GBM samples from the FindMarkers function in Seurat were analyzed using Ingenuity Pathway Analysis (Qiagen).26 An expression log ratio between −1.9624 and 1.7441 was used for the analysis, which included 759 analysis-ready molecules. Four hundred twenty-two molecules had a negative expression log ratio and 337 molecules had a positive expression log ratio. Plots showing significantly different pathways, diseases, and cell functions were made using the ggplot2 package (V3.3.1) in R (V3.6.2).
Regulon Analysis
Gene regulatory networks were identified at the single cell level using SCENIC (v1.2.2).27 First, co-expression patterns were screened between transcription factors and their downstream target genes, and then the top regulon activities were computed in every single cell. This workflow is robust with both raw expression data and normalized data. Significant differences in the key regulons were not observed when input data were normalized. Heatmaps showing regulon activities were plotted using ComplexHeatmap (v2.5.6).28
Lentiviral Production and Transduction of Patient-Derived GBM Cells
Construct overexpressing CRYAB (Dharmacon, #OHS5897-202616161) was purchased from Dharmacon. pLOC CMV-IRES-Dsred-blasticidin was a gift from Oskar Laur (Addgene plasmid # 129428; RRID: Addgene_129428). Lentivirus was produced by co-transfecting pMD2.G (Addgene, #12259), psPAX2 (Addgene, #12260), and the overexpression plasmid into HEK293T cells by Lipofectamine 2K (Invitrogen, #11668019) according to the manufacturer’s instructions. Lipofectamine 2K was removed after 6 h and replaced with serum-free high glucose DMEM (Gibco, #11965092) and 20% fetal bovine serum (FBS) (Gibco, #10082147). Virus was collected 48 h later using Lenti-X Concentrator (Takara Bio, #631231) according to the manufacturer’s instructions. Lentiviral particles were resuspended in serum-free DMEM and stored at −80°C until transduction. For lentiviral transduction, GBM cells were digested in Accutase (BioLegend, #423201) for 2 min at 37°C until in single cell suspension. Dextran (2 ug/ul) and lentivirus were added to the digested GBM cells. Cells were cultured overnight before replacing with the fresh complete neurobasal medium.
Western Blot
Cells were collected and lysed in RIPA buffer (Thermo Scientific, #89901) with protease inhibitors and incubated on ice for 10 min. Lysates were centrifuged at 4°C for 30 min at 14 000 rpm, and the supernatant was collected. The Pierce BCA Protein Assay kit (Thermo Scientific, #23225) was used to determine protein concentration. Equal amounts of protein samples were mixed with SDS Laemmli loading buffer, boiled for 5 min at 95°C, electrophoresed using 4%–20% Mini-PROTEAN TGX Stain-Free Protein Gels (Bio-Rad, #4568093), and transferred onto PVDF membranes. Non-fat dry milk (5% in tris-buffered saline-Tween [TBST]) was used to block the membrane for 1 h. The membrane was cut and incubated with primary antibodies for at least 16 h at 4°C. The membrane was then washed with TBST and incubated with the appropriate secondary antibody at room temperature for 1 h. The signal was developed using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific, #34580). Primary antibodies used were CRYAB (Novus, #NB100-2519) and β-actin (Cell Signaling, #4970S). Secondary antibodies used were Digital anti-Mouse-HRP (Kindle Biosciences, #R1005) and Digital anti-Rabbit-HRP (Kindle Biosciences, #R1006).
Proliferation
Cells were plated in a 48-well plate at a concentration of 5 × 104 cells/ml in 200 μl. Cell concentration was determined at the following time points: 3, 6, 9, 12, and 15 days after seeding.
Mouse In Vivo Studies
Female athymic NCr-nu/nu mice (5–6 weeks old, Charles River Laboratories) were given free access to food and water before all experiments. All animal experiments were approved by the Yale University Institutional Animal Care and Utilization Committee. Tumor-bearing mice were established through intracranial injection of 5 × 104 cells. Mice were sacrificed 5 weeks after tumor inoculation, and their brains were collected. Brains were prepared for histological analysis as described above.
Mouse Model of GBM Resection
Luciferase-expressing tumor cells were inoculated as described above. When the tumors reached a luciferase threshold of 1 × 106 relative light unit (RLU) after 1 min exposure, surgical resection of the tumor was performed. Using a high-power microscope, a micro-dissection was made to remove as much as the tumor as safely possible, while minimizing damage to surrounding normal brain tissue. The extent of resection was determined by the tumor depth. After tumor removal, the surgery area was thoroughly cleaned and washed using PBS containing penicillin and streptomycin. The bone flap was placed back onto the exposed brain, and the skin was sewn with sutures. Mice were monitored until awake and alert, and tumors were imaged using in vivo imaging system (IVIS) before, after, and 10 days after surgery.
Figure Schematics
Schematics (Figures 1A and 6C) were created with BioRender.com.
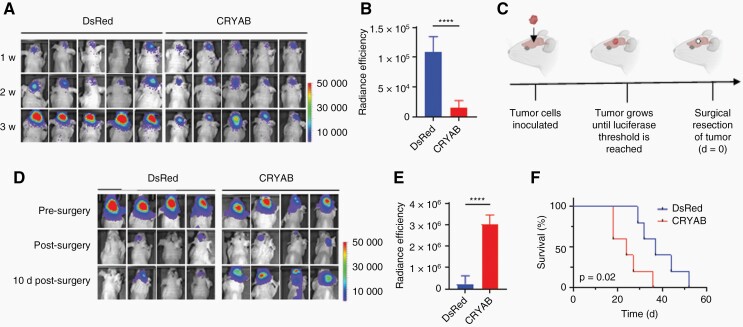
Fig. 6.
Overexpression of CRYAB increases tumor recurrence after surgical resection.
