Abstract
Tight control of lateral root (LR) initiation is vital for root system architecture and function. Regulation of cortical microtubule reorganization is involved in the asymmetric radial expansion of founder cells during LR initiation in Arabidopsis (Arabidopsis thaliana). However, critical genetic evidence on the role of microtubules in LR initiation is lacking and the mechanisms underlying this regulation are poorly understood. Here, we found that the previously uncharacterized microtubule-stabilizing protein TPX2-LIKE5 (TPXL5) participates in LR initiation, which is finely regulated by the transcription factor ELONGATED HYPOCOTYL5 (HY5). In tpxl5 mutants, LR density was decreased and more LR primordia (LRPs) remained in stage I, indicating delayed LR initiation. In particular, the cell width in the peripheral domain of LR founder cells after the first asymmetric cell division was larger in tpxl5 mutants than in the wild-type. Consistently, ordered transverse cortical microtubule arrays were not well generated in tpxl5 mutants. In addition, HY5 directly targeted the promoter of TPXL5 and downregulated TPXL5 expression. The hy5 mutant exhibited higher LR density and fewer stage I LRPs, indicating accelerated LR initiation. Such phenotypes were partially suppressed by TPXL5 knockout. Taken together, our data provide genetic evidence supporting the notion that cortical microtubules are essential for LR initiation and unravel a molecular mechanism underlying HY5 regulation of TPXL5-mediated microtubule reorganization and cell remodeling during LR initiation.
The microtubule-stabilizing protein TPXL5 regulates microtubule reorganization and cell remodeling during LR initiation mediated by the transcription factor HY5.
IN A NUTSHELL.
Background: Tight control of lateral root (LR) initiation is vital for root system architecture. During LR initiation, the specific pairs of founder cells expand more in the central domains and less in the peripheral domains. Such an asymmetric radial expansion promotes the subsequent progression and outgrowth of LRs. However, the underlying cellular mechanisms are largely unclear. The reorganization of cortical microtubules is necessary for plant cell expansion, and is regulated by microtubule-associated proteins (MAPs). Microtubules are involved in the early events of LR initiation in Arabidopsis thaliana. However, the MAPs regulating microtubule reorganization during LR initiation remain unclear.
Question: We wanted to know whether and how specific MAPs mediate LR initiation by regulating microtubule reorganization.
Findings: We found that a microtubule-stabilizing protein TPX2-LIKE5 (TPXL5) participates in LR initiation. In the tpxl5 mutant, the radial expansion in the peripheral domain of LR founder cells after the first asymmetric division was stronger than that in the wild-type, and the ordered transverse cortical microtubule arrays were not well generated. Moreover, the transcription factor ELONGATED HYPOCOTYL5 (HY5) downregulated TPXL5 expression. Mutant hy5 exhibited the opposite phenotype compared to the tpxl5 mutant. Our study demonstrates that TPXL5 positively regulates cortical microtubules reorganization in the peripheral domain, thus promoting the asymmetric radial expansion of founder cells during LR initiation. It also reveals a novel molecular mechanism by which HY5 regulates TPXL5-mediated cortical microtubule reorganization and cell remodeling during LR initiation.
Next steps: In future studies, we will focus on how microtubule organization is specifically regulated in the peripheral and central domains of the founder cells. We will also examine whether there is a feedback mechanism between cortical microtubules and auxin signals in HY5-mediated LR development.
Introduction
The root system of land plants is anchored in the soil to absorb nutrients and water during plant growth and development, and to sense diverse signals enabling plant adaptation to the environment (Motte et al., 2019; Pierik et al., 2021). The overall surface of the root system is promoted and strengthened by root branches. Lateral root (LR) formation leads to a highly branched root system (Osmont et al., 2007; Bellini et al., 2014). The regular pattern of LRs, including the number, length and distribution, is a major determinant of root system architecture and is directly related to biological function (Lynch, 1995). LR formation is triggered by oscillations in gene expression over a confined root region known as the oscillation zone to determine future sites of LR primordium (LRP) initiation (Moreno-Risueno et al., 2010; Santos Teixeira and Ten Tusscher, 2019). In Arabidopsis (Arabidopsis thaliana), LRs originate from founder cells that are specific pairs of pericycle cells adjacent to the protoxylem (xylem pole pericycle cells, XPP cells) (Dubrovsky et al., 2001; Casimiro et al., 2003). Their nuclei migrate toward the common cell wall, reflecting the founder cell identity. These pairs of founder cells undergo an asymmetric, anticlinal division to form single-cell layered LRPs (stage I), leading to LR initiation. In this process, one crucial cellular event is the asymmetric radial expansion of founder cells, which expand more in the central domain than in the peripheral domain. This dynamic process creates a bulge in the central domain, which facilitates the subsequence progression and outgrowth of LRPs. The division planes in stage I LRP cells are reoriented and the following periclinal divisions give rise to a two-cell layered LRP (stage II). The central LRP cells continue to undergo several rounds of divisions, grow through the outer cell layers of the primary root (stage III–VII), and finally emerge from the primary root surface as the LR (stage VIII, also called stage E) (Malamy and Benfey, 1997; Péret et al., 2009; Benkova and Bielach, 2010; Du and Scheres, 2018). Most recent studies of LR development have focused on the mechanisms related to auxin biosynthetic pathways, auxin transport, and auxin signaling pathways. For example, auxin accumulates in founder cells to activate the auxin response module IAA-AUXIN RESPONSE FACTOR7 (ARF7)/ARF19, which regulates the expression of the downstream factors LATERAL ORGAN BOUNDARIES-DOMAIN16 (LBD16), LBD18, and LBD33 to mediate nuclear migration and the asymmetric divisions of founder cells during LR initiation (Lee et al., 2009; Goh et al., 2012; Santos Teixeira and Ten Tusscher, 2019). However, the cellular mechanisms underlying the regulation of the asymmetric radial expansion of founder cells during LR initiation are largely unclear.
The reorganization of cortical microtubules is required for plant cell expansion and plant survival in response to diverse developmental and environmental cues, a process that is regulated by different microtubule-associated proteins (MAPs) (Landrein and Hamant, 2013; Hamada, 2014; Wang and Mao, 2019). Cortical microtubules directly orientate cellulose fibrils to control plant cell expansion. Studies have indicated that an upstream signal, such as a phytohormone or light, directly regulates the gene expression levels and protein levels of MAPs to mediate microtubule organization for plant cell expansion (Wang et al., 2012; Lian et al., 2017). Auxin induces the reorientation of cortical microtubules from perpendicular to parallel to the long cell axis in root cells, and from longitudinal to transverse in light-grown hypocotyl cells (Chen et al., 2014; True and Shaw, 2020). However, the relationship between auxin-induced microtubule reorientation and cell elongation is complicated and remains controversial (Baskin, 2015; Adamowski et al., 2019). Increasing evidence indicates that microtubules provide feedbacks on polar auxin transport and alter the auxin maxima during plant cell growth (Ambrose et al., 2013; Deng et al., 2021). For example, the MAP WAVE-DAMPENED2-LIKE4 (WDL4) positively regulates apical hook opening by modulating proper auxin transport and distribution in differential plant cell growth (Deng et al., 2021). These reports thus demonstrate that the functions and regulations of microtubules in different tissues and organs are sophisticated. Previous studies indicated that microtubules are involved in the early events of LR initiation in Arabidopsis. Confocal imaging showed that cortical microtubules exhibit differential organizations in the central domain and peripheral domain of founder cells. Cortical microtubules form more isotropic arrays in the central domain and more transverse parallel arrays in the peripheral domain. Disruption of microtubules with the microtubule-depolymerizing agent oryzalin results in the overexpansion of founder cells in both central and peripheral domains (Vilches Barro et al., 2019). This evidence suggests that microtubule integrity and organization are required for the asymmetric radial expansion of founder cells. Although pharmacological evidence illustrates the impact of microtubules on LR initiation, crucial genetic evidence is still lacking. Some MAPs participate in LR development, such as KATANIN1 and MAP70-5; however, the MAPs involved in LR initiation remain unidentified (Ovečka et al., 2020; Stockle et al., 2022). In particular, much remains to be learned about the molecular mechanisms underlying the regulation of MAP-microtubule modules in LR initiation.
Targeting Protein for Xklp2 (TPX2) is an evolutionarily conserved MAP that mainly participates in microtubule nucleation and mitotic spindle assembly (Smertenko et al., 2021). Several TPX2-LIKE (TPXL) members have been identified in Arabidopsis, which are classified into group A (TPXL2/3/4/8) and group B (TPXL1/5/6/7) (Boruc et al., 2019; Dvorak Tomaštíková et al., 2020; Smertenko et al., 2021). TPXL proteins co-localize with microtubules, being particularly located at spindle microtubules as in the cases of TPXL2 and TPXL3. Genetic analysis showed that TPXL3 loss-of-function mutants exhibited embryonic lethality (Boruc et al., 2019). However, the physiological processes regulated by TPXL proteins are largely unclear. It is well established that TPX2 acts as an activator of the mitotic kinase Aurora A in humans and Aurora 1 in Arabidopsis (Gruss and Vernos, 2004; Boruc et al., 2019; Dvorak Tomaštíková et al., 2020). A previous study showed that aurora1 aurora2 double mutants display severe defects in LR formation and outgrowth (Van Damme et al., 2011; Lucas et al., 2013), suggesting that plant kinase Auroras play a role in LR development. However, whether and how TPXL proteins are involved in LR development remain unclear.
In this study, we found that a previously uncharacterized microtubule-stabilizing protein TPXL5 positively regulates the reorganization of cortical microtubules in founder cells, especially in the peripheral domain, thus facilitating the asymmetric radial expansion of founder cells during LR initiation. Moreover, the transcription factor ELONGATED HYPOCOTYL5 (HY5) inhibits LR initiation partially by suppressing TPXL5 expression, thereby maintaining proper LR development. These findings revealed the important roles of TPXL5 functioning downstream of HY5 to regulate cortical microtubule reorganization and cell remodeling during LR initiation.
