Significance
Amphioxus diverged from vertebrates around half a billion years ago, but shares the basic vertebrate body plan, making them an important model for understanding the vertebrate origin and innovations. Here we analyze genome sequences of three amphioxus species, and uncover their extraordinary conservation with vertebrates in chromosome composition and conformation. Some amphioxus chromosomes share majorities of gene content with many small-sized chicken microchromosomes, suggesting they have been preserved since their ancient origin in the vertebrate ancestor. Similar to many vertebrates, amphioxus establish the spatial genome topology after the zygotes become broadly activated in transcription, and form two regulatory domains at the Hox gene cluster. We finally reveal different amphioxus species have undergone recent turnovers of sex chromosomes, illuminating their unappreciated diversity.
Keywords: amphioxus, genome, sex chromosomes, microchromosomes, topologically associated domains
Abstract
The slow-evolving invertebrate amphioxus has an irreplaceable role in advancing our understanding of the vertebrate origin and innovations. Here we resolve the nearly complete chromosomal genomes of three amphioxus species, one of which best recapitulates the 17 chordate ancestor linkage groups. We reconstruct the fusions, retention, or rearrangements between descendants of whole-genome duplications, which gave rise to the extant microchromosomes likely existed in the vertebrate ancestor. Similar to vertebrates, the amphioxus genome gradually establishes its three-dimensional chromatin architecture at the onset of zygotic activation and forms two topologically associated domains at the Hox gene cluster. We find that all three amphioxus species have ZW sex chromosomes with little sequence differentiation, and their putative sex-determining regions are nonhomologous to each other. Our results illuminate the unappreciated interspecific diversity and developmental dynamics of amphioxus genomes and provide high-quality references for understanding the mechanisms of chordate functional genome evolution.
First described in 1774, the lesser-known marine invertebrate amphioxus (or lancelets) was already central to comparative embryology and anatomical studies throughout the 18th and 19th centuries. After a short decline in the beginning of the 20th century, the interest into it revived with even more strength before the development of modern biology techniques (1). It was later established that amphioxus diverged from the ancestor of two other chordate subphyla, urochordates (tunicates) and vertebrates about 550 Mya (2, 3). Amphioxus has a vertebrate-like but simpler body plan and underwent much less lineage-specific changes of chromosomes and genomic sequences than urochordates (4). Therefore, it represents the best-known living proxy for the chordate ancestor (5, 6). Amphioxus has one, and the largest reported Hox gene cluster with 15 genes (7), which was reported to form one structural and regulatory unit of topologically associated domain (TAD) (8). By contrast, most vertebrates have at least four Hox gene clusters and up to 13 genes per cluster with a few exceptions (9–13), with the mouse HoxA and HoxD clusters each forming two TADs. Such a fourfold difference of Hox gene cluster numbers provided early evidence for Ohno’s hypothesis of two rounds of whole-genome duplications (WGDs) (the 2R hypothesis) (14, 15) that shaped the genome evolution and regulation of vertebrates since they diverged from other chordates.
Broader understanding beyond individual genes into the scenario and functional consequences of vertebrate WGDs, whose times and timing recently became a subject of debate (16), necessitate high-quality sequence assembly and annotation of genes and cis-regulatory elements of amphioxus (17), as a pre-WGD outgroup. The first draft genome of Florida amphioxus Branchiostoma floridae (Bf) was published over a decade ago and has been frequently used to reconstruct the ancestral vertebrate protokaryotype, with however different estimates of ancestral linkage group number between studies (16, 18–20). A recent work improved the Bf genome into the chromosome level and proposed a refined the 2R hypothesis with 17 ancestral chordate linkage groups: the first WGD occurred in the ancestor of all vertebrates, and the second WGD only occurred in the lineage of jawed vertebrates (21). The duplicated gene products of WGDs in vertebrates (“ohnologues”) seem to have generally a higher number of and more specialized regulatory elements and gene expression between copies, relative to their single-copy orthologs of amphioxus (17). Besides results at the gene level, to address how vertebrates evolved globally more complex regulatory circuits after WGDs requires knowledge of higher-order chromatin organization of amphioxus.
An often-overlooked factor among previous studies using only one species’ genome is the largely unexplored interspecific genomic diversity of amphioxus. It is known that different amphioxus species have different chromosome numbers and exhibit frequent disruptions of gene synteny which may confound the inference of vertebrate ancestral state (22). Moreover, the available amphioxus genome assemblies are either incomplete or fragmented because of the high intraspecific polymorphisms associated with their large effective population size (4). To elucidate the evolution of genes, genomes, and chromatin landscapes of different amphioxus species compared to vertebrates, we resolve here the nearly complete haploid genomes of three Branchiostoma amphioxus species Chinese amphioxus (Branchiostoma belcheri, Bb), Japanese amphioxus (Branchiostoma japonicum, Bj), and Bf.
Results
Haploid Chromosomal Genomes of Three Amphioxus Species.
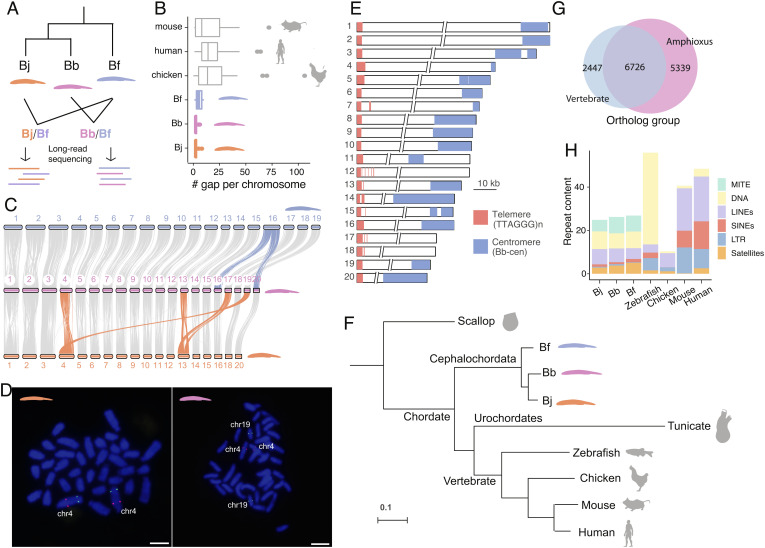
We estimated the genome-wide heterozygosity levels of three amphioxus species and found they range from 3.2 to 4.2%, among the highest in animal species (23) (SI Appendix, Fig. S1). To overcome this great challenge for genome assembly, we devised an interspecific trio sequencing strategy and produced respectively more than 100-fold short and long sequencing reads for the F1 hybrids derived from Bf-Bb or Bf-Bj crosses (Fig. 1A and SI Appendix, Fig. S2). Given an estimated at least 50 MYs’ species divergence time (SI Appendix, Fig. S3), the hybrids contain two haploid parental genomes that have become too diverged in sequences to form cross-species chimeric assembly (SI Appendix, Fig. S1). By mapping short reads derived from the respective parental species, we were able to attribute each assembled contig into one of the four haploid (Bb, Bj, and two Bf) genomes (SI Appendix, Fig. S4). The new haploid amphioxus genomes have an assembled size ranging from 382 to 491 Mb, and an over 200-fold contig N50 length (between 6.4 Mb and 14.2 Mb) compared to the published genomes (4, 17, 22), an over 97% genome completeness (measured by BUSCO (Benchmarking Universal Single-Copy Orthologs)) and a reduced level of false duplications (SI Appendix, Table S1 and Fig. S5 A and B). Using Hi-C data, we anchored more than 98.6% of three species’ contig sequences into chromosomes, with a much lower gap number (on average only 3.8 gaps) per chromosome than those of major vertebrate reference genomes and that of a recently improved Bf genome (21) (Fig. 1B and SI Appendix, Fig. S5C). The three amphioxus genomes show a highly conserved chromosomal synteny to each other (Fig. 1C), with most chromosomes showing a one-to-one homologous relationship except for a few chromosome fusions. And we further confirmed such fusions by mapping the Hi-C reads indicative of long-range linkage relationship between species (SI Appendix, Fig. S6) or by fluorescence in situ hybridization (Fig. 1D).
Fig. 1.
Three haploid genomes of amphioxus species. (A) We performed long-read sequencing of interspecific hybrids between the three amphioxus species and assembled their haploid genomes. Bj: B. japonicum (orange), Bb: B. belcheri (pink), Bf: B. floridae (blue). (B) The amphioxus haploid genomes have a lower gap content (numbers of gaps per chromosome) compared to other vertebrate reference genomes (assessed by January 26, 2022). (C) Pairwise chromosome synteny between amphioxus species. The chromosomes that have experienced interchromosomal changes are highlighted in color. Bj has 18 chromosomes, but we named each of its chromosomes with its homologous chromosome of Bf. Hence the fused chr17 and chr19 are not named here. (D) FISH experiment confirming the chromosome fusion in Bj (chr4, Left) relative to Bb (Right) (E) Most amphioxus chromosomes are telocentric. The 10 kb scale applies to the two tips of the chromosomes only, and the two slash lines represent the gaps between the two chromosomal tips. (F) Phylogenomic tree based on whole-genome alignments of amphioxus vs. other chordate species. (G) A large number of orthologous gene groups (6,726) is shared between amphioxus and vertebrates, but amphioxus species have 5,339 specific gene groups. (H) MITEs (Miniature Inverted-repeat, green) comprise ~6.7% of the amphioxus genomes but are largely absent in vertebrates. In the DNA transposon category (yellow) MITE was excluded.
