Abstract
Acetate and formate are major fermentation products of Escherichia coli. Below pH 7, the balance shifts to lactate; an oversupply of acetate or formate retards growth. E. coli W3110 was grown with aeration in potassium-modified Luria broth buffered at pH 6.7 in the presence or absence of added acetate or formate, and the protein profiles were compared by two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Acetate increased the steady-state expression levels of 37 proteins, including periplasmic transporters for amino acids and peptides (ArtI, FliY, OppA, and ProX), metabolic enzymes (YfiD and GatY), the RpoS growth phase regulon, and the autoinducer synthesis protein LuxS. Acetate repressed 17 proteins, among them phosphotransferase (Pta). An ackA-pta deletion, which nearly eliminates interconversion between acetate and acetyl-coenzyme A (acetyl-CoA), led to elevated basal levels of 16 of the acetate-inducible proteins, including the RpoS regulon. Consistent with RpoS activation, the ackA-pta strain also showed constitutive extreme-acid resistance. Formate, however, repressed 10 of the acetate-inducible proteins, including the RpoS regulon. Ten of the proteins with elevated basal levels in the ackA-pta strain were repressed by growth of the mutant with formate; thus, the formate response took precedence over the loss of the ackA-pta pathway. The similar effects of exogenous acetate and the ackA-pta deletion, and the opposite effect of formate, could have several causes; one possibility is that the excess buildup of acetyl-CoA upregulates stress proteins but excess formate depletes acetyl-CoA and downregulates these proteins.
The growth and survival of Escherichia coli at low pH involves numerous genetic responses (9, 25, 62, 65). Survival in acid enables the pathogenesis of E. coli O157:H7 (49), Shigella flexneri (74), and Vibrio cholerae (52). Acidic stress is intensified by the presence of short-chain fatty acids, high concentrations of which are produced as fermentation products by microflora in the intestine (18). These weak acids permeate the cell membrane in undissociated form and dissociate within the cytoplasm, thus delivering protons to the cytoplasm and depressing internal pH, as well as delivering a corresponding amount of the acid anion to the cytoplasm (41, 47, 57, 59). The extent of this process is determined by the transmembrane pH difference, which is most pronounced in an acid environment. Thus, membrane-permeant organic acids intensify the stress of extracellular acidity, but their presence also induces protective responses against acid (31, 61).
The major fermentation acids excreted by E. coli include acetate, formate, d-lactate, and succinate (10, 21, 40, 58). A high concentration of fermentation acids limits growth (57, 59), and acetate induces the RpoS regulon associated with entry into stationary phase (6, 61). Above pH 7, the favored fermentation products are acetate (with ethanol) and formate (for a review see reference 40). Production of these two acids is maximal in the absence of oxygen or other respiratory electron acceptors (40), but oxygenated cultures also excrete significant amounts of acetate (13, 15) and formate (2, 68), a significant concern for bioreactors (22). As pH falls, E. coli limits internal acidification from metabolism by producing lactate instead of acetate plus formate (14), by reuptake and activation of acetate to acetyl-coenzyme A (CoA) to enter the tricarboxylic acid (TCA) cycle (21, 44, 56), and by conversion of formate to H2 and CO2 (58). The mechanisms of regulation and the responses to high concentrations of different acids remain unclear.
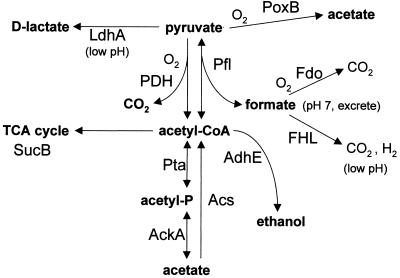
A candidate for participation in the responses to fermentation acids is acetyl-CoA, a key intermediate for interconversion of pyruvate with both acetate and formate (Fig. 1). The conversion between acetyl-CoA and acetate is mediated by two pathways: (i) acetate kinase (AckA) and acetyl phosphotransferase (Pta), which rapidly convert acetyl-CoA via acetyl-phosphate to acetate as an overflow pathway and also convert exogenous acetate back to acetyl-CoA (21, 56), and (ii) acetyl-CoA synthetase (Acs), a high-affinity, low-capacity uptake pathway for acetate which produces acetyl-CoA via an enzyme-bound acetyl-adenylate intermediate (13, 44). The overall modulation of acetyl-CoA and acetyl-phosphate levels involves complex functions of oxygen concentration and carbon source (51, 43).
FIG. 1.
Metabolic pathways connecting pyruvate with fermentation acids in E. coli.
The conversion between pyruvate and acetyl-CoA is mediated aerobically by pyruvate dehydrogenase (PDH), producing CO2, and anaerobically by pyruvate formate lyase (Pfl), producing formate (42, 60). Catalysis by Pfl involves a glycine radical inactivated by oxygen in vitro (73), but recent evidence suggests that in vivo Pfl is both expressed (2) and functional (19) in the presence of some oxygen. Pfl activity is reversible, so it could convert formate to pyruvate, consuming acetyl-CoA (40, 42). E. coli also has several poorly characterized homologs of Pfl, including YfiD (29, 50), which is induced aerobically at low pH (9). The effects of exogenous formate on protein expression, and the relationship between formate and acetate stress, remain unclear.
Some pyruvate is also converted directly to acetate and CO2 by an oxidative enzyme, PoxB (16). This process conserves less energy than the acetyl-CoA pathway; its activity is maximal in early stationary phase, activated by RpoS.
We used the proteomic approach to study steady-state responses of E. coli metabolism to exogenous acetate or formate. One prediction would be that the presence of either acetate or formate might induce certain proteins responding to the stress of internal acidification, such as YfiD (9, 32) or InaA (64). Instead, in cultures grown in complex medium, we found largely opposite responses: most proteins induced by acetate were repressed by formate, and several proteins induced by formate were repressed by acetate. This pattern of responses suggests that the major effects of acetate or formate are particular to the specific anion. Thus, there appears to be a metabolic switch converting the cell between two different growth states, one preferred in the presence of each organic acid.
The role of the acetyl kinase-phosphotransferase pathway in this switch was tested using a strain with ackA-pta deleted, in which excess acetyl-CoA accumulates and inhibits PDH, causing accumulation of pyruvate (15, 36). Most of the proteins that were elevated in this strain (compared to the parent) were the same as those induced by acetate in the parent. One possible explanation, requiring further experiments for confirmation, is that the concentration of acetyl-CoA modulates some of the observed responses to acetate and formate. We also report additional proteins induced by acetate but insensitive to formate, including the oligopeptide permease OppA (24, 38) and the autoinducer synthesis protein LuxS (69).
MATERIALS AND METHODS
Strains and growth conditions.
E. coli K-12 strains included W3110 (66), RM7076 (W3110 ackA-pta), and RM7194 (ack-pta crl::cam). Strain RM7076 was constructed as follows. The ackA-linked locus purF92::Tn10 (Tet) was transduced from strain NK6078 into W3110 using phage P1. Into this construct, Δ(ackA-hisP)862 was transduced from strain TA3516 (48). Transductants containing the deletion were identified by screening pur+ prototrophs for sensitivity to 0.25% alizarin yellow, a pta phenotype. Strain RM7194 was constructed by transduction from LP895 (MC4100 crl920::cam) (55).
For two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (2-D gels), growth media contained LBK broth (10 g of tryptone, 5 g of yeast extract, 7.45 g of KCl) or M63 salts [3 g of KH2PO4, 7 g of K2HPO4, 2 g of (NH4)2SO4, 0.5 ml of FeSO4 at 1 mg/ml, 2 ml of 0.5 M MgSO2, 100 mg of thiamine/ml] with 30 mM glycerol as a carbon source. All LBK and M63 media were buffered with 50 mM MOPS [3-(N-morpholino)propanesulfonic acid] and 50 mM TES (N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid) (62, 64), adjusted for pH using KOH in order to avoid high concentrations of sodium ions, which inhibit growth at high pH (34, 37). Acetic acid (50 mM) or formic acid (20 mM) was included at a concentration that did not substantially retard growth. Cultures in LBK broth were grown overnight and then diluted 200-fold in fresh medium with or without added acid, grown to an optical density at 600 nm (OD600) of 0.4 to 0.5 at 37°C, and oxygenated (rotary aeration in a flask whose capacity was eight times the culture volume); doubling times were 30 to 40 min for W3110 and 40 to 50 min for RM7076. Cultures in M63 were grown to an OD600 of 0.2 to 0.3, with doubling times of 100 to 120 min.