Results
TPXL5 participates in LR initiation
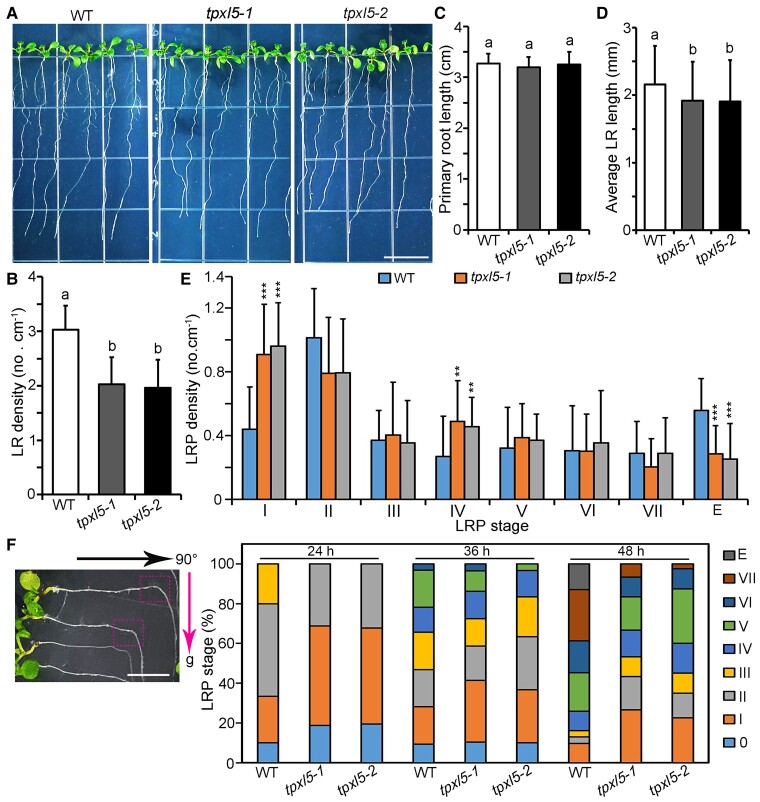
To understand the functions and mechanisms of the MAP-microtubule module in LR development, we examined the LR density in mutants of some MAPs that we have focused on in recent years, including MAP65-1, WDL3, WDL5, WDL7, and TPXL5. Compared with in the wild-type (WT), we found significantly decreased LR density in loss-of-function mutants of TPXL5 (tpxl5-1 and tpxl5-2, created by CRISPR/Cas9), but not in the other mutants (Figure 1, A and B; Supplemental Figure 1, A–D), demonstrating that TPXL5 participates in LR development. This phenotype was complemented by ProTPXL5:TPXL5-GFP (Supplemental Figure 2, A and B), indicating that the decreased LR density in tplx5-1 is caused by TPXL5 dysfunction. Further phenotype analysis showed that the primary root length was similar in the WT and tpxl5 mutants, whereas the average length of LRs was shorter in tpxl5 mutants than in the WT (Figure 1, C and D).
Figure 1.
TPXL5 promotes LR development. A, The LR phenotype of 10-day-old seedlings of the wild-type (WT) and tpxl5 mutants (tpxl5-1 and tpxl5-2). Scale bar = 1 cm. B, C, and D. Quantification of the LR density, primary root length, and LR length, as shown in (A). Values represent mean ± SD (n ≥ 30). Different letters represent significant differences at P < 0.05 (one-way ANOVA). The experiments were independently repeated three times. E, Analysis of the developmental stage distribution of LR primordia (LRPs) from (A). Sds (n = 15) are indicated. 0.01 < **P < 0.05 and ***P < 0.01, as determined by one-way ANOVA. F, Analysis of the developmental stage distribution of LRPs from 5-day-old seedlings of the WT and tpxl5 mutants (tpxl5-1 and tpxl5-2) in LR induction systems. LR synchronization was obtained after a 90° gravitropic stimulus, and LRPs were induced at the root bend site, as indicated by the boxes. The direction of the arrow labeled with g represents the direction of the gravistimulation induction. Graph shows the quantification of the percentage of LRPs in WT, tpxl5-1 and tpxl5-2 mutant seedlings at various developmental stages (n ≥ 30). Here 0 represents no LR initiation or obvious LRPs after gravistimulation. The experiment was repeated three times as different biological replicates.
LR formation involves multiple stages, including the stepwise priming of LRs, the establishment of founder cell identity, the initiation of LRs, the development of LRPs (stages I–VII), and the emergence of LRs (stage E) (Malamy and Benfey, 1997). To determine which stages TPXL5 participates in, we examined the developmental status of LRPs in the WT and tpxl5 mutants. We found that the LRP density in tpxl5 mutants was clearly altered in stages I, IV, and E (emergence) compared with that in the WT (Figure 1E). To further investigate the function of TPXL5 in LRP development, we performed an LR induction experiment to synchronize LR initiation by gravistimulation (Ditengou et al., 2008; Xun et al., 2020). The WT and tpxl5 mutants were grown for 5 days and then placed horizontally with their primary root tip bending 90° toward gravity. The developmental stages of LRPs were observed and analyzed after gravity induction for different time periods. We found that most LRPs in the WT developed to stage II, whereas about 50% of the LRPs in tpxl5 mutants remained in stage I after 24 h of LR induction, indicating slower LRP development in tpxl5 mutants. Consistently, we observed more LRPs remaining in stage I in tpxl5 mutants compared to in the WT after a longer induction for 36 and 48 h (Figure 1F). Taken together, these results demonstrate that TPXL5 positively regulates LRP development, especially the progression beyond stage I.
TPXL5 expression is negatively regulated by HY5
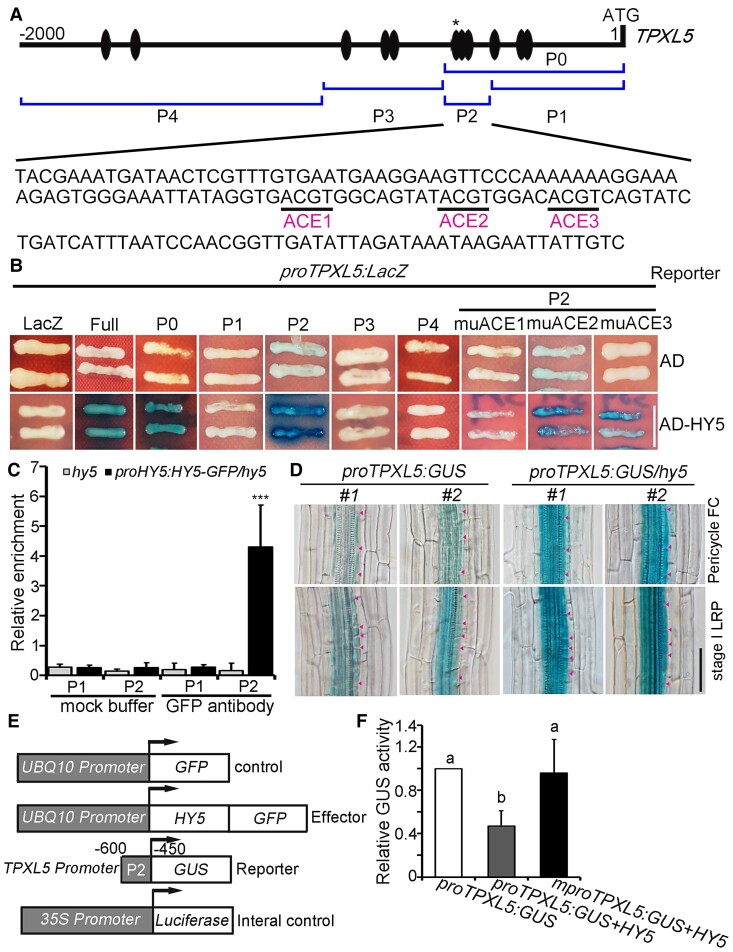
To investigate the regulatory mechanisms of TPXL5-mediated LR development, we generated transgenic seedlings overexpressing TPXL5-MYC and TPXL5-GFP. We found that both seedlings exhibited similar phenotypes with increased LR density (Supplemental Figure 3, A–D), suggesting that the expression level of TPXL5 is critical for its functions during LR development. Previous chromatin immunoprecipitation-DNA chip hybridization (ChIP-chip) analysis suggested that TPXL5 is a putative target of transcription factor HY5 (Lee et al., 2007). We analyzed the sequence of the TPXL5 promoter and found that it contains multiple putative ACGT-containing elements (ACEs) that are generally targeted by HY5, HY5 HOMOLOG (HYH), and PHYTOCHROME-INTERACTING FACTORs (PIFs) (Figure 2A) (Foster et al., 1994; Chattopadhyay et al., 1998; Shin et al., 2007). Yeast one-hybrid assays demonstrated that HY5, but not PIF1, PIF3, PIF4, PIF5, PIF7, or HYH, bound to the TPXL5 promoter (Supplemental Figure 4), indicating that TPXL5 is a HY5 target gene. Further fragmentation assays showed that HY5 bound to the P2 region of the TPXL5 promoter, which contains three ACEs (named ACE1, ACE2, and ACE3) (Figure 2, A and B). We then point-mutated these ACEs (muACE; ACGT was mutated to TTTT) to determine which one is critical for HY5 binding (Li et al., 2010). The results showed that ACE1, but not ACE2 and ACE3, was important for HY5 binding to the P2 region of the TPXL5 promoter (Figure 2B).
Figure 2.
