Abstract
We report the first evidence of a chromosome-encoded toxin-antitoxin locus in spirochetes. This locus has been found in the pathogenic spirochete Leptospira interrogans and exhibits homologies with the pem/chp loci. The L. interrogans chp locus consists of two genes: chpK (for “killer protein”) and its upstream partner chpI (for “inhibitory protein”). Expression of ChpK in Escherichia coli results in the inhibition of bacterial growth. The coexpression of ChpI neutralizes ChpK toxicity. By Southern blot analysis, chp homologs were found in all representative pathogenic strains of L. interrogans.
The recent discovery of toxin-antitoxin modules in many bacteria suggests that programmed cell death may be a general phenomenon in bacteria. Most of these modules were first identified in plasmids, where they constitute a postsegregational killing system (1). Toxin-antitoxin loci are organized into operons in which the first gene encodes the antitoxin and the second gene encodes the toxin. The product of the toxin gene is long lived and toxic, while the product of the second is short lived and counteracts the cell killing activity of the toxin. When the bacteria lose the plasmid, the degradation of the antitoxins by cellular proteases leads to the activation of the toxins and the selective killing of plasmid-free cells (1, 5). However, toxin-antitoxin loci have now been identified in many bacterial chromosomes and may therefore be involved in functions other than plasmid maintenance. The physiological role of chromosome-encoded toxin-antitoxin systems remains unclear. It has been suggested that they may be part of the response to environmental stimuli, such as nutritional stress (5). The overall design of the suicide strategy of these toxin-antitoxin modules is similar: the lethal effect of the toxin is counteracted by the antitoxin.
Homologs of these chromosome-encoded toxin-antitoxin modules have been identified in gram-negative and gram-positive bacteria and in Archaea (5). However, such modules seem to be absent from the genomes of the spirochetes Borrelia burgdorferi and Treponema pallidum (3, 4). In this study, we report the first evidence of a chromosome-encoded toxin-antitoxin system in spirochetes.
Identification of a chromosome-encoded toxin-antitoxin locus in Leptospira interrogans.
The Chinese Human Genome Center is currently sequencing the pathogenic spirochete L. interrogans serovar icterohaemorrhagiae strain Lai by the whole-genome shotgun strategy (2). The genome of Leptospira spp. contains two chromosomes, a 4,400-kb chromosome and a 350-kb chromosome. DNA sequence analyses revealed that the L. interrogans large chromosome contains a 342-bp open reading frame (ORF) that encodes a putative protein of 113 amino acids with 28% identity with PemK of plasmid R100. The pem (for “plasmid emergency maintenance”) locus of R100 consists of two genes, pemI and pemK. PemK inhibits growth of the host cell, while PemI suppresses the killing effect of PemK. By database searching, homologs of unknown functions were also identified, such as Rv1991c of Mycobacterium tuberculosis, ORF136 of Staphylococcus aureus, and a putative protein encoded by the Bacillus subtilis ydcE gene; they all share 35 to 43% identity with the L. interrogans ChpK putative protein. In a recent review on toxin-antitoxin modules (5), a phylogenetic analysis assigned Rv1991c, ORF136, and YdcE to the ChpK proteins (killer proteins); they all are chromosomally encoded and they have an upstream partner that could correspond to the antitoxin gene. The chp locus is a chromosome-borne toxin-antitoxin module that is homologous to the pem operon. Figure 1 shows an alignment of the putative ChpK proteins and Escherichia coli PemK. Additional homologs of unknown functions (putative proteins of 93 to 120 amino acids) were identified in Staphylococcus epidermidis, Bacillus halodurans, Lactobacillus plantarum, and Lactobacillus reuteri. The genetic organization of toxin-antitoxin loci led us to suspect the existence of a second ORF upstream from the pemK homolog of L. interrogans. Indeed, a small ORF of 249 bp was located upstream of the pemK homolog, but the transcribed protein (82 amino acids) does not have homologs in the databases. The pemK homolog translation start codon overlaps the 249-bp ORF translation stop codon (Fig. 1), a strong indication that the two genes constitute an operon. Sequence analysis of the promoter region of this L. interrogans locus reveals a 10-bp inverted repeat (IR) (Fig. 1), which shares the consensus sequence 5′-GTTATAC-3′ with an IR of the E. coli chpB promoter region (7). In E. coli, IRs in the chp/pem promoter regions correspond to specific DNA binding sites of the Chp/Pem proteins (7, 10). Because of the similarities with the chromosomal chp loci, we refer to the L. interrogans pemK homolog as chpK (for “killer protein”) and its upstream partner as chpI (for “inhibitory protein”). The L. interrogans chp system is referred to as chpL (L for Leptospira).
