Significance
Genetic defects in copper transport to cuproenzymes result in infantile disorders for which no effective therapies are currently available. Recent studies have uncovered the therapeutic potential of elesclomol, a copper-transporting drug, in the treatment of copper deficiency disorders. To realize the full potential of this drug, it is necessary to gain a mechanistic understanding of how elesclomol makes copper available to different cellular cuproenzymes. Here, we demonstrate that copper is released from elesclomol within mitochondria via FDX1, and outside of mitochondria in an FDX1-independent manner to make copper available to mitochondrial and nonmitochondrial cuproenzymes, respectively. These modes of copper release are distinct from other currently used copper-transporting drugs and may explain the high potency of elesclomol in rectifying copper deficiency.
Keywords: copper, mitochondria, elesclomol, cytochrome c oxidase, FDX1
Abstract
Recent studies have uncovered the therapeutic potential of elesclomol (ES), a copper-ionophore, for copper deficiency disorders. However, we currently do not understand the mechanism by which copper brought into cells as ES–Cu(II) is released and delivered to cuproenzymes present in different subcellular compartments. Here, we have utilized a combination of genetic, biochemical, and cell-biological approaches to demonstrate that intracellular release of copper from ES occurs inside and outside of mitochondria. The mitochondrial matrix reductase, FDX1, catalyzes the reduction of ES–Cu(II) to Cu(I), releasing it into mitochondria where it is bioavailable for the metalation of mitochondrial cuproenzyme— cytochrome c oxidase. Consistently, ES fails to rescue cytochrome c oxidase abundance and activity in copper-deficient cells lacking FDX1. In the absence of FDX1, the ES-dependent increase in cellular copper is attenuated but not abolished. Thus, ES-mediated copper delivery to nonmitochondrial cuproproteins continues even in the absence of FDX1, suggesting alternate mechanism(s) of copper release. Importantly, we demonstrate that this mechanism of copper transport by ES is distinct from other clinically used copper-transporting drugs. Our study uncovers a unique mode of intracellular copper delivery by ES and may further aid in repurposing this anticancer drug for copper deficiency disorders.
Copper (Cu) is an essential trace element required for the activity and stability of several cuproenzymes involved in a diverse array of physiological processes, including mitochondrial energy generation, iron homeostasis, and collagen cross-linking (1). Genetic mutations that prevent Cu transport across cellular membranes or Cu delivery to cuproenzymes result in lethal human disorders such as Menkes disease and a subset of mitochondrial disorders (1–5). Currently, no Food and Drug Administration-approved therapies are available for the treatment of these disorders (6). The direct administration of hydrophilic Cu salts such as Cu–histidine has shown limited efficacy in clinical trials (7), which is likely due to inefficient Cu delivery across cellular membranes. Thus, there is an unmet need to identify compounds that can safely and effectively transport Cu across biological membranes and restore cellular Cu homeostasis.
Our recent studies have highlighted the therapeutic potential of the Cu ionophore, elesclomol (ES), in treating diseases of Cu deficiency (8, 9). Specifically, we have shown that ES rescues Cu deficiency phenotypes in yeast, zebrafish, and mouse models by delivering Cu to mitochondria and restoring the function of the essential cuproenzyme cytochrome c oxidase (CcO). While such studies highlight the therapeutic potential of ES, the mechanism by which ES-mediates intracellular Cu delivery remains poorly understood. ES binds Cu(II) in the extracellular environment with extremely high affinity (Ka = 1017.1) at physiological pH (10). This raises the question of how and where Cu is released from ES inside the cell. These questions are critical to address since Cu must be released from ES to be made bioavailable to various intracellular cuproenzymes.
Previous studies offer some clues regarding the mechanism of intracellular Cu delivery via ES. It has been shown that ES-transported Cu accumulates in the mitochondria in a bioavailable form (8, 11). More recently, ferredoxin 1 (FDX1), a mitochondrial matrix enzyme involved in steroidogenesis, Fe-S cluster biosynthesis, and lipoylation, was identified as the protein target of ES through a genome-wide CRISPR-Cas9 screen (12–15). Specifically, it was shown that ES–Cu(II) can serve as a neosubstrate of FDX1 and is essential for mediating cuproptosis, a unique form of Cu-dependent cell death (14, 15). These findings have prompted a working model of ES-mediated cytotoxicity in which FDX1 catalyzes the release of Cu from ES in vivo by reducing ES-bound Cu(II) to Cu(I) (16). Here, we test this model in the context of ES-mediated rescue of cuproenzyme deficiencies that are the result of mutations in Cu homeostasis pathways. Using a combination of genetic, biochemical, and cell-biological approaches, we demonstrate that Cu is released from ES inside the mitochondria in an FDX1-dependent manner. Interestingly, we find that Cu is also released outside the mitochondria in an FDX1-independent manner. These modes of Cu transport are unique to ES among other clinically used Cu-transporting drugs we tested and explain its high efficacy in delivering Cu to intracellular cuproenzymes.
Results
FDX1 Is Essential for ES-Mediated Rescue of Mitochondrial Respiration in Cu-Depleted Cells.
