Abstract
Insects display a vast array of eye and body colors. Genes encoding products involved in biosynthesis and deposition of pigments are ideal genetic markers, contributing, for example, to the power of Drosophila genetics. Oncopeltus fasciatus is an emerging model for hemimetabolous insects, a member of the piercing-sucking feeding order Hemiptera, that includes pests and disease vectors. To identify candidate visible markers for O. fasciatus, we used parental and nymphal RNAi to identify genes that altered eye or body color while having no deleterious effects on viability. We selected Of-vermilion for CRISPR/Cas9 genome editing, generating three independent loss-of-function mutant lines. These studies mapped Of-vermilion to the X-chromosome, the first assignment of a gene to a chromosome in this species. Of-vermilion homozygotes have bright red, rather than black, eyes and are fully viable and fertile. We used these mutants to verify a role for Of-xdh1, ortholog of Drosophila rosy, in contributing to red pigmentation using RNAi. Rather than wild-type-like red bodies, bugs lacking both vermilion and xdh1 have bright yellow bodies, suggesting that ommochromes and pteridines contribute to O. fasciatus body color. Our studies generated the first gene-based visible marker for O. fasciatus and expanded the genetic toolkit for this model system.
Subject terms: Developmental biology, Genetics
Introduction
Recent years have witnessed an explosion of genome sequencing and gene annotation for representative species across the tree of life. Understanding the roles these genes play in generating, maintaining, or even limiting biodiversity will require approaches to assess gene function at key branches of the tree. Insects are excellent subjects of investigation because of the large number of species, their diverse habitats, physiology, behaviors and morphologies. Additionally, many can be reared in lab settings to allow for molecular genetic analyses. Most studies of insects to date have focused on the holometabolous insects, including beetles, butterflies and flies, with Drosophila melanogaster among the most powerful genetic model systems. As a close outgroup to holometabolous insects, the order Hemiptera comprises > 80,000 described species1 that share a piercing-sucking mode of feeding but which are otherwise enormously diverse (reviewed in2,3). This order includes agricultural pests such as aphids and harlequin bugs, nuisance pests of humans such as bed bugs and brown marmorated stink bugs, and vectors of serious human disease, such as kissing bugs, which transmit Chagas disease4,5. The milkweed bug Oncopeltus fasciatus (Of) is a promising model system for Hemiptera: a genome sequence is available and multiple tools for molecular genetic analyses have been developed, including RNA interference (RNAi) and CRISPR/Cas9 mutagenesis6–9. However, lacking for this species and Hemiptera more broadly are visible markers that are needed to pursue more rigorous genetic studies.
Pigmentation has long fascinated researchers and has been the target of genetic approaches because it varies extensively, even within species, and is simple and direct to assay, being readily visible externally. Mutations in genes that cause variation in body or eye color often do not impact organismal viability, allowing for isolation and maintenance of mutant lines. This was exploited in the early 1900s to launch the field of Drosophila genetics10,11. At that time, studies of eye color mutants from several different insect species contributed to the “one gene-one enzyme hypothesis” and established an ordered biochemical pathway involved in synthesis of insect pigments12–17. Body color in some insects is based on melanin (reviewed in18). However, the brightly colored eye and body colors of many insects are comprised of two other pigment types: ommochromes (named from the Greek omma—eye, and khroma—color) which derive from tryptophan; and pteridines (named for the Greek pteron—wing, based on their first identification from the orange, yellow and white colors in the wings of Pieridae butterflies (see19)), which are derived from GTP. Overall, ommochrome pigments are in the brown range, with mutations generating red- or white-eyed insects, while pteridines range from bright red to yellow, with mutations often leading to loss of body pigmentation and brown eyes (reviewed in20,21). Biosynthetic pathways leading to ommochrome pigments include products of the karmoisin (kar), vermilion (v), cinnabar (cn) and cardinal (cd) genes22–24 (recently reviewed25), while biosynthesis of pteridine pigments requires gene products of punch (pu) and purple (pr), and includes additional enzymatic steps, such as the product of rosy (ry) which encodes xanthine dehydrogenase, to generate a variety of colors19,26 (recent review27). For each pathway, specific transporters are required for pigment deposition28. The genes white (w), scarlet (st), and brown (bw) are closely related and comprise a subfamily of the ABC-transporter-encoding genes, with White forming a heterodimer with Brown for pteridine precursor transport, and with Scarlet for ommochrome precursor transport29–34. In Bombyx mori, another member of this subfamily, ok was identified and found to be most closely related to Bm-brown; Bm-ok mutants display defects in uric acid deposition in the larval epidermis resulting in a translucent skin35. Although no ok ortholog was found in the Drosophila genome, orthologs have been identified in many other insect genomes.
Hemiptera display diverse colors and patterns that include contributions from melanin (reviewed in18,36) in addition to ommochromes and pteridines. Roles for these latter pigment types in color variation have been investigated recently for a number of species within this order. Brent and Hull (2019)37 combined gene expression analysis and RNAi to document the roles of five ommochrome pathway genes in eye color development in Lygus hesperus (Lh). Knockdown of Lh-bw suggested that the pteridine pathway contributes to eye color while w knock down impacted both eyes and body, with phenotypes including smaller and paler bodies and soft cuticles. Subsequent CRISPR/Cas9 mutagenesis in this species showed that the ommochrome pathway components Lh-cn and Lh-cd are both involved in eye pigmentation, with Lh-cn mutants displaying sex-specific phenotypic differences in adults38. In the brown planthopper, Nilaparvata lugens, CRISPR/Cas9 genome editing suggested that Nl-w and ommochrome pathway member Nl-cn are necessary for wild type eye color39. Orthologs of ABC transporter genes w, st and ok as well as ommochrome pathway member cn were identified in the kissing bug Rhodnius prolixus (Rpro), and their roles in pigmentation tested with RNAi40. Interestingly, nymphal RNAi (nRNAi) knockdown of Rpro-cn resulted in animals with reddish, rather than black, ommatidia, but parental RNAi (pRNAi) caused lethality post-blood meal in first instars. This suggests the Rpro-cn gene is required for functions other than eye color, making it a poor candidate for a visible marker gene. Transporter knockdown revealed a role in ommochrome deposition in the eyes for Rpro-st, with newly added ommatidia displaying bright red color similar to that seen after Rpro-cn nymphal RNAi, and for Rpro-w, which also impacted viability, but no change in eye color was observed after knockdown of either ok ortholog. However, subtle changes in body pigmentation were noted after knockdown of Rpro-st or either of the two Rpro-ok paralogs, suggesting roles for both types of pigment in body color. Results from an extensive study of pigmentation-related genes in several water strider species41 suggested that ommochromes do not contribute to body color but are utilized for eye color in these species, with parental RNAi targeting either cn or st in Limnogonus franciscanus producing embryos with bright red eyes. Pteridines, on the other hand, appear to contribute to both eye and body pigmentation, with pRNAi of several different genes in that pathway producing loss of body coloration and brown eyes. Orthologs of ry were examined in several species where it is expressed and functions in both body and eye pigmentation.
O. fasciatus was one of the first hemipterans for which genetic approaches were taken to study pigmentation42–44. Through an EMS mutagenesis screen, Lawrence and coworkers identified genes impacting both pteridine and ommochrome pathways. One mutation, white body (wb), resulted in a loss of specific pteridines in the body; while no pigment analysis was performed on these mutants, they likely lacked erythropterin, as levels of this pigment were elsewhere found to coincide with the reddening of the body during embryogenesis45. Conversely, red eye (re) mutants had bright red eyes and wild type pteridines, and thus resulted from mutation in ommochrome synthesis. With both pteridine and ommochrome pathways disrupted, wb/wb; re/re mutants displayed loss of both red and brown pigments in the eye, resulting in white eyes and white bodies. In one study, Shelton and Lawrence (1974) made use of these mutants to show that ommatidia added as the compound eye grows through successive molts are recruited from cells of nearby epidermis46. After transplanting eye-proximal epidermal tissue from wild type nymphs into the corresponding area of wb/wb; re/re mutants, they observed mosaic eyes in the adult, mostly white eyes with clusters of wild type ommatidia at the eye margin closest to the graft, showing that these newer ommatidia had formed from transplanted epidermal tissue. The first study of these pathways using RNAi to target candidate genes in O. fasciatus was carried out by Liu47. Separate analyses of ommochrome pathway genes v, cn and st, and pteridine pathway genes bw, pu, pr, as well as w suggested roles for both pigment types in O. fasciatus coloration. Both parental and nymphal RNAi implicated pteridines in eye and body coloration, while nymphal RNAi knockdowns of ommochrome pathway genes generated animals with bright red eye margins but wild type body coloration, similar to the re mutant of Lawrence. Recently, RNAi knockdown demonstrated a role for Of-red Malpighian Tubules (red) in eye color, while also contributing somewhat to body pigmentation. This gene likely encodes a protein involved in intracellular pigment trafficking and, accordingly, knockdown impacted both ommochrome and pteridine pathways48.
