Significance
The pathogenesis of multiple myeloma (MM), a fatal malignancy of bone marrow plasma cells, is initiated by genetic aberrations in germinal center (GC) B cells, including well-defined chromosomal translocations. The latter define distinct MM subgroups with different clinical courses, prognosis, and gene expression profiles. Here, we model two such subgroups in mice, by targeting the expression of the genes involved in the subgroup-specific translocations, together with a secondary, common MM driver mutation, into GC B cells. Strikingly, in both cases, MM-like diseases develop, but their gene expression profiles differ, resembling those of the human MM subgroups. Distinct chromosomal translocations in GC B cells thus govern the development of distinct MM disease entities, for which we present preclinical models.
Keywords: multiple myeloma, chromosomal translocations, translocation subgroups, Cyclin D1/MMSET, conditional mouse models
Abstract
Multiple myeloma (MM), a tumor of germinal center (GC)-experienced plasma cells, comprises distinct genetic subgroups, such as the t(11;14)/CCND1 and the t(4;14)/MMSET subtype. We have generated genetically defined, subgroup-specific MM models by the GC B cell-specific coactivation of mouse Ccnd1 or MMSET with a constitutively active Ikk2 mutant, mimicking the secondary NF-κB activation frequently seen in human MM. Ccnd1/Ikk2ca and MMSET/Ikk2ca mice developed a pronounced, clonally restricted plasma cell outgrowth with age, accompanied by serum M spikes, bone marrow insufficiency, and bone lesions. The transgenic plasma cells could be propagated in vivo and showed distinct transcriptional profiles, resembling their human MM counterparts. Thus, we show that targeting the expression of genes involved in MM subgroup-specific chromosomal translocations into mouse GC B cells translates into distinct MM-like diseases that recapitulate key features of the human tumors, opening the way to a better understanding of the pathogenesis and therapeutic vulnerabilities of different MM subgroups.
Multiple myeloma (MM) is a tumor of antibody-producing plasma cells that expand within the bone marrow (BM). The disease shows a stepwise progression from a premalignant stage (monoclonal gammopathy of undetermined significance, MGUS) to overt MM, commonly defined by the occurrence of CRAB symptoms (i.e., hypercalcemia, renal failure, anemia, bone lesions) (1, 2). Recurrent early MGUS-initiating genetic events include chromosomal translocations into the immunoglobulin heavy chain (IgH) locus (3–5), which typically arise during the germinal center (GC) reaction (6, 7) and frequently affect CCND1, t(11;14), or the histone methyltransferase MMSET, t(4;14), in 20% and 15% of MM cases, respectively (8, 9). The MGUS to MM transition requires additional genetic aberrations, such as c-MYC translocations, aberrant NF-κB or MAPK signaling, and p53 lesions (10, 11). Significantly, the chromosomal translocations are usually already detectable at the MGUS stage of the disease, whereas this is not the case for genetic aberrations resulting in NF-κB activation, qualifying the latter as secondary events in MM pathogenesis (12–14). This complex genetic architecture characterizes MM as a heterogeneous disease, in line with gene expression studies that separate MM patients into 7 to 10 distinct subgroups (15–17). Of these, four are associated with a particular IgH translocation, e.g., t(4;14) with the MMSET group, implying an important role of the early genetic aberrations for subclassification. The significance of the primary events is further supported by recent in-depth single-cell analyses that detect common overexpressed pathways, such as CCND1 and MMSET, shared across subgroups of otherwise highly heterogeneous MM tumors (18). Given that the distinct MM subclasses differ in terms of gene expression, tumor biology, and clinical outcome, new therapeutic strategies have to take into account the heterogeneity of the disease. One approach to delineate and eventually target subgroup-specific disease mechanisms is the creation of genetically defined animal models based on the introduction of subgroup-defining genetic aberrations into the mouse genome. Most of the existing transgenic models do not recapitulate the heterogeneity of human MM as they are either based on the ectopic expression of plasma cell-related genes like XBP1s (19), the overexpression of signaling molecules involved in plasma cell survival such as IL-6 (20) and gp-130 (21), or the enforced expression of genetic events that are associated with the MGUS to MM progression like c-MYC (22) alone or in combination with the NrasQ61R mutation (23)—all of which promote the expansion and malignant transformation of plasma cells but do not consider the molecular characteristics of the various MM subclasses, emphasizing the need for mouse models of genetically distinct MM subgroups (24). While a few transgenic mouse strains are available that carry transgenes involved in MM subgroup-defining chromosomal translocations such as Maf (25), MafB (26), and Ccnd1 (27), these transgenes are expressed from hematopoietic stem and progenitor cells (26) or early B cell stages (25, 27) onward and not targeted to GC B cells, where the respective human IgH translocations take place in MM pathogenesis (6, 7). Here, we describe the generation and characterization of subgroup-specific MM models by the conditional overexpression of mouse Ccnd1 or MMSET together with constitutive NF-κB signaling from the GC B cell stage onward. MMSET/Ikk2ca and Ccnd1/Ikk2ca mice developed an MM-like disease with age presenting with a pronounced and clonally restricted plasma cell expansion, serum M proteins, and pathologies commonly associated with MM. The outgrowing mouse plasma cells demonstrated transcriptional profiles of human (subgroup-specific) MM tumors and could be propagated into secondary hosts, introducing our models as valuable tools to study and assess subgroup-specific therapeutic concepts for MM in the future.
Results
Generation and Characterization of the Conditional MM Mouse Strains.
To establish genetically defined, subgroup-specific models for the two most frequent primary translocation events, t(11;14) and t(4;14), we generated conditional gain-of-function alleles for mouse Ccnd1 and MMSET in the Rosa26 locus (Fig. 1 A and B and SI Appendix, Fig. S1 A–E). To mimic the secondary event necessary for MGUS-MM transition, we combined these with the R26 Ikk2castopF allele (28) that expresses a constitutively active Ikk2 mutant, recapitulating the NF-κB activation commonly seen in human MM, either due to genetic aberrations (29–32) or BM microenvironment-mediated signaling (33) (Fig. 1 A and B). To restrict transgene activation in vivo to GC B cells and their progeny, R26 Ccnd1stopF (hereafter termed Ccnd1), R26 MMSETstopF (hereafter: MMSET), and R26 Ikk2castopF (hereafter: Ikk2ca) mice were crossed to the Cγ1-cre strain (34) (Fig. 1B). Notably, all single- (Ccnd1, MMSET or Ikk2ca) and double-mutant (Ccnd1/Ikk2ca or MMSET/Ikk2ca) mice were able to mount GC and antibody responses upon immunization with the T cell-dependent antigen NP-CGG (4-hydroxy-3-nitrophenylacetyl-chicken-gamma-globulin) (SI Appendix, Fig. S1 F–I). However, while Ccnd1 and MMSET single-mutant mice showed a similar response as BFP control mice, activation of NF-κB signaling significantly enhanced plasma cell differentiation at the expense of the GC reaction (SI Appendix, Fig. S1 H and I)—an expected result that is probably due to upregulation of Prdm1 in Ikk2ca-expressing GC B cells (35). Since the GC B cell-specific induction of the chosen human MM candidate genes is compatible with terminal differentiation of mouse B cells, all (newly generated) transgenic strains were considered appropriate to study myeloma pathogenesis.
