Summary
Microglia play an important role in neuroinflammation and neurodegeneration. Here, we report an approach for generating microglia-containing cerebral organoids derived from human pluripotent stem cells involving the supplementation of growth factors (FGF, EGF, heparin) and 10% CO2 culture conditions. Using this platform, Western Pacific Amyotrophic Lateral Sclerosis and Parkinsonism–Dementia Complex (ALS-PDC) cerebral organoids were generated from patient-derived induced pluripotent stem cells (iPSCs). These ALS-PDC-affected organoids had more reactive astrocytes and M1 microglia, and had fewer M2 microglia than their unaffected counterparts, leading to impaired microglia-mediated phagocytosis. RNA-seq analysis of ALS-PDC and control organoids indicated that the most significant changes were microglia- and astrocyte-related genes (IFITM1/2, TGF-β, and GFAP). The most significantly downregulated pathway was type I interferon signaling. Interferon-gamma supplementation increased IFITM expression, enhanced microglia-mediated phagocytosis, and reduced beta-amyloid accumulation in ALS-PDC-affected network. The results demonstrated the feasibility of using microglia-containing organoids for the study of neurodegenerative diseases.
Subject areas: Neuroscience, Immunology, Cell biology, Stem cells research
Graphical abstract
Highlights
-
•
Report an approach for the generation of microglia-containing cerebral organoids
-
•
Cerebral organoids derived from ALS-PDC patient had fewer M2 state microglia
-
•
Transcriptome analysis indicated that type I interferon signaling was downregulated
-
•
Interferon−γ supplementation promoted IFITM expression and M2 microglia polarization
Neuroscience; Immunology; Cell biology; Stem cells research
Introduction
Microglia are the main immune cells of the central nervous system (CNS), representing 10–15% of the total cell population in the brain.1 Microglia have the capacity for self-renewal and their development is dependent on the interleukin-34 (IL-34)/colony-stimulating factor 1 receptor (CSF1R) and transforming growth factor β (TGF-β) signaling pathways.2,3 The roles of microglia include involvement in astrocyte activation, neuronal development, synaptogenesis, aggregated protein uptake, and the maintenance of extracellular matrix (ECM) organization.4,5,6 Microglia display notable plasticity and can undergo substantial morphological, molecular, and functional changes in response to damage-associated molecular patterns (DAMPs) such as ATP and reactive oxygen species (ROS).7,8
Microglia can be polarized into different phenotypes, the predominant of which are the pro-inflammatory M1 and non-inflammatory, repair promoting M2 states. M1 microglia release pro-inflammatory cytokines, such as tumor necrosis factoralpha (TNF-α), and ROS, which promote NLRP3-mediated neuroinflammation and neuronal cell death.9,10 Several studies have shown that M1 microglia-mediated immune responses are key contributors to the pathogenesis of several neurodegenerative diseases, including Alzheimer’s disease (AD) and Parkinson’s disease (PD).11,12,13
The non-inflammatory, repair promoting M2 state of microglia is primarily associated with the promotion of neurogenesis, the reconstruction of the ECM, and the clearance of aggregated proteins through phagocytosis.14 M2 microglia are associated with the secretion of anti-inflammatory and tissue repair-related cytokines such as IL-4, IL-10, and TGF-β.15 TGF-β can modulate the functional properties of microglia, playing a critical role in microglia homeostasis and preventing microglia from entering the neurotoxic M1 state.16,17,18 However, the cellular mechanisms that govern the M2 phenotype and the role of M2 microglia in neurodegenerative diseases are not well understood, mainly due to the lack of cellular models containing neurons, astrocytes, and microglia.
Stem cell technology has opened a new avenue for investigating the role of microglia in neurodegenerative diseases.19,20,21,22,23,24,25 Several methods for deriving microglia from human iPSCs have been developed to date26; however, most of these involve the differentiation of iPSCs into mesodermal hematopoietic stem cells, which are subsequently directed toward a microglial fate. In this study, we report a novel method for generating 2D and 3D organoids containing neurons, astrocytes, and microglia from these iPSCs using a growth factor-defined culture medium and 10% CO2 culture conditions.
Western Pacific Amyotrophic Lateral Sclerosis and Parkinsonism-Dementia Complex (ALS-PDC) is an environmentally mediated prototypical neurodegenerative disease that exhibits clinical and neuropathological similarities to ALS, PD, and AD.27,28,29 ALS-PDC was first identified among the Chamorro people of Guam. Studies showed that ALS-PDC in Guam was associated with the traditional use of cycad seeds (Cycads spp.) for medicine and food among the Chamorro population.30 One of the cycad toxins has been identified as β-methylamino-L-alanine (BMAA), which is produced by cyanobacteria that live symbiotically in the roots of cycad plants.31 BMAA affects alanine, aspartate, glutamate, and nitrogen metabolism.32,33 No convincing animal model currently exists for the study of ALS-PDC, likely because of differences in inflammatory responses and metabolism between humans and animals.34 In this study, iPSCs were derived from lymphoid cell lines obtained from a patient with ALS-PDC and an age- and gender-matched healthy control were subsequently used to generate microglia-containing cerebral organoids via our differentiation platform. Using these organoids, we found that an imbalance in microglia polarization may play a key role in the etiology of ALS-PDC. Our results further indicated that iPSC-derived cerebral organoids, containing neurons, astrocytes, and microglia, represent an excellent cellular model for researching the impact of genetic and environmental factors on microglia-mediated neurodegenerative diseases as well as a platform for future studies targeting microglial phenotype switching for neurodegenerative disease treatment.
Results
Cerebral organoids containing microglia were generated from human embryonic stem cells under defined culture conditions
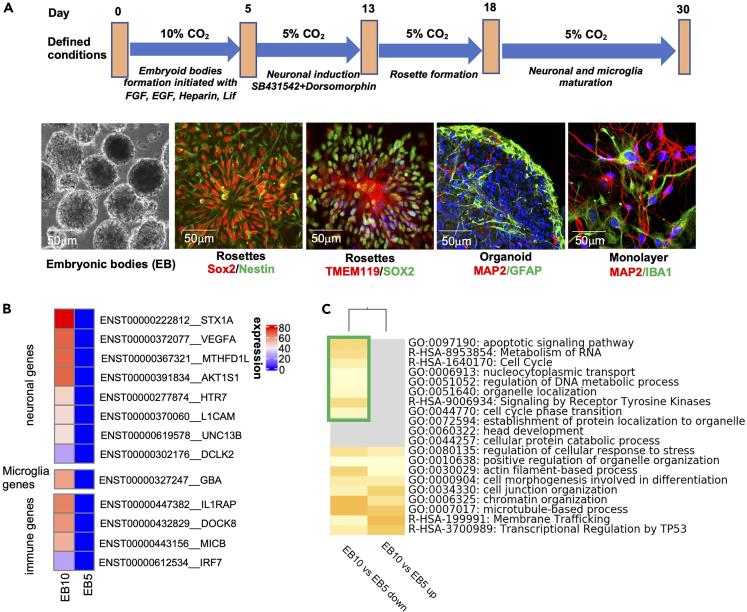
Here, we report a protocol for the generation of microglia-containing cerebral organoids involving the supplementation of growth factors and culturing under 10% CO2 culture conditions. Embryoid bodies (EBs) were generated from H9 human embryonic stem cells (hESCs) cultured in knock out serum replacement (KSR) supplemented with basic fibroblast growth factor (bFGF,10 ng/mL), epidermal growth factor (EGF, 50 ng/mL), leukemia inhibitory factor (LIF, 1,000 Unit/mL), heparin (1 μg/mL), and ROCK inhibitor (10 μM) under 10% CO2 at 37°C for five days. EBs generated under these conditions displayed good morphology with bright edges (Figure 1A). Global gene expression analysis of five-day-old EBs cultured under 5% CO2 and 10% CO2 indicated that, under 10% CO2, the expression of the neuronal related genes STX1A and VEGFA, the microglia related gene GBA, and the immune genes IL1RAP, DOCK8, MICB, and IRF7 were upregulated compared with that under 5% CO2 (Figure 1B). Gene Ontology (GO) enrichment analysis further indicated that the apoptotic signaling pathway and the cell cycle were downregulated when the EBs were cultured under 10% CO2 (Figure 1C). Neural rosettes were derived from the EBs using dual-SMAD inhibition, namely, supplementation of the culture medium with 10 μM SB431542 and 1 μM dorsomorphin for 6 days. Neural rosettes were subjected to immunostaining with the neuroectodermal stem cell markers Sox2 and Nestin and the microglial marker transmembrane protein 119 (TMEM119) (Figure 1A). Rosettes were detached using STEMdiff Neural Rosette Selection Reagent and transferred to 24-well plates for the culture of cerebral organoids in neuronal maintenance medium (NMM) supplemented with IL-34 (100 ng/mL), CSF (5 ng/mL), and TGF-β (50 ng/mL). The rosettes can be replaced in matrigel-coated plates for 2D monolayer culture. The neuronal network was immunostained for the neuronal and microglia markers microtubule-associated protein 2 (MAP2), glial fibrillary acidic protein (GFAP) and ionized calcium-binding adapter molecule 1 (Iba1), respectively (Figure 1A).
