Abstract
Classical microbiology techniques are relatively slow in comparison to other analytical techniques, in many cases due to the need to culture the microorganisms. Furthermore, classical approaches are difficult with unculturable microorganisms. More recently, the emergence of molecular biology techniques, particularly those on antibodies and nucleic acid probes combined with amplification techniques, has provided speediness and specificity to microbiological diagnosis. Flow cytometry (FCM) allows single- or multiple-microbe detection in clinical samples in an easy, reliable, and fast way. Microbes can be identified on the basis of their peculiar cytometric parameters or by means of certain fluorochromes that can be used either independently or bound to specific antibodies or oligonucleotides. FCM has permitted the development of quantitative procedures to assess antimicrobial susceptibility and drug cytotoxicity in a rapid, accurate, and highly reproducible way. Furthermore, this technique allows the monitoring of in vitro antimicrobial activity and of antimicrobial treatments ex vivo. The most outstanding contribution of FCM is the possibility of detecting the presence of heterogeneous populations with different responses to antimicrobial treatments. Despite these advantages, the application of FCM in clinical microbiology is not yet widespread, probably due to the lack of access to flow cytometers or the lack of knowledge about the potential of this technique. One of the goals of this review is to attempt to mitigate this latter circumstance. We are convinced that in the near future, the availability of commercial kits should increase the use of this technique in the clinical microbiology laboratory.
Microbiology in general and clinical microbiology in particular have witnessed important changes during the last few years (82). An issue for microbiology laboratories compared with other clinical laboratories is the relative slowness of definitive reports. Traditional methods of bacteriology and mycology require the isolation of the organism prior to identification and other possible testing. In most cases, culture results are available in 48 to 72 h. Virus isolation in cell cultures and detection of specific antibodies have been widely used for the diagnosis of viral infections (181). These methods are sensitive and specific, but, again, the time required for virus isolation is quite long and is governed by viral replication times. Additionally, serological assays on serum from infected patients are more useful for determining chronic than acute infections. Life-threatening infections require prompt antimicrobial therapy and therefore need rapid and accurate diagnostic tests. Procedures which do not require culture and which detect the presence of antigens or the host's specific immune response have shortened the diagnostic time. More recently, the emergence of molecular biology techniques, particularly those based on nucleic acid probes combined with amplification techniques, has provided speediness and specificity to microbiological diagnosis (139). These techniques have led to a revolutionary change in many of the traditional routines used in clinical microbiology laboratories. Results are offered quickly, the diagnosis of emerging infections has become easier, and unculturable pathogens have been identified (109).
On the other hand, the current organization of clinical microbiology laboratories is now subject to automation and competition, both overshadowed by increasing costs (282, 339). Increased use of automation in clinical microbiology laboratories is best exemplified by systems used for detecting bacteremia, screening of urinary tract infections, antimicrobial susceptibility testing, and antibody detection. To obtain better sensitivity and speed, manufacturers continuously modify all these systems. Nevertheless, the equipment needed for all these approaches is different, and therefore the initial costs, both in equipment and materials, are high.
Flow cytometry (FCM) could be successfully applied to most of these situations. In bacteremia and bacteriuria, FCM would not only rapidly detect organisms responsible for the infection but would also initially identify the type of microorganism on the basis of its cytometric characteristics. Although FCM offers a broad range of potential applications for susceptibility testing, a major contribution would be in testing for slow-growing microorganisms, such as mycobacteria and fungi (108, 163, 262). Results are obtained rapidly, frequently in less than 4 h; when appropriately combined with the classical techniques, FCM may offer susceptibility results even before the microorganism has been identified. The most outstanding contribution offered by FCM is the detection of mixed populations, which may respond to antimicrobial agents in different ways (331).
This technique could also be applied to study the immune response in patients, detect specific antibodies (27, 133), and monitor clinical status after antimicrobial treatments (58, 244). Moreover, when properly applied, FCM can be adjusted to use defined parameters that avoid subjectivity and aid the clinical microbiologist in the interpretation of specific results, particularly in the field of rapid diagnosis.
TECHNICAL BASIS OF FLOW CYTOMETRY
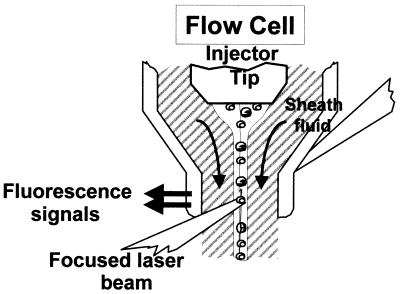
FCM is an analytical method that allows the rapid measurement of light scattered and fluorescence emission produced by suitably illuminated cells. The cells, or particles, are suspended in liquid and produce signals when they pass individually through a beam of light (Fig. 1). Since measurements of each particle or cell are made separately, the results represent cumulative individual cytometric characteristics. An important analytical feature of flow cytometers is their ability to measure multiple cellular parameters (analytical flow cytometers). Some flow cytometers are able to physically separate cell subsets (sorting) based on their cytometric characteristics (cell sorters) (Fig. 2). The scattered light (intrinsic parameters) and fluorescence emissions of each particle are collected by detectors and sent to a computer, where the distribution of the population with respect to the different parameters is represented. Scattered light collected in the same direction as the incident light is related to cell size, and scattered light collected at an angle of 90° gives an idea of the particle complexity. This parameter is related to cell surface roughness and the number of organelles present in the cell. Size and complexity are considered intrinsic parameters since they can be obtained without having to stain the sample. To obtain additional information, samples can be stained using different fluorochromes. Fluorochromes can be classified according to their mechanism of action (127): those whose fluorescence increases with binding to specific cell compounds such as proteins (fluorescein isothiocyanate [FITC]), nucleic acids (propidium iodide [PI]), and lipids (Nile Red); those whose fluorescence depends on cellular physiological parameters (pH, membrane potential, etc.); and those whose fluorescence depends on enzymatic activity (fluorogenic substrates) such as esterases, peroxidases, and peptidases (Table 1). Fluorochromes can also be conjugated to antibodies or nucleotide probes to directly detect microbial antigens or DNA and RNA sequences.
FIG. 1.
Light-scattering and fluorescence signal production at the flow cell analysis point of the flow cytometer. From Purdue Cytometry CD-ROM vol. 1 (adapted with permission of the publisher).
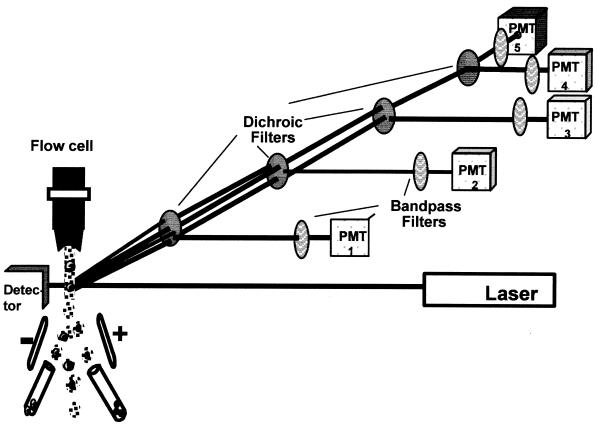
FIG. 2.
Scheme of optic (dichroic mirrors and bandpass filters) and illumination (laser) systems of a flow cytometer with six parameters detected (size, granularity, and four fluorescences) by separate photomultiplier tubes (except size, which can be detected by photodiode or a PMT tube) and sorting capacity. From Purdue Cytometry CD-ROM vol. 1 (adapted with permission of the publisher).
TABLE 1.
Some of the fluorescent molecules used to study microorganisms by flow cytometry
| Dye | Excitation wavelength (λmax) (nm) | Emission wavelength (λmax) (nm) | Ligand or substrate | Applications |
|---|---|---|---|---|
| TOTO-3 | 642 | 660 | DNA, RNA | DNA quantification, cell cycle studies |
| SYTOX Green | 504 | 525 | DNA, RNA | Viability, DNA quantification |
| PI | 536 | 625 | DNA, RNA | Viability, DNA quantification, cell cycle studies |
| Ethidium bromide | 510 | 595 | DNA, RNA | DNA quantification, cell cycle studies |
| Hoechst 33258/33342 | 340 | 450 | DNA (GC pairs) | Cell cycle studies |
| SYTO 13 | 488 | 509 | DNA, RNA | Viability, DNA quantification, cell cycle studies |
| Mithramycin | 425 | 550 | DNA | Cell cycle studies |
| Pyronine Y | 497 | 563 | RNA | RNA quantification |
| FITC | 495 | 525 | Protein | Microbe detection |
| Texas Red (sulforhodamine isothiocyanate) | 580 | 620 | Protein | Microbe detection |
| Oregon Green isothiocyanate | 496 | 526 | Protein | Microbe detection |
| Indo-1 | 340 | 398–485 | Ca2+ | Ca2+ mobilization |
| Fura-2 | 340 | 549 | Ca2+ | Ca2+ mobilization |
| Fluor-3 | 469 | 545 | Ca2+ | Ca2+ mobilization |
| BCECF | 460–510 | 520–610 | pH | Metabolic variations |
| SNARF-1 | 510 | 587–635 | pH | Metabolic variations |
| DIOC6(3) | 484 | 501 | Membrane potential | Antibiotic susceptibility, metabolic variations |
| Oxonol [DiBAC4(3)] | 488 | 525 | Membrane potential | Antibiotic susceptibility, metabolic variations |
| Rhodamine 123 | 507 | 529 | Membrane potential (mitochondria) | Antibiotic susceptibility, metabolic variations |
| Fun-1 | 508 | 525–590 | Yeast vacuolar enzyme activity | Yeast metabolic state |
| Nile Red | 490–550 | 540–630 | Lipids | |
| Lectins | Depends on fluorochrome conjugated | Depends on fluorochrome conjugated | Membrane oligosaccharides | Cell wall composition, microbe detection |
| Fluorescently labeled oligonucleotides | Depends on fluorochrome conjugated | Depends on fluorochrome conjugated | Nucleotide sequences | Microbe identification |
| Calcofluor white | 347 | 436 | Chitin and other carbohydrate polymers | Fungal detection |
| Substrates linked to fluorochromes | Enzyme activities | Metabolic activity | ||
| Antibodies labeled with flurochormes | Antigens | Microbe detection |
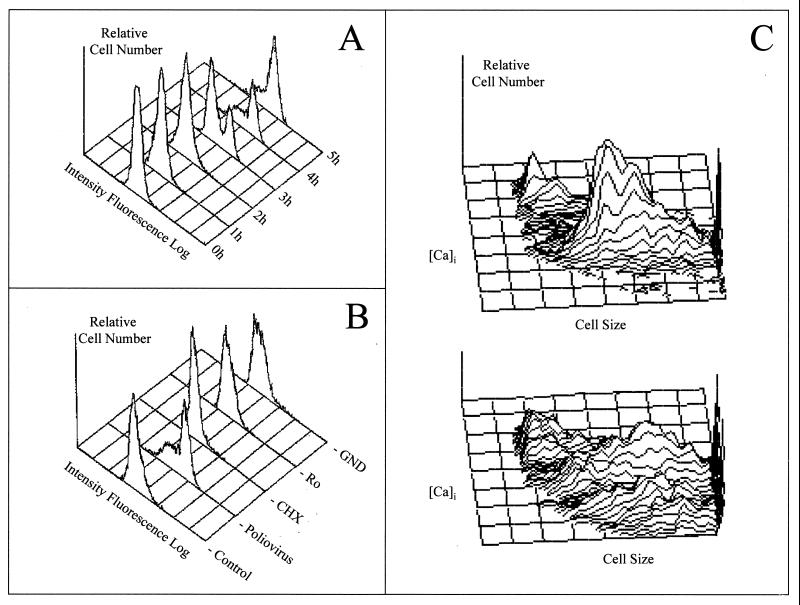
A typical flow cytometer has several parts. (i) The hydraulic system produces the fluid stream, with a liquid sheath surrounding the cell suspension (hydrodynamic focusing). This sheath is responsible for the passage of the particles through the sensing point at a constant velocity. (ii) The illumination system consists of the light that produces the scatter signals and fluorescence emission when the particles pass through it. There are two types of flow cytometers, depending on the illumination source: those with a laser light source, and those with an arc lamp source. Each has it own advantages and disadvantages, but the main difference lies in their fields of application. Arc lamp cytometers are frequently used in microbiological applications due to their better scatter resolution and versatility. In contrast, laser flow cytometers have wider applications in immunology and hematology because they excite fluorochromes associated with cells. Studies comparing the two types of cytometers have concluded that the selection of one rather than the other depends mainly on the range of wavelengths required for the excitation of the selected fluorescent stains (13, 161). Our personal experience supports this opinion and work should aim at developing protocols according to the type of cytometer available. (iii) The optic system focuses incident light on the crossing particles, recovers the scattered light and the fluorescence produced by the fluorochromes present in the cells, and directs both to the appropriate photomultiplier tubes (Fig. 2). (iv) The electronic system transforms the incident light from fluorescence and light scattered into electric pulses (analogic). The magnitudes of these pulses are distributed electronically into channels, permitting the display of histograms of the number of cells plotted against the channel numbers (digital) (68). If the instrument has the capacity to do so, it also controls the cell-sorting process. Fluorescence-activated cell sorting refers to the ability to select a subpopulation from the whole population, following cytometric classification, and to physically separate this particular population. To do this, the machine produces a uniform stream of droplets; a particular droplet containing a cell can be charged, permitting selection of the droplet when it passes through an electrical field produced by deflection plates (Fig. 2). In this way, two populations can be sorted at the same time (positively and negatively charged droplets). A new high-speed sorter machine has been developed with the possibility of sorting four populations at the same time (MoFlo; Cytomation, Freiburg, Germany). (v) The data analysis system consists of software that allows the analysis of the huge amount of information produced by multiparameter data acquisition. The analytical software permits the study and independent analysis of a particular subpopulation. Besides all the statistical information, the data can be represented in several different ways: monoparametric histograms, biparametric histograms, and three-dimensional representations (Fig. 3). There is a growing market of commercial FCM software. Free software can also be downloaded from the Internet, where it is possible to find information about all the fields related to FCM (cytometry network sites, http://nucleus.immunol.washington.edu/ISAC /network_sites.html; JCSMR flow cytometry software, http://jcsmr.anu.edu.au/facslab/facs.html; ISAC WWW home page, http://www10.uniovi.es/ISAC.html).
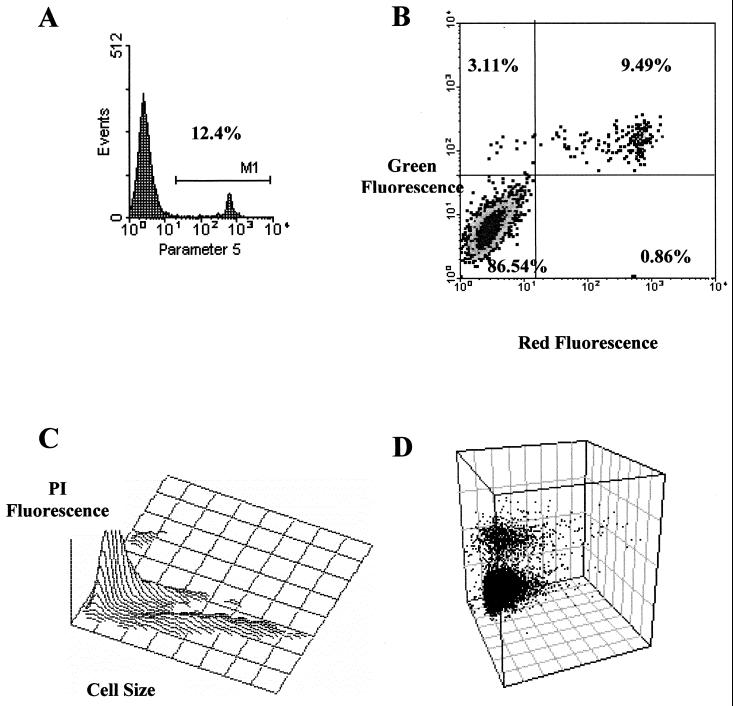
FIG. 3.
The data obtained from a flow cytometer can be displayed in several ways. The most common are the mono- and biparametric histograms (A and B), which usually include a statistical analysis of the results. (A) Monoparametric histogram showing the selected parameter on the x axis and the relative cell number on the y axis. (B) Biparametric histogram showing cells distributed as a function of their signal intensity with respect to each parameter. Cells located in the upper left quadrant are positive for the parameter represented on the y axis, cells located in the upper right quadrant are positive for both parameters, cells located in the lower left quadrant are double negative, while cells in the lower right panel are positive for the parameters on the x axis. (C and D) Three-dimensional representations. The z axis can represent the relative number of cells (C) or a third parameter (D), such as scattered light on the x and y axes and fluorescence signals on the z axis.
FLOW CYTOMETRY AND MICROBIOLOGY, A LONG TIME TOGETHER
From the beginning of FCM (68), the ancestor of modern flow cytometers has been identified with an aerosol particle counter designed to analyze mine dust (124). This apparatus was used in World War II by the U.S. Army in experiments for the detection of bacteria and spores. Gucker et al. (124) reported that the instrument could be used with biological samples (bacteria), as well as particles in air suspension or aerosols. Thus, FCM with an application to microbiology originated many years before the use of flow cytometry as a tool for studying mammalian cells. The original device incorporated a sheath of filtered air to limit the air sample stream to the central portion of the flow chamber. The detector used was a then recently developed device called a photomultiplier tube. Particle counters based on the Coulter orifice principle, in which the difference in electrical conductivity between the cells and the medium in which they are suspended is measured by the change in electrical impedance produced as they pass through an orifice, were later developed. These instruments were widely applied in hematology studies. However, the first real flow cytometer was built by Kamentsky et al. (154), using spectophotometric techniques to detect and measure nucleic acids and light scattering of unstained cervical cells in a flow stream. At the same time, Fulwyler, working at the Los Alamos Scientific Laboratory, described the first flow cytometer with sorting capability (104). This machine worked by measuring cell volumes obtained by the Coulter orifice principle. Fulwyler adapted the ink jet printer principle, using electrostatic deflection of charged droplets, as a cell-sorting mechanism. In fact, sorting capability was introduced to demonstrate the accuracy of the signals obtained by the machine and to ascribe a given distribution of cell volume detected by an electronic signal to a specific cell type. During the 1970s, applications of FCM to research into mammalian cells advanced rapidly, but at that time few instruments were developed for microbiological studies. The subsequent applications to microbiology of FCM techniques that were initially developed to study mammalian cells were due to optical improvements in flow cytometers and newly developed fluorochromes. The development of an arc lamp-based instrument by Steen's group in 1979 (301, 303) allowed the use of FCM for basic research on bacteria. Because of the design of the flow chamber and the use of photomultiplier tubes for detecting scattered light, this instrument was ideal for studying microorganisms (7, 37). The promising tool described by Boye and Steen in 1983 became a “potent illuminating light” in the 1990s (38), as was stated in the book edited by David Lloyd, Flow Cytometry in Microbiology (186a), from which most microbiological cytometrists have learned their trade. In the last years of the 1990s, the applications of FCM in microbiology have significantly increased (9, 28, 103, 148, 291).
General Applications of Flow Cytometry to Microbiology
The applications of FCM to microbiology have been so widespread that discussion of all of them is beyond the scope of this review. For more information, see the excellent reviews by Davey and Kell (68), Porter et al. (263), and McSharry (211) and the “Bible” of flow cytometry by Howard Shapiro, Practical Flow Cytometry (291). Below, we briefly describe some of the applications of FCM in the field of microbiology, focusing on present or future applications in clinical microbiology.
Earlier works by Steen had demonstrated the applicability of dual-parameter analysis to discriminate among different bacteria in the same sample (301, 302, 306). One parameter was light scattered (size), and the other was either fluorescence emission from fluorochromes coupled to cellular components (protein and DNA) or autofluorescence (Fig. 4), or light scattered acquired from another angle (42, 151, 277, 293, 301, 302, 306, 316). However, the use of several fluorochromes for direct staining or through antibody or oligonucleotide conjugates plus size detection is the simplest way to visualize or identify microorganisms by FCM (6, 7, 10, 11, 257, 317, 332).
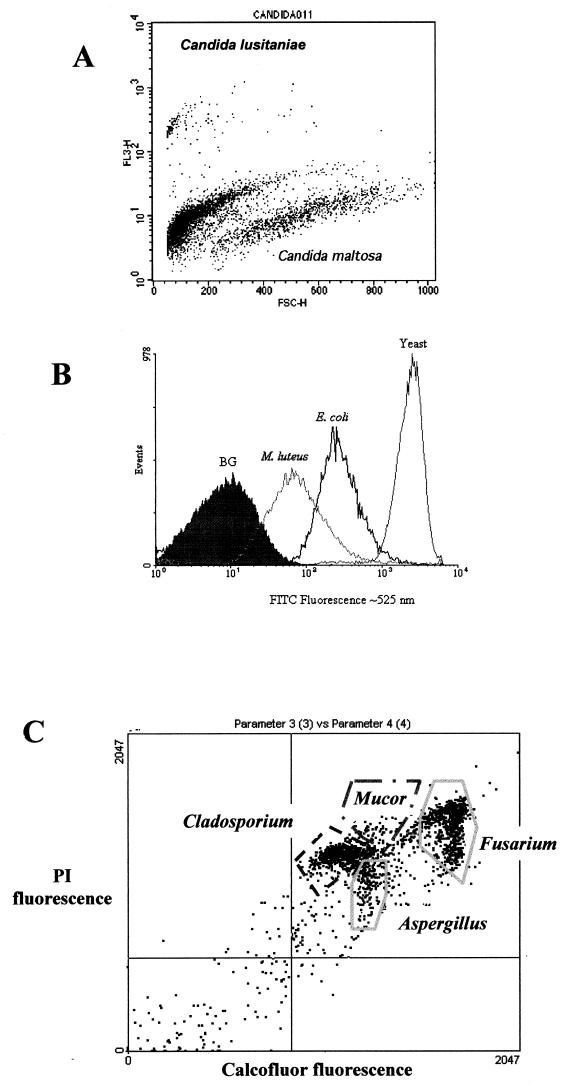
FIG. 4.
(A) Dual-parameter analysis of forward light scatter (size) and red fluorescence signals allowed the discrimination between two species of Candida, based on different fluorochrome staining backgrounds. These yeast species are indistinguishable by monoparametric analysis of forward light scatter or red autofluorescence. However, after addition of PI, they show different basal levels, and if this is plotted against size, it is possible to discriminate them. This kind of analysis permits quantification of both species in mixed cultures. (B) Quantification of different protein amounts (measured as FITC fluorescence) can be used to distinguish different microorganisms such as those represented in the histogram (from Purdue Cytometry CD-ROM, vol 2., ISSN 1091-2037, provided by Hazel M. Davey [adapted with permission of the publisher]). (C) Dual-fluorescence discrimination of fungal spores. Spores from Aspergillus, Mucor, Cladosporium, and Fusarium were fixed and stained with Calcofluor, which binds to chitin in the spore wall, and PI, which stains nucleic acids. As shown, the spores have different amounts of chitin and nucleic acids, permitting their segregation by FCM. Samples shown in panel A were run on a FACScan (Becton-Dickinson) flow cytometer, the ones shown in panel B were run on an EPICS Elite (Coulter) flow cytometer, and those shown in panel C were run on a Bryte-HS (Bio-Rad) flow cytometer.
The simple and rapid assessment of the viability of a microorganism is another important aspect of FCM. The effect of environmental stress or starvation on the membrane potential of bacteria has been studied by several groups using fluorochromes that distinguish among nonviable, viable, and dormant cells (155–157, 188; see references 68, 71, 78, 89, 135, 150, 200–203, 265, and 328 for reviews). Other authors have demonstrated the use of PI as a viability marker in yeasts (72), Pneumocystis carinii (177), and bacteria (89, 230, 264).
FCM has also been used in metabolic studies of microorganisms. This was first accomplished by Thorell (316), using autofluorescence due to NADPH and flavins as metabolic status markers. Other authors studied DNA, protein, peroxide production, and intracellular pH (3, 5, 39, 140, 324, 345). Recently developed fluorochromes and kits (Sytox Green and Live/Dead kits; Molecular Probes, Eugene, Oreg.) have been used for the FCM-based counting of live and dead bacteria and yeasts (176, 178, 311), simplifying staining protocols and making data interpretation easier. Other kits are available for detecting gram-positive and gram-negative bacteria or for studying yeast organelles (127). Recently, Mason et al. (204) described a method which enables the discrimination of gram-positive from gram-negative bacteria on the basis of the fluorescence emitted when the organisms are stained with two fluorochromes. These authors correctly predicted the Gram stain reaction of 45 strains of clinically relevant organisms, including several known to be gram variable. In addition, representative strains of gram-positive anaerobic organisms, which are normally decolorized during the traditional Gram stain procedure, were classified correctly by this method.
FCM also offers the possibility of studying gene expression using reporter genes in yeasts (56, 258, 297, 338) and bacteria (8, 55). The development of gene expression systems based on green fluorescent proteins facilitates this kind of study due to the simplicity of the technique (60, 77, 229, 320).
The sensitivity of FCM allowed Philips and Martin (255) to detect Bacillus spores (254). Using a similar approach, Griffiths et al. (121) and Challier et al. (49) were able to sort spores from Dictyostelium discoideum and Enterocytozoon bieneusi, respectively. These examples show the potential of FCM in the investigation of small microbes.
The interaction between pathogens and phagocytic cells has also been studied by FCM (22, 23). The development of fluorochromes to detect oxidative bursts due to phagocytosis (17, 251, 281) increased the number of studies with different microbes such as Borrelia burgdorferi (17), Staphylococcus spp. (128, 196), Escherichia coli (70, 271), Bordetella pertussis (299), Cryptococcus neoformans (48), Salmonella (272), and yeasts (90, 96, 114).
FCM has been extensively used for studying virus-cell interactions (172, 180, 334). This topic was reviewed in depth by McSharry in 1994 (211). Modulation of the expression of cellular proteins due to viral infection has been studied by FCM for cytomegalovirus (CMV) (116), herpes simplex virus (HSV) (149), adenovirus (168), human immunodeficiency virus (HIV) (53), and hepatitis B virus (HBV) (346). Perturbation of the cell cycle and DNA replication in virus-infected cells have also been studied by FCM for papillomavirus (25), CMV (80) and human HIV-1 (273). This technique has been also used to study the effect of viral infection on intracellular Ca2+ levels ([Ca2+]i) by Irurzun et al. (145) and by Miller et al. (221). FCM has also permitted the demonstration of apoptosis associated with viral infection, including HSV (144, 146), Epstein-Barr virus (EBV) (4), influenza virus (267), measles virus (93), papillomavirus (340), and HIV-1 (129, 194) infections. Furthermore, by means of biotinylated or directly FITC-labeled virus, interactions of EBV (142, 159, 182, 335), echovirus (206), adenovirus (217), influenza virus (228), simian virus 40 (SV40) (20), human T-cell leukemia virus type 1 (HTLV-1) (110), measles virus (226, 232), bovine herpesvirus (326), papillomavirus (268), bunyaviruses (249), poliovirus (102), and HIV-1 (14, 308, 318) with their putative cell receptors have been described. These investigations show that FCM is able to provide solutions to problems arising when working with microorganisms.
