Significance
Post COVID-19 induced pulmonary fibrosis is the most severe of a number of clinical symptoms associated with “long COVID”, a condition which is currently incompletely understood but affects millions given the ongoing COVID-19 pandemic. Using systems biology and mechanistic studies in a humanized mouse model of COVID lung fibrosis, we identify the essential genetic and immunologic perturbations occurring in patients with long COVID lung fibrosis and reveal promising therapeutic targets.
Keywords: long COVID pulmonary fibrosis, innate immunity, immune checkpoint therapy, humanized mouse model
Abstract
COVID-19 remains a global pandemic of an unprecedented magnitude with millions of people now developing “COVID lung fibrosis.” Single-cell transcriptomics of lungs of patients with long COVID revealed a unique immune signature demonstrating the upregulation of key proinflammatory and innate immune effector genes CD47, IL-6, and JUN. We modeled the transition to lung fibrosis after COVID and profiled the immune response with single-cell mass cytometry in JUN mice. These studies revealed that COVID mediated chronic immune activation reminiscent to long COVID in humans. It was characterized by increased CD47, IL-6, and phospho-JUN (pJUN) expression which correlated with disease severity and pathogenic fibroblast populations. When we subsequently treated a humanized COVID lung fibrosis model by combined blockade of inflammation and fibrosis, we not only ameliorated fibrosis but also restored innate immune equilibrium indicating possible implications for clinical management of COVID lung fibrosis in patients.
The coronavirus disease 2019 (COVID-19) outbreak caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to impact human health long after a large proportion of the population has been vaccinated (1–3). Available data indicate immune system activation in the early phase of infections characterized by acute respiratory distress syndrome (ARDS) (4). Post-ARDS survival, a significant portion of patients develop pulmonary inflammation and fibrosis (PIF) (COVID lung fibrosis) (5–8); however, mechanisms aimed at identifying the underlying pathophysiology of long COVID lung fibrosis are scarce (7). A common feature in COVID lung fibrosis is the development of progressive, fibrotic, irreversible interstitial lung disease characterized by declining lung function, increased fibrosis, and increased morbidity and mortality (9). Understanding the immunological mechanisms driving COVID lung fibrosis is an essential step to determine potential treatment targets and design rational strategies to effectively treat COVID lung fibrosis.
Fibrosis is defined by a deposition of excessive amounts of extracellular matrix proteins in and around an inflamed or injured tissue. Instead of structural reorganization, tissue is progressively destroyed, which leads to a loss of organ function (10, 11). Despite intensive research, the etiology of pulmonary fibrosis is still unclear. A lack of understanding of molecular mechanisms, etiology, and progression that underlie this condition makes it a life-threatening disease. In the current paradigm of pulmonary fibrosis pathogenesis, many distinct triggers can contribute to the development of pulmonary fibrotic disease (12, 13). However, regardless of the initiating events, a feature common to all fibrotic diseases is the activation of extracellularmatrix (ECM)-producing myofibroblasts, which are the key mediators of fibrotic tissue remodeling. It is now clear that several pathogenic mechanisms have been implicated in its development: vascular leakage, fibroblast recruitment and activation, myofibroblast differentiation, epithelial-to-mesenchymal transition, and immune cell dysfunction among others. Fibrosis-associated inflammation is a double-edged sword. While a strong early inflammatory response promotes fibrosis, late-onset inflammation inhibits the profibrotic phenotype (10, 14).
While most studies that focused on the immune system of COVID-19 using peripheral blood mononuclear cells (PBMCs) or bronchoalveolar lavage fluid samples demonstrate an immune response characterized by the secretion of proinflammatory cytokines, like IL-6 and TNF-α (15, 16), others have shown recruitment of innate immune cells like myeloid cells (17–20) and changes in adaptive immunity as T cells (21–23) and natural killer (NK) cells (24). Yet, we still lack comprehensive insights into the immunopathology of post–severe COVID-19 human lung tissue. Furthermore, the exact mechanisms governing the pathophysiology of COVID lung fibrosis and questions pertaining to the differential susceptibility of individuals to this condition remain undefined.
In this study, we first analyzed the single-cell transcriptomic profile of COVID-19 lung tissue to identify the innate immune mediators of COVID lung fibrosis. Our results demonstrate unique cellular composition and features of myeloid cells and fibroblasts. Matured neutrophils and monocyte-derived alveolar macrophages (MoAM) are strikingly increased accompanied by elevated IL-6 production in SARS-CoV-2–infected lungs. Of particular interest, we observed that the SARS-CoV-2 infection modulated profibrotic gene signatures like JUN and CD47 in fibroblasts. Using histopathology and mass spectrometry, we further describe an alteration between the innate immunity and fibrotic cell landscape post–COVID-19 fibrotic progression in humans. This pathogenic phenotype can be recapitulated in our COVID lung fibrosis mouse model and a human lung organoid xenograft model. Our conventional experimental models provide support for in vivo evaluation of human immune therapies. Thus, using a systems biologic approach combined with histopathological and therapeutic studies in vivo, we show that a combination therapy involving antibody-mediated blockade of both IL-6 and CD47 can be used to treat pulmonary fibrosis in COVID-19 patients. In all, we provide a plausible mechanism underlying the progression of pulmonary fibrosis in SARS-CoV-2–infected patients and suggest treatment strategies that involve targeting both the inflammatory response pathways and immune suppressive pathways in vivo. Our results are significant because we provide preclinical data to suggest a therapeutic regimen that has currently impaired almost one fourth of the world’s population. Additionally, our results suggest an assay that could potentially be used to prescreen COVID-19 patient samples for the presence of fibrosis that can warrant intervention in clinic.
Results
Longitudinal Analysis of Patients 21 and 103 d after COVID-19 Demonstrates Innate Immune Activation Accompanied by a Profibrotic Program.