Figure 1.
Generation of microglia-containing organoids from human embryonic stem cells (hESCs)
(A) Schematic showing the procedure used for the derivation of organoids containing neurons, astrocytes, and microglia from hESCs. hESCs were differentiated into embryoid bodies by culturing in medium supplemented with growth factors under 10% CO2 and then induced to a neuronal fate by incubation with SB 431542 and dorsomorphin, yielding rosette-forming neuronal and microglia progenitors as determined by Sox2/Nestin and TMEM119/Sox2 staining, respectively. The rosettes were then cultured with our defined medium, leading to the differentiation of the progenitors into a mature neural network (expressing MAP2/GFAP/Iba1) in two weeks.
(B) Global gene expression analysis indicated that, under 10% CO2, the expression of neuronal microglial and immune related genes was upregulated compared with that under 5% CO2.
(C) Enrichment of selected Gene Ontology (GO) terms between 5% and 10% CO2 culture conditions. The green box indicates downregulated genes under 10% CO2 culture conditions.
Generation of cerebral organoids from ALS-PDC-affected and-unaffected iPSCs derived from lymphoblasts
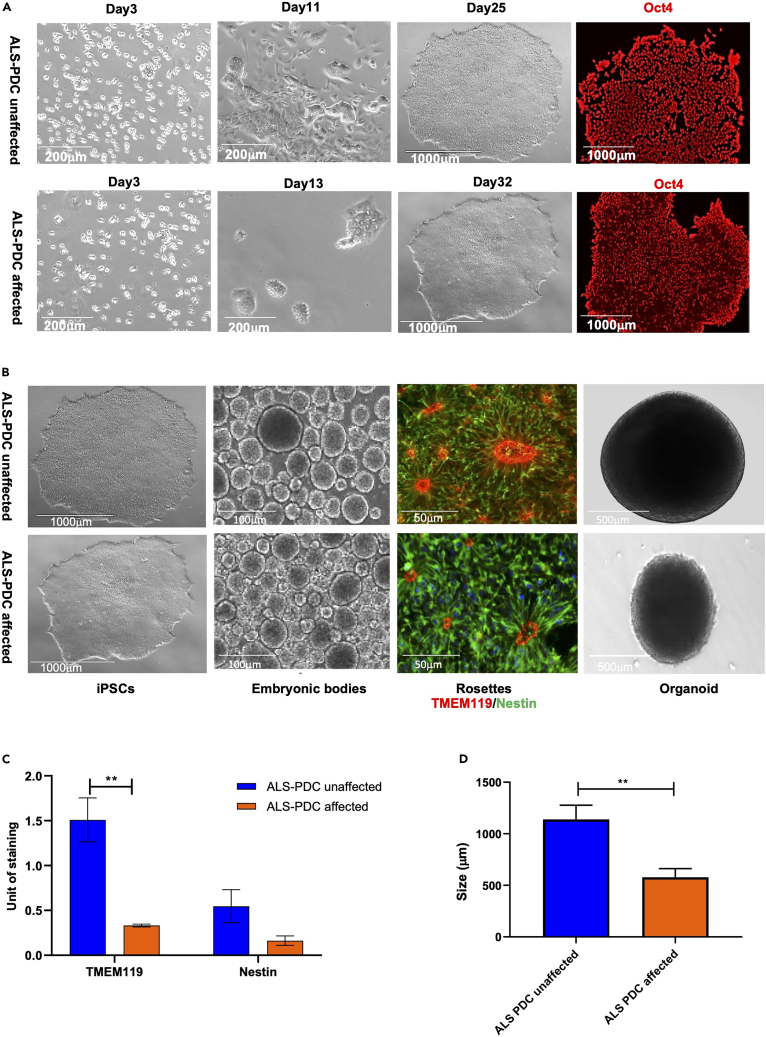
Gender- and age-matched lymphoid cell lines (LCLs) were obtained from a patient with ALS-PDC and an unaffected individual. As shown in Figure 2A, the lymphoid cells were cultured in RPMI 1640 complete medium. After three days of culture, lymphoid cells at the same density (10,000 cells/mL) were transfected with a mixture of episomal plasmids (pCEhOCT3/4, pCE-hSK, pCE-hUL, and pCE-mp53DD) encoding reprogramming factors (OCT3/4, Sox2, KLF4, LMYC, and LIN28) and mouse p53DD (p53 carboxy-terminal dominant-negative fragment) via Neon nucleofection.35 ALS-PDC-unaffected iPSC colonies formed on day 25, whereas ALS-PDC-affected colonies of the same size appeared on day 32. The pluripotency of three ALSPDC-affected lines and three unaffected iPSC lines was assessed via immunostaining for the stem cell factor OCT4 (Figure 2A). RNA-seq results showed that the levels of NANOG, POU5F1, and SOX2 were significantly higher in both affected and unaffected iPSCs than in lymphoid cells (differentially expressed gene [DEG] read numbers: 79, 37, and 1,016 versus 63, 803, and 1,016, respectively). Furthermore, KaryoStat-based analysis showed that the samples originated from a female, and no chromosomal aberrations were detected in either ALS-PDC-affected or-unaffected iPSCs (Figure S1).
Figure 2.
Generation of iPSCs and cerebral organoids from the lymphoid cells of a 57-year-old female Chamorro patient with ALS-PDC (lower images) and those of an age- and gender-matched healthy control subject (upper images)
(A) ALS-PDC-unaffected lymphoid cells at 3, 11, and 25 days and ALS-PDC-affected lymphoid cells at 3, 13, and 32 days following transfection with episomal plasmids. Representative iPSCs were immunostained with an anti-OCT4 antibody.
(B) The procedure employed for the derivation of cerebral organoids containing a neuron-astrocyte-microglia network. iPSCs were differentiated into embryoid bodies by culturing in medium supplemented with growth factors under 10% CO2 and then induced to a neuronal fate by incubation with SB 431542 and dorsomorphin, yielding rosettes containing neuronal and microglia progenitors as determined by Nestin and TMEM119 staining, respectively. Rosettes were picked further allowed to grow into 3D organoids.
(C and D) Quantification of ALS-PDC-affected unaffected neuronal and microglia progenitors and the size of organoids. The data were analyzed using the Student’s t test in GraphPad Prism 8. All values were compared to the ALS-PDC unaffected condition and expressed as means ± SEM.∗∗p< 0.01.
Rosettes, neuronal tube-like structures containing neural progenitor cells (NPCs) and microglia progenitor cells (MPCs), formed on day 18 using our neuronal differentiation platform. Immunostaining results showed that the number of microglial progenitors in ALS-PDC-affected rosettes was significantly reduced compared with that of the ALS-PDC-unaffected controls (Figures 2B and2C). These rosettes were detached with STEMdiff Reagent and grown into organoids. After three months of culture, the average size of thirty the ALS-PDC-affected organoids was 544 μm, half of that of ALS-PDC-unaffected organoids (1,025 μm) (Figures 2D, S2). Cells were observed to detach from ALS-PDC-affected organoids, suggesting that the size difference was because of cell death.
