Abstract
We showed previously that transcription of a plasmid-borne kan allele increases C-to-T mutations in the nontranscribed strand. Using two new plasmid-borne kan alleles, one cmp allele, and a chromosomal kan allele, we found in this study that transcription-induced mutations are not limited to specific genes, alleles, or locations and are likely to be a general property of transcript elongation in Escherichia coli.
Hydrolytic deamination of cytosines in DNA creates uracils (12). If they are not repaired, uracils are replaced by thymine after two rounds of replication, resulting in C-to-T mutations. The double-helical structure of DNA provides excellent protection against the deamination reaction, reducing its rate by 140-fold compared to the rate for single-stranded DNA (8, 11). Based on these observations, we speculated that the nontranscribed (nontemplate) strand may suffer more cytosine deaminations when a gene is being actively transcribed because of the transient single-stranded state of the strand.
To test this idea, we constructed an allele of the kanamycin resistance gene, kan, from Tn5 which reverted by C-to-T mutations in the nontranscribed strand. When this allele, kanS-D94, was introduced into an Escherichia coli strain deficient in excision of uracil from DNA (uracil-DNA glycosylase deficient, genotype ung), it reverted at a higher frequency when it was transcribed from strong promoters. These promoters included the tac promoter (1, 5), a tac-derived promoter in which an UP element from an rRNA gene (9) was inserted upstream of the promoter (UP-tac [4]), and a bacteriophage T7 promoter (6). With these plasmid-borne or mini-F-based constructs, induction of transcription of the kan allele increased the frequency of C-to-T mutations up to 10-fold (4, 5). The absence of Ung in the cells allowed fixation of uracil as thymine through DNA replication, and consequently, the increases in reversion frequency were directly related to increases in cytosine deamination. Using this genetic system, we showed that the frequency of C-to-T mutations increased with increasing levels of isopropyl-β-d-thiogalactopyranoside (IPTG) in growth media (4). We also showed that transcription-induced C-to-T mutations (TIM) increased about twofold in a ung+ strain (5).
Although the results described above support the hypothesis that the nontranscribed strand is more susceptible to C-to-U deamination during transcription, all of the work was done with a single kan allele. It is possible that some special structural feature at the site of the target cytosine, such as a hairpin or a transcriptional pause site, is responsible for the effect observed. Furthermore, much of our previous work on TIM was done with multicopy plasmids, and it could be argued that the transcription elongation complexes for plasmid-borne genes may be different in some way from those for chromosomal genes. We did demonstrate TIM in a mini-F-based genetic system (6) that was almost single copy, but in this case the gene was transcribed by the T7 RNA polymerase. In the present study, we found that TIM can occur at many gene locations, including a gene in the E. coli chromosome.
Construction of new alleles that revert by C-to-T change.
The previously described kan allele contains a mutation at codon 94 that reverts by C-to-T change (18). In this study we used two new plasmid-borne kan alleles that are defective in the ability to confer the kanamycin resistance phenotype on their hosts (phenotype Kans) (Table 1). While one of the new alleles also contains sequence changes at codon 94 (pUP41), the other allele contains changes at codon 212 (plasmid pUP51) (Table 1). We have shown elsewhere (2, 3) that these alleles, which contain Leu-to-Pro codon changes, revert through a C-to-T transition. These alleles contain the mutable cytosine in different sequence contexts (Table 1) and were cloned under the control of the UP-tac promoter. To construct plasmid pUP41, an MscI-NcoI fragment in the pUP31 plasmid was replaced with the corresponding fragment from plasmid pKanS94H. The latter plasmid contains the kanS-H94 allele (3). Similarly, a MscI-BstBI fragment in pUP31 was replaced with the corresponding fragment from plasmid pKanS212D to obtain pUP51. The latter plasmid contains the kanS-D212 allele (2).
TABLE 1.
Sequence context and location of target cytosine
| Gene | Allele | Promotera | Sequenceb | Location(s) |
|---|---|---|---|---|
| kan | kanS-D94 | UP-tac | CTACCAGGC | Plasmid and chromosomal |
| kanS-H94 | UP-tac | CTCCCGGGC | Plasmid | |
| kanS-D212 | UP-tac | CGGCCAGGT | Plasmid | |
| cmp | cmpS-S146P | tac | CGTGCCGGC | Plasmid |
The UP-tac and tac promoters are negatively regulated by the repressor encoded by the lacIq gene present in the same replicon as the antibiotic resistance alleles.
The target cytosine is underlined.
Additionally, we constructed an allele of the chloramphenicol acetyltransferase (CAT) gene that also reverts by a C-to-T change. There is a conserved serine in all CATs (serine 146 in CAT I) that is responsible for transition state stabilization and is essential for catalysis (10). We introduced a T-to-C mutation in the first position of the serine 146 codon (TCA) in cat in plasmid pRW2042 (15) by unique-site elimination mutagenesis (7), which changed the codon to a proline codon (CCG). The resulting plasmid, pCAT146P, is sensitive to chloramphenicol and contains the cmpS-S146P allele under control of the tac promoter.
