Abstract
Three genes, coxB, coxA, and coxC, found in a clone from a gene library of the cyanobacterium Anabaena variabilis strain ATCC 29413, were identified by hybridization with an oligonucleotide specific for aa3-type cytochrome c oxidases. Deletion of these genes from the genome of A. variabilis strain ATCC 29413 FD yielded strain CSW1, which displayed no chemoheterotrophic growth and an impaired cytochrome c oxidase activity. Photoautotrophic growth of CSW1, however, was unchanged, even with dinitrogen as the nitrogen source. A higher cytochrome c oxidase activity was detected in membrane preparations from dinitrogen-grown CSW1 than from nitrate-grown CSW1, but comparable activities of respiratory oxygen uptake were found in the wild type and in CSW1. Our data indicate that the identified cox gene cluster is essential for fructose-dependent growth in the dark, but not for growth on dinitrogen, and that other terminal respiratory oxidases are expressed in this cyanobacterium. Transcription analysis showed that coxBAC constitutes an operon which is expressed from two transcriptional start points. The use of one of them was stimulated by fructose.
Cyanobacteria are prokaryotes capable of oxygenic photosynthesis. However, all cyanobacteria also perform aerobic respiration in the dark. Indeed, cyanobacteria are the only known organisms in which these two most important bioenergetic processes occur in the same compartment. In a natural environment, respiration probably serves as an energy-conserving mechanism during the night, although only a few strains are able to grow heterotrophically in the dark. Some filamentous cyanobacteria can fix dinitrogen in specialized cells termed heterocysts. It has been suggested that in these strains, respiration may have an additional function as a protective mechanism, since nitrogenases, the key enzymes of dinitrogen fixation, are extremely sensitive to oxygen and respiratory activity is considerably higher in heterocysts than in vegetative cells (9).
The respiratory chain in cyanobacteria is intimately linked to photosynthetic electron transport (24), and the only electron transport components not directly involved in photosynthesis are the terminal respiratory oxidases, which act as the terminal acceptors of the electron transport chains and are therefore the key enzymes of cyanobacterial respiration. In many bacteria, there is more than one terminal respiratory oxidase (for a recent review, see reference 21). The best-characterized terminal oxidases are those that are homologous to the aa3-type cytochrome c oxidases of mitochondria. Almost all cyanobacteria, even those that are obligate photoautotrophs, contain membrane-bound enzymes capable of the oxidation of cytochrome c (24). Terminal respiratory oxidases with sequence similarity to the cytochrome bd quinol oxidase, which is found in bacteria, have also been identified in cyanobacteria (reference 16 and the Anabaena website http://www.kazusa.or.jp /cyano/anabaena/). In Synechocystis sp. strain PCC 6803, there are three terminal respiratory oxidases, two homologous of the cytochrome c oxidase of mitochondria and one putative quinol oxidase homologous to Escherichia coli cytochrome bd (15).
We are interested in characterizing the roles of the different branches of the respiratory chain in heterocyst-forming cyanobacteria. For the present work, which is aimed at the isolation of cytochrome c oxidase-encoding genes, we chose Anabaena variabilis strain ATCC 29413, one of the few cyanobacteria that is able to form heterocysts, that grows in the dark with both bound nitrogen and N2 (34), and that can be manipulated by molecular biology tools (28). The variant A. variabilis strain ATCC 29413 FD (5), which displays a higher efficiency than strain ATCC 29413 in conjugative gene transfer from E. coli, was used for construction of the cytochrome c oxidase mutant.
MATERIALS AND METHODS
Strains and growth conditions.
This study was carried out with the heterocyst-forming cyanobacterium A. variabilis strain ATCC 29413 and with its derivative strain ATCC 29413 FD, which can grow at 40°C (5). This strain was grown photoautotrophically, mixotrophically, or chemoheterotrophically at 30°C in BG110 medium (medium BG11 [23] without NaNO3) supplemented with 0.84 g of NaHCO3 per liter (BG110C medium) and bubbled with a mixture of CO2 (1%, vol/vol) and air. When indicated, 8 mM NH4Cl plus 16 mM TES-NaOH buffer (pH 7.5), 17.6 mM NaNO3, or 20 mM fructose was added. For RNA isolation, exponentially growing cells were used. Cells used for the measurement of oxygen uptake and the preparation of membranes were grown at 32°C in a shaker in 0.25% CO2 in air without sparging. Cultures shaken in an air atmosphere at 30°C were used to test the growth phenotype of the CSW1 mutant. E. coli strain DH5α (2) was used for cloning and subcloning purposes.
Gene cloning and inactivation.