With some exceptions, all chromosomal sequences of the three species have been assembled from the telomere at one end to the centromere at the other (Fig. 1E and SI Appendix, Figs. S7 and S8). This is consistent with the reported predominantly telocentric karyotype of amphioxus (24–26), the low levels of recombination rate and nucleotide diversity at centromeric and pericentromeric regions (SI Appendix, Figs. S9 and S10), and is also verified by our fluorescent in situ hybridization (FISH) experiment for Bf (SI Appendix, Fig. S11). The telomeres contain conserved telomeric motifs (TTAGGG)n (27) with an average length of 3.6 kb, and they account for the majority of G-quadruplex content in the genome (SI Appendix, Fig. S12). Our cytogenetic and genomic investigations also confirmed the presence of interstitial telomeric sequences in a few amphioxus chromosomes (SI Appendix, Figs. S7 and S11). The putative centromeric regions consist of species-specific satellite monomers of different sequences and lengths, with inverted repeat structures (SI Appendix, Fig. S13). Our new Bf genome shows a high level of chromosomal synteny with a previous assembly, but contains much more resolved complex regions, including satellite DNA or rDNA arrays (SI Appendix, Figs. S12–S14). This excludes the possible impact of “genomic shock” events, if any, on the Bf or other amphioxus species genomes that might involve TE (Transposable Element) amplification or chromosome rearrangements in the hybrid (28–30).
Our phylogenomic analyses using whole-genome alignments of amphioxus against other chordates and one invertebrate outgroup confirmed amphioxus as the most basal chordate lineage, with a relatively lower genome-wide substitution rate (Fig. 1F). Based on 3,653 single-copy orthologous genes, we estimated that different chordate lineages diverged about 592.5 Mya, and three amphioxus species diverged about 99.9 Mya (Materials and Methods and SI Appendix, Fig. S3 and Table S2). Over 73% vertebrate orthologous gene groups are present in amphioxus genomes (Fig. 1G). The vertebrate-specific genes are enriched for various gene ontology (GO) categories including signaling pathway regulation and muscle functions, while the amphioxus-specific genes are enriched for GOs of tissue regeneration (31) and apoptosis, among many others (SI Appendix, Table S3). We also identified 27,032 conserved sequence elements between vertebrates and amphioxus, and majorities of them (26,955) are located in protein-coding regions. Finally, the amphioxus genomes were found to have a moderate repeat content of about 30% (Fig. 1H), but they contain abundant MITEs (miniature inverted-repeat transposable elements) that are nearly absent in vertebrates. These MITEs seem to have propagated more recently in amphioxus species, relative to other DNA transposons (SI Appendix, Fig. S15).
Reconstructing the Ancestral Karyotypes of Amphioxus, Chordates, and Vertebrates.
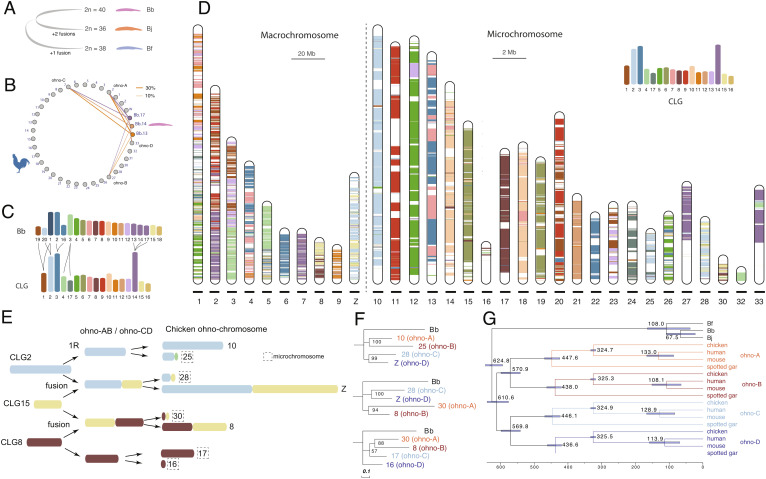
The assembled chromosome number of Bj, Bf, and Bb is respectively 18, 19, and 20, consistent with their reported karyotypes by previous cytogenetic works (27, 32). Based on their whole-genome alignments, we inferred that similar to the karyotype of Bb, the Branchiostoma amphioxus ancestor had 20 linkage groups, which then underwent two chromosome fusions in Bj, and one fusion in Bf after their species divergence (Figs. 1C and 2A and SI Appendix, Fig. S16).
Fig. 2.
Ancestral karyotypes of amphioxus, chordates, and vertebrates. (A) Bb probably best recapitulates the ancestral karyotype of Branchiostoma amphioxus, with Bj and Bf having undergone chromosomal fusions. (B) Genes on chr13, chr14, and chr17 of Bb have their homologous genes located on the same set of chicken chromosomes. Each line connecting chromosomes of Bb and chicken chromosomes is scaled to the proportion of Bb genes that are homologous to the genes of one chicken chromosome. (C) The inferred relationship between Bb chromosomes and CLG. (D) Composition of chicken chromosome by CLG homologous sequences. The colored bands represent the Bb-chicken synteny blocks. A different scale for macrochromosomes (20 Mb) and microchromosomes (2 Mb) was used. (E) Reconstructed 1R and 2R of three CLGs. One color represents one CLG, and when one chromosome is composed with more than one CLG, two or more CLG blocks are linked together. (F) The ohnolog genes were used to construct the phylogeny of ohno-chromosomes (ohno-A, B, C, D), which refer to gene groups derived from WGDs. Bb homologs were used as the outgroup. Bootstrapping values shown placed at the internal nodes. (G) 244 ohnolog gene groups were used to date 1R and 2R. Fossil calibration for the mouse-human nodes: 62 to 101 My, bird-mammal nodes: 306 to 332 My.
Genomic comparison between Bb vs. chicken allows us to reconstruct the karyotype of chordate ancestor. We chose chicken because it is one of the vertebrates that exhibit the lowest rates of lineage-specific chromosomal evolution (20, 21, 33, 34) and gene duplications (35, 36). Consistent with two rounds of WGDs followed by gene loss, one single-copy amphioxus gene typically has between one to four homologs in vertebrates (SI Appendix, Fig. S17). Moreover, genes from one Bb chromosome are more frequently found to have homologs distributed on four different chromosomes in chicken (SI Appendix, Fig. S18), compared to spotted gar or human (SI Appendix, Fig. S19), confirming that chicken has better preserved the ancestral vertebrate karyotype with less interchromosomal rearrangements. We also found several Bb chromosomes share their combination of homologous chicken chromosomes. For instance, Bb chr13, chr14, and chr17 all have their homologous genes located on the chicken chr2 (GGA2), GGA7, 27, and 33 (Fig. 2B). This suggested that these three Bb chromosomes were likely derived from one single chordate ancestral linkage group (CLG) (Fig. 2C). Similarly, Bb chr1 shares its homologous chicken chromosomes exclusively with either Bb chr19 or chr20 (SI Appendix, Fig. S18 and Fig. 2C), suggesting Bb chr1 originated from a translocation between two CLGs. Moreover, we inferred that Bb chr2 and chr16 fused at the vertebrate ancestor prior to the whole-genome duplication, while Bb chr3 was split into two (SI Appendix, Fig. S18 and Fig. 2C). Taken together, we inferred that there was a total of 17 CLGs (Fig. 2C), consistent with previous results (4, 18, 21).
To reconstruct the evolutionary trajectories of how CLGs gave rise to the representative extant vertebrate karyotypes, we mapped the homologs of Bb genes assigned to 17 CLGs (Fig. 2C) across the chromosomes of chicken or spotted gar. Most chicken and gar microchromosomes have homologous Bb genes predominantly derived from one single CLG (Fig. 2D and SI Appendix, Fig. S20). Such striking evolutionary stability of microchromosomes spanning the entire chordate evolution supports the hypothesis that they were likely present at the ancestor of bony vertebrates (21, 37–40). Some chicken microchromosomes (e.g., GGA28 and GGA30), like most macrochromosomes, nevertheless are homologous to two or more CLGs (Fig. 2D). When the same combination of CLGs was found for two different homologous GGAs, e.g., GGA28 and GGAZ (homologous to CLG2 and CLG15), we inferred a fusion or translocation likely occurred between 1R and 2R, as illustrated in Fig. 2E. We identified a total of five such putative post-1R chromosome fusions or translocations (SI Appendix, Fig. S21), whose 2R descendant genes are predicted to be grouped together (Fig. 2F, e.g., GGA28 and GGAZ genes) apart from other ohnologs (GGA10 and GGA25 genes) of the same CLG origin but without undergoing post-1R fusions or translocations. This was broadly supported by the phylogenetic trees (Fig. 2F and SI Appendix, Fig. S21) constructed from chicken ohnolog gene groups (Dataset S1 and SI Appendix, Fig. S22). Extending our phylogenetic reconstructions to 243 chicken paralog groups with at least three ohnologs available, we found among the nine CLGs that gave rise to ohnologs distributed on four GGAs (we termed genes of each of these four GGAs as “ohno linkage group,” ohno-A, B, C, D), six CLGs’ ohnolog trees exhibited a phylogenetic structure that strongly supported the 2R hypothesis (SI Appendix, Fig. S23). That is, ohnologs from two GGAs of the same post-1R origin (ohno-A/B or C/D) were grouped together in their phylogenetic trees. When such ohno linkage groups involve microchromosomes, we revealed that microchromosomes always contain much less ohnologs than the other macrochromosomes of the same post-1R origin (SI Appendix, Fig. S24). This led to our conclusion that microchromosomes possibly originated by asymmetric sequence loss after the 2R in the vertebrate ancestor.
By concatenating chicken ohnologs from the same ohno linkage group (A, B, C, or D), together with their orthologs of human, mouse and gar, we constructed their phylogenetic trees and dated the timing of 1R and 2R (Fig. 2G). The 1R was estimated to occur 547 Mya, in less than 10 My since the divergence of chordate common ancestor (Fig. 2G). In addition, we estimated that jawed vertebrates experienced 2R about 517 Mya (Fig. 2G), 10 My after their divergence from jawless vertebrates (SI Appendix, Fig. S3).
Amphioxus-Specific Gene Duplications.
