Abstract
Background
Ginsenoside Rb2, a major active component of Panax ginseng, has various physiological activities, including anticancer and anti-inflammatory effects. However, the mechanisms underlying the rejuvenation effect of Rb2 in human skin cells have not been elucidated.
Methods
We performed a senescence-associated β-galactosidase staining assay to confirm cellular senescence in human dermal fibroblasts (HDFs). The regulatory effects of Rb2 on autophagy were evaluated by analyzing the expression of autophagy marker proteins, such as microtubule-associated protein 1A/1B-light chain (LC) 3 and p62, using immunoblotting. Autophagosome and autolysosome formation was monitored using transmission electron microscopy. Autophagic flux was analyzed using tandem-labeled GFP-RFP-LC3, and lysosomal function was assessed with Lysotracker. We performed RNA sequencing to identify potential target genes related to HDF rejuvenation mediated by Rb2. To verify the functions of the target genes, we silenced them using shRNAs.
Results
Rb2 decreased β-galactosidase activity and altered the expression of cell cycle regulatory proteins in senescent HDFs. Rb2 markedly induced the conversion of LC3-Ⅰ to LC3-Ⅱ and LC3 puncta. Moreover, Rb2 increased lysosomal function and red puncta in tandem-labeled GFP-RFP-LC3, which indicate that Rb2 promoted autophagic flux. RNA sequencing data showed that the expression of DNA damage-regulated autophagy modulator 2 (DRAM2) was induced by Rb2. In autophagy signaling, Rb2 activated the AMPK-ULK1 pathway and inactivated mTOR. DRAM2 knockdown inhibited autophagy and Rb2-restored cellular senescence.
Conclusion
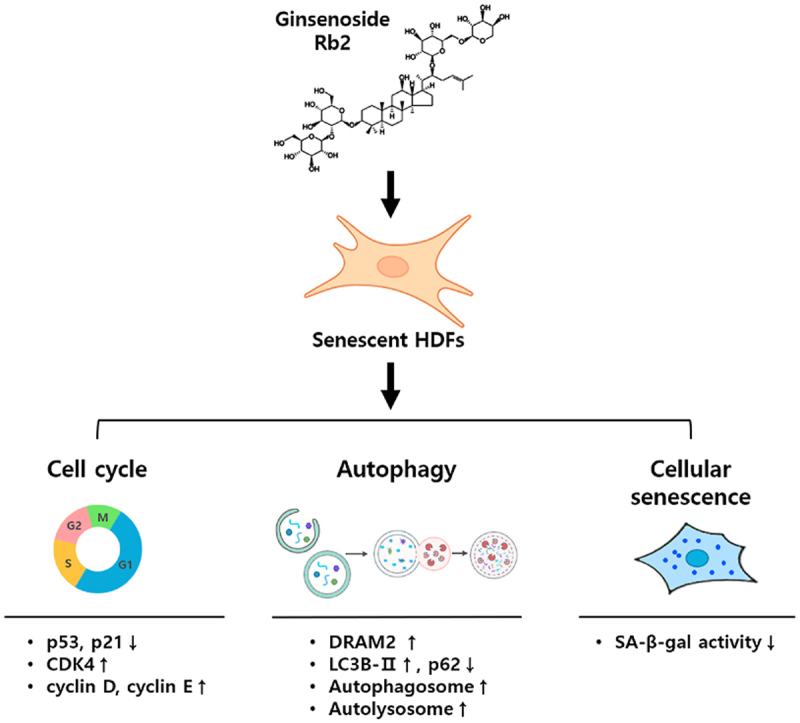
Rb2 reverses cellular senescence by activating autophagy via the AMPK-mTOR pathway and induction of DRAM2, suggesting that Rb2 might have potential value as an antiaging agent.
Keywords: Rb2, Senescence, Autophagy, DRAM2
Graphical abstract
1. Introduction
Cellular senescence is an important hallmark of aging, which is characterized by irreversible cell cycle arrest and permanent proliferation arrest [1]. Hayflick first demonstrated this phenomenon in serially cultured human diploid fibroblasts [2]. Various aging-associated triggers, such as DNA damage, telomere shortening, reactive oxygen species accumulation, and oncogene activation, induce senescence. Senescent cells have several features, including flattened and enlarged morphology, senescence-associated β-galactosidase (SA-β-gal) activity, and upregulated expression of cell cycle inhibitors p16, p21 and p53 [3]. More importantly, autophagy is increased in senescent cells [4] and altered autophagy is found in senescent stem cells [5], indicating that autophagy may be involved in cellular senescence.
Autophagy, a recycling process, which is highly evolutionarily conserved from yeast to mammals, is a crucial mechanism for maintaining cellular homeostasis; it eliminates harmful cytoplasmic contents, including dysfunctional/damaged organelles and misfolded protein [6]. The autophagic degradation pathway is initiated through nucleation of phagophore (nascent autophagosome) by Beclin 1 complex formation, and the phagophore elongation is induced by two ubiquitin-like conjugate systems: (Atg5/Atg12/Atg16, LC3-Ⅱ) and Atg9 [7]. After elongation, the edges of the phagophore are closed to form autophagosomes, which fuse with lysosomes to form autolysosomes, where the autophagic cargo is eventually degraded by resident hydrolases [8]. Various stresses, such as nutrient deprivation and/or caloric restriction (CR), induce autophagy to provide necessary amino acids, which leads to homeostasis regulation [9]. Numerous reports suggest that increased autophagy extends life span and delays aging: thus, autophagy is considered as a novel strategy for rejuvenation. For example, CR induces autophagy by activation of Sirtuin-1, a deacetylase for fundamental autophagic factors, such as Atg5 and Atg7, resulting in the promotion of longevity [10]. Atg5-overexpressing transgenic mice show anti-aging features, such as leanness and enhancement of insulin sensitivity through the activation of autophagy [11]. In addition, astaxanthin, a strong antioxidant, delays brain aging by inducing autophagy in senescence-accelerated mouse prone 10 [12]. Recently, studies have shown that melatonin induces stem cell rejuvenation by restoring autophagy and inhibiting cellular senescence [13]. Therefore, it is necessary to identify the agents and/or regulators of autophagy in aging, particularly in senescence.
The DNA damage-regulated autophagy modulator 2 (DRAM2), also known as TMEM77, harbors six putative transmembrane domains [14]. DRAM2 is localized in lysosomes [14] and cytoplasm [15]. There is accumulating evidence that DMRM2 plays a key role in the autophagic process [16,17]. DRAM2 expression is regulated by several microRNAs, including MIR144∗, miR-27a-3p and miR-125b, which are involved in various diseases, including tuberculosis, cancer, and brain injury [[17], [18], [19]]. However, the role of DRAM2 in cellular senescence has not been fully clarified.
