Abstract
In Escherichia coli and related bacteria, the very-short-patch (VSP) repair pathway uses an endonuclease, Vsr, to correct T · G mismatches that result from the deamination of 5-methylcytosines in DNA to C · G. The products of mutS and mutL, which are required for adenine methylation-directed mismatch repair (MMR), enhance VSP repair. Multicopy plasmids carrying mutS alleles that are dominant negative for MMR were tested for their effects on VSP repair. Some mutS mutations (class I) did not lower VSP repair in a mutS+ background, and most class I mutations increased VSP repair in mutS cells more than plasmids containing mutS+. Other plasmid-borne mutS mutations (class II) and mutS+ decreased VSP repair in the mutS+ background. Thus, MutS protein lacking functions required for MMR can still participate in VSP repair, and our results are consistent with a model in which MutS binds transiently to the mispair and then translocates away from the mispair to create a specialized structure that enhances the binding of Vsr.
The product of gene vsr, which is present in several species of enteric bacteria, is a sequence-specific endonuclease (reviewed in reference 14). Vsr recognizes T · G mismatches in DNA that arise as a result of deamination of 5-methylcytosine in the sequence 5′-CmeC(A or T)GG (meC, 5-methylcytosine) and nicks 5′ to the mismatched T. Following nicking, replacement of T with C is accomplished by DNA polymerase I and DNA ligase. T · G mispairs in related contexts are also subject to very-short-patch (VSP) repair but at a lower frequency (15). The removal of the mispaired T by VSP repair is enhanced by MutS and MutL (8, 11), although these products are not required if Vsr is supplied by a multicopy plasmid (15). MutS, on the other hand, binds to most mismatched base pairs, with highest affinity for T · G and C · A (20). MutL interacts with MutS, and the MutS-MutL complex initiates repair of replication errors by stimulating nicking of one DNA strand by the MutH protein (2, 7). The strand discrimination signal for nicking is adenine methylation within 5′-GATC sites; hence, this is referred to as methyladenine-directed mismatch repair (MMR) (reviewed in reference 18). We describe below experiments in which the effects of multicopy plasmids carrying either wild-type MutS or mutant versions of MutS on VSP repair were studied.
Overproduction of MutS+ is ineffective in restoring VSP repair in mutS and mutL bacteria.
The presence of a mutS+ plasmid reduces the frequency of VSP repair in mutS+ mutL+ bacteria (Table 1, and reference 15). In contrast, introducing a mutS+ plasmid into cells containing chromosomal mutS+ does not disrupt MMR (24). It was also shown that in strain GM31, which is deficient in Vsr, excess MutS essentially abolishes VSP repair (15) (Table 1). We have now found that a mutS+ plasmid increased VSP repair only marginally in a mutS strain (Table 1.) In a mutL background, the amount of repair in the presence of plasmid pMutS+ was substantially lower than when MutS was produced solely by chromosomal mutS+ (Table 1). These results suggest that MutL and MutS serve distinct functions in promoting VSP repair and that the absence of MutL in cells cannot be compensated for by increasing amounts of MutS. Furthermore, just as overproduction of Vsr disrupts MMR (3, 16), overproduction of MutS disrupts VSP repair, suggesting that both proteins must be present in cells at optimal levels for both mismatch repair processes to minimize mutations.
TABLE 1.
Effects of excess MutS+ on VSP repair
| Strain | Relevant genotype or (phenotype) | % cI+a
|
|
|---|---|---|---|
| No plasmid | With pMutS+ | ||
| CC106 | mutS+mutL+vsr+ | 3.2 | 2.3 |
| GM30 | mutS+mutL+vsr+ | 3.1b | 1.6b |
| GM31 | GM30 dcm-6 (Vsr−) | 0.49b | 0.18b |
| SP1 | mutS::Tn10 mutL+vsr+ | 0.83 | 1.1 |
| KMBL3752 | mutS+mutL+vsr+ | 5.0 | 3.9 |
| KMBL3760 | mutS+mutL-101 vsr+ | 0.64 | 0.34 |
The values are percentages of cI+ among N+O+ recombinants in a cross of λNcIam6 O+ with λN+ cICP7 O and are averages of results from two or more experiments. During recombination, heteroduplexes containing T · G mismatches are generated which are repaired by VSP repair to create cI+ phage. Thus, a higher percentage of cI+ indicates more VSP repair (see references 12 and 15). Plasmid pMutS+ has been described previously (21).
Value from reference 15.
Dominant-negative mutS mutations differ with respect to their effect on VSP repair.