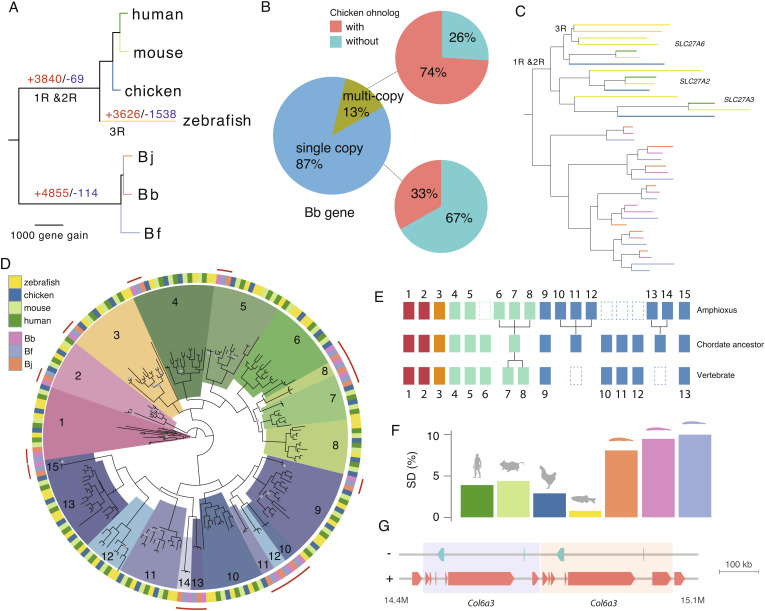
Despite not having undergone WGDs, the three amphioxus species have a comparable number of protein-coding genes (between 22,733 and 26,497) to that of vertebrates (SI Appendix, Fig. S25A). By phylogenetic reconstruction of 8,464 orthologous gene groups whose members are present in both amphioxus and vertebrates, we estimated that the amphioxus ancestor had acquired 4,855 genes (Fig. 3A), some of which may also result from gene loss in the vertebrate ancestor. Interestingly, genes that retained at least two paralogs in vertebrates are more likely to have undergone duplications in amphioxus (P < 1.71e-13, Fisher’s exact test, SI Appendix, Table S4), suggesting convergent gene gains in vertebrates and amphioxus. For example, among the orthologous gene groups that have multi-copy genes in Bb, 74% have multicopy homologs in chicken, but only 33% of the orthologous gene groups with single-copy Bb genes have multiple homologs in chicken (Fig. 3B). We also found cases of recurrent duplication in amphioxus (SI Appendix, Fig. S25B) as demonstrated by a recent study for MRF genes (41). For instance, there are three ohnologs of the Slc27a gene family derived from a single chordate ancestral gene which was independently duplicated multiple times at the ancestor of amphioxus (Fig. 3C).
Fig. 3.
Gene expansion in amphioxus species. (A) Reconstructed gene gains (red) and losses (blue) events during the chordate evolution based on the ortholog gene groups. The branch length is scaled to the number of gene gain. (B) Using Bb as an example, we show duplicated genes in amphioxus more frequently have paralogs in vertebrates. A majority (74%) of the Bb multi-copy genes have chicken paralogs compared with only 33% of Bb single-copy genes. (C) Independent expansion of SLC27A gene copies in vertebrates (due to WGD) and amphioxus (due to gene duplication). Each species (zebrafish and three amphioxus species) is marked with the same color as shown in (A). (D) Phylogenetic tree of Hox genes. The homologous Hox gene (denoted by the number) group of amphioxus and vertebrates was marked in the same color. The grey dots at the internal nodes indicate a bootstrapping value lower than 60. (E) An inferred model of Hox gene evolution in chordates according to the results of (D). Dashed boxes denote gene loss, each aligned column denotes homologous relationship, individual gene duplications are also shown for either amphioxus or vertebrates. (F) Amphioxus has a higher portion of genome derived from segmental duplication compared to vertebrates (G) One example of segmental duplication involving Col6a3 in Bf. The two copies are next to each other highlighted in different background colors.
The other prominent case of convergent gene acquisition in amphioxus and vertebrates is demonstrated by certain members of Hox genes. Amphioxus has one prototypical Hox gene cluster (AmphiHox), whose posterior Hox genes (e.g., Hox14) (42) have an ambiguous orthologous relationship with the vertebrate Hox paralog groups (HPGs), leaving the Hox gene number of chordate ancestor still controversial (7, 43, 44). Our phylogenetic analysis confirmed one-to-one homologous relationships of some Hox (1–5, 9, 15) genes between amphioxus and vertebrates, dating their likely existence to the chordate ancestor (Fig. 3D and SI Appendix, Fig. S26). Other Hox genes likely have undergone gain and loss events independently in the ancestors of the two clades’ (Fig. 3E). For instance, the amphioxus Hox6-8 and the vertebrate HPG8 seem to be acquired after the two chordate clades diverged from each other. The posterior amphioxus Hox genes Hox10-12 and Hox13-14 are respectively grouped with the vertebrate HPG9 and HPG11-13, suggesting amphioxus-specific duplications from an ancestral chordate Hox gene that might have subsequently become lost in the vertebrate ancestor. Similar to HPGs, majorities of amphioxus Hox genes exhibit a temporal ollinearity of expression pattern (45, 46), with the anterior genes expressed in earlier developmental stages than the posterior genes (SI Appendix, Fig. S27).
One major molecular mechanism that contributed to the gene acquisition of amphioxus is segmental duplications, which tend to be of more recent origin and often species-specific (SI Appendix, Fig. S28). Segmental duplications accounted for a higher percentage of the genome in amphioxus vs. vertebrates (9% vs. 3.5%, Fig. 3F); they are on average 7.8 kb long but can be up to 300 kb (Fig. 3G and SI Appendix, Fig. S29). These duplicated segments encompass genes that are enriched for GO categories of G-protein coupled receptor activities, protein tyrosine kinase activities or nucleic acid binding functions (SI Appendix, Table S5). These genes are also frequently enriched for multi-copy ohnologs in vertebrates (47–49). Transcriptional factors or genes involved in early development that are often retained after vertebrate WGDs (50, 51), however, are not enriched in amphioxus segmental duplicates.
Developmental Dynamics of Amphioxus Chromatin Architecture.
Eukaryotic genomes are folded into (active/A or inactive/B) chromatin compartments and to a finer scale of TADs. Such hierarchical three-dimensional (3D) chromatin architectures were previously shown in Drosophila, teleosts, and mammals to be gradually established or reprogrammed during embryonic development (52–54).
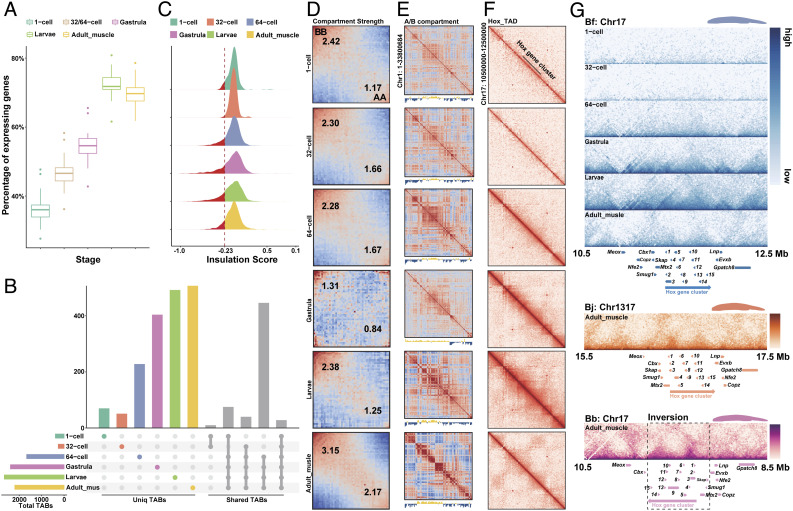
To examine whether this is a broadly conserved feature between invertebrates and vertebrates, we collected time-series population Hi-C data of Bf spanning six developmental stages of 1-cell zygote, 32-cell, 64-cell embryos, gastrula, larvae, and adult muscle tissues (SI Appendix, Table S6). Both the percentage of actively transcribed genes (Fig. 4A) and the total number of TAD boundaries (TAB) (Fig. 4B and SI Appendix, Figs. S30 and S31) display a significant (P < 0.01, Wilcoxon test) increase after zygotic genome activation (ZGA) around the 64-cell stage (55). The strength of TABs measured by insulation scores also becomes generally intensified during development particularly in those strong TABs (Fig. 4C). These patterns are similar to those found in Drosophila and mammals (53, 56) where major TAD structures of zygote genomes emerge after, although do not necessarily depend on ZGA. In contrast to mammals and Drosophila, the amphioxus genome is highly compartmentalized before ZGA. The A/B compartment strength further becomes significantly (P < 0.05, SI Appendix, Fig. S32) increased after embryonic stages, but becomes decreased, i.e., possibly reprogrammed on some chromosomes at the gastrula stage (Fig. 4 D and E and SI Appendix, Figs. S32 and S33).
Fig. 4.
Developmental dynamics of amphioxus chromatin architecture. (A) The percentage of actively transcribed genes (TPM > 1) across five developmental stages of 1-cell zygote, 32-/64-cell, gastrula, larvae, and adult muscle tissues of Bf. (B) The number of TABs at 5 kb resolution across six developmental stages of Bf. The horizontal bars show the number of TABs of each stage. The vertical colored bars show the number of specific TABs of each stage, and the grey bars show the number of shared TABs among six stages. (C) The distribution of insulation scores of TABs across different stages. The smaller the insulation score is, the higher strength the TAB has. (D) Saddle plots of amphioxus Hi-C data binned at 250 kb resolution at six different developmental stages. Bins are sorted by their PC1 value. B-B (inactive-inactive) interactions are in the upper left corner, and preferential A-A (active–active) interactions are in the lower right corner. Numbers in the corners show the strength of AA interactions as compared to AB interaction and BB interactions against BA interactions. (E) Correlation matrix and eigenvector 1(PC1) values value tracks for amphioxus chromosome 1 at 250 kb resolution at six different developmental stages. (F) Distribution of interaction at 15 kb resolution at the Bf Hox cluster. (G) Distribution of TADs at the 15 kb resolution in three different amphioxus Hox regions with the gene tracks.