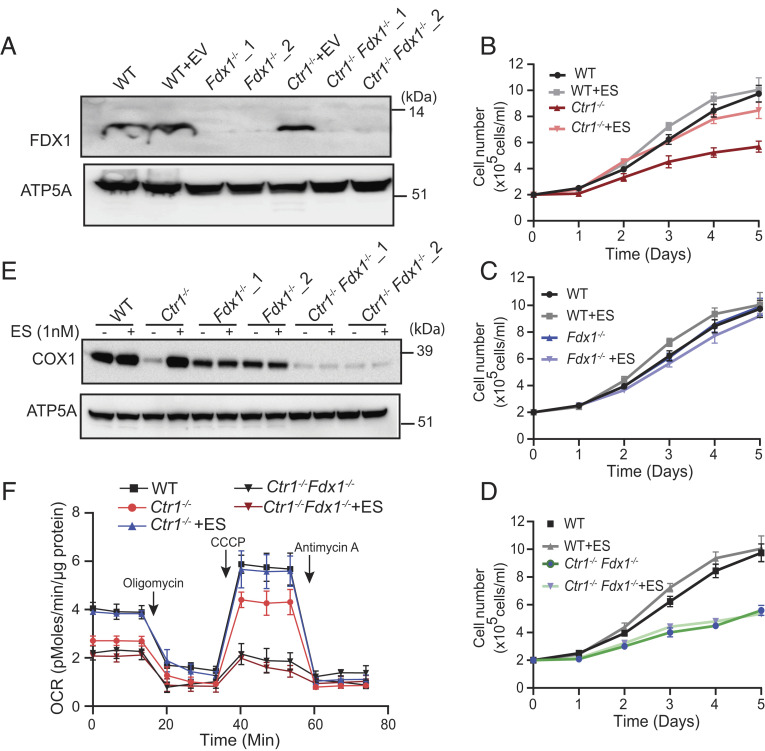
Previous studies reported that ES treatment results in an increase in mitochondrial Cu content and that loss of FDX1 conferred resistance to ES-mediated killing (8, 11, 15). Taken together, these results suggest that FDX1 may play a role in releasing Cu from the ES–Cu complex in vivo. To test this hypothesis, we constructed CRISPR-Cas9-based knockouts of FDX1 in the wild-type (WT) and Cu-deficient Ctr1−/− H9c2 cardiac cell lines that lack the primary cellular Cu importer, CTR1 (SLC31A1) (1), and validated the loss of FDX1 by western blotting (Fig. 1A). Consistent with a previous study (15), we found that Fdx1−/−cells exhibited significant resistance to ES-mediated killing (IC50 of 20 nM in WT vs. 122 nM in Fdx1−/−) (SI Appendix, Fig. S1A).Notably, Ctr1−/−Fdx1−/−double-knockout cells were almost completely resistant to ES-mediated death (SI Appendix, Fig. S1A). However, treatment with ES preloaded with Cu (ES–Cu) significantly increased the sensitivity of all cell lines including Ctr1−/−Fdx1−/−, which suggests that Cu is essential for ES-mediated cytotoxicity (SI Appendix, Fig. S1B). Next, we tested whether ES-mediated Cu transport could rescue the Cu-deficient growth phenotype of Ctr1−/− cells by measuring the rate of cell proliferation in the presence or absence of ES. As expected, Ctr1−/− cells showed a reduced rate of cell proliferation relative to WT cells, which was rescued by 1 nM ES supplementation (Fig. 1B). Fdx1 null cells showed no growth defect (Fig. 1C) and Ctr1−/−Fdx1−/− double-knockout cells phenocopied Ctr1−/− cells (Fig. 1 B and D). Consistent with our model, ES supplementation did not rescue the growth defect in Ctr1−/−Fdx1−/− cells, suggesting that FDX1 is required to release Cu from the ES–Cu complex in mitochondria (Fig. 1D). We further tested this hypothesis by measuring the steady-state levels of COX1, a Cu-containing subunit of mitochondrial CcO, the abundance of which positively correlates with mitochondrial Cu content. Although ES treatment fully restored COX1 abundance in Ctr1−/− cells, the loss of FDX1 in Ctr1−/−Fdx1−/− double-knockout cells prevented ES-mediated restoration of COX1 (Fig. 1E). We found that Fdx1−/− cells have lower basal levels of COX1 as compared with the parental control cells, a finding in line with the recently reported role of FDX1 in CcO biogenesis (17). Consistent with the requirement for FDX1 in restoring COX1 abundance in Cu-deficient cells, we found that ES supplementation was able to increase mitochondrial oxygen consumption rate in Ctr1−/− cells, but not in Ctr1−/−Fdx1−/− cells (Fig. 1F). Together, these results suggest that FDX1 is essential for ES-dependent rescue of mitochondrial cuproenzyme abundance and activity in Cu-deficient cells.
Fig. 1.
Loss of FDX1 prevents ES-mediated rescue of mitochondrial respiration. (A) Western blot-based detection of FDX1 in the mitochondria isolated from wild-type (WT) and Ctr1−/− cells transduced with either empty vector or two different sgRNAs targeting Fdx1. ATP5A was used as a loading control. A representative blot from two independent trials is shown. (B–D) Cells were cultured in the high-glucose media in the presence or absence of 1 nM ES, and cell numbers are plotted over time. Data are expressed as mean ± SD, n = 3. (E) Western blot analysis of COX1 and ATP5A in indicated cells treated with or without 1 nM ES for 72 h. A representative blot from two independent trials is shown. (F) The OCR of WT, Ctr1−/− and Ctr1−/−Fdx1−/− cells treated with or without 1 nM ES for 72 h. Oligomycin, CCCP, and antimycin A were used to measure adenosine triphosphate (ATP)-coupled respiration, maximum respiratory capacity, and mitochondria-specific respiration. Data are shown as mean ± SEM, n = 3.
FDX1 Catalyzes the Release of Cu from the ES–Cu Complex In Vitro.
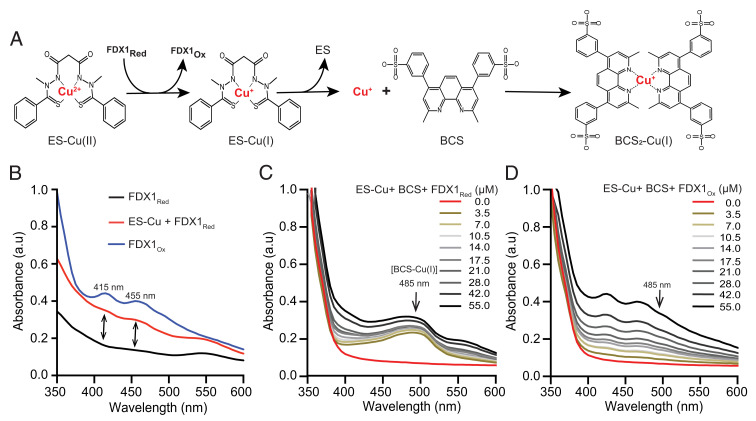
We reasoned that the reduction of the tightly bound Cu(II) in the ES–Cu complex to Cu(I) by FDX1 could facilitate its dissociation from ES. To test this idea, we designed an in vitro assay using purified human FDX1 protein and the colorimetric Cu-specific chelator bathocuproine disulfonate (BCS). Our assay was based on the rationale that if reduced FDX1 (FDX1Red) donates an electron to ES–Cu(II), the resulting ES–Cu(I) complex would be susceptible to Cu(I) chelation by BCS owing to its higher affinity for reduced Cu (Fig. 2A). Thus, the formation of a BCS2–Cu(I) complex, which can be measured spectrophotometrically at 485 nm (18), would provide evidence of FDX1-mediated Cu release from the ES–Cu (Fig. 2A).
Fig. 2.
Reduced FDX1 releases ES-bound Cu in vitro. (A) A schematic representation of in vitro assay to determine FDX1-mediated release of Cu from ES. (B) UV/Vis spectra of 50 µM human FDX1Red /FDX1Ox ± 50 µM ES–Cu(II) in Tris buffer (10 mM Tris, 50 mM NaCl, pH 7.5). (C) UV/Vis spectra of the BCS–Cu(I) complex in the presence of increasing concentrations (0 to 55 µM) of FDX1Red or (D) FDX1Ox protein. FDX1Ox/Red proteins were mixed with 1 mM BCS in Tris buffer, and 20 µM ES–Cu(II) was added to start the reaction. The final dimethyl sulfoxide (DMSO) concentration in the solution was 20% after injection.
