Abstract
A number of transcriptional regulators mediate their effects through direct contact with the ς70 subunit of Escherichia coli RNA polymerase (RNAP). In particular, several regulators have been shown to contact a C-terminal portion of ς70 that harbors conserved region 4. This region of ς contains a putative helix-turn-helix DNA-binding motif that contacts the −35 element of ς70-dependent promoters directly. Here we report the use of a recently developed bacterial two-hybrid system to study the interaction between the putative anti-ς factor Rsd and the ς70 subunit of E. coli RNAP. Using this system, we found that Rsd can interact with an 86-amino-acid C-terminal fragment of ς70 and also that amino acid substitution R596H, within region 4 of ς70, weakens this interaction. We demonstrated the specificity of this effect by showing that substitution R596H does not weaken the interaction between ς and two other regulators shown previously to contact region 4 of ς70. We also demonstrated that AlgQ, a homolog of Rsd that positively regulates virulence gene expression in Pseudomonas aeruginosa, can contact the C-terminal region of the ς70 subunit of RNAP from this organism. We found that amino acid substitution R600H in ς70 from P. aeruginosa, corresponding to the R596H substitution in E. coli ς70, specifically weakens the interaction between AlgQ and ς70. Taken together, our findings suggest that Rsd and AlgQ contact similar surfaces of RNAP present in region 4 of ς70 and probably regulate gene expression through this contact.
Sigma factors are subunits of bacterial RNA polymerase (RNAP) that direct the holoenzymes that contain them to promoters of a specific class (20). In Escherichia coli there are seven different species of ς factors, and ς70 is the principal ς factor (27). The presence of different types of ς factors within a cell with distinct DNA sequence binding specificities provides a mechanism for coordinate regulation of genes that are controlled by promoters of the same class. Competition between different ς factors for the available RNAP core enzyme in part determines which genes are transcribed within a cell at any given time (27). This competition can be influenced by anti-ς factors, which are regulatory proteins that bind ς factors and often prevent their association with the RNAP core enzyme (19, 26). Anti-ς factors ultimately inhibit transcription from the class of promoters recognized by the ς factors that they sequester.
The ς70 subunit of RNAP participates in a number of protein-protein interactions, including interactions with other subunits of the polymerase complex (43, 52) and interactions with transcriptional regulators (18, 23). The regulators that interact with ς70 often contact a region of ς that contains a putative helix-turn-helix DNA-binding motif responsible for contacting the −35 elements of ς70-dependent promoters (18, 23, 37). This DNA-binding region of ς is conserved in members of the ς70 family of proteins and is called region 4 (36).
Recently, Jishage and Ishihama identified a protein in E. coli that was preferentially made by cells during the stationary phase of growth and was associated with the ς70 subunit of RNAP in stationary-phase extracts (28). The protein was named Rsd (which stands for regulator of sigma D) since it was found to associate specifically with ς70 (but not with several alternative ς factors) and was shown to be capable of inhibiting ς70-dependent transcription from certain promoters in vitro (28). The binding site for Rsd on ς70 was mapped to a C-terminal tryptic fragment encompassing conserved region 4 (28). On the basis of these observations and because the synthesis of Rsd coincides with the general shutdown in ς70-dependent transcription that occurs as cells enter the stationary phase of growth, Jishage and Ishihama suggested that Rsd might be an anti-ς factor (28). Subsequent work has shown that consistent with this idea, Rsd may facilitate the replacement of ς70 by the stationary-phase-specific ς factor ς38 in functional RNAP holoenzyme complexes as cells go from the exponential phase to the stationary phase of growth (29).
The sequence of putative anti-ς factor Rsd is similar to the sequence of a regulator of alginate production in Pseudomonas aeruginosa called AlgQ (or AlgR2) (28). Alginate is an important virulence factor that imparts the characteristic mucoid phenotype to P. aeruginosa isolated from the lungs of cystic fibrosis patients (17). P. aeruginosa isolates from other sources are typically nonmucoid and do not exhibit activated expression of genes involved in alginate production (10). However, the production of alginate is believed to promote survival of P. aeruginosa in the special environment of the lungs of cystic fibrosis patients, contributing to resistance to both immune responses and antibiotics (10). AlgQ was originally identified as a positive transcriptional regulator of the key alginate biosynthetic gene algD (8, 32), which is expressed at high levels in mucoid cells. The regulation of the algD gene is complex and involves two different ς factors; transcription initiates from two superimposed promoters, one of which is recognized by RNAP containing ςE (AlgU/AlgT) and the other of which is recognized by RNAP containing ς54 (RpoN) (3, 9, 11, 21, 38, 49, 59). Furthermore, at least one DNA-binding protein, AlgR (AlgR1), is known to bind to specific sites upstream of the algD promoter and activate transcription (31, 41, 42). The mechanism by which AlgQ positively regulates transcription of the algD gene is not known.
We were interested in testing the idea that like Rsd, AlgQ interacts with region 4 of the ς70 subunit of RNAP. In this study we tested this idea explicitly by using a bacterial two-hybrid system. We found that both Rsd and AlgQ can interact with a ς70 moiety encompassing region 4 in vivo, and we identified an amino acid substitution in region 4 that specifically weakens the interaction of Rsd and AlgQ with the ς moiety.
MATERIALS AND METHODS
Plasmids and strains.
E. coli XL1-blue (Stratagene) was used as the recipient strain for all plasmid constructions. E. coli KS1 harbors on its chromosome the lac promoter derivative placOR2-62 driving expression of a linked lacZ reporter gene and has been described previously (14). E. coli SF1 harbors an F′ episome containing the lac promoter derivative plac OR2-55/Cons −35 driving expression of a linked lacZ reporter gene and has also been described previously (13).
Plasmids pACλcI, pACΔcI, pBRα, pBRα-ς38, pBRα-ς70, and pBRα-ς70 (R596H) have all been described previously (13, 14). Plasmid pACλcI-Rsd encodes λcI (residues 1 to 236) fused to Rsd (residues 1 to 158) via three alanine residues. pACλcI-Rsd was made by cloning the appropriate NotI-BamHI-digested PCR product into NotI-BstYI-digested pACλcI32 (24); expression of the cI-rsd fusion gene was therefore under the control of the lacUV5 promoter. Plasmid pACλcI-AlgQ encodes λcI (residues 1 to 236) fused to AlgQ (residues 1 to 160) via three alanine residues, and it was made in a manner similar to the manner in which pACλcI-Rsd was made. Expression of the cI-algQ fusion gene on pACλcI-AlgQ is also under the control of the lacUV5 promoter. Plasmid pACλcI-AsiA encodes λcI (residues 1 to 236) fused to AsiA from bacteriophage T4 (residues 1 to 90) via three alanine residues, and it was made in a manner similar to the manner in which pACλcI-Rsd was made. Plasmid pACΔ-35λcI-AsiA contains the cI-asiA fusion gene from plasmid pACλcI-AsiA under the control of a lacUV5 promoter variant in which the −35 element of the promoter has been deleted. Plasmid pACΔ-35λcI-AsiA, therefore, expresses less of the λcI-AsiA fusion protein than plasmid pACλcI-AsiA expresses under identical conditions. pACΔ-35λcI-AsiA was made by cloning the appropriate HindIII-BstYI fragment from pACλcI-AsiA into plasmid pA3B2 (57) cut with both HindIII and BstYI. Plasmid pBRα-ς70PA encodes residues 1 to 248 of the α subunit of E. coli RNAP fused to residues 532 to 617 of the ς70 subunit of P. aeruginosa RNAP. The hybrid α-ς70PA gene was made by performing PCR and was cloned into HindIII-BamHI-digested pBRα. Expression of the chimeric gene on pBRα-ς70PA is, therefore, under the control of tandem lpp and lacUV5 promoters. pBRα-ς70PA (R600H) is a derivative of pBRα-ς70PA in which the R600H substitution in the ς moiety of the chimera was introduced by PCR.