To explore the formation mechanisms of TADs in amphioxus, we examined the TABs and found that they are enriched for putative binding motifs of chromatin architectural protein CTCF (CCCTC-binding factor) (SI Appendix, Fig. S34), whose transcription level is also specifically increased at ZGA (SI Appendix, Fig. S35). There are disproportionately more (>52%) CTCF-binding site pairs present with convergent forward and reverse orientations at the two TABs of the same TAD (SI Appendix, Fig. S36) (57). These results together suggested that similar to vertebrates, loop extrusion facilitated by CTCF protein might play an important role during the formation of TADs upon ZGA of amphioxus. Another mechanism of TAD formation, i.e., self-organization likely mediated by heterochromatin interactions, could also play a role; however, it requires chromatin profiling data of different embryonic stages before and after ZGA to be tested in the future.
Once established at 64-cell stage, 26.82% of the TABs are overlapped with those in all the later developmental stages, with about 16.88 to 22.95% of TABs only present in one certain stage or tissue (Fig. 4B). This indicates that similar to Drosophila and mammals, substantial numbers of, but not all TADs become stabilized and conserved across stages after ZGA, with many others showing dynamic changes during development. To further illustrate this process, we scrutinized the Hox cluster of Bf, which is encompassed in one single TAD from 1- to 64-cell stages, but becomes segregated into two TADs (SI Appendix, Fig. S30) since the gastrula stage during later development (Fig. 4F). The TABs within the Hox cluster is weak at gastrula and larvae stages, but becomes clearer in the adult tissue (Fig. 4F). This is in contrast to the previous result that characterized the Hox cluster of European amphioxus (B. lanceolatum) as being included in one TAD. This previous work used the 4C technique and pooled samples of different embryonic stages (8). The pooling might have interfered with resolving the Hox cluster TAB, which is supposedly also weak during the embryonic stages of B. lanceolatum. This needs to be verified in the future with samples of separate embryonic stages, as well as adult samples. For the three amphioxus species studied in this work, the Hox TABs in adult muscles seem to be conserved around Hox7. Interestingly, the entire Hox cluster of Bb (together with three neighboring genes) is included in a large genomic inversion (Fig. 4G) that occurred after its divergence from Bj in the last 50 My, with its functional impact on the Bb genome remained to be elucidated in future.
Evolutionary Turnovers of Sex-Determining Regions between Amphioxus Species.
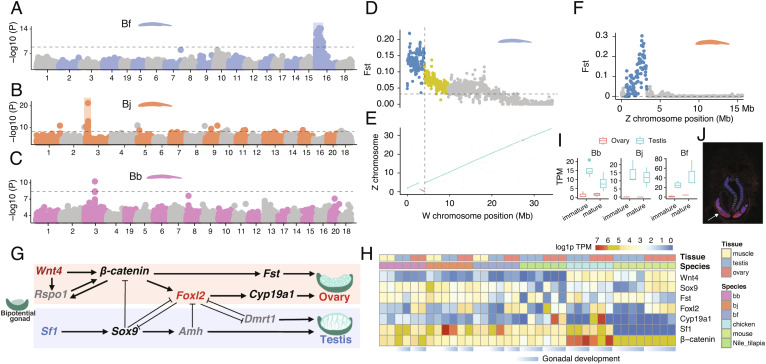
The sex-determination (SD) mechanisms of amphioxus remain largely enigmatic, with no cytogenetic evidence for the existence of differentiated sex chromosome pair in Bf and Bb (24, 58). Using whole-genome resequencing data of between 10 and 48 individuals per sex per species (SI Appendix, Table S7), we identified the sexually differentiated regions (SDR) that harbor female-associated variants, i.e., excessive female heterozygotes, and are not shared between the three amphioxus species (Fig. 5 A–C). In particular, the SDR of Bf is located on Chr16 and harbors 194 genes (SI Appendix, Table S8); and those of Bj and Bb are located at two different genomic loci of Chr3, harboring 35 genes and one gene with unknown function respectively (SI Appendix, Table S9). Bf Chr16 is not homologous with Chr3 of Bj or Bb (Fig. 1C). These SDRs consistently exhibit the highest levels of population differentiation (measured by Fst) between sexes throughout the genome (SI Appendix, Fig. S37), but do not exhibit sexually differentiated patterns of mapped read coverage. These results together indicated that all three amphioxus species have independently evolved female heterogametic sex chromosomes, and the result of Bf is consistent with a recent genetic study (59).
Fig. 5.
Turnovers of SDRs between amphioxus species. (A–C) Genome-wide association study (GWAS) identified the sex-linked regions in amphioxus. The Y axis shows the log10 transformed P value of GWAS. (D) The FST statistics between male and female populations of Bf reveal the evolutionary strata. Each dot represents a 50 kb sliding window. The horizontal dashed lines show the genomic average levels. (E) The synteny plot between the Z and W chromosomes of Bf. The purple lines represent reversed alignments. The vertical dashed line indicates the boundary of stratum 1 as well as the inversion. (F) The FST statistics between male and female populations of Bj. (G) The 10 conserved vertebrate SD pathway genes, genes in grey are absent in amphioxus. Only Foxl2 and Sf1 are sex-biased in amphioxus. (H) The expression profiles of chordate SD-related genes over developing gonads. (I) The candidate Bb SD gene has a conserved testis-biased expression. (J) RNA fluorescence in situ hybridization shows the candidate Bb SD gene has a specific expression in testis.
The homomorphic sex chromosomes of amphioxus are similar to those of many fish and frog species, sharing the feature of rapid evolutionary turnovers between species (60). This is in contrast to the relatively stable and highly differentiated sex chromosomes of most birds and mammals and may be explained by the “fountain-of-youth” hypothesis. It postulates that occasional sex reversal may induce rare recombination between sex chromosomes and prevent them from becoming differentiated (61). Supporting this, we found between 10% and 40% of the phenotypic female or male individuals of the three species exhibit a genotype of the opposite sex in their SDRs (SI Appendix, Fig. S38).
With the advantage of fully assembled sequences of ChrZ of Bb and Bj, and particularly those of both ChrZ and ChrW of Bf (Fig. 1), we further reconstructed the evolutionary history of these species’ SDRs. The SDR of Bf can be divided into two regions which likely have suppressed or reduced homologous recombination between ChrZ/W at different time points [termed “evolutionary strata” (62)]. The older stratum spans 4.1 Mb sequence at one end of Bf ChrW chromosome and exhibits uniformly much higher levels of ChrZ/W pairwise sequence divergence and intersexual Fst than the rest SDR (Fig. 5D and SI Appendix, Fig. S38). The boundary of this stratum aligns with that of chromosomal inversion between ChrZ/W of Bf (Fig. 5E and SI Appendix, Fig. S39), which probably accounted for the recombination suppression in this stratum. In contrast, the Fst values and ChrZ/W sequence divergence levels are not uniform in the rest SDR of Bf (4.1 Mb to 11.5 Mb), suggesting homologous recombination may have been gradually reduced without involving chromosomal inversions (SI Appendix, Fig. S38). To verify the sex-linked region of Chr16 in Bf, we generated a heterozygous female mutant strain of Pitx located on Chr16, and found that its mutant alleles are only carried by their daughters, but the mutant alleles can be found only in sons of male heterozygous mutant strain (SI Appendix, Fig. S40) The SDRs of Bj and Bb do not exhibit a pattern of “evolutionary strata” and seem to have gradually reduced recombination, suggested by their Fst patterns (Fig. 5F and SI Appendix, Figs. S41 and S42).
The SDR of each amphioxus species is expected to harbor respective upstream sex-determining genes, which may constitute the sex-determining pathways together with genes on the other chromosomes. We examined the orthologs of 10 reported vertebrate sex-determining genes and found none of them are present in SDRs of amphioxus. Three upstream SD genes of some vertebrates, Dmrt1 (SI Appendix, Fig. S43), Amh and Rspo1 do not have an ortholog in the amphioxus genome (Fig. 5G); among the rest, only Sf1 and Foxl2 exhibit a testis- or ovary-biased expression pattern in amphioxus (Fig. 5H). The result of Dmrt1 is consistent with a recent study characterizing the Dmrt family genes across bilaterian species, which reported Dmrt1 and its orthologs are only found in vertebrates (63). This suggests that many key genes of the vertebrate sex determination pathway may originate after the WGDs. Among the amphioxus SDR genes, we identified a candidate Bj SD gene that is absent in Bf and Bb (SI Appendix, Fig. S44), and a candidate Bb SD genes (named tesD), on the other hand, are present in Bf and Bj though not in vertebrates, and shows specific or biased expression in amphioxus testis (Fig. 5 I and J). These results together indicated that amphioxus and vertebrates independently evolved their SD pathways.
Conclusions
With three reference-quality genomes of amphioxus, we uncovered their interspecific diversities of genes and chromosomes to an unprecedented resolution. This enabled more direct and accurate reconstruction of ancestral status of the ancestors of both amphioxus and chordates, which was previously based on the draft genome of one amphioxus species. We inferred that there were 20 ancestral linkage groups in the ancestor of Branchiostoma amphioxus, best approximated by the Bb genome; and confirmed there were 17 ancestral linkage groups in the chordate ancestor (18, 21). The phylogenetic analyses of vertebrate ohnologs and their amphioxus orthologs dated the timing of WGDs, and further characterized the rearrangements and asymmetric loss/retention among the duplicated descendants of CLGs that gave rise to the vertebrate ancestral karyotype. These evolutionarily distant comparisons between amphioxus and vertebrates can be attributed to the slow-evolving genomes of the former relative to those of urochordates.
Our analyses also revealed shared or independently evolved genomic features of amphioxus and vertebrates. For example, both clades seem to establish their major TAD architecture after ZGA, and form two TADs within the Hox gene cluster, suggesting these patterns probably originated in their chordate ancestor. In the absence of WGDs, amphioxus species expanded their gene repertoire by segmental duplications or individual gene duplications; and independently evolved their sex determination pathways from each other, and from vertebrates. By the development of rich genomic resources from this and previous works (17, 21, 22), as well as that of gene knockout techniques (64), we expect the resurgence of interest into this classic evo-devo model organism, with more functional insights into its genes to be uncovered in future.
Materials and Methods
Genome Sequencing and Assembly.