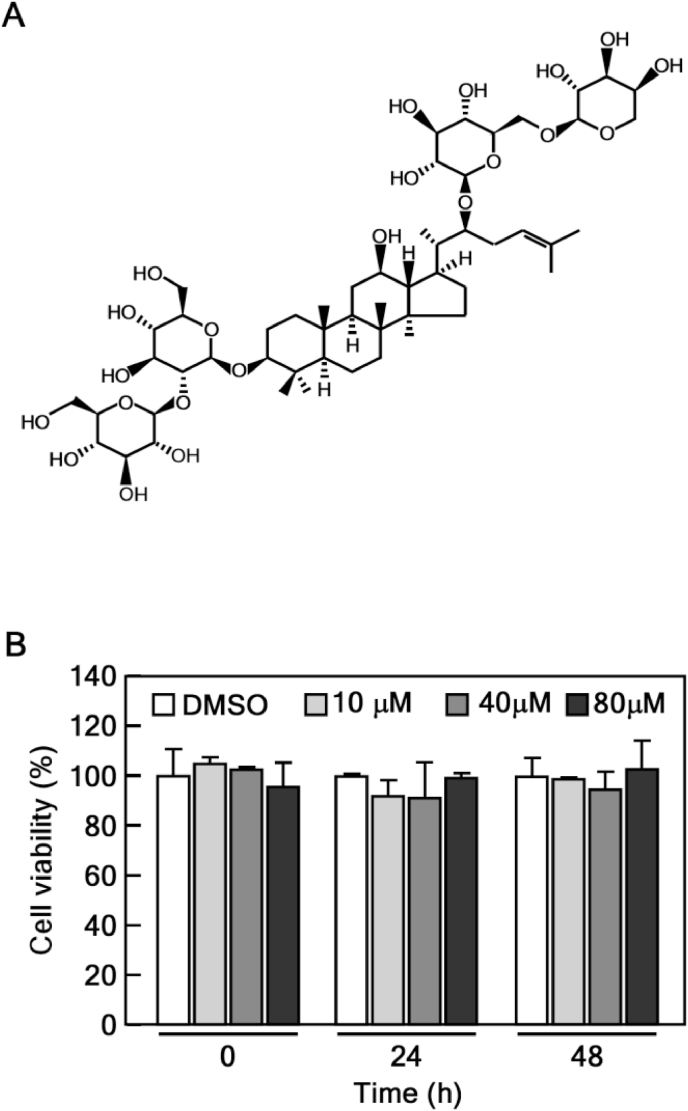
Panax ginseng has been traditionally used as an oriental medicine in East Asia, including Korea, China and Japan. Numerous pharmacological properties of P. ginseng, such as antioxidant [20], anti-inflammatory [21], antitumor [22], and antiaging [23], have been reported in several studies. It is well known that these effects are due to ginsenosides, active components of ginseng. Ginsenosides are categorized into protopanaxadiol (PPD), protopanaxatriol (PPT), oleanolic acid and ocotillol type, according to the chemical structure [24]. Ginsenoside Rb2 (Fig. 1A), most abundantly found in ginseng, is a major PPD-type ginsenoside that differs from other PPDs owing to the sugar moiety in its side chains [25]. Rb2 has various biological and therapeutic effects, including anti-diabetes [26], anti-atherosclerosis [27] and neuroprotective effects [28]. Recently, Rb2 was reported to inhibit endothelial senescence and inflammation by targeting microRNA-216a [29]. However, the rejuvenating effect and precise mechanism of Rb2 in HDFs remain largely unknown.
Fig. 1.
Effects of Rb2 on the viability of old HDFs. (A) The chemical structure of Rb2. (B) Cytotoxicity of Rb2 on old HDFs. Old HDFs were treated with 10, 40 and 80 μM of Rb2. Cell viability was measured at 24 and 48 h using CCK-8 solution.
To elucidate the rejuvenating effects of Rb2 and the underlying molecular mechanisms, we established and used senescent HDFs, which were confirmed by SA-β-gal assay. We investigated the effect of Rb2 on SA-β-gal staining and expression of cell cycle regulatory proteins in senescent HDFs. We also analyzed the regulatory effect of Rb2 on autophagy using tandem sensor RFP-GFP-LC3B, LysoTracker and transmission electron microscopy. Moreover, we conducted RNA-Sequencing to identify the potential target genes involved in Rb2-induced autophagy and rejuvenation.
2. Materials and methods
2.1. Reagents and antibodies
Reagents and antibodies used in this study are listed in the supplementary materials and methods.
2.2. Cell culture and treatment
Primary HDFs purchased from Coriell Institute for Medical Research (AG08498) were cultured in DMEM containing 10% fetal bovine serum (FBS) and 1% Antibiotic-Antimycotic solution. HDFs were subcultured at a ratio of 1:4 when they were about 80%–90% confluent in 100 mm cell culture dishes and maintained until senescence at 37 °C in a 5% CO2 humidified atmosphere; young cells were those in passage number 8–10 and old cells were those in passage number 34–36. Old HDFs were treated with Rb2 in complete medium at the indicated concentrations for 24 h.
2.3. Cell viability assay
To confirm the viability of old HDFs, 3 × 103 cells were seeded in a 96-well plate and cultured for 24 h. The cells were treated with vehicle or 10, 40, and 80 μM of Rb2, respectively, and incubated for 24 and 48 h. After Rb2 treatment for the indicated time, 10 μL of CCK-8 was added to the respective wells and the plates were incubated for 1 h at 37 °C with 5% CO2. The absorbance of solution in each well was measured at 450 nm using a microplate reader (Tecan, Männedorf, Switzerland).
2.4. Immunoblotting
Rb2-treated cells were washed with cold PBS and harvested. The cells were disrupted using Mammalian Protein Extraction Buffer containing a protease inhibitor and phosphatase inhibitor cocktail. Whole cell lysate was obtained by centrifugation at 13,000 rpm for 20 min at 4 °C, and was quantified using the BCA protein assay kit. The same amount of proteins was separated using 8% SDS PAGE gel and 1X ProNATM G-Effect PAGE Running buffer (TransLab, Korea), and then transferred onto a nitrocellulose membrane using 1X ProNATM Cooling-free Wet Transfer buffer (TransLab, Korea). The membrane was blocked with 5% skim milk in 1X TBST for total proteins, or was blocked with 5% BSA in 1X TBST for phosphorylated proteins. The membranes hybridized with the indicated primary antibodies at 4 °C overnight were washed three times with 1X TBST and probed with HRP-conjugated secondary antibody at room temperature for 1 h. The expression of proteins was visualized with the enhanced chemiluminescence detection system using a chemiluminescent imaging system (Azure Biosystems).
2.5. Senescence-associated β-galactosidase staining assay
To confirm cellular senescence, 3 × 104 young and old HDFs were seeded in the wells of 12-well plates. These were treated with 10, 20, and 40 μM of Rb2 for 24 h. SA-β-gal was analyzed using a SA-β-gal staining kit according to the manufacturer's instructions. Briefly, cells were fixed with a fixation solution for 10 min at room temperature and washed two times with 1X PBS. The cells in each well were stained with 500 μL β-galactosidase staining solution for 16 h at 37 °C without humidity and CO2. Stained cells were photographed at × 100 magnification under a microscope with a camera (Nikon Eclipse Ti2–U). The number of SA-β-gal-positive cells was counted in three randomly chosen fields.
2.6. Transmission electron microscopy
Detailed methods are described in the supplementary materials and methods section.
