Abstract
Introduction
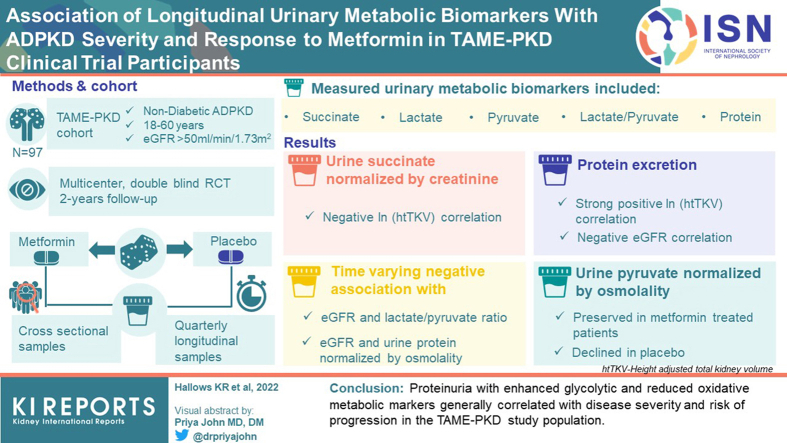
Dysregulated cellular metabolism contributes to autosomal dominant polycystic kidney disease (ADPKD) pathogenesis. The Trial of Administration of Metformin in Polycystic Kidney Disease (TAME-PKD) tested the effects of metformin treatment over 2 years in adult ADPKD patients with mild-moderate disease severity. Metformin was found to be safe and tolerable with an insignificant trend toward reduced estimated glomerular filtration rate (eGFR) decline compared to placebo. Here we tested whether targeted urinary metabolic biomarkers measured in TAME-PKD participants correlated with disease progression, severity, and metformin treatment in cross-sectional and longitudinal analyses.
Methods
Concentrations of total protein, targeted metabolites (lactate, pyruvate, and succinate), and glycolytic enzymes (pyruvate kinase-M2, lactate dehydrogenase-A, and pyruvate dehydrogenase kinase-1) were measured and normalized by creatinine or osmolality in urine specimens and compared with height-adjusted total kidney volume (htTKV) and eGFR at the different study timepoints.
Results
In cross-sectional analyses utilizing placebo group data, urinary succinate normalized by creatinine negatively correlated with ln (htTKV), whereas protein excretion strongly positively correlated with ln (htTKV), and negatively correlated with eGFR. Significant time-varying negative associations occurred with eGFR and the lactate/pyruvate ratio and with urine protein normalized by osmolality, indicating correlations of these biomarkers with disease progression. In secondary analyses, urinary pyruvate normalized by osmolality was preserved in metformin-treated participants but declined in placebo over the 2-year study period with a significant between-arm difference, suggesting time-dependent urinary pyruvate changes may serve as a discriminator for metformin treatment effects in this study population.
Conclusion
Proteinuria with enhanced glycolytic and reduced oxidative metabolic markers generally correlated with disease severity and risk of progression in the TAME-PKD study population.
Keywords: ADPKD, biomarkers, eGFR, metformin, proteinuria
Graphical abstract
ADPKD, the most common genetic kidney disease affecting approximately 1 in 700 individuals, is caused by mutations in the PKD1 or PKD2 genes encoding polycystin 1 and polycystin 2. ADPKD is characterized by a gradual development and enlargement of cysts and overall kidney size that eventually results in kidney failure in half of all patients by the end of the sixth decade of life.1 The only current US Food and Drug Administration-approved ADPKD therapy is tolvaptan,2 which has been shown to slow total kidney volume growth and kidney function (measured by eGFR) decline. Moreover, there are limitations in the tolerability of tolvaptan because of polyuria and potential hepatotoxicity that necessitates close monitoring of liver function tests.3 Of note, as many cellular signaling pathways are dysregulated in ADPKD,4 there is an expanding list of experimental drugs and therapeutic targets under various stages of investigation in preclinical and clinical studies for ADPKD.5
Although critical details of ADPKD pathogenesis remain unclear,4 there has been growing evidence and recognition based on results from a pioneering 2013 study by Rowe et al. that dysregulated metabolism plays a key role.6 Specifically, this study identified a Warburg effect-like shift to excessive aerobic glycolysis, as evidenced by increased levels of certain key glycolytic enzymes (e.g., lactate dehydrogenase-A [LDHA], pyruvate dehydrogenase kinase 1, and the pyruvate kinase M2 isoform), occurs in ADPKD cystic epithelial cells as compared with normal kidney epithelial cells in ADPKD mouse models and in patient kidney tissue.6 These metabolic derangements may contribute to cyst formation and expansion, impaired fatty acid oxidation, and reduced adenosine monophosphate-activated protein kinase activity.6, 7, 8 Indeed, we recently reported that urinary excretion of pyruvate kinase M2 isoform and LDHA correlated with ADPKD severity (greater htTKV or lower eGFR) in baseline urine samples from patients enrolled in the TAME-PKD study, thus providing evidence of upregulated glycolytic flux as a feature of disease severity.9
In preclinical studies, the AMP-activated protein kinase activator, metformin,10 was found to inhibit ADPKD cystic growth and cell proliferation in vitro and improve various disease parameters in both mice models11,12 and a miniature pig model13 of ADPKD. Metformin-induced cellular AMP-activated protein kinase activation may confer beneficial effects in the treatment of diseases such as the metabolic syndrome, diabetes, and polycystic ovary syndrome,14 along with additional pleiotropic effects of metformin that are adenosine monophosphate-activated protein kinase-independent.15 The recently completed TAME-PKD study was a multicenter phase 2 double-blinded, randomized controlled clinical trial whose primary objectives were to test the safety, tolerability, and preliminary efficacy of metformin relative to placebo in adult ADPKD patients with eGFR ≥50 ml/min per 1.73 m2 over a 2-year treatment period. Metformin was found to be safe and tolerable with an insignificant trend toward reduction in eGFR decline as compared with placebo.16
Ongoing and future clinical trials evaluating the efficacy of metformin will require larger enrollment and potentially enrichment for patients and those more at risk for rapid ADPKD progression. There is considerable variability in ADPKD disease severity among affected individuals, even among related patients with the same mutation.17 Therefore, identification of patients at high risk for disease progression is critical for prognostic reasons and for determining those most likely to benefit from new therapies and participation in clinical trials. Current methods to evaluate disease severity and predict progression include measurements of htTKV relative to the patient’s age and type of mutation,1,18 but these assessments may be expensive and not readily accessible. Recently, there has been significant interest in defining noninvasive biomarkers that associate with disease severity and progression along with potential clinical responses to new ADPKD therapies.19
The primary goals of this study were to evaluate, in longitudinal urine samples from TAME-PKD study participants, metabolic biomarkers that may correlate with ADPKD disease severity in cross-sectional (between-subjects) analyses, predict response to metformin, and be impacted by metformin use. We analyzed changes in urinary biomarker levels over time for each participant in longitudinal (within-subjects) analyses and correlated these with metformin treatment status and disease severity or progression over time, as measured by concurrent htTKV and eGFR measurements. In addition, secondary analyses were performed to assess potential differences in the treatment effect on the trajectories of each biomarker over the course of the study.