Wu and Marinus (24) isolated on multicopy plasmids a series of dominant mutS mutations proteins that interfere with MMR. Most of these plasmids produce a high level of spontaneous mutations when they are introduced into mutS+ cells. In Table 2, the dominant-negative mutations of MutS whose sites are known (24) are listed in the order of their positions in the mutS gene. To elucidate the role of MutS in VSP repair, we tested a number of these dominant-negative mutant proteins for their effects on VSP repair in mutS and mutS+ bacteria. It should be noted that none of the mutS plasmids affected phage λ recombination in the VSP repair assay.
TABLE 2.
VSP repair in bacteria containing mutS plasmids
|
mutS plasmid
|
VSP repair in bacteria with indicated genotype
|
Complementing protein(s) | Class | ||||
|---|---|---|---|---|---|---|---|
|
mutS+
|
mutS mutant
|
||||||
| USCe no. | aa change | % cI+a | Relative value | % cI+ | Relative value | ||
| Nonef | 3.17b | 1.00 | 0.83b | 1.00 | |||
| 1 | None (WT)d | 2.26 | 0.71 | 1.07b | 1.29 | ||
| 24 | G117D | 1.16 | 0.37 | NDc | II | ||
| 2 | G117S | 3.31 | 1.04 | 1.29 | 1.57 | MutS | I |
| 22 | A123V | 1.77 | 0.55 | ND | ND | II | |
| 8 | R305H | 4.02 | 1.27 | 0.97b | 1.18 | MutS | I |
| 23 | R350H | 1.62 | 0.51 | ND | II | ||
| 11 | G583D | 0.42 | 0.13 | 0.23 | 0.28 | II | |
| 3 | G614D | 1.12 | 0.35 | 0.25 | 0.30 | II | |
| 5 | G618S | 0.52 | 0.16 | 0.23b | 0.27 | MutS | II |
| 9 | G619D | 0.16 | 0.05 | 0.14 | 0.17 | II | |
| 6 | S621N | 3.23 | 1.02 | 4.51 | 5.50 | MutS MutH | I |
| 7 | D659N | ND | 0.80 | 0.98 | MutS MutH | I | |
| 10 | S668L | 0.18 | 0.06 | 0.19b | 0.23 | II | |
| 14 | T669I | 3.04b | 1.24 | 4.06 | 4.95 | MutS MutL MutH | I |
| 4 | E673K | 3.01 | 0.97 | 1.52b | 1.85 | MutS MutH | I |
| 12 | D693N | 2.89 | 0.89 | 2.08 | 2.50 | MutS MutH | I |
| 21 | E694K | 4.16 | 1.31 | ND | MutS | I | |
| 16 | G698E | 3.20 | 1.01 | 2.14 | 2.58 | MutS MutL MutH | I |
% cI+, the number of cI+ phage/total number of recombinant phages.
Average of values from two or more crosses. The mutS+ strain was CC106, and the mutS strain was SP1.
ND, not determined.
Plasmid with wild-type (WT) mutS.
USC, University of Southern California collection.
No plasmid.
The mutations can be placed into two classes based on their effects on VSP repair in mutS+ bacteria. Class I includes two mutations in the 5′ half of mutS and several mutations at the 3′ end. In the mutS+ background, plasmids with class I mutations either did not reduce VSP repair significantly or produced slightly more repair than was obtained in the absence of a mutS plasmid. This finding contrasts with the reduction in VSP repair caused by the mutS+ plasmid in mutS+ cells (Table 1 and reference 15). In mutS bacteria, plasmids with some of the class I mutations increased VSP repair significantly more than mutS+ plasmids. Notably, mutations at amino acid (aa) 621 or 669 raised cI+ recombinant frequencies to values comparable to those obtained in mutS+ bacteria. We suggest that class I mutations result in a product that retains the ability to facilitate the binding of a T · G mismatch by Vsr but that is deficient in another MMR function(s) not required by VSP repair.
In contrast, class II mutations strongly inhibited VSP repair. In phage crosses made in bacteria carrying plasmids containing class II mutations, the frequency of cI+ recombinants was as low as 0.16% (G619D mutation). A frequency of approximately 0.15% cI+ is expected in the absence of any VSP repair. Thus, class II mutations have a dominant-negative phenotype for VSP repair as well as for MMR and some class II mutations eliminate VSP repair completely.
Overproduction of Vsr increases VSP repair in the presence of dominant-negative mutS alleles.
