Abstract
The general stress response of Bacillus subtilis is controlled by the ςB transcription factor, which is activated in response to diverse energy and environmental stresses. These two classes of stress are transmitted by separate signaling pathways which converge on the direct regulators of ςB, the RsbV anti-anti-ς factor and the RsbW anti-ς factor. The energy signaling branch involves the RsbP phosphatase, which dephosphorylates RsbV in order to trigger the general stress response. The rsbP structural gene lies downstream from rsbQ in a two-gene operon. Here we identify the RsbQ protein as a required positive regulator inferred to act in concert with the RsbP phosphatase. RsbQ bound RsbP in the yeast two-hybrid system, and a large in-frame deletion in rsbQ had the same phenotype as a null allele of rsbP—an inability to activate ςB in response to energy stress. Genetic complementation studies indicated that this phenotype was not due to a polar effect of the rsbQ alteration on rsbP. The predicted rsbQ product is a hydrolase or acyltransferase of the α/β fold superfamily, members of which catalyze a wide variety of reactions. Notably, substitutions in the presumed catalytic triad of RsbQ also abolished the energy stress response but had no detectable effect on RsbQ structure, synthesis, or stability. We conclude that the catalytic activity of RsbQ is an essential constituent of the energy stress signaling pathway.
In Bacillus subtilis and related bacteria, a general stress response controlled by the ςB transcription factor confers multiple stress resistance on nongrowing cells (reviewed in references 17 and 26). The activity of ςB is governed by a signal transduction pathway with two distinct branches. One branch is specific for energy stresses, such as carbon, phosphorus, or oxygen limitation, and the other is specific for environmental stresses, such as acid, ethanol, heat, or salt stress (20, 41, 43, 44). According to the model shown in Fig. 1, each branch terminates with a differentially regulated serine phosphatase: RsbU in the environmental signaling branch and RsbP in the energy signaling branch. When activated by its particular class of stress, either RsbU or RsbP engages the common regulators RsbV and RsbW. RsbV and RsbW together control ςB activity via a partner-switching mechanism in which alternate protein-protein interactions are governed by the phosphorylation state of RsbV (3, 8, 13, 41, 42, 44).
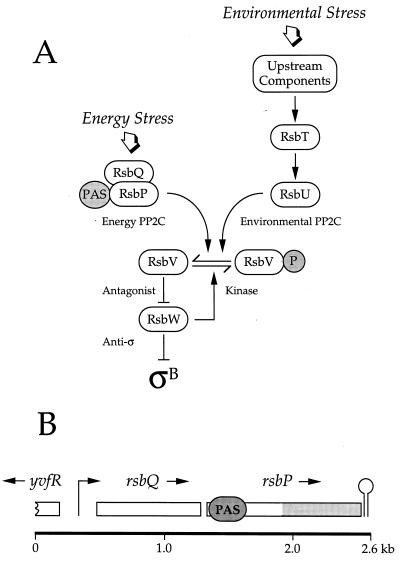
FIG. 1.
Model of the signal transduction network that activates the general stress transcription factor ςB. (A) Two signaling pathways converge on RsbV-P, the antagonist form found in unstressed cells. The energy stress branch terminates with the RsbP phosphatase (Energy PP2C), which contains a PAS domain in its amino-terminal region (41). In contrast the environmental stress branch terminates with the RsbU phosphatase (Environmental PP2C), which is activated by RsbT in response to upstream signaling elements (2, 20, 42–44). When triggered by stress, either the RsbP or the RsbU phosphatase removes the serine phosphate from RsbV-P. Dephosphorylated RsbV then binds the RsbW anti-ς factor, forcing it to release ςB, which can then activate its target genes (3, 8, 13). We show here that RsbQ (formerly called YvfQ) is an essential component of the energy stress branch and propose that it acts in concert with the RsbP phosphatase. (B) Genetic organization of the rsbQ-rsbP operon. The rsbQ and rsbP reading frames are denoted by open rectangles above the kilobase scale. The regions indicated within rsbP code for the PAS domain (PAS) and the PP2C phosphatase domain (shaded). Promoter activity (right-angled arrow) has been proximately located between 0 and 0.7 kb, and a potential terminator sequence (stem-loop) lies downstream from rsbP. Energy signaling does not require increased transcription from this promoter, suggesting that signaling is accomplished by a posttranslational mechanism (41).
The means by which energy and environmental stress signals enter their respective branches of the pathway are unknown, and it is clear that additional signaling components remain to be identified (1, 33, 41, 45). Here we focus on the energy signaling branch. The amino-terminal half of the RsbP phosphatase contains a Per-ARNT-Sim (PAS) domain (41), similar to those found in a wide variety of proteins involved in sensing fluctuations in redox, light, or oxygen (38). In some proteins, such PAS domains function by binding a ligand or a chromophore and in others by controlling protein-protein interactions with other proteins. As part of a search for new elements of the energy signaling branch, we report here that RsbP interacts with RsbQ, a positive regulator essential for energy stress activation of ςB. Moreover, our genetic analysis indicates that the predicted catalytic activity of RsbQ is required for energy stress signaling in B. subtilis.
MATERIALS AND METHODS
Bacterial strains and genetic methods.