To test this hypothesis, we first custom synthesized the ES–Cu complex and confirmed the presence of Cu(II) by electron paramagnetic resonance (EPR) spectroscopy (SI Appendix, Fig. S2A) and Ultraviolet/Visible (UV/Vis) spectroscopy (SI Appendix, Fig. S2B). Using UV/Vis spectrophotometry, we also established that the absorption maxima of 485 nm was unique to BCS2–Cu(I), and was not exhibited by ES, ES–Cu(II), or BCS species (SI Appendix, Fig. S3 A–D). Incubation of equimolar amounts of FDX1Red with ES–Cu(II) led to the appearance of shoulders at 415 and 455 nm in the absorption spectrum consistent with the formation of oxidized FDX1 (FDX1Ox) (Fig. 2B), as has been shown previously (15). This result suggested electron transfer from FDX1Red to ES–Cu(II), leading to the formation of FDX1Ox and ES–Cu(I) species. To check if this reduction of Cu(II) to Cu(I) can release Cu from ES, we coincubated ES–Cu(II) and BCS together with increasing concentrations of FDX1Red in the reaction mixture, which resulted in the development of an absorption peak at 485 nm, indicating formation of the BCS2–Cu(I) complex (Fig. 2C). Consistent with our model, the addition of FDX1Ox in the BCS and ES–Cu(II) reaction mixture did not result in the development of an absorption peak at 485 nm (Fig. 2D). As expected, the BCS–Cu(I) complex was also not observed when FDX1Red was incubated with ES alone (SI Appendix, Fig. S3E). Interestingly, we also observed an absorption peak at 485 nm when FDX1Red was coincubated with CuCl2, which suggests that FDX1 can also directly act on Cu(II) (SI Appendix, Fig. S3 F and G). Together, these results from in vitro experiments demonstrate that FDX1Red can facilitate the reductive release of Cu(I) from the ES–Cu(II) complex.
Fdx1−/− Cells Show Reduced Cu Accumulation upon ES–Cu Treatment.
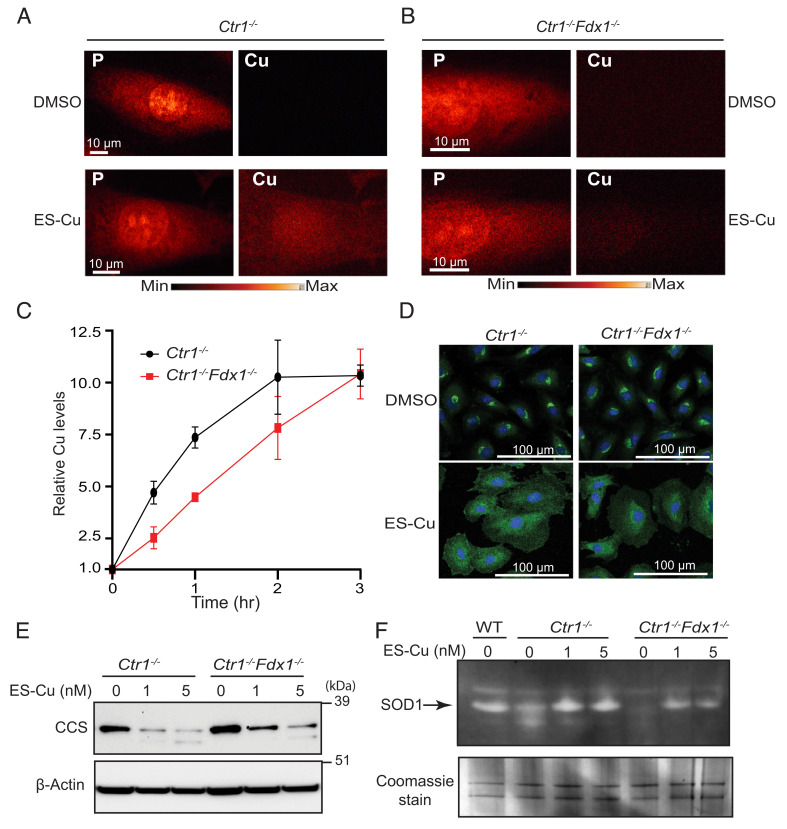
Although our genetic and biochemical data demonstrated a requirement for FDX1 in Cu release from the ES–Cu complex, alternate mechanism(s) of intracellular Cu release from ES were not ruled out by these studies. To directly observe Cu in intact cells, we utilized X-ray fluorescence microscopy (XFM), a label-free technique that is based on the intrinsic fluorescence properties of metals to visualize their intracellular distribution (19). In a typical XFM experiment lighter elements such as phosphorus are used to visualize the nucleus (owing to the high abundance of phosphorus-containing DNA) and the general outline of cells as shown in Fig. 3 A and B for Ctr1−/− and Ctr1−/−Fdx1−/− cells. As expected, there was very little detectable Cu signal in Cu-deficient Ctr1−/− or Ctr1−/−Fdx1−/− cells treated with DMSO (Fig. 3 A and B). We were unable to detect an increase in Cu signal upon treatment with 5 nM ES–Cu, which likely reflects the low sensitivity of XFM to small changes in Cu concentrations (SI Appendix, Fig. S4). Indeed, XFM analysis of cells treated with 100 nM ES–Cu for 30 min revealed an elevated Cu signal in Ctr1−/− cells as compared with Ctr1−/−Fdx1−/− cells (Fig. 3 A and B). These findings suggested that ES–Cu treatment can increase cellular Cu abundance even in the absence of FDX1, albeit to a lesser degree. Next, we utilized inductively coupled plasma-mass spectroscopy (ICP-MS) to quantify the relative magnitude and rate at which ES–Cu stimulates the accumulation of Cu in Ctr1−/− and Ctr1−/−Fdx1−/− cells. While ES–Cu stimulated rapid accumulation of Cu in Ctr1−/− cells which peaked by 2 h, the kinetics of Cu accumulation in the Ctr1−/−Fdx1−/− cells were comparatively slow (Fig. 3C). The decreased accumulation of Cu in Ctr1−/−Fdx1−/− cells is likely because the FDX1-dependent removal of Cu allows ES to reenter the extracellular space where it can facilitate additional cycles of Cu uptake (Fig. 3C) (11). Taken together, these results suggest that the role of FDX1 in ES-mediated intracellular Cu accumulation is rate-limiting, but not essential.
Fig. 3.
Uptake kinetics and intracellular distribution of Cu from the ES–Cu complex. (A and B) X-ray fluorescence microscopy elemental maps for phosphorus (P) and Cu in (A) Ctr1−/− and (B) Ctr1−/−Fdx1−/− cells treated with DMSO or 100 nM ES–Cu for 30 min. Elemental concentration ranges are represented by false coloring from the darkest (lowest concentration) to the brightest (highest concentration). Images are representative of two independent experiments. (C) Relative Cu accumulation in Ctr1−/− and Ctr1−/−Fdx1−/− cells treated with 100 nM ES–Cu for the indicated time intervals. Cu levels were normalized to 0 h. Data are expressed as mean ± SD, n = 3. (D) Immunofluorescence analysis of the steady-state localization and trafficking of endogenous ATP7A (green) in Ctr1−/− and Ctr1−/−Fdx1−/− cells treated with DMSO or 5 nM ES–Cu for 3 h. DAPI was used to stain the nuclei (blue). (E) SDS–PAGE/western blot analysis of CCS protein levels in the indicated cell types treated without or with 1 or 5 nM ES–Cu. β-actin was used as a loading control. (F) In-gel SOD1 activity in the indicated cell types treated with or without 1 or 5 nM ES–Cu. Coomassie stain of the same gel is used as a loading control.