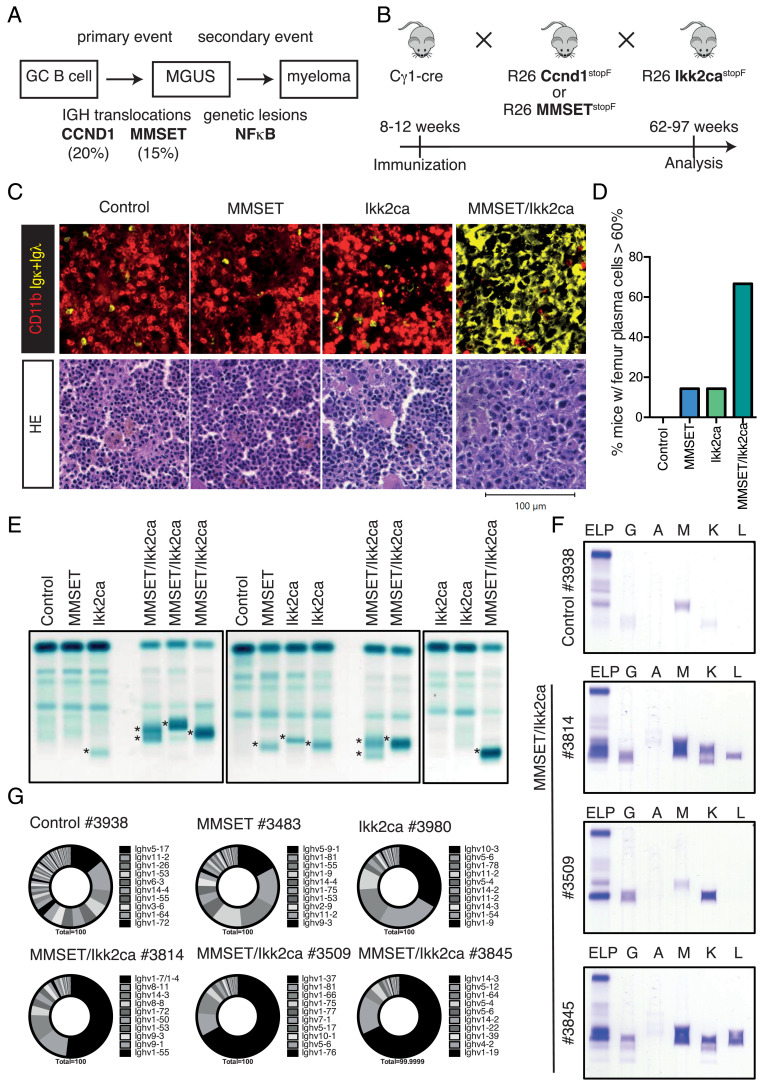
Fig. 1.
Aged MMSET/Ikk2ca mice develop a clonally restricted plasma cell expansion. (A) Simplified scheme of multistep human MGUS-MM development. (B) Outline for the generation of MM subgroup-specific mouse cohorts. Control (i.e., Cγ1-cre only, floxed R26 alleles or Cγ1-cre; R26 BFPstopF mice), single-mutant (Ccnd1, MMSET, or Ikk2ca), and double-mutant (Ccnd1/Ikk2ca or MMSET/Ikk2ca) mice were immunized once with NP-CGG at 8 to 12 wk of age, monitored for tumor development and killed upon onset of disease-defining endpoints (between 62 and 97 wk of age). (C) Upper panel: representative immunofluorescence images of femur sections stained with α-CD11b (red, myeloid cells) and α-Igκ/α-Igλ (yellow, plasma cells). Lower panel: representative HE-stained femur sections. (D) Graph depicts the percentage of mice that demonstrated ≥60% plasma cell infiltration within the femur section by pathologic assessment following HE staining; summary of SI Appendix, Table S1. (E) Representative serum protein electrophoresis (SPEP) of MMSET cohort mice; M spikes are marked with an asterisk. (F) SPEP coupled with immunofixation to determine the immunoglobulin isotype of the detected M proteins. (G) Ighv gene usage in sorted BM plasma cells of MMSET cohort mice. Pie charts show the fraction of individual Ighv genes among all annotated Ighv gene reads as determined by RNA-Seq. The 10 most dominantly expressed Ighv genes are listed (ordered clockwise).
The GC B Cell-Specific Coexpression of MMSET and Ikk2ca Induces a Prominent and Clonally Restricted Plasma Cell Outgrowth with Age.
To analyze whether the GC B cell-specific activation of Ccnd1 or MMSET alone or in combination with Ikk2ca results in MM development, mice were immunized once and then aged to monitor tumor formation (Fig. 1B). Within our t(4;14) cohort, MMSET/Ikk2ca mice reached disease-defining end-points between 72 and 97 wk of age, presenting with splenomegaly (SI Appendix, Fig. S2A) and porous bones (Fig. 2B). HE staining and immunofluorescence analyses of femur sections revealed a pronounced expansion (≥60% infiltration) of large, immunoglobulin light chain-positive plasma cells with abundant cytoplasm and eccentric nuclei in the vast majority of MMSET/Ikk2ca samples, compared to normal femur morphology in most age-matched control and single-mutant mice (Fig. 1 C and D and SI Appendix, Table S1). Similarly, plasma cells dominated in MMSET/Ikk2ca splenic sections (SI Appendix, Fig. S2G and Table S1). This was also seen in flow cytometric analysis, where MMSET/Ikk2ca mice showed a significant increase of spleen and BM CD138+TACI+ plasma cells, most of which expressed both reporters (BFP reporting MMSET and GFP indicating Ikk2ca expression), suggesting synergy between the two transgenes (SI Appendix, Fig. S2 C–F). However, the percentage of plasma cells determined by flow cytometry was lower than that observed in histology (SI Appendix, Fig. S2D and Table S1), most likely reflecting rapid cell death ex vivo, consistent with the strong overall reduction of recovered cells (SI Appendix, Fig. S2B). The observed plasma cell expansion of MMSET/Ikk2ca mice was further corroborated by elevated immunoglobulin serum titers (SI Appendix, Fig. S2H) and an enrichment of antibody-secreting BM cells (SI Appendix, Fig. S2I). With respect to antibody secretion, a characteristic hallmark of MM is the accumulation of a monoclonal (M) immunoglobulin in the serum. In line with this, we detected prominent M spikes in MMSET/Ikk2ca mice in contrast to control and single-mutant animals that showed either no or only faint bands in serum protein electrophoresis (Fig. 1E and SI Appendix, Fig. S3A). Subsequent immunofixation revealed that the M proteins were either of the IgG or IgM isotype and that more than one clone contributed to the plasma cell expansion, as most M proteins precipitated with both Igκ and Igλ antiserum, suggestive of a clonally restricted but not monoclonal plasma cell outgrowth in most MMSET/Ikk2ca mice (Fig. 1F and SI Appendix, Fig. S4C). This finding was supported by a limited Ighv gene repertoire in sorted double-mutant CD138+TACI+ BM plasma cells. In the majority of MMSET/Ikk2ca mice, the respective dominant clone represented ≥50% of all Ighv reads identified by RNA-Seq (Fig. 1G and SI Appendix, Fig. S3 B–D). Given our observation that a substantial fraction of MMSET/Ikk2ca plasma cells dies during ex vivo purification, the Ighv gene analysis of sorted cells might underestimate the real clonal restriction. However, we reliably identified shared clones between sorted plasma cells and snap-frozen femur or spleen sections by RNA-Seq and VDJ-PCR (36) (SI Appendix, Fig. S4A and Table S2), as well as matching immunoglobulin isotype expression patterns of sorted plasma cells and the respective serum M proteins (SI Appendix, Fig. S4 B and C)—suggesting that the molecular analysis of sorted plasma cells is representative of the actual repertoire in double-mutant mice. Thus, the coexpression of MMSET and Ikk2ca from the GC B cell stage onward induces a marked expansion of a few plasma cell clones in aged animals.