(A) Representative IVIS images 1 week, 2 weeks, and 3 weeks after inoculation of the indicated tumor cells without surgical intervention. (B) Quantification of (A) showing that GBM30-DsRed tumors are larger than GBM30-CRYAB tumors 3 weeks after tumor inoculation. n = 5. (C) Schematic of mouse GBM resection model. (D) Representative IVIS images before, after, and 10 days-after surgical resection of tumor in mice inoculated with GBM30-DsRed or GBM30-CRYAB cells. (E) Quantification of (D) showing that GBM30-CRYAB tumors have greater recurrence than that of GBM30-DsRed tumors 10 days post-surgery. n = 4. (F) Survival of mice inoculated with GBM30-DsRed or GBM-CRYAB cells after surgical resection of the tumor. n = 5.
Statistical analysis
All histopathological quantification data were collected as n = 5. All other data were collected in triplicate unless otherwise specified and reported as mean and standard deviation. Statistical analyses for differential gene expression were performed with the default tests and parameters unless otherwise specified. Comparison between two groups was performed using an unpaired, two-tailed Student t-test. One-way ANOVA was used to analyze multiple comparisons by GraphPad Prism 9.1.1 P < .05 (*), .01 (**), .001 (***), and .0001 (****) were considered significant.
scRNA-Seq Data Availability
The scRNA-seq data used in this study are available in the Gene Expression Omnibus under accession GSE125587.
Results
Histopathological and scRNA-Seq Analysis Identify Two Cohorts of GBM Samples With Distinct Phenotypes, Invasive and Nodular
To generate a multi-platform library of GBM data, human histology samples, PDX mouse histology samples, and scRNA-seq data were collected from 10 GBM patients (Figure 1A, Supplementary Table 1). PDX mouse models were generated using patient-derived CSC cultures, which are known to recapitulate histopathology of human GBM.29 All patient samples were classified as isocitrate dehydrogenase (IDH)-wild type; O6-methylguanine-DNA methyltransferase promoter methylation status, epidermal growth factor receptor (amplification and mouse tumorgenicity were determined (Supplementary Table 2). To investigate differences in tumor invasion among GBM samples, GBM tissue was stained with H&E (Supplementary Figure 1A). The distribution of cell nuclei around the tumor boundary was visualized by applying a mask over raw histopathology images that detects, differentiates, and quantifies tumor nuclei based on pixel color and intensity (Supplementary Figure 1B), as described in our previous study.12 Interestingly, while most GBM samples (GBM1, GBM4, GBM5, GBM6, GBM12, and GBM16) had diffuse tumor boundaries, other samples (GBM14, GBM30, and GS5) were nodular with clearly defined tumor edges (Figure 1B). To quantify tumor invasion, cell nuclei percentage within and outside of the tumor boundary was determined. GBM14 was not included because of a missing PDX sample. Compared to other samples, GBM30 and GS5 had significantly lower tumor invasion (Figure 1C). This finding was further verified in vitro (Supplementary Figure 1C).
Single-cell technologies have recently emerged as a useful tool for elucidating the heterogeneity of GBM at both the cellular and transcription levels.10,30 To validate our histopathological findings, we analyzed scRNA-seq data of all 10 GBM samples (Supplementary Table 3). To minimize batch-to-batch variability, we prepared 2 to 3 batches of cDNA library for each patient sample for a total of 26 batches. Each batch contained between 393 and 1695 cells, 1740–3626 median genes per cell, 4408–10 010 median UMIs, and 17 254–23 500 total genes. After quality control filtering, 21 750 cells with expression levels for 24 120 genes were used for downstream analysis. To create a genetic profile of GBM invasion, we visualized all cells with UMAP and grouped them using unbiased, graph-based clustering (Figure 1D, Supplementary Figure 1D). To investigate whether GBM14, GBM30, and GS5 are less invasive on a transcriptomic level, we determined the normalized expression levels of genes related to invasion for each sample (Figure 1E, Supplementary Figure 1E). Consistent with our histopathological findings, GBM14, GBM30, and GS5, had the lowest average invasion-related gene expression (Figure 1F). As expected, tumor invasion quantified from histopathological and scRNA-seq analysis were positively correlated (Supplementary Figure 1F). Next, we performed pathway analysis on differentially expressed genes between invasive (GBM1, GBM4, GBM5, GBM6, GBM12, GBM16) and nodular (GBM14, GBM30, and GS5) samples to determine significantly different pathways. Out of 28 pathways related to cell movement and migration, 24 of them were significantly upregulated in invasive GBM samples, with general migration of cells and cell movement being the most significant (Figure 1G). Cell movement-related pathways that were downregulated in invasive GBM samples were related to cell movement in ovarian cancer cell lines, prostate cancer cell lines, and melanoma cell lines, suggesting that different genes regulate different types of cancer cell invasion, which is consistent with previous studies.31,32 To exclude the possibility that histopathological analysis was segmenting by molecular subtype and not invasiveness, we performed single-sample Gene Set Enrichment Analysis33 on all GBM samples (Supplementary Figure 1G). Nodular samples were classified as classical (GS5), mesenchymal (GBM30), and proneural (GBM14), while invasive samples were classified as classical (GBM5), mesenchymal (GBM1, GBM4, GBM6), and proneural (GBM12, GBM16, GBM24) (Supplementary Figure 1H). Therefore, scRNA-seq, in conjunction with histopathological analysis, confirmed that GBM14, GBM30, and GS5 are significantly less invasive than other GBM samples both phenotypically and transcriptomically. For the remainder of the study, we refer to GBM1, GBM4, GBM5, GBM6, GBM12, GBM16, and GBM24 as invasive, and GBM14, GBM30, and GS5 as nodular.