Bb and Bj were collected from Xiamen Rare Marine Creature Conservation Areas (Fujian, China) and Bf was introduced from Dr. Jr-Kai Yu’s laboratory (SI Appendix, Fig. S1). All of them were cultured as previously described (64, 65). Interspecific hybrids were produced by pooling the sperm of one species, and the eggs of another species except that Bj and Bb cannot be crossed with each other. We extracted high molecular weight genomic DNAs from the muscle tissues of a single individual (male Bj/Bf F1 offspring, Bb/Bf F1 offspring with unidentified sex) using the DNeasy Blood & Tissue Kit (QIAGEN, Valencia, CA), and inspected the DNA quality by Qubit 2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA) and 2100 Agilent Bioanalyzer (Agilent). We prepared the 20 kb SMRTbell™ PacBio libraries and generated sequencing data of ~50G for the two hybrids (Bb/Bf and Bj/Bf) at Annoroad Gene Technology (Beijing, China). We estimated the heterozygosity levels of three species’ genomes using Illumina reads by GenomeScope (66). For the hybrids, the estimated genome size was equivalent to the sum of the haploid genome sizes of the parental species (SI Appendix, Fig. S1).
We used Falcon (67) to assemble the PacBio subreads of two hybrid samples, after discarding raw subreads and corrected reads (preads) shorter than 8 kb. We used the following parameters to avoid collapse of reads derived from different parental species: pa_HPCdaligner_option = -v -dal128 -t8 -e0.75 -M24 -l3200 -k18 -h480 -w8 -s100, ovlp_HPCdaligner_option = -v -dal128 -M24 -k24 -h1024 -e.96 -l2500 -s100. We used the arrow (67) using pilon (1.22) (68) to polish the contigs with the Illumina reads from the same hybrid individual. We aligned the Illumina reads of either parental species to the contigs by bwa-mem with default parameters, and only kept the alignments with a mapping quality higher than 60. For each contig, we calculated the proportion of nucleotide sequences that were mapped by each species’ reads (coverage), without considering the contigs shorter than 20 kb. We assigned a contig to either parental species if the sequencing coverage was larger than 10% for one parental species, while the sequencing coverage for the other was below 1% (SI Appendix, Fig. S2). We then used minimap2 (2.15-r905) (69) to align the PacBio reads of hybrids to the assembly, with the option “--secondary=no,” and partitioned the species-specific haploid reads. These partitioned reads were used for assembling the four haploid assemblies (one Bb, one Bj and two Bf) by Canu (1.6) (70) (“corOutCoverage=200 correctedErrorRate=0.15”) and Falcon (“pa_daligner_option= -k18 -e0.7 -l2000 -h480 -w8 -s100, ovlp_daligner_option=-k24 -e.93 -l2000 -h600 -s100”). Since the read length of Bb/Bf was longer, we increased the “−l” parameter from 2,000 to 2,500 in “pa_daligner_option” and from 2,000 to 3,000 in “ovlp_daligner_option.” The polishing steps were similar to those for the diploid assembly of hybrids. Then contigs of two pipelines were merged: we aligned the Canu contigs against the falcon contigs using the nucmer aligner (MUMmer 3.0) (71) with the option -b 400. When one Falcon contig spanned the boundaries of two Canu contigs, we linked the Canu contigs with a gap of 200 Ns.
Finally, we used the Juicer (1.7.6) (72) and 3D-DNA (180922) (73) to connect the contigs into chromosome-level scaffolds, with the following parameters: --editor-coarse-resolution 500000 --editor-coarse-region 1000000 --editor-saturation-centile 1 -r 0 --editor-repeat-coverage 1 --editor-coarse-stringency 70. We manually curated the chromosome assembly by Juicebox (1.90) (74) and updated the assembly using the “review” module of 3D-DNA. The unanchored scaffolds are highly repetitive, with repeat content as high as 79.0%, 63.5%, and 81.7% for Bb, Bj, and Bf, respectively.
Genome Annotation.
We generated Iso-seq and RNA-seq data from whole-body adult male and female individuals of the three species. We used IsoSeq3 (3.1.0) (75) and Trimmomatic (0.36) (76) for pre-processing the raw reads. Then we generated reference-guided and de novo assembled transcript sequences using Cupcake (5.8) with Iso-Seq reads, and StringTie (1.3.3b) (77) (-m 300 -j 5 -c 8) and Cufflinks (2.2.1) (78) (–multi-read-correct –max-intron-length 30000) and Trinity (2.6.6) (79) (--min_glue 10 --path_reinforcement_distance 30 --min_contig_length 400 --jaccard_clip) with RNA-seq reads. We then used the Mikado (1.2.2) (80) to integrate all transcript sequences. We used RepeatModeler (1.0.10) (81), Tandem Repeat Finder (409) (82) (“2 7 7 80 10 50 500 -d -l 6”) and MITE_Hunter (83) (“-I 86 -n 8 -c 8”) for annotating and classifying the repeat families.
To produce a consensus gene model, we ran MAKER (2.31.10) (84), after masking the annotated repeats. We used the query protein sequences from NCBI RefSeq database (Bb: GCA_001625405.1 and Bf: GCA_000003815.1) and the transcriptome annotations produced by Mikado. The MAKER gene annotation was then used to train SNAP (2013–11–29) (85) (maker2zff -c 0.99 -e 0.99 -o 0.99 -l 800 -x 0.01) and AUGUSTUS (3.3) (86) for ab initio predictions. Gene evidence from protein alignment, StringTie transcripts, ISO-seq transcripts, SNAP and AUGUSTUS predictions, were combined by EvidenceModeler (1.1.1) (87), with the highest weight on the protein alignment and StringTie transcripts (10), intermediate weight on ISO-seq transcripts (5) and the lowest weight on the ab initio predictions. We used the PASApipeline (v2.3.3) (88) to polish the gene models. We used InterProScan (5.35 to 74.0) (89) to annotate GO for the predicted coding genes.
Based on the RepeatMasker results, we inferred that the most abundant and longest satellite sequences were associated with centromeres. The identified centromeric monomer of Bf is consistent with the reported result (90). The recombination rates were estimated with ReLERNN program. The nucleotide diversity was estimated in 100 kb windows using VCFtools (0.1.16) (91, 92). To annotate telomeres, we searched for clusters of (AACCCT)n repeats throughout the genomes using RepeatMasker. We only kept those with a total length of 200 bp (33.3 consecutive AACCCT repeats) to reduce false positives. We used the R package Quadron (93) to predict the G-quadruplexes (G4) throughout the genome with default settings, then calculated the length of G4 elements over 20 kb sliding windows along the chromosomes using bedtools coverage.
For the fluorescence in situ hybridization (FISH) experiment, chromosome preparations were obtained from regenerated tissues from cute tails of a male and a female of individual, using colcemid/hypotonic solution treatment protocols (94). To verify the chromosome fusion in the Bb and BJ, we prepared two probe of fusion regions in the BJ Chr4 (Chr4:18824480-18838055; Chr4:18824480-18838055) and labeled with CY3 and CY5, respectively. FISH experiment was performed as a previously described in method (95). An Olympus BX53 epifluorescence microscope was used to observe metaphase plates with fluorescent signals that were photographed with a cooled CCD camera and visualized using cellSens Dimension 1.9 software (Olympus Corporation, Tokyo, Japan).
Comparative Genomic Analyses.
We included three amphioxus species and four vertebrate species [human (GCF_000001405.39), mouse (GCF_000001635.26), zebrafish (GCF_000002035.6), and chicken (GCF_000002315.6)], with the longest transcript of each gene to infer the orthologous gene groups. We ran OrthoFinder (2.2.7) (96) to group the orthologous genes, diamond (0.9.21) for protein alignment. We used Last (1042) (97) to align genomes of mouse (GRCm38.p4), chicken (GRCg6a), zebrafish (GRCz11), Bb, Bj, and Bf against the human genome (GRCh38.p12), with -uMAM4 for mouse alignment, and more sensitive -uMAM8 for other species, and merged the one-to-one best alignments by Multiz (v11.2) (98).
For reconstructing the chordate phylogeny, we added Ciona intestinalis (GCA_009617815.1) (99) and scallop (Mizuhopecten yessoensis, ASM211388v2) (100), with the latter set as an outgroup. We excluded the alignments in which the sequences were aligned to non-homologous chromosomes among amphioxus for alignment errors. The filtered alignments contained 5,074 loci, with a total size of 276,373 bp. We used IQ-TREE (2.0-rc1, TVMe+R3) (101), to construct the phylogenomic tree, and ran bootstrapping for 100 times. We used the PhastCons from the PHAST package (1.5) (102) to annotate the conserved non-coding elements across the genomes.
Ancestral Karyotype Reconstruction.
