Significance
Plant multitrophic interactions are extremely complex, and the underlying mechanisms are not easy to unravel. Using tomato plants as a model system, we demonstrated that a soil fungus, Trichoderma afroharzianum, widely used as a biocontrol agent of plant pathogens, negatively affects the development and survival of the lepidopteran pest Spodoptera littoralis by altering the gut microbiota and its symbiotic contribution to larval nutrition. Our results indicate that insect-plant interactions can be correctly interpreted only at the metaorganism level, focusing on the broad network of interacting holobionts which spans across the soil and the above-ground biosphere. Here, we provide a new functional framework for studying these intricate trophic networks and their ecological relevance.
Keywords: holobionts, insect–plant interactions, phytophagous insects, insect gut microbiota, soil microbiota
Abstract
Plants generate energy flows through natural food webs, driven by competition for resources among organisms, which are part of a complex network of multitrophic interactions. Here, we demonstrate that the interaction between tomato plants and a phytophagous insect is driven by a hidden interplay between their respective microbiotas. Tomato plants colonized by the soil fungus Trichoderma afroharzianum, a beneficial microorganism widely used in agriculture as a biocontrol agent, negatively affects the development and survival of the lepidopteran pest Spodoptera littoralis by altering the larval gut microbiota and its nutritional support to the host. Indeed, experiments aimed to restore the functional microbial community in the gut allow a complete rescue. Our results shed light on a novel role played by a soil microorganism in the modulation of plant–insect interaction, setting the stage for a more comprehensive analysis of the impact that biocontrol agents may have on ecological sustainability of agricultural systems.
Plants, as primary producers in all ecosystems, fix solar energy, which then flows through natural food webs driven by continuous coevolutionary arm-races among organisms competing for nutritional resources (1). This competition for resources is part of a complex network of multitrophic interactions and drives life evolution, resulting in an amazing diversification of ecological niches. Understanding the dynamics of evolutionary processes and the underpinning mechanisms of resource allocation through competition is crucial for shedding light on the functioning of both natural and anthropic ecosystems, as a basis to develop safe and sound strategies for their sustainable management.
Interspecific competition among closely related species sharing overlapping ecological niches is a well-known and consolidated concept (2, 3). In contrast, far less investigated and understood are the competitions among species belonging to distant phyla or to different kingdoms, including viruses (3), even though competition is the commonest form of interaction existing in nature (4). For example, sharing the same host is the major driving force of peculiar competition strategies among unrelated entities, such as parasitic wasps and viral pathogens attacking the same insect host, which can reinforce its defense barriers against wasps by acquiring viral genes encoding toxins through horizontal transfer (5).
A further layer of complexity is added to these bioevolutionary scenarios by microorganisms associated with their interacting hosts, collectively denoted as holobionts, which are frequently considered as superorganisms controlled by hologenomes (i.e., the host genome and its microbiome) evolving as single units of selection (6). Even though this concept is debated (7), it is certainly true that the host phenotype is partly determined by its microbiota.
Plants provide excellent examples of these intricate interactions among holobionts. Their response to environmental stress agents is strongly conditioned by the root microbiota (8). Indeed, these plant-associated microorganisms can concur in the regulation of the plant hormonal balance underlying the defense response against different types of attackers, finely orchestrated by cross-modulating pathways, targeting different stress agents (9). Ultimately, plant parasites, pathogens, and the root microbiota compete amongst themselves for the nutritional resources made available by plants. More generally, competitive interactions are largely modulated by immune barriers and attack strategies used by competing organisms that are strongly influenced by associated microorganisms.
Root microbiota and endophytic microorganisms have established a broad range of intimate interactions with plants, which can be of mutual benefit and exert a significant impact on higher trophic levels, conferring protection against different biotic stress agents and promoting plant growth (10, 11). Among the many members of the soil microbiota, fungi show an amazing level of diversification of ecological habits and modes of nutrition, offering unparalleled opportunities to study interkingdom competition processes. In particular, the Sordariomycetes, one of the largest classes in the division Ascomycota, are characterized by a wealth of nutritional strategies ranging from saprotrophy to biotrophic interactions with several organisms, including bacteria, plants, animals, and fungi (12). Hypocreales is the largest order in this class, deriving from ancestors (~200 to 170 Mya) originally mycoparasites of wood-decomposing Basidiomycota fungi which during their evolution underwent several intra- and inter-kingdom host shifts, involving fungi, plants, and animals (13–15). The shift in the animal host likely started in the Jurassic (201.3 to ~145 Mya) and culminated during the Cretaceous (~145 to 66 Mya) with the appearance of entomopathogens, in parallel with the high diversification of insects and angiosperms (14).
The genus Trichoderma represents one of the most successful groups in Hypocreales, which includes a plethora of species present both in natural and managed ecosystems, where they play a well-known role as plant decomposers (saprotrophs), plant endophytes, and fungal antagonists/parasites (mycoparasites) (16). These biological characteristics have promoted the wide use of many Trichoderma species as efficient biocontrol agents of fungal plant pathogens and as a source of enzymes for industrial applications (17–19). The direct biocontrol activity of Trichoderma species is associated with the antagonism or parasitism of fungal plant pathogens, whereas indirect biocontrol activity involves the activation of plant defenses, via induced resistance (local or systemic), and a priming effect that stimulates more rapid and intense response upon pathogen or pest attack (13, 17, 18, 20–23). These plant responses are induced either by fungal metabolites released in the soil or modulated by the endophytic behavior of several Trichoderma species, able to enter cortical layers of plant roots and establish a molecular dialogue (18, 23, 24). The resulting Trichoderma–plant interaction, well characterized at the functional and molecular level, is clearly mutualistic, with plants receiving protection against biotic and abiotic stress, increased nutrient availability, and growth promotion from the fungal symbiont, which in turn obtains nutritional benefits (17, 18, 20, 25–27).
Root colonization by Trichoderma can also enhance the direct plant defense response to insects, such as aphids (28, 29), whiteflies (30), thrips (31), and Lepidoptera (25, 29, 32, 33), in addition to tetranychid spider mites (34) and nematodes (35). Moreover, this beneficial fungus reinforces indirect defense barriers, by determining the production of plant volatiles attracting pest natural enemies, including both parasitoids (29, 36, 37) and predators (36, 38). All these studies describe how insect survival and behavior are affected by Trichoderma colonization of the plant, as well as plant transcriptomic and metabolomic changes associated with the observed effects. However, how these plant changes can negatively impact insect survival remains poorly understood. This is a complex phenomenon and likely modulated by a wealth of synergistic defense barriers such as toxic plant secondary metabolites (e.g., phenylpropanoids, flavonoids, anthocyanins, alkaloids, terpenoids, and glucosinolates), and antinutritional enzymes and proteins (e.g., proteinase inhibitors, amino acid catabolizing enzymes, polyphenol oxidases, and peroxidases) (39). Research addressing this latter aspect needs to consider the amazing diversity of insect–microorganism associations and their pivotal role in the regulation of insect physiology and structure/dynamics of their communities, composed of interacting holobionts (40–42). Indeed, the study of plant–insect interactions from this perspective shows high level of complexity and sheds light on the key role played by the interplay among microorganisms associated with plants and insects, a fascinating research area still in its infancy (43).
In this study, we investigate how Trichoderma afroharzianum, a well-known plant-beneficial microorganism widely used in agriculture as a biocontrol agent of plant pathogens, when applied as a seed treatment to tomato plants (Solanum lycopersicum) can negatively affect the development and survival of the lepidopteran pest Spodoptera littoralis feeding on leaves of the treated plants. This experimental system offers unique opportunities for elucidating the underlying functional mechanism, given the significant amount of molecular and functional data already available for tomato plants colonized by Trichoderma, and the suitability of noctuid moth caterpillars for physiological and molecular analyses. Using a multifaceted experimental approach, we demonstrate that T. afroharzianum modulates the fate of this insect–plant interaction by altering the insect gut microbiome, indicating the central importance that microorganisms may have in the regulation of natural food webs, spanning across the soil and the above-ground biosphere.
Results
Trichoderma Colonization of Tomato Plants Has a Negative Impact on Survival and Development of S. littoralis.
