Significance
Mfsd2a is a transporter expressed at the blood–brain barrier that mediates the major pathway by which the brain acquires essential fatty acids from blood. Mfsd2a transports fatty acids conjugated as lysophosphatidylcholine (LPC) in blood circulation. Humans with loss of function mutations in Mfsd2a present with severe microcephaly, indicating a critical role of LPC transport in brain development. A model for Mfsd2a transport posits that Mfsd2a acts as a LPC “flippase,” inverting LPC as it moves through the transporter from the outer to inner leaflet of the plasma membrane. By establishing an in vitro biochemical assay, this study provides the direct proof that Mfsd2a has “flippase” activity, thus defining the biochemical mechanism of how LPC is transported into the brain.
Keywords: Mfsd2a, transporter, flippase, membrane, blood–brain barrier
Abstract
Major Facilitator Superfamily Domain containing 2a (Mfsd2a) is a sodium-dependent lysophosphatidylcholine (LPC) transporter expressed at the blood–brain barrier that constitutes the main pathway by which the brain obtains omega-3 fatty acids, such as docosahexanoic acid. Mfsd2a deficiency in humans results in severe microcephaly, underscoring the importance of LPC transport by Mfsd2a for brain development. Biochemical studies and recent cryo-electron microscopy (cryo-EM) structures of Mfsd2a bound to LPC suggest that Mfsd2a transports LPC via an alternating access mechanism between outward-facing and inward-facing conformational states in which the LPC inverts during transport between the outer and inner leaflet of a membrane. However, direct biochemical evidence of flippase activity by Mfsd2a has not been demonstrated and it is not understood how Mfsd2a could invert LPC between the outer and inner leaflet of the membrane in a sodium-dependent manner. Here, we established a unique in vitro assay using recombinant Mfsd2a reconstituted in liposomes that exploits the ability of Mfsd2a to transport lysophosphatidylserine (LPS) coupled with a small molecule LPS binding fluorophore that allowed for monitoring of directional flipping of the LPS headgroup from the outer to the inner liposome membrane. Using this assay, we demonstrate that Mfsd2a flips LPS from the outer to the inner leaflet of a membrane bilayer in a sodium-dependent manner. Furthermore, using cryo-EM structures as guides together with mutagenesis and a cell-based transport assay, we identify amino acid residues important for Mfsd2a activity that likely constitute substrate interaction domains. These studies provide direct biochemical evidence that Mfsd2a functions as a lysolipid flippase.
Mfsd2a is a sodium-dependent lysophosphatidylcholine (LPC) transporter highly expressed at the blood–brain and blood–eye barriers and is essential for brain and eye acquisition of the omega-3 fatty acid docosahexaenoic acid (DHA) (1–3). Mfsd2a is expressed by endothelial cells of the blood–brain barrier and by several specialized cells in the brain, endothelium of the blood–eye barriers, and in peripheral organs and cells such as alveolar type 2 cells of the lung and in specific immune cell types (1, 4–8). Most prominently, human loss-of-function mutations result in autosomal recessive microcephaly 15, a disease characterized by severe microcephaly, hypomyelination, and intellectual and developmental delay (9–12). Studies using Mfsd2a knockout mice recapitulated many of these phenotypes and indicated that LPC transport is critical for neuron membrane arborization and maintaining normal membrane phospholipid composition (13). Mfsd2a belongs to the Major Facilitator Superfamily (MFS) of secondary transporters. Unlike primary active transporters that utilize adenosine triphosphate (ATP) for translocation of solutes across membranes, many MFS proteins are facilitative transporters or secondary active transporters utilizing the cotransport of sodium ions or protons to drive the uphill transport of solutes (14, 15). Similar to other MFS proteins, Mfsd2a contains twelve transmembrane (TM) helices divided into two semisymmetrical six-transmembrane bundles comprising the N- and C- domains of the protein. Biochemical and structural studies of bacterial and mammalian MFS proteins specific for sugars and other water-soluble compounds have supported an alternating access of transport mechanism, whereby the N- and C-domains undergo rigid body movements to alternate between open and closed conformations at either side of the membrane (14, 15). Because Mfsd2a transports lysolipids, it is considered to be an atypical MFS transporter. There are only a few known lysolipid MFS transporters, namely LplT in bacteria (16), the mammalian sphinogosine-1-phosphate transporters Mfsd2b and Spns2 (17–19), and Spns1 that we recently discovered to be a proton-coupled lysosomal LPC and lysophosphatidylethanolamine (LPE) transporter (20). Structural information on LplT, Mfsd2b, Spns2, and Spns1 is still lacking.
Current information on Mfsd2a lysolipid specificity indicates a requirement for a zwitterionic headgroup and a fatty acyl chain with a minimum chain length of 14 carbons (21). Determination of a mechanism by which Mfsd2a mediates lysolipid transport has been an area of high interest in part because of its physiological importance in human brain development and because it has been proposed as a mechanism to potentially carry small molecules across the largely impermeable blood–barrier endothelium (22–24). Importantly, the transport mechanism is unresolved by which Mfsd2a and the other atypical MFS lysolipid transporters work. Currently, three high-resolution structures of Mfsd2a have been solved using single-particle cryo-EM. One such structure of chicken Mfsd2a (gMfsd2a) was captured in an inward open conformation bound to LPC containing the omega-3 fatty acid linolenic acid (LPC-18:3). The structure revealed the LPC headgroup in an inward-facing cavity positioned in a lateral opening formed between TM5 and TM8 with the fatty acyl tail of the LPC molecule buried in a hydrophobic cleft (25). This gMfsd2a structure likely represents a late step in the transport cycle in which the LPC has inverted from the outer leaflet to the inner leaflet of the membrane and lodged laterally between TM5 and TM8, but prior to exiting into the surrounding membrane. The second high-resolution structure is of mouse Mfsd2a (mMfds2a) that was captured in an outward-open conformation and revealed electron density suggestive of lysolipid located in a large lateral opening formed by TM2-TM11 and TM5-TM8 raising the possibility that the first step of LPC entry into the transporter could be from either opening, although the biochemical evidence to support this is lacking (26). A third structure was recently elucidated of an outward-open occluded conformation of human Mfsd2a (hMfsd2a) in complex with syncytin2 (27). Computational docking of an LPC into this structure suggested that LPC could adopt a bent conformation lying close to parallel relative to the plane of the membrane, suggestive of a step in the transport process in which the LPC is rotating toward an inverted position (27). All three structures suggest that Mfsd2a transports LPC by flipping LPC through the transport cleft with polar headgroup and hydrophobic acyl tail interactions occurring in distinct pockets. However, due to the limited in vitro tools available to study lysolipid transporters, direct biochemical evidence of this flippase mechanism remains speculative from the available structural snapshots of Mfsd2a (28–30). Here, we developed an in vitro assay using purified Mfsd2a reconstituted in liposomes to demonstrate that Mfsd2a exhibits Na+-dependent lysolipid flippase activity with similar qualitative features of LPC transport in vivo.
Results and Discussion
Establishment of an In Vitro Lysophosphatidylserine (LPS) Flippase Assay.
