Abstract
Background
The loss of neurogenic tumor suppressor microRNAs miR-124, miR-128, and miR-137 is associated with glioblastoma’s undifferentiated state. Most of their impact comes via the repression of a network of oncogenic transcription factors. We conducted a high-throughput functional siRNA screen in glioblastoma cells and identify E74 like ETS transcription factor 4 (ELF4) as the leading contributor to oncogenic phenotypes.
Methods
In vitro and in vivo assays were used to assess ELF4 impact on cancer phenotypes. We characterized ELF4’s mechanism of action via genomic and lipidomic analyses. A MAPK reporter assay verified ELF4’s impact on MAPK signaling, and qRT-PCR and western blotting were used to corroborate ELF4 regulatory role on most relevant target genes.
Results
ELF4 knockdown resulted in significant proliferation delay and apoptosis in GBM cells and long-term growth delay and morphological changes in glioma stem cells (GSCs). Transcriptomic analyses revealed that ELF4 controls two interlinked pathways: 1) Receptor tyrosine kinase signaling and 2) Lipid dynamics. ELF4 modulation directly affected receptor tyrosine kinase (RTK) signaling, as mitogen-activated protein kinase (MAPK) activity was dependent upon ELF4 levels. Furthermore, shotgun lipidomics revealed that ELF4 depletion disrupted several phospholipid classes, highlighting ELF4’s importance in lipid homeostasis.
Conclusions
We found that ELF4 is critical for the GBM cell identity by controlling genes of two dependent pathways: RTK signaling (SRC, PTK2B, and TNK2) and lipid dynamics (LRP1, APOE, ABCA7, PLA2G6, and PITPNM2). Our data suggest that targeting these two pathways simultaneously may be therapeutically beneficial to GBM patients.
Keywords: ELF4, glioblastoma, lipid dynamics, miRNA-transcription factor networks, RTK signaling
Key Points.
miR-124, miR-128, and miR-137 regulate a network of oncogenic transcription factors.
A functional screening defined ELF4 as the leading contributor to oncogenic phenotypes.
ELF4 affects GBM cell identity by simultaneously controlling RTK signaling and lipid dynamics.
Importance of the Study.
A functional screen of miR-124, miR-128, and miR-137 shared transcription factor targets in glioblastoma defined ELF4 as the most prominent contributor to oncogenic phenotypes and GBM cell identity. More specifically, genomic and lipidomic analyses established that ELF4 controls a set of highly associated genes linked to RTK signaling and lipid homeostasis. RTK dysfunction is common in GBM, yet RTK inhibitors have not been effective clinically. GBM RTK signaling is dependent upon lipid membrane dynamics, and we found that ELF4 controls the levels of two phospholipid classes (phosphatidylcholine and phosphatidylethanolamine). Based on ELF4’s importance in GBM cell identity, it is evident that GBM cells depend upon both RTK signaling and lipid dynamics for survival. Finally, our findings indicate that combinations of RTK inhibitors with lipid homeostasis inhibitors could provide an alternative strategy to treat GBM.
Glioblastoma multiforme (GBM) is the most common and aggressive form of a brain tumor in adults, with a median survival time of 13 months.1,2 Sequencing efforts have revealed three molecular subtypes driven by differing alterations and pathways, while single-cell analyses identified four cellular subtypes.3–5 Despite these sequencing efforts, targeted therapies against these subtypes remain unrealized. More importantly, GBM tumors display significant transcriptomic plasticity, where cells can shift between molecular subtypes in response to stress and therapy,5 resulting in treatment evasion, recurrence, and ultimately death. Understanding the dynamics of gene regulators, identifying synergistic and antagonistic interactions and their impact on GBM phenotypes and relevant pathways are critical to improving treatment outcomes and preventing relapse.
GBMs display reduced expression of several tumor suppressor microRNAs (miRNAs).6 Of particular interest are pro-neurogenic miRNAs as their absence could contribute to GBM’s poorly differentiated state.7 Activation of differentiation pathways via delivery of miRNA mimics has been explored as potential therapeutic options for GBM.8 Among the many miRNAs that fall in this category, miR-124, miR-128, and miR-137 are among the most studied ones. These three miRNAs share similar expression patterns during neurogenesis and in gliomas, and function synergistically in both normal adult mammalian neural stem cells (NSCs) and glioma stem cells (GSCs).9,10 Ectopic expression of these three miRNAs enhanced NSC differentiation into neurons, while in GBM they inhibited proliferation, reduced cell viability, and induced phenotypic changes. When combined, these three miRNAs produced much more dramatic changes in comparison to individual miRNAs.10 Genomic analysis of miR-124, miR-128, and miR-137 in both NSCs and GSCs revealed that transcription factors (TFs) are preferentially targeted. Moreover, transcription factors regulated by one or more of these miRNAs are associated, forming a highly interconnected miRNA-transcription factor network.9,10 miRNA-TF networks are powerful regulatory systems, often forming feed-forward loops. They play essential roles in numerous functions, ranging from development to death.11 In cancer, miRNA-TF networks can be responsible for a variety of cancer-relevant phenotypes, such as cell cycle progression and metastasis.11
To identify transcription factors in the miRNA-TF network9 contributing to GBM phenotypes the most, we performed a high-throughput siRNA functional screen. E74 like ETS transcription factor 4 (ELF4), which is a target of both miR-124 and miR-128, emerged as the lead candidate. Subsequent analysis in GSCs determined that ELF4 knockdown strongly impacts their growth and viability. Transcriptomic analyses to identify ELF4’s mechanism of action found that ELF4 controls two interlinked pathways: 1) receptor tyrosine kinase (RTK) signaling and 2) lipid dynamics. Following the loss of ELF4, RTK signaling is impaired and phospholipid dynamics are disrupted as indicated by shotgun lipidomics and polar head group studies. Our combined, transcriptomic and lipidomic analyses tie these two pathways and identified important genes in each pathway. This work validates prior observations demonstrating the necessity of RTK signaling and lipid regulation in GBM cells.12,13 More importantly, we identify a single transcription factor, ELF4, that directly connects the two pathways.
Materials and Methods
Cell Lines and Transfection
Glioblastoma cell line U251 was obtained from Uppsala, Sweden. T98G cell line was obtained from the American Type Culture Collection (ATCC). Cells were grown in DMEM (Hyclone) supplemented with 10% fetal bovine serum (GIBCO) and 1% pen/strep (GIBCO). HeLa cells were obtained from the American Type Culture Collection (ATCC) and were cultured in RPMI-1640 (GIBCO) supplemented with 10% FBS and 1% pen/strep. 293T cells were obtained from the ATCC and were cultured in DMEM-High Glucose (HyClone) supplemented with 10% FBS and 1% pen/strep. Cells were passaged no more than 15 times and were tested for mycoplasma contamination using DAPI staining (2 µg/mL; Thermo Fisher).