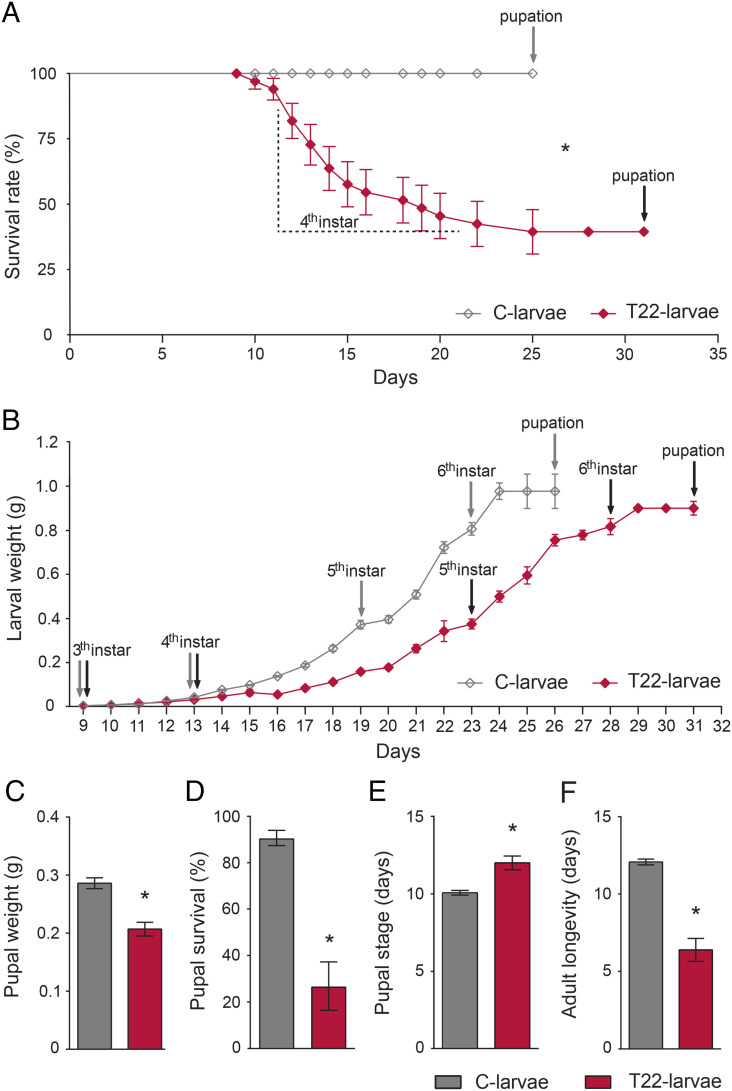
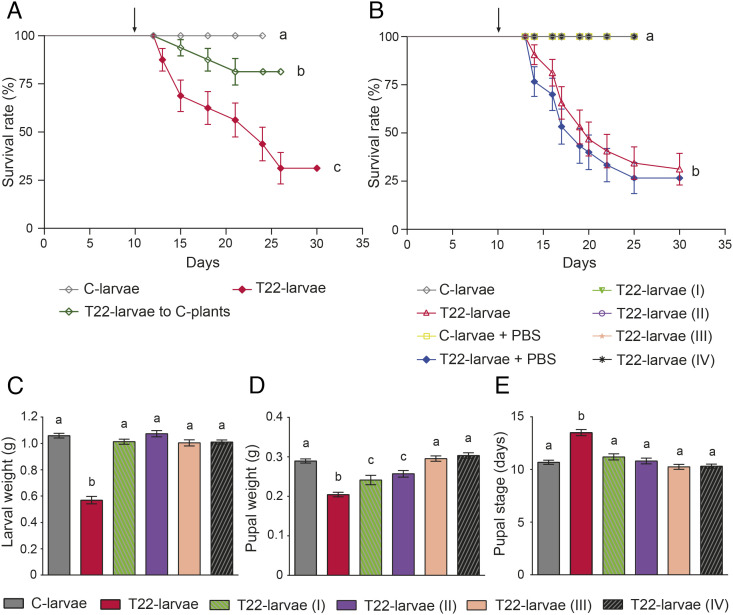
The survival rate of S. littoralis larvae fed on leaves from tomato plants obtained from seeds coated with spores of T. afroharzianum strain T22 (i.e., T22-larvae and T22-plants) was significantly lower than in larvae fed with untreated control plant leaves (i.e., C-larvae and C-plants) (Fig. 1A). The higher mortality induced by T22-plants was evident from day 2 of the fourth instar (Fig. 1A), accompanied by a significant reduction in weight gain over time (Fig. 1B and SI Appendix, Results and Table S1). This resulted in a significant developmental delay since surviving T22-larvae took longer than C-larvae to attain the critical weight needed to pupate (Fig. 1B). Moreover, the weight of T22-pupae (Fig. 1C) and their survival (Fig. 1D) were both significantly lower than in controls. T22-adults emerged significantly later (Fig. 1E) and showed reduced longevity compared to controls (Fig. 1F).
Fig. 1.
Effects on survival and development of S. littoralis larvae feeding on leaves from tomato plants colonized by T. afroharzianum. (A) Survival rate of larvae reared on tomato leaves obtained from plants treated with T. afroharzianum T22 (T22-larvae) or untreated control plants (C-larvae). Here and in the following panels, the asterisk denotes a statistically significant difference (log-rank test: χ2(1) = 29.72, P value < 0.0001). (B) Larval weight from third instar to pupation (statistical indices are reported in SI Appendix, Table S1); T22-larvae took longer than controls to pupate [Student’s t test: t(41) = 14.03, P value < 0.0001]. (C) Weight of day 3 pupae [Student’s t test: t(41) = 4.54, P value < 0.0001]. (D) Pupal survival [log-rank test: χ2(1) = 16.18, P value < 0.0001]. (E) Duration of pupal stage [Student’s t test: t(30) = 4.82, P value < 0.0001]. (F) Adult longevity [Student’s t test: t(28) = 10.67, P value < 0.0001]. The values are means ± SE. The gray arrows and the black arrows indicate the molting occurrence for C-larvae and T22-larvae, respectively.
The delayed growth and development of T22-larvae were associated with levels of food consumption like those observed for C-larvae (SI Appendix, Results and Fig. S1). This indicates that the T22-plant material shows unaltered palatability and excludes the possible occurrence of antifeedant effects. Since the observed negative effects on survival and development could be mediated by the fungal induction of plant defense barriers targeting midgut functionality (39), we focused our attention on the direct impact that Trichoderma colonization of tomato plants may have on larval midgut and/or on the microbial community harbored in its lumen.
T22-Plants Cause Metabolic Redirection and Activation of Antibacterial Immune Genes in Midgut Cells of S. littoralis Larvae.
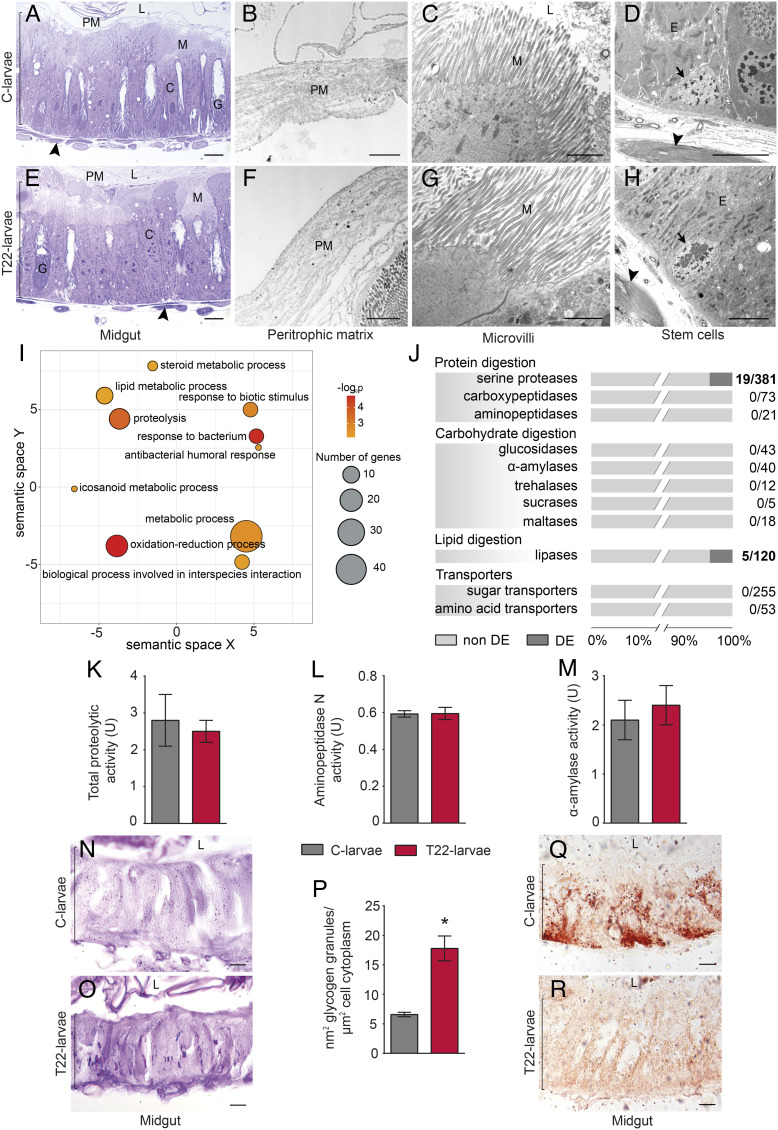
To assess the possible occurrence of any direct impact on the midgut structure of T22-larvae, we carried out a microscopy analysis on the midgut isolated from fourth instar larvae, which were those showing the highest level of mortality during the feeding bioassay (Fig. 1A). T22-larvae and C-larvae showed very similar morphological and ultrastructural features (Fig. 2 A–H), with no signs of evident alterations of the general structure (Fig. 2 A and E), peritrophic matrix (Fig. 2 B and F), and microvilli (Fig. 2 C and G and SI Appendix, Results and Fig. S2 A and C); furthermore, stem cells from both T22- and C-larvae exhibited similar blast morphology with no sign of proliferation (Fig. 2 D and H), usually associated with structural lesions (44).
Fig. 2.