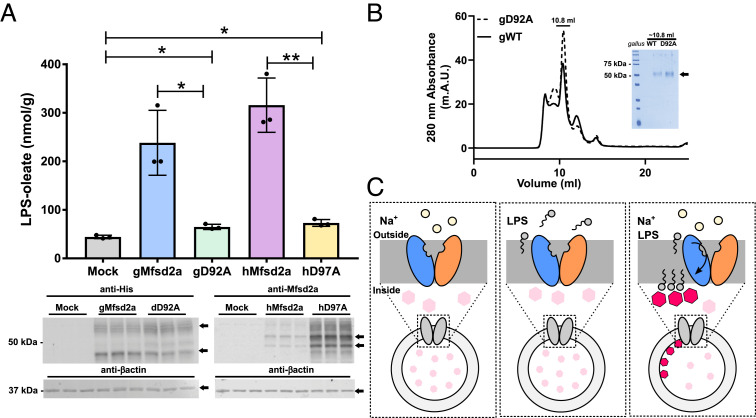
We sought to establish a proteoliposome assay that would directly measure the flipping of lysolipids from the outer to the inner leaflet of a membrane by detecting the location of the charged lipid headgroup. Currently used assays for measuring activity of lipid scramblases and flippases have relied on methods to detect the transport of chemically modified phospholipids. Spin-labeled phospholipids have been used to quantify the membrane location of phospholipid following ATP-dependent flipping (31). A more widely used and robust method for quantifying lipid flipping is the protection of dithionate quenching of fluorophore-conjugated phospholipids and other types of lipids (31–34). One major technical issue with quantifying flipping activity of Mfsd2a using LPC with headgroup-conjugated fluorophores is that the phosphocholine headgroup is essential for transport and modifications to the LPC headgroup could abolish transport (1). To achieve an assay that could be a direct readout for lysolipid flipping without modification to the lysolipid substrate, we wanted to develop an assay that reports directly on the location of the lysolipid headgroup within liposome membranes using a native lysolipid. Probes to detect the phosphocholine headgroup of membrane-embedded PC do not exist and the fact that liposomes are primarily composed of PC excluded the possibility of using LPC in a flippase assay. One promising lipid headgroup binding probe is the small molecule compound PSVue550 that binds specifically to the phosphoserine headgroup of phosphatidylserine (PS) (35, 36). Fortunately, human and chicken Mfsd2a transports LPS (Fig. 1A), raising the possibility that it could be used as a suitable lysolipid in combination with PSVue550 to test for in vitro flippase activity by Mfsd2a. PSVue550 was shown to bind specifically to the phosphoserine headgroup of PS localized in the membrane bilayer in a calcium-independent fashion in apoptotic cells (36). Upon binding to PS headgroups immobilized in the membrane bilayer, PSVue550 exhibits an emission wavelength shift to 610 nm and a corresponding increase in intensity upon excitation at 550 nm, thus having the potential to detect the presence of PS headgroups embedded in an artificially created liposome membrane (36). Mammalian membrane proteins are notoriously difficult to extract from membranes and upon extraction are often unstable resulting in limited success for proteoliposome incorporation and subsequent assay development (29, 30). Hence, we reasoned that for an in vitro flippase assay to be robust, Mfsd2a needs to be thermostable in order to survive the extraction, incorporation, and room temperature assay conditions. Hence, for assay development, we focused on using purified chicken Mfsd2a (gWT) that we previously determined to be thermostable that was used for determining a cryo-EM structure (25). In addition to purified gWT, we produced the sodium binding site mutant D92A (gD92A) to be used as a potential negative control (Fig. 1B). The goal was to incorporate Mfsd2a into palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) liposomes containing 20% cholesterol with PSvue550 encapsulated within the aqueous environment of the proteoliposome and to monitor the fluorescence emission of the encapsulated PSvue550 upon addition of LPS and sodium chloride to the extra-liposomal reaction buffer. We hypothesized that when LPS and sodium are added to Mfsd2a proteoliposomes, PSVue550 fluorescence would increase if LPS is flipped from the outer to the inner leaflet of the proteoliposome due to the interaction of the LPS headgroup with PSVue550 located in the aqueous core of the proteoliposome (Fig. 1C).
Fig. 1.
Experimental setup for a proteoliposome flippase assay. (A) Mass uptake of transported LPS in cells transfected with Mock (empty vector), gMfsd2a, gD92a, hMfsd2a, and hD97A. Data are expressed as mean ± SD. Experiment was performed in triplicate; *P < 0.05, **P < 0.005. The panel below the graph is a western blot analysis showing expression of indicated constructs (arrow); β-actin served as a loading control. (B) FPLC trace of purified G. gallus Mfsd2a (gWT) and sodium-binding mutant gD92A. (C) Experimental setup for the proteoliposome flippase assay which involves the incorporation of PSvue550 into the proteoliposome lumen, depicted as pink hexagons (nonactivated, low fluorescence) and red hexagons (activated, high fluorescence). The three panels depict the experimental conditions used to determine proteoliposome Mfsd2a-mediated LPS transport. The model shows that only in the presence of NaCl and LPS does Mfsd2a mediate flipping of LPS from the outer to the inner leaflet and activates the luminally located PSvue550 (red hexagons).
Mfsd2a-Mediated LPS Flipping in Proteoliposomes.
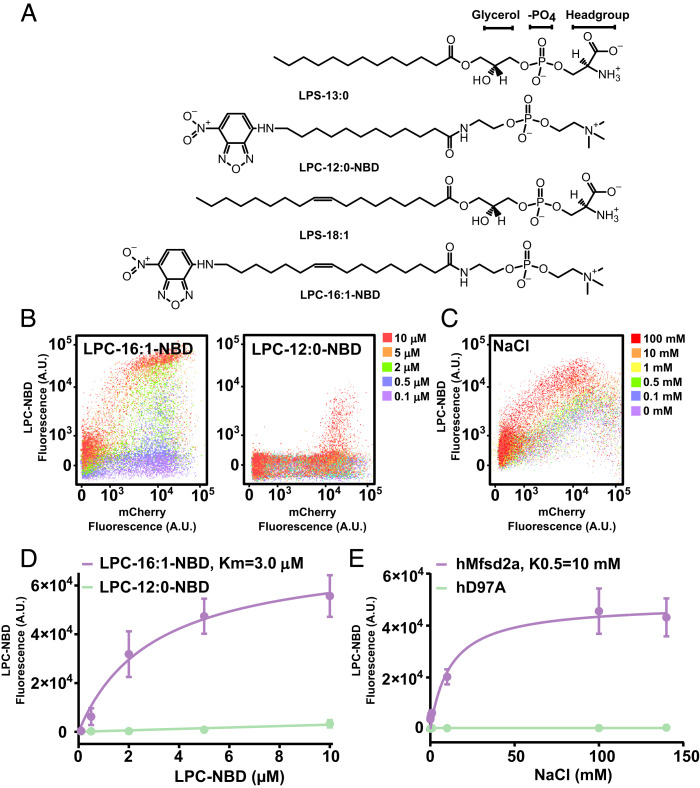
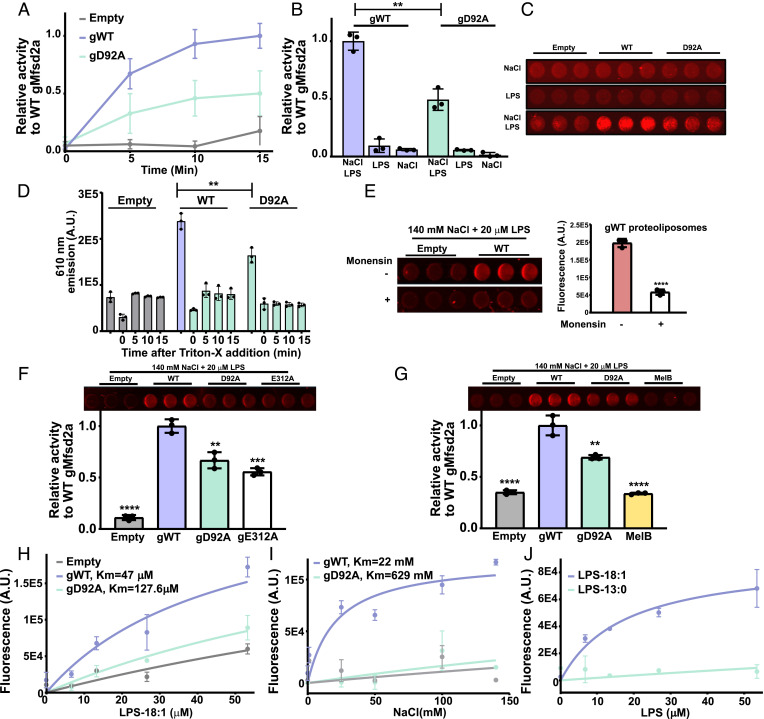