HY5 directly binds to the promoter of TPXL5 and inhibits the expression of TPXL5. A, Diagram of the 2-kb promoter fragments of TPXL5 and the sequences of the P2 fragment. The adenine residue of the translational start codon (ATG) was assigned position +1. The ovals indicate ACE motifs and the asterisk indicates ACE1. P0, P1, P2, P3, and P4 indicate the corresponding promoter subfragments used in the yeast one-hybrid assays shown in (B). B, Yeast one-hybrid assays showing that HY5 binds to the TPXL5 promoter mainly through the ACE1 motif in the P2 fragment. muACE: ACGT was mutated to TTTT. ACE1, ACE2, and ACE3 in P2 fragment of the TPXL5 promoter were mutated separately. An empty vector expressing the AD domain alone was used as a negative control. Scale bar = 5 mm. C, ChIP assays showing that HY5 binds to the promoter of TPXL5 in vivo. Chromatin was immunoprecipitated from hy5 mutant seedlings expressing proHY5:HY5-GFP using anti-GFP mAb-Magnetic Agarose. The immunoprecipitated DNA was analyzed by qPCR using P1 and P2 primers. P1 primers were used to amplify the P1 fragment as a control. P2 primers were used to amplify the P2 fragment containing ACE1. The hy5 mutant and ACTIN were used as negative controls. Values represent mean ± Sd. ***P < 0.01 (one-way ANOVA). The experiments were independently repeated three times. D, TPXL5 is expressed in pericycle founder cells (FCs) and stage I lateral root primordia (LRPs) in transgenic seedlings expressing proTPXL5:GUS in the wild-type (WT) and hy5 mutant background. The seedlings used for GUS staining were 8 days old. Arrowheads indicate the edges of the cells. Scale bar = 50 μm. E, Schematic representation of the constructs used in the transient GUS/LUC assay in Nicotiana benthamiana leaves. The arrows indicate the transcriptional start site. Here −450 to −600 indicates the length of the TPXL5 promoter sequence (P2 fragment) that was fused to the pCAMBIA1391 vector to drive transcription of the GUS reporter gene. muACE: ACGT was mutated to TTTT. 35S:LUC was used as an internal control. F, Transient GUS/LUC assay showed that GUS expression driven by the TPXL5 promoter was decreased by the co-expression of HY5. Luciferase (LUC) was used as an internal control. GUS and LUC activity were quantified, and the GUS/LUC ratio was used to determine promoter activity. Values represent mean ± Sd. Different letters represent significant differences at P < 0.01 (one-way ANOVA). The experiments were independently repeated three times.
To verify whether HY5 binds to the TPXL5 promoter in vivo, we generated transgenic seedlings expressing ProHY5:HY5-GFP in the hy5 mutant background. Phenotype analysis showed that ProHY5:HY5-GFP fully rescued the typical phenotypes of hy5 mutants, including longer hypocotyls and shorter primary roots in light-grown seedlings, and also higher LR density (Supplemental Figure 5, A–E). These results indicated that the HY5-GFP fusion protein could function normally in these seedlings. We then performed ChIP-qPCR with the roots of these seedlings. The results showed that the P2 fragment containing ACE1 was enriched in chromatin immunoprecipitated with the anti-GFP antibody, while the P1 fragment showed no enrichment (Figure 2C). These results confirmed that TPXL5 is a direct target gene of HY5.
To analyze how HY5 regulates TPXL5 during LR initiation, we generated transgenic seedlings expressing ProTPXL5:GUS in the WT and hy5 mutant backgrounds. GUS staining showed that TPXL5 is widely expressed in hypocotyls, primary roots, and LRs, but not in flowers, siliques, and root hair (Supplemental Figure 6, A–G). In addition, close observation showed clear staining in different stages of LRPs, indicating that TPXL5 expression is obviously related to LR development (Supplemental Figure 6H, first and third lines). The staining was almost invisible in outer cell layers in primary roots including the endodermis, cortex and epidermis cells. However, the staining was obvious in vasculature and pericycle cells. In particular, TPXL5 was expressed in stage I LRPs and in pericycle founder cells (Figure 2D). Moreover, the staining was clearly increased in hy5 mutants, indicating that HY5 inhibited TPXL5 expression (Figure 2D). Next, a transient GUS activity assay was performed in Nicotiana benthamiana leaves to analyze the direct effect of HY5 on TPXL5 expression. ProTPXL5:GUS or mProTPXL5:GUS with mutated ACE1 was co-transformed with the construct harboring the HY5 coding sequence driven by the UBQ10 promoter. Relative GUS activity was significantly decreased when HY5 was co-infiltrated with ProTPXL5:GUS, but not with mProTPXL5:GUS, demonstrating the negative effect of HY5 on TPXL5 expression through a target ACE1 in the TPXL5 promoter (Figure 2, E and F). Collectively, these results demonstrated that HY5 directly targets TPXL5 and downregulates TPXL5 expression during LR initiation.
TPXL5 functions as a microtubule stabilizer
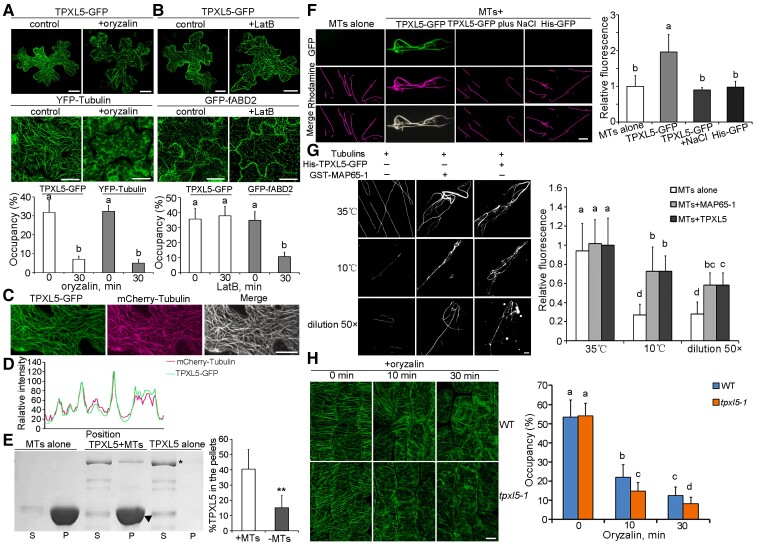
TPXL5 is a potential MAP with unknown functions in microtubules (Dvorak Tomaštíková et al., 2020; Smertenko et al., 2021). Transient expression analysis showed that TPXL5-GFP exhibited a filamentous structure in Arabidopsis pavement cells (Figure 3, A and B). Furthermore, the microtubule-disrupting reagent oryzalin and the actin filament-depolymerizing reagent LatB were used to investigate the characteristics of TPXL5-GFP filamentous structure. YFP-Tubulin5A was used to label microtubules and GFP-fimbrin actin-binding domain 2 (GFP-fABD2) was used to label actin filaments. To quantitatively analyze the changes in these filamentous structures, we calculated the occupancy of GFP/YFP signals to reflect their density as described previously by Higaki et al. (2010) and Dou et al. (2021). The density of TPXL5-GFP filaments and microtubules was clearly decreased by treatment with 50 μM oryzalin for 30 min (Figure 3A). In contrast, TPXL5-GFP filaments were largely unaffected when the cells were treated with 0.5 μM LatB for 30 min, while the density of actin filaments were significantly decreased (Figure 3B). These results suggest that the filamentous localization of TPXL5-GFP is related to microtubules and not to actin filaments. To confirm this result, we transiently expressed TPXL5-GFP in the pavement cells of transgenic Arabidopsis seedlings expressing mCherry-Tubulin. Confocal microscopy revealed that the green fluorescent signal of TPXL5-GFP overlapped with the red fluorescent signal of mCherry-Tubulin. By plotting their signal intensities using ImageJ software, we observed a close association between GFP and mCherry signals (Figure 3, C and D, and Supplemental Figure 7). The results demonstrated that TPXL5 colocalized with cortical microtubules in vivo. Furthermore, an in vitro co-sedimentation assay showed that the His-TPXL5-GFP fusion protein directly bound to paclitaxel-stabilized microtubules in vitro (Figure 3E). This evidence demonstrated the property of TPXL5 as a MAP.
Figure 3.