It has been shown that the frequency of VSP repair is increased when excess Vsr is supplied (13, 15). Therefore, we expected that the addition of a vsr+ plasmid to bacteria carrying plasmids with dominant-negative mutS mutations might also increase VSP repair. In the case of crosses in which one parent carries the cI mutation am6, which is in the optimum context for VSP repair, excess Vsr increased VSP repair in bacteria carrying any of the plasmids with the dominant-negative mutS mutations tested (Table 3). These results are consistent with the idea that MutS enhances the binding of Vsr to the mispair and that a defect in the ability of MutS to perform this function can be compensated for by increasing the cellular concentration of Vsr. Similar results were obtained in crosses to test the VSP repair of cI mutation am9, which produces a T · G mispair in a context that is less favorable for VSP repair (5′-GTAGG). However, in bacteria with plasmids having mutS mutations at aa 619 or 668, excess Vsr did not improve VSP repair at am9 significantly. It is possible that these mutations result in a MutS protein that competes very strongly with Vsr, particularly for mispairs not in the optimum context for Vsr binding.
TABLE 3.
Competition between MutS and Vsr
| Mutation site (aa) or genotype | Class | Frequency of cI+ in mutS bacteria with mismatch in:
|
|||
|---|---|---|---|---|---|
| CTAGG
|
GTAGG
|
||||
| pmutS | pmutS + pvsr+ | pmutS | pmutS + pvsr+ | ||
| None | 3.17a | 6.79b | 0.88a | 2.21b | |
| mutS+ | 2.26 | 5.01 | 0.30 | 1.09 | |
| 614 | II | 1.12 | 4.24 | 0.07 | 0.44 |
| 618 | II | 0.52 | 2.08 | 0.13 | 0.34 |
| 619 | II | 0.16 | 0.37 | 0.06 | 0.08 |
| 621 | I | 3.23 | 6.81 | 0.84 | 2.40 |
| 668 | II | 0.18 | 0.52 | 0.09 | 0.10 |
| 669 | I | 3.94 | 7.28 | 1.12 | 2.74 |
Frequency of cI+ in mutS+ bacteria (CC106) with no plasmids.
Frequency of cI+ in mutS+ bacteria containing pDCM28 (19).
What is the role of MutS in VSP repair?
The role played by MutS in MMR is controversial. A variety of models have been proposed for the MutS function, some of which suggest that MutS remains bound to the mispair during its activation of nicking at 5′-GATC sites by MutH (9, 10, 17). Others have suggested that MutS leaves the mismatch following initial binding (1, 5, 6). Specifically, Allen et al. (1) suggested that MutS mediates the formation of a DNA loop with the mispair at its apex. Loop formation depends on the hydrolysis of ATP. MutL assists in this reaction but is not necessary for it.
The crystal structure of Vsr complexed with DNA shows that Vsr intercalates several of its residues into the DNA helix, deforming base stacking and widening both the major and the minor groove (22). If the models of the crystal structures of both Vsr (22) and MutS (9, 10) bound to DNA are accurate, it is impossible to visualize simultaneous binding of Vsr and MutS at a T · G mismatch. This means that, at least in VSP repair, MutS must leave the mismatch to allow the binding of Vsr. Similarly, a model in which MutS simply translocates away from the mismatch following an ADP-to-ATP exchange (5, 6) does not explain the beneficial effects of MutS on VSP repair. We are forced to conclude that MutS leaves the mismatch following initial binding, leaving behind a specialized structure that is conductive to binding by Vsr. Creating an underwound DNA loop is one such possible way in which MutS could promote Vsr binding to T · G, and Tsutakawa et al. (22) previously proposed such a role for MutS in VSP repair.
We propose further that the products of dominant-negative mutS mutants that have the ability to move the T · G mismatch into a loop but are defective in some subsequent step in MMR retain VSP repair-enhancing activity (class I mutants). However, mutants whose products bind to the mismatch in an irreversible manner prevent access to the mismatch by Vsr and have a dominant-negative effect on VSP repair (class II). Although a majority of class I mutations lie near the 3′ end of mutS and fewer class II mutations lie in this part of the gene, there is no simple correlation between the positions of the mutS mutations and the class to which they belong. The two classes are intermixed in terms of their positions in the gene, and mutations in adjacent or nearby residues often belong to different classes. This is dramatically seen at aa 117, where, depending on the substitution, the mutations can be class I or II (Table 2).