2.7. Determination of autophagy-associated lysosomal activity
For LysoTracker Green DND-26 staining, HDFs were seeded into 4-well chamber slide, cultured overnight, and treated with Rb2 for 24 h. The cells were treated with 100 nM of lysotracker in complete medium for 1 h at 37°C in a 5% CO2 incubator and observed using a fluorescence microscope (Nikon Eclipse Ti2–U).
2.8. Monitoring and measuring autophagy
To measure LC3B puncta, HDFs were seeded into 4-well chamber slide, treated with Rb2 and fixed with 4% formalin for 20 min at RT. The cells were permeabilized with 0.5% Triton X-100/PBS for 10 min and blocked with 3% BSA/Tween-20/PBS at RT for 1 h. After removal of the blocking buffer, the cells were hybridized with primary LC3 antibody in 3%BSA/PBS overnight at 4°C, washed five times with 1% BSA/Tween-20/PBS and incubated with Alexa 488-conjugated secondary antibody in 3%BSA/PBS for 2 h at RT. The LC3 puncta was visualized using fluorescence microscope. To monitor stages of autophagy, we used tandem sensor RFP-GFP-LC3B plasmid. HDFs transfected with RFP-GFP-LC3B plasmid were treated with Rb2 for 24 h, fixed with 4% formalin, and then washed with PBS. Autophagosome or autolysosome were detected under fluorescence microscope.
2.9. shRNA-mediated knockdown assay
For depletion of endogenous DRAM2, shRNA vectors (TRCN0000159300, TRCN0000163085, TRCN0000164262, TRCN0000164485) were purchased from Sigma. To generate viral particles containing with sh-DRAM2, 293T cells were transfected with sh-DRAM2 and packaging plasmids (pMD2.G and psPAX2). After incubation for 48 h, the medium containing virus particles were harvested and filtered through 0.2 μm syringe filters. Viral particles were used for the infection to HDFs with 2 μg/mL of polybrene. The infected cells were selected using 2 μg/mL of puromycin for 2 days to eliminate the noninfected cells.
2.10. Library preparation and sequencing
Detailed methods regarding sample preparation (RNA isolation), sequencing and data analysis are described in the supplementary materials and methods section.
2.11. Statistical analysis
All experiments were independently repeated at least three times. All statistical analyses were performed using the Student's t-test with Graph-Pad Prism (GraphPad, La Jolla, CA). Data are presented as means ± standard deviation. P values less than 0.05 or 0.01 were considered statistically significant.
3. Results
3.1. Rb2 reverses cellular senescence in old HDFs
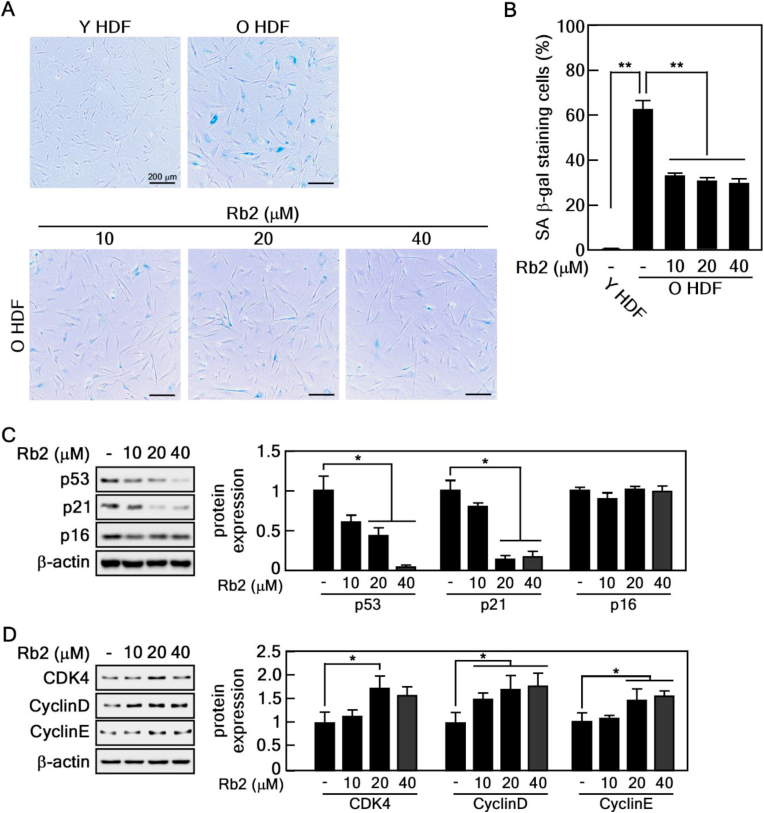
Our research group previously demonstrated that Rg3 rejuvenates the senescent state of old HDFs by inducing peroxiredoxins [30] and via the Akt-mTOR-Sirtuin pathway [31]. Although Rb2 is the most abundant ginsenoside in P. ginseng (Fig. 1A), its effects and the molecular mechanisms underlying rejuvenation remain largely unknown. We first determined the optimal concentrations of Rb2 for treating old HDFs, before assessing the rejuvenating effects of Rb2. Rb2 did not show apparent cytotoxicity up to 80 μM of Rb2 (Fig. 1B). Next, we performed the SA-β-gal assay to verify the replicative senescence of old HDFs. Approximately 60% of old HDFs were stained, whereas less than 1% of young HDFs were stained (Fig. 2A and B). We treated old HDFs with Rb2 and found that Rb2 significantly decreased the number of SA-β-gal-positive cells, compared with that in vehicle-treated control cells (Fig. 2A and B). Because cell cycle arrest and increased expression of cell cycle inhibitors, such as p53, p21 and p16, are distinctive features of senescent cells, we conducted immunoblotting to confirm the changes of the protein expression in Rb2-treated old HDFs. We found that the protein levels of p53 and p21, but not of p16, were gradually decreased under Rb2 treatment (Fig. 2C). It has been well documented that p21, the first identified transcriptional target of p53, is a fundamental regulator of p53-dependent cell cycle arrest [32]. Mechanistically, p21 forms with cyclin/CDK complexes, which are pivotal regulators of cell cycle progression, to suppress its activity. Thus, we assessed the cyclin D, cyclin E and CDK4 protein levels to verify the detailed mechanism underlying the regulatory effect of Rb2 on cell cycle. As expected, the protein expression levels were significantly upregulated by Rb2 treatment (Fig. 2D). These data suggest that Rb2 restores cell cycle arrest and promotes cell cycle progression. Collectively, these results indicate that Rb2 reverses senescence by enhancing cell cycle progression.
Fig. 2.
Rb2 reduces cellular senescence of old HDFs. (A-B) Regulatory effect of Rb2 on cellular senescence was confirmed by SA-β-gal activity assay. Old HDFs treated with Rb2 were stained and observed under light microscopy. Scale bar: 200 μm. Percentage of SA-β-gal staining cells was normalized by counting using optical microscopy images and normalized with total cell number, ∗∗P < 0.01. (C-D) Rb2 regulates expression of cell cycle proteins. Old HDFs treated with 10, 20 and 40 μM of Rb2 for 24 h were harvested for immunoblot analysis. The protein expressions were normalized against β-actin and are shown in graph, ∗P < 0.05.
