Abstract
Citrus cancer, caused by strains of Xanthomonas citri (Xc) and Xanthomonas aurantifolii (Xa), is one of the most economically important citrus diseases. Although our understanding of the molecular mechanisms underlying citrus canker development has advanced remarkably in recent years, exactly how citrus plants fight against these pathogens remains largely unclear. Using a Xa pathotype C strain that infects Mexican lime only and sweet oranges as a pathosystem to study the immune response triggered by this bacterium in these hosts, we herein report that the Xa flagellin C protein (XaFliC) acts as a potent defence elicitor in sweet oranges. Just as Xa blocked canker formation when coinfiltrated with Xc in sweet orange leaves, two polymorphic XaFliC peptides designated flgIII‐20 and flgIII‐27, not related to flg22 or flgII‐28 but found in many Xanthomonas species, were sufficient to protect sweet orange plants from Xc infection. Accordingly, ectopic expression of XaFliC in a Xc FliC‐defective mutant completely abolished the ability of this mutant to grow and cause canker in sweet orange but not Mexican lime plants. Because XaFliC and flgIII‐27 also specifically induced the expression of several defence‐related genes, our data suggest that XaFliC acts as a main immune response determinant in sweet orange plants.
Keywords: Citrus sinensis, citrus canker, Xanthomonas citri, Xanthomonas aurantifolii pathotype C, flagellin C, flgIII‐20, flgIII‐27, FliC
This study unravels the molecular basis underlying immunity against citrus canker bacteria, showing that novel flagellin epitopes elicit a defence response that protects citrus plants from Xanthomonas citri infection.
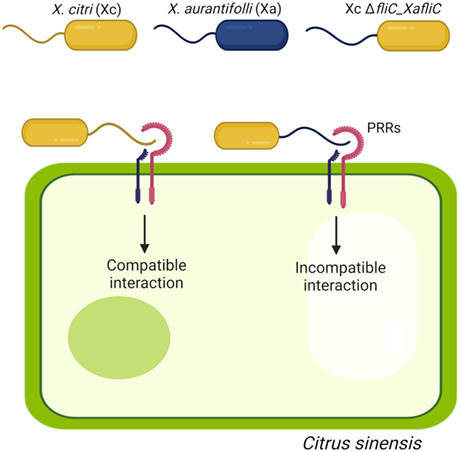
1. INTRODUCTION
Citrus canker, caused by the bacterial pathogen Xanthomonas citri (Xc), also referred to as X. citri pv. citri, is one of the most important citrus diseases and affects all commercial citrus varieties causing great economic losses to the citrus industry worldwide. The disease is characterized by water‐soaked and eruptive lesions that develop on leaves, fruits, and twigs, causing defoliation, premature fruit drop, and general tree decline (Brunings & Gabriel, 2003; Graham et al., 2004).
Different strains of Xc have been isolated from numerous citrus hosts around the world. The Xc Asiatic or type A strains are widely spread in citrus‐producing areas, cause the most severe canker symptoms, and have a broad host range, infecting all commercial orange (Citrus sinensis), lemon (Citrus limon), grapefruit (Citrus paradisi), and mandarin orange (Citrus reticulata) varieties. On the other hand, the related canker pathogen Xanthomonas aurantifolii (Xa), also classified as X. citri pv. aurantifolii, Xanthomonas fuscans pv. aurantifolii, or X. citri pv. fuscans, causes cankers on a limited number of citrus plants and is restricted to certain regions of Brazil and Argentina (Brunings & Gabriel, 2003). In particular, the Xa pathotype C strain ICMP 8435, henceforth called Xa, causes typical canker symptoms on Mexican lime (Citrus aurantifolia) trees only, whereas in sweet oranges it triggers a basal defence response that halts canker development (Cernadas et al., 2008).
The precise molecular mechanisms by which Xc induces cell growth and division in citrus, the hallmarks of canker lesions, are not fully understood. However, numerous studies carried out in the last two decades revealed that transcriptional activator‐like (TAL) effectors of the AvrBs3/PthA family play an essential role in triggering the transcriptional reprogramming of citrus cells, leading to cell hypertrophy and hyperplasia, the main host physiological and morphological changes that favour pathogen growth and spread. Of note, PthA4, the only Xc PthA variant that is essential for full canker symptom development, directly transactivates the citrus LOB1 gene (CsLOB1), which appears to function as a master regulator of downstream genes that control cell division and expansion (Abe & Benedetti, 2016; Al‐Saadi et al., 2007; Brunings & Gabriel, 2003; Duan et al., 2018; Hu et al., 2014; Pereira et al., 2014). Moreover, PthA4 has also been shown to interact with host proteins implicated in transcriptional control, including CsMAF1, a cell growth regulator and negative modulator of RNA polymerase III (Oliveira Andrade et al., 2020; Soprano et al., 2013, 2017; de Souza et al., 2012).
On the other hand, it remains largely unknown how citrus plants fight against Xc infection and what are the main molecular determinants underlying the immune response triggered by Xa in sweet orange plants. Although PthA variants from Xc strains have recently been shown to trigger immune responses on certain citrus hosts (Roeschlin et al., 2019; Teper et al., 2021), none of the PthA‐related effectors of Xa B and C strains, referred to as PthB and PthC, appear to act as avirulence factors on citrus (Al‐Saadi et al., 2007; Brunings & Gabriel, 2003). Accordingly, the ectopic expression of a Xa PthC effector in sweet orange epicotyls did not obviously induce the expression of defence executor genes (Pereira et al., 2014), which is also consistent with the fact that the pthC knockout mutant of a Xa C strain lost its pathogenicity in Mexican lime but not the ability to trigger a hypersensitive response (HR) in grapefruit (Al‐Saadi et al., 2007). These results thus suggest that, at least for Xa C isolates, TAL effectors do not seem to play a critical role in the immune response triggered by this bacterium in sweet orange and grapefruit. Moreover, the observation that X. citri pv. aurantifolii B and C strains have a reduced repertoire of pathogenicity‐related genes, which in principle could explain the narrower host range and lower aggressiveness of these pathogens compared to Xc strains (Fonseca et al., 2019), offers no clues as to the mechanism by which Xa triggers an immune response in sweet oranges. In line with this idea, it was shown that a X. fuscans pv. aurantifolii C strain, which causes canker only in key lime (C. aurantifolia), possesses a type III effector of the XopAG family, named AvrGf2, that is responsible for the HR observed in grapefruit (Gochez et al., 2015). AvrGf2 interacts with grapefruit cyclophilin GfCyp (Gochez et al., 2017); nevertheless, the precise mechanism by which AvrGf2 triggers an HR in grapefruit remains unclear.
Large‐scale transcriptional analyses of sweet orange plants challenged with Xa, relative to Xc, revealed that, in contrast to Xc, Xa strongly up‐regulates a set of defence‐related genes very early during infection, encoding, for instance, the mitogen‐activated protein kinase CsMAPK1, several WRKY factors, and pathogenesis‐related (PR) proteins, consistent with pathogen‐associated molecular pattern (PAMP)‐induced activation of a defence MAPK signalling cascade (Cernadas et al., 2008; de Oliveira et al., 2013). In fact, the augmented expression of CsMAPK1 in transgenic Troyer citrange plants was correlated with a decrease in Xc growth in infected leaves, accompanied by an increase in PR gene expression and reactive oxygen species (ROS) production and a reduction in canker pustule formation (de Oliveira et al., 2013). Thus, the characteristics of the immune response observed in sweet orange plants in response to Xa infection are consistent with a PAMP‐triggered immunity (PTI) type of defence response. Although plants recognize a wide range of bacterial PAMPs, flagellin has been reported as one of the major innate immunity elicitors in many plant–bacterial interactions (Cai et al., 2011; Clarke et al., 2013; Gomez‐Gomez & Boller, 2002; Nicaise et al., 2009; Wang et al., 2015; Zipfel et al., 2004). Because we found variations in the amino acid sequence between the flagellin C (FliC) proteins isolated from Xc (XcFliC) and Xa (XaFliC) and because FliC polymorphism has been reported to affect elicitation as well as evasion of plant defences (Malvino et al., 2022; Sun et al., 2006; Wei et al., 2020), we decided to investigate whether XaFliC could be involved in the immune response triggered by Xa in sweet orange plants.
Here, we present evidence that XaFliC is sufficient to trigger a robust defence response that completely halts the development of canker lesions caused by virulent Xc in sweet oranges but not Mexican lime plants. We also show that two polymorphic peptides of XaFliC, denoted here as flgIII‐20 and flgIII‐27, which are not related to the well‐known flg22 and flgII‐28 peptides, also protected sweet orange leaves from Xc infection. To the best of our knowledge, this is the first description of FliC as a PAMP or molecular determinant of an immune response induced by a citrus canker pathogen.
2. RESULTS
2.1. Xa protects sweet orange leaves from Xc infection
It is well established that Xa triggers a basal defence response in all commercial sweet orange varieties (Brunings & Gabriel, 2003; Cernadas et al., 2008). To verify whether this defence response could prevent Xc infection and canker formation, sweet orange leaves of cultivars Natal and Pera were infiltrated with a mixed suspension of Xa and Xc cells at a 1:1 ratio. In comparison to leaf sectors inoculated with Xa or Xc alone at equivalent cell densities, used as controls for the basal defence response and canker development, respectively, leaf sectors infiltrated with the mixed Xa + Xc suspension showed no canker symptoms, in both Natal and Pera plants (Figure 1a). When Xa was infiltrated 24 h prior to Xc inoculation, the defence response still prevailed and no canker symptoms developed in either Natal or Pera leaves (Figure 1b). However, when Xc was infiltrated 24 h prior to Xa inoculation, canker symptoms developed normally on both citrus hosts (Figure 1c). These results show that the defence response induced by Xa in sweet orange leaves can suppress canker formation if triggered early or before Xc suppresses basal defences and establishes an infection.
FIGURE 1.