In our first attempt to generate a visible eye color marker for O. fasciatus, we chose to target Of-w, given its historic use as a visible marker49. While that approach established CRISPR/Cas9 mutagenesis for O. fasciatus, we were surprised to find lethality in animals homozygous for white null alleles9, although w had been shown to be required for viability in the moth Helicoverpa armigera50 and more recently in L. hesperus38. Here, we used RNAi to assess roles of selected genes encoding candidate visible markers, testing for detrimental consequences of gene knockdown before proceeding to genome editing. Our experiments verified the role of Of-v in eye color47,48 and identified a novel role of the Of-xdh/ry gene in body color. We generated three independent Of-v null mutations, each of which has been maintained in lab colony for multiple generations without discernable impact on development. Thus, Of-v is an ideal visible marker for transgenesis, co-CRISPR, and other genetic tools, adding to the range of genetic studies feasible for this insect.
Results
RNAi targeting ok paralogs fails to reveal clear role in pigmentation
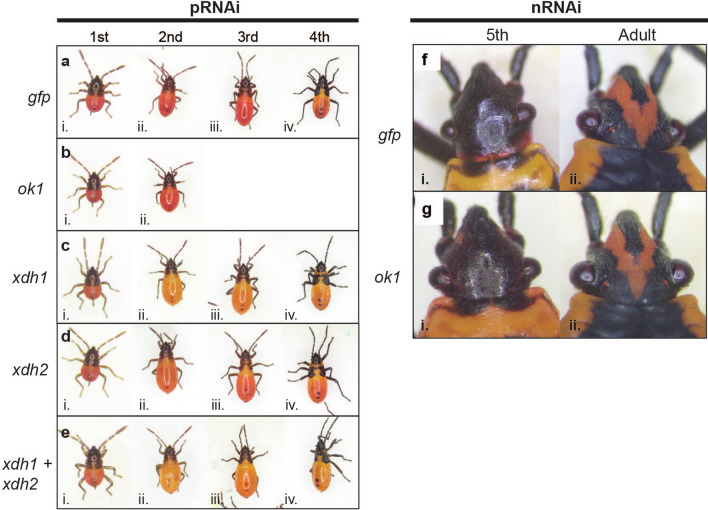
We first investigated two components of the pteridine pigment pathway: two previously identified ok orthologs in the O. fasciatus genome9. Since the ok subfamily of ABC transporter-encoding genes is most closely related to the brown subfamily35, which is required for pteridine pigment transport in many insects (“Introduction”), the two O. fasciatus ok paralogs are good candidates for pteridine transport in this species. To evaluate the roles of these paralogs in O. fasciatus pigmentation, we designed two non-overlapping dsRNAs per gene. Following Of-ok1 pRNAi, no difference in eye or body color in 1st or 2nd instar nymphs was observed compared to gfp controls (Fig. 1A,B). Following Of-ok2 pRNAi, very few embryos hatched despite appearing fully developed. Only 0.87% of embryos hatched following pRNAi with dsRNA-A (n = 344), and 4.96% hatched after pRNAi using dsRNA-B (n = 262), compared with an average hatch rate of 91.52% (n = 544 across four replicates) for gfp controls (Table 1). Co-injection of both Of-ok1 and Of-ok2 dsRNA produced intermediate hatch rates (Table 1), but no change in coloration of hatched nymphs was observed. Furthermore, nRNAi knockdown of Of-ok1 in 4th instar nymphs yielded no discernible change in eye or body coloration in either 5th instar nymphs or adults (Fig. 1G) compared to gfp dsRNA-injected controls (Fig. 1F). In sum, despite being strong candidates for involvement in pteridine pigment transport in O. fasciatus, we found no evidence that either would serve as a useful visible marker gene for this species.
Figure 1.
RNAi targeting pteridine synthesis pathway in Oncopeltus. Photos of nymphs and adults, as indicated. (A) Offspring of females injected with a gfp dsRNA control exhibit wild type phenotypes at (i) first, (ii) second, (iii) third, and (iv) fourth instars. (B) Offspring of females injected with Of-ok1 dsRNA were indistinguishable from controls. pRNAi knockdown of (C) Of-xdh1, (D) Of-xdh2, or (E) Of-xdh1 and Of-xdh2. Knockdown of Of-xdh1 caused a lightening of body color from red (A) to yellow-orange, particular evident in second through fourth instars (Cii–iv; Eii–iv). (F,G) Nymphal RNAi. The eye color of last instar nymphs and adults are similar in controls (Fi,ii) and after nRNAiok1 (Gi,ii).
Table 1.
Summary of hatch rates observed in pRNAi experiments.
| Gene(s) | dsRNA | dsRNA concentration (μM) | Analyzed (n) | # Hatch | Hatch rate (%) |
|---|---|---|---|---|---|
| gfp | A | 5 | 124 | 111 | 89.52 |
| gfp | A | 5 | 156 | 139 | 89.10 |
| Gfp | A | 5 | 142 | 137 | 96.48 |
| Gfp | A | 5 | 122 | 111 | 90.98 |
| Of-ok1 | A | 5 | 262 | 240 | 91.60 |
| Of-ok1 | B | 5 | 236 | 236 | 100.00 |
| Of-ok2 | A | 5 | 344 | 3 | 0.87 |
| Of-ok2 | B | 5 | 262 | 13 | 4.96 |
| Of-ok1 + Of-ok2 | A | 2.5 each | 327 | 155 | 47.40 |
| Of-ok1 + Of-ok2 | B | 5 each | 196 | 114 | 58.16 |
| Of-st | A | 5 | 188 | 179 | 95.21 |
| Of-st | B | 5 | 100 | 90 | 90.00 |
| Of-v | A | 5 | 257 | 246 | 95.72 |
| Of-xdh1 | A | 5 | 190 | 184 | 96.84 |
| Of-xdh1 | B | 5 | 272 | 199 | 73.16 |
| Of-xdh2 | A | 5 | 208 | 182 | 87.50 |
| Of-xdh2 | B | 5 | 189 | 188 | 99.47 |
| Of-xdh1 + Of-xdh2 | A | 2.5 each | 127 | 113 | 88.98 |
| Of-xdh1 + Of-xdh2 | B | 5 each | 87 | 76 | 87.36 |
| gfp (v3 mut) | A | 5 | 255 | 255 | 100.00 |
| Of-xdh1 (v3 mut) | B | 5 | 212 | 204 | 96.23 |
xdh1 contributes to body pigmentation in O. fasciatus
In Drosophila, rosy (ry) mutants display dark red eyes26 resulting from mutation of the xanthine dehydrogenase gene, a component of the pteridine pathway. To further investigate potential components of the O. fasciatus pteridine pathway, we used pRNAi to assess the functions of the Of-xanthine dehydrogenase (xdh) ortholog. BLAST searches of the O. fasciatus official gene set (OGS) followed by phylogenetic analysis (Fig. S1) revealed two orthologs of Dmel-ry, which we refer to as Of-xdh1 (OFAS027123) and Of-xdh2 (OFAS013815) (Figs. S1, S2). However, as we were unable to amplify the region encompassing both dsRNA templates of Of-xdh2 from cDNA, it is possible the OFAS013815 gene model encodes more than one transcript (see “Methods” for more detail). Knockdown of Of-xdh1 by pRNAi produced nymphs with slightly lighter body coloration than gfppRNAi nymphs, appearing yellow-orange rather than red (Fig. 1A,C). This difference was most discernible by the 2nd instar (Fig. 1Cii), likely because the compaction of the body in newly hatched 1st instar nymphs obscures slight differences in pigmentation (Fig. 1Ci). The body color change persisted through the third and fourth nymphal instars (Fig. 1Ciii,iv). Some Of-xdh1 dsRNA-treated nymphs appeared wild type by the fourth instar, indicating that the RNAi effect waned over time in some individuals. However, following Of-xdh2 pRNAi, no clear change in pigmentation relative to gfp controls was observed at any instar, although a marginal decrease in red body color intensity cannot be ruled out (Fig. 1D). Simultaneous knockdown of both Of-xdh1 and Of-xdh2 resulted in nymphs with yellow-orange coloration indistinguishable from the Of-xdh1 single knockdown (Fig. 1E).