2-D gels.
Our detailed methods for 2-D gels and analysis are reported elsewhere (63), and updates are maintained on line (http://www2.kenyon.edu/depts/biology/slonc/labtools/2d_method.html). Protein samples were solubilized in sodium dodecyl sulfate and urea according to the procedure of Genomic Solutions (9, 45, 72). The rehydration solution from AP Biotech replaced buffer 3; this replacement has improved the reproducibility of solubilization. First-dimension IPGphor gel strips (AP Biotech) were run according to the instructions of the manufacturer. For each gel, approximately 50 μg of protein was loaded onto an IPG strip (pH 4 to 7). 2-D gels were performed on the ESA Investigator 2-D electrophoresis system (Genomic Solutions). Thirteen to 15 ml of each culture was chilled, pelleted, and washed with LBK broth. The cell pellets were then treated with urea-sodium dodecyl sulfate sample buffer and DNase or RNase, according to ESA procedures (Genomic Solutions). Samples were applied to 18-cm-long polyacrylamide gel strips with an immobilized gradient (pH 4 to 7) for isoelectric focusing. The gel strips were applied to 2-D gels containing 11.5% acrylamide. For comparative analysis, the gels were silver stained.
For comparison of spot quantities between different growth conditions, gels were scanned and digitized. The pI and molecular weight scales for the gels are based on comparison with E. coli reference gels (reference 72 and the SWISS-2DPAGE database of the Swiss Institute of Bioinformatics [http://www.expasy.ch/]). Spot densities were quantified using Compugen Z3 software, version 1.5, as described elsewhere (http://www.2dgels.com/). The Z3 algorithm for pairwise comparison matches each pair of spots pixel by pixel, discarding values of saturated pixel density as well as values from the edge of the background. A histogram of all spot ratios normalizes them to 1, on the assumption that 90% of all proteins should have the same concentration under the two growth conditions; this assumption corrects for loading differences without requiring summation of all spots and subtraction of background, procedures which introduce error.
For each growth condition, spot densities were obtained from three gel images from independently grown cultures. The differential expression (DE) of the spot densities between two growth conditions (reference condition and comparative condition) was determined for all pairwise comparisons between a set of three reference gels and a set of three comparative gels. A protein spot was considered a candidate for significant induction if seven out of nine pairwise comparisons produced a difference of greater than 50% (63). The log10 of all nine DE values was obtained, and the mean log10 DE (LDE) was reported as the measure of induction or repression. For example, an LDE of +0.5 represents about a threefold increase in spot density of the comparative over the reference gel.
In some cases, significantly induced proteins showed a spot on the comparative gels that was unmatched on one or more of the reference gels (that is, present under the inducing condition but undetectable under the noninducing condition). Because the induction ratios could not be quantified, these strongly induced proteins are indicated as “plus” in the comparative condition compared to the reference condition or as “minus” if repressed (Table 1). Proteins generating DE ratios greater than 10-fold (LDE = 1.0) generally had to be scored plus (induced) or minus (repressed), because of the likelihood that the repressed protein fades into the background in one of the three replicates.
TABLE 1.
Proteins showing differential expression on 2-D gels
| Spot no. | Protein | Differential expressiona
|
|||||
|---|---|---|---|---|---|---|---|
| Acetate in W3110 (Fig. 2A) | ackA-pta/W3110 (Fig. 2B) | Formate in W3110 (Fig. 3A) | Formate in ackA-pta (Fig. 3B) | Acetate in ackA-pta (Fig. 4A) | Acetate in W3110 (glycerol) (Fig. 4B) | ||
| 1 | −0.68 ± 0.37 | −0.41 ± 0.08 | |||||
| 2 | OppAb | + | + | (−) | |||
| 3 | OppA | 0.46 ± 0.12 | 0.43 ± 0.15 | −0.55 ± 0.11 | |||
| 4 | OppA | 0.38 ± 0.20 | 0.45 ± 0.19 | −0.26 ± 0.07 | |||
| 5 | 0.25 ± 0.10 | ||||||
| 6 | + | (+) | (−) | −0.38 ± 0.20 | |||
| 7 | (+) | + | −1.11 ± 0.16 | ||||
| 8 | TnaAc | − | − | −0.70 ± 0.30 | |||
| 9 | 0.79 ± 0.23 | ||||||
| 10 | −0.55 ± 0.11 | ||||||
| 11 | (+) | ||||||
| 12 | (+) | ||||||
| 13 | MalE | −0.81 ± 0.20 | |||||
| 14 | − | ||||||
| 15 | − | ||||||
| 16 | − | ||||||
| 17 | PfkB | 0.40 ± 0.06 | 0.36 ± 0.10 | −0.22 ± 0.12 | |||
| 18 | 0.32 ± 0.10 | ||||||
| 19 | (−) | ||||||
| 20 | 0.29 ± 0.09 | ||||||
| 21 | (+) | (+) | − | ||||
| 22 | 0.45 ± 0.18 | 0.35 ± 0.16 | |||||
| 23 | (−) | ||||||
| 24 | −0.29 ± 0.16 | ||||||
| 25 | OmpC | 0.55 ± 0.33 | (+) | ||||
| 26 | Pta | −0.38 ± 0.08 | − | ||||
| 27 | (+) | 0.32 ± 0.16 | |||||
| 28 | + | ||||||
| 29 | 0.43 ± 0.17 | −0.32 ± 0.17 | |||||
| 30 | ProX | 0.38 ± 0.10 | 0.32 ± 0.21 | −0.68 ± 0.22 | |||
| 31 | − | ||||||
| 32 | 0.46 ± 0.09 | ||||||
| 33 | 0.39 ± 0.09 | ||||||
| 34 | MglB | −0.42 ± 0.12 | −0.49 ± 0.10 | ||||
| 35 | − | (−) | |||||
| 36 | + | + | |||||
| 37 | + | 0.55 ± 0.46 | |||||
| 38 | GatY | 0.53 ± 0.15 | −0.24 ± 0.10 | −0.35 ± 0.20 | 0.40 ± 0.23 | ||
| 39 | 0.71 ± 0.09 | ||||||
| 40 | SucB | −0.32 ± 0.13 | |||||
| 41 | + | −0.61 ± 0.11 | −0.45 ± 0.31 | ||||
| 42 | 0.43 ± 0.18 | ||||||
| 43 | + | (+) | −0.62 ± 0.25 | ||||
| 44 | Lpd | −0.33 ± 0.21 | |||||
| 45 | (+) | ||||||
| 46 | (+) | (+) | −0.47 ± 0.11 | −0.52 ± 0.39 | −0.38 ± 0.12 | −0.34 ± 0.20 | |
| 47 | + | ||||||
| 48 | FliY | 0.28 ± 0.06 | 0.36 ± 0.08 | ||||
| 49 | ArtI | 0.26 ± 0.10 | −0.22 ± 0.06 | ||||
| 50 | + | ||||||
| 51 | − | −0.33 ± 0.15 | 0.42 ± 0.43 | ||||
| 52 | (+) | 0.90 ± 0.45 | |||||
| 53 | + | (+) | (+) | ||||
| 54 | WrbA | 0.29 ± 0.09 | 0.34 ± 0.11 | ||||
| 55 | + | ||||||
| 56 | 0.73 ± 0.52 | ||||||
| 57 | + | ||||||
| 58 | Ppa | 0.23 ± 0.09 | |||||
| 59 | + | 0.46 ± 0.09 | |||||
| 60 | (−) | 0.74 ± 0.10 | + | ||||
| 61 | 0.74 ± 0.17 | −0.32 ± 0.08 | |||||
| 62 | LuxS | 0.40 ± 0.10 | |||||
| 63 | OsmY | 0.45 ± 0.11 | 0.50 ± 0.20 | −0.34 ± 0.14 | −0.35 ± 0.15 | ||
| 64 | DksA | −0.34 ± 0.10 | |||||
| 65 | AroK | −0.39 ± 0.16 | −0.32 ± 0.10 | ||||
| 67 | + | ||||||
| 68 | (+) | 0.30 ± 0.17 | |||||
| 69 | 0.33 ± 0.11 | ||||||
| 70 | −0.31 ± 0.19 | −0.23 ± 0.08 | |||||
| 71 | −0.26 ± 0.10 | −0.31 ± 0.15 | |||||
| 72 | 0.24 ± 0.12 | 0.28 ± 0.14 | −0.37 ± 0.10 | −0.50 ± 0.18 | |||
| 73 | TpiA | 0.32 ± 0.24 | |||||
| 74 | + | 0.51 ± 0.36 | |||||
| 75 | (+) | + | (−) | ||||
| 76 | 0.58 ± 0.43 | ||||||
| 77 | (+) | ||||||
| 78 | YfiD | 0.32 ± 0.11 | 0.39 ± 0.09 | ||||
| 79 | (−) | ||||||
| 80 | 0.43 ± 0.29 | ||||||
| 81 | HdeB | (+) | (+) | (−) | |||
| 82 | HdeA | 0.41 ± 0.13 | 0.42 ± 0.18 | −0.93 ± 0.24 | − | ||
| 83 | GuaB | −0.42 ± 0.25 | −0.50 ± 0.15 | ||||
| 84 | 0.38 ± 0.07 | ||||||
Relative protein expression shown as LDE ± standard deviation (n = 9) determined as described in Materials and Methods. +, spot unmatched on reference gel (i.p., protein induced in the comparative condition compared to the reference condition); −, repressed; (+), (−), spot showing differential expression based on visual inspection, though not quantifiable by the Z3 software. The column headers indicate the comparative gel/the reference gel, with the figure number of the layered view in parentheses.