Both XFM and ICP-MS-based intracellular Cu measurements cannot distinguish between free and ES-bound Cu. Additionally, these techniques required a higher dose of ES–Cu (100 nM) to measure any perturbation in intracellular Cu levels. Therefore, to check whether Cu can be released from ES–Cu in an FDX1-independent manner at a lower dose (5 nM), we tested ATP7A translocation upon the addition of 5 nM ES–Cu. Previous studies have shown that the Golgi membrane-localized ATP7A protein undergoes trafficking from the Golgi to vesicular compartments in response to increased levels of Cu(I) in the cytoplasm (20). Therefore, we utilized immunofluorescence microscopy to test whether FDX1 is required to stimulate ATP7A trafficking in cells treated with the ES–Cu complex. As expected, the ATP7A protein was predominantly localized in the perinuclear region of both Ctr1−/− and Ctr1−/−Fdx1−/− cells treated with vehicle control (Fig. 3D). Upon treatment with 5 nM ES–Cu, a significant fraction of ATP7A protein was localized to cytoplasmic vesicles that extended beyond the perinuclear region in both Ctr1−/− and Ctr1−/−Fdx1−/− cells (Fig. 3D). These observations suggest that Cu(I) is released from ES–Cu(II) in a bioavailable form even in the absence of FDX1. To independently confirm this observation, we measured the abundance of the CCS protein, a Cu metallochaperone for SOD1, the levels of which inversely correlate with the cytosolic Cu levels (21). ES–Cu treatment reduced the abundance of CCS in Ctr1−/−as well as Ctr1−/−Fdx1−/− cells, albeit with lower efficiency (Fig. 3E). We further measured the bioavailability of Cu from the ES–Cu complex in cells containing or lacking FDX1 by measuring the activity of SOD1, the major cytoplasmic cuproenzyme (22). Consistent with our CCS data, we found that ES–Cu treatment could stimulate SOD1 activity in Ctr1−/−and to a lesser extent in Ctr1−/−Fdx1−/− cells (Fig. 3F). Together, these results demonstrate the bioavailability of Cu from ES–Cu even in the absence of FDX1.
To investigate the kinetics of Cu release from the ES–Cu complex, we measured the change in the mitochondrial redox state using a human glutaredoxin-1 fused to redox-sensitive green fluorescent protein 2 (Grx1-roGFP2) ratiometric probe targeted to mitochondria (23). This probe, which consists of a fusion protein containing roGFP2 tethered to the redox enzyme glutaredoxin-1, is used for the measurement of glutathione redox potential in vivo. If the release of Cu(I) from the ES–Cu complex changes the redox potential of mitochondria, this should be accompanied by a change in the fluorescence of Grx1-roGFP2. Although we could not detect changes in the mitochondrial redox state at low concentrations (5 nM) of ES–Cu (SI Appendix, Fig. S5A), at higher concentrations (50 nM), there was a time-dependent perturbation in the mitochondrial redox state in Ctr1−/− cells, and to a lesser extent in Ctr1−/−Fdx1−/− cells (SI Appendix, Fig. S5B). Together with the above results, these observations suggest that the intracellular release of Cu from ES–Cu occurs via two distinct mechanisms: one FDX1-dependent in the mitochondria and the other FDX1-independent outside of mitochondria.
FDX1 Is Not Essential for the Release of Cu from Other Cu-Transporting Drugs.
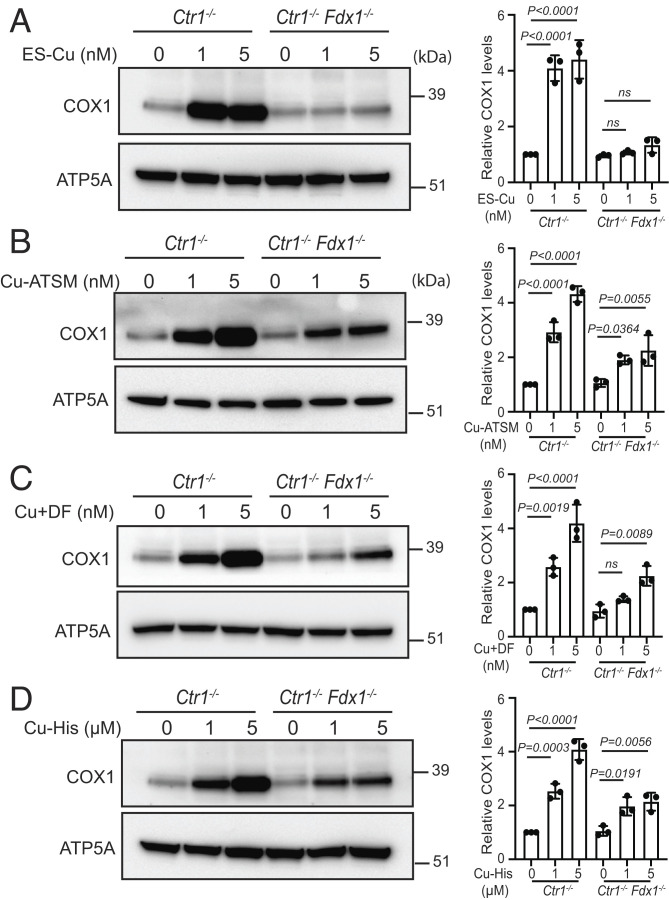
To test whether FDX1 is essential for releasing Cu in mitochondria from other Cu-transporting agents, we measured the rescue of COX1 levels in Ctr1−/− and Ctr1−/−Fdx1−/− cells after treatment with Cu-ATSM [diacetylbis(N(4)-methylthiosemicarbazonato) Cu(II)], Cu-DF (Cu + Disulfiram), and Cu–histidine (Cu(II)–Bis-L-histidine complex). We chose these drugs because they have been tested in clinical trials for amyotrophic lateral sclerosis, cancer, and Menkes disease, respectively (6, 24). As expected, loss of FDX1 prevented the rescue of COX1 levels in Ctr1−/−Fdx1−/− cells when treated with ES–Cu (Fig. 4A). By contrast, each of the other drugs rescued COX1 levels in both Ctr1−/− and Ctr1−/−Fdx1−/− cells (Fig. 4 B–D). However, the magnitude of this rescue was lower in Ctr1−/−Fdx1−/− cells, which suggests that FDX1 can also facilitate the release of Cu from other drugs but compared with ES, they are much less dependent on FDX1 for making Cu bioavailable to COX1. Notably, as compared with hydrophobic Cu-ionophores ES, ATSM, and disulfiram, we find that a 1,000-fold higher concentration of Cu–histidine was required to achieve similar levels of COX1 rescue (Fig. 4 A–D). When comparing two highly efficient Cu-ionophores, ES and ATSM, we find that the kinetics and magnitude of the change in the mitochondrial redox state as measured by Grx1-roGFP2 were much faster and higher in ES–Cu(II)-treated cells (SI Appendix, Fig. S5C). These results demonstrate the high efficiency of ES in making Cu bioavailable to mitochondrial cuproenzyme.