3.2. Rb2 regulates autophagy in senescent HDFs
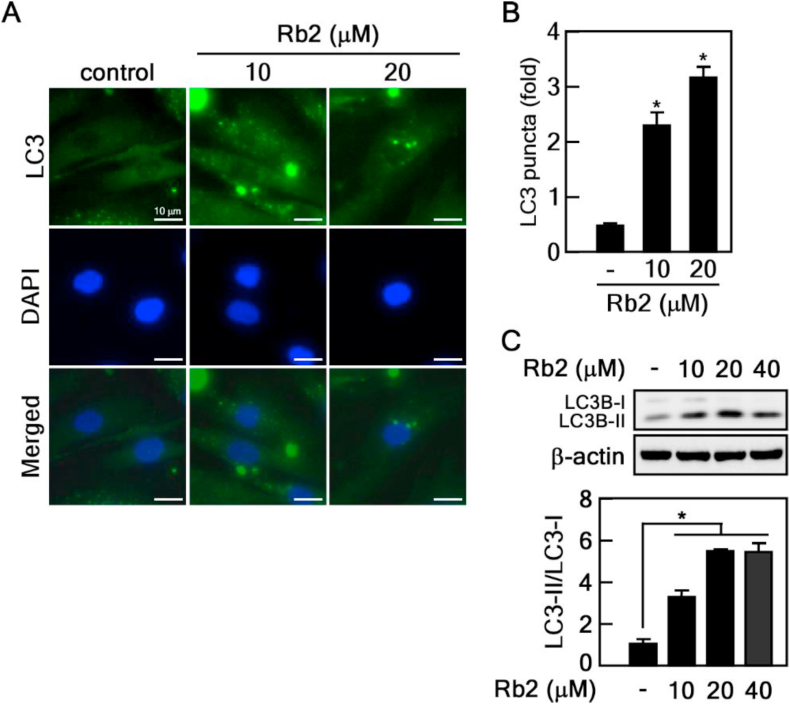
Several studies have reported that reduced autophagy promotes aging, while accelerating autophagy may exert antiaging effects. Therefore, we hypothesized that Rb2 might have regulatory effects on the autophagy pathway. To confirm this hypothesis, we performed immunocytofluorescence assay with an anti-LC3 antibody to evaluate the formation of autophagosomes in Rb2-treated old HDFs. Rb2 treatment significantly increased the LC3 puncta formation (Fig. 3A and B). Next, we analyzed LC3 protein expression using immunoblot assay following treatment of old HDFs with Rb2. The results showed that LC3-Ⅱ protein levels were increased, accompanied with a decrease in LC3-Ⅰ levels (Fig. 3C). Our results support the hypothesis that Rb2 is a regulator of autophagy.
Fig. 3.
Rb2 controls autophagy in senescent HDFs. (A-B) Measuring LC3 puncta. LC3 puncta formation was confirmed using an LC3 specific antibody and visualized by FITC-conjugated secondary antibody in Rb2-treated old HDFs. The number of LC3 puncta in DMSO- or Rb2-treated old HDFs was counted (n = 10), ∗P < 0.05. (C) Immunoblot analysis of LC3 proteins. Old HDFs treated with 10 and 20 μM of Rb2 were harvested for immunoblot analysis. LC3 was detected with the specific antibody. The relative ratio of LC3-Ⅱ and LC3-Ⅰ levels were shown in the graph, ∗P < 0.05.
3.3. Rb2 activates autophagic flux
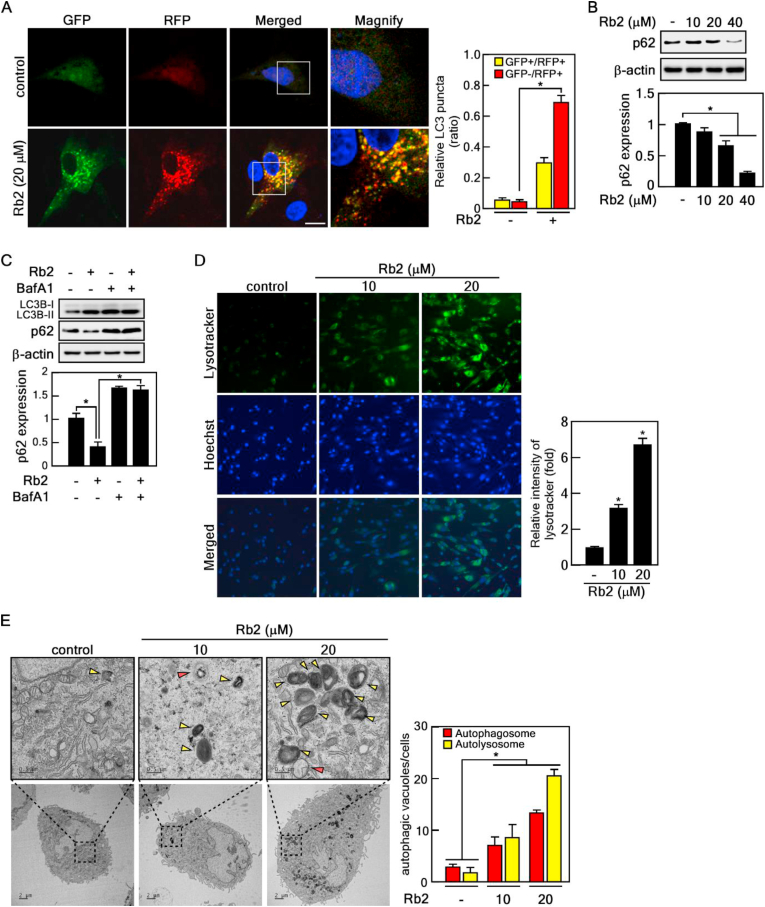
The increased LC3-Ⅱ protein levels reflect autophagosome formation, as well as inhibition of autophagic flux. To determine whether Rb2 activates or suppresses autophagic flux, we performed the GFP-RFP-LC3 tandem reporter assay, which is widely used to monitor the differences between autophagosomes (yellow puncta) and autolysosomes (red puncta). We found that Rb2 induced the formation of autophagosomes and autolysosomes (Fig. 4A). p62, a crucial autophagy marker protein, recognizes ubiquitinated protein aggregates and recruits them to autophagosomes for sequential degradation by autolysosomes [33]. It has been reported that the p62 reduction is associated with an increase of autophagic flux [34]. Therefore, we analyzed the p62 levels in Rb2-treated old HDFs. Immunoblot results showed that p62 protein levels were inhibited by Rb2 treatment (Fig. 4B). To determine whether this decrease in p62 protein level by Rb2 treatment represents autophagy-dependent degradation, we used bafilomycin A1 (BafA1), an autophagy inhibitor that blocks autophagic flux by inhibiting the autophagosome-lysosome fusion and autolysosome-mediated degradation. BafA1 suppressed the LC3-Ⅱ/LC3-Ⅰ protein level increased by Rb2 and recovered the p62 protein level reduced by Rb2 (Fig. 4C), indicating that Rb2 activates autophagic flux. As Rb2 induces autophagosome formation and autophagic flux, we explored whether Rb2 affects the regulation of lysosomal function. Old HDFs were treated with Rb2 and stained with LysoTracker dye, which stains acidic cellular compartments, such as lysosomes and autolysosomes. We observed and analyzed the stained cells under a fluorescence microscope and found that Rb2 significantly enhanced LysoTracker staining (Fig. 4D). The results suggest that Rb2 increases lysosomal activity. To observe the ultrastructure, including autophagosomes and autolysosomes, during the autophagic process, we performed transmission electron microscopy in Rb2-treated HDFs. Autophagic vacuole formation was dramatically induced by Rb2 treatment, compared with that in control cells (Fig. 4E). Collectively, these results indicate that Rb2 promotes autophagosome maturation and autophagic flux.