The lac promoter derivative placCOP-93+OL2-62 was made by the PCR and contains two λ operators; a near-consensus λ operator (TACCACCGGCGGTGATA) and OL2 (CAACACCGCCAGAGATA) are centered 93 and 62 bp, respectively, upstream of the transcriptional start site of the lac core promoter. Plasmid pFW11-COP-93+OL2-62 was constructed by cloning an EcoRI-HindIII-cut PCR product containing placCOP-93+OL2-62 into pFW11 (56) cut with EcoRI and HindIII. Plasmid pFW11-COP-93+OL2-62 was then transformed into strain CSH100, and the promoter-lacZ fusion was recombined onto an F′ episome and mated into strain FW102 (56) to create reporter strain F′93+62. The PCR-amplified regions of all plasmids were sequenced to confirm that no errors had been introduced as a result of the PCR process.
Experimental procedures.
Cells were grown in LB supplemented with kanamycin (50 μg/ml), chloramphenicol (25 μg/ml), carbenicillin (50 μg/ml), and isopropyl-β-d-thiogalactoside (IPTG) at the concentration indicated. Cells were permeabilized with sodium dodecyl sulfate-CHCl3 and assayed for β-galactosidase activity essentially as described previously (39). Assays were performed at least three times in duplicate on separate occasions, and representative data sets are shown below. The values are averages based on one experiment; duplicate measurements differed by less than 10%.
RESULTS
λcI-Rsd activates transcription from a test promoter in the presence of a chimeric α-subunit harboring region 4 of E. coli ς70.
We sought to detect an interaction between Rsd and region 4 of ς70 in vivo by using a bacterial two-hybrid system that we had recently developed (12, 14). This bacterial two-hybrid system is based on the finding that any sufficiently strong interaction between a protein bound upstream of a suitable test promoter and a component of RNAP can activate transcription in E. coli (12, 14). Thus, two proteins that interact with one another can mediate transcriptional activation in E. coli provided that one protein is fused to a DNA-binding protein and the other is fused to a component of RNAP (12, 14).
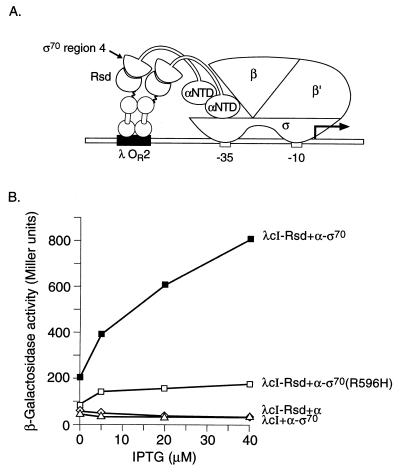
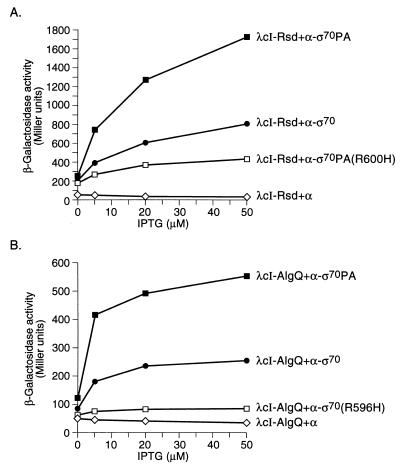
Our strategy for detecting an interaction between Rsd and region 4 of ς70 involved the use of two chimeric proteins, one comprising Rsd fused to the repressor of bacteriophage λ (λcI) and the other comprising a modified form of the α-subunit of RNAP in which the C-terminal domain (CTD) of α has been replaced by a C-terminal fragment of ς70. We reasoned that RNAP containing the resulting α-ς70 chimera would display a target for Rsd that could be contacted by a DNA-bound λcI-Rsd dimer (Fig. 1A). Having fused the entire Rsd protein (residues 1 to 158) to the C terminus of λcI, we placed the gene encoding this chimeric protein on a plasmid vector downstream of the IPTG-inducible lacUV5 promoter, thus creating plasmid pACλcI-Rsd. We used plasmid pBRα-ς70 as a source of the α-ς70 chimera. This plasmid encodes a chimera in which residues 528 to 613 of E. coli ς70 are fused to residues 1 to 248 of the α-subunit of RNAP (13). We introduced plasmids pACλcI-Rsd and pBRα-ς70 into E. coli KS1 (14), which harbors on its chromosome the lac promoter derivative placOR2-62 (bearing a single λ operator centered 62 bp upstream of the transcriptional start site) linked to a lacZ reporter gene. We then tested the ability of the λcI-Rsd chimera to activate transcription from the placOR2-62 test promoter in the presence of the α-ς70 chimera. Figure 1B shows that λcI-Rsd activated transcription from the test promoter up to ∼24-fold in cells containing the α-ς70 chimera compared to control cells containing only wild-type α. An additional control revealed that λcI (lacking the fused Rsd moiety) did not activate transcription from the test promoter in the presence of the α-ς70 chimera (Fig. 1B). We also found that λcI-Rsd did not activate transcription from the test promoter in the presence of an α-ς38 chimera (comprising residues 1 to 248 of α fused to residues 243 to 330 of ς38) encoded by plasmid pBRα-ς38 (13; data not shown).
FIG. 1.
Transcriptional activation by λcI-Rsd in the presence of the α-ς70 chimera. (A) Replacement of the RNAP α-CTD by a C-terminal fragment of E. coli ς70 (residues 528 to 613) permits interaction with the Rsd moiety of a λcI-Rsd chimera bound to DNA. The diagram depicts the test promoter placOR2-62, which bears the λ operator OR2 centered 62 bp upstream from the transcriptional start site of the lac core promoter. In reporter strain KS1 the placOR2-62 test promoter is located on the chromosome and drives expression of a linked lacZ gene. The N-terminal domain of α is designated αNTD. (B) Effect of λcI-Rsd on transcription in vivo from placOR2-62 in the presence of the α-ς70 or α-ς70(R596H) chimera. KS1 cells harboring compatible plasmids expressing the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity.