Escherichia coli DH5α (Gibco BRL, Gaithersburg, Md.) was host for all plasmid constructions, while E. coli BL21(DE3)pLysS (Novagen, Madison, Wis.) was the expression host for protein purification. Standard recombinant DNA methods were as described by Sambrook et al. (32). B. subtilis strains used are shown in Table 1. B. subtilis PB2 and its derivatives were made competent by standard methods (12). We used the four-primer method of site-directed mutagenesis (18) to make three separate alterations within the cloned rsbQ gene. To make the large, in-frame deletion of rsbQ (rsbQΔ2), we removed 546 bp from within the 807-bp coding region, deleting triplets 38 to 219. To make the missense mutations, triplet 96 was changed from serine (TCC) to alanine (GCC) to yield rsbQS96A and triplet 247 was changed from histidine (CAT) to alanine (GCA) to yield rsbQH247A. Each PCR-mutagenized product was cloned into the pCP115 integration vector (27), yielding pMB56 (rsbQΔ2), pMB58 (rsbQH247A), and pMB59 (rsbQS96A). We then introduced the deletion or missense mutations into the homologous copy of rsbQ on the B. subtilis chromosome by means of a two-step allele replacement method (35). Substitution of the rsbQΔ2 allele was confirmed by PCR amplification. Substitution of the missense mutations was confirmed by chromosomal sequencing.
TABLE 1.
Bacillus subtilis strains
| Strain | Genotype | Reference or constructiona |
|---|---|---|
| PB2 | trpC2 | Wild-type Marburg strain |
| PB198 | amyE::ctc-lacZ trpC2 | 10 |
| PB567 | rsbPΔ1::spc amyE::ctc-lacZ trpC2 | 41 |
| PB604 | rsbQΔ2 trpC2 | pMB56→PB2b |
| PB605 | rsbQΔ2 amyE::ctc-lacZ trpC2 | pDH32-ctc (10)→PB604 |
| PB606 | amyE::ctc-lacZ trpC2 pMB64 | pMB64→PB198 |
| PB607 | amyE::ctc-lacZ trpC2 pKV2 | pKV2→PB198 |
| PB631 | rsbQH247A trpC2 | pMB58→PB2b |
| PB632 | rsbQH247A amyE::ctc-lacZ trpC2 | pDH32-ctc→PB631 |
| PB633 | rsbQS96A trpC2 | pMB59→PB2b |
| PB634 | rsbQS96A amyE::ctc-lacZ trpC2 | pDH32-ctc→PB633 |
| PB716 | rsbQΔ2 amyE::ctc-lacZ trpC2 pKV2 | pKV2→PB491 |
| PB719 | rsbPΔ1::spc amyE::ctc-lacZ trpC2 pMB64 | pMB64→PB567 |
Arrow indicates transformation from donor to recipient.
Two-step allele replacement (35).
β-Galactosidase accumulation assays.
All cultures were grown in a shaking water bath at 37°C. For energy stress experiments, cells were either grown into stationary phase in buffered Luria broth (LB) lacking salt (9) or subjected to carbon and energy starvation in a minimal salt medium (5) containing 0.05% glucose. For environmental stress experiments, cells were grown to early logarithmic phase in buffered LB lacking salt. At this point, either NaCl (0.3 M final concentration) or ethanol (4% [vol/vol] final concentration) was added to one of two parallel cultures. For all experiments, samples were collected at the times indicated and treated as described by Miller (22). Cells were washed with Z buffer and permeabilized using sodium dodecyl sulfate and chloroform. Protein concentrations were determined using the Bio-Rad Protein Assay reagent (Bio-Rad, Richmond, Calif.). Activity was defined as ΔA420 × 1,000 per minute per milligram of protein.
Yeast two-hybrid analysis.
We used the Matchmaker Two-Hybrid System (Clontech, Palo Alto, Calif.) to detect possible interactions among RsbP, RsbQ, RsbT, and RsbU. The rsbQ and rsbP reading frames were fused to either the GAL4 DNA binding domain in plasmid pGBT9 or the GAL4 activation domain in plasmid pGAD424, as previously described for rsbT and rsbU (44). These constructs were then paired in yeast SFY526 cells by selecting double transformants on minimal plates. Transcriptional activation of the lacZ reporter gene in these transformants was determined qualitatively using a colony lift filter assay according to the manufacturer's protocol.
Overexpression of rsbQ and rsbP products in B. subtilis.
We used PCR to amplify the individual rsbQ and rsbP reading frames along with their predicted ribosomal binding sites, placing them under control of the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible Pspac promoter in the multicopy expression vector pDG148 (36). Plasmid pMB64, carrying rsbQ, was transformed into B. subtilis PB198 (wild-type) and PB567 (rsbPΔ1::spc). Plasmid pKV2, carrying rsbP, was transformed into PB198 and PB605 (rsbQΔ2). These strains also carried a ςB-dependent ctc-lacZ fusion in single copy at the amyE locus (10) to permit measurement of ςB activity. Expression from the plasmid was induced by adding IPTG (1 mM final concentration) to cultures in the early logarithmic phase of growth.
Construction of overexpression clones and purification of His-tagged proteins from E. coli.
To purify the wild-type, S96A, and H247A forms of RsbQ, we fused the appropriate coding region to a hexahistidine tag in the pET15b expression vector (Novagen). Hexahistidine-tagged proteins were purified from E. coli BL21(DE3)pLysS extracts on nickel-nitrilotriacetic acid metal affinity columns (Qiagen, Valencia, Calif.) according to the manufacturer's protocol.
