Abstract
Mitochondrial diseases (MDs) were a large group multisystem disorders, attributable in part to the dual genomic control. The advent of massively sequencing has improved diagnostic rates and speed, and was increasingly being used as a first-line diagnostic test. Paediatric patients (aged < 18 years) who underwent dual genomic sequencing were enrolled in this retrospective multicentre study. We evaluated the mitochondrial disease criteria (MDC) and molecular diagnostic yield of dual genomic sequencing. Causative variants were identified in 177 out of 503 (35.2%) patients using dual genomic sequencing. Forty-six patients (9.1%) had mitochondria-related variants, including 25 patients with nuclear DNA (nDNA) variants, 15 with mitochondrial DNA (mtDNA) variants, and six with dual genomic variants (MT-ND6 and POLG; MT-ND5 and RARS2; MT-TL1 and NARS2; MT-CO2 and NDUFS1; MT-CYB and SMARCA2; and CHRNA4 and MT-CO3). Based on the MDC, 15.2% of the patients with mitochondria-related variants were classified as “unlikely to have mitochondrial disorder”. Moreover, 4.5% of the patients with non-mitochondria-related variants and 1.43% with negative genetic tests, were classified as “probably having mitochondrial disorder”. Dual genomic sequencing in suspected MDs provided a more comprehensive and accurate diagnosis for pediatric patients, especially for patients with dual genomic variants.
Subject terms: Neurological disorders, Medical genetics
Introduction
Mitochondrial Diseases (MDs) are a group of complex inborn metabolic defects caused by mutations in nuclear DNA (nDNA) or mitochondrial DNA (mtDNA), with a prevalence of approximately 1 in 5,000 and a carrier rate of approximately 1 in 200 individuals1,2. In general, mitochondrial disorders in paediatric patients are mainly caused by nuclear-encoded genes with severe manifestations3. The majority of protein subunits and proteins that maintain mitochondrial structure and function are encoded by approximately 1500 nuclear genes4. Moreover, the expression of mtDNA genes is regulated by nDNA. nDNA variants could exacerbate mtDNA variants, resulting in multiple deletions in mtDNA, which are mostly seen in adults5,6. In addition, even typical phenotypes, such as Leigh syndrome (LS), identifiable by neuroimaging, could be caused by numerous nDNA or mtDNA variants7. Overall, the relationship between nDNA and mtDNA is extremely complicated and nDNA and mtDNA variants are difficult to be distinguished based on clinical data. Due to the clinically and genetically heterogeneous features, selection of genomic tests and interpretation of variants are still challenging.
Respiratory chain enzyme activity assays in muscle biopsy or skin fibroblasts have been widely used for MDs diagnosis. However, biopsy as an invasive method is less acceptable to parents of paediatric patients8,9. Therefore, the diagnosis process of “genotype first” has become popular. Forny et al. recommended histological testing in cases of critical illness to confirm the pathogenicity of variants of unknown significance and clinical MDs, wherein molecular diagnosis was negative, and remained highly suspicious10. With the development of high-throughput sequencing technologies and decreasing costs, whole-exome sequencing and mtDNA sequencing are convenient for molecular diagnosis of MDs. A non-invasive, bigenomic sequencing approach comprising whole exome and mtDNA sequencing has been recommended as the first step in identifying MDs11.
In this study, we retrospectively evaluated the MD criteria (MDC)12 and molecular diagnostic yield of dual genomic sequencing in 503 unrelated paediatric patients from 9 hospitals, aiming to summarize the efficiency of dual genomic sequencing and propose future directions for diagnostic approaches.
Materials and methods
Patient cohort
A total of 503 paediatric patients undergoing dual genomic analysis were enrolled from nine hospitals belonging to the Mitochondrial Consortium Unit between January 2017 and March 2021. These patients had an unexplained neuromuscular system or multisystem progressive disorder, suspected to be a mitochondrial disorder. Clinical data of the patients, including phenotypic information, neuroimaging, routine biochemical test results, and MDC scores, were retrospectively analysed. We assessed their molecular diagnosis based on whether the variants explained the clinical features and clinical matching between our patients and previously reported ones, combining the American College of Medical Genetics (ACMG) classification of variants and segregation pattern. Genes were classified as mitochondria-related if they were associated with MDs documented in the literature13 or non-mitochondria-related when this was not the case.
mtDNA sequencing
Peripheral blood samples were collected, and 3–5 μg of DNA was extracted using the salt extraction method with a DNA extraction kit, according to the manufacturer’s instructions (Mike Bio, China). Mitochondrial amplification was performed using mitochondria-specific primers14. Thereafter, the ultrasonic method was used to disrupt mitochondrial amplification products. Mitochondrial libraries were established using the KAPA HTP Library Preparation Kit (Kapa Biosystems Inc., Woburn, MA, USA). High-throughput sequencing was performed using a NextSeq 500 sequencer (Illumina, San Diego, CA, USA). The average sequencing depth was not less than 4000 × (Supplementary Table S1). After collecting data, bioinformatic analysis of the gene sequence was performed to confirm loci of pathogenic genes. The mtDNA variants were classified according to the mRNA15 and tRNA16 variant classification criteria.
nDNA sequencing
Genomic DNA extracted from whole blood of the patients and their parents was sequenced using Illumina HiSeq X sequencers or Illumina Novaseq platforms with at least Q20 base quality and > 30 × mean nuclear coverage (Supplementary Table S1Sequences were compared to the human reference genome using the NextGENe software (SoftGenetics, State College, Pa, USA) for true and false variant identification. Single-nucleotide variants were retained and annotated by filtering high-frequency variants using population frequency (dbSNP, ExAC, and gnomAD) and literature databases (OMIM, HGMD, ClinVar, and MasterMind). Multiple software (SIFT, Polyphen2, MutationTaster, and AlamutVisual) were used to predict pathogenicity based on amino acid conservation, evolutionary predictions, and splicing site effects. Variants were confirmed by Sanger sequencing and classified according to the ACMG and Genomics guidelines17.
CNV-sequencing
The experimental and analytical methods were performed as previously described18. Genomic DNA was extracted, followed by random fragmentation and short-read sequencing using an Illumina NextSeq500 or NovaSeq6000 sequencer (Illumina, San Diego, CA, USA). The resolution was 25 kb to 100 kb. Sequencing reads were cleaned by removing the reads when the base quality was less than Q20 and were mapped to the reference human genome version GRCh38/hg38. The high-variation regions, indicating highly homologous or repeated regions across different samples, were excluded from further analysis. The interpretation of copy number variations (CNVs) was based on the ACMG and ClinGen19.
Data analysis and statistics
Statistical analysis was performed using GraphPad Prism 8.0 software (San Diego, CA, USA). Statistically significant differences between groups were analyzed using one-way ANOVA. Statistical significance was defined as *P < 0.05, **P < 0.005, ***P < 0.001, and ****P < 0.0001.
Ethics approval and consent to participate//////
Ethical approval for this study was obtained from Xiangya Hospital Ethics Committee. Written informed consent was obtained from participants and parents or legal guardians of any participant under the age of 16. All methods were performed in accordance with the relevant guidelines and regulations.
Consent for publication
This study does not include any personal information leading to the identification of any participants.