APPLICATIONS OF FLOW CYTOMETRY TO CLINICAL MICROBIOLOGY
The isolation of microbes and their identification, the detection of increased levels of antibodies to a particular pathogen in the course of an illness, and direct detection of microbial components (nucleic acids and proteins) in clinical samples obtained from different tissues or body fluids are the main tools for laboratory diagnoses of microbial infections. Effective antimicrobial therapies have indeed been developed because early treatment is crucial in many cases; therefore, rapid diagnosis is essential in the fight against infection.
Direct Detection
Bacteria.
Antibodies are currently changing the way in which we identify microbes, making it easier and faster. Their specificity and the possibility of using fluorochrome-labeled antibodies to specific antigens render them one of the most powerful tools in the identification of pathogens. The main disadvantage of this method is still the limited availability of antibodies directed against particular microbes. Other advantages of using antibodies are that the cells do not need to be cultivable and that the method is simple and fast. In an early work from 1983, Groschel (122) explained somewhat prophetically the use of antibodies in clinical microbiology. FCM in conjunction with fluorescent antibodies has been used to detect surface antigens in Haemophilus (298), Salmonella (57, 207), Mycobacterium (238), Brucella (35), Branhamella catarrhalis (29), Mycoplasma fermentans (50), Pseudomonas aeruginosa (134), Bacteroides fragilis (191, 239) and Legionella (143), among other microorganisms. These examples illustrate the sensitivity and specificity of using antibodies that allow the detection of particular cell types (of as few as 100 cells per ml in 30 min) in heterogeneous populations (57).
The first study detecting of microbes in blood by using FCM was done with ethidium bromide as the detecting fluorochrome (195). Blood cells were lysed, and the remaining bacteria were stained with ethidium bromide; as few as 10 E. coli cells/ml were detected. Using ethidium bromide fluorescence and light-scattered signals, Cohen et al. (58) were able to detect bacteria in 43 clinical specimens from several sources, such as wound exudates, bile, serous-cavity fluids, and bronchial-lavage fluids, in less than 2 h, although they were unable to identify them.
An FCM method for the direct detection of anaerobic bacteria in human feces was described by van der Waaij et al. (322), using PI for discriminating the patient's cells and excluding large particles by forward light scatter. At the same time, fluoresceinated antibodies against human immunoglobulin A (IgA) were added to detect IgA-coated bacteria. This method allows the rapid and highly sensitive assessment of fecal flora by specific IgA-FITC fluorescence without the need to culture the samples.
Another way in which FCM can achieve direct diagnosis is by fluorescent-oligonucleotide detection. By combining rRNA-targeted fluorescent probes and 4′,6-diamidino-2-phenylindole (DAPI) for nucleic acid staining, Wallner et al. (333) showed that it was possible to detect Acinetobacter spp. by FCM. To date, this approach has not been used with clinical samples, perhaps owing to its methodological complexity. Nevertheless, the specificity provided by the oligonucleotide probe to identify the putative infectious agent can be taken advantage of (315), thus promising many future applications.
The use of different-sized fluorescent microspheres coated with antibodies against microbes is a new application of flow cytometry for direct diagnosis (169). This method detects the binding of specific microbes to antibody-coated microspheres by measuring the decrease in the fluorescence emission of the microspheres due to the shading effect of microbes on both the exciting and emitting light. With different-sized fluorescent microspheres, several pathogens can be detected simultaneously in the same sample. This approach could also be used with fungi, parasites, and viruses, as well as in infections produced by combinations of these. In fact, as discussed below, a similar approach has been used for the simultaneous detection of plant viruses (137).
Fungi.
With regard to yeasts, the work by Groshen et al. (122), Chaffin et al. (47), and Han et al. (126) has shown that surface antigens of Candida albicans can be detected by flow cytometry in conjunction with available specific antibodies. As discussed below, this approach can be used for clinical samples.
The possibility of serotyping Candida isolated from clinical samples emerged from the work of Chaffin et al. in 1988 (47) and Brawner and Cutler in 1989 (40). However, it was not until 1996 that Mercure et al. (219) validated the FCM serotyping procedure, using serotype A-specific antisera. According to Mercure et al., the most striking feature of this method is its reliability. Ninety-four strains isolated from patients were analyzed by a slide immunofluorescence assay and FCM. FCM was able to detect the presence of two different strains in a culture that was assumed to be pure and serotyped four strains whose serotypes could not be determined by slide immunofluorescence. Again, it was acknowledged that when a cytometer is available, the procedure is probably more cost-effective than a commercially available kit for Candida serotype determination. Since the origin of the infecting strain(s) is often questioned when clinicians encounter patients with repeated episodes of Candida infections, FCM can help to discriminate among strains, as demonstrated in this work (219).
The diagnosis of onychomycosis based on clinical presentation, culture, and microscopy is hampered by false-negative and false-positive results that confuse treatment outcomes. Using FCM and antibodies directed against yeasts, Pierard et al. (256) identified fungal pathogens and differentiated them from nonpathogenic ones. Furthermore, the authors demonstrated that mixed infections occur, and hence the treatment for such circumstances can be established.
Parasites.
The first applications of FCM to parasites involved a study of the cell cycle and the amounts of DNA of Physarum polycephalum myxamoebae (319) and the characterization of monoclonal antibodies against membrane antigens from Leishmania (337). Specific clinical applications came later, when Flores et al. (99) used monoclonal antibodies, FCM, and immunofluorescence microscopy for the direct identification of Naegleria fowleri and Acanthamoeba spp. in clinical specimens.
Several approaches have been developed in the last few years to detect intracellular parasites, such as Plasmodium (147, 153, 240–242, 250, 319, 325). Such work took advantage of the absence of DNA in erythrocytes. Thus, if the parasite is inside the cell, its DNA can be stained with specific fluorochromes and detected by FCM. The multiparameter analysis permitted by FCM can be used to study other characteristics, such as parasite antigens expressed by the erythrocyte (which can be detected by antibodies conjugated with fluorochromes) (54, 153) or the viability state of the parasitised cell. Furthermore, the technique can be used with either fresh or fixed cells (241, 242). An important study showing the benefits of flow cytometry is that of Dixon et al. (81), who compared normal light microscopy, immunofluorescence microscopy, and FCM in the detection of Giardia lamblia cysts. They showed that when FCM is used in combination with immunofluorescence, a larger number of samples can be analyzed in a relatively short period and, more important, that this technique affords more consistent results than either conventional or immunofluorescence microscopy when samples containing small number of cysts are analyzed.
Viruses.
FCM allows both detection and quantification of infected cells directly in clinical samples or after inoculation and culture of the virus in cell culture.
(i) Detection and quantification of viral antigens.
FCM can detect viral antigens either on the surface of or within infected cells (172). It can rapidly detect and quantify virus-infected cells using antibodies that specifically recognize surface or internal antigens (91, 211, 213, 334); in the latter case, permeabilization of the cells is required. A thorough review of different permeabilization methods, including the advantages and disadvantages of each, for viral antigen and nucleic acid detection has been written by McSharry (211). Direct and indirect fluorescent-antibody methods are used. Direct detection involves the use of fluorescently labeled antibody (labeled with FITC or phycoerythrin). In the indirect fluorescent-antibody method, unlabeled antibody is bound to infected cells, which are then incubated with fluorescence-labeled anti-Ig that binds to the first viral antibody.
As previously stated, FCM is carried out on single cells, and therefore FCM analysis of virus-infected cells is best suited to blood, bronchoalveolar lavage fluid, and urine samples (172). However, it is also possible to analyze cells from tissues that have previously been treated enzymatically (211).
Based on the potential of FCM for multiparametric analysis, there are two key advantages to its use in studying viral infection: (i) its ability to analyze several parameters in single-infected cells at the same time and (ii) its ability not only to detect but also to quantify infected cells. These parameters may be related to particular components or events of the infected cell or components (proteins or nucleic acids) of the virus. For this reason, FCM has been a powerful tool to characterize the mechanisms of viral pathogenesis. Furthermore, FCM allows simultaneous detection of several viruses in a sample by using (i) antibodies to different viral antigens conjugated to different fluorochromes, or (ii) specific viral antibodies conjugated to latex particles of different sizes. As stated above, the presence of different viral antigens is detected by differences in the forward-scattered light as a consequence of the different sized particle used for each antibody. For example, Iannelli et al. (137) simultaneously detected cucumber mosaic, tomato, and potato viruses by using 3-, 6-, and 10-μm-diameter latex particles, respectively. Although this method was aimed at the detection of plant viruses, its basis could be applied to the detection of animal or human viruses in any clinical sample, such as the simultaneous detection of CMV, HSV, and HBV in organs destined for transplantation as well as in transplanted patients and coinfections in HIV-infected individuals.
Flow cytometric analysis has allowed the detection and quantification of SV40 T antigen in infected cells and monitoring the kinetics of T-antigen expression. By means of multiparametric analysis, using PI for the measurement of cellular DNA and FITC-labeled antibody for the detection of viral antigens, Lehman et al. (171, 179) related high levels of SV40 T antigen to the appearance of cells with tetraploid DNA content due to a cell cycle block at G2/M.
Detection of immediate-early, early, and late CMV antigens by monoclonal antibodies permits direct diagnosis and quantification of CMV infection. This is a frequent complication in immunosuppressed patients, including transplant recipients and AIDS patients. In 1988, by means of FCM, Elmendorf et al. (91) detected early CMV antigen 30 min after virus adsorption to fibroblasts. Thus, FCM permits the detection of viral infection earlier than does conventional immunofluorescence microscopy or the detection of cytopathic effects in cell culture (91, 289, 310).
During active infection, CMV disseminates in the blood, and viremia has been described as a major risk factor for the progression to clinical disease, particularly in allogeneic bone marrow transplant recipients (31). Accordingly, quantification of the viral load in persistently infected hosts may provide a method to predict the development of CMV disease and help to differentiate symptomatic infection from asymptomatic shedding. Preventive strategies increasingly use the CMV load as a surrogate marker for disease and initiate antiviral treatment based on the systemic viral load (31). Sensitive techniques, such as the pp65 antigenemia assay or the quantitative PCR assay, allow the detection and quantification of systemic CMV. Both assays provide a good estimation of the systemic CMV burden. Owing to the high sensitivity of these assays, CMV is also detectable in patients with asymptomatic infections. However, patients with disease often have a higher viral load and can therefore be discriminated (31, 314).
The pp65 antigenemia assay determines the systemic CMV load and consists of direct staining of polymorphonuclear leukocytes with monoclonal antibodies against the lower matrix protein pp65 (31, 112, 314). This determination has classically been made by very difficult and time-consuming microscopic observation of immunostained cells (112). Recent studies have evaluated FCM for the direct detection and quantification of CMV antigens in polymorphonuclear leukocytes from transplant recipients (76, 132, 141). Measurement of pp65 CMV antigenemia by FCM overcame these problems owing to its speed and automation and showed it to be a specific and reproducible method, especially when the paraformaldehyde-methanol permeabilization-fixation method and antibody 1C3 (to late antigens) were used (141). Good agreement was found between the degree of DNA load and the level of antigenemia detected by FCM in renal transplant recipients. Although the sensitivity of the method was somewhat lower than that of the slide method, it might be sufficient to predict the disease.
Honda et al. (132) also identified specific CMV-infected cell populations in peripheral blood lymphocytes from CMV-infected patients by FCM. Using monoclonal antibodies directed against immediate-early CMV antigen (see above) or against several cell membrane markers to phenotype infected peripheral blood cells from bone marrow transplant recipients, these authors developed a rapid and quantitative FCM method for the detection of immediate-early CMV antigen. The detection of CMV antigens specifically in the polymorphonuclear leukocytes from transplanted patients with CMV pneumonia suggests that the FCM antigenemia assay would be useful for predicting CMV-associated disease in transplant recipients (132). In summary, FCM offers a rapid and suitable quantification of the CMV viral load. Although systematic comparative evaluations of the CMV viral load using this method are needed, current data are promising.
The detection of cytomegalic endothelial cells in peripheral blood of patients is another means of monitoring active CMV infection. Through enrichment of endothelial cells in the mononuclear fraction by density centrifugation, endothelial cell-specific staining, and fluorescence-activated cell sorting of these cells, a method with 10-fold greater sensitivity than cytocentrifugation of the mononuclear cell fraction alone has recently been developed for quantification of cytomegalic endothelial cells by FCM (158). Belles-Isles et al. (24) have also suggested using FCM to monitor CMV infections by monitoring the CD8+ CD38+ T-cell subset in kidney transplant recipients; this T-cell subpopulation usually increases during active viral infections. Quantification of CD8+ CD38+ T cells by dual-color FCM in 77 kidney transplant recipients during the posttransplantation period detected high levels of CD8+ CD38+ subsets in all patients with CMV disease. Belles-Isles et al. (24) therefore concluded that the percentage of CD8+ CD38+ T cells constitutes an immunologic marker that can serve as a tool for early detection of viral diseases.
Viral antigens have also been detected by FCM for the diagnosis of hepatitis and herpesvirus infections. Quantitative and dynamic analyses of hepatitis virus markers are important in the follow-up of antiviral treatments (32). HBV surface (HBsAg) and HBV core (HbcAg) antigens in peripheral blood mononuclear cells (PBMCs) from HBV patients have been detected by FCM using antibodies (52, 278). In one study, 35 patients with HBV chronic active hepatitis and 38 out of 60 patients with acute hepatitis B (63%) expressed HbsAg in PBMC. In another work, Chemin et al. demonstrated the selective detection of HBsAg and HBcAg on B lymphocytes and natural killer cells from chronically HBV-infected patients (52). Hepatitis C virus (HCV), woodchuck hepatitis virus, and varicella-zoster virus were also detected by FCM in PBMCs using monoclonal antibodies to HCV core antigen (34), polyclonal antibodies to woodchuck hepatitis virus (51), and antibodies to the gpI glycoprotein from varicella-zoster virus (296), respectively. Thus, FCM detection of viral antigens offers a potentially useful automated assay for the clinical diagnosis of multiple blood-borne viruses.
HSV antigens have also been detected in HSV-1, HSV-2, and human herpesvirus 8 (HHV-8)-infected cells by FCM (117, 213, 300, 347). After overnight amplification of clinical samples suspected to contain HSV, FCM detected virus 1 to 3 days before cytopathic effects were detected in cell culture (213). Sensitive FCM assays have also been recently developed to quantitate rotavirus in clinical and environmental samples (2, 19).
Finally, FCM has also been extremely useful in the study of HIV infection (174, 211, 213). By studying HIV-infected cell lines by FCM in 1987, Cory et al. (61) determined the percentage of infected cells and the relative amount of p24 antigen per cell. These and other authors used the same assay to detect and quantify HIV-infected cells in cell cultures by monitoring p24, p17, nef, gp120, gp41, and gp160/gp41 expression (61, 62, 130). This method proved to be more sensitive and accurate for quantitative studies and faster than other methods of HIV-1 detection, such as the reverse transcriptase (RT) assay or determination of syncytium formation.
Detection of HIV antigens on peripheral blood mononuclear cells by FCM is a useful method for monitoring HIV replication in vivo by monitoring the number of circulating CD4+ cells positive for p24 (63, 131, 235), p24 and nef (213), or p18 and p24 (107). In all these works, the percentage of cells expressing these antigens was statistically correlated with the clinical status of the patient. Furthermore, the authors reported an inverse correlation of HIV antigen-positive mononuclear cells and the number of CD4+ cells. Therefore, these assays are useful for rapidly monitoring disease progression in HIV-seropositive individuals and for monitoring the effect of antiviral therapy. Some studies found a lack of correlation between cell-associated antigen detection by FCM and the detection of HIV antigens in sera from HIV-seropositive individuals by the standard antigen capture enzyme-linked immunosorbent assay (ELISA). As explained by McSharry et al. (213), the lack of correlation is due to a masked antigenemia in the presence of immunocomplexes, which could underestimate the amount of antigen present in peripheral blood detected by ELISA. However, in spite of being a good assay for evaluating disease progression, FCM detection of HIV antigens is not sensitive enough to detect the low levels of HIV-infected peripheral blood cells in asymtomatic HIV-1-seropositive individuals (213). This lack of sensitivity can be overcome, as discussed below, by coupling in situ PCR and FCM for the detection of small numbers of peripheral blood cells infected with low levels of HIV in asymptomatic individuals.
(ii) Detection and quantification of viral nucleic acids.
The emergence of PCR and rPCR RT-PCR techniques has allowed the highly sensitive detection of specific viral nucleic acids (DNA or RNA) in virus-infected cells. These methods are indeed the most sensitive for the detection and characterization of viral genomes, especially in the case of rare target viral sequences (123). However, the association between the viral nucleic acid and an individual cell is lost, and therefore no information about productively infected cell populations is obtained by this method. FCM analysis of fluorescent in situ hybridization in cell suspension overcomes this problem (33, 173), since this assay can be coupled with simultaneous cell phenotyping (by using specific antibodies to different cell markers).
FCM detection of in situ hybridization has been used to analyze rare virus-producing cells in peripheral blood samples. HIV-1 RNA in infected cell lines was detected by fixation of the cells in suspension, hybridization with HIV-1 genomic probes labeled with digoxigenin–11-dUTP, detection with fluorescent anti-digoxigenin antibody, and FCM analysis of the fluorescence signals thus generated (33). Link et al. (184) detected CMV antigen pp65 with immunoenzymatic labeling by day 4, whereas CMV-DNA was detected by PCR coupled to FCM detection of in situ hybridization 4 h postinfection on T-lymphoblastoid cells (MOLT-4). This method also detected and quantified mononuclear peripheral blood leukocytes in a patient with active CMV infection (184). Of CMV-DNA-positive mononuclear peripheral blood leukocytes from patients with active CMV infection, 15% were detected by this method, while only 0.9% were found when CMV antigens were analyzed by immunoenzymatic labeling (184). Identification of specific CMV-DNA or RNA by this method, with the possibility of phenotyping cells by FCM, should permit latency studies in CMV infections through the identification of specific cells actively replicating the virus and cells that harbor the virus in a latent state, acting as a reservoir for infection. Furthermore, FCM permits these cells to be sorted for the characterization of latency and reactivation mechanisms.
Two fluorescence in situ hybridization-FCM assays have also been developed for monitoring EBV-infected cells in blood (66). Crouch et al. were able to quantify EBV-infected cells in suspension for both the latent and replicative phases of the virus, using in situ hybridization with two different fluorescently labeled probes (specific for each phase of EBV replication) and FCM (66). This in situ hybridization-FCM assay detected one positive cell out of 9,000, which is sufficient for a diagnosis of EBV-infected cells in transplant recipients with lymphoproliferative disease.
As an alternative to conventional PCR radioactive methods, nonradioisotope FCM detection of viral PCR products was developed (342–344). Following virus-specific PCR amplification incorporating digoxigenin-labeled dUTP, labeled amplicons are hybridized with biotinylated probes, the hybrid DNA is captured using streptavidin-coated beads, and FCM analysis of the binding of FITC-labeled anti-digoxigenin antibodies is performed (342–344). This PCR immunoreactive bead (PCR-IRB) assay has been used for the detection and quantification of HIV-1 (342, 343) and HBV (344) viral genomes. As few as two or three copies of HIV-1 proviral DNA sequences were rapidly detected in PBMC from HIV-1-infected blood donors, a sensitivity comparable to that of the conventional radioactive detection of PCR products (342, 343). The PCR-IRB assay is a very simple, specific, sensitive, and automatic assay for the detection of specific HIV-1 amplicons. Yang et al. (343), testing a panel of 20 pedigreed PBMC specimens, demonstrated a perfect correlation with the results from conventional radioactive assays. By this method, Dorenbaum et al. (84) detected about three copies of proviral HIV DNA using primers for the long terminal repeat sequences. In a double-blind study of blood samples from 14 mother-infant pairs using the PCR-IRB assay, these authors obtained similar results to those found with the commercial Amplicor HIV-1 PCR kit. On testing 20 specimens of blood donors, with or without markers of HBV infection, PCR-IRB detected HBV DNA in a 1,000-fold-higher dilution than the infectious dose needed to produce infection in chimpanzees (344). The PCR-IRB assay proved to be specific and more sensitive than the PCR analyses involving hybridization with radioactive probes for the detection of HBV in blood. Importantly, the FCM assay avoids the use of radioisotopes.
FCM and RT-PCR have detected gene expression in individual cells (111). Muratori et al. (225) developed an in situ RT-PCR technique using fluorescein-labeled HCV specific primers detected by FCM for the quantification and phenotyping of HCV-infected cells in clinical blood samples. Although HCV infects PBMC, the small proportion of circulating infected cells is not easily detectable by conventional RT-PCR. These authors detected HCV in PBMC cells of 50% of patients with chronic hepatitis C tested; the proportion of HCV-infected cells ranged from 0.2 to 8.1%.
Recently, a very sensitive and powerful PCR-driven in situ hybridization assay has been developed (115). This method combines the sensitivity of PCR with the specificity of in situ hybridization, allowing rapid and reproducible detection of single-copy proviral DNA or low-abundance viral mRNA in subsets of cells in suspension. This assay employs PCR- or RT-PCR-driven in situ amplification of viral sequences in fixed cells in suspension with sequence-specific primers and digoxigenin-linked dUTP. The product DNA is hybridized with a fluorescein-labeled oligonucleotide probe, and the cell suspension is then analyzed by FCM (245). This method has been used for the detection of HIV-1 DNA and mRNA sequences in individual cells in both cell lines and cells from HIV-1-infected patients (243–245). The sensitivity and specificity of this technique revealed a linear relationship for the detection of a single copy of intracellular proviral DNA over a wide range of HIV-1-infected cell concentrations (245). Re et al. (274) analyzed the presence of HIV-1 proviral DNA in PBMC from HIV-infected patients at different stages of the disease by a PCR-in situ hybridization FCM assay and correlated the data with p24 antigenemia and virus isolation. p24 antigenemia correlated with the number of CD4-positive cells but was detected in only a very low percentage of patients with a cell count greater than 200 CD4+ T cells per liter. As stated above, detection of HIV antigens is not sensitive enough to detect the low levels of HIV-infected peripheral blood cells in asymtomatic HIV-seropositive individuals. The virus was isolated in most patients with a T-cell count below 500 per liter but only in 4 of 14 patients with a cell count higher than 500 CD4+ T cells per liter. In contrast, the PCR-in situ hybridization FCM assay revealed detectable levels of proviral DNA in all the HIV-1-positive subjects studied, even those with a cell count higher than 500 CD4+ T cells per liter. These data underscore the potential of this assay for detecting small numbers of PBMC infected with low levels of HIV in asymptomatic individuals.
In HIV-1-infected patients, plasma viral RNA levels (viral load) correlate with disease progression (105, 216, 327). Accordingly, evaluation of this marker by RT-PCR is extensively used to monitor the kinetics of HIV infection and the effects of antiretroviral treatments (12). However, the RT-PCR assay does not characterize the cell populations contributing to the plasma viral load. The PCR-driven in situ hybridization method coupled to the FCM assay described above allows simultaneous phenotyping of infected cells and hence quantification of HIV-1 proviral DNA or RNA molecules in specific cell populations. Patterson et al. (244) identified and quantified cell subsets in the peripheral blood of HIV-1 patients expressing HIV-1 RNA by using PCR-driven in situ hybridization coupled to FCM. They found a good correlation between the FCM-determined percentage of HIV RNA-positive cells and the expected percentage of HIV RNA-positive cells on the basis of plasma viral load, with sensitivities of less than 30 copies of RNA per cell and a detection limit of 3.5% HIV-1 RNA-positive cells within a heterogeneous population. Simultaneous immunophenotyping by FCM showed that a significantly higher fraction of patients with a high plasma viral load (more than 20,000 copies/ml) harbored HIV RNA-positive monocytes than did those with a low plasma viral load. Furthermore, the PCR-driven in situ hybridization and FCM assay permits the determination of the presence in HIV-1-infected patients of both latent and transcriptionally active viral infection by detection of proviral DNA or viral mRNA. Patterson et al. (245), testing nine HIV-1-infected patients, observed a significant proportion of PBMCs infected with HIV-1, with most of the cells having viruses in a latent state (the percentage of PBMCs with proviral DNA varied from 4 to 15% whereas the percentage of PBMCs with tat mRNA varied from 1 to 8%).
The above FCM nucleic acid detection techniques have similar sensitivity to conventional PCR, but with the added benefits derived from expression analysis in individual cells (in conventional PCR, nucleic acid expression is not analyzed independently in each cell). Multiparametric analysis of infected cells allows the detection of single-copy proviral DNA or low-abundance viral mRNA (225, 244, 245) in specific subsets of cells that can be phenotyped at the same time. Moreover, FCM is a very useful tool to study the mechanisms of viral latency by association of different stages of the virus cycle and disease progression with the location of the virus in specific cell populations. This can be achieved by using probes to specific mRNAs related to different viral replication stages (66). Knowledge of cell populations in which the virus is either replicating or in a latent state has important implications for our understanding of virus replication in vivo and progression to disease and hence for therapeutic treatments. Furthermore, it should be possible to sort these cells for further analysis. Double staining of viral nucleic acids together with viral proteins or surface markers is also possible (33, 66). In comparison with the detection of the PCR-driven in situ hybridization by fluorescence microscopy (92), in which a large number of microscopic fields must be studied, FCM allows the analysis of thousands of cells in a few seconds. The speed and automation of these assays make them optimal for the rapid diagnosis of viral infections. Since these assays can also determine the relative number of cells bearing viral genomes and the viral load, they could be used to evaluate and monitor antiviral treatments. Amplification of nucleic acid sequences of viruses from cerebrospinal fluid, blood, or tissues, which are difficult to isolate by conventional diagnostic techniques, together with the detection of such nucleic acids by FCM open new possibilities in the diagnosis of viral infections and the characterization of viral pathogenesis.
Serological Diagnosis
Bacteria.
The identification of pathogens by microsphere immunoassays using FCM offers the specificity provided by antibodies coupled with the speed and multiparametric analysis provided by FCM. Although these assays can be used to directly detect microbes, they are more useful for detecting antibodies against microbes in sera obtained from patients. Generally, either a bacterial antigen preparation or the whole organism is attached to polystyrene microspheres with a uniform diameter. The antigen-coated microspheres are incubated with the sera, the putative human antibodies recognize the antigen, and, in a second step, a fluorescence-conjugated antibody against human Igs is used for detection. FCM allows this assay to be completed in a short time with excellent sensitivity and reliability. Using this approach, Best et al. (27) detected the presence of antibodies against Helicobacter pylori in sera from 55 patients. These authors demonstrated that this method was as sensitive and reliable as ELISA but faster and cheaper. The simplicity of the technique and the stability of the coated microspheres make the FCM immunofluorescence assay highly practical for serodiagnosis.