Glioblastoma Stem Cells (GSCs) 3565 (mesenchymal), 3128 (mesenchymal), 1919 (proneural), and 19NS (proneural) were gifts from Drs. Jeremy Rich, Christopher Hubert, and Ichiro Nakano,14,15 and were grown in Neurobasal-A with B27, glutamine, sodium pyruvate, 20 ng/mL of both EGF (ThermoFisher) and hFGF (PeproTech). Every 72 h, GSCs were pulsed with EGF/FGF. Dissociation was performed by incubating GSCs with Accutase (ThermoFisher) at room temperature for 10 min. SMARTpool siRNAs against the transcription factors were obtained from Horizon Discovery (Supplementary Table 1). Glioblastoma cell lines and GSCs were reverse transfected into 96-wells with siRNAs using RNAiMAX (ThermoFisher).
MTS Assay
Glioblastoma cells were plated into 96-well plates and transfected as described above. Quantification of viable cells was assessed using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS; Promega).
Cell Proliferation Assay
Glioblastoma cells were plated into 96-well plates, transfected as described above, and placed into the IncuCyte ZOOM Imaging System (Essen BioScience). Cell confluence was monitored periodically using the Confluence Processing analysis tool (Essen BioScience). Cell growth curves were generated by plotting cell confluence as a function of time.
Caspase-3/-7 Assay
Cells were plated into 96-well opaque plates and transfected as described above. After 48 h, cells were assessed for Caspase-3/-7 activity utilizing the Caspase-Glo® 3/7 Assay (Promega).
mRNA Expression Analysis
Total RNA was isolated from cells treated with specific conditions (eg, siRNA against TFs, siControl/siELF4, Mimic Control/miR-124/miR-128) with TRIzol (ThermoFisher). cDNA was synthesized using a High-Capacity cDNA Synthesis Kit (Thermo Fisher). Primers and TaqMan probes used for knockdown validation and gene expression quantification are listed in (Supplementary Table 2). PowerUp SYBR Green and TaqMan Master Mix were used for qRT-PCR (ThermoFisher) ACTB and 18S were utilized as reference genes. The delta-delta Ct method was used to compare mRNA levels between different conditions.
Luciferase Assay
293T cells were plated into 96-well plates and transfected with the ELF4 3′UTR construct16 using Lipofectamine 3000 (ThermoFisher) along with miRNA mimics or control oligos. Firefly luciferase luminescence was measured 48 h later using the Dual-Glo® Luciferase Assay System (Promega). Sea pansy luminescence was measured as a transfection control.
Serum Response Element Reporter
The serum response element (SRE) reporter was generated by cloning the Luc2 gene into a vector containing six SREs (Addgene: 82686). 293T cells were plated into 96-well plates and transfected 16 h later using Lipofectamine 3000. Stable lines were generated by transfecting cells with linearized SRE plasmid using Lipofectamine 3000. Cells were selected with hygromycin for 2 weeks. A polyclonal population was utilized for knockdown experiments.
Western Blot Analysis
Cells were collected and lysed using the freeze–thaw method in Laemmli buffer. Proteins were separated on SDS-PAGE gels and transferred to PDVF membranes. Membranes were blocked in TBST in 5% milk and probed with the following antibodies: LRP1 (Abcam: ab92544), ABCA7 (Bethyl Laboratories: A304-427A), PITPNM2 (Novus Biologicals: NBP1-80994), SRC (Cell Signaling: 2110), p38 (Cell Signaling: 9212), ELF4 (GeneTex: 103625) p-p38 (Santa Cruz: 7973), p-SRC (R&D: AF2685), Beta-Actin (Abcam: 9227), and Alpha-Tubulin (ThermoFisher: 62204). Anti-rabbit (Cell Signaling) and Anti-mouse (ThermoFisher) HRP-conjugated secondary antibodies were used for development with Immobilon Western Chemiluminescent HRP Substrate (Millipore). Membranes were stripped with LI-COR WesternSure ECL Stripping Buffer and re-blocked for additional probing.
Preparation Xenograft Cell Line
See Supplementary Methods for details.
Xenograft
See Supplementary Methods for details.
RNA-Sequencing Analyses
To identify the transcriptional impact of ELF4, 3565 cells were treated in triplicate with either siELF4 or siControl. About 48 h later, RNA was isolated with TRIzol (ThermoFisher). Samples were sequenced using poly-A selected mRNA at the GCCRI Genome Sequencing Facility (UTHSCSA). Sequencing reads were first processed using Kallisto (version 0.43; parameters: --bootstrap-samples 100 --single --fragment-length 51 --sd 1e-0817; with an index of 31 k-mers and GENCODE (www.gencodegenes.org/; v29) as the reference to the human transcriptome. Gene-level abundance estimates were obtained using the R package tximport.18 Differential gene expression analysis between siELF4 and siControl samples was performed using DESeq2 with default parameters,19 and genes were considered differentially expressed using a threshold of |log2FoldChange| ≥ 0.5 and Benjamini–Hochberg (FDR) adjusted P-value < .05.
Chromatin Immunoprecipitation Sequencing Analyses
To identify genes potentially regulated by ELF4, we first obtained processed ELF4 ChIP-seq data on human HEK293T and K562 cell lines from the ENCODE project (https://www.encodeproject.org/; accession numbers: ENCSR778QLY and ENCSR638QHV), and on T3M-1 Cl-10 cells.20 Next, we defined the promoter regions of genes overexpressed in siELF4 compared with control samples as 5 kb around their transcription start sites (TSSs), which were obtained from Cap Analysis of Gene Expression data at the FANTOM5 project (https://fantom.gsc.riken.jp/5/). Overlaps between gene’s promoter regions and significant chromatin immunoprecipitation sequencing (ChIP-seq) peaks (FDR < 0.05) from all experiments were calculated using bedtools intersect (VERSION 2.26; default parameters).21 We considered potential ELF4 targets those genes with promoter regions overlapping significant Chip-seq peaks. As a negative control, we also overlapped ChIP-seq peaks with promoter regions of genes overexpressed in control in comparison to siELF4 samples.
Gene Set Enrichment Analysis and Network Interaction Assessment
See Supplementary Methods for details.
Analysis of Patient Survival
See Supplementary Methods for details.
Sample Preparation for Lipid Analysis
Lipid extracts were isolated by modified Bligh–Dyer, as previously described.22 Chilled extraction buffer (methanol: water [1:1] with 10 mM ammonium bicarbonate) and a proportionate volume of chloroform were added to frozen cell pellets. See Supplementary Methods for details.
Lipidomics
Untargeted lipidomics analysis was performed on a high-resolution Hybrid Quadrupole-Orbitrap mass spectrometer (Q Exactive, Thermo Scientific, Waltham, MA, USA) equipped with an automated chip-based nanoelectrospray ionization (nESI) source (TriVersa NanoMate, Advion, Ithaca, NY). See Supplementary Methods for details.