Fig. 4.
FDX1 is specific for Cu release from ES–Cu complex. Western blot analysis of COX1 protein levels in the indicated cells treated with or without (A) 1 or 5 nM ES–Cu, (B) 1 or 5 nM Cu-ATSM, (C) 1 or 5 nM CuCl2+ disulfiram, and (D) 1 or 5 µM Cu–histidine for 72 h. ATP5A was used as a loading control. The relative abundance of COX1 was quantified by densitometry analysis using ImageJ software. Data were normalized to protein levels in untreated Ctr1−/− cells and expressed as mean ± SD (n = 3). Statistical significance was assessed by one-way ANOVA with Tukey’s multiple comparison test using GraphPad Prism 9 software. ns: not significant; each dot on the bar chart represents individual data point.
Discussion
Perturbations in intracellular Cu homeostasis cause devastating genetic diseases in humans, as evidenced by the lethal effects of Cu deficiency in Menkes disease and Cu toxicity in Wilson disease (2, 25). Pharmacological agents that cross biological membranes and restore cellular Cu homeostasis by increasing or decreasing Cu bioavailability could be of great therapeutic benefit (16, 24). In support of this idea, we recently showed that ES could prevent postnatal lethality and suppress Cu deficiency phenotypes in genetic models of mitochondrial disorders and Menkes disease (8, 9). While these studies clearly established the ability of ES to make Cu bioavailable to the mitochondrial cuproenzyme CcO, the mechanism of Cu release from the ES–Cu complex and its delivery to other nonmitochondrial cuproenzymes has remained unclear. Here, we show that Cu(I) is released from the ES–Cu(II) complex inside and outside mitochondria via mechanisms that are FDX1-dependent and -independent, respectively. In this manner, Cu is made bioavailable to mitochondrial as well as cytosolic cuproenzymes. Unlike other Cu-binding ionophores tested, these mechanisms of Cu release appear to be unique to ES and may account for its unusually high efficiency as a Cu delivery agent compared with currently used Cu-transporting drugs.
Elesclomol was originally developed as a chemotherapy agent after it was discovered to selectively kill rapidly proliferating cancer cells (11, 26). A more recent study showed that ES–Cu-mediated cytotoxicity occurs via a novel mechanism termed cuproptosis, a nonapoptotic process that targets protein lipoylation mediated by FDX1 (14). Consistent with FDX1 being a target of ES (15), we show that FDX1 is essential for releasing Cu in mitochondria and making it bioavailable to CcO (Fig. 1). Moreover, the ability of ES to restore CcO function in several Cu-deficient eukaryotic model organisms (8) is consistent with FDX1 being an evolutionarily conserved mitochondrial protein.
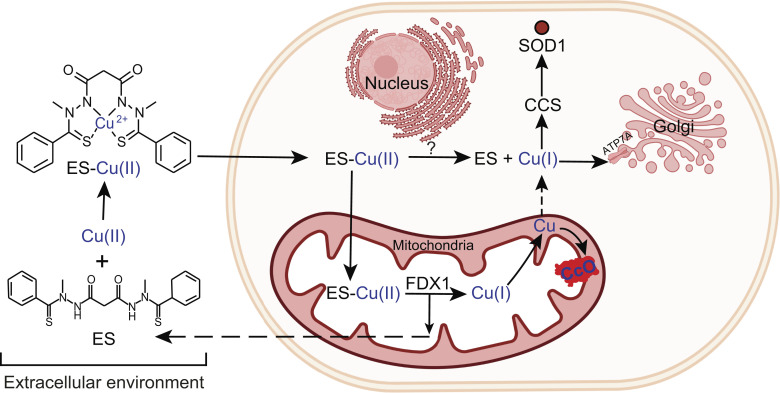
Surprisingly, we found that even in the absence of FDX1, Cu can be released from the ES–Cu(II) complex and made bioavailable outside mitochondria (Fig. 3). Although unexpected, this observation is consistent with our recent study in yeast showing that ES can circumvent the loss of CTR1-mediated Cu uptake by making Cu available to Ccc2, a Golgi-localized Cu pump that metallates the lumenal multicopper oxidase Fet3, which is required for cellular iron import (27). Such findings raise the obvious question—what are the reductive mechanism(s) that release Cu from ES outside of mitochondria? A previous study has shown that ES–Cu-induced cuproptosis is attenuated by hypoxia and by inhibitors of the electron transport chain under normoxic conditions (14), which suggests that oxygen concentration may directly or indirectly play a role in Cu(I) release from ES–Cu. A recent study uncovering a novel Cu(II) reductase activity of histones H3-H4 tetramer (28) offers a possibility that they may be involved in releasing Cu from ES–Cu in a manner akin to FDX1. However, our XFM data showing low levels of Cu in the nucleus upon ES–Cu treatment in Ctr1−/− Fdx1−/− as compared with Ctr1−/− cells (Fig. 3 A and B) suggests that it is unlikely. Our data also show that some of the Cu released in mitochondria by FDX1 very likely makes its way to the cytoplasm because we observed a greater decrease in the levels of CCS in Ctr1−/− as compared with Ctr1−/− Fdx1−/− cells (Fig. 3E). Based on these results, we propose a model of ES-mediated intracellular Cu distribution that depicts mitochondrial and nonmitochondrial sites of Cu release to make Cu bioavailable to cuproenzymes localized in different subcellular compartments (Fig. 5). Our study provides a mechanistic framework to understand the ES-mediated Cu trafficking in cells that may further help in repurposing this investigational anticancer drug for the disorders of Cu metabolism.
Fig. 5.
A model depicting ES-mediated intracellular Cu distribution. ES binds Cu(II) in the extracellular environment forming an electrically neutral, square planar ES–Cu complex that can pass through the lipid membranes and enter the mitochondria where Cu(I) is released from ES–Cu by the action of FDX1 protein. The released Cu(I) is now bioavailable for the metalation of cytochrome c oxidase (solid arrow) and to a certain extent to other cuproenzymes present outside the mitochondria (broken arrow). Cu is also released from ES–Cu outside of the mitochondria where it is bioavailable to cytosolic cuproproteins—CCS and SOD1 (solid arrows) and other subcellular compartment.
Materials and Methods
Compounds.
Elesclomol and elesclomol-copper were purchased from Accel Pharmtech; disulfiram, Cu-ATSM, carbonyl cyanide 3-cholorophenylhydrazone (CCCP), oligomycin, antimycin A, BCS, CuCl, and CuCl2 were purchased from Sigma.
Mammalian Cell Culture Conditions.
The rat H9c2 WT and knockout cell lines were cultured in high-glucose Dulbecco's Modified Eagle Medium (DMEM) media supplemented with 10% fetal bovine serum (FBS) (Sigma), 1 mM sodium pyruvate and Pen/Strep (Life Technologies). Cells were cultured under 5% CO2 at 37 °C and were treated with indicated concentrations of ES or ES–Cu.