Structural, transcriptional, and functional changes in S. littoralis larval midgut. (A–H) Morphological and ultrastructural features of the midgut in C-larvae (A–D) and T22-larvae (E–H); optical microscopy images (A and E); Transmission Electron Microscopy images of the peritrophic matrix (B and F), microvilli of the columnar cells (C and G), and stem cells (D and H); arrow: stem cell; arrowhead: muscles; bracket and E: epithelium; C: columnar cell; G: goblet cell; L: lumen; M: microvilli; PM: peritrophic matrix; bars: 10 µm (A and E), 2 µm (B, C, and F–H), and 5 µm (D). (I) Semantic clustering of enriched biological process Gene Ontology (GO) terms associated with T22-larvae upregulated genes resulting from midgut transcriptomic analyses; circle color indicates the −log10 P value of the enriched GO terms, while the size is proportional to the number of Differentially Eexpressed (DE) genes associated with the GO term. (J) Proportion of differentially expressed genes in the midgut (DE; in black) and non-DE genes (in gray); numbers on the right represent the ratio between DE genes and the total genes listed within the category. (K–M) Activity of digestive enzymes in C-larvae and T22-larvae; (K) total proteolytic activity [Welch’s t test: t(2.58) = 0.44, P value = 0.7]; l aminopeptidase N activity [Welch’s t test: t(7.41) = −0.07, P value = 0.94]; (M) α-amylase activity [Welch’s t test: t(7.79) = −0.58, P value = 0.58]. (N and O) Glycogen deposits, visible as purple/violet spots, in the midgut epithelium of C-larvae (N) and T22-larvae (O), evidenced with Periodic Acid-Schiff staining; bracket: epithelium, L: lumen; bars: 10 µm. (P) Quantification of glycogen deposits in the midgut epithelium of C-larvae and T22-larvae [Welch’s t test: t(4.24) = −5.28, P value < 0.01]. (Q and R) Accumulation of lipid droplets, visible as red spots, in the midgut epithelium of C-larvae (Q) and T22-larvae (R) evidenced with Oil Red O staining; bracket: epithelium, L: lumen; bars: 10 µm. In K–M and P, the values are means ± SE, asterisk indicates statistically significant difference.
Transcriptional changes in the midgut cells of fourth instar T22-larvae, compared to C-larvae, revealed the differential expression of 85 genes, with 5 downregulated and 80 upregulated (SI Appendix, Results and Dataset S1A). The Gene Onthology (GO) terms enrichment analysis of upregulated genes (Fig. 2I) highlighted biological processes associated with immune response activation (GO:0009617 response to bacterium; GO:0019731 antibacterial humoral response; GO:0009607 response to biotic stimulus). Other enriched biological processes associated with upregulated genes in T22-larvae included oxidative processes (GO:0055114 oxidation-reduction process) along with lipid and protein metabolic processes (GO:0006629 lipid metabolic process; GO:0006508 proteolysis). Enriched molecular function GO terms were also associated with lipid and protein degradation (e.g., GO:0004252 serine-type endopeptidase activity; GO:0052689 carboxylic ester hydrolase activity; SI Appendix, Results, Fig. S3, and Results 1).
To analyze possible alterations in specific midgut functions in response to T. afroharzianum, we investigated the variation in the expression of genes associated with digestive processes and membrane transporters involved in nutrient absorption. The analysis of 1,022 genes, subdivided into 11 functional categories (SI Appendix, Results and Dataset S1B), clearly indicated that the expression patterns remained largely unchanged in T22-larvae compared to C-larvae, except for a few serine proteases (19 out of 381) and lipase (5 out of 120) genes, which were upregulated in T22-larvae (Fig. 2J, SI Appendix, Results, and Dataset S1B). However, these limited transcriptional changes of genes associated with serine proteases, the major family of endopeptidases in Lepidoptera (45), did not influence the activity profile of enzymes involved in the initial phase of protein digestion (i.e., total proteolytic activity; Fig. 2K). Moreover, in agreement with the expression pattern, the activity of aminopeptidase N, one of the most abundant exopeptidase families present in the midgut brush border of insect larvae responsible for the final phase of protein digestion (46), and the activity of α-amylase, the major enzyme that acts in the first step of maltopolysaccharide digestion (47), also did not show any differences (Fig. 2 L and M). Although a few genes encoding lipases were differentially expressed in the midgut of T22-larvae, no difference in lipase activity was detected between C-larvae and T-22 larvae, with activity being under the limit of detection in both samples.
Transcriptional changes indicated the possible occurrence of metabolic alterations (Fig. 2I and SI Appendix, Results and Fig. S3); we therefore performed histochemical analyses to assess the potential impact of T22-plants on the storage of nutrients, namely glycogen and acylglycerols, because of the metabolic redirection induced in T22-larvae. Glycogen deposits in the midgut cells of T22-larvae were not only significantly higher compared to C-larvae but also significantly larger (Fig. 2 N–P; SI Appendix, Results and Fig. S2 B and D for the specificity of glycogen staining). However, in contrast, the accumulation of lipid droplets was significantly greater in midgut cells of C-larvae, being absent in the T22-larvae (Fig. 2 Q and R).
Collectively, these data indicate that midgut cells exposed to T22-plant material show neither structural alterations nor reduction in digestion capability but exhibit a significant metabolic redirection, characterized by an extensive use of intracellular proteins and lipid stores and an increased accumulation of glycogen, as a likely consequence of changes in the nutritional profile of the biochemical milieu in the gut lumen. Moreover, another evident alteration is the activation of antibacterial immune genes, suggesting the occurrence of a change in the microbiota composition with a possible switch of commensal species to a pathogenic lifestyle.
T22-Plants Induce Structural and Transcriptional Alterations in the Gut Microbiota of S. littoralis Larvae.
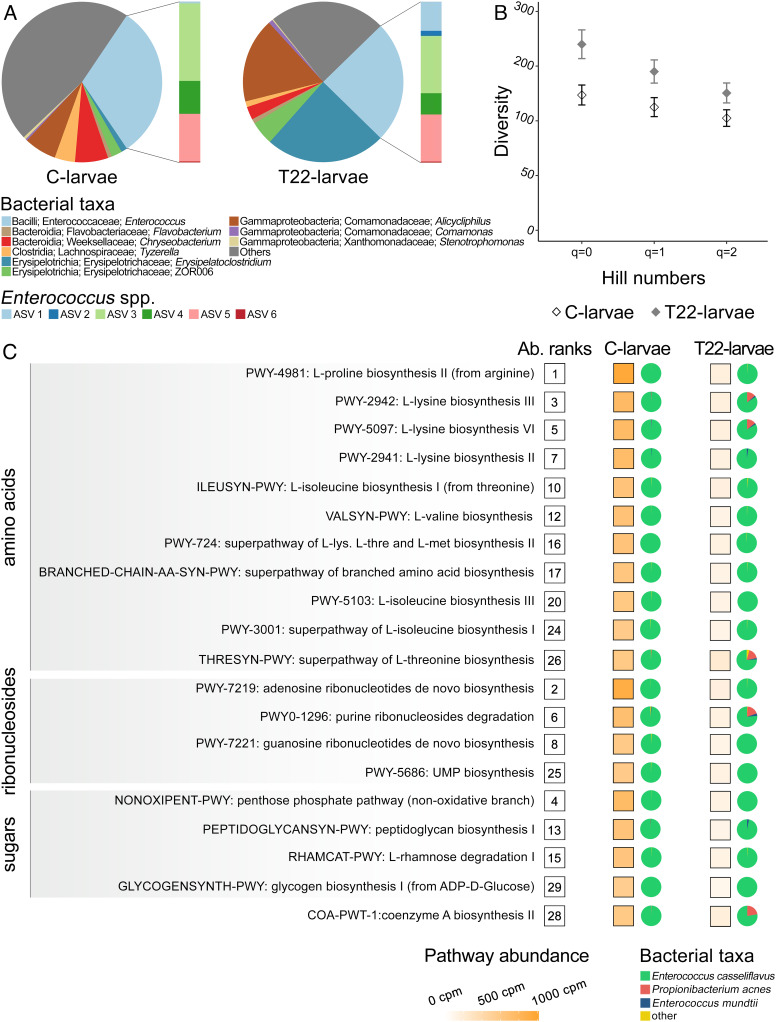
Given the key role played by the gut microbiota on host fitness (e.g., supply of essential nutrients), we analyzed the impact of T22-plant leaves on the microbiota of S. littoralis fourth instar larvae. In T22-larvae, we observed a taxonomic shift in the composition of the microbiota and an increase of its diversity (Fig. 3 A and B, SI Appendix, Results and Results 2, and Dataset S1C). Two bacterial genera, Erysipelatoclostridium (Erysipelotrichia; Erysipelatoclostridiaceae) and Alicycliphilus (Gammaproteobacteria; Comamonadaceae) represented only a negligible fraction of the C-larvae microbiota (1% and 6%, respectively), while reaching an average relative abundance of ~24% and ~17% in the microbiota of T22-larvae (Fig. 3A). Overall, the relative abundance of Enterococcus, the dominant bacterial genus in the microbiota of S. littoralis (48, 49), remained unchanged between T22-larvae and C-larvae (Fig. 3A and SI Appendix, Results 2). Among the five most abundant Amplicon Sequence Variants (ASVs) assigned to Enterococcus, which accounted for 97.8% of the total Enterococcus reads, two ASVs (ASV 1 and ASV 2, 1 Single Nucleotide Polymorphisms (SNPs) of difference), identified as Enterococcus mundtii (confidence 0.98), were more represented in T22-larvae than in C-larvae (Fig. 3A). Two further ASVs (ASV 3 and ASV 4, 1 SNP of difference) were more abundant in the control group, while ASV 5 had similar abundance in both groups (Fig. 3A). However, these last three ASVs can only be assigned to Enterococcus casseliflavus or to Enterococcus gallinarum based on the variable 3 and 4 (V3-V4) 16S ribosomal RNA (rRNA) gene region (1 or 2 SNPs of difference). The observed microbiota composition of C-larvae is consistent with previous studies in which a relatively simple microbiota, at least in terms of the core taxa, was detected (50, 51).
Fig. 3.
Compositional and functional alterations in the midgut microbiota of S. littoralis. (A) 16S rRNA metagenomics of S. littoralis microbiota; pie charts represent the taxonomic composition of the microbiota expressed as taxa relative abundance, while the associated bar plots represent the relative abundance of ASVs attributed to the Enterococcus genus. (B) Microbiota diversity of C-larvae and T22-larvae inferred by 16S rRNA metagenomics and expressed as Hill’s numbers. (C) Metatranscriptomics pathways significantly associated with T22-larvae; on the right of each pathway, the following information are reported: i) the overall abundance of the pathway in C-larvae, numbers in boxes express the rank value; for C-larvae and T22-larvae; ii) the pathway abundance expressed as copies per million in a white to orange scale color and iii) pie charts representing the relative contribution of bacterial taxa to a specific pathway. Abbreviation: Ab.: abundance.