Fig. 4.
Rb2 activates autophagic flux in senescent HDFs. (A) Rb2 induces autophagic flux. Old HDFs transfected with the RFP-GFP-LC3 plasmid were treated with 20 μM of Rb2 for 24 h. The cells were analyzed with a confocal microscope. Quantification of yellow and red puncta (n = 10), ∗P < 0.05. (B) Rb2 decreased p62 protein level. Old HDFs treated with 10, 20 and 40 μM of Rb2 were harvested for immunoblot. p62 was detected with the specific antibody. The relative expression of p62 was shown in the graph, ∗P < 0.05. (C) Immunoblot analysis of autophagic flux. Old HDFs were treated with 20 μM of Rb2 in the presence or absence of 20 nM of BafA1. LC3 and p62 bands were normalized with β-actin and are shown in the graph, ∗P < 0.05. (D) Rb2 activated lysosomal function. Old HDF cells treated with 10 and 20 μM of Rb2 were stained with lysosome staining dye (LysoTracker green). The fluorescence intensity was measured using ImageJ software and the lysotracker intensity was normalized with Hoechst, ∗P < 0.05. (E) Rb2 increased formation of ultrastructure of autophagy. Red arrow: autophagosome, yellow arrow: autolysosome. The number of autophagosomes and autolysosomes per area was quantified (n = 10).
3.4. Rb2 specifically modulates DRAM2 expression and mTOR signaling pathway
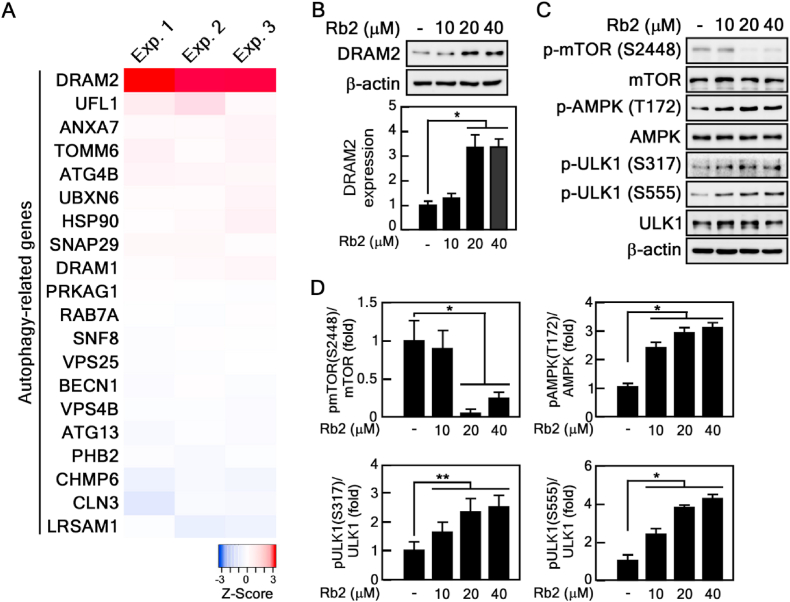
Because our data demonstrated that Rb2 enhances autophagic flux (Fig. 4), we further studied how Rb2 affects autophagy and which signaling pathways are involved in Rb2-induced autophagy. To this end, we conducted RNA-Seq to identify Rb2 target genes associated with the regulation of autophagy in Rb2-treated old HDFs. Among 25730 human genes, we analyzed 247 autophagy-associated genes exported from QuickGO (Gene ontology annotation, https://www.ebi.ac.uk/QuickGO/annotations). Interestingly, the expression of DRAM2 mRNA was increased by Rb2 treatment in triplicate experiments (Fig. 5A). Next, we conducted immunoblot assay to confirm the alteration in DRAM2 protein expression under Rb2 treatment and found that Rb2 increased DRAM2 protein levels (Fig. 5B). Previous studies have reported that DRAM1 induces autophagy by inhibiting the Akt-mTOR signaling pathway [35], a major regulatory pathway in autophagy, and DRAM2 is closely related to DRAM1 [36]. Thus, we hypothesized that Rb2 may affect the mTOR signaling pathway by regulating DRAM2 expression. To confirm this hypothesis, we performed immunoblot assay to assess the mTOR signaling pathway in Rb2-treated old HDFs. Rb2 inhibited the phosphorylation of mTOR at Ser-2448 (Fig. 5C and D), indicating that the mTOR activity was suppressed by Rb2 treatment. Since mTOR is a substrate of AMP-activated protein kinase (AMPK) and the AMPK-mTOR-ULK1 pathway plays a key role in autophagy, we hypothesized that Rb2 might activate AMPK and ULK1. To test this hypothesis, we analyzed the profile of phosphorylated proteins under Rb2 treatment. Rb2 increased the phosphorylation of AMPK at Thr-172, and of ULK1 at Ser-317 and Ser-555, which are important for initiation of autophagy (Fig. 5C and D). These results collectively demonstrate that Rb2 induces DRAM2 expression and regulates the AMPK-mTOR-ULK1 signaling pathway.
Fig. 5.
Rb2 specifically modulates DRAM2 expression and AMPK-mTOR-ULK1 signaling pathway. (A) Analysis of RNA-Seq using mRNA extracted from Rb2-treated old HDFs (n = 3). (B) Rb2 induced DRAM2 protein level. The relative expression of DRAM2 was shown in the graph, ∗P < 0.05. (C-D) Rb2 affected mTOR-AMPK pathway. Cell lysates from Rb2-treated old HDFs were blotted with the indicated antibodies for proteins. The total protein expression was normalized with β-actin, and phospho-protein expression was normalized with the specific total protein are shown in graph, ∗P < 0.05, ∗∗P < 0.01.