Construction of Knockout Cell Lines.
We used lentiCRISPRv2 plasmid (Addgene #52961) to construct CRISPR/Cas9-based Fdx1 knockouts in WT and Ctr1−/− rat H9c2 cell lines. Three guide RNA sequences targeting Fdx1 were identified using (https://portals.broadinstitute.org/gppx/crispick/public). LentiCRISPRv2 plasmid vector backbone was digested using BsmBI restriction enzyme followed by T4 ligation of the digested vector with the phosphorylated and annealed oligo pairs (SI Appendix, Fig. S6). The ligated constructs were amplified in the Stbl3 Escherichia coli strain and sequence verified. Lentiviral particles were prepared using standard protocols. Transduction using the lentivirus was performed in H9c2 WT and Ctr1-KO cells using 8 μg/mL polybrene (EMD Millipore). After 24-h incubation, the virus-containing medium was removed, and a fresh culture medium containing blasticidin was added and selected for 72 h. Cells were then trypsinized, diluted, and plated at a density of 1 cell per well in 96-well plates. Clonal population was established in the selection medium containing blasticidin. Disruption of the Fdx1 gene was confirmed by genomic DNA sequencing and western blotting.
Cell Viability Assay.
Cells were seeded at a density of 5,000 cells/well in a 96-well plate in high-glucose DMEM growth media supplemented with 10% FBS (Sigma) and 1 mM sodium pyruvate and incubated at 37 °C and 5% CO2 for ~12 to 16 h. Spent media from each well were replaced using growth media containing indicated concentrations of ES or ES–Cu, and the plates were incubated for 72 h. Cell viability was assessed using the CellTiter-Glo-Cell viability assay (Promega) according to the manufacturer's instructions.
Cell Proliferation Assay.
Cells were seeded at a density of 5 × 105 cells/well in 6-well plates. After 12 to 16 h, growth media were replaced with fresh growth media containing either DMSO or 1 nM ES and incubated at 37 °C in 5% CO2 incubator. After the indicated time points, cells were trypsinized and counted using a hemocytometer.
Sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS–PAGE) Immunoblotting.
Cells were harvested, washed with phosphate-buffered saline (PBS), and lysed on ice in radioimmunoprecipitation assay (RIPA) extraction buffer (Boston BioProducts, BP-115), supplemented with a 1× cOmplete protease inhibitor cocktail (Roche), for 30 min and centrifuged at 14,000 × g for 15 min at 4 °C. Clear supernatants were collected, and protein concentration was determined using the bicinchoninic acid (BCA) assay (Pierce). Proteins were fractionated in 12% NuPAGE Bis-Tris gel (Life Technologies) followed by western blotting using the following antibodies: FDX1, 1:500 (12592-1-AP; Proteintech); COX1, 1:1,000 (ab14705; Abcam); ATP5A, 1: 20,000 (ab14748; Abcam); CCS, 1:1,000 (sc-55561; Santacruz), and β-Actin,1:20,000 (A2228, Sigma).
Oxygen Consumption Rate Measurements.
Oxygen consumption rate (OCR) measurements were carried out as previously described (29) with minor modifications. Briefly, H9c2 WT and knockout cardiomyocytes were treated with DMSO or 1 nM ES for 48 h in high-glucose DMEM growth media supplemented with 10% FBS and 1 mM sodium pyruvate. The cells were then seeded in XF24-well cell culture microplates (Agilent Technologies) at 10,000 cells/well in 250-µL growth media with DMSO or 1 nM ES and incubated at 37 °C in 5% CO2 incubator for ~20 h. Before measurements, 525 µL of the prewarmed growth medium was added to each well, and cells were further incubated at 37 °C for 30 min in a non-CO2 incubator. OCR measurements were carried out in intact cells using Seahorse XF24 Extracellular Flux Analyzer (Agilent Technologies). Mix, wait, and measure durations were set to 2, 2, and 2 min, respectively. For the mitochondrial stress test, oligomycin, carbonyl cyanide 3-cholorophenylhydrazone (CCCP), and antimycin A were sequentially injected to achieve final concentrations of 0.5, 10, and 1 µM, respectively. Immediately after the assay, cells were washed thrice with 500 µL PBS, and protein was extracted using 25 µL RIPA lysis buffer. Protein concentration in each well was measured by the BCA assay and was used to normalize OCR values.
Elesclomol–Cu EPR.
Powder EPR spectra were recorded on a continuous wave X-band Bruker 300E spectrometer at the frequency of 9.36 GHz at room temperature using microcrystalline solid samples. The spectra were recorded at a modulation amplitude of 1.0 G, modulation frequency of 100 kHz, and microwave power of 25 dB.
ICP-MS.
Cellular Cu levels were measured by inductively coupled plasma mass spectrometer (ICP-MS) using a Perkin Elmer, NexION 300D instrument. Briefly, cells were harvested and washed twice with 1 mL 0.9% NaCl containing 100 μM ethylenediamine tetraacetic acid (EDTA) prepared in ultrapure metal-free water (Trace SELECT; Sigma) followed by two more washes with 0.9% NaCl to eliminate EDTA. After washing, cell pellets were weighed and digested with 40% (w/v) nitric acid (Trace SELECT; Sigma) at 90 °C for 18 h, followed by 4-h digestion with 0.75% H2O2 (Sigma Supelco). The samples were then diluted in ultrapure water to 5 to 10 mL before injecting.
X-ray Fluorescence Microscopy.
Samples were prepared as described previously with slight modifications (30). Briefly, sterilized silicon nitride membranes (SiN, 1.5 × 1.5 mm, Norcada) were treated with 0.01% Poly-L-lysine solution (Sigma-Aldrich). Cells were seeded directly on SiN membranes placed in 6-well culture plates and incubated at 37 °C, 5% CO2 overnight. Cells were then treated using fresh growth media containing 100 nM ES–Cu for 30 min. Used media were discarded, and cells were washed twice with PBS and fixed with 4% paraformaldehyde for 30 min, followed by sequential washing with PBS, isotonic 100 mM ammonium acetate, nanopure water, and air-dried. XFM data were collected on beamline 9-ID-B [Bionanoprobe] (31) and beamline 2-ID-D in July 2021 and November 2021 at the Advanced Photon Source part of the Argonne National Laboratory, Argonne, IL. The incident X-ray energy was tuned to 10 keV using a Si-monochromator, and the monochromatic beam was focused to 80 × 80 nm using a Fresnel zone plate (Bionanoprobe) or 300 × 300 nm (beamline 2-ID-D). The sample was placed at 15° to the incident X-ray beam, and the resulting X-ray fluorescence was collected at 90° using an energy dispersive detector. Elemental maps were generated by extracting, background subtracting, and fitting the fluorescence counts for each element at each pixel using the program XRF-MAPS (https://github.com/AdvancedPhotonSource/XRF-Maps). The fluorescent photon counts were translated into µg/cm2 using calibrated X-ray standards (AXO products).
Real-Time Redox Measurements.