TPXL5 directly binds to and stabilizes microtubules (MTs). A, Ballistics-mediated transient expression of UBQ10:TPXL5-GFP in Arabidopsis (Arabidopsis thaliana) (Col-0) leaf epidermal cells. TPXL5-GFP exhibits a filamentous pattern in pavement cells. The TPXL5-GFP filaments and microtubules (labeled by YFP-Tubulin5A) were treated with 50 μM oryzalin for 30 min. Scale bar = 20 μm. The graph shows the densities of the filaments. Values represent mean ± Sd (n ≥ 15 cells). Different letters represent significant differences at P < 0.01 (one-way ANOVA). B, The TPXL5-GFP filaments and actin filaments (labeled by GFP-fABD2) were treated with 0.5 μM LatB for 30 min. Scale bar = 20 μm. The graph shows the densities of the filaments. Values represent mean ± Sd (n ≥ 15 cells). Different letters represent significant differences at P < 0.01 (one-way ANOVA). C, Ballistics-mediated transient expression of UBQ10:TPXL5-GFP in leaf epidermal cells of mCherry-Tubulin6B transgenic seedlings. TPXL5-GFP colocalizes with the microtubule marker mCherry-Tubulin6B in Arabidopsis pavement cells. Scale bar = 20 μm. D, Plot of the relative signal intensities along the line drawn in (C). It shows a strong association between the spatial localization of TPXL5-GFP and mCherry-Tubulin6B. E, His-TPXL5-GFP fusion proteins were co-sedimented with paclitaxel-stabilized microtubules in vitro. The supernatant (S) and pellet (P) fractions are labeled. Symbol * represents TPXL5 fusion proteins, ▴ represents tubulins. The graph shows the percentage of TPXL5 in the pellets. t-test, **P < 0.01. Values represent mean ± Sd. The experiment was repeated three times with similar results. F, Fluorescent images showing the effect of TPXL5 on the microtubules in vitro. Rhodamine-labeled microtubules were stabilized by paclitaxel and incubated with His-TPXL5-GFP or His-GFP protein for 10 min before the addition of 200 mM NaCl. The graph shows the relative fluorescence intensity of rhodamine-labeled microtubules. Values represent mean ± Sd (n ≥ 10). The experiment was repeated three times with similar results. Scale bar = 10 μm. G, Fluorescent images showing the microtubule-stabilizing activity of TPXL5 in vitro. Rhodamine-labeled tubulins were incubated with TPXL5 fusion proteins for 40 min. The system was then treated at 10°C for 30 min or diluted (50×) for 60 min. GST-MAP65-1 was used as a positive control. The graph shows the relative fluorescence intensity of rhodamine-labeled microtubules. Values represent mean ± Sd (n ≥ 10). Different letters represent significant differences at P < 0.01 (one-way ANOVA). The experiment was repeated three times with similar results. Scale bar = 10 μm. H, The microtubule-stabilizing activity of TPXL5 in vivo. Cortical microtubules in epidermal cells in the middle regions of hypocotyls were observed in wild-type (WT) and tpxl5-1 seedlings expressing YFP-Tubulin. The seedlings were grown in the light for 4 days and treated with 10 μM oryzalin for 0, 10, and 30 min. Scale bar = 20 μm. The graph shows the densities of the cortical microtubules. Values represent mean ± Sd (n ≥ 30 cells). Different letters represent significant differences at P < 0.01 (one-way ANOVA). The experiments were independently repeated three times.
To test the effect of TPXL5 on microtubules in vitro, we incubated His-TPXL5-GFP with paclitaxel-stabilized microtubules polymerized from rhodamine-labeled tubulins, and His-GFP was used as a control. Confocal imaging revealed that His-TPXL5-GFP induced the formation of relatively large microtubule bundles, but not His-GFP alone. When NaCl was added to the system to strip His-TPXL5-GFP from microtubules, the large bundles dispersed into single filaments (Figure 3F), further confirming the bundling activity of TPXL5. Additional in vitro assays showed that TPXL5 could stabilize the microtubules when low temperature and dilution treatment induced microtubule depolymerization (Figure 3G). These results together demonstrated that TPXL5 can bundle and stabilize microtubules in vitro.
We then investigated the microtubule-stabilizing activity of TPXL5 in vivo. Phenotypic analysis showed that hypocotyl elongation of tpxl5 mutants was clearly suppressed compared with that of WT when the plants were grown in the light for 7 days, which could be complemented by ProTPXL5:TPXL5-GFP (Supplemental Figure 2C). Statistical analysis indicated that the length of hypocotyl epidermal cells was decreased in tpxl5 mutant seedlings, particularly in the middle region (Supplemental Figure 2D). To observe the microtubule organization and stability in hypocotyl epidermal cells, we generated transgenic seedlings expressing 35S:YFP-Tubulin in the WT and tpxl5 mutant backgrounds. The seedlings were grown in the light for 4 days, when the hypocotyls are in a rapid elongation stage (Liu et al., 2013). Confocal microscopy showed that the parallel arrays of cortical microtubules in WT hypocotyl epidermal cells were generally transversely and obliquely oriented relative to the hypocotyl growth axis, while the microtubule organization was more random in tpxl5 mutants, with a higher portion of oblique and longitudinal arrays (Supplemental Figure 2E). These results were consistent with the decrease in hypocotyl cell elongation in tpxl5 mutants. Moreover, WT and tpxl5 seedlings expressing 35S:YFP-Tubulin were treated with microtubule-disrupting drug oryzalin. We used the density of cortical microtubules in hypocotyl epidermal cells to assess the effects of oryzalin on the stability of cortical microtubules (Higaki et al., 2010; Dou et al., 2021). Before treatment, the density of cortical microtubules in WT hypocotyl epidermal cells was similar to that in tpxl5 mutants. After treatment with 10 μM oryzalin for 10 min, the density of cortical microtubules was decreased in WT cells. Moreover, this effect was obviously enhanced in tpxl5 mutant cells. Longer oryzalin treatment (30 min) resulted in disruption of most cortical microtubules in tpxl5 mutant cells, whereas more cortical microtubules were observed in WT cells (Figure 3H). These results confirmed that TPXL5 functions as a microtubule stabilizer.
TPXL5 regulates the asymmetric radial expansion of LR founder cells by arranging microtubule arrays
Cortical microtubule reorganization is concomitant to the asymmetric radial expansion of founder cells during LR initiation. We found that TPXL5 is a microtubule-stabilizing protein and participates in LR initiation. Thus we reasoned that TPXL5 might regulate cortical microtubule reorganization and founder cell remodeling to promote LR initiation. To confirm this hypothesis, we generated transgenic seedlings expressing 35S:GFP-MBD in the WT and tpxl5 mutant backgrounds. Phenotype analysis showed that GFP-MBD did not affect the density of LRs (Supplemental Figure 8). We performed an LR induction experiment by gravistimulation and found that pericycle cells in the induction regions had similar cell widths before the first asymmetric cell division (ACD) in the WT and tpxl5 mutant (Supplemental Figure 9, A and C). We further quantified the anisotropy (whether microtubules are well ordered, where 0 represents no order and 1 represents perfectly ordered) of cortical microtubules in these cells using FibrilTool (Boudaoud et al., 2014; Vilches Barro et al., 2019). We found that the cortical microtubules exhibited anisotropic arrays that were oriented in a spiral along the long cell axis in both the WT and tpxl5 mutant (Supplemental Figure 9, B and D), which is consistent with the previous work (Vilches Barro et al., 2019). Taken together, these pieces of evidence indicate that TPXL5 does not participate in the regulation of microtubule arrangement and cell expansion in founder cells prior to the first ACD.
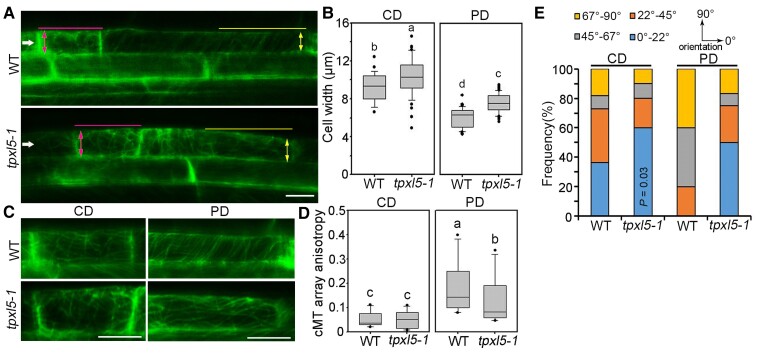
Previous studies have shown that cortical microtubules in LR founder cells become more isotropic in the central domain and form a well-organized array transverse to the long cell axis in the peripheral domain, which together facilitate the asymmetric radial expansion of founder cells (Vilches Barro et al., 2019). We further analyzed the cell expansion and microtubule organization in the central and peripheral domains of LR founder cells after the first ACD. The results showed that the cell width in both central and peripheral domains in tpxl5 mutants were clearly larger than those in the WT (Figure 4, A and B). This result indicated that the founder cells in tpxl5 mutants were more “expansive” during LR initiation, and especially the radial expansion of the peripheral domain was less constrained. Such an altered asymmetric radial expansion of founder cells may delay the LRP progression and result in more LRPs remaining in stage I in tpxl5 mutants. Then, we checked the cortical microtubule arrangements and orientations (stacks were rotated so that the longitudinal axis of cells was horizontal [0°]) in founder cells after the first ACD. In the central domain, cortical microtubules organized in similar isotropic arrays in both the tpxl5 mutant and WT (Figure 4, C and D, left panel). Accordingly, cortical microtubules exhibited similar orientation patterns, although the portion of cortical microtubules orientated from 0° to 22° was clearly increased in tpxl5 mutants compared with in the WT (Figure 4E, left panel).
Figure 4.
Cortical microtubule reorganization and asymmetric radial expansion of lateral root (LR) founder cells in the tpxl5-1 mutant are clearly altered during LR initiation. A, Confocal images of the founder cells (shootward end) after the first ACD in the wild-type (WT) and tpxl5-1 mutant were visualized. Six-day-old WT and tpxl5-1 mutant seedlings expressing 35S:GFP-MBD under continuous white light were induced by gravistimulation (180°) for 15 h. The filled arrowheads indicate the founder cells after the first ACD (LR primordium (LRP) stage I). The lines mark the zone in which the orientation and anisotropy of the cortical microtubule arrays were quantified in the central domain (CD) and peripheral domain (PD), respectively. The double arrowheads mark the cell width in the CD and PD, respectively. Scale bar = 10 μm. B, Quantification of founder cell radial expansion. Cell width was measured in the CD and PD from (A). n ≥ 30 cells from 4 independent experiments. The box and whisker plots generated by SigmaPlot version 10.0 show minimum and maximum values, respectively. The line in the box indicates the median value, and the boundaries represent the 25th percentile (upper) and 75th percentile (lower). The outliers were indicated by points. Different letters represent significant differences at P < 0.01 (one-way ANOVA). C, Larger views showing the arrangement of cortical microtubules in the CD and PD from A. Scale bar = 10 μm. D, Quantification of the anisotropy (0 = no order; 1 = order) of cortical microtubule arrays in the CD and PD from (C), respectively. The results were obtained from 12 cells from eight individual roots from four independent experiments for the WT and from 11 cells from nine individual roots from four independent experiments for the tpxl5-1 mutant. The box and whisker plots show minimum and maximum values. The line in the box indicates the median value, and the boundaries represent the 25th percentile (upper) and 75th percentile (lower). The outliers were indicated by points. Different letters represent significant differences at P < 0.05 (one-way ANOVA). E, Quantification of the orientation (0°–90°, respective to the long axis of the cell) of cortical microtubule arrays in the CD and PD from (C), respectively. The results were obtained from the same cells from (D). P = 0.03 indicates significant difference between the frequency of microtubules from 0° to 22° in WT and that in tpxl5-1.