Substitution R596H in the ς moiety of the α-ς70 chimera weakens the interaction between ς and Rsd
Our ability to link the protein-protein interaction between Rsd and its target on ς70 to transcriptional activation provided us with a useful genetic tool for dissecting this interaction. We were particularly interested in identifying mutant forms of ς70 that were specifically defective in the ability to interact with Rsd. We therefore introduced a variety of amino acid substitutions into the ς moiety of the α-ς70 chimera and tested the effects of these substitutions on the ability of the λcI-Rsd fusion protein to mediate transcriptional activation from the test promoter. Figure 1B shows that substitution R596H in the ς moiety of the α-ς70 chimera strongly reduced the magnitude of λcI-Rsd-dependent activation (to a factor of ∼5). Substitution R596A in the ς moiety of the α-ς70 chimera had a nearly identical effect on the magnitude of the activation (data not shown). In contrast, substitutions L573A, E591A, E591Q, H600A, and H600R did not decrease the magnitude of λcI-Rsd-dependent activation (data not shown).
Substitution R596H in the ς moiety of the α-ς70 chimera does not compromise the ability of a superactivating variant of λcI to stimulate transcription from an appropriate test promoter.
To determine whether the R596H substitution affects the interaction of ς70 with Rsd specifically, we tested its effect on the ability of another protein to interact with the ς moiety of the chimera. We showed recently that the λcI protein (a transcriptional activator as well as a repressor) can interact specifically with the ς moiety of the α-ς70 chimera and stabilize its binding to a promoter −35 element (13). The in vivo assay which we designed to detect the interaction between λcI and region 4 of ς70 is shown in Fig. 2A. In this experimental setup a DNA-bound λcI dimer activates transcription from test promoter placOR2-55/Cons-35 by stabilizing the binding of the ς moiety of the α-ς70 chimera to the ectopic −35 element present upstream of the core promoter elements (13) (Fig. 2A). Transcriptional activation from this test promoter is dependent not only on the protein-protein interaction between λcI and the tethered ς moiety but also on the protein-DNA interaction between the tethered ς moiety and the ectopic −35 element (13).
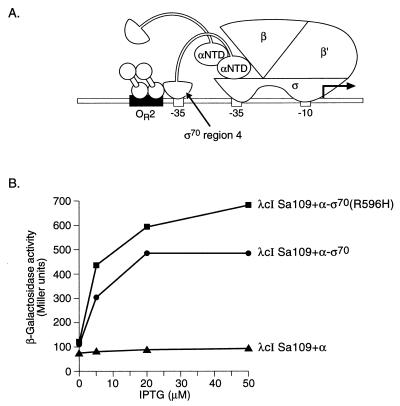
FIG. 2.
Transcriptional activation by λcI superactivator in the presence of the α-ς70 chimera. (A) λcISa109 stabilizes the binding of region 4 of ς70 to an ectopic −35 element. The diagram depicts the test promoter placOR2-55/Cons-35, which bears an ectopic −35 element and the λ operator OR2 centered 45.5 and 55 bp, respectively, upstream from the transcriptional start site of the lac core promoter. In reporter strain SF1 the placOR2-55/Cons-35 test promoter is located on an F′ episome and drives expression of a linked lacZ gene. (B) Effect of substitution R596H in the ς moiety of the α-ς70 chimera on the ability of λcISa109 to activate transcription in vivo from placOR2-55/Cons-35. SF1 cells harboring compatible plasmids expressing the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity.
We tested whether the R596H substitution in the α-ς70 chimera has an effect on the ability of a superactiviating variant of λcI to activate transcription from test promoter placOR2-55/Cons-35 (Fig. 2A). Reporter strain SF1 carries promoter placOR2-55/Cons-35 fused to the lacZ gene in single copy on an F′ episome (13). We assayed the ability of λcI superactivator 109 (λcISa109) (5, 13) to activate transcription from this reporter in the presence of the α-ς70 chimera with or without the R596H substitution in the ς moiety. λcISa109 activated transcription a maximum of ∼5.5-fold from the test promoter in the presence of the α-ς70 chimera and a maximum of ∼7.5-fold in the presence of the chimera harboring the R596H substitution (Fig. 2B). (Although the R596H substitution has previously been shown to inhibit the ability of wild-type λcI to activate transcription from PRM [35], we have observed that this substitution does not inhibit the ability of λcISa109 to activate transcription from PRM [see below].) We concluded that substitution R596H in the ς moiety of the α-ς70 chimera does not result in a general defect in the ability of the ς moiety to interact with other proteins.
We also tested the effect of the R596A substitution in the ς moiety of the α-ς70 chimera on the ability of λcISa109 to activate transcription from the same test promoter. Unlike the R596H substitution, the R596A substitution significantly reduced the magnitude of the activation by λcISa109 (data not shown). Using another assay, however, we were able to determine that this reduction in activation was not due to a general defect caused by the R596A substitution (see below).
λcI-AlgQ activates transcription from a test promoter in the presence of a chimeric α-subunit harboring region 4 of P. aeruginosa ς70.
Jishage and Ishihama (28) noted that Rsd exhibits 31% identity with the alginate regulatory protein AlgQ (also known as AlgR2) from P. aeruginosa. AlgQ was originally identified as a positive regulator of alginate production in P. aeruginosa (8, 32) but has subsequently been shown to regulate several other gene products in this organism (see below).
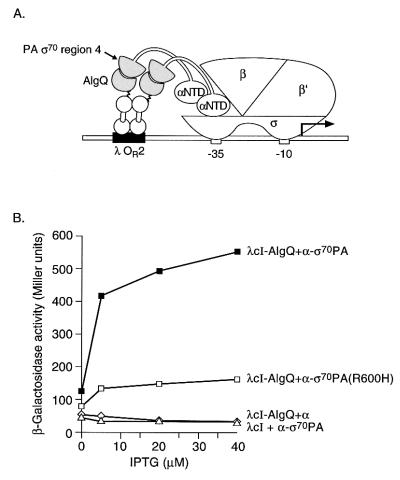
Very little is known about how AlgQ mediates its effects on gene expression. In order to test the hypothesis that AlgQ, like Rsd, can interact with region 4 of ς70, we made a cI-algQ fusion gene analogous to the cI-rsd fusion gene. We also made a fusion gene encoding a protein analogous to the α-ς70 chimera that contained region 4 of ς70 from P. aeruginosa. We fused the entire AlgQ protein (residues 1 to 160) to the C terminus of λcI. We placed the gene encoding this chimeric protein on a plasmid vector downstream of the IPTG-inducible lacUV5 promoter, creating plasmid pACλcI-AlgQ. We then fused residues 532 to 617 of P. aeruginosa ς70 (equivalent to residues 528 to 613 of E. coli ς70) to residues 1 to 248 of α. The resulting chimera was called α-ς70PA. The hybrid α-ς70PA gene encoding this chimera was placed on a plasmid vector downstream of tandemly arranged lpp and lacUV5 promoters, creating plasmid pBRα-ς70PA. We introduced plasmids pACλcI-AlgQ and pBRα-ς70PA into E. coli KS1. We then tested the ability of the λcI-AlgQ chimera to activate transcription from placOR2-62 in the presence of the α-ς70PA chimera (Fig. 3A). Figure 3B shows that λcI-AlgQ activated transcription from the test promoter a maximum of ∼17-fold in cells containing the α-ς70PA chimera compared to control cells containing only wild-type α. An additional control revealed that λcI without the fused AlgQ moiety did not activate transcription from the test promoter in the presence of the α-ς70PA chimera (Fig. 3B).