Drug Combination Assay
GSCs were plated onto geltrex-coated plates. The following day cells were treated with different combinations of Lovastatin (Cayman Chemical: 10010338) and Dasatinib (Cayman Chemical: 11498). About 168 h later, MTS assay was performed as described earlier.
Statistical Analysis
siRNA screen was performed utilizing technical triplicates for each of the and a Dunnett’s test was used for comparison with siControl. Differences in proliferation were identified utilizing multiple t-testing adjusted with a Bonferroni correction. qRT-PCR measurements were performed with biological and technical triplicates, with differences in expression assessed by Student’s t-test. Luciferase assays were performed with biological and technical triplicates, with differences in activity assessed by Student’s t-test.
Results
Phenotypic Screening of Transcription Factors Targeted by miR-124, miR-128, and miR-137
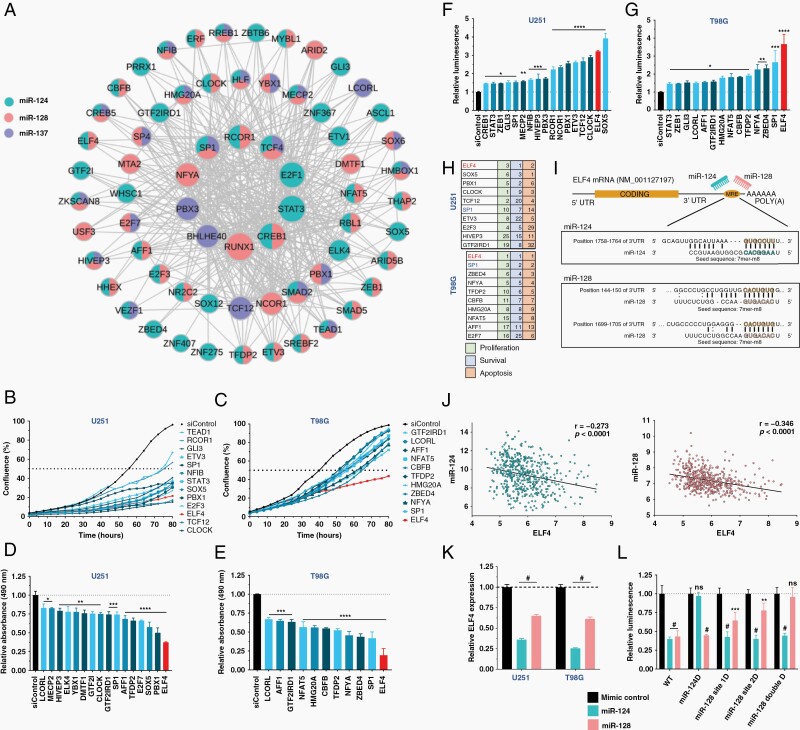
The pro-neurogenic and tumor suppressor miRNAs, miR-124, miR-128, and miR-137, are often downregulated in GBM. We have previously shown that they work synergistically to promote neuronal differentiation and repression of a network of transcription factors (TFs; Figure 1A) is an important component of their effect.9,10 To evaluate the contribution of each TF in this network to GBM phenotypes, we conducted a high-throughput siRNA screen in two GBM cell lines, U251 and T98G. We measured the impact of their knockdown (KD) on proliferation via live-cell imaging, cell viability based on an MTS assay and apoptosis using the caspase-3/-7 assay (Figure 1B–G). To rank the importance of the TFs in GBM, we generated a score for all assays and considered their impact on patient survival using data from the TCGA (Supplementary Table 3 and Supplementary Figure 1A). We summarized the top-10 hits in each cell line considering the results of the three different assays (Figure 1H). E74 like ETS transcription factor 4 (ELF4) emerged as our lead candidate, appeared as the top hit in both cell lines. Based on GTEx and TCGA datasets, ELF4 shows higher expression in GBM in comparison to low-grade glioma (LGG) and normal brain. Similarly, analysis of the CGGA dataset indicated that ELF4 expression is higher in secondary and recurrent glioma in comparison to primary tumors (Supplementary Figure 1B–C). ELF4 expression shows increased expression in the more aggressive GBM sub-group, mesenchymal, in comparison to proneural and classical (Supplementary Figure 1D). Finally, ELF4 displays ahigh expression correlation with several glioma stem cell and neuronal stem cell markers including CD44, CD36, CD15, CD70, S100A4, and ALDH1A3 in TCGA GBM samples, suggesting a potential role in stemness and tumor initiation (Supplementary Figure 1E).23
Figure 1.
miR-124, -128, and -137 regulated transcription factors drive the glioblastoma phenotype. (A) TF network regulated by the three miRNAs. (B–G) U251 and T98G cells were reverse transfected with siRNAs against each TF. Only siRNAs that caused a significant difference from siControl are shown. (B, C) Cell proliferation was monitored using a live-cell imager (IncuCyte). (D, E) Cell viability was measured using an MTS assay 48 h after transfection. (F, G) Caspase-3/-7 activity in GBM cells 48 h after knockdown. (H) Summary table of screening results displaying top-10 TFs based on their rank in each assay. (I) miR-124/-128 binding sites in ELF4’s 3′UTR. (J) Correlation between ELF4 and miR-124/-128 RNA expression in the TCGA GBM cohort. (K) ELF4 expression 48 h after miRNA transfection. (L) 293T cells cotransfected with ELF4 3′UTR luciferase reporter and miRNA mimics. miRNA binding sites in (I) were deleted. Luminescence was measured after 48 h. Student’s t-test was used to determine statistical significance in qPCR and luciferase experiments, **P < .01, ***P < .001, #P < .0001. Significant differences in screening assays were determined by performing a Dunnett’s multiple comparison test against siControl (P-adjusted < .05). Full results in (Supplementary Table 4).
Previously, we demonstrated that ELF4 is a target of miR-12416; however, the ELF4 3′UTR also possesses miR-128 binding sites. We find that ELF4 expression is negatively correlated with the two miRNAs in GBM tumors. Transfection of GBM cells with miR-128 mimics led to a reduction in ELF4 expression. ELF4’s 3′UTR has two predicted binding sites for miR-128. Luciferase assays conducted with ELF4 3′UTR reporter constructs showed that both binding sites are functional since their mutation impaired miR-128 mediated regulation (Figure 1I–L).