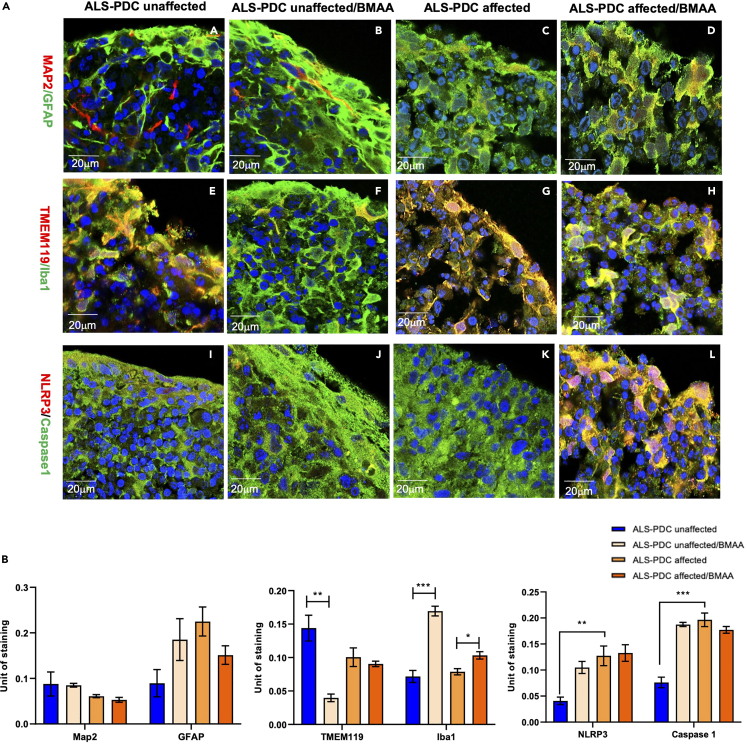
ALS-PDC-affected organoids had a greater number of reactive astrocytes and M1 microglia and exhibited an exacerbated inflammatory response to acute BMAA exposure
Three-month-old ALS-PDC-affected and-unaffected cerebral organoids were treated with BMAA at the concentration 100 μm. BMAA is a neurotoxin amino acid produced by cyanobacteria found in cycad plants for two weeks with untreated organoids serving as controls. Following treatment, the organoids were fixed and sectioned into 10-μm-thick slices for immunostaining. Neurons and astrocytes were detected using anti-MAP2 and anti-GFAP antibodies, respectively. Resting microglia were detected using anti-transmembrane protein (TMEM119) antibody and reactive microglia were detected with an antibody targeting Iba1. Compared with controls, ALS-PDC-affected organoids microglial phenotypes differed significantly between the two groups of organoids (Figure 3A). Furthermore, the results showed that exposure to BMAA increased GFAP, Iba1, and caspase-1 expression in ALS-PDC-unaffected organoids, relative to that in affected organoids. These results suggested that BMAA treatment induced microglia and astrocyte activation. In addition, ALS-PDC-affected inflammatory response exhibited significantly different from ALS-PDC unaffected one (Figures 3A and 3B). The statistical analysis showed that NLRP3/caspase1 expression is higher in ALS-PDC affected organoids before BMAA exposure and exhibited an exacerbated inflammatory response to BMAA exposure.
Figure 3.
Comparison of neuronal, astrocytic, and microglial phenotypes based on marker gene expression both in 3D cerebral organoids derived from ALS-PDC- affected and-unaffected iPSCs, along withβ-methylamino-L-alanine (BMAA) treatment
(A) Immunostaining of ALS-PDC-affected and-unaffected organoids using antibodies targeting MAP2/GFAP, TMEM119/Iba1, and NLRP3/caspase-1 as listed on the left. Images were taken with a Zeiss LSM 880 using a 60× objective. Scale bars, 20 μm.
(B) Quantification of MAP2/GFAP, TMEM119/Iba1, and NLRP3/caspase-1 expression in ALS-PDC-affected, -unaffected and BMAA treated samples. The data were analyzed using the Student’s t test in GraphPad Prism 8. All values were compared to the ALS-PDC-unaffected condition and expressed as means ± SEM. ∗p< 0.05, ∗∗p< 0.01. ∗∗∗p< 0.001.
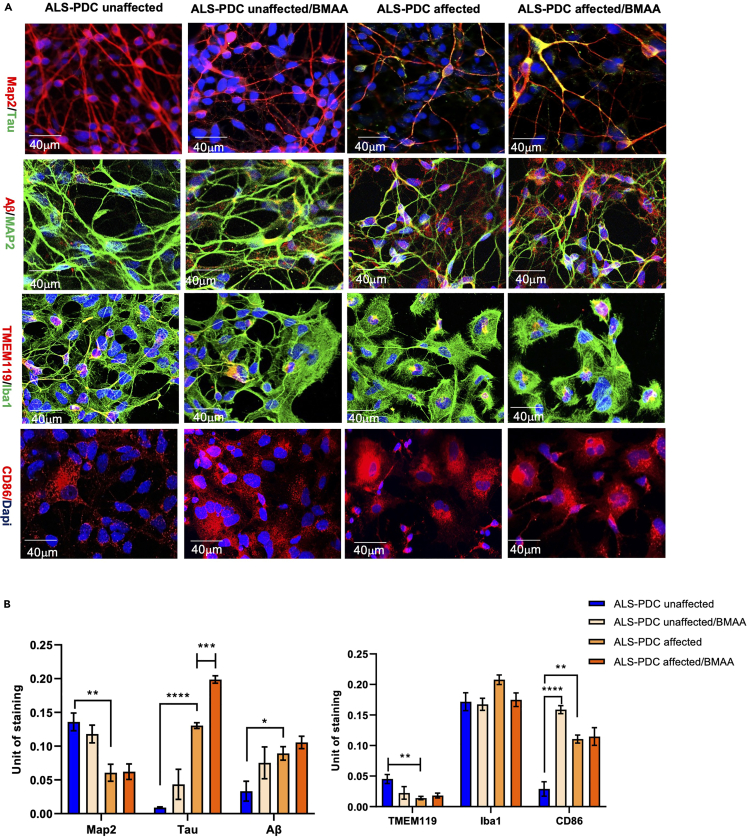
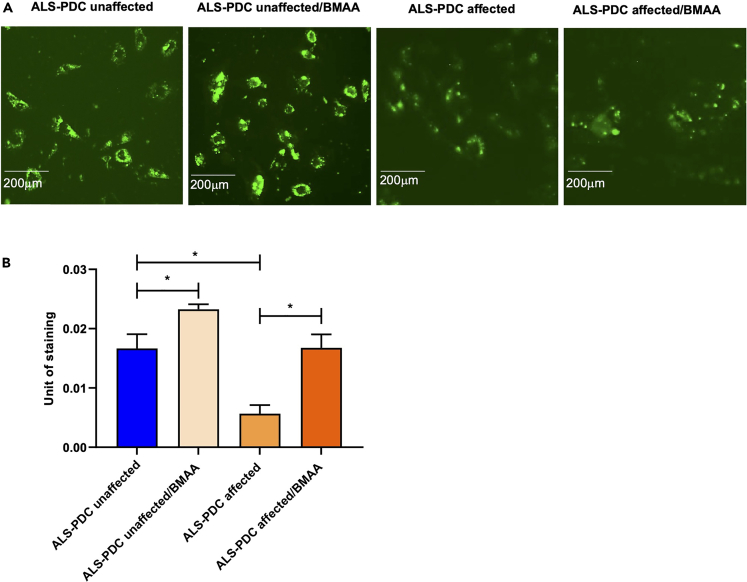
The accumulation of Tau and Aβ and microglial subpopulations in neuronal network were further examined in 2D cultures via immunostaining for MAP2/Tau, MAP2/Aβ, TMEM119/Iba1, and the M1 microglia marker CD86. The results showed that neurons derived from ALS-PDC-affected iPSCs displayed greater intraneural tau protein accumulation as well as increased accumulation of beta-amyloid protein in the extracellular space. Importantly, this phenotype could be recapitulated in ALS-PDC-unaffected 2D cultures by exposure to BMAA. Furthermore, microglia derived from ALS-PDC-affected iPSCs were less ramified and presented a greater diversity of ameboid shapes reminiscent of an M1-positive microglial phenotype. In contrast, microglia derived from healthy control iPSCs displayed a more ramified morphology with elongated dendrites and changed to a large brush shape in response to BMAA exposure (Figures 4A and 4B). Beta-amyloid uptake by microglia was examined via incubation with HiLyte Fluor 488-labeled β-Amyloid (1–40) peptide (AS-60491-01; 1 μM final concentration). The results showed that BMAA exposure upregulated the phagocytic capacity of ALS-PDC-unaffected microglia and impaired that of microglia derived from ALS-PDC-affected iPSCs (Figures 5 A and 5B).
Figure 4.