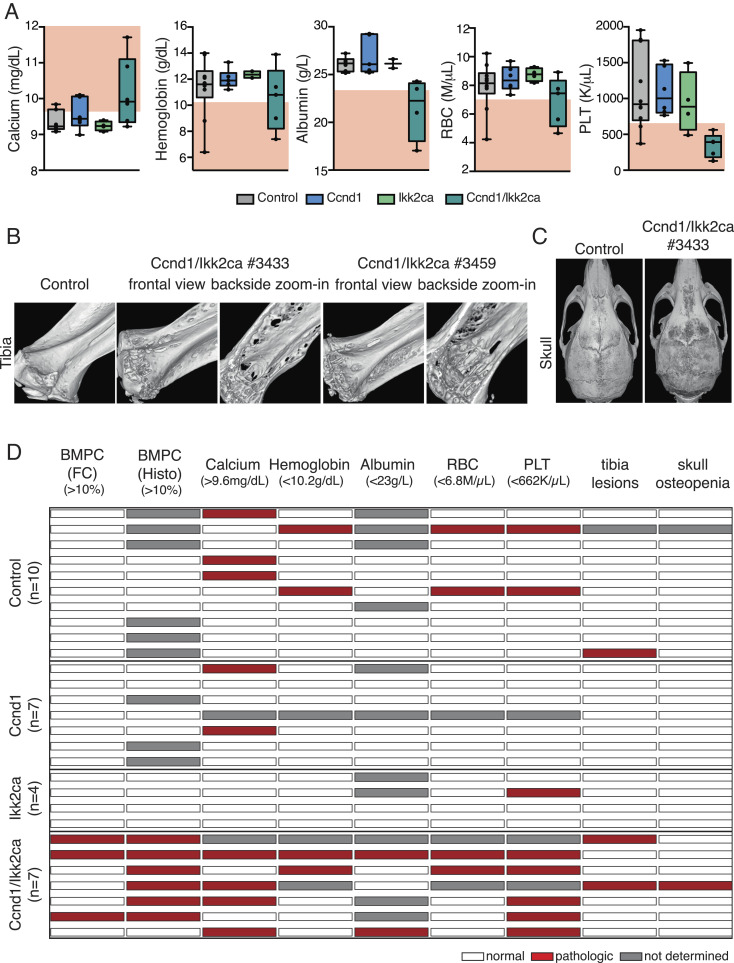
Fig. 2.
The plasma cell outgrowth in aged MMSET/Ikk2ca mice is accompanied with MM-like pathologies. (A) Graphs depicting serum calcium, hemoglobin, serum albumin, red blood cell (RBC), and platelet (PLT) values of MMSET cohort mice. The pathologic range of either parameter (calcium > 9.6 mg/dL; hemoglobin < 10.2 g/dL; albumin* < 23 g/L; RBC < 6.8 M/µL; PLT < 662 K/µL) is marked in red (reference: Jackson Laboratory; 78 wk old C57BL/6J; *the lower limit for albumin had to be adjusted for our cohort to discern healthy and diseased mice for animals >78 wk). (B and C) Representative 3D images reconstructed from µCT scans of tibiae (B) and skulls (C). Tibiae are shown in frontal view (B; Left) and as zoomed-in image (B; Right) to demonstrate that lesions destroyed the cortex. (D) Heatmap summarizing the assessment of selected MM-associated parameters. Columns include from Left to Right: ≥10% of BM plasma cells (BMPC) measured by flow cytometry (FC); ≥10% BMPC in the femur biopsy (histology); serum calcium, hemoglobin, serum albumin, RBC, PLT, tibia lesions, and skull osteopenia. For blood parameters, the pathologic range is indicated in brackets (same reference values as before). Each row represents an individual mouse, ordered by genotype. White marks indicate that the parameter is within the normal range, red marks indicate that the parameter is within the defined pathologic range, and gray marks indicate that the parameter was not measured. For the µCT analysis, red marks indicate mice with ≥3 lesions within the tibiae and diffuse osteopenia of the skull.
A Major Fraction of MMSET/Ikk2ca Mice Shows MM-Associated Disease Phenotypes.
Next, we analyzed the MMSET/Ikk2ca cohort animals for the presence of disease parameters. With regard to classical CRAB criteria, most MMSET/Ikk2ca mice demonstrated increased serum calcium levels (>9.6 mg/dL; 11/15 mice) and signs of anemia such as decreased hemoglobin titers (<10.2 g/dL; 6/15 mice) and a reduction of red blood cells (RBC < 6.8 M/µL; 11/15 mice) (Fig. 2A). Moreover, almost all MMSET/Ikk2ca mice suffered from thrombocytopenia [platelets (PLT) < 662 K/µL; 14/15 mice], which along with the decrease of RBCs indicates disturbed hematopoiesis in the BM (Fig. 2A). In addition, a major fraction of MMSET/Ikk2ca mice presented with hypoalbuminemia (<23 g/L; 10/15 mice) (Fig. 2A), suggestive of an advanced disease stage (37). Another important feature of human MM is the occurrence of bone lesions. µ-computer tomographic (CT) analysis revealed that a substantial fraction of MMSET/Ikk2ca mice showed defined lesions within the tibia (8/16 mice), which destroyed the cortical bone (Fig. 2 B and D). Furthermore, we frequently detected extended areas of lower bone density in the skull of MMSET/Ikk2ca animals, reminiscent of osteopenia (6/16 mice) (Fig. 2 C and D). Thus, the prominent plasma cell outgrowth in MMSET/Ikk2ca mice is accompanied by the development of pathologies commonly associated with human MM. In summary, based on the defined parameters, 14 of 16 MMSET/Ikk2ca mice developed an MM-like disease, one MMSET/Ikk2ca mouse showed a prominent plasma cell outgrowth but could not be further analyzed for hematology and clinical chemistry, and only one mouse did not develop the disease (Fig. 2D). Importantly, longitudinal serum protein electrophoresis was performed for six of the 16 MMSET/Ikk2ca mice and demonstrated the occurrence of the respective M proteins already several weeks before the mice became sick, strongly suggesting the presence of an asymptomatic MGUS phase in MMSET/Ikk2ca animals (SI Appendix, Fig. S5).
The Conditional Activation of Ccnd1 and Ikk2ca Induces a Similar Plasma Cell Phenotype as in Aged MMSET/Ikk2ca Mice.
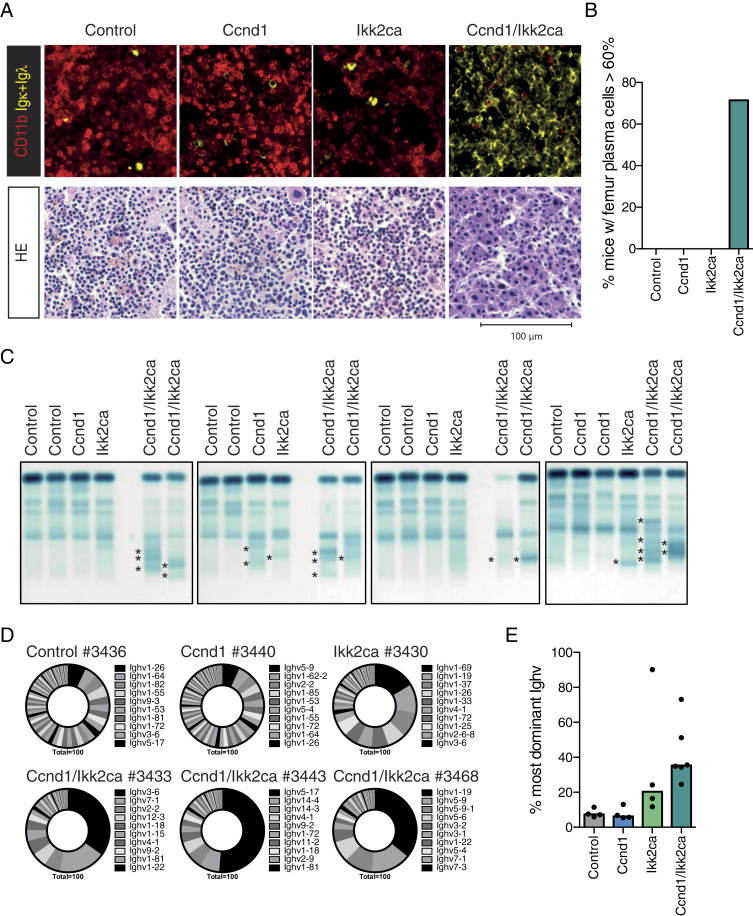
Similar to MMSET/Ikk2ca mice, also Ccnd1/Ikk2ca animals developed a pronounced plasma cell phenotype between 62 and 91 wk of age (Fig. 3 and SI Appendix, Fig. S6). The plasma cell outgrowth was confirmed by histology of femur and spleen sections, demonstrating ≥60% BM plasma cells in five of seven double-mutant mice (Fig. 3 A and B and SI Appendix, Fig. S6G and Table S3), increased immunoglobulin serum titers (SI Appendix, Fig. S6H), and an enrichment of antibody-secreting cells within the long bone-derived BM (SI Appendix, Fig. S6I). As for MMSET/Ikk2ca mice, flow cytometry confirmed a significant expansion of double transgene-expressing (BFP+GFP+) CD138+TACI+ cells, but greatly underestimated their frequency compared to histology (SI Appendix, Fig. S6 C–F). In line with an oligoclonal plasma cell outgrowth, Ccnd1/Ikk2ca mice developed multiple serum M spikes of various Ig isotypes (Fig. 3C and SI Appendix, Fig. S8 B and C), which was further confirmed by the presence of a restricted Ighv gene repertoire determined by RNA-Seq (Fig. 3 D and E and SI Appendix, Fig. S7) and the detection of dominant clones by VDJ-PCR (36) (SI Appendix, Fig. S8A and Table S4). Thus, also the GC B cell-specific activation of Ccnd1 as a primary genetic event and constitutive NF-κB signaling as a secondary genetic lesion induce the expansion of plasma cell clones with age.