Invasive GBM Are Less Dense and Less Proliferative Than That of Nodular GBM
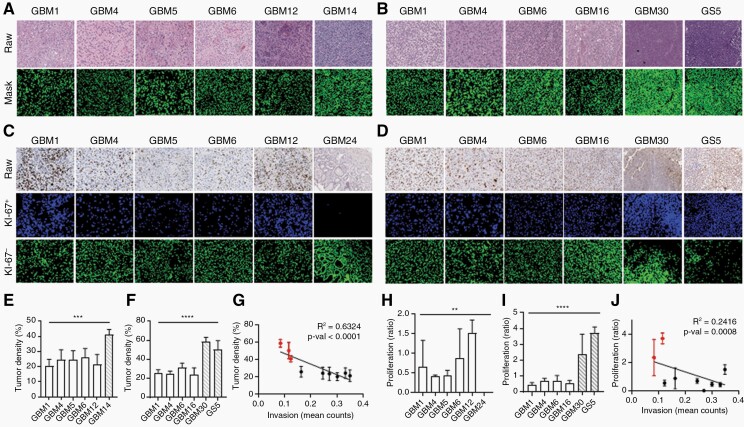
To characterize differences between invasive and nodular GBM, tumor density for all samples was determined by quantifying cell nuclei percentage in areas with the densest cellularity identified from H&E staining. Among human H&E samples, GBM1, GBM4, GBM5, GBM6, and GBM12 had similar tumor densities, while GBM14 (a nodular sample) was significantly denser (Figure 2A and E). Similarly, for PDX mouse H&E samples, GBM30 and GS5 (both nodular samples) were significantly denser than that of GBM1, GBM4, GBM6, and GBM16 (Figure 2B and F). GBM1, GBM4, and GBM6 had comparable levels of tumor density in both human and mouse H&E samples (Supplementary Figure 2A), confirming the histological fidelity of PDX models for human GBM. Next, linear regression analysis was performed, and tumor invasion was found to be significantly negatively correlated with tumor density, with nodular samples (GBM14, GBM30, and GS5) having the highest density and lowest invasion (Figure 2G).
Fig. 2.
Invasive GBM has low cell density and low proliferative rate.
(A, B) H&E staining of (A) human and (B) PDX mouse GBM tissue. (Mask) Cell nuclei are shown in green. Data is reported as percent of positive pixels within a specified window. (E) Quantification of (A). (F) Quantification of (B). (G) Correlation between tumor invasion and density. (C, D) Ki-67 staining of (C) human and (D) PDX mouse GBM tissue. Data is reported as a ratio of Ki-67+ cells34 to Ki-67− cells within a specified window. (H) Quantification of (C). (I) Quantification of (D). (J) Correlation between tumor invasion and proliferation. (E, F and H, I) White: invasive samples. Striped: nodular samples. n = 5. (G, J) Red: nodular samples. Black: invasive samples.
To determine how cell proliferation is related to tumor invasion, GBM tissue was stained for Ki-67, a cell proliferation marker (Supplementary Figure 2B and C). Cell proliferation was determined by overlaying a mask onto raw histopathology images stained for Ki-67 and detecting cell nuclei stained brown and blue to identify Ki-67+ and Ki-67− cells, respectively. Cell proliferation was reported as the ratio of Ki-67+ to Ki-67− cells in areas with the highest Ki-67+ signal. All human samples had similar levels of cell proliferation ranging from a ratio of 0 to 1.5 (Figure 2C and H). Among PDX mouse samples, GBM1, GBM4, GBM6, and GBM16 had similar levels of proliferation to that of the human samples; however, GBM30 and GS5 (both nodular samples) had significantly higher cell proliferation (Figure 2D and I). Similar to that of tumor density, linear regression analysis determined that cell proliferation was significantly negatively correlated with tumor invasion, with nodular samples (GBM30 and GS5) having the highest proliferation and lowest invasion (Figure 2J).
A Subset of CSCs Contributes Significantly to GBM Invasion
CSCs greatly contribute to GBM malignancy due to their ability to self-renew, proliferate, and invade surrounding brain tissue.6 Although PROM1 (CD133) was the first reported for CSC identification in GBM,8 many other CSC-related markers have since been reported.9,35–37 Due to their heterogeneity and plasticity, subpopulations of CSCs are known to have different phenotypes within the tumor microenvironment.38
We sought to determine whether a subset of CSCs contributes more to tumor invasion in GBM. We stained PDX mouse GBM samples for SOX2, a canonical CSC marker in GBM,37 and quantified SOX2 expression as a ratio of SOX2+ cells to SOX2− cells (Figure 3A). GBM1, GBM4, GBM6, and GBM16 (invasive samples) had higher SOX2 expression, while GBM30 (nodular) had significantly less SOX2 expression. SOX2 expression quantified using histopathological analysis was consistent with that of scRNA-seq data (Figure 3B). However, GBM14 and GS5 had comparable SOX2 expression to invasive samples even though histopathology analysis identifies them as nodular, suggesting that SOX2 expression alone is not enough to determine the invasiveness of GBM samples.
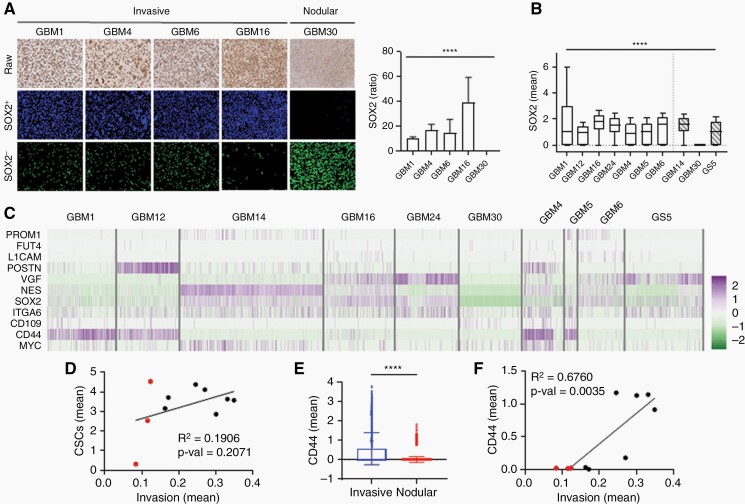
Fig. 3.
CD44 is more highly expressed in invasive GBM than other CSC-related genes.