Trypsin proteolysis.
Purified wild-type and mutant RsbQ proteins were subjected to limited trypsin proteolysis as follows. Trypsin (l-1-tosylamido-2-phenyl ethyl chloromethyl ketone [TPCK]-treated type XIII from bovine pancreas; Sigma, St. Louis, Mo.) was used to digest a 500-fold-molar excess of the target protein in a buffer containing 25 mM Tris (pH 8.0), 50 mM NaCl, and 1 mM CaCl2. Digestions were performed at 25°C and were stopped at 2-min intervals by adding phenylmethylsulfonyl fluoride (1 mM final concentration), removing the mixtures to ice, and then freezing them at −80°C. Samples were electrophoresed on sodium dodecyl sulfate (SDS)–15% polyacrylamide gels and were visualized by Coomassie blue staining.
Detection of RsbQ by Western blotting.
Polyclonal antibody was raised by injecting rabbits with the purified, His-tagged RsbQ. To test antibody specificity, extracts of PB2 and PB604 (rsbQΔ2) were prepared from early-stationary-phase cells. For determining RsbQ levels in vivo, strains PB2, PB633 (rsbQS96A), and PB631 (rsbQH247A) were grown in buffered LB lacking salt, and samples were taken at the indicated times during the logarithmic and stationary growth phases. Cells were harvested by centrifugation, washed in 1 ml of sonication buffer (10 mM Tris, pH 7.4, and 5 mM MgCl2), and resuspended in a minimal volume (50 to 100 μl) of the same buffer containing 10 μg of lysozyme. Following a 10-min incubation at 37°C, cells were broken by sonication and centrifuged to remove cell debris from the extracts. Approximately 100 μg of total cell protein per lane (as determined by Bio-Rad Protein Assay) was separated on an SDS–12% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Bio-Rad). Blots were blocked and washed in phosphate-buffered saline–Tween (PBST) (10 mM Na2HPO4, pH 7.2; 137 mM NaCl; 2.7 mM KCl; 3% [vol/vol] Tween 20) containing 5% (wt/vol) nonfat dry milk (BLOTTO) and were then exposed to the rabbit polyclonal anti-RsbQ antiserum for 1 h at 25oC. Following four washes with BLOTTO and PBST, blots were incubated at 25°C for 1 h with the secondary antibody, a goat anti-rabbit immunoglobulin G peroxidase conjugate (Sigma). Washes were repeated as before, with the addition of a final wash in PBS. Bound antibody was detected using the ECL Plus Western blotting detection kit (Amersham Pharmacia Biotech, Piscataway, N.J.) according to the manufacturer's instructions.
RESULTS
rsbQ is required to activate ςB in response to energy stress.
As shown in Fig. 1B, rsbQ is located in an operon together with rsbP, the structural gene for the serine phosphatase which activates ςB under conditions of energy stress. Although the promoter for the rsbQP operon has not been located precisely, fusion analysis found that energy signaling does not require increased operon transcription, leading to the notion that signaling is largely accomplished via a posttranslational mechanism (41). Moreover, because a similar arrangement of rsbQ and rsbP homologues is also found in Streptomyces coelicolor, we inferred that both gene products might affect a common cellular process (41). To determine whether RsbQ is involved in ςB regulation, we made a large in-frame deletion within the rsbQ coding region and substituted it for the chromosomal copy via a two-step allele replacement procedure. The resulting null mutant also contained a single-copy transcriptional fusion between the ςB-dependent ctc promoter and an E. coli lacZ reporter gene to provide an assay for ςB activity.
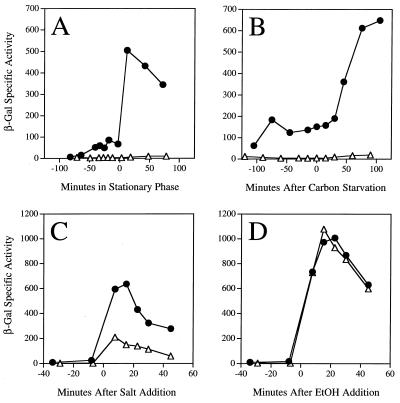
As shown in Fig. 2, the strain bearing the rsbQΔ2 null allele was unable to induce expression of the reporter fusion in two different energy stress assays: entry into stationary phase in buffered LB (Fig 2A) and glucose limitation in a minimal medium (Fig. 2B). In contrast, the rsbQΔ2 null mutant retained the ability to respond to signals of environmental stress. Logarithmically growing cells were challenged by the addition of either 0.3 M salt (Fig. 2C) or 4% ethanol (Fig. 2D). Response to ethanol stress was similar in the mutant and wild-type strains, whereas response to salt stress was decreased about threefold by the rsbQΔ2 null allele. Although it appears that rsbQ plays a role in response to salt stress, perhaps due to an effect on the levels of ςB in unstressed cells, rsbQ function is clearly not required for the environmental stress response as it is for the energy stress response.
FIG. 2.