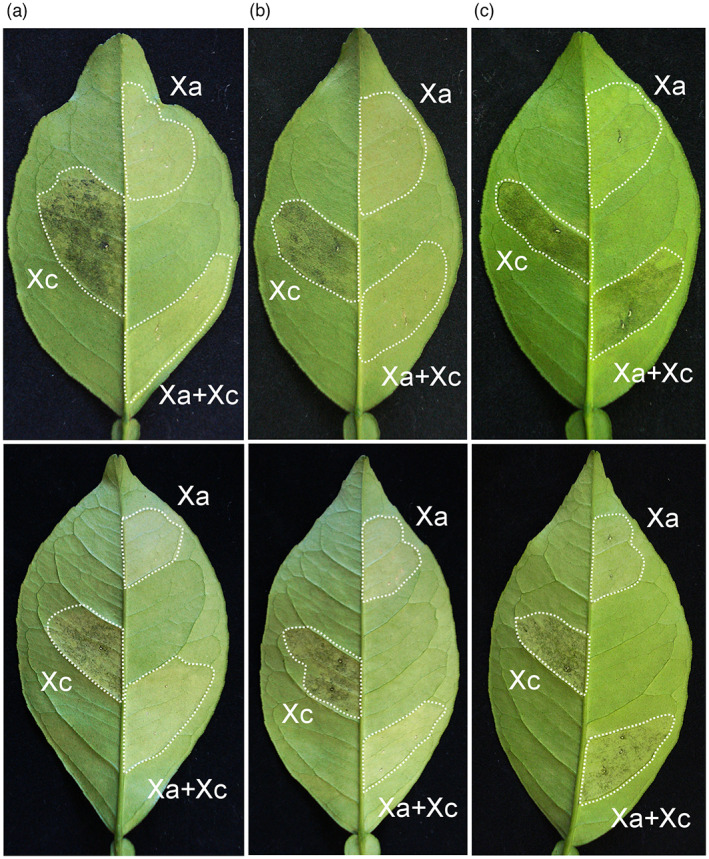
Xanthomonas aurantifolii (Xa) protects sweet orange leaves from Xanthomonas citri (Xc) infection. (a) Leaves of Natal (upper panel) and Pera (bottom panel) plants infiltrated with a suspension of Xc or Xa (OD600 = 0.1) or with a mixed suspension of Xa and Xc cells at a 1:1 ratio (Xa + Xc). Xa blocks canker development when simultaneously inoculated with Xc. (b) Twenty‐four hours after Xa infiltration, Xc was inoculated in the same leaf area (Xa + Xc) and canker symptoms did not develop. (c) Twenty‐four hours after Xc infiltration, Xa was inoculated in the same leaf area (Xa + Xc) and canker symptoms developed normally, indicating that Xc suppresses the Xa‐triggered defence if established first. Infiltrated leaf sectors are indicated by the dashed lines and those inoculated with Xc or Xa separately served as controls. Canker symptoms were evaluated 10 days postinoculation. Three leaves of each citrus variety were infiltrated and the images presented are representative of the phenotype observed in all infiltrated leaves
2.2. Xa‐induced protection against Xc infection is time‐dependent
The results shown in Figure 1 suggest that Xa‐induced protection of sweet orange leaves against Xc infection is time‐dependent. To further evaluate this response and test whether spray inoculation of Xa could also protect plants from Xc infection, leaves of Natal plants were sprayed with a Xa suspension, and after different time intervals Xc was infiltrated into the leaves. We found that spray application of Xa prevented canker formation only when Xc was infiltrated 24 h after spraying, whereas no protection was observed when Xc was infiltrated 4 h or 5 days after Xa application (Figure 2). These results suggest that the defence response triggered by Xa has a short time frame of hours after bacterial spraying, which is consistent with previous transcriptome analysis of Pera leaves challenged with this pathogen (Cernadas et al., 2008). Moreover, Xa‐induced protection appears to require bacterial penetration into the leaf mesophyll, as canker symptoms developed normally in leaves sprayed with Xa 4 h before Xc infection (Figure 2), but not in leaves coinfiltrated or previously infiltrated with Xa (Figure 1a,b).
FIGURE 2.
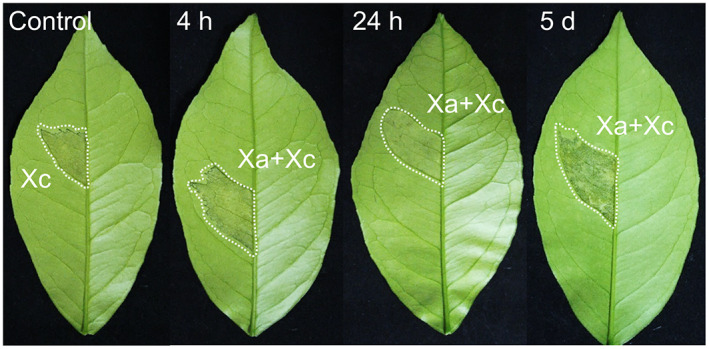
Xanthomonas aurantifolii (Xa)‐induced protection against Xanthomonas citri (Xc) infection is time‐dependent. Natal leaves were sprayed with a suspension of Xa (OD600 = 0.1) and after 4 h, 24 h, or 5 days, Xc (OD600 = 0.1) was infiltrated into the leaves (areas surrounded by dashed lines). Canker symptoms were recorded 10 days after Xc inoculation. Xa prevented canker formation when Xc was inoculated 24 h after Xa application. No protection was observed when Xc was inoculated 4 h or 5 days after the Xa treatment, indicating that the Xa‐induced protection response has a short time frame of a few hours and requires bacterial penetration into the leaf mesophyll to occur. The images presented are representative of the phenotype observed in most infiltrated leaves
2.3. Amino acid polymorphisms between Xa and Xc FliC proteins
The observation that bacterial penetration into the leaf mesophyll was critical for Xa‐induced protection against Xc infection suggested that FliC, a major innate immunity elicitor in many plant–bacterial interactions, might be involved in this process. To investigate this possibility, we compared the amino acid sequences of FliC from Xa (XaFliC) and Xc (XcFliC). Because the genome of the Xa strain used in this study (ICMP 8435, Cernadas et al., 2008) is not available, flagella of Xa and Xc were extracted and proteins associated with this preparation were resolved by SDS‐PAGE and identified by mass spectrometry (Figure 3). These analyses confirmed that the major approximately 50 kDa bands, observed at higher levels in Xa samples (Figure 3a), correspond to FliC proteins. All the peptides identified by mass spectrometry (Table S1) showed perfect matches to the FliC proteins of Xc and Xa strains deposited at the NCBI database with the reference sequences WP_003482972 and WP_007962409, respectively.
FIGURE 3.
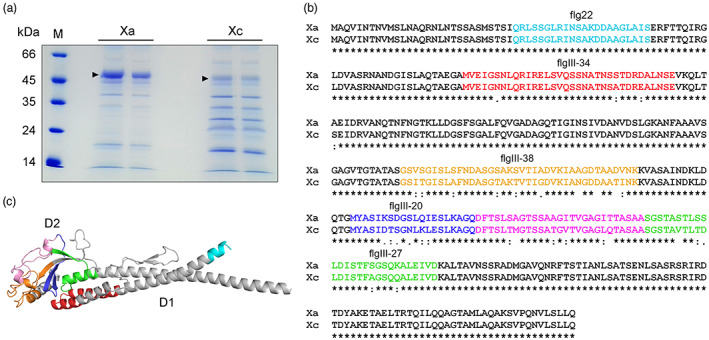
The FliC proteins from Xanthomonas aurantifolii (Xa) and Xanthomonas citri (Xc) display several amino acid polymorphisms. (a) SDS‐PAGE of flagellum preparations showing the major approximately 50 kDa band corresponding to the FliC proteins from Xa and Xc (arrowheads). M indicates the molecular marker. (b) Protein sequence alignment performed with Clustal Omega showing that the amino acid polymorphisms between XaFliC (Xa) (GenBank accession WP_007962409) and XcFliC (Xc) (GenBank accession WP_003482972) lie outside of the flg22 region (coloured in cyan). Regions coloured in red (flgIII‐34), orange (flgIII‐38), blue (flgIII‐20), magenta, and green (flgIII‐27) represent the polymorphic peptides between the two FliC proteins. The flgIII‐34 sequence includes the Pseudomonas syringae flgII‐28 peptide. (c) Structural model of XaFliC based on the crystal structure of the Sphingomonas sp. A1 flagellin (PDB code 3K8V) showing that the corresponding polymorphic regions depicted in (b) are located mostly in the globular D2 domain
Protein sequence alignment revealed a number of amino acid polymorphisms between XaFliC and XcFliC (Figure 3b). Notably, all amino acid changes found between XaFliC and XcFliC lie outside the flg22 region (Figure 3b). Moreover, according to our structural models, most of the FliC polymorphic regions, hereinafter referred to as the flgIII‐20, flgIII‐27, flgIII‐34, and flgIII‐38 peptides, are located in the FliC D2 domain (Figure 3b,c). The only exception is flgIII‐34, which corresponds to the Pseudomonas syringe pv. tomato flgII‐28, which is recognized by the tomato FLS3 receptor (Cai et al., 2011; Clarke et al., 2013) (Figure 3c).
2.4. Polymorphic peptides from XaFliC protects citrus from Xc infection
The amino acid polymorphisms found between XaFliC and XcFliC suggested that a specific region of the XaFliC polypeptide could evoke the immune response triggered by Xa in sweet orange plants. To test this hypothesis, the five peptides spanning the FliC polymorphic regions (Figure 3b) were synthesized and coinfiltrated with Xc in Natal leaves. However, the peptide located between flgIII‐20 and flgIII‐27 (Figure 3b) was insoluble in aqueous solutions and thus could not be tested.
We found that while none of the XcFliC peptides affected canker development in sweet orange leaves, the XaFliC peptides flgIII‐20 and flgIII‐27 fully protected Natal leaves from Xc infection, as no canker symptoms developed in the leaf sectors coinfiltrated with them (Figure 4a). Similar results were observed in Troyer citrange leaves (Figure 4b). On the other hand, flgIII‐20 and flgIII‐27 derived from XaFliC only partially protected Mexican lime leaves from Xc infection (Figure 4c). Likewise, canker symptoms elicited by Xa in Mexican lime were slightly attenuated by the XaFliC flgIII‐20 and flgIII‐27 peptides (Figure 4d). Because all tested peptides did not inhibit Xc or Xa growth in culture medium (Figure S1), the results indicate that flgIII‐20 and flgIII‐27 derived from XaFliC elicit a robust defence response in Natal and Troyer, but not Mexican lime plants. In addition, to rule out the possibility that the flg22 peptide, which is identical between XaFliC and XcFliC, does not contribute to the defence response triggered by Xa in sweet oranges, Xc was infiltrated in the presence and absence of flg22 in Natal, Pera, and Valencia leaves. The results confirm that flg22 does not protect sweet orange leaves from Xc infection (Figure S2).