We generated whole genome alignments between amphioxus species by minimap2 (2.15-r905) (70) (-x asm20) and visualized the alignments by D-Genies online tool (103) (SI Appendix, Fig. S16). We selected 7269 orthologous gene groups (orthogroups) in which Bb genes are located within the same chromosome. A total of 1,799 orthogroups contained more than one gene in chicken which were informative for reconstructing the chordate ancestral karyotype. For each Bb chromosome (i), we asked which chicken chromosome (j) its homologous genes belong to, and counted the gene number for each chicken chromosome (Ckij). Then we calculated the relative abundance of genes of a chicken chromosome for a given Bb chromosome (nCKij):
We included 33 chicken chromosomes, and retained a chicken chromosome when the nCKij value was larger than 4%, for a given Bb chromosome (i). Then we visualized the nCKij values for every Bb chromosome with a network-style graph (Fig. 2B and SI Appendix, Fig. S17), using the igraph R package. We used 244 orthogroups that retained three or four chicken ohnologs and performed coding sequence alignments using MAFFT (v7.294b) (104). Then we constructed the phylogenetic tree using concatenated sequence alignments of the same CLG using IQ-TREE, with 1,000 times bootstrapping. Based on the phylogenetic relationships, we assigned the four ohno-chromosomes derived from a single CLG as ohno-A, ohno-B, ohno-C, and ohno-D. For each ohno-chromosome group, we further included the orthologous genes of human, mouse, and spotted gar of the chicken gene in that group (Dataset S1 and SI Appendix, Fig. S21). Then all the coding sequences of three amphioxus species and vertebrates were aligned with MAFFT (7.427) (104) and GUIDANCE2 (2.02) (105) pipeline, producing concatenated alignments with 409,659 nucleotide sites. We then used BASEML (4.9j) (106) to estimate the overall mutation rate with the time calibration on the root node (575 My for the vertebrate and amphioxus split (107). The topology “((bf,(bb,bj)),((((chicken-Ohn_A,(human-Ohn_A,mouse-Ohn_A)),gar-Ohn_A),((chicken-Ohn_B,(human-Ohn_B,mouse-Ohn_B)),gar-Ohn_B)),(((chicken-Ohn_C,(human-Ohn_C,mouse-Ohn_C)),gar-Ohn_C),((chicken-Ohn_D,(human-Ohn_D,mouse-Ohn_D)),gar-Ohn_D))))” was used. General reversible substitution model and discrete gamma rates were estimated by maximum likelihood approach under the strict clock. The divergence time was then estimated using MCMCtree (4.9j) (clock 3) (108), with three soft-bound calibration time points (SI Appendix, Table S2): 514-636.1 My for the vertebrate and Branchiostoma species split, and 65.6 to 64.6 My for the human and mouse split, 318 to 332.9 My for the chicken and mammal split, 378.2 to 422.4 My for the zebrafish-spotted gar split according to the Fossil Calibration database (109). We used priors of G (1, 8.49) for the overall substitution rates (rgene_gamma), G (1, 4.5) for the rate-drift parameter (sigma2_gamma). The MCMC chains were first run for 500,000 as burn-in, and then were sampled every 400 generations until a total of 20,000 samples were collected. The out tree file was visualized and trimmed by FigTree v1.4.4.
Gene Evolution.
We used SDquest (0.1) (110) to identify segmental duplications (SDS) in all amphioxus species and other studied vertebrates including human (hg38), mouse (mm10), chicken (galGal6), and zebrafish (danRer11). We excluded the sex chromosomes of human, mouse, and chicken, and alternate-loci scaffolds of zebrafish as these sequences may confound the identification of SDS. We only kept SDS that are longer than 1,000 kb and show a sequence similarity level of at least 70%. For studying gene gain and loss, we selected 8,464 orthologous gene groups that contain at least one vertebrate species and one amphioxus species as the input for Notung (2.9.1) (111) gene family reconstruction. We identified 200 orthogroups that had more than one gene copy in all amphioxus species, but had single-copy genes in vertebrates. The mean copy number of the expanded gene families were 3.6, 3.8, and 4.8 for Bb, Bj, and Bf, respectively. To elucidate the evolution of the Hox genes across chordate species, protein, and CDS sequences of chicken, mouse, human, and zebrafish Hox genes were downloaded from NCBI, and aligned to those of amphioxus species by MAFFT (v7.407), with alignment polishing by trimAl (v1.4.rev15) (112). We used IQ-TREE to infer the phylogeny, and the AVX+FMA model was selected automatically by IQ-TREE. We used EvolView online tool (https://www.evolgenius.info/evolview) to visualize our phylogenetic tree. RNA-seq data of multiple Bf developmental stages were downloaded from NCBI SRA (PRJDB3785) for estimating the Hox gene expression level using HISAT2 (2.0.4) and featureCounts (v1.5.2).
3D Genome Analyses.
In situ Hi-C libraries were constructed from the muscle and embryonic tissues of B. floridae, and the adult muscle tissues of B. japonicum and B. belcheri as described before (113). Hi-C data were mapped to the genomes using bwa-mem (0.7.17-r1188) with parameters “-A 1 -B 4 -E 50 -L 0.” The quality control including valid pairs and cis/trans ratio of Hi-C data was finished by using pairtools (0.3.0) (https://pairtools.readthedocs.io/en/latest/) and the estimated resolution was calculated by HiCRes (2.0) (114).
Then we used HiCExplorer (2.2.1) suite (115) to generate the coordinates of TADs and the TAD insulation score of each bin (--thresholdComparisons = 0.01, --delta = 0.01). To investigate the overlaps of TABs between stages, we combined the TABs of all stages into one set, and extended each boundary for 5 kb of both sides to form 15 kb windows and merged adjacent windows when their distance was not longer than 10 kb. This generated a set of boundaries that existed in at least one developmental stage. We then compared the boundaries of each stage to this common set, and defined conservation of boundaries as an overlap of at least 15 kb in size. We used cooltools (0.3.2) (https://cooltools.readthedocs.io/en/latest/) and to call A/B compartments with a 250 kb resolution. Compartment strength was calculated as AA × BB/AB2 for each chromosome. Saddle plot was also obtained by cooltools. We used FIMO (Find Individual Motif Occurrences) (116) to search for human CTCT motif (MA0139.1) in the amphioxus genomes and identified 62,987 putative CTCF motifs. To test whether the CTCF motif was enriched in the TAD boundary, we used bedtools intersect to identify the CTCF motifs located in the 15 kb TABs (5 kb boundary extended by 5 kb of both sides) of the six developmental stages. In addition, we also checked whether the TABs contain more CTCF motifs than by chance, we randomly selected 15 kb windows across the genome and calculated the proportion of windows that contain CTCF motifs (SI Appendix, Fig. S45). The pairings of convergent CTCF sites at domain boundaries are considered as a hallmark of the conserved role of CTCF/cohesion in TAD formation (117). The enrichment pattern of putative CTCF binding sites (SI Appendix, Fig. S34) and the distribution pattern of convergent CTCF site pairs (SI Appendix, Fig. S36) were similar for TAD results derived from different TAD-calling bin sizes.
Sex Chromosome Analyses.
Pitx mutants were generated and detected using the TALEN method as described before (118). The TALEN pair used for mutant generation are Fw3: 5′-GCAACCGTTCGACGAC-3′ and Rv3 5′-TGTAGGCCGGCGAGTA-3′ which are from the third coding exon of the gene. A Tat restriction site was included in the target site for genotyping and primer pair used for genotyping are Pitx-TALEN-PCR-F2 (5′-AGGTCTGGTTCAAGAACCG-3′) and Pitx-TALEN-PCR-R4 (5′-TCACGGTAAGCGTAAGGCTG-3′). Two different mutant stains were generated. The founder of stain 1 is a female, which was crossed with a wild-type male to generate F1 offspring, from which a female heterozygote was further crossed with a wild type to generate F2 descendants. In contrast, the founder of strain 2 was a male and an F1 heterozygous male was used to generate its F2 descendants.
We g-enerated re-sequencing Illumina data of multiple individuals of both male and female (on average 25 individuals of each sex) at a coverage larger than 20X (SI Appendix, Table S7). The raw reads were mapped to the genome using bwa-mem (0.7.16a), and we used GATK (3.8) (119) pipeline to call variants. We filtered the SNPs with the following criteria: QD < 2.0 || FS > 60.0 || MQRankSum < -12.5 || RedPosRankSum < -8.0 || SOR > 3.0 || MQ < 40.0, and used the biallelic SNPs (bcftools -m2 -M2) to screen for sex-linked variants. We further excluded the variants that have minor allele frequency less than 0.05 and missing rate larger than 10%. We used Beagle (28Sep18.793) (120) to do the imputation for the variants and used SHAPEIT (v2.r904) (121) to produce a more accurate set of phased genotypes on the variants. A total of 4,954,852, 7,213,889, and 12,016,687 high-quality phased SNPs in Bj, Bb, and Bf, respectively, were used to perform whole-genome association analysis for the sex trait (male or female) with EMMAX (version 8.22) (122). The genome-wide significance thresholds of all tested traits were evaluated with the formula P = 0.05/n (where n is the effective number of independent SNPs). We calculated the FST values between male and female populations using VCFtools (0.1.13) (80). SNPs with more than two alleles were removed. The FST values were estimated in a 10 kb sliding window with an overlapping size of 5 kb. For Bb whose sex-determining region is much smaller, we used 5 kb windows instead of 10 kb. We defined the nonrecombining regions of the sex chromosomes by the sex-linked SNPs identified through the whole-genome association tests. We evaluated the extent of sex chromosome differentiation with two measures: 1) FST and 2) the difference between male and female SNP density. We collected transcriptomes of immature (identifiable but not functionally mature) and mature gonads for studying the candidate sex-determining genes of amphioxus. After aligning and filtering the reads, we used featureCounts (1.6.2) (123) to count the reads mapped to the annotated transcripts and normalized the counts with the TPM (transcripts per million) method.
We chose 10 conserved vertebrate SD pathway genes: Wnt4, Sf1, β-catenin, Rspo1, Sox9, Amh, Foxl2, Fst, Cyp19a1, and Dmrt1 to check their presence or absence in the amphioxus genomes. We first checked the orthogroups that contain those SD genes and whether amphioxus is present in these orthogroups. If amphioxus is absent in the orthogroups, we searched the coding sequences of the SD genes against the amphioxus genomes by BLAST. The absence of Dmrt1 in amphioxus is consistent with a recent study (SI Appendix, Fig. S43) (63).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
Q. Zhou is supported by the National Natural Science Foundation of China (32170415 and 32061130208), Natural Science Foundation of Zhejiang Province (LD19C190001), European Research Council Starting Grant (grant agreement 677696) and start-up funds from Zhejiang University. L. Xu. is supported by the Erwin Schrödinger Fellowship (J4477-B) from the Austrian Science Fund and start-up funds from Southwest University. Z.H. is supported by Natural Science Foundation of Fujian Province of China (2022N0012) and the 13th Five-Year Plan for the Marine Innovation and Economic Development Demonstration Projects (FZHJ14 and FZHJ11) from Fujian Provincial Fund. W.C. is supported by the Research Foundation of the Education Bureau of Fujian Province (JT180104). Q. Zhou is supported by the Natural Science Foundation of Fujian Province (2019J01277). J.-K.Y. is supported by intramural funding from the Cellular and Organismic Biology, Academia Sinica, and grants from the Ministry of Science and Technology, Taiwan (105-2628-B-001-003-MY3; 108-2311-B-001-035-MY3). The Life Science Compute Cluster (LiSC) of University of Vienna provided the computational resources.