FIG. 3.
Transcriptional activation by λcI-AlgQ in the presence of the α-ς70PA chimera. (A) Replacement of the RNAP α-CTD by a C-terminal fragment of P. aeruginosa ς70 (residues 532 to 617) permits interaction with the AlgQ moiety of a λcI-AlgQ chimera bound to DNA. The diagram depicts the test promoter placOR2-62, which bears the λ operator OR2 centered 62 bp upstream from the transcriptional start site of the lac core promoter. In reporter strain KS1 the placOR2-62 test promoter is located on the chromosome and drives expression of a linked lacZ gene. (B) Effect of λcI-AlgQ on transcription in vivo from placOR2-62 in the presence of the α-ς70PA or α-ς70PA(R600H) chimera. KS1 cells harboring compatible plasmids expressing the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity.
Substitution R600H in the ς moiety of the α-ς70PA chimera weakens the interaction between ς and AlgQ.
The ς70 subunits from E. coli and P. aeruginosa are very similar to one another, exhibiting ∼83% identity over the length of the ς fragments (86 amino acids) that we used in our experiments (36). We wanted to test whether substitution R600H in ς70 from P. aeruginosa, which corresponds to the R596H substitution in E. coli ς70, had any effect on the ability of λcI-AlgQ to activate transcription in the presence of the α-ς70PA chimera. To do this, we made a version of the α-ς70PA chimera harboring substitution R600H [α-ς70PA(R600H)] and assayed the ability of λcI-AlgQ to activate transcription in KS1 cells expressing this chimera. Figure 3B shows that λcI-AlgQ activated transcription from the reporter gene a maximum of ∼5-fold in the presence of the α-ς70PA(R600H) chimera, compared to a maximum of ∼17-fold with the chimera derived from the wild-type form of P. aeruginosa ς70.
Substitution R600H in the ς moiety of the α-ς70PA chimera does not compromise the ability of a superactivating variant of λcI to stimulate transcription from an appropriate test promoter.
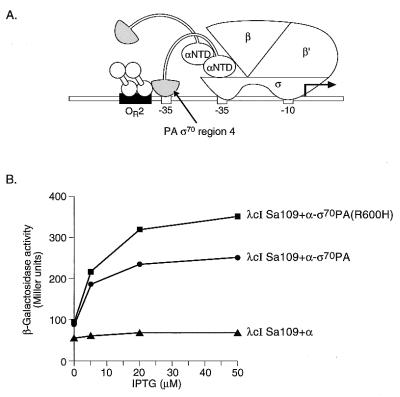
In order to assess whether the effect of the R600H substitution was specific for the interaction between ς70PA and AlgQ, we first tested whether λcISa109 could interact with the ς moiety of the α-ς70PA chimera. We found that λcISa109 stimulated transcription from test promoter placOR2-55/Cons-35 up to ∼3.5-fold specifically in the presence of the α-ς70PA chimera (Fig. 4). Furthermore, introduction of the R600H substitution into the ς moiety of the α-ς70PA chimera did not abrogate the stimulatory effect of λcISa109 (instead it resulted in a modest increase in the observed activation), suggesting that the effect of the R600H substitution is specific for the λcI-AlgQ chimera (Fig. 4B).
FIG. 4.
Transcriptional activation by λcI superactivator in the presence of the α-ς70PA chimera. (A) λcISa109 stabilizes the binding of region 4 of ς70 from P. aeruginosa to an ectopic −35 element. The diagram depicts the test promoter placOR2-55/Cons-35, which bears an ectopic −35 element and the λ operator OR2 centered 45.5 and 55 bp, respectively, upstream from the transcriptional start site of the lac core promoter. In reporter strain SF1 the placOR2-55/Cons-35 test promoter is located on an F′ episome and drives expression of a linked lacZ gene. (B) Effect of substitution R600H in the ς moiety of the α-ς70PA chimera on the ability of λcISa109 to activate transcription in vivo from placOR2-55/Cons-35. SF1 cells harboring compatible plasmids expressing the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity.
The E. coli protein Rsd can interact with region 4 of ς70 from P. aeruginosa, and the P. aeruginosa protein AlgQ can interact with region 4 of ς70 from E. coli.
Given the high degree of similarity between the regions of ς70 from E. coli and P. aeruginosa used in our experiments, we thought that each regulator might be able to contact region 4 of ς70 from either E. coli or P. aeruginosa. We explicitly tested whether Rsd could interact with region 4 of ς70 from P. aeruginosa and also whether AlgQ could interact with region 4 of ς70 from E. coli. Figure 5A shows that the λcI-Rsd chimera activated transcription from the test promoter in KS1 cells a maximum of ∼52-fold in the presence of the α-ς70PA chimera, compared to ∼24-fold in the presence of the E. coli α-ς70 chimera. We also found that introduction of substitution R600H into the α-ς70PA chimera reduced the ability of λcI-Rsd to stimulate transcription from the test promoter to a factor of ∼13 (Fig. 5A).
FIG. 5.
Rsd can interact with region 4 of ς70 from P. aeruginosa, and AlgQ can interact with region 4 of ς70 from E. coli. (A) Effect of λcI-Rsd on transcription in vivo from placOR2-62 in the presence of the α-ς70PA, α-ς70PA(R600H), or α-ς70 chimera. KS1 cells harboring compatible plasmids expressing the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity. (B) Effect of λcI-AlgQ on transcription in vivo from placOR2-62 in the presence of the α-ς70, α-ς70(R596H), or α-ς70PA chimera. KS1 cells harboring compatible plasmids expressing the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity.
Figure 5B shows that the λcI-AlgQ chimera activated transcription from the test promoter in KS1 cells a maximum of ∼7-fold in the presence of the α-ς70 chimera, compared to ∼15-fold in the presence of the P. aeruginosa α-ς70PA chimera. Introduction of substitution R596H into the ς moiety of the E. coli α-ς70 chimera reduced the ability of λcI-AlgQ to activate transcription from the test promoter to a factor of ∼2 (Fig. 5B).
λcI-AsiA activates transcription from a test promoter in the presence of either α-ς chimera.
We took advantage of another protein known to interact with region 4 of E. coli ς70 to test further the specificity of the effect of the R596H substitution on the interaction of ς70 with either Rsd or AlgQ. The AsiA protein encoded by bacteriophage T4 is an anti-ς factor that has been shown to interact specifically and very strongly with region 4 of E. coli ς70 (1, 7, 22, 40, 50, 51). AsiA is thought to work by interacting with ς70 that is free in solution rather than with ς70 that is already complexed with the RNAP core enzyme (22). The interaction between AsiA and E. coli ς70 inhibits the activity of RNAP holoenzyme (containing the AsiA-ς70 complex) by preventing region 4 of ς from contacting the −35 element of ς70-dependent promoters (7, 51).