Fig. 3.
Ccnd1/Ikk2ca mice show an oligoclonal plasma cell outgrowth with age. (A) Upper panel: representative immunofluorescence images of femur sections stained with α-CD11b (red, myeloid cells) and α-Igκ/α-Igλ (yellow, plasma cells). Lower panel: representative HE-stained femur sections. (B) Graph shows the fraction of mice that demonstrated ≥60% plasma cell infiltration within the femur section by pathologic assessment following HE staining; summary of SI Appendix, Table S3. (C) Representative serum protein electrophoresis (SPEP) of Ccnd1 cohort mice; M spikes are marked with an asterisk. (D) Ighv gene usage in sorted CD138+TACI+ BM plasma cells of Ccnd1 cohort mice. Pie charts show the fraction of individual Ighv genes among all annotated Ighv gene reads as determined by RNA-Seq. The 10 most dominantly expressed Ighv genes are listed (ordered clockwise). (E) Graph illustrating the percentage of the most dominant Ighv gene per mouse of the indicated genotypes.
Ccnd1/Ikk2ca Animals Also Present with MM-Associated Pathologies.
With regard to clinical parameters, Ccnd1/Ikk2ca mice presented with similar disease phenotypes as MMSET/Ikk2ca animals. A major fraction demonstrated increased serum calcium levels (4/6 mice) with two mice reaching hypercalcemia values of ≥11 mg/dL (Fig. 4A). Furthermore, anemia was observed in 2 of 5 Ccnd1/Ikk2ca animals as judged by decreased hemoglobin levels and RBC counts (Fig. 4A). Interestingly, as for MMSET/Ikk2ca mice, two prominent features of Ccnd1/Ikk2ca animals were thrombocytopenia (5/5 mice), which likely indicates progressive BM failure, and hypoalbuminemia (2/4 mice) (Fig. 4A). Moreover, two Ccnd1/Ikk2ca mice exhibited tibia lesions, accompanied in one case by skull osteopenia (Fig. 4 B and C). Thus, also the coactivation of Ccnd1 and Ikk2ca from the GC B cell stage onward translated into an MM-like disease with age in six of seven double-mutant mice (Fig. 4D). Longitudinal serum protein electrophoresis revealed the existence of M spikes some weeks prior to the final analysis, indicative of an indolent MGUS stage also in Ccnd1/Ikk2ca animals (SI Appendix, Fig. S9).
Fig. 4.
Ccnd1/Ikk2ca mice present with MM-associated pathologies. (A) Graphs depicting serum calcium, hemoglobin, serum albumin, red blood cell (RBC), and platelet (PLT) values of Ccnd1 cohort mice (as in Fig. 2A). (B and C) 3D images reconstructed from µCT scans of tibiae (B) and skulls (C). Tibiae are shown in frontal view (B, Left) and as zoomed-in image (B, Right) to demonstrate that lesions destroyed the cortical bone. (D) Heatmap summarizing the assessment of selected MM-associated parameters. Columns include from Left to Right: ≥10% of BM plasma cells (BMPC) measured by flow cytometry (FC); ≥10% BMPC in the femur biopsy (histology); serum calcium, hemoglobin, serum albumin, RBC, PLT, tibia lesions, and skull osteopenia. For blood parameters the pathologic range is indicated in brackets (as in Fig. 2D). Each row represents an individual mouse, ordered by genotype. White marks indicate that the parameter is within the normal range, red marks indicate that the parameter is within the defined pathologic range and gray marks indicate that the parameter was not measured. For the µCT analysis, red marks indicate mice with ≥3 lesions within the tibiae and diffuse osteopenia of the skull.
The Transgenic Mouse Plasma Cells Share Gene Expression Signatures with Human MM Tumor Cells and Exhibit Subgroup-Specific Gene Expression Profiles.
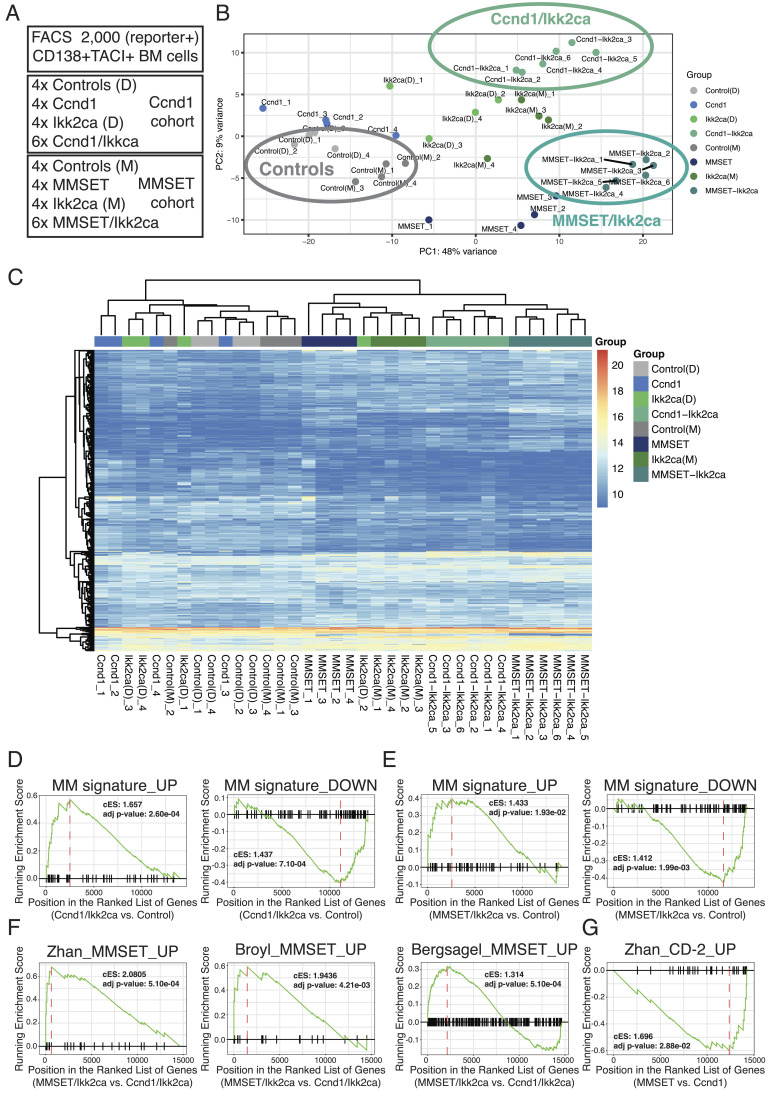
To link the new mouse models to human MM, we sorted BM plasma cells from aged control, single-mutant (Ccnd1, MMSET, or Ikk2ca), or double-mutant (Ccnd1/Ikk2ca or MMSET/Ikk2ca) mice and performed RNA-Seq (Fig. 5A). Principal component (PC) analysis based on the top 500 genes showing the highest variability in expression across all samples accurately clustered the samples according to genotype (Fig. 5B). Double-transgenic cells were clearly separated from control BM plasma cells by PC1, suggestive of an aberrant disease-related gene expression profile. Moreover, PC2 further divided double-mutant cells into Ccnd1/Ikk2ca and MMSET/Ikk2ca populations, reminiscent of the emergence of distinct MM subgroups upon gene expression profiling of human MM samples (38) (Fig. 5B). Likewise, unsupervised hierarchical clustering positioned all double-transgenic BM plasma cells into one branch, with clearly defined sub-branches for Ccnd1/Ikk2ca and MMSET/Ikk2ca samples (Fig. 5C). We then employed gene set enrichment analysis (GSEA) using the tmod algorithm (39) to determine whether human MM signature genes are enriched in double-mutant mouse plasma cells. To this end, we combined two published lists of genes differentially up- or down-regulated in MGUS/MM cells compared to normal human plasma cells into an MM signature_UP or MM signature_DOWN module, respectively (40, 41). Notably, the MM signature_UP was significantly enriched among genes up-regulated in Ccnd1/Ikk2ca or MMSET/Ikk2ca BM plasma cells in comparison to their respective controls, whereas the MM signature_DOWN was significantly enriched among the down-regulated genes (Fig. 5 D and E), suggesting that the outgrowing mouse plasma cells share a transcriptional profile with human MGUS/MM cells. It should be noted that these signatures were also enriched in single-mutant plasma cells (SI Appendix, Fig. S10 A and B), suggestive of a premalignant MGUS stage in these mice, which occasionally showed faint M proteins (Figs. 1E and 3C and SI Appendix, Fig. S3A) in the absence of overt disease (Figs. 2D and 4D). To investigate whether the transgenic plasma cells match to their respective human MM subgroup counterparts, we performed GSEA within the genes differentially expressed between MMSET/Ikk2ca versus Ccnd1/Ikk2ca and MMSET versus Ccnd1 samples, respectively (Fig. 5 F and G and SI Appendix, Fig. S10C), using previously described MM subgroup-specific gene sets (15–17). Strikingly, all three MMSET signatures were significantly enriched among MMSET-specific genes, linking MMSET/Ikk2ca (and also MMSET single mutant) plasma cells to the human t(4;14) subgroup (Fig. 5F and SI Appendix, Fig. S10C). For Ccnd1-specific genes, only the comparison of MMSET versus Ccnd1 single-mutant plasma cells resulted in a significant enrichment, namely the CD-2 signature (16), suggesting that transgenic Ccnd1-expressing plasma cells show transcriptional features of the human t(11;14) subgroup (Fig. 5G).