FIGURE 4.
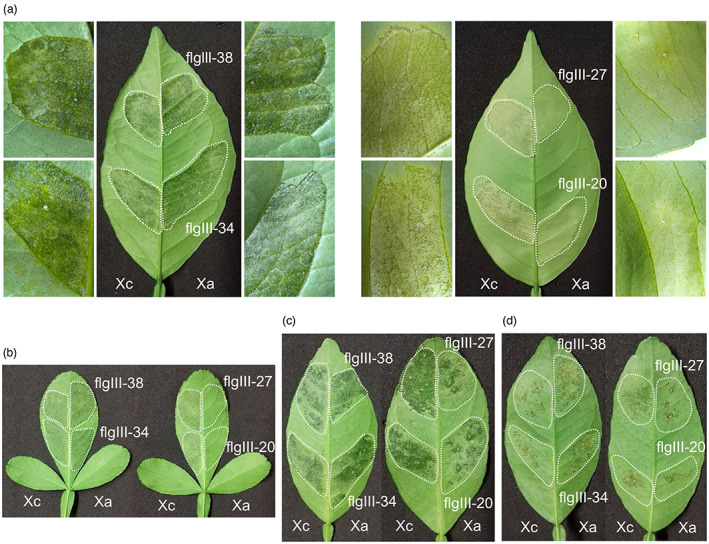
Polymorphic peptides from XaFliC, but not XcFliC, protected citrus from Xanthomonas citri (Xc) infection. (a) Natal leaves were infiltrated with a water suspension of Xc (OD600 = 0.05) in the presence of the XcFliC (Xc) or XaFliC (Xa) peptides at a final concentration of 70 μM (areas surrounded by dashed lines). Canker symptoms were recorded 10 days after bacterial inoculation. Only peptides flgIII‐20 and flgIII‐27 inhibited canker formation. Details of the infiltrated leaf sectors at higher magnification (10×) are shown on the left and right. (b) Same experiment performed with Troyer citrange showing that only peptides flgIII‐20 and flgIII‐27 protected leaves from Xc infection. (c) Peptides flgIII‐20 and flgIII‐27 partially protected Mexican lime leaves from Xc infection. (d) Peptides flgIII‐20 and flgIII‐27 attenuated canker development caused by Xanthomonas aurantifolii (Xa) in Mexican lime leaves. Three leaves of each citrus variety were infiltrated and the images presented are representative of the phenotype observed in most infiltrated leaves
2.5. XaFliC elicits canker resistance in sweet orange plants
To confirm the role of XaFliC as a major defence response elicitor in sweet orange plants, a Xa fliC mutant, defective in XaFliC production (XaΔfliC), was generated and further complemented with the XaFliC gene, as revealed by western blot (Figure 5a) and cell motility assays (Figure 5b). We found that contrary to wild‐type Xa, the XaΔfliC mutant did not protect Natal, Pera, or Valencia leaves from Xc infection (Figure 5c). Although complementation of the XaΔfliC mutant with XaFliC did not restore the protective role of Xa in most of the sweet orange varieties tested, except in Natal plants (Figure 5c), possibly due to low levels of complementation, as revealed by the motility assay (Figure 5b), the results do show that XaFliC contributes to the defence response triggered by Xa in sweet oranges. Interestingly, disruption of XaFliC was not sufficient to cause canker on sweet oranges (Figure S3); however, the reduction in leaf yellowing induced by the XaΔfliC mutant, observed in Natal, Pera, and Sorocaba leaves (Figure S3), suggests that the XafilC mutation attenuated the defence response triggered by Xa in these hosts.
FIGURE 5.
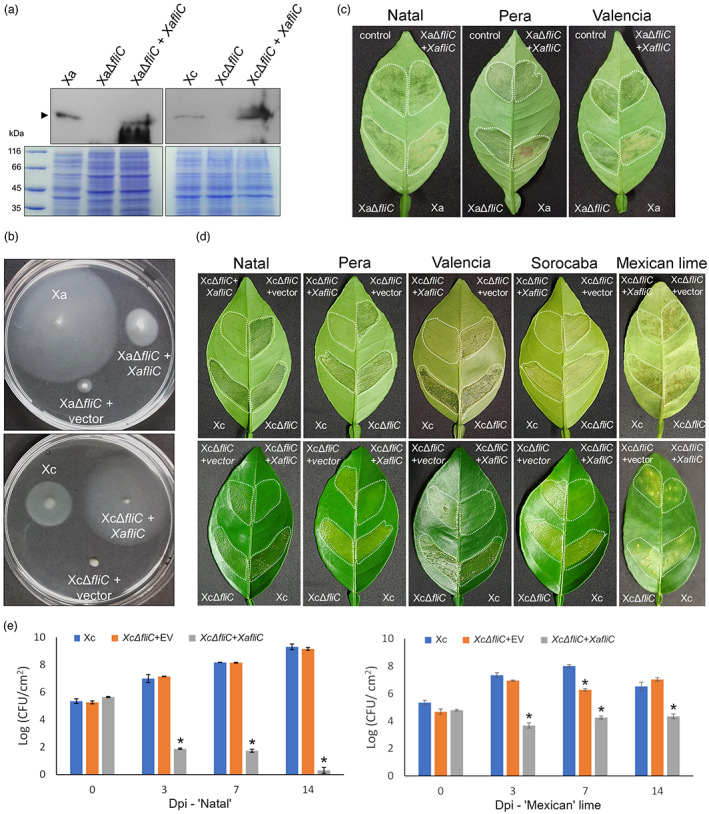
XaFliC is the main elicitor of canker resistance in sweet oranges. (a) Western blot analysis showing the expression of XaFliC (arrow) in wild‐type Xanthomonas aurantifolii (Xa) and Xanthomonas citri (Xc), fliC mutants XaΔfliC and XcΔfliC, and corresponding fliC mutants complemented with XaFliC (XaΔfliC + XaFliC, XcΔfliC + XaFliC). Blots were probed with anti‐XaFliC serum. (b) Motility assays performed on soft agar showing that disruption or deletion of FliC in Xa and Xc significantly reduces bacterial motility. Complementation of these mutants with XaFliC only partially restored Xa motility, whereas in Xc, XaFliC expression significantly increased cell motility. (c) Leaves of Natal, Pera, and Valencia plants infiltrated with Xc (control) or with a mixed suspension (OD600 = 0.1) of Xc + Xa (Xa) or Xc + XaΔfliC transformed with empty vector or vector expressing XaFliC (at a 1:1 ratio), showing that Xa but not XaΔfliC blocks canker development when simultaneously inoculated with Xc. (d) Canker symptoms developed on Natal, Pera, Valencia, and Sorocaba sweet oranges and in Mexican lime leaves 10 days after they were infiltrated with a water suspension (OD600 = 0.1) of Xc or the corresponding XcΔfliC mutants (areas surrounded by dashed lines). Expression of XaFliC in the XcΔfliC mutant was sufficient to trigger a robust defence response in all sweet orange varieties tested, but not in Mexican lime. Three leaves of each citrus variety were infiltrated and the images presented are representative of the phenotype observed in most infiltrated leaves. (e) Bacterial counts on Natal and Mexican lime leaves infiltrated with wild‐type Xc or XcΔfliC transformed with XaFliC or the empty vector (EV), used as control, determined at 0, 3, 7, and 14 days post‐infection (dpi). While in Natal leaves the XcΔfliC mutant expressing XaFliC showed a sharp reduction in growth within 14 days of bacterial inoculation, relative to Xc or the XcΔfliC mutant carrying the empty vector, this complemented strain reached much higher titres in Mexican lime leaves at 3, 7, and 14 dpi. Error bars represent standard deviations of three independent biological samples, and asterisks indicate statistically significant differences between the means at α = 0.05 relative to Xc
Next, we expressed the XaFliC gene in the Xc FliC‐deletion mutant (XcΔfliC). Western blot (Figure 5a) and bacterial motility assays (Figure 5b) confirmed that complementation of the XcΔfliC mutant with the XaFliC gene not only restored but even increased bacterial motility, relative to wild‐type Xc or the XcΔfliC mutant transfected with empty vector, used as control (Figure 5b).
Plant inoculations with these bacterial mutants revealed that deletion of FliC in Xc did not alter canker development. Surprisingly, however, the expression of XaFliC in the XcΔfliC mutant was sufficient to elicit a robust defence response in several sweet orange varieties, but not in Mexican lime, although canker symptoms produced by this mutant in Mexican lime were slightly reduced in comparison to wild‐type Xc or XcΔfliC transfected with empty vector (Figure 5d). In line with these results, bacterial counts on Natal leaves revealed that leaf sectors infiltrated with XcΔfliC expressing XaFliC showed a marked reduction in bacterial growth at 3, 7, and 14 days after bacterial inoculation, compared to leaf sectors infiltrated with Xc or XcΔfliC carrying the empty vector, used as control (Figure 5e). Conversely, the XcΔfliC mutant expressing XaFliC reached much higher titres in Mexican lime leaves at 3 to 14 days postinoculation, although it grew more slowly than Xc or XcΔfliC carrying the empty vector (Figure 5e), which is consistent with the fact that this mutant also induced less severe canker symptoms in lime relative to Xc or XcΔfliC carrying the empty vector (Figure 5d).
Together, these results confirm that XaFliC is a major elicitor of the immune response triggered by Xa in sweet orange plants.
2.6. FLS2 and WRKY22 induction in sweet orange is dependent on XaFliC expression
In a previous gene expression study using differential display and microarray analyses, we found that Xa induces the expression of several sweet orange genes involved in basal defence within 48 h of bacterial inoculation (Cernadas et al., 2008). To broaden our understanding of the transcriptional changes associated with the defence response triggered by Xa in sweet orange plants, we performed RNA sequencing (RNA‐seq) analysis of Natal leaves infiltrated with Xa, Xc, or water, at 24 and 48 h after bacterial inoculation.
The RNA‐seq data not only corroborated our previous gene expression studies (Cernadas et al., 2008; Pereira et al., 2014), but also revealed a larger number of innate immunity‐related genes being predominantly or exclusively up‐regulated by Xa, relative to Xc infection, including genes encoding several leucine‐rich repeat–receptor‐like kinases (RLKs), genes encoding transcription factors of the WRKY, BHLH, and MYB families, and genes associated with oxidative burst, the HR, and senescence (Data S1). Interestingly, while Xc preferentially induced the expression of genes associated with canker formation, including CsLOB1, Xa rapidly induced the expression of FLS2 and WRKY22, suggesting that flagellin perception was predominantly activated by this bacterium (Data S1).