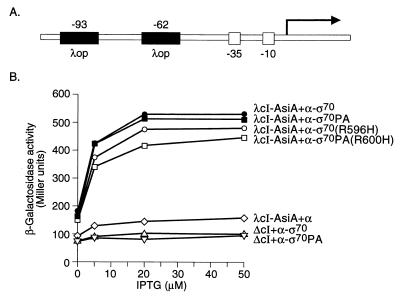
We therefore sought to detect the interaction between AsiA and region 4 of ς70 by fusing AsiA to λcI and measuring the ability of the resulting λcI-AsiA chimera to activate transcription from placOR2-62 in the presence of either the α-ς70 chimera or the α-ς70PA chimera. We fused the entire AsiA protein (residues 1 to 90) to the C terminus of λcI. We placed the gene encoding this chimeric protein on a plasmid vector downstream of the IPTG-inducible lacUV5 promoter, creating plasmid pACλcI-AsiA. Initial experiments demonstrated that the λcI-AsiA chimera was extremely toxic to cells at the levels provided by the expression vector pACλcI-AsiA (data not shown). For subsequent experiments we constructed a different expression vector, pACΔ-35λcI-AsiA, in which the chimeric cI-asiA gene was under control of an IPTG-inducible variant of the lacUV5 promoter that lacked the normal −35 element and therefore directed the synthesis of smaller amounts of the fusion protein. Since pACΔ-35λcI-AsiA made insufficient levels of λcI-AsiA to saturate the λ operator present on the placOR2-62 promoter construct, we also constructed a new test promoter bearing two relatively strong λ operators in the upstream region (Fig. 6A; also see Materials and Methods). Figure 6B shows that the λcI-AsiA chimera activated transcription from the modified test promoter in the presence of the α-ς70 chimera derived from ς70 of either E. coli or P. aeruginosa (similar findings with the α-ς70 chimera from E. coli have been obtained by S. Pande and D. Hinton [submitted for publication]). The data also show that substitution R596H in the E. coli ς moiety or the corresponding R600H substitution in the P. aeruginosa ς moiety had only a modest effect on the ability of the λcI-AsiA chimera to activate transcription from placCOP-93+OL2-62. Similarly, the R596A substitution in the E. coli ς moiety did not reduce the stimulatory effect of the λcI-AsiA chimera (data not shown). Since these substitutions do not appear to cause nonspecific defects in the abilities of the ς moieties to interact with other proteins, we suggest that residue R596 of E. coli ς70 is at or near the contact surface for Rsd and the equivalent residue of P. aeruginosa ς70, R600, is at or near the contact surface for AlgQ (see Discussion). Furthermore, the finding that substitution R596A in E. coli ς70 results in a strong defect in the ability of ς to interact with Rsd suggests that the arginine side chain may make an energetically significant contact with Rsd.
FIG. 6.
Transcriptional activation by λcI-AsiA in the presence of the α-ς chimeras. (A) Schematic diagram of test promoter placCOP-93+OL2-62 used to determine the effects of substitutions in the α-ς chimeras on transcriptional activation by λcI-AsiA. The test promoter placCOP-93+OL2-62 bears both a consensus λ operator and OL2 centered 93 and 62 bp, respectively, upstream from the transcriptional start site of the lac core promoter. In reporter strain F′93+62 the placCOP-93+OL2-62 test promoter is located on an F′ episome and drives expression of a linked lacZ gene. (B) Effect of λcI-AsiA on transcription in vivo from placCOP-93+OL2-62 in the presence of the α-ς chimeras. F′93+62 cells harboring compatible plasmids expressing the indicated proteins were grown in the presence of different concentrations of IPTG and assayed for β-galactosidase activity. ΔcI refers to the fact that no λcI was made by control plasmid pACΔcI.
DISCUSSION
Rsd and AlgQ can interact with region 4 of ς70 in vivo.
We found that the E. coli Rsd protein and the P. aeruginosa AlgQ protein can interact in vivo with a C-terminal fragment of the ς70 subunit of E. coli and P. aeruginosa RNAP, respectively. This fragment of ς70 encompasses conserved region 4, which contains a DNA-binding domain that mediates recognition of the −35 element of ς70-dependent promoters (18). Furthermore, each protein can also interact with the corresponding ς70 fragment from the heterologous organism. We also identified a single amino acid substitution (R596H and the corresponding substitution R600H in E. coli and P. aeruginosa ς70, respectively) that inhibits the binding of both Rsd and AlgQ to either ς70 fragment. The interchangeability of the E. coli and P. aeruginosa ς70 fragments in our experiments suggests that regulators from P. aeruginosa or E. coli that interact with region 4 of ς70 might in general be expected to function in either organism. In fact, a previous in vitro study showed that the λcI protein (which contacts region 4) can activate transcription from the λ promoter PRM by the P. aeruginosa RNAP (16).
Analysis of the interaction between Rsd and region 4 of ς70.
Our bacterial two-hybrid results for Rsd and region 4 of ς70 are consistent with the biochemical data of Jishage and Ishihama (28). These authors found that Rsd could interact with ς70 in vitro but not with ς38 (or several other alternative ς factors), and they showed that Rsd could interact with a tryptic fragment of ς70 composed of residues ∼500 to 613. We found that Rsd can interact in vivo with an 86-amino-acid C-terminal fragment of ς70 that contains region 4 but cannot interact with the equivalent 88-amino-acid C-terminal fragment of ς38. Jishage et al. (30) have recently reported that alanine substitutions at two positions flanking residue 596 (L595 and L598) disrupt the association of Rsd with ς70 in vitro. However, in contrast with our results, they reported that substitution R596A in ς70 did not prevent association of Rsd with ς70 (in a glutathione S-transferase pulldown assay). To better compare the results of Jishage et al. with our results, we introduced substitutions L595A and L598A into the ς moiety of the α-ς70 chimera and tested their effects on the interaction with Rsd in vivo. We found that both the L595A substitution and the L598A substitution strongly inhibited the interactions with Rsd and that their effects were more severe than that of the R596A substitution (data not shown). We suggest, therefore, that the in vivo assay may be more sensitive than the in vitro assay, allowing us to detect the less severe effect of the R596A substitution. Moreover, a structural model of region 4 of ς70 based on the crystal structure of the NarL protein (see below) suggests that the side chain of residue 595, at least, is likely to be partially buried. Substitutions at position 595 and possibly also at position 598 may affect the interaction with Rsd indirectly; consistent with this possibility, we found that substitutions L595A and L598A both completely eliminated the ability of λcISa109 to activate transcription from our artificial test promoter in the presence of the α-ς70 chimera (data not shown).