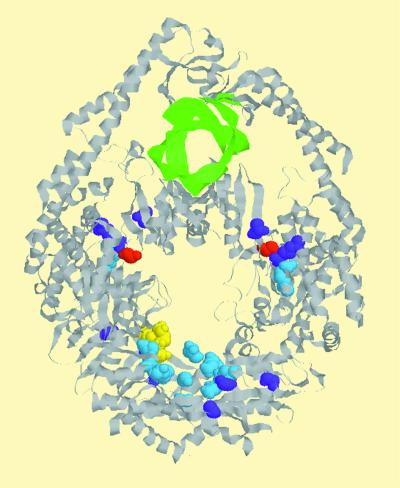
The distribution of all tested dominant-negative mutations along the crystal structure of MutS shows that they line the surface of the clamp within the dimer (Fig. 1). They are seen to cluster in two groups, with those of one group surrounding the bound ADP molecule and with those of the other residing in a segment that connects the DNA-binding domain with the ATP-binding domain. These mutations may affect ATP binding, ATP hydrolysis, the communication between the binding sites for ATP and DNA, or the interaction between MutS and MutL. Therefore, biochemical work is necessary to identify the functional defects in these mutant proteins.
FIG. 1.
The figure was drawn using coordinates kindly provided by T. Sixma (Netherlands Cancer Institute, Amsterdam, The Netherlands) for E. coli MutS bound to DNA containing a T · G mismatch (10). The color scheme is as follows: gray indicates the MutS protein, green ribbons are DNA, and yellow indicates ADP. Residues in which class I mutations lie are shown in light blue, while residues for class II mutations are shown in purple. Mutations in residue 117 can either be class I or II (see the text), and this residue is shown in red.
To explain the enhancement of VSP repair in mutS bacteria by some of the mutS plasmids, one can suppose that the mutations affect regions of the protein where MutS normally interacts with MutL. Consequently, these plasmid increase the amount of MutL available to enhance Vsr binding to the mismatch. There is evidence that MutL can interact with MutS that is not DNA bound (23). It has been pointed out that in both wild-type and mutL bacteria, overproduction of MutS+ reduced VSP repair. Thus, the class I mutS mutations may fail to reduce the amount of available MutL. The dominant-negative effect on MMR would be attributable to the failure of MutS to interact with MutL, an action which is required to activate MutH. It should be noted that the mutagenic phenotype produced by the mutS mutations that strongly enhance VSP repair in mutS bacteria is complemented by MutL+ or by MutH+ (24).
In conclusion, our findings are compatible with the following model: MutS binds to a T · G mispair and translocates the mispair into a specialized DNA structure such as a loop. The affinity of Vsr for the mismatch is increased when the mismatch is present in a loop. The ability of Vsr to interfere with MMR (3, 16) suggests a direct interaction of Vsr with an MMR protein. Complementation of the mutagenic effects of Vsr overproduction by MutL and MutH, but not MutS, indicates that Vsr can reduce the MutL available for MMR, presumably by forming a complex that can help Vsr bind to its specific mismatch (4). It is possible that MutH and Vsr compete for the same site on MutL; hence, increasing the amount of MutH in cells can overcome the mutagenic effects of Vsr. Thus, MutS and MutL may enhance VSP repair by independent mechanisms, MutS by bending DNA and MutL by forming a transient complex with Vsr.
Acknowledgments
We thank M. Marinus, M. Radman, J. P. Claverys, and P. Modrich for providing bacterial strains or plasmids.
The work in the laboratory of A.S.B. is supported by National Institutes of Health grant GM 53273.