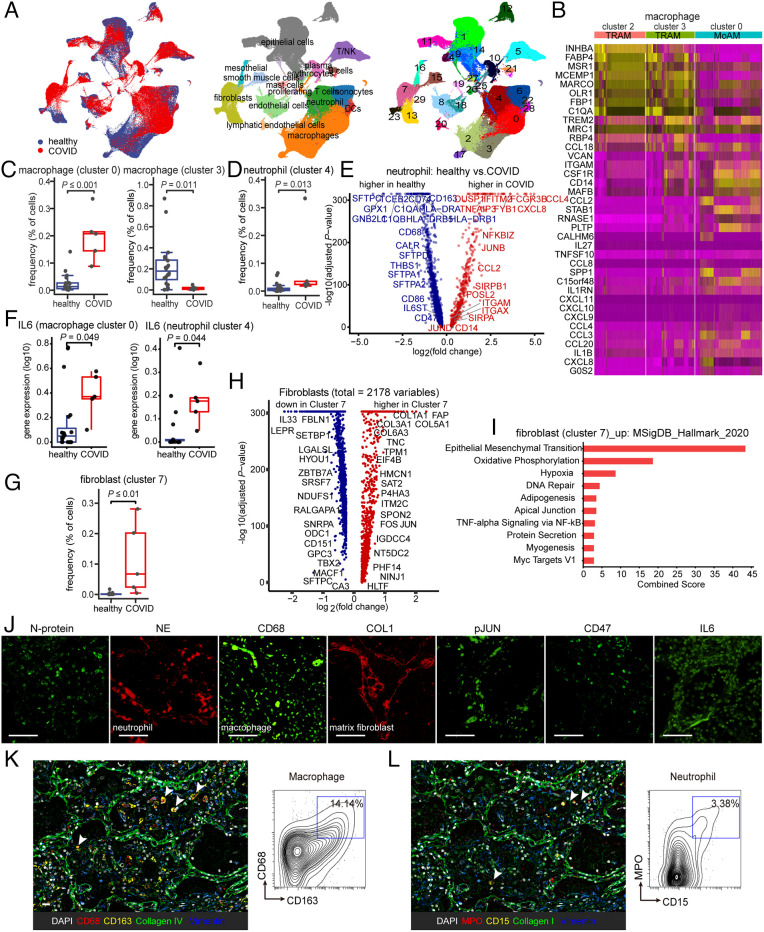
Coronavirus disease 2019 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Although most of the patients infected with SARS-CoV-2 are either asymptomatic or have mild symptoms and fully recover, a significant number of patients suffer from long COVID syndrome, a number of conditions which have been linked to COVID-19. Micro-CT (7) and further pathological analysis (25) showed clinical features that resembled end-stage pulmonary fibrosis in such patients. Owing to the existing conundrum underlying SARS-CoV-2–mediated pulmonary fibrosis, we analyzed lung tissues from independent patient cohorts published in GEO GSE149878 (26), GSE122960 (27), GSE163919, and GSE158127 (25). We studied the alignment of cell populations in lung tissue from five patients with COVID and 20 control patients with hypoxic brain injury, but otherwise, healthy lungs and the clinical demographic data are shown in Table 1. In total, high-quality single-cell transcriptomes of 251,612 cells from 5 COVID human lungs and 20 healthy human control lungs were assessed, and their population structure was visualized using Uniform Manifold Approximation and Projection (UMAP). Unsupervised cluster analysis separated cell populations into 29 clusters (Fig. 1A and featured genes are shown in SI Appendix, Fig. S1 and Dataset S1). Total leukocytes comprising neutrophils, macrophages, and T cells were significantly changed in all COVID lungs and fibroblasts which showed a very distinct distribution (SI Appendix, Fig. S2). The annotation resolved multiple clusters corresponding to macrophages subdividing clusters 0, 2, and 3 as alveolar macrophages (AM) and cluster 17 as interstitial macrophages (SI Appendix, Fig. S3A). Further clustering of the AM-related genes allowed us to identify clusters 2 and 3 as tissue-resident alveolar macrophages (TRAM). Conversely, cluster 0 is characterized as monocyte-derived alveolar macrophages (MoAM) by gradually increasing expression of alveolar macrophage maturation markers (Fig. 1B and SI Appendix, Fig. S3B). Cluster 0 is noted highly expressed in COVID lungs, whereas cluster 3 is enriched in healthy lungs (Fig. 1C, P value showing a significant difference between healthy and COVID patients). All these results indicated that the macrophage compartment is particularly affected in the lungs of long COVID; most alveolar macrophages are lost, but additionally, monocytes undergo recruitment and differentiate into alveolar macrophages during this ongoing lung injury.
Table 1.
Clinical characteristics of human lung specimens used in this study
| GEO accession no. | Age (yrs) | Symptoms | Pathology | Diagnosis/Cause of death | Gender | Race | Tissue |
|---|---|---|---|---|---|---|---|
| GSE149878 | 66 | hypoxia | Long COVID lung fibrosis | Long COVID, Death at Day 37 | M | As | lung |
| GSE163919 | 19 | hypoxia | Long COVID lung fibrosis | Long COVID, Lung tx at Day 70 | unknown | K | lung |
| GSE158127 | 28 | hypoxia | Long COVID lung fibrosis | Long COVID, Lung tx at Day 21 | F | His | lung |
| GSE158127 | 62 | hypoxia | Long COVID lung fibrosis | Long COVID, Lung tx at Day 103 | M | K | lung |
| GSE158127 | 43 | hypoxia | Long COVID lung fibrosis | Long COVID, Lung tx at Day 90 | M | K | lung |
| GSE122960 | 63 | neurologic Sx | no fibrosis | Stroke | F | AA | lung |
| GSE122960 | 55 | neurologic Sx | no fibrosis | Intracranial hemorrhage | M | As | lung |
| GSE122960 | 29 | neurologic Sx | no fibrosis | Anoxic brain injury | F | AA | lung |
| GSE122960 | 57 | neurologic Sx | no fibrosis | Anoxic brain injury | F | AA | lung |
| GSE122960 | 49 | neurologic Sx | no fibrosis | Intracranial hemorrhage | F | K | lung |
| GSE122960 | 22 | neurologic Sx | no fibrosis | Anoxic brain injury | F | AA | lung |
| GSE122960 | 47 | neurologic Sx | no fibrosis | Intracranial hemorrhage | F | K | lung |
| GSE122960 | 21 | neurologic Sx | no fibrosis | Head trauma | M | AA | lung |
| GSE122960 | 41 | neurologic Sx | no fibrosis | Stroke | M | K | lung |
| GSE122960 | 57 | neurologic Sx | no fibrosis | Intracranial hemorrhage | M | K | lung |
| GSE122960 | 64 | neurologic Sx | no fibrosis | Intracranial hemorrhage | M | AA | lung |
| GSE122960 | 49 | neurologic Sx | no fibrosis | Intracranial hemorrhage | F | K | lung |
| GSE122960 | 43 | neurologic Sx | no fibrosis | Anoxic brain injury | F | K | lung |
| GSE122960 | 50 | neurologic Sx | no fibrosis | Intracranial hemorrhage | M | His | lung |
| GSE122960 | 51 | neurologic Sx | no fibrosis | Stroke | M | AA | lung |
| GSE122960 | 26 | neurologic Sx | no fibrosis | Anoxic brain injury | F | AA | lung |
| GSE122960 | 54 | neurologic Sx | no fibrosis | Hyperlipidemia | M | K | lung |
| GSE122960 | 57 | neurologic Sx | no fibrosis | Stroke | F | K | lung |
| GSE122960 | 52 | neurologic Sx | no fibrosis | Stroke | M | K | lung |
| GSE122960 | 25 | neurologic Sx | no fibrosis | Head trauma | F | K | lung |
*As = Asian, K = White/Caucasian, His = Hispanic, AA = African American.