The characteristics of FCM allow the detection of more than one antigen at the same time. Thus, simultaneous detection of multiple antibodies using different-sized particles coated with different antigens or microbes would make it possible to detect multi-infection diseases. Furthermore, using several fluorochrome-conjugated antibodies against human Igs and differently sized microspheres, FCM can gather information from each different-sized microsphere and particular fluorescence signals. Using Brucella abortus-coated microspheres and Staphylococcus aureus fixed cells, dual antibody detection in sera and milk from cows has been reported by Iannelli et al. (138). In this assay, antibodies against the two bacteria are identified on the basis of the altered size of B. abortus cells due to the microspheres.
Another use of the microsphere fluoroimmunoassay by FCM is to detect bacterial toxins. In cases where the suspicion of Clostridium difficile infection is high, it is necessary to confirm the presence of toxin A in patient samples. Renner (275) reported a microsphere fluoroimmunoassay for C. difficile toxin A using microspheres of two different sizes. The largest one was coated with polyclonal antibody against toxin A, and the smaller one, which was fluorescent, was coated with monoclonal antibody against toxin A. In the first step, large microspheres were added to the stool samples, and after incubation, smaller fluorescent microspheres were added. FCM measurement allowed the separation and washing steps to be omitted by gating the light scattered by the larger microspheres and measuring only the associated fluorescence from the smaller particles. Renner compared this method with the cytotoxin assay and culture of the organism from patients with C. difficile-associated gastrointestinal disease. The results showed that the fluoroimmunoassay was less sensitive than the cytotoxin assay and culture but had the same specificity, with the advantage of being rapid, and, as the author stated “in laboratories with a flow cytometer, this offers an alternative method for the laboratory diagnosis of C. difficile-associated gastrointestinal disease.” Tapp and Stotzky (313), using a similar approach to detect and track the fate of toxins from Bacillus thuringiensis, concluded that FCM is more sensitive and rapid than dot blot ELISA and that it is possible to process many samples easily.
The detection of antibodies with borreliacidal activity in sera from patients with Lyme disease can help in both early and late serodiagnosis. Using FCM, it is easy to detect the loss of viability of Borrelia burgdorferi incubated with sera from patients with Lyme disease, using fluorochromes that detect the damage caused by antibodies. Callister et al. (43) used acridine orange to demonstrate this effect.
The above work demonstrates the potential of FCM as a routine technique in clinical microbiology laboratories for detecting the presence of antibodies against microbes in patient sera and for reliably checking the presence of toxins in clinical samples.
Fungi.
The use of FCM and the antibodies present in patient sera to detect fungal pathogens was first described by Libertin et al. (183) in 1984. Pneumocystis carinii cysts in lung homogenates from biopsy specimens were detected by these authors using sera from patients and experimentally infected rats. Bergbrant (26), using FCM to monitor antibodies against Pityrosporum ovale in sera from patients with seborrheic dermatitis, demonstrated that there was no relationship between this microorganism and the illness.
Although the tools to directly detect antibodies against fungi in patient sera do exist, no work validating the FCM procedure has yet been published.
Parasites.
The presence in patient sera of antibodies to any particular parasite can permit an FCM-based diagnosis of parasitism. Martins-Filho et al. (199), using FCM on serum from patients chronically infected with Trypanosoma cruzi, developed a sensitive method for the immunodetection of anti-trypomastigote membrane-bound antibodies. They were also able to monitor the treatment in order to establish its effectiveness. A similar assay was developed by Cozon et al. (64) with Toxoplasma gondii, using fixed tachyzoites and specific conjugates for different human Ig heavy chains. They were able to quantify the amounts of IgM, IgG, and IgA antibodies in patient sera by measuring the amount of fluorescence bound to tachyzoites. The authors stated that the method might offer a major improvement in cost-effectiveness per sample (especially when a large number of tests is used) compared with routine immunofluorescence assays by fluorescence microscopy and further stressed that it could be fully automated.
Viruses.
Some methods have been routinely used to detect specific antibodies to viral antigens. Among these techniques are ELISA, complement fixation, indirect immunofluorescence microscopy, and Western blotting. In addition, the detection and quantification of antibodies to viral antigens can be carried out by FCM. This technique has been used to detect and quantify antibodies to CMV (209), HSV-1 and HSV-2 (46, 209), HCV (187, 210, 279), and HIV-1 (100, 118, 133, 290, 294).
Most of these viral antibody quantifications use a microsphere-based immunoassay and FCM. In this assay, polystyrene microspheres attached to viral antigens are used as a support for viral antibody detection by FCM. For the simultaneous detection of two or more viruses, different-sized microspheres, each coated with a specific viral antigen, are used. The assay has the advantage of simultaneous detection of multiple antibodies with high analytic sensitivity. Simultaneous detection and quantification of antibodies to CMV and HSV was achieved by McHugh et al. (209) using this method. Using particles of different sizes coated with p31, gp120, p24, and gp41 antigens from HIV-1, Scillian et al. (290) were able to detect and quantify the specific antibodies. An FCM immunofluorescence assay (FIFA) with high sensitivity and specificity was developed by Sligh et al. (294) to detect antibodies to HIV-1 by using HIV-1-infected cell lines. The cells are incubated with the sera to be tested, and incubation with an FITC-conjugated anti-human Ig and FCM allows the quantification of HIV-1 antibodies in the sera. Based on this assay, Folghera et al. (100) developed a FIFA for the quantitative determination of HIV p24 in HIV-1-infected cells and used the reduction in HIV-1 p24 antigen expression in these cells to determine the neutralizing-antibody titers in human sera (100). This method also allowed the rapid detection and monitoring of antibodies to native and recombinant human HIV-1 envelope glycoproteins following gp160 immunization (118). A new serological assay, the recombinant FIFA, was later described (133) for the early detection of HIV-1 antibodies. In this assay, antibodies in sera are evaluated by FCM for binding to the HIV-1 recombinant insoluble forms of proteins Gag-p45, Gag-gp41, and gp160 expressed in insect cells by a baculovirus expression system. The sensitivity of this method permits earlier detection of antibodies after initial infection than for enzyme immunoassays, with a reduction in the “window” period, i.e., the time between initial infection and the time of seroconversion, a parameter which is critical in infection from blood transfusions (133).
The humoral immune response to HCV has been evaluated in patients with chronic hepatitis (187). Antibodies to HCV core and NS3 antigens have been quantified using immunoassay beads and FCM (210) in blood donors. The microsphere assay resulted in increased sensitivity (fivefold higher than that of reference methods) of HCV detection and resolved a significant proportion of indeterminate samples. A fast FCM assay that measures the neutralization of the binding of recombinant HCV E2 envelope protein by antibodies to human cells has also been described (279). This method permits study of the natural immunity to HCV and should be useful in the development and validation of vaccination protocols.
To conclude, the investigations of Best et al. (27) and Iannelli et al. (138), among others, offer the possibility of performing FCM serodiagnosis in an elegant, rapid, cheap, and precise manner. The technique is simple and can be used for many pathogens (including viruses), with an additional possible advantage of detecting more than one microorganism in a single sample. The use of FCM in clinical microbiology laboratories would allow detection times and costs to be reduced. At present, however, it is not in general use and the setting up of such protocols can be fairly time-consuming.
ANTIMICROBIAL EFFECTS AND SUSCEPTIBILITY TESTING BY FLOW CYTOMETRY
The first experiments in which FCM was used to study the effects of antimicrobial agents in prokaryotes were carried out at the beginning of the 1980s (136, 302, 304, 306). In the 1990s, there were interesting advances in this field from microbiology laboratories, and the number of scientific articles addressing the antimicrobial responses of bacteria (including mycobacteria), fungi, and parasites to antimicrobial agents increased considerably. The development of FCM in combination with fluorochromes permits the assessment of individual viability and functional capacity (membrane potential and metabolic pathways) within microbial populations. This allows investigators to explore the possibility of performing susceptibility testing within the applications of FCM. Also, the introduction of this technique in clinical laboratories for routine susceptibility testing has been proposed, since this approach can be performed reliably in just a few hours. Several examples of the study of the antimicrobial effect and susceptibility testing by FCM are shown in Table 2.
TABLE 2.
Antimicrobial effects and susceptibility testing by FCM in bacteria
| Antibiotic | Organism(s) | Time to results (h) | Dye | Parameter(s) studied | Reference(s) |
|---|---|---|---|---|---|
| Amoxicillin | E. coli | 1–2 | PI + FITC | DNA and protein content | 86 |
| Ampicillin | E. coli | 1–3 | PI | Morphology, membrane effects | 108 |
| E. coli, K. pneumoniae | 1 | Ethidium bromide + mithramycin | Light scattering, morphology, DNA content | 330 | |
| E. coli | 0.5 | Oxonol | Light scattering, membrane effects | 200 | |
| Azithromycin | E. coli | <1 | Oxonol | Viable cells | 150 |
| R. tsutsugamushi | 72 | FITC | Stained bacterial cells | 160 | |
| Benzylpenicillin | S. aureus, MRSAa | 2–4 | Oxonol | Membrane effects | 310 |
| E. coli | 4 | Mithramycin + ethidium bromide + FITC | Morphology, DNA and protein content | 37 | |
| Cefamandole | E. coli | <0.5 | Ethidium bromide | Light scattering, DNA content | 198 |
| Cefazoline | E. coli | <0.5 | Ethidium bromide | Light scattering, DNA content | 198 |
| Cefotaxime | E. coli | 1–3 | PI | Light scattering, morphology, membrane effects | 108 |
| Ceftazidime | E. coli | 0.5–1 | Ethidium bromide + mithramycin | Light scattering, morphology, DNA content | 331 |
| Cefuroxime | E. coli | <1 | Oxonol | Viable cells | 150 |
| Chloramphenicol | E. coli | <2 | Mithramycin | DNA content | 302 |
| E. coli | 1–2 | PI + FITC | DNA and protein content | 86 | |
| Ciprofloxacin | E. coli | 1–3 | PI | Light scattering, morphology, membrane effects | 108 |
| E. coli | <6 | PI + cyanoditodyl tetrazolium chloride | Membrane effects, viable cells | 201 | |
| E. coli | 0.5–1 | Ethidium bromide + mithramycin | Light scattering, morphology, DNA content | 331 | |
| E. coli | 1–2 | PI + FITC | DNA and protein content | 86 | |
| Mycobacterium spp. | 6–24 | FDA | Viable cells, metabolic status | 36 | |
| E. coli | 0.5–1 | Oxonol | Light scattering, membrane effects, viable cells | 150, 200 | |
| Clarithromycin | Mycobacterium spp. | 6–24 | FDA | Cell viability, metabolic status | 36 |
| Doxycycline | C. trachomatis | 48 | FITC-antibody | Antibody binding | 75 |
| R. tsutsugamushi | 72 | FITC | Stained bacterial cells | 160 | |
| E. coli | <2 | Mithramycin | DNA content | 302 | |
| Erythromycin | C. trachomatis | 48 | FITC-antibody | Antibody binding | 75 |
| Mycobacterium spp. | 6–24 | FDA | Cell viability, metabolic status | 36 | |
| E. coli | <2 | Mithraymcin | DNA content | 302 | |
| Ethambutol | M. tuberculosis | 24 | FDA | Viable cells, metabolic status | 231 |
| M. tuberculosis | ≤24 | FDA | Viable cells, metabolic status | 164 | |
| Formaldehyde | E. coli | 1 | PI SYTO-13 | Membrane effects, DNA content | 59 |
| Gentamicin | E. coli | 1–3 | PI | Light scattering, morphology, membrane effects | 108 |
| E. coli | 0.5–1 | Ethidium bromide + mithramycin | Light scattering, morphology, DNA content | 330 | |
| E. coli | 0.5 | Oxonol | Light scattering, membrane effects | 200 | |
| E. coli | 1 | Acridine orange | Membrane effects, nucleic acid staining | 202 | |
| Gramicidin | E. coli | 1 | Propidium iodide + SYTO-13 | Membrane effects, DNA content | 125 |
| Isoniazid | Mycobacterium spp. | 24–72 | Auramine | Microencapsulated bacteria colony growth | 283 |
| M. tuberculosis | 24 | FDA | Viable cells, metabolic status | 231 | |
| M. tuberculosis | ≤24 | FDA | Viable cells, metabolic status | 164 | |
| Kanamycin | Mycobacterium spp. | 6–24 | FDA | Viable cells, metabolic status | 36 |
| Mecilinam | E. coli | 1–3 | PI | Light scattering, morphology, membrane effects | 108 |
| E. coli | 1–2 | PI + FITC | DNA and protein content | 86 | |
| Methicillin | S. aureus, MRSA | 2–4 | Oxonol | Membrane effects | 310 |
| Moxalactam | E. coli | <0.5 | Ethidium bromide | Light scattering, DNA content | 198 |
| Ofloxacin | C. trachomatis | 48 | FITC-antibody | Antibody binding | 75 |
| Oxacillin | S. aureus | 2 | DIOC5(3) | Membrane effects | 237 |
| Penicillin | S. aureus | 2 | DIOC5(3) | Membrane effects | 237 |
| Rifampin | Mycobacterium spp. | 6–24 | FDA | Viable cells, metabolic status | 36 |
| M. tuberculosis | ≤24 | FDA | Viable cells, metabolic status | ||
| Mycobacterium spp. | 24–72 | Auramine | Microencapsulated bacteria colony growth | 283 | |
| M. tuberculosis | 24 | FDA | Viable cells, metabolic status | 231 | |
| Streptomycin | M. tuberculosis | 24 | FDA | Viable cells, metabolic status | 231 |
| E. coli | <2 | Mithraymcin | DNA content | 302 | |
| Surfactants | E. coli | 1 | Rhodamine + SYTO-13, oxonol + SYTO-17 | Membrane effects, DNA content | 59, 125 |
| Thrimethoprim | E. coli | 1–2 | PI + FITC | DNA and protein content | 86 |
| Tobramycin | Mycobacterium spp. | 6–24 | FDA | Viable cells, metabolic status | 36 |
| Vancomycin | S. aureus, MRSA | 2–4 | Oxonol | Membrane effects | 310 |
| Enterococcus spp., E. coli | 0.5–2 | Baclight kit (Molecular Probes.) | Live and dead cells | This work |
MRSA, methicillin-resistant S. aureus.
Available data clearly demonstrate the utility of this technique to also assay viral susceptibility. Since the pioneer works of Rosenthal et al. (280) and Pauwels et al. (247) on the use of FCM to study the effect of antiviral agents on herpesvirus and HIV, FCM has been used by several groups, particularly those studying these viruses and CMV. The detection and quantification of viral antigens and viral nucleic acids in combination with antibodies or nucleic acid probes, respectively, or any other cellular parameter associated with viral infection have been used in FCM protocols for the in vitro evaluation of antiviral-drug activities. In addition, the possibility of on-line monitoring of the antiviral treatments ex vivo has also been explored. The latter approach has also been applied to bacterial and fungal infections; rapid and sensitive protocols for the evaluation of microbial responses to antimicrobial agents are also available.
In the following sections, the possible uses of FMC in antimicrobial susceptibility testing are discussed according to the type of pathogen involved.
Antibacterial Agents
Standardized methods for performing in vitro susceptibility testing of bacteria in clinical microbiology laboratories are widespread (98). Qualitative and/or quantitative results are given on the basis of the size of the zone of inhibition or MIC. Despite the automation of MIC-based broth microdilution systems, an 18- to 24-h incubation period is usually needed before antimicrobial activity can be quantified. Recently, fluorogenic, turbidometric, and colorimetric technology has reduced susceptibility testing to 4 to 6 h (83, 98). However, unless the bacterial inoculum is quantified, only the bacteriostatic effect, i.e., the MIC, is generally tested by clinical laboratories. The MBC, which reflects the bactericidal effect of antimicrobial drugs, is rarely determined. This requires calculation of bacterial counts, generally expressed as CFU, which involves bacterial culture dilutions and subcultures. In addition, neither MIC nor MBC determinations consider the heterogeneity of bacterial populations. Similarly, postantibiotic and subinhibitory concentration effects (106), which offer pharmacodynamic data based on the antimicrobial activity, are rarely determined in clinical laboratories since these determinations (190) are tedious and time-consuming.
FCM has proved to be very useful for studying the physiological effects of antimicrobial agents on bacterial cells due to their effect on certain metabolic parameters (membrane potential, cell size, and amount of DNA). In addition, FCM is a reliable approach for susceptibility testing, offering results in terms of the bactericidal or bacteriostatic effect (86, 108, 198, 262, 331).
The studies by Martinez et al. (198) and Steen et al. (302) in 1982 on the antimicrobial effects on bacteria assayed by FCM are now considered classic. They demonstrated that the effect of β-lactam antibiotics on E. coli can be detected by measurements of light scattering and DNA content after 10 min of incubation with the drug. Using FCM, Steen et al. (302) showed the effect of several antibiotics that inhibit protein synthesis (chloramphenicol, erythromycin, doxycycline, and streptomycin) on E. coli with the fluorescent DNA probe mithramycin (305). Since then, FCM has been used to measure the effects of different antimicrobial agents on bacteria (Table 2).
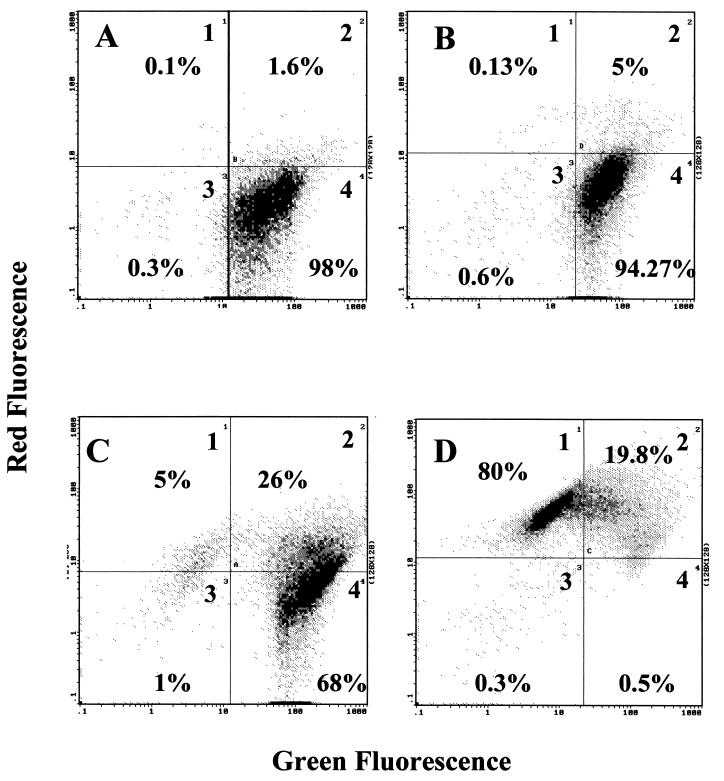
Realizing the potential of FCM in this field, several companies have developed new products to rapidly perform antibiotic susceptibility testing in clinical microbiology laboratories. We have used one of these products (Bac-light; Molecular Probes) to analyze the reliability of this test in clinical microbiology. We worked with two strains of Enterococcus, one sensitive to vancomycin (E. faecalis ATCC 29212) and other resistant (E. faecium U2A1), with a vanA element responsible for its vancomycin resistance phenotype. In addition, as a control we included an E. coli isolate showing intrinsic resistance to vancomycin. The above-mentioned products are kits with two fluorochromes that permit the detection of live and dead cells. Bacterial cells were incubated with and without vancomycin for 2 h at the respective MICs of this drug (Fig. 5). Antimicrobial effects were detected by measuring the variations in fluorescence due to dead cells within 3 h of incubation with the antibiotic. Another advantage of FCM (Fig. 5), as mentioned above, is the visualization of the heterogeneity of the response of the cells to the antimicrobial agent. This heterogeneity is detected by determining the presence of subpopulations that are less susceptible to the antimicrobial agent treatment. Davies and Hixson have recently reported similar results (69). Therefore, it is feasible to use FCM and fluorescence probes to perform rapid testing of the susceptibility of bacteria to a panel of antimicrobial agents before choosing the treatment. Moreover, resistant subpopulations can be effectively detected, representing an interesting advantage of FCM that can be suitably exploited in clinical studies (331).
FIG. 5.
Antimicrobial susceptibility testing by FCM using the Bac/live kit (Molecular Probes). Antimicrobial susceptibility is shown by the decrease in green fluorescence (live cells) and the increase in red fluorescence (dead cells). (A) Distribution of E. coli cultures without antimicrobial agent incubation; therefore, the cells appear mainly in quadrant 4 (positive for green fluorescence). (B) An E. coli culture was incubated for 2 h with vancomycin at 1.024 g/ml. The distribution is similar to that seen in panel A. Accordingly, this strain was not affected by antimicrobial agent treatment, although a small percentage of cells was positive for the red fluorescence, meaning that the cells were sensitive to vancomycin. The same protocol was applied to E. faecium (C) and E. faecalis (D) cultures. As described in the text, E. faecium is vancomycin resistant and E. faecalis is vancomycin sensitive. (C) One subpopulation of E. faecium lies in quadrant 1 (positive for red fluorescence only), another is in quadrant 2 (positive for red and green fluorescence), and the majority is in quadrant 4 (positive for green fluorescence only). This means that the behavior of the E. faecium population is not homogeneous in the presence of vancomycin, perhaps due to the loss of the element responsible for vancomycin resistance. (D) After 2 h of incubation, most E. faecalis cells appear in quadrant 1 and the rest appear in quadrant 2. Therefore, almost all cells are positive for red fluorescence and are dead. However, a small population (0.5%) is still present in quadrant 4, meaning that this population is less sensitive to vancomycin than the rest (over 99% of the cells).
Measurement of bacterial susceptibility.
By using fluorescence probes, FCM measures the light scattering of cells in suspension and fluorescence of cellular contents or metabolic activity. Thus, the activity of antimicrobial agents can also be described in terms of changes in cell shape and in fluorescence (262, 329, 330). In this sense, Walberg et al. (329, 330), and others (108, 200) showed that light scattering is a useful parameter of antimicrobial effects, irrespective of their mechanisms of action. This was demonstrated with two different drugs: ceftazidime, which acts on the bacterial envelope, and ciprofloxacin, whose target is a gyrase. Nevertheless, the same authors (329, 330) used both fluorescence detection and light-scattering analysis when susceptibility testing was performed directly in clinical samples in order to distinguish bacterial cells from debris and other components.
Two major categories of fluorochromes have been used to study antimicrobial effects and short-term antimicrobial activity: (i) DNA- and RNA-labeling dyes and (ii) metabolic or protein-binding probes, which include membrane potential-sensitivity dyes (262) (Table 2). The choice of these substances in a particular protocol is related to their excitation and emission properties and to the mechanism of action of the antimicrobial. Mithramycin with ethidium bromide (DNA-staining dyes) were the first probes used for susceptibility testing by FCM (302) and are still in use (331). These probes estimate viability in an indirect way. After staining, the antibacterial activity in antimicrobial-exposed cells is measured as a result of the ploidy in organisms undergoing filamentation (unable to generate independent cells after division). This has been performed with β-lactam antibiotics, which affect cell membrane integrity but induce an increase in nucleic acid contents in growing but not dividing cells. DNA ploidy has also been studied by Durodie et al. (86) in E. coli with sub-MIC concentrations of β-lactam antibiotics.
PI, a fluorochrome that intercalates into double-stranded nucleic acid, is a more appropriate stain than ethidium bromide for susceptibility testing by FCM (262). Because PI is a nonpermeable dye that is excluded by viable cells, measurement of the loss of viability produced by antimicrobial agents or other compounds is possible. For example, Gant et al. (108) studied the effect of several classes of antimicrobial agents, such as an aminoglycoside (gentamicin), β-lactams (ampicillin, mecillinam, and cefotaxime), and a DNA gyrase inhibitor (ciprofloxacin), on the integrity of the outer membrane of E. coli as measured by the uptake of PI. This method allows differentiation among antibiotics with different cell wall enzyme targets due to its effects on cell light-scattering characteristics and its effect on the integrity of the outer cell membrane of E. coli, measured simultaneously.
Other new nucleic acid-staining fluorochromes that provide more intense fluorescence and resolution have also been used to test antimicrobial activity (189, 203). SYTO-13 and SYTO-17 label both DNA and RNA (127). These stains were used by Comas and Vives-Rego (59) in combination with oxonol to assess the effects of gramicidin, formaldehyde, and surfactants on E. coli viability and membrane potential. Another fluorochrome is acridine orange (202), which stains DNA and RNA at the same time but with different wavelength emissions. Therefore, variations in the relative amount of each nucleic acid in the presence of antimicrobials can be evaluated. Also, acridine orange penetration depends on plasma membrane integrity, and so the method can also be used to measure alterations in plasma membrane permeability produced by the action of antimicrobial agents.
Bacterial cells respond to different antimicrobial agents by decreasing or increasing their membrane potential. Moreover, cell death and/or membrane damage brings about a collapse of the electrical potential. Membrane potential can be effectively measured by FCM using potential-sensitive probes (262). These dyes are normally positively charged and accumulate inside the cell, rendering it fluorescent. As the cell depolarizes, the dye is redistributed or the electronic environment changes and the fluorescence produced by each fluorochrome molecule decreases. Some of these probes belong to the carbocyamide group of dyes, which includes 3,3′-dipentyloxocarbocyanine iodine [DiOC5]. This dye was previously used to measure membrane potential in mammalian cells by FCM (291). Ordoñez and Wehman (237) applied DiOC5 to assess the susceptibility of S. aureus to penicillin G and oxacillin. The results were comparable to those obtained by conventional susceptibility tests but were obtained 90 min after addition of the antimicrobial, as opposed to the usual overnight period.
Rhodamine 123 and oxonol [bis(1,3-dibutylbarbituric acid)trimethine oxonol] are other membrane potential probes used to assess drug effects by FCM (59, 150, 310). Their accumulation inside bacterial cells depends on the difference in charge between the two sides of the plasma membrane. Rhodamine 123 is a cationic lipophilic dye. Because it is not readily taken up by gram-negative organisms (157, 205), it is used only for gram-positive organisms. Rhodamine 123 is accumulated in the cytosol by cells with an active transmembrane electrochemical potential. In contrast, oxonol is an anionic lipophilic dye that can enter the cell only when the membrane has become depolarized. Therefore, fluorescence-emitting cells have a low membrane potential; the higher the decrease in membrane potential, the higher the intensity of oxonol fluorescence (67, 78, 157, 189, 203, 237).
Since the same fluorochrome can be used in different studies, versatility is an essential property. Oxonol has been used for susceptibility testing of different classes of antimicrobial agents in both gram-positive and gram-negative organisms (Table 2). Mason et al. (200) used this dye to analyze membrane damage caused by gentamicin, a protein synthesis inhibitor, and ciprofloxacin, a DNA gyrase inhibitor. Suller et al. (310) used it to study the effect of penicillin, methicillin, and vancomycin, all of which interfere with bacterial envelope synthesis. In addition, Jepras et al. (150) used oxonol to study the action of azithromycin, cefuroxime, and ciprofloxacin on E. coli. Another advantage of oxonol is that it can be added directly to the liquid culture, avoiding any pretreatment steps that may cause perturbations in bacteria and interfere with the effect of the antimicrobials. Furthermore, oxonol is nontoxic to bacterial cells, unlike cationic dyes (79).
Measurement of antimycobacterial drug susceptibility.