Methods
Urine Specimen Collection and Preparation
As previously reported,9,16,20,21 this study was conducted ethically in accordance with the World Medical Association Declaration of Helsinki. All study subjects have given their written informed consent, and the TAME-PKD study protocol was approved by the Institutional Review Boards at each of the study sites. Eligible patients in the TAME-PKD study population included nondiabetic adults (N = 97) aged from 18 years to 60 years, with eGFR ≥50 ml/min per 1.73 m2 and ADPKD.20 Although all baseline urine specimens were collected in the morning after overnight fasting, most of the later longitudinal samples (87.7%) were nonfasting and collected later in the day, as necessitated by constraints of patient scheduling and convenience. Clean catch spot urine samples were obtained by standard methods in sterile containers and then processed into 1.8 ml aliquots before storing within 3 hours at −80 ˚C. They were then shipped on dry ice to the University of Southern California for biomarker analyses and additional aliquots were kept at a Biorepository at the University of Maryland for longterm storage. In preparation for analyses, samples were thawed and centrifuged at 1000 x g for 20 minutes at 4 °C. The supernatant was then used to perform biomarker assays.
Urinary Biomarker Analyses
For each longitudinal urine sample, measurements of urine osmolality and the concentrations of creatinine, total protein, key metabolites (lactate, pyruvate, and succinate), and the candidate glycolytic enzymes pyruvate kinase M2 isoform, LDHA, and pyruvate dehydrogenase kinase 1 were performed essentially as described previously in detail.9 Briefly, all enzymatic assay measurements were fit to standard curves at appropriate dilutions so that the levels fell within a linear range of the standard fit curve. The measured absorbance or fluorescence was used to calculate the concentration of the biomarker in the urine samples by subtracting the sample background control readings from the sample readings and then comparing the sample signals to those of a standard curve. The mean value of replicate measurements for each sample was normalized to the urine creatinine and to the urine osmolality measured for the same sample.
The following summarizes how each of the analytes were quantitated in the specimens from each patient at the different time points as described.9 Creatinine, lactate, pyruvate, and succinate were measured from urine samples using colorimetric or fluorometric enzymatic assay kits. Urinary pyruvate kinase M was measured using an enzyme-linked immunosorbent assay kit with a colorimetric readout. Urine osmolality of each sample was measured with a vapor pressure osmometer. Total urinary protein was isolated by the methanol-chloroform precipitation method and then quantitated by dilution and absorbance using the Bradford technique.22 LDHA and pyruvate dehydrogenase kinase 1 standards from normal control urine and patient samples were loaded into individual wells, separated by sodium dodecyl-sulfate polyacrylamide gel electrophoresis, and then transferred to a nitrocellulose membrane. As described previously,9 the membrane was blocked in blocking buffer and first stained for total protein, imaged, and then destained and reblocked. Membranes were then subjected to immunoblotting for LDHA and pyruvate dehydrogenase kinase 1, whose protein levels were detected and quantified by densitometry of the relevant bands in comparison to densitometric quantitation of standards run on the same gel. For immunoblots of patient samples, all available longitudinal patient samples were run on the same gel to allow more reliable direct comparisons in the longitudinal analyses.
Statistical Analyses
Descriptive statistics are presented as mean ± SD or median (range) for continuous variables and frequency (percentage) for categorical variables. Because of the skewed nature of some outcomes, natural log transformations were applied before analyses. Our primary objective was to identify both prognostic predictive biomarkers of htTKV and eGFR as well as assess the impact of metformin on longitudinal biomarkers.
Prognostic Analysis
The analysis for prognostic biomarkers was restricted to participants taking placebo and utilized a Laird and Ware linear mixed model23 for each of htTKV and eGFR with the following predictors: time (months since baseline visit), a time-dependent covariate for the biomarker of interest, clinical site, and random effects for the intercept and slope. In addition, we reparametrized the model such that cross-sectional (between-subjects) and longitudinal (within-subjects) effects on htTKV and eGFR trajectories could be estimated and compared.
Predictive Analysis
The analysis for predictive biomarkers utilized the same Laird and Ware mixed model for each of htTKV and eGFR with the following predictors: time, time-by-study arm interaction, baseline biomarker of interest, time-by-biomarker interaction, and the 3-way interaction of these predictors, and clinical site. A significant 3-way interaction denoted subgroups of participants who would benefit differently from metformin therapy.
Secondary Analysis
Secondary analyses were performed to assess the impact of metformin treatment on the trajectories of each normalized biomarker over the course of the study. Similar to the primary results of the TAME-PKD trial, the 24-month mean change for each biomarker was compared between the metformin and placebo arms using the Laird and Ware linear mixed model.23 Natural log-transformed outcomes were modeled as a function of time, the interaction between time and the study arm, and clinical site. If the modeling allowed, the intercept and slope were allowed to vary. A significant interaction between time and the study arm indicated an impact on biomarker trajectories because of metformin. Statistical analyses were performed using SAS version 9.4 (SAS Institute, Cary, NC). P-values less than 0.05 were considered statistically significant.
Results
Study Overview
Urinary biomarker specimens from the 97 subjects enrolled in the study were collected at baseline (0), 1, 3, 6, 9, 12, 15, 18, 21, and 24 months after randomization and treatment initiation on the study drug (metformin or placebo). A total of 826 specimens were obtained and analyzed (85% of the maximum expected of 970) (see Supplementary Table S1 and Supplementary Figure S1). This shortfall primarily resulted from missed in-person patient visits, most commonly because of COVID-19 precautions during the pandemic and/or failure to collect urine sample at a study visit, and patient dropout from the study (∼15%). The demographic and clinical characteristics of the study population at baseline have been described previously9 and are presented along with the baseline biomarker values, summarized by randomized arms, in Supplementary Table S2.
Prognostic Analyses
None of the time-varying urinary metabolic biomarkers normalized to either urinary creatinine or osmolality significantly correlated with changes in ln(htTKV). However, urinary creatinine itself did have a significant positive correlation with ln(htTKV) over time (Table 1). Both urinary creatinine and osmolality also had significant negative correlations with eGFR over time (Table 2). The relevance of these changes is unclear because these are markers of urinary concentration, not total excretion or glomerular filtration rate. Importantly, there were also significant time-varying negative correlations of the urinary lactate/pyruvate ratio and total protein normalized by osmolality with eGFR (Table 2). These latter findings suggest that to the extent that eGFR changed over time among participants in this study, both the lactate/pyruvate ratio and the total protein excretion normalized by urinary osmolality (Uosm) tended to move in the opposite direction (i.e., with a decline in eGFR, there was an associated increase in these parameters in the study population). In the case of proteinuria, our findings confirm previous reports in other study populations that proteinuria serves as an important negative prognostic biomarker for ADPKD disease progression.24,25 Of note, further analysis of urine protein staining from aliquots of participants’ baseline urine specimens subjected to sodium dodecyl-sulfate polyacrylamide gel electrophoresis and transfer to nitrocellulose membrane reveal that the most abundant proteins present in most participants’ urine samples were albumin and uromodulin (Supplementary Figure S2). However, overall and relative protein expression levels of these 2 major proteins were found to be highly variable across the different study participants. In general, there appeared to be relatively less low-molecular weight proteinuria in these polycystic kidney disease patient samples.