Comparison of Tau and Aβ, and microglial phenotypes derived from ALS-PDC- affected and-unaffected iPSCs, with β-methylamino-L-alanine (BMAA) treatment in 2D culture
(A) Intraneural tau accumulation, the distinct microglial morphology, and M1 microglia abundance in both ALS-PDC-affected and-unaffected organoids were assessed in 2D culture using antibodies against MAP2, tau, Aβ, TMEM119, Iba1, and M1. Images were obtained with a Zeiss LSM 880 using a 20× objective. Scale bar, 40 μm.
(B) Quantification of MAP2, Tau, and Ab in ALS-PDC affected, unaffected and BMAA treated neuronal networks. The data were analyzed using the Student’s t test in GraphPad Prism 8. All values were compared to the ALS-PDC-unaffected condition and expressed as means ± SEM. ∗p< 0.05, ∗∗p< 0.01. ∗∗∗p< 0.001,∗∗∗∗p< 0.0001.
Figure 5.
Comparison of microglia phagocytic capacity of ALS-PDC-affected and ALS-PDC-unaffected neuronal network
(A) Microglia derived from ALS-PDC-affected iPSCs displayed reduced uptake of HiLyte Fluor 488-labeled beta-amyloid (1–40). BMAA treatment upregulated beta-amyloid uptake in ALS-PDC-unaffected microglia, but impaired that of microglia derived from ALS-PDC affected iPSCs.
(B) Quantification of microglia phagocytosis in ALS-PDC-affected, -unaffected and BMAA treated samples. The data were analyzed using the Student’s t test in GraphPad Prism 8. All values were compared to the ALS-PDC-unaffected condition and expressed as means ± SEM. ∗p< 0.05, ∗∗p< 0.01. ∗∗∗p< 0.001,∗∗∗∗p< 0.0001.
Transcriptome analysis revealed that the expression of genes related to metabolism, the immune response, and the type I interferon signaling pathway was significantly reduced in ALS-PDC-affected organoids compared with that in control organoids
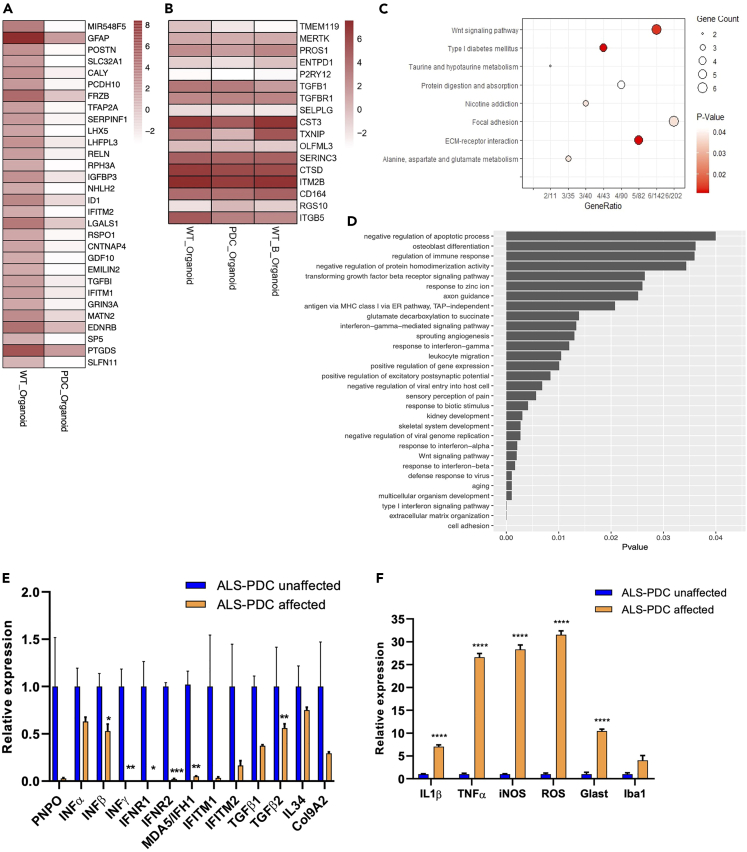
To further understand the molecular mechanism involved in ALS-PDC etiology, we next undertook an RNA-seq analysis in three-month-old ALS-PDC-affected and-unaffected cerebral organoids. DEGs were defined as those showing a >2-fold up- or down-regulation (pvalue: <0.05). Four genes were found to be upregulated and 133 downregulated. Most of these genes encoded microglia- or immunity-related proteins such as interferon inducible transmembrane proteins (IFITM 1/2), and TGF-β (Figure 6A). BMAA treatment reduced TMEM119 and TGF-β expression (Figure 6B). KEGG pathway analysis of organoid DEGs showed that the most downregulated pathways were taurine, alanine, aspartate, and glutamate metabolism; protein digestion and absorption; and extracellular matrix (ECM)-receptor interaction (Figure 6C). GO enrichment analysis of DEGs between ALS-PDC-affected and-unaffected organoids indicated that the most significantly downregulated genes were associated with the type I interferon signaling pathway, cell adhesion, and ECM organization (Figure 6D). To validate these results, we next evaluated the expression levels of selected genes through reverse transcription-quantitative PCR (RT-qPCR), focusing on those encoding metabolic enzymes and factors involved in the type I interferon signaling pathway. The results indicated that ALS-PDC-affected organoids expressed very low levels of pyridoxine/pyridoxine 5′-phosphate oxidase (PNPO), INFG, interferon production regulators 1/2 (IFNR1/2), IFITM1/2, interferon-induced helicase C domain (1MDA5/IFH1), IL34, TGFB, and COL9A2 (Figure 6E) whereas the expression levels of IL1B, TNFA, iNOS, and IBA1 and ROS production (Figure 6F) were significantly upregulated.
Figure 6.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) enrichment analysis of differentially expressed genes (DEGs) as determined by RNA-seq
(A) The top 30 DEGs between ALS-PDC-affected and-unaffected cerebral organoids.
(B) and among ALS-PDC-unaffected, ALS-PDC-affected, and ALS-PDC-unaffected/BMAA-treated cerebral organoids.
(C) Significantly downregulated KEGG pathways between ALS-PDC-affected and-unaffected organoids.
(D) GO term enrichment analysis showing the downregulated biological processes between ALS-PDC-affected and-unaffected organoids.
(E and F) The expression of selected genes was validated by RT-qPCR. The data were analyzed using the Student’s t test in GraphPad Prism 8. All values were compared to the ALS-PDC-unaffected condition and expressed as means ± SEM. ∗p< 0.05, ∗∗p< 0.01. ∗∗∗p< 0.001,∗∗∗∗p< 0.0001.
IFN-γ supplementation increased the expression of the interferon response protein (IFITM), promoted ALS-PDC-affected M2 microglia polarization, and restored phagocytosis
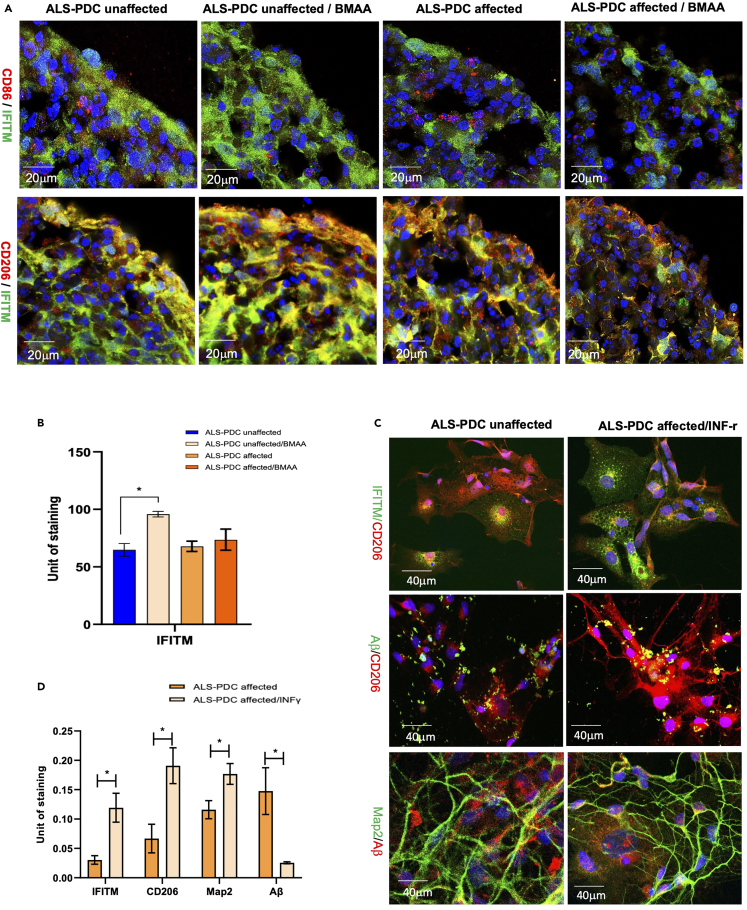
Our data indicated that type I interferon signaling pathway deficiency and microglia polarization might play a significant role in ALS-PDC etiology. Because IFITM has been shown to be involved in microglia polarization,36 we further examined the IFITM expression profile both in ALS-PDC affected and-unaffected organoids employing co-immunostaining with the M1 microglia marker CD86 and the M2 microglia marker mannose receptor (CD206). We found that IFITM was co-expressed with CD206, but not CD86. In addition, BMAA treatment increased IFITM expression and the abundance of M2 microglia in ALS-PDC-unaffected organoids but not in ALS-PDC-affected organoids (Figures 7A and 7B). Supplementation of the culture medium with IFN-γ (10 ng/mL) increased IFITM expression and M2 microglia numbers. M2 microglia Beta-amyloid uptake was examined via incubation with HiLyte Fluor 488-labeled β-Amyloid (1–40) peptide and coimmunostained with M2 microglia marker. The results indicated IFN-r increased the IFITM expression, the number of M2 microglia and the levels of phagocytosis, thereby reducing Aβ accumulation in the extracellular space in the ALS-PDC-affected neuronal network (Figures 7C and 7D).