Protein identified based on position (http://www.expasy.ch/).
Protein identified based on position and on inducibility at high pH (9).
A few proteins were determined to be induced or repressed based on visual inspection, although the software could not effectively quantify them because of the irregular shape of the spot or because of overlap between spots. These are identified in Table 1.
In addition to the pairwise comparisons, each set of three replicate gel images was digitally combined into a composite image. The composite images representing each of two growth conditions or strain constructs (reference condition and comparative condition) were overlaid by Z3 so that a spot showing only in the comparative gel (or the border of a spot showing increased induction) appears pink, whereas a spot showing only in the reference gel appears green (see Fig. 2 to 4). This layered view provides a global “snapshot” of the gel comparison. Most of the spots that appear pink or green on the layered view generate significantly positive or negative LDE values, respectively, in the pairwise comparisons. In some cases, however, a spot which appears pink or green is present on only one of the original three gels of a composite. We did not count these proteins as induced in Table 1.
FIG. 2.
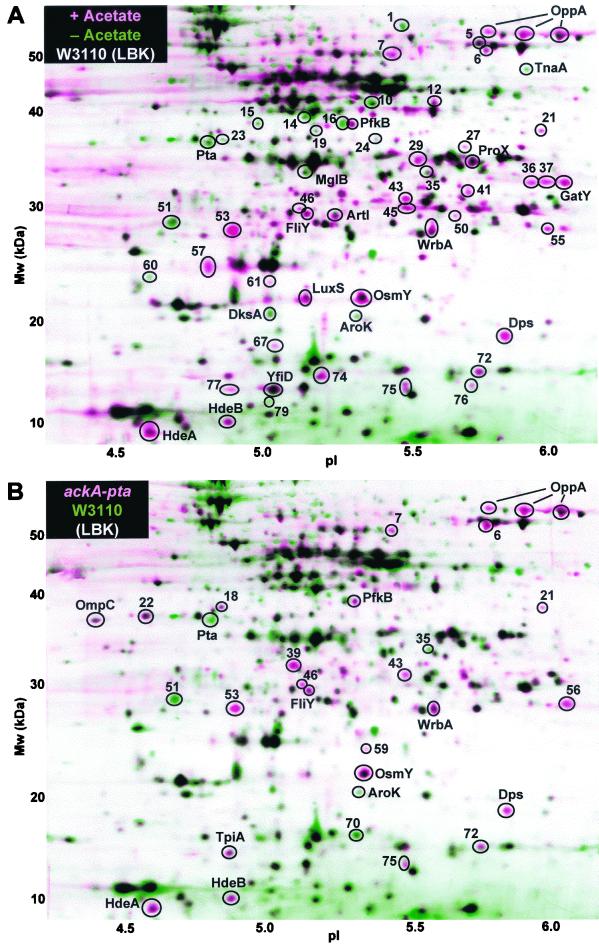
Proteins differentially expressed with acetic acid or in the ackA-pta strain. The layered view superimposes two composite images representing different growth conditions. Each composite image is based on three 2-D gels from replicate cultures. All E. coli cultures were grown at 37°C to an OD600 of 0.4 in LBK broth buffered with 50 mM MOPS and 50 mM TES, pH 6.7. (A) Strain W3110 was grown in buffered LBK broth with (pink) or without (green) 50 mM acetic acid. (B) Strain W3110 ackA-pta (pink) or W3110 (green) was grown in buffered LBK broth. In the layered views, pink and green indicate proteins induced and repressed, respectively, in the presence of acetic acid (A) or proteins constitutively induced or repressed in the ackA-pta strain (B), based on pairwise comparison of individual gels (summarized in Table 1). The horizontal axis represents the approximate pH range of the isoelectric focusing first dimension. The vertical axis represents the molecular mass in kilodaltons. Proteins in the 2-D gels were silver stained. For N-terminal sequence identification, the proteins were electroblotted and stained with Coomassie blue.
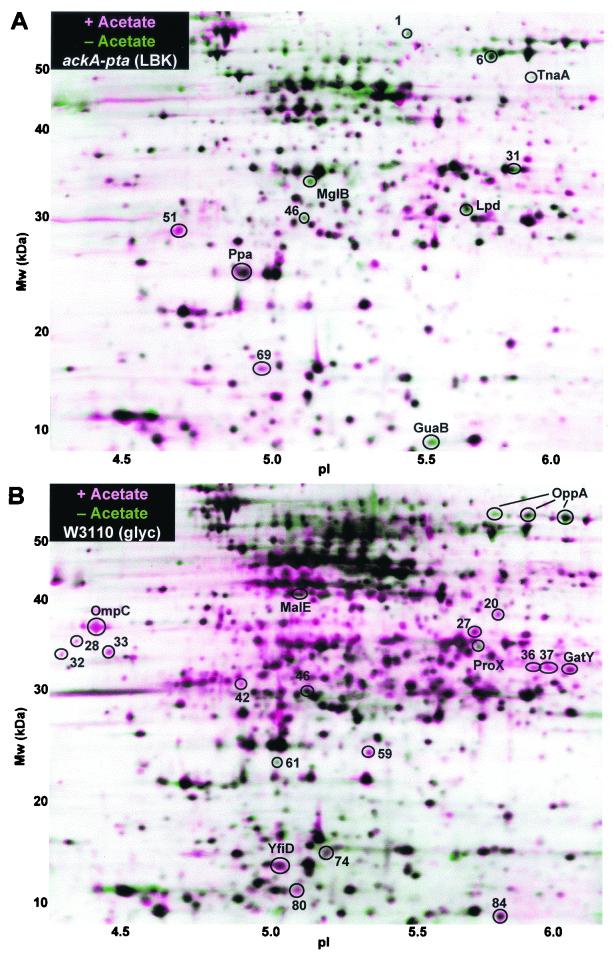
FIG. 4.
Proteins differentially expressed with (pink) or without (green) 50 mM acetic acid. (A) Strain W3110 ackA-pta was grown in LBK broth–50 mM MOPS–50 mM TES, pH 6.7, to an OD600 of 0.4. (B) Strain W3110 was grown in glycerol M63 salts–50 mM MOPS–50 mM TES, pH 6.7, to an OD600 of 0.2. Other growth and gel conditions were as for Fig. 2.