To validate the RNA‐seq data and reveal which genes among the most induced by Xa show XaFliC‐dependent expression, RNA samples from Natal leaves infiltrated with Xc, XcΔfliC, XcΔfliC complemented with XaFliC, or water were analysed by reverse transcription‐quantitative real‐time PCR (RT‐qPCR), 24 h after bacterial inoculation (Figure 6). We found that among the seven RLK genes predominantly induced by Xa (Data S1), only three (FLS2, RLK15, and RLKC) showed a significant XaFliC‐dependent up‐regulation (Figure 6a). Likewise, among the six Xa‐induced transcription factor‐encoding genes tested, WRKY22 and MYB102 were the most highly induced in response to XaFliC expression (Figure 6b). Moreover, several Xa‐induced genes related to oxidative burst (respiratory burst oxidase homologue D [RBOHB], glutathione‐S‐transferase [GST], and peroxidase 52 [PRX52]) and the HR and senescence (senescence‐associated gene 13 [SAG13], accelerated cell death 2 [ACD2], plant U‐box protein 24 [PUB24], cytochrome P450 3A [CYP3A], and cysteine‐rich repeat protein 38 [CRR38]) also showed XaFliC‐dependent up‐regulation (Figure 6c,d, respectively). This pattern of gene regulation is thus consistent with the fact that XaFliC triggers a basal defence response in sweet oranges.
FIGURE 6.
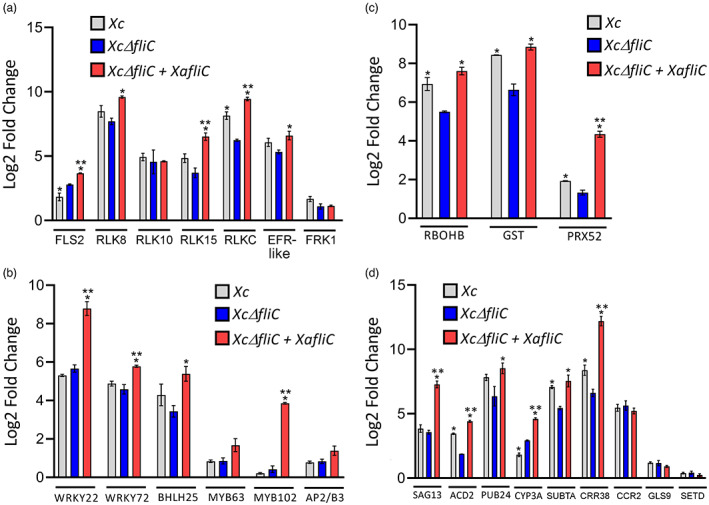
XaFliC‐dependent expression of FLS2, WRKY22, and other defence‐related genes in sweet orange. Reverse transcription‐quantitative PCR analysis of Natal leaves challenged with Xanthomonas citri (Xc), XcΔfliC, XcΔfliC complemented with XaFliC, or water as control, 24 h after bacterial inoculation. (a) Relative expression levels of RLK genes showing XaFliC‐dependent expression of Xanthomonas aurantifolii (Xa)‐induced genes, including FLS2. (b) Relative expression levels of transcription factor‐encoding genes showing that the WRKY22 and MYB102 homologues are the most highly up‐regulated in response to XaFliC expression. (c) Relative expression levels of oxidative burst‐associated genes showing significant induction of the putative peroxidase gene PRX52 in leaves infiltrated with XcΔfliC complemented with XaFliC relative to leaves infiltrated with Xc or XcΔfliC. (d) Relative expression levels of hypersensitive response (HR)‐ and senescence‐related genes showing XaFliC‐dependent expression of the SAG13, ACD2, CYP3A, and CRR38 homologues. Error bars represent standard deviations of three independent biological samples, whereas asterisks denote statistically significant differences between the means at the α = 0.05 level relative to XcΔfliC (*) or Xc (**)
To know whether flgIII‐20 and flgIII‐27 from XaFliC could also induce the expression of the citrus genes up‐regulated by XaFliC (Figure 6), we performed RT‐qPCR analysis of Natal leaves infiltrated with these peptides. Considering that flg22 induces rapid activation of gene expression in plant cells with maximum transcriptional changes occurring for instance in Arabidopsis at 3 h after treatment (Czékus et al., 2021; Denoux et al., 2008), we inspected the mRNA levels of the XaFliC‐induced genes (Figure 6) in Natal leaves 3 h after leaves had been infiltrated with flgIII‐20 or flgIII‐27 from XaFliC and XcFliC, or with flg22 or water as controls. Although flgIII‐20 did not significantly change the expression levels of the XaFliC‐induced genes analysed, except for a small induction of FLS2, WRKY22, and MYB102 caused by flgIII‐20 from XaFliC only, we found that flgIII‐27 from XaFliC, but not XcFliC, specifically induced the expression of FLS2, RLK15, RLKC, FRK1, CRR38, bHLH25, MYB102, SAG13, and ACD2, relative to water infiltration (Figure 7). In addition, except for FLS2, all these genes were more strongly up‐regulated in response to flgIII‐27 than in response to flg22 (Figure 7). Surprisingly, WRKY22 and WRKY72 were equally induced by flgIII‐27 from XaFliC and XcFliC (Figure 7).
FIGURE 7.
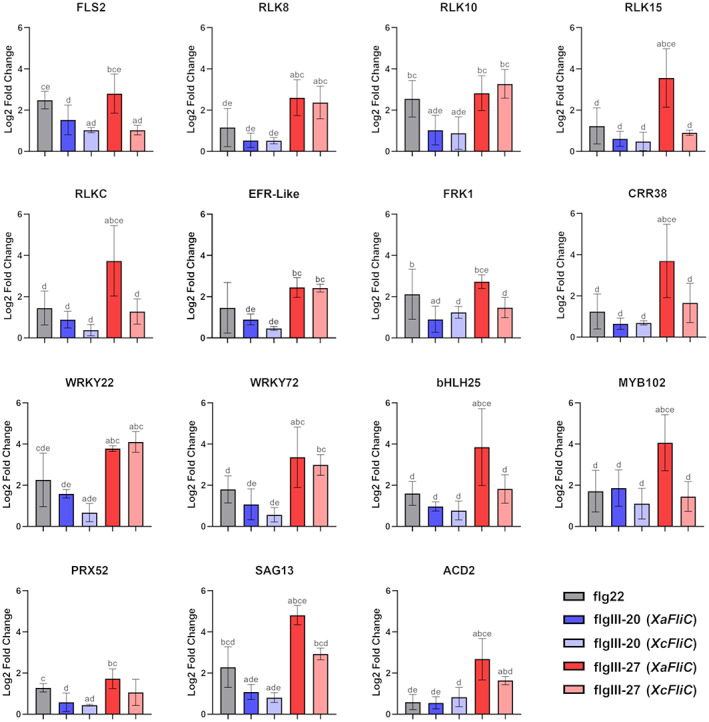
Gene expression analysis of XaFliC‐induced genes in response to treatment with flg peptides. Reverse transcription‐quantitative PCR analysis of Natal leaves infiltrated with 70 μM flgIII‐20 or flgIII‐27 from XaFliC and XcFliC, or with flg22 or water, used as controls, 3 h postinfiltration. While flgIII‐20 did not significantly change the expression levels of most XaFliC‐induced genes, flgIII‐27 from XaFliC, but not XcFliC, specifically induced the expression of FLS2, RLK15, RLKC, FRK1, CRR38, bHLH25, MYB102, SAG13, and ACD2, relative to the water control. Error bars denote standard deviations of two independent biological samples with three technical replicates of each treatment. Statistically significant differences among the groups are indicated by the letters a, b, c, d, and e, which represent flg22, flgIII‐20 (XaFliC), flgIII‐20 (XcFliC), flgIII‐27 (XaFliC), and flgIII‐27 (XcFliC) treatments, respectively, where a indicates, for instance, that the analysed group is statistically different (p < 0.05) from the flg22 treatment group
2.7. Amino acid polymorphisms of flgIII‐20 and flgIII‐27 are commonly found in the Xanthomonas genus
To analyse whether the polymorphisms observed between XaFliC and XcFliC are conserved among Xc and Xa strains or also found in other Xanthomonas species/pathovars, we initially performed BlastP searches using XaFliC as query. Notably, we found that the amino acid sequence of XaFliC is fully conserved in all the designated X. aurantifolii, X. citri pv. aurantifolii, X. fuscans pv. aurantifolii, and X. citri pv. fuscans strains, but not in the X. citri pv. citri or Xanthomonas axonopodis pv. citri strains. The NCBI accession WP_007962409 refers to 109 proteins identical to XaFliC belonging to several Xanthomonas species/pathovars most closely related to Xa, including X. citri pv. phaseoli var. fuscans and Xanthomonas phaseoli pv. dieffenbachiae. Likewise, BlastP searches using XcFliC as query revealed 410 identical proteins belonging mainly to X. citri, X. citri pv. citri, and X. axonopodis pv. citri strains and other Xanthomonas species, but none belonging to X. aurantifolii strains (NCBI multispecies accession number WP_003482972).
Next, we performed BlastP searches using only the XaFliC region spanning the flgIII‐20 and flgIII‐27 peptides. The resulting sequence alignment comprising mostly nonidentical sequences sharing 98% to 65% identity to XaFliC is shown in Figure 8. This protein alignment confirms that the amino acid polymorphism found between XaFliC and XcFliC is shared among many Xanthomonas species/pathovars, suggesting that the FliC regions spanning flgIII‐20 and flgIII‐27 are subject to selective pressure. This assumption is supported by the type of amino acid changes observed at specific positions in these regions. For instance, in flgIII‐20, a K‐to‐D change at position 6 and an S/T‐to‐N change at position 10 distinguish a large group of Xanthomonas species/pathovars, including Xa, from the X. citri, Xanthomonas passiflorae, Xanthomonas manihotis, and Xanthomonas malvacearum group (Figure 8). It is expected that only these two nonconserved amino acid changes could potentially alter recognition or affinity of flgIII‐20 by a pattern recognition receptor (PRR). Likewise, the nonconserved changes from S to V at position 6 and from S/G/A to T at position 9 in flgIII‐27 also distinguish the Xa‐related group from the Xc‐related group (Figure 8). Furthermore, two serine residues, at positions 10 and 17 in flgIII‐27, are replaced by N/D and A, respectively, between these two groups of Xanthomonas, except in Xanthomonas fragariae, which has an asparagine residue at position 10 (Figure 8). Because serine and threonine residues are prone to phosphorylation and glycosylation (Nothaft & Szymanski, 2010; Shi et al., 2014), such substitutions may represent important changes in the FliC structure and consequently receptor recognition.
