Significance
One of the most important physiological damage-control mechanisms is detoxification. Because of their roles in removing potentially harmful substances, detoxification systems are highly promiscuous in the molecules they recognize. While evolutionarily protective, this presents a roadblock to drug development. To understand how drug molecules will react during physiological exposure, we must identify the mechanisms of promiscuity in detoxification systems. In this study, we have performed structural, biochemical, chemical, and cellular experiments to characterize a structural mechanism of promiscuity in pregnane X receptor, a major transcriptional regulator of drug metabolizing enzymes.
Keywords: nuclear receptor, promiscuity, drug design
Abstract
Ligand-binding promiscuity in detoxification systems protects the body from toxicological harm but is a roadblock to drug development due to the difficulty in optimizing small molecules to both retain target potency and avoid metabolic events. Immense effort is invested in evaluating metabolism of molecules to develop safer, more effective treatments, but engineering specificity into or out of promiscuous proteins and their ligands is a challenging task. To better understand the promiscuous nature of detoxification networks, we have used X-ray crystallography to characterize a structural feature of pregnane X receptor (PXR), a nuclear receptor that is activated by diverse molecules (with different structures and sizes) to up-regulate transcription of drug metabolism genes. We found that large ligands expand PXR’s ligand-binding pocket, and the ligand-induced expansion occurs through a specific unfavorable compound-protein clash that likely contributes to reduced binding affinity. Removing the clash by compound modification resulted in more favorable binding modes with significantly enhanced binding affinity. We then engineered the unfavorable ligand-protein clash into a potent, small PXR ligand, resulting in marked reduction in PXR binding and activation. Structural analysis showed that PXR is remodeled, and the modified ligands reposition in the binding pocket to avoid clashes, but the conformational changes result in less favorable binding modes. Thus, ligand-induced binding pocket expansion increases ligand-binding potential of PXR but is an unfavorable event; therefore, drug candidates can be engineered to expand PXR’s ligand-binding pocket and reduce their safety liability due to PXR binding.
Promiscuity in cellular detoxification pathways is an evolutionary trait that allows cells to maintain chemical homeostasis, even when presented with diverse endobiotics and xenobiotics (1–4). However, due to their negative influence on drug pharmacokinetics and toxicity, these pathways contain perhaps the most infamous promiscuous proteins related to human physiology. Metabolic enzymes modify diverse small molecules to protect the body from toxicological harm and maintain physiological homeostasis, and, in the process, alter drug activities by disabling active parental molecules or creating metabolites with new activities (5). Like the detoxification enzymes themselves, transcription factors that control expression of drug metabolism-related genes show great ligand-binding promiscuity. These “xenobiotic receptors” sense exogenous substances and respond by up-regulating detoxification programs. The nuclear receptor pregnane X receptor (PXR) is a master transcriptional regulator of drug metabolism and disposition gene networks (6–8). Among various functions, PXR has a central role in controlling the expression of the cytochrome P450 3A (CYP3A) family of enzymes, which metabolizes more than half of clinically applied drugs (9). Importantly, direct inhibition of CYP3A activity or induction of CYP3A expression through mechanisms such as PXR activation are the dominant sources of drug–drug interactions (10). Circumventing small molecule-mediated PXR activation and interactions with downstream metabolic enzymes is a major roadblock to drug development, as these events lead to drug–drug interactions and decreased drug efficacy. Thus, assessing and designing out the PXR liability of small molecules is an integral part of drug development pipelines (11).
Mechanistically, structure–activity relationships (SAR) are challenging to perform with promiscuous ligand binders because of the allowed conformational heterogeneity in both the proteins and bound ligands. Various structural models have been proposed (12), but the mechanisms of promiscuity differ from protein to protein, and a single protein may have promiscuity mechanisms specific to each ligand. Therefore, a holistic view of promiscuity is difficult to achieve and requires the study of diverse protein–ligand partners. The immense chemical space of PXR ligands is exhibited in SI Appendix, Table S1, in which all previously cocrystallized ligands are displayed. Importantly, PXR has been crystallized with a large variety of chemicals, resulting in some understanding of its promiscuity mechanisms. PXR is known to allow multiple binding poses of ligands (13), incorporate multiple ligands in the binding pocket simultaneously (14, 15), and expand its binding pocket to accommodate large ligands (16). The diversity of PXR ligands can be explained, in part, by PXR’s ligand-binding pocket volume, which is large even when compared with its partner xenobiotic receptor, constitutive androstane receptor (CAR; 1,200 to 1,600 Å3 for PXR versus ~600 Å3 for CAR) (17). However, although previous studies have illustrated PXR’s structural response to ligands of various shapes and sizes, no studies have yet focused on PXR-ligand changes related to analogs of single scaffolds. Thus, in the current work, we characterize a conformational mechanism that governs PXR ligand promiscuity and demonstrate how this structural feature can be exploited across analogs of multiple chemical scaffolds to modulate PXR binding.
The large macrocyclic antibiotic rifampicin was the first discovered human PXR agonist, and rifampicin’s ability to up-regulate CYP3A4 was the very reason the human PXR homolog was initially discovered and characterized (6, 8, 18). A rifampicin-PXR ligand binding domain (LBD) cocrystal structure previously showed that rifampicin binding causes expansion of, and disorder in, the ligand-binding pocket (16). We performed SAR and cocrystallography of selected rifampicin analogs and identified a key chemical appendage responsible for both the induced PXR LBD structural disorder and comparatively weak PXR binding affinity of rifampicin. Removal of rifampicin’s 3-position (4-methylpiperazin-1-yl)iminomethyl appendage “unlocks” the macrolide binding pose, allowing a more favorable position within the pocket that can exist without unfavorable binding pocket expansion. We used these observations to rationally design analogs of one of the most potent PXR agonists, T0901317, that were predicted to enlarge the binding pocket as rifampicin does. The analogs showed a marked decrease in PXR binding affinity that correlated with the length of the added chemical appendage, and we confirmed by cocrystallography that the reduction in affinity was due to unfavorable changes in binding mode and PXR conformation. PXR’s binding pocket inflation represents a mechanism that enables it to accommodate diversified compounds, augmenting its role as a detoxification program regulator. However, our results show that while the PXR ligand-binding pocket can expand to incorporate large ligands, ligand-induced expansion reduces a ligand’s binding affinity, an unfavorable attribute that can be utilized in drug development to minimize the PXR liability of small molecules. Our studies support the feasibility to control the function of a promiscuous protein by engineering its ligands.
Results
Rifampicin Analogs Exhibit Differential Binding Affinity for PXR LBD.
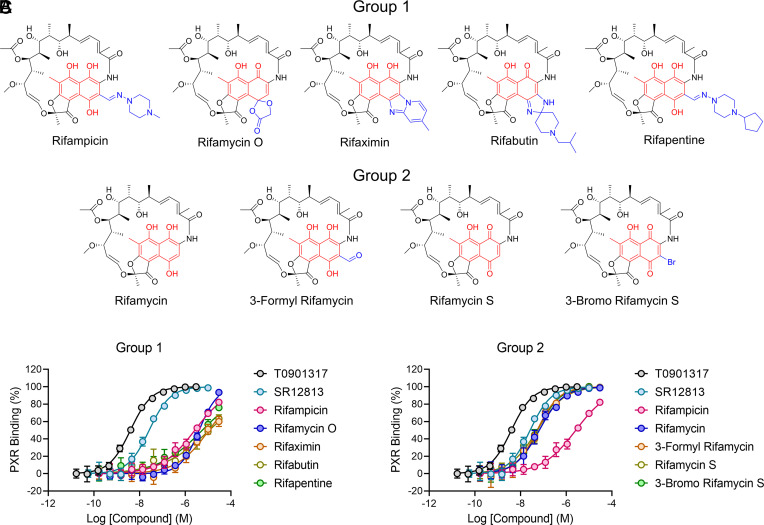
Because rifampicin is the prototypical PXR agonist and has long been a benchmark for assessing CYP3A4 induction through PXR, we performed SAR for the binding affinity of rifampicin to PXR LBD. Rifampicin and eight analogs were chosen for the analysis, including additional clinically approved drugs such as rifaximin and rifapentine (Fig. 1). To measure ligand binding, we used a time-resolved fluorescence resonance energy transfer (TR-FRET) assay that measures displacement of a fluorescent PXR ligand from PXR LBD (19). T0901317 and SR12813 were used for comparison because they are two of the most potent and commonly used PXR agonists (20–24). Interestingly, rather than a gradient of binding affinities, the test compounds clustered into two distinct groups (Fig. 1 B and C and Table 1). Group 1 analogs bound with micromolar affinity roughly equal to that of rifampicin, while group 2 analogs bound with nanomolar affinity roughly equal to that of SR12813.