Fig. 1.
Immunological features in the lungs of COVID patients display fibrosis and innate immune activation. (A) UMAP plots demonstrating the major cell types and associated cluster in the lungs of five patients with COVID and 20 healthy lungs. Cells are colored according to donor origin, cell type classification, and clusters (for patient demographics, see Table 1). (B) Heat map comparing the expression of alveolar macrophage-related genes in two subsets of tissue-resident alveolar macrophages (TRAM, clusters 2 and 3) and one subset of monocyte-derived alveolar macrophages (MoAM, cluster 0). (C) Frequencies of macrophage subsets (clusters 0 and 3) which were presenting a significant difference between healthy and COVID cohorts. Each dot represents one individual patient. (D) The frequency of neutrophils (cluster 4) demonstrates a qualitative increase in COVID lung tissues. Each dot represents one individual patient. (E) Associations of genes with COVID status were determined using differential expression analysis for transcripts and linear regression in log-likelihood tests for neutrophil-related genes. The adjusted P values are plotted relative to the log2 fold change of the mean values between COVID and healthy lung cohorts. Blue indicates genes highly expressed in the healthy lung cohort and red genes highly expressed in the COVID lung cohort. (F) IL-6 is expressed in monocyte-derived alveolar macrophages (MoAM, cluster 0) and neutrophil (cluster 4) and in COVID lungs. Each dot represents one individual patient. (G–I) Cluster 7 represents a cluster of fibroblasts uniquely enriched in COVID which demonstrates elevated expression of AP1 and fibroblast and epithelial-to-mesenchymal transition-related genes (fibroblasts are represented in clusters 7, 13, and 23). Each dot represents one individual patient. (J) Representative confocal images of lung tissue from COVID patients demonstrate positive staining of cells for SARS-CoV-2 N-protein, as well as indicating the presence of infiltrating neutrophils and macrophages by neutrophil elastase (NE) and CD68 staining, collagen accumulation with COL1, and activation of IL-6–pJUN–CD47 axis. (Scale bar, 100 µm.) (K and L) Representative CODEX images of COVID lung tissues; selected markers collagen IV, collagen I, and vimentin indicate fibrosis, CD68, and CD163 indicate macrophage (K), and myeloperoxidase (MPO) and CD15 indicate the presence of neutrophilic/myeloid cells (L). Contour plots display the double-positive populations. (Scale bar, 100 µm.) Data are expressed as mean ± SD of five COVID lung and 20 healthy lung cohorts. Data were analyzed by the two-tailed unpaired t test; P values were labeled.
Next, we further examined the unique transcriptional state of Cluster 4 which emerged specifically in COVID lungs and corresponded to increased neutrophil populations (Fig. 1D). To describe the differential transcriptional programming in COVID lungs relative to healthy samples, we identified differentially expressed genes between the two groups. Our results show an enrichment of gene members of the AP1 family, neutrophil maturation-related chemokine and cytokine genes (Fig. 1E). The Elsevier pathway database analysis showed curated pathways that are up-regulated in cluster 4 (combined score > 75). Not surprisingly, we found that these pathways were associated with neutrophil activation (SI Appendix, Fig. S3C).
Previous studies of patients with severe COVID-19 described high levels of inflammatory cytokines in their blood (28). Here, we compared the IL-6 expression levels in COVID in all clusters and found the clusters that differed significantly are clusters 0, 3, and 4 that comprise alveolar macrophages (MoAM), tissue-resident alveolar macrophages (TRAM), and neutrophils (neutrophils) (Fig. 1F and SI Appendix, Fig. S4). These results further underscore the important role of increased MoAM and matured neutrophils as well as the injured TRAM in the lung in COVID.
Next, we analyzed single-cell RNA-seq data and probed divergent fibroblasts in lung tissue: clusters 7, 13, and 23 (SI Appendix, Fig. S3D). Of note, a majority of the fibroblasts in COVID-19 lung tissues clustered together, mainly in cluster 7 (Fig. 1G, P value showing a significant difference between healthy and COVID patients). Among all the fibroblasts, cluster 7 comprises increased gene expression such as AP1 family members JUN (SI Appendix, Fig. S3E) in addition to genes involved in extracellular matrix like collagen and fibroblast activation (Fig. 1H). Furthermore, evaluating the Molecular Signature Database, we identified the epithelial-to-mesenchymal transition pathway to be most significantly enriched in fibroblast cluster 7 (Fig. 1I). Overall, evaluating single-cell transcriptomic studies of COVID-19 lung samples helped elucidate a complex dysregulation of the pulmonary immune response and profibrotic pathways leading to irreversible lung tissue damage and progressive fibrosis in severe COVID-19 patients.
Last, we validated differentially expressed markers obtained in our scRNA-seq analysis using immunofluorescence staining on COVID lung tissue (SI Appendix, Fig. S3F) with markers of SARS-CoV-2 infection N-protein, the neutrophil elastase (NE) protein for neutrophils, CD68 that marks macrophages, CD31 for endothelium, TTF1 for epithelium, CD11b for monocytes, CD20 for B cells, CD3 for T cells and collagen I that is a marker of matrix fibroblast tissue, mesothelin for active fibroblast precursors, smooth muscle actin (SMA) for vessels, and FSP1 for fibroblasts. Meanwhile, striking expression of phospho-JUN (pJUN), CD47, and IL-6 was noted in the SARS-CoV-2–infected lungs (Fig. 1J and SI Appendix, Fig. S3G). By using multiplexed immunostaining (codetection by indexing, CODEX; Fig. 1 K and L), we confirmed the presence of a profibrotic macrophage subset, indicated by CD68 and CD163 double-positive cells, and activated neutrophils, indicated by CD15 and myeloperoxidase (MPO) double-positive cells, in the fibrotic area positive for collagen I, collagen IV, and vimentin.
Taken together, our data suggest that the SARS-CoV-2 infection elicits an innate immune activation accompanied by a profibrotic program. This inflammatory milieu consisting of IL-6 released by MoAM and matured neutrophils and the overexpression of AP1 in active fibroblasts could be a potential mechanism describing the progression of fibrosis and inflammation in SARS-CoV-2–infected patients via interactions with the innate immune system.
A Mouse Model Recapitulates COVID Lung Fibrosis in Patients.