Figure 7.
ALS-PDC-affected organoids showed a reduced abundance of M2 microglia whereas IFN-γ supplementation promoted IFITM expression, restored M2 microglia numbers, and decreased Aβ accumulation
(A) BMAA treatment increased the expression of IFITM and M2 microglia numbers in ALS-PDC-unaffected but not ALS-PDC-affected organoids. Scale bar, 20 μm.
(B) Analysis of IFITM fluorescence intensity.
(C) IFN-γ treatment increased the expression of IFITM, promoted M2 microglia polarization, increased Aβ uptake and reduced Aβ accumulation in the extracellular space.
(D) Quantification of IFITM, CD206, MAP2 and Ab between ALS-PDC-affected, and ALS-PDC-affected INFr treated samples. The data were analyzed using the Student’s t test in GraphPad Prism 8. ∗p< 0.05.
Discussion
The metabolic and immune systems are fundamental for survival.37 Microglia play a key role in the integration of metabolic regulation and immune responses, highlighting the importance of microglia homeostasis and proper function to CNS health. Studies have demonstrated that microglia are highly plastic in their response to environmental insults. Activated microglia participate in defense, phagocytosis, uptake and clearance of aggregated proteins, and tissue regeneration. Sustained chronic inflammatory states because of nutritional imbalance or neurotoxin-containing foods can negatively influence microglia homeostasis and the risk for microglia-associated disorders.38 Despite the critical role of microglia in brain physiology and pathology, little is currently known about the cellular mechanisms involved in the shifting of microglial phenotypes and their impact on neurodegenerative diseases, mainly because of a lack of a cellular model that contains the necessary cell type diversity like CNS. In this study, we reported a protocol for generation of organoids containing microglia from iPSCs. Ormel group had published a protocol that shows microglia generation in cerebral organoids.39 The difference between our method and Ormel group is that our embryoid bodies were generated under 10% CO2 culture conditions. Global gene expression analysis of five-day-old EBs cultured under 10% CO2 compared with 5% CO2 indicated that under 10% CO2 increased the expression of the neuronal and microglia related genes, and down regulate the apoptotic signaling pathway. In addition, Ormel et al. protocol indicated that without using dual-SMAD inhibition could promote microglia production. In contrast, we do use dual-SMAD inhibitors in our differentiation protocol. Neuronal progenitors’ rosettes generated with our protocol displayed nice morphology including microglia progenitors which were stained with TMEM119. One explanation to the different of these two protocols is that Dual-SMAD inhibitors were unable to inhibit all mesoderm-derived progenitors. We do see some other cell types expressing mesodermal marker brachyury during our differentiation process; that is why we only picked up rosettes for 3D organoid generation to avoid the presentation of other cell linages. The origin of microglia has been a long-standing matter of debate. Some thought it was derived from mesodermal pial. Other believed that they are from neuroectodermal.40 Further research is needed to clarify microglia origin.
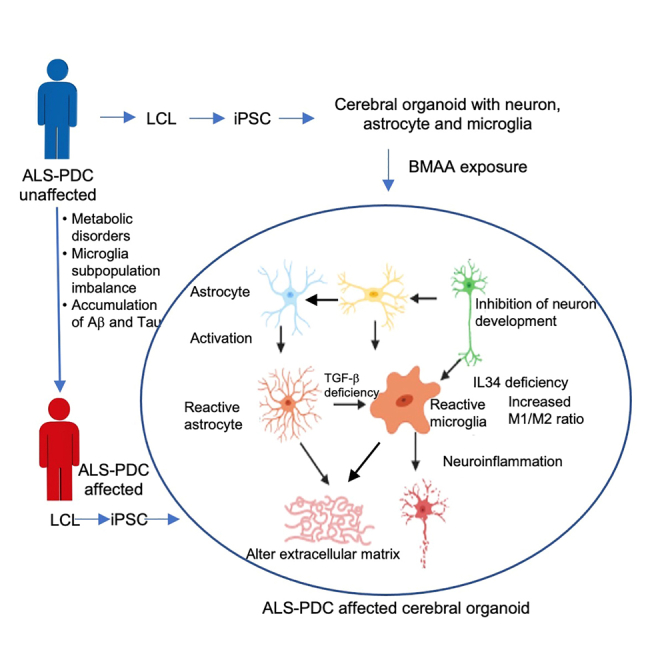
Cerebral organoids containing neurons, astrocytes, and microglia were generated from patient-derived ALS-PDC-affected iPSCs using this directed differentiation approach to model the etiological role of cycad toxin BMAA in ALS-PDC. Cerebral organoids similarly derived from a healthy individual were used as controls. Our results indicated that ALS-PDC-affected organoids had greater numbers of reactive astrocytes and M1 microglia, and fewer resting and M2 microglia when compared with control organoids.
Transcriptome analysis suggested that the genes most significantly affected by BMAA were associated with metabolic pathways, the regulation of neuron development, the immune response, and the interferon signaling pathway. The results obtained through RT-qPCR and immunostaining assays confirmed that the metabolic gene such as PNPO and interferon signaling pathway and M2 microglia polarization were impaired in ALS-PDC-affected organoids. We further examined the role of the interferon signaling pathway and the interferon-induced protein IFITM in M2 microglia polarization. The results showed that supplementation with IFN-γ promoted M2 microglia polarization and increased the levels of microglia-mediated phagocytosis, suggesting that the interferon I signaling pathway plays a significant role in M2 microglia polarization. Numerous studies have established IFNs as critical antiviral molecules in the innate immune system. IFNs can be produced by most cell types, including those of the CNS. Nevertheless, the relationship between IFNs, metabolic disorder, and M2 microglia polarization in neurodegenerative diseases remains poorly understood.36 Future studies should focus on employing gene knockout and single-cell gene expression methodology to identify the molecular mechanisms underlying the interferon-mediated microglia phenotypic switch.
In summary, we have developed a platform for obtaining patient-derived organoids containing neurons, astrocytes, and microglia for disease modeling. This system can recapitulate human brain development and neurodegeneration in response to exposure to the neurotoxin BMAA exposure. Our results suggested that the etiology of ALS-PDC was because of metabolic disorders that promoted a shift in the microglial phenotype toward a more pro-inflammatory M1 and less phagocytosis and repair-promoting M2 state because of a deficiency in the interferon pathway. An imbalance in the microglial population leads to an exacerbated inflammatory response and a reduction in aggregated protein uptake in ASL-PDC-affected organoids, with a consequent increase in tau protein and beta-amyloid peptide accumulation and neuronal cell death. Our results further suggested that IFN-γ supplementation can restore M2 microglial levels and enhance the uptake of beta-amyloid protein in ASL-PDC-affected organoids. Our directed differentiation protocol provides an excellent platform for further characterizing the biological roles of microglia and their interactions with neurons, astrocytes, and ECM components in both healthy and pathophysiological conditions.