Fig. 1.
Rifampicin analogs cluster into two distinct groups for PXR LBD binding affinity. (A) Chemicals are divided into two groups: 1) compounds that bind PXR LBD with affinity equal to rifampicin and 2) compounds that bind PXR LBD with affinity greater than rifampicin. The 1,4-dihydroxynaphthalene core is red, and the 3- and 4- position appendages are blue. (B and C) PXR LBD binding assays are shown for the indicated compounds.
Table 1.
PXR LBD binding activities of rifampicin analogs
| Compound | Group | PXR LBD binding [IC50 ± SD (µM)] | FC* |
|---|---|---|---|
| T0901317 | NA | 0.004 ± 0.0003 | 0.0015 |
| SR12813 | NA | 0.026 ± 0.003 | 0.0096 |
| Rifampicin | 1 | 2.7 ± 0.4 | 1 |
| Rifamycin O | 1 | 4.1 ± 0.4 | 1.5 |
| Rifaximin | 1 | 11.4 ± 3.3 | 4.2 |
| Rifabutin | 1 | 11.8 ± 4.4 | 4.4 |
| Rifapentine | 1 | 5.7 ± 1.6 | 2.1 |
| Rifamycin | 2 | 0.067 ± 0.009 | 0.025 |
| 3-Formyl Rifamycin | 2 | 0.054 ± 0.01 | 0.020 |
| Rifamycin S | 2 | 0.054 ± 0.014 | 0.020 |
| 3-Bromo Rifamycin S | 2 | 0.057 ± 0.011 | 0.021 |
*FC, fold change, calculated by dividing a compound’s IC50 value by that of rifampicin.
Rifampicin Analog Binding Affinity Is Correlated with the Size of the 3-Position Appendage at Its 1,4-Dihydroxynaphthalene Core.
The two-group clustering of binding affinities suggested that a single specific difference exists between compounds of group 1 versus group 2, and upon inspection of the chemical structures, a clear correlation between activity and structures could be observed. Rifampicin contains a bulky substituent in the 3-position of the substituted 1,4-dihydroxynaphthalene core (Fig. 1A), and all other group 1 analogs likewise have bulky moieties in the 3- or 4-positions. However, in the more potent group 2 binders, these bulky appendages are absent.
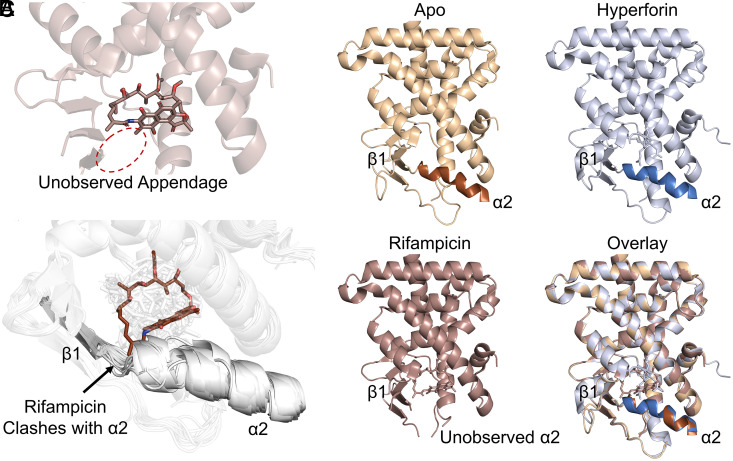
To gain insight into the potential impacts of the appendage on PXR binding, we analyzed the previously solved cocrystal structure of PXR LBD in complex with rifampicin (PDB ID 1SKX) (16). The appendage, however, is not present in the structure due to lack of observable electron density (Fig. 2A). In this previous report, Chrencik et al. found that the 3-position substituent clashed with regions on the floor of the ligand-binding pocket, resulting in disordered expansion of the pocket and lack of electron density for both the rifampicin appendage and PXR residues comprising the pocket floor (16). This phenomenon can be observed by comparing the rifampicin-PXR LBD complex with the apo and hyperforin-PXR LBD structures, which have well-defined floors (Fig. 2B). Additional rifampicin atoms other than the appendage can be seen to clash with the native conformation of the loop connecting alpha helix 2 (α2) to beta strand 1 (β1), explaining the absence of α2 from the electron density (Fig. 2C and SI Appendix, Table S1). Analysis of all PXR LBD crystal structures to date (n = 49, SI Appendix, Table S1) shows that while this region has some amount of flexibility to allow binding of various ligands, rifampicin is the only ligand to have significant overlap with the native α2-β1 loop configuration (Fig. 2C).
Fig. 2.
Rifampicin clashes with the native PXR α2-β1 loop conformation. (A) PXR LBD is shown in complex with rifampicin (PDB ID 1SKX). The location of the unobserved 3-position substituent is indicated with a dotted oval. (B) Structures of apo PXR LBD (PDB ID 1ILG) or PXR LBD in complex with hyperforin (PDB ID 1M13, Right) or rifampicin (PDB ID 1SKX, Left) are shown. Alpha helix 2 (α2) and beta strand 1 (β1) are indicated, and α2 is not observed in the rifampicin-bound structure. (C) All previously reported PXR LBD structures are overlaid. Ligands are shown as sticks, and protein is represented as cartoon. Rifampicin is shown in chocolate color and is the only ligand to clash with the native α2-β1 loop conformation shown in gray.
Crystal Structure of PXR LBD Bound to the High-Affinity Binder Rifamycin S Reveals Loss of Ligand-Protein Clash and Gain of Favorable Interactions.
In Fig. 1, four rifampicin analogs were found to have higher binding affinity than rifampicin for PXR LBD (rifamycin, 3-formyl rifamycin, rifamycin S, and 3-bromo rifamycin S). Based on our comprehensive evaluation of all PXR LBD structures (Fig. 2C), we hypothesized that the affinity shift may be related to two chemical and structural features. First, the lack of a bulky 3-position substituent would allow compound binding without significant clashes occurring with the ligand-binding pocket floor. Second, the high-affinity analogs would bind in such a way that avoids additional clashes between the protein and macrocycle.