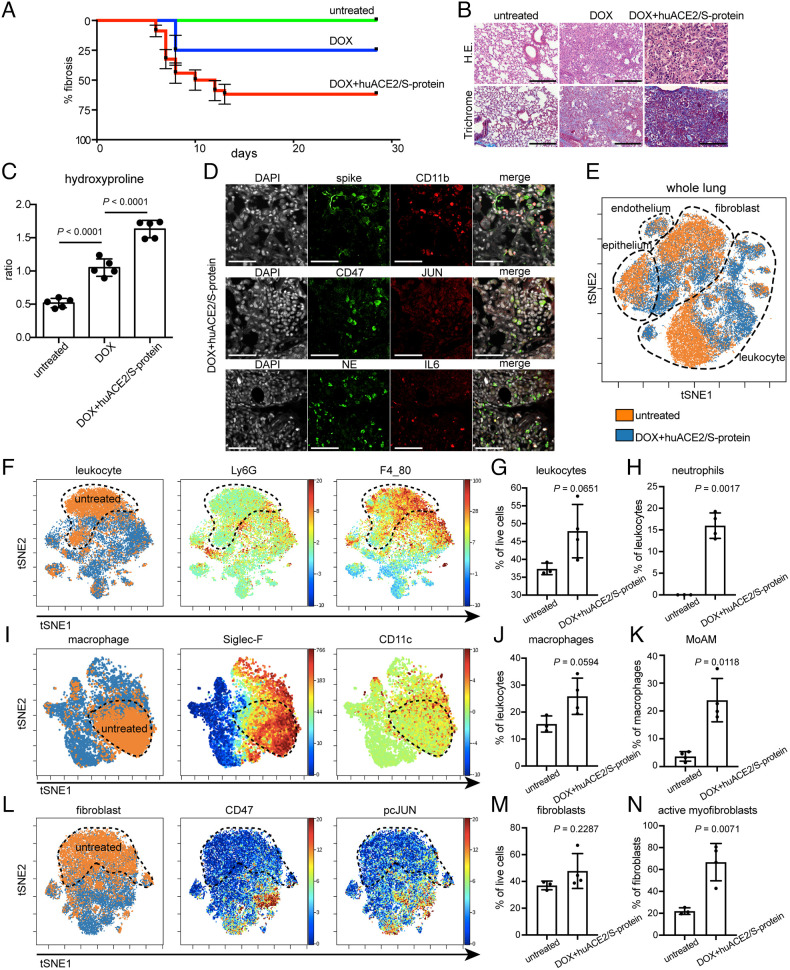
To recapitulate the pathogenesis of lung fibrosis after COVID-19 infection, we generated a mouse model which developed lung fibrosis in response to pseudotyped SARS-CoV-2 transduction. The main impediment to the infection of mouse cells with SARS-CoV-2 is the lack of appropriate receptors to initiate viral infection (29). To overcome this, we cotransduced mice with human ACE2 lentivirus and a SARS-CoV-2 pseudovirus in the lung via aerosol and induced JUN with doxycycline (30) and confirmed that mice transduced with SARS-CoV-2 developed pronounced fibrosis in the lung compared to doxycycline-alone treated and untreated lungs (Fig. 2 A and B). Kaplan–Meier plots and histopathological analysis indicated that aerosol of SARS-CoV-2 pseudovirus contributed to 75% fibrosis at 4 wk, while doxycycline JUN induction contributed to only 25% at the same time point (Fig. 2A); however, huACE2 and SARS-CoV-2 pseudovirus are insufficient to induce fibrosis in every mouse. These findings were supported by a hydroxyproline assay which also confirmed exacerbated fibrosis by SARS-CoV-2 (Fig. 2C) along with an increased frequency of macrophages and neutrophils and highly expressed profibrotic markers CD47, JUN, and IL-6 (Fig. 2D) within the mouse lungs. Furthermore, we performed cytometry by time of flight (CyTOF) analyses of the entire lung tissue obtained from the mice exposed to Dox+huACE2/S-protein, and the gating strategy was shown in SI Appendix, Fig. S5. t-distributed stochastic neighbor embedding (tSNE) plots revealed that leukocyte and fibroblast populations drastically differed from healthy mouse lung tissue (Fig. 2E). Further investigations into specific cell type populations revealed leukocyte subpopulations to be significantly increased in the mouse lungs exposed to Dox+huACE2/S-protein relative to untreated healthy controls (Fig. 2 F and G), especially the neutrophils as indicated by Ly6G expression (Fig. 2 F and H). Of note, remodeling of macrophage and fibroblast distribution was observed (Fig. 2 I–K). Of interest, we found no significant difference between the frequency of macrophage and fibroblast cell populations between the fibrotic mouse lungs exposed to Dox+huACE2/S-protein and healthy control lungs. However, a striking increase in the MoAM population was observed (Fig. 2K) demonstrated by expression of Siglec-F and CD11c (Fig. 2I). In addition, we found subpopulations of fibroblasts commonly associated with pathogenic fibrosis to be elevated post–SARS-CoV-2 infection (Fig. 2 L–N) and were characterized by the expression of CD47 and pcJUN (Fig. 2L). Taken together, our analyses of single-cell lung tissue data in combination with histopathological characterization model and explain the onset of fibrosis after COVID-19 as a disordered structure of the infected lung, extensive immune infiltration, and profibrotic fibroblast activation.
Fig. 2.
Innate immune infiltrates and fibrosis in mice indicating the transition to COVID lung fibrosis. (A) Kaplan–Meier analysis depicting onset and % fibrosis in untreated (5 mice), doxycycline-treated (5 mice), and doxycycline plus human ACE2/S-protein–treated (10 mice) mice. (B) Histology demonstrating a drastic exacerbation of lung fibrosis with combined aerosol treatment of doxycycline for JUN induction and human ACE2/S-protein compared to doxycycline aerosol alone and untreated. (Scale bar, 100 µm.) (C) Hydroxyproline assay of mouse lung tissues confirmed that the addition of human ACE2/S-protein to doxycycline for JUN induction drastically increased the fibrosis also quantitatively (P < 0.0001). 5 mice per group were analyzed. (D) Representative immunofluorescence stains demonstrate positive cells for SARS-CoV-2 spike protein, macrophages, and monocytes (CD11b), neutrophils (neutrophil elastase, NE), JUN, CD47, and IL-6. (Scale bar, 100 µm.) (E) tSNE of single-cell CyTOF data of lung tissues of 4 mice in transition to lung exposed to Dox+huACE2/S-protein (blue) and three healthy lungs (orange). (F) tSNE plots represent leukocytes in the lungs of mice in transition to COVID and healthy controls. The healthy control is labeled with a dotted line. Ly6G and F4_80 expression is used to determine the abundance of neutrophils and macrophages. (G) The statistical analysis of the frequency of leukocytes in all live cells as indicated. (H) Quantification of neutrophil frequency in the lung as indicated. (I) tSNE alignment of macrophage clusters, Siglec-F, and CD11c abundance labeling heterogeneous macrophages. (J) The abundance of lung macrophages as indicated. (K) Dot plot shows the relative contributions of monocyte-derived alveolar macrophage (MoAM) per mouse. (L) tSNE of CyTOF profiling protein expression in fibroblasts as indicated. (M) Dot plots of percentages of fibroblast gated on live cells as indicated. (N) Activated myofibroblasts were drastically increased in the mouse lungs in transition to COVID. (Scale bar, 100 µm.) Data are expressed as mean ± SD of four mouse lungs in transition to COVID and three healthy control lungs. Data were analyzed by one-way ANOVA (C) and the two-tailed unpaired t test (G, H, J, K, M, and N); P values are indicated and deemed statistically significant if P < 0.05.