We first set out to determine the Km of transport for LPC conjugated by nitrobenzoxadiazole (NBD) (LPC-NBD) (Fig. 2A) and for sodium by Mfsd2a in cells, which would set an in vivo benchmark to compare with transport data derived from purified Mfsd2a in proteoliposomes. Utilizing our established single-cell–based transport assay (25) that quantifies the amount of fluorescent LPC-NBD uptake by Mfsd2a (Fig. 2 B and C), we obtained an apparent Km of 3 μM for LPC-16:1-NBD and 244 μM for LPC-12:0-NBD (Fig. 2D), in agreement with a minimal LPC acyl chain length requirement of 14 carbons for high capacity transport (21). The stimulation constant for NaCl was calculated to be 10 mM (Fig. 2 C and E). However, the curve fitting was poor for NaCl, and the data appear to indicate an all-or-nothing sodium activation of LPC transport, a characteristic that is very similar to sodium activation of MelB (37). Using the proteoliposome assay setup described in Fig. 1C, we performed a time course experiment using 50 μM LPS-oleate (LPS-18:1) and 140 mM NaCl, including the sodium binding site mutant gD92A and empty liposomes as negative controls. In the presence of LPS and NaCl, fluorescence was significantly increased over time relative to controls, indicative of sodium-dependent flippase activity (Fig. 3 A–C and Movies S1 and S2). It is worth noting that the time-resolved reaction rate in this assay is on the order of minutes, which is significantly slower than the time-resolved reaction rate in seconds for dithionate fluorescence quenching (38–40). The likely cause for the relatively slow rate of reaction is that the rate of flipping in our experimental setup would be governed by the rate of LPS binding to the proteolipososme membrane, Mfsd2a-mediated flipping of LPS, and binding of LPS with PSVue550. While the rate of LPS absorption to proteoliposomes is not known, the binding affinity of PSVue550 to PS is approximately 10 µM in vesicles highly enriched in PS (containing 1:1 POPC:POPS) (35). The published studies using dithionate fluorescence quenching were in experimental setups whereby the fluorescent lipid was pre-incorporated into liposomes (28, 29, 32, 33, 38, 39); thus, the rate of lipid absorption into the liposome membrane is not a factor contributing to the rate of lipid flipping with dithionate fluorescence quenching as it is in our experimental setup. The addition of the detergent Triton-X100 to disrupt proteoliposomes resulted in complete loss in PSVue550 fluorescence (Fig. 3D), indicating that PSVue550 does not detect freely rotating LPS headgroups in solution, and supporting the conclusion that PSVue550 is detecting the membrane immobilized LPS headgroup that has flipped from the outer to inner leaflet. Consistent with sodium-dependent Mfsd2a-mediated lysolipid flippase activity, addition of monensin, a sodium-specific ionophore, at the start of the flippase reaction abolished Mfsd2a activity (Fig. 3E).
Fig. 2.
Transport parameters of LPC-NBD using a single-cell assay. (A) Chemical structures of lysolipids used in this study, LPS-13:0, LPS-18:1, LPC-12:0-NBD, and LPC-16:1-NBD. (B) Single-cell flow data for LPC-NBD (16:1 and 12:0) uptake in a dose–response setup for HEK293 cells transfected with hMfsd2a-mCherry. The LPC-NBD dose is indicated in the legend. (C) Flow data for 2 μM LPC-NBD (16:1) uptake in HEK293 cells transfected with hMfsd2a-mCherry in response to increasing concentrations of NaCl. (D) Curve fitting of data in panel B was used to derive the indicated Km for the transport of LPC-16:1-NBD. (E) Curve fitting of data in panel C. Each data point shown in D and E represents the mean ± SD.
Fig. 3.
Demonstration of flippase activity in proteoliposomes. (A) Relative time-dependent activity of WT gMfsd2a (gWT), gD92A mutant, and empty liposomes (Empty). (B) Graphical representation of the 15 min timepoint shown in panel A; NaCl and LPS are added to the reactions as indicated at 140 mM NaCl and 50 μM LPS. (C) A representative image of wells from a 96-well plate at the 15 min time point shows NaCl and LPS-dependent fluorescence produced by WT gMfsd2a is greater than gD92A and empty liposomes. (D) Proteoliposomes incubated with 140 mM NaCl and 50 μM LPS for 15 min then treated with 1% Triton-X and fluorescence was quantified as a function of time. (E) Dissipation of the sodium gradient using 30 μM of monensin abolished Mfsd2a activity in proteoliposomes. Left panel showing fluorescence emitted from gWT proteoliposomes treated with and without monensin and quantified in the Right panel. (F) Comparison of activity of gWT to gD92A, gE312A, and empty liposomes at the 15 min time point using same conditions as described in panel B. Top panel showing fluorescence image of the reaction wells. (G) Comparison of gWT relative to gD92A and MelB, with Top panel showing fluorescence image of the reaction wells. (H) Dose–response experiment with indicated proteoliposomes with increasing concentrations of LPS-18:1. (I) Dose–response experiment with indicated proteoliposomes with increasing concentrations of NaCl. (J) Dose–response experiment using LPS-18:1 and LPS-13:0. All experiments were performed in technical triplicate with data represented as mean ± SD. **P < 0.005, ***P < 0.002, ****P < 0.0001.
It is notable that the sodium-binding site mutant gD92A showed significant activity in this assay at higher LPS concentrations, although significantly lower than gWT (Fig. 3 B–D). Moreover, the sodium binding site mutants gD92A and hD97A expressed in cells exhibited residual transport activity (Fig. 1A and SI Appendix, Fig. S3). Residual LPC transport activity of hD97A was previously reported (1) suggesting that the flippase assay is sensitive and specific. Furthermore, we purified (SI Appendix, Fig. S2) and tested the effect of gE312A on fippase activity. E312 side chain is predicted to be involved in sodium binding that exhibits low activity in cells when mutated to alanine (25). In agreement with in vivo findings, gE312A showed similarly reduced flippase activity as gD92A relative to gWT (Fig. 3F). As an independent test of the specificity of this flippase assay, we tested the activity of the melibiose transporter from Salmonella typhimurium (MelB) in this assay. MelB is a Na+-coupled MFS protein that is ~25% similar to Mfsd2a that transports sugars and not lipids (21, 41). MelB did not exhibit LPS flippase activity (Fig. 3G), further supporting the conclusion that our assay is sensitive and specific for measuring LPS flipping activity of Mfsd2a.