Limitations of the study
We have a limited number of ALS-PDC affected samples because of their availability. The goal of this paper is to report a novel approach for the generation of microglia-containing cerebral organoids derived from human pluripotent stem cells, in a hope to provide a differentiation platform for the study of the role of microglia in neurological diseases.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti- microtubule associated protein 2 (Map2) | Santa Cruz Biotechnology | Cat #sc20172; RRID:AB_2250101 |
| Mouse monoclonal anti- ionized calcium binding adapter molecule 1 (Iba1) | Abcam | Cat # ab15690; RRID:AB_2224403 |
| Mouse monoclonal anti-interferon inducible transmembrane protein (IFITM) | Santa Cruz Biotechnology | Cat # sc-374026; RRID:AB_10116884 |
| Rabbit monoclonal anti CD86 | Abcam | Cat # ab239075; RRID:AB_2921417 |
| Mouse monoclonal anti- mannose receptor (CD208) | Abcam | Cat # ab8918; RRID:AB_306860 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant human interlukin-34 (IL34) | BioLegend | Cat # 577906 |
| Recombinant human colonystimulating factor (CSF) | BioLegend | Cat # 574804 |
| Recombinant human transforming growth factor beta 1(TGF-β) | Life Technologies Inc | Cat # PIRP8600 |
| SB431542 | Tocris Bioscience Inc. | Cat # 1614/1 |
| Dorsomorphin | Tocris Bioscience Inc. | Cat # 3093/10 |
| Deposited data: accession code PRJNA75348 | ||
| ALS-PDC unaffected organoid 1 raw and analyzed data | This paper | SRR15404813 |
| ALS-PDC unaffected organoid 2 raw and analyzed data | This paper | SRR15409003 |
| ALS-PDC affected organoid 1 raw and analyzed data | This paper | SRR15409909 |
| ALS-PDC affected organoid 2 raw and analyzed data | This paper | SRR15410515 |
| Oligonucleotides | ||
| Primers for PCR | This paper | Supplemental table |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Yiling Hong (Yhong@westernu.edu).
Materials availability
This study generated two new iPSC cell lines. There are restrictions to the availability of iPSCs due to additional documents needed for the approval.
Method details
Generation of patient-specific iPSCs from ALS-PDC-affected lymphoid cell lines
Gender- and age-matched ALS-PDC-affected and-unaffected LCLs were gifts from Drs. Teequ Siddique and Glen Kisby. Lymphoid cells were cultured in RPMI 1640 complete medium until confluence. As previously reported,35 patient-specific iPSCs were generated from lymphoid cells by transfection with a combination of episomal plasmids (equal amounts [∼2.5 μg] of pCE-hOCT3/4, pCE-hSK, pCE-hUL, and pCE-mp53DD) (Addgene Inc.). Co-transfection into ∼1×106 lymphoid cells was performed using Neon nucleofection. The transfected cells were then plated onto matrigel-coated six-well plates and maintained in TeSr-E7 xeno-free medium (Stem Cell Tech Inc., Catalog # 05919). Twelve days after transfection, this medium was slowly replaced with mTeSR-1 medium (Stem Cell Tech Inc., Catalog # 85851). Approximately 20–30 days after transfection, colonies that exhibited a morphology resembling that of human human embryonic stem cells (hESCs) were plated onto Matrigel-coated six-well plated in mTesr-1 medium. Soon after, putative iPSC colonies were transferred into Matrigel-coated T25 flasks and grown in the same medium. The colonies were passaged every four days. Pluripotency was determined by immunostaining with an anti-OCT4 antibody (Santa Cruz, Catalog # sc-9081) and RNA-seq analysis. Karyotyping of the iPSCs was undertaken using a KaryoStat assay. Genomic DNA (gDNA) was purified using the Genomic DNA Purification Kit (Catalog #K0512) and quantified using the Qubit dsDNA BR Assay Kit (Catalog #Q32850). Total gDNA (250 ng) was used to prepare the CytoScan HT-CMA 96 array for the KaryoStat assay according to the manufacturer’s instructions. This array is used to detect genome-wide copy number variation and single nucleotide polymorphisms. The assay was performed, and the data were analyzed by Thermo Fisher Scientific (Quote No. CB16785).
Differentiation of cerebral organoids containing a neuron-astrocyte-microglia network from ALS-PDC-affected and-unaffected iPSCs
H9 human embryonic stem cells were ordered from the University of Wisconsin (WiCell Research Institute) and iPSCs of ALS-PDC-affected patient and an unaffected control were expanded and enzymatically dissociated with collagenase IV (2 mg/mL: Life Technologies Inc., Catalog # 1710419). Dissociated cells were transferred into a low-adhesion suspension culture plate (Olympus, Inc.) and cultured in knock out serum replacer (KSR (Life Technologies Inc., Catalog # 10828010) supplemented with 10 ng/mL bFGF (Life Technologies Inc., Cat # PHG0360), 50 ng/mL EGF (R&D Systems Inc., Cat #P01133), 1,000 units/mL LIF (Millipore Inc., Cat # GF342), 1 μg/mL heparin (Sigma-Aldrich Inc., Cat# 375095), and 10 μM ROCK inhibitor (Tocris Bioscience Inc., Cat # 4009/10) at 37°C and under 10% CO2 for five days to derive EBs containing the three germ layers. These EBs were further induced to a neural fate by culturing in neuronal induction medium (NIM), which is a 1:1 combination of KSR and NMM (Life Technologies Inc., Cat # A1647801) supplemented with 10 μM SB431542 (Tocris Bioscience Inc., Cat # 1614/1) and 1 μM dorsomorphin (Tocris Bioscience Inc., Cat # 3093/10) for 10 days. NMM is a 1:1 mixture of DMEM/F12 and Neurobasal-A medium (Life Technologies Inc., Cat# 10888022) containing N-2 supplement (Life Technologies Inc., Cat# A1370701), B-27 supplement (Life Technologies Inc., Cat # 1750444), 5 μg/mL insulin (Tocris Bioscience Inc., Cat # 3435/10), 1 mM GlutaMAX, 100 μM nonessential amino acids, and 100 μM β-mercaptoethanol (Life Technologies Inc., Cat # 21985023). Over the 10 days of incubation, KSR medium was gradually replaced with NMM medium by increasing the NMM: KSR ratio by 25% every two days, i.e., five changes of medium were required for complete replacement. After neuronal induction, the EBs containing neuronal and microglia progenitors were collected by centrifugation at 400 × g for 5 min, gently suspended in Gentle Cell Dissociation Reagent (Stem Cell Technologies Inc., Cat# 07174), incubated for 7 minat 37°C, and then broken down into smaller fragments using a 5-mL polystyrene serological pipette. The cells were subsequently plated in matrigel-coated plates and cultured in NMM at 37°C with 5% CO2. After three to five days, neuroepithelial sheets containing neural rosettes with neuronal and microglia progenitors had formed. For organoid generation, the neural rosettes were detached via incubation with STEMdiff Neural Rosette Selection Reagent (Stem Cell Technologies Inc., Cat # 5832) for 30 minat 37°C. Individual rosettes were transferred into separate wells of a low-adhesion, 24-well suspension culture plate. The organoids were maintained in NMM supplemented with 100 ng/mL IL34 (BioLegend, Cat# 577906), 5 ng/mL CSF (BioLegend, Cat# 574804), and 50 ng/mL TGF-β (Life Technologies Inc., Cat # PIRP8600).
For the generation of a 2D monolayer neuronal culture, neuronal rosettes were treated with Gentle Cell Dissociation Reagent at 37°C for 10 min and plated at a density of approximately 1 × 104 cells in Matrigel-coated plates with coverslips. After culture in NMM supplemented with IL-34 (100 ng/mL), CSF (5 ng/mL), and TGF-β (50 ng/mL) for two weeks, the progenitors developed into mature neurons, astrocytes, and microglia.
Cycad toxin (BMAA) exposure
For BMAA dosing assessment, neuronal rosettes were dissociated and seeded in 24-well plates at a density of 50,000 cells/cm2 and cultured in NMM for two weeks. Mature neuronal network and organoids were exposed to varying concentrations (1, 10, 100, and 1,000 μM BMAA; Sigma Cat # B-107) of BMAA for three days. The 100 μM concentration was selected for BMAA exposure experiments (14 days) for cerebral organoids. The neurotoxin was reintroduced into the wells whenever the culture medium was replaced.