Anti-IL-6/CD47 Antibody Therapy Could Be an Effective Strategy for the Treatment of Post–COVID-19 Pulmonary Inflammation and Fibrosis.
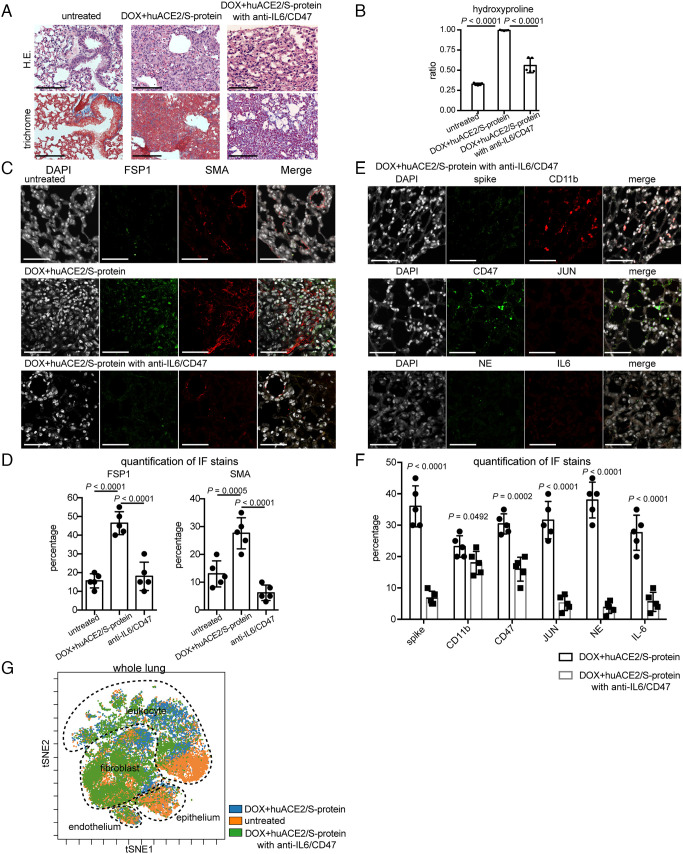
Our results underscore the importance of the IL-6-JUN-CD47 nexus as a potential therapeutic target for resolving fibrosis in response to SARS-CoV-2 infection. To determine the effect of this therapeutic target, anti-IL-6 and anti-CD47 antibodies were administered in mice after 13 d of DOX+huACE2/S-protein aerosol. We assessed single-agent treatment and found it insufficient to reduce fibrosis in these models. These findings are also in line with our previous observation in a model of idiopathic pulmonary fibrosis (IPF) lung fibrosis (14). Histopathological analyses performed after 2 wk of treatment revealed significant restoration of normal lung morphology and less extracellular matrix/collagen deposition (Fig. 3A). A hydroxyproline assay was also used to calculate the extent of fibrosis development revealing a significant decrease in fibrosis (Fig. 3B). Dramatic improvement was further verified by assessing the immunolocalization of FSP1 and SMA, which showed a decrease in activated fibroblasts posttreatment (Fig. 3 C and D), suggesting a mitigation of the immune dysregulation (Fig. 3 E and F). We further confirmed these results by CyTOF analysis of major population abundance after IL-6/CD47 blockade (Fig. 3G).
Fig. 3.
Mice in transition to COVID lung fibrosis and mice treated with anti-CD47/anti-IL-6 demonstrate improved fibrosis. (A) H&E and trichrome demonstrating lung histology after anti-CD47/anti-IL-6 treatment in mice in transition to COVID fibrosis. (Scale bar, 100 µm.) (B) Hydroxyproline assay calculated less extent of collagen accumulation. (C) Representative immunofluorescence images of FSP1 and SMA demonstrating that inhibiting CD47 and IL-6 improves lung fibrosis. (Scale bar, 100 µm.) (D) Quantification of expression of FSP1 and SMA in mice in transition to COVID with anti-CD47/anti-IL-6 treatment displaying decreased numbers of activated fibroblasts. (E) Confocal imaging of lung tissues showing attenuation of immune infiltration and fibrosis in mice in transition to COVID after anti-CD47/anti-IL-6 treatment. (Scale bar, 100 µm.) (F) Quantitation of marker expression in mouse lung tissues with or without anti-CD47/anti-IL-6 treatment. Data are shown as means ± SD from five different fields. (G) tSNE projection of CyTOF analysis probes global immune cellular changes after anti-CD47/anti-IL-6 treatment in the mouse lungs in transition to COVID. (Scale bar, 100 µm.) Data are shown as mean ± SD of five different mouse lungs per each group. Data were analyzed by one-way ANOVA (B and D) and multiple t tests (F); P values are indicated.
Humanized Mice Carrying Human Lung Organoids Transduced with huACE2/S-Protein Model of Long COVID Lung Fibrosis Which Is Alleviated by Combined Blockade of Inflammation and Fibrosis.
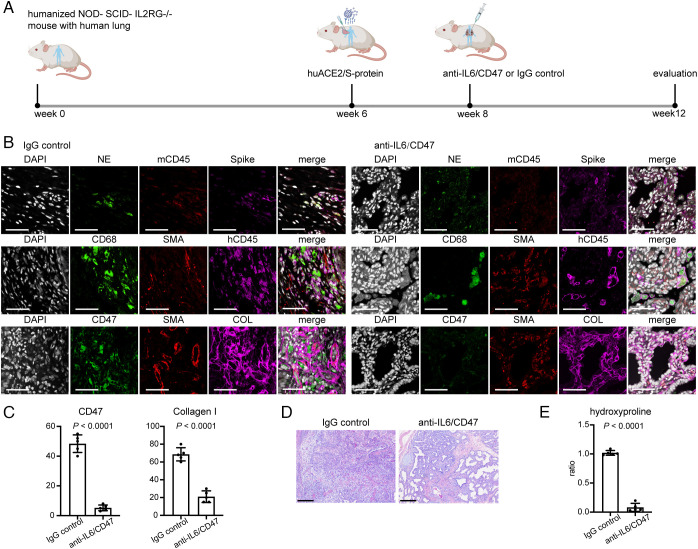
We then tested our therapeutic strategy in a humanized mouse model of long COVID lung fibrosis. Irradiated 4-m-old NOD-SCID-IL2RG-/- (NSG) mice were reconstituted by implanting a sandwich of human fetal thymus–liver–thymus tissue under the kidney capsule. Two pieces of human lung tissue (~2 to 4 mm3) were simultaneously implanted subcutaneously into the left and right flanks. Following tissue implantation, mice received bone marrow grafts from human CD34+ HSC via tail vein injections. After 6 wk, human chimerism was assessed by peripheral blood flow analysis at ~20%. Then, SARS-CoV-2 pseudovirus and bleomycin were administered after efficient engraftment of human lung organoids to model fibrosis in long COVID. Treatment with anti-IL-6/CD47 commenced on week 8 after establishment, and evaluation was carried out until week 12 (Fig. 4A). Immunofluorescent staining revealed successful humanization of mice evident from human CD45 positive staining of tissue, spike protein staining positive for SARS-CoV-2 transduction, NE and CD68 positive staining depicting neutrophil and macrophage infiltration and α-SMA, and collagen stains suggesting fibrotic expansion. After treatment, the antifibrotic effect of the combinatorial therapy on anti-CD47 and anti-IL-6 was sufficient to resolve fibrosis in this humanized mouse model of COVID-19 (Fig. 4 B and C). In addition, lung tissue obtained from this humanized mouse model shares features reminiscent of human COVID lung fibrosis. The lungs showed infiltration by neutrophils and macrophages, interstitial expansion by fibrosis, and bronchiolization of alveoli (Fig. 4D). All these pathological alterations were ameliorated post-IL-6/CD47 treatment. Measurement of hydroxyproline content also confirmed less collagen deposition posttreatment (P < 0.0001) (Fig. 4E). Overall, our data suggest CD47/IL-6 inhibition as a beneficial strategy to clinically manage COVID lung fibrosis.