Results
Dual genome test results
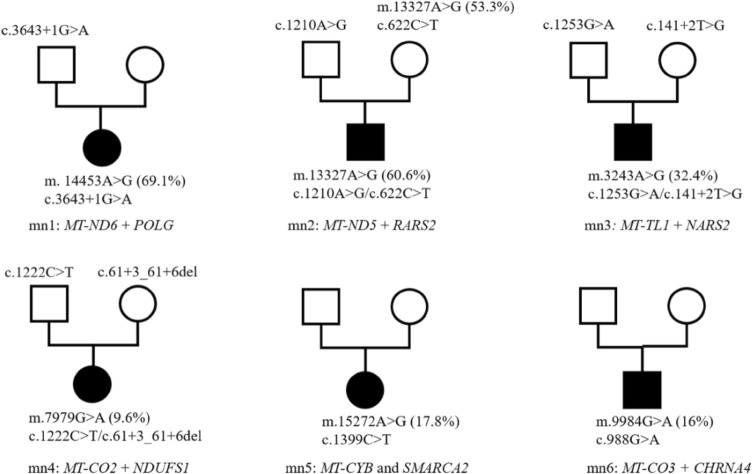
Among the 503 patients, mitochondria-related variants were identified in 46 (8.5%) (Table 1). In addition, 25 patients presented with variants in nDNA-encoded MDs genes, 15 with mtDNA variants, and six harboured both mitochondria-related nDNA and mtDNA variants. The nuclear-encoded MDs genes included POLG (n = 2), GTPBP3 (n = 2), ETFDH3 (n = 2), HIBCH (n = 2), AARS2, ACAD9, AIFM1, CARS2, COQ4, DNM1L, GFM1, HSD17B10, LIPT1, NAXE, OPA3, NDUFAF5, PANK2, PDHB, TWNK, WARS2, and PDHA1. Furthermore, mtDNA genes included MT-TL1(n = 7), MT-ATP6(n = 4), MT-ND1, MT-ND3, MT-ND4, MT-CO2 and a deletion m.10947-15362del. Four patients harboured dual genomic variants related to the mitochondria (Fig. 1), MT-ND6 and POLG; MT-ND5 and RARS2; MT-TL1 and NARS2; and MT-CO2 and NDUFS1. Two patients presented with mtDNA and non-mitochondria-related nDNA variants (Fig. 1): MT-CYB and MARCA2 and CHRNA4 and MT-CO3.
Table 1.
Summary of clinical features of patients with mtDNA or/and nDNA variants.
| Patient | Gender | Gene | Inheritance pattern | Variant | Zygosity | Origin | Age of onset (years) | Lactate (mmol/L) | MRI | Clinical features | ACMG | Evidence | Final clinical diagnosis | Novel mutation |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| nDNA | ||||||||||||||
| n6 | F | HIBCH | AR | c.1118A > G (p.N373S); c.810-4A > G | com.het | parental | 10.2 | 8.57 | Normal | Psychological and behavioral disorder | VUS/VUS | PM2 + PP3/PM2 + PP3 | Behavioral abnormality | Y/Y |
| n10 | M | PANK2 | AR | c.1355A > G (p.D452G) | hom | parental | 1 | 3.38 | Basal ganglia involvement | DD, ataxia, retinitis pigmentosa | VUS | PS1 + PP4 | Pantothenate kinase-associated neurodegeneration | N |
| n14 | F | AARS2 | AR | c.2682 + 5G > A; c.331G > C (p.A111P) | com.het | parental | 1 | 2.9 | Normal | Seizure, myoclonus, developmental delay, ataxia | VUS/VUS | PM2 + PP3/PM2 + PP3 | Epileptic encephalopathy | N/N |
| n22 | F | GFM1 | AR | c.2167 T > C (p.C723R); c.539delG (p.G180Afs*11) | com.het | parental | 7 | 5.83 | Basal ganglia involvement | Muscle weakness, dystonia | VUS/P | PM2 + PM3 + PP4/PVS1 + PS1 + PM2 | Leigh syndrome | Y/N |
| n26 | M | AIFM1 | XLR | c.1084A > C (p.K362Q) | hem | maternal | at birth | 9.39 | Thin corpus callosum | DD, hypertonia, microcephaly | VUS | PM2 + PP2 | GDD | Y |
| n33 | F | HSD17B10 | XLD | c.628C > G (p.P210A) | het | de novo | at birth | 10.22 | Normal | DD | LP | PS2 + PM2 + PP3 | HSD10 mitochondrial disease | Y |
| n20 | M | CARS2 | AR | c.323 T > G (p.F108C); c.1036C > T (p.R346W) | com.het | parental | 2 | 2.74 | Cortical atrophy | Seizure, psychological and behavioral disorder | VUS/VUS | PM2 + PP3/PM2 + PP3 | Epileptic encephalopathy | Y/Y |
| n54 | M | HIBCH | AR | c.958A > G (p.K320E); c.439-2A > G | com.het | parental | 1.5 | 6.3 | Basal ganglia involvement | Developmental regression, myoclonus, feeding difficulties | VUS/LP | PM2_P + PP3/ PVS1_M + PM3 + PP3 | 3-hydroxyisobutryl-CoA hydrolase deficiency | Y/Y |
| n59 | F | POLG | AR | c.2890C > T (p.R964C); c.2584G > A (p.A862T) | com.het | parental | 13 | 2.74 | Cortical abnormalities | Seizure, tremer, liver disorder | VUS/VUS | PM3 + PM2_P + PP3/ PM3 + PM2_P + PP3 | Alpers syndrome | N/N |
| n62 | M | COQ4 | AR | c.550 T > C (p.W184R); c.743 T > C (p.L248P) | com.het | parental | 2 | 3.31 | Basal ganglia involvement | DD, hypertonia, feeding difficulties | VUS/VUS | PM3 + PM2_P_PP3/PM2_P + PP3 | Primary coenzyme Q10 deficiency | Y/Y |
| n89 | F | OPA3 | AR | c.123C > G (p.I41M) | hom | parental | 1 | 1.23 | Cerebellar atrophy | DD, seizure, ataxia | VUS | PM2_P + PP3 | DD, seizure, ataxia | N |
| n98 | F | GTPBP3 | AR | c.187C > T (p.R63*); c.776A > G (p.N259S) | com.het | parental | at birth | 6.94 | Bilateral thalamus and left cortical involvement | DD, seizure | P/VUS | PVS1 + PM2.PP3/PM2 + PM3 + PP3 | Combined oxidative phosphorylation deficiency 23 | N/N |
| n108 | F | PDHA1 | XLD | c.901C > G(p.R301G) | het | de novo | 0.5 | 3 | Basal ganglia involvement | Muscle weakness | P | PS3 + PM2_P + PP3 + PP1_S | Pyruvate dehydrogenase E1-alpha deficiency | Y |
| n111 | M | WARS2 | AR | c.751 T > C (p.F251L) | hom | parental | 1 | 4.68 | Na | DD, seizure, tic disorder, ataxia, liver disorder | VUS | PM2_P + PP3 | NEMMLAS | Y |
| n115 | M | LIPT1 | AR | c.302G > A (p.S101N); c.316G > A (p.V106I) | com.het | parental | 4.6 | 1.5 | Involvement of brainstem and thalamus | DD, gastrointestinal disorder | VUS/VUS | PM2_P + PP3/ PM2_P + PP3 | Lipoyltransferase 1 deficiency | Y |
| n116 | M | ACAD9 | AR | c.1693-1G > A; c.1237G > A (p.E413K) | com.het | parental | 0.3 | 1.9 | Normal | Cardiovascular disorder, respiratory distress | VUS/LP | PVS1_M + PM2_P/PS3 + PM3 + PM2_P + PP3 | Mitochondrial complex I deficiency | Y |
| n126 | F | PDHB | AR | c.97-3C > G | hom | parental | 2 | 8.67 | White matter abnormalities | Ptosis, DD, seizure | VUS | PM2_P + PP3 | Pyruvate dehydrogenase E1-beta deficiency | Y |
| n129 | F | DNM1L | AD | c.1207C > T (p.R403C) | het | de novo | 4.4 | 3.3 | Involvement of brainstem and thalamus | Seizure, seizure status, coma | P | PS3 + PS2_M + PM2_P + PP1_M + PP3 | Encephalopathy, lethal, due to defective mitochondrial peroxisomal fission 1 | N |