3.5. DRAM2 depletion downregulates Rb2-induced rejuvenation by inhibiting autophagy
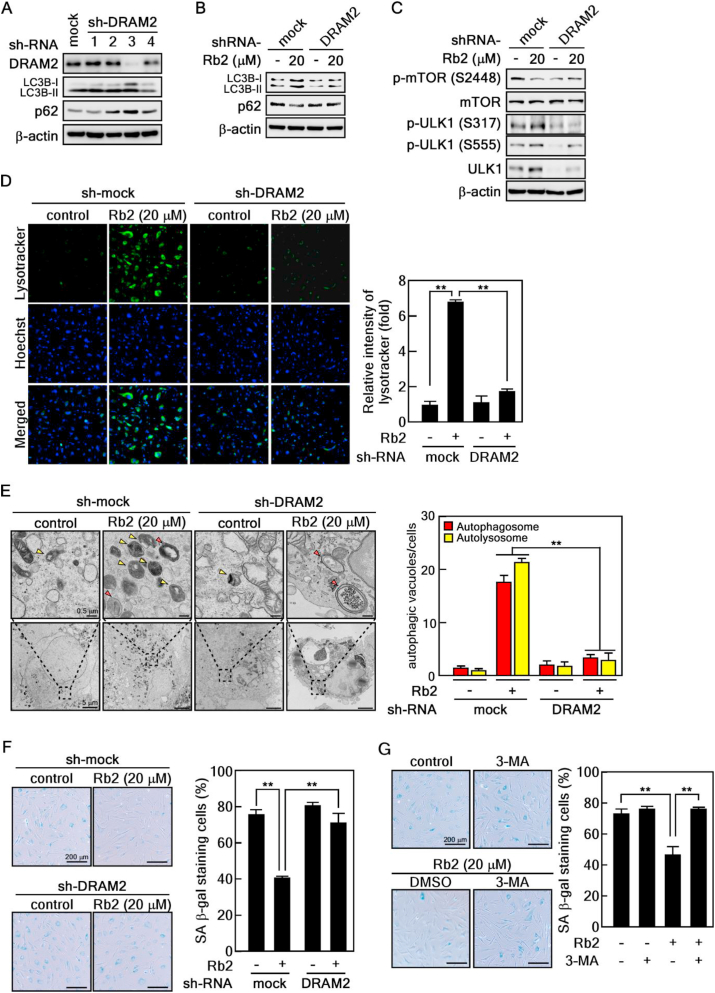
We show that Rb2 induced DRAM2 expression as well as autophagy (Fig. 3, Fig. 4, Fig. 5). Therefore, we hypothesized that DRAM2 knockdown might suppress Rb2-induced autophagy. To validate this hypothesis, we first generated stable old HDFs expressing DRAM2-specific shRNA. Importantly, DRAM2 depletion largely increased the ratio of LC3-Ⅰ/LC3-Ⅱ and p62 protein levels (Fig. 6A, relative density summarized in supplementary Fig. 1A). Among the four DRAM2 shRNAs, we selected number 3, which showed the highest DRAM2-knockdown efficiency and used it for subsequent experiments. To confirm whether the expression of autophagy marker proteins altered by Rb2 treatment is affected by DRAM2 depletion, DRAM2-knockdown cells were treated with Rb2. We found that the increased LC3 protein level following Rb2 treatment was decreased by DRAM2 knockdown, and the decreased p62 protein level following Rb2 treatment was restored by DRAM2 depletion (Fig. 6B, relative density summarized in supplementary Fig. 1B). Furthermore, DRAM2 depletion increased Rb2-induced reduction in the phosphorylation of mTOR whereas it disturbed the Rb2-induced phosphorylation of ULK1 (Fig. 6C, relative density summarized in supplementary Fig. 1C), which correlated with our previous results that Rb2-induced autophagy is associated with DRMA2 expression and the mTOR pathway in Fig. 5B and C. Additionally, we examined the lysosomal activity using LysoTracker in DRAM2-knockdown old HDFs under Rb2 treatment. As expected, DRAM2 depletion largely suppressed Lysotracker-stained cells induced by Rb2 (Fig. 6D). Consistently, we observed that the elevated autophagic vacuoles by Rb2 were remarkably inhibited in DRAM2-knockdown old HDFs (Fig. 6E). These data indicated that DRAM2 plays an important role in the activation of autophagy induced by Rb2. Next, we hypothesized that DRAM2 might affect the rejuvenation via the regulation of autophagy, because Rb2 inhibits senescence of old HDFs (Fig. 2). To confirm this hypothesis, we performed the SA-β-gal assay in DRAM2-knockdown cells under Rb2 treatment. Importantly, DRAM2 depletion did not inhibit SA-β-gal staining upon Rb2 treatment, compared with that in Rb2-treated control cells (Fig. 6F). To further understand the relationship between autophagy and rejuvenation of old HDFs by Rb2, we treated old HDFs with 3-methyladenine (3-MA), commonly used for autophagy inhibitor, in the presence or absence of Rb2. We found that the inhibition of autophagy by 3-MA treatment increased the number of SA-β-gal- stained cells, compared with that in Rb2-treated control cells (Fig. 6G). Collectively, these results demonstrate that DRAM2 knockdown inhibits autophagy and rejuvenation of senescent HDFs.
Fig. 6.
DRAM2 depletion inhibits autophagy and rejuvenation induced by Rb2. (A) Immunoblot analysis of cell lysate extracted from old HDFs infected with shRNA lentiviral vectors against for DRAM2. (B–C) Immunoblot analysis of DRAM2-knockdown cells treated with Rb2. DRAM2-knockdown cells treated with 20 μM of Rb2 were harvested for immunoblot. (D) DRAM2 depletion inhibited lysosomal function. Old HDFs infected with shRNA lentiviral vectors against for DRAM2 were treated with 20 μM of Rb2 and stained with lysosome staining dye (Lysotracker green). The fluorescence intensity was measured using ImageJ software and the Lysotracker intensity was normalized with Hoechst, ∗∗P < 0.01. (E) DRAM2 depletion inhibited formation of ultrastructure of autophagy induced by Rb2. Red arrow: autophagosome, yellow arrow: autolysosome. The number of autophagosomes and autolysosomes per area was quantified (n = 10). (F-G) Representative images of SA-β-gal activity assay. Old HDFs infected with shRNA lentiviral vectors against for DRAM2 were treated with 20 μM of Rb2 for 24 h and stained (F). Old HDFs treated with 20 μM of Rb2 in the presence or absence of 3-MA were stained (G). The SA-β-gal-stained cells were observed under light microscopy. Scale bar: 200 μm. Percentage of SA-β-gal staining cells was normalized by counting using optical microscopy images and normalized with total cell number, ∗∗P < 0.01.
4. Discussion
In the present study, we found that the SA-β-gal activity was markedly increased in old HDFs compared with that in young HDFs and, old HDFs showed enlarged and flattened morphology, indicating features of cellular senescence. Notably, Rb2 largely inhibited the increased SA-β-gal activity in old HDFs, suggesting that Rb2 may have an inhibitory effect on cellular senescence (Fig. 2A). This result agrees with those of a previous study showing that Rb2 abrogates the SA-β-gal activity in miR-216a-induced senescent HUVECs [29]. As cellular senescence is a form of irreversible cell cycle arrest, we explored the effect of Rb2 on the expression of cell cycle regulatory proteins, including p16, p21 and p53. Interestingly, Rb2 reduced the levels of p53 and p21, but not of p16 (Fig. 2C and D). These results suggest that the restorative effect of Rb2 on cellular senescence relies on the p53-p21 pathway. Although we did not investigate the precise mechanisms, by which Rb2 selectively affects the p53-p21 pathway, further studies are required to understand the effect of Rb2 on cell cycle. The regulatory effects of ginsenosides on cell cycle have been previously reported. For instance, Rh2 increases G1-phase arrest of cell cycle by inducing the expression of p21 and p27 mRNAs and proteins [37]. More recently, Rb1 was reported to exhibit anti-aging benefits through restoration of cell cycle progression via reduction of the p53-p21 axis in aging mice [38].