Fig. 4.
Fibrosis decreased in human lung tissues after treatment with blocking antibodies against CD47 and IL-6 in a humanized mouse model of COVID lung fibrosis. (A) Evaluation of therapeutic effects of CD47 and IL-6 inhibition on humanized mice grafted with human lung organoids infected with SARS-CoV-2 and treated with IgG or anti-IL-6/anti-CD47 blocking antibodies. (B) Representative immunofluorescence images of humanized COVID mouse treated with or without anti-CD47/anti-IL-6 antibodies highlighting the change of human immune cell infiltrates and extracellular matrix deposition. (Scale bar, 100 µm.). (C) Quantification of CD47 and collagen I expression in humanized mouse infected with SARS-CoV-2 without and with anti-CD47/anti-IL-6 treatment. (D) Representative images of histopathology demonstrating normalization of the lung structure in humanized COVID mouse model after anti-CD47/anti-IL-6 treatment. (Scale bar, 100 µm.). (E) Measurement of hydroxyproline content decreased with anti-CD47/anti-IL-6 treatment. (Scale bar, 100 µm.) Data are shown as mean ± SD of five different human lung tissue pieces per each group. Data were analyzed by the two-tailed unpaired t test; P values were labeled.
Discussion
As the pandemic continues, COVID-19 is now defined as a multiorgan disease with a broad spectrum of manifestations (31). A majority of patients with COVID-19 have reported continuing debilitating symptoms months after the onset of disease now summarized as long COVID. The COVID-19 sequelae display varying symptoms ranging from fatigue, muscle aches, and breathlessness to pulmonary inflammation and fibrosis (PIF) (32). Although ARDS seems to be the main predictor of the onset of long COVID after acute COVID-19 infection, underlying pathological causes remain unknown. Clinically pertinent questions like why some individuals recover from COVID-19–induced pulmonary insult, whereas others develop progressive pulmonary fibrosis due to accumulation of profibrotic fibroblasts and excessive deposition of collagen remain unanswered. Given these observations, the burden of pulmonary fibrosis after acute COVID-19 recovery poses a significant challenge to global health. It is also essential to prevent a second wave of late mortality associated with this pandemic with a specific focus on older and severe ARDS populations. Designing therapeutic interventions is dependent on our understanding of the immunological response elicited during infection.
Exacerbated immune responses play a major role in the pathophysiology of SARS-CoV-2, leading to severe lung injury and respiratory failure (15). In this study, by analyzing scRNA-seq data from severe COVID-19 lung tissues in comparison with a healthy cohort, we addressed the contribution of immune compartments and the important role of the profibrotic modulator JUN–CD47–IL-6 axis in promoting pulmonary fibrosis postacute COVID-19 infection. We show that neutrophils and monocytes and macrophages are the most prevalent innate immune subsets present in these lungs and show a correlation with SARS-CoV-2–associated fibrotic progression, suggesting that these are the immediate effector cells responsible for the chronic inflammation in COVID lung fibrosis.
Several studies have reported that severe COVID-19 progression is associated with a dysregulated release of proinflammatory cytokines commonly called the “cytokine storm” (33). Specifically, IL-6, TNF-α, vascular endothelial growth factor (VEGF), and IL-1α are enhanced in the bronchoalveolar lavage and plasma. Among these cytokines, IL-6 is thought to be required for pathogen recognition and subsequent translation into an emergency granulopoiesis response to inflammatory milieu. Previous literature shows IL-6 signaling through the JanusKinase (JAK)–signal transducer and activator of transcription (STAT) pathway to stimulate neutrophil precursor proliferation and differentiation (34, 35). Also, our previous studies suggest IL-6 as an integral component in the initiation and progression of fibrosis via the JUN and CD47 axis (14). Although previous scRNA-seq analyses of PBMCs failed to identify circulating cells producing IL-6 (15), our analysis concludes IL-6 to be produced by inflammatory cells as matured neutrophils and monocyte-derived alveolar macrophages (MoAM) in the lung tissue. Meanwhile, we identified that JUN and CD47 expression is up-regulated in SARS-CoV-2–infected lung tissues, especially in activated fibroblasts.
Mitigation of immune dysregulation is therefore viewed as a major therapeutic avenue for the treatment and prevention of COVID lung fibrosis. Inflammatory myeloid cells as neutrophil and macrophage are demonstrated that affect fibrosis progression accompanied by a dysregulation of the profibrotic modulating JUN–CD47–IL-6 axis. Thereafter, we investigated the efficiency of combined CD47/IL-6 blockade for lung fibrosis in the COVID lung fibrosis mouse model and humanized COVID-19 mouse model. Overall, the robust improvement of fibrosis in a combinatorial therapy of CD47 and IL-6 inhibition highlights the importance of JUN, CD47, and IL-6 as potential therapeutic targets for resolving fibrosis in COVID lung fibrosis.
Materials and Methods
Animal Studies.
Animal trials were approved by the Stanford Administrative Panel on Laboratory Animal Care (nos. 30911 and 30912), and mice were kept in the facilities of the Stanford Veterinary Service Center on a regular diet, and experiments were performed according to the approved protocol.