Fluorescein diacetate (FDA) (a nonfluorescent diacetyl fluorescein ester that becomes fluorescent upon hydrolysis by cytoplasmic esterases) staining and FCM have been used by Norden et al. (231) for the FCM susceptibility testing of Mycobacterium tuberculosis. In contrast to the delay in obtaining data in current antimycobacterial drug susceptibility testing, results were available within 24 h. In addition, using FDA, Bownds et al. (36) demonstrated that mycobacterial multiplication was not required to measure antimycobacterial drug activity, as is the case with traditional methods. Mycobacterium avium, Mycobacterium gordonae, Mycobacterium fortuitum, and Mycobacterium marinum treated with different concentrations of ciprofloxacin, kanamycin, and other tuberculostatic agents were easily differentiated from untreated cells after 6 and 24 h of incubation (36). Using the same protocol, Kirk et al. (164) determined the susceptibilities of 35 clinical isolates of M. tuberculosis to several concentrations of isoniazid, rifampin, and ethambutol in 24 h. To avoid manipulator infection risks, Moore et al. (222) have developed a variation of the above method using paraformaldehide to inactivate Mycobacterium. In this case, the time needed to obtain susceptibility results increased to 72 h.
A different approach has been explored to use FCM to detect mycobacterial growth inhibition under antimicrobial pressure based on mycobacterial microencapsulation in agarose gel microdrops. FCM was used to monitor colony growth by evaluating the intensity of auramine staining after different times of incubation in the presence or absence of rifampin and isoniazid. With this system, growth inhibition was demonstrated after 1 to 3 days, and, indeed, the presence of an isoniazid-resistant subpopulation was observed (283).
Intracellular bacterial pathogens.
Antimicrobial susceptibility testing by FCM has also been used for intracellular pathogens such as Rickettsia tsutsugamushi (160). Conventional susceptibility methods for this organism include direct microscopic counting of Giemsa-stained cells and mouse protection assays. Both methods are slow, labor-intensive, and expensive in comparison with FCM screening. Kelly et al. (160) evaluated the in vitro effectiveness of doxycycline and azithromycin against both doxycycline-sensitive and -resistant R. tsutsugamushi strains. Inhibitory concentrations were assayed by measuring the reduction in the number of infected cells determined by fluorescence emission of stained R. tsutsugamushi. This method allowed screening of numerous isolates for their susceptibility to a variety of antibiotics that was faster than conventional techniques. The correlation between FCM screening and conventional susceptibility testing was high.
Recently, Dessus-Babus et al. (75) developed a specific and reproducible technique for the susceptibility testing of another intracellular pathogen, Chlamydia trachomatis. The results for susceptibility to doxycycline, ofloxacin, and erythromycin obtained by FCM were compared with those obtained using the standard method of determining the minimal chlamydicidal concentration by microscopie examination. The assay was performed on 13 clinical isolates after infection of McCoy cells and incubation for 25 h with and without the antimicrobial agent. An FITC-conjugated monoclonal antibody to the major outer membrane protein of Chlamydia was used to stain chlamydial inclusions. Calculation of the 50% inhibitory concentration (IC50) by FCM allowed a more objective and precise evaluation of antimicrobial activity than did reading of microscopic inhibitory concentrations. The values obtained were reproducible and could be applied to compare the results obtained with different drugs.
Postantibiotic effect.
The presence of a transient inhibition of bacterial growth after prior exposure to antimicrobial agents (postantibiotic effect) was first described by Bigger (30). Although this effect is responsible for a persistent inhibition of bacterial cells after short-term exposure to an antimicrobial agent (190), the mechanism(s) through which this effect is exerted remains obscure (106). These data are generally used to define antimicrobial dosing, e.g., single daily doses, as in the case of the new fluoroquinolones and aminoglycosides (101, 307). Traditionally, studies of the postantibiotic effect have been based on bacterial counts (190), and only a few authors have explored the possibility of using other approaches (65). By detecting changes in the impedance of the culture media, Baquero et al. (18) were able to measure bacterial growth. This parameter is related to the metabolic activity of the cell population and may be considered equivalent to measuring viable cells in a given population. Other authors have used optical density, carbon dioxide generation, intracellular ATP measurements, and the rate of [3H]thymidine incorporation to assess microbial growth (65). Recently, FCM has been used to successfully determine the postantibiotic effect (119, 309). This technique has the advantage of studying the heterogeneity of bacterial populations and particular metabolic events during the postantibiotic-effect period. Gottfredsson et al. (119) studied microbial sizes and nucleic acid contents in E. coli and P. aeruginosa populations during the exposure of these microorganisms to antimicrobial agents as well as during the postantibiotic effect period (after drug removal). Aliquots of bacterial growth were stained with PI at regular time intervals during and after drug exposure. Different effects could be observed during the post-antibiotic period. In general, the postantibiotic effect induced by β-lactam antibiotics and ciprofloxacin is characterized by the appearance in the population of enlarged cells with increased nucleic acid contents whereas rifampin induces a decrease in the size and nucleic acid contents of the microorganism. Both effects would have been missed if only bacterial counts had been monitored. Suller et al. (310) also reported the heterogeneity of bacterial populations during the postantibiotic-effect phase in S. aureus cultures previously exposed to methicillin. Despite the observation of very few variations in the numbers of CFU during this phase, a continuous decline in membrane potential and respiratory activity was found using FCM. Again, these effects would have been missed if FCM had not been available.
Antifungal Agents
Interest in antifungal agents and resistance to these antimicrobials has redoubled during the last decade. Invasive fungal infections have emerged as major causes of morbidity and mortality due to the increasing numbers of immunocompromised and HIV patients. Nevertheless, antifungal susceptibility testing in clinical laboratories is currently performed to a far lesser extent than is susceptibility testing of antibacterial agents. Unlike bacteria, standardized methods and interpretable guidelines have only recently been proposed for antifungal agents, and even then they remain controversial (253). Different definitions of breakpoints for susceptibility testing may occur as a result of the absence of consensus-standardized MIC techniques for different antifungal drugs (165), hindering the interpretation of results and protocol standardization. The broth dilution methods for yeast susceptibility testing of the National Committee for Clinical Laboratory Standards (NCCLS) are labor-intensive and require 48 to 72 h of incubation. Alternative methods including the E-test (253), the Bioscreen microdilution method (323), and a colorimetric system (94) have also been proposed.
The FCM assay has been proposed as a reliable alternative method to study antifungal activity. Moreover, FCM has also been used for yeast susceptibility testing (120, 163, 234, 252, 259–262, 270, 336). The results obtained with FCM agreed with those obtained by NCCLS methods and were available in only 24 to 32 h or even in 9 h (163, 233). Moreover, the definition of end points for antifungal treatment by FCM is probably easier (252).
There are several studies based on FCM. In 1990, Pore et al. (259) developed an FCM assay to study the susceptibility of C. albicans to ketoconazole, amphotericin B, and 5-fluorocytosine. Using both PI and rose bengal, the time required for susceptibility determinations decreased from 24 to 48 h to only 9 h. Sensitivity and reliability were greater than in the standard NCCLS broth dilution method. The assay could also be applied to other pathogenic yeasts such as Candida tropicalis, Candida parapsilosis, Candida lusitaniae, Candida krusei, Candida guilliermondii, Torulopsis glabrata, and Rhodotorula rubra (262) and Crytococcus neoformans (261). O'Gorman and Hopfer (233) used ethidium bromide and FCM to study drug susceptibility in yeasts, measuring the effects of amphotericin B on Candida species after 8 h of incubation. Carter et al. (45) performed similar studies using different fluorochromes to detect dead and living cells (PI and FDA) and changes in membrane potential (oxonol) and intracellular ion concentrations (Indo-1 and Fluo-3). Unfortunately, no relationship between the antifungal agents and the ion concentration could be established, since the ion-sensitive probes were unable to stain yeast cells. The authors reported alterations in oxonol fluorescence after 20 min of incubation with amphotericin B, although with other fluorochromes, such as PI, the response time was longer (8 h). Differences in the detection times of the various fluorochromes depend on the parameter measured by the fluorochrome. Since amphotericin B mainly affects the plasma membrane potential, oxonol will first detect this effect. By increasing the amphotericin B incubation time, the plasma membrane will be affected to a greater extent, which will permit the entrance of PI.
In 1994, using FDA, Green et al. (120) showed that FCM was a better and faster method than conventional ones for determining and comparing the antifungal activities of compounds with different modes of action. Since then, several studies have shown that FCM can be effectively used for antifungal susceptibility testing. Ordóñez and Wehman (237) measured the variations in the membrane potential of different Candida species with DIOC5 when cells were incubated with amphotericin B. Sensitivity or resistance was determined after 30 min in the presence of the antifungal agent. Moreover, two recent articles have reported the effectiveness of PI in testing antifungal susceptibility by FCM (163, 270). The results of both studies were comparable to those obtained with the reference NCCLS macrodilution method during a period ranging from 2 to 24 h. The antifungals studied were amphotericin B, fluconazole, and itraconazole. Recently, Peyron et al. (252) have compared FCM with the NCCLS broth dilution method for amphotericin B. Oxonol was used to measure the variations in membrane potential. From the dose-response curve obtained for each isolate, the concentrations at which the fluorescence intensity was reduced by 50, 80, and 90%, i.e., the IC50, IC80, and IC90, respectively, were calculated. Regression analysis revealed that the best agreement with the M27-T NCCLS document was obtained with the IC80 end point. Thus, FCM can be effectively used for routine susceptibility testing since this end point is currently used in clinical laboratories to distinguish susceptible from resistant isolates. Moreover, the results were highly reproducible and were obtained within 1 h.
Also, susceptibility studies of Pneumocystis carinii have been performed using FCM and PI (223). Although significant advances have been made in susceptibility testing for filamentous fungi (73), to our knowledge FCM has not yet been used.
Factors Affecting Bacterial and Fungal Susceptibility Testing
Several authors have underscored the importance of different factors that can affect bacterial susceptibility testing by FCM. Although such factors do not imply a clear limitation in the FCM assay, they may give misleading results. Knowledge of these factors is critical for the implementation of methods to avoid false resistance and/or false susceptibility results. These factors have been studied with bacteria, but similar considerations could also be applied to other organisms, such as fungi or parasites.
The organism itself can affect susceptibility testing by FCM. Because different bacterial species and occasionally even different strains of the same species show different abilities to take up membrane potential-sensitive dyes, this has been suggested to affect susceptibility testing by FCM (201). The ability of rhodamine 123 to stain cells differs between gram-negative and gram-positive organisms (205), possibly due to the highly charged nature of gram-negative bacterial lipopolysaccharide. Moreover, enhanced efflux mechanisms resulting in extrusion of the internalized dye may decrease the amount of dye present inside the cells of certain bacteria and thus give misleading results (95, 151, 220).
Dye concentrations and staining times can also affect susceptibility testing by FCM (59, 108, 201). For example, the labeling properties of oxonol in E. coli are related to dye concentrations (201). The effects of valinomycin were better defined at oxonol concentrations of 10 than 0.5 mg/liter. Nevertheless, a high dye concentration can produce cytotoxic effects (259). Therefore, staining protocols for susceptibility testing should be carefully established and appropriate controls should be used.
Another important issue is the cell preparation. For instance, permeabilization is used in certain FCM protocols. This procedure allows certain nonpermeable dyes to cross the cytoplasmic membrane, while permeable dyes cross more readily. Therefore, incubation times must be changed. Pretreating gram-negative bacteria with 1 mM EDTA improves the uptake of the membrane potential dye rhodamine 123, allowing the detection of antibiotic effects on the membrane potential of gram-negative bacteria. However, such pretreatment can affect the physiological parameters of bacterial viability and indeed the membrane potential (79, 205). A fixation step is mandatory in DNA staining of bacteria with nonpermeant stains. This can be achieved by 70% ethanol treatment (37), but two steps of centrifugation are required, thus increasing the processing time and loss of cells. An alternative to ethanol fixation is the cold shock procedure, which has been used with E. coli cells (151). Permeabilization of exponentially growing E. coli involves placing the cells in ice-cold EDTA (10 mm) in phosphate-buffered saline (pH 7.4) containing 4 g of azide per liter. The cold shock procedure was recently used by Walberg et al. (331) in combination with ethidium bromide and mithramycin. This seemed to be as efficient as ethanol fixation for detecting the effects of β-lactams and quinolones but not for detecting the effect of gentamicin.
The use of surfactants has also been proposed to render cells permeable to fluorescence dyes but can affect the FCM results (59). Because these agents may exert biocidal activity, they could interfere with the antibacterial effect of the antimicrobial agent being tested. Also, these agents may generate independent particles (debris) which are detectable by the cytometer as light scatter signals (59). Sometimes these signals are proportional to the surfactant concentration and can be attributed to micelle formation and cellular aggregation. Nevertheless, by using cationic surfactants with null biocidal activity, this pitfall has been circumvented (59).
The antimicrobial drug concentration should be carefully controlled since different concentrations may give different results. In early studies, Steen and coworkers used concentrations exceeding the MIC values for bacteria (302, 303) but other authors used lower and even sub-MIC concentrations (87, 198). Over the last few years, antimicrobial effects have been studied at clinically achievable concentrations (58, 200, 237, 329, 330). Gant et al. (108) tested antimicrobials by FCM at concentrations equivalent to conventional MICs. In this study, the exposure time of bacteria to the antimicrobial agents prior to the FCM assay was also critical for the demonstration of the inhibitory action.
The inoculum and bacterial growth phase are other factors that may affect FCM results. Comas and Vives-Rego (59) used stationary-phase instead of exponential-phase cells mainly for two reasons: (i) stationary-phase inocula are used in conventional antimicrobial assays, and (ii) stationary-phase cells remain stable longer than do exponential- or dead-phase cells. These authors observed a progressive decline in rhodamine 123 uptake during the growth phase, whereas exponential or stationary populations did not take up oxonol, which was incorporated only in the dead-phase. In general, bacterial and yeast inocula are slightly higher in FCM susceptibility assays than in conventional assays. With this approach, resistant subpopulations can be better detected.
Like any other technique, FCM requires work and time in the setting up of protocols. The factors affecting the results are the same as those affecting conventional susceptibility protocols plus those intrinsic to FCM, such as aggregation, cell size, and autofluorescence. Once these are delineated, FCM can be routinely used in antimicrobial susceptibility testing. The possibility of using simple commercial kits, such as the one described in Fig. 5, will encourage the use of FCM in clinical microbiology laboratories.
Antiparasite Susceptibility
Drug resistance among parasites is an increasingly recognized phenomenon, particularly among different Plasmodium species (88, 242). Clinical failure, rather than laboratory results, is taken as indicative of a drug resistance process. Unlike bacteria and yeasts, testing of parasites for susceptibility to antimicrobial drugs is not generally performed in clinical laboratories due to technical difficulties and the absence of standardized methods. Interestingly, different authors have used FCM to study the effect of antiparasitic drugs and explore the possibility of routine susceptibility testing. For example, Azas et al. (15) used FCM to assess the effects of pentamidine, allopurinol, and amphotericin B on Leishmania infantum metabolism with PI to measure viability and DIOC5 to detect membrane potential alterations. Moreover, using FCM and rhodamine 123, Maarouf et al. (192) demonstrated that paromomycin rapidly affects the mitochondrial activity of Leishmania donovani, lowering rhodamine 123 fluorescence. They suggested that the decrease in rhodamine 123 fluorescence could be used to detect sensitivity to drugs in this parasite. An FCM multiparameter approach was developed by Basselin et al. (21) to study the differences between pentamidine-resistant and -nonresistant Leishmania. They detected variations in mitochondrial membrane potential and lipid metabolism in pentamidine-resistant cells.
Pattanapanyasat et al. (242), working with a drug-resistant Plasmodium strain, described an antiparasite drug susceptibility method using FCM. Using several iron chelators as antiparasite drugs, FCM measured their effects on Plasmodium viability by detecting parasites inside the red blood cells. They demonstrated the importance of iron for parasite growth and showed that FCM provides a simple and reliable means of antimalarial drug susceptibility testing. Analyzing environmental samples, Bamdad et al. (16) used FCM to test the uptake and efflux of several polycyclic aromatic hydrocarbons in Tetrahymena spp. in an attempt to obtain evidence of a resistance mechanism.
As stated above, FCM has already been used to study the effects of antiparasite drugs. Despite this, its application in routine practise does not seem to be as common as in the study of antibacterial and antifungal agents, perhaps for socioeconomic reasons rather than because of scientific criteria.
Antiviral Susceptibility
Although antiviral in vitro susceptibility testing is not routine in the clinical microbiology laboratory, the development of rapid and accurate tests is essential for the screening of new antiviral compounds and antiviral susceptibility studies. From a clinical point of view, the emergence of antiviral resistance increases the necessity for rapid, automated, and objective assays. Different methods commonly used to test viral susceptibility include plaque reduction assays, virus yield reduction assays, assays based on inhibition of the virus-induced cytopathic effect, and assays for the detection of viral nucleic acids and viral antigens. These conventional methods are labor-intensive and time-consuming. Because viral antigens, nucleic acids, or any viral parameter related to viral infection can be quantified by FCM, the antiviral susceptibilities of clinical isolates or laboratory strains can be rapidly determined. FCM also permits rapid screening and assessment of new potential antiviral compounds and their mechanisms of action. In a recent review, McSharry (212) has described the applications of FCM to (i) the screening of antiviral compounds active against human CMV, HSV, and HIV and (ii) the performance of drug susceptibility testing for clinical isolates. Since both FCM studies of antiviral susceptibility in clinical isolates or laboratory strains and studies of antiviral activities or antiviral mechanisms of action are based on the same principles and techniques, we will review both in this section by virus type.
HSV.
Rosenthal et al. (280) developed a rapid and easy FCM method to screen the anti-HSV activities of several antiviral agents. The method was based on HSV-induced changes in cellular DNA content measured by the binding of PI to the DNA. The lower the antiviral activity, the larger was the DNA amount and the more intense was the PI fluorescence (280). The activity of antiviral drugs against HSV-1 (300), HHV-6 (276) and varicella-zoster virus (295, 296) has also been assayed by FCM by measuring the inhibition of specific viral antigen expression for each drug with antibodies against virus antigens. For example, Pavic et al. (248) developed a quantitative FCM method measuring gpC expression to determine the susceptibility of HSV-1 to acyclovir, ganciclovir, and foscarnet; this was done by measuring the amount of antibody against gpC binded to infected cells. The results were compared with those obtained by the standard cytopathic effect inhibitory assay. A very interesting conclusion was reached: while both methods gave identical qualitative results, the range of IC50 detected for acyclovir-resistant strains by FCM was much broader than that detected by the cytopathic effect inhibitory assay for the same isolate. Therefore, it is possible to detect resistant viral subpopulations within phenotypically characterized sensitive populations by using FCM. In view of the emergence of drug-resistant viral subpopulations after long-term treatment with acyclovir, this finding is of great interest in the clinical setting, since it should be possible to detect not only resistance but also the emergence of resistance in a patient.
CMV.
FCM has been used for rapid in vitro evaluation of anti-CMV agents (185, 227, 295). Neyts et al. (227) used FDA to measure the cytoplasmic esterase activity after CMV infection. Differences in FDA fluorescence allowed these authors to differentiate between CMV-infected and -uninfected cells and to evaluate several antiviral agents. Snoeck et al. (295) monitored the anti-CMV activity of several phosphonyl methoxyalkylpurines and pyrimidines by FCM and DNA hybridization. Recently developed by McSharry et al. (214, 215) and Kesson et al. (162), an FCM assay differentiated gancyclovir-sensitive and -resistant strains of human CMV clinical isolates and laboratory strains by quantification of human CMV immediate-early and late antigens with specific monoclonal antibodies. In all these studies, the accuracy of the FCM assay was confirmed by parallel testing by the plaque reduction assay (PRA). There was an excellent correlation between IC50 determined by FCM and the values determined by PRA, although the FCM assay gave a broader range of IC50 than the PRA did (214, 215). Furthermore, the effect of ganciclovir was detectable by FCM at 96 h postinfection instead of the 7 days required for PRA.
HIV.
In 1987 Pauwells et al. (247) used an FCM assay to quantitatively measure the inhibition of viral antigen expression by 3′-azido-2′,3′-dideoxythymidine (AZT) and 2′,3′-dideoxycytidine (ddCyd) in HIV-1-infected human T4 lymphocyte cells. The anti HIV-1 activity of a broad range of compounds has subsequently been studied using FCM (113, 166, 193, 208, 218, 224, 236, 284–288, 341). Using FCM and MT-4 cells, Schols et al. (287) monitored the cell death produced by HIV-1 infection using FDA and also observed the effect of AZT. A dose-dependent viral inhibition was observed by FCM that was comparable to the results obtained by the more tedious and time-consuming conventional methods. FCM determination was able to monitor the effects of antiretroviral treatment regardless of the status of p24 antigen expression in infected cells (62, 208). FCM has also evaluated the effect of inhibitors of HIV-1 proteases and other antiretroviral drugs on the expression of cell-associated viral proteins in treated peripheral blood lymphocytes and cell lines (166, 218, 341). AZT and 2′,3′-dideoxyinosine (ddI) inhibit the establishment of HIV-1 infection in CD8+ cells derived from peripheral blood lymphocytes as shown by FCM analysis using anti-glycoprotein 120 and anti-CD8 monoclonal antibodies to quantify the percentage of CD8+ infected cells (CD8+ gp120+) (218).
Using a coculture system of chronically HIV-1 infected monocytes and noninfected CD4+ T lymphocytes, it is possible to use FCM to study viral dissemination and hence the inhibition of dissemination in the presence of antiviral compounds (224). Recently, Macchi et al. (193), using a similar coculture system of PBMC isolated from normal adults and the chronically HTLV-1-infected MT-2 cell line, measured the levels of viral DNA and RNA by PCR and viral core p19 expression by FCM. The inhibitory effect of AZT on virus transmission to PBMC cells was demonstrated (193). Coculturing uninfected JM cells and HIV-1 chronically infected H9 cells and using two-fluorescence FCM analyses, Bridges et al. (41) demonstrated the dose-dependent effect of the drug 6-O-butanoyl castanospermine on virus production as measured by the decreases in cell conjugate formation between normal and chronically infected cells.
A different way to study the effect of antiviral agents is to quantify the expression of viral genes using gene reporters. A green fluorescent protein reporter system in which the gene encoding this protein is under the control of the HIV-1 long terminal repeat promoter was described for the rapid and simple quantification of HIV infection (85). In the presence of HIV-1, expression of the reporter gene is strongly induced, and FCM measurement of the relative fluorescence in cells allows reliable quantification of infected cells. Recently, Gervaix et al. (113) obtained a stable T-cell line expressing a similar green fluorescent protein construct. This stable cell line offers a very rapid, easy, and accurate method for monitoring HIV-1 infection and evaluating new antiretroviral compounds. Rapid determination of drug susceptibility to HIV-1 RT and protease inhibitors as well as other antiretroviral drugs was also performed by these authors.
Poliovirus.
The infection of human fibroblasts by poliovirus leads to morphological changes related to cell size and granularity in infected cells, which can be detected by FCM. The analysis of this induced cytopathic effect, as stated above, has been extensively used to monitor viral infection, and FCM allows perfect monitoring of these changes. Furthermore, [Ca2+]i can be monitored simultaneously. As shown in Fig. 6, [Ca2+]i increases from 2 to 3 h postinfection and 90% of the cells increase their [Ca2+]i by the 5 h postinfection. These effects on cell size and the increase in [Ca2+]i in infected cells must be a consequence of viral gene expression, since such modifications are not observed when cells are incubated with cycloheximide, guanidine, or Ro 09-0179 compounds (145). This is a good example of multiparametric analysis of viral infection and also highlights the potential of this technique for studying antiviral activity.
FIG. 6.
FCM analysis of poliovirus-infected HeLa cells. (A) Kinetics of intracellular free calcium during poliovirus infection. Poliovirus infection was performed at a multiplicity of infection of 50. After 1 h of adsorption at the indicated times postinfection, poliovirus-infected cells were detached from the plates, incubated with 6 μM fluo-3 AM, and analyzed in a FACScan flow cytometer. (B) Effects of guanidine, cycloheximide, and Ro 09-179 on [Ca2+]i during poliovirus infection. Guanidine (GND) (500 μM), Ro 09-179 (Ro) (1 μg/ml), or cycloheximide (CHX) (50 μM) was added to the infected cells after 1 h of poliovirus adsorption. The [Ca2+]i was monitored using FCM 4 h postinfection. Nontreated poliovirus-infected cells (Poliovirus) and nontreated uninfected cells (Control) are also shown. (C) Simultaneous FCM analysis of cytopathic effect and [Ca2+]i on poliovirus-infected cells. Noninfected (top) and poliovirus-infected (bottom) HeLa cells were analyzed 4 h postinfection for cytosolic free calcium (as described in panel A) and cell size (measured as forward light scatter). Adapted from reference 145 with permission of the publisher.
Antiviral drugs: mechanisms of action.
Understanding the mechanisms of action of antiviral drug is important for the design of new antiviral compounds and the improvement of existing ones. Some examples of FCM used in studies of antiviral mechanisms of action with HIV are explained below. Since FCM has permitted the study of virion-cell binding through specific receptors, it allows the characterization of antiviral drugs that inhibit this process, as demonstrated by Schols et al. (284). They used human anti-HIV serum in an FCM assay to detect the HIV-1 virion binding to the cell membrane, analyzing several compounds for their ability to inhibit it. While AZT did not affect viral binding, heparin, dextran sulfate, and pentosan polysulfate completely inhibited the process. Suramin, an inhibitor of RT, partially blocked HIV-1 binding. Subsequently, these authors showed that these compounds which inhibited HIV-1 binding to MT-4 cells block the interaction between the gp120 and the CD4 receptor (286). Using an FCM assay based on a DNA-staining protocol which reveals differences in DNA contents in HUT-78 cells persistently infected with HIV-1 and CD4+ MOLT-4 cells, these compounds were shown to suppress syncytium formation and destruction of CD4+ cells. Of note, Oravecz et al. (236) later studied the role of cell surface proteoglycans in chemokine-mediated anti-HIV-1 activity in T cells and macrophages. FCM revealed that digestion of cell surface proteoglycans, including heparin sulfate, prevented the binding of RANTES to PM1(CD21H) cells (a human T-cell line selected for high CD26 expression). Finally, using FCM, Schols et al. (288) measured the effect of AMD3100 (a prototype of bicyclams that is a new selective inhibitor of HIV-1 and HIV-2 replication) on the binding of a specific CXCR4 monoclonal antibody to SUP-T1 cells. The drug inhibited this binding, indicating an interaction with the CXC chemokine receptor CXCR4 (coreceptor for T-tropic viruses). However, AMD3100 did not inhibit the binding of CC chemokine macrophage inflammatory protein, a ligand of the chemokine CCR5 receptor (coreceptor for M-tropic viruses).
Summarizing, a close correlation is always found between the FCM assay and conventional susceptibility methods. Furthermore, FCM decreases the time required for antimicrobial susceptibility studies and also allows multiparametric analysis, meaning that more information is obtained from the same sample. Finally, it can be used to gain further insight into the mechanisms of action of antimicrobial agents.