We next performed a dose–response assay with increasing concentrations of LPS-oleate. Increasing LPS-oleate concentration from 0 to 50 µM while holding NaCl constant at 140 mM showed a significant dose-dependent flipping activity relative to the gD92A and empty liposomes (Fig. 3H and Movie S2). We calculated the Km for gWT-mediated LPS flipping to be 47 μM. This value is higher than the Km measured for cell-based transport of LPC-NBD, estimated to be 3 μM (Fig. 2D). The higher Km for LPS in this in vitro assay could be attributed to reduced affinity of Mfsd2a for LPS relative to LPC. To determine whether Mfsd2a LPS flipping activity exhibited a NaCl dose–response, we kept the LPS concentration fixed at 50 μM with increasing NaCl from 0 to 140 mM. In this assay, LPS-oleate flipping across the membrane exhibited a dose–response relationship in the presence of increasing NaCl concentration with an estimated Km for Na+ to be 22 mM (Fig. 3I and Movie S3). The increased Km for Na+ in this assay relative to findings in cells (Fig. 2E) could be a consequence of lack of maintenance of a sodium gradient. While the Km for LPS and sodium are increased in this in vitro flippase assay relative to the cell-based assay, the overall transport characteristics of sodium dependence of LPS transport and dose–response relationships are qualitatively similar to in vivo transport.
We next wanted to determine whether the same acyl chain length constraints hold true for LPS in our flippase activity assay as observed in the cell-based assay (Fig. 2B). We examined flippase activity using LPS-18:1 and LPS-13:0, as LPS-12:0 was not commercially available. Only in the presence of increasing concentrations of LPS-18:1 but not LPS-13:0, flippase activity of Mfsd2a was observed (Fig. 3J). These data are in line with previously reported lysolipid acyl chain requirements of 14 carbons for transport by Mfsd2a ((1) and Fig. 2D) and provide an additional control demonstrating the specificity of this flippase assay. These data further support the conclusion that Mfsd2a is a lysolipid flippase.
Refinement of a Model for Mfsd2a-Mediated Lysolipid Transport.
The mode of lysolipid entry into the transporter and its interaction with charged residues lining the central core of Mfsd2a remains to be understood. Using the currently available high-resolution structures and our single-cell transport assay, we further attempt to understand the first step of lysolipid entry into the outward-open structure of Mfsd2a.
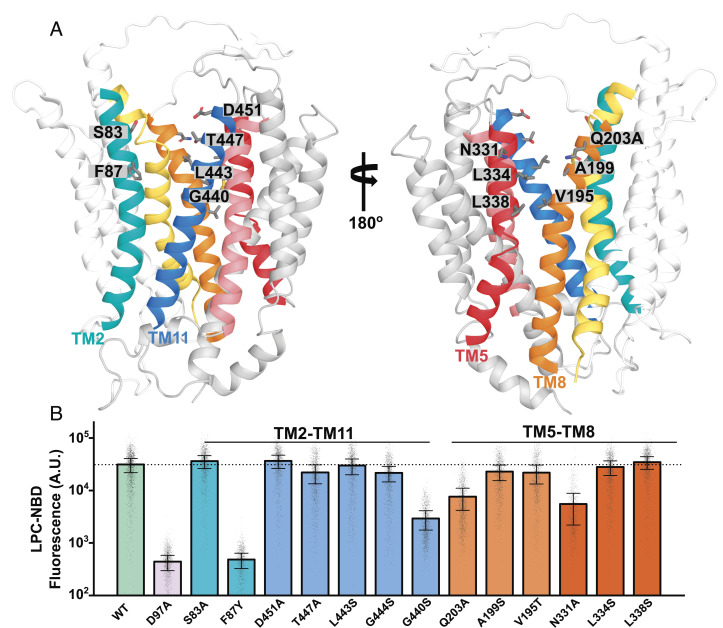
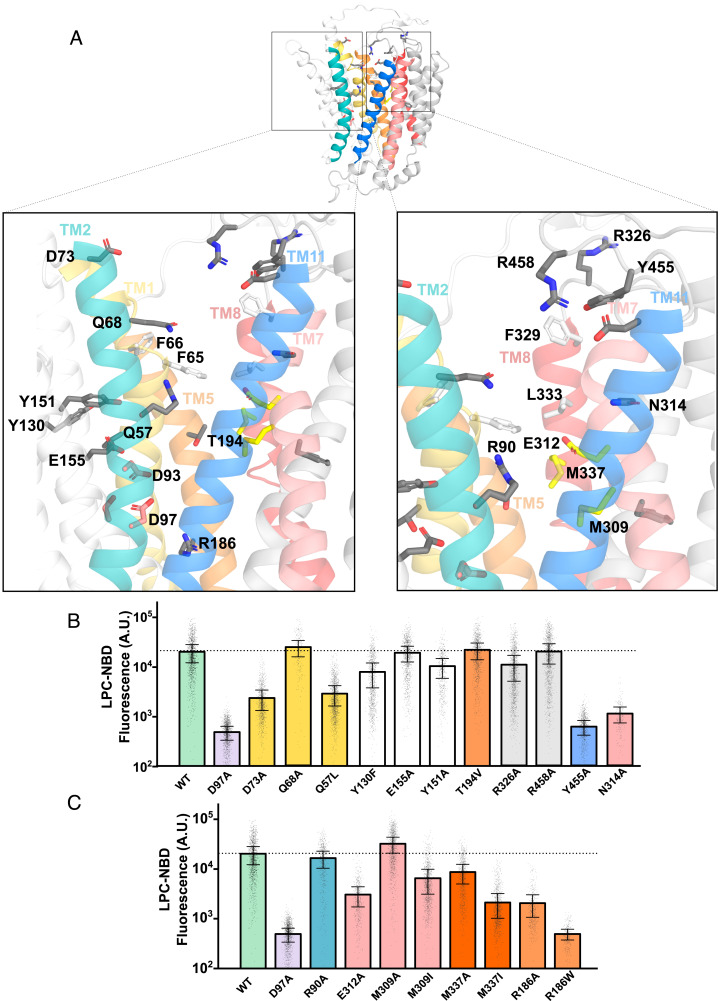
The recently elucidated outward-open mMfsd2a structure showed two unassignable densities at the two lateral openings formed by TM5-TM8 and TM2-TM11 suggesting them to be sites of lysolipid entry (26). The authors of that report were able to fit a LPC-like molecule into the density observed at TM5-TM8, suggesting that LPC might enter Mfsd2a at this lateral opening. Lateral openings into the membrane bilayer at TM2-TM11 and TM5-TM8 have also been reported to be important in other lipid transporters, such as LtaA that transports diacylglycerol-linked-gentiobiose in gram-positive bacteria using a proton gradient, where it has been shown by molecular modeling that the diacyl chains of phospholipids can intrude into the proposed substrate translocation pathway (32). The outward-open mMfsd2a structure appears remarkedly similar to the LtaA structure, with a rmsd ~9.8 Å, with an exception that the distances between the two lateral openings are smaller in Mfsd2a than found in LtaA (SI Appendix, Fig. S6A). This structural difference in the size of the lateral openings might account for the fact that Mfsd2a transports a single-acyl chain phospholipid while LtaA transports a larger diacylglycerol glycolipid (38). Despite these similarities, the functional importance of the residues lining TM2-TM11 and TM5-TM8 near the extracellular face of Mfsd2a has not been studied. Residues lining the two lateral openings begin with polar residues, such as S83 and D451 at TM2-TM11 and Q203 and N331 at TM5-TM8 (Fig. 4A). These residues are followed by aliphatic and aromatic residues that could allow interaction with the LPC acyl tail predicted to reside in the membrane bilayer. Mutating these polar residues residing at TM2-TM11 (S83 and D451) and TM5-TM8 (Q203 and N331) to alanine partially reduced LPC transport (Fig. 4B). We next tested if Mfsd2a could tolerate a reduction in the size of the lateral openings by the substitution of a single bulky tryptophan residue at F87, G440, G444, V195, A199, L334, and L338. These modifications at both TM2-TM11 and TM5-TM8 abrogated transport (SI Appendix, Fig. S5), indicating that size of the lateral openings is important. It is possible that tryptophan substitutions at F87, G440, G444, V195, A199, and L334 sterically interfered with alternating access. To test the importance of the hydrophobic character of these residues lining the lateral openings near the extracellular face of Mfsd2a, we substituted aliphatic and aromatic residues lining TM2-TM11 (F87, L443, G440, and G444) and TM5-TM8 (A199, V195, L334, and L338) with serine. Remarkedly, only the introduction of F87Y and G440S lining TM2-TM11, but not V195S, L334S, and L338S at TM5-TM8, disrupted LPC transport (Fig. 4 A and B). These findings raise the possibility that the acyl chain of LPC might enter through or interact within the region at F87 and G440 at TM2-TM11 (Fig. 4 A and B). This conclusion is similar to what has been shown for LtaA, where the lateral opening formed by TM2-TM11 was demonstrated to have a more important role in transport than the lateral opening at TM5-TM8 (38).