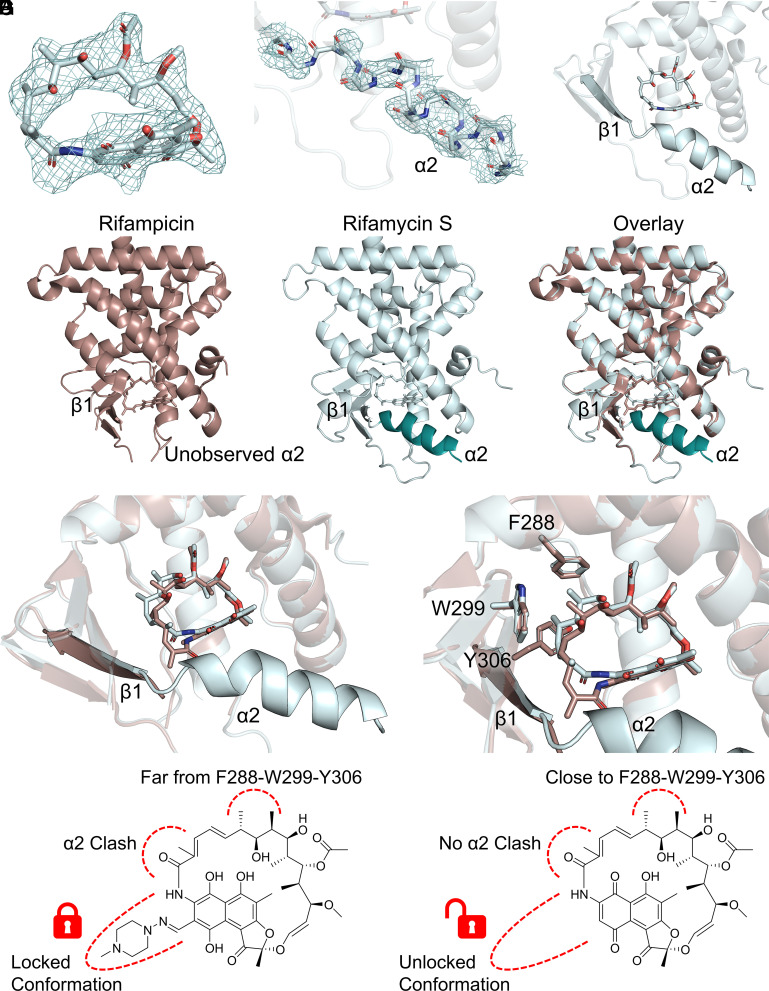
To fully understand the mechanism of increased binding affinities, we solved the crystal structure of rifamycin S-bound PXR LBD to a resolution of 2.25 Å (SI Appendix, Table S2). The binding location of rifamycin S in the structure was well defined, with the refined Fo–Fc map showing clear density for the macrolide ring (Fig. 3A). Unlike the rifampicin–PXR LBD complex, the rifamycin S-bound structure had clear density for helix 2 (Fig. 3B), and this correlated with the absence of a rifamycin S clash with the α2-β1 loop (Fig. 3C). Alpha helix 2 was present at the same location as in other PXR LBD structures (Fig. 3D). Rifamycin S was twisted in the pocket from rifampicin’s position, resulting in increased distance between the ligand and α2 (Fig. 3E). Interestingly, the shift away from α2 resulted in rifamycin S binding deeper in the F288-W299-Y306 aromatic cage, which has been shown to be vital for PXR ligand binding (Fig. 3F). Therefore, the enhanced binding affinity of group 2 rifampicin analogs appears to be due to 1) no clash with the ligand-binding pocket floor and 2) increased hydrophobic interactions of the compound with the F288-W299-Y306 subpocket. Although the rifampicin appendage is not observed in the structure, it is likely that its physical presence restrains rifampicin to a conformation that clashes with the α2-β1 loop; removal of the appendage frees the macrocycle to bind in a more favorable position (Fig. 3G).
Fig. 3.
Rifamycin S has a more favorable binding position than rifampicin in the PXR ligand-binding pocket. (A) The rifamycin S binding mode in PXR LBD is well defined. The 2Fo–Fc map is contoured in mesh at 1.0 RMSD and carved around rifamycin S at 2 Å. (B) α2 in the rifamycin S-PXR LBD cocrystal structure is well-defined. The α2 main chain is shown as sticks with the 2Fo–Fc map contoured in mesh at 1.0 RMSD and carved around main chain atoms at 2 Å. (C) Rifamycin S does not clash with α2 in the cocrystal structure. (D) Comparison of overall PXR LBD structures bound to rifamycin S or rifampicin (PDB ID 1SKX). The rifamycin S-bound LBD has a clearly placed α2. (E) Overlay of rifamycin S and rifampicin positions. Rifamycin S is shifted away from α2 compared with rifampicin. (F) Overlay of rifamycin S and rifampicin positions. Rifamycin S binds deeper in the F288-W299-Y306 subpocket than rifampicin. (G) Schematic of structural changes between rifampicin-bound (Left) and rifamycin S-bound (Right) PXR LBD.
Structural Findings from the Rifamycin S-PXR LBD Complex Can Be Applied to Different Ligand Scaffolds.
An inherent complexity of conducting SAR on promiscuous ligand-binding proteins is the specificity of the SAR findings to a single chemical scaffold. We found that removing a ligand-PXR α2 clash enhanced the ligand’s binding affinity (Fig. 3). Therefore, we would predict that engineering a chemical clash with the α2-β1 region will reduce ligand-binding affinity. To concurrently test this prediction and whether the rifampicin analog results can be applied to additional chemical space, we chose to design analogs of the highly potent PXR agonist T0901317.
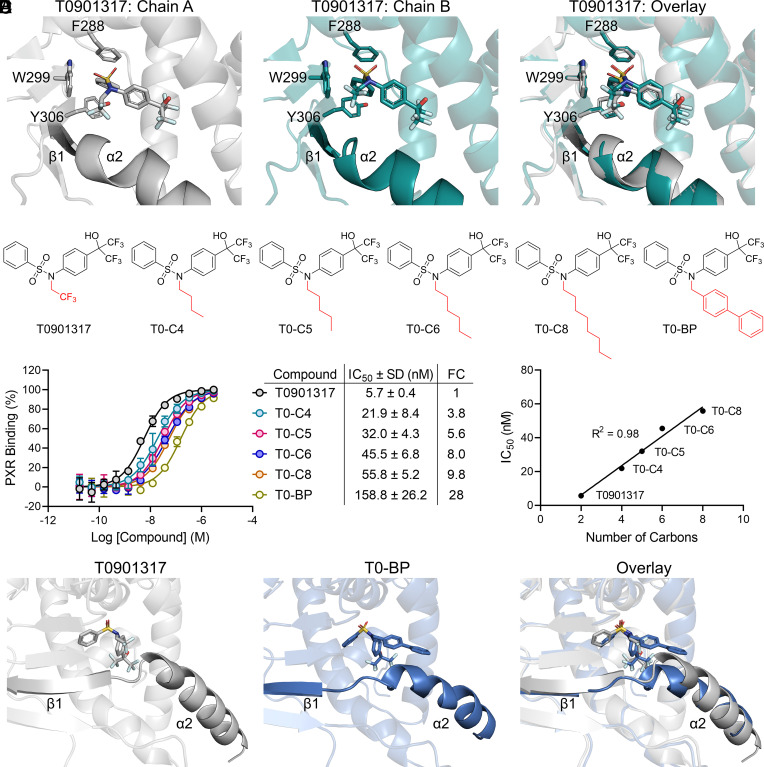
The published crystal structure of T0901317 in complex with PXR LBD (PDB ID 2O9I) has a PXR LBD dimer with T0901317 bound to each monomer (Fig. 4A). T0901317 is placed in slightly different poses in the two chains, but in both cases the N-trifluoroethyl group is oriented toward the α2-β1 loop (Fig. 4A). This positioning made T0901317 an attractive candidate to test the possibility to design a α2-β1 clash, and we synthesized five analogs with extensions of the N-trifluoroethyl group (Fig. 4B). Four analogs were carbon chain extensions (T0-C4, T0-C5, T0-C6, and T0-C8), and the fifth compound was a biphenyl substitution (T0-BP). When assayed for PXR LBD binding, affinity of the analogs was reduced in a stepwise manner, with T0-BP being 28-fold less potent than the parental T0901317 (Fig. 4C). Interestingly, upon inspection of the carbon chain extension analogs, there was a linear correlation between binding potency and carbon chain length (Fig. 4D). Although the strategy was not able to completely abolish PXR binding, it is noteworthy that we achieved nearly a 30-fold reduction in binding affinity of one of the most potent PXR agonists currently known.
Fig. 4.