Cerebral organoid processing and immunohistochemistry
Organoids were fixed in 4% PFA for 24 h and then transferred to 30% sucrose solution at room temperature. Before sectioning, the organoids were transferred into polyethylene Molding Cup Trays (Polysciences Inc.) filled with OCT compound (Tissue Tek Inc.) and flash-frozen in liquid nitrogen. The frozen OCT blocks containing the organoids were then sectioned into 10-μM-thick slices using a cryostat (Leica, Model CM 1950, Germany). For immunocytochemistry, the slices were washed three times with PBS, permeabilized in 0.1% Triton X-100 (Sigma-Aldrich) for 15 minat room temperature, blocked with 2% BSA, and incubated with primary antibodies against the mature neuron marker MAP2 (Santa Cruz, Cat# sc20172); the astrocyte marker GFAP (Thermo Fisher, Cat# MA5-12023); the microglia markers TMEM119 (Abcam, Cat#185333), Iba1 (Abcam, Cat#15690), IFITM1 (Cat# sc-374026), CD86 (Abcam, Cat# ab239075), and CD208 (Abcam, Cat# ab8918); beta-amyloid (1–42) (Abcam, Cat# ab10148); tau (Abcam, Cat# ab80579); and the inflammasome markers NLRP3 (Thermo Fisher, Cat# PA5-79740) and caspase-1 (Santa Cruz, Cat# sc-392736) overnight at 4°C in a humid chamber follow the instructions provided by companies. The next day, the slices were washed three times with PBST (1× PBS containing 0.1% Tween 20) and twice with PBS, incubated for 2 hat room temperature with fluorescent secondary antibodies (Life Technologies Inc., Cat# A-11029 and A11010), washed three times with PBST and once with PBS, mounted in Fluoromount-G medium containing DAPI, and cover-slipped. Immunohistochemistry images were captured with a Zeiss LSM 880 confocal microscope with Airyscan. Data were collected from three independent images and analyzed using ImageJ software (https://imagej.nih.gov/ij/) (ANOVA and Student’s t test; a pvalue < 0.05 was considered significant). Correlations were determined by calculating Pearson coefficients. All statistical analyses were conducted in GraphPad Prism (GraphPad Software, Inc.).
Beta-amyloid phagocytic assay
Neuronal network containing microglia derived from different ALS-PDC affected and unaffected iPSCs were incubated with Amyloid-beta (1–40), HiLyte Fluor™488-labeled (AS-60491-01) at 1 μM for overnight at 37 C. Following the incubation, the cells were washed 3X with PBS and fixed in 4% paraformaldehyde (PFA) and cell with internalized substrates were visualized using an EVOS inverted microscope. At least 10 cells from each group were analyzed from three independent experiments. Data were prepared and analyzed by De Novo Software. Tukey’s post-hoc test was used to assess significant differences between the groups.
RNA-seq and data analysis
Total RNA was extracted from ALS-PDC-affected and-unaffected organoids using TRIzol Reagent (Invitrogen Inc.) according to the manufacturer’s instructions. RNA-seq library construction, sequencing, and analysis were performed at the City of Hope Integrative Genomics Core according to routine procedures. Libraries were prepared with the Kapa RNA mRNA Hyper-Prep Kit (Kapa Biosystems Inc.) following the manufacturer’s protocol. Reads were aligned against the human genome (hg19) using TopHat2.41 Read counts were quantified using htseq-count42 with the UCSC known gene annotation package [TxDb.Hsapiens.UCSC.hg19.knownGene (Hsu et al., 2006)]. Fold-change was calculated from Fragments Per Kilobase per Million mapped reads (FPKM)43-normalized expression values, which were also used for visualization following log2 transformation. Aligned reads were counted using GenomicRanges.44 Before pvalue calculation, genes were filtered to only include transcripts with an FPKM expression level of 0.1 (after a rounded log2 transformation) in at least 50% of samples45 and genes longer than 150 bp. The general scripts are a modified version of a template for RNA-Seq gene expression analysis (https://github.com/cwarden45/RNAseq_templates/tree/master/TopHat_Workflow). RNA-Seq data for this study are deposited in NCBI underBioproject #: PRJNA 75348.
For the ALS-PDC-affected vs-unaffected comparison (n = 2), pvalues were calculated from raw counts using DESeq2,46while the false discovery rate (FDR) values were calculated using the method of Benjamini and Hochberg.47 DEGs were defined as those with a fold-change >2.0 (unadjusted pvalue: <0.05; FDR: <0.25). GO enrichment was calculated using goseq.48 Gene symbol GO mapping was defined within goseq for the genome reference “hg19”. A histogram was created from the results using plotting −log10 (pvalue) for gene.49 Selected genes were validated by RT-qPCR on a Bio-Rad CFX 96 system (Bio-Rad Inc.) using SYBR Premix according to the manufacturer’s instructions. All primer sequences are shown in Table S1. The rich factor is the ratio of DEG numbers annotated in a pathway to the numbers of all the genes annotated in that pathway.
Quantification and statistical analysis
Immunohistochemistry images were captured with a Zeiss LSM 880 confocal microscope with Airyscan. Data were collected from three independent images and analyzed using ImageJ software (https://imagej.nih.gov/ij/) presented as mean ± standard error of the mean (S.E.M.) and analyzed using Student’s t test. A pvalue < 0.05 was considered significant. All statistical analyses were conducted in GraphPad Prism (GraphPad Software, Inc.).
Acknowledgments
We would like to thank Drs. Teequ Siddique and Glen Kisby for the LCLs, the City of Hope Integrative Genomics Core for providing support for RNA-seq and data analysis, and the support of Western University of Health Sciences through an intramural grant.
Author contributions
Y.H. conceived and designed the project, analyzed the data, and wrote the manuscript; X.D. generated the iPSC lines and performed cell culture, RNA isolation, RT-qPCR, and immunohistochemistry; L.C. developed the procedure for the generation of organoids from iPSCs and performed immunohistochemistry; C.X., M.C., and J.L. performed cell cultures and immunohistochemistry; J.S.A. performed qPCR and data analysis; J-C. L. analyzed data. Q.Q.L. co-mentored students, analyzed the data, and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location.
Published: February 24, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106267.
Supplemental information
Data and code availability
RNA-seq data had been deposited to NCBI. Bioproject # is PRJNA75348 and are publicly available as of the date of publication. This paper does not report original code, and any additional information required to reanalyze the data reported in this paper is available from the lead contactupon request.