MONITORING OF INFECTIONS AND ANTIMICROBIAL THERAPY
The evaluation of patient responses to antimicrobial treatments is usually based on clinical findings. Nevertheless, different microbiological studies are frequently performed to assess the efficacy of antimicrobial treatments (152). In vitro susceptibility tests provide information about the concentration at which a specific microorganism is susceptible. However, in vitro results may not correlate with the response to in vivo therapy since other factors affect this susceptibility (152). Among these factors are the immunological status of the patient, the viral or bacterial load, and the possibility of selecting antibiotic-resistant strains as a consequence of the antimicrobial treatment. The last point is particularly interesting in viral infections. Virus-infected patients can shed multiple viral subpopulations with potentially different susceptibilities, which hinders the determination of the correct treatment. For these reasons, monitoring of antimicrobial treatments ex vivo will in the future be a powerful tool to evaluate the response to treatment or to show whether the treatment should be modified. This is especially interesting for infections in which resistant strains might emerge during the course of treatment, particularly in HIV infection, for which simultaneous multitherapies have been developed. From the therapeutic point of view, it would be desirable to detect, as early as possible, evolving resistant strains in a patient in order to decide on the most suitable treatment.
As mentioned above, Cohen and Sahar (58) described a FCM method to detect bacteria in body fluids and exudates (wound exudates, bile, serous-cavity fluids, and bronchial-lavage fluids). These authors also demonstrated that the antimicrobial effects of amikacin could be detected directly in bacteria present in body fluids. A total of 43 clinical samples were prepared for FCM by centrifugation, filtration, resuspension, and the addition of growth medium including 100 mg of amikacin per liter. Light scattering and nucleic acid staining with ethidium bromide were used to assess the bacterial response. The results obtained in less than 2 h were in agreement with those obtained by conventional methods (plate counts and turbidometry). Susceptibility to amikacin was detected in 1 h in 92% of 13 bacterium-positive specimens. Furthermore, with respect to yeasts, Martin et al. (197) developed a PI-based FCM bioassay that measures amphotericin B concentrations in human serum, detecting its fungicidal effect on C. albicans cells in only 5 h.
More recently, Walberg et al. (331) studied antibiotic sensitivity testing in polymicrobial infections. They designed an in vitro model for the FCM detection of heterogeneous drug responses in exponentially growing E. coli and Klebsiella pneumoniae cells. Bacteria were incubated with ampicillin, fixed, and stained with a nucleic acid-specific fluorochrome. Discrimination of heterogeneous populations was demonstrated, as well as the differentiation between susceptible and resistant subpopulations. The detection of resistant subpopulations is crucial in the monitoring of antimicrobial treatments, since these populations may become predominant under antibiotic pressure. Preliminary results have also been obtained with other members of the Enterobacteriaceae and Pseudomonas aeruginosa (331). Although this in vitro model has not yet been used for the ex vivo assessment of infections, this approach should make it possible to monitor polymicrobial infections and their respective treatments.
The viral load is commonly used as a marker of disease progression (105), and antiretroviral therapy has therefore been proposed in asymptomatic patients with a plasma viral RNA load of more than 30,000 copies/ml (44). Inhibition of viral replication induced by anti-HIV drugs is generally associated with an increase in the number of CD4+ cells and a reduction in plasma HIV-1 RNA levels and disease progression. FCM has been used to monitor HIV-1 antiretroviral therapy, mostly by measuring the viral load by PCR and evaluating CD4 counts by FCM in plasma PBMC and lymph node mononuclear cells (170, 266, 269, 312).
Other constituents of the peripheral blood of AIDS patients have been used to monitor the effect of antiviral agents ex vivo by FCM. For example, the number of dendritic cells in the peripheral blood of AIDS patients decreases progressively and, quantification by FCM of the increase in dendritic-cell numbers in patients given AZT therapy can be used to monitor the response to antiviral treatments (246). Fluorescence-labeled human leukocyte antigen tetrameric complexes are obtained in vitro by refolding of the heavy chain, and hence β2-microglobulin and specific peptides can be used to quantify HIV-1-specific cytotoxic T lymphocytes in HIV-infected patients by FCM (233). An inverse correlation between HIV-specific cytotoxic T-lymphocyte proportion and plasma viral load, as well as a decrease in the number of HIV-1-specific cytotoxic T lymphocytes, was found after 6 months of treatment of patients with a triple combination of antiretroviral therapy.
Measurement of the viral load in peripheral blood by PCR-driven in situ hybridization and FCM (244, 245), phenotyping of virus-infected cells, and simultaneous quantitation of CD4+ cells offer a very practical method of using FCM to monitor antiretroviral treatments in HIV-1-infected patients. The relationship between disease progression and the predominant infection of specific cell subpopulations remains to be fully elucidated, but FCM opens new possibilities for the rapid monitoring of these parameters in antiretrovirus-treated patients.
CONCLUDING REMARKS AND FUTURE PERSPECTIVES
Cohen and Sahar (58) pointed out in 1989 that despite the rapid and automatic analysis provided by FCM, the introduction of this technique in clinical microbiology laboratories was limited by the cost of the instrumentation and the need for well-trained operators. Since then, FCM devices have become user-friendly, the number of applications has been expanded, instrument software has been improved, and costs are gradually decreasing. Furthermore, another key characteristic of FCM is its versatility: the same machine can be used to perform direct diagnosis, antimicrobial susceptibility testing, serum antibody detection, and even microbial identification.
Today's flow cytometers allow the analysis of several cell functions in a rapid and multiparametric fashion (several thousand cells in few seconds and a minimum of five parameters per cell). At the same time, it is possible to perform qualitative and quantitative analyses of samples. Thus, FCM is a suitable technique for the study of heterogeneous populations (68, 186) under circumstances of infections due to the presence of multiple microorganisms (292) or for the study of the effects of treating microorganisms with antimicrobial agents (119, 309, 330), since each particle is analyzed individually. As stated above, its main advantages are simplicity, reliability, speed, and the possibility of detecting simultaneous infections.
Monitoring different parameters in clinical samples during the course of an illness allows an assessment of the effect of therapy in patients. This information is very important since the effectiveness of antimicrobial therapy can be measured on-line by FCM. By monitoring the patient's progress via FCM, it should also be possible to detect emerging resistant strains, hence permitting suitable modifications in treatment.
The use of flow cytometers in clinical microbiology should therefore be more of a current issue than a future perspective. For microbial diagnosis, FCM has been used primarily for the detection of viruses in blood and bacteria in urine samples (a flow cytometer specific for urine has been developed) (97, 175), saving time and labor; however, when high sensitivity is needed, visual microscopic review should be performed (167). FCM should be extended to other human samples, particularly sterile body fluids and those with low microbial counts.
To our knowledge, heterogeneity has not yet been studied in the clinical setting. In cases of mixed infections or infections produced by different bacterial subpopulations, such as the persistence of different P. aeruginosa morphotypes in bronchial secretions from cystic fibrosis patients (74) or the presence of H. pylori subpopulations with different pathogenic determinants in the gastric mucosa (321), FCM can help to provide the correct treatment. FCM may also be useful for discriminating microbial variants of recognized clinical impact, i.e., small-colony variants, which show different morphologies and responses to antimicrobial agents from the parent strains (R. Cantón, personal communication).
To optimize the cost-benefit ratio of FCM in clinical microbiology laboratories, automation is necessary. This implies adapting flow cytometers to robotics for automatic sample analysis and developing software to automate analysis of the results. This kind of software is currently used in hematological studies, and hence its adaptation to susceptibility testing, for example, should not be too difficult. The robotics are more complex, but if clinical laboratories create the need, companies should show interest in developing the necessary instruments.
The increasing availability of commercial kits to test antimicrobial susceptibility by FCM in bacteria and fungi (127) points to the future importance of this application. Moreover, the ability of FCM to measure individual cells, in contrast to traditional susceptibility testing methods that analyze the entire population, would lend impetus to the spread of this technique. FCM antimicrobial susceptibility protocols will probably become crucial for the detection of resistant subpopulations, a potential clinical concern. This additional information provided by FCM could be useful to clinicians for the selection of the appropriate antimicrobial therapy in very short times, reducing the time during which the patient is without treatment. In addition, this approach can be beneficial in preventing the emergence and dissemination of resistance. From this review, many technical approaches based on FCM currently in use in basic research can be “exported” to clinical microbiology. Nevertheless, prior to the routine application of FCM to susceptibility testing, the standardization of FCM protocols is mandatory. This would involve the establishment of susceptibility criteria with specific fluorochromes based on NCCLS criteria. The work of Ramani et al. (270), Peyron et al. (252), and Kirk et al. (164), among others, in which FCM susceptibility criteria are compared with NCCLS criteria demonstrates that the use of FCM in clinical microbiology is more than a mere possibility. Today's ubiquitous presence of flow cytometers should help clinical microbiologists to progressively incorporate FCM into their standard protocols.
ACKNOWLEDGMENTS
We thank Rafael Rotger for critical reading of the manuscript; Virginia Piqueras, Fernando Martínez, Anabel Moreno, and Amalia Vázquez for technical assistance; and N. Skinner for language corrections.
This work was partially supported by grants FISS97/0047-01, CAM PR182/96, and SAF97-0134 (CICYT).
Footnotes
We dedicate this review to Luis Carrasco.
REFERENCES
- 1.Reference deleted.
- 2.Abad F X, Pinto R M, Bosch A. Flow cytometry detection of infectious rotaviruses in environmental and clinical samples. Appl Environ Microbiol. 1998;64:2392–2396. doi: 10.1128/aem.64.7.2392-2396.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Agar D W, Bailey J E. Cell cycle operation during batch growth of fission yeast populations. Cytometry. 1982;3:123–128. doi: 10.1002/cyto.990030210. [DOI] [PubMed] [Google Scholar]
- 4.Akbar A N, Borthwick N, Salmon M, Gombert W, Bofill M, Shamsadeen N, Pilling D, Pett S, Grundy J E, Janossy G. The significance of low bcl-2 expression by CD45RO T cells in normal individuals and patients with acute viral infections. The role of apoptosis in T cell memory. J Exp Med. 1993;178:427–438. doi: 10.1084/jem.178.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alberghina L, Porro D. Quantitative flow cytometry: analysis of protein distributions in budding yeast. A mini-review. Yeast. 1993;9:815–823. doi: 10.1002/yea.320090802. [DOI] [PubMed] [Google Scholar]
- 6.Allman R. Characterization of fungal spores using flow cytometry. Mycol Res. 1992;96:1016–1018. [Google Scholar]
- 7.Allman R, Hann A C, Manchee R, Lloyd D. Characterization of bacteria by multiparameter flow cytometry. J Appl Bacteriol. 1992;73:438–444. doi: 10.1111/j.1365-2672.1992.tb05001.x. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez A M, Ibanez M, Rotger R. Beta-galactosidase activity in bacteria measured by flow cytometry. BioTechniques. 1993;15:974–976. [PubMed] [Google Scholar]
- 9.Álvarez-Barrientos A, O'Connor Blasco J, Sánchez Pérez M. La citometría de flujo y el estudio de microorganismos de importancia industrial y clínica. Ind Farm. 1996;5:87–90. [Google Scholar]
- 10.Amann R, Binder B J, Olson R, Chisholm S, Devereux R, Stahl D. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Amann R, Ludwig L, Scheleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreoni M, Sarmati L, Ercoli L, Nicastri E, Giannini G, Galluzzo C, Pirillo M F, Vella S. Correlation between changes in plasma HIV RNA levels and in plasma infectivity in response to antiretroviral therapy. AIDS Res Hum Retrovir. 1997;13:555–561. doi: 10.1089/aid.1997.13.555. [DOI] [PubMed] [Google Scholar]
- 13.Arvinte T, Schulz B, Cudd A, Nicolau C. Low-pH association of proteins with the membranes of intact red blood cells. I. Exogenous glycophorin and the CD4 molecule. Biochim Biophys Acta. 1989;981:51–60. doi: 10.1016/0005-2736(89)90081-3. [DOI] [PubMed] [Google Scholar]
- 14.Ayehunie S, Garcia-Zepeda E A, Hoxie J A, Horuk R, Kupper T S, Luster A D, Ruprecht R M. Human immunodeficiency virus-1 entry into purified blood dendritic cells through CC and CXC chemokine coreceptors. Blood. 1997;90:1379–1386. [PubMed] [Google Scholar]
- 15.Azas N, Di G C, Delmas F, Gasquet M, Timon-David P. Assessment of amphotericin B susceptibility in Leishmania infantum promastogotes by flow cytometry membrane potential assay. Cytometry. 1997;28:165–169. doi: 10.1002/(sici)1097-0320(19970601)28:2<165::aid-cyto10>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 16.Bamdad M, Reader S, Groliere C A, Bohatier J, Denizeau F. Uptake and efflux of polycyclic aromatic hydrocarbons by Tetrahymena pyriformis: evidence for a resistance mechanism. Cytometry. 1997;28:170–175. doi: 10.1002/(sici)1097-0320(19970601)28:2<170::aid-cyto11>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 17.Banfi E, Cinco M, Perticarari S, Presani G. Rapid flow cytometric studies of Borrelia burgdorferi phagocytosis by human polymorphonuclear leukocytes. J Appl Bacteriol. 1989;67:37–45. doi: 10.1111/j.1365-2672.1989.tb04952.x. [DOI] [PubMed] [Google Scholar]
- 18.Baquero F, Culebras C, Patron C, Pérez-Díaz J C, Medrano J C, Vicente M F. Postantibiotic effect of imipenem on gram-positive and gram-negative microorganisms. J Antimicrob Chemother. 1986;18:47–59. doi: 10.1093/jac/18.supplement_e.47. [DOI] [PubMed] [Google Scholar]
- 19.Barardi C R, Emslie K R, Vesey G, Williams K L. Development of a rapid and sensitive quantitative assay for rotavirus based on flow cytometry. J Virol Methods. 1998;74:31–38. doi: 10.1016/s0166-0934(98)00061-5. [DOI] [PubMed] [Google Scholar]
- 20.Basak S, Turner H, Compans R W. Expression of SV40 receptors on apical surfaces of polarized epithelial cells. Virology. 1992;190:393–402. doi: 10.1016/0042-6822(92)91225-j. [DOI] [PubMed] [Google Scholar]
- 21.Basselin M, RobertGero M. Alterations in membrane fluidity, lipid metabolism, mitochondrial activity, and lipophosphoglycan expression in pentamidine-resistant Leishmania. Parasitol Res. 1998;84:78–83. doi: 10.1007/s004360050361. [DOI] [PubMed] [Google Scholar]
- 22.Bassoe C F. Processing of Staphylococcus aureus and zymosan particles by human leukocytes measured by flow cytometry. Cytometry. 1984;5:86–91. doi: 10.1002/cyto.990050113. [DOI] [PubMed] [Google Scholar]
- 23.Bassoe C F, Laerum O D, Glette J, Hopen G, Haneberg B, Solberg C O. Simultaneous measurement of phagocytosis and phagosomal pH by flow cytometry: role of polymorphonuclear neutrophilic leukocyte granules in phagosome acidification. Cytometry. 1983;4:254–262. doi: 10.1002/cyto.990040311. [DOI] [PubMed] [Google Scholar]
- 24.Belles-Isles M, Houde I, Lachance J G, Noel R, Kingma I, Roy R. Monitoring of cytomegalovirus infections by the CD8+CD38+ T-cell subset in kidney transplant recipients. Transplantation. 1998;65:279–282. doi: 10.1097/00007890-199801270-00026. [DOI] [PubMed] [Google Scholar]
- 25.Belyavskyi M, Westerman M, DiMichele L, Wilson V G. Perturbation of the host cell cycle and DNA replication by the bovine papillomavirus replication protein E1. Virology. 1996;219:206–219. doi: 10.1006/viro.1996.0238. [DOI] [PubMed] [Google Scholar]
- 26.Bergbrant I M. Seborrhoeic dermatitis and Pityrosporum ovale: cultural, immunological and clinical studies. Acta Dermatol Venereol Suppl. 1991;167:1–36. [PubMed] [Google Scholar]
- 27.Best L M, Veldhuyzen van Zanten S J, Bezanson G S, Haldane D J, Malatjalian D A. Serological detection of Helicobacter pylori by a flow microsphere immunofluorescence assay. J Clin Microbiol. 1992;30:2311–2317. doi: 10.1128/jcm.30.9.2311-2317.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Betz J W, Aretz W, Hartel W. Use of flow cytometry in industrial microbiology for strain improvement programs. Cytometry. 1984;5:145–150. doi: 10.1002/cyto.990050208. [DOI] [PubMed] [Google Scholar]
- 29.Bhushan R, Kirkham C, Sethi S, Murphy T F. Antigenic characterization and analysis of the human immune response to outer membrane protein E of Branhamella catarrhalis. Infect Immun. 1997;65:2668–2675. doi: 10.1128/iai.65.7.2668-2675.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bigger J W. The bactericidal action of penicillin Staphylococcus aureus. Ir J Med Sci. 1944;227:533–568. [Google Scholar]
- 31.Boeckh M, Boivin G. Quantitation of cytomegalovirus: methodologic aspects and clinical applications. Clin Microbiol Rev. 1998;11:533–554. doi: 10.1128/cmr.11.3.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bonino F, Colloredo M G, Bellati G, Ideo G, Oliveri F, Colombatto P, Brunetto M R. Problems in diagnosing viral hepatitis. Gut. 1993;34:S36–S38. doi: 10.1136/gut.34.2_suppl.s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borzi R M, Piacentini A, Monaco M C, Lisignoli G, Degrassi A, Cattini L, Santi S, Facchini A. A fluorescent in situ hybridization method in flow cytometry to detect HIV-1 specific RNA. J Immunol Methods. 1996;193:167–176. doi: 10.1016/0022-1759(96)00070-1. [DOI] [PubMed] [Google Scholar]
- 34.Bouffard P, Hayashi P H, Acevedo R, Levy N, Zeldis J B. Hepatitis C virus is detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells of infected patients. J Infect Dis. 1992;166:1276–1280. doi: 10.1093/infdis/166.6.1276. [DOI] [PubMed] [Google Scholar]
- 35.Bowden R A, Cloeckaert A, Zygmunt M S, Bernard S, Dubray G. Surface exposure of outer membrane protein and lipopolysaccharide epitopes in Brucella species studied by enzyme-linked immunosorbent assay and flow cytometry. Infect Immun. 1995;63:3945–3952. doi: 10.1128/iai.63.10.3945-3952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bownds S E, Kurzynski T A, Norden M A, Dufek J L, Schell R F. Rapid susceptibility testing for nontuberculosis mycobacteria using flow cytometry. J Clin Microbiol. 1996;34:1386–1390. doi: 10.1128/jcm.34.6.1386-1390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boye E, Steen H E. Flow cytometry of bacteria: a promising tool in experimental and clinical microbiology. J Gen Microbiol. 1983;129:973–980. doi: 10.1099/00221287-129-4-973. [DOI] [PubMed] [Google Scholar]
- 38.Boye E, Lobner-Olesen A. Flow cytometry: illuminating microbiology. New Biol. 1990;2:119–125. [PubMed] [Google Scholar]
- 39.Bracey D, Holyoak C D, NebevonCaron G, Coote P J. Determination of the intracellular pH (pH(i)) of growing cells of Saccharomyces cerevisiae: the effect of reduced-expression of the membrane H+-ATPase. J Microbiol Methods. 1998;31:113–125. [Google Scholar]
- 40.Brawner D L, Cutler J E. Oral Candida albicans isolates from nonhospitalized normal carriers, immunocompetent hospitalized patients, and immunocompromised patients with or without acquired immunodeficiency syndrome. J Clin Microbiol. 1989;27:1335–1341. doi: 10.1128/jcm.27.6.1335-1341.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bridges C G, Brennan T M, Taylor D L, McPherson M, Tyms A S. The prevention of cell adhesion and the cell-to-cell spread of HIV-1 in vitro by the alpha-glucosidase 1 inhibitor, 6-O-butanoyl castanospermine (MDL 28574) Antiviral Res. 1994;25:169–175. doi: 10.1016/0166-3542(94)90105-8. [DOI] [PubMed] [Google Scholar]
- 42.Bronk B V, Van de Merwe W P, Stanley M. In vivo measure of average bacterial cell size from a polarized light scattering function. Cytometry. 1992;13:155–162. doi: 10.1002/cyto.990130208. [DOI] [PubMed] [Google Scholar]
- 43.Callister S M, Jobe D A, Schell R F, Pavia C S, Lovrich S D. Sensitivity and specificity of the borreliacidal-antibody test during early Lyme disease: a “gold standard”? Clin Diagn Lab Immunol. 1996;3:399–402. doi: 10.1128/cdli.3.4.399-402.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Carpenter C C, Fischl M A, Hammer S M, Hirsch M S, Jacobsen D M, Katzenstein D A, Montaner J S, Richman D D, Saag M S, Schooley R T, Thompson M A, Vella S, Yeni P G, Volberding P A. Antiretroviral therapy for HIV infection in 1996. Recommendations of an international panel. International AIDS Society-USA. JAMA. 1996;276:146–154. [PubMed] [Google Scholar]
- 45.Carter E A, Frank E P, Hunter P A. Cytometric evaluation of antifungal agents. In: Lloyd D, editor. Flow cytometry in microbiology. London, United Kingdom: Springer-Verlag; 1993. pp. 111–120. [Google Scholar]
- 46.Caruso A, Tinti M, Peroni L, Cabibbo E, De R C, Manca N, Turano A. Flow cytometric indirect immunofluorescence assay with high sensitivity and specificity for the detection of antibodies to HSV-1 and HSV-2. Eur J Epidemiol. 1993;9:547–552. doi: 10.1007/BF00209534. [DOI] [PubMed] [Google Scholar]
- 47.Chaffin W L, Ringler L, Larsen H S. Interactions of monospecific antisera with cell surface determinants of Candida albicans. Infect Immun. 1998;56:3294–3296. doi: 10.1128/iai.56.12.3294-3296.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaka W, Scharringa J, Verheul A F, Verhoef J, Van Strijp A G, Hoepelman I M. Quantitative analysis of phagocytosis and killing of Cryptococcus neoformans by human peripheral blood mononuclear cells by flow cytometry. Clin Diagn Lab Immunol. 1995;2:753–759. doi: 10.1128/cdli.2.6.753-759.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Challier S, Brown S, Ombrouck C, Desportes-Livage I, De Nay D, Gentilini M. Flow cytometry as a possible method of isolation of spores of the microsporidian Enterocytozoon bieneusi. J Eukaryot Microbiol. 1994;41:27S–27S. [PubMed] [Google Scholar]
- 50.Cheek R F, Olszak I, Madoff S, Preffer F I. In vitro detection of Mycoplasma fermentans binding to B-lymphocytes in fresh peripheral blood using flow cytometry. Cytometry. 1997;28:90–95. doi: 10.1002/(sici)1097-0320(19970501)28:1<90::aid-cyto11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 51.Chemin I, Baginski I, Vermot-Desroches C, Hantz O, Jacquet C, Rigal D, Trepo C. Demonstration of woodchuck hepatitis virus infection of peripheral blood mononuclear cells by flow cytometry and polymerase chain reaction. J Gen Virol. 1992;73:123–129. doi: 10.1099/0022-1317-73-1-123. [DOI] [PubMed] [Google Scholar]
- 52.Chemin I, Vermot-Desroches C, Baginski I, Saurin J C, Laurent F, Zoulim F, Bernaud J, Lamelin J P, Hantz O, Rigal D. Selective detection of human hepatitis B virus surface and core antigens in some peripheral blood mononuclear cell subsets by flow cytometry. J Clin Lab Immunol. 1992;38:63–71. [PubMed] [Google Scholar]
- 53.Chen B K, Gandhi R T, Baltimore D. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J Virol. 1996;70:6044–6053. doi: 10.1128/jvi.70.9.6044-6053.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Choudhury A, Goel A, Raje M, Vohra H, Varshney G C. Recognition of the parasite infested cell surface determinants by homologous antiserum raised against infected cell membranes. Parasitol Res. 1997;83:746–754. doi: 10.1007/s004360050334. [DOI] [PubMed] [Google Scholar]
- 55.Chung J D, Conner S, Stephanopoulos G. Flow cytometric study of differentiating cultures of Bacillus subtilis. Cytometry. 1995;20:324–333. doi: 10.1002/cyto.990200408. [DOI] [PubMed] [Google Scholar]