Since we have noticed variation in body coloration within our O. fasciatus lab population and wanted to ensure that the lighter body coloration observed after xdh1 pRNAi was indeed due to specific knockdown of this gene and not lighter variants already present in the population, we performed another round of Of-xdh1 pRNAi to be scored according to a single-blind protocol. In this experiment, adults were injected with either gfp or Of-xdh1 dsRNA by one researcher, and second instar offspring from both treatments were individually placed in tubes labeled with a random number to be analyzed by a second researcher, described in more detail in the “Methods”. The second researcher scored each individual using a four-point scale (yellow-orange, orange, orange-red, or red). Of 188 gfp pRNAi offspring scored, 97.8% were scored as orange-red or red; just 4 (2%) were scored as orange. Of 193 Of-xdh1 pRNAi bugs scored, none were scored as red, just one (0.5%) was scored as orange-red, and 99.4% were scored as yellow-orange or orange. Thus, Of-xdh1 knockdown produced a definitive and consistent reduction in body pigmentation outside the range of variation seen in wild type populations. Overall, these results demonstrate that Of-xdh1 is required for dark red pigments in the O. fasciatus body, consistent with a role in the pteridine synthesis pathway, and does not appear to be required for viability, making this a potentially useful marker in this species. Interestingly, we did not observe any effect of Of-xdh1 on eye color.
Of-v and Of-st are required for brown pigmentation in adult compound eyes
We next investigated roles of genes expected to function in ommochrome pigmentation pathways. The tryptophan 2,3-dioxygenase-coding gene vermilion (v) and ABC transporter-encoding gene scarlet (st) are involved in ommochrome pigment synthesis and transport in Drosophila and other insects (see “Introduction”). Furthermore, previous work showed that RNAi targeting either Of-v or Of-st results in loss of dark brown pigmentation in the eyes47,48. We employed both pRNAi and nRNAi to assess the function of v and st in O. fasciatus. Following pRNAi targeting either Of-v or Of-st, no change in eye or body coloration was observed relative to gfp controls in 1st or 2nd instar nymphs (Fig. 2A–C). However, nRNAi targeting Of-v or Of-st in 4th instar nymphs resulted in a loss of brown pigments at the medial edge of the compound eyes in 5th instar nymphs (Fig. 2Ei,Fi). The effect was even more pronounced after individuals molted to adulthood (Fig. 2Eii,Fii). This partial change in eye color is attributed to the gradual addition of newly formed ommatidia after each molt. No change in body coloration following knockdown of Of-v or Of-st was observed. In short, Of-v and Of-st both appear to play roles in ommochrome synthesis or transport, respectively, with loss of these transcripts resulting in clear phenotypic differences late in development. In addition, no impact on viability was observed after RNAi knockdown for either gene (Table 1), making these genes good candidates for use as eye color markers in this species.
Figure 2.
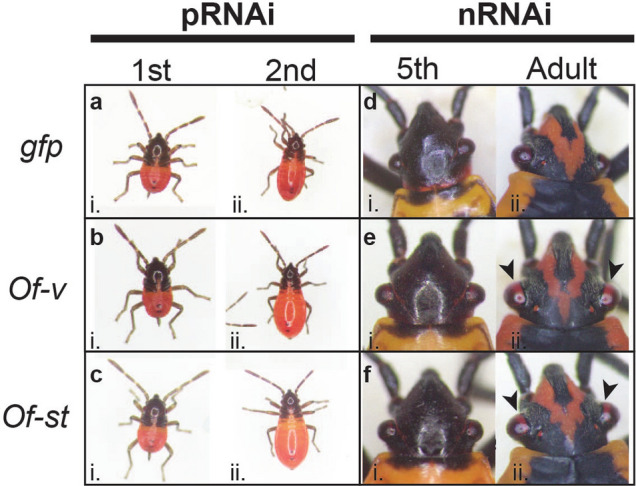
RNAi knockdown of ommochrome pathway genes Of-st and Of-v results in loss of brown eye pigmentation. Photos of nymphs and adults, as indicated. (A) Offspring of females injected with a gfp dsRNA control exhibit wild type phenotypes at first (i), second (ii) instars. Offspring of females injected with (B) Of-v dsRNA or (C) Of-st dsRNA. Fourth instar nymphs were injected with (D) gfp, (E) Of-v, or (F) Of-st dsRNA, shown after molting to fifth instar (i) and adulthood (ii). While gfp dsRNA-treated individuals display wild type black eyes, those treated with either Of-v or Of-st dsRNA display clear loss of brown eye pigment in the new ommatidia along the medial border of the adult compound eye (Eii,Fii, arrowheads).
Loss-of-function red-eyed Of-vermilion mutants are viable
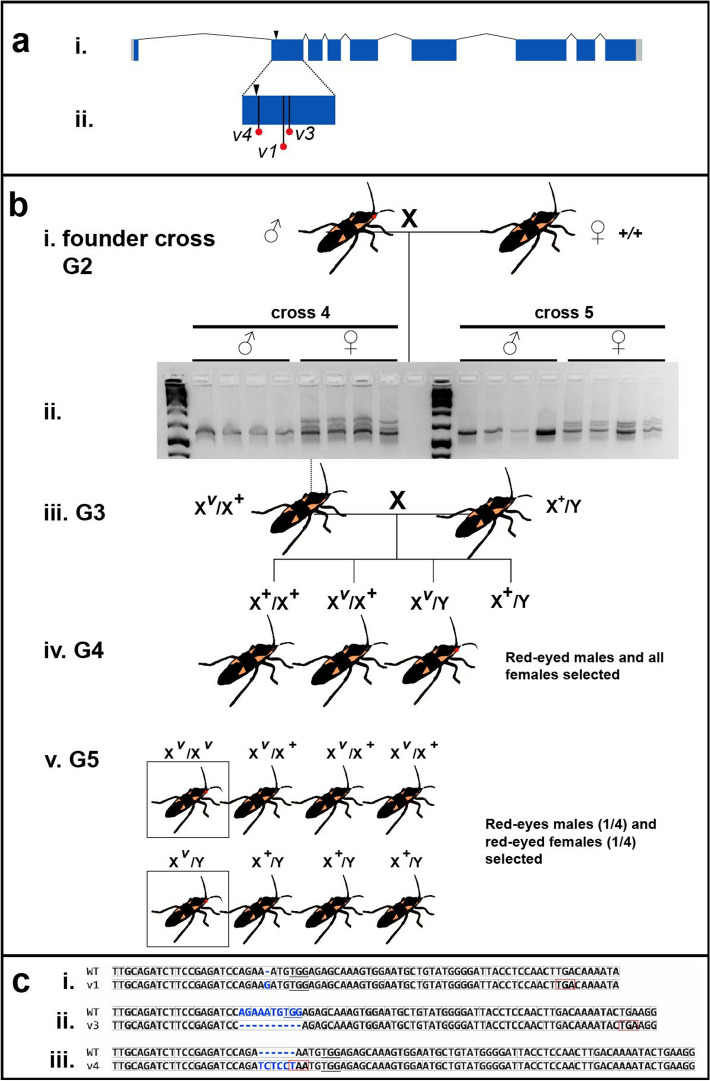
Given that RNAi knockdown of Of-v did not appear to negatively impact viability or fertility, we chose it as a candidate for generating a stable eye color marker for O. fasciatus. We used CRISPR/Cas9 to generate an Of-v null mutant, with tools we developed previously for mutagenesis in O. fasciatus9. We designed a single guide RNA (gRNA) to direct Cas9 to a site within exon 2 of Of-v (Fig. 3A arrowhead; see “Methods”). Of 1177 embryos injected with Of-v gRNA and Cas9 protein, 779 (66%) hatched. Since it takes at minimum four months from the time of O. fasciatus embryo injection to generation of homozygous individuals when reared at 25 °C, we sought to increase the developmental rate by rearing the G0s at 29 °C and 75% relative humidity. At their final nymphal instar, we observed an unusually high number of dead G0 nymphs, all of which had swollen black abdomens, a presentation we had not previously seen in our lab populations (Fig. S3). While we cannot be sure of the cause, the symptoms we observed closely matched those described for a disease of O. fasciatus populations infected with the bacterium Pseudomonas aeruginosa51, namely: paralysis, darkening of the body, bad smell, and very quick death following the onset of symptoms. The disease caused by P. aeruginosa was likewise associated with populations raised at high temperature (30 °C) and humidity.
Figure 3.
Generation of Of-v mutant lines using CRISPR/Cas9. (A) An Of-v gRNA was designed to guide Cas9 to generate a double-stranded break (arrowhead) in Of-v exon 2. (i) Of-v gene structure, with coding DNA sequence shown in dark blue and untranslated regions shown in light gray. (ii) Premature stop codons found in exon 2 of all mutant alleles. (B) Crossing scheme to generate homozygous lines from a heteroallelic Of-v mutant stock. (i) Five single crosses were set up between a single red-eyed Of-v G2 male founder (derived from the heteroallelic Of-v population) and three wild-type virgin females. (ii) Of-v exon 2 was amplified from genomic DNA isolated from the progeny of these crosses and a heteroduplex mobility assay performed; all females gave rise to heteroduplex bands, while no heteroduplex bands were found in male samples, consistent with Of-v X linkage; (iii) One G3 female offspring from each founder cross was mated with one or more wild type males; (iv) Male G4 offspring had either red or black eyes, while all female G4s had black eyes. All red-eyed males were selected to mate with female siblings; (v) In the G5, all red-eyed males and red-eyed virgin females were selected to establish homozygous lines. The expected genotypes and phenotypes are shown in their expected proportions. (C) Alignment between the wild-type Of-v allele and the three mutant alleles for which homozygous lines were successfully established: (i) v1, single nucleotide insertion at the predicted double-stranded break site; (ii) v3, 10 bp deletion; (iii) v4, 6 bp insertion. Insertions or deletions, blue. Reading frame, light gray boxes. Introduced stop codons, red boxes. PAM site, underline.