References
- 1.Harry G.J. Microglia during development and aging. Pharmacol. Ther. 2013;139:313–326. doi: 10.1016/j.pharmthera.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Y., Szretter K.J., Vermi W., Gilfillan S., Rossini C., Cella M., Barrow A.D., Diamond M.S., Colonna M. IL-34 is a tissue-restricted ligand of CSF1R required for the development of Langerhans cells and microglia. Nat. Immunol. 2012;13:753–760. doi: 10.1038/ni.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E., et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cowan M., Petri W.A., Jr. Microglia: immune regulators of neurodevelopment. Front. Immunol. 2018;9:2576. doi: 10.3389/fimmu.2018.02576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nayak D., Roth T.L., McGavern D.B. Microglia development and function. Annu. Rev. Immunol. 2014;32:367–402. doi: 10.1146/annurev-immunol-032713-120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paolicelli R.C., Bolasco G., Pagani F., Maggi L., Scianni M., Panzanelli P., Giustetto M., Ferreira T.A., Guiducci E., Dumas L., et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–1458. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 7.Hickman S., Izzy S., Sen P., Morsett L., El Khoury J. Microglia in neurodegeneration. Nat. Neurosci. 2018;21:1359–1369. doi: 10.1038/s41593-018-0242-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hickman S.E., Kingery N.D., Ohsumi T.K., Borowsky M.L., Wang L.C., Means T.K., El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemonnot A.L., Hua J., Ulmann L., Hirbec H. Microglia in Alzheimer disease: well-known targets and new opportunities. Front. Aging Neurosci. 2019;11:233. doi: 10.3389/fnagi.2019.00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Y., Le W. Differential roles of M1 and M2 microglia in neurodegenerative diseases. Mol. Neurobiol. 2016;53:1181–1194. doi: 10.1007/s12035-014-9070-5. [DOI] [PubMed] [Google Scholar]
- 11.Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. A unique microglia type associated with restricting development of Alzheimer's disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 12.Moehle M.S., West A.B. M1 and M2 immune activation in Parkinson's Disease: foe and ally? Neuroscience. 2015;302:59–73. doi: 10.1016/j.neuroscience.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Gomez J.A., Kavanagh E., Engskog-Vlachos P., Engskog M.K.R., Herrera A.J., Espinosa-Oliva A.M., Joseph B., Hajji N., Venero J.L., Burguillos M.A. Microglia: agents of the CNS pro-inflammatory response. Cells. 2020;9:1717. doi: 10.3390/cells9071717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathys H., Adaikkan C., Gao F., Young J.Z., Manet E., Hemberg M., De Jager P.L., Ransohoff R.M., Regev A., Tsai L.H. Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 2017;21:366–380. doi: 10.1016/j.celrep.2017.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lana D., Ugolini F., Giovannini M.G. An overview on the differential interplay among neurons-astrocytes-microglia in CA1 and CA3 Hippocampus in hypoxia/ischemia. Front. Cell. Neurosci. 2020;14:585833. doi: 10.3389/fncel.2020.585833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brionne T.C., Tesseur I., Masliah E., Wyss-Coray T. Loss of TGF-beta 1 leads to increased neuronal cell death and microgliosis in mouse brain. Neuron. 2003;40:1133–1145. doi: 10.1016/s0896-6273(03)00766-9. [DOI] [PubMed] [Google Scholar]
- 17.Noh M.Y., Lim S.M., Oh K.W., Cho K.A., Park J., Kim K.S., Lee S.J., Kwon M.S., Kim S.H. Mesenchymal stem cells modulate the functional properties of microglia via TGF-beta secretion. Stem Cells Transl. Med. 2016;5:1538–1549. doi: 10.5966/sctm.2015-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tzavlaki K., Moustakas A. TGF-Beta signaling. Biomolecules. 2020;10:487. doi: 10.3390/biom10030487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begum A.N., Guoynes C., Cho J., Hao J., Lutfy K., Hong Y. Rapid generation of sub-type, region-specific neurons and neural networks from human pluripotent stem cell-derived neurospheres. Stem Cell Res. 2015;15:731–741. doi: 10.1016/j.scr.2015.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dutta D., Heo I., Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol. Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 21.Fujimori K., Tezuka T., Ishiura H., Mitsui J., Doi K., Yoshimura J., Tada H., Matsumoto T., Isoda M., Hashimoto R., et al. Modeling neurological diseases with induced pluripotent cells reprogrammed from immortalized lymphoblastoid cell lines. Mol. Brain. 2016;9:88. doi: 10.1186/s13041-016-0267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelava I., Lancaster M.A. Dishing out mini-brains: current progress and future prospects in brain organoid research. Dev. Biol. 2016;420:199–209. doi: 10.1016/j.ydbio.2016.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoon S.J., Elahi L.S., Pașca A.M., Marton R.M., Gordon A., Revah O., Miura Y., Walczak E.M., Holdgate G.M., Fan H.C., et al. Reliability of human cortical organoid generation. Nat. Methods. 2019;16:75–78. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 26.Muffat J., Li Y., Yuan B., Mitalipova M., Omer A., Corcoran S., Bakiasi G., Tsai L.H., Aubourg P., Ransohoff R.M., Jaenisch R. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miklossy J., Steele J.C., Yu S., McCall S., Sandberg G., McGeer E.G., McGeer P.L. Enduring involvement of tau, beta-amyloid, alpha-synuclein, ubiquitin and TDP-43 pathology in the amyotrophic lateral sclerosis/parkinsonism-dementia complex of Guam (ALS/PDC) Acta Neuropathol. 2008;116:625–637. doi: 10.1007/s00401-008-0439-2. [DOI] [PubMed] [Google Scholar]
- 28.Spencer P.S., Palmer V., Kisby G. The ALS/PDC syndrome of Guam and the cycad hypothesis. Neurology. 2009;72:474–476. author reply 475-476. [PubMed] [Google Scholar]
- 29.Spencer P.S., Palmer V.S., Kisby G.E. Seeking environmental causes of neurodegenerative disease and envisioning primary prevention. Neurotoxicology. 2016;56:269–283. doi: 10.1016/j.neuro.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Z.X., Anderson D.W., Mantel N., Román G.C. Motor neuron disease on Guam: geographic and familial occurrence, 1956-85. Acta Neurol. Scand. 1996;94:51–59. doi: 10.1111/j.1600-0404.1996.tb00039.x. [DOI] [PubMed] [Google Scholar]
- 31.Kisby G.E., Ellison M., Spencer P.S. Content of the neurotoxins cycasin (methylazoxymethanol beta-D-glucoside) and BMAA (beta-N-methylamino-L-alanine) in cycad flour prepared by Guam Chamorros. Neurology. 1992;42:1336–1340. doi: 10.1212/wnl.42.7.1336. [DOI] [PubMed] [Google Scholar]
- 32.Delcourt N., Claudepierre T., Maignien T., Arnich N., Mattei C. Cellular and molecular aspects of the beta-N-Methylamino-l-alanine (BMAA) mode of action within the neurodegenerative pathway: facts and controversy. Toxins. 2017;10:6. doi: 10.3390/toxins10010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engskog M.K.R., Ersson L., Haglöf J., Arvidsson T., Pettersson C., Brittebo E. Beta-N-Methylamino-L-alanine (BMAA) perturbs alanine, aspartate and glutamate metabolism pathways in human neuroblastoma cells as determined by metabolic profiling. Amino Acids. 2017;49:905–919. doi: 10.1007/s00726-017-2391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seok J., Warren H.S., Cuenca A.G., Mindrinos M.N., Baker H.V., Xu W., Richards D.R., McDonald-Smith G.P., Gao H., Hennessy L., et al. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proc. Natl. Acad. Sci. USA. 2013;110:3507–3512. doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barrett R., Ornelas L., Yeager N., Mandefro B., Sahabian A., Lenaeus L., Targan S.R., Svendsen C.N., Sareen D. Reliable generation of induced pluripotent stem cells from human lymphoblastoid cell lines. Stem Cells Transl. Med. 2014;3:1429–1434. doi: 10.5966/sctm.2014-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wee Y.S., Weis J.J., Gahring L.C., Rogers S.W., Weis J.H. Age-related onset of obesity corresponds with metabolic dysregulation and altered microglia morphology in mice deficient for Ifitm proteins. PLoS One. 2015;10:e0123218. doi: 10.1371/journal.pone.0123218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paludan S.R., Pradeu T., Masters S.L., Mogensen T.H. Constitutive immune mechanisms: mediators of host defence and immune regulation. Nat. Rev. Immunol. 2021;21:137–150. doi: 10.1038/s41577-020-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.El Khoury J. Neurodegeneration and the neuroimmune system. Nat. Med. 2010;16:1369–1370. doi: 10.1038/nm1210-1369. [DOI] [PubMed] [Google Scholar]
- 39.Ormel P.R., Vieira de Sá R., van Bodegraven E.J., Karst H., Harschnitz O., Sneeboer M.A.M., Johansen L.E., van Dijk R.E., Scheefhals N., Berdenis van Berlekom A., et al. Microglia innately develop within cerebral organoids. Nat. Commun. 2018;9:4167. doi: 10.1038/s41467-018-06684-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ginhoux F., Garel S. The mysterious origins of microglia. Nat. Neurosci. 2018;21:897–899. doi: 10.1038/s41593-018-0176-3. [DOI] [PubMed] [Google Scholar]
- 41.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R13–R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mortazavi A., Williams B.A., McCue K., Schaeffer L., Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. https://www.nature.com/nmeth/journal/v5/n7/suppinfo/nmeth.1226_S1.html [DOI] [PubMed] [Google Scholar]
- 44.Lawrence M., Huber W., Pagès H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warden C., Yuan Y., Wu X. Optimal calculation of RNA-seq fold-change values. Int. J. Comput. Bioinforma. Silico Model. 2013;2:285–292. [Google Scholar]
- 46.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B. 1995;57:289–300. [Google Scholar]
- 48.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young M.D., Wakefield M.J., Smyth G.K., Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14. doi: 10.1186/gb-2010-11-2-r14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data had been deposited to NCBI. Bioproject # is PRJNA75348 and are publicly available as of the date of publication. This paper does not report original code, and any additional information required to reanalyze the data reported in this paper is available from the lead contactupon request.