Amino acid substitution R596H has been isolated previously. It was first identified based on its effect on expression of the arabinose operon (25, 53). Expression of the ara genes is positively controlled by two regulators, the global regulator cyclic AMP receptor protein (CRP) and the operon-specific regulator AraC (15). CRP is active only when it is complexed with the small molecule effector cyclic AMP, and consequently mutant strains that are not able to synthesize cyclic AMP (cya mutants) cannot induce expression of CRP-dependent operons, including the arabinose operon. A mutation in the ς70 (rpoD) gene specifying the R596H substitution was isolated as a suppressor that restored expression of the arabinose operon in a cya mutant background (25). It has been suggested that the R596H substitution enhances the ability of AraC to interact productively with RNAP (25, 37). This same amino acid substitution was subsequently isolated based on its ability to suppress the effect of a λcI positive control mutation at the λ promoter PRM. Suppressor mutations in the rpoD gene were sought that would reverse the activation defect of a λcI mutant bearing substitution D38N in its activating region, and a single mutation specifying the R596H change was obtained (35). Although the R596H substitution in ς70 reduces the ability of wild-type λcI to activate transcription from PRM, we have shown that this substitution actually enhances the ability of λcISa109 (which bears a non-wild-type residue at position 38) to stimulate transcription from PRM (unpublished data). Evidently, certain amino acid-amino acid combinations at position 38 of λcI and position 596 of ς70 permit efficient activation, while others do not.
Taken together, the data obtained in previous studies and data obtained in this study suggest that R596 of ς70 is exposed on the surface of the polypeptide such that it can be contacted by interacting proteins. This suggestion is supported by the results of structural modeling. Region 4 of ς70 contains a putative helix-turn-helix motif, and the structure of this DNA-binding domain has been modeled based on the three-dimensional crystal structures of two related helix-turn-helix proteins, the E. coli NarL protein and the bacteriophage 434 Cro protein (4, 37). In both structural models, residue 596 is solvent exposed and accessible when the protein domain is bound to DNA.
AlgQ and regulation of gene expression in P. aeruginosa.
Our findings with AlgQ may be relevant to understanding the mechanism by which it regulates gene expression in P. aeruginosa. The finding that AlgQ, like Rsd, can interact with a C-terminal fragment of the ς70 subunit of RNAP provides strong support for the proposal that it is a functional homolog of Rsd. This conclusion is reinforced by our finding that the interactions of Rsd and AlgQ with region 4 of ς70 are both weakened by the same substitution in ς70.
The algQ (algR2) gene was originally identified as a regulatory gene implicated in activation of the algD promoter in experimentally derived mucoid strains of P. aeruginosa (8, 32). In particular, a putative algQ mutation was found to eliminate transcription from the algD promoter in a laboratory strain of P. aeruginosa (8). Furthermore, overexpression of algQ was shown to reverse the nonmucoid phenotype of spontaneous Alg− mutants derived from mucoid cystic fibrosis isolates of P. aeruginosa (32). Interestingly, expression of algQ was also shown to mediate strong activation of the algD promoter in E. coli cells grown under high-osmolarity conditions (32). Activation of the algD promoter has been shown to occur by at least two different mechanisms involving one of two alternative sigma factors, ςE and ς54 (3, 9, 11, 21, 38, 44, 49, 59). In particular, mutations that activate ςE (encoded by the algU gene) lead to increased expression of algD (reviewed in reference 9), but algD expression can also be activated by a ς54-dependent pathway (3). Our results support the idea that the effect of AlgQ on algD expression is likely to be indirect since there is no evidence that the ς70 form of RNAP can recognize the algD promoter. Possibly AlgQ functions as an anti-ς factor, increasing the amount of RNAP core that is available to bind the relevant alternative ς factor (either ςE or ς54), thereby increasing the occupancy of the algD promoter. It is interesting that algD gene expression is negatively controlled by an anti-ς factor (the product of the mucA gene), which is specific for ςE (44, 48, 58). Thus, most mucoid cystic fibrosis isolates of P. aeruginosa have been found to bear mutations in the mucA gene, which result in constitutive alginate production (2).
Our finding that AlgQ can bind to ς70 from E. coli as well as to ς70 from P. aeruginosa could be relevant to its ability to activate the algD promoter in E. coli (32). Nevertheless, it is possible that the role of AlgQ in activation of the algD promoter is unrelated to its ability to bind to ς70 in either P. aeruginosa or E. coli. Although AlgQ was originally reported to have a kinase activity (45), the subsequent finding that it regulates production of a kinase (nucleoside diphosphate kinase) which has a similar molecular weight suggests that AlgQ is not itself a kinase (47).
The regulatory effects of AlgQ are not limited to alginate production. For example, AlgQ regulates production of a variety of secretable virulence factors, up-regulating a neuraminidase and a siderophore and down-regulating extracellular proteases and a rhamnolipid biosurfactant (6, 46, 54). AlgQ also regulates production of Ndk (see above) and succinyl coenzyme A synthetase, an enzyme of the tricarboxylic acid cycle that forms a complex with Ndk in P. aeruginosa (33, 46, 47). Finally, characterization of an algQ null mutant revealed a dramatic loss of viability in the stationary phase of growth, as well as reductions in the intracellular concentrations of GTP, ppGpp, and inorganic polyphosphate (34). Although the molecular basis for these regulatory effects has not been defined, the pleiotropic nature of AlgQ-dependent phenotypes and our results support the suggestion that AlgQ is a global regulator of transcription in P. aeruginosa. In addition to its similarity to Rsd, AlgQ exhibits 58% identity with PfrA, a positive regulator of siderophore biosynthetic genes in Pseudomonas putida (54). Interestingly, both PfrA and a putative member of the extracytoplasmic function family of alternative ς factors (PfrI) are required for transcriptional activation of siderophore biosynthetic genes under iron limitation conditions in P. putida (54, 55).
Two-hybrid assay for the interaction of transcriptional regulators with region 4 of ς70.
The two-hybrid system that we used to study the interactions of Rsd and AlgQ with region 4 of ς70 should facilitate studies of other regulators that interact with this region of ς70 from E. coli, P. aeruginosa, or other bacteria. Use of the α-ς70 chimeras, in particular, could facilitate genetic analysis of these interactions by providing a convenient vehicle for mutagenesis of region 4 of ς70. Whereas isolation and analysis of rpoD mutations are complicated by the fact that ς70 is an essential protein that exerts global effects on cellular transcription, our α-ς chimeras exert their effects at specifically designed test promoters. Moreover, mutant chimeras can be assayed to determine their abilities to interact with a number of different regulators so that the specificities of their effects can be assessed.
ACKNOWLEDGMENTS
We thank Arne Rietsch for providing the P. aeruginosa PAO1 chromosomal DNA and Debbie Hinton for the gift of plasmid pEG-AsiA as a source of the asiA gene. We also thank Debbie Hinton for communicating results prior to publication and Cliff Boucher for helpful discussions.
This work was supported by National Institutes of Health grant GM44025, by an established investigatorship from the American Heart Association (to A.H.), and by a Charles A. King Trust postdoctoral fellowship (to S.L.D.).