RsbQ function is required for energy stress signaling. Effect of the rsbQΔ2 allele on β-galactosidase accumulation from a ςB-dependent transcriptional fusion in response to entry into stationary phase in buffered LB medium (A), carbon and energy starvation in a minimal medium containing 0.05% glucose (B), addition of 0.3 M NaCl to logarithmically growing cells in buffered LB (C), or addition of 4% ethanol (EtOH) to logarithmically growing cells in buffered LB (D). Time zero indicates when the stress was imposed on wild-type PB198 (●) and on the PB605 mutant bearing the rsbQΔ2 allele (▵).
The phenotype elicited by the rsbQΔ2 null allele was essentially the same as that previously reported for the rsbPΔ1::spc null allele (41). This raised the possibility that the in-frame deletion in rsbQ was polar on rsbP gene expression. To address this concern, we performed a complementation experiment in the rsbQ null background. ςB activation in response to stationary-phase stress was restored by insertion of a single copy of rsbQ+ at the thr locus, in trans to the rsbQΔ2 allele at the rsbQ locus (data not shown). We therefore conclude that rsbQ encodes a positive regulator which plays an essential role in transmitting energy stress signals to ςB.
Activation of ςB by RsbP requires RsbQ function in vivo.
In the parallel environmental signaling pathway (Fig. 1), the RsbU phosphatase is activated by direct protein-protein interaction with RsbT, the product of the gene immediately upstream from rsbU. RsbT therefore functions as a regulatory subunit of the RsbU environmental signaling phosphatase (21, 44). To probe whether a similar relationship exists between RsbQ and the RsbP energy-signaling phosphatase, we first asked if the two proteins could interact to activate the yeast two-hybrid system. As shown in Table 2, RsbQ and RsbP together activated transcription of the yeast reporter fusion. Moreover, this interaction was specific in that there was no detectable activation when RsbQ was paired with RsbU or when RsbP was paired with RsbT.
TABLE 2.
Activation of the yeast two-hybrid system by RsbQ paired with RsbP, RsbT, or RsbUa
| Fusion protein in pGAD424 | Activation result for different fusion proteins in pGBT9
|
|||
|---|---|---|---|---|
| RsbQ (0)b | RsbP (+) | RsbT (0) | RsbU (0) | |
| RsbQ | ND | ++ | ND | 0 |
| RsbP | ++ | ND | 0 | 0 |
| RsbT | ND | + | ND | ++ |
| RsbU | 0 | ND | ++ | ND |
++, strong blue color in colony lift assay; +, weak blue; 0, no blue detected; ND, not done.
The symbol shown in parentheses indicates the basal activation elicited by the protein of interest that was fused to the DNA binding domain carried by pGBT9 and tested in the absence of a companion activating domain carried by pGAD424.
We next used overexpression experiments to further characterize the relationship between RsbQ and RsbP. We moved each gene into a multicopy plasmid that placed its expression under control of the LacI-repressible, IPTG-inducible Pspac promoter. pMB64 (bearing the rsbQ coding region) was introduced into the wild-type strain and also into a strain harboring the rsbPΔ1::spc null allele, whereas pKV2 (bearing the rsbP coding region) was introduced into the wild-type strain and also into a strain harboring the rsbQΔ2 null allele. To permit measurement of ςB activity, these four strains each contained a single-copy ctc-lacZ reporter fusion.
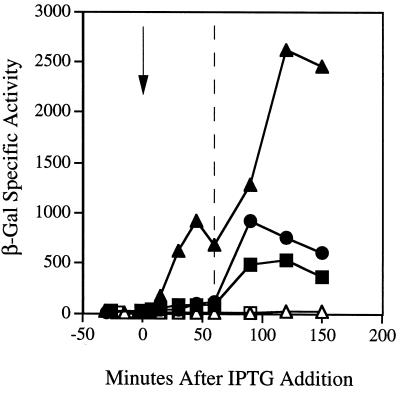
As shown in Fig. 3, overexpression of rsbQ during early logarithmic growth did not immediately induce expression of the reporter fusion in wild-type cells. Induction occurred only when cells encountered the energy stress of stationary phase, and the magnitude of this induction was reduced compared to the wild-type control in which rsbQ was not overexpressed. Moreover, overexpression of rsbQ could not compensate for the loss of rsbP function in the strain bearing the rsbPΔ1::spc null allele. The phenotype manifested in this latter strain was the same as that of the rsbP null mutant without overexpression of rsbQ: ctc-lacZ was not induced by energy stress. The failure of rsbQ overexpression to induce ςB activity in logarithmically growing cells stands in marked contrast to the results obtained by Yang et al. (44) in their study of the environmental stress pathway. These authors found a strong, rsbU-dependent induction of ςB activity immediately upon overexpression of the rsbT regulator. From the experiments shown in Fig. 3, we conclude that RsbQ does not act as a regulatory subunit for its cognate phosphatase, as does RsbT.
FIG. 3.
Overexpression of either RsbP or RsbQ cannot compensate for the loss of the other. At time zero (arrow) IPTG was added to logarithmically growing cells to induce overexpression of either rsbP (carried in pKV2) or rsbQ (carried in pMB64). Samples were periodically removed for assay of β-galactosidase activity both before and after cells had entered stationary phase in buffered LB medium (indicated by the dashed line at 60 min). ●, wild-type PB198; ▪, wild-type PB606 with rsbQ overexpressed; ▴, wild-type PB607 with rsbP overexpressed; □, PB719 mutant bearing the rsbPΔ1::spc allele with rsbQ overexpressed; ▵, PB716 mutant bearing the rsbQΔ2 allele with rsbP overexpressed.