Identification of proteins.
For protein identification, 200 μg of cell protein was loaded onto each IPG strip. The slab gel proteins were transferred to an Immobilon-P polyvinylidene difluoride membrane (Millipore) using the ESA Investigator graphite electroblotter, type II (Genomic Solutions). The membrane was stained with Coomassie blue, and protein spots were excised for sequence determination. Protein spots cut from the transfer membrane were washed three times in 10% methanol and then dried and stored at −70°C.
N-terminal peptide sequence analysis was performed by the Biopolymer Facility at the University of California at Davis (Table 2). Samples were processed using a model 470A or 477A ABI sequencer with on-line high-pressure liquid chromatography and capable of picomole analysis. Peptide sequences were matched against all known and putative E. coli proteins in the Swiss-Prot database, using the ExPASy Fasta program (http://www.expasy.ch/).
TABLE 2.
Identified proteins and peptide sequences
| Spot no. | Protein | Protein name | N-terminal sequence |
|---|---|---|---|
| 3 | OppA | Periplasmic oligopeptide-binding protein | ADVPAGV |
| 4 | OppA | Periplasmic oligopeptide-binding protein | XDVPAGVTLA |
| 8 | TnaA | Tryptophanase | See reference 8 |
| 13 | MalE | Maltose-binding periplasmic protein | KIEEGKLVIXIN |
| 17 | PfkB | 6-Phosphofructokinase isozyme 2 | MVRIYTLTLAP |
| 26 | Pta | Phosphate acetyltransferase | KAGKRIVLPE |
| 25 | OmpC | Outer membrane protein C | AEVYNKDGNKLD |
| 30 | ProX | Glycine betaine-binding periplasmic protein | ADLPGKGITVNP |
| 34 | MglB | d-Galactose-binding periplasmic protein | DTRIGVTI |
| 38 | GatY | Tagatose-bisphosphate aldolase | YVVSTKQMLN |
| 40 | SucB | Dihydrolipoamide succinyltransferase | VDILVPDLPE |
| 44 | Lpd | Dihydrolipoamide dehydrogenase | KFNLMLETKVT |
| 48 | FliY | Cysteine-binding periplasmic protein | DEGLLNKVKER |
| 49 | ArtI | Arginine-binding periplasmic protein 1 | AETIRFATEA |
| 54 | WrbA | Trp repressor binding protein | AKVLVLYYSXYG |
| 58 | Ppa | Inorganic pyrophosphatase | LLNVPAXKDL |
| 63 | OsmY | Hyperosmotically inducible protein Y | ENNAQTTNESAG |
| 62 | LuxS | Autoinducer-2 synthesis protein | PLLDSFTVDDHR |
| 64 | DksA | DnaK suppressor protein | QEGQNRK |
| 65 | AroK | Shikimate kinase I | EKRNIFLVGP |
| 66 | Dps | DNA protection during starvation | STAKLVKSKAT |
| 73 | TpiA | Triosephosphate isomerase | KFAVLKEQGLTP |
| 78 | YfiD | Pyruvate formate-lyase homolog | MITGIQITKAAN |
| 81 | HdeB | Extreme-acid periplasmic chaperone | ANESAKDMTCQE |
| 82 | HdeA | Extreme-acid periplasmic chaperone | ADAQKAADNKKP |
| 83 | GuaB | Inosine-5′-monophosphate dehydrogenase | LRIAKEALTFD |
Extreme-acid survival.
Survival of log-phase cultures exposed to pH 2.5 was determined by a previously described procedure, with slight modification (65). Overnight cultures were diluted 200-fold in Luria broth (LB), pH 6.7 (10 g of Bacto Tryptone, 5 g of Bacto yeast extract, 5 g of NaCl, and 100 mM MOPS buffer adjusted to pH 6.7 with NaOH) and grown with aeration at 37°C for approximately 2 h, at which point the OD600 was typically 0.15 to 0.20 (≈1 × 108 to 2 × 108 cells/ml). An aliquot of each culture was diluted 1,000-fold into LB, pH 2.5 (as above but adjusted to pH 2.5 with HCl in place of the MOPS buffer). At the same time, another aliquot of each culture was diluted and plated to obtain the input CFU. After 1 h at room temperature, the acid challenge was terminated by the addition of sufficient 1 N NaOH to return the pH to 6.7. Survivors were assayed as CFU; the geometric mean of three trials is reported. For wild-type cells, whose survival is very low, the log-phase culture was concentrated 10-fold in water and then diluted 100-fold in acid LB and further processed as described above to obtain meaningful numbers of survivors.
RESULTS
Quantitative measurement of protein expression levels.
E. coli K-12 strains were cultured for at least four doublings in order to observe steady-state levels of protein expression during growth in the presence of acetate or formate. Protein expression levels with several different pairs of growth conditions and/or strain constructs were compared (Table 1). The quantification of relative expression levels was based on pairwise comparisons of all three reference gels and all three comparative gels. Note, however, that because of the inherent limitations of the sensitivity ranges of both the silver stain and the gel scanner, the actual protein concentration ratios for matched spots could be significantly greater than the LDE values for spot density.
For a given protein spot, we defined significant DE based on DE ratios showing a 50% increase (DE ≥ 1.5 or ≤ 0.67) in seven out of nine pairwise comparisons between the sets of three comparative and three reference gels. These criteria produced no significant spot differences when two gel sets with equivalent growth conditions were compared (data not shown). The software detected 80% of the spots that appeared to show induction based on visual inspection of individual gel images. The remaining 20% of the spots showed irregularities in shape that interfered with the detection algorithm.
Acetate-induced proteins during growth in LBK broth.
The effect of acetate on steady-state protein levels was tested by growth of cultures to mid-log phase in buffered LBK broth in the presence or absence of 50 mM acetic acid (Fig. 2A). Several periplasmic transporters showed elevated expression levels during growth with acetate (Table 1). The periplasmic oligopeptide binding protein OppA (38, 39) transports short peptides for a nutrient and recycles cell wall components (27). OppA appeared as a “train” of three different spots, an appearance in 2-D gels that is poorly understood but common for highly expressed proteins (9, 28). All three OppA spots showed clear induction in LBK broth but were strongly repressed in glycerol minimal medium. Other transporters induced by acetate included ArtI, FliY, OsmY, and ProX. On the other hand, acetate repressed the d-galactose-binding protein MglB.
Acetate induced several members of the RpoS growth phase regulon (11, 33, 46), including Dps, HdeA, HdeB, OsmY, PfkB, and WrbA. This induction is consistent with previous reports that acetate induces RpoS (6, 61). The most strongly induced member of this regulon was the DNA-binding protein Dps (3), which is also regulated by the oxidative stress regulator OxyR during log phase (4). Acetate also induced metabolic enzymes, including tagatose-bisphosphate aldolase (GatY) and the pyruvate formate-lyase homolog YfiD, which are induced by low pH (9). The autoinducer synthesis protein LuxS (69) was induced.
Seventeen proteins were repressed during growth with acetate, including Pta, MglB, DksA, AroK, and TnaA. We have not discerned any functional or regulatory commonalities in this group so far. The repression of Pta by acetate may be associated with some of the similarities in the pattern of gene expression seen in the ackA-pta strain (see below).
Protein expression levels in an ackA-pta deletion strain.
The Ack and Pta enzymes provide the major route for introduction of exogenous acetate into metabolism, and it seemed likely that they would also be required for gene expression effects of acetate. Therefore, we observed protein expression in a mutant with ack and pta deleted. One hypothesis would be that in the absence of acetate, the mutant and parent would show similar protein profiles and that the response of the mutant to acetate, if any, would be attenuated in comparison to that of the parent. In fact, however, the protein profile of the mutant, in the absence of added acetate, appeared remarkably similar to that of the parent in the presence of acetate (Fig. 2B). The ackA-pta deletion strain showed increased basal levels of 16 of the same proteins induced in the parent by exogenous acetate, including most of the transporters and the RpoS regulon (Table 1). Three proteins repressed by acetate in the parent, including shikimate kinase (AroK), were also repressed by ackA-pta.