Transcription-induced reversion of plasmid-borne antibiotic resistance alleles.
The newly constructed plasmids were introduced into an ung strain of E. coli, BH156 (13), by transformation, and several independent colonies for each plasmid were grown in parallel cultures in Luria-Bertani (LB) media. The effects of transcription on the frequency of acquisition of resistance to the appropriate antibiotic were determined by using previously described procedures (5). Briefly, each culture was diluted and split into two cultures, and these two cultures were then grown for 3 h. For each pair of cultures one of the cultures was grown in the absence of an inducer, and the other was grown in the presence of IPTG (150 μM for plasmids with the UP-tac promoter and 1 mM for the tac promoter). Following growth, cells were harvested and plated onto LB medium plates to determine the total number of viable cells and onto LB medium with kanamycin (50 μg/ml) or chloramphenicol (40 μg/ml) to determine the number of kanamycin-resistant (Kanr) or chloramphenicol-resistant (Cmpr) revertants. The revertant frequency was the ratio of the number of Kanr or Cmpr cells to the total number of cells.
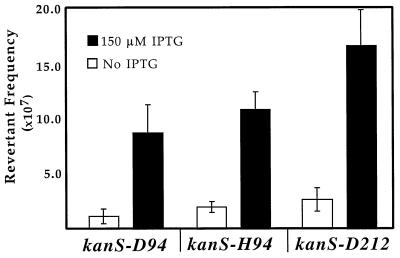
As Fig. 1 shows, all of the kan alleles reverted at a higher frequency when cells were grown in the presence of the inducer, IPTG. The Kanr revertant frequency in the presence of IPTG was five- to sevenfold greater than the frequency in the uninduced culture and was greater than the increase observed with the weaker tac promoter (fourfold) (5). It should be noted that the UP-tac promoter is a very strong promoter and that concentrations of IPTG higher than 150 μM significantly inhibited cell growth (4). Consequently, we were unable to determine the effects of fully induced UP-tac promoter on TIM.
FIG. 1.
Transcription-induced reversion of kan alleles. For each plasmid the mean revertant frequency based on seven or more independent cultures is shown. The error bars indicate standard deviations.
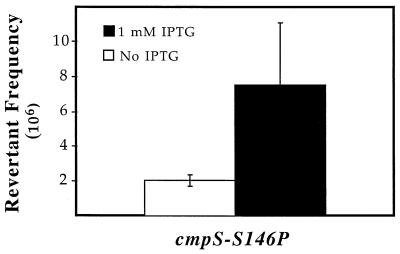
Transcription of the cmp allele also increased the frequency of Cmpr revertants (Fig. 2). In these experiments the selective plates contained 100 μM IPTG in addition to chloramphenicol. This was necessary because the tac promoter has a lower basal level of expression than the UP-tac promoter, and consequently, expression of Cmpr revertants requires partial induction of transcription on plates. Under these conditions, full induction of the tac promoter during growth in liquid cultures resulted in an approximately fourfold increase in the frequency of revertants (Fig. 2). The increase in mutation frequency was smaller for the cmp allele than for the kan alleles because as noted previously, even upon complete induction the tac promoter does not transcribe as frequently as the UP-tac promoter in the presence of 150 μM IPTG (4). These results show that TIM is not dependent on the gene used for the reversion assay or the sequence surrounding the mutable cytosine.
FIG. 2.
Transcription-induced reversion of cmp allele. The mean revertant frequencies based on nine independent cultures are shown. The error bars indicate standard deviations.
TIM in a chromosomal gene.
To demonstrate that TIM can also occur in a chromosomal gene transcribed by the E. coli RNA polymerase, kanS-D94 expressed from the UP-tac promoter and the lacIq gene were cloned into bacteriophage λ. This was accomplished by releasing these genes from pUP31 as a BstZ17I-StuI fragment and ligating the fragment at the BamHI site of λD69 DNA (14). The DNA ligation mixture was packaged by using Gigapack III Gold packaging extract (Stratagene, La Jolla, Calif.) according to manufacturer's instructions, and the resulting phage plaques were screened with the red plaque test (16) and by PCR. We obtained two classes of recombinant λ phages containing the kan allele in the two possible orientations with respect to phage promoter PI (λ-UP71 and λ-UP81). It is known (17) that in one orientation residual transcription from PI results in a higher basal level of transcription of cloned genes. To maintain low basal transcription levels and background reversion frequency, the construct in which the direction of kanS-D94 transcription was opposite that of promoter PI (λUP81) was selected for further work. This phage was lysogenized into the chromosome of BH156 by using helper and selection phages from a λDE3 lysogenization kit (Novagen, Madison, Wis.), and the resulting lysogen was used in reversion assays.