Table 1.
Association of time-varying urinary biomarkers with ln (height-adjusted total kidney volume)
| Biomarker | % Change (95% CI) | P-value |
|---|---|---|
| Creatinine (mg/dl) | 0.038 (0.006, 0.070) | 0.0200a |
| Osmolality (mmol/kg) | 0.004 (−0.003, 0.011) | 0.2445 |
| Lactate (μM)/Ucrea | −0.295 (−1.422, 0.844) | 0.6071 |
| Lactate (μM)/Uosm | 1.999 (−18.289, 27.326) | 0.8600 |
| Lactate/Pyruvate | −0.181 (−0.896, 0.539) | 0.6184 |
| Succinate (μM)/Ucrea | −0.728 (−1.854, 0.412) | 0.2077 |
| Succinate (μM)/Uosm | −6.150 (−21.727, 12.526) | 0.4899 |
| Pyruvate (μM)/Ucrea | −2.975 (−7.869, 2.179) | 0.2501 |
| Pyruvate (μM)/Uosm | −20.016 (−68.056, 100.268) | 0.6308 |
| Total Protein (mg/dl)/Ucrea | −0.743 (−4.059, 2.688) | 0.6645 |
| Total Protein (mg/dl)/Uosm | −3.597 (−45.338, 70.017) | 0.8984 |
| PKM2 (ng/ml)/Ucrea | −0.729 (−3.975, 2.626) | 0.6635 |
| PKM2 (ng/ml)/Uosm | −5.146 (−40.031, 50.030) | 0.8198 |
| LDHA (ng/ml)/Ucrea | −1.250 (−12.983, 12.065) | 0.8442 |
| LDHA (ng/ml)/Uosm | −31.609 (−83.760, 188.012) | 0.6017 |
| PDK1 (pg/ml)/Ucrea | −0.019 (−0.148, 0.111) | 0.7747 |
| PDK1 (pg/ml)/Uosm | −0.076 (−2.989, 2.923) | 0.9593 |
CI, confidence interval; LDHA, lactate dehydrogenase-A; PDK1, pyruvate dehydrogenase kinase 1; PKM2, pyruvate kinase M2; Ucrea, urinary creatinine concentration; Uosm, urinary osmolality.
Significant P values.
Table 2.
Association of time-varying urinary biomarkers with eGFR
| Biomarker | Mean Change (95% CI) | P-value |
|---|---|---|
| Creatinine (mg/dl) | −0.056 (−0.076, −0.036) | <0.0001a |
| Osmolality (mmol/kg) | −0.005 (−0.009, −0.001) | 0.0147a |
| Lactate (μM)/Ucrea | −0.270 (−0.692, 0.152) | 0.2091 |
| Lactate (μM)/Uosm | −8.364 (−17.507, 0.780) | 0.0729 |
| Lactate/Pyruvate | −0.407 (−0.794, −0.021) | 0.0388a |
| Succinate (μM)/Ucrea | 0.384 (−0.234, 1.003) | 0.2223 |
| Succinate (μM)/Uosm | −3.553 (−13.278, 6.172) | 0.4729 |
| Pyruvate (μM)/Ucrea | 1.870 (−0.242, 3.982) | 0.0825 |
| Pyruvate (μM)/Uosm | 0.805 (−30.386, 31.996) | 0.9595 |
| Total Protein (mg/dl)/Ucrea | −0.140 (−2.710, 2.431) | 0.9150 |
| Total Protein (mg/dl)/Uosm | −54.083 (−98.638, −9.528) | 0.0175a |
| PKM2 (ng/ml)/Ucrea | 1.881 (−0.568, 4.330) | 0.1318 |
| PKM2 (ng/ml)/Uosm | −10.697 (−41.326, 19.933) | 0.4927 |
| LDHA (ng/ml)/Ucrea | 0.185 (−8.768, 9.138) | 0.9676 |
| LDHA (ng/ml)/Uosm | −84.184 (−171.014, 2.645) | 0.0574 |
| PDK1 (pg/ml)/Ucrea | 0.042 (−0.061, 0.145) | 0.4231 |
| PDK1 (pg/ml)/Uosm | −0.196 (−2.123, 1.731) | 0.8415 |
CI, confidence interval; eGFR, estimated glomerular filtration rate; LDHA, lactate dehydrogenase-A; PDK1, pyruvate dehydrogenase kinase 1; PKM2, pyruvate kinase M2; Ucrea, urinary creatinine concentration; Uosm, urinary osmolality.
Significant P values.
We also analyzed and compared cross-sectional (between-subjects) and longitudinal (within-subjects) correlations of the various biomarkers with disease severity as assessed by htTKV and eGFR among all samples from study participants in the placebo group. For several biomarkers, there were significant differences between the cross-sectional and longitudinal effects. Specifically, in cross-sectional analyses, urinary succinate excretion normalized by urinary creatinine concentration (succinate/UCrea) negatively correlated with ln(htTKV) (P = 0.008), whereas total protein excretion positively correlated with ln(htTKV) when normalized either by Ucrea (P = 0.0024) or by Uosm (P = 0.0007) (Table 3). Conversely, there was a strong negative correlation of total protein excretion when normalized by either Ucrea (P = 0.0009) or Uosm (P < 0.0001) with eGFR in the cross-sectional analysis (Table 4).
Table 3.