(A) SOX2 staining of PDX mouse GBM tissue. Data is reported as a ratio of SOX2+ cells34 to SOX2− cells within a specified window. (B) Box plot of normalized SOX2 expression from scRNA-seq data. Nodular GBM samples have a wide range of SOX2 expression. (C) Heatmap of normalized counts for canonical CSC genes in GBM samples. (D) Correlation between tumor invasion and known CSC-related genes. Red: nodular samples. Black: invasive samples. (E) Bar graph of normalized CD44 expression in invasive and nodular GBM. Invasive GBM have higher CD44 expression. (F) Correlation between invasion and CD44 expression in GBM samples. Red: nodular samples. Black: invasive samples.
To investigate how other CSC genes contribute to GBM invasion, we analyzed all 10 GBM samples and determined the normalized expression levels of canonical CSC-related genes38 (Figure 3C). Interestingly, most CSC markers were widely expressed across GBM samples, regardless of histopathological invasion (Supplementary Figure 3A). Next, we calculated the stemness score, which was defined as the sum of all CSC-related gene expressions, for each sample. Both invasive and nodular samples had similar stemness scores (Supplementary Figure 3B). Notably, GBM14 had the highest stemness score, even though its histopathological phenotype suggests it is nodular (Supplementary Figure 3C). Indeed, there was a non-significant correlation between tumor invasion and total CSC expression (Figure 3D), suggesting that only a subset of CSCs contribute to GBM invasion.
To determine if a subset of CSCs contributes more strongly to tumor invasion, we looked at individual CSC gene expression to identify any differences between invasive and nodular GBM samples. CSC genes PROM1, FUT4, L1CAM, POSTN, ITGA6, CD109, and MYC had low expression in both invasive and nodular groups, while NES and SOX2 had similar expression regardless of invasive profile (Supplementary Figure 3D). VGF, a neuropeptide, had high expression in invasive samples, but also high expression in GS5, a nodular sample. Interestingly, CD44, a transmembrane glycoprotein known to be related to tumor progression,39 was highly expressed in invasive but not nodular samples (Figure 3E). Next, linear regression analysis was performed to determine the correlation between individual CSC gene expression and tumor invasion. While the correlation for most CSC genes was non-significant (Supplementary Figure 3E), CD44 expression was significantly correlated with tumor invasion. These results suggest that CD44 contributes more to tumor invasion than other CSC-related genes (Figure 3F).
Patient-Derived scRNA-Seq Data Reveals Transcriptional Dynamics of GBM Invasion
To determine molecular characteristics of GBM invasion, we identified differentially expressed genes between invasive (GBM1, GBM4, GBM5, GBM6, GBM12, GBM16, GBM24) and nodular (GBM14, GBM30, GS5) GBM (Supplementary Table 4). Four hundred thirty-seven genes were identified to be upregulated in invasive GBM, and of these, 111 genes had a significant fold change (log2FC > 0.5) (Figure 4A). Many of the genes upregulated in invasive GBM, such as CRYAB, S100 calcium-binding protein B (S100B), and adrenomedullin (ADM), have previously been reported to contribute to GBM invasion.40–42 In particular, prior studies have found that CRYAB is related to apoptosis in gliomas,43 and promotes invasion in gastric44 and colorectal45 cancers. Consistent with our previous observations that a subset of CSCs contributes to GBM invasion, CD44 was also significantly upregulated in invasive GBM. Many of the genes downregulated in invasive GBM were related to ribosomal RNA processing, such as NOP56 and FBL, and cellular metabolism, such as CKB, suggesting that invasive GBM cells rely on alternative protein and energy production mechanisms to invade surrounding brain tissue.
Fig. 4.
Cellular and molecular profile of invasion in GBM.
(A) Volcano plot showing differentially expressed genes between invasive and nodular GBM samples. Positive fold-change means gene is upregulated in invasive GBM. log2FC > 0.5 was considered significant. Q-value < 0.05 was considered significant. (B) Differentially expressed regulons between invasive and nodular GBM samples. Positive fold-change means regulon is upregulated in invasive GBM. (C) Canonical pathways significantly upregulated in invasive GBM samples. (D) Cellular functions significantly upregulated in invasive GBM samples.
Next, we examined differentially expressed regulons between invasive and nodular GBM to determine whether specific transcription factors contribute to GBM invasion (Figure 4B and Supplementary Figure 4A). Regulons related to Jun proto-oncogene (JUN) and FBJ murine osteosarcoma viral oncogene homolog B (FOSB), both components of the activator protein 1 (AP-1) transcription factor complex,46 were highly expressed in invasive GBM. AP-1 has previously been reported to contribute to GBM invasion through interleukin-8 upregulation,47 and is known to increase GBM resistance to radiotherapy and chemotherapy.48 In addition, early growth response protein 1 (EGR1) and CCAAT/enhancer-binding protein delta (CEBPD) were also upregulated in invasive GBM. EGR1 has been reported to contribute to GBM stemness and progression,49 while CEBPD has been reported to contribute to stemness of CSCs in GBM after inflammatory stimulation.50 Interestingly, expression of both EGR1 and CEBPD are significantly positively correlated with CD44 (Supplementary Figure 4B), suggesting that the CD44 subset of CSCs may regulate GBM invasion through regulation of these transcription factors.
We performed pathway analysis to dissect differentially expressed pathways between invasive and nodular GBM (Supplementary Table 5). As expected, pathways related to glioma invasiveness, actin cytoskeleton, and Rho signaling (RhoA, Cdc42) were upregulated in invasive GBM (Figure 4C). The Rho family of GTPases is known to contribute to GBM invasion through rearrangement of actin cytoskeleton, cell adhesion, and invasion.7 Rho GDP-dissociation inhibitor (RhoGDI), an inhibitor of Rho GTPases, was significantly downregulated in invasive GBM, further supporting GBM invasion is mediated through Rho GTPase-related signaling. Also consistent with previous findings,39 integrin-related signaling (integrin signaling, integrin-linked kinase signaling) was found to be significantly upregulated in invasive GBM.