| n130 | M | NAXE | AR | c.473G > C (p.C158S); c.490C > A (p.P164T) | com.het | parental | 2 | Na | White matter abnormalities | DD, seizure | VUS/VUS | PM2_P + PP3/ PM2_P + PP3 | Encephalopathy, progressive, early-onset, with brain edema and/or leukoencephalopathy | Y |
| n155 | M | POLG | AR | c.2890C > T (p.R964C); c.2584G > A (p.A862T) | com.het | parental | 5 | Na | Cortical abnormalities | Seizure, muscle weakness, headache | VUS/VUS | PM3 + PM2_P + PP3/ PM3 + PM2_P + PP3 | Alpers syndrome | Y |
| n167 | M | GTPBP3 | AR | c.413C > T (p.A138V); c.509_510delAG (p.E170Gfs*42) | com. het | parental | at birth | 26 | Na | respiratory failure | VUS/LP | PM3 + PM2_P + PP3/ PVS1 + PM2_P | Combined oxidative phosphorylation deficiency 23 | Y |
| n179 | M | ETFDH | AR | c.886G > T (p.G296C); c.1773_1774delAT (p.C592*) | com.het | parental | 0.5 | 2 | Normal | Muscle weakness, gastrointestinal disorder, impaired vision | VUS/VUS | PM2_P + PP3/ PVS1_M + PM2_P | Multiple acyl-CoA dehydrogenase deficiency |
N/ N |
| n180 | M | NDUFAF5 | AR | c.752 T > G (p.M251R); c.155A > C (p.K52T) | com.het | parental | 0.6 | 4.61 | White matter abnormalities | Developmental regression | LP/LP | PM3_S + PM2_P + PP3/ PM3_S + PM2_P + PP3 | Cavitating Leukoencephalopathy | N |
| 2n4 | M | ETFDH | AR | c.250G > A (p.A84T); c.2 T > G (p.M1?) | com.het | parental | 0.5 | Na | Na | Muscle weakness, liver disorder, gastrointestinal disorder | LP/VUS | PS3 + PM2 + PP3 + PPM2 + PP3 | Combined oxidative phosphorylation deficiency 23 | N/N |
| 2n5 | M | TWNK | AD | c.1421G > C (p.W474S) | het | maternal | at birth | 5 | Na | Ptosis | LP | PM2_P + PP3 + PM5 + PM6 | Congenital myasthenic syndrome | Y |
| CHRNB1 | AD | c.1394 T > C (p.M465T) | het | maternal | VUS | - | Y | |||||||
| mtDNA | ||||||||||||||
| m1 | M | MT-TL1 | maternal | m.3243A > G (71.2%) | 71.20% | maternal (23.6%) | 14 | Na | Occipital cortex involvement with calcification | Seizure, headache, vomiting | P | PM2 + PP3-B + PS5 + PS2 + PS3 | MELAS | N |
| m3 | F | MT-TL1 | maternal | m.3243A > G | 66.20% | maternal (24.0%) | 6.3 | 6.47 | Cortical and basal ganglia involvement | Seizure, headache, vomiting | P | PM2 + PP3-B + PS5 + PS2 + PS3 | MELAS | N |
| m4 | F | MT-TL1 | maternal | m.3243A > G | 71.70% | maternal (24.4%) | 4.7 | 9.33 | Cortical, basal ganglia and thalamus involvement | Seizure, behavioral disorder | P | PM2 + PP3-B + PS5 + PS2 + PS3 | MELAS | N |
| m6 | F | MT-ND3 | maternal | m.10197G > A | 99.60% | de novo | 0.5 | 3.5 | Basal ganglia, thalamus and brainstem involvement | Developmental regression, seizure | LP | PP3-B + PP4 + PM9 + PM8 | Leigh syndrome | N |
| m8 | M | MT-TL1 | maternal | m.3243A > G | 66.10% | maternal (24.4%) | 1 | 6.5 | Basal ganglia involvement | Seizure, ID, diabetes | P | PM2 + PP3-B + PS5 + PS2 + PS3 | Leigh syndrome | N |
| m9 | M | MT-ATP6 | maternal | m.9176 T > C | 99.50% | maternal (88.2%) | 8 | 4.49 | Basal ganglia and brainstem involvement | Ptosis, muscle weakness, dysuria, tachycardia, tachypnea | P | PP3-A1 + PP3-B + PS1 + PM5 + PM9 + PM10 + PP4 | Leigh syndrome | N |
| m10 | M | MT-TL1 | maternal | m.3243A > G | 72.40% | de novo | 12.6 | 7.33 | Cortical abnormalities | Seizure | P | PM2 + PP3-B + PS5 + PS2 + PS3 | MELAS | N |
| m12 | M | MT-TL1 | maternal | m.3243A > G | 74.20% | maternal (10.2%) | 8.7 | 4.73 | Cortical abnormalities | Headache, vomiting, impaired vision | P | PM2 + PP3-B + PS5 + PS2 + PS3 | MELAS | N |
| m13 | M | MT-ND1 | maternal | m.3761C > A | 81.40% | 0% | 0.6 | 5.31 | Ventriculomegaly | West syndrome | LP | PM2 + PM9 | West syndrome | N |
| m14 | M | mtDNA (tissue) | maternal | m.10947-15362del | / | Na | 11 | 3.68 | Na | Ptosis, growth restriction, skeletal muscle biopsy showing ragged red fibers | P | PM2 + PVS1 + PP4 | KSS | Y |
| m16 | F | MT-TL1 | maternal | m.3243A > G | 66.40% | de novo | 8 | 6.69 | Basal ganglia involvement | Developmental delay | P | PM2 + PP3-B + PS5 + PS2 + PS3 | Leigh syndrome | N |
| m17 | M | MT-CO2, MT-ATP6 | maternal | m.7929G > A; m.9035 T > C | 13.9%, 99.5% | de novo, de novo | 1 | 3.3 | Basal ganglia and brainstem involvement | Muscle weakness, hearing loss | VUS/LP | PM2 + PM9/PM2 + PM9 + PP4 | Leigh syndrome | Y |
| m19 | M | MT-ATP6 | maternal | m.9176 T > C | 99.80% | maternal (99.2%) | 7 | 2.07 | Abnormal signal foci around the midbrain aqueduct | ptosis, dysuria | P | PP3-A1 + PP3-B + PS1 + PM5 + PM9 + PM10 + PP4 | Leigh syndrome | N |
| mn6 | M | MT-ATP6 | maternal | m.8993 T > C | 99.50% | maternal (10.0%) | 1.7 | 3.3 | Basal ganglia involvement | muscle weakness, easy fatigability | P | PM2 + PP3-B + PS3 + PM5 + PS2 | Leigh syndrome | N |
| mn9 | M | MT-ND4 | maternal | m.11778G > A | 99.40% | maternal (99.4%) | 0.6 | 3 | Normal | Vision loss | P | PP3-A1 + PP3-B + PS1 + PS3 | LHON | N |
| mtDNA + nDNA | ||||||||||||||
| mn1 | F | MT-ND6 | maternal | m.14453A > G | 69.10% | de novo | 4.9 | 6.2 | Cortical and basal ganglia involvement | Seizure | P | PM2 + PP3-B + PS4_M + PS2 | MD | N |
| POLG | AD/AR | c.3643 + 1G > A | het | paternal | VUS | PM2_P + PVS1_M | Y | |||||||
| mn2 | M | MT-ND5 | maternal | m.13327A > G | 60.60% | maternal (53.3%) | at birth | 4.13 | Ventriculomegaly | DD, seizure, microcephalus | VUS | BS1 + BS4 | EIEE | Y |
| RARS2 | AR |
c.1210A > G (p.M404V); c.622C > T (p.Q208*) |
com. het | parental | VUS/LP | PM2_P + PP3-B2(+ PP4) / PM2_P + PVS1(+ PP4) | Y/Y | |||||||