Mitochondria-targeted Grx1-roGFP2 plasmid was procured from Addgene (#64977). H9C2 Ctr1−/− and Ctr1−/−Fdx1−/− cells were transfected using Lipofectamine 3000 (Thermo Fisher) according to the manufacturer’s instructions. Cells were then seeded into an imaging chamber for 48 h before imaging. Cells showing clear membrane boundary and nucleus with vigorous Grx1-roGFP2 fluorescent intensity in the brightfield and fluorescence micrographs were selected for analysis. For redox measurements, 5 or 50 nM ES–Cu was added at the start of the experiment, followed by continuous imaging for 45 min. All micrographs were collected on an inverted microscope (IX 83, Olympus). The Grx1-roGFP2-transfected samples were excited sequentially with the 405 nm and 488 nm laser under epiillumination at each timestamp. A 40× objective (N1492800, Olympus) was used to collect the GFP emission at ~520 nm. A 525/50-nm bandpass filter (Lot# 344306, Chroma) was used for imaging on the scientific complementary metal-oxide-semiconductor (CMOS) camera (Prime 95B, Photometrics). The fluorescence intensities of all cells were corrected by subtracting the background reading using a cell-free area in each micrograph for subsequent redox responses analysis under various treatments.
FDX1 Expression and Purification.
A plasmid encoding human FDX1 was transformed in E. coli BL21(DE3) cells (32, 33). The cells were grown in Luria-Bertani media (Becton Dickinson Biosciences) at 37 °C and induced with 0.4 mM β-D-1-thiogalactopyranoside, 1 mM L-cysteine, and 0.1 mg/mL ferric ammonium citrate at OD600 of 0.6. The temperature was decreased to 25 °C, and cells were harvested 16 h later. Cells were lysed by sonication (Branson Sonifier 450), and the soluble fraction was loaded onto an anion exchange column (26/20 POROS 50HQ, Applied Biosystems). FDX1 was eluted with a linear gradient from 0 to 1,000 mM NaCl in 50 mM Tris (pH 7.5). Fractions containing FDX1 were pooled, concentrated, and loaded onto a HiPrep 26/60 Sephacryl S100 HR (Cytiva) size-exclusion column equilibrated in 50 mM Tris (pH 7.4) and 50 mM NaCl. Brown fractions containing holo-FDX1 were concentrated, frozen in liquid nitrogen, and stored at −80 °C. The sample was determined to be greater than 95% pure as estimated by SDS–PAGE analysis. The concentration of FDX1 was determined using a Bradford assay.
FDX1-Dependent Reduction of ES–Cu(II) In Vitro.
Purified [2Fe-2S]2+-FDX1 was reduced with 10 mM titanium (III) citrate to produce [2Fe-2S]+-FDX1. Titanium (III) citrate stock solution (83 mM, pH 8.0) was prepared as previously described (34). The reduction of FDX1 was monitored using peak attenuation at 415 and 455 nm with UV/Vis spectroscopy. Excess titanium citrate was removed using a PD-10 desalting column (Sephadex G-25 M, Cytiva). To ensure that FDX1 remained reduced, experiments were conducted in an anaerobic glovebox (mBraun, 22 °C, < 0.5 ppm O2; monitored by Teledyne Model 3110 Oxygen Gas Analyzer). All materials were extensively degassed and incubated in the glovebox for at least 24 h prior. In Cu(I) trapping experiments, absorbance scans were carried out in a Tecan M200 plate reader using bottom-read absorbance measurements. Corning 96-well half-area plates (white flat bottom, polystyrene) with a nonbinding surface were used. The reduction of ES–Cu(II) by [2Fe-2S]+- FDX1 was monitored spectrophotometrically using a Cu(I)-specific chelator, BCS. BCS (1 mM) and 20 µM ES–Cu(II) with increasing amounts of FDX1Red was mixed with 1 mM BCS in 10 mM Tris, 50 mM NaCl pH 7.5 solution. ES–Cu(II) (20 µM in 100% DMSO) was used to initiate the reaction and was incubated for 5 min prior to absorbance scans.
Immunofluorescence Microscopy.
Cells were grown in 24-well trays for 24 h on sterile glass coverslips. The following day, cells were incubated with 5 nM ES–Cu complex or basal medium for 3 h. Cells were then washed twice with 1 mL ice-cold PBS and fixed for 10 min at 25 °C using 4% paraformaldehyde. Cells were then permeabilized with 0.1% Triton X-100 in PBS for 6 min, blocked for 2 h with 1% bovine serum albumin in PBS, and incubated with the anti-ATP7A antibody (1:2,000) overnight at 4 °C. The next day, cells were washed 3× with PBS followed by 1-h incubation with Alexa488 anti-rabbit antibody (1:1,000). Nuclei were stained with DAPI (ThermoFisher). Finally, specimens were analyzed using a STELLARIS 5 confocal microscope (Leica Microsystems).
In-Gel SOD1 Activity Assay.
Sod1 activity was measured using an in-gel assay as described previously (35). Briefly, cells were solubilized using 4X volume of cell pellets in cell lysis buffer (20 mM potassium phosphate, pH 7.4, 1 mM EDTA, 0.5% Triton X-100, 4 mM PMSF with protease inhibitor cocktail) by incubating on ice for 15 min. The supernatant was collected after centrifugation at 20,000 × g for 15 min followed by protein concentration measurement by BCA assay. Then, 20 mg of protein supplemented with NativePAGE sample buffer (4X) was separated using a 4 to 16% native PAGE gel. The gel was stained for Sod1 activity as described previously (35).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. John Markley for providing the FDX1 plasmid, Deepika Das for the FDX1 purification protocol, and Dr. Byung-Eun Kim for providing H9c2 Ctr1−/− cells. We also thank Adriana Okonkwo for assistance with cell culture experiments and Vishav Sharma for help with ChemDraw. Research reported in this publication was supported by the National Institute of General Medical Sciences of the NIH awards R01GM143630 and R01GM111672 to V.M.G., R21GM129592 to M.R., R35GM133505 to T.-Y.C., R01GM096100 to D.P.B. and National Institute of Diabetes and Digestive and Kidney Diseases award DK131190 and National Cancer Institute award CA262664 to M.J.P. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Use of the Advanced Photon Source part of the Argonne National Laboratory is supported by the Department of Energy, Office of Basic Energy Sciences, under contract no. DEAC02-06CH11357. The authors have research support to disclose: The corresponding author’s university (Texas A&M) has entered into a licensing agreement with Engrail Therapeutics for the development of elesclomol:copper as a therapeutic agent for the disorders of copper metabolism.
Author contributions
M.Z. and V.M.G. designed research; M.Z., A.N.S., Y.Z., S.S., S.C., L.L., T.L., V.S., and M.R. performed research; M.Z. contributed new reagents/analytic tools; M.Z., A.N.S., Y.Z., S.S., S.C., L.L., T.L., V.S., M.J.P., T.-Y.C., M.R., D.P.B., and V.M.G. analyzed data; and M.Z., M.J.P., T.-Y.C., M.R., D.P.B., and V.M.G. wrote the paper.