Addition of acetate to the ackA-pta strain affected the expression of only 11 proteins, less than a third of the number induced or repressed in the parent (see Fig. 4A and Table 1). One protein repressed was the pyruvate dehydrogenase component Lpd (30), possibly a homeostatic mechanism to limit the overaccumulation of acetyl-CoA predicted for this mutant.
Extreme-acid survival.
Acetate and other short-chain organic acids induce extreme-acid resistance (31) and the RpoS regulon (6, 61). Since the ackA-pta strain showed elevated expression of the RpoS regulon, we tested whether it also showed constitutive extreme-acid resistance. Log-phase cultures of the ackA-pta mutant showed 13% survival after extreme-acid treatment, compared to 0.012% survival of the parental W3110. The acid resistance of ackA-pta was lowered 25-fold by a crl::cam insertion, which inactivates a gene required for most RpoS-dependent phenomena, although its mode of action is unknown (55). Thus, the ackA-pta strain showed constitutive extreme-acid resistance dependent upon RpoS.
Formate-induced proteins.
Formic acid, like acetic acid, is a membrane-permeant acidic product of fermentation (2, 40, 58). The effect of 20 mM formate on the protein profiles of strain W3110 is shown in Fig. 3A.
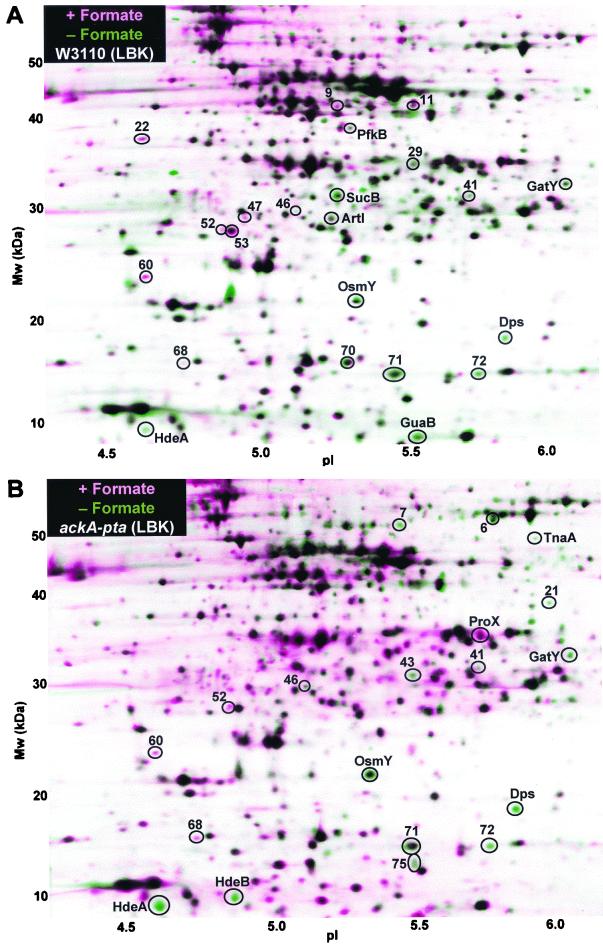
FIG. 3.
Proteins differentially expressed with (pink) or without (green) 20 mM formic acid. (A) Strain W3110; (B) strain W3110 ackA-pta. Other growth and gel conditions were as for Fig. 2.
Only one protein (no. 53) showed positive induction by both acetate and formate. One protein repressed by acetate, TnaA, was also repressed by formate in the ackA-pta strain, consistent with the known repression of TnaA at low pH (9). By contrast, 10 proteins induced by acetate were repressed by formate, including transporters, metabolic enzymes, and members of the RpoS regulon. In the ackA-pta strain, which constitutively elevated 14 acetate stress proteins, 11 of these were repressed by formate (Fig. 3B and Table 1). Thus, the major effect of formate appeared to be opposite to the effect of acetate or the ack-pta deletion.
A small number of proteins responded to formate but not to acetate; for example SucB, a component of 2-ketoglutarate dehydrogenase (5, 35). This component of the TCA cycle is repressed anaerobically, the condition under which formate predominates.
Acetate addition under conditions of low acetate production.
In glycerol minimal medium, acetate production is low, and deletion of ackA-pta does not affect the growth rate (15, 48). The inclusion of acetate during growth of W3110 in glycerol medium caused elevation of only six of the proteins induced by acetate in LBK broth, including GatY and YfiD. Other proteins induced by acetate in LBK broth, including OppA and ProX, were repressed by acetate in glycerol medium. Seven proteins were induced by acetate only in glycerol medium, whereas ProX and MalE were repressed.
The ackA-pta strain produces almost no acetate in glycerol medium. The protein profile of this strain grown on glycerol appeared essentially the same as that of the parent (data not shown). The lack of effect of ackA-pta is consistent with the very low flux through the ackA-pta pathway in wild-type cells in this medium (51).
DISCUSSION
Several previous studies using proteomic or microarray approaches have revealed E. coli proteins induced by membrane-permeant weak acids. 2-D gel experiments show that benzoate induces heat shock and universal stress proteins (45) and that propionate induces AhpC, GatY, ManX, and YfiD (9). Acetate-induced proteins in E. coli O157 probed with an E. coli K-12 microarray include members of the RpoS regulon, as well as various transporters and metabolic enzymes (6). Growth on glucose, which generates high acetate, elevates expression of the RpoS regulon, as well as two other genes encoding proteins we found to be induced by acetate or constitutive in the ackA-pta strain: OmpC and TpiA (70).
Our present study utilized improved proteomic methods and more sensitive N-terminal sequence detection than did previous 2-D gel studies (9). In comparison with microarray studies, 2-D gels can screen only a limited subset of cellular proteins (53, 72), but they also detect modulation of proteins whose predominant regulation appears to be translational, such as OppA (38, 71).
Responses to acetate.
Growth with exogenous acetate increased the expression of several amino acid transporters, as well as the peptide transporter OppA. A possible function of these transporters could be to enhance uptake of alternative energy sources if the cell perceives acetate uptake as a sign of decreasing availability of carbohydrates. Enhanced amino acid catabolism increases the survival of emerging mutants in stationary phase (75). Another possible advantage of ArtI (arginine uptake) and OppA could be to obtain amino acids for decarboxylation, producing alkaline amines; amino acid decarboxylation is one strategy for protection from acidification (67). This finding is consistent with the induction of ArtI and OppA during growth below pH 5 (D. Stancik, L. Stancik, and J. Slonczewski, unpublished data). The induction of OppA by acetate may have pharmaceutical significance, because OppA expression increases sensitivity to aminoglycoside antibiotics (39).
A group of transporters and the components of the RpoS regulon showed similar elevation of expression during growth with acetate and elevated basal levels in the ackA-pta strain. The elevation of the RpoS regulon was consistent with the extreme-acid survival of exponential-phase cultures of the ackA-pta strain; survival below pH 3 normally is highly dependent upon RpoS. Several of the acetate-induced members of the RpoS regulon are known to enhance acid resistance, including the DNA-binding protein Dps (17) and the extreme-acid periplasmic chaperone, HdeA (26).
The ackA-pta strain shows diminished efflux, as well as uptake, of acetate. This perturbation of the fermentation pathways involves substantial increase of acetyl-CoA due to the loss of the major pathway for dumping unneeded acetyl-CoA (15). The increase of acetyl-CoA in the ackA-pta strain may be similar to the elevation of acetyl-CoA caused by growth of the parental strain in medium with acetate. With respect to acetyl-phosphate, however, the two situations are opposed: the wild type accumulates acetyl-phosphate in the presence of excess acetate, whereas the ackA-pta mutant makes no acetyl-phosphate (51). Acetyl-phosphate has been identified as a positive effector of RpoS turnover, through phosphorylation of the proteolysis cofactor, RssB (12). In an ackA-pta mutant, the absence of acetyl-phosphate produces an ≈3-fold elevation in the growth-phase level of RpoS. In wild-type cells grown with added acetate, however, the acetyl-phosphate level increases; this would be expected to increase turnover of RpoS and reduce its steady-state level. Thus, we consider acetyl-phosphate an unlikely mediator for the induction of RpoS by acetate.