| mn3 | M | MT-TL1 | maternal | m.3243A > G | 32.40% | de novo | 2 | 1.67 | Normal | Developmental regression, seizure, hearing loss | P | PM2 + PP3-B + PS5 + PS2 + PS3 | MD | N |
| NARS2 | AR | c.1253G > A (p.R418H); c.141 + 2 T > G | com. het | parental | VUS/LP | PM2_P + PP3/ PVS1 + PM2_P | N/Y | |||||||
| mn4 | F | MT-CO2 | maternal | m.7979G > A | 9.60% | de novo | at birth | 6.52 | Basal ganglia and brainstem involvement, ventriculomegaly | Muscle weakness, growth restriction | VUS | PM2 + BP4 | Leigh syndrome | Y |
| NDUFS1 | AR | c.1222C > T(p.R408C); c.61 + 3_61 + 6delGAGT | com.het | parental | VUS/VUS | PM3_S + PM2_P + PP3/ PM2_P + PP3 | N/Y | |||||||
| mn5 | M | MT-CYB | maternal | m.15272A > G | 17.80% | de novo | 1.9 | 6.81 | Hippocampus involvement | Development regression, seizure, dysarthria | VUS | PP3-B | Epileptic encephalopathy | N |
| SMARCA2 | AD | c.1399C > T (p.R467W) | het | de novo | LP | PS2 + PM2_P + PP3 | Y | |||||||
| mn6 | F | CHRNA4 | AD | c.988G > A (p.V330M) | het | paternal | 2.1 | 1.31 | Normal | seizure | VUS | PM2_P + PP3 | Epilepsy | Y |
| MT-CO3 | maternal | m.9984G > A | 16% | de novo | VUS | PP7 + PS6 | N | |||||||
F, female; M, male; Na, not available; hom. homozygote; het, heterozygote; com. het. compound heterozygote; hem, hemizygote; DD, development delay; ID, intellectual disorder; NEMMLAS, neurodevelopmental disorder, mitochondrial, with abnormal movements and lactic acidosis, with or without seizures; GDD, global developmental delay; MELAS, mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes; LHON, Leber's hereditary optic neuropathy; MD, mitochondrial disease; EIEE, early onset epileptic encephalopathy; KSS, Kearns-Sayre syndrome; ACMG, American College of Medical Genetics and Genomics; P, pathogenic; LP, likely pathogenic; VUS, variant of uncertain significance.
Family history: n111, father had seizure; 2n5, mother, mother's sister, father, two father's sisters, and grandmother have ptosis; m8, mother had diabetes.
Figure 1.
Family pedigree of six patients with dual genomic variants.
131 patients (26.0%) were found to have non-mitochondria-related variants, including 13 patients with pathogenic CNVs (Supplementary Table S2 and S3). Dual genomic analysis failed to identify definitive causative variants in 326 patients (64.8%). A few patients harboured low level of heteroplasmic mtDNA variants in blood, such as m.4358G > A (8.60%), m.10437G > A (blood 3.00%, urine 0%), m.594insC (21.80%), m.15867A > G (9.20%), m.5703G > A (2.50%), and m.3250 T > C (6.60%). Some variants of uncertain significance, such as PMP22 (c.235 T > C) in infantile spasms, KCNH5 (c.976G > T) in epilepsy, and SEMA3E, PLEC in suspected MDs.
Patient demographics and clinical presentations
There was a male preponderance in 299 boys (59.4%) and 204 girls (40.6%) were included. A male to female ratio of 2.75 was observed in patients with mtDNA variants (Table 2). The median age of onset was 6.3 years (range, 0.5–14 years) in patients with mtDNA variants and one year (range, 0–13 years) in those with mitochondria-related nDNA variants. The patients with non-mitochondria-related variants (median, 0.5 years) and negative genetic test (median, one year) had an even younger age at first presentation than did those with mtDNA defects. The first symptoms included seizures; developmental delay or regression; muscle weakness; failure to thrive; ptosis; vomiting; increased alanine transaminase/aspartate transaminase; ataxia; headache; and visual signs. Seizures and developmental delay were the two most common symptoms in mitochondrial and non-mitochondrial patients in our cohort.
Table 2.
Summary of demographics and clinical presentations in this study.
| Groups | Mitochondria-related | Non-mitochondria-related | Negative | ||
|---|---|---|---|---|---|
| mtDNA | nDNA | mtDNA + nDNA | |||
| Gender | |||||
| Male | 11 | 15 | 3 | 66 | 204 |
| Female | 4 | 10 | 3 | 65 | 122 |
| Median age of onset (years) | 6.3 | 1.0 | 1.2 | 0.5 | 1.0 |
| Serum lactate (Mean ± SD, mmol/L) | 5.0 ± 2.0 | 5.7 ± 5.4 | 4.4 ± 2.5 | 3.3 ± 3.28 | 2.80 ± 2.02 |
The serum lactate level in patients with mtDNA, mitochondria-related nDNA, and dual genomic variants were 5.0 ± 2.0 mmol/L, 5.7 ± 5.4 mmol/L, and 4.4 ± 2.5 mmol/L, respectively. The patients with non-mitochondria-related variants and negative genetic tests showed a lower lactate level of 3.3 ± 3.28 mmol/L and 2.80 ± 2.02 mmol/L, respectively (Table 1). Compared with the patients with negative genetic result, those with mtDNA (P = 0.0076) or mitochondria-related nDNA (P = 0.0483) variants showed significantly higher lactate level. In addition, the patients with mitochondria-related nDNA variants had significantly higher lactate level than that with non-mitochondria-related nDNA variants (P = 0.0076).
Neuroimaging data were available in 386 patients, 177 of whom exhibited normal brain magnetic resonance imaging (MRI) findings. The most frequent neuroradiological presentations included white matter (14.0%), basal ganglia (9.2%), cortical abnormalities (6.1%), cerebral atrophy (5.6%), cerebellar atrophy (3.9%), corpus callosum (3.6%), and brainstem and thalamic (2.9%) involvements. Less common presentations include optic atrophy, ventriculomegaly, and cysts. Basal ganglia characteristics were observed in the most mitochondrial patients (32.6%).