Fig. 4.
Testing the importance of residues lining the extracellular facing half of TM2-TM11 and TM5-TM8 in LPC transport. (A) The model of the outward-open gMfsd2a structure (PDB: 798N). TM2 is in teal, TM11 in blue, TM5 in red, and TM8 in orange. Gray sticks represent residues that were mutated. Residues correspond to human Mfsd2a numbering. (B) Quantification of transport of indicated mutants using a single-cell Flow cytometry assay. The location of residues lining TM2-TM11 and TM5-TM8 are indicated above the graph. Data are represented as the mean ± SD, with each dot representing a single analyzed cell with approximately 1,000 cells analyzed per construct. The dotted line represents the average transport level of WT. See SI Appendix, Fig. S4 for primary flow data.
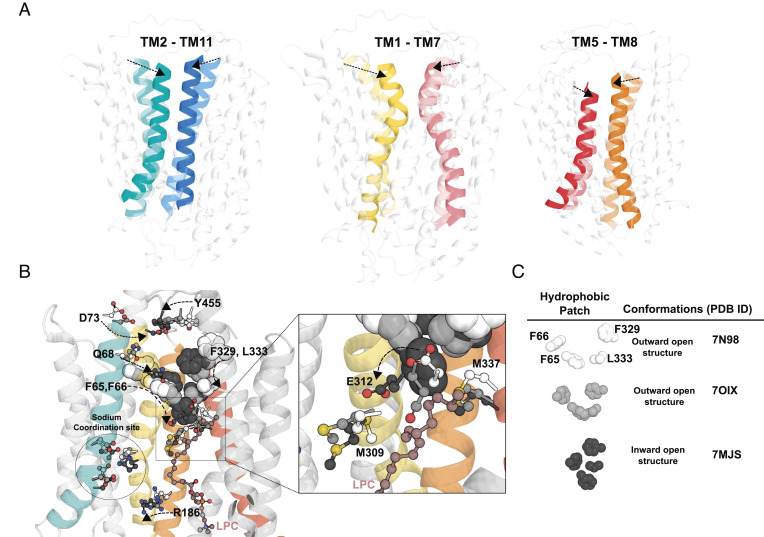
On closer examination of the central cavity, the charged and hydrophilic residues are mostly located at the N-terminal domain near to the cytoplasmic side that shows limited similarity to LtaA, where the central charged residue E32 in LtaA plays an important role for proton-dependent transport (38). However, Mfsd2a is a sodium-coupled transporter and the central cavity of the outward-open mMfsd2a structure shares similarities with sodium-binding and substrate translocation residues of MelB (with a rmsd ~4.07 Å), where the sodium binding site of hMfsfd2a consists of D93, D97, T154, and other charged residues such as K436, R186, and R90 (MelB corresponding residues: D55, D59, T121, K377, R149, and R52) (SI Appendix, Fig. S6B) (42). To determine whether other residues in the central cavity are required for transport activity, we mutated R326, R458, Y455, Q68, N314A, Y130F, E155A, Y151A, T194V, Q57, F308, R90, E312, M309, M337, and R186 to alanine (Fig. 5A). In our mutagenesis of polar residues along the transporter cavity only D73A and Y455A are located at the extracellular face, N314A and E312A are at the middle of the transporter, and R186A is located in proximity to the sodium-binding site and cytoplasmic face showed significant abrogation of LPC transport (Fig. 5 B and C), suggesting that these residues could be involved in sodium or lipid headgroup interactions. E312A in the outward-open and inward-open structures is in close proximity to M309 and M337, respectively (Fig. 5A). A similarly positioned charged residue in the C-domain of LtaA and MelB has not been observed (SI Appendix, Fig. S6). The methyl-thio group of methionine can coordinate with glutamic acid to neutralize the negative charge on the carboxyl group by proton donation (43). Overlaying the three determined structures of Mfsd2a revealed that the position of E312 coordinating with M337 and M309 changes between the outward-open and inward-open structures. M337 is closer to E312 in the outward-open structure and E312 turns toward M309 in the inward-open structure (Fig. 6 A and B). Mutation of E312 to alanine reduced LPC transport in cells and LPS flipping in proteoliposomes (Figs. 3F and 5C), whilst alanine mutations of M337 also resulted in partial disruption of LPC transport (Fig. 5C). Mutation of M337 to isoleucine, which places an aliphatic residue at position 337 that cannot donate a proton to E312, despite isoleucine having similar bond length to methionine, partially disrupted transport (Fig. 5C). Notably, E312 lies directly below a quad of hydrophobic residues, F65, F66, F329, and L333, previously shown to be important in LPC transport (25). The positions of these hydrophobic residues also change greatly between the outward-open and inward-open structures (Fig. 6B). These findings suggest that E312 likely communicates substrate binding (sodium and/or lipid headgroup) to conformational changes between M309 and M337 (Fig. 6B). Moving further down the transport cleft toward the cytoplasmic surface, we find R186 that was previously found to be mutated to tryptophan in a patient with microcephaly (44). Consistent with the predicted pathogenicity of R186W, we found R186W to be nonfunctional (Fig. 5C). Given the functional importance of residues lining TM1 (F65, F66, and D73), TM7 (E312 and N314), and TM8 (L333 and F329), it is possible that these TMs act as gating helices important for the conformational transition from the outward-open to the inward-open states. Such gating helices are also observed in other MFS proteins such as the GLUT1-3 transporters (45).