The eyes of healthy G0s, sequestered from the diseased population, were inspected for the expected red-eyed mosaic phenotype, which would result from biallelic hits in the injected animals. Mosaicism was observed in many individuals. In our efforts to isolate potentially infected bugs from the rest of our O. fasciatus lab population, we were unable to tabulate the frequencies of G0 phenotypes. Rather, all G0s with a red-eyed mosaic phenotype were allowed to mate to each other, producing many red-eyed G1s, which were selected and maintained as a heteroallelic Of-v mutant population.
Of-vermilion X-linkage and establishment of homozygous lines
Having established a population of red-eyed mutant bugs with many different v mutant alleles, we sought to establish single-allele homozygous lines. Five G2 males, offspring of the red-eyed G1 heteroallelic Of-v stock described above, were individually outcrossed to wild type virgins (“founder crosses”, Fig. 3Bi). When founder offspring reached the 5th instar or early adulthood, a single leg was removed for genomic DNA isolation, allowing us to genotype individuals before crossing them. We found that samples from all female offspring of crosses 3–5 showed the presence of a mutant allele, while no male samples produced heteroduplex bands, consistent with Of-v X-linkage (Fig. 3Bii). To test for X-linkage, a single red-eyed virgin female from the heteroallelic Of-v population was outcrossed to a wild-type male. When all progeny of this cross were in their final nymphal instar or adults, their eye color was examined. All female offspring (37/37) were found to have wild-type black eyes, and all male offspring (30/30) were found to have red eyes, consistent with X-linkage (data not shown).
Accordingly, to generate homozygous lines, a single virgin presumably heterozygous G3 female offspring from each of the original five founder crosses was outcrossed to one or more wild-type males (Fig. 3Biii). All of these crosses produced both red- and black-eyed G4 male progeny (Fig. 3Biv). Red-eyed males and all females from each cross were selected and crossed to each other. Finally, the G5 red-eyed progeny were selected to produce homozygous lines (Fig. 3Bv).
From the original five founder crosses, three homozygous Of-v mutant lines were established: v1, v3, and v4. Sequencing of Of-v exon 2 from each line showed all three alleles result in premature stop codons within exon 2 (Fig. 3Aii, red dots; Fig. 3C). The wild type Of-v allele has a 1167 bp long coding DNA sequence (CDS). Allele v1 has a single guanine insertion at the expected double-stranded break (DSB) site, causing a frameshift and introduction of a stop codon at position 87 of the CDS (Fig. 3Ci). Allele v3 has a 10 bp deletion, removing bases from both sides of the expected DSB, as well as introducing a stop codon at CDS position 98 (Fig. 3Cii). We also observed a single substitution in the intron preceding exon 2 in which a guanine was present where a thymine was observed in 8/8 wild-type samples. As this is 61 bp away from the expected DSB site, the difference most likely represents a polymorphism present in the G0 individual before Cas9 cleavage. Allele v4 has a 6 bp (TC|TCC|T) insertion immediately before AA (Fig. 3Ciii), resulting in a TAA stop codon at that site (Fig. 3Ciii, red box).
Scoring Of-v mutants for use as genetic tool
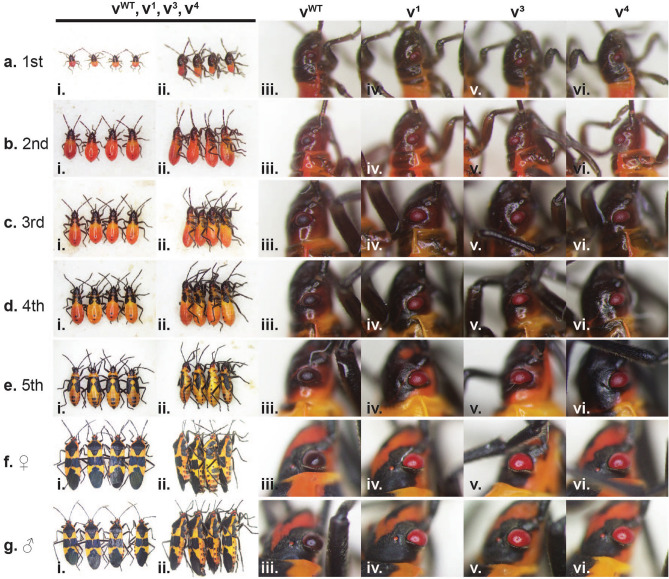
All three Of-v mutant lines are viable and fertile, making Of-v useful as an eye color marker for this species. To determine at which instars the Of-v mutant phenotype can reliably be scored, we photographed individuals from each of our three Of-v mutant lines at every nymphal instar and as adults to document and compare the phenotype to wild-type bugs over the lifespan. During the first instar (Fig. 4A), the Of-v lines are almost indistinguishable from wild type, though the body color of Of-v mutants is slightly lighter (Fig. 4Ai, dorsal view; ii, lateral view). Consistent with pRNAi results, the eye color of wild type first instar nymphs is not clearly different from that of the Of-v lines (Fig. 4Aiii-vi), perhaps because ommochromes have not yet been deposited in the eye. By the second instar, there is a clear difference in eye color between wild type (Fig. 4Biii) and Of-v mutant lines (Fig. 4Biv–vi). Any differences in body coloration become less apparent as development proceeds, while differences in eye pigmentation become much more pronounced as the compound eyes enlarge in subsequent instars (Fig. 4C–E). In adulthood, male and female Of-v mutants have bright red compound eyes in stark contrast to the black eyes of wild type adults (Fig. 4F,G). Overall, we noticed no differences in phenotype between the three Of-v strains. We have maintained each Of-v line for multiple generations and have noticed no difference in their developmental rate or overall health compared to our wild type population. Thus, these mutants provide a reliable visible marker for the O. fasciatus research community for transgenesis and other genetic manipulations.
Figure 4.
Phenotypes of homozygous Of-v lines across lifespan. Photographs of whole animals or heads of different individuals are shown in (A–G). (A) First instar nymphs, (B) second instar nymphs, (C) third instar nymphs, (D) fourth instar nymphs, (E) fifth instar nymphs, (F) adult females, (G) adult males. (i) Dorsal view of each genotype in the following order: vWT, v1, v3, v4. (ii) Lateral view of each genotype in the same order as shown in i. (iii–vi) Close-up view of right compound eye from individuals shown in i–ii, with genotypes indicated at top. Wild-type and v phenotypes are essentially indistinguishable during the first instar (A), but differences in coloration become apparent as wild-type eyes darken by the second instar (B), while v compound eyes remain red for the rest of nymphal development (C–E) and appear bright red in adults (F,G).
Because of issues with disease described above, we were not able to determine the frequency of mutagenesis in our initial Of-v CRISPR experiment. To determine basic statistics of CRISPR/Cas9 efficiency and germline mutation rates, we conducted a second round of Of-v CRISPR injections, making use of our new Of-v mutant lines for crosses (Fig. S4). Wild type embryos were injected with Cas9 protein and an Of-v gRNA as described above. We characterized the phenotype of mosaic G0s, and crossed these to v4 virgins. G1 offspring from each cross were then analyzed for red or black eyes. We observed a progressive increase in germline mutation rates with the severity of somatic knockout seen in the mosaic G0s. G0s that appeared wild type produced a germline mutation rate of 66.18 ± 9.12%, while G0s with one red eye had a germline mutation rate of 74.56 ± 9.37%, and those with two red eyes had a rate of 90.41 ± 4.33% (Table 2). Importantly, these results suggest that Of-v co-mutation may be a helpful strategy to concentrate screening efforts when trying to isolate non-visible mutations at other loci.
Table 2.