Modification in the composition of the gut-associated bacterial community observed in T22-larvae was coupled with alterations in the microbiota transcriptional profile (Fig. 3C and SI Appendix, Results 3). The activity of the microbiota harbored in the midgut of C-larvae was characterized by the prevalence of pathways associated with the biosynthesis and degradation of purine ribonucleosides, sugars degradation (rhamnose, galactose, and stachyose), and biosynthesis of glycogen, peptidoglycan, and amino acids (Ile, Leu, Lys, Met, Pro, Thr, and Val; SI Appendix, Results and Fig. S4). Noteworthily, most of the metabolic pathways consistently transcribed in the microbiota of C-larvae were associated with biosynthesis of essential amino acids for the host (Ile, Leu, Lys, Met, Thr, and Val). A clear separation between C-larvae and T22-larvae was further highlighted by both principal component analysis (PCA) and cluster analyses on abundances of metabolic pathways (SI Appendix, Results and Fig. S5). In the PCA, the first principal component accounted for 83.9% of the variance, allowing a clear-cut separation of the two groups, while in the cluster analyses the samples grouped into two well-separated sets, corresponding to C-larvae and T22-larvae. In the microbiota associated with T22-larvae, a strong perturbation of the transcriptional activity was observed for 20 pathways (Fig. 3C). Notably, these pathways were among the most highly expressed in C-larvae and were consistently associated with the biosynthesis of amino acids and the metabolism of sugars and ribonucleotides. Moreover, the entirety of the pathways undergoing changes in abundance was mainly attributed to E. casseliflavus (Fig. 3C), supporting the assignment of ASVs 3, 4, and 5 to this taxon. These results highlight the important nutritional support provided by E. casseliflavus to S. littoralis, through the supply of six essential amino acids (see above), thus having a more conspicuous role for the host than E. mundtii (52, 53).
The overall analysis of the hologenomic domain shows an evident functional complementarity between the host and microbiota domains. The substantial reduction in the supply of amino acids by the gut microbiota in T22-larvae results in a reduced support to the energy metabolism, which, in the gut of Lepidoptera is largely dependent on amino acids and lipids (54–56). Therefore, the increased expression of genes associated with lipid and protein metabolism, as well as with the degradation of host intracellular proteins and lipids (Fig. 2I and SI Appendix, Results and Fig. S3), and the observed increased degradation of lipid stores (Fig. 2R) could likely be the consequence of a shortage of amino acids due to impairment of the microbiota. Moreover, the reduced glycogen biosynthesis by the gut microbiota (Fig. 3C) may favor the uptake of the resulting monosaccharide excess present in the lumen by the gut cells and the downstream accumulation of glycogen in the midgut of T22-larvae (Fig. 2O).
Trichoderma Colonization of Tomato Plants Significantly Alters the Midgut Metabolome of S. littoralis.
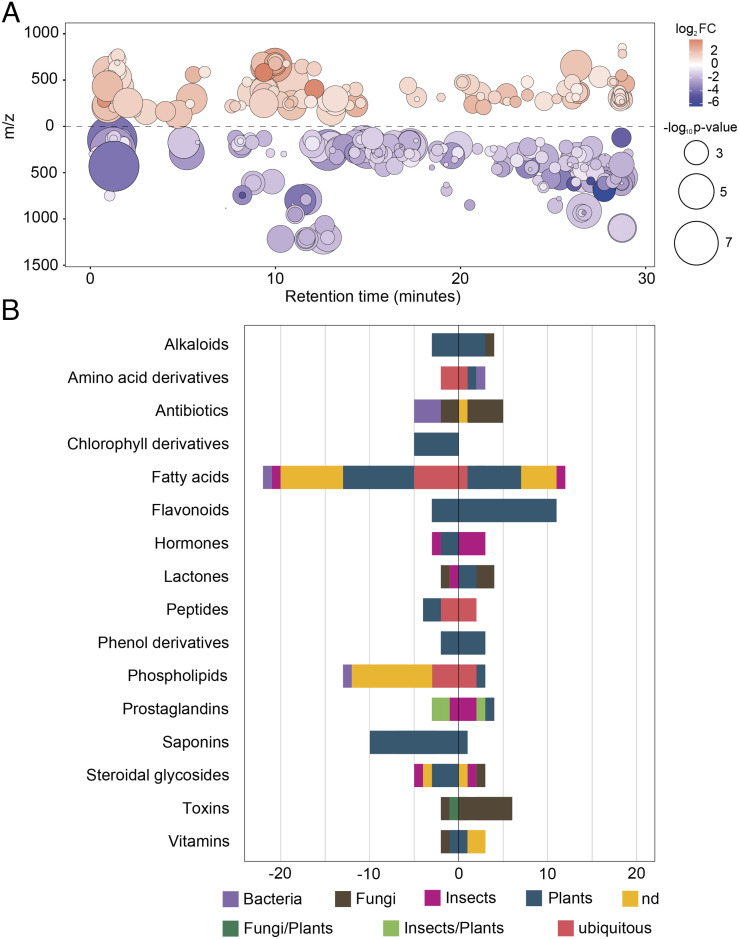
To study the changes induced by T22-plant leaves on the biochemical milieu of the midgut, metabolomics analyses of midguts isolated from T22-larvae and C-larvae were carried out. From the 1,469 features (i.e., ions with a unique m/z and retention time) detected, 337 differentially accumulated metabolites (DAMs) were observed in T22-larvae compared to C-larvae (with 210 and 127 down- and over-accumulated, respectively; Fig. 4A, SI Appendix, Results and Results 4, and Dataset S1D). Overall, the Liquid Chromatography-ElectroSpray Ionization-High Resolution Mass Spectrometry (LC-ESI-HRMS) analysis highlighted a stark disparity in the metabolites present in the midgut of the two groups, with features of plant origin (e.g., flavonoids, fatty acids, saponins, and alkaloids) accounting for the larger part of DAMs (Fig. 4B). The most represented classes of plant origin were flavonoids and fatty acids. In T22-larvae, flavonoids were the most over-accumulated metabolites (11 features), followed by fatty acids involved in the jasmonic acid pathway (5 features), while all the other fatty acids showed a clear reduction in accumulation (22 features) (Fig. 4B). The resulting general lower abundance of fatty acids in the midgut of T22-larvae is consistent with the reduced lipid content in midgut cells (Fig. 2R), suggesting that this change may have both a direct effect on larvae as well as on the gut microbiota. An additional change, which is nutritionally relevant, is the reduction in amino acid content, corresponding with the reduced abundance of amino acid-associated pathways highlighted by the metatranscriptomics analysis (Fig. 3C); this finding further indicates reduced nutritional support by the midgut microbiota to the insect host. However, the correspondence between metabolomics and the gut microbiota transcriptomics was not observed for nucleosides (even though guanine is less prevalent in T22-larvae) or sugar metabolites (SI Appendix, Results and Dataset S1D).
Fig. 4.
Feeding on T22-plant leaves affects the midgut metabolome of S. littoralis. (A) Metabolomic cloud plot showing the 337 features with P value ≤ 0.05 and fold change ≥ 1.5 identified by untargeted LC-ESI-HRMS analysis. Dot color intensity is positively correlated with P value, while its radius with fold-change. Position on the x-axis represents retention time. Position on the y-axis represents mass-to-charge ratio. Light red and purple colors represent over-accumulated and down-accumulated metabolites, respectively. (B) Class and origin of annotated DAMs in T22-larvae (only classes with five or more metabolites were reported). Values on the x-axis represent the number of metabolites down- (−) or over- (+) accumulated in T22-larvae.
The over-accumulation in T22-larvae of metabolites acting as toxins or having antibiotic properties was also observed (Fig. 4B), indicating the presence of molecules that could concur in the induction of the midgut dysbiosis, such as aphidicolin-like, acetylneomycin-like, and aibekacin-like metabolites (SI Appendix, Results and Dataset S1D). Among metabolites of insect and plant origin, it is interesting to note the differential accumulation of prostaglandins and prostaglandin-like compounds in the midgut of T22-larvae (Fig. 4B), which are mediators of both systemic and gut immune responses (57). This evidence, together with the upregulation of immune genes (Fig. 2I), indicates the occurrence of an immune alteration associated with gut dysbiosis.
Rescue of T22-Larvae by Restoring the Midgut Microbiota Composition.
To provide direct experimental evidence supporting the potential functional link between the fitness reduction of T22-larvae (Fig. 1) and the alteration of the gut microbiota composition and functionality (Fig. 3 A and C), we performed rescue experiments to recover insect development and survival as “proof of concept.” We first verified that a significant rescue effect could be obtained when T22-larvae were transferred to C-plant leaves on day 2 of the third instar (Fig. 5A and SI Appendix, Results and Fig. S6). To demonstrate that these effects could be attributed to changes in the midgut microbiota, we attempted to rescue T22-larvae through in vivo manipulations of the midgut microbiota. Starting from day 2 of third instar, T22-larvae were fed with T22-plant leaves overlaid with the whole midgut microbiota obtained from fourth instar C-larvae [T22-larvae (I)] and with three different amounts of E. casseliflavus cells [T22-larvae (II to IV)] (SI Appendix, Methods 8), the bacterium that accounts for most of the changes in the metatranscriptome (Fig. 3C). The administration of healthy midgut microbiota and E. casseliflavus cells resulted in a complete rescue in terms of survival and development of T22-larvae (Fig. 5B). The same rescue pattern was observed in terms of larval weight (Fig. 5C and SI Appendix, Results, Fig. S7, and Table S2), pupal weight (Fig. 5D), and duration of the pupal stage (Fig. 5E).