Lastly, we linked significant pathways to cellular functions that are upregulated in invasive GBM (Supplementary Table 6). Functions related to nucleic acid metabolism were upregulated in invasive GBM (Figure 4D). Conversely, functions related to cell proliferation, protein synthesis, and cell cycle progression were downregulated, which further supports our histopathological observations.
Collectively, our analysis identified differentially expressed genes, regulons, pathways, and cell functions between invasive and nodular GBM, establishing a molecular landscape of GBM invasion.
CRYAB and CD44 Are Co-localized in Highly Invasive GBM Samples
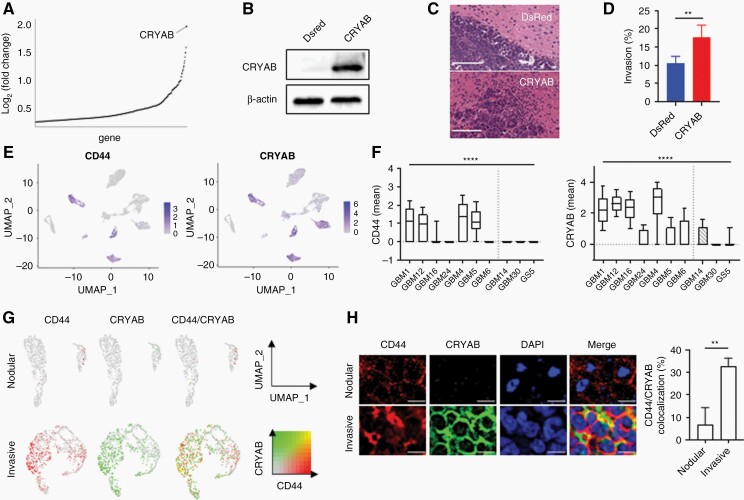
CRYAB, a member of the small heat shock protein family, was identified as one of the most significantly upregulated genes in invasive GBM (Figure 5A). Interestingly, TCGA GBM patients expressing higher levels of CRYAB had longer overall survival compared to those expressing lower levels of CRYAB (Supplementary Figure 5A). We tested if CRYAB genetically drives GBM invasion through overexpression in GBM30, the most nodular tumor (Figure 1C and F). To establish GBM30 cells that overexpress CRYAB, cDNA open reading frames encoding for CRYAB were cloned into a lentiviral express vector and transduced into GBM30 cells. Control cells were prepared using the same procedure but with cDNA open reading frames encoding for DsRed. The resulting cells were designated as GBM30-CRYAB and GBM30-DsRed, respectively. Western blot analysis confirmed that CRYAB was overexpressed in GBM30-CRYAB and not GBM30-DsRed cells (Figure 5B). To determine whether CRYAB can increase GBM migration in vivo, we inoculated GBM30-CRYAB or GBM30-DsRed cells in immunodeficient nude mice. Mice were sacrificed 5 weeks after inoculation, and their brains were collected and sliced for H&E staining. Mice inoculated with GBM30-DsRed cells had a nodular tumor with distinct tumor boundaries, which was similar to that of untreated GBM30 cells (Figure 5C). On the other hand, mice inoculated with GBM30-CRYAB cells had tumors with more diffuse tumor boundaries, demonstrating that overexpression of CRYAB increases GBM migration in vivo (Figure 5D). Overexpression of CRYAB did not affect SOX2 expression in GBM30 cells (Supplementary Figure 5B), further supporting that only a subset of CSCs contributes to GBM invasion. In addition, GBM30-CRYAB cells had significantly slower proliferation compared to that of GBM30-DsRed control cells (Supplementary Figure 5C), which is consistent with our finding that invasive GBM is less proliferative than that of nodular GBM.
Fig. 5.
CRYAB and CD44 are expressed in the same subpopulation of cells and contribute to GBM invasion.
(A) CRYAB is the most upregulated gene in invasive GBM compared to that of nodular GBM. (B) Western blot demonstrating overexpression of CRYAB in GBM30-CRYAB cells. (C, D) Overexpression of CRYAB increases tumor invasiveness of GBM30. Scale bar = 100 µm. n = 3. (E) UMAP of GBM samples colored by CD44 (left) and CRYAB (right) expression. (F) Box plot of mean CD44 (left) and CRYAB (right) expression from scRNA-seq data. Invasive GBM samples express both CD44 and CRYAB. (G) UMAP of nodular GBM (GBM1) and invasive GBM (GBM30) to examine intra-tumoral heterogeneity of CD44- and CRYAB-expressing cells. Color threshold = 0.1. H: Confocal imaging of nodular GBM (GBM1) and invasive GBM (GBM30) to determine co-localization of CD44 and CRYAB. Correlation was determined by Pearson R value. n = 3. Scale bar = 10 µm.
To examine how CRYAB affects cells on the transcriptomic level, we compared differentially expressed genes, pathways, and cellular functions between GBM30 cells with and without CRYAB expression. Growth differentiation factor 15 (GDF15), which has been reported to promote CSC growth in GBM,51 was the most significantly upregulated gene in CRYAB-expressing cells (Supplementary Figure 5d). Interestingly, genes from the S100 protein family were upregulated in invasive GBM (S100B) and CRYAB-expressing GBM30 cells (S100A10), suggesting that this family of genes may regulate GBM invasion, which has been previously reported.41 In addition, we found that cellular functions related to cellular movement and invasion were upregulated in CRYAB-expressing GBM30 cells, while functions related to cell death and apoptosis were downregulated (Supplementary Figure 5E). Similar to that of invasive GBM, functions related to protein synthesis were downregulated. Furthermore, functions related to DNA repair, such as metabolism, repair, and fragmentation of DNA, were all downregulated in CRYAB-expressing cells, suggesting that CRYAB alters DNA repair pathways to promote tumor invasion. We found that pathways related to Rho and integrin signaling were upregulated in CRYAB-expressing cells, similar to that of invasive GBM (Supplementary Figure 5F). In addition, CD44 and interleukin-related genes (IL1A, IL1B, IL6, IL8) all had significantly positive correlations (Supplementary Figure 5H).