FIG. 1.
Genetic structure of the chp locus of L. interrogans. (A) Schematic representation of the chp locus. The arrows for chpI and chpK show their transcription orientation. IRs are boxed. (B) Sequence alignment of the putative ChpK proteins of L. interrogans (Lint), M. tuberculosis (Mtub), B. subtilis (Bsub), and S. aureus (Saur) and of the PemK protein of E. coli (Ecol). Residues conserved in at least three homologous proteins are shaded.
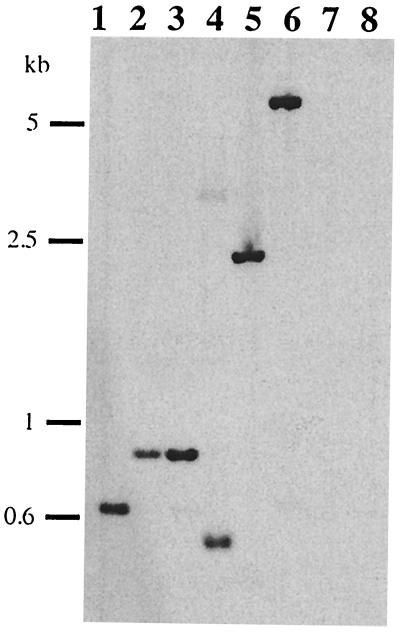
Southern blot analysis showed that chpK was present in a single copy in L. interrogans serovar icterohaemorrhagiae strain Lai (Fig. 2, lane 1). In addition, total genomes of five different serovars of the pathogenic species L. interrogans and two strains of the saprophytic species Leptospira meyeri and Leptospira biflexa were investigated for the presence of chpK homologs. Results revealed a single hybridizing band in all strains tested except saprophytic species (Fig. 2). This discrepancy may be due to the phylogenetic distance between pathogenic and saprophytic Leptospira species. In conclusion, the chpL locus is conserved and distributed in all the pathogenic strains tested.
FIG. 2.
Southern blot analyses of Leptospira spp. Southern blotting of EcoRI-digested DNA was performed as previously described (8), and blots were probed at 50°C with the radiolabeled chpK probe from L. interrogans serovar icterohaemorrhagiae strain Lai. The chpK probe was amplified by PCR with primers PkA (5′-TTC ATT TGG AAG TGA GCC TG-3′) and PkB (5′-AAT CTA AGC CTG TAA CCA AC-3′). Lane 1, L. interrogans serovar icterohaemorrhagiae strain Lai; lane 2, L. interrogans serovar copenhageni strain Wijnberg; lane 3, L. interrogans serovar icterohaemorrhagiae strain Verdun; lane 4, L. interrogans serovar grippotyphosa strain Moskva V; lane 5, L. interrogans serovar canicola strain Hond Utrecht IV; lane 6, L. interrogans serovar sejroe strain M84; lane 7, L. biflexa serovar patoc strain Patoc1; lane 8, L. meyeri serovar semaranga strain Veldrat.
Expression of the toxin and the antitoxin in E. coli.
To see whether the putative chpK gene of L. interrogans encodes a killer protein, we tested the effect of its expression (alone or together with the putative chpI gene) in E. coli. E. coli XL10 (Stratagene) was routinely used during vector constructions. For expression of recombinant proteins in E. coli, we used the pET system and the E. coli BL21(DE3) strain (Novagen) containing a chromosomal copy of the T7 RNA polymerase gene. Briefly, cloned genes are under the control of a strong promoter from bacteriophage T7. Expression of the cloned gene is induced by the T7 RNA polymerase, whose expression is inducible by isopropyl-β-d-thiogalactopyranoside (IPTG). The cloned gene may therefore have an extremely low transcriptional activity in an uninduced state, which is important for expression of proteins potentially toxic to the host cell.