Fig. 5.
The transgenic mouse plasma cells express human MGUS/MM signature genes. (A) Overview of RNA-Seq samples. (B and C) Principal component analysis (B) and unsupervised hierarchical clustering (C) of control (Cγ1-cre or floxed R26 alleles), single-mutant (Ccnd1, MMSET, or Ikk2ca), and double-mutant (Ccnd1/Ikk2ca or MMSET/Ikk2ca) BM plasma cells using the top 500 genes showing the highest variability in expression across all samples. (D–G) Graphical representations of gene set enrichment analysis results employing the tmod algorithm. (D and E) The x-axis shows the ranked gene list (ordered from most up- to most down-regulated) when comparing Ccnd1/Ikk2ca (D) or MMSET/Ikk2ca (E) to control BM plasma cells. The tested gene sets represent genes differentially up- (MM signature_UP) or down-regulated (MM signature_DOWN) in human MGUS/MM cells versus normal human plasma cells (40, 41). (F) The x-axis shows the ranked gene list (ordered from most up- to most down-regulated) when comparing MMSET/Ikk2ca versus Ccnd1/Ikk2ca BM plasma cells. The tested gene sets represent genes specifically up-regulated in the human t(4;14)/MMSET subgroup compared to the other MM subgroups (15–17). (G) The x-axis shows the ranked gene list (ordered from most up- to most down-regulated) when comparing MMSET versus Ccnd1 BM plasma cells. The tested gene set represents genes specifically up-regulated in the human t(11;14)-associated CD-2 subgroup compared to the other MM subgroups (16).
The Outgrowing Double-Mutant Plasma Cells Can Be Propagated in Secondary Hosts.
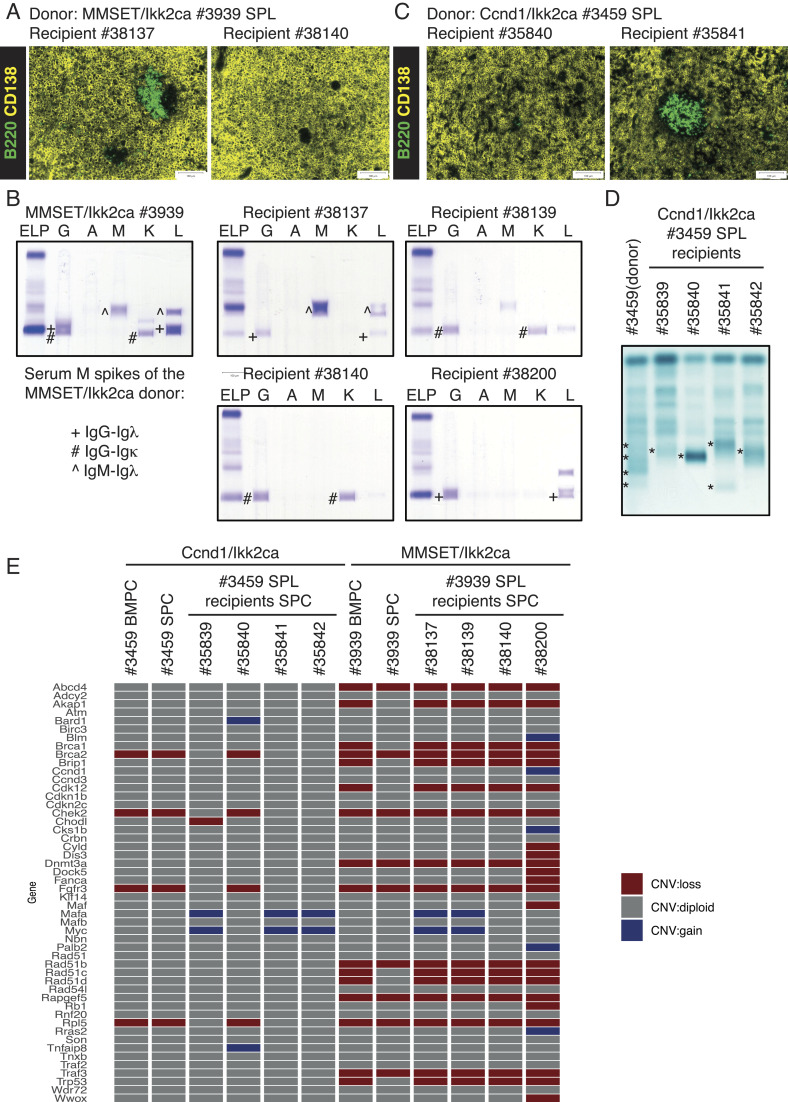
Having shown that our MM mouse models share clinical, phenotypic, and molecular features with the human disease, we set out to establish a system to propagate the expanded plasma cells in vivo. The first sets of experiments were based on the transfer of total spleen cells from MMSET/Ikk2ca and Ccnd1/Ikk2ca donor mice into sublethally irradiated Rag2-Il2r- recipients (SI Appendix, Fig. S11A). More precisely, all four recipients of one MMSET/Ikk2ca donor spleen (#3939) presented with a prominent plasma cell outgrowth in the spleen (Fig. 6A and SI Appendix, Fig. S11 B, C, and G) and serum M spikes (SI Appendix, Fig. S11D) within 34 wk. Strikingly, although the donor mouse had demonstrated at least three serum M proteins, three of four recipient mice showed only one of the three spikes (Fig. 6B), implying continuing clonal selection of the transferred plasma cells. This finding was also corroborated by whole-exome sequencing (WES), which revealed unique but also shared somatic mutations especially between recipient #38200 and the donor (#3939) plasma cells as well as between recipients #38139 and #38140 (SI Appendix, Fig. S11E) reflecting the M spike pattern, further confirming the effective propagation of donor plasma cell clones and the acquisition of additional genetic aberrations. Successful propagation of plasma cells could also be shown for a second MMSET/Ikk2ca donor spleen (#3814) (SI Appendix, Fig. S11 F and G). Likewise, the transfer of total spleen cells from a Ccnd1/Ikk2ca donor (#3459) into irradiated immunodeficient hosts resulted in a massive plasma cell outgrowth within 31 wk (Fig. 6C and SI Appendix, Fig. S12 A, B, and E). This was again accompanied by clonal selection as shown by SPEP that demonstrated only one major M protein in recipients’ sera, whereas the donor mouse had presented with multiple spikes (Fig. 6D), as well as by WES that revealed in addition to a limited set of shared mutations a number of uniquely acquired aberrations within the recipients’ plasma cells (SI Appendix, Fig. S12C). However, we noticed that the transfer of total splenocytes occasionally resulted in a rapid outgrowth of double-transgenic CD19+ B cells, a phenomenon that also rarely occurred in primary Ccnd1/Ikk2ca mice (3 of 13), preventing propagation of the more slowly expanding plasma cells. Thus, a more stringent strategy was designed based on the transplantation of sorted CD138+TACI+CD19- splenic plasma cells. Using this strategy, two recipient mice of a second Ccnd1/Ikk2ca donor (#3443) showed a prominent plasma cell outgrowth in the spleen and BM (SI Appendix, Fig. S12 D and E) approximately 30 wk after the transfer, validating this more stringent approach and suggesting that the CD138+TACI+CD19- plasma cell population indeed contains the propagating activity.