Somatic and germline mutation rates observed in second Of-v CRISPR experiment.
| G0 phenotype | Number | Percent of all G0s | Mean germline mutation rate | SEM germline mutation rate |
|---|---|---|---|---|
| WT (total) | 21 | 30.43 | 66.18 | 9.12 |
| WT (males) | 9 | 13.04 | 45.93 | 16.04 |
| WT (females) | 12 | 17.39 | 81.38 | 8.67 |
| 1 red eye (total) | 14 | 20.29 | 74.56 | 9.37 |
| 1 red eye (males) | 5 | 7.25 | 63.90 | 20.82 |
| 1 red eye (females) | 9 | 13.04 | 89.81 | 4.96 |
| 2 red eyes (total) | 34 | 49.28 | 90.41 | 4.33 |
| 2 red eyes (males) | 24 | 34.78 | 89.09 | 5.69 |
| 2 red eyes (females) | 10 | 14.49 | 93.56 | 5.80 |
xdh1 knockdown in v3 mutant produces bright yellow body color
Given that we observed a lighter body color after Of-xdh1 pRNAi (Fig. 1C) and after Of-v mutation (Fig. 4A–C), we wondered if both genes might contribute to body pigmentation in an additive manner. To test this, we performed Of-xdh1 pRNAi in an Of-v mutant background. Of-v3 females were injected with either Of-xdh1 dsRNA or gfp dsRNA as a negative control. Interestingly, v3; Of-xdh1 pRNAi offspring had bright yellow bodies (Fig. 5B), lighter than the v3; gfppRNAi controls (Fig. 5A), and even lighter than the yellow-orange pigmentation observed after xdh1 pRNAi knockdown in a wild type background (compare to Fig. 1C). Since Of-v mutants do not synthesize ommochromes and thus have red eyes throughout the lifespan (Fig. 4), any effect of Of-xdh1 knockdown on eye color that may have been masked by the presence of ommochromes in a wild type background should be more apparent in the Of-v3 background. However, no distinguishable effect of the knockdown on Of-v3 eye pigmentation was observed, consistent with our previous observation that Of-xdh1 knockdown in a wildtype background had no effect on eye pigmentation. The yellow body phenotype was marginally detectable in 1st instar nymphs (Fig. 5Bi) but was clear in 2nd instars (Fig. 5Bii) and lasted through the 4th instar (Fig. 5Biv). These results suggest that both Of-v and Of-xdh1 contribute independently to O. fasciatus body coloration and that Of-xdh1 would be useful as a visible body color marker for O. fasciatus.
Figure 5.
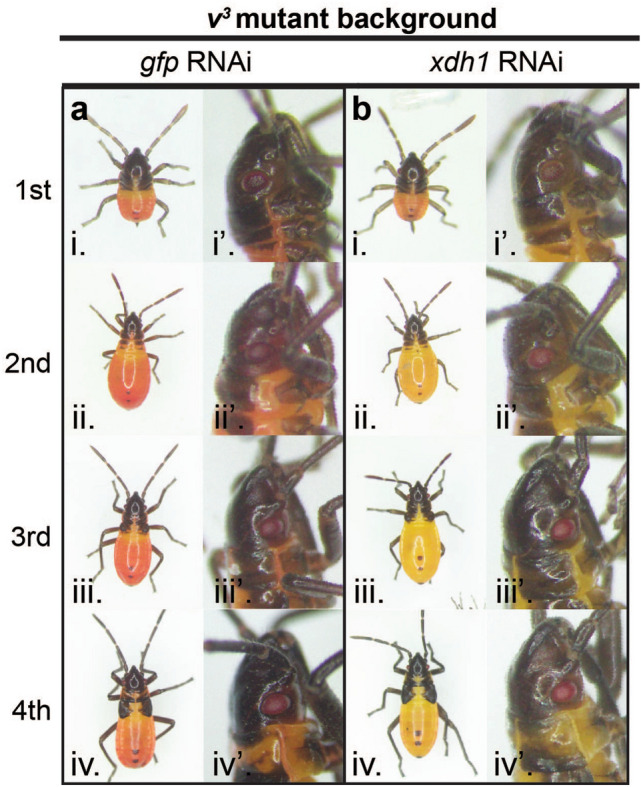
Of-xdh1 knockdown in Of-v3 mutant background produces bright yellow body color. Offspring of v3 mothers treated with gfp (A) or Of-xdh1 (B) dsRNA were observed at first (i), second (ii), third (iii), and fourth (iv) instars. While the knockdown did not appear to produce an effect in the eyes of pRNAi offspring (i’–iv’), body pigmentation was greatly reduced in Of-xdh1 treated individuals, resulting in a bright yellow color (Bi–iv).
Discussion
Here we have expanded the repertoire of genetic tools available for O. fasciatus, an emerging model system for hemimetabolous insects. Among pigmentation mutants, those that display loss of eye pigmentation are particularly valuable, given the established efficacy of the 3XP3 synthetic enhancer-promoter construct, which can direct gene expression in eyes across a range of phylogenetically diverse insects52. Combined with a fluorescent protein-encoding gene or rescue allele, this becomes a powerful tool for screening low-frequency genetic events, including gene replacement using CRISPR/Cas9 or transposon-mediated transgenesis. Even in the absence of validated transgenes, pigmentation mutants are useful for tracking germline mutation in the case of CRISPR/Cas9 co-conversion, allowing alleles that may not produce visible phenotypes to be more easily detected.
In investigating genes that would be useful as visible markers, we identified functions for both ommochrome and pteridine synthesis pathways in eye and body color of these bugs. This contrasts with holometabolous insects such as the beetle Tribolium castaneum, where mutations in ommochrome pathway genes generate white-eyed animals, suggesting no role for pteridines in eye color in that species53,54. Based on our previous experience that loss-of-function mutation of Of-w results in lethality in homozygotes9, we pre-screened candidate genes by RNAi before performing CRISPR/Cas9 mutagenesis. Similar to previous work in O. fasciatus47 and R. prolixus40, we did not observe roles for the Of-ok paralogs in eye pigmentation, but also observed no change in body pigmentation following Of-ok1 knockdown, in contrast to the functions of these genes in R. prolixus. However, we did observe an unexpected role for Of-ok2 in embryonic viability. In Bombyx mori, where ok is involved in transporting uric acid to the larval epidermis, both ok and white mutants display translucent larval skin55, leading Wang et al. (2013) to hypothesize that Bmor-White and Bmor-Ok form a heterodimer responsible for uric acid transport in this species. A similar heterodimer forming between Ofas-White and Ofas-Ok2 could explain the lethality seen here after Of-ok2 RNAi and after Of-w mutation9, perhaps resulting from the inability to transport waste products like uric acid. Knockdown of Of-xdh1—ortholog of rosy, a gene required for pteridine pigment synthesis in Drosophila—revealed a subtle but consistent change in body color, with loss of red pigmentation, while no role was identified for Of-xdh2 (Fig. 1). Within the ommochrome pathway, our studies confirmed roles of Of-st, encoding a putative ommochrome transporter, and Of-v, encoding tryptophan 2,3-dioxygenase, in eye color (Fig. 2). Neither parental nor nymphal RNAi appeared to impact viability, making both of these genes potentially useful eye color markers.
Accordingly, we used CRISPR/Cas9 genome editing to generate three independent loss-of-function mutations via non-homologous end joining in Of-v (Figs. 3, 4), each of which has been maintained as a viable colony of bright red-eyed homozygotes for multiple generations without any apparent impact on viability or fertility. As in Drosophila, this gene is located on the X-chromosome of O. fasciatus (Fig. 3). Thus, although the phenotype appears identical to that of Lawrence’s re mutants, they are not allelic as re behaved as an autosomal recessive44. We further made use of Of-v mutants to test the efficiency of CRISPR/Cas9 mutagenesis in this species, showing that G0 individuals exhibiting somatic mutation displayed germline transmission rates as high as 90%, making Of-v an excellent marker for co-CRISPR56. Finally, using RNAi to knock down Of-xdh1 in Of-v homozygotes, we revealed a role for the ommochrome pathway in body coloration, as the body color of these double “mutants” was bright yellow (Fig. 5), compared to the yellow-orange color of Of-xdh1RNAi animals (Fig. 1). Of-xdh1 does not appear to be required for viability, making this another potentially useful O. fasciatus marker. In contrast to our findings, studies in water striders suggested that ommochromes play no role in body pigmentation but function only for eye color41. Conversely, rosy plays roles in body and eye pigmentation in water striders41, but we found it to function only in body coloration in O. fasciatus. These differences may reflect species variation or stage-specific utilization of the different pathways, which remain to be examined.
Methods
O. fasciatus rearing
An O. fasciatus wild type population was maintained at room temperature (23–24 °C) in clear plastic storage containers (15.25 × 11 × 11 in.) and exposed to a 16 h:8 h light:dark cycle. All RNAi experiments were performed at 25 °C. As described in the “Results”, subpopulations were briefly maintained at 29 °C and 75% relative humidity, which appeared to promote disease (Fig. S3). A paper towel and cotton ball wick placed atop a 32 oz. plastic container full of water provided a constant source of water and raw organic sunflower seeds were provided as food. Single crosses were performed either using Drosophila vials as described9 or using one gallon plastic storage containers.