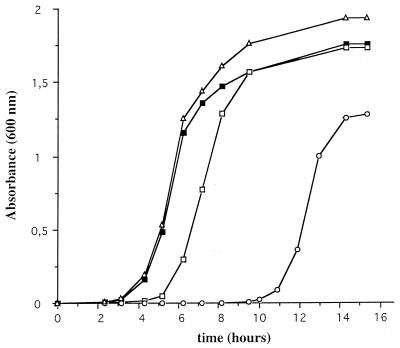
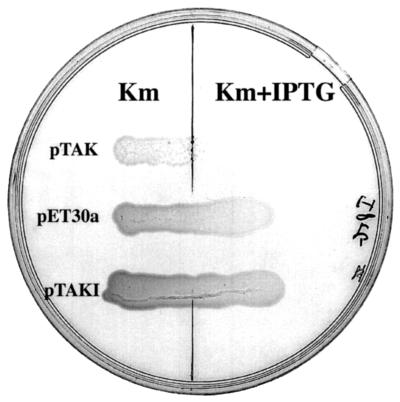
The coding region of the chpK gene was PCR amplified from L. interrogans (strain Lai) by using the sense primer 5′-GGA ATT CCA TAT GAT TCG TGG TG-3′ and the antisense primer 5′-TAA CGG GAT CCA GGT TTG GGA G-3′. NdeI and BamHI restriction sites, which were added at the 5′ ends of the sense and antisense primers, respectively, are underlined. After gel purification, NdeI-BamHI double digestion, and another step of DNA purification, the PCR product was inserted into the NdeI-BamHI sites of the polylinker of the E. coli expression vector pET30a (Novagen) to generate the plasmid pTAK. Plasmid constructs were checked by DNA sequence analysis. Transformation efficiencies of pET30a and pTAK were similar in E. coli BL21(DE3). Inoculation of E. coli transformants in Luria-Bertani (LB) liquid media supplemented with 50 μg of kanamycin per ml, but without the IPTG inducer, showed dramatic growth differences, suggesting background expression of the cloned chpK gene. The leaky activity of the uninduced promoter of pET30a is sufficient to inhibit the growth of cells harboring pTAK, at least for the first 10 h (Fig. 3). After 10 h, cells start to grow and enter the exponential phase. Interestingly, under similar conditions, the same period (10 h) was found in three independent experiments. Restriction analysis of plasmids extracted from overnight cultures of cells harboring pTAK did not show obvious DNA rearrangements (data not shown). When overnight cultures were reinoculated in fresh liquid media, cells harboring pTAK still exhibited a much longer lag period than the wild-type strain, suggesting that toxin resistance was not stably maintained. Cells harboring pTAK were unable to grow on LB solid media supplemented with the IPTG inducer (Fig. 4), suggesting that there was a lethal effect of L. interrogans ChpK in E. coli. The L. interrogans chpK gene may therefore encode a toxin protein.
FIG. 3.
Effect of the expression of the L. interrogans ChpK in the presence or absence of ChpI on growth of E. coli cells harboring pET30a (filled square), pTAK (chpK with an AUG start codon) (open circles), pTGK (chpK with a GUG start codon) (open squares), and pTAKI (chpK and chpI) (open triangles) in LB liquid media. Growth was determined by measuring the optical density at 600 nm.
FIG. 4.
Growth of E. coli expressing L. interrogans ChpK in the presence or absence of 100 μM IPTG. Cells harboring pET30a, pTAK (chpK), and pTAKI (chpK chpL) are indicated. Plates supplemented with 50 μg of kanamycin (Km) per ml were incubated for 24 h at 37°C.
As suggested by Gerdes (5), we also replaced the AUG start codon of the putative toxin gene with GUG by amplifying the L. interrogans chpK gene with the sense primer 5′-GGA ATT CAT ATG TGA TTC GTG GTG-3′ and the previously used antisense primer to generate pTGK. This change is supposed to reduce the level of gene expression 5- to 10-fold. This was indeed the case: growth of cells harboring pTGK were only slightly inhibited, and the lag period was 2 h longer than that of normally growing cells (Fig. 3). Cells harboring pTGK did not exhibit any obvious defect during exponential growth, and they reached stationary phase with the same optical density as cells harboring pET30a. This indicates that even when the AUG start codon is replaced, we still detect a low expression of the toxin protein ChpK in an uninduced state, leading to a slight inhibition of cell growth in comparison to cells harboring pET30a.