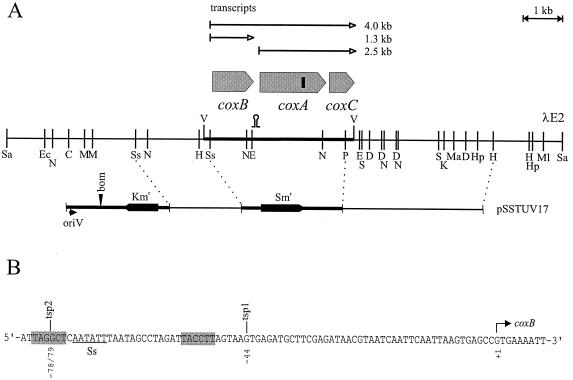
A gene library consisting of partially Sau3AI-restricted total DNA from wild-type A. variabilis strain ATCC 29413 cloned into the BamHI site of λEMBL3A was used to isolate a clone (called λE2) hybridizing with oligonucleotide C (5′-AACATRTGRTGNGCCCA-3′) (22). A physical map of the insert of λE2 was prepared (Fig. 1), and sequences of subclones were determined on both sides of the oligonucleotide C hybridization site until the complete coxBAC locus was covered. For the inactivation of the coxBAC locus in A. variabilis ATCC 29413 FD, plasmid pSSTUV17 was constructed. This plasmid consists of the following (Fig. 1A, left to right): the HindIII-PvuII fragment from pUC19 (positions 449 to 630 [36]); the 2,704-bp blunted BamHI-HindIII fragment from plasmid pRL25 (35) containing oriV and the bom site from pBR322 and the kanamycin resistance cassette from transposon Tn5; the 1.9-kb SspI-SspI 5′ flanking region of the A. variabilis coxBAC locus; the HindIII (blunted)-FspI fragment from transposon Tn5 (positions 1195 to 888 [1]); the FspI-Sau96I fragment (positions 262 to 175), the 192-bp Sau96I-Sau96I fragment (positions 4344 to 4361 and 1 to 174), and the Sau96I-XhoII (blunted) fragment (positions 3409 to 3228), all from pBR322 (33); the PvuII-HindIII fragment from pUC18 (positions 309 to 399); the 2.0-kb HindIII-BamHI streptomycin-spectinomycin resistance cassette C.S3 (8); the BamHI-PstI fragment from pUC19 (positions 418 to 439); and the 3.8-kb PstI-HindIII 3′-flanking region of the A. variabilis coxBAC locus. In effect, pSSTUV17 contains a 9.2-kb chromosomal region of A. variabilis in which the coxBAC locus has been almost completely supplanted by a streptomycin-spectinomycin resistance cassette. The kanamycin resistance cassette in the vector part of pSSTUV17 is useful to distinguish between single and the desired double recombinants.
FIG. 1.
(A) The coxBAC locus of A. variabilis strain ATCC 29413. The restriction sites in λE2 are CelII (C), DraI (D), Eco47III (E), EcoRI (Ec), HindIII (H), HpaI (Hp), KpnI (K), MunI (M), MamI (Ma), MluNI (Ml), NheI (N), PstI (P), ScaI (S), SspI (Ss), SalI (Sa), and VspI (V). The small vertical bar in the coxA gene represents the binding site for oligonucleotide C (see Materials and Methods). pSSTUV17 is the plasmid used for construction of the CSW1 mutant. bom, basis of mobility (10); oriV, origin of replication in E. coli, not active in Anabaena. The hairpin denotes the position of a putative cutting site for RNase III (see Discussion). (B) Sequence of the cox locus around the SspI site (Ss) close to the putative translational start of coxB containing the two tsp (see Fig. 5). The hatched hexamers show putative −10 and −35 regions for tsp1. Similar sequences for tsp2 could not be identified.
A. variabilis mutant construction.
Conjugation of plasmid pSSTUV17 to A. variabilis strain ATCC 29413 FD, grown at 40°C, was carried out essentially as described previously (7). The triparental mating involved conjugal plasmid pRL443 and helper plasmids pRL528 and pRL591-W45 (6). Exconjugants were selected and further grown in medium containing streptomycin and spectinomycin at 2 μg ml−1 each. Segregation of the mutant chromosomes was confirmed by PCR using the oligonucleotides cox4 (5′-CACGCGGGTCATCAAGCGCC-3′; forward primer, internal to coxB) and cox3 (5′-CAATACGAGCCGGGATATTGGG-3′; reverse primer, internal to coxA) and the total DNA isolated from strains ATCC 29413 FD and CSW1.
Cytochrome c oxidase and O2 uptake activities.