REFERENCES
- 1.Allen D J, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith J D. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au K G, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. J Biol Chem. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 3.Doiron K M J, Viau S, Koutroumanis M, Cupples C G. Overexpression of vsr in Escherichia coli is mutagenic. J Bacteriol. 1996;178:4294–4296. doi: 10.1128/jb.178.14.4294-4296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drotschmann K, Aronshtam A, Fritz H J, Marinus M G. The Escherichia coli MutL protein stimulates binding of Vsr and MutS to heteroduplex DNA. Nucleic Acids Res. 1998;26:948–953. doi: 10.1093/nar/26.4.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gradia S, Acharya S, Fishel R. The human mismatch recognition complex hMSH2-hMSH6 functions as a novel molecular switch. Cell. 1997;91:995–1005. doi: 10.1016/s0092-8674(00)80490-0. [DOI] [PubMed] [Google Scholar]
- 6.Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R. hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell. 1999;3:255–261. doi: 10.1016/s1097-2765(00)80316-0. [DOI] [PubMed] [Google Scholar]
- 7.Hall M C, Matson S W. The Escherichia coli MutL protein physically interacts with MutH and stimulates the MutH-associated endonuclease activity. J Biol Chem. 1999;274:1306–1312. doi: 10.1074/jbc.274.3.1306. [DOI] [PubMed] [Google Scholar]
- 8.Jones M, Wagner R, Radman M. Mismatch repair of deaminated 5-methyl-cytosine. J Mol Biol. 1987;194:155–159. doi: 10.1016/0022-2836(87)90724-8. [DOI] [PubMed] [Google Scholar]
- 9.Junop M S, Obmolova G, Rausch K, Hsieh P, Yang W. Composite active site of an ABC ATPase: MutS uses ATP to verify mismatch recognition and authorize DNA repair. Mol Cell. 2001;7:1–12. doi: 10.1016/s1097-2765(01)00149-6. [DOI] [PubMed] [Google Scholar]
- 10.Lamers M H, Perrakis A, Enzlin J H, Winterwerp H H, de Wind N, Sixma T K. The crystal structure of DNA mismatch repair protein MutS binding to a G · T mismatch. Nature. 2000;407:711–717. doi: 10.1038/35037523. [DOI] [PubMed] [Google Scholar]
- 11.Lieb M. Bacterial genes mutL, mutS, and dcm participate in repair of mismatches at 5-methylcytosine sites. J Bacteriol. 1987;169:5241–5246. doi: 10.1128/jb.169.11.5241-5246.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieb M. Specific mismatch correction in bacteriophage lambda crosses by very short patch repair. Mol Gen Genet. 1983;191:118–125. doi: 10.1007/BF00330898. [DOI] [PubMed] [Google Scholar]
- 13.Lieb M. Spontaneous mutation at a 5-methylcytosine hotspot is prevented by very short patch (VSP) mismatch repair. Genetics. 1991;128:23–27. doi: 10.1093/genetics/128.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lieb M, Bhagwat A S. VSP repair: reducing the cost of cytosine methylation. Mol Microbiol. 1996;20:467–473. doi: 10.1046/j.1365-2958.1996.5291066.x. [DOI] [PubMed] [Google Scholar]
- 15.Lieb M, Rehmat S. Very short patch repair of T:G mismatches in vivo: importance of context and accessory proteins. J Bacteriol. 1995;177:660–666. doi: 10.1128/jb.177.3.660-666.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Macintyre G, Doiron K M J, Cupples C G. The Vsr endonuclease of Escherichia coli: an efficient DNA repair enzyme and a potent mutagen. J Bacteriol. 1997;179:6048–6052. doi: 10.1128/jb.179.19.6048-6052.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Obmolova G, Ban C, Hsieh P, Yang W. Crystal structures of mismatch repair protein MutS and its complex with a substrate DNA. Nature. 2000;407:703–710. doi: 10.1038/35037509. [DOI] [PubMed] [Google Scholar]
- 18.Rasmussen L J, Samson L, Marinus M G. Dam-directed DNA mismatch repair. In: Nickoloff J A, Hoekstra M F, editors. DNA damage and repair. Vol. 1. Totowa, N.J: Human Press; 1998. pp. 205–228. [Google Scholar]
- 19.Sohail A, Lieb M, Dar M, Bhagwat A S. A gene required for very short patch repair in Escherichia coli is adjacent to the DNA cytosine methylase gene. J Bacteriol. 1990;172:4214–4221. doi: 10.1128/jb.172.8.4214-4221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Su S S, Lahue R S, Au K G, Modrich P. Mispair specificity of methyl-directed DNA mismatch correction in vitro. J Biol Chem. 1988;263:6829–6835. [PubMed] [Google Scholar]
- 21.Su S S, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proc Natl Acad Sci USA. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsutakawa S E, Jingami H, Morikawa K. Recognition of a TG mismatch: the crystal structure of very short patch repair endonuclease in complex with a DNA duplex. Cell. 1999;99:615–623. doi: 10.1016/s0092-8674(00)81550-0. [DOI] [PubMed] [Google Scholar]
- 23.Wu T H, Marinus M G. Deletion mutation analysis of the mutS gene in Escherichia coli. J Biol Chem. 1999;274:5948–5952. doi: 10.1074/jbc.274.9.5948. [DOI] [PubMed] [Google Scholar]
- 24.Wu T-H, Marinus M G. Dominant negative mutator mutations in the mutS gene of Escherichia coli. J Bacteriol. 1994;176:5393–5400. doi: 10.1128/jb.176.17.5393-5400.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]