In the peripheral domain, the anisotropy of cortical microtubules was significantly decreased in tpxl5 mutants compared with in the WT, suggesting less-ordered microtubule organization in tpxl5 mutants (Figure 4, C and D, right panel). Indeed, we observed more well-organized microtubule arrays transverse to the cell long axis in the WT compared with in the tpxl5 mutant (Figure 4C). Orientation analysis of the cortical microtubules was consistent with this observation. Moreover, a high frequency of microtubule arrays from 0° to 22° was observed in the tpxl5 mutant but not in the WT (Figure 4E, right panel). Notably, the microtubule orientation patterns in the peripheral domain and central domain were clearly different in the WT, while they were similar in the tpxl5 mutant (Figure 4E). Thus, the differential microtubule organization in the founder cells was less obvious in the tpxl5 mutant, consistent with the altered asymmetric radial expansion of founder cells in the tpxl5 mutant.
Taken together, this evidence supported the notion that TPXL5 is required for the asymmetric radial expansion of founder cells through regulating the anisotropy and orientation of cortical microtubules, particularly in the peripheral domain.
HY5 negatively regulates LR initiation and controls the microtubule arrangement in founder cells
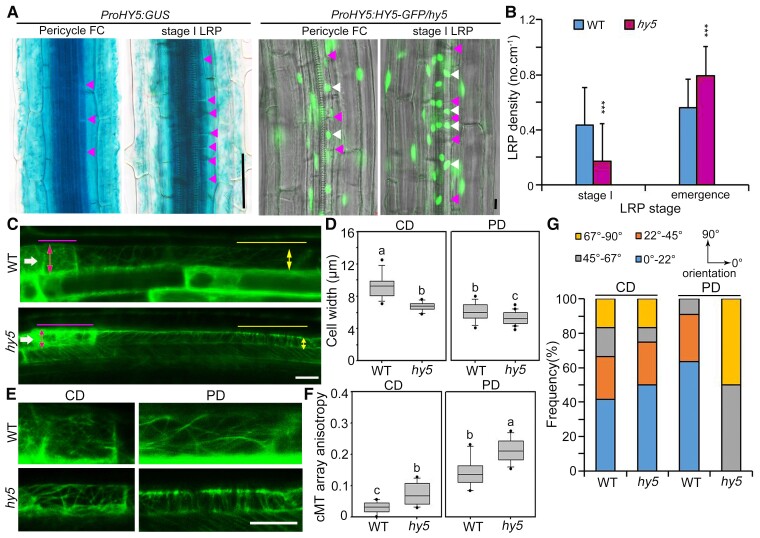
Previous studies showed significantly increased LR density in hy5 mutants (Oyama et al., 1997; Cluis et al., 2004), indicating that HY5 participates in LR formation. We identified TPXL5 as a direct target gene of HY5 and a positive regulator of LR initiation. To further investigate whether HY5 is involved in LR initiation, we first analyzed the expression pattern of HY5 during LR development using ProHY5:GUS transgenic seedlings. GUS staining showed that HY5 was broadly expressed in diverse root tissues during LR development, with the strongest expression in vasculature tissue (Supplemental Figure 6H). In particular, HY5 was highly expressed in pericycle founder cells and stage I LRPs during LR initiation (Figure 5A, left panels). Confocal observation using hy5 mutant seedlings expressing proHY5:HY5-GFP confirmed that HY5-GFP was expressed in the nucleus in the pericycle founder cells and stage I LRP cells (Figure 5A, right panels). Furthermore, we examined the developmental stages of LRPs in the hy5 mutant. The density of LRPs in the emergence stage was increased in the hy5 mutant (Figure 5B, right panel), which is consistent with previous studies (Cluis et al., 2004; van Gelderen et al., 2018a, 2018b, 2021). Moreover, the density of stage I LRPs was significantly decreased in the hy5 mutant (Figure 5B, left panel), showing that fewer stage I LRPs remained in the hy5 mutant. This phenomenon suggests that LRP development in the hy5 mutant was accelerated through stage I and that HY5 negatively regulates LR initiation.
Figure 5.
HY5 negatively regulates LR initiation and controls the microtubule arrangement in founder cells. A, HY5 is expressed in pericycle founder cells (FCs) and stage I LR primordia (LRPs) in the roots. GUS staining showed the expression pattern of HY5 in 8-day-old transgenic seedlings expressing proHY5:GUS. Confocal images from 8-day-old proHY5:HY5-GFP transgenic seedlings in the hy5 mutant background showed the expression pattern of HY5-GFP. GFP signal represents the expression of HY5 in the nucleus. The cell edges and nuclei in founder cells and stage I LRPs are indicated by arrowheads. Scale bar = 50 μm. B, Analysis of LRP density in stage I and stage E in 10-day-old wild-type (WT) and hy5 mutant seedlings. Sds (n = 11) are indicated. ***P < 0.05, as determined by one-way ANOVA. C, Confocal image of the founder cells (shootward end) after the first ACD in WT and hy5 mutant seedlings expressing 35S:GFP-MBD. Six-day-old seedlings under continuous white light were induced by gravistimulation (180°) for 8 h. The filled arrowheads indicate the founder cells after the first ACD. The lines mark the zone in which the orientation and anisotropy of the cortical microtubule array were quantified in the central domains (CD) and peripheral domains (PD), respectively. The double arrowheads mark the cell width in the CD and PD, respectively. Scale bar = 10 μm. D, Quantification of founder cell radial expansion. Cell widths were measured in the CD and PD from (C). Multiple cells (n ≥ 13) from 4 independent experiments were analyzed. The box and whisker plots generated by SigmaPlot version 10.0 show minimum and maximum values, respectively. The line in the box indicates the median value, and the boundaries represent the 25th percentile (upper) and 75th percentile (lower). The outliers were indicated by points. Different letters represent significant differences at P < 0.05 (one-way ANOVA). E, Larger views showing the arrangement of cortical microtubules in the CD and PD from (C). Scale bar = 10 μm. F and G. Quantification of anisotropy (0 = no order; 1 = order) and orientation (0°–90°, relative to the long axis of the cell) of cortical microtubule organization in the CD and PD from (E). The results were obtained from 10 cells from 10 individual roots from four independent experiments for the WT and from 12 cells from 10 individual roots in four independent experiments for the hy5 mutant. The box and whisker plots show minimum and maximum values. The line in the box indicates the median value, and the boundaries represent the 25th percentile (upper) and 75th percentile (lower). The outliers were indicated by points. Different letters represent significant differences at P < 0.05 (one-way ANOVA).
Next, we generated transgenic seedlings overexpressing GFP-MBD in the hy5 mutant background to analyze the asymmetric radial expansion and microtubule organization of founder cells during LR initiation. We performed an LR induction experiment by gravistimulation and found that pericycle cells in the induction regions had similar cell widths before the first ACD in the WT and hy5 mutant (Supplemental Figure 10, A and C). Accordingly, the anisotropy of cortical microtubules in these cells was similar in the hy5 mutant and WT (Supplemental Figure 10, B and D), suggesting that HY5 did not regulate cell expansion or microtubule arrangement in founder cells before the first ACD.
Then, we measured the cell width of LR founder cells after the first ACD and found that the cell widths in both central and peripheral domains were clearly decreased in the hy5 mutant (Supplemental Figure 11, A and B), indicating that radial expansion of founder cells in the hy5 mutant is more constrained than that in the WT during LR initiation. Statistical analysis revealed that the cortical microtubules in the central domain in the hy5 mutant were more ordered than those in the WT, and those in the peripheral domain were similarly well-organized in both the WT and hy5 mutant (Supplemental Figure 11, C and D). Surprisingly, the cortical microtubules in the peripheral domain in the hy5 mutant were more parallel to the long axis of the cell (Supplemental Figure 11, C and E). However, this is not in agreement with a previous study showing that ordered microtubule arrays transverse to the cell long axis in the peripheral domain facilitate the inhibition of radial expansion (Vilches Barro et al., 2019). LR development is accelerated in the hy5 mutant according to our results and a previous study (Cluis et al., 2004). Thus, we observed cell expansion and microtubule organization early after gravistimulation (8 h, compared with 15 h for above microscopic observations of the cortical microtubules in the founder cells). In both central and peripheral domains of founder cells after the first ACD, the cell width was decreased in the hy5 mutant and the microtubule arrays were more ordered (Figure 5, C–G). Importantly, we observed well-organized arrays transverse to the cell long axis in the peripheral domain in the hy5 mutant, supporting the restricted expansion (Figure 5E). Results of the orientation analysis of the cortical microtubules were consistent with this observation, showing a high frequency of cortical microtubules orientated from 67° to 90° in the hy5 mutant but not in the WT (Figure 5G). We also checked the tpxl5 mutant at the same time frame. The tpxl5 mutant showed opposite changes to those in the hy5 mutant during cell expansion and microtubule orientation in the peripheral domains, including larger cell widths than those in the WT and more ordered microtubule arrays parallel to the cell long axis (Supplemental Figure 12).
Taken together, these results demonstrate that HY5 is required for the asymmetric radial expansion of founder cells by controlling microtubule arrangement and that HY5 negatively regulates LR initiation.