FIGURE 8.

Amino acid polymorphisms in flgIII‐20 and flgIII‐27 are found in several Xanthomonas species and pathovars. Protein sequence alignment of Xanthomonas FliC proteins showing that the amino acid polymorphisms found in flgIII‐20 and flgIII‐27 between Xanthomonas aurantifolii (Xa) and X. citri (Xc) are also present in several species/pathovars of Xanthomonas. The alignment, performed with ClustalW, includes the FliC regions spanning the flgIII‐20 and flgIII‐27 peptides from the following Xanthomonas species/pathovars: Xa (WP_007962409), X. citri pv. glycines WP_016848727, X. campestris pv. mirabilis WP_218495416, X. euvesicatoria WP_218195708, X. euvesicatoria pv. allii MCP3041826, X. citri pv. thirumalacharii WP_223574663, X. campestris pv. paulliniae WP_218557524, X. arboricola WP_055854457, X. perforans MCF5971967, X. dyei WP_104616676, X. prunicola WP_101364107, X. oryzae WP_134953848, X. oryzae pv. oryzicola AEQ96507, X. fragariae WP_254223128, X. hortorum WP_115038939, X. pisi WP_046964182, X. floridensis WP_064510802, X. cucurbitae WP_104603719, X. cannabis WP_047696158.1, X. bromi WP_065467843, X. vesicatoria WP_005994319, X. campestris WP_218559892, X. vasicola WP_010373629, X. axonopodis pv. passiflorae MBV6815034, X. axonopodis pv. manihotis WP_017154654, X. citri pv. malvacearum WP_003482972.1, and Xc (WP_003482972). A K‐to‐D change at position 6 and an S/T‐to‐N change at position 10 in flgIII‐20 distinguish a large group of Xanthomonas species/pathovars, including Xa, from the citri, passiflorae, manihotis, and malvacearum group. The nonconserved changes from S to V at position 6 and from S/G/A to T at position 9 in flgIII‐27 also distinguish the Xa‐related group from the Xc‐related group. Two serine residues at positions 10 and 17 in flgIII‐27 are replaced by N/D and A, respectively, between the Xa‐ and Xc‐related groups, except in X. fragariae, which has an asparagine residue at position 10
3. DISCUSSION
Understanding how plants perceive and defend themselves from pathogen attack is of paramount importance for crop protection and food security. In this study, we unravelled the molecular basis underlying the immune response triggered by Xa in sweet orange plants, which for a long time remained a mystery to plant pathologists.
We show here that the XaFliC protein is sufficient to induce a robust defence response in several sweet orange varieties and Troyer citrange plants, protecting them against Xc infection. We also show that two polymorphic XaFliC peptides, flgIII‐20 and flgIII‐27, which belong to the structural domain D2, inhibited Xc‐induced canker development more pronouncedly in orange cultivars than in Mexican lime, indicating that XaFliC acts as a PAMP in sweet oranges.
The identification of XaFliC as a major elicitor that triggers an immune response in sweet oranges during Xa infection is in line with the fact that none of the PthA effectors from Xa strains act as avirulence factors in sweet orange cultivars (Al‐Saadi et al., 2007; Brunings & Gabriel, 2003), which thus characterize this defence mechanism as PAMP‐ and not effector‐triggered immunity. Additionally, although amino acid and glycosylation polymorphisms in FliC proteins have been well documented as plant defence evasion mechanisms exploited by many bacterial pathogens (Buscaill et al., 2019; Hirai et al., 2011; Malvino et al., 2022; Sun et al., 2006; Wang et al., 2015; Wei et al., 2020), to the best of our knowledge, this is the first description of amino acid polymorphisms among FliC proteins of citrus canker bacteria determining the host range. Furthermore, such polymorphisms were found in peptide regions other than flg22 and flgII‐28, the only known FliC peptides and the best‐characterized PAMPs recognized by FLS receptors (Cai et al., 2011; Clarke et al., 2013; Gomez‐Gomez & Boller, 2002; Hind et al., 2016; Zipfel et al., 2004). Importantly, we found no FliC protein belonging to X. aurantifolii strains carrying the amino acid substitutions of flgIII‐20 and flgIII‐27 of XcFliC. Likewise, no FliC protein belonging to X. citri strains carries the amino acid substitutions of flgIII‐20 and flgIII‐27 of XaFliC. Nevertheless, the polymorphisms found in flgIII‐20 and flgIII‐27 between Xa and Xc are found in many Xanthomonas species/pathovars. Furthermore, the nonconserved amino acid exchanges found in flgIII‐20 and flgIII‐27 between XaFliC and XcFliC are expected to significantly alter the affinity to or the recognition by a PRR. Thus, our data suggest that these regions are under selective pressure and that the flgIII‐20 and flgIII‐27 polymorphisms could contribute to plant immunity evasion. Interestingly, the region covering the FliC D2 domain of the rice pathogen Acidovorax avenae, carrying the corresponding flgIII‐20 and flgIII‐27 peptides, also induced an immune response in Oryza sativa, Brachypodium distachyon, and Asparagus persicus (Katsuragi et al., 2015; Murakami et al., 2022).
Consistent with its role as a PAMP, ectopic expression of XaFliC in the XcΔfliC mutant led to the up‐regulation of several defence‐related genes also induced by Xa in sweet orange plants. Many of such genes that displayed a robust induction in a XaFliC‐dependent manner, including the citrus homologues FLS2, WRKY22, ACD2, PRX52, and SAG13, are typically induced in incompatible plant–pathogen interactions. For instance, the Arabidopsis ACD2 gene, which encodes the red chlorophyll catabolite enzyme, not only delays plant cell death, but also modulates the onset of HR affecting pathogen survival during bacterial infection (Yao & Greenberg, 2006). This type of response is reminiscent of what is normally observed in sweet orange leaves infected with Xa, where necrosis or extensive plant cell death is atypical (Cernadas et al., 2008). Moreover, the XaFliC‐induced PRX52 gene encodes a protein related to C. sinensis CsPRX25, an apoplastic peroxidase implicated in ROS homeostasis and cell wall lignification that confers resistance against Xc when overexpressed in transgenic citrus (Li et al., 2020). This is particularly important because lignification of the cell wall is a hallmark of Xa infection in sweet orange leaves (Cernadas et al., 2008). Furthermore, in addition to SAG13, a gene that is rapidly induced during HR in response to avirulent Pseudomonas syringae and flg22 (Dhar et al., 2020), the concomitant up‐regulation of the transcription factor genes bHLH25 and MYB102 by XaFliC is of particular interest because these transcriptional factors can form heterodimers to regulate distinct cellular processes, including defence (Pireyre & Burow, 2015).
In line with the results of gene induction mediated by XaFliC, FLS2, WRKY22, and MYB102 showed a slight induction in response to flgIII‐20, whereas FLS2, RLK15, RLKC, FRK1, CRR38, bHLH25, MYB102, SAG13, and ACD2 were greatly up‐regulated by flgIII‐27, thus supporting the idea that flgIII‐20 and flgIII‐27 from XaFliC, but not XcFliC, act as PAMPs in sweet oranges. In fact, activation of most XaFliC‐induced genes was more pronounced in response to flgIII‐27 than in response to flg22.
The observation that activation of FLS2 by flg22 leads to rapid degradation with subsequent de novo mRNA accumulation and synthesis of the receptor (Smith et al., 2014) suggests that up‐regulation of PRR genes in plants can occur in response to receptor activation after ligand binding. Based on this premise and on our gene expression data, we consider that the citrus FLS2‐1, RLK15, and RLKC proteins could represent potential PRR candidates to recognize flgIII‐20 or flgIII‐27. For instance, the citrus FLS2 gene, which is up‐regulated in response to XaFliC, flgIII‐20, and flgIII‐27, corresponds to the FLS2‐1 allele (GenBank XP_006478775) firstly characterized in Duncan grapefruit and shown to be induced by Xc flg22 (Hao et al., 2016; Shi et al., 2016, 2018). However, considering that the FLS2‐1 protein has not yet been demonstrated to bind flg22 with high affinity and that the Arabidopsis FLS2 receptor recognizes an array of flg22 variants to tune the immune response against pathogens and commensal bacteria (Colaianni et al., 2021; Parys et al., 2021; Stringlis & Pieterse, 2021), one cannot rule out the possibility that FLS2‐1 could sense flgIII‐20 or flgIII‐27. Likewise, the citrus RLK15, RLKC, and CRR38 proteins, which were more pronouncedly induced by flgIII‐27 than by flg22, also remain as possible receptor candidates for flgIII‐27 perception. In particular, citrus CRR38 belongs to the subfamily of plant RLKs that contain two C‐X8‐C‐X2‐C motives and are referred to as cysteine‐rich receptor‐like secreted proteins (Vaattovaara et al., 2019). However, CRR38 harbours a predicted transmembrane helix at the N‐terminus, suggesting that it might be surface‐localized and attached to the cell membrane. CRR38 is related to Arabidopsis CRK28, a membrane receptor induced by flg22 and whose enhanced expression increases disease resistance against P. syringae. Notably, CRK28 is associated with BAK1 and was detected in the FLS2–BAK1 immune complex (Yadeta et al., 2017). Therefore, because plant CRK receptors are thought to sense a diverse set of ligands (Vaattovaara et al., 2019), it is possible that CRR38 could act in conjunction with FLS2‐1, RLK15, or RLKC to sense flgIII‐27. In fact, the citrus RLKC protein belongs to the family of lectin receptor‐like kinases recently found to play crucial roles in PAMP recognition and plant defence (Luo et al., 2017; Sun et al., 2020; Wang & Bouwmeester, 2017). Although further studies will be needed to demonstrate whether any of these PRR candidates recognizes the XaFliC polymorphic peptides to trigger the immune response in sweet oranges, we believe this work provides a better understanding of the molecular players involved in such defence mechanism, which may support the development of new approaches to protect citrus plants from bacterial pathogens.