ELF4’s Regulatory Landscape
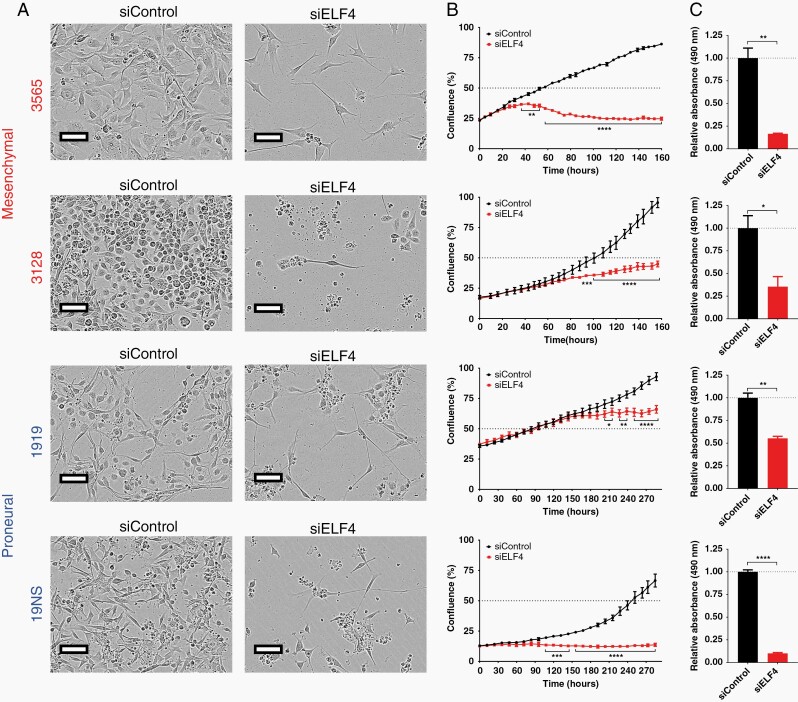
To expand on the regulatory impact of ELF4 expression, we conducted additional analyses in two mesenchymal and two proneural glioma stem cells (GSCs) lines. Decreasing ELF4 expression via siRNA transfection resulted in significant proliferation delay and more importantly, produced morphological changes featuring long neurite protrusions, suggesting a more differentiated phenotype (Figure 2).
Figure 2.
ELF4 is critical for glioma stem cell phenotype. GSCs were reverse transfected with siRNAs onto a Geltrex matrix. (A) GSCs displayed long-term morphological changes following transient knockdown of ELF4 (3565 and 3128: 160 h later; 1919 and 19NS: 286 h later). (B) Proliferation was monitored over time using a live-cell imager (IncuCyte). Statistical significance was determined by performing multiple t-tests with P-adjusted value corrected by the Holm–Šídák method (*P < .05; **P < .01; ***P < .001; and ****P < .0001). (C) MTS assay following the end of live-cell imaging. Student’s t-test was used to determine statistical significance *P < .05, **P < .01, ****P < .0001.
ELF4 levels have shown to impact the aggressiveness of a PDGF-driven mouse model of GBM.24 We sought to determine whether ELF4 played a role in tumor progression using an orthotopic xenograft model. We inoculated NCR-SCID mice with U251-luciferase cells expressing tet-inducible shRNAs against ELF4. Administration of doxycycline started 11 days after implantation. ELF4 knockdown caused an increase in survival (median difference = 14 days; Supplementary Figure 2A). More importantly, we observed differences in tumor volume based on bioluminescence between control and ELF4 knockdown tumors during the first 2 weeks after we initiated Dox treatment. This result suggests that a reduction in ELF4 levels caused a delay in tumor initiation (Supplementary Figure 2B). Finally, we noticed that tumors in doxycycline-treated mice were smaller based on Ki67 staining (Supplementary Figure 2C).
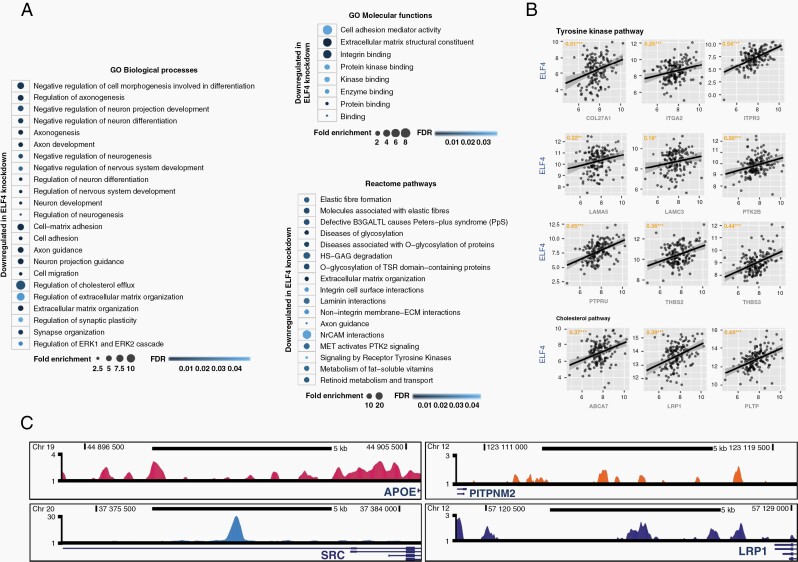
Next, we performed a transcriptome analysis to identify changes in gene expression driven by ELF4 knockdown in 3565 GSCs (Supplementary Table 4).14 Gene ontology and pathway analyses of downregulated genes highlight an enrichment in connected categories, including regulation of neuronal differentiation, extracellular matrix organization, receptor signaling, and lipid transportation (Figure 3A, Supplementary Table 4). Analysis of publicly available ELF4 ChIP-seq datasets indicated that over 30% of downregulated genes have ELF4 binding peaks in their promoter region (Supplementary Table 5). We compared the downregulated set to a previous study performed on neuroblastoma cells.16 Circa 25% of the genes downregulated in GBM cells after ELF4 knockdown also displayed a decrease in expression in neuroblastoma cells. Most of these shared genes are linked to differentiation, development, and migration (Supplementary Table 6). The comparison between the upregulated sets showed almost no overlap. However, an integrated analysis of enriched gene ontology terms pointed out commonalities between the two sets with respect to development, differentiation, and morphogenesis. Moreover, network analysis with genes upregulated upon ELF4 knockdown in GBM and/or neuroblastoma cells showed that these genes are highly interconnected, suggesting that despite differences in gene sets, similar pathways were activated (Supplementary Figure 3).
Figure 3.
ELF4’s regulatory landscape. (A) Enriched gene ontology (GO) and reactome terms following ELF4 knockdown in 3565 cells. Gene set was analyzed using PANTHER (http://pantherdb.org; Supplementary Table 3). (B) Downregulated genes, in tyrosine kinase and cholesterol pathways, whose expression is strongly correlated with ELF4 in TCGA GBM patients. (C) ELF4 binding sites in the promoters of relevant genes in K562 cells (fold change over control; Supplementary Table 5).
ELF4 Regulates Receptor Signaling
Our analysis identified new regulatory roles for ELF4, in particular its impact on two linked processes, receptor tyrosine kinase (RTK) signaling and lipid homeostasis. The regulatory impact of ELF4 on the expression of several genes associated with these two pathways was supported not only by ChIP-seq data but also observed a high expression correlation between ELF4 and these genes in TCGA glioblastoma samples (Figure 3B–C). When analyzed as a group, we also observed a strong expression correlation among genes in these two pathways supporting their connection (Supplementary Figure 4).