TPXL5 knockout weakens the acceleration of LR initiation in the hy5 mutant
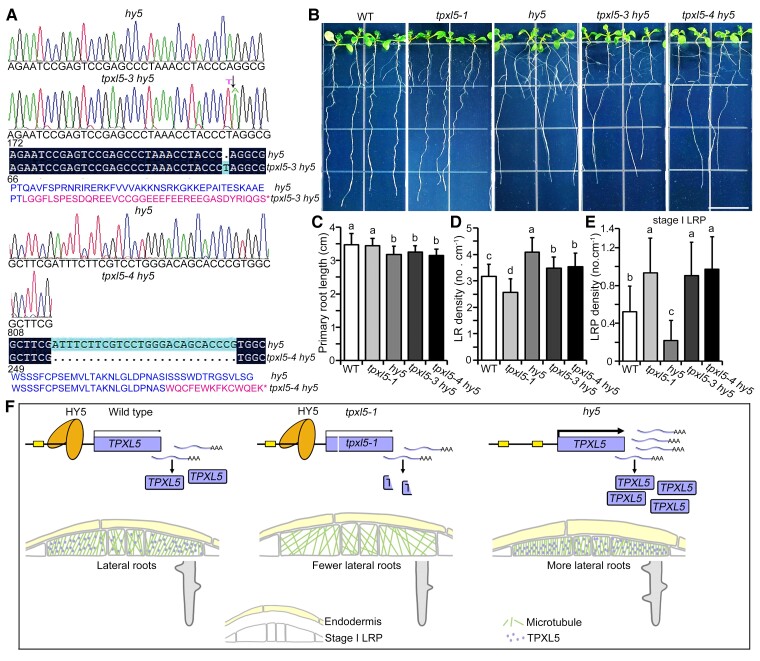
Because HY5 downregulates TPXL5 expression during LR initiation, we hypothesized that knockout of TPXL5 would partially counteract the acceleration of LR initiation in the hy5 mutant. We tried to generate a tpxl5 hy5 double mutant by crossing. TPXL5 and HY5 are located on chromosome 5. Unfortunately, we failed to obtain the double mutant combination, probably caused by a lack of recombination. Therefore, we generated two tpxl5 hy5 double mutant lines using the CRISPR/Cas9 system (tpxl5-3 hy5 and tpxl5-4 hy5; Figure 6A). Phenotype analysis showed similar primary root lengths in the tpxl5 hy5 double mutants and hy5 mutant (Figure 6, B and C), whereas the LR density in tpxl5 hy5 double mutants was clearly decreased compared with that in the hy5 mutant (Figure 6D). This indicated that TPXL5 functions genetically downstream of HY5 during LR development, but not during primary root growth. Furthermore, we examined the density of stage I LRPs in tpxl5 hy5 double mutants. We found that the lower density of stage I LRPs in the hy5 mutant was mostly recovered in tpxl5 hy5 double mutants (Figure 6E). These results indicated that HY5 controls LR initiation and development partially by repressing TPXL5 expression.
Figure 6.
Knockout of TPXL5 partially suppresses the increase of LR density in the hy5 mutant. A, Schematic diagrams showing the sequencing results of the tpxl5 hy5 double mutants generated by CRISPR/Cas9. TPXL5 translation is early terminated in the hy5 mutant background. The tpxl5-3 hy5 and tpxl5-4 hy5 mutants encode terminators at positions 106 and 287, respectively, compared with the amino acid sequence of TPXL5. Normal and mutated amino acid sequences of TPXL5 were shown. Symbol * represents terminator. B, The LR phenotype of 10-day-old seedlings of wild-type (WT), tpxl5-1, hy5, tpxl5-3 hy5, and tpxl5-4 hy5 seedlings. Scale bar = 1 cm. C and D, Quantification of the primary root length and density of LRs as shown in (B). Values represent mean ± Sd (n > 20). Different letters represent significant differences at P < 0.01 (one-way ANOVA). The experiments were independently repeated three times. E, Analysis of stage I LR primordia (LRPs) in the seedlings from (B). Sds (n = 15) are indicated. Different letters represent significant differences at P < 0.05 (one-way ANOVA). The experiments were independently repeated three times. F, Working model for the function of TPXL5 during HY5-mediated LR initiation. In the WT, HY5 inhibits the expression of TPXL5 and thereby regulates microtubule arrangement and asymmetric radial expansion in the peripheral domains of LR founder cells, properly controlling the initiation and development of LRs. In tpxl5 mutants, the anisotropy and orientation of cortical microtubule arrays were clearly altered, which resulted in abnormal radial expansion in the peripheral domains of LR founder cells, thereby inhibiting the initiation and development of LRs. In the hy5 mutant, the expression of TPXL5 was increased, which led to more transverse microtubule arrays and suppression of asymmetric radial expansion in the peripheral domain, promoting the initiation and development of LRs.
Discussion
Precise regulation of LR development is important for root system architecture and root functions, especially in response to diverse environmental and stress cues. In this study, we found that the microtubule-stabilizing protein TPXL5 participates in LR initiation by altering the microtubule reorganization and asymmetric radial expansion of founder cells. Furthermore, the transcription factor HY5 controls LR initiation partially by suppressing TPXL5 expression, thereby maintaining proper LR development. Our results shed new light on the microtubule reorganization in LR initiation and suggest a hypothetical model for the function of TPXL5 and HY5 in LR initiation (Figure 6F).
Diverse upstream signals regulate MAP-microtubule modules to directly mediate plant cell expansion, which have been well investigated in hypocotyls (Wang et al., 2012; Lian et al., 2017). However, the roles and regulatory mechanisms of cortical microtubules in LR initiation remain unclear. A previous study hypothesized that cortical microtubules do not function as the driver of the initial differential growth between the central and peripheral domains, but as an amplifier (Vilches Barro et al., 2019). In this study, we provide critical genetic evidence supporting the notion that proper regulation of cortical microtubule reorganization is required for the asymmetric radial expansion of founder cells. During LR initiation, the organization of cortical microtubules and radial expansion in founder cells after the first ACD are clearly altered in tpxl5 and hy5 mutants compared with those in WT. Accordingly, LR density and LR initiation are abnormal in tpxl5 and hy5 mutants. Knockout of TPXL5 partially restored the phenotype of the hy5 mutant, including fewer LRPs in stage I and the increased LR density. Thus, the tight control of TPXL5 expression by HY5 and the subsequent regulation of microtubule organization are important for proper LR initiation and further development.
If the LR primordium was pressurized and the periphery was more constrained than the central domain, the central domain would expand more than the peripheral domain, creating a bulge (Lucas et al., 2013; Schutz et al., 2021). Confocal imaging and pharmacological assays suggest that cortical microtubules become more isotropic in the central domain after the first ACD, which is suggested to facilitate the radial expansion. By contrast, cortical microtubules exhibit more well-organized transverse arrays in the peripheral domain, which is believed to facilitate the inhibition of radial expansion (Vilches Barro et al., 2019). In this study, the organization of cortical microtubules was found to be significantly altered in the peripheral domain in tpxl5 and hy5 mutants, but not in the central domain, demonstrating the importance of TPXL5 and HY5 for the re-establishment of organized microtubule arrays in the peripheral domain after the first ACD during LR initiation. This is consistent with a previous report showing that microtubule destabilization through expressing a mutated version of the atypical tubulin kinase PROPYZAMIDE-HYPERSENSITIVE 1 (PHS1ΔP) in founder cells resulted in greater expansion of the peripheral domain but not the central domain (Vilches Barro et al., 2019). Accordingly, we found elevated radial expansion in the peripheral domain and more LRPs remaining in stage I in tpxl5 mutants, suggesting delayed LR development. In contrast, the radial expansion in the peripheral domains was significantly suppressed in hy5 mutants, and fewer LRPs remained in stage I, which is consistent with previous reports showing that LR development is accelerated in the hy5 mutant (Cluis et al., 2004). Taken together, these lines of evidence support the notion that it is not important for the reinforcement of the central region to change in any particular way, only that the central region be less heavily restricted than the periphery, which adds to our understanding of how cells may transition from a callus-like proliferative state in which all cells are expanding at roughly equal rates to the formation of distinct, individualized primordia.
Confocal imaging showed the differential arrangements of cortical microtubules between the central and peripheral domains of founder cells. In this study, we also found enhanced microtubule stability and bundling in the peripheral domain compared with in the central domain (Supplemental Figure 13). This is consistent with the functions of TPXL5 on microtubules and supports the notion that TPXL5 mainly affects cortical microtubule organization in the peripheral domains. However, the expression patterns of TPXL5 were comparable between these two domains and so were those of its negative regulator HY5. This suggests that the regulatory module HY5-TPXL5 may not participate in establishing the initial differences but does facilitate the maintenance of the differences in microtubule organization and stability between these two domains in founder cells, thereby regulating proper LR initiation. This evidence suggests that some unknown microtubule regulators may be differentially expressed or regulated between the central and peripheral domains. Future study should be performed to investigate how microtubule organization and stability are specifically regulated in the two domains of founder cells.