Isolated membranes from A. variabilis for determination of in vitro cytochrome c oxidase activity were prepared from 200-ml cultures at an optical density at 730 nm of about 3. After harvesting, the cells were resuspended in 10 ml of HEPES buffer (10 mM HEPES [pH 7.4], 5 mM NaCl) supplemented with 20% (mass/vol) sucrose and 10 mg of lysozyme and incubated for 30 min at 37°C. The cells were centrifuged in a JA-20 rotor for 5 min, resuspended in 6 ml of ice-cold HEPES buffer per g (wet weight), and incubated on ice for 1 h. After the addition of 1 mM phenylmethylsulfonyl fluoride (100 mM in ethanol) and 0.005% (mass/vol) DNase I, the cells were passed through a French press at 11,000 lb/in2 three consecutive times and centrifuged at 0°C and 14,000 rpm in a JA-20 rotor for 10 min. Further processing depended on the nitrogen source used for growth of the cells. In the case of BG110 medium-grown cells, the supernatant contained very little membrane material, and the pellet, which contained no detectable unbroken cells, was used directly. In the case of the cells grown with bound nitrogen, the supernatant was centrifuged in an SW41Ti rotor at 0°C at 40,000 rpm for 50 min and the supernatant was discarded. In both cases, the membrane pellets were resuspended on ice in HEPES buffer in a Potter homogenizer to a protein concentration of 2 to 5 mg ml−1. The assay medium contained, in a total volume of 3 ml, membranes at 150 to 500 μg of protein ml−1 in HEPES buffer and 10 or 20 μM horse heart cytochrome c prereduced with ascorbic acid. The difference between A550 and A540 was monitored in a stirred cuvette at 30°C in a Varian Cary 5 spectrophotometer. Rates of horse heart cytochrome c oxidation were determined using E550s of 29.5 and 8.4 cm−1 mM−1 for reduced and oxidized cytochrome c, respectively (31). Chlorophyll (Chl) content was determined by the method of Mackinney (18). For the determination of protein content, 2 to 10 μl of membranes in HEPES buffer was mixed with an equal volume of 20% (mass/vol) sodium dodecyl sulfate and vortexed for 10 min and then the protein concentration was determined by the method of Chang (4). Respiratory O2 uptake activity in the dark was determined as described earlier (20).
Gene expression analysis.
The total DNA (3) and RNA (by using the glass bead method [14] as modified by García-Domínguez and Florencio [13]) from A. variabilis strain ATCC 29413 FD and its derivative CSW1 were isolated as previously described. DNA fragments were purified from agarose gels with the Geneclean II kit (Bio 101 Inc.). For Northern analysis, 70 μg of RNA was loaded per lane and was electrophoresed in 1% agarose–denaturing formaldehyde gels. Transfer and fixation to Hybond-N Plus membranes (Amersham Pharmacia) were carried out using 0.1 M NaOH. Hybridization was performed at 65°C according to the recommendations of the manufacturers of the membranes. The coxB probe was a 1.1-kb EcoRI-AseI fragment from λE2 containing most of the coxB gene. The coxAC probe was a 1.8-kb BstXI-PstI fragment from λE2 containing most of the coxAC genes. The fragments used as probes were 32P labeled with a Ready to Go DNA labeling kit (Amersham/Pharmacia) using [α-32P]dCTP. Images of radioactive filters were obtained and quantified using a Cyclone storage phosphor system and OptiQuant image analysis software (Packard). The oligonucleotide used for primer extension analysis of the coxB transcript was CB1 (5′-GGCTTGCCAGGGTTAGCCCG-3′; complementary to positions +58 to +39 relative to the putative translation start of coxB). Oligonucleotide labeling and primer extension reactions were carried out as described previously (19). Images of radioactive gels were obtained and quantified as described above.
Nucleotide sequence accession number.
The sequence data for the coxBAC genes have been submitted to the EMBL database under accession number Z98264.
RESULTS
Cloning and sequencing of coxBAC.
To isolate by hybridization the A. variabilis cox genes, oligonucleotide C (22) was used as a probe. This oligonucleotide is specific for a very highly conserved region of subunit I of aa3-type cytochrome c oxidases. Clone λE2 from a λEMBL3A library containing a 14,300-bp insert was identified and characterized (Fig. 1). The 3,954-bp VspI fragment was sequenced and found to contain open reading frames whose putative products were homologous to the coxB, coxA, and coxC gene products. When compared with well-characterized cytochrome c oxidases from other organisms, amino acid sequence identities were highest with Synechocystis sp. strain PCC 6803 (71.0% for CoxA, 54.0% for CoxB, and 64.9% for CoxC) (16). The identities with the Paracoccus denitrificans cytochrome c oxidase (22) were 45.0% for CoxA, 31.9% for CoxB, and 34.5% for CoxC.
Inactivation of coxBAC and growth phenotype of the mutant.