We then performed the reciprocal experiment to determine the dependence of rsbP upon rsbQ. As shown in Fig. 3, overexpression of rsbP during early logarithmic growth caused a rapid and significant increase in ςB activity in wild-type cells. This activation occurred well before the energy stress of stationary phase, whereupon a further increase in ςB activity could be seen. However, this result required the presence of least one copy of rsbQ. In the absence of rsbQ function, overexpression of rsbP had no apparent effect. The phenotype manifested in this experiment was therefore the same as that of the rsbQΔ2 null mutant without overexpression of rsbP: ctc-lacZ was not induced by energy stress. We conclude that overexpression of either rsbQ or rsbP is not sufficient to compensate for the loss of the other regulator.
Serine 96 and histidine 247 of RsbQ are important for energy stress signaling.
In order to suggest the in vivo function of RsbQ, we first considered the Cluster of Orthologous Groups of proteins (COG) to which it belonged. The COG database (http://www.ncbi.nlm.nih.gov/COG) is a phylogenetic classification of proteins encoded by 44 complete microbial genomes representing 30 lineages, and it allows detection of both close and distant relationships (37). RsbQ (formerly called YvfQ) is a member of COG 0596—predicted hydrolases or acyltransferases (α/β hydrolase superfamily). Examples of the α/β hydrolase superfamily are widely distributed and manifest unusually diverse catalytic activities, functioning variously as proteases, lipases, peroxidases, epoxide hydrolases, dehalogenases, and esterases (23, 25). While their primary amino acid sequences are not highly conserved, members of the α/β hydrolase superfamily share a common three-dimensional core structure containing the catalytic domain. Within this domain is a catalytic triad in an invariant order: nucleophilic residue/acidic residue/histidine residue (23, 25). The residues comprising this triad can be broadly separated in the primary sequence but are brought together in the tertiary structure.
Comparison of the predicted RsbQ sequence to well-characterized α/β hydrolases yielded good candidates for the catalytic triad residues. Based on the alignments shown in Fig. 4, we chose serine 96 as the potential catalytic nucleophile for RsbQ and histidine 247 as the potential catalytic histidine. In contrast, the assignment of aspartate 219 as the acidic residue was less certain due to other nearby candidates. We therefore focused our analysis on serine 96 and histidine 247.
FIG. 4.
Partial alignment of RsbQ with well-characterized members of the α/β hydrolase superfamily. The crystal structures have been solved and the residues comprising the catalytic triads have been suggested or established for BpoA2, a bromoperoxidase from Streptomyces aureofaciens (16); DhlA, a haloalkane halidohydrolase from Xanthobacter autotrophicus (14, 28, 29, 39, 40); and EchA, an epoxide hydrolase from Agrobacterium radiobacter (24, 31). RsbQ and these other family members are aligned by the Reverse Position Blast algorithm (4) against the consensus for the α/β superfamily found in the Conserved Domain Database, maintained by the National Library of Medicine (www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml). RsbQ was 23.5% identical to the α/β hydrolase fold consensus over a 226-residue overlap (E = 3 × 10−15). The two regions surrounding the proposed catalytic triad residues are shown here. Residues in boldface indicate the positions conserved in at least three of the four proteins. (A) Residues surrounding the proposed nucleophile S96 of RsbQ, shown here in larger font together with the S→A substitution. The catalytic nucleophiles in the other proteins and in the consensus are also identified by the larger font. (B) Residues surrounding the proposed acidic residue D219 and the proposed histidine residue H247 of RsbQ, shown here in larger font together with the H→A substitution at H247. The catalytic acidic and histidine residues in the other proteins and in the consensus are also identified by the larger font.
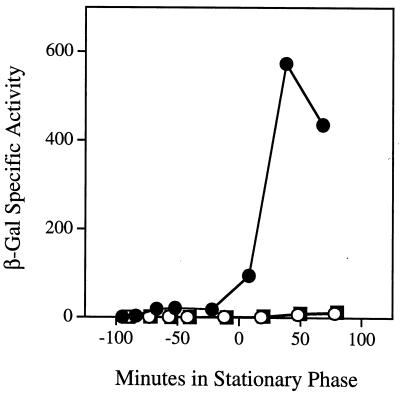
If serine 96 and histidine 247 comprised part of a catalytic triad in RsbQ and if the enzymatic activity of RsbQ was important for its signaling function, we would predict that changing either residue would affect ςB activation in response to energy stress. We therefore constructed two mutant versions of rsbQ, one in which an alanine replaced serine 96 (RsbQS96A) and another in which an alanine replaced histidine 247 (RsbQH247A). These mutant versions were substituted for the wild-type copy of rsbQ on the chromosome. The resulting mutant strains also contained a ctc-lacZ reporter fusion to measure ςB activity. As shown in Fig. 5, replacement of either serine 96 or histidine 247 with alanine completely abolished induction of the reporter fusion in response to energy stress. These results suggest that the catalytic activity of RsbQ is required for energy stress signaling. However, to strengthen this conclusion, it was necessary to test the possibility that the S96A and H247A alterations might only indirectly perturb catalytic activity as a result of their primary effects on RsbQ structure, synthesis, or stability.
FIG. 5.