Mitochondrial disease criteria
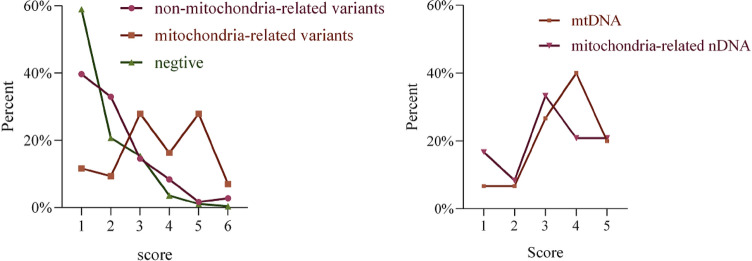
According to the MDC, 15.2%, 52.2% and 32.6% of the patients with mtDNA or mitochondria-related nDNA variants, were classified into “unlikely to have mitochondrial disorder” (score 1), “possibly having mitochondrial disorder” (score 2 to 4) and “probably having mitochondrial disorder” (score 5 to 7), respectively (Fig. 2). A comparison of the MDC scores between mtDNA and mitochondria-related nDNA variants is shown in Fig. 2. 42.6% and 18.7% of patients whose causative variants were not detected were determined as “unlikely to have mitochondrial disorder” and “possibly having mitochondrial disorder”. In patients with non-mitochondria-related variants, the corresponding percentages were 39.0% and 39.0%. A few confirmed mitochondrial cases had low MDC scores due to an organ-specific presentation, such as Leber hereditary optic neuropathy (LHON), progressive external ophthalmoplegia, cerebellar ataxia, or a single clinical symptom, such as epileptic encephalopathy without characteristic MRI or lactate level findings. The importance of the parts that might have been missed because of low scores should be noted.
Figure 2.
Percentages distribution of different groups referring to MDC scores. Left, comparison of three groups including negative, mitochondria-related and non-mitochondria-related nDNA variants. Right, comparison between mtDNA and mitochondria-related nDNA variants.
In addition, there were a few noteworthy genes in the patients with high MDC scores. A few cases of non-mitochondria-related variants were evaluated as probably having mitochondrial disorders, such as variants in MMACHC, MCEE, FOLR1 and G6PC genes, which were grouped as causative genes of metabolic diseases affecting multiple systems.
A few cases of high MDC scores in which confirmed causative variants could not be detected require further study. An eight-year-old girl presenting with developmental delay, vomiting, and abnormal behaviour whose examination showed hypoglycaemia, high serum lactate (5.04 mmol/L), bilateral basal ganglia involvement, and a de novo variant c.760G > C (p.E254Q) of SEMA3E gene was identified. A girl with a ventricular sepal defect presented with seizures after fever, and examination showed CK 5833U/L, CKMB 369.9U/L, LDH 1876U/L, and bilateral basal ganglia involvement on brain MRI. However, no mitochondria-related variant other than the PLEC gene with c.10947C > A (p.F3649L) and c.7603C > T (p.R2535C) was identified.
Discussion
A two-step next-generation sequencing approach, by which patients were evaluated based on whole exome sequencing after excluding pathogenic variants in the mtDNA, was frequently used in previous cohort20,28. Simultaneous sequencing of the dual genome, including mtDNA genome-wide sequencing combined with whole-exon sequencing of trios, is not common in paediatric cohorts of MDs. Recently, whole-genome sequencing has been used to determine the gene-disease association of MDs21,22. Dual genomic sequencing was a choice before the widespread use of whole genome sequencing, which was helpful for comprehensive diagnosis and establishment of a mitochondrial disease knowledge base23.
MDs in children is mainly caused by nDNA variation, accounting for 75–95%24,25. In China, mtDNA variations accounted for the main part (63%) of MDs in children in a previous study26. In addition, similar proportion was found in our previous local cohort27. The explanation might be the selection bias of the detection method and limitation of targeted gene panel sequencing in previous years. Because conditional MDs such as LS, mitochondrial encephalomyopathy with lactate acidosis and stroke-like episodes (MELAS), and LHON are relatively easy to catch the attention of physicians, targeted gene panel or mtDNA hotspot tests are unlikely to detect novel genes or variants. In this study, dual genomic sequencing provided more accurate data that nDNA variants (25/46) accounted for a larger proportion than did mtDNA variants (15/46) in MDs.
Here, we describe the findings in 503 routine diagnostic nDNA sequencing combined with mtDNA genome-wide sequencing in patients with a suspected MD. Overall, a molecular diagnosis was established in 177 patients (34.5%) in this cohort, which was similar to a finding in a previous study with more stringent inclusion criteria28. Similarly, 31% of participants with suspected MDs under 18 years old received a molecular diagnosis by using whole genome sequencing22. In a previous study, 5.3%, 37.2%, and 16.8% of 113 patients with suspected MD were identified with mtDNA, mitochondria-related nDNA, and non-mitochondria-related nDNA variants, respectively29. In 177 patients with molecular diagnosis, a lower percentage of mitochondria-related (26%) and higher percentage of non-mitochondria-related (74%) variants were detected in this cohort. This ratio was similar with a previous study22, wherein 37% (28/75) of patients with a definite diagnosis were in genes known to cause primary mitochondrial disease, including four mtDNA variants and 24 diagnoses in nuclear-mitochondrial genes, whereas 63% (47/75) were in non-mitochondrial genes. A precise comparison of overall diagnostic rates with previous studies is difficult, given the existence of several biases that affect the diagnostic rate, including prior mtDNA/nDNA genetic screening, population characteristics, and phenotyping accuracy. This cohort consists of patients from nine hospitals. The clinical suspicion for a mitochondrial disease of the patients varied from relatively low to very high and therefore the cohort represents the heterogeneous group of suspected mitochondrial patients. First and foremost, this is a retrospective cohort study, and it is not possible to control for factors that led to the selection of certain genetic testing methods. The diagnostic rate varied depending on the presenting clinical phenotype30. The highest diagnostic rates were achieved when patients with suspected MD presented with clear clinical phenotypes. It is well known that the percentage of detected MD-related genes could increase with increased MDC scores29. The limited diagnostic rate could be associated with relative low MDC scores in our cohort, since the diagnosis process of “genotype first” was performed widely, lacking of biochemical or histopathological supports. Median age of onset was 0.9 (0–13) years in this cohort. Genetic tests were performed rapidly and broadly in infants and children with a suspected hereditary disease30.
Non-mitochondrial disorders were more common than mitochondrial disorders and had features resembling mitochondrial diseases (often referred to as phenocopies). These could be broadly classified as developmental disorders with intellectual disability, metabolic disorders, myopathies, cardiomyopathies, epileptic encephalopathies, leukodystrophies, ciliopathies, amyloidosis, and other neurogenetic disorders, including basal ganglia calcification and neurodegeneration with iron accumulation. The MDC system was built by summarising known typical MDs and guiding the selection of diagnostic methods. A high MDC score is a hallmark of an MD diagnosis. Similarly in the study, there is a correlation between the degree of MD diagnosis and MDC score.