Cross-sectional and longitudinal associations of urinary biomarkers with ln (height-adjusted total kidney volume)
| Biomarker | Difference P-valuea |
% Change (95% CI) | P-value |
|---|---|---|---|
| Creatinine (mg/dl) | 0.1380 | ||
| Cross-sectional effect | 0.360 (−0.071, 0.793) | 0.0997 | |
| Longitudinal effect | 0.038 (0.005, 0.071) | 0.0246b | |
| Osmolality (mmol/kg) | 0.3981 | ||
| Cross-sectional effect | 0.045 (−0.051, 0.140) | 0.3540 | |
| Longitudinal effect | 0.004 (−0.003, 0.011) | 0.2394 | |
| Lactate (μM)/Ucrea | 0.5796 | ||
| Cross-sectional effect | −13.386 (−47.942, 44.110) | 0.5722 | |
| Longitudinal effect | -0.285 (−1.432, 0.876) | 0.6264 | |
| Lactate (μM)/Uosm | 0.2842 | ||
| Cross-sectional effect | −95.290 (−99.985, 1358.902) | 0.2887 | |
| Longitudinal effect | 2.673 (−18.130, 28.762) | 0.8178 | |
| Lactate/Pyruvate | 0.3353 | ||
| Cross-sectional effect | −13.907 (−36.650, 17.002) | 0.3304 | |
| Longitudinal effect | −0.159 (−0.891, 0.578) | 0.6687 | |
| Succinate (μM)/Ucrea | 0.0095b | ||
| Cross-sectional effect | −20.878 (−33.222, −6.252) | 0.0079b | |
| Longitudinal effect | −0.635 (−1.787, 0.530) | 0.2809 | |
| Succinate (μM)/Uosm | 0.0623 | ||
| Cross-sectional effect | −85.651 (−98.066, 6.457) | 0.0573 | |
| Longitudinal effect | −5.083 (−21.162, 14.275) | 0.5787 | |
| Pyruvate (μM)/Ucrea | 0.7563 | ||
| Cross-sectional effect | −17.758 (−72.655, 147.350) | 0.7222 | |
| Longitudinal effect | −2.561 (−10.118, 5.633) | 0.5264 | |
| Pyruvate (μM)/Uosm | 0.7293 | ||
| Cross-sectional effect | −93.459 (−100.000, 13484225) | 0.7072 | |
| Longitudinal effect | −20.078 (−68.687, 103.989) | 0.6365 | |
| Total Protein (mg/dl)/Ucrea | 0.0023b | ||
| Cross-sectional effect | 909.537 (137.604, 4189.341) | 0.0024b | |
| Longitudinal effect | −0.904 (−4.276, 2.588) | 0.6042 | |
| Total Protein (mg/dl)/Uosm | 0.0006b | ||
| Cross-sectional effect | 2.5993 × 108 (77536.75, 8.702 × 1011) | 0.0007b | |
| Longitudinal effect | −9.759 (−49.443, 61.073) | 0.7260 | |
| PKM2 (ng/ml)/Ucrea | 0.1726 | ||
| Cross-sectional effect | 225.658 (−42.143, 1733.028) | 0.1753 | |
| Longitudinal effect | −0.731 (−4.050, 2.702) | 0.6698 | |
| PKM2 (ng/ml)/Uosm | 0.1148 | ||
| Cross-sectional effect | 4384830 (−93.818, 3.11 × 1012) | 0.1169 | |
| Longitudinal effect | −5.518 (−40.909, 51.069) | 0.8110 | |
| LDHA (ng/ml)/Ucrea | 0.1783 | ||
| Cross-sectional effect | 1394.614 (−72.911, 82364.38) | 0.1810 | |
| Longitudinal effect | −1.508 (−13.511, 12.162) | 0.8173 | |
| LDHA (ng/ml)/Uosm | 0.2959 | ||
| Cross-sectional effect | 3202585 (−99.997, 3.027 × 1015) | 0.3172 | |
| Longitudinal effect | −35.548 (−85.303, 182.638) | 0.5571 | |
| PDK1 (pg/ml)/Ucrea | 0.2341 | ||
| Cross-sectional effect | −4.040 (−10.404, 2.777) | 0.2322 | |
| Longitudinal effect | −0.018 (−0.150, 0.114) | 0.7881 | |
| PDK1 (pg/ml)/Uosm | 0.9217 | ||
| Cross-sectional effect | −3.340 (−50.837, 90.046) | 0.9198 | |
| Longitudinal effect | −0.088 (−3.057, 2.971) | 0.9539 |
CI, confidence interval; LDHA, lactate dehydrogenase-A; PDK1, pyruvate dehydrogenase kinase 1; PKM2, pyruvate kinase M2; Uosm, urinary osmolality; Ucrea, urinary creatinine concentration.
Test for difference between cross-sectional and longitudinal effects.
Significant P values.
Table 4.
Cross-sectional and longitudinal associations of urinary biomarkers with eGFR
| Biomarker | Difference P-valuea |
Mean change (95% CI) | P-value |
|---|---|---|---|
| Creatinine (mg/dl) | 0.1303 | ||
| Cross-sectional effect | −0.150 (−0.277, −0.023) | 0.0217b | |
| Longitudinal effect | −0.055 (−0.075, −0.035) | <0.0001b | |
| Osmolality (mmol/kg) | 0.8715 | ||
| Cross-sectional effect | −0.002 (−0.030, 0.025) | 0.8586 | |
| Longitudinal effect | −0.005 (−0.009, −0.001) | 0.0274b | |
| Lactate (μM)/Ucrea | 0.2457 | ||
| Cross-sectional effect | 8.028 (−6.236, 22.291) | 0.2626 | |
| Longitudinal effect | −0.287 (−0.707, 0.133) | 0.1799 | |
| Lactate (μM)/Uosm | 0.1287 | ||
| Cross-sectional effect | 113.735 (−46.715, 274.185) | 0.1601 | |
| Longitudinal effect | −9.056 (−18.171, 0.059) | 0.0515 | |
| Lactate/Pyruvate | 0.4119 | ||
| Cross-sectional effect | 3.142 (−5.584, 11.868) | 0.4717 | |
| Longitudinal effect | −0.436 (−0.821, −0.050) | 0.0268b | |
| Succinate (μM)/Ucrea | 0.6654 | ||
| Cross-sectional effect | 1.508 (−3.723, 6.739) | 0.5644 | |
| Longitudinal effect | 0.391 (−0.229, 1.011) | 0.2151 | |
| Succinate (μM)/Uosm | 0.4912 | ||
| Cross-sectional effect | 15.905 (−44.425, 76.235) | 0.5988 | |
| Longitudinal effect | −4.276 (−14.090, 5.538) | 0.3921 | |
| Pyruvate (μM)/Ucrea | 0.2675 | ||
| Cross-sectional effect | 18.734 (−11.762, 49.231) | 0.2222 | |
| Longitudinal effect | 1.842 (−0.265, 3.948) | 0.0864 | |
| Pyruvate (μM)/Uosm | 0.1037 | ||
| Cross-sectional effect | 328.152 (−73.346, 729.649) | 0.1067 | |
| Longitudinal effect | −0.677 (−31.796, 30.441) | 0.9659 | |
| Total Protein (mg/dl)/Ucrea | 0.0008b | ||
| Cross-sectional effect | −71.137 (−111.192, −31.082) | 0.0009b | |
| Longitudinal effect | 0.332 (−2.240, 2.903) | 0.8000 | |
| Total Protein (mg/dl)/Uosm | <0.0001b | ||
| Cross-sectional effect | −530.282 (−749.266, −311.298) | <0.0001b | |
| Longitudinal effect | −37.636 (−82.823, 7.550) | 0.1023 | |
| PKM2 (ng/ml)/Ucrea | 0.7401 | ||
| Cross-sectional effect | −6.327 (−56.420, 43.766) | 0.8002 | |
| Longitudinal effect | 1.944 (−0.527, 4.414) | 0.1227 | |
| PKM2 (ng/ml)/Uosm | 0.1634 | ||
| Cross-sectional effect | −281.878 (−668.279, 104.524) | 0.1486 | |
| Longitudinal effect | −11.984 (−43.350, 19.381) | 0.4529 | |
| LDHA (ng/ml)/Ucrea | 0.4837 | ||
| Cross-sectional effect | −39.747 (−155.612, 76.118) | 0.4929 | |
| Longitudinal effect | 0.730 (−8.286, 9.746) | 0.8736 | |
| LDHA (ng/ml)/Uosm | 0.5487 | ||
| Cross-sectional effect | −267.240 (−873.747, 339.267) | 0.3800 | |
| Longitudinal effect | −88.729 (−177.500, 0.042) | 0.0501 | |
| PDK1 (pg/ml)/Ucrea | 0.7146 | ||
| Cross-sectional effect | 0.400 (−1.578, 2.378) | 0.6852 | |
| Longitudinal effect | 0.040 (−0.062, 0.143) | 0.4409 | |
| PDK1 (pg/ml)/Uosm | 0.6573 | ||
| Cross-sectional effect | −4.575 (−23.928, 14.779) | 0.6364 | |
| Longitudinal effect | −0.336 (−2.286, 1.614) | 0.7348 |
CI, confidence interval; eGFR, estimated glomerular filtration rate; LDHA, lactate dehydrogenase-A; PDK1, pyruvate dehydrogenase kinase 1; PKM2, pyruvate kinase M2; Ucrea, urinary creatinine concentration; Uosm, urinary osmolality.