Fig. 6.
The transgenic mouse plasma cells can be propagated in vivo and exhibit genetic aberrations that are associated with human MM. Total spleen cells of a MMSET/Ikk2ca (#3939) and a Ccnd1/Ikk2ca (#3459) donor mouse were transferred into sublethally irradiated (3 Gy) Rag2-Il2r-immunodeficient recipients, which were analyzed 30 to 34 wk after the transplantation. (A and C) Representative immunofluorescence images of two MMSET/Ikk2ca #3939 recipient spleens (A) and two Ccnd1/Ikk2ca #3459 recipient spleens (C) stained with α-B220 (green, B cells) and α-CD138 (yellow, plasma cells). (B) SPEP coupled to immunofixation of the MMSET/Ikk2ca donor #3939 and its four recipients to determine the immunoglobulin isotypes of the M proteins (corresponding heavy and light chains are marked by the same symbol; G—IgG, A—IgA, M—IgM, K—Igκ, L—Igλ). (D) SPEP of the Ccnd1/Ikk2ca donor #3459 and its corresponding four recipients; M spikes are marked (*). (E) Double-mutant (BFP+GFP+) plasma cells were sorted from all four Ccnd1/Ikk2ca #3459 and MMSET/Ikk2ca #3939 recipients’ spleens (SPC) as well as the donors’ spleen (SPC) and bone marrow (BMPC) and subjected to WES to determine copy number variations (CNVs) (reference: myeloid cells sorted from the donor mice BM). The graph depicts genes shown to be frequently affected by CNVs in human MM (42) and their copy number status (loss—red, gain—blue, diploid—gray) in the respective mouse plasma cells.
Given that the expanded mouse plasma cells can be successfully transplanted into secondary hosts with increased clonal selection, we analyzed the MMSET/Ikk2ca #3939 and Ccnd1/Ikk2ca #3459 donor and respective recipient plasma cells for the acquisition of genetic lesions that are associated with the human disease. A prominent feature of human MM genomes is the presence of copy number variations (CNVs), e.g., monosomy of chromosome 13, amplifications in 1q, and deletions in 1p, 12p, 14q, 16q, and 22q (43, 44). In agreement with this, our mouse plasma cells also demonstrated large-scale chromosomal aberrations, which resulted in the loss or gain of genes known to be affected by recurrent CNVs in human MM, such as the amplification of MYC or the deletion of the tumor suppressor genes TRAF3 and BRCA2 (Fig. 6E) (42), further linking our MM mouse models to the human malignancy.
Discussion
Taken together, our study reports genetically defined mouse models for distinct human MM subgroups by the GC B cell-specific coactivation of either Ccnd1 or MMSET, the two genes most frequently affected by a primary IgH translocation, with constitutive NF-κB signaling as a secondary event. Most double-transgenic mice reached a myeloma-defining event (2), based on ≥60% femur plasma cell infiltration and/or the presence of CRAB criteria as well as thrombocytopenia and hypoalbuminemia, which have prognostic importance in human MM (45). Most importantly, RNA-Seq of double-mutant plasma cells confirmed the presence of a human MGUS/MM gene expression signature and linked MMSET and Ccnd1 transgene expression to their respective human t(4;14) and t(11;14) subgroup counterparts, mirroring the heterogeneity of human MM. This implies that the primary genetic events (here: deregulated MMSET or Ccnd1 expression) dictate the transcriptional phenotype of the tumor cells independently of secondary genetic aberrations (i.e., the Ikk2ca allele or randomly acquired genetic lesions)—a finding that is in line with the published human MM subgroup classifications for which unsupervised hierarchical clustering reveals the existence of transcriptionally distinct MM entities that align to defined primary genetic lesions (16, 17). Interestingly, though, only very few single mutant mice presented with a full disease phenotype, highlighting the low oncogenic potential of the primary genetic aberrations on their own and the importance of both primary and secondary mutations for the development of overt MM.
Despite their resemblance to human MM, some aspects of the disease are still not fully recapitulated in the present mouse models. Thus, likely due to the large fraction of mutated GC B cells and slow rate of expansion, the expanding plasma cell clones are oligoclonal, in contrast to the monoclonality of human MM and a MYC-based MM mouse model, where MYC activation is restricted to very few cells (22). Other limitations include the substantial fraction of IgM isotype expression and the overall low degree of somatic hypermutation within the VDJ sequences of the expanding plasma cell clones (SI Appendix, Tables S2 and S4). These are likely consequences of early Ikk2ca activation by the Cγ1-cre transgene, since NF-κB activation actively drives B cells out of the GC, promoting plasma cell differentiation (35). An expansion of IgM-expressing plasma cells is usually associated with Waldenström’s macroglobulinemia (WM), an indolent lymphoplasmacytic lymphoma (46). However, the occurrence of IgM MM has also been described, differing from WM in gene expression (47) and clinical manifestation (48). In line with this, Ccnd1/Ikk2ca and MMSET/Ikk2ca BM plasma cells were clearly distinguished from WM cells by GSEA using MM- and WM-specific gene sets (49) (SI Appendix, Fig. S10 D and E).
Our studies are also limited by their restriction to a single secondary mutation in the scenario of MM pathogenesis, namely activation of the canonical NF-κB signaling pathway through the Ikk2ca transgene (28). Although most genetic aberrations targeting NF-κB in MM appear to cluster in the alternative pathway (29), a number of genetic and functional studies indicate a frequent coactivation and extensive crosstalk between the two NF-κB branches, with a high (and maybe dominant) functional impact of the canonical pathway in MM cells (50, 51). In addition, the use of the Ikk2ca transgene had proven to be a reliable experimental strategy to induce a robust shift toward plasma cell differentiation in previous tumor models (35, 52). While the Ikk2ca transgene clearly served its function in the present context, the impact of other secondary mutations remains to be studied and is addressed by ongoing work.
In summary, the genetically distinct MM models described here exhibit a prominent outgrowth of selected plasma cell clones with the onset of MM-associated pathologies, particularly the emergence of bone lesions. The expanded plasma cells can be propagated in vivo, opening the way to the identification of MM subgroup-specific vulnerabilities, subtype-oriented drug screening approaches, or selective cell transfer experiments to address the long-standing question of the identity of MM-initiating cells (53). Thus, our MM models do not only recapitulate basic features of the human disease but also reflect the heterogeneity of human MM, offering the opportunity to study and target subgroup-specific disease mechanisms in a defined genetic system.
Materials and Methods
Mice, Immunization, and Tumor Cohorts.