Author contributions
Z.H., L. Xu, G. Lin, Q. Zhang, and Q. Zhou designed research; Z.H., L. Xu, C.C., Y. Zhou, J.L., Z.X., Z.Z., W.K., G. Li, G. Lin, Q. Zhang, and Q. Zhou performed research; Y. Zhou, J.-K.Y., G. Li, and Q.Z. contributed new reagents/analytic tools; Z.H., L. Xue, J.L., W.K., W.C., S.P., D.C., C.S., X.W., Y.H., C.X., Y. Yan, Y. Yang, T.X., W.H., X.H., Y. Zhang, Y.C., C.B., C.H., L. Xue, S.X., Z.Y., Y.J., and Q.Z. analyzed data; J.-K.Y. comment on the paper; and Z.H., L. Xu, C.C., E.D.J., G.L., and Q. Zhou wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Contributor Information
Luohao Xu, Email: luohaox@gmail.com.
Gang Lin, Email: lgffz@fjnu.edu.cn.
Qiujin Zhang, Email: qjzhang@fjnu.edu.cn.
Qi Zhou, Email: zhouqi1982@zju.edu.cn.
Data, Materials, and Software Availability
1) Genomic Reads, Genome Assembly and Annotation, Sequencing Data; 2) SNP VCF files, genome annotation files, whole-genome alignments, and Hox gene alignment; 3) Scripts used data have been deposited in 1) GenBank, 2) Dryad (124), 3) Github (125): 1) PRJNA603158 (126), PRJNA603159 (127), PRJNA647830 (128), PRJNA602496 (129); 2) SNP VCF files: https://datadryad.org/stash/downloads/file_stream/2070006, https://datadryad.org/stash/downloads/file_stream/2070005 , https://datadryad.org/stash/downloads/file_stream/2070032; Genome annotation files: https://datadryad.org/stash/downloads/file_stream/2069997, https://datadryad.org/stash/downloads/file_stream/2069996, https://datadryad.org/stash/downloads/file_stream/2069995; whole-genome alignments: https://datadryad.org/stash/downloads/file_stream/2070001; Hox gene alignment https://datadryad.org/stash/downloads/file_stream/2070002; 3) https://github.com/lurebgi/amphioxusGenome. All study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Holland N. D., Holland L. Z., The ups and downs of amphioxus biology: A history. Int. J. Dev. Biol. 61, 575–583 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Bourlat S. J., et al. , Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature 444, 85–88 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Delsuc F., Brinkmann H., Chourrout D., Philippe H., Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature 439, 965–968 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Putnam N. H., et al. , The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Bertrand S., Escriva H., Evolutionary crossroads in developmental biology: Amphioxus. Development 138, 4819–4830 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Holland P., The dawn of amphioxus molecular biology–a personal perspective. Int. J. Dev. Biol. 61, 585–590 (2017). [DOI] [PubMed] [Google Scholar]
- 7.Holland L. Z., et al. , The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1100–1111 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acemel R. D., et al. , A single three-dimensional chromatin compartment in amphioxus indicates a stepwise evolution of vertebrate Hox bimodal regulation. Nat. Genet. 48, 336–341 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Powers T. P., Amemiya C. T., Evidence for a Hox14 paralog group in vertebrates. Curr. Biol. 14, R183–R184 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Venkatesh B., et al. , Survey sequencing and comparative analysis of the elephant shark (Callorhinchus milii) genome. PLoS Biol. 5, e101 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuraku S., et al. , Noncanonical role of Hox14 revealed by its expression patterns in lamprey and shark. Proc. Natl. Acad. Sci. U.S.A. 105, 6679–6683 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oulion S., et al. , Evolution of Hox gene clusters in gnathostomes: Insights from a survey of a shark (Scyliorhinus canicula) transcriptome. Mol. Biol. Evol. 27, 2829–2838 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Feiner N., Ericsson R., Meyer A., Kuraku S., Revisiting the origin of the vertebrate Hox14 by including its relict sarcopterygian members. J. Exp. Zool. B Mol. Dev. Evol. 316, 515–525 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Ohno S., Evolution by Gene Duplication (Springer, 1970). [Google Scholar]
- 15.Holland P. W., Garcia-Fernàndez J., Williams N. A., Sidow A., Gene duplications and the origins of vertebrate development. Dev. 1994 (Supplement), 125–133 (1994). [PubMed] [Google Scholar]
- 16.Smith J. J., Keinath M. C., The sea lamprey meiotic map improves resolution of ancient vertebrate genome duplications. Genome Res. 25, 1081–1090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marletaz F., et al. , Amphioxus functional genomics and the origins of vertebrate gene regulation. Nature 564, 64–70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sacerdot C., Louis A., Bon C., Berthelot C., Roest Crollius H., Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 19, 166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohn M., et al. , Reconstruction of a 450-My-old ancestral vertebrate protokaryotype. Trends Genet. 22, 203–210 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Smith J. J., et al. , The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 50, 270–277 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simakov O., et al. , Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 4, 820–830 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S., et al. , Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat. Commun. 5, 5896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang G., et al. , The oyster genome reveals stress adaptation and complexity of shell formation. Nature 490, 49–54 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Saotome K., Ojima Y., Chromosomes of the lancelet Branchiostoma belcheri Gray. Zool. Sci. 18, 683–686 (2001). [Google Scholar]
- 25.Wang C., Zhang S., Zhang Y., The karyotype of amphioxus Branchiostoma belcheri tsingtauense (Cephalochordata). J. Mar. Biol. Assoc. U. K. 83, 189–191 (2003). [Google Scholar]
- 26.Colombera D., Male chromosomes in two populations of Branchiostoma lanceolatum. Experientia 30, 353–355 (1974). [DOI] [PubMed] [Google Scholar]
- 27.Castro L. F., Holland P. W., Fluorescent in situ hybridisation to amphioxus chromosomes. Zoolog. Sci. 19, 1349–1353 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Graves J. A., Gene amplification in a mouse embryo? Double minutes in cell lines independently derived from a Mus musculus X M. caroli fetus. Chromosoma 89, 138–142 (1984). [DOI] [PubMed] [Google Scholar]
- 29.O’Neill R. J., O’Neill M. J., Graves J. A., Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393, 68–72 (1998). [DOI] [PubMed] [Google Scholar]
- 30.O’Neill R. J., Eldridge M. D., Graves J. A., Chromosome heterozygosity and de novo chromosome rearrangements in mammalian interspecies hybrids. Mamm. Genome 12, 256–259 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Somorjai I. M., Somorjai R. L., Garcia-Fernandez J., Escriva H., Vertebrate-like regeneration in the invertebrate chordate amphioxus. Proc. Natl. Acad. Sci. U.S.A. 109, 517–522 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Q., Li G., Sun Y., Wang Y., Chromosome preparation and preliminary observation of two amphioxus species in Xiamen. Zoolog. Res. 30, 131–136 (2009). [Google Scholar]
- 33.Braasch I., et al. , The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 48, 427–437 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uno Y., et al. , Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS One 7, e53027 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blomme T., et al. , The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 7, R43 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang G., et al. , Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346, 1311–1320 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burt D. W., Origin and evolution of avian microchromosomes. Cytogenet. Genome Res. 96, 97–112 (2002). [DOI] [PubMed] [Google Scholar]
- 38.O’Connor R. E., et al. , Patterns of microchromosome organization remain highly conserved throughout avian evolution. Chromosoma 128, 21–29 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohno J. M. S., Stenius C., Christian L., Kittrell W. A., Atkin N. B., Microchromosomes in holocephalian, chondrostean and holostean fishes. Chromosoma 26, 35–40 (1969). [DOI] [PubMed] [Google Scholar]
- 40.Waters P. D., et al. , Microchromosomes are building blocks of bird, reptile, and mammal chromosomes. Proc. Natl. Acad. Sci. U.S.A. 118, e2112494118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aase-Remedios M. E., Coll-Llado C., Ferrier D. E. K., More than one-to-four via 2R: Evidence of an independent amphioxus expansion and two-gene ancestral vertebrate state for MyoD-related myogenic regulatory factors (MRFs). Mol. Biol. Evol. 37, 2966–2982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrier D. E., Minguillon C., Holland P. W., Garcia-Fernandez J., The amphioxus Hox cluster: Deuterostome posterior flexibility and Hox14. Evol. Dev. 2, 284–293 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Pascual Anaya J., D’Aniello S., Kuratani S., Garcia Fernàndez J., E volution of Hoxgene clusters in deuterostomes. BMC Dev. Biol. 13, 1–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Amemiya C. T., et al. , The amphioxus Hox cluster: Characterization, comparative genomics, and evolution. J. Exp. Zool. B. Mol. Dev. Evol. 310, 465–477 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Pascual-Anaya J., et al. , Broken colinearity of the amphioxus Hox cluster. Evodevo 3, 28 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pascual-Anaya J., et al. , Hagfish and lamprey Hox genes reveal conservation of temporal colinearity in vertebrates. Nat. Ecol. Evol. 2, 859–866 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Semyonov J., Park J. I., Chang C. L., Hsu S. Y., GPCR genes are preferentially retained after whole genome duplication. PLoS One 3, e1903 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brunet F. G., Volff J. N., Schartl M., Whole genome duplications shaped the receptor tyrosine kinase repertoire of jawed vertebrates. Genome Biol. Evol. 8, 1600–1613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li J. T., et al. , The fate of recent duplicated genes following a fourth-round whole genome duplication in a tetraploid fish, common carp (Cyprinus carpio). Sci. Rep. 5, 8199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh P. P., Arora J., Isambert H., Identification of ohnolog genes originating from whole genome duplication in early vertebrates, based on synteny comparison across multiple genomes. PLoS Comput. Biol. 11, e1004394 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McGrath C. L., Gout J. F., Johri P., Doak T. G., Lynch M., Differential retention and divergent resolution of duplicate genes following whole-genome duplication. Genome Res. 24, 1665–1675 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ke Y., et al. , 3D Chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170, 367–381.e20 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Hug C. B., Grimaldi A. G., Kruse K., Vaquerizas J. M., Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169, 216–228.e19 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Chen X., et al. , Key role for CTCF in establishing chromatin structure in human embryos. Nature 576, 306–310 (2019). [DOI] [PubMed] [Google Scholar]