Gene isolation
To identify the O. fasciatus ortholog of v, a BLAST search against the O. fasciatus official gene set (OGS)8 was conducted using the Dmel-v amino acid sequence as a query. Only one hit had an e-value less than 1.4, that of OFAS025214 (e-value = 2e−129), suggesting that there is only one Of-v ortholog. To confirm the identity of OFAS025214, a phylogenetic tree was constructed from an alignment of this gene model to amino acid sequences of several insect v orthologs. The OFAS025214 gene model clustered with Lhes-v, the only other heteropteran v ortholog included in this analysis, suggesting that OFAS025214 is indeed the Of-v ortholog. To identify the Of-xdh ortholog, the O. fasciatus OGS was searched using BLAST and the Dmel-ry amino acid sequence as query. Several hits had significant e-values, so sequences for all gene models with e-values below 8e−71 were collected and subjected to phylogenetic analysis. The resulting gene tree showed two gene models—OFAS027123 and OFAS013815—clustering with other insect xdh/ry orthologs, whereas the remaining O. fasciatus gene models clustered with Dmel-aox, which was included in the analysis as an outgroup, suggesting that there are two Of-xdh paralogs. The O. fasciatus orthologs of st, ok1, and ok2 were analyzed previously9. Primers for all sequences were designed based on the O. fasciatus genome sequence (see Table S1). For Of-v, we amplified the coding DNA sequence using primers 30 and 31, and the PCR product was blunt-end ligated into SmaI-cut pUC19, and subsequently Sanger sequenced. We Sanger sequenced fragments from all other genes spanning the two dsRNA templates (Fig. S2), except for Of-xdh2. For this gene model (OFAS013815), we were able to amplify two non-overlapping fragments, but we were not able to amplify a fragment uniting these two regions. This may suggest that the two regions are not part of the same transcript; more work is needed to determine if the OFAS013815 gene model represents a single gene.
Phylogenetic analysis
To construct gene trees, amino acid sequences were aligned in MUSCLE57followed by phylogenetic inference using MrBayes 3.1.158 within the TOPALi v2.5 framework59 with the following parameters: runs = 2, generations = 100,000, sample freq. = 10, burnin = 25%. Tree formatting was done in MEGA X 10.1.660.
Double-stranded RNA (dsRNA) synthesis
All dsRNA templates were PCR-amplified using Q5 high-fidelity DNA polymerase (NEB) from O. fasciatus first instar complementary DNA (cDNA). In some cases, this PCR product was inserted into the pGEM T-easy vector before dsRNA synthesis, while in others the PCR product was used directly as template for dsRNA synthesis. A dsRNA template targeting turbo gfp (tGFP) was amplified from plasmid pBac[3xP3-DsRed; UAS-Tc'hsp_p-tGFP-SV40] (Addgene 86453) for use as a negative control. All primers used to amplify dsRNA templates have T7 RNA polymerase promoter sequences at the 5’ end allowing synthesis of both RNA strands in a single reaction. PCR products were used as templates in an RNA transcription reaction using the MEGAscript T7 Transcription kit (Invitrogen) at 37 °C overnight. 1 ul kit-provided TURBO DNase was then added and the reaction was incubated at 37 °C for 15 min. The RNA was denatured by heating to 95 °C for 3 min in a heat block; the heat block was then turned off, and the RNA strands were allowed to anneal as the heat block temperature slowly decreased. The dsRNA was precipitated with ethanol and lithium chloride, and resuspended in water. For each gene except Of-v and tGFP, two non-overlapping dsRNAs were made (denoted dsRNA A and B, Fig. S2). All primer sequences are listed in Table S1.
Parental RNA interference (pRNAi)
For pRNAi, each dsRNA was diluted to 5 uM in injection buffer (5 mM KCl, 0.1 mM phosphate buffer pH 6.8) with green food coloring (McCormick) diluted 1:50. Each adult female was injected with 3 μl injection mix between the 4th and 5th abdominal sternites using a glass needle pulled from borosilicate glass capillary tubes (World Precision Instruments). Each injected female was paired with a male and kept individually in a Drosophila vial supplied with raw sunflower seeds, damp cotton as a source of water, and dry cotton for egg laying. Vials were laid on their sides to keep seeds from contacting the wet cotton. Embryos were collected daily over 7 days after injection and incubated at 25 °C for approximately one week until hatching. Offspring were checked every day, and photographed within 24 h of hatching or molting to the next instar, after melanin deposition. For Of-xdh1 + xdh2 and Of-ok1 + ok2 double injections, either 2.5 μM of each dsRNA (dsRNA A, Fig. S2) or 5 μM of each dsRNA (dsRNA B, Fig. S2) were used.
After an initial xdh1 knockdown experiment produced a subtle phenotype, we carried out a blind experiment to ensure that this phenotype was indeed due to xdh1 knockdown and not to lighter colored variants in our population. In this experiment, phenotypes of offspring of Of-xdh1 or gfp dsRNA-injected females were scored by a researcher who had no knowledge of the treatment each bug had received. Two researchers independently injected five adult females each with Of-xdh1-B dsRNA or gfp dsRNA (as described above). The first four egg clutches laid by each female were discarded as we have often observed that the first several clutches fail to display knockdown phenotypes, likely due to their position within the ovariole at the time of injection. Each additional clutch of embryos was placed in a Drosophila vial provided with sunflower seeds and damp cotton and incubated at 25 °C for approximately 7 days until hatching. Nymphs were allowed to molt into 2nd instar as we had noticed the lighter coloration seen after xdh1 knockdown is more apparent at this stage. Each researcher selected 100 gfp pRNAi and 100 Of-xdh1 pRNAi nymphs derived from their independent injections and placed each nymph in a tube labeled with a random number from 1 to 200. Each researcher gave the other researcher (the “analyzer”) the 200 nymphs to score on a 4-point scale from light orange to dark red. Any individuals found by the analyzer to be dead or at the wrong instar were excluded from analysis. The analyzer had no information about any of the nymphs aside from their random labels. Finally, the score sheets of the two analyzers were combined, treatment groups were unblinded, and scores assigned to individuals from each RNAi treatment group were tabulated and compared.
Nymphal RNA interference (nRNAi)
For nRNAi, 4th instar nymph injections were performed as described above for pRNAi, except that only 1 μl of the 5 μM dsRNA injection mix was injected into each nymph and individuals were injected between the 5th and 6th abdominal sternites. After injection, nymphs were kept in small cages provided with damp cotton and sunflower seeds, incubated at 25 °C, and were photographed after molting to 5th instar and/or adulthood.
CRISPR/Cas9 mutagenesis of Of-vermilion
The Of-v exon 2 sequence was submitted to CHOPCHOP61 to identify potential gRNA target sites. Each of the suggested gRNAs was aligned to the O. fasciatus genome assembly using blastn -task blastn-short, and the resulting alignments were used to evaluate the potential of off-target Cas9 cleavage. A gRNA was selected that produced no alignments in the PAM plus seed region for any genomic region other than Of-v exon 2. This gRNA was synthesized in vitro following the recommendations of https://www.crisprflydesign.org/grnatranscription/. Briefly, the gRNA sequence was introduced using primer 24, which was used with primer 23 to amplify a gRNA scaffold sequence in plasmid pCFD361 using Phusion polymerase. This PCR product was added as template to a T7 RNA polymerase transcription reaction using the Megascript T7 kit (Invitrogen). Following 3 h of RNA transcription at 37 °C, TURBO DNase was added to the reaction and incubated for 15 min at 37 °C. The RNA was purified using the Monarch RNA Cleanup kit (NEB). To form ribonucleoprotein complexes prior to injection, the Of-v gRNA was diluted in injection buffer (5 mM KCl, 0.1 mM phosphate buffer pH 6.8) and heated to 70 °C for 2 min, then transferred to ice. Cas9-NLS protein (PNA Bio) was added to the gRNA and incubated at room temperature for 5 min. The final injection mix contained 80 ng/μl Of-v gRNA, 300 ng/μl Cas9 protein, and green food coloring (McCormick) diluted 1:50. Embryo injection was performed as described in Reding and Pick (2020). Briefly, O. fasciatus embryos were collected over a ~ 3.5 h period, washed in 1:50 commercial bleach: water for 3 min, then aligned into the grooves of 3% agar sheets. Embryos were injected at ~ 3 to 7 h AEL using a Narishige IM-400 microinjector attached to a micromanipulator. Sheets of injected embryos were moved to petri dishes and allowed to develop at 25 °C until hatching.
Heteroduplex mobility assay
When sampling 5th instar or adult bugs, genomic DNA was prepared as described in62, except that a single mesothoracic leg was used instead of whole individuals and only 25 μl of squishing buffer was used, which was then diluted 1:2 with water after DNA extraction. When sampling embryos, a single embryo was squished in 6 μl squishing buffer. Primers 25 and 26 were used to amplify a 581 bp fragment around Of-v exon 2, and the heteroduplex mobility assay was performed following standard methods9,63.