- 56.Cid V J, Alvarez A M, Santos A I, Nombela C, Sánchez M. Yeast exo-beta-glucanases can be used as efficient and readily detectable reporter genes in Saccharomyces cerevisiae. Yeast. 1994;10:747–756. doi: 10.1002/yea.320100606. [DOI] [PubMed] [Google Scholar]
- 57.Clarke R G, Pinder A C. Improved detection of bacteria by flow cytometry using a combination of antibody and viability markers. J Appl Microbiol. 1998;84:577–584. doi: 10.1046/j.1365-2672.1998.00384.x. [DOI] [PubMed] [Google Scholar]
- 58.Cohen C Y, Sahar E. Rapid flow cytometric bacterial detection and determination of susceptibility to amikacin in body fluids and exudates. J Clin Microbiol. 1989;27:1250–1256. doi: 10.1128/jcm.27.6.1250-1256.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comas J, Vives-Rego J. Assessment of the effects of gramicidin, formaldehyde, and surfactants on Escherichia coli by flow cytometry using nucleic acid and membrane potential dyes. Cytometry. 1997;29:58–64. doi: 10.1002/(sici)1097-0320(19970901)29:1<58::aid-cyto6>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 60.Cormack B P, Bertram G, Egerton M, Gow N, Falkow S, Brown A. Yeast-enhanced green fluorescent protein (yEGFP)a reporter of gene expression in Candida albicans. Microbiology. 1997;143:303–311. doi: 10.1099/00221287-143-2-303. [DOI] [PubMed] [Google Scholar]
- 61.Cory J M, Ohlsson-Wilhelm B M, Brock E J, Sheaffer N A, Steck M E, Eyster M E, Rapp F. Detection of human immunodeficiency virus-infected lymphoid cells at low frequency by flow cytometry. J Immunol Methods. 1987;105:71–78. doi: 10.1016/0022-1759(87)90415-7. [DOI] [PubMed] [Google Scholar]
- 62.Cory J M, Ohlsson-Wilhelm B M, Steck M E, Smithgall M D, Rozday V, Eyster M E, Rapp F. Kinetics of infected cell appearance as a determinant of number of human immunodeficiency virus-1 infectious units. AIDS Res Hum Retrovir. 1989;5:97–106. doi: 10.1089/aid.1989.5.97. [DOI] [PubMed] [Google Scholar]
- 63.Costigliola P, Tumietto F, Ricchi E, Chiodo F. Detection of circulating p24 antigen-positive CD4+ cells during HIV infection by flow cytometry. AIDS. 1992;6:1121–1125. doi: 10.1097/00002030-199210000-00009. [DOI] [PubMed] [Google Scholar]
- 64.Cozon G, Roure C, Lizard G, Greenland T, Larget-Piet D, Gandilhon F, Peyron F. An improved assay for the detection of Toxoplasma gondii antibodies in human serum by flow cytometry. Cytometry. 1993;14:569–575. doi: 10.1002/cyto.990140518. [DOI] [PubMed] [Google Scholar]
- 65.Craig W A. The postantibiotic effect. Clin Microbiol Newsl. 1998;13:121–124. [Google Scholar]
- 66.Crouch J, Leitenberg D, Smith B R, Howe J G. Epstein-Barr virus suspension cell assay using in situ hybridization and flow cytometry. Cytometry. 1997;29:50–57. [PubMed] [Google Scholar]
- 67.Davey H M, Kaprelyants A S, Kell D B. Flow cytometry analysis using rhodamine 123 of Micrococcus luteus at low growth rate in chemostat culture. In: Lloyd D, editor. Flow cytometry in microbiology. London, United Kingdom: Springer-Verlag; 1993. pp. 83–89. [Google Scholar]
- 68.Davey H M, Kell D B. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol Rev. 1996;60:641–696. doi: 10.1128/mr.60.4.641-696.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Davies D, Hixson C. Study of the effect of Ampicillin concentration and exposure time on E. coli using flow cytometry and the FAST-2 kit. Cytometry. 1998;9(Suppl.):39–39. [Google Scholar]
- 70.de Boer E C, Bevers R F, Kurth K H, Schamhart D H. Double fluorescent flow cytometric assessment of bacterial internalization and binding by epithelial cells. Cytometry. 1996;25:381–387. doi: 10.1002/(SICI)1097-0320(19961201)25:4<381::AID-CYTO10>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 71.Deere D, Porter J, Edwards C, Pickup R. Evaluation of the suitability of bis(1,3-dibutylbarbituric acid) trimethine oxonol, (diBAC4(3)-), for the flow cytometric assessment of bacterial viability. FEMS Microbiol Lett. 1995;130:165–169. doi: 10.1111/j.1574-6968.1995.tb07714.x. [DOI] [PubMed] [Google Scholar]
- 72.de la Fuente J, Alvarez A, Nombela C, Sanchez M. Flow cytometric analysis of Saccharomyces cerevisiae autolytic mutants and protoplasts. Yeast. 1992;8:39–45. doi: 10.1002/yea.320080104. [DOI] [PubMed] [Google Scholar]
- 73.Denning D W, Radford S A, Oakley K L, Hall L, Johnson E M, Warnock D W. Correlation between in-vitro susceptibility testing to itraconazole and in-vivo outcome of Aspergillus fumigatus infection. J Antimicrob Chemother. 1997;40:401–414. doi: 10.1093/jac/40.3.401. [DOI] [PubMed] [Google Scholar]
- 74.Deretic V, Schurr M J, Yu H. Pseudomonas aeruginosa mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 1995;3:351–356. doi: 10.1016/s0966-842x(00)88974-x. [DOI] [PubMed] [Google Scholar]
- 75.Dessus-Babus S, Belloc F, Bébéar C M, Poutiers F, Lacombe F, de Barbeyrac B. Antibiotic susceptibility testing for Chlamydia trachomatis using flow cytometry. Cytometry. 1998;31:37–44. [PubMed] [Google Scholar]
- 76.Detrick B, Hooks J J, Keiser J, Tabbara I. Detection of cytomegalovirus proteins by flow cytometry in the blood of patients undergoing hematopoietic stem cell transplantation. Exp Hematol. 1999;27:569–575. doi: 10.1016/s0301-472x(98)00076-9. [DOI] [PubMed] [Google Scholar]
- 77.Dhandayuthapani S, Via L E, Thomas C A, Horowitz P M, Deretic D, Deretic V. Green fluorescent protein as a marker for gene expression and cell biology of mycobacterial interactions with macrophages. Mol Microbiol. 1995;17:901–912. doi: 10.1111/j.1365-2958.1995.mmi_17050901.x. [DOI] [PubMed] [Google Scholar]
- 78.Diaper J P, Edwards C. Survival of Staphylococcus aureus in lakewater monitored by flow cytometry. Microbiology. 1994;140:35–42. doi: 10.1099/13500872-140-1-35. [DOI] [PubMed] [Google Scholar]
- 79.Diaper J P, Tither K, Edwards C. Rapid assessment of bacterial viability by flow cytometry. Appl Microbiol Biotechnol. 1992;22:1–5. doi: 10.1007/BF00174481. [DOI] [PubMed] [Google Scholar]
- 80.Dittmer D, Mocarski E S. Human cytomegalovirus infection inhibits the G1/S transition. J Virol. 1997;71:1629–1634. doi: 10.1128/jvi.71.2.1629-1634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dixon B R, Parenteau M, Martineau C, Fournier J. A comparison of conventional microscopy, immunofluorescence microscopy and flow cytometry in the detection of Giardia lamblia cysts in beaver fecal samples. J Immunol Methods. 1997;202:27–33. doi: 10.1016/s0022-1759(96)00239-6. [DOI] [PubMed] [Google Scholar]
- 82.Doern G V. Introduction. Diagnostic technologies in clinical microbiology. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 1st ed. Washington, D.C.: ASM Press; 1995. pp. 14–43. [Google Scholar]
- 83.Doern G V, Vaoutour R, Gaudet M, Levy B. Clinical impact of rapid in vitro susceptibility testing and bacterial identification of rapid in vitro susceptibility testing and bacterial identification. J Clin Microbiol. 1994;32:1757–1762. doi: 10.1128/jcm.32.7.1757-1762.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dorenbaum A, Venkateswaran K S, Yang G, Comeau A M, Wara D, Vyas G N. Transmission of HIV-1 in infants born to seropositive mothers: PCR-amplified proviral DNA detected by flow cytometric analysis of immunoreactive beads. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:35–42. doi: 10.1097/00042560-199705010-00006. [DOI] [PubMed] [Google Scholar]
- 85.Dorsky D I, Wells M, Harrington R D. Detection of HIV-1 infection with a green fluorescent protein reporter system. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13:308–313. doi: 10.1097/00042560-199612010-00002. [DOI] [PubMed] [Google Scholar]
- 86.Durodie J, Coleman K, Simpson I N, Loughborough S H, Winstanley D W. Rapid detection of antimicrobial activity using flow cytometry. Cytometry. 1995;21:374–377. doi: 10.1002/cyto.990210409. [DOI] [PubMed] [Google Scholar]
- 87.Durodie J, Coleman K, Wilkinson M J. Characterization of bacterial size and ploidy using flow cytometry and image analysis. In: Lloyd D, editor. Flow cytometry. London, United Kingdom: Springer-Verlag; 1993. pp. 55–109. [Google Scholar]
- 88.Dvorak J A. Analysis of the DNA of parasitic protozoa by flow cytometry. Methods Mol Biol. 1993;21:191–204. doi: 10.1385/0-89603-239-6:191. [DOI] [PubMed] [Google Scholar]
- 89.Edwards C. Assessment of viability of bacteria by flow cytometry. In: Al-Rubeai M, Emery A N, editors. Flow cytometry applications in cell culture. New York, N.Y: Marcel Dekker, Inc; 1996. pp. 291–310. [Google Scholar]
- 90.Egido J M, Vinuelas J. Flow cytometric quantitation of phagocytosis in heparinized complete blood with latex particles and Candida albicans. Rev Soc Bras Med Trop. 1997;30:441–446. doi: 10.1590/s0037-86821997000600001. [DOI] [PubMed] [Google Scholar]
- 91.Elmendorf S, McSharry J, Laffin J, Fogleman D, Lehman J M. Detection of an early cytomegalovirus antigen with two-color quantitative flow cytometry. Cytometry. 1988;9:254–260. doi: 10.1002/cyto.990090311. [DOI] [PubMed] [Google Scholar]
- 92.Embretson J, Zupancic M, Beneke J, Till M, Wolinsky S, Ribas J L, Burke A, Haase A T. Analysis of human immunodeficiency virus-infected tissues by amplification and in situ hybridization reveals latent and permissive infections at single-cell resolution. Proc Natl Acad Sci USA. 1993;90:357–361. doi: 10.1073/pnas.90.1.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Esolen L M, Park S W, Hardwick J M, Griffin D E. Apoptosis as a cause of death in measles virus-infected cells. J Virol. 1995;69:3955–3958. doi: 10.1128/jvi.69.6.3955-3958.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Espinel-Ingroff A, Rodriguez-Tudela J L, Martinez-Suarez J V. Comparison of two alternative microdilution procedures with the National Committee for Clinical Laboratory Standards reference macrodilution method M27-P for in vitro testing of fluconazole-resistant and -susceptible isolates of Candida albicans. J Clin Microbiol. 1995;33:3154–3158. doi: 10.1128/jcm.33.12.3154-3158.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eyler Y L, Lantz L M, Lewis A M J. Flow cytometric detection of DNA tumor virus nuclear oncogene products in unfixed cells: saponin FACS of viral oncogene products. J Virol Methods. 1994;46:23–27. doi: 10.1016/0166-0934(94)90013-2. [DOI] [PubMed] [Google Scholar]
- 96.Fattorossi A, Nisini R, Pizzolo J G, D'Amelio R. New, simple flow cytometry technique to discriminate between internalized and membrane-bound particles in phagocytosis. Cytometry. 1989;10:320–325. doi: 10.1002/cyto.990100311. [DOI] [PubMed] [Google Scholar]
- 97.Fenili D, Pirovano B. The automation of sediment urinalysis using a new urine flow cytometer (UF-100) Clin Chem Lab Med. 1998;36:909–917. doi: 10.1515/CCLM.1998.158. [DOI] [PubMed] [Google Scholar]
- 98.Ferraro M J, Jorgensen J H. Instrument-based antibacterial susceptibility testing. In: Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 1st ed. Washington, D.C.: ASM Press; 1995. pp. 1379–1384. [Google Scholar]
- 99.Flores B M, Garcia C A, Stamm W E, Torian B E. Differentiation of Naegleria fowleri from Acanthamoeba species by using monoclonal antibodies and flow cytometry. J Clin Microbiol. 1990;28:1999–2005. doi: 10.1128/jcm.28.9.1999-2005.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Folghera S, Fiorentini S, Martinelli F, Ravizzola G, Gargiulo F, Terlenghi L, De F M, Caruso A, Turano A. Development of a flow cytometric assay for the detection and measurement of neutralizing antibodies against human immunodeficiency virus. New Microbiol. 1994;17:21–28. [PubMed] [Google Scholar]
- 101.Freeman C D, Nicolau D P, Belleveau P P, Nightingale P P. Once-daily dosing of aminoglycosides: review and recommendations for clinical practice. J Antimicrob Chemother. 1997;39:677–683. doi: 10.1093/jac/39.6.677. [DOI] [PubMed] [Google Scholar]
- 102.Freistadt M S, Fleit H B, Wimmer E. Poliovirus receptor on human blood cells: a possible extraneural site of poliovirus replication. Virology. 1993;195:798–803. doi: 10.1006/viro.1993.1433. [DOI] [PubMed] [Google Scholar]
- 103.Frelat G, Laplace-Builhe C, Grunwald D. Microbial analysis by flow cytometry: present and future. In: Yen A, editor. Flow cytometry: advanced research and clinical applications. Boca Raton, Fla: CRC Press, Inc.; 1989. pp. 256–279. [Google Scholar]
- 104.Fulwyler M J. Electronic separation of biological cells by volume. Science. 1965;150:910–911. doi: 10.1126/science.150.3698.910. [DOI] [PubMed] [Google Scholar]
- 105.Furtado M R, Kingsley L A, Wolinsky S M. Changes in the viral mRNA expression pattern correlate with a rapid rate of CD4+ T-cell number decline in human immunodeficiency virus type 1-infected individuals. J Virol. 1995;69:2092–2100. doi: 10.1128/jvi.69.4.2092-2100.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fuursted K. Postantibiotic effect: with a highlight on its mechanism. Antimicrob Infect Dis Newsl. 1995;14:21–24. [Google Scholar]
- 107.Gadol N, Crutcher G J, Busch M P. Detection of intracellular HIV in lymphocytes by flow cytometry. Cytometry. 1994;15:359–370. doi: 10.1002/cyto.990150412. [DOI] [PubMed] [Google Scholar]
- 108.Gant V A, Warnes G, Phillips I, Savidge G F. The application of flow cytometry to the study of bacterial responses to antibiotics. J Med Microbiol. 1993;39:147–154. doi: 10.1099/00222615-39-2-147. [DOI] [PubMed] [Google Scholar]
- 109.Gao S J, More P S, Phil M. Molecular approaches to the identification of unculturable infectious agents. Emerg Infect Dis. 1996;2:159–167. doi: 10.3201/eid0203.960301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gavalchin J, Fan N, Lane M J, Papsidero L, Poiesz B J. Identification of a putative cellular receptor for HTLV-I by a monoclonal antibody, Mab 34-23. Virology. 1993;194:1–9. doi: 10.1006/viro.1993.1228. [DOI] [PubMed] [Google Scholar]
- 111.Gaynor E M, Mirsky M L, Lewin H A. Use of flow cytometry and RT-PCR for detecting gene expression by single cells. BioTechniques. 1996;21:286–291. doi: 10.2144/96212rr02. [DOI] [PubMed] [Google Scholar]
- 112.Gerna G, Revello M G, Percivalle E, Morini F. Comparison of different immunostaining techniques and monoclonal antibodies to the lower matrix phosphoprotein (pp65) for optimal quantitation of human cytomegalovirus antigenemia. J Clin Microbiol. 1992;30:1232–1237. doi: 10.1128/jcm.30.5.1232-1237.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gervaix A, West D, Leoni L M, Richman D D, Wong-Staal F, Corbeil J. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc Natl Acad Sci USA. 1997;94:4653–4658. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Giaimis J, Lombard Y, Poindron P, Muller C D. Flow cytometry distinction between adherent and phagocytized yeast particles. Cytometry. 1994;17:173–178. doi: 10.1002/cyto.990170210. [DOI] [PubMed] [Google Scholar]
- 115.Gibellini D E, Re M C, Furlini G, La P M. Flow cytometry analysis of an in situ PCR for the detection of human immunodeficiency virus type-1 (HIV-1) proviral DNA. Methods Mol Biol. 1997;71:113–122. doi: 10.1385/0-89603-395-3:113. [DOI] [PubMed] [Google Scholar]
- 116.Gilbert M J, Riddell S R, Li C R, Greenberg P D. Selective interference with class I major histocompatibility complex presentation of the major immediate-early protein following infection with human cytomegalovirus. J Virol. 1993;67:3461–3469. doi: 10.1128/jvi.67.6.3461-3469.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Goolsby C, Gay H, Docherty J J, Todd P. Flow cytometric detection of herpes simplex virus type 2 infected and transformed cells by immunoenzymatic and by indirect immunofluorescence staining. Cytometry. 1988;9:126–130. doi: 10.1002/cyto.990090205. [DOI] [PubMed] [Google Scholar]
- 118.Gorse G J, Frey S E, Newman F K, Belshe R B. Detection of binding antibodies to native and recombinant human immunodeficiency virus type 1 envelope glycoproteins following recombinant gp160 immunization measured by flow cytometry and enzyme immunoassays. The AIDS Vaccine Clinical Trials Network. J Clin Microbiol. 1992;30:2606–2612. doi: 10.1128/jcm.30.10.2606-2612.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gottfredsson M, Erlendsdottir H, Sigfusson A, Gudmundsson S. Characteristics and dynamics of bacterial populations during postantibiotic effect determined by flow cytometry. Antimicrob Agents Chemother. 1998;42:1005–1011. doi: 10.1128/aac.42.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Green L, Petersen B, Steimel L, Haeber P, Current W. Rapid determination of antifungal activity by flow cytometry. J Clin Microbiol. 1994;32:1088–1091. doi: 10.1128/jcm.32.4.1088-1091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Griffiths K R, Champion A C, Vesey G, Williams K L. Isolation of glycosylation mutants in Dictyostelium discoideum using flow cytometry. Cytometry. 1996;25:133–143. doi: 10.1002/(SICI)1097-0320(19961001)25:2<133::AID-CYTO2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 122.Groschel D H. Methods, reagents and media: new technology in clinical microbiology. Infect Control. 1983;4:232–233. [PubMed] [Google Scholar]
- 123.Guatelli J C, Gingeras T R, Richman D D. Nucleic acid amplification in vitro: detection of sequences with low copy numbers and application to diagnosis of human immunodeficiency virus type 1 infection. Clin Microbiol Rev. 1989;2:217–226. doi: 10.1128/cmr.2.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Gucker F, O'Konski C O, Pickard H B, Pitts J. A photoelectric counter for colloidal particles. Am J Chem. 1947;69:2422–2431. doi: 10.1021/ja01202a053. [DOI] [PubMed] [Google Scholar]
- 125.Guindulain T, Comas J, Vives-Rego J. Use of nucleic acid dyes SYTO-13, TOTO-1, and YOYO-1 in the study of Escherichia coli and marine prokaryotic populations by flow cytometry. Appl Environ Microbiol. 1997;63:4608–4611. doi: 10.1128/aem.63.11.4608-4611.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Han Y, Kanbe T, Cherniak R, Cutler J E. Biochemical characterization of Candida albicans epitopes that can elicit protective and nonprotective antibodies. Infect Immun. 1997;65:4100–4107. doi: 10.1128/iai.65.10.4100-4107.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Haugland R P. Handbook of fluorescent probes and research chemicals. Eugene, Oreg: Molecular Probes Inc.; 1996. [Google Scholar]
- 128.Heinzelmann M, Herzig D O, Swain B, Mercer-Jones M A, Bergamini T M, Polk H C J. Phagocytosis and oxidative-burst response of planktonic Staphylococcus epidermidis RP62A and its non-slime-producing variant in human neutrophils. Clin Diagn Lab Immunol. 1997;4:705–710. doi: 10.1128/cdli.4.6.705-710.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Herbein G, Van L C, Lovett J L, Verdin E. Distinct mechanisms trigger apoptosis in human immunodeficiency virus type 1-infected and in uninfected bystander T lymphocytes. J Virol. 1998;72:660–670. doi: 10.1128/jvi.72.1.660-670.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heynen C A, Holzer T J. Evaluation of a flow cytometric model for monitoring HIV antigen expression in vitro. J Immunol Methods. 1992;152:25–33. doi: 10.1016/0022-1759(92)90085-8. [DOI] [PubMed] [Google Scholar]
- 131.Holzer T J, Heynen C A, Novak R M, Pitrak D L, Dawson G J. Frequency of cells positive for HIV-1 p24 antigen assessed by flow cytometry. AIDS. 1993;7(Suppl. 2):S3–S5. doi: 10.1097/00002030-199311002-00002. [DOI] [PubMed] [Google Scholar]
- 132.Honda J, Okubo Y, Imamura Y, Kusaba M, Saruwatari N, Oizumi K. Flow cytometric detection of cytomegalovirus antigen in peripheral blood cells after bone marrow transplantation. Br J Haematol. 1997;99:415–417. doi: 10.1046/j.1365-2141.1997.3863200.x. [DOI] [PubMed] [Google Scholar]
- 133.Hu Y W, Birch P, Balaskas E, Zeibdawi A, Scalia V, Theriault-Valin S A, Gill P, Aye M T. Flow cytometric immunofluorescence assay for detection of antibodies to human immunodeficiency virus type 1 using insoluble precursor forms of recombinant polyproteins as carriers and antigens. J Clin Microbiol. 1996;34:1412–1419. doi: 10.1128/jcm.34.6.1412-1419.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hughes E E, Matthews-Greer J M, Gilleland H E J. Analysis by flow cytometry of surface-exposed epitopes of outer membrane protein F of Pseudomonas aeruginosa. Can J Microbiol. 1996;42:859–862. doi: 10.1139/m96-109. [DOI] [PubMed] [Google Scholar]
- 135.Humphreys M J, Allman R, Lloyd D. Determination of the viability of Trichomonas vaginalis using flow cytometry. Cytometry. 1994;15:343–348. doi: 10.1002/cyto.990150410. [DOI] [PubMed] [Google Scholar]
- 136.Hutter K J, Oldiges H. Alterations of proliferating microorganims by flow cytometric measurements after heavy metal intoxication. Ecotoxicol Environ Saf. 1980;4:57. doi: 10.1016/0147-6513(80)90008-1. [DOI] [PubMed] [Google Scholar]
- 137.Iannelli D, D'Apice L, Cottone C, Viscardi M, Scala F, Zoina A, Del Sorbo S G, Spigno P, Capparelli R. Simultaneous detection of cucumber mosaic virus, tomato mosaic virus and potato virus Y by flow cytometry. J Virol Methods. 1997;69:137–145. doi: 10.1016/s0166-0934(97)00149-3. [DOI] [PubMed] [Google Scholar]
- 138.Iannelli D, D'Apice L, Fenizia D, Serpe L, Cottone C, Viscardi M, Capparelli R. Simultaneous identification of antibodies to Brucella abortus and Staphylococcus aureus in milk samples by flow cytometry. J Clin Microbiol. 1998;36:802–806. doi: 10.1128/jcm.36.3.802-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ieven M, Goossens H. Relevance of nucleic acid amplification techniques for diagnosis of respiratory tract infections in the clinical laboratory. Clin Microbiol Rev. 1997;10:242–256. doi: 10.1128/cmr.10.2.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Imai T, Ohno T. The relationship between viability and intracellular pH in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol. 1995;61:3604–3608. doi: 10.1128/aem.61.10.3604-3608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Imbert-Marcille B M, Robillard N, Poirier A S, Coste-Burel M, Cantarovich D, Milpied N, Billaudel S. Development of a method for direct quantification of cytomegalovirus antigenemia by flow cytometry. J Clin Microbiol. 1997;35:2665–2669. doi: 10.1128/jcm.35.10.2665-2669.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Inghirami G, Nakamura M, Balow J E, Notkins A L, Casali P. Model for studying virus attachment: identification and quantitation of Epstein-Barr virus-binding cells by using biotinylated virus in flow cytometry. J Virol. 1988;62:2453–2463. doi: 10.1128/jvi.62.7.2453-2463.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ingram M, Cleary T J, Price B J, Castro A. Rapid detection of Legionella pneumophila by flow cytometry. Cytometry. 1982;3:134–147. doi: 10.1002/cyto.990030212. [DOI] [PubMed] [Google Scholar]
- 144.Inoue Y, Yasukawa M, Fujita S. Induction of T-cell apoptosis by human herpesvirus 6. J Virol. 1997;71:3751–3759. doi: 10.1128/jvi.71.5.3751-3759.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Irurzun A, Arroyo J, Alvarez A, Carrasco L. Enhanced intracellular calcium concentration during poliovirus infection. J Virol. 1995;69:5142–5146. doi: 10.1128/jvi.69.8.5142-5146.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ito M, Watanabe M, Kamiya H, Sakurai M. Herpes simplex virus type 1 induces apoptosis in peripheral blood T lymphocytes. J Infect Dis. 1997;175:1220–1224. doi: 10.1086/593672. [DOI] [PubMed] [Google Scholar]
- 147.Janse C J, Van Vianen P H. Flow cytometry in malaria detection. Methods Cell Biol. 1994;42(B):295–318. doi: 10.1016/s0091-679x(08)61081-x. [DOI] [PubMed] [Google Scholar]
- 148.Jansson J K, Prosser J I. Quantification of the presence and activity of specific microorganisms in nature. Mol Biotechnol. 1997;7:103–120. doi: 10.1007/BF02761746. [DOI] [PubMed] [Google Scholar]
- 149.Jennings S R, Rice P L, Kloszewski E D, Anderson R W, Thompson D L, Tevethia S S. Effect of herpes simplex virus types 1 and 2 on surface expression of class I major histocompatibility complex antigens on infected cells. J Virol. 1985;56:757–766. doi: 10.1128/jvi.56.3.757-766.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jepras R I, Carter J, Pearson S C, Paul F E, Wilkinson M J. Development of a robust flow cytometric assay for determining numbers of viable bacteria. Appl Environ Microbiol. 1995;61:2696–2701. doi: 10.1128/aem.61.7.2696-2701.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jernaes M W, Steen H B. Staining of Escherichia coli for flow cytometry: influx and efflux of ethidium bromide. Cytometry. 1994;17:302–309. doi: 10.1002/cyto.990170405. [DOI] [PubMed] [Google Scholar]
- 152.Johnson C C. In vitro testing: correlations of bacterial susceptibility, body fluids levels, and effectiveness of antibacterial therapy. In: Conan V, editor. Antibiotics in laboratory medicine. Baltimore, Md: The Williams & Wilkins Co.; 1996. pp. 813–834. [Google Scholar]