Moreover, a growing number of new nDNA and mtDNA variants have been found in patients with atypical clinical presentations or isolated symptoms. Developmental, neurological, and metabolic abnormalities are common manifestations in other metabolic diseases, leading to a high MDC score. In this cohort, most of the diagnoses were non-mitochondrial disorders, including developmental disorders, epilepsies, myopathies, and other multisystem disorders. In a previous study, 34 patients had a probable or definite mitochondrial disorder according to the modified MDC score (between 5 and 12), of these, 18% having a non-mitochondrial disorder22. Regarding MD mimics, combination of nDNA and mtDNA sequencing was able to detect variants in a more comprehensive perspective, thereby allowing diagnosis and differentiation of patients with other treatable conditions, enabling appropriate care and treatment for their respective diseases, while ruling out MD. As observed in our cohort, methylmalonic acidemia (MMACHC, MCEE), glycogen storage disease (G6PC), cerebral folate deficiency (FOLR1), and MDs had many overlapping features. As sequencing costs decreased and genetic sequencing became readily available, genetic tests were more acceptable than invasive muscle biopsies. After genetic tests giving a direction, these genetic metabolic diseases could be confirmed by further diagnostic tests, such as gas chromatography and mass spectrometry and liquid chromatography-tandem mass spectrometry. SEMA3E and PLEC were identified in two patients with high MDC scores. Based on the existing research, these two genes could not completely account for the symptoms observed in two patients.
The mtDNA variants had an older age of onset and higher lactate levels in this cohort. However, these were not definite guidelines for the selection of genomic tests. The distributions of MDC scores in mtDNA and mitochondria-related nDNA variants were not significantly different. Selecting mtDNA or nDNA sequences based on clinical features or MDC scores was difficult. Regarding patients with dual genomic variants, selecting candidate variants and interpreting their disease-causing roles were even more challenging. Nuclear-mitochondrial intergenomic communication disorders, which result in loss or instability of the mitochondrial genome and, in turn, impaired maintenance of qualitative and quantitative mtDNA integrity.
In the mn1 patient (MDC: 5), dual genomic variants of MT-ND6 (m.14453A > G, 69.1%) and POLG (c.3643 + 1G > A) were tested. DNA polymerase errors have a prominent role in mtDNA mutation, arising through spontaneous errors of DNA replication or through unrepaired damage to mtDNA that introduces miscoding lesions22. It is already known that the POLG gene, responsible for the replication of mtDNA, could lead to depletion of mtDNA and/or accumulation of multiple mtDNA deletions31. And in a mice model with Polg mutation, the frequency of mutations was found to be 500-fold higher in heterozygous mice and 2,500-fold higher in homozygous mice than in aged wild-type mice22. However, regarding this patient, c.3643 + 2G > A had been reported in trans with another pathogenic variant in a child with Alpers syndrome32. Therefore, c.3643 + 1G > A was suspected to have an autosomal recessive inheritance. Since m.14453A > G, with a heteroplasmy of 69.1%, was a reported cause of LS and MELAS, and classified as “pathogenic”. mtDNA variant might be considered to be finally responsibe in this patient.
Guan et al. identified that mutant cell lines harbouring both m.7511A > G and YARS2 mutations exhibited a greater deficiency in mitochondrial function33, and overexpression of HARS2 in cytoplasmic hybrid cells carrying the m.12201 T > C mutation reversed mitochondrial dysfunctions34. It is possible that an interaction occurred between nDNA and mtDNA variants, rather than a simple single-gene disorder, since all NARS2, RARS2, HARS2 and EARS2 belong to a large gene family of aminoacyl tRNA synthetases that function as a translation of mtDNA. Although we tried to analyse the major causative genes between mtDNA and nDNA, some cases were still ambiguous, such as NARS2 and MT-TL1 (m.3243A > G) in the mn3 patient (MDC: 2). For m.3243A > G, it has been reported that patients with higher heteroplasmy levels of this point mutation exhibit Mitochondrial Encephalomyopathy, Lactic Acidosis and Stroke-Like Episodes (MELAS), while those with low heteroplasmy levels mainly have maternally inherited diabetes and deafness (MIDD). A study conducted a long-term hearing evaluation in patients with MELAS or MIDD who harbored the m.3243A > G mutation of mitochondrial DNA. They identified the age of onset of hearing loss was correlated with the heteroplasmy, and the mean age of onset of hearing loss was 28.6 years. However, hearing loss occurred earlly in our patient, and he was treated with cochlear implantation at three years old. Mutations in NARS2 are associated with combined oxidative phosphorylation deficiency 24 and autosomal recessive deafness 94. Seizures and hearing impairments were most common in the clinical findings of NARS2 patients35. A previous study reported a patient with novel NARS2 variants, causing infantile-onset severe epilepsy. The patient had continuous bilateral clonic and myoclonic seizures and paroxysmal episodes of upward eye deviation. He had normal serum and CSF lactate levels. And initially the brain MRI was normal, repeated MR imaging showed progressive cortical and periventricular brain atrophy. However, Finsterer commented that investigations of the mDNA should be performed to tell if the NARS2 variants altered the mtDNA sequence or resulted in mtDNA depletion. As observed in our patient, developmental regression, seizure, and hearing loss could be explaind perfectly by NARS2 variants, however, investigations of the mDNA found another variant, m.3243A > G, which was not observed in his mother. We could not tell if m.3243A > G was caused by NARS2 variants, and if these clinical manifestation was attributed to m.3243A > G. The mn2 patient (MDC: 4) harboured MT-ND5 (m.13327A > G, 60.60%) and RARS2 (c.1210A > G, c.622C > T). However, m.13327A > G was interpretated to be a benign variant based on the modified ACMG guidelines (BS1, BS4)15; RARS2 was more likely to cause early onset epileptic encephalopathy in this patient. Similarly.
The mn4 patient (MDC: 4) harbored a mtDNA variant, m.7979G > A, and two variants in NDUFS1 (c.1222C > T, c.61 + 3_61 + 6delGAGT). The m.7979G > A is located in MT-CO2, encoding a subunit of Complex IV, and NDUFS1 encodes a Fe-S protein operating within complex I. Variants in MT-CO2 and NDUFS1 are associated with Leigh syndrome. The clinical manifestation of muscle weakness, growth restriction, and brain MRI with basal ganglia and brainstem involvement, can be explained by the two genes. A overlapping effect could not be comfirmed. NDUFS1 was assessed as major pathogenic variants, however, the m.7979G > A, as a noval variant, can not be fully ruled out though at a low-level heteroplasmy.
In regard to the other two patients with mtDNA and non-mitochondria-related nDNA variants, judging whether they could be diagnosed with MDs was difficult. The patient with MT-CYB (m.15272A > G, 17.8%) and de novo SMARCA2 (c.1399C > T) variants presented with psychomotor regression, seizure, dysarthria, and high serum lactate 6.81 mmol/L. Although this patient had auricular deformity, he lacked distinctive facial appearance of Nicolaides-Baraitser syndrome or Blepharophimosis-impaired intellectual development syndrome caused by SMARCA2 variants36. The m.15272A > G was reclassified as “uncertain significance” according to modified ACMG15. MT-CYB is associated with Leigh syndrome, however, this patient did not harbored typical clinical features of Leigh syndrome. Existing clinical data and low-level heteroplasmy of m.15272A > G could not perfectly be connected. Another patient harboured MT-CO3 (m.9984G > A, 16%) and CHRNA4 (c.988G > A) variants. m.9984G > A was reported in a patient with suspected MD37 and reclassified as “uncertain significance” by Wong et al.15. CHRNA4 is assosited with nocturnal frontal lobe epilepsy. Seizure, as the main presentation in our patient, did not provide definitive evidence.