Fig. 5.
Rescue experiments on S. littoralis larvae. (A) Survival rate of S. littoralis larvae was affected by switching from T22- to C-plant leaves (arrow indicates when T22-larvae were transferred to control leaves), showing a significant rescue of the survival curve [log-rank test: χ2(1) = 8.84, P value < 0.003], which approached that of C-larvae but still resulted significantly lower [log-rank test: χ2(1) = 4.918, P value < 0.0266] [log-rank test among the three groups: χ2(2) = 27.15, P value < 0.0001]. (B) The complete rescue of survival rate of T22-larvae was obtained when larvae were daily offered T22-plant leaves overlaid with healthy midgut microbiota (5.2 × 106 bacterial cells) (T22-larvae I) and three different amounts of E. casseliflavus cells (5.2 × 106, 1.3 × 106, and 6.5 × 105 cells for T22 larvae II, III, and IV, respectively) [log-rank test: χ2(7) = 99.99, P value < 0.0001]; arrow indicates when T22-larvae were transferred to T22-plant leaves overlaid with bacteria. (C) Larval weight before pupation [Kruskal–Wallis χ2(5) = 78.38, P value < 0.0001], (D) pupal weight [Kruskal–Wallis χ2(5) = 74.43, P value < 0.0001], and (E) duration of pupal stage were restored when larvae were offered T22-plant leaves overlaid with bacteria [one-way ANOVA: F(5,159) = 18.51, P value < 0.0001]; in C–E, the values are means ± SE. Different letters indicate mean values that are statistically different.
Collectively, these results exclude a direct effect of plant/fungal toxins on the insect and demonstrate that i) the alteration of the midgut microbiota caused by T. afroharzianum plant colonization is of key importance in the induced plant resistance against insects, and ii) E. casseliflavus is a key player for the survival of the insect host.
Discussion
The fungal genus Trichoderma is composed of a vast number of species (58), widely distributed in almost all geographic areas, both in natural and anthropogenic environments. The remarkable biodiversity and multifaceted ecological roles (13) make it difficult to provide a comprehensive description of all significant biological traits. However, the reconstruction of the evolution of nutritional mode supports multiple interkingdom host jumps which generated different lineages attributed to nutritional roles as mycoparasites and saprotrophs or as plant mutualists (26, 27), that eventually conferred benefits to plants (13, 17). Members of the genus Trichoderma have been widely used over the past decades as biocontrol agents of plant pathogens and plant growth promoters in agriculture, as well as model organisms for the industrial production of cellulolytic enzymes (18, 59).
The mechanisms underlying the interaction between Trichoderma and plants have been studied in great detail; they involve an intricate cross-talk modulating different pathways that generate responses enabling the plant to face various biotic or abiotic stress agents, therefore to modulate its ability to defend and/or grow (13). Recent studies have demonstrated that root colonization by Trichoderma can induce plant resistance barriers against insects (25, 28–33). In-depth metabolic reprogramming of the plant generates both indirect (i.e., the induction of volatiles attracting insect natural enemies) and direct (i.e., molecules with a negative impact on insect physiology and fitness) defense barriers (13).
The multifaceted experimental approach we have used in this study demonstrates that these direct defense barriers do not always exert a direct negative effect on insect physiology. Here, we show that the metabolic changes induced in tomato plants by T22 colonization, associated with enhanced resistance to biotic stress agents and growth promotion (28, 29, 32, 60), can mediate gut dysbiosis in S. littoralis larvae feeding on T22-plant leaves. Notably, the dysbiosis affected the relative abundance of symbiotic Enterococcus bacteria and the role of E. casseliflavus in providing nutritional support to the host (i.e., mainly essential amino acids and sugars), that results in a negative impact on insect development and survival (Fig. 6). It has been recently demonstrated that bacteria of the genus Enterococcus can improve the performance of a S. littoralis congener when reared on suboptimal diets (61). Indeed, our study indicates that the larvae fed on tomato leaves from T22-plants did not suffer structural damage to the gut and alterations in the digestive capacity. In fact, the negative impact on insect development and survival could be fully rescued by oral administration of the functional S. littoralis gut microbiota obtained from larvae fed on control plant leaves or alternatively from the administration of the E. casseliflavus bacteria alone.
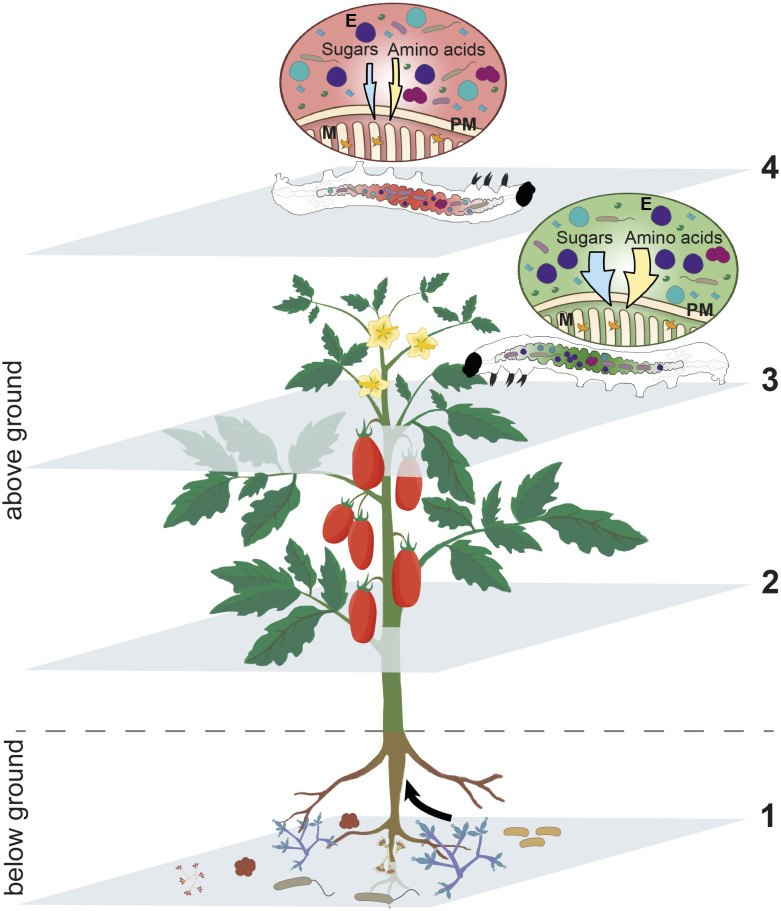
Fig. 6.
Schematic representation of the interactions among the plant S. lycopersicum, the fungus T. afroharzianum strain T22, and the phytophagous insect S. littoralis and its gut microbiome. The colonization of S. lycopersicum roots by the fungus T. afroharzianum strain T22 (1) systemically conditions the plant (2), generating a dysbiosis of the gut microbiome in S. littoralis larvae feeding on tomato leaves with neither structural damages to the midgut epithelium and peritrophic matrix nor alterations in the digestive capacity of the insect. This dysbiosis, among others, affects symbiotic bacteria of the genus Enterococcus and the functional capability of E. casseliflavus to nutritionally support the insect host with sugars and amino acids (3), with a consequent negative impact on S. littoralis development and survival (4). E: E. casseliflavus bacterial cells; M: microvilli; PM: peritrophic matrix; orange, green, and light blue shapes: insect digestive enzymes.
The functional basis of the gut dysbiosis could be associated with changes in the midgut metabolome due to toxins or compounds of fungal and/or plant origin, such as saponins and flavonoids both known to have antimicrobial (62, 63) and insecticide activity (64, 65), that can have a strong negative impact on midgut microbiota. Interestingly, the analysis of the T22-larvae midgut samples indicated a remarkable reduction of several fatty acids and hydroxy-fatty acids, in particular of plant origin, which are important membrane components and carbon suppliers for bacteria (66). Therefore, this marked alteration of their profile might have an impact on specific components of the gut microbiota, which may contribute to the observed dysbiosis.
The reduced accumulation of plant-derived fatty acids could be partly due to the effect of the Trichoderma treatment to tomato plants which causes a shift in the plant biosynthesis processes, redirecting the upstream precursors of fatty acids in favor of the eicosanoid pathway supporting the enhanced biosynthesis of jasmonic acid and other oxylipins (67), such as plant prostaglandins (68). In our study, over- and down-accumulated prostaglandin levels, both of plant and insect origin, were noted in the gut metabolome, indicating the possibility of plant contribution to the induction of an immune disguise causing a dysbiosis and a concurrent activation of the insect immune reaction against an altered gut microbial community. Indeed, prostaglandins are involved in several physiological functions and, particularly, in the modulation of the immune response (57). It has been recently reported that prostaglandins trigger the expression of Duox and the production of antimicrobial reactive oxygen species in the gut of Spodoptera exigua larvae (69). Moreover, the metabolic alteration observed in the microbiota of T22-larvae, and mainly attributed to E. casseliflavus, is compatible with an unhealthy status of this bacterium. This evidence, together with the increased midgut microbiota diversity, may indicate a possible reduced capability of E. casseliflavus to keep under control the expansion of opportunistic bacteria, a role reported for E. mundtii associated with the same lepidopteran host (49). Therefore, the loss of this regulation ability by E. casseliflavus could contribute to dysbiosis induction, with the consequent activation of a gut immune response, that is supported by the upregulation of antibacterial immune genes.