In the absence of IPTG, the kanS-D94 lysogen reverted to Kanr at a low frequency (mean, 2.9 × 10−8; nine cultures). Upon induction of the UP-tac promoter, the mutation frequency increased more than 10-fold (mean, 3.1 × 10−7). It should be noted that in these experiments the presence of the UP-tac promoter in the chromosome rather than a multicopy plasmid allowed the use of 1 mM IPTG for induction of transcription without affecting cell growth. Presumably, the higher IPTG concentration resulted in higher levels of transcription of the kan gene and a larger increase in the revertant frequency. The results obtained clearly show that TIM does not depend on having the test gene on an extrachromosomal genetic element.
Concluding remarks.
We demonstrated in this study that cytosines in four different sequence contexts are more susceptible to deamination when they are present in the nontranscribed strand of an actively transcribed gene. The sequences used were chosen for the convenience of the reversion assay and without regard to their ability to form any secondary structure. Consequently, the target cytosines in these alleles are flanked by different sequences on both sides (Table 1), and it seems unlikely that all of the sequences adopt some common secondary structure that would cause the cytosines to mutate in a transcription-dependent fashion. Additionally, we found that for one kan allele increases in the number of mutations occurred in response to transcription regardless of whether the gene was present in a multicopy plasmid or in the chromosome. When combined with the finding that TIM also occur in a kan gene transcribed by the T7 RNA polymerase and when the gene is in a mini-F (6), these results show that TIM occur independent of the sequence context of the mutable C, the gene in which the cytosine is present, the replicon that carries the allele, and the RNA polymerase that transcribes the gene. In other words, TIM is likely to be a general property of transcript elongation in E. coli.
Acknowledgments
We thank R. Weisberg (National Cancer Institute, Frederick, Md.) and M. Gottesman (Columbia University, New York, N.Y.) for providing λD69 phage and strains for the red plaque test, respectively.
This work was supported by the NIH grant GM57200 to A.S.B.
REFERENCES
- 1.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 2.Bandaru B. Ph.D. thesis. Detroit, Mich: Wayne State University; 1996. [Google Scholar]
- 3.Bandaru B, Wyszynski M, Bhagwat A S. HpaII methyltransferase is mutagenic in Escherichia coli. J Bacteriol. 1995;177:2950–2952. doi: 10.1128/jb.177.10.2950-2952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beletskii A, Bhagwat A S. Correlation between transcription and C to T mutations in the non-transcribed DNA strand. Biol Chem. 1998;379:549–551. [PubMed] [Google Scholar]
- 5.Beletskii A, Bhagwat A S. Transcription-induced mutations: increase in C to T mutations in the nontranscribed strand during transcription in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:13919–13924. doi: 10.1073/pnas.93.24.13919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beletskii A, Grigoriev A, Joyce S, Bhagwat A S. Mutations induced by bacteriophage T7 RNA polymerase and their effects on the composition of T7 genome. J Mol Biol. 2000;300:1057–1065. doi: 10.1006/jmbi.2000.3944. [DOI] [PubMed] [Google Scholar]
- 7.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–88. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 8.Frederico L A, Kunkel T A, Shaw B R. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 9.Gourse R L, Ross W, Gaal T. Ups and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol. 2000;37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 10.Lewendon A, Murray I A, Shaw W V, Gibbs M R, Leslie A G. Evidence for transition-state stabilization by serine-148 in the catalytic mechanism of chloramphenicol acetyltransferase. Biochemistry. 1990;29:2075–2080. doi: 10.1021/bi00460a016. [DOI] [PubMed] [Google Scholar]
- 11.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 12.Lindahl T, Nyberg B. Heat-induced deamination of cytosine residues in deoxyribonucleic acid. Biochemistry. 1974;13:3405–3410. doi: 10.1021/bi00713a035. [DOI] [PubMed] [Google Scholar]
- 13.Lutsenko E, Bhagwat A S. Principal causes of hot spots for cytosine to thymine mutations at sites of cytosine methylation in growing cells. A model, its experimental support and implications. Mutat Res. 1999;437:11–20. doi: 10.1016/s1383-5742(99)00065-4. [DOI] [PubMed] [Google Scholar]
- 14.Mizusawa S, Ward D F. A bacteriophage lambda vector for cloning with BamHI and Sau3A. Gene. 1982;20:317–322. doi: 10.1016/0378-1119(82)90200-1. [DOI] [PubMed] [Google Scholar]
- 15.Rahmouni A R, Wells R D. Direct evidence for the effect of transcription on local DNA supercoiling in vivo. J Mol Biol. 1992;223:131–144. doi: 10.1016/0022-2836(92)90721-u. [DOI] [PubMed] [Google Scholar]
- 16.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. [Google Scholar]
- 17.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 18.Wyszynski M, Gabbara S, Bhagwat A S. Cytosine deaminations catalyzed by DNA cytosine methyltransferases are unlikely to be the major cause of mutational hot-spots at sites of cytosine methylation in E. coli. Proc Natl Acad Sci USA. 1994;91:1574–1578. doi: 10.1073/pnas.91.4.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]