Plasmid pSSTUV17 contains a DNA fragment of 9,200 bp from the λE2 clone in which the 3,805-bp fragment containing all of coxB and coxA and most of coxC was replaced by a streptomycin-spectinomycin resistance cassette. The vector part is not replicative in A. variabilis, can be mobilized by conjugative plasmid RP4 (and derivatives) to A. variabilis, and carries a kanamycin resistance gene. After conjugation to A. variabilis strain ATCC 29413 FD and selection for resistance to streptomycin-spectinomycin, a number of colonies were obtained. Four colonies were transferred to liquid medium and tested for growth in the dark and for the presence of wild-type chromosomes by PCR analysis using oligonucleotides from the deleted DNA region as primers. One clone showed no growth in the dark, contained no wild-type chromosome, and was called strain CSW1. This strain was also sensitive to kanamycin, confirming integration of the streptomycin-spectinomycin resistance cassette by double recombination. The direct isolation of a segregated double recombinant was probably facilitated by the long flanking A. variabilis DNA fragments in pSSTUV17.
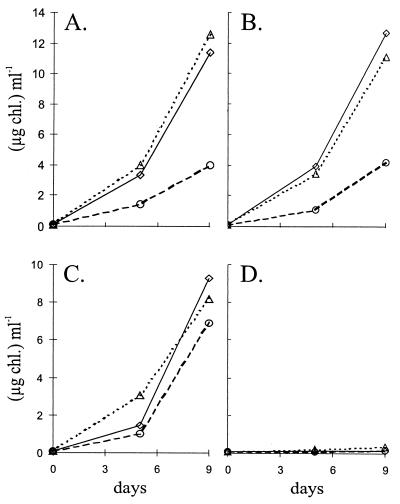
The growth of the mutant was tested in light and darkness using different nitrogen sources. While the wild-type strain grew in the dark independent of the nitrogen source used, no growth was obtained with the CSW1 mutant in any nitrogen source (Fig. 2C and D). In the light, however, growth rates were similar for the two strains (Fig. 2A and B). This shows that the mutated gene cluster is essential for chemoheterotrophic growth but not for dinitrogen fixation.
FIG. 2.
Growth phenotype of the A. variabilis CSW1 mutant. Cells grown in BG11 medium (with antibiotics in the case of CSW1) were washed and inoculated with 0.1 μg of Chl ml−1 in medium with nitrate (◊), ammonium (▵), or dinitrogen (○) as the nitrogen source and were incubated in the light (A and B) or in the dark with 20 mM fructose (C and D). Results are shown for wild-type strain ATCC 29413 FD (A and C) and mutant strain CSW1 (B and D).
Cytochrome c oxidation and respiratory activities.
Cytochrome c oxidase activity of membranes prepared from the two Anabaena strains is presented in Table 1. Since the protein-to-Chl ratio of the membranes was found to depend highly on the growth conditions, activities based on both Chl and protein contents are given. In cells grown photoautotrophically with nitrate (BG11), CSW1 showed about one-fifth of the activity of the wild type. Therefore, the coxBAC genes encode a cytochrome c oxidase. However, a significant activity remained in the mutant, indicating the presence of at least one other cytochrome c oxidase in A. variabilis. Chemoheterotrophically grown wild-type cells displayed a significantly increased cytochrome c oxidase activity, presumably due to the induction of CoxBAC by fructose (see below). When dinitrogen was used instead of nitrate as the N source in the light, higher cytochrome c oxidase activities in membranes were observed, especially on a Chl basis, and these activities were somewhat higher in CSW1 than in the wild type.
TABLE 1.
Horse heart cytochrome c oxidation by isolated membranes of Anabaena strainsa
| Growth medium (condition) | Strain | hh-cytox activityb (mean ± SD)
|
|
|---|---|---|---|
| nmol h−1 (mg of Chl)−1 | nmol h−1 (mg of protein)−1 | ||
| BG11 (light) | ATCC 29413 FD | 810 ± 144 | 140 ± 14 |
| CSW1 | 199 ± 2 | 29 ± 1 | |
| BG11 + fructose (dark) | ATCC 29413 FD | 7,241 | 298 |
| BG110 (light) | ATCC 29413 FD | 4,166 | 192 |
| CSW1 | 5,591 | 299 | |
For experimental details, see Materials and Methods.
hh-cytox, horse heart cytochrome c oxidase. Inhibition with 5 μM KCN was always >88%.
Respiratory O2 uptake (given in micromoles of O2 h−1 milligrams of Chl−1) was measured in cells grown photoautotrophically with nitrate. Respiration rates were similar with endogenous electron sources (5.87 ± 0.87 [mean ± standard deviation] for wild-type cells and 5.00 ± 0.50 for CSW1) and with 10 mM exogenous fructose (7.13 ± 1.50 for wild-type cells and 6.13 ± 0.47 for CSW1). All respiratory activities were 100% inhibitable by 1 mM KCN, showing the absence of a KCN-insensitive terminal oxidase, an enzyme found in plants but so far not in cyanobacteria (32).
Expression of coxBAC.