A few cases of the coexistence of mutations in nDNA and mtDNA have been reported in a previous study. However, the lack of functional data has confused the digenic mechanisms. The relationship between nDNA and mtDNA was determined by observing attributing phenotypes, such as KRT10 and MT-ND638, MT-ND1/MT-ND4 and OPA1, MT-ND1/MT-ND4 and RTN4IP1, ND4 and TMEM126A39. A single case could not easily distinguish the contribution of two genes, particularly both causative genes of similar MDs, as in our patients. By observing the co-segregation phenomenon and calculating the minor allele frequency of variants and prevalence of disease, a study further explored the digenic mechanisms and defined the contribution of TRMU and EARS2 heterozygous variants to the clinical manifestation of the disease caused by m.14674 T > C40. Currently, to identify recurrence in similarly affected, unrelated patients, further studies are required to determine its likely clinical significance.
Kerr et al. found that whole-exome sequencing provided a second diagnosis in two patients who already had a pathogenic variant in mtDNA. Notably, many variants purported to be causal of disease may, in some cases, be of unknown significance or benign polymorphisms11. Recently, whole-genome sequencing has been recommended for early diagnosis in a patient’s local secondary or tertiary care centre and before invasive tests such as muscle biopsy22. At present, whole genomic sequencing is not easy to use for clinical diagnosis due to the massive amount of data it yields, and it is difficult to select candidate variants and interpret their disease-causing roles. The combination of nDNA and mtDNA sequencing is alternative for suspected MD patients.
This study had several limitations. This was a preliminary study, and the overall cohort was not followed up consistently. Variants of unknown significance in nDNA and mtDNA genes needed further tests and functional studies to verify. Our findings highlighted the importance of dual genomic sequencing for pediatric patients before widespread use of whole genome sequencing. Then, in combination with biochemical testing and muscle histology studies correctly, molecular diagnosis would be more definitive. Next, follow-up of cases and data reanalysis based on dual genomic sequencing, combined with transcriptome sequencing, could significantly improve the predictive probability of MDs and identify underlying pathogenic genes8.
Conclusion
This study identified six patients with dual genomic variants in mtDNA and nDNA and demonstrated variants of non-mitochondria-related nDNA and variants of unknown significance in patients with high MDC scores. The MDC could guide the implementation of dual genomic sequencing in routine diagnostics. Esipecially, for patients assessed as“possibly having mitochondrial disorder” and “probably having mitochondrial disorder”, dual genomic sequencing provided a more comprehensive research strategy for identifiying patients with dual genomic variants and informing the risk of genetic transmission.
Supplementary Information
Acknowledgements
The authors thank all the patients. This work was supported by the National Natural Science Foundation of China (Grant number 82071462).
Author contributions
Material preparation and data collection were performed by T.H.W., L.Y., F.H., Y.H.C., X.L.L., J.T. H., J.Y.Y., W.X. O.Y., Y.Y.S., Y.N.X., X.M., H.M.Y., R.N.R., J.X. and Z.H.C. Statistical analysis were performed by T.H.W. and J.P. Writing of the first draft were performed by T.H.W. and G.Z.L. Study design was performed by F.H. Manuscript revision was performed by J.P. and V.W.Z.
Funding
This work was supported by the National Natural Science Foundation of China (Grant number 82071462).
Data availability
The analyzed clinical and genetic data generated during the study are available from the corresponding author on reasonable request. The mtDNA and nDNA variants of patiens with mitochondrial disease are deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar; accession numbers: SCV002760197 - SCV002760205, SCV002761197 - SCV002761232). The raw sequencing data are not publicly available due to restriction of human data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-31134-5.
References
- 1.Ferreira CR, van Karnebeek C, Vockley J, Blau N. A proposed nosology of inborn errors of metabolism. Genet. Med. 2019;21:102–106. doi: 10.1038/s41436-018-0022-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smeitink JA, Zeviani M, Turnbull DM, Jacobs HT. Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Gibson K, Halliday JL, Kirby DM, Yaplito-Lee J, Thorburn DR, Boneh A. Mitochondrial oxidative phosphorylation disorders presenting in neonates: Clinical manifestations and enzymatic and molecular diagnoses. Pediatrics. 2008;122:1003–1008. doi: 10.1542/peds.2007-3502. [DOI] [PubMed] [Google Scholar]
- 4.Rath S, Sharma R, Gupta R, Ast T, Chan C, Durham TJ, et al. MitoCarta3.0: an updated mitochondrial proteome now with sub-organelle localization and pathway annotations. Nucleic Acids Res. 2021;49:D1541–D1547. doi: 10.1093/nar/gkaa1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saneto RP, Sedensky MM. Mitochondrial disease in childhood: mtDNA encoded. Neurotherapeutics. 2013;10:199–211. doi: 10.1007/s13311-012-0167-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, et al. Human mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 7.Rahman J, Noronha A, Thiele I, Rahman S. Leigh map: A novel computational diagnostic resource for mitochondrial disease. Ann. Neurol. 2017;81:9–16. doi: 10.1002/ana.24835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stenton SL, Prokisch H. Genetics of mitochondrial diseases: Identifying mutations to help diagnosis. Ebiomedicine. 2020;56:102784. doi: 10.1016/j.ebiom.2020.102784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wortmann SB, Mayr JA, Nuoffer JM, Prokisch H, Sperl W. A guideline for the diagnosis of pediatric mitochondrial disease: The value of muscle and skin biopsies in the genetics era. Neuropediatrics. 2017;48:309–314. doi: 10.1055/s-0037-1603776. [DOI] [PubMed] [Google Scholar]
- 10.Forny P, Footitt E, Davison JE, Lam A, Woodward CE, Batzios S, et al. Diagnosing mitochondrial disorders remains challenging in the omics era. Neurol Genet. 2021;7:e597. doi: 10.1212/NXG.0000000000000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerr M, Hume S, Omar F, Koo D, Barnes H, Khan M, et al. MITO-FIND: A study in 390 patients to determine a diagnostic strategy for mitochondrial disease. Mol. Genet. Metab. 2020;131:66–82. doi: 10.1016/j.ymgme.2020.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Morava E, van den Heuvel L, Hol F, de Vries MC, Hogeveen M, Rodenburg RJ, et al. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67:1823–1826. doi: 10.1212/01.wnl.0000244435.27645.54. [DOI] [PubMed] [Google Scholar]
- 13.Thompson K, Collier JJ, Glasgow R, Robertson FM, Pyle A, Blakely EL, et al. Recent advances in understanding the molecular genetic basis of mitochondrial disease. J. Inherit. Metab. Dis. 2020;43:36–50. doi: 10.1002/jimd.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang W, Cui H, Wong LJ. Comprehensive one-step molecular analyses of mitochondrial genome by massively parallel sequencing. Clin. Chem. 2012;58:1322–1331. doi: 10.1373/clinchem.2011.181438. [DOI] [PubMed] [Google Scholar]