Crosses to establish homozygous Of-v lines
All G0s were inspected as young adults or last instar nymphs to identify mosaic animals, reflecting biallelic gene targeting events. As described in the “Results”, G0s with at least one red eye were selected and allowed to mate with each other. All G1s with two red eyes—presumably homozygous for Of-v mutation—were selected to establish a heteroallelic Of-v mutant population. Five G2 red-eyed males drawn from this population were each crossed to three wild-type virgins (Fig. 3B). A heteroduplex mobility assay was performed on embryos resulting from these crosses. Since this assay is not sensitive enough to detect very small indels, it was expected that not all samples would give rise to heteroduplex bands; indeed no heteroduplex bands were observed in samples of offspring from two of the five crosses. Offspring from three of the five founder crosses produced heteroduplex bands in all female fifth instar or adult samples and in none of the male samples, suggesting that Of-v is X-linked. Crosses proceeded as described in the Results and illustrated in Fig. 3B. To characterize each line’s mutation, genomic DNA was isolated as described above from two individuals per line, Of-v exon 2 was independently PCR-amplified from each sample using primers 25 and 26, and PCR products were Sanger sequenced.
Second Of-v CRISPR experiment
Wild type embryos were injected with 300 ng/μl Cas9 protein (PNA Bio), 500 ng/μl pBac[3xP3-EGFP;Tc'hsp5'-Gal4Delta-3'UTR] (Addgene #86449), 80 ng/μl of the Of-v gRNA described above (see “CRISPR/Cas9 mutagenesis of Of-vermilion”), and 80 ng/μl of a gRNA designed to target the ampicillin resistance gene of the injected plasmid for Cas9-mediated linearization. This AmpR gRNA was synthesized in vitro as described above for the Of-v gRNA, except that primers 23 and 29 (Table S1) were used to amplify the gRNA template. The AmpR gRNA was injected to allow whole plasmid integration through non-homologous end joining at the Of-v locus, potentially tagging an Of-v mutation with a 3xP3 > egfp fluorescent eye marker.
The phenotypes of adult G0s were recorded, before being crossed to v4 virgins. 99 G0 crosses were set up; 30 G0s either died before mating or were infertile. G1s were CO2-anaesthetized and the eye phenotype of each was characterized as wild-type or v under a brightfield microscope; the presence of GFP in the eyes was assessed using a fluorescent microscope equipped with a GFP filter. Unfortunately, we observed no GFP+ G1 offspring, suggesting that either no plasmid incorporation occurred or that the reduction in eye pigmentation in Of-v mutants is not sufficient to allow GFP visualization. Germline mutation rate was differentially calculated for G0 males and G0 females, since male v G1 offspring of male G0s obtained their mutant v allele from their v mothers, and thus are not informative of v mutations that may have occurred in the paternal germline. Germline mutation rate for male G0s was calculated as the number of v female G1s/total female G1s; for G0 females the calculation was v G1s/total G1s.
Supplementary Information
Acknowledgements
This work was supported by the National Institutes of Health (grant R01GM113230 to L.P.). K.R. was supported by a National Sciences Foundation Graduate Research Program Fellowship. pBac[3xP3-DsRed; UAS-Tc'hsp_p-tGFP-SV40] (Addgene 86453) and pBac[3xP3-EGFP;Tc'hsp5'-Gal4Delta-3'UTR] (Addgene 86449) were gifts from Gregor Bucher. We thank Patricia Graham and Matthew D. Fischer for comments on the manuscript.
Author contributions
K.R. and M.L. carried out experiments. K.R., M.L. and L.P. analyzed the data and wrote the manuscript. L.P. obtained funding.
Data availability
Mutant strains and materials are available upon request. The gene sequences generated and/or analysed during the current study are available in the GenBank repository: vermilion, OQ151805; ok1, OQ151806; ok2, OQ151807; scarlet, OQ151808; xdh1, OQ151809; xdh2, OQ151810; v_exon2_WT, OQ151811; v_exon2_v1, OQ151812; v_exon2_v3, OQ151813; v_exon2_v4, OQ151814.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-31145-2.
References
- 1.Stork NE. How many species of insects and other terrestrial arthropods are there on Earth? Annu. Rev. Entomol. 2018;63:31–45. doi: 10.1146/annurev-ento-020117-043348. [DOI] [PubMed] [Google Scholar]
- 2.Panfilio KA, Angelini DR. By land, air, and sea: Hemipteran diversity through the genomic lens. Curr. Opin. Insect Sci. 2018;25:106–115. doi: 10.1016/j.cois.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Jockusch EL, Fisher CR. Something old, something new, something borrowed, something red: The origin of ecologically relevant novelties in Hemiptera. Curr. Opin. Genet. Dev. 2021;69:154–162. doi: 10.1016/j.gde.2021.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Nunes-da-Fonseca R, Berni M, Tobias-Santos V, Pane A, Araujo HM. Rhodnius prolixus: From classical physiology to modern developmental biology. Genesis. 2017 doi: 10.1002/dvg.22995. [DOI] [PubMed] [Google Scholar]
- 5.Pacheco ID, Walling LL, Atkinson PW. Gene editing and genetic control of Hemipteran pests: Progress, challenges and perspectives. Front. Bioeng. Biotechnol. 2022;10:900785. doi: 10.3389/fbioe.2022.900785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hughes CL, Kaufman TC. RNAi analysis of deformed, proboscipedia and sex combs reduced in the milkweed bug Oncopeltus fasciatus: Novel roles for Hox genes in the hemipteran head. Development. 2000;127:3683–3694. doi: 10.1242/dev.127.17.3683. [DOI] [PubMed] [Google Scholar]
- 7.Chipman AD. Oncopeltus fasciatus as an evo-devo research organism. Genesis. 2017 doi: 10.1002/dvg.23020. [DOI] [PubMed] [Google Scholar]
- 8.Panfilio KA, et al. Molecular evolutionary trends and feeding ecology diversification in the Hemiptera, anchored by the milkweed bug genome. Genome Biol. 2019;20:64. doi: 10.1186/s13059-019-1660-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reding K, Pick L. High-efficiency CRISPR/Cas9 mutagenesis of the white gene in the milkweed bug Oncopeltus fasciatus. Genetics. 2020;215:1027–1037. doi: 10.1534/genetics.120.303269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morgan TH. Sex limited inheritance in Drosophila. Science. 1910;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- 11.Sturtevant, A. H. The use of mosaics in the study of the developmental effect of genes. In Proceedingsofthe6thInternationalCongressofGenetics. Vol. 1. 304–307 (1932).