Ligands designed to clash with the α2-β1 loop have decreased PXR binding affinity. (A) Comparison of T0901317 binding poses in the two PXR LBD chains of PDB ID 2O9I. (B) Chemical structures of T0901317 and synthesized analogs. (C) PXR LBD binding assays were performed for the indicated compounds. Fold change (FC) was calculated by dividing a compound’s IC50 value by that of T0901317. (D) The binding IC50s are plotted against the number of carbons on the N-position of the respective T0901317 analog. T0-BP was excluded from the analysis. (E) Comparison of T0901317-bound (PDB ID 2O9I, chain A) and T0-BP-bound PXR LBD.
The binding results suggest that our structure-guided approach to modulate ligand proximity to the α2-β1 region can be applied to various ligand scaffolds. To confirm that the differences in analog binding affinity were due to PXR-ligand structural changes, we cocrystallized PXR LBD with T0-BP and T0-C6 (Fig. 4E and SI Appendix, Tables S3 and S4 and Figs. S1–S3). Both compounds were well defined in the pocket (SI Appendix, Fig. S1); however, due to its flexibility, apparent lack of stabilizing interactions, and substantial available surrounding space for it to freely move, we did not observe electron density for the T0-C6 carbon chain (SI Appendix, Fig. S1B). As predicted, the biphenyl rings of T0-BP occupied space that would normally be occupied by α2, and the helix was displaced downward compared with T0901317-bound PXR LBD (Fig. 4E). Strikingly, when the α2-β1 region of T0-BP-bound PXR LBD was compared with the α2-β1 region of all other PXR LBD structures, we found that the helix and beta strand were displaced more by T0-BP than by any other previously cocrystallized ligand (SI Appendix, Fig. S2). T0-C6-induced structural changes were more subtle, but we observed three significant differences between T0901317-bound and T0-C6-bound PXR LBD (SI Appendix, Fig. S3). First, the L209 sidechain in the α2-β1 loop flipped to accommodate the carbon chain. Second, α2 was shifted away from the compound binding position due to its being dislodged by the carbon chain. Third, the C terminus of α11 was displaced in T0-C6-bound versus T0901317-bound PXR LBD. These three events likely occur in succession upon binding of the extended T0901317 analog: Binding requires extra space that is afforded by the L209 flip and α2 displacement, and the α2 movement then forces repositioning of α11. These observations indicate that extending the N-position arm of T0901317 toward the α2-β1 loop results in unfavorable PXR conformational changes to incorporate the ligand.
Incorporation of the α2 Clash Translates to Reduced Cellular Activity.
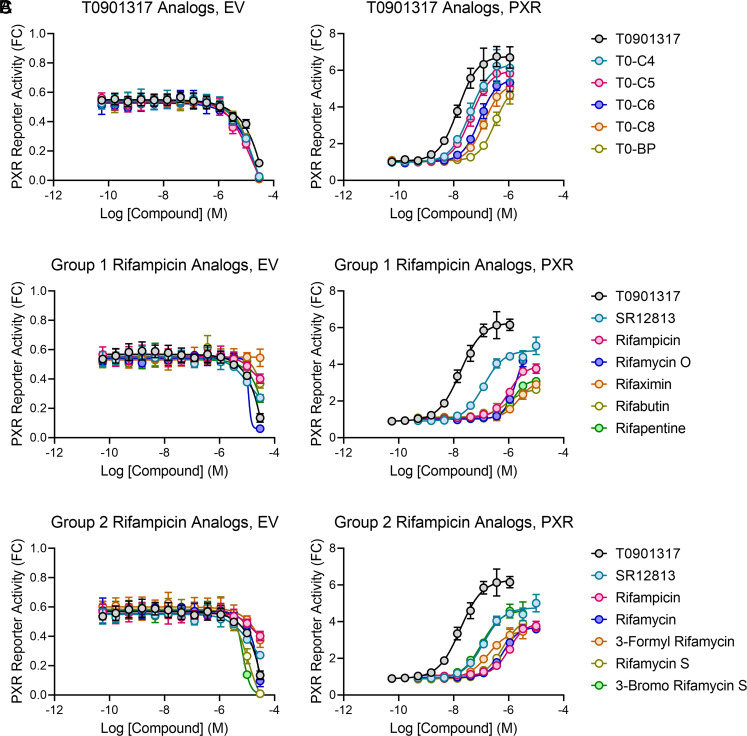
We have proposed a structural feature that can be used to manipulate PXR ligand binding and validated the proposal in biochemical experiments. However, the biological impacts remain to be seen. Therefore, we tested the rifampicin and T0901317 analogs in cellular reporter assays that measure the expression of firefly luciferase under the control of a PXR-responsive CYP3A4 promoter. We used 293T cells that do not express PXR and cotransfected the reporter with either PXR-expressing plasmid or an empty vector (EV) to control for reporter activity changes unrelated to PXR. High concentrations of compounds tend to induce nonspecific signals due to cytotoxicity and other nonspecific mechanisms. To identify an appropriate range of compound concentrations for assessing PXR activation, we first tested the EV-transfected cells using a large range of compound concentrations (Fig. 5 A–C, Left). The concentrations that nonspecifically reduced reporter signal were excluded from analysis in the PXR-transfected cells.
Fig. 5.
Structural predictions and biochemical results translate to cellular PXR activity. 293T cells were cotransfected with a PXR-responsive luciferase reporter and either an empty vector (EV) or PXR-expressing plasmid. Cells were treated for 24 h with the indicated compounds and assayed for luciferase activity. Compounds were used in threefold dilution series starting at 30 µM, and concentrations that exhibited signal reduction in the EV wells were excluded from analysis in the PXR-transfected samples. Results are shown as fold change (FC) relative to the averaged dimethyl sulfoxide (DMSO) controls of PXR-transfected cells, and compounds are split based on their previous designations: (A) T0901317 analogs, (B) group 1 rifampicin analogs, and (C) group 2 rifampicin analogs.
The T0901317 analogs showed an excellent correlation between binding potencies (Fig. 4C) and cellular activity (Fig. 5 A, Right), thereby validating our structural prediction in a cellular model. The group 1 rifampicin analog trends were also as expected, with all analog activities equal to or slightly less than rifampicin (Fig. 5 B, Right). The group 2 rifampicin analogs were less consistent with the PXR binding potency but still exhibited the expected trend (Figs. 1 C and 5 C, Right). Cellular activities of rifamycin and rifamycin S were marginally better than rifampicin, 3-formyl rifamycin activity was more significantly enhanced over rifampicin, and 3-bromo rifamycin S had the expected activity equal to that of SR12813 (Fig. 5C). The inconsistencies may be attributed to various factors, including cellular permeability, differential cellular metabolism, or altered non-PXR protein binding profiles. Indeed, unlike rifampicin, several of the analogs showed significant reduction in reporter expression in the absence of PXR, indicating transcriptional or cytotoxic effects. Nevertheless, together, the data indicate that designing a ligand clash with the α2-β1 area reduces PXR binding and subsequent transcriptional transactivation.
Groups 1 and 2 Rifampicin Analogs Have Distinct Binding Requirements.
Studies of promiscuity may involve two general approaches: 1) Ligands are modified to impart enhanced or diminished specificity in their protein-binding profiles and 2) proteins are modified to obtain enhanced or diminished specificity in their ligand-binding profiles. Thus far, we have identified a PXR structural feature that regulates PXR’s ligand-binding promiscuity and synthesized ligands to unfavorably clash with the feature (approach 1). Next, we conducted PXR mutagenesis to alter PXR ligand selectivity (approach 2).