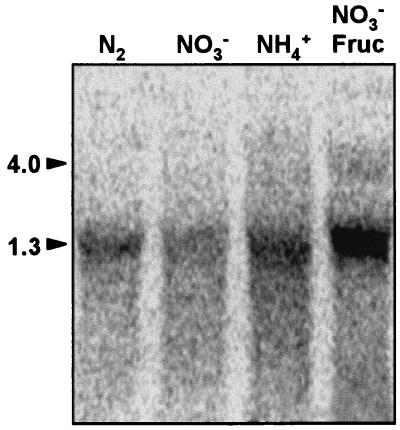
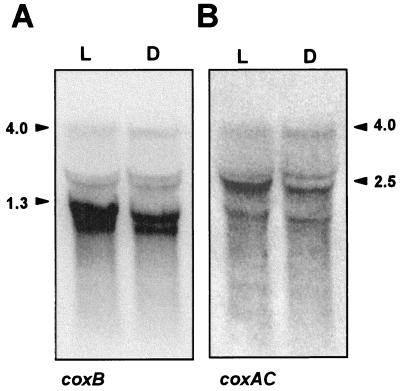
Northern analysis was performed with a probe of coxB using RNA isolated from A. variabilis strain ATCC 29413 FD cells grown autotrophically with nitrate, ammonium, or dinitrogen as the nitrogen source as well as from cells grown mixotrophically (with fructose in the light) and with nitrate as the nitrogen source (Fig. 3). An abundant transcript of 1.3 kb was observed with all the RNA preparations. The addition of fructose to the medium induced this transcript as well as a transcript of about 4 kb that might also be present at a low level in the absence of fructose (see the NH4+ sample in Fig. 3). Further Northern analysis was carried out with probes of coxB and coxAC using RNA isolated from cells grown in the presence of nitrate and fructose both in the light and in the dark. No additional induction by growth in darkness took place (Fig. 4). The 4- and 1.3-kb hybridization transcripts were again observed with the coxB probe; hybridization signals above the 1.3-kb band might represent degradation products of the 4-kb transcript and/or material excluded from the zone of the gel containing a high amount of rRNA. The 4-kb transcript was also observed with the coxAC probe, which additionally produced an increased hybridization signal at about 2.5 kb that might represent a specific mRNA species. Since the length of the cox gene cluster is 3,729 bp, detection of a 4-kb transcript with the two probes indicates cotranscription of the three genes, which would thus constitute an operon. The 1.3-kb transcript detected with the coxB probe might cover coxB (1,068 bp), and the putative 2.5-kb transcript detected with the coxAC probe might cover coxA and coxC (together, 2,476 bp).
FIG. 3.
Northern blot analysis of expression of the coxB gene in A. variabilis strain ATCC 29413 FD. RNA was isolated from cells grown in the light with nitrate (NO3−), ammonium (NH4+), or nitrate plus fructose (NO3− Fruc) or in medium lacking combined nitrogen (N2). Hybridization to a probe of the coxB gene was carried out as described in Materials and Methods. Samples contained 70 μg of RNA. Arrowheads point to the coxB transcripts of 4.0 and 1.3 kb.
FIG. 4.
Northern blot analysis of expression of the coxB (A) and coxAC (B) genes in A. variabilis strain ATCC 29413 FD grown in the light or in darkness. RNA was isolated from cells grown with nitrate plus fructose in the light (L) or under dark conditions (D). Hybridization of the filter with probes of the coxB (A) and coxAC (B) genes was carried out in that order as described in Materials and Methods. Samples contained 70 μg of RNA. Arrowheads point to the different transcripts.
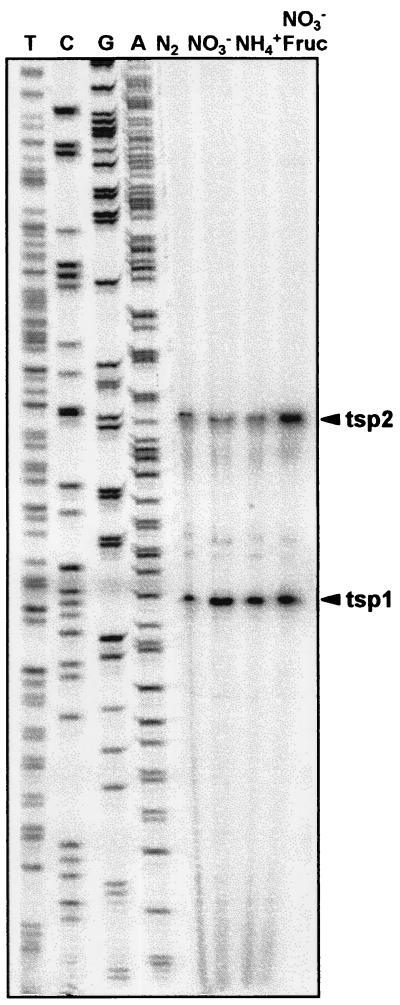
Primer extension analysis using a primer from the 5′ region of coxB (CB1) and the RNA preparations shown in Fig. 3 indicated the presence of two putative transcription start points (tsp) for the operon, a constitutive tsp located at −44 with respect to the putative initiation coxB codon (tsp1), and a tsp at −78/−79 (tsp2) whose use was increased in the presence of fructose (Fig. 5). Quantification of the −78/−79 band showed that its intensity was similar for the RNA from the three autotrophic cultures but was double that of the mixotrophic culture.