- 153.Jouin H, Goguet de la Salmoniere Y O, Behr C, Huyin Q D, Michel J C, Sarthou J L, Pereira D S, Dubois P. Flow cytometry detection of surface antigens on fresh, unfixed red blood cells infected by Plasmodium falciparum. J Immunol Methods. 1995;179:1–12. doi: 10.1016/0022-1759(94)00265-x. [DOI] [PubMed] [Google Scholar]
- 154.Kamentsky L M, Melamed M R, Derman H. Spectrophotometer: new instrument for ultrarapid cell analysis. Science. 1965;150:630–631. doi: 10.1126/science.150.3696.630. [DOI] [PubMed] [Google Scholar]
- 155.Kaprelyants A S, Gottschal J C, Kell D B. Dormancy in non-sporulating bacteria. FEMS Microbiol Rev. 1993;104:271–286. doi: 10.1111/j.1574-6968.1993.tb05871.x. [DOI] [PubMed] [Google Scholar]
- 156.Kaprelyants A S, Kell D B. Dormancy in stationary-phase cultures of Micrococcus luteus: flow cytometry analysis of starvation and resuscitation. Appl Environ Microbiol. 1993;59:3187–3196. doi: 10.1128/aem.59.10.3187-3196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaprelyants A, Kell D B. Rapid assessment of bacterial viability and vitality using rhodamine 123 and flow cytometry. J Appl Bacteriol. 1992;72:410–422. [Google Scholar]
- 158.Kas-Deelen A M, Harmsen M C, de Maar E F, van Son W J, The T H. A sensitive method for quantifying cytomegalic endothelial cells in peripheral blood from cytomegalovirus-infected patients. Clin Diagn Lab Immunol. 1998;5:622–626. doi: 10.1128/cdli.5.5.622-626.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kearns-Jonker M, Monell-Torrens E, Abbasi F, Holers V M, Notkins A L, Sigounas G. EBV binds to lymphocytes of transgenic mice that express the human CR2 gene. Virus Res. 1997;50:85–94. doi: 10.1016/s0168-1702(97)00052-x. [DOI] [PubMed] [Google Scholar]
- 160.Kelly D J, Salata K F, Strickman D, Hershey J N. Rickettsia tsutsugamushi infection in cell culture: antibiotic susceptibility determined by flow cytometry. Am J Trop Med Hyg. 1995;53:602–606. doi: 10.4269/ajtmh.1995.53.602. [DOI] [PubMed] [Google Scholar]
- 161.Kesson A M, Fear W R, Williams L, Chang J, King N J, Cunningham A L. HIV infection of placental macrophages: their potential role in vertical transmission. J Leukoc Biol. 1994;56:241–246. doi: 10.1002/jlb.56.3.241. [DOI] [PubMed] [Google Scholar]
- 162.Kesson A M, Zeng F, Cunningham A L, Rawlinson W D. The use of flow cytometry to detect antiviral resistance in human cytomegalovirus. J Virol Methods. 1998;71:177–186. doi: 10.1016/s0166-0934(97)00214-0. [DOI] [PubMed] [Google Scholar]
- 163.Kirk S M, Callister S M, Lim L C, Schell R F. Rapid susceptibility testing of Candida albicans by flow cytometry. J Clin Microbiol. 1997;35:358–363. doi: 10.1128/jcm.35.2.358-363.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kirk S M, Schell R F, Moore A V, Callister S M, Mazurek G H. Flow cytometric testing of susceptibilities of Mycobacterium tuberculosis isolates to ethambutol, isoniazid, and rifampin in 24 hours. J Clin Microbiol. 1998;36:1568–1573. doi: 10.1128/jcm.36.6.1568-1573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Klepser M E, Ernst E J, Pfaller M A. Update on antifungal resistance. Trends Microbiol. 1997;5:372–375. doi: 10.1016/S0966-842X(97)01108-6. [DOI] [PubMed] [Google Scholar]
- 166.Kort J J, Bilello J A, Bauer G, Drusano G L. Preclinical evaluation of antiviral activity and toxicity of Abbott A77003, an inhibitor of the human immunodeficiency virus type 1 protease. Antimicrob Agents Chemother. 1993;37:115–119. doi: 10.1128/aac.37.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kouri T T, Kahkonen U, Malminiemi K, Vuento R, Rowan R M. Evaluation of Sysmex UF-100 urine flow cytometer vs chamber counting of supravitally stained specimens and conventional bacterial cultures. Am J Clin Pathol. 1999;112:25–35. doi: 10.1093/ajcp/112.1.25. [DOI] [PubMed] [Google Scholar]
- 168.Kring S C, Spindler K R. Lack of effect of mouse adenovirus type 1 infection on cell surface expression of major histocompatibility complex class I antigens. J Virol. 1996;70:5495–5502. doi: 10.1128/jvi.70.8.5495-5502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kulaga H, Anderson P E, Cain M, Stopa P J. Identification of pathogenic agents via microsphere-based immunoassays on a flow cytometer. Cytometry. 1998;9(Suppl.):55–56. [Google Scholar]
- 170.Lafeuillade A, Tamalet C, Poggi C, Pellegrino P, Tourres C, Izopet J. Antiretroviral effect of zidovudine-didanosine combination on blood and lymph nodes. AIDS. 1997;11:67–72. doi: 10.1097/00002030-199701000-00010. [DOI] [PubMed] [Google Scholar]
- 171.Laffin J, Fogleman D, Lehman J M. Correlation of DNA content, p53, T antigen, and V antigen in simian virus 40-infected human diploid cells. Cytometry. 1989;10:205–213. doi: 10.1002/cyto.990100212. [DOI] [PubMed] [Google Scholar]
- 172.Laffin J, Lehman J M. Detection of intracellular virus and viral products. Methods Cell Biol. 1994;41:543–557. doi: 10.1016/s0091-679x(08)61739-2. [DOI] [PubMed] [Google Scholar]
- 173.Lalli E, Gibellini D, Santi S, Facchini A. In situ hybridization in suspension and flow cytometry as a tool for the study of gene expression. Anal Biochem. 1992;207:298–303. doi: 10.1016/0003-2697(92)90015-y. [DOI] [PubMed] [Google Scholar]
- 174.Landay A, Ohlsson-Wilhelm B, Giorgi J V. Application of flow cytometry to the study of HIV infection. AIDS. 1990;4:479–497. doi: 10.1097/00002030-199006000-00001. [DOI] [PubMed] [Google Scholar]
- 175.Langlois M R, Delanghe J R, Steyaert S R, Everaert K C, de Buyzere M L. Automated flow cytometry compared with an automated dipstick reader for urinalysis. Clin Chem. 1999;45:118–122. [PubMed] [Google Scholar]
- 176.Langsrud S, Sundheim G. Flow cytometry for rapid assessment of viability after exposure to a quaternary ammonium compound. J Appl Bacteriol. 1996;81:411–418. doi: 10.1111/j.1365-2672.1996.tb03527.x. [DOI] [PubMed] [Google Scholar]
- 177.Lapinsky S E, Glencross D, Car N G, Kallenbach J M, Zwi S. Quantification and assessment of viability of Pneumocystis carinii organisms by flow cytometry. J Clin Microbiol. 1991;29:911–915. doi: 10.1128/jcm.29.5.911-915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lebaron P, Parthuisot N, Catala P. Comparison of blue nucleic acid dyes for flow cytometric enumeration of bacteria in aquatic systems. Appl Environ Microbiol. 1998;64:1725–1730. doi: 10.1128/aem.64.5.1725-1730.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Lehman J M, Friedrich T D, Laffin J. Quantitation of simian virus 40 T-antigen correlated with the cell cycle of permissive and non-permissive cells. Cytometry. 1993;14:401–410. doi: 10.1002/cyto.990140409. [DOI] [PubMed] [Google Scholar]
- 180.Lehman J M, Jacobberger J W. Virus-cell interactions analyzed with flow cytometry. In: Melamed M R, Lindmo T, Mendelsohn M L, editors. Flow cytometry and sorting. New York, N.Y: Wiley-Liss, Inc.; 1990. pp. 623–631. [Google Scholar]
- 181.Lennette D A. Collection and preparation of specimens for virological examination. In: Murray P R, Baron E J, Pfaller M A, Tenover F, Yolken R, editors. Manual of clinical microbiology. 1st ed. Washington, D.C.: ASM Press; 1995. pp. 868–875. [Google Scholar]
- 182.Li Q X, Young L S, Niedobitek G, Dawson C W, Birkenbach M, Wang F, Rickinson A B. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature. 1992;356:347–350. doi: 10.1038/356347a0. [DOI] [PubMed] [Google Scholar]
- 183.Libertin C R, Woloschak G E, Wilson W R, Smith T F. Analysis of Pneumocystis carinii cysts with a fluorescence-activated cell sorter. J Clin Microbiol. 1984;20:877–880. doi: 10.1128/jcm.20.5.877-880.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Link H, Battmer K, Kleine H D. Detection of cytomegalovirus-infected cells by flow cytometry and fluorescence in suspension hybridisation (FLASH) using DNA probes labeled with biotin by polymerase chain reaction. J Med Virol. 1992;37:143–148. doi: 10.1002/jmv.1890370213. [DOI] [PubMed] [Google Scholar]
- 185.Lipson S M, Soni M, Biondo F X, Shepp D H, Kaplan M H, Sun T. Antiviral susceptibility testing-flow cytometric analysis (AST-FCA) for the detection of cytomegalovirus drug resistance. Diagn Microbiol Infect Dis. 1997;28:123–129. doi: 10.1016/s0732-8893(97)00040-0. [DOI] [PubMed] [Google Scholar]
- 186.Lloyd D, editor. Flow cytometry in microbiology. London, United Kingdom: Springer-Verlag; 1993. Flow cytometry: a technique waiting for microbiologists; pp. 1–10. [Google Scholar]
- 186a.Lloyd D. Flow cytometry in microbiology. London, United Kingdom: Springer-Verlag; 1993. [Google Scholar]
- 187.Lohr H F, Elste C, Dienes H P, Michel G, Braun H B, Meyer zum Buschenfelde K H, Gerken G. The quantitative humoral immune response to the hepatitis C virus is correlated with disease activity and response to interferon-alpha. J Hepatol. 1996;25:292–300. doi: 10.1016/s0168-8278(96)80114-0. [DOI] [PubMed] [Google Scholar]
- 188.Lopez-Amoros R, Castel S, Comas-Riu J, Vives-Rego J. Assessment of E. coli and Salmonella viability and starvation by confocal laser microscopy and flow cytometry using rhodamine 123, DiBAC4(3), propidium iodide, and CTC. Cytometry. 1997;29:298–305. doi: 10.1002/(sici)1097-0320(19971201)29:4<298::aid-cyto6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 189.Lopez-Amoros R, Comas J, Vives-Rego J. Flow cytometric assesment of Escherichia coli and Salmonella typhimurium starvation survival in seawater using rhodamine 123, propidium iodide, and oxonol. Appl Environ Microbiol. 1995;65:2521–2526. doi: 10.1128/aem.61.7.2521-2526.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Lorian V, Ernst J, Amaral L. The post-antibiotic effect defined by bacterial morphology. J Antimicrob Chemother. 1989;23:485–491. doi: 10.1093/jac/23.4.485. [DOI] [PubMed] [Google Scholar]
- 191.Lutton D A, Patrick S, Crockard A D, Stewart L D, Larkin M J, Dermott E, McNeill T A. Flow cytometric analysis of within-strain variation in polysaccharide expression by Bacteroides fragilis by use of murine monoclonal antibodies. J Med Microbiol. 1991;35:229–237. doi: 10.1099/00222615-35-4-229. [DOI] [PubMed] [Google Scholar]
- 192.Maarouf M, de Kouchkovsky Y, Brown S, Petit P X, Robert-Gero M. In vivo interference of paromomycin with mitochondrial activity of Leishmania. Exp Cell Res. 1997;232:339–348. doi: 10.1006/excr.1997.3500. [DOI] [PubMed] [Google Scholar]
- 193.Macchi B, Faraoni I, Zhang J, Grelli S, Favalli C, Mastino A, Bonmassar E. AZT inhibits the transmission of human T cell leukaemia/lymphoma virus type I to adult peripheral blood mononuclear cells in vitro. J Gen Virol. 1997;78:1007–1016. doi: 10.1099/0022-1317-78-5-1007. [DOI] [PubMed] [Google Scholar]
- 194.Maldarelli F, Sato H, Berthold E, Orenstein J, Martin M A. Rapid induction of apoptosis by cell-to-cell transmission of human immunodeficiency virus type 1. J Virol. 1995;69:6457–6465. doi: 10.1128/jvi.69.10.6457-6465.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Mansour J D, Robson J A, Arndt C W, Schulte T H. Detection of Escherichia coli in blood using flow cytometry. Cytometry. 1985;6:186–190. doi: 10.1002/cyto.990060303. [DOI] [PubMed] [Google Scholar]
- 196.Martin E, Bhakdi S. Flow cytometric assay for quantifying opsonophagocytosis and killing of Staphylococcus aureus by peripheral blood leukocytes. J Clin Microbiol. 1992;30:2246–2255. doi: 10.1128/jcm.30.9.2246-2255.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Martin E, Schlasius U, Bhakdi S. Flow cytometric assay for estimating fungicidal activity of amphotericin B in human serum. Med Microbiol Immunol. 1992;181:117–126. doi: 10.1007/BF00202051. [DOI] [PubMed] [Google Scholar]
- 198.Martinez O V, Gratzner H G, Malinin T I, Ingram M. The effect of some beta-lactam antibiotics on Escherichia coli studied by flow cytometry. Cytometry. 1982;3:129–133. doi: 10.1002/cyto.990030211. [DOI] [PubMed] [Google Scholar]
- 199.Martins-Filho O A, Pereira M E, Carvalho J F, Cancado J R, Brener Z. Flow cytometry, a new approach to detect anti-live trypomastigote antibodies and monitor the efficacy of specific treatment in human Chagas' disease. Clin Diagn Lab Immunol. 1995;2:569–573. doi: 10.1128/cdli.2.5.569-573.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Mason D J, Allman R, Stark J M, Lloyd D. Rapid estimation of bacterial antibiotic susceptibility with flow cytometry. J Microsc. 1994;176:8–16. doi: 10.1111/j.1365-2818.1994.tb03494.x. [DOI] [PubMed] [Google Scholar]
- 201.Mason D J, Gant V A. The application of flow cytometry to the estimation of bacterial antibiotic susceptibility. J Antimicrob Chemother. 1995;36:441–443. doi: 10.1093/jac/36.2.441. [DOI] [PubMed] [Google Scholar]
- 202.Mason D J, Lloyd D. Acridine orange as an indicator of bacterial susceptibility to gentamicin. FEMS Microbiol Lett. 1997;153:199–204. doi: 10.1111/j.1574-6968.1997.tb10482.x. [DOI] [PubMed] [Google Scholar]
- 203.Mason D J, Lopez-Amoros R, Allman R, Stark J M, Lloyd D. The ability of membrane potential dyes and calcafluor white to distinguish between viable and non-viable bacteria. J Appl Bacteriol. 1995;78:309–315. doi: 10.1111/j.1365-2672.1995.tb05031.x. [DOI] [PubMed] [Google Scholar]
- 204.Mason D J, Shanmuganathan S, Mortimer F C, Gant V A. A fluorescent Gram stain for flow cytometry and epifluorescence microscopy. Appl Environ Microbiol. 1998;64:2681–2685. doi: 10.1128/aem.64.7.2681-2685.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Matsuyama T. Staining of living bacteria with rhodamine 123. FEMS Microbiol Lett. 1984;21:153–157. [Google Scholar]
- 206.Mbida A D, Pozzetto B, Sabido O, Akono Y, Grattard F, Habib M, Gaudin O G. Competition binding studies with biotinylated echovirus 11 in cytofluorimetry analysis. J Virol Methods. 1991;35:169–176. doi: 10.1016/0166-0934(91)90132-j. [DOI] [PubMed] [Google Scholar]
- 207.McClelland R G, Pinder A C. Detection of low levels of specific Salmonella species by fluorescent antibodies and flow cytometry. J Appl Bacteriol. 1994;77:440–447. doi: 10.1111/j.1365-2672.1994.tb03447.x. [DOI] [PubMed] [Google Scholar]
- 208.McGrath M S, Santulli S, Gaston I. Effects of GLQ223 on HIV replication in human monocyte/macrophages chronically infected in vitro with HIV. AIDS Res Hum Retrovir. 1990;6:1039–1043. doi: 10.1089/aid.1990.6.1039. [DOI] [PubMed] [Google Scholar]
- 209.McHugh T M, Miner R C, Logan L H, Stites D P. Simultaneous detection of antibodies to cytomegalovirus and herpes simplex virus by using flow cytometry and a microsphere-based fluorescence immunoassay. J Clin Microbiol. 1988;26:1957–1961. doi: 10.1128/jcm.26.10.1957-1961.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.McHugh T M, Viele M K, Chase E S, Recktenwald D J. The sensitive detection and quantitation of antibody to HCV by using a microsphere-based immunoassay and flow cytometry. Cytometry. 1997;29:106–112. [PubMed] [Google Scholar]
- 211.McSharry J J. Uses of flow cytometry in virology. Clin Microbiol Rev. 1994;7:576–604. doi: 10.1128/cmr.7.4.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.McSharry J J. Antiviral drug susceptibility assays: going with the flow. Antiviral Res. 1999;43:1–21. doi: 10.1016/s0166-3542(99)00039-x. [DOI] [PubMed] [Google Scholar]
- 213.McSharry J J, Costantino R, Robbiano E, Echols R, Stevens R, Lehman J M. Detection and quantitation of human immunodeficiency virus-infected peripheral blood mononuclear cells by flow cytometry. J Clin Microbiol. 1990;28:724–733. doi: 10.1128/jcm.28.4.724-733.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.McSharry J J, Lurain N S, Drusano G L, Landay A L, Notka M, O'Gorman M R, Weinberg A, Shapiro H M, Reichelderfer P S, Crumpacker C S. Rapid ganciclovir susceptibility assay using flow cytometry for human cytomegalovirus clinical isolates. Antimicrob Agents Chemother. 1998;42:2326–2331. doi: 10.1128/aac.42.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.McSharry J M, Lurain N S, Drusano G L, Landay A, Manischewitz J, Nokta M, O'Gorman M, Shapiro H M, Weinberg A, Reichelderfer P, Crumpacker C. Flow cytometric determination of ganciclovir susceptibilities of human cytomegalovirus clinical isolates. J Clin Microbiol. 1998;36:958–964. doi: 10.1128/jcm.36.4.958-964.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Mellors J W, Rinaldo C R J, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 217.Mentel R, Dopping G, Wegner U, Seidel W, Liebermann H, Dohner L. Adenovirus-receptor interaction with human lymphocytes. J Med Virol. 1997;51:252–257. [PubMed] [Google Scholar]
- 218.Mercure L, Brenner B J, Phaneuf D, Tsoukas C, Wainberg M A. Effect of 3′-azido-3′-deoxythymidine and 2′,3′-dideoxyinosine on establishment of human immunodeficiency virus type 1 infection in cultured CD8+ lymphocytes. Antimicrob Agents Chemother. 1994;38:986–990. doi: 10.1128/aac.38.5.986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mercure S, Senechal S, Auger P, Lemay G, Montplaisir S. Candida albicans serotype analysis by flow cytometry. J Clin Microbiol. 1996;34:2106–2112. doi: 10.1128/jcm.34.9.2106-2112.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Midgley M. The phosphonium ion efflux system of E. coli: relationship to the ethidium efflux system and energetics study. J Gen Microbiol. 1986;132:3187–3193. doi: 10.1099/00221287-132-11-3187. [DOI] [PubMed] [Google Scholar]
- 221.Miller C L, Longnecker R, Kieff E. Epstein-Barr virus latent membrane protein 2A blocks calcium mobilization in B lymphocytes. J Virol. 1993;67:3087–3094. doi: 10.1128/jvi.67.6.3087-3094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Moore A V, Kirk S M, Callister S M, Mazurek G H, Schell R F. Safe determination of susceptibility of Mycobacterium tuberculosis to antimycobacterial agents by flow cytometry. J Clin Microbiol. 1999;37:479–483. doi: 10.1128/jcm.37.3.479-483.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Morley P J. Evaluation of anti-Pneumocystis agents using flow cytometry. J Eukaryot Microbiol. 1994;41:105S–106S. [PubMed] [Google Scholar]
- 224.Murakami T, Nakashima H, Ito M, Yamamoto N. A novel coculture system for evaluating anti-HIV drugs. AIDS. 1992;6:1279–1285. doi: 10.1097/00002030-199211000-00007. [DOI] [PubMed] [Google Scholar]
- 225.Muratori L, Gibellini D, Lenzi M, Cataleta M, Muratori P, Morelli M C, Bianchi F B. Quantification of hepatitis C virus-infected peripheral blood mononuclear cells by in situ reverse transcriptase-polymerase chain reaction. Blood. 1996;88:2768–2774. [PubMed] [Google Scholar]
- 226.Naniche D, Varior-Krishnan G, Cervoni F, Wild T F, Rossi B, Rabourdin-Combe C, Gerlier D. Human membrane cofactor protein (CD46) acts as a cellular receptor for measles virus. J Virol. 1993;67:6025–6032. doi: 10.1128/jvi.67.10.6025-6032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Neyts J, Snoeck R, Schols D, Himpens B, De Clercq E. Sensitive, reproducible and convenient fluorometric assay for the in vitro evaluation of anti-cytomegalovirus agents. J Virol Methods. 1991;35:27–38. doi: 10.1016/0166-0934(91)90082-b. [DOI] [PubMed] [Google Scholar]
- 228.Nichols J E, Mock D J, Roberts N J J. Use of FITC-labeled influenza virus and flow cytometry to assess binding and internalization of virus by monocytes-macrophages and lymphocytes. Arch Virol. 1993;130:441–455. doi: 10.1007/BF01309672. [DOI] [PubMed] [Google Scholar]
- 229.Niedenthal R K, Riles L, Johnston M, Hegemann J H. Green fluorescent protein as a marker for gene expression and subcellular localization in budding yeast. Yeast. 1996;12:773–786. doi: 10.1002/(SICI)1097-0061(19960630)12:8%3C773::AID-YEA972%3E3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 230.Niven G W, Mulholland F. Cell membrane integrity and lysis in Lactococcus lactis: the detection of a population of permeable cells in post-logarithmic phase cultures. J Appl Microbiol. 1998;84:90–96. doi: 10.1046/j.1365-2672.1997.00316.x. [DOI] [PubMed] [Google Scholar]
- 231.Norden M A, Kurzynski T A, Bownds S E, Callister S M, Schell R F. Rapid susceptibility testing of Mycobacterium tuberculosis (H37Ra) by flow cytometry. J Clin Microbiol. 1995;33:1231–1237. doi: 10.1128/jcm.33.5.1231-1237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nussbaum O, Broder C C, Moss B, Stern L B, Rozenblatt S, Berger E A. Functional and structural interactions between measles virus hemagglutinin and CD46. J Virol. 1995;69:3341–3349. doi: 10.1128/jvi.69.6.3341-3349.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ogg G S, Jin X, Bonhoeffer S, Dunbar P R, Nowak M A, Monard S, Segal J P, Cao Y, Rowland-Jones S L, Cerundolo V, Hurley A, Markowitz M, Ho D D, Nixon D F, McMichael A J. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science. 1998;279:2103–2106. doi: 10.1126/science.279.5359.2103. [DOI] [PubMed] [Google Scholar]
- 234.O'Gorman M R G, Hopfer R L. Amphotericin B susceptibility testing of Candida species by flow cytometry. Cytometry. 1991;12:743–747. doi: 10.1002/cyto.990120808. [DOI] [PubMed] [Google Scholar]
- 235.Ohlsson-Wilhelm B M, Cory J M, Kessler H A, Eyster M E, Rapp F, Landay A. Circulating human immunodeficiency virus (HIV) p24 antigen-positive lymphocytes: a flow cytometric measure of HIV infection. J Infect Dis. 1990;162:1018–1024. doi: 10.1093/infdis/162.5.1018. [DOI] [PubMed] [Google Scholar]
- 236.Oravecz T, Pall M, Wang J, Roderiquez G, Ditto M, Norcross M A. Regulation of anti-HIV-1 activity of RANTES by heparan sulfate proteoglycans. J Immunol. 1997;159:4587–4592. [PubMed] [Google Scholar]
- 237.Ordoñez J V, Wehman N M. Rapid flow cytometric antibiotic susceptibility assay for Staphylococcus aureus. Cytometry. 1993;14:811–818. doi: 10.1002/cyto.990140714. [DOI] [PubMed] [Google Scholar]
- 238.Ozanne V, Ortalo-Magne A, Vercellone A, Fournie J J, Daffe M. Cytometric detection of mycobacterial surface antigens: exposure of mannosyl epitopes and of the arabinan segment of arabinomannans. J Bacteriol. 1996;178:7254–7259. doi: 10.1128/jb.178.24.7254-7259.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Patrick S, Stewart L D, Damani N, Wilson K G, Lutton D A, Larkin M J, Poxton I, Brown R. Immunological detection of Bacteroides fragilis in clinical samples. J Med Microbiol. 1995;43:99–109. doi: 10.1099/00222615-43-2-99. [DOI] [PubMed] [Google Scholar]
- 240.Pattanapanyasat K, Thaithong S, Kyle D E, Udomsangpetch R, Yongvanitchit K, Hider R C, Webster H K. Flow cytometric assessment of hydroxypyridinone iron chelators on in vitro growth of drug-resistant malaria. Cytometry. 1997;27:84–91. doi: 10.1002/(sici)1097-0320(19970101)27:1<84::aid-cyto11>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 241.Pattanapanyasat K, Udomsangpetch R, Webster H K. Two-color flow cytometric analysis of intraerythrocytic malaria parasite DNA and surface membrane-associated antigen in erythrocytes infected with Plasmodium falciparum. Cytometry. 1993;14:449–454. doi: 10.1002/cyto.990140415. [DOI] [PubMed] [Google Scholar]
- 242.Pattanapanyasat K, Webster H K, Udomsangpetch R, Wanachiwanawin W, Yongvanitchit K. Flow cytometric two-color staining technique for simultaneous determination of human erythrocyte membrane antigen and intracellular malarial DNA. Cytometry. 1992;13:182–187. doi: 10.1002/cyto.990130212. [DOI] [PubMed] [Google Scholar]
- 243.Patterson B K, Goolsby C, Hodara V, Lohman K L, Wolinsky S M. Detection of CD4+ T cells harboring human immunodeficiency virus type 1 DNA by flow cytometry using simultaneous immunophenotyping and PCR-driven in situ hybridization: evidence of epitope masking of the CD4 cell surface molecule in vivo. J Virol. 1995;69:4316–4322. doi: 10.1128/jvi.69.7.4316-4322.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Patterson B K, Mosiman V L, Cantarero L, Furtado M, Bhattacharya M, Goolsby C. Detection of HIV-RNA-positive monocytes in peripheral blood of HIV- positive patients by simultaneous flow cytometric analysis of intracellular HIV RNA and cellular immunophenotype. Cytometry. 1998;31:265–274. doi: 10.1002/(sici)1097-0320(19980401)31:4<265::aid-cyto6>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 245.Patterson B K, Till M, Otto P, Goolsby C, Furtado M R, McBride L J, Wolinsky S M. Detection of HIV-1 DNA and messenger RNA in individual cells by PCR-driven in situ hybridization and flow cytometry. Science. 1993;260:976–979. doi: 10.1126/science.8493534. [DOI] [PubMed] [Google Scholar]
- 246.Patterson S, Helbert M, English N R, Pinching A J, Knight S C. The effect of AZT on dendritic cell number and provirus load in the peripheral blood of AIDS patients: a preliminary study. Res Virol. 1996;147:109–114. doi: 10.1016/0923-2516(96)80224-x. [DOI] [PubMed] [Google Scholar]
- 247.Pauwels R, De Clercq E, Desmyter J, Balzarini J, Goubau P, Herdewijn P, Vanderhaeghe H, Vandeputte M. Sensitive and rapid assay on MT-4 cells for detection of antiviral compounds against the AIDS virus. J Virol Methods. 1987;16:171–185. doi: 10.1016/0166-0934(87)90002-4. [DOI] [PubMed] [Google Scholar]