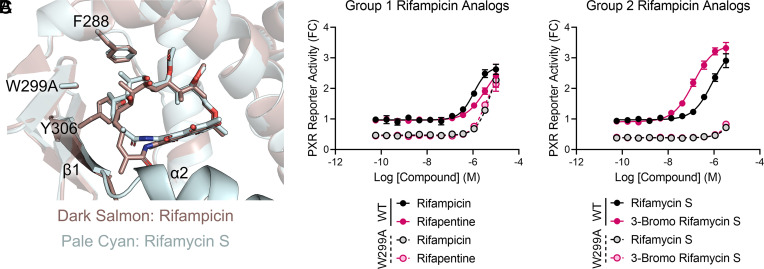
In addition to changes at the α2 region, our rifamycin S-PXR LBD structure indicated that rifamycin S binds deeper in the F288-W299-Y306 subpocket than rifampicin (Fig. 3F), suggesting that the higher affinity group 2 analogs may have increased reliance on the aromatic cage for binding. The subpocket was previously identified as a key element in PXR ligand binding (25), and we have found that mutations in the region have ligand-dependent outcomes (26). Specifically, the W299A mutation weakens the aromatic cage, thereby impacting ligand binding (Fig. 6A), but the specific effects vary among ligands. The small ligand T0901317 is ~100-fold less potent in activating W299A mutant PXR compared with wild-type (WT), but the relatively large rifampicin is minimally affected by the mutation (26). To test our structure-based prediction that the group 2 analogs may more heavily rely on the W299 region than rifampicin and group 1 analogs, we performed reporter assays with WT and W299A PXR. Like rifampicin, rifabutin activity changed very little from WT to W299A (Fig. 6B). However, the group 2 analogs rifamycin S and 3-bromo rifamycin S were nearly completely inactive for W299A PXR (Fig. 6C). For 3-bromo rifamycin S, the mutation resulted in >24-fold reduction in activity (EC50 = 137 ± 40 nM for WT and EC50 > 3.3 µM for W299A; 3.3 µM was the highest concentration before reporter signal reduction was observed in Fig. 5B). Therefore, we found that removal of rifampicin’s 3-position moiety improves ligand potency but alters the protein–ligand interactions in a manner that can be exploited by modifying PXR residues.
Fig. 6.
Group 2 rifampicin analogs have increased reliance on the F288-W299-Y306 aromatic cage. (A) Overlay of rifampicin (dark salmon) and rifamycin S (pale cyan) positions in wild-type (WT) PXR LBD (from Fig. 3F) with W299 modeled as A299. (B and C) 293T cells were cotransfected with a PXR-responsive luciferase reporter and either a WT or W299A mutant PXR-expressing plasmid. Cells were treated for 24 h with the indicated compounds and assayed for luciferase activity. Results are shown as fold change (FC) relative to the averaged DMSO controls of WT PXR-transfected cells.
Discussion
Promiscuity is often discussed in the context of enzymatic substrates and reactions, but xenobiotic receptors are also excellent models of ligand-binding promiscuity. Among the 48 human nuclear receptors, the xenobiotic receptors PXR and CAR are unique in their evolutionary emergence by positive rather than negative selection, suggesting a significant fitness benefit in their current states (27). Because of their roles in transcriptionally regulating detoxification programs, PXR and CAR must be able to respond to a variety of chemical ligands. PXR, in particular, is highly efficient in this regard due to its large binding pocket volume (1,200 to 1,600 Å3) (17) that allows multiple binding poses of ligands (13), simultaneous incorporation of multiple ligands in the binding pocket (14, 15), and further expansion of its binding pocket to accommodate large ligands (16). Additionally, because of its role in regulating CYP3A expression, analysis of PXR activation is standard in drug development pipelines and is in the United States Food and Drug Administration guidance (https://www.fda.gov/regulatory-information/search-fda-guidance-documents/in-vitro-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter-mediated-drug-interactions) (11). Because of PXR’s enormous ligand diversity, it is not possible to predict the PXR activation potential of small molecule drug candidates. All molecules must be experimentally tested, and substantial medicinal chemistry effort is required to remove the PXR liability of lead compounds. Therefore, studying the mechanisms of PXR promiscuity is of great importance in drug development.
Although PXR is well described as being promiscuously activated by diverse molecules, certain selectivity exists among chemicals. This selectivity is clear in two observations: 1) PXR proteins from different species are activated by different compounds (28) and 2) drug molecules can be chemically altered to escape PXR binding (11). Structural studies are required to reveal the mechanisms of promiscuity and selectivity, but no such studies have been performed with structurally related compounds to divulge the differences between strong and weak binders. Structure-guided reduction in PXR binding is performed in pharmaceutical development, but reduction in PXR activity empirically through medicinal chemistry is the usual method of removing the PXR liability of small molecules. Campaigns that use a crystallographic approach generally begin with a PXR LBD cocrystal structure with a single potent PXR agonist; the structure is then used to guide medicinal chemistry efforts with no further structural characterization. Introducing a molecule clash with the F288-W299-Y306 aromatic cage is the most common result of these structure-guided programs (11). Thus, we have performed a crystallographic comparison of structurally related chemicals that bind and activate PXR with different potencies and used our findings to characterize a PXR feature that can be used to enhance or diminish PXR binding by small molecules.
Rifampicin was the first discovered human PXR agonist, and nearly three decades later, it remains one of the most common experimental PXR inducers (6, 8, 18). However, with a micromolar binding affinity (IC50 = 2.7 µM in PXR LBD binding assay, Table 1 and Fig. 1), rifampicin is orders of magnitude less potent than other PXR agonists, such as T0901317 (IC50 = 4 nM, Table 1 and Fig. 1). A previously reported rifampicin-PXR LBD cocrystal structure showed that rifampicin binding causes expansion of, and disorder in the floor of, the ligand-binding pocket (16). With a molecular weight (MW) of 823 Da, rifampicin is substantially larger than T0901317 (MW = 481 Da), invoking the question of whether ligand size is a limiting factor of PXR activity. However, with MW of 775 Da, 3-bromo rifamycin S had a binding IC50 of 57 nM, ~50-fold enhanced over rifampicin. This affinity was only ~twofold worse than SR12813 (IC50 = 26 nM, Table 1 and Fig. 1), which has a MW of 505 Da. Therefore, size is not directly correlated with activity. After testing multiple rifampicin analogs, we found a clear correlation between binding affinity and presence of a bulky substituent on the dihydroxynaphthalene core (Fig. 1). Our crystal structure of PXR LBD bound to rifamycin S revealed that a compound lacking the bulky moiety on the dihydroxynaphthalene core no longer induces structural disorder in PXR LBD (Fig. 3). We may then conclude that although PXR has structural flexibility in its ligand-binding pocket to accommodate large ligands, usage of this expansion feature is unfavorable.
While the 3-position appendage seems to be the driving factor for the decreased potency of group 1 rifampicin analogs versus group 2 analogs, it is a different part of the molecule that physically clashes with the α2-β1 region of PXR LBD (Fig. 2C). Therefore, the dihydroxynaphthalene core appendage likely restrains the macrolide conformation, resulting in the α2-β1 clash. Appendage removal allowed the molecule (rifamycin S) to shift away from α2-β1, resulting in a more favorable binding mode (Fig. 3 E–G). To test whether a α2-β1 clash itself could reduce ligand potency, we modified the most potent PXR ligand T0901317 to result in a structurally guided α2-β1 clash (Fig. 4). Using this approach, we achieved a 28-fold reduction in binding potency that translated to significantly reduced cellular activity (Figs. 4C and 5C). Certain PXR ligands have been shown to be conformationally flexible within the binding pocket, likely due to the large pocket size relative to the small ligands (13, 29). Although not discussed in the original report, T0901317 appears to be flexible based on the presence of positive difference density around the molecule (PDB ID 2O9I) (30). The flexibility of PXR LBD combined with its large pocket volume make SAR difficult, as modified small molecules may bind in different poses than the parental compounds. Therefore, while our results indicate that the modified T0901317 analogs indeed clash with α2-β1 and displace it, as predicted, we cannot rule out the possibility that some analogs are instead forced to bind in modes that are less favorable than the parental T0901317 mode. However, in either case, the desired result is achieved by extending the molecule toward the α2-β1 region. Additional examples of this exist in the literature, although loss of binding is not specifically discussed with respect to α2-β1. For example, we previously described a thalidomide-linked PXR ligand with nearly absent PXR binding (31), and, based on crystallographic analysis of a precursor of this ligand (32), loss of binding is likely due to the rigid thalidomide group being incompatible with any available α2-β1 position.