Fig. 5.
Testing the importance of hydrophilic residues lining the central core of Mfsd2a for LPC transport. (A) The model of the outward-open gMfsd2a structure (PDB: 798N). TM2 is in teal and TM11 in blue, TM5 in orange and TM8 in pink, and TM1 in yellow and TM7 in light pink. Residues mutated are shown in gray sticks; hydrophobic residues previously shown to be important for transport (F65, F66, F329, and L333) are shown in white sticks; residues E312, M309, and M337 are shown in yellow sticks. Residues are labeled with human Mfsd2a numbering. (B and C) Quantification of transport of indicated mutants using a single-cell Flow cytometry assay. Data are represented as the mean ± SD, with each dot representing a single analyzed cell with approximately 1,000 cells analyzed per construct. The dotted line represents the average transport level of WT. See SI Appendix, Fig. S4 for primary flow data.
Fig. 6.
Conformational changes during LPC flipping. (A) Movement of the TM2-TM11, TM1-TM7, and TM5-TM8 from the superposition of the three determined cryo-EM structures, outward-open (Left panel, Protein Data Bank (PDB): 798N), outward-open occluded (Middle panel, PDB: 7OIX), and inward-open (Right panel, PDB: 7MJS) conformations. Left panel showing TM2 and TM11, Middle panel showing TM1 and TM7, Right panel showing TM5 and TM8, with dotted arrows showing conformational changes in the indicated TMs from the outward-open structure to inward-open structure. (B) Leftmost structure showing residues of TM1 (D73, Q68, F65, and F66) and TM7 (F329, L333, and E312) are seen to move toward each other from the outward-open structure 7N98 to the inward-open structure 7MJS (see dotted line arrows). As the residues from both TM1 and TM7 move toward each other to form the inward-open structure, E312 is seen to move from M337 to M309 (see enlarged area indicated by a square). (C) Changes in the relative positions of the four hydrophobic residues F65, F66, F329, and L333 from the outward-open structure 7N98 to the occluded structure 7OIX resulting in the formation of a hydrophobic plug in the inward-open structure 7MJS. Residues D73, Q68, Y455, E312, M309, and M337 are represented in sticks and colored white for outward-open structure 7N98, light gray for outward-open occluded structure 7OIX and dark grey for inward-open structure 7MJS (see Figure Legend). An LPC molecule that is found in the inward-open structure, 7MJS, is represented in maroon ball and sticks.
Insights into the flippase mechanism used by Mfsd2a might be gleaned from structural information on LtaA which was predicted to transport via a trap-and-flip mechanism (32). In LtaA (SI Appendix, Fig. S6A), charged residues (E32, R35, N44, and N162) are mostly confined in the N-domain and aliphatic residues (L237, L296, L300, L309, and I316) forming the hydrophobic core within the transporter. Mfsd2a also displays a similar amphipathic transport cavity and might adopt a similar transport mechanism as LtaA. Based on Mfsd2a cryo-EM structures together with our functional studies, polar residues closer to the extracellular side, namely Q68, D73, and Y455, could be important in sodium ion and LPC headgroup binding while we would predict that the LPC acyl chain interacts with the hydrophobic residue F87 of TM2. In the outward open structure, the hydrophobic patch comprising residues F65, F66, F329, and L333 could form an acyl chain binding site to capture the LPC from the surrounding membrane (Fig. 6 B and C and SI Appendix, Fig. S7). Charged residue E312 lies directly below this hydrophobic patch in the outward open structure and could be the next charged residue to orient the LPC headgroup or sodium ion toward the designated sodium-binding site composed of D93, D97, and K436 (1, 21, 41, 46). Indeed, E312 and the sodium-binding site have been observed to have significant interactions with sodium ions in Molecular Dynamic modeling studies (25). The hydrophobic patch of residues that were seen to form a plug in the inward-open structure (Fig. 6 B and C) (25) could be a site that holds in place the LPC acyl tail while sodium moves with the LPC headgroup downward constituting the inversion or flipping step in the LPC transport process (SI Appendix, Fig. S7). This hydrophobic domain within the core of Mfsd2a consists of a greater number of aromatic residues (F65, F66, F329, and L333) than LtaA (Fig. 6 B and C and SI Appendix, Fig. S6A), that could be important features for substrate selectivity between these two transporters (25, 26, 32).
The mechanisms by which MFS proteins transport lysolipids are currently understudied relative to other well-known MFS proteins that transport hydrophilic substates, such as GlpT, LacY, MelB, and GLUTs (41, 43, 46–48). Demonstration of substrate transport using proteoliposomes has been achieved for many of these transporters that supports the underlying mechanism of rocker-switch alternating access, although variations to this mechanism have been described (14, 15, 45, 49) Based on cryo-EM structures of Mfsd2a and mutagenesis studies, sodium-dependent LPC transport mediated by Mfsd2a has been proposed to be via a flippase type mechanism (1, 21, 25, 26) (SI Appendix, Fig. S7). Crucially, direct biochemical evidence that Mfsd2a is a flippase was lacking. Proteoliposome-based assays are difficult to develop for membrane proteins, and Mfsd2a poses a particular technical challenge because its LPC ligand is amphipathic and spontaneously associates with lipid bilayers making it difficult to measure transport over background membrane binding. In the current study, we overcame these challenges by development of a unique fluorescence-based assay that detects the movement of the LPS headgroup of a native LPS as it transits from the outer membrane leaflet to the inner leaflet of proteoliposomes. This assay recapitulates the transport characteristics of Mfsd2a measured in cell-based transport assays, namely its sodium dependency and lysolipid acyl chain length constraints, and provides direct biochemical evidence that Mfsd2a functions as a lysolipid flippase. It is possible that our assay could be adapted to study the biochemical activity of Spns1, Spns2, and Mfsd2b-MFS proteins that might also function as lipid flippases (17–20).
Materials and Methods
Purification of Gallus gallus Mfsd2a Mutants in Insect Cells.
G. gallus Mfsd2a (gMfsd2a), codon optimized for expression in insect cells, was subcloned into a pFastbac vector containing a TEV cleavage site followed by a His-Tag containing 10 Histidines as previously described (25). Insect cell viruses for gMfsd2a WT and its mutants were generated as described previously to induce protein production in SF9 cells (25).
LPS-Oleate Transport Assay Using Mass Spectrometry.
LPS-oleate transport was conducted on Mfsd2a-transfected HEK293 cells, and lipids were extracted and analyzed with mass spectrometry as described in SI Appendix, Methods. Expression of Mfsd2a in transfected cells was analyzed by western blot.
Single-Cell Transport Assay.