To investigate the relationship between CD44 and CRYAB, we visualized all cells with UMAP colored by CD44 and CRYAB expression, respectively (Figure 5E). Interestingly, three (GBM1, GBM12, GBM4) out of the four CD44-expressing GBM samples (CD44normalized_mean > 0.5) also had high expression of CRYAB (Figure 5F). Indeed, linear regression analysis confirmed that CD44 and CRYAB are significantly positively correlated (Supplementary Figure 6A). Next, we examined intra-tumoral expression of CD44 and CRYAB in all GBM samples. We found that CD44 and CRYAB are co-expressed within the same subpopulation of cells at both the mRNA (Figure 5G, Supplementary Figure 6B) and protein (Figure 5H) levels. Indeed, knockdown of CD44 in GBM30-CRYAB cells (GBM30-CRYAB-sgCD44) significantly reduced invasion compared to that of GBM30-CRYAB-sgGFP controls (Supplementary Figure 6C). Collectively, these results suggest that CD44 and CRYAB are co-expressed within invasive GBM samples, and cells expressing CD44 and CRYAB can be potential targets for GBM invasion intervention.
CRYAB Slows Tumor Growth but Promotes Rapid Postoperative Recurrence
To determine how overexpression of CRYAB affects GBM progression in vivo, we inoculated luciferase-expressing GBM30-DsRed or GBM30-CRYAB cells in immunodeficient nude mice. Consistent with our in vitro observations, GBM30-DsRed cells proliferated more quickly than that of GBM30-CRYAB cells (Figure 6A). Three weeks after tumor inoculation, GBM30-DsRed tumors were significantly larger than that of GBM30-CRYAB tumors (Figure 6B).
Currently, surgical resection of the tumor is the first step in standard GBM treatments.2 To mimic the standard of care used for GBM patients clinically, we developed a mouse GBM resection model (Figure 6C). Luciferase-expressing GBM30-DsRed or GBM30-CRYAB cells were inoculated in immunodeficient nude mice. When the tumor grew to a certain size, as determined by a radiance efficiency range between 1 × 106 and 1 × 107 after 1 min exposure, surgical resection of the tumor was performed, and mice were imaged post-surgery to monitor tumor recurrence. After surgical resection of the tumor, mice inoculated with GBM30-CRYAB cells had quicker tumor recurrence than that of mice inoculated with GBM30-DsRed cells due to increased tumor invasion (Figure 6D). Ten days after surgery, GBM30-CRYAB tumors were significantly larger than that of GBM30-DsRed tumors (Figure 6E). Mouse survival was also monitored to determine how tumor invasion affects overall survival after surgical resection. Mice inoculated with GBM30-CRYAB cells had significantly shorter survival (t = 24 days) than those inoculated with GBM30-DsRed cells (t = 37 days) (Figure 6F). Furthermore, CRYAB expression was significantly higher in both recurrent GBM mouse models (Supplementary Figure 6D) and recurrent GBM patient samples (Supplementary Figure 6E) compared to that of nodular samples. Collectively, these data show that GBM invasiveness is associated with slow tumor growth but rapid post-operative recurrence and expression of CRYAB.
Discussion
Tumor invasion, a hallmark of GBM, contributes significantly to treatment failure and recurrence.3,4 However, the biology of GBM invasion and how CSCs contribute, remain poorly understood. Previous studies have used transgenic mouse models52 or GBM cell lines53 to study GBM invasion; unfortunately, these models may not be able to accurately recapitulate the aggressive nature of GBM seen clinically or lack the tumor microenvironment. Others have attempted to identify potential molecular targets or markers of invasion by comparing genomic profiles of normal brain tissue and GBM tissue or low-grade astrocytoma tissue and GBM tissue.54 These studies may be highlighting genetic differences between GBM and other tissues but not necessarily features related to invasion.
In this study, we used an alternative approach to profile GBM invasion through integrative analysis of human histology, PDX model, and scRNA-seq data from 10 GBM patients. Our analysis identified two cohorts of GBM, invasive and nodular, which are distinct at both the cellular and molecular levels, but, apparently, not age and sex (Supplementary Table 2). Of samples where, patient survival data was available (GBM16, GBM24, and GBM30), the patient with the shortest survival (GBM16, t = 54 days) had the highest expression of invasion-related genes and CSC markers compared to the other 2 samples (Supplementary Figure 6F). We show that, compared to non-invasive counterparts, invasive GBM is less dense and proliferative. Additional studies are needed to determine whether cellular proliferation outpaces migration in nodular tumors. We found that consistent with previous reports,7,55 CSCs correlate with GBM invasion. However, among them, only the CD44+ subset contributes significantly to the invasive phenotype, likely through its interaction with the hyaluronic acid-enriched brain extracellular matrix.39 The analysis further established a transcriptomic landscape of GBM invasion with differentially expressed genes, regulons, pathways, and cell functions between invasive and nodular GBM. Consistent with previous findings,7 we found that the Rho family of GTPases plays a major role in GBM invasion.
Lastly, we investigated CRYAB, one of the most prominent invasive genes that was previously shown to be highly expressed in the infiltrative edge of GBM.40 We found that overexpression of CRYAB increased invasion of nodular GBM in vivo and switched the molecular phenotype at both the cellular and molecular levels. We demonstrated that CD44 and CRYAB are co-expressed in invasive GBM samples, and that knockout of CD44 reduces invasion in the context of CRYAB overexpression. In addition, we showed that GBM invasiveness, although associated with slow tumor growth, causes rapid postoperative recurrence.
In summary, through integrative analysis of histological and scRNA-seq data from GBM patients, we reveal the cellular and molecular landscape of GBM invasion. We identified and validated CRYAB as a major GBM invasion driver. These findings are significant as they may guide the development of more effective therapies by targeting GBM invasion through genetic regulation of major molecular targets, such as CD44 and CRYAB, or targeted elimination of a subset of cells, such as those with co-expression of CD44 and CRYAB.