Physiologically, orally administered drugs are at their highest concentrations in organs where PXR resides (liver and intestine), and even low-affinity drugs can activate PXR and experience a first pass effect. Therefore, complete abolition of PXR agonism is required. Medicinal chemistry campaigns to avoid metabolic events while retaining target potency are difficult and time-consuming. Introducing the α2-β1 clash is an approach that can be used in isolation or in combination with other strategies such as the F288-W299-Y306 clash to achieve full loss of PXR activity. Our findings indicate that while PXR is a wildly promiscuous ligand binder, certain elements that contribute to promiscuity also limit the efficiency of ligands at the edges of PXR’s binding range. The flexibility of the ligand-binding pocket floor extends the binding potential of PXR, but ligands that bind in this manner do so with a penalty.
Materials and Methods
General Chemistry Methods and Synthesis.
DMSO, rifampicin, SR12813, and T0901317 were purchased from MilliporeSigma. BODIPY FL vindoline was synthesized as previously reported (19). Rifaximin and 3-formyl rifamycin were purchased from Cayman Chemical. Rifabutin and rifamycin S were purchased from MedChemExpress. Rifamycin, rifamycin O, and 3-bromo rifamycin S were purchased from Toronto Research Chemicals. Rifapentine was purchased from AstaTech. The manufacturer-determined purity of all compounds was >95%. Synthesis and evaluation of the T0901317 analogs is described in SI Appendix.
TR-FRET PXR Competitive Binding Assay.
The TR-FRET PXR competitive binding assay was performed as previously described (19). The assay buffer composition was 50 mM Tris (pH 7.5), 20 mM MgCl2, 0.1 mg/mL bovine serum albumin, and 0.05 mM dithiothreitol. BODIPY FL vindoline (15 µL/well, 133.3 nM in assay buffer) was dispensed into 384-well low-volume black assay plates. An Echo 555 Acoustic Liquid Handler (Labcyte) then dispensed 60 nL/well compound stocks or DMSO. Lastly, 5 µL/well 20 nM Tb-anti-GST (Thermo Fisher Scientific) and 20 nM GST-PXR LBD (Thermo Fisher Scientific) in assay buffer were added. The final concentrations of the assay components (in a 20 µL final assay volume per well) were: 100 nM BODIPY FL vindoline, 5 nM Tb-anti-GST, 5 nM GST-PXR LBD, and 0.3% DMSO. DMSO alone (0.3%) and 3 µM T0901317 (diluted from 60 nL of 1 mM stock to a 20 µL assay volume) were included in each plate to serve as negative and positive controls, respectively. The plates were shaken at 900 rpm (80 × g) on an IKA MTS 2/4 digital microtiter shaker (IKA Works) for 1 min then centrifuged at 1,000 rpm (201 × g) for 30 s in an Eppendorf 5810 centrifuge equipped with an A-4-62 swing-bucket rotor. The plates were protected from light exposure and incubated for 1 h at room temperature. After incubation, the TR-FRET signal from each well was collected with a PHERAstar FS Microplate Reader (BMG Labtech). The percent inhibition for each well was calculated using the following equation,
Protein Purification, Crystallization, and Structure Determination: Rifamycin S.
PXR LBD was expressed and purified as previously described (32), with modifications. Codon-optimized sequences for His-tagged PXR LBD (residues 130 to 434) and untagged mouse SRC-1 (residues 623 to 710) were cloned into pETDuet-1 (Novagen), which allows coexpression of two genes from separate inducible T7 promoters. The plasmid was transformed into TurboCells Competent E. coli BL21(DE3) (Genlantis), grown in terrific broth at 37 °C to an OD600 of 3 to 4, and induced overnight at 16 °C with 500 µM isopropyl β-D-thiogalactoside (IPTG). Cells were pelleted by centrifugation at 4,000 × g and resuspended in lysis buffer [20 mM Tris (pH 7.5), 250 mM NaCl, 5% (v/v) glycerol, 10 mM imidazole] supplemented with EDTA-free SIGMAFAST protease inhibitor cocktail tablets (MilliporeSigma) and 1 mg/mL lysozyme (Thermo Fisher Scientific). The suspension was sonicated and then centrifuged at 20,000 × g for 1 h, and the supernatant was applied to a 5 mL HisTrap FF column (Cytiva). The column was washed with 50 mL lysis buffer, and bound proteins were eluted with lysis buffer containing 500 mM imidazole. Elution fractions were collected and analyzed by SDS–PAGE for protein amount and purity. Selected fractions were pooled, and a 2:1 molar ratio of SRC-1 peptide was added to stabilize PXR LBD (N-CPSSHSSLTERHKILHRLLQEGSPS-C, prepared by the Macromolecular Synthesis Section at St. Jude Children’s Research Hospital). The PXR LBD/peptide mixture was concentrated to ≤10 mL in an Amicon Ultra-15 centrifugal filter unit with 10 kDa cutoff (MilliporeSigma), filtered through a 0.22-µm syringe filter, and loaded onto a HiLoad 26/600 Superdex 200 pg size exclusion column (Cytiva) equilibrated with storage buffer [20 mM Tris (pH 7.8), 250 mM NaCl, 5% (v/v) glycerol, 5 mM DTT, 1 mM EDTA]. Elution fractions were collected and analyzed by SDS–PAGE, pure fractions were pooled, and a 2:1 molar ratio of SRC-1 peptide was again added. The PXR LBD/peptide mixture was concentrated to 3 mg/mL, aliquoted, flash frozen in liquid nitrogen, and stored at −80 °C.
PXR LBD (3 mg/mL, 83 µM) was mixed with 2 mM rifamycin S and incubated for 1.5 h at 4 °C. This mixture contained 2% DMSO from the compound dilution. Hanging drop trays were set with 1 µL protein-ligand complex and 1 µL reservoir solutions containing 50 or 100 mM imidazole (pH 7 to 7.6) and 10 to 16% isopropanol. Crystals grew in various conditions within 3 to 5 d and were cryoprotected in respective mother liquors containing 2 mM rifamycin S, 2% DMSO, and 20% (+/−)-2-methyl-2,4-pentanediol (MPD, Hampton Research). The data presented were collected from a single crystal grown in 100 mM imidazole (pH 7.2) with 14% isopropanol.
X-ray diffraction data were collected to a resolution of 2.25 Å at FMX Beamline 17-ID-2 at the National Synchrotron Light Source II at Brookhaven National Laboratory. Frames were processed with XDS (33). The crystals belonged to space group P43212 with one molecule in the asymmetric unit. The structure was solved by molecular replacement in Phaser (34) using PDB ID 1M13 as the search model (35). The search model was stripped of solvent and ligand prior to molecular replacement. Rifamycin S was placed with Phenix LigandFit (36, 37). Iterative cycles of model building and refinement were performed in Coot (38) and Phenix (39). The data collection and refinement statistics are shown in SI Appendix, Table S2. All crystallographic figures were made in PyMOL (Schrödinger). The structure is deposited as PDB ID 8E3N.
Protein Purification, Crystallization, and Structure Determination: T0-BP.
Protein was purified as above and stored at 4 mg/mL. PXR LBD (4 mg/mL, 110 µM) was mixed with 2 mM T0-BP and incubated for 1 h at room temperature. This mixture contained 2% DMSO from the compound dilution. Hanging drop trays were set with 1 µL protein-ligand complex and 1 µL reservoir solutions containing 50 mM imidazole (pH 6.8 to 7.8) and 8 to 14% isopropanol. Crystals grew in various conditions within 1 to 3 d and were cryoprotected in respective mother liquors containing 1 mM T0-BP, 1% DMSO, and 40% ethylene glycol. The data presented were collected from a single crystal grown in 50 mM imidazole (pH 7.0) with 12% isopropanol.