To determine whether the expression of the putative L. interrogans chpI gene can suppress the inhibitory function of L. interrogans ChpK, we introduced L. interrogans chpI into pTAK. As described above, the chpI coding region was PCR amplified from L. interrogans by using the sense primer 5′-GGA ATT CCA TAT GAA GAC GGC G-3′ and the antisense primer 5′-TAA CGG GAT CCA GAA CTG GAC G-3′. The amplified product was digested with NdeI and BamHI and inserted into the NdeI-BamHI restriction sites of pET30a (Novagen) to generate the plasmid pTAI. A SphI-BamHI fragment containing the chpI gene under the control of the pET30a promoter was then released from pTAI, blunt ended, and inserted into the unique FspI site of pTAK to generate pTAKI. Growth of the cells harboring pTAKI, which carries both chpK and chpI genes under the control of pET30a promoters, was not inhibited (Fig. 3); the growth curve was similar to that of cells harboring pET30a. Growth was also restored in the presence of IPTG (Fig. 4). This shows that L. interrogans ChpI suppresses the inhibitory function of L. interrogans ChpK and restores normal growth. It is likely that, in both L. interrogans and E. coli, the two proteins interact directly and that toxicity of ChpK is due to the loss of that interaction.
Conclusions.
Our data clearly demonstrate that the chpL system of L. interrogans belong to the toxin-antitoxin family: the expression of the chpK gene is toxic to E. coli cells, and the product of the chpI gene counteracts the toxin in E. coli. Our results also suggest that the molecular target of the ChpK protein is conserved between E. coli and L. interrogans. Similarly, another study has recently shown that a chromosome-encoded toxin-antitoxin from E. coli (the relBE locus, which does not belong to the chp/pem family) was able to inhibit the growth of yeast cells and that RelE and RelB interact in yeast (6). At present, the cellular target of ChpK proteins remains unknown. On the basis of a study on the pem locus, one can hypothesize that ChpK proteins inhibit DNA replication of the host cell (9). Further study of L. interrogans ChpK and ChpI in E. coli could help in understanding the cellular target of these toxins and their physiological role in bacteria.
Nucleotide sequence accession number.
The GenBank accession number for the L. interrogans chpL locus is AF395875.
Acknowledgments
M. Picardeau and S. Ren contributed equally to this work.
We thank K. Gerdes for critical reading of the manuscript, C. Le Dantec for her participation in part of this work, and C. Buchrieser for help with Artemis software. We also thank Xiugao Jiang and Jianguo Xu (The Institute of Epidemiology and Microbiology, Chinese Academy of Preventive Medicine), Yan Shen (Chinese National Human Genome Center, Beijing), Yumei Wen (Fudan University), Zhu Chen (Chinese Human Genome Center, Shanghai), and Guoping Zhao (Shanghai Institute of Biology Sciences).
This work received support from the Institut Pasteur, the National Natural Science Foundation of China, the Shanghai Commission for Science and Technology, and Programme de Recherches Avancées Franco-Chinois (PRA B00-05).
REFERENCES
- 1.Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann R, Adams M, White O, Clayton R, Kirkness E, Kerlavage A, Bult C, Tomb J, Dougherty B, Merrick J, et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 3.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature. 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 4.Fraser C M, Norris S J, Weinstock C M, White O, Sutton G G, Dodson R, Gwinn M, Hickey E K, Clayton R, Ketchum K A, et al. Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science. 1998;281:375–388. doi: 10.1126/science.281.5375.375. [DOI] [PubMed] [Google Scholar]
- 5.Gerdes K. Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol. 2000;182:561–572. doi: 10.1128/jb.182.3.561-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kristoffersen P, Jensen G B, Gerdes K, Piskur J. Bacterial toxin-antitoxin gene system as containment control in yeast cells. Appl Environ Microbiol. 2000;66:5524–5526. doi: 10.1128/aem.66.12.5524-5526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Picardeau M, Brenot A, Saint Girons I. First evidence for gene replacement in Leptospira spp. Inactivation of L. biflexa flaB results in non-motile mutants deficient in endoflagella. Mol Microbiol. 2001;40:189–199. doi: 10.1046/j.1365-2958.2001.02374.x. [DOI] [PubMed] [Google Scholar]
- 9.Ruiz-Echevarria M J, Gimenez-Gallego G, Sabariegos-Jareno R, Diaz-Orejas R. Kid, a small protein of the parD stability system of plasmid R1, is an inhibitor of DNA replication acting at the initiation of DNA synthesis. J Mol Biol. 1995;247:568–577. doi: 10.1006/jmbi.1995.0163. [DOI] [PubMed] [Google Scholar]
- 10.Tsuchimoto S, Ohtsubo E. Autoregulation by cooperative binding of the PemI and PemK proteins to the promoter region of the pem operon. Mol Gen Genet. 1993;237:81–88. doi: 10.1007/BF00282787. [DOI] [PubMed] [Google Scholar]