- 55.Yang K. Y., et al. , Transcriptome analysis of different developmental stages of amphioxus reveals dynamic changes of distinct classes of genes during development. Sci. Rep. 6, 23195 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dixon J. R., et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao S. S., et al. , A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang C., Zhang S., Chu J. J. H., G-banding patterns of the chromosomes of amphioxus Branchiostoma belcheri tsingtauense. Hereditas 141, 2–7 (2004). [DOI] [PubMed] [Google Scholar]
- 59.Shi C., et al. , A ZZ/ZW sex chromosome system in cephalochordate amphioxus. Genetics 214, 617–622 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vicoso B., Molecular and evolutionary dynamics of animal sex-chromosome turnover. Nat. Ecol. Evol. 3, 1632–1641 (2019). [DOI] [PubMed] [Google Scholar]
- 61.Perrin N., Sex reversal: A fountain of youth for sex chromosomes? Evolution 63, 3043–3049 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Lahn B. T., Page D. C., Four evolutionary strata on the human X chromosome. Science 286, 964–967 (1999). [DOI] [PubMed] [Google Scholar]
- 63.Mawaribuchi S., Ito Y., Ito M., Independent evolution for sex determination and differentiation in the DMRT family in animals. Biol. Open 8, bio041962 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li G., et al. , Cerberus-nodal-lefty-pitx signaling cascade controls left-right asymmetry in amphioxus. Proc. Natl. Acad. Sci. U.S.A. 114, 3684–3689 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Q.-J., et al. , Continuous culture of two lancelets and production of the second filial generations in the laboratory. J. Exp. Zool. B Mol. Dev. Evol. 308B, 464–472 (2007). [DOI] [PubMed] [Google Scholar]
- 66.Ranallo-Benavidez T. R., Jaron K. S., Schatz M. C., GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 11, 1432 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chin C. S., et al. , Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 13, 1050–1054 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang J., et al. , Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9, 464–472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H., Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34, 3094–3100 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Koren S., et al. , Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kurtz S., et al. , Versatile and open software for comparing large genomes. Genome Biol. 5, 1–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durand N. C., et al. , Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 3, 95–98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dudchenko O., et al. , De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 356, 92–95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Durand N. C., et al. , Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 3, 99–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng D., et al. , Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS One 10, e0132628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bolger A. M., Lohse M., Usadel B., Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pertea M., et al. , StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trapnell C., et al. , Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Venturini L., Caim S., Kaithakottil G. G., Mapleson D. L., Swarbreck D., Leveraging multiple transcriptome assembly methods for improved gene structure annotation. Gigascience 7, giy093 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flynn J. M., et al. , RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. U.S.A. 117, 9451–9457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Benson G., Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han Y., Wessler S. R., MITE-Hunter: A program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 38, e199 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cantarel B. L., et al. , MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Korf I., Gene finding in novel genomes. BMC Bioinformatics 5, 59 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stanke M., Schoffmann O., Morgenstern B., Waack S., Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics 7, 62 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Haas B. J., et al. , Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Haas B. J., et al. , Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 31, 5654–5666 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jones P., et al. , InterProScan 5: Genome-scale protein function classification. Bioinformatics 30, 1236–1240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Melters D. P., et al. , Comparative analysis of tandem repeats from hundreds of species reveals unique insights into centromere evolution. Genome Biol. 14, 1–20 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu J. D., et al. , Sex chromosomes in the spiny eel (Mastacembelus aculeatus) revealed by mitotic and meiotic analysis. Cytogenet. Genome Res. 98, 291–297 (2002). [DOI] [PubMed] [Google Scholar]
- 92.Danecek P., et al. , The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sahakyan A. B., et al. , Machine learning model for sequence-driven DNA G-quadruplex formation. Sci. Rep. 7, 14535 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Q., Li G., Sun Y., Wang Y., Chromosome preparation and preliminary observation of two Amphioxus species in Xiamen. Zoolog. Res. 30, 131–136 (2009). [Google Scholar]
- 95.O’Connor R. E., et al. , Reconstruction of the diapsid ancestral genome permits chromosome evolution tracing in avian and non-avian dinosaurs. Nat. Commun. 9, 1883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Emms D. M., Kelly S., OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kielbasa S. M., Wan R., Sato K., Horton P., Frith M. C., Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blanchette M., et al. , Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 14, 708–715 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Satou Y., et al. , A nearly complete genome of ciona intestinalis type A (C. robusta) reveals the contribution of inversion to chromosomal evolution in the Genus Ciona. Genome Biol. Evol. 11, 3144–3157 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wang S., et al. , Scallop genome provides insights into evolution of bilaterian karyotype and development. Nat. Ecol. Evol. 1, 120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Minh B. Q., et al. , IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic Era. Mol. Biol. Evol. 37, 1530–1534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Siepel A., et al. , Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Cabanettes F., Klopp C., D-GENIES: Dot plot large genomes in an interactive, efficient and simple way. PeerJ 6, e4958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sela I., Ashkenazy H., Katoh K., Pupko T., GUIDANCE2: Accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 107.Benton M. J., Donoghue P. C., Asher R. J., Calibrating and constraining molecular clocks. J. Vertebr. Paleontol. 35, 86 (2009). [Google Scholar]
- 108.Yang Z., Rannala B., Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol. Biol. Evol. 23, 212–226 (2006). [DOI] [PubMed] [Google Scholar]
- 109.Benton M. J., Donoghue P. C., Paleontological evidence to date the tree of life. Mol. Biol. Evol. 24, 26–53 (2007). [DOI] [PubMed] [Google Scholar]
- 110.Pu L., Lin Y., Pevzner P. A., Detection and analysis of ancient segmental duplications in mammalian genomes. Genome Res. 28, 901–909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen K., Durand D., Farach-colton M., NOTUNG: A program for dating gene duplications. J. Comput. Biol. 7, 429–447 (2000). [DOI] [PubMed] [Google Scholar]
- 112.Capella-Gutierrez S., Silla-Martinez J. M., Gabaldon T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shi J., et al. , Chromosome conformation capture resolved near complete genome assembly of broomcorn millet. Nat. Commun. 10, 464 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Marchal C., Singh N., Corso-Díaz X., Swaroop A., HiCRes: A computational method to estimate and predict the resolution of HiC libraries 2. biorxiv [Preprint] (2020), 10.1101/2020.09.22.307967 (Accessed 22 September 2020). [DOI] [PMC free article] [PubMed]
- 115.Ramírez F., et al. , High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9, 189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Grant C. E., Bailey T. L., Noble W. S., FIMO: Scanning for occurrences of a given motif. Bioinformatics 27, 1017–1018 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rowley M. J., et al. , Evolutionarily conserved principles predict 3D chromatin organization. Mol. Cell 67, 837–852.e7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li G., et al. , Mutagenesis at specific genomic loci of amphioxus Branchiostoma belcheri using TALEN method. J. Genet. Genom. 41, 215–219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.DePristo M. A., et al. , A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Browning S. R., Browning B. L., Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 81, 1084–1097 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Delaneau O., Zagury J.-F., Marchini J., Improved whole-chromosome phasing for disease and population genetic studies. Nat. Methods 10, 5–6 (2013). [DOI] [PubMed] [Google Scholar]
- 122.Kang H. M., et al. , Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42, 348–354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2013). [DOI] [PubMed] [Google Scholar]
- 124.Huang Z., et al. , Three amphioxus reference genomes reveal gene and chromosome evolution of chordates. DRYAD. https://datadryad.org/stash/dataset/dryad.9p8cz8wkx. Accessed 31 January 2023. [DOI] [PMC free article] [PubMed]
- 125.Huang Z., et al. , AmphioxusGenome. GitHub. https://github.com/lurebgi/amphioxusGenome.git. Accessed 25 October 2021.
- 126.Huang Z., et al. , Branchiostoma floridae x Branchiostoma belcheri isolate: bbbf Genome sequencing and assembly. NCBI. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA603158. Deposited 25 January 2020.
- 127.Huang Z., et al. , Branchiostoma floridae x Branchiostoma belcheri isolate: bbbf Genome sequencing and assembly. NCBI. https://www.ncbi.nlm.nih.gov/sra/PRJNA603159. Deposited 25 January 2020.
- 128.Huang Z., et al. , Branchiostoma floridae x Branchiostoma japonicum Genome sequencing and assembly. NCBI. https://www.ncbi.nlm.nih.gov/genome/?term=PRJNA647830. Deposited 7 December 2020.
- 129.Huang Z., et al. , Genome sequencing and assembly of three amphioxuses. NCBI. https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA602496. Deposited 12 January 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
1) Genomic Reads, Genome Assembly and Annotation, Sequencing Data; 2) SNP VCF files, genome annotation files, whole-genome alignments, and Hox gene alignment; 3) Scripts used data have been deposited in 1) GenBank, 2) Dryad (124), 3) Github (125): 1) PRJNA603158 (126), PRJNA603159 (127), PRJNA647830 (128), PRJNA602496 (129); 2) SNP VCF files: https://datadryad.org/stash/downloads/file_stream/2070006, https://datadryad.org/stash/downloads/file_stream/2070005 , https://datadryad.org/stash/downloads/file_stream/2070032; Genome annotation files: https://datadryad.org/stash/downloads/file_stream/2069997, https://datadryad.org/stash/downloads/file_stream/2069996, https://datadryad.org/stash/downloads/file_stream/2069995; whole-genome alignments: https://datadryad.org/stash/downloads/file_stream/2070001; Hox gene alignment https://datadryad.org/stash/downloads/file_stream/2070002; 3) https://github.com/lurebgi/amphioxusGenome. All study data are included in the article and/or SI Appendix.