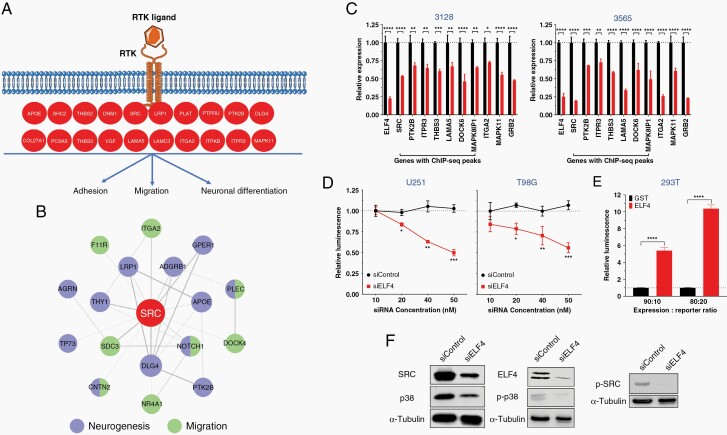
Dysfunction of RTK signaling is a hallmark of GBM with genetic or expression alterations observed in various receptors such as EGFR, PDGFRα, and MET.4 We validated ELF4’s impact on RTK genes by qRT-PCR (Figure 4A). To determine whether ELF4 loss impacted MAPK signaling, we generated U251 and T98G cell lines stably expressing a serum response element (SRE) reporter that indicates MAPK activity based on luciferase activity. Upon ELF4 KD, luminescence decreased suggesting MAPK signaling was hindered (Figure 4B). Additionally, transgenic ELF4 expression in 293T cells resulted in dramatic increases in MAPK signaling (Figure 4C). Western confirmed ELF4’s regulatory impact on MAPK signaling (Figure 4D). Overall, these results suggest that ELF4 directly regulates RTK-MAPK signaling.
Figure 4.
(A) qPCR for RTK-related genes in 3565 and 3128 cells 48 h after transfection. Student’s t-test was used to determine statistical significance between siControl and siELF4, *P < .05, **P < .01, ***P < .001, ****P < .0001. (B) U251 and T98G cells stably expressing a construct containing firefly luciferase under the control of serum response element promoter (SRE-Luciferase), were reverse transfected with different concentrations of siRNA. About 24 h later, luciferase activity was measured. Statistical significance was determined by performing multiple t-tests with P-adjusted value corrected by the Holm–Šídák method (*P < .05; **P < .01; ***P < .001). (C) HEK 293T cells were cotransfected with either GST (control) or ELF4 expression construct along with the SRE-luciferase construct, and luminescence was measured 24 h later. Student’s t-test was used to determine statistical significance between GST and ELF4, ****P < .0001. (D) Immunoblot analysis 48 h after transfection.
ELF4 Maintains Lipid Dynamics
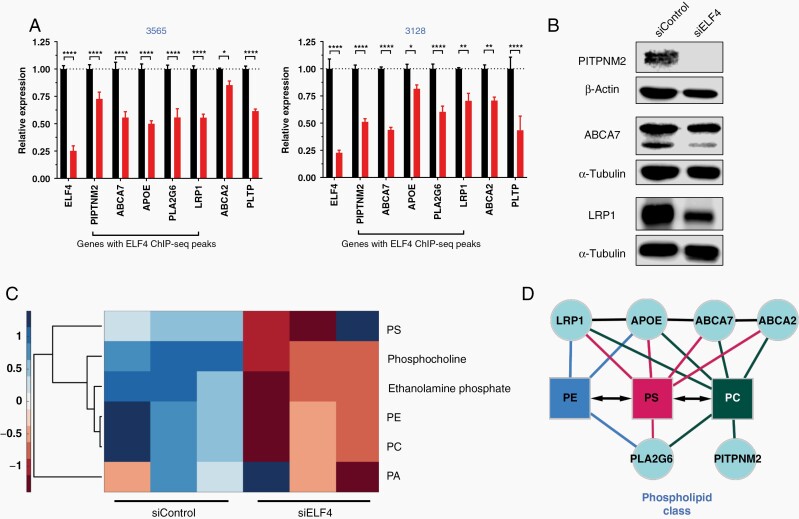
Receptors, as membrane-bound proteins, are impacted by disturbances in the lipid bilayer. Membrane signaling in GBM cells is particularly dependent upon lipid metabolism, as recent studies have implicated coordination between the two pathways.12,13 Our RNA-seq analysis identified several genes responsible for lipid efflux, suggesting ELF4 directly regulates lipid homeostasis. We first confirmed the downregulation of several lipid efflux genes at the RNA level and protein levels (Figure 5A and B). Several of these genes, PITPNM2, APOE, PLA2G6, and ABCA7 possess ELF4 binding sites in their promoter region, indicating direct regulation (Figure 3C, Supplementary Table 5). To characterize ELF4’s impact on lipid dynamics, we opted to perform an unbiased shotgun lipidomic analysis in the same GSCs used in the RNA-seq experiments. ELF4 appears critical for maintaining specific phospholipid levels, as ELF4 KD resulted in a significant global decrease in phosphocholine (PC) and phosphatidylethanolamine (PE) levels (Figure 5C, Supplementary Table 7). Untargeted polar metabolomics also shows a similar decreasing trend in corresponding polar headgroups phosphocholine and ethanolamine phosphate. Additionally, metabolite and protein–protein interaction analyses further implicate these genes in lipid homeostasis (Figure 5D).
Figure 5.
ELF4 controls lipid dynamics. (A) qPCR for RTK-related genes in 3565 and 3128 cells 48 h after transfection. Student’s t-test was used to determine statistical significance between siControl and siELF4, *P < .05, **P < .01, ***P < .001, ****P < .0001. (B) Immunoblot analysis 48 h after transfection. (C) Heatmap displaying major phospholipid classes and corresponding polar head groups based on lipidomics analysis (Supplementary Table 6). (D) Network analysis. Protein–protein interactions based on STRING using Text mining, Experiments, and Databases with a 0.4 confidence score. Protein interactions with phospholipids based on literature.