The activation of plant defense compounds plays a key role in the modulation of gut microbiota (43) and insect health. Our study demonstrates the important role of plant metabolites induced by T. afroharzianum, and/or produced by the fungus itself, in the alteration of microbiota structure and functionality which modulates a complex network of multitrophic interactions, having important evolutionary and ecological implications. These findings describe an interesting case of interkingdom relationships between a nonpathogenic soil fungus colonizing the plant and a phytophagous insect interacting with the same host plant as its nutritional resource (Fig. 6). Once the symbiotic fungus establishes a mutualistic relationship with the plant (20), i.e., endophytic status (18, 70), it acquires nourishment and protection, while the plant increases its primary metabolism (71) and resistance to both pathogens (21, 72) and pests (32) which can be considered as competitors for food resources. Even though the functional details on how Trichoderma induces plant changes remain to be studied, the present investigation provides insights into the ecology of these beneficial fungi, shedding light on a yet undescribed multitrophic interaction, which can be extremely important in the modulation of energy flow in natural food webs.
Similar interkingdom interactions have likely driven the evolution of other plant-associated soil fungi, which have acquired the ability to infect and kill insects, giving rise to new lineages of endophytic insect pathogens characterized by multifunctional lifestyles (73). In this context, the entomopathogenic Metarhizium is an excellent example of a fungus with genotypic plasticity allowing a multifunctional lifestyle, ranging from insect pathogen to plant colonizer or saprotroph depending upon exposure to different environmental conditions (74–76).
The regulation of pest insects and plant pathogens by soil fungi (e.g., refs. 17, 26, and 32) clearly suggests a central role of soil microbiota in controlling the allocation of plant resources in natural food webs, for the orchestration of nutrient cycling, determining the quantum resources recaptured in the soil and made available to plants (77–79). The soil microbiota not only provides an essential ecosystem service by decomposing plant biomass, directly providing nutrients to the plant, but also regulates the development and survival of plant pathogens and parasites sharing the same host, for optimal energy allocation. The competition between kingdoms is extremely frequent and not always well understood (4), but these soil biota-driven pathways of resource allocation in natural food webs through interkingdom interaction are of key importance for Earth’s health: they are pacemakers of the global energy budget.
This case study demonstrates that insect–plant interplays are part of a much broader and complex network of interactions, finely modulated by microorganisms spanning across below-ground and above-ground ecological communities, including those associated with other living organisms (Fig. 6). Both direct and indirect plant responses to microbial interactions can largely modulate plant–insect relations through partially hidden connections in the food web and further substantiate the concept that interactions among living organisms can be fully understood only when analyzed at the metaorganism level (6, 7). This is an aspect that should be carefully considered when studying the mechanisms of plant resistance and insect–plant coevolutionary pathways. Indeed, insect population diversity in terms of susceptibility to plant resistance traits may be attributed to different assemblages of gut microbiota, which are known to vary in a context-specific manner (80). This concept is further corroborated by the recent findings that plant resistance can trigger insect gut dysbiosis, with resident commensal bacteria switching to a pathogenic lifestyle (81). The possible occurrence of a similar phenomenon in the present study appears to be supported by the observed alteration in the composition of the microbiota and the enhanced expression of immune genes in S. littoralis midgut. It will be interesting to investigate how the microbial community in the haemocoel of T22-larvae changes over time and correlates to variations in mortality rates. Collectively, these fragments of emerging information clearly indicate that microorganims are key players in the modulation of insect physiological pathways required to face the challenging use of plant tissues as a food substrate.
The intricate interactions among living organisms and the key role played by associated microorganisms make it imperative that we investigate the potential impact of plant protection products on plant and animal microbiota, of both target and nontarget organisms, in any risk assessment study. For example, the results of this study suggest that potential negative impacts by Trichoderma and other plant symbionts on higher trophic levels (i.e., insect predators and parasitoids, and pollinators) cannot be ruled out. On the contrary, a positive effect could be the longer developmental time of surviving larvae, which increases their exposure to the action of natural antagonists, enhancing the impact of natural biocontrol (82). These aspects should be all carefully considered in developing new risk assessment protocols, based on rigorous modeling approaches, inspired by a holistic vision of the risk posed by any pest control strategy (83). Plant health, in its broadest sense, is crucial for primary production and food safety/security and can only be pursued if the complexity of natural and managed ecosystems is well understood, so that management strategies can be developed with a genuine one-health approach that should always guide our sustainable presence on Earth.
The discovery of how a network of microbial interactions, interconnecting the below-ground and above-ground environments, contributes to the control of the global energy budget in nature sheds light on the hitherto poorly understood interkingdom interactions and the important role they may have in the evolution of natural communities.
Materials and Methods
Insect Bioassays on Leaves of Tomato Plants Colonized by T. afroharzianum Strain T22.
S. lycopersicum cv “Dwarf San Marzano” plants colonized by T. afroharzianum strain T22 (T22-plants) and control plants (C-plants) were obtained by seed coating with spores as described by Coppola et al. (29). Plants were grown in the same environmental chamber at 25 ± 1 °C, photoperiod 16:8 light/dark, and irrigated three times a week.
Bioassays were performed on S. littoralis larvae alimented from hatching with subapical leaves of 4-wk-old T22-plants (T22-larvae) or of C-plants (C-larvae) to assess their impact on insect survival and development. Details on S. littoralis colony, rearing procedures, protocols for bioassays, parameters recorded during the experiments, and statistical analyses are reported in SI Appendix, Methods 1.
Effects of T22-Plant Leaves on the Midgut of S. littoralis Larvae.
Midgut microscopy.
Day 2 fourth instar T22-larvae and C-larvae were anesthetized on ice and then dissected to isolate the midgut with the enclosed intestinal content. Light and transmission electron microscopy observations were carried out to assess any structural, ultrastructural, and histochemical (i.e., glycogen and lipid deposits) change of the midgut caused by the ingestion of T-22 plant leaves. Protocols for midgut sample preparations for microscopy analyses, as well as the method used to quantify glycogen deposits in C-larvae and T22-larvae midgut cells, are reported in SI Appendix, Methods 2.
Midgut transcriptomics.
Day-2 fourth instar C-larvae and T22-larvae were cold-anesthetized and surface-sterilized as previously described (84) and then dissected under a horizontal-flow hood to isolate the midgut. After the removal of fat body and tracheal residues, the midgut epithelium was separated from the peritrophic matrix and its content and then washed with sterile phosphate-buffered saline (PBS). Each sample was immediately placed in TRIzol™ (Life Technologies) and stored at −80 °C until RNA extraction, which was performed according to the manufacturer’s instructions. For both the midgut epithelium and the peritrophic matrix content, the RNA extracted from three fourth instar larvae was pooled at equimolar concentration and used as a biological replicate in the high-throughput sequencing analyses (midgut epithelium transcriptomics, microbiota targeted metagenomics, and metatranscriptomics); three replicates per group were used for midgut epithelium and microbiota metatranscriptomics and five for microbiota targeted metagenomics. Details on RNA quantification, library preparation, sequencing, and bioinformatics analyses can be found in SI Appendix, Methods 3.
Digestive enzyme assays.
Day 2 fourth instar C-larvae and T22-larvae were anesthetized on ice and dissected, as described above, to separate the midgut epithelium from the peritrophic matrix and its content. The peritrophic matrix with its content was centrifuged at 15,000 × g for 10 min at 4 °C, and the supernatant (i.e., the midgut juice) was stored at −80 °C until use. The midgut epithelium was washed in PBS, lightly blotted on filter paper, weighed, and stored in liquid nitrogen until use.
Enzymatic assays were performed at 25 °C on midgut juice samples (total proteolytic activity, α-amylase activity, and lipase activity) and on midgut tissue homogenates (aminopeptidase N activity) using experimental conditions under which product formation was linearly associated with enzyme concentration. The protein concentration in each sample was determined using Coomassie Protein Assay Reagent (Thermo Fisher Scientific). Detailed protocols for enzymatic assays are reported in SI Appendix, Methods 4.
Taxonomic and Functional Characterization of the Midgut Microbiota.
Taxonomy.
In 16S rRNA metagenomic experiments, five replicates of endoperitrophic-derived RNA of fourth instar C-larvae and T22-larvae were processed (see Midgut transcriptomics). For each replicate, 500 µg of the total RNA was treated with ribonucleases-free deoxyribonuclease (Promega) according to the manufacturer’s instructions. deoxyribonuclease-treated RNAs were checked for DNA contamination through PCR using Bakt 341F/805R primers targeting the hypervariable V3 to V4 regions of the 16S rRNA gene (85), and no positive amplifications were obtained. For each sample, ~200 ng of deoxyribonuclease-treated RNA was reverse-transcribed into complementary DNA by using GoScript™ Reverse Transcriptase (Promega) and random primers according to the manufacturer’s instructions. V3 to V4 16S rRNA libraries were obtained using Bakt 341F/805R primers (85). Library preparation, NGS sequencing, bioinformatics, and statistical analyses are described in SI Appendix, Methods 5.
Transcriptomics.
Three replicates of endoperitrophic-derived RNA from fourth instar C-larvae and T22-larvae (see Midgut transcriptomics) were used as templates in library preparation for the transcriptomics of the endoperitrophic microbiota. Each indexed library was prepared from 500 ng of purified RNA with the Universal Prokaryotic RNA-Seq (Tecan) according to the manufacturer’s instructions. Library preparation, NGS sequencing, and bioinformatic and statistical analyses are described in SI Appendix, Methods 6.
Midgut Metabolomics.
For untargeted metabolomics analysis, the midguts from six day-2 fourth instar C-larvae and 6 T22-larvae were collected, weighted (~0.06 g each), and subjected to LC-ESI-HRMS. A detailed description of the extraction method, parameters used for the analysis of mass spectrometry data, and MS chromatograms is reported in SI Appendix, Methods 7.
Rescue Bioassays.