4. EXPERIMENTAL PROCEDURES
4.1. Bacterial strains and generation of fliC mutants
X. citri (strain 306) and X. aurantifolii pathotype C (strain ICMP 8435) were grown in LBON medium supplemented with 100 mg/L ampicillin for 48 h at 28°C (Cernadas et al., 2008). XcΔfliC and XaΔfliC mutants were generated as previously described (Andrade et al., 2014). Briefly, DNA fragments of approximately 1 kb flanking the XcFliC coding region were amplified using a high‐fidelity polymerase (Phusion; Thermo Scientific) and cloned into the HindIII site of the pNPTS138 vector. To generate the XaFliC knockout mutant, a 0.5‐kb DNA fragment encoding only the central region of the XaFliC gene was amplified and cloned into the HindIII site of pNPTS138. The resulting constructs pNPTS‐XcΔfliC and pNPTS‐ XaΔfliC were used to transform Xc and Xa cells by electroporation. The wild‐type copies of XcFliC and XaFliC were replaced with the respective deleted versions of the genes after two recombination events. All mutant clones were confirmed using PCR. To complement the XcΔfliC and XaΔfliC mutants, the XaFliC coding sequence was amplified by PCR using genomic DNA extracted from wild‐type Xa as template. The amplified fragment was cloned into pUFR053 at the BamHI and HindIII restriction sites to obtain the recombinant plasmid pUFR053_XaFliC, which was used for genetic complementation. The construction was transferred into both mutant strains using electroporation, and cells were selected using 10 μg/ml gentamicin. The oligonucleotides used for producing the fliC mutant constructs are listed in Table S2.
4.2. Plant growth and bacterial inoculations
The sweet orange (C. sinensis) varieties Natal, Pera, Valencia, and Sorocaba and the Mexican lime (C. aurantifolia) cultivar Galego were obtained from certified nurseries and kept in a greenhouse. Plant leaves were infiltrated with water suspensions of Xc, Xa, the corresponding fliC mutants, or mixed suspensions of Xa and Xc or Xc and XaΔfliC at a 1:1 ratio (OD600 = 0.1). Single colonies of Xc, Xa, or corresponding fliC mutants were suspended in sterile water to a final OD600 of 0.05 or 0.1 for leaf infiltration. Citrus leaves were also infiltrated with a water suspension of Xc (OD600 = 0.05) in the presence of FliC peptides at a final concentration of 70 μM. Canker symptoms were typically recorded 10 to 15 days after bacterial inoculation.
4.3. Bacterial growth analysis
The growth of Xc and corresponding XcΔfliC mutants complemented with XaFliC or with the empty vector were analysed in Natal and Mexican lime leaves. Discs of 0.5 cm in diameter were removed from leaf sectors infiltrated with a water suspension of bacterial cells (OD600 = 0.2) at different time intervals after bacterial inoculation and macerated in 1 ml of sterile water. Tenfold serial dilutions of the bacterial suspensions were plated on LBON supplemented with ampicillin, and bacterial colonies were counted from three independent leaf extractions. Statistical analyses were performed with one‐way analysis of variance (ANOVA) followed by Bonferroni's test for multiple comparisons (p < 0.05).
4.4. FliC identification and peptide mapping
Bacterial cells were grown in liquid LBON supplemented with ampicillin for 18 h at 28°C under mild agitation (100 rpm). Cells were collected by centrifugation (5000 × g) at 4°C for 5 min and suspended in 25 mM Tris–HCl, 150 mM NaCl, pH 7.0. The suspensions were homogenized in a cell mixer (Politron PT‐MR 1600 E) for 5 min on ice. Bacterial cells were recovered by centrifugation and the suspensions containing bacterium flagellum were precipitated with acetone (at a 1:1 ratio) at −20°C overnight. The suspensions were centrifuged (5000 × g) at 4°C for 10 min and the protein pellets were suspended in SDS‐PAGE sample buffer and resolved on 10% SDS‐PAGE gels. Protein bands were extracted from the gels and digested with trypsin, and the cleaved peptides were identified by mass spectrometry, as previously described (Soprano et al., 2017). The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (Perez‐Riverol et al., 2022) with the dataset identifier PXD037760.
4.5. Protein sequence alignment and molecular modelling
Protein sequence alignments were performed with Clustal Omega using default parameters, whereas 3D structural models of the FliC proteins were generated with SWISS‐MODEL using the crystal structure of the Sphingomonas sp. A1 flagellin (PDB code 3K8V) as a template model. The structures were visualized and compared using PyMOL (Schrodinger, LLC, 2015).
4.6. Bacterial motility assays
Bacterial motility assays were performed as previously described (Andrade et al., 2020). Briefly, cells grown in LBON plus 1.5% (wt/vol) agar for 48 h at 28°C were stabbed into 0.3% (wt/vol) agar nutrient broth medium plates. Motility was evaluated and bacteria were photographed after 3 days of incubation at 28°C. The assays were performed with three independent replicates.
4.7. FliC purification and antibody production
The XaFliC gene was cloned into the pET28a vector for the expression of recombinant 6×His‐tagged XaFliC in Escherichia coli BL21(DE3) cells. Bacterial cells were grown in Luria‐Bertani medium supplemented with kanamycin (50 mg/L) at 37°C and protein expression was induced with 0.1 mM IPTG for 3 h under agitation (200 rpm). Bacterial cells were suspended in 50 mM Tris–HCl, pH 8.0, 300 mM NaCl, 5 mM imidazole, lysed with lysozyme (1 mg/ml) and sonication, and centrifuged at 18,000 × g at 4°C for 30 min. The recombinant protein in the soluble fraction was purified by affinity chromatography and eluted with 50 mM Tris–HCl, pH 8.0, 300 mM NaCl, 250 mM imidazole. Fractions containing the recombinant XaFliC were pooled, concentrated, and fractionated on a Superdex 200 10/30 column equilibrated with 50 mM Tris–HCl, pH 8.0, 300 mM NaCl. Fractions containing purified XaFliC were concentrated and used to immunize white rabbits for antibody production (Rhea Biotech, Campinas, Brazil).
4.8. Western blot analyses
Xc and Xa cells were suspended in SDS‐PAGE sample buffer and incubated at 90°C for 5 min for cell disruption. Cell debris was pelleted by centrifugation at 15,000 × g for 5 min and the supernatants were resolved by 10% SDS‐PAGE. The proteins were transferred to PVDF membranes, which were blocked with skimmed milk and probed with the primary anti‐XaFliC antibodies (1:1000) overnight at 4°C under mild agitation. After washing with phosphate‐buffered saline, the membranes were incubated with horseradish peroxidase‐conjugated secondary anti‐rabbit IgG (GE Healthcare) at 1:3000 dilution for 3 h at room temperature with mild agitation. Protein bands were detected by chemiluminescence with Super Signal West Pico Chemiluminescent substrate (Thermo Scientific).
4.9. RNA‐seq and gene expression analysis
Natal leaves of similar size and age were infiltrated with water suspensions of Xc or Xa (OD600 = 0.1), or water as control. The infiltrated leaf sectors were collected at 24 and 48 h after bacterial infiltration and immediately frozen in liquid nitrogen. Leaf tissues were ground in liquid nitrogen and total RNA was extracted from three independent leaves using TRIzol reagent (Invitrogen). The quality and quantity of the RNA samples were verified by NanoDrop measurements and agarose gels. The RNA samples were treated with DNase I to remove traces of DNA and subjected to HISEQ2500 Illumina sequencing at the LaCTAD facility of the State University of Campinas. Reads of each sample (28,000,000 on average) were aligned with the C. sinensis transcripts of the Phytozome database using the RSEM program (Li & Dewey, 2011), which allowed the mapping of more than 20,000,000 reads per sample, and the output files were recorded in bam format (Li et al., 2009; Mortazavi et al., 2008). The expression value of each transcript (FPKM) was calculated using RSEM according to Mortazavi et al. (2008). The RNA‐seq data were deposited in the GEO database with the following accession numbers: GSE202469 and GSM6122945 to GSM6122959.
For RT‐qPCR analyses, leaves of Natal plants were infiltrated with Xc, XcΔfliC, XcΔfliC complemented with the XaFliC gene, or water as control. Total RNA was extracted from the infiltrated leaf sectors 24 h after bacterial inoculation. In addition, Natal leaves were infiltrated with water suspensions of the flg peptides at a final concentration of 70 μM, and total RNA was extracted 3 h after peptide inoculation. RNA extraction followed by mRNA purification and RT‐qPCR were performed as described previously (Andrade et al., 2020). The oligonucleotides used for the amplification of the citrus genes are listed in Table S2. Gene expression values were normalized using the GAPDH gene as endogenous control and three independent biological replicates were evaluated for all treatments. Fold change values were calculated using the 2−ΔΔCt method (Livak & Schmittgen, 2001). Statistical analyses were conducted with one‐way ANOVA followed by Dunnett's and Tukey's tests for multiple comparisons (p < 0.05).
Supporting information
Figure S1 The polymorphic FliC peptides do not inhibit Xanthomonas citri (Xc) growth in culture medium. Xc was incubated with the polymorphic XaFliC and XcFliC peptides at a final concentration of 70 μM for 30 min at room temperature and then plated on LBON medium. Bacterial cells grew normally in the presence of the peptides after 48 h incubation at 28°C
Figure S2 The flg22 peptide does not protect sweet orange leaves from Xanthomonas citri (Xc) infection. Natal, Pera, and Valencia leaves were infiltrated with a water suspension of Xc (OD600 = 0.05) in the presence or absence of the flg22 peptide at a final concentration of 70 μM (areas surrounded by dashed lines). Canker symptoms were recorded 10 days after bacterial inoculation
Figure S3 Deletion of the FliC gene in Xanthomonas aurantifolii (Xa) is not sufficient to induce canker symptoms on sweet oranges. Natal, Pera, Valencia, and Sorocaba leaves were infiltrated with a water suspension (OD600 = 0.1) of Xa or the corresponding XaΔfliC mutants (areas surrounded by dashed lines). While no cancer pustule developed in the leaf sectors infiltrated with the XaΔfliC mutant, a reduction in the yellowing of the leaf is observed in these leaf sectors, compared to leaf sectors infiltrated with Xa, particularly in Natal, Pera, and Sorocaba leaves
Table S1 XcFliC and XaFliC peptides identified by mass spectrometry
Table S2 Oligonucleotides used in this study
Data S1 Major citrus genes preferentially or exclusively up‐regulated by Xanthomonas aurantifolii (Xa) in Natal leaves, relative to water inoculation or Xanthomonas citri (Xc) infection at 24 and 48 h postinoculation (hpi); citrus genes differentially induced by Xc in Natal leaves, relative to water inoculation, at 24 and 48 hpi
ACKNOWLEDGEMENTS
We acknowledge the use of the LNBio facilities LPP and MAS for protein production and mass spectrometry analysis, respectively. We thank Jackeline Zanella, Romenia Domingues, and Bianca Pauletti for technical help. We also thank Dr Marcelo Falsarella Carazzolle for helping us with the initial analysis of the RNA‐seq data. This work was supported by the São Paulo Research Foundation – FAPESP (grants 2020/02547‐0, 2018/08535‐4, and 2014/50880‐0 and fellowship 2013/22507‐0 to H.M.S. and fellowship 2017/18570‐9 to M.O.A.), the Ministry of Science, Technology and Innovation, and the Conselho Nacional de Desenvolvimento Científico e Tecnológico – CNPq (fellowships 303238/2016‐0 to C.E.B. and 164832/2017‐3 to M.O.A.).