X-ray diffraction data were collected to a resolution of 2.3 Å at AMX Beamline 17-ID-1 at the National Synchrotron Light Source II at Brookhaven National Laboratory. Frames were processed with XDS (33). The crystals belonged to space group P43212 with one molecule in the asymmetric unit. The structure was solved by molecular replacement in Phaser (34) using PDB ID 1M13 as the search model (35). The search model was stripped of solvent and ligand prior to molecular replacement. T0-BP was placed with Phenix LigandFit (36, 37). Iterative cycles of model building and refinement were performed in Coot (38) and Phenix (39). The data collection and refinement statistics are shown in SI Appendix, Table S3. All crystallographic figures were made in PyMOL (Schrödinger). The structure is deposited as PDB ID 8FPE.
Protein Purification, Crystallization, and Structure Determination: T0-C6.
For the T0-C6-PXR LBD cocrystal structure, we used a PXR LBD tethered to a 33-amino acid SRC-1 peptide (40–42) and optimized the expression, purification, and crystallization in-house. A codon-optimized sequence for His-tagged PXR LBD (residues 130 to 434) tethered to SRC-1 (residues 678 to 710) with SGGSGG linker was cloned into pET3a (Novagen). The plasmid was transformed into TurboCells Competent E. coli BL21(DE3) (Genlantis), grown in terrific broth + 0.2% glucose at 37 °C to an OD600 of 3 to 4, and induced overnight at 16 °C with 200 µM IPTG. Cells were pelleted by centrifugation at 4,000 × g and resuspended in lysis buffer [25 mM Tris (pH 7.9), 500 mM NaCl, 5% (v/v) glycerol, 1 mM DTT, 10 mM imidazole] supplemented with EDTA-free SIGMAFAST protease inhibitor cocktail tablets. The suspension was lysed by passage through a microfluidizer at 18,000 psi and then centrifuged at 20,000 × g for 1 h, and the supernatant was applied to a 5 mL HisTrap FF column. The column was washed with 50 mL lysis buffer, and bound proteins were eluted with a 100 mL linear gradient from lysis buffer to lysis buffer containing 250 mM imidazole. Elution fractions were collected and analyzed by SDS–PAGE for protein amount and purity. Selected fractions were pooled and diluted to 125 mM NaCl by addition of lysis buffer without NaCl. The protein was applied to a 5 mL HiTrap SP HP column (Cytiva), and the flow-through contained the PXR LBD-SRC-1. The protein was concentrated to ≤10 mL in a Amicon Ultra-15 centrifugal filter unit with 10 kDa cutoff, filtered through a 0.22-µm syringe filter, and loaded onto a HiLoad 26/600 Superdex 200 pg size exclusion column equilibrated with storage buffer [25 mM Tris-HCl (pH 8.0), 100 mM NaCl, 5% (v/v) glycerol, 2 mM DTT]. Elution fractions were collected and analyzed by SDS-PAGE, and pure fractions were pooled, concentrated to 12.4 mg/mL, aliquoted, flash frozen in liquid nitrogen, and stored at −80 °C.
PXR LBD-SRC-1 (12.4 mg/mL, 308 µM) was mixed with 2 mM T0-C6 and incubated for 1.5 h at room temperature. This mixture contained 2% DMSO from the compound dilution. Hanging drop trays were set with 1 µL protein-ligand complex and 1 µL reservoir solutions containing 100 mM Bis-Tris (pH 5.5 to 6.8) and 9 to 15% MPD. Crystals grew in various conditions within 1 to 2 d, with the chosen crystal growing in 9% MPD at pH 5.8. The crystal was cryoprotected in 100 mM Bis-Tris (pH 5.8), 1 mM T0-C6, 1% DMSO, and 25% MPD.
X-ray diffraction data from a single crystal were collected to a resolution of 2.37 Å at AMX Beamline 17-ID-1 at the National Synchrotron Light Source II at Brookhaven National Laboratory. Frames were processed with XDS (33). The crystals belonged to space group P212121 with two molecules in the asymmetric unit. The structure was solved by molecular replacement in Phaser (34) using PDB ID 3CTB as the search model (35). The search model was stripped of solvent prior to molecular replacement. T0-C6 was placed with Phenix LigandFit (36, 37). Iterative cycles of model building and refinement were performed in Coot (38) and Phenix (39). The data collection and refinement statistics are shown in SI Appendix, Table S4. All crystallographic figures were made in PyMOL (Schrödinger). The structure is deposited as PDB ID 8EQZ.
PXR Transactivation Assays.
293T/17 cells were obtained from the American Type Culture Collection (ATCC, cat. # CRL-11268) and maintained in Dulbecco's Modified Eagle Medium (DMEM, ATCC) with 10% fetal bovine serum (FBS; HyClone). Cells were incubated in a humidified atmosphere at 37 °C with 5% CO2 and routinely verified to be mycoplasma free by using the MycoProbe Mycoplasma Detection Kit (R&D Systems). Cell counts were obtained with a Countess II Automated Cell Counter using trypan blue staining.
PXR transactivation assays were performed similarly as previously described, with modifications (43). 293T cells (1 × 106/well in 2 mL DMEM + 10% FBS) were reverse transfected in 6-well plates with pGL3-CYP3A4-luc (2 µg/well) (44, 45) and 100 ng/well of either empty vector (pcDNA3) or pcDNA3-FLAG-PXR (WT or W299A) using FuGENE HD (Promega). Twenty-four hours after transfection, cells were trypsinized and suspended in phenol red-free DMEM (Thermo Fisher Scientific) supplemented with 5% charcoal/dextran-treated FBS (HyClone), and 5,000 cells/well in 25 µL volume were added to PerkinElmer CulturPlate-384 plates. An Echo 555 Acoustic Liquid Handler was used to dispense 75 nL/well of DMSO or stock compounds into the wells, for a final DMSO concentration of 0.3%. After 24 h, a luciferase assay was performed using the steadylite plus Reporter Gene Assay System and EnVision microplate reader (PerkinElmer).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
Research reported in this publication was supported by ALSAC and NIH National Institute of General Medical Sciences [Grant R35GM118041]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This work utilized the FMX and AMX beamlines, which are primarily supported by the NIH National Institute of General Medical Sciences through a Center Core P30 Grant (P30GM133893), and by the U.S. Department of Energy Office of Biological and Environmental Research (KP1607011). As part of the National Synchrotron Light Source II, a national user facility at Brookhaven National Laboratory, work performed at the Center for BioMolecular Structures is supported in part by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences Program under contract number DE-SC0012704.
Author contributions
W.L., A.D.H., S.P., Y.L., and T.C. designed research; W.L., A.D.H., S.P., and Y.L., performed research; Y.L. contributed new reagents/analytic tools; W.L., A.D.H., Y.L., J.S., and D.J.M. analyzed data; and A.D.H. and T.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Andrew D. Huber, Email: andrew.huber@stjude.org.
Taosheng Chen, Email: taosheng.chen@stjude.org.
Data, Materials, and Software Availability
This manuscript includes three X-ray crystal structures that have been deposited in the Protein Data Bank with IDs 8E3N (46), 8FPE (47), and 8EQZ (48).