Plant.
Feeding bioassays, performed as described above (see Insect Bioassays on Leaves of Tomato Plants Colonized by T. afroharzianum Strain T22 and SI Appendix, Methods 1), were carried out on two experimental groups made of 16 synchronous T22-larvae, which, on day 2 of the third instar, were transferred to C-plant leaf disks to assess any rescue effect. T22-larvae and C-larvae acted as controls (two groups of 16 individuals for each experimental condition). During the bioassays, insect survival and development were daily recorded (SI Appendix, Methods 1).
Midgut microbiota.
Midgut microbiota was obtained by isolating the peritrophic matrix and its content (see Midgut transcriptomics) from 10 fourth instar C-larvae, on day 2. Details for midgut microbiota isolation, quantification, and E. casseliflavus identification are reported in SI Appendix, Methods 8. T22-larvae at day 2 of the third instar (two groups of 16 individuals for each experimental condition) were used in the feeding bioassay and daily offered with T22-plant 4-cm2 leaf disks overlaid with different bacterial suspensions, to assess their capacity to mitigate the negative effects of plant material, as detailed in SI Appendix, Methods 8.
Data, Materials, and Software Availability
The raw data, the Markdown file describing the whole pipeline of the analyses, and the intermediate files are available at the following link: https://figshare.com/s/b6db24859a65c391e629 (86). Raw reads were uploaded to Sequence Read Archive (SRA) under the BioProject PRJNA784009 (87). All other data are included in the manuscript and/or SI Appendix.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank Angharad Gatehouse (Newcastle University, UK) and David Giron (Institut de Recherche sur la Biologie de l’Insecte, IRBI, -National Center for Scientific Research, CNRS-, FR; Université François-Rabelais de Tours, FR) for their comments on the manuscript. This work was supported by: i) the European Union Horizon 2020 Research and Innovation Program (EcoStack; grant agreement No. 773554); ii) the Ministry of University and Research, Projects of National Relevance 2017 (PROSPECT, grant No. 2017JLN833); and iii) the European Union Next-GenerationEU [PIANO NAZIONALE DI RIPRESA E RESILIENZA (PNRR)—MISSIONE 4 COMPONENTE 2, INVESTIMENTO 1.4—D.D. 1032 17/06/2022, CN00000022]. This manuscript reflects only the authors’ views and opinions; neither the European Union nor the European Commission can be considered responsible for them.
Author contributions
M.C., M.M., and F.P. designed research; I.D.L., G.M., D.B., A.B., M.G.D.L., M.S., E.B., M. Bonelli, S.F., G.D., M.C.D., S.L.W., G.T., R.R., M.C., M.M., and F.P. performed research; I.D.L., G.F., G.M., M. Brunetti, D.B., A.B., M.G.D.L., M.S., E.B., S.F., G.D., M.C.D., S.L.W., G.T., R.R., M.C., M.M., and F.P. data analyses and interpretation; M.L., M.M., and F.P. funds acquisition; and M.L., M.C., M.M., and F.P. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Morena Casartelli, Email: morena.casartelli@unimi.it.
Matteo Montagna, Email: matteo.montagna@unina.it.
Francesco Pennacchio, Email: f.pennacchio@unina.it.
Supporting Information
References
- 1.Barnes A. D., et al. , Energy flux: The link between multitrophic biodiversity and ecosystem functioning. Trends Ecol. Evol. 33, 186–197 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gause G. F., The Struggle for Existence (The Williams and Wilkins Co., 1934). [Google Scholar]
- 3.Burns J. H., Strauss S. Y., More closely related species are more ecologically similar in an experimental test. Proc. Natl. Acad. Sci. U.S.A. 108, 5302–5307 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hochberg M. E., Lawton J. H., Competition between kingdoms. Trends Ecol. Evol. 5, 367–371 (1990). [DOI] [PubMed] [Google Scholar]
- 5.Gasmi L., et al. , Horizontally transmitted parasitoid killing factor shapes insect defense to parasitoids. Science 373, 535–541 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Rosenberg E., Zilber-Rosenberg I., Microbes drive evolution of animals and plants: The hologenome concept. mBio 7, e01395 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas A. E., Werren J. H., Holes in the hologenome: Why host-microbe symbioses are not holobionts. mBio 7, e02099 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berendsen R. L., Pieterse C. M., Bakker P. A., The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Pieterse C. M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S. C., Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Busby P. E., Ridout M., Newcombe G., Fungal endophytes: Modifiers of plant disease. Plant Mol. Biol. 90, 645–655 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Santoyo G., Moreno-Hagelsieb G., del Carmen Orozco-Mosqueda M., Glick B. R., Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Zhang N., et al. , An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98, 1076–1087 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Woo S. L., Hermosa R., Lorito M., E., Monte Trichoderma: A multipurpose, plant-beneficial microorganism for eco-sustainable agriculture. Nat. Rev. Microbiol. (2022), online (ahead of print), 10.1038/s41579-022-00819-5. [DOI] [PubMed]
- 14.Sung G. H., Poinar G. O. Jr., Spatafora J. W., The oldest fossil evidence of animal parasitism by fungi supports a Cretaceous diversification of fungal-arthropod symbioses. Mol. Phylogenet. Evol. 49, 495–502 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Spatafora J. W., Sung G. H., Sung J. M., Hywel-Jones N. L., White J. F. Jr., Phylogenetic evidence for an animal pathogen origin of ergot and the grass endophytes. Mol. Ecol. 16, 1701–1711 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Druzhinina I. S., et al. , Trichoderma: The genomics of opportunistic success. Nat. Rev. Microbiol. 9, 749–759 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Harman G. E., Howell C. R., Viterbo A., Chet I., Lorito M., Trichoderma species–opportunistic, avirulent plant symbionts. Nat. Rev. Microbiol. 2, 43–56 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Lorito M., Woo S. L., Harman G. E., Monte E., Translational research on Trichoderma: From ‘omics to the field. Annu. Rev. Phytopathol. 48, 395–417 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Mukherjee P. K., Horwitz B. A., Herrera-Estrella A., Schmoll M., Kenerley C. M., Trichoderma research in the genome era. Annu. Rev. Phytopathol. 51, 105–129 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Hermosa R., et al. , The contribution of Trichoderma to balancing the costs of plant growth and defense. Int. Microbiol. 16, 69–80 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Conrath U., Beckers G. J., Langenbach C. J., Jaskiewicz M. R., Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Martínez-Medina A., et al. , Shifting from priming of salicylic acid- to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 213, 1363–1377 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Moran-Diez M. E., et al. , Trichoderma and the plant heritable priming responses. J. Fungi 7, 318 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mendoza-Mendoza A., et al. , Molecular dialogues between Trichoderma and roots: Role of the fungal secretome. Fungal Biol. Rev. 32, 62–85 (2018). [Google Scholar]
- 25.Contreras-Cornejo H. A., Macías-Rodríguez L., del-Val E., Larsen J., The root endophytic fungus Trichoderma atroviride induces foliar herbivory resistance in maize plants. Appl. Soil. Ecol. 124, 45–53 (2018). [Google Scholar]
- 26.Chaverri P., Samuels G. J., Evolution of habitat preference and nutrition mode in a cosmopolitan fungal genus with evidence of interkingdom host jumps and major shifts in ecology. Evolution 67, 2823–2837 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Kubicek C. P., et al. , Evolution and comparative genomics of the most common Trichoderma species. BMC Genomics 20, 485 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coppola M., et al. , Transcriptome and metabolome reprogramming in tomato plants by Trichoderma harzianum strain T22 primes and enhances defense responses against aphids. Front. Physiol. 10, 745 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coppola M., et al. , Trichoderma atroviride P1 colonization of tomato plants enhances both direct and indirect defense barriers against insects. Front. Physiol. 10, 813 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jafarbeigi F., Samih M. A., Alaei H., Shirani H., Induced tomato resistance against Bemisia tabaci triggered by salicylic acid, β-aminobutyric acid, and Trichoderma. Neotrop. Entomol. 49, 456–467 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Muvea A. M., et al. , Colonization of onion roots by endophytic fungi and their impacts on the biology of Thrips tabaci. PLoS One 9, e108242 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Lelio I., et al. , Temperature differentially influences the capacity of Trichoderma species to induce plant defense responses in tomato against insect pests. Front. Plant Sci. 12, 678830 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Papantoniou D., et al. , Cascading effects of root microbial symbiosis on the development and metabolome of the insect herbivore Manduca sexta L. Metabolites 11, 731 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pappas M. L., Samaras K., Koufakis I., Broufas G. D., Beneficial soil microbes negatively affect spider mites and aphids in pepper. Agronomy 11, 1831 (2021). [Google Scholar]
- 35.Medeiros H., et al. , Tomato progeny inherit resistance to the nematode Meloidogyne javanica linked to plant growth induced by the biocontrol fungus Trichoderma atroviride. Sci. Rep. 7, 40216 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Battaglia D., Tomato below ground-above ground interactions: Trichoderma longibrachiatum affects the performance of Macrosiphum euphorbiae and its natural antagonists. Mol. Plant Microbe Interact. 26, 1249–1256 (2013). Erratum in: Mol. Plant Microbe Interact. 26, 1499 (2013). [DOI] [PubMed] [Google Scholar]