- 248.Pavic I, Hartmann A, Zimmermann A, Michel D, Hampl W, Schleyer I, Mertens T. Flow cytometric analysis of herpes simplex virus type 1 susceptibility to acyclovir, ganciclovir, and foscarnet. Antimicrob Agents Chemother. 1997;41:2686–2692. doi: 10.1128/aac.41.12.2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Pekosz A, Griot C, Nathanson N, Gonzalez-Scarano F. Tropism of bunyaviruses: evidence for a G1 glycoprotein-mediated entry pathway common to the California serogroup. Virology. 1995;214:339–348. doi: 10.1006/viro.1995.0043. [DOI] [PubMed] [Google Scholar]
- 250.Perraut R, Mercereau-Puijalon O, Mattei D, Bourreau E, Bonnefoy S, Bonnemains B, Gysin J, Michel J C, Da S L. Immunogenicity and efficacy trials with Plasmodium falciparum recombinant antigens identified as targets of opsonizing antibodies in the naive squirrel monkey Saimiri sciureus. Am J Trop Med Hyg. 1997;56:343–350. doi: 10.4269/ajtmh.1997.56.343. [DOI] [PubMed] [Google Scholar]
- 251.Perticarari S, Presani G, Mangiarotti M A, Banfi E. Simultaneous flow cytometric method to measure phagocytosis and oxidative products by neutrophils. Cytometry. 1991;12:687–693. doi: 10.1002/cyto.990120713. [DOI] [PubMed] [Google Scholar]
- 252.Peyron F, Favel A, Guiraud-Dauriac H, El Mzibri M, Chastin C, Dumenil G, Regli P. Evaluation of a flow cytofluorometric method for rapid determination of amphotericin B susceptibility of yeast isolates. Antimicrob Agents Chemother. 1997;41:1537–1540. doi: 10.1128/aac.41.7.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Pfaller M A, Herwaldt L A. The clinical microbiology laboratory and infection control: emerging pathogens, antimicrobial resistance, and new technology. Clin Infect Dis. 1997;25:858–870. doi: 10.1086/515557. [DOI] [PubMed] [Google Scholar]
- 254.Phillips A P, Martin K L. Immunofluorescence analysis of Bacillus spores and vegetative cells by flow cytometry. Cytometry. 1983;4:123–131. doi: 10.1002/cyto.990040205. [DOI] [PubMed] [Google Scholar]
- 255.Phillips A P, Martin K L. Dual-parameter scatter-flow immunofluorescence analysis of Bacillus spores. Cytometry. 1985;6:124–129. doi: 10.1002/cyto.990060207. [DOI] [PubMed] [Google Scholar]
- 256.Pierard G E, Arrese J E, De Donker P, Pierard-Franchimont C. Present and potential diagnostic techniques in onychomycosis. J Am Acad Dermatol. 1996;34:273–277. doi: 10.1016/s0190-9622(96)80122-8. [DOI] [PubMed] [Google Scholar]
- 257.Pinder A C, Purdy P W, Poulter S A, Clark D C. Validation of flow cytometry for rapid enumeration of bacterial concentrations in pure cultures. J Appl Bacteriol. 1990;69:92–100. doi: 10.1111/j.1365-2672.1990.tb02916.x. [DOI] [PubMed] [Google Scholar]
- 258.Plovins A, Alvarez A M, Ibanez M, Molina M, Nombela C. Use of fluorescein-di-beta-d-galactopyranoside (FDG) and C12-FDG as substrates for beta-galactosidase detection by flow cytometry in animal, bacterial, and yeast cells. Appl Environ Microbiol. 1994;60:4638–4641. doi: 10.1128/aem.60.12.4638-4641.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Pore R S. Antibiotic susceptibility testing of Candida albicans by flow cytometry. Curr Microbiol. 1990;20:323–328. [Google Scholar]
- 260.Pore R S. Ketoconazole susceptibility of yeast by the FCTS method. Curr Microbiol. 1991;23:45–50. [Google Scholar]
- 261.Pore R S. Amphotericine B synergy test by the FCTS. Curr Microbiol. 1992;24:171–177. [Google Scholar]
- 262.Pore R S. Antibiotic susceptibility testing by flow cytometry. J Antimicrob Chemother. 1994;34:613–627. doi: 10.1093/jac/34.5.613. [DOI] [PubMed] [Google Scholar]
- 263.Porter J, Deere D, Hardman M, Edwards C, Pickup R. Go with the flow—use of flow cytometry in environmental microbiology. FEMS Microbiol Ecol. 1997;24:93–101. [Google Scholar]
- 264.Porter J, Deere D, Pickup R, Edwards C. Fluorescent probes and flow cytometry: new insights into environmental bacteriology. Cytometry. 1996;23:91–96. doi: 10.1002/(SICI)1097-0320(19960201)23:2<91::AID-CYTO1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 265.Porter J, Edwards C, Pickup R W. Rapid assessment of physiological status in Escherichia coli using fluorescent probes. J Appl Bacteriol. 1995;79:399–408. doi: 10.1111/j.1365-2672.1995.tb03154.x. [DOI] [PubMed] [Google Scholar]
- 266.Portnoi D, Stall A M, Schwartz D, Merigan T C, Herzenberg L A, Basham T. Zidovudine (azido dideoxythymidine) inhibits characteristic early alterations of lymphoid cell populations in retrovirus-induced murine AIDS. J Immunol. 1990;144:1705–1710. [PubMed] [Google Scholar]
- 267.Price G E, Smith H, Sweet C. Differential induction of cytotoxicity and apoptosis by influenza virus strains of differing virulence. J Gen Virol. 1997;78:2821–2829. doi: 10.1099/0022-1317-78-11-2821. [DOI] [PubMed] [Google Scholar]
- 268.Qi Y M, Peng S W, Hengst K, Evander M, Park D S, Zhou J, Frazer I H. Epithelial cells display separate receptors for papillomavirus VLPs and for soluble L1 capsid protein. Virology. 1996;216:35–45. doi: 10.1006/viro.1996.0032. [DOI] [PubMed] [Google Scholar]
- 269.Rabin L, Hincenbergs M, Moreno M B, Warren S, Linquist V, Datema R, Charpiot B, Seifert J, Kaneshima H, McCune J M. Use of standardized SCID-hu Thy/Liv mouse model for preclinical efficacy testing of anti-human immunodeficiency virus type 1 compounds. Antimicrob Agents Chemother. 1996;40:755–762. doi: 10.1128/aac.40.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ramani R, Ramani A, Wong S J. Rapid flow cytometric susceptibility testing of Candida albicans. J Clin Microbiol. 1997;35:2320–2324. doi: 10.1128/jcm.35.9.2320-2324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Raponi G, Keller N, Overbeek B P, Rozenberg-Arska M, van Kessel K P, Verhoef J. Enhanced phagocytosis of encapsulated Escherichia coli strains after exposure to sub-MICs of antibiotics is correlated to changes of the bacterial cell surface. Antimicrob Agents Chemother. 1990;34:332–336. doi: 10.1128/aac.34.2.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Raybourne R B, Bunning V K. Bacterium-host cell interactions at the cellular level: fluorescent labeling of bacteria and analysis of short-term bacterium-phagocyte interaction by flow cytometry. Infect Immun. 1994;62:665–672. doi: 10.1128/iai.62.2.665-672.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Re F, Braaten D, Franke E K, Luban J. Human immunodeficiency virus type 1 Vpr arrests the cell cycle in G2 by inhibiting the activation of p34cdc2-cyclin B. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Re M C, Furlini G, Gibellini D, Vignoli M, Ramazzotti E, Lolli E, Ranieri S, La P M. Quantification of human immunodeficiency virus type 1-infected mononuclear cells in peripheral blood of seropositive subjects by newly developed flow cytometry analysis of the product of an in situ PCR assay. J Clin Microbiol. 1994;32:2152–2157. doi: 10.1128/jcm.32.9.2152-2157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Renner E D. Development and clinical evaluation of an amplified flow cytometric fluoroimmunoassay for Clostridium difficile toxin A. Cytometry. 1994;18:103–108. doi: 10.1002/cyto.990180209. [DOI] [PubMed] [Google Scholar]
- 276.Reymen D, Naesens L, Balzarini J, Holy A, Dvorakova H, De Clercq E. Antiviral activity of selected acyclic nucleoside analogues against human herpesvirus 6. Antiviral Res. 1995;28:343–357. doi: 10.1016/0166-3542(95)00058-5. [DOI] [PubMed] [Google Scholar]
- 277.Robertson B R, Button D K. Characterizing aquatic bacteria according to population, cell size, and apparent DNA content by flow cytometry. Cytometry. 1989;10:70–76. doi: 10.1002/cyto.990100112. [DOI] [PubMed] [Google Scholar]
- 278.Roisman F R, Castello A, Fainboim H, Morelli A, Fainboim L. Hepatitis B virus antigens in peripheral blood mononuclear cells during the course of viral infection. Clin Immunol Immunopathol. 1994;70:99–103. doi: 10.1006/clin.1994.1016. [DOI] [PubMed] [Google Scholar]
- 279.Rosa D, Campagnoli S, Moretto C, Guenzi E, Cousens L, Chin M, Dong C, Weiner A J, Lau J Y, Choo Q L, Chien D, Pileri P, Houghton M, Abrignani S. A quantitative test to estimate neutralizing antibodies to the hepatitis C virus: cytofluorimetric assessment of envelope glycoprotein 2 binding to target cells. Proc Natl Acad Sci USA. 1996;93:1759–1763. doi: 10.1073/pnas.93.5.1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Rosenthal K S, Hodnichak C M, Summers J L. Flow cytometric evaluation of anti-herpes drugs. Cytometry. 1987;8:392–395. doi: 10.1002/cyto.990080408. [DOI] [PubMed] [Google Scholar]
- 281.Rothe G, Valet G. Phagocytosis, intracellular pH, and cell volume in the multifunctional analysis of granulocytes by flow cytometry. Cytometry. 1988;9:316–324. doi: 10.1002/cyto.990090408. [DOI] [PubMed] [Google Scholar]
- 282.Rubin F A, Rothman S W. Extinction or revitalization: clinical microbiologists look to the future. ASM News. 1997;63:530–531. [Google Scholar]
- 283.Ryan C, Nguyen B T, Sullivan S J. Rapid assay for mycobacterial growth and antibiotic susceptibility using gel microdrop encapsulation. J Clin Microbiol. 1995;33:1720–1726. doi: 10.1128/jcm.33.7.1720-1726.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Schols D, Baba M, Pauwels R, de Clercq E. Flow cytometric method to demonstrate whether anti-HIV-1 agents inhibit virion binding to T4+ cells. J Acquir Immune Defic Syndr. 1989;2:10–15. [PubMed] [Google Scholar]
- 285.Schols D, Pauwels R, Baba M, Desmyter J, de Clerck E. Syncytium formation and destruction of bystander CD4+ cells cocultured with T cells persistently infected with human immunodeficiency virus as demonstrated by flow cytometry. J Gen Virol. 1989;70:2397–2408. doi: 10.1099/0022-1317-70-9-2397. [DOI] [PubMed] [Google Scholar]
- 286.Schols D, Pauwels R, Desmyter J, de Clerck E. Flow cytometric method to monitor the destruction of CD4+ cells following their fusion with HIV-infected cells. Cytometry. 1990;11:736–743. doi: 10.1002/cyto.990110611. [DOI] [PubMed] [Google Scholar]
- 287.Schols D, Pauwels R, Vanlangendonck F, Balzarini J, de Clerck E. A highly reliable, sensitive, flow cytometric/fluorometric assay for the evaluation of the anti-HIV activity of antiviral compounds in MT-4 cells. J Immunol Methods. 1988;114:27–32. doi: 10.1016/0022-1759(88)90149-4. [DOI] [PubMed] [Google Scholar]
- 288.Schols D, Proost P, Van D J, de Clerck E. RANTES and MCP-3 inhibit the replication of T-cell-tropic human immunodeficiency virus type 1 strains (SF-2, MN, and HE) J Virol. 1997;71:7300–7304. doi: 10.1128/jvi.71.10.7300-7304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Schols D, Snoeck R, Neyts J, de Clerck E. Detection of immediate early, early and late antigens of human cytomegalovirus by flow cytometry. J Virol Methods. 1989;26:247–254. doi: 10.1016/0166-0934(89)90107-9. [DOI] [PubMed] [Google Scholar]
- 290.Scillian J J, McHugh T M, Busch M P, Tam M, Fulwyler M J, Chien D Y, Vyas G N. Early detection of antibodies against rDNA-produced HIV proteins with a flow cytometric assay. Blood. 1989;73:2041–2048. [PubMed] [Google Scholar]
- 291.Shapiro H M. Practical flow cytometry. 3rd ed. New York, N.Y: Wiley-Liss; 1995. [Google Scholar]
- 292.Siegman-Igra Y, Kulka T, Schwarz D, Konforti N. The significance of polymicrobial growth in urine: contamination or true infection. Scand J Infect Dis. 1993;25:85–91. [PubMed] [Google Scholar]
- 293.Skarstad K, Steen H B, Boye E. Cell cycle parameters of slowly growing Escherichia coli B/r studied by flow cytometry. J Bacteriol. 1983;154:656–662. doi: 10.1128/jb.154.2.656-662.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Sligh J M, Roodman S T, Tsai C C. Flow cytometric indirect immunofluorescence assay with high sensitivity and specificity for detection of antibodies to human immunodeficiency virus (HIV) Am J Clin Pathol. 1989;91:210–214. doi: 10.1093/ajcp/91.2.210. [DOI] [PubMed] [Google Scholar]
- 295.Snoeck R, Schols D, Andrei G, Neyts J, de Clerck E. Antiviral activity of anti-cytomegalovirus agents (HPMPC, HPMPA) assessed by a flow cytometric method and DNA hybridization technique. Antiviral Res. 1991;16:1–9. doi: 10.1016/0166-3542(91)90053-t. [DOI] [PubMed] [Google Scholar]
- 296.Snoeck R, Schols D, Sadzot-Delvaux C, Cloes J M, Andrei G, de Clerck E, Piette J, Rentier B. Flow cytometric method for the detection of gpI antigens of varicella zoster virus and evaluation of anti-VZV agents. J Virol Methods. 1992;38:243–254. doi: 10.1016/0166-0934(92)90114-s. [DOI] [PubMed] [Google Scholar]
- 297.Srienc F, Campbell J L, Bailey J E. Flow cytometry analysis of recombinant Saccharomyces cerevisiae populations. Cytometry. 1986;7:132–141. doi: 10.1002/cyto.990070203. [DOI] [PubMed] [Google Scholar]
- 298.Srikumar R, Chin A C, Vachon V, Richardson C D, Ratcliffe M J, Saarinen L, Kayhty H, Makela P H, Coulton J W. Monoclonal antibodies specific to porin of Haemophilus influenzae type b: localization of their cognate epitopes and tests of their biological activities. Mol Microbiol. 1992;6:665–676. doi: 10.1111/j.1365-2958.1992.tb01514.x. [DOI] [PubMed] [Google Scholar]
- 299.Steed L L, Akporiaye E T, Friedman R L. Bordetella pertussis induces respiratory burst activity in human polymorphonuclear leukocytes. Infect Immun. 1992;60:2101–2105. doi: 10.1128/iai.60.5.2101-2105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Steele-Mortimer O A, Meier-Ewert H, Loser R, Hasmann M J. Flow cytometric analysis of virus-infected cells and its potential use for screening antiviral agents. J Virol Methods. 1990;27:241–252. doi: 10.1016/0166-0934(90)90092-t. [DOI] [PubMed] [Google Scholar]
- 301.Steen H B. Simultaneous separate detection of low angle and large angle light scattering in an arc lamp-based flow cytometer. Cytometry. 1986;7:445–449. doi: 10.1002/cyto.990070509. [DOI] [PubMed] [Google Scholar]
- 302.Steen H B, Boye E, Skarstad K, Bloom B, Godal T, Mustafa S. Applications of flow cytometry on bacteria: cell cycle kinetics, drug effects, and quantitation of antibody binding. Cytometry. 1982;2:249–257. doi: 10.1002/cyto.990020409. [DOI] [PubMed] [Google Scholar]
- 303.Steen H B, Lindmo T. Flow cytometry: a high-resolution instrument for everyone. Science. 1979;204:404. doi: 10.1126/science.441727. [DOI] [PubMed] [Google Scholar]
- 304.Steen H B, Skarstad K, Boye E. Flow cytometry of bacteria: cell cycle and effects of antibiotics. Ann N Y Acad Sci. 1986;486:329–338. doi: 10.1111/j.1749-6632.1986.tb42050.x. [DOI] [PubMed] [Google Scholar]
- 305.Steen H B, Skarstad K, Boye E. DNA measurements of bacteria. In: Darzynkiewicz Z, Crissman H, editors. Flow cytometry. London, United Kingdom: Academic Press, Ltd.; 1990. pp. 519–526. [Google Scholar]
- 306.Steen H, Boye E. Growth of Escherichia coli studied by dual-parameter flow cytometry. J Bacteriol. 1981;145:1091–1094. doi: 10.1128/jb.145.2.1091-1094.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Stein G E. Pharmacokinetics and pharmacodynamics of newer fluoroquinolones. Clin Infect Dis. 1996;23(Suppl. 1):S19–S24. doi: 10.1093/clinids/23.supplement_1.s19. [DOI] [PubMed] [Google Scholar]
- 308.Stoiber H, Frank I, Spruth M, Schwendinger M, Mullauer B, Windisch J M, Schneider R, Katinger H, Ando I, Dierich M P. Inhibition of HIV-1 infection in vitro by monoclonal antibodies to the complement receptor type 3 (CR3): an accessory role for CR3 during virus entry? Mol Immunol. 1997;34:855–863. doi: 10.1016/s0161-5890(97)00108-9. [DOI] [PubMed] [Google Scholar]
- 309.Suller M T E, Lloyd D. Flow cytometric assessment of the postantibiotic effect of methicillin on Staphylococcus aureus. Antimicrob Agents Chemother. 1998;42:1195–1199. doi: 10.1128/aac.42.5.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Suller M T E, Stark J M, Lloyd D. A flow cytometric study of antibiotic-induced demage and evaluation as rapid antibiotic susceptibility test for methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1997;40:77–83. doi: 10.1093/jac/40.1.77. [DOI] [PubMed] [Google Scholar]
- 311.Swarts A J, Hastings J W, Roberts R F, von Holy A. Flow cytometry demonstrates bacteriocin-induced injury to Listeria monocytogenes. Curr Microbiol. 1998;36:266–270. doi: 10.1007/s002849900307. [DOI] [PubMed] [Google Scholar]
- 312.Tamalet C, Lafeuillade A, Fantini J, Poggi C, Yahi N. Quantification of HIV-1 viral load in lymphoid and blood cells: assessment during four-drug combination therapy. AIDS. 1997;11:895–901. doi: 10.1097/00002030-199707000-00009. [DOI] [PubMed] [Google Scholar]
- 313.Tapp H, Stotzky G. Monitoring the insecticidal toxins from Bacillus thuringiensis in soil with flow cytometry. Can J Microbiol. 1997;43:1074–1078. doi: 10.1139/m97-153. [DOI] [PubMed] [Google Scholar]
- 314.The T H, van den Berg A P, Vlieger A M, van de Giessen G, van der Son W J. Direct detection of cytomegalovirus in peripheral blood leukocytes—a review of the antigenemia assay and polymerase chain reaction. Transplantation. 1992;54:193–198. doi: 10.1097/00007890-199208000-00001. [DOI] [PubMed] [Google Scholar]
- 315.Thomas J C, Desrosiers M, St-Pierre Y, Lirette P, Bisaillon J G, Beaudet R, Villemur R. Quantitative flow cytometric detection of specific microorganisms in soil samples using rRNA targeted fluorescent probes and ethidium bromide. Cytometry. 1997;27:224–232. doi: 10.1002/(sici)1097-0320(19970301)27:3<224::aid-cyto3>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 316.Thorell B. Flow cytometric analysis of cellular endogenous fluorescence simultaneously with emission from exogenous fluorochromes, light scatter and absorption. Cytometry. 1981;2:39–43. doi: 10.1002/cyto.990020109. [DOI] [PubMed] [Google Scholar]
- 317.Trask B J, van den Engh G J, Elgershuizen J H. Analysis of phytoplankton by flow cytometry. Cytometry. 1982;2:258–264. doi: 10.1002/cyto.990020410. [DOI] [PubMed] [Google Scholar]
- 318.Trischmann H, Davis D, Lachmann P J. Lymphocytotropic strains of HIV type 1 when complexed with enhancing antibodies can infect macrophages via Fc gamma RIII, independently of CD4. AIDS Res Hum Retrovir. 1995;11:343–352. doi: 10.1089/aid.1995.11.343. [DOI] [PubMed] [Google Scholar]
- 319.Turner H M, Mohberg J, Goodman E M. A flow cytometry study of the cell cycle and of ploidy levels in Physarum polycephalum myxamoebae and plasmodia. Microbios. 1981;32:29–36. [PubMed] [Google Scholar]
- 320.Valdivia R H, Falkow S. Fluorescence-based isolation of bacterial genes expressed within host cells. Science. 1997;277:2007–2011. doi: 10.1126/science.277.5334.2007. [DOI] [PubMed] [Google Scholar]
- 321.van der Ende A, Rauws E A, Feller M, Mulder C J, Tytgat G N, Dankert J. Heterogeneous Helicobacter pylori isolates from members of a family with a history of peptic ulcer diseases. Gastroenterology. 1996;111:638–647. doi: 10.1053/gast.1996.v111.pm8780568. [DOI] [PubMed] [Google Scholar]
- 322.van der Waaij L A, Mesander G, Limburg P C. Direct flow cytometry of anaerobic bacteria in human feces. Cytometry. 1994;16:270–279. doi: 10.1002/cyto.990160312. [DOI] [PubMed] [Google Scholar]
- 323.van Eldere J, Joosten L, Verhaeghe V, Surmont I. Fluconazole and amphotericin B antifungal susceptibility testing by National Committee for Clinical Laboratory Standards broth macrodilution method compared with E-test and semiautomated broth microdilution test. J Clin Microbiol. 1996;34:842–847. doi: 10.1128/jcm.34.4.842-847.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Vanoni M, Vai M, Popolo L, Alberghina L. Structural heterogeneity in populations of the budding yeast Saccharomyces cerevisiae. J Bacteriol. 1983;156:1282–1291. doi: 10.1128/jb.156.3.1282-1291.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.van Vianen P H, Thaithong S, Reinders P P, van Gnger A, Tanke H J, van der Kaay H J, Mons B. Automated flow cytometric analysis of drug susceptibility of malaria parasites. Am J Trop Med Hyg. 1990;43:602–607. doi: 10.4269/ajtmh.1990.43.602. [DOI] [PubMed] [Google Scholar]
- 326.Varthakavi V, Minocha H C. Identification of a 56 kDa putative bovine herpesvirus 1 cellular receptor by anti-idiotype antibodies. J Gen Virol. 1996;77:1875–1882. doi: 10.1099/0022-1317-77-8-1875. [DOI] [PubMed] [Google Scholar]
- 327.Volberding P A. HIV quantification: clinical applications. Lancet. 1996;347:71–73. doi: 10.1016/s0140-6736(96)90205-6. [DOI] [PubMed] [Google Scholar]
- 328.Votyakova T V, Mukamolova G V, ShteinMargolina V A, Popov V I, Davey H M, Kell D B, Kaprelyants A S. Research on the heterogeneity of a Micrococcus luteus culture during an extended stationary phase: subpopulation separation and characterization. Microbiology. 1998;67:71–77. [Google Scholar]
- 329.Walberg M, Gaustad P, Steen H B. Rapid flow cytometric assessment of mecillinam and ampicillin bacterial susceptibility. J Antimicrob Chemother. 1996;37:1063–1075. doi: 10.1093/jac/37.6.1063. [DOI] [PubMed] [Google Scholar]
- 330.Walberg M, Gaustad P, Steen H B. Rapid assessment of ceftazidime, ciprofloxacin, and gentamicin susceptibility in exponentially-growing E. coli cells by means of flow cytometry. Cytometry. 1997;27:169–178. [PubMed] [Google Scholar]
- 331.Walberg M, Gaustad P, Steen H B. Rapid discrimination of bacterial species with different ampicillin susceptibility levels by means of flow cytometry. Cytometry. 1997;29:267–272. [PubMed] [Google Scholar]
- 332.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 333.Wallner G, Fuchs B, Spring S, Beisker W, Amann R. Flow sorting of microorganisms for molecular analysis. Appl Environ Microbiol. 1997;63:4223–4231. doi: 10.1128/aem.63.11.4223-4231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Wattre P. Contribution of flow cytometry in virology. Ann Biol Clin (Paris) 1993;51:1–6. . (In French.) [PubMed] [Google Scholar]
- 335.Wells A, Steen H, Saeland S, Godal T, Klein G. A microassay for quantitatively detecting the Epstein-Barr virus receptor on single cells utilizing flow cytometry. J Virol Methods. 1981;3:127–136. doi: 10.1016/0166-0934(81)90047-1. [DOI] [PubMed] [Google Scholar]
- 336.Wenisch C, Linnau K F, Parschalk B, Zedtwitz-Leibenstein K, Georgopoulos A. Rapid susceptibility testing of fungi by flow cytometry using vital staining. J Clin Microbiol. 1997;35:5–10. doi: 10.1128/jcm.35.1.5-10.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Williams K M, Sacci J B, Anthony R L. Characterization and quantitation of membrane antigens of New World Leishmania species by using monoclonal antibodies in Western blot and flow microfluorometric assays. J Protozool. 1986;33:490–497. doi: 10.1111/j.1550-7408.1986.tb05648.x. [DOI] [PubMed] [Google Scholar]
- 338.Wittrup K D, Bailey J E. A single-cell assay of beta-galactosidase activity in Saccharomyces cerevisiae. Cytometry. 1988;9:394–404. doi: 10.1002/cyto.990090418. [DOI] [PubMed] [Google Scholar]
- 339.Woods G L. Automation in clinical microbiology. Am J Clin Pathol. 1992;98:S22–S30. [PubMed] [Google Scholar]
- 340.Xu C, Meikrantz W, Schlegel R, Sager R. The human papilloma virus 16E6 gene sensitizes human mammary epithelial cells to apoptosis induced by DNA damage. Proc Natl Acad Sci USA. 1995;92:7829–7833. doi: 10.1073/pnas.92.17.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Yang D W, Ohta Y, Yamaguchi S, Tsukada Y, Haraguchi Y, Hoshino H, Amagai H, Kobayashi I. Sulfated colominic acid: an antiviral agent that inhibits the human immunodeficiency virus type 1 in vitro. Antiviral Res. 1996;31:95–104. doi: 10.1016/0166-3542(96)00957-6. [DOI] [PubMed] [Google Scholar]
- 342.Yang G, Garhwal S, Olson J C, Vyas G N. Flow cytometric immunodetection of human immunodeficiency virus type 1 proviral DNA by heminested PCR and digoxigenin-labeled probes. Clin Diagn Lab Immunol. 1994;1:26–31. doi: 10.1128/cdli.1.1.26-31.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Yang G, Olson J C, Pu R, Vyas G N. Flow cytometric detection of human immunodeficiency virus type 1 proviral DNA by the polymerase chain reaction incorporating digoxigenin- or fluorescein-labeled dUTP. Cytometry. 1995;21:197–202. doi: 10.1002/cyto.990210212. [DOI] [PubMed] [Google Scholar]
- 344.Yang G, Ulrich P P, Aiyer R A, Rawal B D, Vyas G N. Detection of hepatitis B virus in plasma using flow cytometric analyses of polymerase chain reaction-amplified DNA incorporating digoxigenin-11-dUTP. Blood. 1993;81:1083–1088. [PubMed] [Google Scholar]
- 345.Yurkow E J, McKenzie M A. Characterization of hypoxia-dependent peroxide production in cultures of Saccharomyces cerevisiae using flow cytometry: a model for ischemic tissue destruction. Cytometry. 1993;14:287–293. doi: 10.1002/cyto.990140309. [DOI] [PubMed] [Google Scholar]
- 346.Zhou D X, Taraboulos A, Ou J H, Yen T S. Activation of class I major histocompatibility complex gene expression by hepatitis B virus. J Virol. 1990;64:4025–4028. doi: 10.1128/jvi.64.8.4025-4028.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Zoeteweij J P, Eyes S T, Orenstein J M, Kawamura T, Wu L, Chandran B, Forghani B, Blauvelt A. Identification and rapid quantification of early- and late-lytic human herpesvirus 8 infection in single cells by flow cytometric analysis: characterization of antiherpesvirus agents. J Virol. 1999;73:5894–5902. doi: 10.1128/jvi.73.7.5894-5902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]