Synergism Between RTK and Lipid Homeostasis Inhibitors
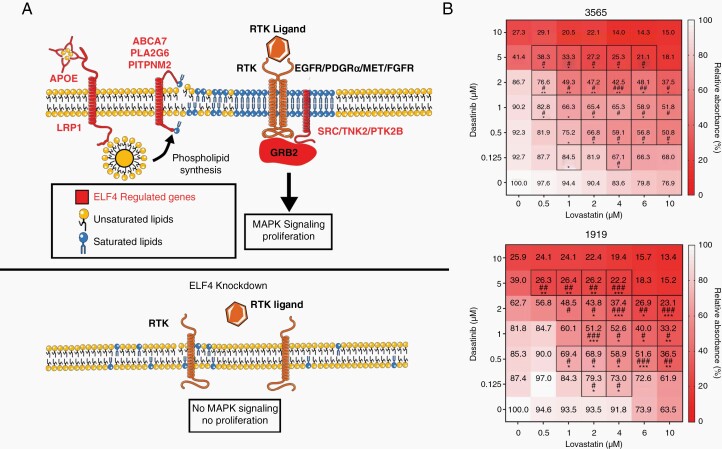
RTK signaling and lipid homeostasis are two codependent pathways that feed one another, with one set of genes increasing phospholipid pools, and another being directly responsible for RTK signaling. As a positive regulator of interconnected genes, ELF4 functions as a bridge factor between these two pathways (Figure 6A). Based on ELF4’s mechanism of action in GBM, we tested the concept of targeting these two codependent pathways simultaneously by combining an RTK inhibitor (Dasatinib) with an inhibitor of lipid homeostasis (Lovastatin). We treated mesenchymal and proneural GSCs with different combinations and measured viability 7 days later using an MTS assay (Figure 6B).25 We found that the two drugs synergized (based on either Bliss or Loewe’s model of synergy), suggesting that targeting these two pathways simultaneously is detrimental to GSCs.25
Figure 6.
Model of ELF4 in GBM reveals synergy between SRC and lipid inhibitors (A) Model of ELF4 in GBM. Normal RTK signaling and lipid dynamics occurs in the presence of ELF4. Knockdown of ELF4 results in disrupted RTK signaling and lipid dynamics and no proliferation. (B) Dose–response matrices for mesenchymal and proneural GSCs 168 h after treatment with different doses of dasatinib and/or lovastatin. Viability was measured with an MTS assay. Boxed in combinations are considered synergistic based on either Bliss (*) or Loewe’s (#) model of synergy which were calculated by Combenfit.25 */#P < .05, **/##P < .01, ***/###P < .001.
Discussion
Glioblastoma remains lethal cancer despite advances in genomics that improved classification and identified prognostic markers and oncogenic drivers.3,4,26 Understanding the regulatory networks contributing to cancer phenotypes is critical in the identification of novel options for therapeutic intervention.27 The tumor suppressor miRNAs miR-124, miR-128, and miR-137 work synergistically as agents promoting neuronal differentiation; acting in part by repressing a network of transcription factors. In glioblastoma, the absence of these three miRNAs leads to increased expression of this set of transcription factors followed by activation of oncogenic pathways.10 Our screening revealed ELF4 as one of the most critical members of this network. ELF4’s importance in cell cycle progression has been demonstrated in different cell types including neuroblastoma, hematopoietic, and glioma cells where ELF4 interacts with several cell cycle kinases.16,24,28 Our analysis established a new oncogenic route for ELF4, demonstrating that this transcription factor regulates two connected processes, GBM receptor signaling and lipid homeostasis, which ultimately influences cell proliferation.
Receptor tyrosine kinases (RTKs) are a highly conserved family of cell surface receptors that form dimers to respond to external ligands.29 They are involved in a wide range of processes, including proliferation, survival, metabolism, and differentiation.30 In GBM, aberrant RTK signaling is seen in approximately 67% of tumors, with alterations appearing in RTKs such as EGFR, PDGFRA/B, MET, and FGFR2/3.4 Single-cell analyses of GBM tumors further highlighted the importance of RTK signaling, as three of the four distinct cellular subtypes identified are driven by either EGFR, PDGFR or NF1 alterations.5 GBMs are highly dependent upon RTK signaling; however, as membrane-bound proteins, RTKs are themselves dependent upon proper lipid dynamics. The cell membrane is a heterogeneous lipid bilayer that dynamically varies in composition to fit the cell needs, forming lipid rafts or membrane microdomains. These rafts act as signaling focal points, being enriched in receptors and different lipid classes.31 In GBM cells, EGFR signaling is highly dependent upon these lipid rafts which are stabilized by several saturated phosphatidylcholines species.12 Likewise, EGFR signaling in glioma stem cells relies upon a pool of polyunsaturated fatty acids.13 Lipid homeostasis is essential for proper RTK signaling, ELF4 simultaneously controls both by regulating several highly connected genes in each pathway.
RTK signaling feeds into several signaling cascades, including the dynamic mitogen-activated protein kinase (MAPK) pathway, which can regulate cell proliferation, differentiation, and death.32,33 MAPK dysregulation is seen in a variety of cancers including GBM.33 ELF4 knockdown resulted in the downregulation of several MAPK-related genes, such as p38, GRB2, SHC2, PTK2B, TNK2, and SRC. Interestingly, many of these MAPK-related genes fall into two protein classes, being either adaptors or non-receptor tyrosine kinases. For example, GRB2 and SHC2 are adaptors that work together to prime RTKs for quick activation upon ligand stimulation.34 On the other hand, non-RTKs like PTK2B, TNK2, and SRC are of equal importance even though they lack extracellular domains, as they aid in RTK dimerization and propagate RTK signaling on the cytoplasmic side of the membrane.35,36 These proteins display promiscuity, in that they facilitate the function of various RTKs such as EGFR, PDGFR, and MET.35,36 For example, EGFR and MET dimerization are dependent upon SRC activity,36 while TNK2 (Ack1) is stimulated by both EGF and PDGF and regulates EGFR trafficking.37,38 Furthermore, PTK2B (FAK2 and PYK2) works with SRC to phosphorylate EGFR and activate MAPK activity.39,40 GBMs can be driven by several different oncogenic RTKs, yet ELF4 appears to be essential for GBM identity. Our data suggest that this essentiality is likely due to ELF4’s regulation of promiscuous signaling adaptors and non-RTKs, which facilitate the signaling of any RTK that a certain GBM cell may be dependent upon.
Lipid dynamics directly impact receptor functionality, and alterations in lipid species affect downstream RTK signaling.12,13 In addition to finding that ELF4 regulates RTK-relevant genes, we identified multiple lipid efflux genes to be direct targets of ELF4. Lipidomics revealed that ELF4 knockdown decreases phosphatidylcholine (PC) and phosphatidylethanolamine (PE) pools, suggesting that the downregulated lipid-associated genes are responsible for maintaining phospholipid levels. Among the genes identified, APOE, ABCA7, PITPNM2, PLA2G6, and LRP1 stand out. APOE and LRP1, for example, work together to transport lipids into the cell.41 ABCA7, PITPNM2, and PLA2G6 emerge as key ELF4 targets due to their ability to regulate phospholipid dynamics. ABCA7 is an ATP-binding cassette transporter that flips phospholipids, including PC and PE, to the exocytoplasmic leaflet of membranes.42 PLA2G6 on the other hand is a calcium-independent phospholipase A2 that regulates phospholipid membrane turnover and directly hydrolyzes PC.43 Finally, PITPNM2 is a membrane-associated phosphatidylinositol (PI) transfer protein that transfers PI and PC between membranes.44 Interestingly PITPNM2 is phosphorylated by ELF4-regulated non-RTK PT2KB further supporting the codependency between RTK signaling and lipid dynamics.30 Supporting ELF4’s role in lipid homeostasis, is a study where transgenic overexpression of ELF4 in osteoblasts caused abnormal adipogenesis in the bone marrow with cells accumulating lipid droplets.45 Additionally, SERPINE1 (PAI-1), a gene that regulates both RTK signaling and lipid homeostasis,46,47 decreases following ELF4 knockdown further connecting the two processes. In summary, we found that ELF4 depletion results in the downregulation of multiple lipid efflux genes which coincides with a global decrease in several important phospholipid classes, implicating ELF4 as an important regulator of lipid dynamics in GBM cells.