- 12.Beadle GW, Ephrussi B. Development of eye colors in Drosophila: Transplantation experiments with suppressor of vermilion. Proc. Natl. Acad. Sci. USA. 1936;22:536–540. doi: 10.1073/pnas.22.9.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beadle GW, Ephrussi B. The differentiation of eye pigments in Drosophila as studied by transplantation. Genetics. 1936;21:225–247. doi: 10.1093/genetics/21.3.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beadle GW, Ephrussi B. Development of eye colors in Drosophila: Diffusible substances and their interrelations. Genetics. 1937;22:76–86. doi: 10.1093/genetics/22.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ephrussi B, Beadle GW. Development of eye colors in Drosophila: Production and release of cn substance by the eyes of different eye color mutants. Genetics. 1937;22:479–483. doi: 10.1093/genetics/22.5.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ephrussi B, Beadle GW. Development of eye colors in Drosophila: Transplantation experiments on the interaction of vermilion with other eye colors. Genetics. 1937;22:65–75. doi: 10.1093/genetics/22.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ephrussi B. Chemistry of “eye color hormones” of Drosophila. Q. Rev. Biol. 1942;17:327–338. doi: 10.1086/394662. [DOI] [Google Scholar]
- 18.Popadic A, Tsitlakidou D. Regional patterning and regulation of melanin pigmentation in insects. Curr. Opin. Genet. Dev. 2021;69:163–170. doi: 10.1016/j.gde.2021.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Ziegler I, Harmsen R. The biology of pteridines in insects. Adv. Insect Physiol. 1970;6:139–203. doi: 10.1016/S0065-2806(08)60112-5. [DOI] [Google Scholar]
- 20.Kikkawa H. Mechanism of pigment formation in Bombyx and Drosophila. Genetics. 1941;26:587–607. doi: 10.1093/genetics/26.6.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summers KM, Howells AJ, Pyliotis NA. Biology of eye pigmentation in insects. Adv. Insect Physiol. 1982;16:119–166. doi: 10.1016/S0065-2806(08)60153-8. [DOI] [Google Scholar]
- 22.Sullivan DT, Kitos RJ, Sullivan MC. Developmental and genetic studies on kynurenine hydroxylase from Drosophila melanogaster. Genetics. 1973;75:651–661. doi: 10.1093/genetics/75.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryall RL, Howells AJ. Ommochrome biosynthetic pathway of Drosophila melanogaster: Variations in levels of enzyme activities and intermediates during adult development. Insect Biochem. 1974;4:47–61. doi: 10.1016/0020-1790(74)90041-9. [DOI] [Google Scholar]
- 24.Howells AJ, Summers KM, Ryall RL. Developmental patterns of 3-hydroxykynurenine accumulation in white and various other eye color mutants of Drosophila melanogaster. Biochem. Genet. 1977;15:1049–1059. doi: 10.1007/BF00484496. [DOI] [PubMed] [Google Scholar]
- 25.Figon F, Casas J. Ommochromes in invertebrates: Biochemistry and cell biology. Biol. Rev. 2019;94:156–183. doi: 10.1111/brv.12441. [DOI] [PubMed] [Google Scholar]
- 26.Reaume AG, Knecht DA, Chovnick A. The rosy locus in Drosophila melanogaster: Xanthine dehydrogenase and eye pigments. Genetics. 1991;129:1099–1109. doi: 10.1093/genetics/129.4.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim H, Kim K, Yim J. Biosynthesis of drosopterins, the red eye pigments of Drosophila melanogaster. IUBMB Life. 2013;65:334–340. doi: 10.1002/iub.1145. [DOI] [PubMed] [Google Scholar]
- 28.Ewart GD, Howells AJ. ABC transporters involved in transport of eye pigment precursors in Drosophila melanogaster. Methods Enzymol. 1998;292:213–224. doi: 10.1016/s0076-6879(98)92017-1. [DOI] [PubMed] [Google Scholar]
- 29.Mount SM. Sequence similarity. Nature. 1987;325:487. doi: 10.1038/325487c0. [DOI] [PubMed] [Google Scholar]
- 30.Dreesen TD, Johnson DH, Henikoff S. The brown protein of Drosophila melanogaster is similar to the white protein and to components of active transport complexes. Mol. Cell Biol. 1988;8:5206–5215. doi: 10.1128/mcb.8.12.5206-5215.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tearle RG, Belote JM, McKeown M, Baker BS, Howells AJ. Cloning and characterization of the scarlet gene of Drosophila melanogaster. Genetics. 1989;122:595–606. doi: 10.1093/genetics/122.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pepling M, Mount SM. Sequence of a cDNA from the Drosophila melanogaster white gene. Nucleic Acids Res. 1990;18:1633. doi: 10.1093/nar/18.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mackenzie SM, et al. Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim. Biophys. Acta. 1999;1419:173–185. doi: 10.1016/s0005-2736(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 34.Dermauw W, Van Leeuwen T. The ABC gene family in arthropods: Comparative genomics and role in insecticide transport and resistance. Insect Biochem. Mol. Biol. 2014;45:89–110. doi: 10.1016/j.ibmb.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 35.Wang L, et al. Mutation of a novel ABC transporter gene is responsible for the failure to incorporate uric acid in the epidermis of ok mutants of the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2013;43:562–571. doi: 10.1016/j.ibmb.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 36.Liu J, Lemonds TR, Marden JH, Popadic A. A pathway analysis of melanin patterning in a hemimetabolous insect. Genetics. 2016;203:403–413. doi: 10.1534/genetics.115.186684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brent CS, Hull JJ. RNA interference-mediated knockdown of eye coloration genes in the western tarnished plant bug (Lygus hesperus Knight) Arch. Insect Biochem. Physiol. 2019;100:e21527. doi: 10.1002/arch.21527. [DOI] [PubMed] [Google Scholar]
- 38.Heu CC, et al. CRISPR-mediated knockout of cardinal and cinnabar eye pigmentation genes in the western tarnished plant bug. Sci. Rep. 2022;12:4917. doi: 10.1038/s41598-022-08908-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue WH, et al. CRISPR/Cas9-mediated knockout of two eye pigmentation genes in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) Insect Biochem. Mol. Biol. 2018;93:19–26. doi: 10.1016/j.ibmb.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 40.Berni M, et al. Atypical strategies for cuticle pigmentation in the blood-feeding hemipteran Rhodnius prolixus. Genetics. 2022 doi: 10.1093/genetics/iyac064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vargas-Lowman A, et al. Cooption of the pteridine biosynthesis pathway underlies the diversification of embryonic colors in water striders. Proc. Natl. Acad. Sci. USA. 2019;116:19046–19054. doi: 10.1073/pnas.1908316116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abbott CE. Inherited melanism in the large milkweed bug, Oncopeltus fasciatus (Heteropters: Lygaeidae) Ann. Entomol. Soc. Am. 1968;61:542. doi: 10.1093/aesa/61.2.542. [DOI] [Google Scholar]
- 43.Smissman EE, Orme JPR. A yellow mutant strain of the large milkweed bug, Oncopeltus fasciastus, that lacks erythropterin. Ann. Entomol. Soc. Am. 1969;62:246. doi: 10.1093/aesa/62.1.246. [DOI] [Google Scholar]
- 44.Lawrence PA. Some new mutants of large milkweed bug Oncopeltus fasciatus Dall. Genet. Res. 1970;15:347–350. doi: 10.1017/S0016672300001713. [DOI] [Google Scholar]
- 45.Forrest HS, Menaker M, Alexander J. Studies in the milkweed bug Oncopeltus fasciatus (Dallas) J. Insect Physiol. 1966;12:1411–1421. doi: 10.1016/0022-1910(66)90155-7. [DOI] [Google Scholar]
- 46.Shelton PMJ, Lawrence PA. Structure and development of Ommatidia in Oncopeltus fasciatus. J. Embryol. Exp. Morph. 1974;32:337–353. [PubMed] [Google Scholar]
- 47.Liu, J. UnravelingtheMolecularMechanismsofAposematicPigmentationinOncopeltusfasciatus. Ph.D.Thesis (2016).
- 48.Francescutti CM, Martin A, Hanly JJ. Knockdowns of red Malphigian tubules reveal pigmentation roles in the milkweed bug. J. Exp. Zool. B Mol. Dev. Evol. 2022;338:382–387. doi: 10.1002/jez.b.23123. [DOI] [PubMed] [Google Scholar]
- 49.Green MM. 2010: A century of Drosophila genetics through the prism of the white gene. Genetics. 2010;184:3–7. doi: 10.1534/genetics.109.110015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan SA, Reichelt M, Heckel DG. Functional analysis of the ABCs of eye color in Helicoverpa armigera with CRISPR/Cas9-induced mutations. Sci. Rep. 2017;7:40025. doi: 10.1038/srep40025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dorn A, Romer F. Structure and function of prothoracic glands and oenocytes in embryos and last larval instars of Oncopeltus fasciatus Dallas (Insecta, Heteroptera) Cell Tissue Res. 1976;171:331–350. doi: 10.1007/BF00224658. [DOI] [PubMed] [Google Scholar]
- 52.Berghammer AJ, Klingler M, Wimmer EA. A universal marker for transgenic insects. Nature. 1999;402:370–371. doi: 10.1038/46463. [DOI] [PubMed] [Google Scholar]
- 53.Grubbs N, Haas S, Beeman RW, Lorenzen MD. The ABCs of eye color in Tribolium castaneum: Orthologs of the Drosophila white, scarlet, and brown genes. Genetics. 2015;199:749–759. doi: 10.1534/genetics.114.173971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lorenzen MD, Brown SJ, Denell RE, Beeman RW. Cloning and characterization of the Tribolium castaneum eye-color genes encoding tryptophan oxygenase and kynurenine 3-monooxygenase. Genetics. 2002;160:225–234. doi: 10.1093/genetics/160.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kômoto N, Quan G, Sezutsu H, Tamura T. A single-base deletion in an ABC transporter gene causes white eyes, white eggs, and translucent larval skin in the silkworm w-3(oe) mutant. Insect Biochem Mol Biol. 2009;39(2):152–156. doi: 10.1016/j.ibmb.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 56.Kim H, et al. A co-CRISPR strategy for efficient genome editing in Caenorhabditis elegans. Genetics. 2014;197:1069–1080. doi: 10.1534/genetics.114.166389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 59.Milne I, et al. TOPALi v2: A rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 2009;25:126–127. doi: 10.1093/bioinformatics/btn575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Port F, Chen H, Lee T, Bullock S. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci. 2014;111(29):E2967–E2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gloor G, Engels W. Single-fly DNA preps for PCR. Drosophila Inf. Newslett. 1991;1:312–314. [Google Scholar]
- 63.Bhattacharya D, Van Meir EG. A simple genotyping method to detect small CRISPR-Cas9 induced indels by agarose gel electrophoresis. Sci. Rep. 2019;9:4437. doi: 10.1038/s41598-019-39950-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mutant strains and materials are available upon request. The gene sequences generated and/or analysed during the current study are available in the GenBank repository: vermilion, OQ151805; ok1, OQ151806; ok2, OQ151807; scarlet, OQ151808; xdh1, OQ151809; xdh2, OQ151810; v_exon2_WT, OQ151811; v_exon2_v1, OQ151812; v_exon2_v3, OQ151813; v_exon2_v4, OQ151814.