Husbandry.
lsl-rtTA JUN mice were kept in the facilities of the Veterinary Service Center at the Stanford University and had a B6/129 background (36). Nod.Scid.IL2RG-/- (NSG) mice were purchased from the Jackson Laboratory. Mice were kept on a standard diet. Female and male mice were used. When different sexes were used for individual experiments, groups were sex-matched. Mice were not backcrossed and between 6 and 12 wk of age during experiments.
Lung Fibrosis Model (JUN Mouse Model).
Doxycycline was administered by intranasal aerosol to mice anesthetized with isoflurane as follows: three doses of 100 µL of a 2 M doxycycline on days 0, 1, and 3. For lung fibrosis studies, antibody or placebo treatments started on day 13, continued for 2 wk, and the end point reached on day 27.
Humanized Mouse.
To establish a humanized mouse, the hematopoietic system was reconstructed by transplanting the human fetal liver and thymus systemically and under the mouse kidney capsule of lethally irradiated NODSCID IL2Rg-/- mice. Human fetal lung tissues obtained from Advanced Bioscience Resources were implanted in small pieces (~2 to 4 mm3) in the right and left dorsal flanks subcutaneously caudal to the scapula as previously described (37, 38). Six weeks after HSC engraftment, human donor chimerism was assessed by flow cytometric immune phenotyping of human myeloid and B and T lymphoid cells in the peripheral blood. Successful engraftment of lung tissue was assessed by volumetric measurements. Shortly after, lung tissues were transduced with single and/or combined virus concomitant with a single dose of bleomycin injected once i.p. at 2 U/kg body weight. At 8 wk, humanized mice were randomized into single or combined anti-CD47, anti-IL-6, or IgG control treatment arms as detailed below.
SARS-CoV-2 Spike Pseudovirus and Human ACE2 Lentivirus Production and Transduction.
Plasmid constructs were kindly provided by Drs. Crawford and Bloom (30). The packaging and tittering methods are described in detail (30). Pseudotyped lentiviral particles with SARS-CoV-2 spike and SARS-CoV-2 receptor ACE2 were effectively expressed and used in this study. For JUN mouse model, 30 μL of pseudovirus supernatant per mouse was delivered via inhalation of aerosol three times on days 0, 1, and 3. For humanized mice, 30 μL of pseudovirus supernatant per mouse was injected into the grafted human lung tissues subcutaneously on days 0, 1, and 3.
In Vivo Antibody Blockade.
For CD47 antibody blockade experiments, mice were injected intraperitoneally (IP) with a dose of 500 μg CD47 antibody (Clone MIAP410, BioXCell) diluted in 100 μL PBS at 8 wk. The same dose was then given every other day for 4 wk. For IL-6 antibody blockade experiments, mice were injected intraperitoneally with 20 mg/kg dose of an anti-IL-6 monoclonal antibody (Clone MP5-20F3, BioXCell) twice a week for 4 wk.
Human Study.
Lung samples were obtained from tissue blocks of autopsies consented for by the decedent's legal next of kin. Autopsy consents included research permission and were collected under IRB 39881.
Tissue Fixation and Hematoxylin Staining.
We kept harvested tissue in 10% formalin overnight. Tissue was then submitted to the Stanford Human Pathology/Histology Service Center for paraffin embedding and cutting. We deparaffinized and rehydrated the tissue slides with xylene and a descending ethanol row. After washing the slides in PBS, we incubated them in hematoxylin (American MasterTech) for 4 min, then in bluing reagent (Thermo Fisher Scientific) for 2 min, and in Harleco® (Millipore Sigma) for 2 min. Slides were dehydrated with ethanol and xylene (Millipore Sigma) and mounted with Permount® (Thermo Fisher Scientific).
Trichrome Staining.
We used a One-Step Trichrome Stain Kit (American MasterTech). After deparaffinization and rehydration, the tissue was incubated in Bouin’s Fluid overnight, followed by Modified Mayer’s Hematoxylin for 7 min and One-Step Trichrome Stain for 5 min. Slides were dehydrated with ethanol and xylene and covered with Permount® (Thermo Fisher Scientific).
Hydroxyproline Assay.
We determined the hydroxyproline content with a Hydroxyproline Assay Kit (Millipore Sigma) according to the manufacturer’s specifications. All samples were performed in biological replicates, and standards were in technical duplicates.
Single-Cell RNA-seq Analysis and Data Accessibility.
scRNA-seq expression datasets of lung tissues (20 normal and five long COVID patients) were obtained from four public datasets (GSE149878, GSE122960, GSE163919, and GSE158127), and the clinical profiles of all patients are presented in Table 1. For each dataset, the low-quality cells were removed with parameters: cell total Unique molecular identifiers (UMIs) <200 UMIs and percentage of mitochondrial genes >15%. After filtering, a total of 251612 cells were left for further analysis. The filtered expression datasets were integrated with Seurat v4 1 by following the reciprocal principal component analysis (PCA) workflow to remove batch effects across different donors. In brief, the filtered individual dataset was first normalized by the LogNormalize function; then, the top 2,000 variable genes identified using the FindVariableFeatures function with the vst method. To identify the aligning anchors, we used the “rpca” method for reduction and k.anchor equal to 20 in the FindIntergrationAnchors function. After integrating the expression datasets, the first 20 dimensions’ principal component analysis (PCA) were used for UMAP visualization and clustering.
Mass Cytometry (CyTOF).
Samples were processed as previously described (14). Formaldehyde-fixed cell samples were incubated with a cell surface antibody cocktail for 1 h at room temperature, washed once with PBS containing 0.5% BSA, permeabilized with methanol on ice for 15 min, washed twice with PBS containing 0.5% BSA, and then incubated with metal-conjugated antibodies against intracellular molecules for 1 h. Cells were washed twice with PBS containing 0.5% BSA and then incubated at room temperature for 20 min with an iridium-containing DNA intercalator (Fluidigm) in PBS containing 2% paraformaldehyde. After intercalation/fixation, the cell samples were washed once with PBS containing 0.5% BSA and twice with water before measurement on a CyTOF mass cytometer (Fluidigm). Normalization for detector sensitivity was performed by using Four Element Calibration Beads (Fluidigm). After measurement and normalization, the individual files were analyzed by first gating out doublets, debris, and dead cell based on cell length, DNA content, and cisplatin staining. tSNE maps and FlowSOM were generated with software tools available at https://www.beckman.com/flow-cytometry/software. org/ by considering all included markers.
Statistics.
Statistical analyses were performed using Prism software (GraphPad Software). Statistical significance was determined by the unpaired Student t test for comparisons between two groups and one-way ANOVA with Tukey’s post hoc test for multigroup comparisons (n.s., P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). In statistical graphs, points indicate individual samples, and results represent the mean ± SD unless indicated otherwise.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (CSV)
Acknowledgments
We would like to acknowledge Pauline Chu for her assistance with processing of histological specimens, the Stanford Stem Cell FACS Core for flow cytometry experiments, Stanford Shared Fluorescence-activated Sell Sorting Facility for CyTOF experiments, Stanford Hospital for autopsy service, Dr. Serena Tan for pathology review, and Drs. Tushar Desai and Tal Raveh for reading parts of the manuscript. Drs. Crawford and Bloom for sharing the viral constructs. Funding sources for this project include Boehringer-Ingelheim (SI Appendix), Stinehart-Reed Foundation, Scleroderma Research Foundation, and Ludwig Institute at the Stanford University.