HY5 is a key negative regulator of plant photomorphogenesis, and the hy5 mutant exhibits multiple developmental phenotypes, including longer hypocotyls and shorter primary roots in light-grown seedlings and also increased LR density (Oyama et al., 1997; Gangappa and Botto, 2016; Zhang et al., 2017, 2019). Although TPXL5 is a direct target gene of HY5, their functions do not fully overlap. TPXL5 participates in hypocotyl elongation and LR initiation but not in the elongation of primary roots. In addition, we found that HY5 is expressed more broadly than TPXL5 during LR development. For example, HY5 but not TPXL5 is clearly expressed in the endodermis, which plays a role in the differential expansion of pericycle cells (Stockle et al., 2022), indicating the complex functions of HY5 in root development. So far, the molecular mechanisms of HY5-regulated LR development are largely unclear. Studies showed that HY5 inhibits the expression of ARF19, which is involved in LR initiation, likely by directly binding its promoter (Lee et al., 2007). Moreover, HY5 reduces auxin concentration in the cortex overlaying LRPs by decreasing the abundance of PIN-FORMED3 (PIN3) and LIKE-AUXIN3 (LAX3), thereby inhibiting LR emergence (van Gelderen et al., 2018a, 2018b). These results revealed a connection between HY5 and auxin signaling during LR development. In this study, we found that HY5-regulated founder cell expansion and microtubule organization partially through inhibiting TPXL5 expression, especially in the peripheral domain of founder cells after the first ACD during LR initiation. However, the relationship between auxin-dependent and microtubule-dependent mechanisms during HY5-mediated LR development is still unknown. Previous studies showed that perturbing auxin signaling leads to the inhibition of asymmetric radial expansion and cortical microtubule reorganization in founder cells (Vilches Barro et al., 2019), suggesting that auxin signaling is required for microtubule reorganization during LR initiation. However, GUS staining showed that IAA treatment did not change TPXL5 expression level strongly in the pericycle founder cells and stage I LRPs but did enhance TPXL5 expression during subsequent LR development and emergence (Supplemental Figure 14). This suggests that auxin regulation of microtubule reorganization during LR initiation may not involve altering TPXL5 expression. In addition, increasing evidence shows that microtubules and MAPs are capable of altering auxin distribution and thereby mediating plant cell growth (Li et al., 2012; Ambrose et al., 2013; Deng et al., 2021). Thus, whether a feedback mechanism between the cortical microtubules and auxin signaling exists in HY5-mediated LR development will be investigated in a future study.
Materials and methods
Plant material and growth conditions
Arabidopsis (Arabidopsis thaliana) Columbia ecotype (Col-0) was used in this study. All plant seeds were surface-sterilized for 15 min, washed with sterile water, and treated in the dark at 4°C for 3 days. Seeds were sown on Murashige and Skoog (MS) medium under continuous white light at 22°C.
The tpxl5 mutant lines were obtained using an egg cell-specific promoter-controlled CRISPR/Cas9 system (Wang et al., 2015). The following primers were constructed for the mutant tpxl5-1: TPXL5-BsF-guide1: 5′-ATATATGGTCTCGATTGAAAGCAGGGACTCCGGTGAGTT-3′; TPXL5-F0-guide1: 5′-TGGAAAGCAGGGACTCCGGTGAGTTTTAGAGCTAGAAATAGC-3′; TPXL5-R0-guide2: 5′-ACCTTCTTCGCAGATTTCATGAGAATCTCTTAGTCGACTCTAC-3′; TPXL5-BsR-guide2: 5′-ATTATTGGTCTGAAACTTCTTCGCAGATTTCATGAGC-3′. The following primers were constructed for the mutant tpxl5-2 and double mutant tpxl5 hy5: TPXL5-BsF-guide3: 5′-ATATATGGTCTCGATTGGAGCCCTAAACCTACCCAGGGTT-3′; TPXL5-F0-guide3: 5′-TGGAGCCCTAAACCTACCCAGGGTTTTAGAGCTAGAAATAGC-3′; TPXL5-R0-guide4: 5′-ACCCGGGTGCTGTCCCAGGACGAAATCTCTTAGTCGACTCTAC-3′; TPXL5-BsR-guide4: 5′-ATTATTGGTCTGAAACCGGGTGCTGTCCCAGGACGAC-3′.
The T-DNA insertion mutant line of HY5 (SALK_096651) was described previously (Yu et al., 2016) and was kindly provided by Prof. Dongtao Ren of China Agricultural University. 35S:GFP-MBD transgenic plants were reported previously (Granger and Cyr, 2001). For proTPXL5:GUS and proHY5:GUS transgenic plants, the promoters of TPXL5 (2 kb) and HY5 (1 kb) from genomic DNA were inserted into a pCAMBIA1391 vector. For proTPXL5:TPXL5-GFP transgenic tpxl5-1 mutant plants, the native promoter and CDS of TPXL5 were PCR-amplified and reconstructed into the pCAMBIA1300 vector. For Super:TPXL5-MYC and UBQ10:TPXL5-MYC transgenic plants, the CDS of TPXL5 was PCR-amplified and reconstructed into the Super1300 and pCAMBIA1390 vectors. For proHY5:HY5-GFP transgenic hy5 mutant plants, the native promoter and CDS of HY5 were PCR-amplified and reconstructed into the pCAMBIA1300 vector. Constructs of all transgenic plants were transformed into Arabidopsis plants by floral dipping with Agrobacterium strain GV3101. Homozygous transgenic lines were used for further analyses. To generate the tpxl5 hy5 double mutant, two fragments were selected from the first exon of TPXL5 as targets of Cas9 and constructs were transformed into the hy5 mutant by floral dipping with Agrobacterium strain GV3101. The 35S:GFP-MBD/tpxl5-1, 35S:GFP-MBD/hy5, and 35S:YFP-Tubulin5A/tpxl5-1 plants were generated by crossing. Genetic materials are listed in Supplemental Table 1. Primers are listed in Supplemental Data Set S1.
Phenotypic analysis
For LR phenotype analysis, seeds were sown on a half-strength MS Basal Salt (Caisson Labs, MSP01-50LT) plate with 0.8% agar, 0.5% MES, pH 5.8, and 1% sucrose (w/v). The plates were vertically grown in the chamber (continuous white light, 116 μmol m−2s−1, 22°C) for 5 days and then the seedlings that were growing consistently were selected and transferred onto fresh medium for 5 days. The primary root and LR lengths of the seedlings were measured with ImageJ software (http://rsb.info.nih.gov/ij/) and the number of LRs was counted manually. The LR densities (number of LRs/length of primary root) of the indicated genotypes were analyzed according to a previous report (Lee et al., 2009).
For hypocotyl phenotype analysis, seeds were sown on a MS (Phytotech, M519) plate with 0.8% agar, 0.5% MES, pH 5.8, and 1% sucrose (w/v). The plates were placed flat against the light in the growth chamber (continuous white light, 116 μmol m−2s−1, 22°C). The hypocotyl lengths of seedlings were measured after 7 days with ImageJ software.
Histochemical GUS activity analysis
For the expression patterns of TPXL5 and HY5, seedlings were vertically grown in continuous white light for 10 days. To test the histochemical localization of GUS activity in LRPs, 8-day-old seedlings grown vertically on half-strength MS Basal Salt plates were subjected to GUS histochemical staining analysis using staining buffer (0.1 M phosphate-buffered saline [PBS], 10 mM EDTA, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 0.1% [v/v] Triton X-100, and 2 mM X-Gluc) for 4–6 h at 37°C in the dark, as previously described (Wang et al., 2007).
RNA extraction and RT-PCR analysis
Total RNA was extracted from 6- or 7-day-old seedlings with an RNA purification kit (BioTeke). Genomic DNA was removed from total RNA (1 g) using RNase-free DNase I (TaKaRa) and reverse transcription was performed with M-MLV reverse transcriptase (TaKaRa). PCR analysis was performed to detect TPXL5 and HY5 expression. 18S rRNA was used as the reference control. The primers used for PCR were as follows: TPXL5-F: 5′-ATGGAGTCGTTGACATTGAAG-3′; TPXL5-R: 5′-CTAGAGAAACTGATATTGGAA-3′, HY5-F: 5′-ATGCAGGAACAAGCGACTAGC-3′; HY5-R: 5′-TCAAAGGCTTGCATCAGCATT-3′.
GUS/LUC assay
GUS and LUC assays were performed as previously described (Zhao et al., 2016). To generate the UBQ10:HY5-GFP construct, the coding sequence of HY5 was amplified by PCR and cloned into the pCAMBIA1390 vector under the UBQ10 promoter. To generate the proTPXL5:GUS construct, the P2 region (−450 bp to −600 bp upstream ATG) of the TPXL5 promoter was amplified by PCR and cloned into the pCAMBIA1391 vector. For the mproTPXL5:GUS construct, ACE1 in the P2 region was mutated from ACGT to TTTT. The constructed vectors were transformed into Agrobacterium strain GV3101. proTPXL5:GUS together with UBQ10:HY5-GFP or UBQ10 empty vector were co-infiltrated into 4- to 5-week-old tobacco leaves using syringes. 35S:LUC was co-transformed as the internal control. GUS and LUC activities were detected after 3 days. The GUS/LUC ratio was used to measure the binding activity of HY5 to the TPXL5 promoter: GUS/LUC = GUS*2*6/(LUC/107).
ChIP assay
The hy5 mutant and proHY5:HY5-GFP transgenic hy5 seedlings were grown for 10 days under continuous white light, and then the roots were collected for a ChIP assay as previously described (Lee et al., 2007). Roots of seedlings were first cross-linked with 1% formaldehyde and then subjected to nuclei isolation and sonication. After incubation, HY5-GFP was immunoprecipitated by anti-GFP mAb-Magnetic Agarose (MBL, D153-10) and DynabeadsTM Protein A (Invitrogen). Both immunoprecipitated DNA and nonimmunoprecipitated DNA was analyzed by qPCR using an ABI 7500 RT-PCR detection system. The primer sequences used to detect HY5-targeted TPXL5 promoter fragments (P1 and P2) were as follows:
P1-F: 5′-TCTAACGGACAAGATTAGGAAAAT-3′;
P1-R: 5′-GGCTAGTCTTTTTGGTGCTTTTTT-3′;
P2-F: 5′-GGAAAAGAGTGGGAAATTATAGCT-3′;
P2-R: 5′-ATCAACCGTTGGATTAAATGATCA-3′.
P1 was used as a negative control while ACTIN2 was used as a loading control.