Half a million cells were seeded into a well of 12 wells for 24 h. Cells were transfected with 2 μg of plasmid and Lipofectamine 2000 (Life Technologies) as per the manufacturer’s protocol for 6 h and replaced with fresh Dulbecco's Modified Eagle Medium (DMEM) (Gibco) High glucose, 10% FBS, and 1% penicillin–streptomycin. Transfected cells were allowed to grow for another 24 h prior to transport assay. Cells were washed once with charcoal stripped DMEM with no fetal bovine serum (FBS), and then, increasing concentrations of LPC-NBD (16:1 and 12:0) were added to the transfected cells and incubated in a 37 °C incubator with 5% CO2 for 30 min. Single cells were then obtained by trypsinization with Tryple (GIBCO) and reaction neutralized with PBS containing 0.5% fatty-acid free BSA and 2% FBS and analyzed with BD Fortessa. Single cells were gated and analyzed for mCherry and NBD-positive populations. See SI Appendix, Methods for more details.
Proteoliposome Preparation.
Briefly, 20 mg 1-palmitoyl-2-oleoly-sn-glycero-3phosphocholine (POPC) and 20 mol percent cholesterol (Avanti) were thoroughly dried down and reconstituted in 1 mL of 100 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), KOH, pH 7.5 buffer at 37 c shaking incubator at 220 rpm for 1.5 h. Then, 1.5% b-octylglucoside (Anatrace) was added to the lipid mix and dialyzed with a 100-kDa molecular weight cutoff dialysis tubing (Spectra-Por) overnight at 4 °C. In addition, 0.25 μM PSVue550 (Mtarget) was added to liposomes destabilized with 0.11% Triton-X and nutated at room temperature for 20 min, followed by dropwise addition of 50 μg of purified proteins that had been buffer exchanged by seven consecutive additions with 20 mM HEPES, KOH, pH 7.5, 140 mM KCl buffer containing 0.05% dodecylmaltoside using 50 kDa molecular weight cut-off Amicon (Merck) and incubated for 30 min at room temperature. For more information, see SI Appendix, Methods.
Flippase Assay.
Each well was preloaded with reaction buffer, with the different assay conditions and mixed well. Proteoliposomes were then added into each well of a 96-well glass-bottomed plate (Perkin-Elmer) and scanned on the Azure Biosystems Imager using 550 nm excitation and 610 nm emission, and the reaction was followed by scanning at the start of the reaction (0 time point) followed by every 5 min for 15 min. The images were quantified using the Azure bio-imager software AzureSpot and plotted with Prism (Graphpad). To calculate the relative activity to WT gMFSD2A, obtained intensity is normalized to the highest intensity (fmax) obtained by gWT proteoliposome in the experiments. For dose experiments, the highest intensity is subtracted from baseline and plotted. Curves were fitted to the Michaelis–Menten equation to derive Vmax and the transport constant, Km.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
WT Mfsd2a flippase activity: 5-minute scans at 550 nm and 610 nm emission for a total of 15 minutes for gWT proteoliposomes under the following three conditions: leftmost three vertical lanes containing 140 mM NaCl, middle three vertical lanes containing 140 mM NaCl and 25 μM LPS, rightmost containing only 25 μM LPS. Images are in three 700 ms loops encompassing four time points (0, 5,10,15 min).
Specificity of Mfsd2a flippase activity: 5-minute scans at 550 nm and 610 nm emission for a total of 15 minutes for empty liposomes, gWT and gD92A proteoliposomes in wells containing 50 μM LPS-oleate and 140 mM of NaCl. Images are in three 700 ms loops at four time points (0, 5,10,15 min).
Sodium-dependent Mfsd2a flippase activity: 5-minute scans at 550 nm excitation and 610 nm emission for a total of 15 minutes for gWT proteolipsomes in 50 mM NaCl (wells 1, 2, 3), 25 mM NaCl (wells 4, 5, 6), 1 mM NaCl (wells 7, 8, 9), 0 mM NaCl (for wells 10,11,12). Images are in three 700 ms loops of 4 time points (0, 5,10,15 min).
Acknowledgments
Lysophosphatidylcholine-NBD (LPC-NBD) (16:1 and 12:0) was a gift from Travecta Therapeutics. We are grateful to Joanne Ong Wei En from Duke-National University of Singapore (NUS) Laboratory for Translational and Molecular Imaging for assisting in the use of the Azure Biosystems and Lim Ze Ming from the Duke-NUS Flow Facility for assistance in the use of the BDFortessa. This work was supported by grants from the National Research Foundation, Ministry of Health, and Ministry of Education, Singapore (NRF-NRFI2017-05, MOH-000217, MOE-T2EP30220-0001, and MOE2019-T2-1-031 to D.L.S.), a Goh Cardiovascular Research Award, and a grant from the Human Frontier Science Program (LT000733/2020-L to A.C.Y.K.), and a grant from the NIH, USA (R01GM122759 to L.G.).
Author contributions
G.-L.C. and D.L.S. designed research; G.-L.C., B.C.T., R.Y.J.L., M.H., C.-F.C., B.H.W., A.C.Y.K., and M.D. performed research; M.R.W., L.G., and F.T. contributed new reagents/analytic tools; G.-L.C., B.H.W., M.D., F.T., and D.L.S. analyzed data; and G.-L.C. and D.L.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. S.-Y.L. is a guest editor invited by the Editorial Board.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Nguyen L. N., et al. , Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature 509, 503–506 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Wong B. H., et al. , Mfsd2a is a transporter for the essential omega-3 fatty acid DHA in eye and important for photoreceptor cell development. J. Biol. Chem. 291, 10501–10514 (2016), 10.1074/jbc.M116.721340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Quek D. Q., Nguyen L. N., Fan H., Silver D. L., Structural insights into the transport mechanism of the human sodium-dependent lysophosphatidylcholine transporter Mfsd2a. J. Biol. Chem. 291, 9383–9394 (2016), 10.1074/jbc.M116.721035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piccirillo A. R., et al. , The lysophosphatidylcholine transporter MFSD2A is essential for CD8(+) memory T cell maintenance and secondary response to infection. J. Immunol. 203, 117–126 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong B. H., et al. , Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid (DHA) in eye and is important for photoreceptor cell development. J. Biol. Chem. 291, 10501–10514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong B. H., et al. , The lipid transporter Mfsd2a maintains pulmonary surfactant homeostasis. J. Biol. Chem. 298, 101709 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger J. H., Charron M. J., Silver D. L., Major facilitator superfamily domain-containing protein 2a (MFSD2A) has roles in body growth, motor function, and lipid metabolism. PLoS One 7, e50629 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabula Sapiens C., et al. , The Tabula Sapiens: A multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alakbarzade V., et al. , A partially inactivating mutation in the sodium-dependent lysophosphatidylcholine transporter MFSD2A causes a non-lethal microcephaly syndrome. Nat. Genet. 47, 814–817 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Guemez-Gamboa A., et al. , Inactivating mutations in MFSD2A, required for omega-3 fatty acid transport in brain, cause a lethal microcephaly syndrome. Nat. Genet. 47, 809–813 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harel T., et al. , Homozygous mutation in MFSD2A, encoding a lysolipid transporter for docosahexanoic acid, is associated with microcephaly and hypomyelination. Neurogenetics 19, 227–235 (2018), 10.1007/s10048-018-0556-6. [DOI] [PubMed] [Google Scholar]