Cγ1-cre, R26 Ikk2castopF, and R26 BFPstopF alleles have been described (28, 34, 54). A similar targeting strategy as for the Ikk2ca allele was applied to generate R26 MMSETstopF and R26 Ccnd1stopF alleles (28). In short, a construct encoding for either mouse Ccnd1 cDNA (NM_007631.3) or mouse MMSET-II cDNA (Nsd2; NM_001081102.2) preceded by a loxP-flanked STOP cassette was integrated into the mouse Rosa26 locus. Transgene transcription is controlled by a CAG promoter and expression can be detected by the enhanced blue fluorescent protein (EBFP), which is placed under control of an internal ribosomal entry site (IRES) downstream of the respective cDNAs. The linearized targeting vector was transfected into the Artemis B6/3 C57BL/6 ES cell line and targeted clones were isolated using positive (NeoR) selection. Correct integration was verified by Southern blot of EcoRI- or PacI-digested genomic DNA from mouse ES cells and founder mouse tails using a Rosa26-specific probe (external Rosa26 probe A) (28) or Neo-specific (BamHI/PstI-digested fragment from NeoR) probe, respectively.
Mice were bred and maintained under specific pathogen-free conditions. Eight- to 12-wk-old mice, indiscriminately of their sex, were immunized intraperitoneally with 100 µg alum-precipitated 4-hydroxy-3-nitrophenylacetyl-hapten conjugated to chicken gamma globulin (NP-CGG, Ratio10-19, LGC Biosearch Technologies Cat#N-5055B) and either analyzed after 10 to 14 d (SI Appendix, Fig. S1 F–I) or monitored for tumor formation and analyzed when reaching disease-defining humane endpoints (between 62 and 97 wk of age). Disease-defining endpoints in this study included the presence of at least one of the following phenotypes: splenomegaly, a weak general condition, slow movement, weight loss, breathing problems, and diarrhea. Cohort mice included heterozygous control (Cγ1-cre only, R26 floxed, or Cγ1-cre; R26 BFPstopF reporter), single-mutant (Cγ1-cre; R26 Ccnd1stopF, Cγ1-cre; R26 MMSETstopF or Cγ1-cre; R26 Ikk2castopF), and double-mutant (Cγ1-cre; R26 Ccnd1stopF; R26 Ikk2castopF or Cγ1-cre; R26 MMSETstopF; R26 Ikk2castopF) mice. On the day of analysis, cohort mice were killed by cardiac puncture to obtain a sufficient blood sample for hematology and clinical chemistry. To this end, mice were first injected subcutaneously with 0.1 mg per kg body weight Buprenorphin (Buprenovet sine), followed by deep anesthesia with 4 Vol.-% isoflurane and subsequent puncture of the left ventricle. Hematologic and clinical serum parameters were measured on the ProCyte Dx (IDEXX) and AU480 Chemistry Analyser (Beckman Coulter), respectively. For the adoptive transfer, 1.15 × 106 thawed total spleen cells of either MMSET/Ikk2ca or Ccnd1/Ikk2ca mice were injected intravenously into Rag2- (55) Il2rg- (56) recipients, which had been irradiated with 3 Gy 1 d before. For the selective transplantation of plasma cells, BFP+GFP+ CD138+TACI+CD19- cells were sorted from thawed spleen suspensions of double-mutant donor animals and 1–2 × 105 sorted cells were injected along with 1 × 106 Rag2-Il2rg- total SPL cells as described above. Breeding, maintenance, and experimental animal procedures were approved by the Landesamt für Gesundheit und Soziales Berlin (G0374/13, G0196/19, G0029/15, G0308/19).
Data Presentation and Statistical Analysis.
Single dots represent the number of biological replicates from independent mice. Unless noted otherwise, bars or lines represent median values. Prism software (GraphPad Prism, RRID: SCR_002798) version 8 was used for pair-wise comparisons between control and single mutant or respective double-mutant samples using nonparametric, unpaired, two-tailed Mann–Whitney U test. Asterisks indicate statistical significance for P values < 0.05 (single), <0.01 (double), <0.001 (triple), and < 0.0001 (quadruple). For representation of blood parameters, box-and-whisker graphs are shown with boxes extending from the 25th to 75th percentile with a line at the median and whiskers plotted down to the minimum and up to the maximum value.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank all members of the M. Janz and S. Mathas and K. Rajewsky groups for their feedback and discussion; D. Calado for support and advice; K. Petsch, C. Salomon, J. Pempe, and the MDC animal caretakers for their outstanding technical help; H.P. Rahn und K. Rautenberg for excellent FACS-related support; R. Kühn and the MDC Transgenic Core Facility for the generation of R26 Ccnd1stopF and R26 MMSETstopF alleles; W. Schneider for assessment of mouse kidney biopsies, and J. Altmüller and the MDC Genomics Facility for performing the RNA-sequencing as well as WES library preparation and sequencing. This work was supported by the Clinical Research Cooperation Program MDC/Charité/ECRC (C12/03) to K.R. and M.J., the Deutsche Krebshilfe (grant #70112800) to K.R. and M.J., the Mildred Scheel MD student program (Deutsche Krebshilfe; #70113398) to M.W., and the Berlin School of Integrative Oncology (BSIO) to C.F.D.
Author contributions
W.W., K.R., and M.J. designed research; W.W., C.F.D., H.N., P.L., M.W., B.W., B.W.-W., T.Y., and I.A. performed research; W.W., C.F.D., E.B., H.N., P.L., M.W., B.W., B.W.-W., T.Y., A.H., D.B., S.M., I.A., A.R., K.R., and M.J. analyzed data; and W.W., K.R., and M.J. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: R.D.-F., Columbia University Irving Medical Center; and S.L.N., The Walter & Eliza Hall Institute of Medical Research.
Contributor Information
Klaus Rajewsky, Email: klaus.rajewsky@mdc-berlin.de.
Martin Janz, Email: mjanz@mdc-berlin.de.
Data, Materials, and Software Availability
The RNA-Seq data are available in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo; RRID:SCR_005012) under the accession number GSE183177 (57). The WES data have been uploaded to the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/docs/; RRID:SCR_004891; ID BioProject PRJNA881497) (58). The materials required to use any of the newly generated mouse strains will be available through K.R. and M.J. under a material transfer agreement from the MDC. Additional information is provided in the SI Appendix, Supporting Information Materials and Methods.