FIG. 5.
Primer extension analysis of expression of the coxB gene in A. variabilis strain ATCC 29413 FD. Primer extension assays were carried out with RNA isolated from cells grown with ammonium (NH4+), nitrate (NO3−), or nitrate plus fructose (NO3− Fruc) or in medium lacking combined nitrogen (N2). Assays were carried out using oligonucleotide CB1 (see Materials and Methods). The sequencing ladder shown was generated with the same oligonucleotide on a subclone of λE2. Arrowheads point to the putative tsp.
DISCUSSION
The coxA, coxB, and coxC genes of the facultatively chemoheterotrophic heterocyst-forming cyanobacterium A. variabilis strain ATCC 29413 have been isolated and found to be clustered together in the order coxB-coxA-coxC, a usual arrangement for these genes in many bacteria. Deletion of this gene cluster from the A. variabilis genome rendered a mutant strain, CSW1, that showed a low cytochrome c oxidase activity when grown in bound nitrogen and a lack of growth with fructose in the dark. Therefore, this cox cluster encodes a cytochrome c oxidase essential for chemoheterotrophic growth. Interestingly, in Synechocystis sp. strain PCC 6803, removal of the related coxBAC genes also leads to loss of chemoheterotrophic (light-activated dark) growth (20).
Removal of the CoxBAC cytochrome c oxidase from A. variabilis strain ATCC 29413 does not lead to loss of respiratory activity in cells grown photoautotrophically in nitrate. Therefore, CoxBAC cannot be the only terminal respiratory oxidase in this strain. Indeed, the existence of additional terminal respiratory oxidases can be inferred from our data. One of them is a cytochrome c oxidase since, compared to the wild type, CSW1 has lower but not negligible cytochrome c oxidase activity. Furthermore, since the total respiratory activity is not significantly different in CSW1 and the wild type, at least one other terminal respiratory oxidase that is not a cytochrome c oxidase may be present. The CoxBAC cytochrome c oxidase, however, is apparently the only respiratory terminal oxidase able to support chemoheterotrophic growth. Not a single colony was observed when 2 × 109 cells of strain CSW1 were plated on a total of 47 plates (BG11 medium plus 20 mM fructose) and incubated for 1 month in complete darkness at 30°C. Thus, not only is strain CSW1 incapable of chemoheterotrophic growth, but also no simple mutation in one of the genes coding for the other respiratory terminal oxidases yields a strain able to grow chemoheterotrophically.
Diazotrophic growth under autotrophic conditions was unaffected in the CSW1 mutant. This finding and the increase in cytochrome c oxidase activity in diazotrophically grown cells suggest the participation of a terminal respiratory oxidase(s) different from CoxBAC in the increased respiratory activity that has been observed in heterocysts (9). A search for cox homologues in the recently available genome sequences of two other heterocyst-forming cyanobacteria, Anabaena sp. strain PCC 7120 (Kazusa DNA Research Institute [www.kazusa.or.jp/cyano/]) and Nostoc punctiforme strain ATCC 29133. (Department of Energy Joint Genome Institute [www.jgi.doe.gov/]) showed the presence of three and four sets of genes, respectively, with high sequence similarity to cox genes.
The A. variabilis coxA, coxB, and coxC genes can be transcribed as an operon, coxBAC, since a 4-kb transcript covering the three genes was observed. Additionally, a very abundant transcript of 1.3 kb and a possible transcript of 2.5 kb that might cover coxB and coxAC, respectively, were detected. The four-gene zwf (glucose-6-phosphate dehydrogenase) operon of N. punctiforme has also been shown to originate a number of transcripts covering different genes or combinations of genes (26). Some of these transcripts show different patterns of expression, and the authors of that study suggested the presence of promoters internal to the operon. In the A. variabilis coxBAC operon, however, a sequence, 5′-AGAGATGACCAA-3′, strongly similar to RNase III cleavage sites (17) and located in a strand of the stem of a putative mRNA stem-loop structure, is detected in the region between the coxB and coxA open reading frames. The length from the coxB tsp to the possible cutting site is approximately 1,250 to 1,285 nucleotides. Therefore, it is possible that the 1.3- and 2.5-kb mRNAs of the coxBAC operon result from processing of the 4-kb transcript (Fig. 1).