REFERENCES
- 1.Adelman K, Orsini G, Kolb A, Graziani L, Brody E N. The interaction between the AsiA protein of bacteriophage T4 and the ς70 subunit of Escherichia coli RNA polymerase. J Biol Chem. 1997;272:27435–27443. doi: 10.1074/jbc.272.43.27435. [DOI] [PubMed] [Google Scholar]
- 2.Boucher J C, Yu H, Mudd M H, Deretic V. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect Immun. 1997;65:3838–3846. doi: 10.1128/iai.65.9.3838-3846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boucher J C, Schurr M J, Deretic V. Dual regulation of mucoidy in Pseudomonas aeruginosa and sigma factor antagonism. Mol Microbiol. 2000;36:341–351. doi: 10.1046/j.1365-2958.2000.01846.x. [DOI] [PubMed] [Google Scholar]
- 4.Busby S, Ebright R H. Promoter structure, promoter recognition and transcription activation in prokaryotes. Cell. 1994;79:743–746. doi: 10.1016/0092-8674(94)90063-9. [DOI] [PubMed] [Google Scholar]
- 5.Bushman F D, Shang C, Ptashne M. A single glutamic acid residue plays a key role in the transcriptional activation function of lambda repressor. Cell. 1989;58:1163–1171. doi: 10.1016/0092-8674(89)90514-x. [DOI] [PubMed] [Google Scholar]
- 6.Cacalano G, Kays M, Saiman L, Prince A. Production of the Pseudomonas aeruginosa neuraminidase is increased under hyperosmotic conditions and is regulated by genes involved in alginate expression. J Clin Invest. 1992;89:1866–1874. doi: 10.1172/JCI115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colland F, Orsini G, Brody E N, Buc H, Kolb A. The bacteriophage T4 AsiA protein: a molecular switch for sigma 70-dependent promoters. Mol Microbiol. 1998;27:819–829. doi: 10.1046/j.1365-2958.1998.00729.x. [DOI] [PubMed] [Google Scholar]
- 8.Deretic V, Konyecsni W M. Control of mucoidy in Pseudomonas aeruginosa: transcriptional regulation of algR and identification of the second regulatory gene, algQ. J Bacteriol. 1989;171:3680–3688. doi: 10.1128/jb.171.7.3680-3688.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deretic V, Schurr M J, Boucher J C, Martin D W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deretic V, Schurr M J, Yu H. Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 1995;3:351–356. doi: 10.1016/s0966-842x(00)88974-x. [DOI] [PubMed] [Google Scholar]
- 11.DeVries C A, Ohman D E. Mucoid-to-nonmucoid conversion in alginate-producing Pseudomonas aeruginosa often results from spontaneous mutations in algT, encoding a putative alternate sigma factor, and shows evidence for autoregulation. J Bacteriol. 1994;176:6677–6687. doi: 10.1128/jb.176.21.6677-6687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dove S L, Hochschild A. Conversion of the ω subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dove S L, Huang F W, Hochschild A. Mechanism for a transcriptional activator that works at the isomerization step. Proc Natl Acad Sci USA. 2000;97:13215–13220. doi: 10.1073/pnas.97.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dove S L, Joung J K, Hochschild A. Activation of prokaryotic transcription through arbitrary protein-protein contacts. Nature. 1997;386:627–630. doi: 10.1038/386627a0. [DOI] [PubMed] [Google Scholar]
- 15.Gallegos M T, Schleif R, Bairoch A, Hofman K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J G, Gussin G N. RNA polymerases from Pseudomonas aeruginosa and Pseudomonas syringae respond to Escherichia coli activator proteins. J Bacteriol. 1991;173:394–397. doi: 10.1128/jb.173.1.394-397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Govan J R W, Deretic V. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev. 1996;60:539–574. doi: 10.1128/mr.60.3.539-574.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross C A, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 19.Helmann J D. Anti-sigma factors. Curr Opin Microbiol. 1999;2:135–141. doi: 10.1016/S1369-5274(99)80024-1. [DOI] [PubMed] [Google Scholar]
- 20.Helmann J D, Chamberlin M J. Structure and function of bacterial sigma factors. Annu Rev Biochem. 1988;57:839–872. doi: 10.1146/annurev.bi.57.070188.004203. [DOI] [PubMed] [Google Scholar]
- 21.Hershberger C D, Ye R W, Parsek M R, Xie Z-D, Chakrabarty A M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative ς factor (ςE) Proc Natl Acad Sci USA. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinton D M, Vuthoori S. Efficient inhibition of Escherichia coli RNA polymerase by the bacteriophage T4 AsiA protein requires that AsiA binds first to free ς70. J Mol Biol. 2000;304:731–739. doi: 10.1006/jmbi.2000.4113. [DOI] [PubMed] [Google Scholar]
- 23.Hochschild A, Dove S L. Protein-protein contacts that activate and repress prokaryotic transcription. Cell. 1998;92:597–600. doi: 10.1016/s0092-8674(00)81126-5. [DOI] [PubMed] [Google Scholar]
- 24.Hu J, Kornacker M G, Hochschild A. Escherichia coli one- and two-hybrid systems for the analysis and identification of protein-protein interactions. Methods. 2000;20:80–94. doi: 10.1006/meth.1999.0908. [DOI] [PubMed] [Google Scholar]
- 25.Hu J C, Gross C A. Mutations in the sigma subunit of E. coli RNA polymerase which affect positive control of transcription. Mol Gen Genet. 1985;199:7–13. doi: 10.1007/BF00327502. [DOI] [PubMed] [Google Scholar]
- 26.Hughes K T, Mathee K. The anti-sigma factors. Annu Rev Microbiol. 1998;52:231–286. doi: 10.1146/annurev.micro.52.1.231. [DOI] [PubMed] [Google Scholar]
- 27.Ishihama A. Functional modulation of Escherichia coli RNA polymerase. Annu Rev Microbiol. 2000;54:499–518. doi: 10.1146/annurev.micro.54.1.499. [DOI] [PubMed] [Google Scholar]
- 28.Jishage M, Ishihama A. A stationary phase protein in Escherichia coli with binding activity to the major ς subunit of RNA polymerase. Proc Natl Acad Sci USA. 1998;95:4953–4958. doi: 10.1073/pnas.95.9.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jishage M, Ishihama A. Transcriptional organization and in vivo role of the Escherichia coli rsd gene, encoding the regulator of RNA polymerase sigma D. J Bacteriol. 1999;181:3768–3776. doi: 10.1128/jb.181.12.3768-3776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jishage M, Dasgupta D, Ishihama A. Mapping of the Rsd contact site on the sigma 70 subunit of Escherichia coli RNA polymerase. J Bacteriol. 2001;183:2952–2956. doi: 10.1128/JB.183.9.2952-2956.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kato J, Chakrabarty A M. Purification of the regulatory protein AlgR1 and its binding in the far upstream region of the algD promoter in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1991;88:1760–1764. doi: 10.1073/pnas.88.5.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kato J, Chu L, Kitano K, DeVault J D, Kimbara K, Chakrabarty A M, Misra T K. Nucleotide sequence of a regulatory region controlling alginate synthesis in Pseudomonas aeruginosa: characterization of the algR2 gene. Gene. 1989;84:31–38. doi: 10.1016/0378-1119(89)90136-4. [DOI] [PubMed] [Google Scholar]