Supplementary Material
Contributor Information
Ann T Chen, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Yang Xiao, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Xiangjun Tang, Department of Neurosurgery, Yale University, New Haven, CT, USA.
Mehdi Baqri, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Xingchun Gao, Department of Neurosurgery, Yale University, New Haven, CT, USA.
Melanie Reschke, Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT, USA.
Wendy C Sheu, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Gretchen Long, Department of Neurosurgery, Yale University, New Haven, CT, USA.
Yu Zhou, Department of Neurosurgery, Yale University, New Haven, CT, USA.
Gang Deng, Department of Neurosurgery, Yale University, New Haven, CT, USA.
Shenqi Zhang, Department of Neurosurgery, Yale University, New Haven, CT, USA.
Yanxiang Deng, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Zhiliang Bai, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Dongjoo Kim, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Anita Huttner, Department of Pathology, Yale University, New Haven, CT, USA.
Russell Kunes, Department of Statistics, Columbia University, New York, NY, USA.
Murat Günel, Department of Neurosurgery, Yale University, New Haven, CT, USA.
Jennifer Moliterno, Department of Neurosurgery, Yale University, New Haven, CT, USA.
W Mark Saltzman, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Rong Fan, Department of Biomedical Engineering, Yale University, New Haven, CT, USA.
Jiangbing Zhou, Department of Biomedical Engineering, Yale University, New Haven, CT, USA; Department of Neurosurgery, Yale University, New Haven, CT, USA.
Funding
This work was partially supported by NIH Grants NS095817 and CA245313.
Conflict of interest statement. The authors declare that there is no conflict of interest.
Authorship statement. Conception and design: A.T. C., Y.X., R.F., and J.Z. Collection and assembly of data: A.T.C., Y.X., X.T., M.B., X.G., M.R., W.C.S., G.L., Y.Z., G.D., S.Z., Y.D., Z.B., and D.K. Manuscript writing: All authors. Final approval of the manuscript: All authors. Accountable for all aspects of the study: All authors.
References
- 1. Holland EC. Glioblastoma multiforme: the terminator. Proc Natl Acad Sci USA. 2000;97(12):6242–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tan AC, Ashley DM, Lopez GY, et al. Management of glioblastoma: state of the art and future directions. CA Cancer J Clin. 2020;70(4):299–312. [DOI] [PubMed] [Google Scholar]
- 3. D’Alessio A, Proietti G, Sica G, Scicchitano BM. Pathological and molecular features of glioblastoma and its peritumoral tissue. Cancers (Basel). 2019;11(4):469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haj A, Doenitz C, Schebesch K-M, et al. Extent of resection in newly diagnosed glioblastoma: impact of a specialized neuro-oncology care center. Brain Sci. 2017;8(1):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aum DJ, Kim DH, Beaumont TL, et al. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg Focus. 2014;37(6):E11. [DOI] [PubMed] [Google Scholar]
- 6. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Volovetz J, Berezovsky AD, Alban T, et al. Identifying conserved molecular targets required for cell migration of glioblastoma cancer stem cells. Cell Death Dis. 2020;11(2):152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. [DOI] [PubMed] [Google Scholar]
- 9. Bao S, Wu Q, Li Z, et al. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68(15):6043–6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao Y, Kim D, Dura B, et al. Ex vivo dynamics of human glioblastoma cells in a microvasculature-on-a-chip system correlates with tumor heterogeneity and subtypes. Adv Sci. 2019;6(8):1801531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou J, Patel TR, Sirianni RW, et al. Highly penetrative, drug-loaded nanocarriers improve treatment of glioblastoma. Proc Natl Acad Sci USA. 2013;110(29):11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Reschke M, DiRito JR, Stern D, et al. A digital pathology tool for quantification of color features in histologic specimens. Bioeng Transl Med. 2021;7(1):e10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Macosko Evan Z, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Satija R, Farrell JA, Gennert D, Schier AF, Regev A. Spatial reconstruction of single-cell gene expression data. Nat Biotechnol. 2015;33(5):495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krishnan H, Rayes J, Miyashita T, et al. Podoplanin: an emerging cancer biomarker and therapeutic target. Cancer Sci. 2018;109(5):1292–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang Z, Xie Q, Hu C-L, et al. CHL1 is expressed and functions as a malignancy promoter in glioma cells. Front Mol Neurosci. 2017;10:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Darmanis S, Sloan SA, Croote D, et al. Single-cell RNA-Seq analysis of infiltrating neoplastic cells at the migrating front of human glioblastoma. Cell Rep. 2017;21(5):1399–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klank RL, Decker Grunke SA, Bangasser BL, et al. Biphasic dependence of glioma survival and cell migration on CD44 expression level. Cell Rep. 2017;19(3):668. [DOI] [PubMed] [Google Scholar]
- 19. Che Mat MF, Abdul Murad NA, Ibrahim K, et al. Silencing of PROS1 induces apoptosis and inhibits migration and invasion of glioblastoma multiforme cells. Int J Oncol. 2016;49(6):2359–2366. [DOI] [PubMed] [Google Scholar]
- 20. Bhat KP, Salazar KL, Balasubramaniyan V, et al. The transcriptional coactivator TAZ regulates mesenchymal differentiation in malignant glioma. Genes Dev. 2011;25(24):2594–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan MSY, Sandanaraj E, Chong YK, et al. A STAT3-based gene signature stratifies glioma patients for targeted therapy. Nat Commun. 2019;10(1):3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vollmann-Zwerenz A, Leidgens V, Feliciello G, Klein CA, Hau P. Tumor cell invasion in glioblastoma. Int J Mol Sci. 2020;21(6):1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Braig S, Wallner S, Junglas B, Fuchshofer R, Bosserhoff A. CTGF is overexpressed in malignant melanoma and promotes cell invasion and migration. Br J Cancer. 2011;105(2):231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang X, Chen T, Zhang J, et al. Notch1 promotes glioma cell migration and invasion by stimulating β-catenin and NF-κB signaling via AKT activation. Cancer Sci. 2012;103(2):181–190. [DOI] [PubMed] [Google Scholar]