Our results highlight the importance of ELF4 in GBM cell identity, by directly regulating RTK signaling and lipid homeostasis. While pharmacological inhibition of transcription factors remains a challenge, our characterization of ELF4’s mechanism of action in GBM suggests that targeting two codependent pathways simultaneously may mimic ELF4 inhibition. RTK inhibitors have been tested in GBM, yet have yielded mixed results, impart due to the different RTKs expressed and heterogeneity of GBMs.48 Oncogenic SRC emerged as an important target of ELF4. Supporting this hypothesis is a pan-cancer study of high ELF4 tumors, which found that ELF4 high expressing cells were only sensitive to SRC-family inhibitors out of 397 anti-cancer drugs tested, indicating that high ELF4 expressing cells are dependent upon SRC signaling.49 Although phase-II trials for SRC inhibitors in GBM failed,50 our results indicate that combining SRC inhibitor along with an inhibitor of lipid efflux genes, such as ABCA7, PITPNM2, and/or PLA2G6, will likely be synergistic, mimicking genetic knockdown of ELF4. This approach is supported by our finding that Dasatinib and Lovastatin synergize in vitro. Alternatively, neurogenic miRNAs that target ELF4, such as miR-124 and miR-128, may prove useful in treating GBM.10 Many transcription factors have been implicated in the regulation of RTK signaling (eg, c-MYC) or lipid homeostasis (eg, PPARgamma), yet ELF4 emerges as one of the only transcription factors to simultaneously control two codependent pathways.
Supplementary Material
Acknowledgments
RNA-seq data were generated at the Genome Sequencing Facility, which is supported by UT Health San Antonio, NIH-NCI P30 CA054174 (Mays Cancer Center, UT Health San Antonio), NIH Shared Instrument grant 1S10OD021805-01, and CPRIT Core Facility Award RP160732. We thank Servier Medical Art for the Graphic templates.
Contributor Information
Adam Kosti, Greehey Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA; Department of Cell Systems and Anatomy, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA.
Jennifer Chiou, Department of Nutritional Sciences, Dell Pediatric Research Institute, Dell Medical School, The University of Texas at Austin, Austin, Texas, USA.
Gabriela D A Guardia, Centro de Oncologia Molecular, Hospital Sirio-Libanes, São Paulo, Brazil.
Xiufen Lei, Greehey Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA; Department of Nutritional Sciences, Dell Pediatric Research Institute, Dell Medical School, The University of Texas at Austin, Austin, Texas, USA.
Henriette Balinda, Department of Medicine, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA.
Tesha Landry, Greehey Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA.
Xiyuan Lu, Greehey Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA; Department of Nutritional Sciences, Dell Pediatric Research Institute, Dell Medical School, The University of Texas at Austin, Austin, Texas, USA.
Mei Qiao, Greehey Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA.
Andrea Gilbert, Department of Pathology, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA.
Andrew Brenner, Department of Medicine, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA.
Pedro A F Galante, Centro de Oncologia Molecular, Hospital Sirio-Libanes, São Paulo, Brazil; Departamento de Bioquimica, Instituto de Química, Universidade de São Paulo, São Paulo, Brazil.
Stefano Tiziani, Department of Nutritional Sciences, Dell Pediatric Research Institute, Dell Medical School, The University of Texas at Austin, Austin, Texas, USA.
Luiz O F Penalva, Greehey Children’s Cancer Research Institute, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA; Department of Cell Systems and Anatomy, University of Texas Health Science Center at San Antonio, San Antonio, Texas, USA.
Funding
This work was supported by grants from the National Institutes of Health [1 R21 NS113344-01A1], the Owens Foundation and the Joe and Teresa Lozano School of Medicine at UTHSCSA to L.O.F.P., and Serrapilheira Foundation and Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) [2018/15579-8] to P.A.F.G. A.K. was supported by [2R01 HG006015S1] and the Greehey Foundation. G.D.A.G. was supported by a fellowship from FAPESP [2017/19541-2].
Conflict of interest statement. The authors have no conflicts to report.
Author contributions
A.K. performed most of the biological experiments, contributed to data analysis, figure generation, and manuscript writing. J.C. and X.L. performed the metabolite extraction and lipidomics analysis. G.D.A.G. performed the genomic analyses and studies with patient data. M.Q. contributed to the siRNA screening experiments. A.B. designed in vivo experiments. H.B. performed in vivo experiments and conducted data analysis. X.L. and T.L. generated cell lines, conducted in vitro experiments and help with bioinformatic analyses. A.G. conducted pathological analyses of tumor samples. P.A.F.G., S.T., and L.O.F.P. designed the experiments, analyzed the data, and wrote the manuscript. All authors contributed to the manuscript.