Residues S96 and H247 of RsbQ are required for energy stress signaling. β-Galactosidase accumulation from a ςB-dependent transcriptional fusion as cells were grown to stationary phase in buffered LB medium. ●, wild-type PB198; ○, PB634 mutant bearing the rsbQS96A allele; ▪, PB632 mutant bearing the rsbQH247A allele.
The S96A and H247A alterations do not detectably affect RsbQ structure or accumulation.
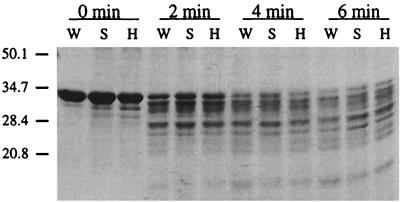
To address the possibility that the S96A and H247A alterations might affect RsbQ structure, we used limited trypsin proteolysis to compare the conformation of mutant and wild-type RsbQ proteins. His-tagged proteins were overexpressed in E. coli and purified as described in Materials and Methods. As shown in Fig. 6, the mutant proteins were not substantially altered in their trypsin susceptibility relative to the wild type. This implies that the native conformation of RsbQ was not materially disrupted by the S96A or H247A substitutions.
FIG. 6.
The S96A and H247A substitutions do not affect trypsin susceptibility of RsbQ. His-tagged versions of wild-type RsbQ (W), the S96A substitution (S), and the H247A substitution (H) were purified by metal affinity chromatography and subjected to limited trypsin digestion. Samples were removed at 2-min intervals and digestion was terminated by addition of phenylmethylsulfonyl fluoride. Protein fragments were separated by SDS-polyacrylamide gel electrophoresis and stained with Coomassie blue. The mobilities of the marker protein are given on the left, labeled with their masses (in kilodaltons).
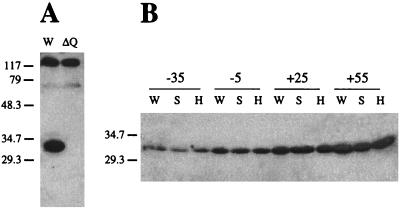
To determine whether the S96A and H247A substitutions affected synthesis or stability of the mutant proteins, we used Western blotting experiments to compare steady-state, in vivo levels of mutant and wild-type RsbQ. Polyclonal antibody against RsbQ was raised by injecting rabbits with the His-tagged protein purified from E. coli; the specificity of this antibody is shown in Fig. 7A. When tested against a whole-cell extract of wild-type cells that had been subjected to SDS-polyacrylamide gel electrophoresis, the anti-RsbQ antibody detected three proteins with distinctly different mobilities. One of these proteins had the mobility expected for RsbQ and was missing from an extract of the rsbQΔ2 null mutant. Figure 7B shows that the signals from this protein were the same in the wild type and the two substitution mutants at multiple points in the growth curve. We infer from these results that the alterations in the mutant proteins had no significant effect on synthesis or stability of RsbQ in vivo.
FIG. 7.
The S96A and H247A substitutions do not affect steady-state levels of RsbQ in vivo. Polyclonal antibody raised against purified RsbQ was used to probe Western blots of whole-cell extracts separated by SDS-polyacrylamide gel electrophoresis. (A) The anti-RsbQ antibody detected three clear signals in the wild-type PB2 extract (W). One of these signals was absent in the extract from the PB604 mutant bearing the rsbQΔ2 allele (ΔQ); this signal had the mobility expected for RsbQ (29.9 kDa). Marker protein masses (in kilodaltons) are shown on the left. (B) The wild-type PB2 (W), the PB633 mutant bearing the rsbQS96A allele (S), and the PB631 mutant bearing the rsbQH247A allele (H) were grown in buffered LB medium. The time before or after entry into stationary phase at which each group of three samples was taken is indicated in minutes at the top. At each time point, the anti-RsbQ antibody detected a signal of similar strength in all three strains. This signal increased in magnitude (relative to total cell protein) as cells entered stationary phase. Only that portion of the gel containing the RsbQ signal is shown; mobilities of the 29.3- and 34.7-kDa marker proteins are given on the left.
From the sum of these analyses, we conclude that the loss of energy stress signaling caused by the RsbQS96A and RsbQH247A substitutions most likely resulted from altered catalysis and not from a disruption of protein structure, synthesis, or stability. We therefore propose that RsbQ possesses an enzymatic activity that plays an essential role in the energy stress signaling pathway which activates the general stress response.
DISCUSSION
Bacteria use a variety of mechanisms to sense and respond to impending energy stress. These include control of individual enzyme activities by energy charge (6) as well as control of regulatory proteins by reaction centers which most frequently sense oxygen tension or electron flux (7, 38). Some of these reaction centers are provided by PAS domains, and in the best-characterized examples the PAS domain binds a chromophore specific for the parameter sensed, such as heme in the oxygen sensors FixL and Dos (11, 15) and flavin adenine dinucleotide in the redox sensors NifL and Aer (30, 34). In eukaryotic signaling pathways, PAS domains are also known to mediate ligand binding or protein-protein interactions (38), but these aspects of PAS function have not yet been demonstrated in prokaryotic systems.
Here we report that RsbQ is a positive regulator required for energy stress activation of the ςB transcription factor. Moreover, RsbQ interacts in the yeast two-hybrid system with the RsbP energy-signaling phosphatase, which contains a PAS domain in its amino-terminal region (41). We presently have no additional biochemical evidence to corroborate this interaction, nor have we detected a hypothetical ligand or chromophore which the PAS domain of RsbP might bind. However, we have shown that the predicted catalytic activity of RsbQ is important for its signaling role, and on the basis of our genetic analysis we can propose a model of RsbQ function.