Test for difference between cross-sectional and longitudinal effects.
Significant P values.
Predictive Analyses
An analysis for predictive biomarkers was used to determine whether individual biomarkers predicted a differential response to metformin treatment versus placebo with respect to time-dependent changes in htTKV or eGFR. However, there were no statistically significant correlations found via 3-way interaction P-values for any of the metabolic biomarkers that were predictive of changes over time in either ln(htTKV) (Table 5) or eGFR (Table 6).
Table 5.
ln (height-adjusted total kidney volume)–metformin versus placebo: baseline urinary biomarkers (modifier)
| Biomarker | Metformin vs. placebo: annual % change (95% CI) | 3-way interaction P-value |
|---|---|---|
| Creatinine (mg/dl) | −0.021 (−0.113, 0.071) | 0.6505 |
| Osmolality (mmol/kg) | 0.013 (−0.005, 0.032) | 0.1465 |
| Lactate (μM)/Ucrea | −1.731 (−11.696, 9.358) | 0.7459 |
| Lactate (μM)/Uosm | −35.098 (−80.078, 111.443) | 0.4684 |
| Lactate/Pyruvate | 2.634 (−1.764, 7.230) | 0.2409 |
| Succinate (μM)/Ucrea | −0.140 (−3.675, 3.524) | 0.9384 |
| Succinate (μM)/Uosm | −12.988 (−40.859, 28.018) | 0.4753 |
| Pyruvate (μM)/Ucrea | −5.153 (−21.536, 14.650) | 0.5802 |
| Pyruvate (μM)/Uosm | −79.460 (−98.241, 139.838) | 0.2037 |
| Total Protein (mg/dl)/Ucrea | −11.113 (−31.047, 14.584) | 0.3586 |
| Total Protein (mg/dl)/Uosm | −57.656 (−90.050, 80.207) | 0.2411 |
| PKM2 (ng/ml)/Ucrea | −5.517 (−30.269, 28.022) | 0.7110 |
| PKM2 (ng/ml)/Uosm | −68.739 (−96.647, 191.490) | 0.3030 |
| LDHA (ng/ml)/Ucrea | −11.219 (−60.069, 97.392) | 0.7677 |
| LDHA (ng/ml)/Uosm | −74.751 (−99.839, 3858.614) | 0.5894 |
| PDK1 (pg/ml)/Ucrea | −0.312 (−1.479, 0.868) | 0.5985 |
| PDK1 (pg/ml)/Uosm | −5.524 (−14.503, 4.399) | 0.2609 |
CI, confidence interval; LDHA, lactate dehydrogenase-A; PDK1, pyruvate dehydrogenase kinase 1; PKM2, pyruvate kinase M2; Ucrea, urinary creatinine concentration; Uosm, urinary osmolality.
Table 6.
eGFR–metformin versus placebo: baseline urinary biomarkers (modifier)
| Biomarker | Metformin vs. placebo: annual mean change (95% CI) | 3-way interaction P-value |
|---|---|---|
| Creatinine (mg/dl) | −0.005 (−0.054, 0.044) | 0.0398a |
| Osmolality (mmol/kg) | −0.002 (−0.012, 0.008) | 0.3524 |
| Lactate (μM)/Ucrea | −1.915 (−7.500, 3.669) | 0.9419 |
| Lactate (μM)/Uosm | −27.757 (−88.693, 33.179) | 0.9972 |
| Lactate/Pyruvate | −2.069 (−4.365, 0.228) | 0.1210 |
| Succinate (μM)/Ucrea | −0.090 (−2.020, 1.840) | 0.9926 |
| Succinate (μM)/Uosm | 2.349 (−18.305, 23.003) | 0.9986 |
| Pyruvate (μM)/Ucrea | 2.741 (−7.443, 12.926) | 0.6934 |
| Pyruvate (μM)/Uosm | 22.068 (−110.382, 154.518) | 0.9775 |
| Total Protein (mg/dl)/Ucrea | 7.158 (−6.507, 20.824) | 0.6720 |
| Total Protein (mg/dl)/Uosm | 28.013 (−52.118, 108.143) | 0.3262 |
| PKM2 (ng/ml)/Ucrea | −1.508 (−17.585, 14.570) | 0.9935 |
| PKM2 (ng/ml)/Uosm | −21.580 (−141.637, 98.477) | 0.9622 |
| LDHA (ng/ml)/Ucrea | −23.458 (−63.787, 16.870) | 0.8668 |
| LDHA (ng/ml)/Uosm | −94.700 (−342.825, 153.425) | 0.6813 |
| PDK1 (pg/ml)/Ucrea | −0.106 (−0.746, 0.533) | 0.3331 |
| PDK1 (pg/ml)/Uosm | 0.021 (−5.372, 5.414) | 0.8319 |
CI, confidence interval; eGFR, estimated glomerular filtration rate; LDHA, lactate dehydrogenase-A; PDK1, pyruvate dehydrogenase kinase 1; PKM2, pyruvate kinase M2; Ucrea, urinary creatinine concentration; Uosm, urinary osmolality.
Significant P values.