Andrade, M.d.O. , da Silva, J.C. , Soprano, A.S. , Shimo, H.M. , Leme, A.F.P. & Benedetti, C.E. (2023) Suppression of citrus canker disease mediated by flagellin perception. Molecular Plant Pathology, 24, 331–345. Available from: 10.1111/mpp.13300
Maxuel de Oliveira Andrade and Jaqueline Cristina da Silva contributed equally to this work.
Contributor Information
Maxuel de Oliveira Andrade, Email: maxuel.andrade@lnbr.cnpem.br.
Celso Eduardo Benedetti, Email: celso.benedetti@lnbio.cnpem.br.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. The RNA‐seq data are openly available in the GEO repository at https://www.ncbi.nlm.nih.gov/geo/ with the following reference numbers: GSE202469 and GSM6122945 to GSM6122959. The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium at http://www.proteomexchange.org via the PRIDE partner repository (Perez‐Riverol et al., 2022) with the dataset identifier PXD037760.
REFERENCES
- Abe, V.Y. & Benedetti, C.E. (2016) Additive roles of PthAs in bacterial growth and pathogenicity associated with nucleotide polymorphisms in effector‐binding elements of citrus canker susceptibility genes. Molecular Plant Pathology, 17, 1223–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Saadi, A. , Reddy, J.D. , Duan, Y.P. , Brunings, A.M. , Yuan, Q. & Gabriel, D.W. (2007) All five host‐range variants of Xanthomonas citri carry one pthA homolog with 17.5 repeats that determines pathogenicity on citrus, but none determine host‐range variation. Molecular Plant‐Microbe Interactions, 20, 934–943. [DOI] [PubMed] [Google Scholar]
- Andrade, M.O. , Farah, C.S. & Wang, N. (2014) The post‐transcriptional regulator rsmA/csrA activates T3SS by stabilizing the 5' UTR of hrpG, the master regulator of hrp/hrc genes, in Xanthomonas. PLoS Pathogens, 10, e1003945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade, M.O. , Pang, Z. , Achor, D.S. , Wang, H. , Yao, T. , Singer, B.H. et al. (2020) The flagella of ‘Candidatus Liberibacter asiaticus’ and its movement in planta. Molecular Plant Pathology, 21, 109–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunings, A.M. & Gabriel, D.W. (2003) Xanthomonas citri: breaking the surface. Molecular Plant Pathology, 4, 141–157. [DOI] [PubMed] [Google Scholar]
- Buscaill, P. , Chandrasekar, B. , Sanguankiattichai, N. , Kourelis, J. , Kaschani, F. , Thomas, E.L. et al. (2019) Glycosidase and glycan polymorphism control hydrolytic release of immunogenic flagellin peptides. Science, 364, eaav0748. [DOI] [PubMed] [Google Scholar]
- Cai, R. , Lewis, J. , Yan, S. , Liu, H. , Clarke, C.R. , Campanile, F. et al. (2011) The plant pathogen Pseudomonas syringae pv. t omato is genetically monomorphic and under strong selection to evade tomato immunity. PLoS Pathogens, 7, e1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cernadas, R.A. , Camillo, L.R. & Benedetti, C.E. (2008) Transcriptional analysis of the sweet orange interaction with the citrus canker pathogens Xanthomonas axonopodis pv. citri and Xanthomonas axonopodis pv. aurantifolii . Molecular Plant Pathology, 9, 609–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke, C.R. , Chinchilla, D. , Hind, S.R. , Taguchi, F. , Miki, R. , Ichinose, Y. et al. (2013) Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. The New Phytologist, 200, 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaianni, N.R. , Parys, K. , Lee, H.S. , Conway, J.M. , Kim, N.H. , Edelbacher, N. et al. (2021) A complex immune response to flagellin epitope variation in commensal communities. Cell Host & Microbe, 29, 635–649.e9. [DOI] [PubMed] [Google Scholar]
- Czékus, Z. , Kukri, A. , Hamow, K.Á. , Szalai, G. , Tari, I. , Ördög, A. et al. (2021) Activation of local and systemic defence responses by flg22 is dependent on daytime and ethylene in intact tomato plants. International Journal of Molecular Sciences, 22, 8354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoux, C. , Galletti, R. , Mammarella, N. , Gopalan, S. , Werck, D. , De Lorenzo, G. et al. (2008) Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Molecular Plant, 1, 423–445. Erratum in: Molecular Plant, 2009 2, 838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhar, N. , Caruana, J. , Erdem, I. , Subbarao, K.V. , Klosterman, S.J. & Raina, R. (2020) The Arabidopsis SENESCENCE‐ASSOCIATED GENE 13 regulates dark‐induced senescence and plays contrasting roles in defense against bacterial and fungal pathogens. Molecular Plant‐Microbe Interactions, 33, 754–766. [DOI] [PubMed] [Google Scholar]
- Duan, S. , Jia, H. , Pang, Z. , Teper, D. , White, F. , Jones, J. et al. (2018) Functional characterization of the citrus canker susceptibility gene CsLOB1. Molecular Plant Pathology, 19 8), 1908–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, N.P. , Patane, J.S.L. , Varani, A.M. , Felestrino, E.B. , Caneschi, W.L. , Sanchez, A.B. et al. (2019) Analyses of seven new genomes of Xanthomonas citri pv. aurantifolii strains, causative agents of citrus canker B and C, show a reduced repertoire of pathogenicity‐related genes. Frontiers in Microbiology, 10, 2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gochez, A.M. , Minsavage, G.V. , Potnis, N. , Canteros, B.I. , Stall, R.E. & Jones, J.B. (2015) Functional XopAG homologue in Xanthomonas fuscans pv. aurantifolii strain C limits host range. Plant Pathology, 64, 1207–1214. [Google Scholar]
- Gochez, A.M. , Shantharaj, D. , Potnis, N. , Zhou, X. , Minsavage, G.V. , White, F.F. et al. (2017) Molecular characterization of XopAG effector AvrGf2 from Xanthomonas fuscans ssp. aurantifolii in grapefruit. Molecular Plant Pathology, 18, 405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez‐Gomez, L. & Boller, T. (2002) Flagellin perception: a paradigm for innate immunity. Trends in Plant Science, 7, 251–256. [DOI] [PubMed] [Google Scholar]
- Graham, J.H. , Gottwald, T.R. , Cubero, J. & Achor, D.S. (2004) Xanthomonas axonopodis pv. citri: factors affecting successful eradication of citrus canker. Molecular Plant Pathology, 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Hao, G. , Pitino, M. , Duan, Y. & Stover, E. (2016) Reduced susceptibility to Xanthomonas citri in transgenic citrus expressing the FLS2 receptor from Nicotiana benthamiana . Molecular Plant‐Microbe Interactions, 29, 132–142. [DOI] [PubMed] [Google Scholar]
- Hind, S.R. , Strickler, S.R. , Boyle, P.C. , Dunham, D.M. , Bao, Z. , O'Doherty, I.M. et al. (2016) Tomato receptor FLAGELLIN‐SENSING 3 binds flgII‐28 and activates the plant immune system. Nature Plants, 2, 16128. [DOI] [PubMed] [Google Scholar]
- Hirai, H. , Takai, R. , Iwano, M. , Nakai, M. , Kondo, M. , Takayama, S. et al. (2011) Glycosylation regulates specific induction of rice immune responses by Acidovorax avenae flagellin. Journal of Biological Chemistry, 286, 25519–25530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Zhang, J. , Jia, H. , Sosso, D. , Li, T. , Frommer, W.B. et al. (2014) Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proceedings of the National Academy of Sciences of the United States of America, 111, E521–E529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi, Y. , Takai, R. , Furukawa, T. , Hirai, H. , Morimoto, T. , Katayama, T. et al. (2015) CD2‐1, the C‐terminal region of flagellin, modulates the induction of immune responses in rice. Molecular Plant‐Microbe Interactions, 28, 648–658. [DOI] [PubMed] [Google Scholar]
- Li, B. & Dewey, C.N. (2011) RSEM: accurate transcript quantification from RNA‐seq data with or without a reference genome. BMC Bioinformatics, 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. et al. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Qin, X. , Qi, J. , Dou, W. , Dunand, C. , Chen, S. et al. (2020) CsPrx25, a class III peroxidase in Citrus sinensis, confers resistance to citrus bacterial canker through the maintenance of ROS homeostasis and cell wall lignification. Horticulture Research, 7, 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak, K.J. & Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(−ΔΔC(T)) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Luo, X. , Xu, N. , Huang, J. , Gao, F. , Zou, H. , Boudsocq, M. et al. (2017) A lectin receptor‐like kinase mediates pattern‐triggered salicylic acid signaling. Plant Physiology, 174, 2501–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvino, M.L. , Bott, A.J. , Green, C.E. , Majumdar, T. & Hind, S.R. (2022) Influence of flagellin polymorphisms, gene regulation, and responsive memory on the motility of Xanthomonas species that cause bacterial spot disease of solanaceous plants. Molecular Plant‐Microbe Interactions, 35, 157–169. [DOI] [PubMed] [Google Scholar]
- Mortazavi, A. , Williams, B.A. , McCue, K. , Schaeffer, L. & Wold, B. (2008) Mapping and quantifying mammalian transcriptomes by RNA‐seq. Nature Methods, 5, 621–628. [DOI] [PubMed] [Google Scholar]
- Murakami, T. , Katsuragi, Y. , Hirai, H. , Wataya, K. , Kondo, M. & Che, F.S. (2022) Distribution of flagellin CD2‐1, flg22, and flgII‐28 recognition systems in plant species and regulation of plant immune responses through these recognition systems. Bioscience, Biotechnology, and Biochemistry, 86, 490–501. [DOI] [PubMed] [Google Scholar]