Autophagy plays a pivotal role in maintaining homeostasis and in protecting cells by eliminating unnecessary cellular substances, including damaged organelles and protein aggregates [6]. Because of the fundamental function of autophagy, it has been considered as an anti-aging mechanism [39], including through anti-senescence. In our in vitro experiments, we provide crucial evidence to elucidate that Rb2 exhibits the rejuvenation effects as a positive regulator of autophagy. Rb2 induced autophagic flux by increasing the formation of autophagosomes and autolysosomes, as well as the lysosomal activity, in senescent HDFs (Fig. 4). These results agree with a previous report that Rb2 increases autophagic flux in hepato-carcinoma cells and primary mouse hepatocytes [40]. In autophagy signaling, the mTOR-AMPK pathway regulates the initiation of autophagy through the ULK1 complex, consisting of ULK1, Atg13 and FIP200, which is required for the formation of photophores and autophagosomes [41]. Mechanistically, mTOR inactivates ULK1 via phosphorylation [42], and AMPK directly phosphorylates ULK1, leading to its activation and inducing ULK1 complex formation [43]. Our data showed that Rb2 activated AMPK and abolished mTOR activity (Fig. 3C), indicating that Rb2 may promote ULK1 complex formation. Notably, the reduction of SA-β-gal stain cells mediated by Rb2 was abrogated by 3-MA treatment (Fig. 6G), suggesting that autophagy mediates Rb2-induced restoration of cellular senescence.
DRAM2 is a crucial regulator of autophagy from its initiation [44] to autophagosomes maturation [17]. Our data also show that DRAM2 depletion inhibited the LC3 conversion and restores p62 protein levels in old HDFs (Fig. 6A). Interestingly, we identified DRAM2 as a target gene involved in Rb2-induced autophagy using RNA-Seq (Fig. 5A). Although other autophagy-related genes were altered by Rb2 treatment, DRAM2 gene expression was induced more than two-fold, compared with that of other genes (Supplementary Table). Moreover, Rb2 dose-dependently increased DRAM2 protein expression (Fig. 5B). DRAM2 knockdown suppressed Rb2-induced autophagy by inhibiting the mTOR pathway and the formation of autophagic vacuoles (Fig. 6C–E). Notably, DRAM2 depletion abrogated Rb2-induced restoration of senescence (Fig. 6F). We revealed the function of DRAM2 in cellular senescence. Recently, a study regarding the role of DRAM2 in non-small cell lung cancer demonstrate that DRAM2 overexpression induces CDK4 and cyclin D protein expression and suppresses p53 and p21 protein expression [15]. Furthermore, DRAM2 knockdown by siRNA shows opposite results [15]. These reports allow us to speculate that the alteration of cell cycle regulatory proteins expression by Rb2 (Fig. 2C and D) might be associated with DRAM2, because Rb2 treatment increased DRAM2 expression (Fig. 5A and B). Taken together, we demonstrate that Rb2 induces DRAM2 expression and regulates the AMPK-mTOR pathway to promote autophagy, resulting in the restoration of cellular senescence.
5. Conclusions
In summary, the present study demonstrates that ginsenoside Rb2 can restore cellular senescence through the activation of autophagy. Mechanistically, Rb2 regulates the AMPK-mTOR pathway to enhance autophagy. Importantly, DRAM2 was identified as a target gene of Rb2 and its regulatory function in cellular senescence has been revealed for the first time. Collectively, our results provide new insights and evidence that Rb2 may be a useful agent for rejuvenating human skin cells.
Author contributions
CJL and JSC designed the study and wrote the manuscript; KEY, SBN, MSJ, JSP and GEL performed the experiments and analyzed the data; YYC and BCJ advised the experimental design and data interpretation.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This research was supported by Korea Basic Science Institute grants (C280320 and C230110) and Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2021R1F1A1057701).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2022.11.004.
Contributor Information
Cheol-Jung Lee, Email: veritas0613@kbsi.re.kr.
Jong-Soon Choi, Email: jschoi@kbsi.re.kr.
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- 1.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayflick L., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 3.Wang A.S., Dreesen O. Biomarkers of cellular senescence and skin aging. Front Genet. 2018;9:247. doi: 10.3389/fgene.2018.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gewirtz D.A. Autophagy and senescence: a partnership in search of definition. Autophagy. 2013;9:808–812. doi: 10.4161/auto.23922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.García-Prat L., Martínez-Vicente M., Perdiguero E., Ortet L., Rodríguez-Ubreva J. Autophagy maintains stemness by preventing senescence. Nature. 2016;529:37–42. doi: 10.1038/nature16187. [DOI] [PubMed] [Google Scholar]
- 6.Chun Y., Kim J. Autophagy: an essential degradation Program for cellular homeostasis and life. Cells. 2018;7 doi: 10.3390/cells7120278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanida I., Ueno T., Kominami E. LC3 conjugation system in mammalian autophagy. Int J Biochem Cell Biol. 2004;36:2503–2518. doi: 10.1016/j.biocel.2004.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hollenstein D.M., Kraft C. Autophagosomes are formed at a distinct cellular structure. Curr Opin Cell Biol. 2020;65:50–57. doi: 10.1016/j.ceb.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sakata T., Saito A., Sugimoto H. In situ measurement of autophagy under nutrient starvation based on interfacial pH sensing. Sci Rep. 2018;8:8282. doi: 10.1038/s41598-018-26719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morselli E., Maiuri M.C., Markaki M., Megalou E., Pasparaki A. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pyo J.O., Yoo S.M., Ahn H.H., Nah J., Hong S.H. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. doi: 10.1038/ncomms3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu M., Liang X., Zhang X., Yang M., Ye Q. Astaxanthin delays brain aging in senescence-accelerated mouse prone 10: inducing autophagy as a potential mechanism. Nutr Neurosci. 2022:1–11. doi: 10.1080/1028415X.2022.2055376. [DOI] [PubMed] [Google Scholar]
- 13.Tan Y.Z., Xu X.Y., Dai J.M., Yin Y., He X.T. Melatonin induces the rejuvenation of long-term ex vivo expanded periodontal ligament stem cells by modulating the autophagic process. Stem Cell Res Ther. 2021;12:254. doi: 10.1186/s13287-021-02322-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park S.M., Kim K., Lee E.J., Kim B.K., Lee T.J. Reduced expression of DRAM2/TMEM77 in tumor cells interferes with cell death. Biochem Biophys Res Commun. 2009;390:1340–1344. doi: 10.1016/j.bbrc.2009.10.149. [DOI] [PubMed] [Google Scholar]
- 15.Wudu M., Ren H., Hui L., Jiang J., Zhang S. DRAM2 acts as an oncogene in non-small cell lung cancer and suppresses the expression of p53. J Exp Clin Cancer Res. 2019;38:72. doi: 10.1186/s13046-019-1068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoon J.H., Her S., Kim M., Jang I.S., Park J. The expression of damage-regulated autophagy modulator 2 (DRAM2) contributes to autophagy induction. Mol Biol Rep. 2012;39:1087–1093. doi: 10.1007/s11033-011-0835-x. [DOI] [PubMed] [Google Scholar]