Two putative tsp were detected upstream from coxB, located at −44 (tsp1) and −78/−79 (tsp2). Whereas tsp2 was induced by fructose, tsp1 was constitutive. Because they showed different regulation patterns, the RNA originating from tsp1 does not appear to correspond to a degradation product of the tsp2 transcript. Induction by fructose of one of the tsp is consistent with the observed induction by fructose of transcripts detected by Northern analysis and of cytochrome c oxidase activity. Induction by fructose has also been shown for some of the transcripts in the N. punctiforme zwf operon (26) as well as for a fructose transport activity in Nostoc sp. strain ATCC 29150 (25). A regulatory system mediating induction by fructose that is yet to be characterized may therefore be relatively common among heterocyst-forming cyanobacteria. With regard to the constitutive tsp1, sequences that may represent a ς70-type promoter are found upstream from it: TACCTT, centered at −8.5, and TAGGCT, centered at −34.5 with respect to the tsp (Fig. 1). A number of genes from heterocyst-forming cyanobacteria have been observed to be expressed from multiple promoters. They are genes which are, or may be, expressed both in vegetative cells and heterocysts; some examples are glnA encoding glutamine synthetase (12, 29), petH encoding ferredoxin:NADP+ oxidoreductase (30), zwf (27), and argD encoding N-acetylornithine aminotransferase (11). It is currently unknown whether the A. variabilis coxBAC operon is expressed in heterocysts, although our data indicate that it is not required for heterocyst function in the light. Utilization of the constitutive promoter would ensure a basal level of expression of the operon, while the use of the inducible promoter would permit a higher expression under mixotrophic or chemoheterotrophic conditions. Enhancement of the expression of coxBAC by fructose is consistent with its essential function in chemoheterotrophic growth with fructose as the substrate.
ACKNOWLEDGMENTS
We thank Himadri Pakrasi for a λEMBL3A gene library of A. variabilis strain ATCC 29413. Preliminary sequence data obtained from the DOE Joint Genome Institute (JGI) at http://www.jgi.doe.gov/ is acknowledged.
This work was supported in part by Austria-Spain Acciones Integradas. Work in Seville, Spain, was supported by grants PB97-1137 and PB98-0481 from the Ministerio de Ciencia y Tecnología, Madrid, Spain. Work in Vienna, Austria, was supported by Human Frontier Science Program grant no. RG-51/97.
REFERENCES
- 1.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 2.Bethesda Research Labs. BRL pUC host: E. coli DH5α competent cells. Focus. 1986;8(2):9. [Google Scholar]
- 3.Cai Y, Wolk C P. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J Bacteriol. 1990;172:3138–3145. doi: 10.1128/jb.172.6.3138-3145.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang Y-C. Efficient precipitation and accurate quantitation of detergent-solubilized membrane proteins. Anal Biochem. 1992;205:22–26. doi: 10.1016/0003-2697(92)90573-p. [DOI] [PubMed] [Google Scholar]
- 5.Currier T C, Wolk C P. Characteristics of Anabaena variabilis influencing plaque formation by cyanophage N-1. J Bacteriol. 1979;139:88–92. doi: 10.1128/jb.139.1.88-92.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elhai J, Vepritskiy A, Muro-Pastor A M, Flores E, Wolk C P. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J Bacteriol. 1997;179:1998–2005. doi: 10.1128/jb.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elhai J, Wolk C P. Conjugal transfer of DNA to cyanobacteria. Methods Enzymol. 1988;167:747–754. doi: 10.1016/0076-6879(88)67086-8. [DOI] [PubMed] [Google Scholar]
- 8.Elhai J, Wolk C P. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene. 1988;68:119–138. doi: 10.1016/0378-1119(88)90605-1. [DOI] [PubMed] [Google Scholar]
- 9.Fay P. Oxygen relations of nitrogen fixation in cyanobacteria. Microbiol Rev. 1992;56:340–373. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Finnegan J, Sherratt D. Plasmid ColE1 conjugal mobility: the nature of bom, a region required in cis for transfer. Mol Gen Genet. 1982;185:344–351. doi: 10.1007/BF00330810. [DOI] [PubMed] [Google Scholar]
- 11.Floriano B, Herrero A, Flores E. Analysis of expression of the argC and argD genes in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol. 1994;176:6397–6401. doi: 10.1128/jb.176.20.6397-6401.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frías J E, Flores E, Herrero A. Requirement of the regulatory protein NtcA for the expression of nitrogen assimilation and heterocyst development genes in the cyanobacterium Anabaena sp. PCC 7120. Mol Microbiol. 1994;14:823–832. doi: 10.1111/j.1365-2958.1994.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 13.García-Domínguez M, Florencio F J. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol. 1997;35:723–734. doi: 10.1023/a:1005846626187. [DOI] [PubMed] [Google Scholar]