- 15.Wong LC, Chen T, Schmitt ES, Wang J, Tang S, Landsverk M, et al. Clinical and laboratory interpretation of mitochondrial mRNA variants. Hum. Mutat. 2020;41:1783–1796. doi: 10.1002/humu.24082. [DOI] [PubMed] [Google Scholar]
- 16.Wong LC, Chen T, Wang J, Tang S, Schmitt ES, Landsverk M, et al. Interpretation of mitochondrial tRNA variants. Genet. Med. 2020;22:917–926. doi: 10.1038/s41436-019-0746-0. [DOI] [PubMed] [Google Scholar]
- 17.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qi Q, Jiang Y, Zhou X, Meng H, Hao N, Chang J, et al. Simultaneous Detection of CNVs and SNVs Improves the Diagnostic Yield of Fetuses with Ultrasound Anomalies and Normal Karyotypes. Genes (Basel). 2020 doi: 10.3390/genes11121397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riggs ER, Andersen EF, Cherry AM, Kantarci S, Kearney H, Patel A, et al. Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen) Genet. Med. 2020;22:245–257. doi: 10.1038/s41436-019-0686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Puusepp S, Reinson K, Pajusalu S, Murumets Ü, Õiglane-Shlik E, Rein R, et al. Effectiveness of whole exome sequencing in unsolved patients with a clinical suspicion of a mitochondrial disorder in Estonia. Mol. Genet. Metab. Rep. 2018;15:80–89. doi: 10.1016/j.ymgmr.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riley LG, Cowley MJ, Gayevskiy V, Minoche AE, Puttick C, Thorburn DR, et al. The diagnostic utility of genome sequencing in a pediatric cohort with suspected mitochondrial disease. Genet. Med. 2020;22:1254–1261. doi: 10.1038/s41436-020-0793-6. [DOI] [PubMed] [Google Scholar]
- 22.Schon KR, Horvath R, Wei W, Calabrese C, Tucci A, Ibañez K, et al. Use of whole genome sequencing to determine genetic basis of suspected mitochondrial disorders: cohort study. BMJ. 2021;375:e66288. doi: 10.1136/bmj-2021-066288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, Song J, Chuai Y, Chen F, Song C, Shu M, et al. The mining and construction of a knowledge base for gene-disease association in mitochondrial diseases. Sci. Rep. 2021;11:23909. doi: 10.1038/s41598-021-03249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham BH. Diagnostic challenges of mitochondrial disorders: complexities of two genomes. Methods Mol. Biol. 2012;837:35–46. doi: 10.1007/978-1-61779-504-6_3. [DOI] [PubMed] [Google Scholar]
- 25.Thorburn DR. Mitochondrial disorders: Prevalence, myths and advances. J Inherit Metab Dis. 2004;27:349–362. doi: 10.1023/B:BOLI.0000031098.41409.55. [DOI] [PubMed] [Google Scholar]
- 26.Fang F, Liu Z, Fang H, Wu J, Shen D, Sun S, et al. The clinical and genetic characteristics in children with mitochondrial disease in China. Sci. China Life Sci. 2017;60:746–757. doi: 10.1007/s11427-017-9080-y. [DOI] [PubMed] [Google Scholar]
- 27.Wu T, He F, Xiao N, Han Y, Yang L, Peng J. Phenotype-genotype analysis based on molecular classification in 135 children with mitochondrial disease. Pediatr. Neurol. 2022;132:11–18. doi: 10.1016/j.pediatrneurol.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 28.Wortmann SB, Koolen DA, Smeitink JA, van den Heuvel L, Rodenburg RJ. Whole exome sequencing of suspected mitochondrial patients in clinical practice. J. Inherit. Metab. Dis. 2015;38:437–443. doi: 10.1007/s10545-015-9823-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pronicka E, Piekutowska-Abramczuk D, Ciara E, Trubicka J, Rokicki D, Karkucińska-Więckowska A, et al. New perspective in diagnostics of mitochondrial disorders: Two years' experience with whole-exome sequencing at a national paediatric centre. J. Transl. Med. 2016;14:174. doi: 10.1186/s12967-016-0930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis RL, Kumar KR, Puttick C, Liang C, Ahmad KE, Edema-Hildebrand F, et al. Use of whole-genome sequencing for mitochondrial disease diagnosis. Neurology. 2022;99:e730–e742. doi: 10.1212/WNL.0000000000200745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rahman S, Copeland WC. POLG-related disorders and their neurological manifestations. Nat. Rev. Neurol. 2019;15:40–52. doi: 10.1038/s41582-018-0101-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mousson DCB, Chassagne M, Mayençon M, Padet S, Crehalet H, Clerc-Renaud P, et al. POLG exon 22 skipping induced by different mechanisms in two unrelated cases of Alpers syndrome. Mitochondrion. 2011;11:223–227. doi: 10.1016/j.mito.2010.07.011. [DOI] [PubMed] [Google Scholar]
- 33.Fan W, Zheng J, Kong W, Cui L, Aishanjiang M, Yi Q, et al. Contribution of a mitochondrial tyrosyl-tRNA synthetase mutation to the phenotypic expression of the deafness-associated tRNA(Ser(UCN)) 7511A>G mutation. J. Biol. Chem. 2019;294:19292–19305. doi: 10.1074/jbc.RA119.010598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong S, Wang X, Meng F, Cui L, Yi Q, Zhao Q, et al. Overexpression of mitochondrial histidyl-tRNA synthetase restores mitochondrial dysfunction caused by a deafness-associated tRNA(His) mutation. J. Biol. Chem. 2020;295:940–954. doi: 10.1074/jbc.RA119.010998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vafaee-Shahi M, Farhadi M, Razmara E, Morovvati S, Ghasemi S, Abedini SS, et al. Novel phenotype and genotype spectrum of NARS2 and literature review of previous mutations. Ir. J. Med. Sci. 2022;191:1877–1890. doi: 10.1007/s11845-021-02736-7. [DOI] [PubMed] [Google Scholar]
- 36.Cappuccio G, Sayou C, Tanno PL, Tisserant E, Bruel AL, Kennani SE, et al. De novo SMARCA2 variants clustered outside the helicase domain cause a new recognizable syndrome with intellectual disability and blepharophimosis distinct from Nicolaides-Baraitser syndrome. Genet. Med. 2020;22:1838–1850. doi: 10.1038/s41436-020-0898-y. [DOI] [PubMed] [Google Scholar]
- 37.Debray FG, Lambert M, Chevalier I, Robitaille Y, Decarie JC, Shoubridge EA, et al. Long-term outcome and clinical spectrum of 73 pediatric patients with mitochondrial diseases. Pediatrics. 2007;119:722–733. doi: 10.1542/peds.2006-1866. [DOI] [PubMed] [Google Scholar]
- 38.Kalinska-Bienias A, Pollak A, Kowalewski C, Lechowicz U, Stawinski P, Gergont A, et al. Coexistence of mutations in keratin 10 (KRT10) and the mitochondrial genome in a patient with ichthyosis with confetti and Leber's hereditary optic neuropathy. Am. J. Med. Genet. A. 2017;173:3093–3097. doi: 10.1002/ajmg.a.38403. [DOI] [PubMed] [Google Scholar]
- 39.Li JK, Li W, Gao FJ, Qu SF, Hu FY, Zhang SH, et al. Mutation screening of mtDNA combined targeted exon sequencing in a cohort with suspected hereditary optic neuropathy. Transl. Vis. Sci. Technol. 2020;9:11. doi: 10.1167/tvst.9.8.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hathazi D, Griffin H, Jennings MJ, Giunta M, Powell C, Pearce SF, et al. Metabolic shift underlies recovery in reversible infantile respiratory chain deficiency. Embo J. 2020; 39:e105364. http://doi.org/10.15252/embj.2020105364 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The analyzed clinical and genetic data generated during the study are available from the corresponding author on reasonable request. The mtDNA and nDNA variants of patiens with mitochondrial disease are deposited in ClinVar (https://www.ncbi.nlm.nih.gov/clinvar; accession numbers: SCV002760197 - SCV002760205, SCV002761197 - SCV002761232). The raw sequencing data are not publicly available due to restriction of human data.