- 12.Scala M., et al. , Biallelic MFSD2A variants associated with congenital microcephaly, developmental delay, and recognizable neuroimaging features. Eur. J. Hum. Genet. 28, 1509–1519 (2020), 10.1038/s41431-020-0669-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan J. P., et al. , The lysolipid transporter Mfsd2a regulates lipogenesis in the developing brain. PLoS Biol. 16, e2006443 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan N., Structural biology of the major facilitator superfamily transporters. Annu. Rev. Biophys. 44, 257–283 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Drew D., Boudker O., Shared molecular mechanisms of membrane transporters. Annu. Rev. Biochem. 85, 543–572 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Lin Y., Deepak R., Zheng J. Z., Fan H., Zheng L., A dual substrate-accessing mechanism of a major facilitator superfamily protein facilitates lysophospholipid flipping across the cell membrane. J. Biol. Chem. 293, 19919–19931 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vu T. M., et al. , Mfsd2b is essential for the sphingosine-1-phosphate export in erythrocytes and platelets. Nature 550, 524–528 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi N., et al. , MFSD2B is a sphingosine 1-phosphate transporter in erythroid cells. Sci. Rep. 8, 4969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawahara A., et al. , The sphingolipid transporter spns2 functions in migration of zebrafish myocardial precursors. Science 323, 524–527 (2009). [DOI] [PubMed] [Google Scholar]
- 20.He M., et al. , Spns1 is a lysophospholipid transporter mediating lysosomal phospholipid salvage. Proc. Natl. Acad. Sci. U.S.A. 119, e2210353119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quek D. Q., Nguyen L. N., Fan H., Silver D. L., Structural insights into the transport mechanism of the human sodium-dependent lysophosphatidylcholine transporter MFSD2A. J. Biol. Chem. 291, 9383–9394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pardridge W. M., A historical review of brain drug delivery. Pharmaceutics 14, 1283 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pardridge W. M., Blood-brain barrier delivery for lysosomal storage disorders with IgG-lysosomal enzyme fusion proteins. Adv. Drug Deliv. Rev. 184, 114234 (2022). [DOI] [PubMed] [Google Scholar]
- 24.Pardridge W. M., The blood-brain barrier: Bottleneck in brain drug development. NeuroRx 2, 3–14 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cater R. J., et al. , Structural basis of omega-3 fatty acid transport across the blood-brain barrier. Nature 595, 315–319 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wood C. A. P., et al. , Structure and mechanism of blood-brain-barrier lipid transporter MFSD2A. Nature 596, 444–448 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maria Martinez-Molledo E. N. N. R., Structural insights into the lysophospholipid brain uptake mechanism and its inhibition by syncytin-2. Nat. Struct. Mol. Biol. 29, 604–612 (2022). [DOI] [PubMed] [Google Scholar]
- 28.Pomorski T., Menon A. K., Lipid flippases and their biological functions. Cell Mol. Life Sci. 63, 2908–2921 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rush J. S., Role of flippases in protein glycosylation in the endoplasmic reticulum. Lipid. Insights 8, 45–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang X., et al. , Engineered XylE as a tool for mechanistic investigation and ligand discovery of the glucose transporters GLUTs. Cell Discov. 5, 14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seigneuret M., Devaux P. F., ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: Relation to shape changes. Proc. Natl. Acad. Sci. U.S.A. 81, 3751–3755 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lambert E., Mehdipour A. R., Schmidt A., Hummer G., Perez C., Evidence for a trap-and-flip mechanism in a proton-dependent lipid transporter. Nat. Commun. 13, 1022 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helenius J., et al. , Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415, 447–450 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Watanabe R., Sakuragi T., Noji H., Nagata S., Single-molecule analysis of phospholipid scrambling by TMEM16F. Proc. Natl. Acad. Sci. U.S.A. 115, 3066–3071 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiVittorio K. M., et al. , Synthetic peptides with selective affinity for apoptotic cells. Org. Biomol. Chem. 4, 1966–1976 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Rice D. R., Clear K. J., Smith B. D., Imaging and therapeutic applications of zinc(ii)-dipicolylamine molecular probes for anionic biomembranes. Chem. Commun. (Camb) 52, 8787–8801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hariharan P., Guan L., Thermodynamic cooperativity of cosubstrate binding and cation selectivity of Salmonella typhimurium MelB. J. Gen Physiol. 149, 1029–1039 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang B., et al. , Structure of a proton-dependent lipid transporter involved in lipoteichoic acids biosynthesis. Nat. Struct. Mol. Biol. 27, 561–569 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menon I., et al. , Opsin is a phospholipid flippase. Curr. Biol. 21, 149–153 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vishwakarma R. A., et al. , New fluorescent probes reveal that flippase-mediated flip-flop of phosphatidylinositol across the endoplasmic reticulum membrane does not depend on the stereochemistry of the lipid. Org. Biomol. Chem. 3, 1275–1283 (2005). [DOI] [PubMed] [Google Scholar]
- 41.Ethayathulla A. S., et al. , Structure-based mechanism for Na(+)/melibiose symport by MelB. Nat. Commun. 5, 3009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guan L., Hariharan P., X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB. Commun. Biol. 4, 931 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaback H. R., Sahin-Toth M., Weinglass A. B., The kamikaze approach to membrane transport. Nat. Rev. Mol. Cell Biol. 2, 610–620 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Khuller K., et al. , MFSD2A-associated primary microcephaly - Expanding the clinical and mutational spectrum of this ultra-rare disease. Eur. J. Med. Genet 64, 104310 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Drew D., North R. A., Nagarathinam K., Tanabe M., Structures and general transport mechanisms by the major facilitator superfamily (MFS). Chem. Rev. 121, 5289–5335 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katsube S., Liang R., Amin A., Hariharan P., Guan L., Molecular basis for the cation selectivity of salmonella typhimurium melibiose permease. J. Mol. Biol. 434, 167598 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deng D., et al. , Crystal structure of the human glucose transporter GLUT1. Nature 510, 121–125 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N., Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301, 616–620 (2003). [DOI] [PubMed] [Google Scholar]
- 49.Law C. J., Maloney P. C., Wang D. N., Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 62, 289–305 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
WT Mfsd2a flippase activity: 5-minute scans at 550 nm and 610 nm emission for a total of 15 minutes for gWT proteoliposomes under the following three conditions: leftmost three vertical lanes containing 140 mM NaCl, middle three vertical lanes containing 140 mM NaCl and 25 μM LPS, rightmost containing only 25 μM LPS. Images are in three 700 ms loops encompassing four time points (0, 5,10,15 min).
Specificity of Mfsd2a flippase activity: 5-minute scans at 550 nm and 610 nm emission for a total of 15 minutes for empty liposomes, gWT and gD92A proteoliposomes in wells containing 50 μM LPS-oleate and 140 mM of NaCl. Images are in three 700 ms loops at four time points (0, 5,10,15 min).
Sodium-dependent Mfsd2a flippase activity: 5-minute scans at 550 nm excitation and 610 nm emission for a total of 15 minutes for gWT proteolipsomes in 50 mM NaCl (wells 1, 2, 3), 25 mM NaCl (wells 4, 5, 6), 1 mM NaCl (wells 7, 8, 9), 0 mM NaCl (for wells 10,11,12). Images are in three 700 ms loops of 4 time points (0, 5,10,15 min).
Data Availability Statement
All study data are included in the article and/or SI Appendix.