Competing interests
The authors have organizational affiliations to disclose: Vishal M. Gohil serves as a consultant to Engrail Therapeutics for their effort in developing elesclomol–copper as a therapeutic agent for Menkes disease. The authors have patent filings to disclose: S.S. and V.M.G. are inventors on the patent US 2021/0290571 A1 submitted by Texas A&M University entitled “Compositions for the Treatment of Copper Deficiency and Methods of Use.”
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Garza N. M., Swaminathan A. B., Maremanda K. P., Zulkifli M., Gohil V. M., Mitochondrial copper in human genetic disorders. Trends Endocrinol. Metab. 34, 21–33 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaler S. G., Inborn errors of copper metabolism. Handb. Clin. Neurol. 113, 1745–1754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Papadopoulou L. C., et al. , Fatal infantile cardioencephalomyopathy with COX deficiency and mutations in SCO2, a COX assembly gene. Nat. Genet. 23, 333–337 (1999). [DOI] [PubMed] [Google Scholar]
- 4.Valnot I., et al. , Mutations of the SCO1 gene in mitochondrial cytochrome c oxidase deficiency with neonatal-onset hepatic failure and encephalopathy. Am. J. Hum. Genet. 67, 1104–1109 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baertling F., et al. , Mutations in COA6 cause cytochrome c oxidase deficiency and neonatal hypertrophic cardiomyopathy. Hum. Mutat. 36, 34–38 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Horn N., Møller L. B., Nurchi V. M., Aaseth J., Chelating principles in Menkes and Wilson diseases: Choosing the right compounds in the right combinations at the right time. J. Inorg. Biochem. 190, 98–112 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Kaler S. G., et al. , Neonatal diagnosis and treatment of Menkes disease. N. Engl. J. Med. 358, 605–614 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soma S., et al. , Elesclomol restores mitochondrial function in genetic models of copper deficiency. Proc. Natl. Acad. Sci. U.S.A. 115, 8161–8166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guthrie L., et al. , Elesclomol alleviates Menkes pathology and mortality by escorting Cu to cuproenzymes in mice. Science 368, 620–625 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yadav A. A., Patel D., Wu X., Hasinoff B. B., Molecular mechanisms of the biological activity of the anticancer drug elesclomol and its complexes with Cu(II), Ni(II) and Pt (II). J. Inorg. Biochem. 126, 1–6 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Nagai M., et al. , The oncology drug elesclomol selectively transports copper to the mitochondria to induce oxidative stress in cancer cells. Free Radic. Biol. Med. 52, 2142–2150 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Sheftel A. D., et al. , Humans possess two mitochondrial ferredoxins, Fdx1 and Fdx2, with distinct roles in steroidogenesis, heme, and Fe/S cluster biosynthesis. Proc. Natl Acad. Sci. U.S.A. 107, 11775–11780 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai K., Tonelli M., Frederick R. O., Markley J. L., Human mitochondrial ferredoxin 1 (FDX1) and ferredoxin 2 (FDX2) both bind cysteine desulfurase and donate electrons for iron–sulfur cluster biosynthesis. Biochemistry 56, 487–499 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tsvetkov P., et al. , Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 375, 1254–1261 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsvetkov P., et al. , Mitochondrial metabolism promotes adaptation to proteotoxic stress. Nat. Chem. Biol. 15, 681–689 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gohil V. M., Repurposing elesclomol, an investigational drug for the treatment of coppermetabolismdisorders. Exp. Opin. Investig. Drugs 30, 1–4 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulz V., et al. , Functional spectrum and specificity of mitochondrial ferredoxins FDX1 and FDX2. Nat. Chem. Biol. 19, 206–217 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hasinoff B. B., Yadav A. A., Patel D., Wu X., The cytotoxicity of the anticancer drug elesclomol is due to oxidative stress indirectly mediated through its complex with Cu(II). J. Inorg. Biochem. 137, 22–30 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Leary S. C., Ralle M., Advances in visualization of copper in mammalian systems using X-ray fluorescence microscopy. Curr. Opin. Chem. Biol. 55, 19–25 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petris M. J., et al. , Ligand-regulated transport of the Menkes copper P-type ATPase efflux pump from the Golgi apparatus to the plasma membrane: A novel mechanism of regulated trafficking. EMBO J. 15, 6084–6095 (1996). [PMC free article] [PubMed] [Google Scholar]
- 21.Bertinato J., L’Abbe M. R., Copper modulates the degradation of copper chaperone for Cu, Zn superoxide dismutase by the 26S proteosome. J. Biol. Chem. 278, 35071–35078 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Robinett N. G., Peterson R. L., Culotta V. C., Eukaryotic copper-only superoxide dismutases (SODs): A new class of SOD enzymes and SOD-like protein domains. J. Biol. Chem. 293, 4636–4643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gutscher M., et al. , Real-time imaging of the intracellular glutathione redox potential. Nat. Methods 5, 553–559 (2008). [DOI] [PubMed] [Google Scholar]
- 24.Hunsaker E. W., Franz K. J., Emerging opportunities to manipulate metal trafficking for therapeutic benefit. Inorg. Chem. 58, 13528–13545 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Członkowska A., et al. , Wilson disease. Nat. Rev. Dis. Primers 4, 1–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirshner J. R., et al. , Elesclomol induces cancer cell apoptosis through oxidative stress. Mol. Cancer Ther. 7, 2319–2327 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Garza N. M., Zulkifli M., Gohil V. M., Elesclomol elevates cellular and mitochondrial iron levels by delivering copper to the iron import machinery. J. Biol. Chem. 298, 102139 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Attar N., et al. , The histone H3–H4 tetramer is a copper reductase enzyme. Science 369, 59–64 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gohil V. M., et al. , Nutrient-sensitized screening for drugs that shift energy metabolism from mitochondrial respiration to glycolysis. Nat. Biotechnol. 28, 249–255 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pek R. H., et al. , Hemozoin produced by mammals confers heme tolerance. eLife 8, e49503 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen S., et al. , The bionanoprobe: Synchrotron-based hard x-ray fluorescence microscopy for 2D/3D trace element mapping. Micros. Today 23, 26–29 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xia B., Cheng H., Bandarian V., Reed G. H., Markley J. L., Human ferredoxin: Overproduction in Escherichia coli, reconstitution in vitro, and spectroscopic studies of iron-sulfur cluster ligand cysteine-to-serine mutants. Biochemistry 35, 9488–9495 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Pagani S., Bonomi F., Cerletti P., Enzymic synthesis of the iron-sulfur cluster of spinach ferredoxin. Eur. J. Biochem. 142, 361–366 (1984). [DOI] [PubMed] [Google Scholar]
- 34.Seefeldt L. C., Ensign S. A., A continuous, spectrophotometric activity assay for nitrogenase using the reductant titanium (III) citrate. Anal. Biochem. 221, 379–386 (1994). [DOI] [PubMed] [Google Scholar]
- 35.Flohe L., Otting F., Superoxide dismutase assays. Methods Enzymol. 105, 93–104 (1984). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix.