Significance
Ultraviolet (UV) light is the primary etiological agent for skin cancer because it induces lesions in DNA. Previous studies have shown that the formation of the major class of UV damage [cyclobutane pyrimidine dimers (CPD) lesions] is significantly modulated by chromatin. UV also induces less common 6-4 photoproducts (6-4PP) and atypical thymine–adenine (TA) photoproducts in DNA, but the effects of chromatin-bound proteins on their formation are unclear. Here, we report maps of these rare and atypical photoproducts across the yeast genome. Our data indicate that a number of DNA-bound proteins significantly alter the formation of 6-4PP and TA photoproducts, in a manner distinct from that of CPD lesions. We further show that 6-4PP hotspots promote UV mutagenesis.
Keywords: 6-4 photoproducts, thymine–adenine photoproduct, TA photoproduct, nucleosome, transcription factor binding site
Abstract
Ultraviolet (UV) light induces different classes of mutagenic photoproducts in DNA, namely cyclobutane pyrimidine dimers (CPDs), 6-4 photoproducts (6-4PPs), and atypical thymine–adenine photoproducts (TA-PPs). CPD formation is modulated by nucleosomes and transcription factors (TFs), which has important ramifications for Ultraviolet (UV) mutagenesis. How chromatin affects the formation of 6-4PPs and TA-PPs is unclear. Here, we use UV damage endonuclease-sequencing (UVDE-seq) to map these UV photoproducts across the yeast genome. Our results indicate that nucleosomes, the fundamental building block of chromatin, have opposing effects on photoproduct formation. Nucleosomes induce CPDs and 6-4PPs at outward rotational settings in nucleosomal DNA but suppress TA-PPs at these settings. Our data also indicate that DNA binding by different classes of yeast TFs causes lesion-specific hotspots of 6-4PPs or TA-PPs. For example, DNA binding by the TF Rap1 generally suppresses CPD and 6-4PP formation but induces a TA-PP hotspot. Finally, we show that 6-4PP formation is strongly induced at the binding sites of TATA-binding protein (TBP), which is correlated with higher mutation rates in UV-exposed yeast. These results indicate that the formation of 6-4PPs and TA-PPs is modulated by chromatin differently than CPDs and that this may have important implications for UV mutagenesis.
Exposure to ultraviolet (UV) light induces mutagenic lesions in cellular DNA, including cyclobutane pyrimidine dimers (CPDs), 6-4 photoproducts (6-4PPs), and atypical thymine–adenine photoproducts (TA-PPs) (1–4). CPDs and 6-4PPs form between neighboring pyrimidine bases, with CPDs comprising 75 to 90% of UV damage and 6-4PPs comprising 10 to 20% of UV damage (1). Although CPD and 6-4PP formation both involve creating covalent bonds between neighboring pyrimidine bases, the necessary photochemistry is quite different (SI Appendix, Fig. S1A). CPDs form in a rapid UV-induced [2+2] cycloaddition reaction between the C5–C6 double bonds of neighboring pyrimidines. In contrast, 6-4PPs are created in a slower two-step reaction that proceeds through a UV-induced oxetane (or azetidine) intermediate (1). TA-PPs also form via a distinct photochemical reaction (SI Appendix, Fig. S1B), in this case between neighboring thymine and adenine bases (3, 5), but the abundance and genomic distribution of TA-PPs in UV-irradiated cells are unknown. To what extent these differences in photochemistry influence the formation of these distinct UV photoproducts in chromatin is also not well understood.
The efficiency of CPD formation is modulated by distortions in the DNA structure caused by DNA-bound proteins (6, 7), which has important ramifications for UV mutagenesis in skin cancers (8). For example, the wrapping of DNA around the histone octamer induces a ~10 base pair (bp) periodicity to CPD formation in nucleosomes, with elevated CPD formation at outward rotational settings, where the DNA minor groove faces away from the histone octamer, and suppressed CPD formation at inward rotational settings (6, 7, 9–12). Similarly, the binding of many transcription factors (TFs), including yeast Reb1 and Abf1, suppresses CPD formation (10), while binding by the ETS family of TFs in human cells induces a DNA conformation that is more susceptible to CPD formation (13–15). Modulation of CPD formation by DNA-bound proteins has important ramifications for UV mutagenesis, as somatic mutation rates in human skin cancers are elevated at ETS binding sites (8, 14–16) and are modulated by the rotational setting of nucleosomal DNA (11, 12).
The formation of 6-4PPs is also thought to be modulated by DNA-bound proteins but in a manner distinct from that of CPDs. Previous studies have indicated that, unlike CPD formation, 6-4PP formation is elevated in linker DNA relative to nucleosomal DNA and shows weak or nonexistent rotational periodicity within nucleosomes (17, 18). This has been attributed to the fact that 6-4PPs cause a greater distortion in the DNA structure than CPDs, so that 6-4PPs may not be readily accommodated or form in the tightly wrapped nucleosomal DNA (6). However, a recent bioinformatics analysis of a genome-wide High sensitivity-damage-sequencing (HS-Damage-seq) map of 6-4PPs (19) suggests that 6-4PPs may show rotational periodicity in nucleosomes in UV-irradiated human cells (11). In addition, 6-4PP formation has also been reported to be modulated by TFs, including TATA-binding protein (TBP) in yeast (20) and in vitro (21) and Nuclear Transcription Factor Y in human cells (19, 22). However, whether 6-4PPs hotspots at these or other TF binding sites are associated with elevated UV mutation rates is unknown.
UV light also induces rare atypical TA-PPs at thymine–adenine sequences (3, 4). We have recently shown that TA-PPs make significant contributions to UV mutagenesis in yeast and may also induce somatic mutations in human skin cancers (23). It has been reported that DNA binding by TBP may modulate TA photoproduct formation in vitro (21). However, the extent to which DNA-bound proteins or nucleosomes modulate TA photoproduct formation in cellular DNA is unclear.
Methods for mapping genome-wide distribution of CPD lesions have proven to be powerful tools for elucidating how DNA-bound proteins modulate CPD formation (10, 13–14, 15). Here, we use a method called UV damage endonuclease-sequencing (UVDE-seq) (24) to characterize the genome-wide formation of 6-4PPs and TA photoproducts at single-nucleotide resolution in UV-irradiated yeast cells.
Results
UVDE-seq Method for Mapping Rare and Atypical UV Photoproducts.
To map 6-4PPs and atypical UV photoproducts at single-nucleotide resolution across the genome, we used the UVDE-seq method (Fig. 1A). This method is similar to our published CPD-seq method for mapping CPD lesions (10, 25) but differs in two important aspects (23, 24). First, the ultraviolet damage endonuclease (UVDE) enzyme was used to cleave DNA lesions, due to its broad substrate specificity in cleaving a variety of UV photoproducts, including cis-syn and trans-syn CPDs, 6-4PPs, and TA-PPs, as well as other DNA lesions (e.g., cisplatin adducts) (23, 26–28). Second, CPD photolyase was used to remove CPD lesions prior to UVDE cleavage, in order to specifically detect non-CPD photoproducts. This step was optimized using T4 endonuclease V (T4 endoV) digestion, which cleaves specifically at CPD lesions, and alkaline gel electrophoresis. Alkaline gel analysis revealed little, if any, cleavage of the UV-irradiated genomic DNA (gDNA) by T4 endoV following CPD photolyase treatment (SI Appendix, Fig. S2, lane 8), confirming CPD removal by photolyase. In contrast, UVDE still readily digested the CPD photolyase-treated DNA (SI Appendix, Fig. S2, lane 9), reflecting UVDE cleavage of 6-4PPs and other atypical UV photoproducts (e.g., TA-PP, see below).
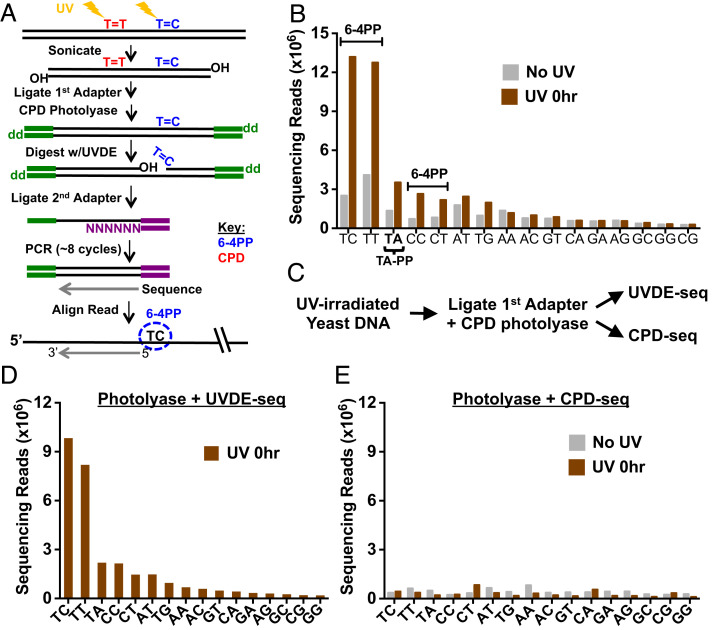
Fig. 1.
UVDE-seq method for mapping UV-induced 6-4PPs and atypical lesion formation across the eukaryotic genome. (A) Schematic of the UVDE-seq method. Induction of damage with UVC light induces both CPD lesions (indicated as T=T) and 6-4PPs (indicated as T=C). Treatment with CPD photolyase repairs CPDs, leaving only 6-4PPs and atypical lesions to be cleaved by UVDE. The first (trP1) adapter is colored green, and the second (A) adapter is in purple. (B) UVDE-seq reads mapping putative lesions at different dinucleotide sequences. Enrichment of UVDE-seq reads was observed at canonical dipyrimidine sequences (TC, TT, CC, and CT) as well as atypical TA sequences in comparison to unexposed (No UV) samples. (C) Schematic of control library preparation. Samples were UV-irradiated and processed through UVDE-seq library preparation including CPD photolyase treatment. Samples were then split, and half were treated with UVDE to map 6-4PPs, while the others were treated with T4 endonuclease V and Ape1 to map CPD lesions as described in our published CPD-seq method (10). (D and E) Analysis of reads mapped to putative lesion sites following UVDE-seq (D) and CPD-seq (E) library preparation (see C for more details) for UV-irradiated yeast cells (UV 0 h) and unirradiated controls (No UV). CPD-seq reads were generally not enriched relative to the No UV control at lesion-forming dipyrimidines, confirming CPD photolyase treatment is sufficient for the removal of all CPD lesions prior to UVDE treatment.
The UVDE-seq method was used to map 6-4PPs and potentially other UV photoproducts in wild-type (WT) yeast cells immediately following UV irradiation (i.e., 0 h). Analysis of the UVDE-seq data revealed that reads are specifically enriched at dipyrimidine sequences in the UV-irradiated cells (UV 0 h) relative to the No UV control (Fig. 1B and SI Appendix, Fig. S3A), consistent with UV-induced 6-4PP formation at dipyrimidine sequences. Moreover, the frequency of UVDE-seq reads associated with each dipyrimidine sequence matches the expected propensity of 6-4PP formation (e.g., TC > TT > CC > CT) observed in previous studies (1). Notably, UVDE-seq reads are also enriched at TA lesion sites following UV irradiation (Fig. 1B and SI Appendix, Fig. S3A), consistent with our previous publication and a prior report suggesting that UVDE cleaves at atypical thymine–adenine (TA) photoproducts (23, 28).
To confirm that UVDE-seq reads at dipyrimidine sequences reflect 6-4PPs, and not the more abundant CPDs, we prepared genomic DNA from UV-irradiated yeast cells and processed this DNA through the CPD photolyase step of the UVDE-seq method (Fig. 1A). Part of the DNA was then continued through the remainder of the UVDE-seq protocol, while the other part was then processed using our published CPD-seq method using T4 endonuclease V to map CPD lesions (Fig. 1C). Analysis of the resulting UVDE-seq data revealed significant enrichment at dipyrimidine and TA sequences (Fig. 1D), consistent with previous experiments. In contrast, there was little to no enrichment of CPD-seq reads at dipyrimidine sequences following the CPD photolyase treatment (Fig. 1E), confirming that CPD lesions were completely removed by the photolyase treatment during the UVDE-seq protocol. These control experiments confirm that UVDE-seq specifically detects 6-4PPs at dipyrimidine sequences.
Rotational Positioning of DNA in Nucleosomes Regulates 6-4PP Formation in Yeast.
While it is well-established that the rotational setting of the DNA in nucleosomes modulates the formation of CPD lesions, the impact of nucleosomes on UV-induced 6-4PP formation is less clear. It has been previously suggested that 6-4PPs are elevated in linker DNA and are only weakly modulated in nucleosomes (6, 17, 18), but a recent bioinformatics analysis of human HS-Damage-seq data (19) indicated that 6-4PPs are elevated at outward rotational settings in nucleosomes (11). To investigate the impact of rotational and translational positioning of nucleosomal DNA on 6-4PP formation, we analyzed the frequency of 6-4PPs mapped using our UVDE-seq data in yeast among a set of ~10,000 strongly positioned nucleosomes (10, 29). To specifically analyze 6-4PPs, we only analyzed UVDE-seq reads associated with 6-4PP-forming dipyrimidine sequences (Fig. 1B); UVDE-seq reads associated with TA dinucleotides or other nondipyrimidine sequences were excluded from this analysis. To account for effects of DNA sequence bias in nucleosomal DNA, we also used UVDE-seq to analyze 6-4PP formation on isolated yeast genomic DNA that was UV irradiated in vitro (SI Appendix, Fig. S3B). The cellular UVDE-seq data (UV 0 h) was then normalized using these naked DNA controls, in order to remove the effects of DNA sequence bias on 6-4PP formation.
This analysis revealed that 6-4PP formation is modulated by the rotational setting of DNA in strongly positioned nucleosomes (Fig. 2A). Of note, 6-4PP enrichment relative to the naked DNA control is specifically increased at outward rotational settings in nucleosomal DNA (outward rotational settings are indicated by dashed lines in Fig. 2A) and decreased at inward rotational settings. This pattern of 6-4PP formation in nucleosomes has a periodicity of 10.1 bp, corresponding to the helical repeat length of DNA, and a signal-to-noise ratio (SNR) of 129 (Fig. 2A), which quantifies the strength of the periodicity in nucleosomes (11, 30). This periodic pattern of 6-4PP formation is essentially absent in a control set of ~7,500 weakly positioned nucleosomes (SNR ~ 10.2; Fig. 2B), indicating that the rotational periodicity in 6-4PP formation is dependent upon the strength of nucleosome positioning. In strongly positioned nucleosomes, peaks of 6-4PP enrichment at outward rotational settings precisely coincide with peaks of CPD enrichment (Fig. 2C), based on analysis of our published yeast CPD-seq data (10). However, the magnitude of damage enrichment at outward rotational settings is weaker for 6-4PPs than for CPDs (Fig. 2C).
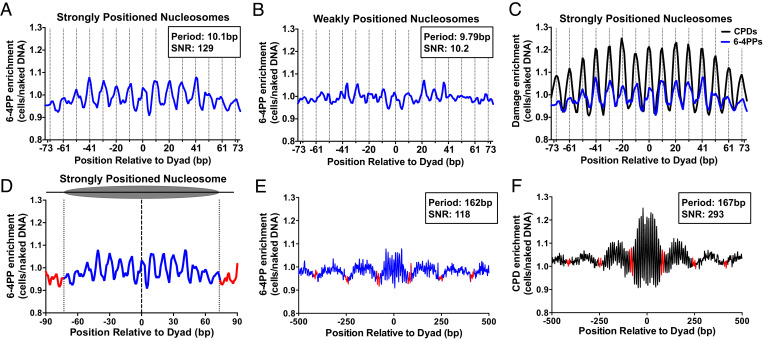
Fig. 2.
Rotational positioning of DNA in nucleosomes regulates 6-4PP formation in yeast and human cells. (A and B) Rotational periodicity of 6-4PPs within ~10,000 strongly positioned (A) and ~7,500 weakly positioned (B) yeast nucleosomes, derived from a published map of yeast nucleosome positions (29). Dashed lines correspond to outward rotational settings. UVDE-seq reads for 6-4PPs in UV-irradiated yeast cells (UV 0 h) were normalized to UVDE-seq reads for 6-4PPs from naked DNA controls, in order to account for DNA sequence biases in nucleosomal DNA and calculate 6-4PP enrichment. The peak periodicity and signal-to-noise ratio (SNR), which quantifies the strength of the periodicity (11, 30), are shown for 6-4PP enrichment in nucleosomes. (C) 6-4PPs data from panel A plotted against CPD enrichment within strongly positioned yeast nucleosomes, using published CPD-seq data (10). (D) Analysis of translational periodicity of 6-4PP enrichment in nucleosomal DNA (blue) relative to linker DNA (red) in strongly positioned (29). Dashed lines at −73 and +73 represent the border between nucleosomal and linker DNA. (E and F) Translational periodicity of E 6-4PP or F CPD enrichment in a 1,000-bp window centered on strongly positioned nucleosomes (29). Red indicates likely location of linker DNA. Peak translational periodicity and its SNR was calculated using mutperiod (30).
In contrast, 6-4PP formation is not elevated in linker DNA immediately adjacent to either strongly or weakly positioned nucleosomes (Fig. 2D and SI Appendix, Fig. S4). To further investigate this question, we analyzed the effects of translational positioning on 6-4PP enrichment in a 1,000-bp window centered on each strongly positioned nucleosome. This analysis indicated that 6-4PP formation is not elevated in linker DNA (red line segments in Fig. 2E) but is actually slightly higher in nucleosome core DNA (blue line segments), presumably due to 6-4PP enrichment at outward rotational settings. Analysis of published CPD-seq data showed a similar pattern of elevated damage levels in nucleosomal DNA relative to linker regions (Fig. 2F). We also examined 6-4PP formation in nucleosomal DNA and nucleosome-free regions (NFRs) near the transcription start site (TSS) of yeast genes. 6-4PP enrichment in UV-irradiated cells (relative to the naked DNA control) is elevated at positioned nucleosomes but also shows a small peak in damage induction in the NFR (SI Appendix, Fig. S5A), which can be attributed to 6-4PP induction at TBP-bound TATA sites (see below).
One possible caveat is that 6-4PPs are rapidly repaired by the global genomic-nucleotide excision repair (GG-NER) pathway in yeast, being removed within minutes after UV irradiation (31). Although the UV 0 h cells were harvested and frozen immediately after UV irradiation, it is possible that some 6-4PPs were repaired during cell harvesting and freezing, particularly in accessible linker DNA. To test this possibility, we repeated the UV 0 h UVDE-seq experiments in a GG-NER-deficient rad16∆ mutant strain (SI Appendix, Fig. S6A). Analysis of 6-4PP formation in strongly positioned nucleosomes (relative to the naked DNA control) showed a rotational periodicity of 6-4PP formation in rad16∆ mutant cells (SI Appendix, Fig. S6B) similar to that observed in WT cells. Moreover, 6-4PP formation was slightly elevated in nucleosomal DNA relative to linker DNA in rad16∆ cells (SI Appendix, Fig. S6 C and D), similar to WT cells. This analysis indicates that the lack of 6-4PP induction in linker DNA is not a consequence of repair, since essentially the same pattern of 6-4PP formation is observed in GG-NER-deficient cells.
TA Photoproducts Show a Unique Pattern of Damage Formation in Nucleosomes.
UV light also induces the formation of atypical TA photoproducts (3, 4), but the impact of DNA-bound proteins and chromatin on their formation is poorly understood. To accurately map TA-PPs in yeast chromatin, we developed an improved version of our method, which we call UVDE-High Sensitivity-seq (UVDE-HS-seq), which reduced background reads associated with aberrant DNA cleavage during the CPD photolyase step (Materials and Methods). Analysis of UVDE-HS-seq reads revealed that they are specifically enriched at dipyrimidine sequences and TA dinucleotides in the UV-irradiated cells (UV 0 h) data (SI Appendix, Fig. S7 A and B), consistent with our previous UVDE-seq data (Fig. 1B). Importantly, the No UV control showed relatively lower read counts than those from the previous iteration of UVDE-seq, most notably in T-containing dinucleotide sequences. This reflects a reduction in background, which is important for accurately mapping the low abundance TA photoproducts across the genome.
Analysis of UVDE-HS-seq reads associated with 6-4PPs-forming dipyrimidine sequences revealed a nearly identical pattern of 6-4PP formation in nucleosomes as our original UVDE-seq analysis (SI Appendix, Fig. S7 C–F). However, analysis of TA-PP formation using the UVDE-HS-seq method revealed a unique pattern of damage formation in strongly positioned nucleosomes (Fig. 3A). Unlike CPDs and 6-4PPs, TA-PP formation is decreased at outward rotational settings in nucleosomes (Fig. 3A, dashed lines indicate outward rotational settings), even after normalizing to the naked DNA control (Fig. 3B). Instead, TA-PP formation is elevated at inward rotational settings in strongly positioned nucleosomes, resulting in a ~10.2 bp periodicity of TA-PP formation in nucleosomes (SNR of 53.8).
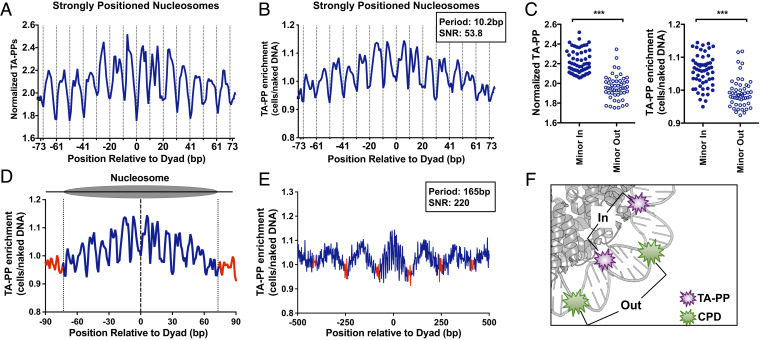
Fig. 3.
TA photoproducts show a unique pattern of damage formation in nucleosomes. (A) Rotational periodicity of cellular TA-PPs within all (~10,000) strongly positioned yeast nucleosomes (29). Data plotted as number of UVDE-seq reads at TA sequences normalized by the number of TA dinucleotides. Dashed lines correspond to outward rotational settings. (B) Rotational periodicity of TA-PP enrichment (TA-PP in UV-irradiated cells relative to the UV-irradiated naked DNA control) in strongly positioned yeast nucleosomes (29). Peak periodicity of TA-PP enrichment and its signal-to-noise ratio (SNR) are indicated. (C) Normalized TA-PP (Left) and TA-PP enrichment (Right) are elevated at inward rotational settings relative to outward rotational settings. Rotational settings derived from a previous study (32). ***P < 0.0001, based on Mann–Whitney test. (D) Translational periodicity of normalized TA-PPs in nucleosomal (blue) and linker DNA (red) in ~10,000 strongly positioned nucleosomes (29). Dashed lines at −73 and +73 represent the border between nucleosomal and linker DNA. (E) Translational periodicity of TA-PPs across multiple yeast nucleosomes, with a 1,000-bp window centered on each strongly positioned nucleosome. TA-PP enrichment from UV-irradiated cells is depicted. Regions of linker DNA are shown in red. Peak translational periodicity of TA-PPs, and its SNR are indicated. (F) Model showing TA-PP formation (purple explosion) is elevated at inward rotational settings, while CPD formation (green explosion) is elevated at outward rotation settings in nucleosomes. Nucleosome image was generated from PDB ID 1id3 and visualized using PyMOL.
To confirm that TA-PP formation is modulated by rotational setting in nucleosomes, we compared the normalized TA-PP formation at all positions in nucleosomes associated with an outward rotational setting (minor-out) to those positions associated with an inward rotational setting (minor-in), using previously published rotational positioning data in nucleosomes (32). This analysis revealed that in UV-irradiated cells, TA-PP formation is significantly elevated at minor-in positions relative to minor-out (P < 0.0001, Left panel of Fig. 3 C). Moreover, TA-PP enrichment in UV-irradiated cells relative to the naked DNA control is also higher at minor-in positions relative to minor-out (P < 0.0001; Right panel of Fig. 3 C). This periodic pattern of TA-PP formation is largely absent within weakly positioned nucleosomes (SI Appendix, Fig. S8A). Taken together, these data indicate that positioned nucleosomes promote TA-PP formation at inward rotational settings and suppress TA-PP formation at outward settings.
TA-PP enrichment is also elevated at positions near the central nucleosome dyad positions (Fig. 3B). Analysis of TA-PP formation in linker DNA adjacent to strongly positioned nucleosomes revealed that TA-PP enrichment in linker DNA is lower than that within nucleosomes (Fig. 3D). This pattern of TA-PP formation is not apparent when analyzed among a control set of ~7,500 weakly positioned nucleosomes (SI Appendix, Fig. S8B), indicating that it is dependent on the strength of nucleosome positioning. Analysis of TA-PP formation across a 1,000-bp window centered on strongly positioned nucleosomes (Fig. 3E) confirmed that TA-PP enrichment is elevated in nucleosome core DNA relative to linker DNA with a periodicity of 165 bp, corresponding to the known nucleosome repeat length of ~160 to 165bp in yeast (33). TA-PP enrichment is also lower in NFRs upstream of TSS of yeast genes relative to neighboring nucleosomes (SI Appendix, Fig. S5B). Taken together, these findings suggest the following conclusions: 1) Neither 6-4PPs nor TA-PPs are elevated in linker DNA and 2) 6-4PPs are elevated at outward rotational settings in nucleosomes, similar to CPDs, but TA-PPs are suppressed at minor out positions (Fig. 3F).
Transcription Factor Binding Modulates 6-4PP and TA-PP Formation in Yeast.
Since TF binding can affect the formation of UV-induced CPDs and 6-4PPs in human cells (13–15, 19, 22, 34), we investigated to what extent TF binding modulates the formation of 6-4PPs in yeast. We analyzed the enrichment of 6-4PPs in UV-irradiated cells relative to the naked DNA control (using UVDE-HS-seq data) in the binding sites of 32 distinct TFs identified by high-resolution ChIP-exo data from yeast (35, 36). This analysis identified four TFs (i.e., Hap3, Hap5, Lys14, and TBP) that strongly induce 6-4PPs and two TFs (i.e., Rap1 and Reb1) that strongly suppress 6-4PP formation (Fig. 4A). Closer inspection of 6-4PP enrichment at 339 Reb1 binding sites (identified by ChIP-exo) confirmed that 6-4PP formation is primarily suppressed by Reb1 binding (Fig. 4B). In contrast, there is little, if any, modulation of 6-4PP formation at a control set of low occupancy Reb1 sites (SI Appendix, Fig. S9A), even though these sites have a nearly identical DNA motif (10, 37). Taken together, these data indicate that 6-4PP modulation is specifically associated with Reb1 binding. Unlike Reb1, DNA binding by both the Hap3 and Hap5 TFs strongly induces 6-4PP formation in yeast (Fig. 4A). Since these TFs bind DNA as a complex (along with Hap2), we analyzed 6-4PP formation at all Hap2/3/5 binding sites identified by ChIP-exo (36). The results confirmed that 6-4PP formation is strongly induced (~sevenfold) at Hap2/3/5 binding sites in UV-irradiated cells relative to the naked DNA control (Fig. 4C).
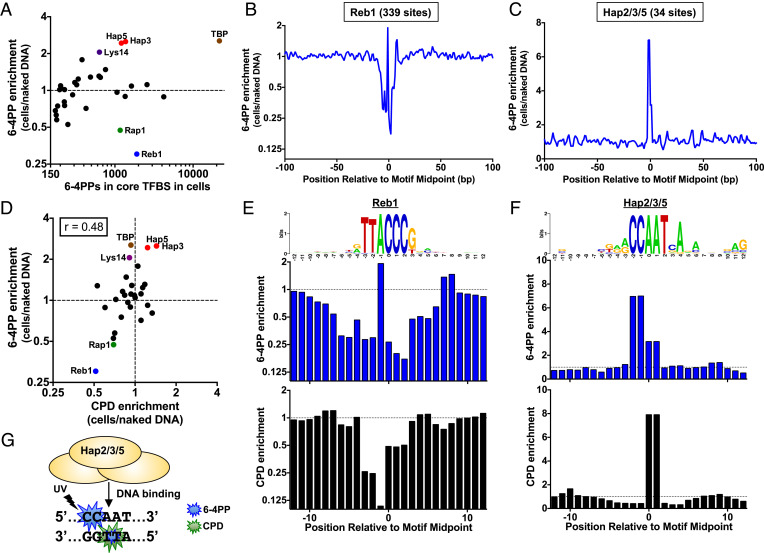
Fig. 4.
DNA binding by yeast TFs Hap2/3/5 and TBP induce 6-4PP formation, while Reb1 suppresses 6-4PP. (A) Plot of 6-4PP enrichment (6-4PPs in UV-irradiated cells relative to the UV-irradiated naked DNA control) relative to total count of 6-4PPs at core binding sites (within 4 bp of binding site midpoint) of 32 different TFs. TF binding site data are derived from published ChIP-exo experiments (35, 36). Only TF binding site classes with at least a count of 150 6-4PPs in both the cellular and naked DNA experiments are depicted. (B and C) Formation of 6-4PPs within (B) Reb1 and (C) Hap2/Hap3/Hap5 binding sites across the yeast genome. Enrichment of each class of photoproducts in UV-irradiated cells relative to the naked DNA control is plotted. (D) Same as panel A, except that 6-4PP enrichment is compared to CPD enrichment at the same binding sites. Pearson correlation coefficient (r) calculated using Graphpad Prism. (E and F) Closer examination of 6-4PP (Middle) and CPD (Bottom) enrichment within (E) Reb1 and (F) Hap2/Hap3/Hap5 binding sites. Top panel is the consensus sequence of each set of binding sites, created using weblogo (38). (G) Schematic showing that 6-4PP formation (blue explosion) is elevated at CC dinucleotide (and to a lesser extent at the TT dinucleotide) in Hap2/Hap3/Hap5 binding sites, while CPD formation (green explosion) is elevated only at the TT dinucleotide on the opposite DNA strand.
Since Reb1 and other TFs in yeast have previously been shown to modulate the formation of UV-induced CPD lesions (10), we used our published CPD-seq data to compare CPD and 6-4PP enrichment at each class of TF binding sites. This analysis revealed a weak but significant correlation (r = 0.48, P = 0.019) between overall CPD and 6-4PP enrichment among the different classes of TF binding sites (Fig. 4D). However, closer inspection of TFs that had a similar level of CPD and 6-4PP enrichment overall (e.g., Reb1 or Hap3/5) revealed key differences in the detailed patterns of UV damage formation (SI Appendix, Fig. S9 B and C and Fig. 4 E and F). Although both 6-4PP and CPD formation are decreased in Reb1 sites, closer inspection revealed that 6-4PP formation is primarily suppressed at CC dinucleotides at the 3′ end of the Reb1-bound TTACCC motif (Fig. 4E), while CPD formation is primarily suppressed at the 5’TT dinucleotide (10). Similarly, although both 6-4PP and CPD formation are elevated at Hap2/3/5 binding sites, 6-4PP formation is primarily induced (~sevenfold) at a conserved CC dinucleotide on the plus strand of the binding motif (CCAAT), while CPD formation is primarily induced (~eightfold) at a TT dinucleotide on the minus strand of the binding motif (Fig. 4 F and G). 6-4PP formation is also induced at this TT dinucleotide, although to a lesser extent (~threefold; see Fig. 4 F and G). Analysis of individual Hap2/3/5 binding sites revealed that most binding sites have both elevated CPDs and 6-4PPs in UV-irradiated yeast relative to the naked DNA control (SI Appendix, Fig. S10). Additionally, overall CPD and 6-4PP enrichment differed markedly at a number of other TF binding sites, including those of TBP and Lys14 (Fig. 4D).
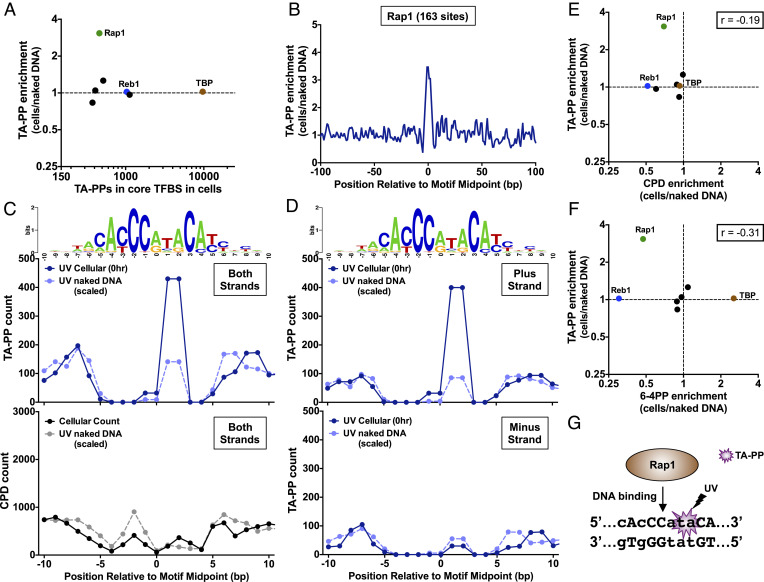
We performed a similar analysis of TA-PP formation using the UVDE-HS-seq data, focusing just on 7 classes of TFs with a sufficient number of binding sites to reliably detect patterns of TA-PP (Fig. 5A). While we did not observe any significant change in overall TA-PP formation at the binding sites of most TFs, this analysis revealed that TA-PPs are induced (~threefold) at the center of Rap1 binding sites (Fig. 5 A and B). Closer inspection revealed that TA-PPs are induced at a TA dinucleotide at positions +1/+2 in the Rap1 binding motif, a sequence that occurs in approximately half (88 out of 163) of Rap1 binding sites (Fig. 5C). There is also enrichment of TA-PPs at the neighboring −1/0 position in the binding motif, although only 2 out of 163 binding sites have a TA dinucleotide at this position. Similar analysis of the CPD-seq data at Rap1 binding sites indicates that CPD levels are also induced (~twofold) at positions +2/+3 in the binding motif (Fig. 5 C, Lower), partially overlapping with the location of the TA-PP hotspot. However, CPD levels elsewhere in the Rap1 binding motif are generally suppressed. Since TA dinucleotides are palindromes, TA-PP can form on either DNA strand. Analysis of strand-specific TA-PP formation (Fig. 5D) revealed that TA-PPs are specifically induced (~fivefold) at the TA dinucleotide on the plus strand (e.g., 5′-CACCCATACA-3′) and are suppressed (~0.5-fold) at the TA dinucleotide on the minus strand (e.g., 5′-TGTATGGGTG-3′). Comparison of overall TA-PP enrichment with CPD or 6-4PP enrichment at different classes of binding sites (Fig. 5 E and F) revealed no significant correlation (r = −0.19 and r = −0.31, respectively, P > 0.05), suggesting that TA-PP formation at TF binding sites is distinct from canonical photoproducts. In summary, these findings indicate that DNA binding by Rap1, an essential TF that regulates ribosomal protein expression and telomeric silencing (39), results in a strand-specific induction of TA-PPs (Fig. 5G).
Fig. 5.
Rap1 specifically induces TA-PP formation in its binding sites. (A) Plot of TA-PP enrichment (TA-PP in UV-irradiated cells relative to the UV-irradiated naked DNA control) relative to total count of TA-PPs at core binding sites (within 4 bp of binding site midpoint) of 32 different TFs. TF binding site data are derived from published ChIP-exo experiments (35, 36). Only TF binding site classes with at least a count of 150 TA-PPs in both the cellular and naked DNA experiments are depicted. (B) Formation of TA-PPs within Rap1 binding sites across the yeast genome. Enrichment of TA-PPs in UV-irradiated cells relative to the naked DNA control is plotted. (C) Closer examination of count of TA-PPs (Middle) and CPDs (Bottom) for both UV-irradiated cellular and scaled naked DNA samples at Rap1 binding sites. Top panel is the consensus sequence of binding sites, created using weblogo (38). (D) Same as panel C, except that counts of TA-PPs on plus and minus strands are plotted separately, as the lesion is able to form on either strand of a TA sequence. (E and F) Same as panel A, except that TA-PP enrichment is compared to E CPD or F 6-4PP enrichment at the same binding sites. Pearson correlation coefficient (r) is calculated using Graphpad Prism. (G) Model of how Rap1 binding to DNA specifically induces a TA-PP hotspot on the plus strand (i.e., top strand) of its binding motif. Weakly conserved positions of the binding motif are shown with lowercase letters, while strongly conserved positions are indicated with uppercase letters.
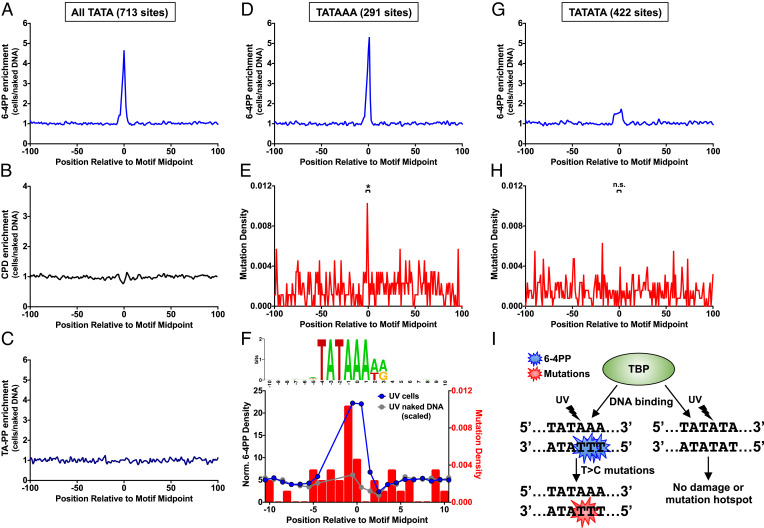
6-4PP Hotspot at TBP-Bound TATA Sites Coincides with a Mutation Hotspot in UV-Irradiated Yeast.
Previous studies have indicated that CPD hotspots at ETS and CTCF binding sites in human cells coincide with mutation hotspots in skin cancers (14, 15, 34, 40, 41); however, whether 6-4PP or TA-PP hotspots are also associated with mutation hotspots is unknown. To test this hypothesis, we analyzed a data set of ~46,000 single-nucleotide variants identified by genome sequencing of yeast cells repeatedly exposed to UV light (23). While we identified 6-4PP or TA-PP hotspots at Rap1, Lys14, Hap2/3/5, and TBP binding sites (Figs. 4 and 5), we focused only on TBP binding sites, as there are too few Rap1, Lys14, and Hap2/3/5 binding sites to reliably measure UV mutation frequency. Analysis of UV photoproduct formation at 713 TBP-bound TATA sites, derived from ChIP-exo data in yeast (35), revealed a striking peak of 6-4PP enrichment in UV-irradiated cells relative to the naked DNA control (Fig. 6A). In contrast, there is no apparent enrichment of CPDs or TA-PPs (Fig. 6 B and C). Notably, TBP binding sites coincide with, and can potentially account for, the peak of 6-4PP formation in NFRs of yeast genes (SI Appendix, Fig. S5C). High-resolution analysis of damage formation at TATA sites revealed that 6-4PPs are primarily induced at TT dinucleotides on the opposite strand at positions −1/0 and 0/+1 relative to the TATA motif midpoint (SI Appendix, Fig. S11).
Fig. 6.
TBP binding induces a 6-4PP hotspot at TATA sites, which coincides with a mutation hotspot in UV-irradiated yeast cells. (A–C) Formation of A 6-4PPs, (B) CPDs (10), and (C) TA-PPs within all TBP-bound TATA sites derived from yeast ChIP-exo data (35). UV photoproduct enrichment was calculated from the ratio of sequencing reads in UV-irradiated yeast cells (UV 0 h) relative to the UV-irradiated naked DNA control. (D) Same as panel A, except for subset of TATA sites containing dipyrimidines on the bottom strand at positions −1/0 and 0/+1 (i.e., TATAAA). (E) Same as panel D, except for density of single nucleotide mutations in UV-exposed yeast cells. Mutation data from ref. 23. Mutation density is significantly enriched in TATA motif relative to expected mutation density based on sequence context, *P < 0.001 based on chi-square test. (F) High-resolution analysis of 6-4PP formation data at yeast TBP-bound TATAAA sites. Top panel is the consensus sequence of subset of TATA sites containing dipyrimidines on the bottom strand at positions −1/0 and 0/+1 (i.e., TATAAA), created using weblogo (38). Half-integer positions were assigned to the average position of the two damaged bases for each 6-4PPs and normalized to the number of lesion-forming dipyrimidine sequences at each position. Normalized 6-4PPs levels are plotted for both UV-irradiated yeast cells (blue) and the scaled naked DNA control (gray). The gap in data points between positions −4 to −2 in motif corresponds to positions where there are no lesion-forming dipyrimidine sequences in the TATA motif. (G) Same as panel A, except for subset of TATA sites not containing dipyrimidines on the bottom strand at positions −1/0 and 0/+1 (i.e., TATATA). (H) Same as panel G, except for density of single-nucleotide mutations in UV-exposed yeast cells. Mutation data from ref. 23. N.S. indicates P > 0.05 based on chi-square test. (I) Model of how TBP binding to DNA specifically induces a 6-4PP hotspot on the strand opposite to its binding motif. This 6-4PP hotspot likely contributes to a T > C mutation hotspot present in TBP binding sites in yeast.
Only a subset (291 out of 713) of TBP-bound TATA sites have lesion-forming dipyrimidines at these positions in the TATA motif, and 6-4PP formation is specifically enriched (~fivefold, Fig. 6D) at this subset of motifs (i.e., TATAAA consensus). The peak of 6-4PP formation coincides with a peak of mutation density in the UV-exposed yeast (Fig. 6E), and mutations in this subset of TATA sites are significantly elevated relative to the expected mutation frequency based on DNA sequence context (P < 0.001). In contrast, a control set of 2,182 sites matching the TATAAA consensus, but located inside of genes (and therefore unlikely to be bound by TBP), did not show elevated mutation density in UV-irradiated yeast (P > 0.05). High-resolution analysis of TBP-bound TATAAA sites revealed that elevated 6-4PP density at positions −1/0 and 0/+1 in UV-irradiated cells (but not UV-irradiated naked DNA) coincides with significantly higher mutation density at positions −1 and 0 (P < 0.05, Fig. 6F). Twelve out of the 13 mutations at positions −1 and 0 are T>C substitutions occurring at the 3′ position of TT dinucleotide. These mutation spectra match the experimentally derived spectra of a chemically synthesized TT 6-4PPs in yeast (42), consistent with the model that this mutation hotspot is caused by 6-4PPs. In contrast, 6-4PP enrichment and UV mutation density are not significantly elevated at TATA sites lacking a dipyrimidine at positions −1/0 or 0/+1 (i.e., TATATA sequences; Fig. 6 G and H). Taken together, these findings indicate that TBP binding induces a 6-4PP hotspot at a subset of TATA sites, which coincides with an elevated mutation rate in UV-exposed yeast (Fig. 6I).
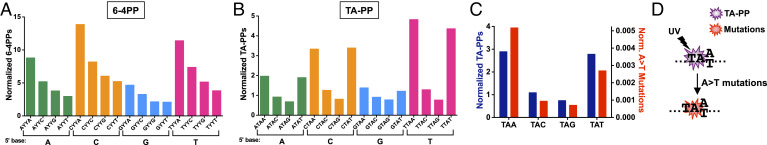
TA-PPs Are Elevated at TAW Sequences, which Coincide with Elevated Frequency of UV-Induced A>T Substitutions.
Previous studies have indicated that the formation of CPDs and 6-4PPs is also modulated by the adjacent sequence context (28, 43–45); however, whether the formation of atypical TA-PPs is similarly modulated by neighboring DNA bases is unknown. Analysis of our UVDE-HS-seq data for UV-irradiated yeast, normalized for the genomic frequency of each sequence context, indicates that 6-4PP lesions are elevated when a C or T base is present on the 5′ side of the lesion-forming dipyrimidine (YY) and an A base is present on the 3′ side (Fig. 7A), while a 5′ G base or a 3′ T base suppresses 6-4PP formation. These findings are consistent with a previous report that analyzed preferred sequence contexts adjacent to 6-4PPs (28). Our data indicate that the observed sequence context preferences are present in 6-4PPs occurring at different dipyrimidine sequences (i.e., TC, TT, and CC dipyrimidines), with the exception of CT sequences (SI Appendix, Fig. S12). Analysis of 6-4PP formation in UV-irradiated naked DNA shows a similar trend (SI Appendix, Fig. S13A).
Fig. 7.
Flanking DNA sequence context modulates 6-4PP and TA-PP formation. (A) Frequency of 6-4PP UVDE-seq reads in different sequence contexts. 6-4PP UVDE-seq reads were normalized by the frequency of each sequence context in the yeast genome. YY = dipyrimidine sequences (i.e., TT, TC, CT, and CC). (B) Same as panel A, except for UVDE-seq reads occurring at TA dinucleotides (i.e., TA-PPs). (C) Plot compares the frequency of TA-PPs in the indicated sequence contexts relative to the frequency of A>T substitutions in UV-irradiated yeast in the same sequence contexts. Both TA-PPs and A>T mutations were normalized by the frequency of the indicated sequence context in the yeast genome. (D) Model that elevated TA-PP formation at TA[A/T] (i.e., TAW) sequences induces A>T substitutions in the same sequence context in UV-irradiated yeast.
Similar analysis of TA-PP formation showed distinct preferences for neighboring sequence context. The normalized UVDE-HS-seq data indicated that TA-PPs are elevated when a 5′ pyrimidine base (C or T) is present, but suppressed with a 5′ G base, similar to our 6-4PP data (Fig. 7B) and previous studies of CPD formation (e.g., refs. 28 and 45). However, TA-PPs are highly enriched when the 3′ base is T or A base (Fig. 7B), while 6-4PPs are disfavored at lesion-forming sites with a 3′ T base (Fig. 7A). Normalized TA-PPs form threefold more frequently at TA[A/T] (i.e., TAW) sequences than TA[C/G] (i.e., TAS) sequences. In contrast, 6-4PPs form only ~1.2-fold more frequently at YY[A/T] sequences than YY[C/G] sequences. Similar patterns of TA-PP sequence preferences are apparent in the UV-irradiated naked DNA (SI Appendix, Fig. S13B), indicating that the preferred sequence contexts are not due to modulation by cellular DNA-bound proteins but rather reflect intrinsic sequence preferences of TA-PP formation.
Since previous studies have suggested that TA-PPs cause A>T substitutions at the 3′ adenine base (23, 46), we wondered whether UV-induced A>T substitutions would show similar flanking sequence preferences. To test this hypothesis, we analyzed a data set of 7,052 A>T substitutions identified by genome sequencing of yeast cells repeatedly exposed to UV light (23). This analysis indicates that UV-induced A>T mutations are enriched at both TAA and TAT sequence contexts (underlined base is mutated), even after normalizing for the genomic frequency of each sequence context, mirroring the 3′ sequence preference of UV-induced TA-PPs (Fig. 7C). In total, UV-induced A>T mutations are 4.7-fold more frequent at TAW sequences than TAS sequences. Analysis of normalized A>T mutation frequency in different tetranucleotide sequence contexts indicated that mutation frequency is highly correlated with normalized TA-PP frequency (SI Appendix, Fig. S14; P < 0.0001). Taken together, these findings indicate that TA-PPs show an intrinsic preference to form at TAW sequences, which correlates with elevated A>T substitutions at these same sequence contexts in UV-irradiated yeast (Fig. 7D).
Discussion
While previous studies have demonstrated that nucleosomes and other DNA-bound proteins modulate the formation of UV-induced CPD lesions (7, 8, 47), the impact of chromatin on the formation of less abundant 6-4PPs and atypical TA-PPs across the genome was previously unclear. Here, we describe genome-wide maps of 6-4PPs and TA-PPs in yeast, which reveal that both of these UV photoproducts are significantly modulated in chromatin. Our data indicate that 6-4PPs are elevated at outward rotational settings in strongly positioned nucleosomes, although to a lesser extent than CPDs, but are not elevated in adjacent linker DNA. In contrast, TA-PPs show the opposite pattern in nucleosomes, being suppressed at outward rotation settings and elevated at inward settings (Fig. 3F). Our data further reveal that a number of TFs specifically induce 6-4PP formation, most notably Hap2/3/5 and TBP, in a manner distinct from CPDs. The yeast TF Rap1 specifically induces TA-PP formation in a strand-specific manner, indicating that atypical TA-PPs can also be modulated by TF binding. We show that the 6-4PP hotspot associated with a subset of TBP binding sites precisely coincides with a mutation hotspot in UV-irradiated yeast. This finding, coupled with our analysis of sequence preferences of TA-PPs and UV-induced A>T substitutions, indicates that induction of these less abundant photoproducts can have important ramifications for UV mutagenesis.
Although it has long been thought that 6-4PP formation is elevated (up to ~sixfold) in linker DNA relative to nucleosomal DNA (6, 17), our data indicate that 6-4PPs are not specifically elevated in linker DNA in either wild-type or repair-deficient yeast. A key experimental difference is that the previous study analyzed 6-4PPs levels in chromatin digested with nuclease after UV irradiation (17). It is likely that the presence of 6-4PPs lesions destabilizes weakly positioned nucleosomes (48), which could give the appearance of elevated damage levels in nonnucleosomal DNA (i.e., even though the lesion originally formed in a nucleosome). Alternatively, this discrepancy could be a consequence of differences in linker length or chromatin organization (i.e., linker histones) in yeast and human cells. This finding has important ramifications for our understanding of 6-4PP repair since it suggests that a substantial portion of 6-4PP lesions must be recognized and repaired by the NER machinery in the context of nucleosomes. Our data also indicate that 6-4PPs are enriched at outward rotational settings in nucleosomes, albeit to a lesser extent than CPDs. These findings suggest that 6-4PP formation in nucleosomes largely mimics that of CPD formation, potentially as a consequence of the same type of structural distortion in nucleosomal DNA. As somatic mutations in skin cancers are also elevated at outward rotational settings in human nucleosomes (11, 12), it is possible that elevated 6-4PP formation may contribute to increased mutation rates at these positions in nucleosomes.
6-4PP formation is also modulated by the binding of TFs, being suppressed at some binding site classes (e.g., Reb1, Rap1, etc.) and strongly induced at others (e.g., Hap2/3/5, Lys14, TBP, etc.). While CPDs are similarly affected at some of these binding site classes (e.g., Reb1, Hap2/3/5), closer inspection revealed that the pattern of 6-4PP formation at these binding sites is largely distinct from that of CPDs. These findings indicate that CPD formation does not predict how 6-4PP formation will respond to TF binding and thus highlights the importance of mapping UV-induced 6-4PPs along with CPDs. CPD hotspots associated with TF binding sites in human cells (e.g., ETS, CTCF) have been linked to mutation hotspots in skin cancers. Whether 6-4PP hotspots also result in elevated mutation rates was previously unclear. Our analysis of a compendium of UV-induced mutations in yeast (23) indicates that the 6-4PP hotspot at TBP binding sites (20, 21) coincides with a mutation hotspot in UV-exposed yeast (Fig. 6I). These mutations are likely caused by 6-4PPs, as CPDs (and TA-PPs) are not induced at TBP binding sites, and because the mutation signature (TT>TC substitutions) matches the published signature of 6-4PPs in yeast (42). It will be important to determine whether 6-4PP hotspots in human cells are also associated with elevated UV mutagenesis.
While UV-induced thymine–adenine photoproducts were first detected nearly 40 y ago (4), the impact of chromatin or other DNA-bound proteins on TA-PP formation is almost entirely unknown. Our genome-wide map of TA-PPs indicates that their formation is affected by both nucleosomes and certain TFs (e.g., Rap1). TA-PP formation in nucleosomes shows a strikingly different pattern than canonical UV photoproducts, as they form more readily at inward rotational settings and less readily at outward settings, exactly opposite the pattern observed for CPDs and 6-4PPs (Fig. 3F). Similarly, TA-PPs are strongly induced at Rap1 binding sites (Fig. 5G), while CPDs and 6-4PPs are generally suppressed. Since it has recently been reported that as many as 12% of all UV-induced mutations in yeast may arise due to mutagenic bypass of TA-PPs (23), these findings have potentially important implications for UV mutagenesis in chromatin. Indeed, our analysis indicates that the intrinsic preference for TA-PPs to form at TAW sequence contexts can explain a similar enrichment of UV-induced A>T substitutions in these sequence contexts across the yeast genome. It will be important to determine in future studies to what extent these patterns of TA-PP formation affect UV mutagenesis in human skin cancers.
Materials and Methods
UVDE-seq and UVDE-HS-seq Library Preparation and Sequencing.
We adopted the emRiboSeq protocol (49) and our CPD-seq protocol (10, 14) to create the UVDE-seq method to accurately map 6-4PP formation, as previously described (23, 24). Briefly, cells were grown in yeast extract peptone dextrose medium, exposed to 600 J/m2 UVC light, and pelleted, and genomic DNA (gDNA) was isolated. Cells used for naked DNA samples were collected as described but not exposed to UVC. Following gDNA isolation, naked samples were spotted onto a microscope cover slide on ice and exposed to 400 J/m2 or 500 J/m2 UVC light. All samples were then sonicated, and DNA fragments were treated with end-repair and dA-tailing modules (New England Biolabs), and a double-stranded trP1 adapter was ligated to both ends of the fragments. Adapter and primer sequences (used to confirm ligation) are listed in SI Appendix, Materials and Methods. Following trP1 ligation confirmation, free 3′-OH groups were blocked with Terminal Transferase and either dideoxy ATP or dideoxy GTP. Samples were then treated with Escherichia coli CPD photolyase under 365 nm UV light for 2 h at room temperature. Those libraries prepared using the UVDE-HS-seq method were treated with uracil DNA glycosylase inhibitor during CPD photolyase treatment, under the same conditions. DNA fragments were purified and treated with Thermus thermophilus UVDE for 45 min at 55 °C. 5′ phosphate groups were removed using shrimp alkaline phosphatase, and DNA was subsequently denatured at 95 °C for 5 min and snap-cooled on ice. A second double-stranded adapter, the A adapter, was then ligated to the 3′-OH created immediately downstream from the cleaved UV lesion and PCR confirmed. DNA containing the biotin label on the A adapter was purified using Streptavidin beads, while the DNA strand lacking the biotin label was removed using 0.15 M NaOH. The remaining single-stranded DNA was then used as a template for second-strand synthesis, and libraries were PCR amplified using primers complimentary to the trP1 and A adapters. Finally, samples were combined and submitted for Ion Proton sequencing (Life Technologies). The resulting UVDE-seq reads were mapped to the yeast (SacCer3) genome using bowtie2 (50), and the corresponding dinucleotide damage site was identified and counted, as previously described (10, 14). More detail can be found in the SI Appendix, Materials and Methods.
UVDE-seq Control Library Preparation and Sequencing.
A control library to confirm that there was no contamination of CPDs in our UVDE-seq libraries was performed as described above, but with the following modifications. Libraries from UV-irradiated wild-type yeast (BY4741) were prepared and carried through CPD photolyase incubation. Following CPD photolyase treatment and ethanol precipitation of genomic DNA, one-half of the library was treated with T4 endonuclease V and APE1 to cleave CPD lesions (as described in ref. 10), and the other was treated with UVDE to cleave CPD, 6-4PP, and atypical lesions. Following enzyme treatment, the samples were purified using Ampure XP beads and continued with library preparation. Sequenced reads were then aligned to the yeast genome using bowtie2 (50), and dinucleotide damage sites were counted. CPD- and 6-4PP-associated control reads are shown in Fig. 1 D and E. We also sequenced a No UV control sample for the UVDE protocol, but this sample was not analyzed further because of aberrantly high sequencing depth for this sample.
UVDE-seq Data Analysis.
UVDE-seq sequencing reads were trimmed of barcode sequences and the 3′ nucleotide of the sequencing read, and then aligned to the SacCer3 genome using the bowtie2 software (50). The resulting alignment files were processed with SAMtools (51) and BEDtools (52), and custom Perl scripts were used to identify the dinucleotide sequence immediately upstream of the 5′ end of each sequencing read. The dinucleotide sequence on the opposite strand was extracted as a putative 6-4PP (or TA-PP) lesion. To analyze UVDE-seq reads specifically associated with 6-4PPs, reads associated with lesion-forming dipyrimidine sequences (i.e., TC, TT, CT, and CC) were identified and used for subsequent analysis. To analyze UVDE-seq reads associated with TA-PPs, reads associated with thymine–adenine (TA) dinucleotides were retained for analysis. Typically, both positions in the dipyrimidine nucleotide were counted as lesion sites, except for high-resolution analysis in which the 6-4PP was assigned to a half-integer position between the two nucleotides comprising the lesion (i.e., a lesion formed in a TT sequence at positions 12 and 13 would have a half-integer position of 12.5). Analysis of the enrichment of UVDE-HS-seq reads associated with 6-4PPs and TA-PPs in different sequence contexts was determined using BEDtools (52) and custom Perl scripts, after normalizing for the genomic frequency of each sequence context across the yeast genome (SacCer3).
CPD-seq Data Analysis.
CPD-seq data mapping CPD lesions in yeast or UV-irradiated naked DNA were derived from a previous publication (10) and generally analyzed as described previously.
Analysis of UV Photoproduct Formation in Nucleosomes.
Analysis of 6-4PP or TA-PP formation in yeast nucleosomes was performed using a previously published high-resolution map of yeast nucleosome dyad positions (29). Strongly positioned nucleosomes in this map were defined as those having a nucleosome score of greater than 5, while weakly positioned nucleosomes had a score less than 1, as previously described (10). Custom perl scripts were used to determine the enrichment of 6-4PPs in UV-irradiated yeast cells relative to the UV-irradiated naked DNA control and scaled so that there were an equivalent number of lesions in both data sets. CPD enrichment was determined in a similar manner using published CPD-seq data (10). TA-PP formation in nucleosomes was analyzed in a similar manner, except that we also analyzed normalized TA-PP density in UV-irradiated yeast, in which TA-PP counts were normalized to the number of TA dinucleotides overlapping with each position in the nucleosome.
For nucleosome analysis, peak rotational and translational periodicity and their corresponding signal-to-noise ratio (SNR) were determined using the mutperiod software package (30). See SI Appendix, Materials and Methods for more details regarding this analysis.
Analysis of UV Photoproduct Formation in TFBS.
We analyzed CPD, 6-4PP, and TA-PP formation at published yeast TF binding sites identified by ChIP-exo (36). Analysis of photoproduct formation at TF binding sites was performed using custom Perl scripts, as previously described (10, 14, 34). We also analyzed UV damage formation at a set of 713 TBP-bound TATA sites, derived from published ChIP-exo data (35), and a set of low-occupancy control Reb1 binding sites (10, 53). Sequence logos were generated using weblogo (38), with default background nucleotide probabilities. Additional details are described in the SI Appendix, Materials and Methods.
UVDE Cleavage Optimization by Alkaline Gel Electrophoresis.
To verify the cleavage of UV-induced lesions by UVDE, we performed a global DNA cleavage assay, essentially as previously described (54). Briefly, wild-type (BY4741) cells were grown to an OD600 of ~0.8, pelleted, and resuspended in water. Cells were then exposed to 600 J/m2 UVC light, pelleted, and frozen at −80 °C. Genomic DNA (gDNA) was isolated using a phenol:chloroform:isoamylalcohol extraction and RNaseA treated. Samples were then treated solely or with varying combinations of E. coli CPD photolyase, T4 endonuclease V, and T. thermophilus UVDE and run on a 1.2% alkaline gel at 30 V for 20 h (SI Appendix, Fig. S2). The gel was subsequently washed, stained with SYBR gold, and imaged using the Typhoon FLA7000 imager and ImageQuant software.
Analysis of UV-Induced Mutations in Yeast.
We analyzed 45,747 single-nucleotide variants derived from whole-genome sequencing of UV-exposed wild-type, rad16∆, and rad26∆ diploid yeast (23). Significant enrichment of mutations within 5 bp of the TBP binding site midpoint was determined by chi-square analysis, comparing the observed mutation count within binding sites and outside the binding sites relative to the expected mutation count based on sequence context. Analysis was performed both for TATA sites containing dipyrimidines at positions −1/0 and 0/+1 relative to the motif midpoint and TATA sites not containing dipyrimidines at these positions. As a control, we identified 2,182 sites that matched the TATAAA site consensus, but were located at least 100 bp in side of a gene (i.e., at least 100 bp from transcription start and end site of the gene), and compared the observed to expected mutation count as described above.
For analysis of UV-induced A>T mutations, we analyzed the normalized frequency of only A>T single nucleotide substitutions (23) in different trinucleotide and tetranucleotide sequence contexts. Correlation with TA-PP frequency was determined using the Spearman correlation coefficient function in Graphpad Prism.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We are grateful to Dr. Michael Smerdon for helpful comments and suggestions. We thank Wei Wei Du and Mark Wildung for assistance with Ion Torrent sequencing. We thank Dr. Alexander Brown for purifying the UVDE enzyme and Marian Laughery for generating the UV mutation data in yeast. This work was supported by funding from National Institute of Environmental Health Sciences grants R21ES027937 (to S.A.R. and J.J.W.), R21ES029655 (to J.J.W.), R01ES028698 (to J.J.W.), and R01ES032814 (to S.A.R. and J.J.W.). K.A.B. was supported by a training grant (T32GM008336) from the National Institute of General Medical Sciences and later a Poncin Fellowship.
Author contributions
K.A.B. and J.J.W. designed research; K.A.B. performed research; P.M. and S.A.R. contributed new reagents/analytic tools; K.A.B., B.M.-H., S.S., and J.J.W. analyzed data; B.M.-H. edited paper; P.M. assisted with bioinformatics analysis and edited paper; S.A.R. helped direct research and edited paper; and K.A.B. and J.J.W. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The UVDE-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accessions GSE144679 and GSE190471. The UVDE-HS-seq data were deposited in GEO under accession GSE214217. Software code is available at https://github.com/bmorledge-hampton19/Yeast_UVDE-seq_Analysis.
Supporting Information
References
- 1.Friedberg E. C., et al. , DNA Repair and Mutagenesis (ASM Press, Washington, D.C., ed. 2, 2006), p. 1118, p. xxvii. [Google Scholar]
- 2.Pfeifer G. P., Besaratinia A., UV wavelength-dependent DNA damage and human non-melanoma and melanoma skin cancer. Photochem. Photobiol. Sci. 11, 90–97 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao X., Nadji S., Kao J. L., Taylor J. S., The structure of d(TpA), the major photoproduct of thymidylyl-(3’5’)-deoxyadenosine. Nucleic Acids Res. 24, 1554–1560 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bose S. N., Davies R. J., Sethi S. K., McCloskey J. A., Formation of an adenine-thymine photoadduct in the deoxydinucleoside monophosphate d(TpA) and in DNA. Science 220, 723–725 (1983). [DOI] [PubMed] [Google Scholar]
- 5.Davies R. J., et al. , High-resolution crystal structure of the intramolecular d(TpA) thymine-adenine photoadduct and its mechanistic implications. Nucleic Acids Res. 35, 1048–1053 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao P., Wyrick J. J., Roberts S. A., Smerdon M. J., UV-induced DNA damage and mutagenesis in chromatin. Photochem. Photobiol. 93, 216–228 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao P., Wyrick J. J., Organization of DNA damage, excision repair, and mutagenesis in chromatin: A genomic perspective. DNA Repair 81, 102645 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts S. A., Brown A. J., Wyrick J. J., Recurrent noncoding mutations in skin cancers: UV damage susceptibility or repair inhibition as primary driver? Bioessays 41, e1800152 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gale J. M., Nissen K. A., Smerdon M. J., UV-induced formation of pyrimidine dimers in nucleosome core DNA is strongly modulated with a period of 10.3 bases. Proc. Natl. Acad. Sci. U.S.A. 84, 6644–6648 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mao P., Smerdon M. J., Roberts S. A., Wyrick J. J., Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 113, 9057–9062 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pich O., et al. , Somatic and germline mutation periodicity follow the orientation of the DNA minor groove around nucleosomes. Cell 175, 1074–1087.e1018 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Brown A. J., Mao P., Smerdon M. J., Wyrick J. J., Roberts S. A., Nucleosome positions establish an extended mutation signature in melanoma. PLoS Genet. 14, e1007823 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Premi S., et al. , Genomic sites hypersensitive to ultraviolet radiation. Proc. Natl. Acad. Sci. U.S.A. 116, 24196–24205 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao P., et al. , ETS transcription factors induce a unique UV damage signature that drives recurrent mutagenesis in melanoma. Nat. Commun. 9, 2626 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott K., et al. , Elevated pyrimidine dimer formation at distinct genomic bases underlies promoter mutation hotspots in UV-exposed cancers. PLoS Genet. 14, e1007849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fredriksson N. J., et al. , Recurrent promoter mutations in melanoma are defined by an extended context-specific mutational signature. PLoS Genet. 13, e1006773 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell D. L., Nguyen T. D., Cleaver J. E., Nonrandom induction of pyrimidine-pyrimidone (6–4) photoproducts in ultraviolet-irradiated human chromatin. J. Biol. Chem. 265, 5353–5356 (1990). [PubMed] [Google Scholar]
- 18.Gale J. M., Smerdon M. J., UV induced (6–4) photoproducts are distributed differently than cyclobutane dimers in nucleosomes. Photochem. Photobiol. 51, 411–417 (1990). [DOI] [PubMed] [Google Scholar]
- 19.Hu J., Adebali O., Adar S., Sancar A., Dynamic maps of UV damage formation and repair for the human genome. Proc. Natl. Acad. Sci. U.S.A. 114, 6758–6763 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aboussekhra A., Thoma F., TATA-binding protein promotes the selective formation of UV-induced (6–4)-photoproducts and modulates DNA repair in the TATA box. EMBO J. 18, 433–443 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Gross M. L., Taylor J. S., Use of a combined enzymatic digestion/ESI mass spectrometry assay to study the effect of TATA-binding protein on photoproduct formation in a TATA box. Biochemistry 40, 11785–11793 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Tornaletti S., Pfeifer G. P., UV light as a footprinting agent: Modulation of UV-induced DNA damage by transcription factors bound at the promoters of three human genes. J. Mol. Biol. 249, 714–728 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Laughery M. F., et al. , Atypical UV photoproducts induce non-canonical mutation classes associated with driver mutations in melanoma. Cell Rep. 33, 108401 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bohm K. A., Sivapragasam S., Wyrick J. J., Mapping atypical UV photoproducts in vitro and across the S. cerevisiae genome. STAR Protocols 3, 101059 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao P., Wyrick J. J., Genome-wide mapping of UV-induced DNA damage with CPD-Seq. Methods Mol. Biol. 2175, 79–94 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Avery A. M., et al. , Substrate specificity of ultraviolet DNA endonuclease (UVDE/Uve1p) from Schizosaccharomyces pombe. Nucleic Acids Res. 27, 2256–2264 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paspaleva K., et al. , Crystal structure of the DNA repair enzyme ultraviolet damage endonuclease. Structure 15, 1316–1324 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Bryan D. S., Ransom M., Adane B., York K., Hesselberth J. R., High resolution mapping of modified DNA nucleobases using excision repair enzymes. Genome Res. 24, 1534–1542 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brogaard K., Xi L., Wang J. P., Widom J., A map of nucleosome positions in yeast at base-pair resolution. Nature 486, 496–501 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morledge-Hampton B., Wyrick J. J., Mutperiod: Analysis of periodic mutation rates in nucleosomes. Comput. Struct. Biotechnol. J. 19, 4177–4183 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li W., Adebali O., Yang Y., Selby C. P., Sancar A., Single-nucleotide resolution dynamic repair maps of UV damage in Saccharomyces cerevisiae genome. Proc. Natl. Acad. Sci. U.S.A. 115, E3408–E3415 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui F., Zhurkin V. B., Structure-based analysis of DNA sequence patterns guiding nucleosome positioning in vitro. J. Biomol. Struct. Dyn. 27, 821–841 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cui F., Cole H. A., Clark D. J., Zhurkin V. B., Transcriptional activation of yeast genes disrupts intragenic nucleosome phasing. Nucleic Acids Res. 40, 10753–10764 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivapragasam S., et al. , CTCF binding modulates UV damage formation to promote mutation hot spots in melanoma. EMBO J. 40, e107795 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rhee H. S., Pugh B. F., Genome-wide structure and organization of eukaryotic pre-initiation complexes. Nature 483, 295–301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rossi M. J., et al. , A high-resolution protein architecture of the budding yeast genome. Nature 592, 309–314 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan M., et al. , High-resolution mapping demonstrates inhibition of DNA excision repair by transcription factors. Elife 11, e73943 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Azad G. K., Tomar R. S., The multifunctional transcription factor Rap1: A regulator of yeast physiology. Front. Biosci. (Landmark edition) 21, 918–930 (2016). [DOI] [PubMed] [Google Scholar]
- 40.Stark B., Poon G. M. K., Wyrick J. J., CTCF puts a new twist on UV damage and repair in skin cancer. Mol. Cell Oncol. 8, 2009424 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elliott K., Larsson E., Non-coding driver mutations in human cancer. Nat. Rev. Cancer 21, 500–509 (2021). [DOI] [PubMed] [Google Scholar]
- 42.Bresson A., Fuchs R. P., Lesion bypass in yeast cells: Pol eta participates in a multi-DNA polymerase process. EMBO J. 21, 3881–3887 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brash D. E., Haseltine W. A., UV-induced mutation hotspots occur at DNA damage hotspots. Nature 298, 189–192 (1982). [DOI] [PubMed] [Google Scholar]
- 44.Mitchell D. L., Jen J., Cleaver J. E., Sequence specificity of cyclobutane pyrimidine dimers in DNA treated with solar (ultraviolet B) radiation. Nucleic Acids Res. 20, 225–229 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Law Y. K., Forties R. A., Liu X., Poirier M. G., Kohler B., Sequence-dependent thymine dimer formation and photoreversal rates in double-stranded DNA. Photochem. Photobiol. Sci. 12, 1431–1439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X., Taylor J. S., Mutation spectra of TA*, the major photoproduct of thymidylyl-(3’5’)-deoxyadenosine, in Escherichia coli under SOS conditions. Nucleic Acids Res. 24, 1561–1565 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gonzalez-Perez A., Sabarinathan R., Lopez-Bigas N., Local determinants of the mutational landscape of the human genome. Cell 177, 101–114 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Duan M. R., Smerdon M. J., UV damage in DNA promotes nucleosome unwrapping. J. Biol. Chem. 285, 26295–26303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding J., Taylor M. S., Jackson A. P., Reijns M. A., Genome-wide mapping of embedded ribonucleotides and other noncanonical nucleotides using emRiboSeq and EndoSeq. Nat. Protoc. 10, 1433–1444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., et al. , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quinlan A. R., Hall I. M., BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kasinathan S., Orsi G. A., Zentner G. E., Ahmad K., Henikoff S., High-resolution mapping of transcription factor binding sites on native chromatin. Nat. Methods 11, 203–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hodges A. J., Plummer D. A., Wyrick J. J., NuA4 acetyltransferase is required for efficient nucleotide excision repair in yeast. DNA Repair 73, 91–98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
The UVDE-seq data have been deposited in the Gene Expression Omnibus (GEO) database under accessions GSE144679 and GSE190471. The UVDE-HS-seq data were deposited in GEO under accession GSE214217. Software code is available at https://github.com/bmorledge-hampton19/Yeast_UVDE-seq_Analysis.