References
- 1. Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 2. Weller M, Felsberg J, Hartmann C, et al. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J Clin Oncol. 2009;27(34):5743–5750. [DOI] [PubMed] [Google Scholar]
- 3. Verhaak RG, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brennan CW, Verhaak RG, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Piwecka M, Rolle K, Belter A, et al. Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Mol Oncol. 2015;9(7):1324–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ahir BK, Ozer H, Engelhard HH, Lakka SS. MicroRNAs in glioblastoma pathogenesis and therapy: a comprehensive review. Crit Rev Oncol Hematol. 2017;120:22–33. [DOI] [PubMed] [Google Scholar]
- 8. Bhaskaran V, Nowicki MO, Idriss M, et al. The functional synergism of microRNA clustering provides therapeutically relevant epigenetic interference in glioblastoma. Nat Commun. 2019;10(1):442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Santos MC, Tegge AN, Correa BR, et al. miR-124, -128, and -137 orchestrate neural differentiation by acting on overlapping gene sets containing a highly connected transcription factor network. Stem Cells. 2016;34(1):220–232. [DOI] [PubMed] [Google Scholar]
- 10. Kosti A, Barreiro R, Guardia GDA, et al. Synergism of proneurogenic miRNAs provides a more effective strategy to target glioma stem cells. Cancers (Basel). 2021;13(2). Article No.: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arora S, Rana R, Chhabra A, Jaiswal A, Rani V. miRNA-transcription factor interactions: a combinatorial regulation of gene expression. Mol Genet Genomics. 2013;288(3–4):77–87. [DOI] [PubMed] [Google Scholar]
- 12. Bi J, Ichu TA, Zanca C, et al. Oncogene amplification in growth factor signaling pathways renders cancers dependent on membrane lipid remodeling. Cell Metab. 2019;30(3):525–538.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gimple RC, Kidwell RL, Kim LJY, et al. Glioma stem cell-specific superenhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov. 2019;9(9):1248–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hubert CG, Rivera M, Spangler LC, et al. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76(8):2465–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mao P, Joshi K, Li J, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110(21):8644–8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kosti A, Du L, Shivram H, et al. ELF4 is a target of miR-124 and promotes neuroblastoma proliferation and undifferentiated state. Mol Cancer Res. 2020;18(1):68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34(5):525–527. [DOI] [PubMed] [Google Scholar]
- 18. Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ando M, Kawazu M, Ueno T, et al. Mutational landscape and antiproliferative functions of ELF transcription factors in human cancer. Cancer Res. 2016;76(7):1814–1824. [DOI] [PubMed] [Google Scholar]
- 21. Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wu H, Southam AD, Hines A, Viant MR. High-throughput tissue extraction protocol for NMR- and MS-based metabolomics. Anal Biochem. 2008;372(2):204–212. [DOI] [PubMed] [Google Scholar]
- 23. Hassn Mesrati M, Behrooz AB, A YA, Syahir A. Understanding glioblastoma biomarkers: knocking a mountain with a hammer. Cells. 2020;9(5). Article No.: 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bazzoli E, Pulvirenti T, Oberstadt MC, et al. MEF promotes stemness in the pathogenesis of gliomas. Cell Stem Cell. 2012;11(6):836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Di Veroli GY, Fornari C, Wang D, et al. Combenefit: an interactive platform for the analysis and visualization of drug combinations. Bioinformatics. 2016;32(18):2866–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Correa BR, de Araujo PR, Qiao M, et al. Functional genomics analyses of RNA-binding proteins reveal the splicing regulator SNRPB as an oncogenic candidate in glioblastoma. Genome Biol. 2016;17(1):125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guardia GDA, Correa BR, Araujo PR, et al. Proneural and mesenchymal glioma stem cells display major differences in splicing and lncRNA profiles. NPJ Genom Med. 2020;5:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lacorazza HD, Yamada T, Liu Y, et al. The transcription factor MEF/ELF4 regulates the quiescence of primitive hematopoietic cells. Cancer Cell. 2006;9(3):175–187. [DOI] [PubMed] [Google Scholar]
- 29. Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141(7):1117–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lev S, Hernandez J, Martinez R, et al. Identification of a novel family of targets of PYK2 related to drosophila retinal degeneration B (rdgB) protein. Mol Cell Biol. 1999;19(3):2278–2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18(6):361–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morrison DK. MAP kinase pathways. Cold Spring Harb Perspect Biol. 2012;4(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Braicu C, Buse M, Busuioc C, et al. A comprehensive review on MAPK: a promising therapeutic target in cancer. Cancers (Basel). 2019;11(10). Article No.: 1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Belov AA, Mohammadi M. Grb2, a double-edged sword of receptor tyrosine kinase signaling. Sci Signal. 2012; 5(249):pe49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saito Y, Haendeler J, Hojo Y, Yamamoto K, Berk BC. Receptor heterodimerization: essential mechanism for platelet-derived growth factor-induced epidermal growth factor receptor transactivation. Mol Cell Biol. 2001;21(19):6387–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mueller KL, Yang ZQ, Haddad R, Ethier SP, Boerner JL. EGFR/Met association regulates EGFR TKI resistance in breast cancer. J Mol Signal. 2010;5:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones S, Cunningham DL, Rappoport JZ, Heath JK. The non-receptor tyrosine kinase Ack1 regulates the fate of activated EGFR by inducing trafficking to the p62/NBR1 pre-autophagosome. J Cell Sci. 2014;127(Pt 5):994–1006. [DOI] [PubMed] [Google Scholar]
- 38. Galisteo ML, Yang Y, Urena J, Schlessinger J. Activation of the nonreceptor protein tyrosine kinase Ack by multiple extracellular stimuli. Proc Natl Acad Sci USA. 2006;103(26):9796–9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dikic I, Tokiwa G, Lev S, Courtneidge SA, Schlessinger J. A role for Pyk2 and Src in linking G-protein-coupled receptors with MAP kinase activation. Nature. 1996;383(6600):547–550. [DOI] [PubMed] [Google Scholar]
- 40. Liu J, Liao Z, Camden J, et al. Src homology 3 binding sites in the P2Y2 nucleotide receptor interact with Src and regulate activities of Src, proline-rich tyrosine kinase 2, and growth factor receptors. J Biol Chem. 2004;279(9):8212–8218. [DOI] [PubMed] [Google Scholar]
- 41. Bu G. Apolipoprotein E and its receptors in Alzheimer’s disease: pathways, pathogenesis and therapy. Nat Rev Neurosci. 2009;10(5):333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quazi F, Molday RS. Differential phospholipid substrates and directional transport by ATP-binding cassette proteins ABCA1, ABCA7, and ABCA4 and disease-causing mutants. J Biol Chem. 2013;288(48): 34414–34426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Engel LA, Jing Z, O’Brien DE, Sun M, Kotzbauer PT. Catalytic function of PLA2G6 is impaired by mutations associated with infantile neuroaxonal dystrophy but not dystonia-parkinsonism. PLoS One. 2010;5(9):e12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vihtelic TS, Goebl M, Milligan S, O’Tousa JE, Hyde DR. Localization of drosophila retinal degeneration B, a membrane-associated phosphatidylinositol transfer protein. J Cell Biol. 1993;122(5):1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baek K, Cho JY, Hwang HR, et al. Myeloid Elf-1-like factor stimulates adipogenic differentiation through the induction of peroxisome proliferator-activated receptor gamma expression in bone marrow. J Cell Physiol. 2012;227(11):3603–3612. [DOI] [PubMed] [Google Scholar]
- 46. Freytag J, Wilkins-Port CE, Higgins CE, et al. PAI-1 mediates the TGF-beta1+EGF-induced “scatter” response in transformed human keratinocytes. J Invest Dermatol. 2010;130(9):2179–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Levine JA, Oleaga C, Eren M, et al. Role of PAI-1 in hepatic steatosis and dyslipidemia. Sci Rep. 2021;11(1):430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pearson JRD, Regad T. Targeting cellular pathways in glioblastoma multiforme. Signal Transduct Target Ther. 2017;2:17040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kafita D, Daka V, Nkhoma P, et al. High ELF4 expression in human cancers is associated with worse disease outcomes and increased resistance to anticancer drugs. PLoS One. 2021;16(4):e0248984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Galanis E, Anderson SK, Twohy EL, et al. A phase 1 and randomized, placebo-controlled phase 2 trial of bevacizumab plus dasatinib in patients with recurrent glioblastoma: Alliance/North Central Cancer Treatment Group N0872. Cancer. 2019;125(21):3790–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.