- Nicaise, V. , Roux, M. & Zipfel, C. (2009) Recent advances in PAMP‐triggered immunity against bacteria: pattern recognition receptors watch over and raise the alarm. Plant Physiology, 150, 1638–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft, H. & Szymanski, C.M. (2010) Protein glycosylation in bacteria: sweeter than ever. Nature Reviews. Microbiology, 8, 765–778. [DOI] [PubMed] [Google Scholar]
- de Oliveira, M.L. , de Lima Silva, C.C. , Abe, V.Y. , Costa, M.G. , Cernadas, R.A. & Benedetti, C.E. (2013) Increased resistance against citrus canker mediated by a citrus mitogen‐activated protein kinase. Molecular Plant‐Microbe Interactions, 26, 1190–1199. [DOI] [PubMed] [Google Scholar]
- Oliveira Andrade, M. , Sforca, M.L. , Batista, F.A.H. , Figueira, A.C.M. & Benedetti, C.E. (2020) The MAF1 phosphoregulatory region controls MAF1 interaction with the RNA polymerase III C34 subunit and transcriptional repression in plants. The Plant Cell, 32, 3019–3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parys, K. , Colaianni, N.R. , Lee, H.S. , Hohmann, U. , Edelbacher, N. , Trgovcevic, A. et al. (2021) Signatures of antagonistic pleiotropy in a bacterial flagellin epitope. Cell Host & Microbe, 29, 620–634.e9. [DOI] [PubMed] [Google Scholar]
- Pereira, A.L. , Carazzolle, M.F. , Abe, V.Y. , de Oliveira, M.L. , Domingues, M.N. , Silva, J.C. et al. (2014) Identification of putative TAL effector targets of the citrus canker pathogens shows functional convergence underlying disease development and defense response. BMC Genomics, 15, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Riverol, Y. , Bai, J. , Bandla, C. , Hewapathirana, S. , García‐Seisdedos, D. , Kamatchinathan, S. et al. (2022) The PRIDE database resources in 2022: a hub for mass spectrometry‐based proteomics evidences. Nucleic Acids Research, 50(D1), D543–D552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pireyre, M. & Burow, M. (2015) Regulation of MYB and bHLH transcription factors: a glance at the protein level. Molecular Plant, 8, 378–388. [DOI] [PubMed] [Google Scholar]
- Roeschlin, R.A. , Uviedo, F. , Garcia, L. , Molina, M.C. , Favaro, M.A. , Chiesa, M.A. et al. (2019) PthA4(AT), a 7.5‐repeats transcription activator‐like (TAL) effector from Xanthomonas citri ssp. citri, triggers citrus canker resistance. Molecular Plant Pathology, 20, 1394–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrodinger, LLC (2015) The PyMOL molecular graphics system, Version 1.8. Available at: https://pymol.org/2/ [Accessed 20 January 2023]. [Google Scholar]
- Shi, L. , Pigeonneau, N. , Ravikumar, V. , Dobrinic, P. , Macek, B. , Franjevic, D. et al. (2014) Cross‐phosphorylation of bacterial serine/threonine and tyrosine protein kinases on key regulatory residues. Frontiers in Microbiology, 5, 495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Q. , Febres, V. J. , Jones, J. B. & Moore, G. A. (2016). A survey of FLS2 genes from multiple citrus species identifies candidates for enhancing disease resistance to Xanthomonas citri ssp. citri . Horticulture Research, 3, 16022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Q. , Febres, V.J. , Zhang, S. , Yu, F. , McCollum, G. , Hall, D.G. et al. (2018) Identification of gene candidates associated with Huanglongbing tolerance, using ‘Candidatus Liberibacter asiaticus’ flagellin 22 as a proxy to challenge citrus. Molecular Plant‐Microbe Interactions, 31, 200–211. [DOI] [PubMed] [Google Scholar]
- Smith, J.M. , Salamango, D.J. , Leslie, M.E. , Collins, C.A. & Heese, A. (2014) Sensitivity to Flg22 is modulated by ligand‐induced degradation and de novo synthesis of the endogenous flagellin‐receptor FLAGELLIN‐SENSING2. Plant Physiology, 164, 440–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano, A.S. , Abe, V.Y. , Smetana, J.H. & Benedetti, C.E. (2013) Citrus MAF1, a repressor of RNA polymerase III, binds the Xanthomonas citri canker elicitor PthA4 and suppresses citrus canker development. Plant Physiology, 163, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano, A.S. , Giuseppe, P.O. , Shimo, H.M. , Lima, T.B. , Batista, F.A.H. , Righetto, G.L. et al. (2017) Crystal structure and regulation of the citrus pol III repressor MAF1 by auxin and phosphorylation. Structure, 25, 1360–1370.e4. [DOI] [PubMed] [Google Scholar]
- de Souza, T.A. , Soprano, A.S. , de Lira, N.P. , Quaresma, A.J. , Pauletti, B.A. , Paes Leme, A.F. et al. (2012) The TAL effector PthA4 interacts with nuclear factors involved in RNA‐dependent processes including a HMG protein that selectively binds poly(U) RNA. PLoS One, 7, e32305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringlis, I.A. & Pieterse, C.M.J. (2021) Evolutionary “hide and seek” between bacterial flagellin and the plant immune system. Cell Host & Microbe, 29, 548–550. [DOI] [PubMed] [Google Scholar]
- Sun, W. , Dunning, F.M. , Pfund, C. , Weingarten, R. & Bent, A.F. (2006) Within‐species flagellin polymorphism in Xanthomonas campestris pv. campestris and its impact on elicitation of Arabidopsis FLAGELLIN SENSING2‐dependent defenses. The Plant Cell, 18, 764–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y. , Qiao, Z. , Muchero, W. & Chen, J.G. (2020) Lectin receptor‐like kinases: the sensor and mediator at the plant cell surface. Frontiers in Plant Science, 11, 596301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teper, D. , Xu, J. , Pandey, S.S. & Wang, N. (2021) PthAW1, a transcription activator‐like effector of Xanthomonas citri subsp. citri, promotes host‐specific immune responses. Molecular Plant‐Microbe Interactions, 34, 1033–1047. [DOI] [PubMed] [Google Scholar]
- Vaattovaara, A. , Brandt, B. , Rajaraman, S. , Safronov, O. , Veidenberg, A. , Luklová, M. et al. (2019) Mechanistic insights into the evolution of DUF26‐containing proteins in land plants. Communications Biology, 2, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Y. & Bouwmeester, K. (2017) L‐type lectin receptor kinases: new forces in plant immunity. PLoS Pathogens, 13, e1006433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Sun, Z. , Wang, H. , Liu, L. , Lu, F. , Yang, J. et al. (2015) Rice OsFLS2‐mediated perception of bacterial flagellins is evaded by Xanthomonas oryzae pvs. oryzae and oryzicola . Molecular Plant, 8, 1024–1037. [DOI] [PubMed] [Google Scholar]
- Wei, Y. , Balaceanu, A. , Rufian, J.S. , Segonzac, C. , Zhao, A. , Morcillo, R.J.L. et al. (2020) An immune receptor complex evolved in soybean to perceive a polymorphic bacterial flagellin. Nature Communications, 11, 3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadeta, K.A. , Elmore, J.M. , Creer, A.Y. , Feng, B. , Franco, J.Y. , Rufian, J.S. et al. (2017) A cysteine‐rich protein kinase associates with a membrane immune complex and the cysteine residues are required for cell death. Plant Physiology, 173, 771–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, N. & Greenberg, J.T. (2006) Arabidopsis ACCELERATED CELL DEATH2 modulates programmed cell DEATH. The Plant Cell, 18, 397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Robatzek, S. , Navarro, L. , Oakeley, E.J. , Jones, J.D. , Felix, G. et al. (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature, 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The polymorphic FliC peptides do not inhibit Xanthomonas citri (Xc) growth in culture medium. Xc was incubated with the polymorphic XaFliC and XcFliC peptides at a final concentration of 70 μM for 30 min at room temperature and then plated on LBON medium. Bacterial cells grew normally in the presence of the peptides after 48 h incubation at 28°C
Figure S2 The flg22 peptide does not protect sweet orange leaves from Xanthomonas citri (Xc) infection. Natal, Pera, and Valencia leaves were infiltrated with a water suspension of Xc (OD600 = 0.05) in the presence or absence of the flg22 peptide at a final concentration of 70 μM (areas surrounded by dashed lines). Canker symptoms were recorded 10 days after bacterial inoculation
Figure S3 Deletion of the FliC gene in Xanthomonas aurantifolii (Xa) is not sufficient to induce canker symptoms on sweet oranges. Natal, Pera, Valencia, and Sorocaba leaves were infiltrated with a water suspension (OD600 = 0.1) of Xa or the corresponding XaΔfliC mutants (areas surrounded by dashed lines). While no cancer pustule developed in the leaf sectors infiltrated with the XaΔfliC mutant, a reduction in the yellowing of the leaf is observed in these leaf sectors, compared to leaf sectors infiltrated with Xa, particularly in Natal, Pera, and Sorocaba leaves
Table S1 XcFliC and XaFliC peptides identified by mass spectrometry
Table S2 Oligonucleotides used in this study
Data S1 Major citrus genes preferentially or exclusively up‐regulated by Xanthomonas aurantifolii (Xa) in Natal leaves, relative to water inoculation or Xanthomonas citri (Xc) infection at 24 and 48 h postinoculation (hpi); citrus genes differentially induced by Xc in Natal leaves, relative to water inoculation, at 24 and 48 hpi
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The RNA‐seq data are openly available in the GEO repository at https://www.ncbi.nlm.nih.gov/geo/ with the following reference numbers: GSE202469 and GSM6122945 to GSM6122959. The mass spectrometry proteomic data have been deposited to the ProteomeXchange Consortium at http://www.proteomexchange.org via the PRIDE partner repository (Perez‐Riverol et al., 2022) with the dataset identifier PXD037760.