It is possible that a common process leads to increased activity of RpoS in the wild type growing in the presence of acetate and in the basal activity level of RpoS in the ackA-pta mutant. This process could explain the acetate induction of the RpoS regulon observed previously (6, 61), as well as our observed induction of several transporters, metabolic enzymes, and unidentified proteins (Tables 1 and 3). A factor that might contribute to acetate induction, requiring further experimentation for confirmation, is the elevated level of acetyl-CoA. Acetyl-CoA might act, for example, by binding to a regulatory protein, analogous to the FadR regulator (20) or the PaaX regulator (23), or it might participate in a protein acetylation reaction, such as the regulatory acetylation of RpoS itself, possibly in a reaction involving the Crl protein (55).
TABLE 3.
Functions of proteins oppositely regulated by acetate and formate
| Functional role | Protein | Expressiona
|
|||
|---|---|---|---|---|---|
| Acetate in W3110 | ackA-pta/W3110 | Formate in W3110 | Formate in ackA-pta | ||
| Arginine uptake | ArtI | + | − | ||
| Cystine uptake | FliY | + | + | ||
| Galactitol catabolism | GatY | + | − | − | |
| Osmotic periplasmic protein | OsmY | + | + | − | − |
| DNA-binding protein | Dps | + | + | − | − |
| Extreme-acid periplasmic chaperone | HdeA | + | + | − | − |
| Extreme-acid periplasmic chaperone | HdeB | + | + | − | |
| Sugar catabolism | PfkB | + | + | − | |
+, increased expression in comparative gel; −, decreased expression in comparative gel. The column headers indicate the comparative gel/the reference gel.
High levels of acetate conversion to acetyl-CoA could also function as a general indicator of high cell density, leading to stationary-phase response. This would be consistent with the induction of LuxS by acetate. Proteins dependent on LuxS appear most strongly in cultures grown with acetate below pH 7 (L. M. Maurer and J. L. Slonczewski, unpublished data). The role of acetate and acetyl-CoA in these observations is being investigated further.
In glycerol minimal medium (Fig. 4B and Table 1), the responses to acetate were very different from those seen in LBK. The elevation of the RpoS regulon and most of the transporters was eliminated; in fact OppA and ProX were repressed. Thus, in glycerol medium, the proteins induced by acetate appear to be mediated by factors other than those acting in complex media. This result may indicate that acetyl-CoA levels remain low during glycerol metabolism, even in the presence of exogenous acetate. The ackA-pta strain grown on glycerol showed a protein profile essentially the same as that of the parent, indicating that absence of the acetate activation system had little effect on protein expression.
Response to formate.
A striking number of proteins induced during growth with acetate, including the RpoS regulon, showed decreased steady-state expression during growth with formate (Table 3). E. coli has several routes for elimination of formate produced by fermentation: direct excretion (10, 40), oxidation of formate by oxygen or other electron acceptors to form CO2 (1, 8), or anaerobic conversion by formate hydrogen lyase (FHL) to CO2 and H2 (40). However, high levels of formate could also drive the reversible Pfl, using up acetyl-CoA to generate pyruvate. If most of this pyruvate is then excreted or converted to lactate, the acetyl-CoA concentration would fall. Pfl activity has generally been thought to require strict anaerobiosis, but recent evidence shows its activity in the presence of limited oxygen (2, 19). If sufficient Pfl activity occurs to deplete acetyl-CoA, this could explain the repression of the TCA cycle, as indicated by the downregulation of a TCA enzyme, 2-ketoglutarate dehydrogenase (SucB), by formate (Table 1). The depletion of acetyl-CoA by formate, and the elevation of acetyl-CoA by acetate or by an ackA-pta defect, could explain the pattern of expression summarized in Table 3; again, the possible role of acetyl-CoA requires further experimentation to confirm.
Expression effects mediated by formate could be involved in the phenomenon of formate protection from antimicrobial peptides (7). The absence of RpoS participation in formate protection is consistent with our finding of lowered steady-state activity of the RpoS regulon in cultures grown with formate.
Our general conclusion is that different acidic fermentation products induce very different genetic and metabolic responses. Few proteins are consistently induced or repressed by both acetate and formate in LBK broth. Protein no. 53 was induced by acetate or by formate. ProX was induced by acetate in the wild type and by formate in the ackA-pta strain; TnaA was repressed under both conditions. Two of the proteins induced by acetate but repressed or unaffected by formate (GatY and YfiD) are known to be induced by propionate (9). YfiD, a homolog of Pfl (29, 50), is also induced during conversion of acetyl-CoA to poly(3-hydroxybutyrate) (32); this would make sense if YfiD is an enzyme that replenishes acetyl-CoA. YfiD is also repressed in gel-entrapped cells (54), a condition whose metabolic state remains poorly characterized. The significance of these responses, and extension to other fermentation products, including lactate and succinate, is being studied further.
ACKNOWLEDGMENTS
This work was supported by grant MCB-9982437 from the National Science Foundation.
We thank David Clark and Alan Wolfe for very helpful discussions. We thank Thomas Silhavy for providing strain LP895.
REFERENCES
- 1.Abaibou H, Pommier J, Benoit S, Giordano G, Mandrand-Berthelot M A. Expression and characterization of the Escherichia coli fdo locus and a possible physiological role for aerobic formate dehydrogenase. J Bacteriol. 1995;177:7141–7149. doi: 10.1128/jb.177.24.7141-7149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexeeva S, de Kort B, Sawers G, Hellingwerf K J, de Mattos M J. Effects of limited aeration and of the ArcAB system on intermediary pyruvate catabolism in Escherichia coli. J Bacteriol. 2000;182:4934–4940. doi: 10.1128/jb.182.17.4934-4940.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almirón M, Link A J, Furlong D, Kolter R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992;6:2646–2654. doi: 10.1101/gad.6.12b.2646. [DOI] [PubMed] [Google Scholar]
- 4.Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and ςs in stationary phase. Mol Microbiol. 1994;13:265–272. doi: 10.1111/j.1365-2958.1994.tb00421.x. [DOI] [PubMed] [Google Scholar]
- 5.Amarasingham C R, Davis B D. Regulation of α-ketoglutarate dehydrogenase formation in Escherichia coli. J Biol Chem. 1965;240:3664–3668. [PubMed] [Google Scholar]
- 6.Arnold C N, McElhanon J, Lee A, Leonhart R, Siegele D A. Global analysis of Escherichia coli gene expression during the acetate-induced acid tolerance response. J Bacteriol. 2001;183:2178–2186. doi: 10.1128/JB.183.7.2178-2186.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker H C, Kinsella N, Jaspe A, Friedrich T, O'Connor C D. Formate protects stationary-phase Escherichia coli and Salmonella cells from killing by a cationic antimicrobial peptide. Mol Microbiol. 2000;35:1518–1529. doi: 10.1046/j.1365-2958.2000.01820.x. [DOI] [PubMed] [Google Scholar]
- 8.Benoit S, Abaibou H, Mandrand-Berthelot M A. Topological analysis of the aerobic membrane-bound formate dehydrogenase of Escherichia coli. J Bacteriol. 1998;180:6625–6634. doi: 10.1128/jb.180.24.6625-6634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blankenhorn D, Phillips J, Slonczewski J L. Acid- and base-induced proteins during aerobic and anaerobic growth of Escherichia coli revealed by two-dimensional gel electrophoresis. J Bacteriol. 1999;181:2209–2216. doi: 10.1128/jb.181.7.2209-2216.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Böck A, Sawers G. Fermentation. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 262–282. [Google Scholar]