We first considered the possibility that RsbQ might hydrolyze a small molecule and thereby produce a direct activator of the RsbP phosphatase. However, simply overexpressing RsbQ in growing cells did not trigger the energy stress response (Fig. 3), so this model would also require that RsbQ activity is somehow regulated by energy stress. Such energy stress regulation was not apparent even when the cells that were overexpressing RsbQ subsequently entered stationary phase. In contrast, overexpressing the RsbP phosphatase in growing cells did immediately induce a mock energy stress response, and this response was intensified upon subsequent challenge with an authentic energy stress. We interpret these results to indicate that in unstressed cells the RsbP phosphatase has a low intrinsic activity which normally sets the steady-state level of functioning ςB, a necessary feature of its autocatalytic induction mechanism (19). Overexpression of RsbP would rapidly increase the total amount of this intrinsic phosphatase activity, and, therefore, of functioning ςB. Moreover, we presume that RsbP activity in vivo is stimulated by energy stress (41). Therefore, induction of ςB activity would occur upon energy stress in wild-type cells, in which RsbP levels are normal, and also in cells in which RsbP levels have been artificially elevated.
We infer that the catalytic activity of RsbQ is required for RsbP phosphatase activity—both for the low intrinsic activity as well as for the activity stimulated by energy stress (Fig. 3). We therefore propose that the target of the RsbQ activity is RsbP itself. In this view, the RsbP phosphatase would be synthesized in an inactive form and converted to an active form by RsbQ. RsbQ could conceivably modify the covalent structure of RsbP by means of its presumed hydrolytic or acyltransferase activity or perhaps modify the hypothetical chromophore which binds the PAS domain of RsbP.
The relationship of the RsbQ and RsbP energy stress regulators is clearly different from that of the RsbT and RsbU environmental stress regulators. RsbT lies directly on the signaling pathway, and it passes the environmental stress signal to the RsbU phosphatase via a specific protein-protein interaction (21, 44). In contrast, RsbQ does not appear to lie directly on the energy signaling pathway but instead acts catalytically to render the RsbP phosphatase competent to receive the signal. This difference in the relationship between the terminal regulators of the energy and environmental branches suggests that the upstream sections of these two signaling pathways will also prove to function by distinctly different mechanisms.
ACKNOWLEDGMENTS
We thank Tania Baker for her helpful discussions and Valley Stewart for his critical comments on the manuscript.
This research was supported by Public Health Service grant GM42077 from the National Institute of General Medical Sciences. Kamni Vijay was a predoctoral trainee supported in part by Public Health Service training grant GM07377 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Akbar S, Gaidenko T A, Kang C M, O'Reilly M, Devine K M, Price C W. New family of regulators in the environmental signaling pathway which activates the general stress transcription factor ςB of Bacillus subtilis. J Bacteriol. 2001;183:1329–1338. doi: 10.1128/JB.183.4.1329-1338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signalling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. doi: 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 3.Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. doi: 10.1006/jmbi.1996.0390. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anagnostopulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atkinson D E. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry. 1968;7:4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- 7.Beinert H, Kiley P J. Fe-S proteins in sensing and regulatory functions. Curr Opin Chem Biol. 1999;3:152–157. doi: 10.1016/S1367-5931(99)80027-1. [DOI] [PubMed] [Google Scholar]
- 8.Benson A K, Haldenwang W G. Bacillus subtilis ςB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boylan S A, Rutherford A, Thomas S M, Price C W. Activation of Bacillus subtilis transcription factor ςB by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992;174:3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delgado-Nixon V M, Gonzalez G, Gilles-Gonzalez M A. Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry. 2000;39:2685–2691. doi: 10.1021/bi991911s. [DOI] [PubMed] [Google Scholar]
- 12.Dubnau D, Davidoff-Abelson R. Fate of transforming DNA following uptake by competent Bacillus subtilis. I. Formation and properties of the donor-recipient complex. J Mol Biol. 1971;56:209–221. doi: 10.1016/0022-2836(71)90460-8. [DOI] [PubMed] [Google Scholar]
- 13.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilis anti-ς factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Franken S M, Rozeboom H J, Kalk K H, Dijkstra B W. Crystal structure of haloalkane dehalogenase: an enzyme to detoxify halogenated alkanes. EMBO J. 1991;10:1297–1302. doi: 10.1002/j.1460-2075.1991.tb07647.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gilles-Gonzalez M A, Gonzalez G, Perutz M F. Kinase activity of oxygen sensor FixL depends on the spin state of its heme iron. Biochemistry. 1995;34:232–236. doi: 10.1021/bi00001a027. [DOI] [PubMed] [Google Scholar]