Secondary Analyses
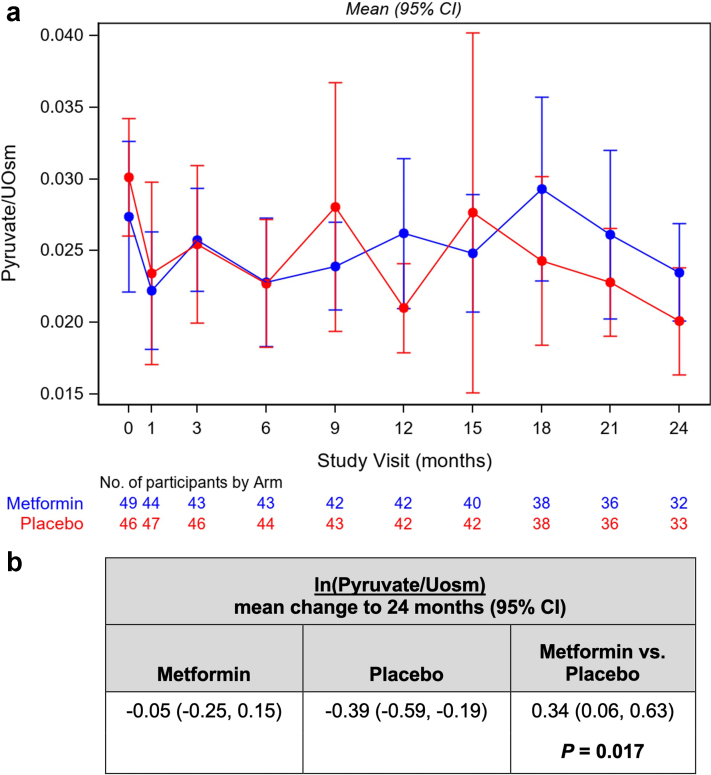
Secondary analyses were performed to investigate changes in each of the normalized biomarkers over the course of the study as a function of treatment group. Although most biomarkers did not differ significantly as a function of treatment group, there was a significant difference in the change of pyruvate excretion (normalized by urine osmolality) in metformin-treated versus placebo-treated subjects. Specifically, whereas there was no significant change over 24 months of ln(Pyruvate/Uosm) in the metformin treatment group (−0.05 [95% class interval −0.25, 0.15]), there was a significant reduction of ln(Pyruvate/Uosm) in the placebo group (−0.39 [95% class interval −0.59, −0.19]) with a between-arm difference of metformin versus placebo of 0.34 (95% class interval 0.06, 0.63; P = 0.017). Therefore, pyruvate excretion in the placebo group declined whereas there was no significant change in metformin-treated subjects. Changes in Pyruvate/Uosm by study visit are shown in Figure 1.
Figure 1.
Changes in urinary pyruvate normalized to urinary osmolality by study visit. (a) Graph depicting time course of changes in urinary pyruvate to osmolality ratio for metformin-treated subjects (blue) and placebo group subjects (red) throughout the 24-month study period. Data shown are means (± 95% CI) at each of the study visits with the number of samples at each visit for the 2 treatment arms indicated at the bottom of the graph. (b) Comparison of the mean change (and 95% CI) in ln (pyruvate/Uosm) over the 24-month treatment period for the metformin and placebo groups. Statistical analysis revealed no significant change over 24 months of ln(Pyruvate/Uosm) in the metformin treatment group (−0.05 [95% CI −0.25, 0.15]), whereas there was a significant reduction of ln (Pyruvate/Uosm) in the placebo group of −0.39 (95% CI −0.59, −0.19), with a between-arm difference of metformin versus placebo of 0.34 (95% CI 0.06, 0.63; P = 0.017). CI, confidence interval;
Discussion
ADPKD progression follows a variable course even across related patients with the same or similar mutations.17 Accordingly, with the current pipeline of emerging ADPKD therapies, there is considerable interest in risk stratification and the identification of new biomarkers that may inform disease severity and the potential for disease progression to prioritize usage in patients most likely to benefit from these therapies. Our recent analysis of baseline biomarkers for the TAME-PKD study confirmed in our study population with mild-to-moderate disease that proteinuria, a significant predictor of ADPKD disease progression,24, 25, 26 correlates with disease severity as assessed by decreasing eGFR and increasing htTKV.9 Moreover, pyruvate kinase M2 isoform and LDHA, key urinary glycolytic enzymes that are markers for excessive aerobic glycolysis and found to be elevated in ADPKD preclinical studies,6 were correlated with disease severity at baseline in the TAME-PKD study population.9 These findings support the notion that dysregulated metabolism with increased glycolytic flux is an important correlate of disease severity in urine samples from ADPKD patients.
In the current study using urine samples obtained from participants longitudinally over the course of the TAME-PKD clinical trial, we sought to determine whether any of the targeted metabolic biomarkers are useful in indicating disease severity, progression rate, and/or response to metformin treatment. An important goal was to identify possible subgroups of participants who may or may not benefit from metformin therapy.
Summary and Implications of Findings
In the prognostic analyses utilizing all placebo group data, none of the time-varying urinary metabolic biomarkers significantly correlated with changes in ln(htTKV) (Table 1). However, urine creatinine itself had a significant positive correlation with ln(htTKV) over time, although there was no significant cross-sectional correlation between urine creatinine and ln(htTKV) (Table 3). Both time-varying Ucrea and Uosm had negative correlations with eGFR (Table 2). The underlying factors accounting for these time-varying correlations are unclear. Of note, Ucrea and Uosm were highly correlated with one another in all study participant samples (Supplementary Figure S3) as previously observed,9 suggesting that both of these parameters may reasonably serve as normalization factors to apply to spot urine sample measurements to facilitate comparisons across different samples.
Importantly, there were negative correlations in both time-varying urinary protein excretion normalized by osmolality and lactate/pyruvate ratio with eGFR changes (Table 2). These negative correlations with changes in eGFR suggest that proteinuria and lactate/pyruvate ratio may serve as prognostic biomarkers for ADPKD progression in our study population. Although not significant, there were trends toward negative correlations (P < 0.10) of lactate/Uosm, and LDHA/Uosm with eGFR changes over time (Table 2). Together, these findings suggest that worsening kidney function in ADPKD patients over time correlates with greater glycolytic metabolism in kidney tubular epithelium.
Our further characterization of the proteinuria in study participant urine samples revealed that the most abundant proteins present were albumin and uromodulin (Supplementary Figure S2). To the extent that the appearance of these proteins was greater in individual urine samples, their excess presence may reflect the following: (i) greater perturbations in the glomerular filtration barrier27; and (ii) enhanced tubular production of uromodulin in the thick ascending limb of the loop of Henle, respectively. Increased uromodulin production is associated with injury in the thick ascending limb of the loop of Henle,28 which could reflect more severe cystic disease leading to localized ischemic tubular injury. In addition, there is decreased uromodulin protein degradation in ADPKD kidneys relative to control kidneys,29 which could contribute to an increase in detectable full-length uromodulin in ADPKD urine samples. Nevertheless, overall and relative protein expression levels of these 2 major proteins were found to be highly variable across the different study participants, suggesting potential heterogeneity of glomerular and tubular disease features across different study participants. Of note, there appeared to be relatively less low-molecular weight proteinuria in most of these ADPKD participants’ samples, suggesting that proximal tubular resorptive capacity was generally not compromised to the extent that overflow proteinuria was apparent.30
In cross-sectional analyses utilizing all placebo group samples available, there was a significant negative correlation between urinary excretion of succinate, a tricarboxylic acid cycle intermediate important for oxidative metabolism, and ln(htTKV) (Table 3). This result suggests that ADPKD patients with greater kidney volumes tend to have reduced oxidative metabolic flux, supporting the notion that reduced mitochondrial oxidative function correlates with more severe ADPKD disease activity, as previously suggested.7,8 Moreover, in cross-sectional analyses of urinary protein excretion, there was a strong positive correlation with htTKV (Table 3) and a strong negative correlation with eGFR (Table 4). Robust correlations of proteinuria with established indicators of ADPKD disease severity in our full-study samples confirm previous cross-sectional results using the TAME-PKD baseline visit samples9 and the conclusions of earlier studies that proteinuria is a biomarker of more severe disease.24, 25, 26
Our predictive analysis did not identify any individual biomarkers that could predict a differential response to metformin treatment versus placebo with respect to changes in either htTKV or eGFR over time (Tables 5 and 6).