- 11.Bohannon D E, Connell N, Keener J, Tormo A, Espinosa-Urgel M, Zambrano M M, Kolter R. Stationary-phase-inducible “gearbox” promoters: differential effects of katF mutations and role of ς70. J Bacteriol. 1991;173:4482–4492. doi: 10.1128/jb.173.14.4482-4492.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouché S, Klauck E, Fischer D, Lucassen M, Jung K, Hengge-Aronis R. Regulation of RssB-dependent proteolysis in Escherichia coli: a role for acetyl phosphate in a response regulator-controlled process. Mol Microbiol. 1998;27:787–795. doi: 10.1046/j.1365-2958.1998.00725.x. [DOI] [PubMed] [Google Scholar]
- 13.Brown T D, Jones-Mortimer M C, Kornberg H L. The enzymic interconversion of acetate and acetyl-coenzyme A in Escherichia coli. J Gen Microbiol. 1977;102:327–336. doi: 10.1099/00221287-102-2-327. [DOI] [PubMed] [Google Scholar]
- 14.Bunch P K, Mat-Jan F, Lee N, Clark D P. The IdhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology. 1997;143:187–195. doi: 10.1099/00221287-143-1-187. [DOI] [PubMed] [Google Scholar]
- 15.Chang D E, Shin S, Rhee J S, Pan J G. Acetate metabolism in a pta mutant of Escherichia coli W3110: importance of maintaining acetyl coenzyme A flux for growth and survival. J Bacteriol. 1999;181:6656–6663. doi: 10.1128/jb.181.21.6656-6663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang Y-Y, Wang A-Y, Cronan J E., Jr Expression of Escherichia coli pyruvate oxidase (PoxB) depends on the sigma factor encoded by the rpoS (katF) gene. Mol Microbiol. 1994;11:1019–1028. doi: 10.1111/j.1365-2958.1994.tb00380.x. [DOI] [PubMed] [Google Scholar]
- 17.Choi S H, Baumler D J, Kaspar C W. Contribution of dps to acid stress tolerance and oxidative stress tolerance in Escherichia coli O157:H7. Appl Environ Microbiol. 2000;66:3911–3916. doi: 10.1128/aem.66.9.3911-3916.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummings J H, Pomare E W, Branch W J, Naylorr C P E, MacFarlane G T. Short chain fatty acids in human large intestine, portal, hepatic, and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Graef M R, Alexeeva S, Snoep J L, Teixeira de Mattos M J. The steady-state internal redox state (NADH/NAD) reflects the external redox state and is correlated with catabolic adaptation in Escherichia coli. J Bacteriol. 1999;181:2351–2357. doi: 10.1128/jb.181.8.2351-2357.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiRusso C C, Tsvetnitsky V, Hojrup P, Knudsen J. Fatty acyl-CoA binding domain of the transcription factor FadR. Characterization by deletion, affinity labeling, and isothermal titration calorimetry. J Biol Chem. 1998;273:33652–33659. doi: 10.1074/jbc.273.50.33652. [DOI] [PubMed] [Google Scholar]
- 21.el-Mansi E M, Holms W H. Control of carbon flux to acetate excretion during growth of Escherichia coli in batch and continuous cultures. J Gen Microbiol. 1989;135:2875–2883. doi: 10.1099/00221287-135-11-2875. [DOI] [PubMed] [Google Scholar]
- 22.Enfors S O, Jahic M, Rozkov A, Xu B, Hecker M, Jurgen B, Kruger E, Schweder T, Hamer G, O'Beirne D, Noisommit-Rizzi N, Reuss M, Boone L, Hewitt C, McFarlane C, Nienow A, Kovacs T, Tragardh C, Fuchs L, Revstedt J, Friberg P C, Hjertager B, Blomsten G, Skogman H, Hjort S, Hoeks F, Lin H Y, Neubauer P, van der Lans R, Luyben K, Vrabel P, Manelius A. Physiological responses to mixing in large scale bioreactors. J Biotechnol. 2001;85:175–195. doi: 10.1016/s0168-1656(00)00365-5. [DOI] [PubMed] [Google Scholar]
- 23.Ferrández A, García J L, Díaz E. J. Biol. Chem. 275:12214–12222. 2000. Transcriptional regulation of the divergent paa catabolic operons for phenylacetic acid degradation in Escherichia coli. [DOI] [PubMed] [Google Scholar]
- 24.Foster J W. Microbial responses to acid stress. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 99–115. [Google Scholar]
- 25.Foster J W, Hall H K. Adaptive acidification tolerance response of Salmonella typhimurium. J Bacteriol. 1990;172:771–778. doi: 10.1128/jb.172.2.771-778.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gajiwala K S, Burley S K. HdeA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J Mol Biol. 2000;295:605–612. doi: 10.1006/jmbi.1999.3347. [DOI] [PubMed] [Google Scholar]
- 27.Goodell E W, Higgins C F. Uptake of cell wall peptides by Salmonella typhimurium and Escherichia coli. J Bacteriol. 1987;169:3861–3865. doi: 10.1128/jb.169.8.3861-3865.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gooley A A, Packer N H. The importance of protein co- and post-translational modifications in proteome projects. In: Wilkins M R, Williams K L, Appel R D, Hochstrasser D F, editors. Proteome research: new frontiers in functional genomics. Berlin, Germany: Springer; 1997. pp. 63–91. [Google Scholar]
- 29.Green J, Baldwin M L, Richardson J. Downregulation of Escherichia coli yfiD expression by FNR occupying a site at −93.5 involves the AR1-containing face of FNR. Mol Microbiol. 1998;29:1113–1123. doi: 10.1046/j.1365-2958.1998.01002.x. [DOI] [PubMed] [Google Scholar]
- 30.Guest J R, Angier S J, Russell G C. Structure, expression, and protein engineering of the pyruvate dehydrogenase complex of Escherichia coli. Ann N Y Acad Sci. 1989;573:76–99. doi: 10.1111/j.1749-6632.1989.tb14988.x. [DOI] [PubMed] [Google Scholar]
- 31.Guilfoyle D E, Hirshfield I N. The survival benefit of short-chain organic acids and the inducible arginine and lysine decarboxylase genes for Escherichia coli. Lett Appl Microbiol. 1996;22:393–396. doi: 10.1111/j.1472-765x.1996.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 32.Han M J, Yoon S S, Lee S Y. Proteome analysis of metabolically engineered Escherichia coli producing poly(3-hydroxybutyrate) J Bacteriol. 2001;183:301–308. doi: 10.1128/JB.183.1.301-308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hengge-Aronis R. The general stress response in Escherichia coli. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 1996. pp. 161–178. [Google Scholar]
- 34.Ishikawa T, Hama H, Tsuda M, Tsuchiya T. Isolation and properties of a mutant of Escherichia coli possessing defective Na+/H+ antiporter. J Biol Chem. 1987;262:7443–7446. [PubMed] [Google Scholar]
- 35.Iuchi S, Lin E C. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc Natl Acad Sci USA. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kakuda H, Hosono K, Shiroishi K, Ichihara S. Identification and characterization of the ackA (acetate kinase A)-pta (phosphotransacetylase) operon and complementation analysis of acetate utilization by an ackA-pta deletion mutant of Escherichia coli. J Biochem. 1994;116:916–922. doi: 10.1093/oxfordjournals.jbchem.a124616. [DOI] [PubMed] [Google Scholar]
- 37.Karpel R, Alon T, Glaser G, Schuldiner S, Padan E. Expression of a sodium proton antiporter (NhaA) in Escherichia coli is induced by Na+ and Li+ ions. J Biol Chem. 1991;266:21753–21759. [PubMed] [Google Scholar]
- 38.Kashiwagi K, Yamaguchi Y, Sakai Y, Kobayashi H, Igarashi K. Identification of the polyamine-induced protein as a periplasmic oligopeptide binding protein. J Biol Chem. 1990;265:8387–8391. [PubMed] [Google Scholar]
- 39.Kashiwagi K, Miyaji A, Ikeda S, Tobe T, Sasakawa C, Igarashi K. Increase of sensitivity to aminoglycoside antibiotics by polyamine-induced protein (oligopeptide-binding protein) in Escherichia coli. J Bacteriol. 1992;174:4331–4337. doi: 10.1128/jb.174.13.4331-4337.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kessler D, Knappe J. Anaerobic dissimilation of pyruvate. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 199–204. [Google Scholar]