- 16.Hecht H J, Sobek H, Haag T, Pfeifer O, van Pee K H. The metal-ion-free oxidoreductase from Streptomyces aureofaciens has an α/β hydrolase fold. Nat Struct Biol. 1994;1:532–537. doi: 10.1038/nsb0894-532. [DOI] [PubMed] [Google Scholar]
- 17.Hecker M, Völker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the ςB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 18.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 19.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate ς factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςB in response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang C M, Vijay K, Price C W. Serine kinase activity of a Bacillus subtilis switch protein is required to transduce environmental stress signals but not to activate its target PP2C phosphatase. Mol Microbiol. 1998;30:189–196. doi: 10.1046/j.1365-2958.1998.01052.x. [DOI] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 23.Nardini M, Dijkstra B W. α/β hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 24.Nardini M, Ridder I S, Rozeboom H J, Kalk K H, Rink R, Janssen D B, Dijkstra B W. The X-ray structure of epoxide hydrolase from Agrobacterium radiobacter AD1. An enzyme to detoxify harmful epoxides. J Biol Chem. 1999;274:14579–14586. [PubMed] [Google Scholar]
- 25.Ollis D L, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken S M, Harel M, Remington S J, Silman I, Schrag J, et al. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 26.Price C W. Protective function and regulation of the general stress response in Bacillus subtilis and related gram-positive bacteria. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: American Society for Microbiology; 2000. pp. 179–197. [Google Scholar]
- 27.Price C W, Doi R H. Genetic mapping of rpoD implicates the major sigma factor of Bacillus subtilis RNA polymerase in sporulation initiation. Mol Gen Genet. 1985;201:88–95. doi: 10.1007/BF00397991. [DOI] [PubMed] [Google Scholar]
- 28.Pries F, Kingma J, Krooshof G H, Jeronimus-Stratingh C M, Bruins A P, Janssen D B. Histidine 289 is essential for hydrolysis of the alkyl-enzyme intermediate of haloalkane dehalogenase. J Biol Chem. 1995;270:10405–10411. doi: 10.1074/jbc.270.18.10405. [DOI] [PubMed] [Google Scholar]
- 29.Pries F, Kingma J, Pentenga M, van Pouderoyen G, Jeronimus-Stratingh C M, Bruins A P, Janssen D B. Site-directed mutagenesis and oxygen isotope incorporation studies of the nucleophilic aspartate of haloalkane dehalogenase. Biochemistry. 1994;33:1242–1247. doi: 10.1021/bi00171a026. [DOI] [PubMed] [Google Scholar]
- 30.Repik A, Rebbapragada A, Johnson M S, Haznedar J O, Zhulin I B, Taylor B L. PAS domain residues involved in signal transduction by the Aer redox sensor of Escherichia coli. Mol Microbiol. 2000;36:806–816. doi: 10.1046/j.1365-2958.2000.01910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rink R, Fennema M, Smids M, Dehmel U, Janssen D B. Primary structure and catalytic mechanism of the epoxide hydrolase from Agrobacterium radiobacter AD1. J Biol Chem. 1997;272:14650–14657. doi: 10.1074/jbc.272.23.14650. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Scott J M, Haldenwang W G. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor ςB. J Bacteriol. 1999;181:4653–4660. doi: 10.1128/jb.181.15.4653-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Söderback E, Reyes-Ramirez F, Eydmann T, Austin S, Hill S, Dixon R. The redox- and fixed nitrogen-responsive regulatory protein NIFL from Azotobacter vinelandii comprises discrete flavin and nucleotide-binding domains. Mol Microbiol. 1998;28:179–192. doi: 10.1046/j.1365-2958.1998.00788.x. [DOI] [PubMed] [Google Scholar]
- 35.Stahl M L, Ferrari E. Replacement of the Bacillus subtilis subtilisin structural gene with an in vitro-derived deletion mutation. J Bacteriol. 1984;158:411–418. doi: 10.1128/jb.158.2.411-418.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 37.Tatusov R L, Natale D A, Garkavtsev I V, Tatusova T A, Shankavaram U T, Rao B S, Kiryutin B, Galperin M Y, Fedorova N D, Koonin E V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taylor B L, Zhulin I B. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev. 1999;63:479–506. doi: 10.1128/mmbr.63.2.479-506.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verschueren K H, Franken S M, Rozeboom H J, Kalk K H, Dijkstra B W. Refined X-ray structures of haloalkane dehalogenase at pH 6.2 and pH 8.2 and implications for the reaction mechanism. J Mol Biol. 1993;232:856–872. doi: 10.1006/jmbi.1993.1436. [DOI] [PubMed] [Google Scholar]
- 40.Verschueren K H, Seljee F, Rozeboom H J, Kalk K H, Dijkstra B W. Crystallographic analysis of the catalytic mechanism of haloalkane dehalogenase. Nature. 1993;363:693–698. doi: 10.1038/363693a0. [DOI] [PubMed] [Google Scholar]
- 41.Vijay K, Brody M S, Fredlund E, Price C W. A PP2C phosphatase containing a PAS domain is required to convey signals of energy stress to the ςB transcription factor of Bacillus subtilis. Mol Microbiol. 2000;35:180–188. doi: 10.1046/j.1365-2958.2000.01697.x. [DOI] [PubMed] [Google Scholar]
- 42.Voelker U, Voelker A, Haldenwang W G. Reactivation of the Bacillus subtilis anti-ςB antagonist, RsbV, by stress- or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 45.Zhang S, Scott J M, Haldenwang W G. Loss of ribosomal protein L11 blocks stress activation of the Bacillus subtilis transcription factor ςB. J Bacteriol. 2001;183:2316–2321. doi: 10.1128/JB.183.7.2316-2321.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]