- 33.Kavanaugh-Black A, Connolly D M, Chugani S A, Chakrabarty A M. Characterization of nucleoside-diphosphate kinase from Pseudomonas aeruginosa: complex formation with succinyl-CoA synthetase. Proc Natl Acad Sci USA. 1994;91:5883–5887. doi: 10.1073/pnas.91.13.5883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H-Y, Schlictman D, Shankar S, Xie Z, Chakrabarty A M, Kornberg A. Alginate, inorganic polyphosphate, GTP and ppGpp synthesis co-regulated in Pseudomonas aeruginosa: implications for stationary phase survival and synthesis of RNA/DNA precursors. Mol Microbiol. 1998;27:717–725. doi: 10.1046/j.1365-2958.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Moyle H, Susskind M M. Target of the transcriptional activation function of phage λ cI protein. Science. 1994;263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 36.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lonetto M A, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase ς70 subunit. J Mol Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
- 38.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 40.Minakhin L, Camarero J A, Holford M, Parker C, Muir T W, Severinov K. Mapping the molecular interface between the ς70 subunit of E. coli RNA polymerase and T4 AsiA. J Mol Biol. 2001;306:631–642. doi: 10.1006/jmbi.2001.4445. [DOI] [PubMed] [Google Scholar]
- 41.Mohr C D, Leveau J H J, Krieg D P, Hibler N S, Deretic V. AlgR-binding sites within the algD promoter make up a set of inverted repeats separated by a large intervening segment of DNA. J Bacteriol. 1992;174:6624–6633. doi: 10.1128/jb.174.20.6624-6633.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohr C D, Hibler N S, Deretic V. AlgR, a response regulator controlling mucoidy in Pseudomonas aeruginosa, binds to the FUS sites of the algD promoter located unusually far upstream from the mRNA start site. J Bacteriol. 1991;173:5136–5143. doi: 10.1128/jb.173.16.5136-5143.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owens J T, Miyake R, Murakami K, Chmura A J, Fujita N, Ishihma A, Meares C F. Mapping the ς70 subunit contact sites on Escherichia coli RNA polymerase with a ς70-conjugated chemical protease. Proc Natl Acad Sci USA. 1998;95:6021–6026. doi: 10.1073/pnas.95.11.6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rowen D W, Deretic V. Membrane-to-cytosol redistribution of ECF sigma factor AlgU and conversion to mucoidy in Pseudomonas aeruginosa isolates from cystic fibrosis patients. Mol Microbiol. 2000;36:314–327. doi: 10.1046/j.1365-2958.2000.01830.x. [DOI] [PubMed] [Google Scholar]
- 45.Roychoudhury S, Sakai K, Chakrabarty A M. AlgR2 is an ATP/GTP-dependent protein kinase involved in alginate synthesis by Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1992;89:2659–2663. doi: 10.1073/pnas.89.7.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schlictman D, Kubo M, Shankar S, Chakrabarty A M. Regulation of nucleoside diphosphate kinase and secretable virulence factors in Pseudomonas aeruginosa: roles of algR2 and algH. J Bacteriol. 1995;177:2469–2474. doi: 10.1128/jb.177.9.2469-2474.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlictman D, Kavanaugh-Black A, Shankar S, Chakrabarty A M. Energy metabolism and alginate biosynthesis in Pseudomonas aeruginosa: role of the tricarboxylic acid cycle. J Bacteriol. 1994;176:6023–6029. doi: 10.1128/jb.176.19.6023-6029.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schurr M J, Yu H, Martinez-Salazar J M, Boucher J C, Deretic V. Control of AlgU, a member of the ςE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schurr M J, Yu H, Martinez-Salazar J M, Hibler N S, Deretic V. Biochemical characterization and posttranslational modification of AlgU, a regulator of stress response in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1995;216:874–880. doi: 10.1006/bbrc.1995.2703. [DOI] [PubMed] [Google Scholar]
- 50.Severinov K, Muir T. Expressed protein ligation: a novel method to study protein-protein interactions in transcription. J Biol Chem. 1998;273:16205–16209. doi: 10.1074/jbc.273.26.16205. [DOI] [PubMed] [Google Scholar]
- 51.Severinova E, Severinov K, Darst S A. Inhibition of Escherichia coli RNA polymerase by bacteriophage T4 AsiA. J Mol Biol. 1998;279:9–18. doi: 10.1006/jmbi.1998.1742. [DOI] [PubMed] [Google Scholar]
- 52.Sharp M M, Chan C L, Lu C Z, Marr M T, Nechaev S, Merritt E W, Severinov K, Roberts J W, Gross C A. The interface of sigma with core RNA polymerase is extensive, conserved, and functionally specialized. Genes Dev. 2000;13:3015–3026. doi: 10.1101/gad.13.22.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silverstone A E, Goman M, Scaife J G. ALT: a new factor involved in the synthesis of RNA by Escherichia coli. Mol Gen Genet. 1972;118:223–234. doi: 10.1007/BF00333459. [DOI] [PubMed] [Google Scholar]
- 54.Venturi V, Ottevanger C, Leong J, Weisbeek P J. Identification and characterization of a siderophore regulatory gene (pfrA) of Pseudomonas putida WCS358: homology to the alginate regulatory gene algQ of Pseudomonas aeruginosa. Mol Microbiol. 1993;10:63–73. doi: 10.1111/j.1365-2958.1993.tb00904.x. [DOI] [PubMed] [Google Scholar]
- 55.Venturi V, Ottevanger C, Bracke M, Weisbeek P. Iron regulation of siderophore biosynthesis and transport in Pseudomonas putida WCS358: involvement of a transcriptional activator and of the Fur protein. Mol Microbiol. 1995;15:1081–1093. doi: 10.1111/j.1365-2958.1995.tb02283.x. [DOI] [PubMed] [Google Scholar]
- 56.Whipple F W. Genetic analysis of prokaryotic and eukaryotic DNA-binding proteins in E. coli. Nucleic Acids Res. 1998;26:3700–3706. doi: 10.1093/nar/26.16.3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whipple F W, Kuldell N H, Cheatham L A, Hochschild A. Specificity determinants for the interaction of λ repressor and P22 repressor dimers. Genes Dev. 1994;8:1212–1223. doi: 10.1101/gad.8.10.1212. [DOI] [PubMed] [Google Scholar]
- 58.Xie Z D, Hershberger C D, Shankar S, Ye R W, Chakrabarty A M. Sigma factor–anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu H, Schurr M J, Deretic V. Functional equivalence of Escherichia coli ςE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J Bacteriol. 1995;177:3259–3268. doi: 10.1128/jb.177.11.3259-3268.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]