- 41.Kihara M, Macnab R M. Cytoplasmic pH mediates pH taxis and weak-acid repellent taxis of bacteria. J Bacteriol. 1981;145:1209–1221. doi: 10.1128/jb.145.3.1209-1221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knappe J, Blaschkowski H P, Grobner P, Schmitt T. Pyruvate formate-lyase of Escherichia coli: the acetyl-enzyme intermediate. Eur J Biochem. 1974;50:253–263. doi: 10.1111/j.1432-1033.1974.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 43.Kumari S, Beatty C M, Browning D F, Busby S J W, Simel E J, Hovel-Miner G, Wolfe A J. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 2000;182:4173–4179. doi: 10.1128/jb.182.15.4173-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumari S, Tishel R, Eisenbach M, Wolfe A J. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lambert L A, Abshire K, Blankenhorn D, Slonczewski J L. Proteins induced in Escherichia coli by benzoic acid. J Bacteriol. 1997;179:7595–7599. doi: 10.1128/jb.179.23.7595-7599.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 47.Lee I S, Slonczewski J L, Foster J W. A low-pH-inducible, stationary-phase acid tolerance response in Salmonella typhimurium. J Bacteriol. 1994;176:1422–1426. doi: 10.1128/jb.176.5.1422-1426.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LeVine S M, Ardeshir F, Ames G F. Isolation and characterization of acetate kinase and phosphotransacetylase mutants of Escherichia coli and Salmonella typhimurium. J Bacteriol. 1980;143:1081–1085. doi: 10.1128/jb.143.2.1081-1085.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin J, Smith M P, Chapin K C, Baik H S, Bennett G N, Foster J W. Mechanisms of acid resistance in enterohemorrhagic Escherichia coli. Appl Environ Microbiol. 1996;62:3094–3100. doi: 10.1128/aem.62.9.3094-3100.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marshall F A, Messenger S L, Wyborn N R, Guest J R, Wing H, Busby S J, Green J. A novel promoter architecture for microaerobic activation by the anaerobic transcription factor FNR. Mol Microbiol. 2001;39:747–753. doi: 10.1046/j.1365-2958.2001.02262.x. [DOI] [PubMed] [Google Scholar]
- 51.McCleary W R, Stock J B. Acetyl phosphate and the activation of two-component response regulators. J Biol Chem. 1994;269:31567–31572. [PubMed] [Google Scholar]
- 52.Miller V L, Taylor R K, Mekalanos J J. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell. 1987;48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 53.Neidhardt F C, VanBogelen R A. Proteomic analysis of bacterial stress responses. In: Storz G, Hengge-Aronis R, editors. Bacterial stress responses. Washington, D.C.: ASM Press; 2000. pp. 445–452. [Google Scholar]
- 54.Perrot F, Hebraud M, Charlionet R, Junter G A, Jouenne T. Protein patterns of gel-entrapped Escherichia coli cells differ from those of free-floating organisms. Electrophoresis. 2000;21:645–653. doi: 10.1002/(SICI)1522-2683(20000201)21:3<645::AID-ELPS645>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 55.Pratt L A, Silhavy T J. Crl stimulates RpoS activity during stationary phase. Mol Microbiol. 1998;29:1225–1236. doi: 10.1046/j.1365-2958.1998.01007.x. [DOI] [PubMed] [Google Scholar]
- 56.Pruss B M, Wolfe A J. Regulation of acetyl phosphate synthesis and degradation, and the control of flagellar expression in Escherichia coli. Mol Microbiol. 1994;12:973–984. doi: 10.1111/j.1365-2958.1994.tb01085.x. [DOI] [PubMed] [Google Scholar]
- 57.Roe A J, McLaggan D, Davidson I, O'Byrne C, Booth I R. Perturbation of anion balance during inhibition of growth of Escherichia coli by weak acids. J Bacteriol. 1998;180:767–772. doi: 10.1128/jb.180.4.767-772.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rossman R, Sawers G, Bock A. Mechanism of regulation of the formate-hydrogenlyase pathway by oxygen, nitrate, and pH: definition of the formate regulon. Mol Microbiol. 1991;5:2807–2814. doi: 10.1111/j.1365-2958.1991.tb01989.x. [DOI] [PubMed] [Google Scholar]
- 59.Russell J B, Diez-Gonzalez F. The effects of fermentation acids on bacterial growth. Adv Microb Physiol. 1998;39:205–234. doi: 10.1016/s0065-2911(08)60017-x. [DOI] [PubMed] [Google Scholar]
- 60.Sawers G, Böck A. Anaerobic regulation of pyruvate formate-lyase from Escherichia coli K-12. J Bacteriol. 1988;170:5330–5336. doi: 10.1128/jb.170.11.5330-5336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schellhorn H E, Stones V L. Regulation of katF and katE in Escherichia coli K-12 by weak acids. J Bacteriol. 1992;174:4769–4776. doi: 10.1128/jb.174.14.4769-4776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Slonczewski J L, Foster J W. pH-regulated genes and survival at extreme pH. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1539–1552. [Google Scholar]
- 63.Slonczewski, J. L., and C. Kirkpatrick. Proteomic analysis of Escherichia coli and Helicobacter pylori, using two-dimensional gel electrophoresis. Methods Enzymol., in press. [DOI] [PubMed]
- 64.Slonczewski J L, Gonzalez T N, Bartholomew F M, Holt N J. Mu d-directed lacZ fusions regulated by low pH in Escherichia coli. J Bacteriol. 1987;169:3001–3006. doi: 10.1128/jb.169.7.3001-3006.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Small P, Blankenhorn D, Welty D, Zinser E, Slonczewski J L. Acid and base resistance in Escherichia coli and Shigella flexneri: role of rpoS and growth pH. J Bacteriol. 1994;176:1729–1737. doi: 10.1128/jb.176.6.1729-1737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith M W, Neidhardt F C. Proteins induced by anaerobiosis in Escherichia coli. J Bacteriol. 1983;154:336–343. doi: 10.1128/jb.154.1.336-343.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stim K P, Bennett G N. Nucleotide sequence of the adi gene, which encodes the biodegradative acid-induced arginine decarboxylase of Escherichia coli. J Bacteriol. 1993;175:1221–1234. doi: 10.1128/jb.175.5.1221-1234.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Suppmann B, Sawers G. Isolation and characterization of hypophosphite-resistant mutants of Escherichia coli: identification of the FocA protein, encoded by the pfl operon, as a putative formate transporter. Mol Microbiol. 1994;11:965–982. doi: 10.1111/j.1365-2958.1994.tb00375.x. [DOI] [PubMed] [Google Scholar]
- 69.Surette M G, Miller M B, Bassler B L. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci USA. 1999;96:1639–1644. doi: 10.1073/pnas.96.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tao H, Bausch C, Richmond C, Blattner F R, Conway T. Functional genomics: expression analysis of Escherichia coli growing on minimal and rich media. J Bacteriol. 1999;181:6425–6440. doi: 10.1128/jb.181.20.6425-6440.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Urbanowski M L, Stauffer L T, Stauffer G V. The gcvB gene encodes a small untranslated RNA involved in expression of the dipeptide and oligopeptide transport systems in Escherichia coli. Mol Microbiol. 2000;37:856–868. doi: 10.1046/j.1365-2958.2000.02051.x. [DOI] [PubMed] [Google Scholar]
- 72.VanBogelen R A, Abshire K Z, Pertsemlidis A, Clark R L, Neidhardt F C. Gene-protein database of Escherichia coli K-12, edition 6. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2067–2117. [Google Scholar]
- 73.Wagner A F, Frey M, Neugebauer F A, Schafer W, Knappe J. The free radical in pyruvate formate-lyase is located on glycine-734. Proc Natl Acad Sci USA. 1992;89:996–1000. doi: 10.1073/pnas.89.3.996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Waterman S R, Small P L. Identification of ςS-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol Microbiol. 1996;21:925–940. doi: 10.1046/j.1365-2958.1996.00058.x. [DOI] [PubMed] [Google Scholar]
- 75.Zinser E R, Kolter R. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J Bacteriol. 1999;181:5800–5807. doi: 10.1128/jb.181.18.5800-5807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]