- 37.Coppola M., Trichoderma harzianum enhances tomato indirect defense against aphids. Insect Sci. 24, 1025–1033 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Contreras-Cornejo H. A., et al. , Alterations of foliar arthropod communities in a maize agroecosystem induced by the root-associated fungus Trichoderma harzianum. J. Pest Sci. 94, 363–374 (2021). [Google Scholar]
- 39.Erb M., Reymond P., Molecular interactions between plants and insect herbivores. Annu. Rev. Plant Biol. 70, 527–557 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Engel P., Moran N. A., The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev. 37, 699–735 (2013). [DOI] [PubMed] [Google Scholar]
- 41.McLean A. H., Parker B. J., Hrček J., Henry L. M., Godfray H. C., Insect symbionts in food webs. Philos. Trans. R. Soc. Lond. B Biol. Sci. 371, 20150325 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dicke M., Cusumano A., Poelman E. H., Microbial symbionts of parasitoids. Annu. Rev. Entomol. 65, 171–190 (2020). [DOI] [PubMed] [Google Scholar]
- 43.Mason C. J., Jones A. G., Felton G. W., Co-option of microbial associates by insects and their impact on plant-folivore interactions. Plant Cell Environ. 42, 1078–1086 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Tian A., Wang B., Jiang J., Injury-stimulated and self-restrained BMP signaling dynamically regulates stem cell pool size during Drosophila midgut regeneration. Proc. Natl. Acad. Sci. U.S.A. 114, E2699–E2708 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srinivasan A., Giri A. P., Gupta V. S., Structural and functional diversities in Lepidopteran serine proteases. Cell Mol. Biol. Lett. 11, 132–154 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adang M. J., “Chapter 81” in Handbook of Proteolytic Enzymes, Rawlings N. D., Salvesen G. S., Eds. (Academic Press, Boston, 2013), vol. 1, pp. 405–409. [Google Scholar]
- 47.Da Lage J. L., The amylases of insects. Int. J. Insect Sci. 10, 1–14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen B., et al. , Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 6, 29505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shao Y., et al. , Symbiont-derived antimicrobials contribute to the control of the lepidopteran gut microbiota. Cell Chem. Biol. 24, 66–75 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Tang X., et al. , Complexity and variability of gut commensal microbiota in polyphagous lepidopteran larvae. PLoS One 7, e36978 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shao Y., Arias-Cordero E., Guo H., Bartram S., Boland W., In vivo Pyro-SIP assessing active gut microbiota of the cotton leafworm, Spodoptera littoralis. PLoS One 9, e85948 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen B., et al. , Draft genome sequence of Enterococcus mundtii SL 16, an indigenous gut bacterium of the polyphagous pest Spodoptera littoralis. Front. Microbiol. 7, 1676 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mazumdar T., et al. , Transcriptomics reveal the survival strategies of Enterococcus mundtii in the gut of Spodoptera littoralis. J. Chem. Ecol. 47, 227–241 (2021). [DOI] [PubMed] [Google Scholar]
- 54.Parenti P., Giordana B., Sacchi V. F., Hanozet G. M., Guerritore A., Metabolic activity related to the potassium pump in the midgut of Bombyx mori larvae. J. Exp. Biol. 116, 69–78 (1985). [Google Scholar]
- 55.Toprak U., Hegedus D., Doğan C., Güney G. A., Journey into the world of insect lipid metabolism. Arch. Insect Biochem. Physiol. 104, e21682 (2020). [DOI] [PubMed] [Google Scholar]
- 56.Zhao X., Karpac J., The Drosophila midgut and the systemic coordination of lipid-dependent energy homeostasis. Curr. Opin. Insect Sci. 41, 100–105 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim Y., Stanley D., Eicosanoid signaling in insect immunology: New genes and unresolved issues. Genes 12, 211 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cai F., Druzhinina I. S., In honor of John Bissett: Authoritative guidelines on molecular identification of Trichoderma. Fungal Divers. 107, 1–69 (2021). [Google Scholar]
- 59.Brotman Y., Kapuganti J. G., Viterbo A., Trichoderma. Curr. Biol. 20, R390–R391 (2010). [DOI] [PubMed] [Google Scholar]
- 60.De Palma M., et al. , Transcriptome reprogramming, epigenetic modifications and alternative splicing orchestrate the tomato root response to the beneficial fungus Trichoderma harzianum. Hortic. Res. 6, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen B., et al. , Enterococcal symbionts of caterpillars facilitate the utilization of a suboptimal diet. J. Insect Physiol. 138, 104369 (2022). [DOI] [PubMed] [Google Scholar]
- 62.Górniak I., Bartoszewski R., Króliczewski J., Comprehensive review of antimicrobial activities of plant flavonoids. Phytochem. Rev. 18, 241–272 (2019). [Google Scholar]
- 63.Zaynab M., et al. , Saponin toxicity as key player in plant defense against pathogens. Toxicon 193, 21–27 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Singh B., Kaur A., Control of insect pests in crop plants and stored food grains using plant saponins: A review. LWT 87, 93–101 (2018). [Google Scholar]
- 65.Schnarr L., Segatto M. L., Olsson O., Zuin V. G., Kümmerer K., Flavonoids as biopesticides—Systematic assessment of sources, structures, activities and environmental fate. Sci. Total Environ. 824, 153781 (2022). [DOI] [PubMed] [Google Scholar]
- 66.Cronan J. E., Thomas J., Bacterial fatty acid synthesis and its relationships with polyketide synthetic pathways. Methods Enzymol. 459, 395–433 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Christie W. W., Harwood J. L., Oxidation of polyunsaturated fatty acids to produce lipid mediators. Essays Biochem. 64, 401–421 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Groenewald E. G., Van der Westhuizen A. J., Prostaglandins and related substances in plants. Bot. Rev. 63, 199–220 (1997). [Google Scholar]
- 69.Sajjadian S. M., Kim Y., PGE2 upregulates gene expression of dual oxidase in a lepidopteran insect midgut via cAMP signalling pathway. Open Biol. 10, 200197 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Harman G. E., Doni F., Khadka R. B., Uphoff N., Endophytic strains of Trichoderma increase plants’ photosynthetic capability. J. Appl. Microbiol. 130, 529–546 (2021). [DOI] [PubMed] [Google Scholar]
- 71.Vitti A., et al. , Trichoderma harzianum T-22 induces systemic resistance in tomato infected by cucumber mosaic virus. Front. Plant Sci. 7, 1520 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adnan M., et al. , Plant defense against fungal pathogens by antagonistic fungi with Trichoderma in focus. Microb. Pathog. 129, 7–18 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Barelli L., Moonjely S., Behie S. W., Bidochka M. J., Fungi with multifunctional lifestyles: Endophytic insect pathogenic fungi. Plant Mol. Biol. 90, 657–664 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Wang C. S., Hu G., St Leger R. J., Differential gene expression by Metarhizium anisopliae growing in root exudate and host (Manduca sexta) cuticle or hemolymph reveals mechanisms of physiological adaptation. Fungal Genet. Biol. 42, 704–718 (2005). [DOI] [PubMed] [Google Scholar]
- 75.Wang C., St Leger R. J., Developmental and transcriptional responses to host and nonhost cuticles by the specific locust pathogen Metarhizium anisopliae var. acridum. Eukaryot. Cell 4, 937–947 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pava-Ripoll M., et al. , The rhizosphere-competent entomopathogen Metarhizium anisopliae expresses a specific subset of genes in plant root exudate. Microbiology 157, 47–55 (2011). [DOI] [PubMed] [Google Scholar]
- 77.Crowther T. W., et al. , The global soil community and its influence on biogeochemistry. Science 365, eaav0550 (2019). [DOI] [PubMed] [Google Scholar]
- 78.van Bruggen A. H., et al. , One health—Cycling of diverse microbial communities as a connecting force for soil, plant, animal, human and ecosystem health. Sci. Total Environ. 664, 927–937 (2019). [DOI] [PubMed] [Google Scholar]
- 79.Pausas J. G., Bond W. J., On the three major recycling pathways in terrestrial ecosystems. Trends Ecol. Evol. 35, 767–775 (2020). [DOI] [PubMed] [Google Scholar]
- 80.Hochberg M. E., Lawton J. H., Competition between kingdoms. Trends Ecol. Evol. 5, 367–371 (1990). [DOI] [PubMed] [Google Scholar]
- 81.Paniagua Voirol L. R., Frago E., Kaltenpoth M., Hilker M., Fatouros N. E., Bacterial symbionts in Lepidoptera: Their diversity, transmission, and impact on the host. Front. Microbiol. 9, 556 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mason C. J., Peiffer M., St Clair A., Hoover K., Felton G. W., Concerted impacts of antiherbivore defenses and opportunistic Serratia pathogens on the fall armyworm (Spodoptera frugiperda). Oecologia 198, 167–178 (2022). [DOI] [PubMed] [Google Scholar]
- 83.Pennacchio F., Strand M. R., Evolution of developmental strategies in parasitic hymenoptera. Annu. Rev. Entomol. 51, 233–258 (2006). [DOI] [PubMed] [Google Scholar]
- 84.Topping C. J., Aldrich A., Berny P., Overhaul environmental risk assessment for pesticides. Science 367, 360–363 (2020). [DOI] [PubMed] [Google Scholar]
- 85.Montagna M., et al. , Metamicrobiomics in herbivore beetles of the genus Cryptocephalus (Chrysomelidae): Toward the understanding of ecological determinants in insect symbiosis. Insect Sci. 22, 340–352 (2015). [DOI] [PubMed] [Google Scholar]
- 86.Di Lelio I., et al. , Figshare, Workflow_analyses_code_data, https://figshare.com/s/b6db24859a65c391e629, 2022-05-18. [Google Scholar]
- 87.Di Lelio I., et al. , NCBI BioProject, PRJNA784009, https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA784009, 27-11-2021. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
The raw data, the Markdown file describing the whole pipeline of the analyses, and the intermediate files are available at the following link: https://figshare.com/s/b6db24859a65c391e629 (86). Raw reads were uploaded to Sequence Read Archive (SRA) under the BioProject PRJNA784009 (87). All other data are included in the manuscript and/or SI Appendix.