- 25. Fumagalli A, Heuninck J, Pizzoccaro A, et al. The atypical chemokine receptor 3 interacts with Connexin 43 inhibiting astrocytic gap junctional intercellular communication. Nat Commun. 2020;11(1):4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Almanzar N, Antony J, Baghel AS, et al. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020;583(7817):590–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Aibar S, González-Blas CB, Moerman T, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. [DOI] [PubMed] [Google Scholar]
- 29. Lee J, Kotliarova S, Kotliarov Y, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 30. Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu H, Liu S, Zhang G, et al. Oncogenic BRAF-mediated melanoma cell invasion. Cell Rep. 2016;15(9):2012–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Barrett CS, Millena AC, Khan SA. TGF-β effects on prostate cancer cell migration and invasion require FosB. Prostate. 2017;77(1):72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Q, Hu B, Hu X, et al. Tumor evolution of glioma-intrinsic gene expression subtypes associates with immunological changes in the microenvironment. Cancer Cell. 2017;32(1):42–56.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Esensten JH, Bluestone JA, Lim WA. Engineering therapeutic T cells: from synthetic biology to clinical trials. Annu Rev Pathol. 2017;12:305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells. 2009;27(10):2383–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trépant AL, Bouchart C, Rorive S, et al. Identification of OLIG2 as the most specific glioblastoma stem cell marker starting from comparative analysis of data from similar DNA chip microarray platforms. Tumour Biol. 2015;36(3):1943–1953. [DOI] [PubMed] [Google Scholar]
- 37. Berezovsky AD, Poisson LM, Cherba D, et al. Sox2 promotes malignancy in glioblastoma by regulating plasticity and astrocytic differentiation. Neoplasia. 2014;16(3):193–206, 206.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bhaduri A, Di Lullo E, Jung D, et al. Outer radial glia-like cancer stem cells contribute to heterogeneity of glioblastoma. Cell Stem Cell. 2020;26(1):48–63.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xiao W, Zhang R, Sohrabi A, et al. Brain-mimetic 3D culture platforms allow investigation of cooperative effects of extracellular matrix features on therapeutic resistance in glioblastoma. Cancer Res. 2018;78(5):1358–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goplen D, Bougnaud S, Rajcevic U, et al. αB-crystallin is elevated in highly infiltrative apoptosis-resistant glioblastoma cells. Am J Pathol. 2010;177(4):1618–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang H, Zhang L, Zhang IY, et al. S100B promotes glioma growth through chemoattraction of myeloid-derived macrophages. Clin Cancer Res. 2013;19(14):3764–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lim SY, Ahn S-H, Park H, et al. Transcriptional regulation of adrenomedullin by oncostatin M in human astroglioma cells: implications for tumor invasion and migration. Sci Rep. 2014;4:6444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lee JS, Kim HY, Jeong NY, et al. Expression of αB-crystallin overrides the anti-apoptotic activity of XIAP. Neuro Oncol. 2012;14(11):1332–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen D, Cao G, Qiao C, et al. Alpha B-crystallin promotes the invasion and metastasis of gastric cancer via NF-κB-induced epithelial-mesenchymal transition. J Cell Mol Med. 2018;22(6):3215–3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shi C, Yang X, Bu X, Hou N, Chen P. Alpha B-crystallin promotes the invasion and metastasis of colorectal cancer via epithelial-mesenchymal transition. Biochem Biophys Res Commun. 2017;489(4):369–374. [DOI] [PubMed] [Google Scholar]
- 46. van Dam H, Castellazzi M. Distinct roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene. 2001;20(19):2453–2464. [DOI] [PubMed] [Google Scholar]
- 47. Ahn S-H, Park H, Ahn Y-H, et al. Necrotic cells influence migration and invasion of glioblastoma via NF-κB/AP-1-mediated IL-8 regulation. Sci Rep. 2016;6(1):24552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dhandapani KM, Mahesh VB, Brann DW. Curcumin suppresses growth and chemoresistance of human glioblastoma cells via AP-1 and NFκB transcription factors. J Neurochem. 2007;102(2):522–538. [DOI] [PubMed] [Google Scholar]
- 49. Knudsen AM, Eilertsen I, Kielland S, et al. Expression and prognostic value of the transcription factors EGR1 and EGR3 in gliomas. Sci Rep. 2020;10(1):9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang S-M, Lin H-Y, Chen Y-L, et al. CCAAT/enhancer-binding protein delta regulates the stemness of glioma stem-like cells through activating PDGFA expression upon inflammatory stimulation. J Neuroinflammation. 2019;16(1):146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu S, Yang N, Guan Y, et al. GDF15 promotes glioma stem cell-like phenotype via regulation of ERK1/2–c-Fos–LIF signaling. Cell Death Discov. 2021;7(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jun HJ, Appleman VA, Wu H-J, et al. A PDGFRα-driven mouse model of glioblastoma reveals a stathmin1-mediated mechanism of sensitivity to vinblastine. Nat Commun. 2018;9(1):3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sha Z, Zhou J, Wu Y, et al. BYSL promotes glioblastoma cell migration, invasion, and mesenchymal transition through the GSK-3β/β-catenin signaling pathway. Front Oncol. 2020;10:565225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gupta MK, Polisetty RV, Sharma R, et al. Altered transcriptional regulatory proteins in glioblastoma and YBX1 as a potential regulator of tumor invasion. Sci Rep. 2019;9(1):10986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ortensi B, Setti M, Osti D, Pelicci G. Cancer stem cell contribution to glioblastoma invasiveness. Stem Cell Res Ther. 2013;4(1):18. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.