Supporting Information
References
- 1.Hoffmann A. A., Willi Y., Detecting genetic responses to environmental change. Nat. Rev. Genet 9, 421–432 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Copley S. D., Evolution of efficient pathways for degradation of anthropogenic chemicals. Nat. Chem. Biol. 5, 559–566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Copley S. D., An evolutionary biochemist’s perspective on promiscuity. Trends Biochem. Sci. 40, 72–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Scholtes C., Giguere V., Transcriptional control of energy metabolism by nuclear receptors. Nat. Rev. Mol. Cell Biol. 23, 750–770 (2022). [DOI] [PubMed] [Google Scholar]
- 5.Guengerich F. P., Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 8, E101–E111 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehmann J. M., et al. , The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J. Clin. Invest. 102, 1016–1023 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kliewer S. A., et al. , An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92, 73–82 (1998). [DOI] [PubMed] [Google Scholar]
- 8.Bertilsson G., et al. , Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc. Natl. Acad. Sci. U.S.A. 95, 12208–12213 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhai Q., van der Lee M., van Gelder T., Swen J. J., Why we need to take a closer look at genetic contributions to CYP3A activity. Front Pharmacol. 13, 912618 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu J., Wang Y., Ragueneau-Majlessi I., Pharmacokinetic drug-drug interactions with drugs approved by the US FOod and drug administration in 2020: Mechanistic understanding and clinical recommendations. Drug Metab. Dispos. 50, 1–7 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Hall A., Chanteux H., Menochet K., Ledecq M., Schulze M. E. D., Designing out PXR activity on drug discovery projects: A review of structure-based methods, empirical and computational approaches. J. Med. Chem. 64, 6413–6522 (2021). [DOI] [PubMed] [Google Scholar]
- 12.Khersonsky O., Tawfik D. S., Enzyme promiscuity: A mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79, 471–505 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Watkins R. E., et al. , The human nuclear xenobiotic receptor PXR: Structural determinants of directed promiscuity. Science 292, 2329–2333 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Delfosse V., et al. , Synergistic activation of human pregnane X receptor by binary cocktails of pharmaceutical and environmental compounds. Nat. Commun. 6, 8089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delfosse V., et al. , Mechanistic insights into the synergistic activation of the RXR-PXR heterodimer by endocrine disruptor mixtures. Proc. Natl. Acad. Sci. U.S.A. 118, e2020551118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chrencik J. E., et al. , Structural disorder in the complex of human pregnane X receptor and the macrolide antibiotic rifampicin. Mol. Endocrinol. 19, 1125–1134 (2005). [DOI] [PubMed] [Google Scholar]
- 17.Buchman C. D., Chai S. C., Chen T., A current structural perspective on PXR and CAR in drug metabolism. Expert Opin. Drug Metab. Toxicol. 14, 635–647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blumberg B., et al. , SXR, a novel steroid and xenobiotic-sensing nuclear receptor. Genes Dev. 12, 3195–3205 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin W., et al. , Development of BODIPY FL vindoline as a novel and high-affinity pregnane X receptor fluorescent probe. Bioconjug Chem. 25, 1664–1677 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz J. R., et al. , Role of LXRs in control of lipogenesis. Genes Dev. 14, 2831–2838 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitro N., Vargas L., Romeo R., Koder A., Saez E., T0901317 is a potent PXR ligand: implications for the biology ascribed to LXR. FEBS Lett. 581, 1721–1726 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Berkhout T. A., et al. , SR-12813 lowers plasma cholesterol in beagle dogs by decreasing cholesterol biosynthesis. Atherosclerosis 133, 203–212 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Berkhout T. A., et al. , The novel cholesterol-lowering drug SR-12813 inhibits cholesterol synthesis via an increased degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J. Biol. Chem. 271, 14376–14382 (1996). [DOI] [PubMed] [Google Scholar]
- 24.Jones S. A., et al. , The pregnane X receptor: A promiscuous xenobiotic receptor that has diverged during evolution. Mol. Endocrinol. 14, 27–39 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Ngan C. H., et al. , The structural basis of pregnane X receptor binding promiscuity. Biochemistry 48, 11572–11581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huber A. D., et al. , Mutation of a single amino acid of pregnane X receptor switches an antagonist to agonist by altering AF-2 helix positioning. Cell Mol. Life Sci. 78, 317–335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iyer M., Reschly E. J., Krasowski M. D., Functional evolution of the pregnane X receptor. Expert Opin. Drug Metab. Toxicol. 2, 381–397 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore L. B., et al. , Orphan nuclear receptors constitutive androstane receptor and pregnane X receptor share xenobiotic and steroid ligands. J. Biol. Chem. 275, 15122–15127 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Watkins R. E., Davis-Searles P. R., Lambert M. H., Redinbo M. R., Coactivator binding promotes the specific interaction between ligand and the pregnane X receptor. J. Mol. Biol. 331, 815–828 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Xue Y., et al. , Crystal structure of the PXR-T1317 complex provides a scaffold to examine the potential for receptor antagonism. Bioorg Med. Chem. 15, 2156–2166 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huber A. D., et al. , SJPYT-195: A designed nuclear receptor degrader that functions as a molecular glue degrader of GSPT1. ACS Med. Chem. Lett. 13, 1311–1320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin W., et al. , SPA70 is a potent antagonist of human pregnane X receptor. Nat Commun 8, 741 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCoy A. J., et al. , Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watkins R. E., et al. , 2.1 A crystal structure of human PXR in complex with the St. John’s wort compound hyperforin. Biochemistry 42, 1430–1438 (2003). [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger T. C., Adams P. D., Moriarty N. W., Cohn J. D., Ligand identification using electron-density map correlations. Acta Crystallogr. D Biol. Crystallogr. 63, 101–107 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Terwilliger T. C., Klei H., Adams P. D., Moriarty N. W., Cohn J. D., Automated ligand fitting by core-fragment fitting and extension into density. Acta Crystallogr. D Biol. Crystallogr. 62, 915–922 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P., Lohkamp B., Scott W. G., Cowtan K., Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liebschner D., et al. , Macromolecular structure determination using X-rays, neutrons and electrons: Recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivaprakasam P., et al. , Structure-based amelioration of PXR transactivation in a novel series of macrocyclic allosteric inhibitors of HIV-1 integrase. Bioorg Med. Chem. Lett. 30, 127531 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Gong H., et al. , Identification of bicyclic hexafluoroisopropyl alcohol sulfonamides as retinoic acid receptor-related orphan receptor gamma (RORgamma/RORc) inverse agonists. Employing structure-based drug design to improve pregnane X receptor (PXR) selectivity. Bioorg Med. Chem. Lett. 28, 85–93 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Duan J. J., et al. , Structure-based discovery of Phenyl (3-Phenylpyrrolidin-3-yl)sulfones as selective, orally active RORgammat inverse agonists. ACS Med. Chem. Lett. 10, 367–373 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang Y. M., et al. , Serine 350 of human pregnane X receptor is crucial for its heterodimerization with retinoid X receptor alpha and transactivation of target genes in vitro and in vivo. Biochem. Pharmacol. 96, 357–368 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin W., et al. , Cyclin-dependent kinase 2 negatively regulates human pregnane X receptor-mediated CYP3A4 gene expression in HepG2 liver carcinoma cells. J. Biol. Chem. 283, 30650–30657 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goodwin B., Hodgson E., Liddle C., The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol. Pharmacol. 56, 1329–1339 (1999). [DOI] [PubMed] [Google Scholar]
- 46.Huber A. D., et al., Crystal structure of pregnane X receptor ligand binding domain complexed with rifamycin S. Worldwide Protein Data Bank (wwPDB). 10.2210/pdb8E3N/pdb5. Deposited 17 August 2022. [DOI] [Google Scholar]
- 47.Huber A. D., et al., Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-BP. Worldwide Protein Data Bank (wwPDB). 10.2210/pdb8FPE/pdb5. Deposited 4 January 2023. [DOI] [Google Scholar]
- 48.Huber A. D., et al., Crystal structure of pregnane X receptor ligand binding domain complexed with T0901317 analog T0-C6. Worldwide Protein Data Bank (wwPDB). 10.2210/pdb8EQZ/pdb. Deposited 11 October 2022. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
This manuscript includes three X-ray crystal structures that have been deposited in the Protein Data Bank with IDs 8E3N (46), 8FPE (47), and 8EQZ (48).