Supporting Information
References
- 1.Landgren O., et al. , Monoclonal gammopathy of undetermined significance (MGUS) consistently precedes multiple myeloma: A prospective study. Blood 113, 5412–5417 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rajkumar S. V., et al. , International myeloma working group updated criteria for the diagnosis of multiple myeloma. Lancet Oncol. 15, e538–e548 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Avet-Loiseau H., et al. , 14q32 translocations and monosomy 13 observed in monoclonal gammopathy of undetermined significance delineate a multistep process for the oncogenesis of multiple myeloma. Cancer Res. 59, 4546–4550 (1999). [PubMed] [Google Scholar]
- 4.Fonseca R., et al. , Genomic abnormalities in monoclonal gammopathy of undetermined significance. Blood 100, 1417–1424 (2002). [PubMed] [Google Scholar]
- 5.Kaufmann H., et al. , Both IGH translocations and chromosome 13q deletions are early events in monoclonal gammopathy of undetermined significance and do not evolve during transition to multiple myeloma. Leukemia 18, 1879–1882 (2004). [DOI] [PubMed] [Google Scholar]
- 6.González D., et al. , Immunoglobulin gene rearrangements and the pathogenesis of multiple myeloma. Blood 110, 3112–3121 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Walker B. A., et al. , Characterization of IGH locus breakpoints in multiple myeloma indicates a subset of translocations appear to occur in pregerminal center B cells. Blood 121, 3413–3419 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Chesi M., et al. , Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood 88, 674–681 (1996). [PubMed] [Google Scholar]
- 9.Chesi M., et al. , The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92, 3025–3034 (1998). [PubMed] [Google Scholar]
- 10.Morgan G. J., Walker B. A., Davies F. E., The genetic architecture of multiple myeloma. Nat. Rev. Cancer 12, 335–348 (2012). [DOI] [PubMed] [Google Scholar]
- 11.Manier S., et al. , Genomic complexity of multiple myeloma and its clinical implications. Nat. Rev. Clin. Oncol. 14, 100–113 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Mikulasova A., et al. , The spectrum of somatic mutations in monoclonal gammopathy of undetermined significance indicates a less complex genomic landscape than that in multiple myeloma. Haematologica 102, 1617–1625 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dutta A. K., et al. , Subclonal evolution in disease progression from MGUS/SMM to multiple myeloma is characterised by clonal stability. Leukemia 33, 457–468 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oben B., et al. , Whole-genome sequencing reveals progressive versus stable myeloma precursor conditions as two distinct entities. Nat. Commun. 12, 1861 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergsagel P. L., et al. , Cyclin D dysregulation: An early and unifying pathogenic event in multiple myeloma. Blood 106, 296–303 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhan F., et al. , The molecular classification of multiple myeloma. Blood 108, 2020–2028 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Broyl A., et al. , Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood 116, 2543–2553 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Ledergor G., et al. , Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat. Med. 24, 1867–1876 (2018). [DOI] [PubMed] [Google Scholar]
- 19.Carrasco D. R., et al. , The differentiation and stress response factor XBP-1 drives multiple myeloma pathogenesis. Cancer Cell 11, 349–360 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suematsu S., et al. , Generation of plasmacytomas with the chromosomal translocation t(12;15) in interleukin 6 transgenic mice. Immunology 89, 232–235 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scherger A. K., et al. , Activated gp130 signaling selectively targets B cell differentiation to induce mature lymphoma and plasmacytoma. JCI Insight 4, e128435 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chesi M., et al. , AID-dependent activation of a MYC transgene induces multiple myeloma in a conditional mouse model of post-germinal center malignancies. Cancer Cell 13, 167–180 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen Z., et al. , Expression of NrasQ61R and MYC transgene in germinal center B cells induces a highly malignant multiple myeloma in mice. Blood 137, 61–74 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pisano M., et al. , Laboratory mice – A driving force in immunopathology and immunotherapy studies of human multiple myeloma. Front. Immunol. 12, 667054 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morito N., et al. , A novel transgenic mouse model of the human multiple myeloma chromosomal translocation t(14;16)(q32;q23). Cancer Res. 71, 339–348 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Vicente-Duenas C., et al. , A novel molecular mechanism involved in multiple myeloma development revealed by targeting MafB to haematopoietic progenitors. EMBO J. 31, 3704–3717 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiancette R., et al. , A myeloma translocation-like model associating CCND1 with the immunoglobulin heavy-chain locus 3’ enhancers does not promote by itself B-cell malignancies. Leuk. Res. 34, 1043–1051 (2010). [DOI] [PubMed] [Google Scholar]
- 28.Sasaki Y., et al. , Canonical NF-κB activity, dispensable for B cell development, replaces BAFF-receptor signals and promotes B cell proliferation upon activation. Immunity 24, 729–739 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Keats J. J., et al. , Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell 12, 131–144 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Annunziata C. M., et al. , Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12, 115–130 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chapman M. A., et al. , Initial genome sequencing and analysis of multiple myeloma. Nature 471, 467–472 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker B. A., et al. , Mutational spectrum, copy number changes, and outcome: Results of a sequencing study of patients with newly diagnosed myeloma. J. Clin. Oncol. 33, 3911–3920 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hideshima T., Anderson K. C., Signaling pathway mediating myeloma cell growth and survival. Cancers 13, 1–17 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Casola S., et al. , Tracking germinal center B cells expressing germ-line immunoglobulin g1 transcripts by conditional gene targeting. Proc. Natl. Acad. Sci. U.S.A. 103, 7396–7401 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calado D. P., et al. , Constitutive canonical NF-κB activation cooperates with disruption of BLIMP1 in the pathogenesis of activated B cell-like diffuse large cell lymphoma. Cancer Cell 18, 580–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehlich A., Martin V., Müller W., Rajewsky K., Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 4, 573–583 (1994). [DOI] [PubMed] [Google Scholar]
- 37.Kim J. E., et al. , Serum albumin level is a significant prognostic factor reflecting disease severity in symptomatic multiple myeloma. Ann. Hematol. 89, 391–397 (2010). [DOI] [PubMed] [Google Scholar]
- 38.Szalat R., Avet-Loiseau H., Munshi N. C., Gene expression profiles in myeloma: Ready for the real world? Clin. Cancer Res. 22, 5434–5442 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weiner J., Domaszewska T., tmod: An R package for general and multivariate enrichment analysis. PeerJ Prepr. 4, e2420v1 (2016). [Google Scholar]
- 40.Zhan F., et al. , Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood 99, 1745–1757 (2002). [DOI] [PubMed] [Google Scholar]
- 41.López-Corral L., et al. , Transcriptome analysis reveals molecular profiles associated with evolving steps of monoclonal gammopathies. Haematologica 99, 1365–1372 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walker B. A., et al. , Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood 132, 587–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Avet-Loiseau H. Â., Daviet A., Saunier S. Â., R., Âgis Bataille, Chromosome 13 abnormalities in multiple myeloma are mostly monosomy 13. Br. J. Haematol. 111, 1116–1117 (2000). [DOI] [PubMed] [Google Scholar]
- 44.Avet-Loiseau H., et al. , Prognostic significance of copy-number alterations in multiple myeloma. J. Clin. Oncol. 27, 4585–4590 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Greipp P. R., et al. , International staging system for multiple myeloma. J. Clin. Oncol. 23, 3412–3420 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Yun S., et al. , Waldenström macroglobulinemia: Review of pathogenesis and management. Clin. Lymphoma Myeloma Leuk. 17, 252–262 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atrash S., et al. , Clinical presentation and gene expression profiling of immunoglobulin M multiple myeloma compared with other myeloma subtypes and Waldenström Macroglobulinemia. J. Glob. Oncol. 4, 1–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schuster S. R., et al. , IgM multiple myeloma: Disease definition, prognosis, and differentiation from Waldenstrom’s macroglobulinemia. Am. J. Hematol. 85, 853–855 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gutiérrez N. C., et al. , Gene expression profiling of B lymphocytes and plasma cells from Waldenström’s macroglobulinemia: Comparison with expression patterns of the same cell counterparts from chronic lymphocytic leukemia, multiple myeloma and normal individuals. Leukemia 21, 541–549 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Demchenko Y. N., et al. , Classical and/or alternative NF-B pathway activation in multiple myeloma. Blood 115, 3541–3552 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cormier F., et al. , Frequent engagement of RelB activation is critical for cell survival in multiple myeloma. PLoS One 8, e59127 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbosa R., et al. , Co-activation of NF-κB and MYC renders cancer cells addicted to IL6 for survival and phenotypic stability. bioRxiv [Preprint] (2020). 10.1101/2020.04.12.038414 (Accessed 13 April 2020). [DOI]
- 53.Karadimitris A., et al. , Myeloma propagating cells, drug resistance and relapse significance statement. Stem Cells 33, 3205–3211 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Sommermann T., et al. , Functional interplay of Epstein-Barr virus oncoproteins in a mouse model of B cell lymphomagenesis. Proc. Natl. Acad. Sci. U.S.A. 117, 14421–14432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hao Z., Rajewsky K., Homeostasis of peripheral B cells in the absence of B cell influx from the bone marrow. J. Exp. Med. 194, 1151–1163 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiSanto J. P., Müller W., Guy-Grand D., Fischer A., Rajewsky K., Lymphoid development in mice with a targeted deletion of the interleukin 2 receptor g chain. PNAS 92, 377–381 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Janz M., Rajewsky K., Winkler W., Blanc E., RNA-Seq of transgenic plasma cells from the described subgroup-specific MM mouse models. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE183177/. Deposited 1 September 2021.
- 58.Winkler W., Blanc E., Whole exome sequencing of transgenic mouse plasma cells. Sequence Read Archive. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA881497/. Deposited 17 September 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The RNA-Seq data are available in the Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo; RRID:SCR_005012) under the accession number GSE183177 (57). The WES data have been uploaded to the NCBI Sequence Read Archive (SRA) database (https://www.ncbi.nlm.nih.gov/sra/docs/; RRID:SCR_004891; ID BioProject PRJNA881497) (58). The materials required to use any of the newly generated mouse strains will be available through K.R. and M.J. under a material transfer agreement from the MDC. Additional information is provided in the SI Appendix, Supporting Information Materials and Methods.