- 14.Golden S S, Brusslan J, Haselkorn R. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 1987;153:215–231. doi: 10.1016/0076-6879(87)53055-5. [DOI] [PubMed] [Google Scholar]
- 15.Howitt C A, Vermaas W F. Quinol and cytochrome oxidases in the cyanobacterium Synechocystis sp. PCC 6803. Biochemistry. 1998;37:17944–17951. doi: 10.1021/bi981486n. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 17.Krinke L, Wulff D L. The cleavage specificity of RNase III. Nucleic Acids Res. 1990;18:4809–4815. doi: 10.1093/nar/18.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mackinney G. Absorption of light by chlorophyll solutions. J Biol Chem. 1941;140:315–322. [Google Scholar]
- 19.Muro-Pastor A M, Valladares A, Flores E, Herrero A. The hetC gene is a direct target of the NtcA transcriptional regulator in cyanobacterial heterocyst development. J Bacteriol. 1999;181:6664–6669. doi: 10.1128/jb.181.21.6664-6669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pils D, Gregor W, Schmetterer G. Evidence for in vivo activity of three distinct respiratory terminal oxidases in the cyanobacterium Synechocystis sp. strain PCC6803. FEMS Microbiol Lett. 1997;152:83–88. doi: 10.1111/j.1574-6968.2001.tb10844.x. [DOI] [PubMed] [Google Scholar]
- 21.Poole R K, Cook G M. Redundancy of aerobic respiratory chains in bacteria? Routes, reasons and regulation. Adv Microb Physiol. 2000;43:165–224. doi: 10.1016/s0065-2911(00)43005-5. [DOI] [PubMed] [Google Scholar]
- 22.Raitio M, Jalli T, Saraste M. Isolation and analysis of the genes for cytochrome c oxidase in Paracoccus denitrificans. EMBO J. 1987;6:2825–2833. doi: 10.1002/j.1460-2075.1987.tb02579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 24.Schmetterer G. Cyanobacterial respiration. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 409–435. [Google Scholar]
- 25.Schmetterer G, Flores E. Uptake of fructose by the cyanobacterium Nostoc sp. ATCC 29150. Biochim Biophys Acta. 1988;942:33–37. [Google Scholar]
- 26.Summers M L, Meeks J C. Transcriptional regulation of zwf, encoding glucose-6-phosphate dehydrogenase, from the cyanobacterium Nostoc punctiforme strain ATCC 29133. Mol Microbiol. 1996;22:473–480. doi: 10.1046/j.1365-2958.1996.1371502.x. [DOI] [PubMed] [Google Scholar]
- 27.Summers M L, Wallis J G, Campbell E L, Meeks J C. Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J Bacteriol. 1995;177:6184–6194. doi: 10.1128/jb.177.21.6184-6194.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thiel T. Molecular genetic techniques for analysis of cyanobacteria. In: Bryant D A, editor. The molecular biology of cyanobacteria. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 581–611. [Google Scholar]
- 29.Tumer N E, Robinson S J, Haselkorn R. Different promoters for the Anabaena glutamine synthetase gene during growth using molecular or fixed nitrogen. Nature. 1983;306:337–342. [Google Scholar]
- 30.Valladares A, Muro-Pastor A M, Fillat M F, Herrero A, Flores E. Constitutive and nitrogen-regulated promoters of the petH gene encoding ferredoxin:NADP+ reductase in the heterocyst-forming cyanobacterium Anabaena sp. FEBS Lett. 1999;449:159–164. doi: 10.1016/s0014-5793(99)00404-4. [DOI] [PubMed] [Google Scholar]
- 31.Van Gelder B F, Slater E C. The extinction coefficient of cytochrome c. Biochim Biophys Acta. 1962;58:593–595. doi: 10.1016/0006-3002(62)90073-2. [DOI] [PubMed] [Google Scholar]
- 32.Vanlerberghe G C, McIntosh L. Alternative oxidase: from gene to function. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:703–734. doi: 10.1146/annurev.arplant.48.1.703. [DOI] [PubMed] [Google Scholar]
- 33.Watson N. A new revision of the sequence of plasmid pBR322. Gene. 1988;70:399–403. doi: 10.1016/0378-1119(88)90212-0. [DOI] [PubMed] [Google Scholar]
- 34.Wolk C P, Shaffer P W. Heterotrophic micro- and macrocultures of a nitrogen-fixing cyanobacterium. Arch Microbiol. 1976;110:145–147. doi: 10.1007/BF00690221. [DOI] [PubMed] [Google Scholar]
- 35.Wolk C P, Cai Y, Cardemil L, Flores E, Hohn B, Murry M, Schmetterer G, Schrautemeier B, Wilson R. Isolation and complementation of mutants of Anabaena sp. strain PCC 7120 unable to grow aerobically on dinitrogen. J Bacteriol. 1988;170:1239–1244. doi: 10.1128/jb.170.3.1239-1244.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]