- 17.Kim J.K., Lee H.M., Park K.S., Shin D.M., Kim T.S. MIR144∗ inhibits antimicrobial responses against Mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2. Autophagy. 2017;13:423–441. doi: 10.1080/15548627.2016.1241922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai S., Tian B., Li A., Yao Q., Zhang G., Li F. MicroRNA-125b promotes tumor growth and suppresses apoptosis by targeting DRAM2 in retinoblastoma. Eye (Lond) 2016;30:1630–1638. doi: 10.1038/eye.2016.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Lu C., Yao W., Xu L., Zhou J., Zheng B. Dexmedetomidine inhibits inflammatory response and autophagy through the circLrp1b/miR-27a-3p/Dram2 pathway in a rat model of traumatic brain injury. Aging (Albany NY) 2020;12:21687–21705. doi: 10.18632/aging.103975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S.K., Hyun S.H., In G., Park C.K., Kwak Y.S. The antioxidant activities of Korean Red Ginseng (Panax ginseng) and ginsenosides: a systemic review through in vivo and clinical trials. J Ginseng Res. 2021;45:41–47. doi: 10.1016/j.jgr.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han S.Y., Kim J., Kim E., Kim S.H., Seo D.B. AKT-targeted anti-inflammatory activity of Panax ginseng calyx ethanolic extract. J Ginseng Res. 2018;42:496–503. doi: 10.1016/j.jgr.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shin H.R., Kim J.Y., Yun T.K., Morgan G., Vainio H. The cancer-preventive potential of Panax ginseng: a review of human and experimental evidence. Cancer Causes Control. 2000;11:565–576. doi: 10.1023/a:1008980200583. [DOI] [PubMed] [Google Scholar]
- 23.Peng X., Hao M., Zhao Y., Cai Y., Chen X. Red ginseng has stronger anti-aging effects compared to ginseng possibly due to its regulation of oxidative stress and the gut microbiota. Phytomedicine. 2021;93 doi: 10.1016/j.phymed.2021.153772. [DOI] [PubMed] [Google Scholar]
- 24.Peng D., Wang H., Qu C., Xie L., Wicks S.M., Xie J. Ginsenoside Re: its chemistry, metabolism and pharmacokinetics. Chinese Medicine. 2012;7:2. doi: 10.1186/1749-8546-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Washida D., Kitanaka S. Determination of polyacetylenes and ginsenosides in Panax species using high performance liquid chromatography. Chem Pharm Bull (Tokyo) 2003;51:1314–1317. doi: 10.1248/cpb.51.1314. [DOI] [PubMed] [Google Scholar]
- 26.Dai S., Hong Y., Xu J., Lin Y., Si Q., Gu X. Ginsenoside Rb2 promotes glucose metabolism and attenuates fat accumulation via AKT-dependent mechanisms. Biomed Pharmacother. 2018;100:93–100. doi: 10.1016/j.biopha.2018.01.111. [DOI] [PubMed] [Google Scholar]
- 27.Wang S., Yang S., Chen Y., Chen Y., Li R. Ginsenoside Rb2 alleviated atherosclerosis by inhibiting M1 macrophages polarization induced by MicroRNA-216a. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.764130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D.H., Kim D.W., Jung B.H., Lee J.H., Lee H. Ginsenoside Rb2 suppresses the glutamate-mediated oxidative stress and neuronal cell death in HT22 cells. J Ginseng Res. 2019;43:326–334. doi: 10.1016/j.jgr.2018.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y., Wang S., Yang S., Li R., Yang Y. Inhibitory role of ginsenoside Rb2 in endothelial senescence and inflammation mediated by microRNA-216a. Mol Med Rep. 2021;23 doi: 10.3892/mmr.2021.12054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jang I.S., Jo E., Park S.J., Baek S.J., Hwang I.H. Proteomic analyses reveal that ginsenoside Rg3(S) partially reverses cellular senescence in human dermal fibroblasts by inducing peroxiredoxins. J Ginseng Res. 2020;44:50–57. doi: 10.1016/j.jgr.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang K.E., Jang H.J., Hwang I.H., Hong E.M., Lee M.G. Stereoisomer-specific ginsenoside 20(S)-Rg3 reverses replicative senescence of human diploid fibroblasts via Akt-mTOR-Sirtuin signaling. J Ginseng Res. 2020;44:341–349. doi: 10.1016/j.jgr.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engeland K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022;29(5):946–960. doi: 10.1038/s41418-022-00988-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W.J., Ye L., Huang W.F., Guo L.J., Xu Z.G. p62 links the autophagy pathway and the ubiquitin-proteasome system upon ubiquitinated protein degradation. Cell Mol Biol Lett. 2016;21:29. doi: 10.1186/s11658-016-0031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshii S.R., Mizushima N. Monitoring and measuring autophagy. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu T., Zhu Z., Wu J., She H., Han R. DRAM1 regulates autophagy and cell proliferation via inhibition of the phosphoinositide 3-kinase-Akt-mTOR-ribosomal protein S6 pathway. Cell Commun Signal. 2019;17:28. doi: 10.1186/s12964-019-0341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Prey J., Skommer J., Wilkinson S., Ryan K.M. Analysis of DRAM-related proteins reveals evolutionarily conserved and divergent roles in the control of autophagy. Cell Cycle. 2009;8:2260–2265. doi: 10.4161/cc.8.14.9050. [DOI] [PubMed] [Google Scholar]
- 37.Chung K.S., Cho S.H., Shin J.S., Kim D.H., Choi J.H. Ginsenoside Rh2 induces cell cycle arrest and differentiation in human leukemia cells by upregulating TGF-β expression. Carcinogenesis. 2013;34:331–340. doi: 10.1093/carcin/bgs341. [DOI] [PubMed] [Google Scholar]
- 38.Yu S., Xia H., Guo Y., Qian X., Zou X. Ginsenoside Rb1 retards aging process by regulating cell cycle, apoptotic pathway and metabolism of aging mice. J Ethnopharmacol. 2020;255 doi: 10.1016/j.jep.2020.112746. [DOI] [PubMed] [Google Scholar]
- 39.Gelino S., Hansen M. Autophagy - an emerging anti-aging mechanism. J Clin Exp Pathol. 2012;Suppl 4 doi: 10.4172/2161-0681.s4-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang Q., Wang T., Yang L., Wang H.Y. Ginsenoside Rb2 alleviates hepatic lipid accumulation by restoring autophagy via induction of Sirt1 and activation of AMPK. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18051063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong P.M., Puente C., Ganley I.G., Jiang X. The ULK1 complex: sensing nutrient signals for autophagy activation. Autophagy. 2013;9:124–137. doi: 10.4161/auto.23323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganley I.G., Lam du H., Wang J., Ding X., Chen S., Jiang X. ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J Biol Chem. 2009;284:12297–12305. doi: 10.1074/jbc.M900573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mack H.I., Zheng B., Asara J.M., Thomas S.M. AMPK-dependent phosphorylation of ULK1 regulates ATG9 localization. Autophagy. 2012;8:1197–1214. doi: 10.4161/auto.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.El-Asrag M.E., Sergouniotis P.I., McKibbin M., Plagnol V., Sheridan E. Biallelic mutations in the autophagy regulator DRAM2 cause retinal dystrophy with early macular involvement. Am J Hum Genet. 2015;96:948–954. doi: 10.1016/j.ajhg.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.