In the secondary analyses, changes in urinary pyruvate levels normalized by Uosm over time could serve as a discriminator for the effects of treatment with metformin versus placebo in this study population. Specifically, urinary pyruvate excretion was relatively preserved over time in metformin-treated participants whereas it declined significantly in the placebo group (Figure 1). It is noteworthy that this effect occurred despite the lack of significant effect of metformin on the primary outcome measures of changes in eGFR or htTKV in the primary TAME-PKD study.16 This effect could reflect an overall relative enhancement in the conversion of pyruvate to lactate in placebo group subjects over time, which was not present in the metformin-treated subjects. Of note, time-varying changes in the lactate/pyruvate ratio negatively correlated with eGFR (Table 2). Therefore, given that eGFR tended to decline more in the placebo group than in the metformin group over the course of the study,16 these results taken altogether would be consistent with decreased lactate formation in metformin-treated subjects and thus perhaps decreased glycolytic flux, although this was not measured directly.
Limitations
There are several limitations of the current study. First, there was considerable longitudinal variability in biomarker values, which likely relates in part to variability in urine collection times and fasting status at the different study visits (see Methods). This is in contrast to our previously reported baseline biomarker measurements, which were all fasting morning samples.9 This limitation resulted from the practical need to allow flexibility in the scheduling of follow-up study visit times for participants’ convenience. Second, the overall power to detect potential differences in the various biomarker analyses is limited by the numbers of study participants and samples obtained in this phase 2 trial, thus many of the conclusions herein should be considered exploratory. In particular, the power to detect significant differences in time-varying data is limited because this analysis was restricted only to the placebo group of study participants (N = 48; 417 samples total) (Supplementary Table S1 and Supplementary Figure S1). Third, the statistical power to detect any correlations of urinary biomarkers with ln(htTKV) is more limited than with eGFR because of the fewer longitudinal measurements of htTKV relative to eGFR over the course of the study (up to 5 imaging vs. 10 blood specimen longitudinal data points per patient, respectively). Nevertheless, the cross-sectional analysis in this longitudinal study is consistent with and generally confirms our previous findings in the baseline TAME-PKD biomarkers study.9 Specifically, urinary total protein and glycolytic enzyme levels generally positively correlated with htTKV and negatively correlated with eGFR.
Perspectives and Significance
A major goal of this study was to define surrogate biomarkers for ADPKD disease severity, risk of progression, and response to metformin based on the emerging notion that metabolic dysregulation is an underlying driver of disease severity and progression.8 We have uncovered additional evidence that reduced oxidative and enhanced glycolytic flux, as evidenced by changes in excretion of key metabolites and glycolytic enzymes, is a feature associated with ADPKD. Moreover, we have confirmed proteinuria as a robust biomarker of ADPKD disease severity and risk of progression in this study population, as measured by both htTKV and eGFR. Although severe proteinuria is relatively uncommon in ADPKD, greater proteinuria is a significant predictor of future ADPKD progression.24, 25, 26 Our findings support this idea because time-varying protein excretion negatively correlates with eGFR changes. Therefore, proteinuria and the urinary lactate/pyruvate ratio seem to be useful predictive biomarkers for progression in early ADPKD.
In the longterm, we expect that identification of additional surrogate biomarkers will play a growing role in ADPKD disease management, because they may represent relatively inexpensive and noninvasive tools to monitor disease severity, progression potential, and response to new therapies, including metformin and others that may be used alone or in combination with metformin.
Disclosure
The authors declare no relevant conflicts of interest. KRH has received research support from Otsuka Pharmaceutical and Esperion Therapeutics and is a consultant to Maze Therapeutics, Otsuka, Bayer AG, Poxel SA, and Aurinia Pharmaceuticals. KTB is a consultant to Kadmon, Otsuka, and Sanofi. RDP has received research funding from Otsuka, Kadmon Corporation, Sanofi-Genzyme, and Reata. RDP is a consultant to Palladiobio, Reata, Sanofi-Genzyme, and Otsuka. SLS has received research funding from Otsuka, Kadmon Corporation, Palladio Biosciences, Sanofi, and Reata. TJW has received research funding from Otsuka, Kadmon Corporation, Palladio Biosciences, Sanofi, and Reata.
Acknowledgments
The authors thank Daniel Rivera and Pei-Yin Ho at the University of Southern California for excellent technical assistance on biomarker measurements. We also thank the study coordinators, administrators, and other support staff at each of the study sites: Susan Spillane and Linda Whiting at the University of Pittsburgh; Charalett Diggs and Ashley Hargrove at the University of Maryland; and Carly Tucker, Margaret Reilly, Victoria Himaras and Raabia Malik at Tufts Medical Center. Finally, we express our gratitude to the study participants and PKD community who are instrumental for the success of this clinical trial.
Funding
This study was supported by the U.S. Department of Defense Congressionally Directed Medical Research Program (W81XWH-15-1-0663), by the National Center for Advancing Translational Sciences, National Institutes of Health, Award Numbers UL1TR002544 and 1UL1TR003098. This work also utilized resources developed by the Baltimore Polycystic Kidney Disease Research Center Clinical and Translational Core P30 DK090868.
Footnotes
Figure S1. Number of samples for each participant over time.
Figure S2. Representative image of urine protein staining from aliquots of 20 baseline participants’ specimens subjected to SDS-PAGE and transfer to nitrocellulose membrane.
Figure S3. Scatter plot of the association between ln(urinary osmolality) and ln(urinary creatinine) for each sample.
Table S1. Distribution of the number of samples for each participant.
Table S2. Baseline characteristics by randomized arm.
Supplementary Material
Figure S1. Number of samples for each participant over time.
Figure S2. Representative image of urine protein staining from aliquots of 20 baseline participants’ specimens subjected to SDS-PAGE and transfer to nitrocellulose membrane.
Figure S3. Scatter plot of the association between ln(urinary osmolality) and ln(urinary creatinine) for each sample.
Table S1. Distribution of the number of samples for each participant.
Table S2. Baseline characteristics by randomized arm.