Author contributions
L.C., S.-Y.C., and G.W. designed research; L.C., C.M.D.S., T.L., I.L., S.-Y.C., and G.W. performed research; S.-Y.C. contributed new reagents/analytic tools; L.C., Z.F., Y.G., V.C., and G.W. analyzed data; and L.C., C.M.D.S., I.W., and G.W. wrote the paper.
Competing interests
The authors have patent filings to disclose: I.W. and G.W. have patents that have been licensed, and it is possible that they could receive royalties from them. No current consultation with companies in this field. I.W. is a founder and director of Bitterroot Bio, Inc, Pheast, Inc, and 48 Bio, Inc, none of which has programs in this field.
Footnotes
Reviewers: J.V.G., University of North Carolina at Chapel Hill; M.L.W., Dartmouth College Geisel School of Medicine; and P.J.W., University of California San Francisco.
Contributor Information
Irving Weissman, Email: irv@stanford.edu.
Gerlinde Wernig, Email: gwernig@stanford.edu.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Huang C., et al. , Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395, 497–506 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laing A. G., et al. , A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 26, 1623–1635 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Lucas C., et al. , Impact of circulating SARS-CoV-2 variants on mRNA vaccine-induced immunity. Nature 600, 523–529 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D., et al. , Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 323, 1061–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tale S., et al. , Post-COVID-19 pneumonia pulmonary fibrosis. QJM 113, 837–838 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwensen H. F., et al. , Fatal pulmonary fibrosis: A post-COVID-19 autopsy case. J. Clin. Pathol. 28, 206879 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Spagnolo P., et al. , Pulmonary fibrosis secondary to COVID-19: A call to arms? Lancet Respir. Med. 8, 750–752 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bazdyrev E., et al. , Lung fibrosis after COVID-19: Treatment prospects. Pharmaceuticals (Basel) 14, 807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown K. K., et al. , The natural history of progressive fibrosing interstitial lung diseases. Eur. Respir. J. 55, 2000085 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wynn T. A., Ramalingam T. R., Mechanisms of fibrosis: Therapeutic translation for fibrotic disease. Nat. Med. 18, 1028–1040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson M. S., Wynn T. A., Pulmonary fibrosis: Pathogenesis, etiology and regulation. Mucosal Immunol. 2, 103–121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wynn T. A., Cellular and molecular mechanisms of fibrosis. J. Pathol. 214, 199–210 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wynn T. A., Integrating mechanisms of pulmonary fibrosis. J. Exp. Med. 208, 1339–1350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui L., et al. , Activation of JUN in fibroblasts promotes pro-fibrotic programme and modulates protective immunity. Nat. Commun. 11, 2795 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arunachalam P. S., et al. , Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 369, 1210–1220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas C., et al. , Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 584, 463–469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathew D., et al. , Deep immune profiling of COVID-19 patients reveals distinct immunotypes with therapeutic implications. Science 369, eabc8511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao M., et al. , Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 26, 842–844 (2020). [DOI] [PubMed] [Google Scholar]
- 19.Schulte-Schrepping J., et al. , Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182, 1419–1440.e23 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee S., et al. , Virus-induced senescence is a driver and therapeutic target in COVID-19. Nature 599, 283–289 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Grant R. A., et al. , Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature 590, 635–641 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poon M. M. L., et al. , SARS-CoV-2 infection generates tissue-localized immunological memory in humans. Sci. Immunol. 6, eabl9105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hasenkrug K. J., et al. , Recovery from acute SARS-CoV-2 infection and development of anamnestic immune responses in T cell-depleted rhesus macaques. mBio 12, e0150321 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsieh W. C., et al. , NK cell receptor and ligand composition influences the clearance of SARS-CoV-2. J. Clin. Invest. 131, e146408 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bharat A., et al. , Lung transplantation for patients with severe COVID-19. Sci. Transl. Med. 12, eabe4282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu G., et al. , Persistent viral activity, cytokine storm, and lung fibrosis in a case of severe COVID-19. Clin. Transl. Med. 10, e224 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reyfman P. A., et al. , Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 199, 1517–1536 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del Valle D. M., et al. , An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 26, 1636–1643 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munoz-Fontela C., et al. , Animal models for COVID-19. Nature 586, 509–515 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crawford K. H. D., et al. , Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses 12, 513 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bolouri H., et al. , The COVID-19 immune landscape is dynamically and reversibly correlated with disease severity. J. Clin. Invest. 131, e143648 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nalbandian A., et al. , Post-acute COVID-19 syndrome. Nat. Med. 27, 601–615 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R., The COVID-19 cytokine storm; What we know so far. Front. Immunol. 11, 1446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fielding C. A., et al. , IL-6 regulates neutrophil trafficking during acute inflammation via STAT3. J. Immunol. 181, 2189–2195 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Hashizume M., Higuchi Y., Uchiyama Y., Mihara M., IL-6 plays an essential role in neutrophilia under inflammation. Cytokine 54, 92–99 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Wernig G., et al. , Unifying mechanism for different fibrotic diseases. Proc. Natl. Acad. Sci. U.S.A. 114, 4757–4762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wahl A., et al. , Precision mouse models with expanded tropism for human pathogens. Nat. Biotechnol. 37, 1163–1173 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wahl A., et al. , SARS-CoV-2 infection is effectively treated and prevented by EIDD-2801. Nature 591, 451–457 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (CSV)
Data Availability Statement
scRNA-seq expression datasets of lung tissues (20 normal and five long COVID patients) were obtained from four public datasets (GSE149878, GSE122960, GSE163919, and GSE158127), and the clinical profiles of all patients are presented in Table 1. For each dataset, the low-quality cells were removed with parameters: cell total Unique molecular identifiers (UMIs) <200 UMIs and percentage of mitochondrial genes >15%. After filtering, a total of 251612 cells were left for further analysis. The filtered expression datasets were integrated with Seurat v4 1 by following the reciprocal principal component analysis (PCA) workflow to remove batch effects across different donors. In brief, the filtered individual dataset was first normalized by the LogNormalize function; then, the top 2,000 variable genes identified using the FindVariableFeatures function with the vst method. To identify the aligning anchors, we used the “rpca” method for reduction and k.anchor equal to 20 in the FindIntergrationAnchors function. After integrating the expression datasets, the first 20 dimensions’ principal component analysis (PCA) were used for UMAP visualization and clustering.
All study data are included in the article and/or SI Appendix.