References
- 1.Chebib F.T., Torres V.E. Autosomal dominant polycystic kidney disease: core curriculum 2016. Am J Kidney Dis. 2016;67:792–810. doi: 10.1053/j.ajkd.2015.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres V.E., Chapman A.B., Devuyst O., et al. Tolvaptan in patients with autosomal dominant polycystic kidney disease. N Engl J Med. 2012;367:2407–2418. doi: 10.1056/NEJMoa1205511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Watkins P.B., Lewis J.H., Kaplowitz N., et al. Clinical pattern of tolvaptan-associated liver injury in subjects with autosomal dominant polycystic kidney disease: analysis of clinical trials database. Drug Saf. 2015;38:1103–1113. doi: 10.1007/s40264-015-0327-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergmann C., Guay-Woodford L.M., Harris P.C., et al. Polycystic kidney disease. Nat Rev Dis Primers. 2018;4:50. doi: 10.1038/s41572-018-0047-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weimbs T., Shillingford J.M., Torres J., et al. Emerging targeted strategies for the treatment of autosomal dominant polycystic kidney disease. Clin Kidney J. 2018;11(suppl 1):i27–i38. doi: 10.1093/ckj/sfy089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rowe I., Chiaravalli M., Mannella V., et al. Defective glucose metabolism in polycystic kidney disease identifies a new therapeutic strategy. Nat Med. 2013;19:488–493. doi: 10.1038/nm.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menezes L.F., Lin C.C., Zhou F., Germino G.G. Fatty acid oxidation is impaired in an orthologous mouse model of autosomal dominant polycystic kidney disease. EBioMedicine. 2016;5:183–192. doi: 10.1016/j.ebiom.2016.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Padovano V., Podrini C., Boletta A., Caplan M.J. Metabolism and mitochondria in polycystic kidney disease research and therapy. Nat Rev Nephrol. 2018;14:678–687. doi: 10.1038/s41581-018-0051-1. [DOI] [PubMed] [Google Scholar]
- 9.Hallows K.R., Althouse A.D., Li H., et al. Association of baseline urinary metabolic biomarkers with ADPKD severity in TAME-PKD clinical trial participants. Kidney360. 2021;2:795–808. doi: 10.34067/kid.0005962020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou G., Myers R., Li Y., et al. Role of AMP-activated protein kinase in mechanism of metformin action. In vitro. J Clin Invest. 2001;108:1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takiar V., Nishio S., Seo-Mayer P., et al. Activating AMP-activated protein kinase (AMPK) slows renal cystogenesis. Proc Natl Acad Sci U S A. 2011;108:2462–2467. doi: 10.1073/pnas.1011498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pastor-Soler N.M., Li H., Pham J., et al. Metformin improves relevant disease parameters in an autosomal dominant polycystic kidney disease mouse model. Am J Physiol Ren Physiol. 2022;322:F27–F41. doi: 10.1152/ajprenal.00298.2021. [DOI] [PubMed] [Google Scholar]
- 13.Lian X., Wu X., Li Z., et al. The combination of metformin and 2-deoxyglucose significantly inhibits cyst formation in miniature pigs with polycystic kidney disease. Br J Pharmacol. 2019;176:711–724. doi: 10.1111/bph.14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jayasena C.N., Franks S. The management of patients with polycystic ovary syndrome. Nat Rev Endocrinol. 2014;10:624–636. doi: 10.1038/nrendo.2014.102. [DOI] [PubMed] [Google Scholar]
- 15.Miller R.A., Chu Q., Xie J., et al. Biguanides suppress hepatic glucagon signalling by decreasing production of cyclic AMP. Nature. 2013;494:256–260. doi: 10.1038/nature11808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrone R.D., Abebe K.Z., Watnick T.J., et al. Primary results of the randomized trial of metformin administration in polycystic kidney disease (TAME PKD) Kidney Int. 2021;100:684–696. doi: 10.1016/j.kint.2021.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang Y.H., Conklin J., Chan W., et al. Refining genotype-phenotype correlation in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 2016;27:1861–1868. doi: 10.1681/ASN.2015060648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lanktree M.B., Iliuta I.A., Haghighi A., et al. Evolving role of genetic testing for the clinical management of autosomal dominant polycystic kidney disease. Nephrol Dial Transplant. 2019;34:1453–1460. doi: 10.1093/ndt/gfy261. [DOI] [PubMed] [Google Scholar]
- 19.Kawano H., Muto S., Ohmoto Y., et al. Exploring urinary biomarkers in autosomal dominant polycystic kidney disease. Clin Exp Nephrol. 2015;19:968–973. doi: 10.1007/s10157-014-1078-7. [DOI] [PubMed] [Google Scholar]
- 20.Seliger S.L., Abebe K.Z., Hallows K.R., et al. A randomized clinical trial of metformin to treat autosomal dominant polycystic kidney disease. Am J Nephrol. 2018;47:352–360. doi: 10.1159/000488807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seliger S.L., Watnick T., Althouse A.D., et al. Baseline characteristics and patient-reported outcomes of ADPKD patients in the multicenter TAME-PKD clinical trial. Kidney360. 2020;1:1363–1372. doi: 10.34067/KID.0004002020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 23.Laird N.M., Ware J.H. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. doi: 10.2307/2529876. [DOI] [PubMed] [Google Scholar]
- 24.Chapman A.B., Johnson A.M., Gabow P.A., Schrier R.W. Overt proteinuria and microalbuminuria in autosomal dominant polycystic kidney disease. J Am Soc Nephrol. 1994;5:1349–1354. doi: 10.1681/ASN.V561349. [DOI] [PubMed] [Google Scholar]
- 25.Gansevoort R.T., Meijer E., Chapman A.B., et al. Albuminuria and tolvaptan in autosomal-dominant polycystic kidney disease: results of the TEMPO 3:4 Trial. Nephrol Dial Transplant. 2016;31:1887–1894. doi: 10.1093/ndt/gfv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schrier R.W., Brosnahan G., Cadnapaphornchai M.A., et al. Predictors of autosomal dominant polycystic kidney disease progression. J Am Soc Nephrol. 2014;25:2399–2418. doi: 10.1681/ASN.2013111184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tojo A., Kinugasa S. Mechanisms of glomerular albumin filtration and tubular reabsorption. Int J Nephrol. 2012;2012:481520. doi: 10.1155/2012/481520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen Y., Parikh C.R. Current concepts and advances in biomarkers of acute kidney injury. Crit Rev Clin Lab Sci. 2021;58:354–368. doi: 10.1080/10408363.2021.1879000. [DOI] [PubMed] [Google Scholar]
- 29.Kistler A.D., Serra A.L., Siwy J., et al. Urinary proteomic biomarkers for diagnosis and risk stratification of autosomal dominant polycystic kidney disease: a multicentric study. PLoS One. 2013;8 doi: 10.1371/journal.pone.0053016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Topham P. Proteinuric renal disease. Clin Med (Lond) 2009;9:284–287. doi: 10.7861/clinmedicine.9-3-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.