Yeast one-hybrid assay
The PIF1, PIF3, PIF4, PIF5, PIF7, HY5, and HYH coding sequences were amplified by PCR and cloned into the pB42AD vector (Clontech). The promoter of TPXL5 was amplified by PCR and cloned into the pLacZ vector (Clontech). AD-fused protein and LacZ reporter plasmids were co-transformed into the EGY48 yeast strain, and transformants were selected and grown on SD/-Trp-Ura dropout media. Yeast transformation and growth were performed based on the Yeast Protocols Handbook (Clontech).
Microtubule co-sedimentation assay
Porcine brain tubulins were purified as previously described (Castoldi and Popov et al., 2003) and used for sedimentation assays. Tubulin assembly and co-sedimentation of microtubules with His-TPXL5-GFP fusion proteins were performed. For microtubule co-sedimentation assays, the purified proteins and tubulins were centrifuged at 100,000 × g for 15 min at 4°C before use. Prepolymerized and paclitaxel-stabilized microtubules (4 μM) were incubated with TPXL5 fusion protein (6 μM) in PEM buffer (1 mM MgCl2, 1 mM EGTA, and 100 mM PIPES-KOH, pH 6.9) plus 20 μM paclitaxel at room temperature for 30 min. After centrifugation at 5,900 × g for 20 min, the supernatant (S) and pellet (P) were subjected to SDS-PAGE.
Low-temperature and dilution assays
Low-temperature and dilution assays were performed as described previously (Liu et al., 2013; Sun et al., 2015). Briefly, 20 μM rhodamine-labeled tubulin and His-TPXL5-GFP protein or GST-MAP65-1 protein (3 μM) were added to PEM buffer (1 mM MgCl2, 1 mM EGTA, and 100 mM PIPES-KOH, pH 6.9) containing 1 mM GTP. The samples were incubated at 35°C for 40 min to allow tubulin assembly. For the low-temperature assay, the samples were immediately transferred to 10°C for 30 min. For the dilution assay, the samples described above were diluted 50-fold with prewarmed PEM buffer and incubated for 60 min at 35°C. The samples were fixed with 1% (v/v) glutaraldehyde for observation and imaging using a confocal microscope (LSM 880, Carl Zeiss; 40× objective).
Ballistics-mediated transient expression in leaf epidermal cells
Ballistics-mediated transient expression assays were performed as previously described (Fu et al., 2002). The experiments used a transiently expressed UBQ10:TPXL5-GFP construct to visualize the subcellular localization of TPXL5-GFP in the leaf epidermal cells of mCherry-Tubulin transgenic seedlings (Chen et al., 2016). We used 1 μg UBQ10:TPXL5-GFP plasmid for particle bombardment. GFP and mCherry signal were detected after bombardment for 6–8 h using a confocal microscope (LSM 880, Carl Zeiss, 40× objective). Filamentous structures of TPXL5-GFP in the leaf epidermal cells were visualized after a 30-min treatment with 50 μM oryzalin (Sigma-Aldrich; 10 mM in DMSO) and 0.5 μM LatB (Sigma-Aldrich; 1 mM in DMSO).
Gravistimulation-induced LR assay
Five-day-old seedlings were grown on half-strength MS medium under continuous white light. To analyze the developmental stages of LRPs, the seedlings were gravistimulated (90°) for 24, 36, and 48 h, according to a previous report (Xun et al., 2020).
Differential-interference contrast microscopy and LRP analysis
For LRP observation, seedlings were treated with a gradient of ethanol solutions (Malamy and Benfey, 1997) prior to microscopic analysis. The seedlings were mounted in 50% glycerol and photographed using an OLYMPUS BX51 DIC (differential-interference contrast) microscope (20× objective). The developmental stages of LRPs were counted manually as previously described (Malamy and Benfey, 1997).
Confocal microscopy and radial expansion analysis
Analyses of the cortical microtubules and radial expansion of founder cells during LR initiation were performed as previously described (Vilches Barro et al., 2019). Six-day-old seedlings grown on half-strength MS Basal Salt medium under continuous white light were induced by gravistimulation (180°). For 35S:GFP-MBD/tpxl5-1 and 35S:GFP-MBD/hy5 seedlings, gravistimulation was performed for 8 or 15 h as indicated in the figure legends. Cortical microtubule arrays and radial expansion in pericycle cells and LR founder cells after the first ACD were observed using a confocal microscope (LSM 880, Carl Zeiss, 40× objective) in 35S:GFP-MBD, 35S:GFP-MBD/tpxl5-1, and 35S:GFP-MBD/hy5 seedlings. To observe HY5-GFP localization, confocal imaging of proHY5:HY5-GFP transgenic hy5 seedlings was performed with a confocal microscope (LSM 880, Carl Zeiss, 40× objective). GFP was excited at 488 nm.
For radial expansion, the confocal microscope collected the maximum face of a single slice image of pericycle cells and LR founder cells after the first ACD. Then, the cell widths of the two domains were measured with ImageJ software.
Cortical microtubule organization analysis
The anisotropy and average orientation of cortical microtubules were quantified in the two domains with ImageJ software using FibrilTool macros (Boudaoud et al., 2014; Vilches Barro et al., 2019; Stockle et al., 2022). Anisotropy analysis describes the microtubule organization of arbitrary units, with 0 indicating lowest anisotropy (not ordered) and 1 indicating highest anisotropy (ordered). The z-stacks were rotated so that the longitudinal axis of cells was horizontal (0°), and the absolute value of the cortical microtubule orientation (from 0° to 90°) was used to represent the direction of the fibrils.
Oryzalin treatment
To observe cortical microtubules in epidermal cells in the middle regions of the hypocotyls, we used 4-day-old seedlings of the WT and tpxl5 mutant expressing 35S:YFP-Tubulin5A grown in the light. Seedlings were treated with 0 μM oryzalin and 10 μM oryzalin for 10 and 30 min. Cortical microtubules in hypocotyl epidermal cells were observed using a confocal microscope (LSM 880, 40× objective; Carl Zeiss) and z-stacks were collected.
To observe cortical microtubules in the central and peripheral domains of LR founder cells after the first ACD, 6-day-old seedlings grown on half-strength MS Basal Salt medium under continuous white light were induced by gravistimulation (180°) for 15 h. The seedlings were then treated with 5 μM oryzalin for 0, 10, and 20 min. Cortical microtubules in LR founder cells after the first ACD were observed using confocal microscopy (LSM 880, Carl Zeiss, 40× objective).
Microtubule density analysis
For hypocotyl epidermal cells, the density of cortical microtubules was defined as the occupancy of the GFP/YFP signal in the cells (Higaki et al., 2010; Dou et al., 2021). Image J software was used to process the images through the menu “Process-Binary-Skeletonize”. The pixel numbers constituting the skeletonized microtubules (NMT) and the whole image (Ntotal) were measured. The density of microtubules (100%) was calculated as 100 × NMT/Ntotal. The values were analyzed using one-way ANOVA. The density of TPXL5-GFP-labeled filaments and GFP-fABD2-labeled actin filaments in leaf epidermal cells were analyzed in a similar way as described above. For the density of cortical microtubules in LR founder cells, the pixel numbers constituting the cell region in each image (Ncell) was used and the density was calculated as 100 × NMT/Ncell.
Statistical analysis
All statistical significance calculations were performed using IBM SPSS Statistics software. One-way ANOVAs were analyzed with a least significant difference (L) test at significance levels of 0.01 < P < 0.05 and P < 0.01. All the statistical data were included in Supplemental Data Set S2.
Accession numbers
Sequence data of genes described in this article can be found in the Arabidopsis Genome Initiative under the following accession numbers: AT5G15510, TPXL5; AT5G11260, HY5; AT3G17609, HYH; AT2G20180, PIF1; AT1G09530, PIF3; AT2G43010, PIF4; AT3G 59060, PIF5; AT5G61270, PIF7.
Supplemental data
The following supplemental materials are available in the online version of this article.
Supplemental Figure S1. CRISPR/Cas9 system was used to generate tpxl5 mutants.
Supplemental Figure S2. Lower density of LR and shorter hypocotyl phenotype in tpxl5-1 are complemented by proTPXL5:TPXL5-GFP.
Supplemental Figure S3. Overexpression of TPXL5 increases LR density.
Supplemental Figure S4. PIFs and HYH do not bind to the promoter of TPXL5.
Supplemental Figure S5. The hy5 mutant phenotypes can be complemented by the expression of proHY5:HY5-GFP.
Supplemental Figure S6. Expression patterns of TPXL5 and HY5 in Arabidopsis (Arabidopsis thaliana).
Supplemental Figure S7. Co-localization analysis between TPXL5-GFP and mCherry-Tubulin.
Supplemental Figure S8. GFP-MBD does not affect LR development.
Supplemental Figure S9. Microtubule rearrangement and radial expansion were not clearly affected in the pericycle cells before the first ACD in the tpxl5-1 mutant.
Supplemental Figure S10. Microtubule rearrangement and radial expansion were not clearly affected in the pericycle cells before the first ACD in the hy5 mutant.
Supplemental Figure S11. Microtubule rearrangement and radial expansion were altered in the founder cells after the first ACD in the hy5 mutant under 15 h induction.
Supplemental Figure S12. Microtubule rearrangement and radial expansion were altered in the founder cells after the first ACD in the tpxl5-1 mutant under 8 h induction.
Supplemental Figure S13. Cortical microtubules are more stable in the peripheral domain than in the central domain in LR founder cells.
Supplemental Figure S14. Auxin does not strongly alter TPXL5 expression in the pericycle founder cells and stage I LRPs.
Supplemental Table S1 Genetics module.
Supplemental Data Set S1. List of primers used in this study.
Supplemental Data Set S2. Supplemental statistical analyses.
Supplementary Material
Acknowledgments
We thank Prof. Dongtao Ren (China Agricultural University) and Prof. Zhaosheng Kong (University of Chinese Academy of Sciences) for generously providing the hy5 mutant and mCherry-Tubulin6B transgenic seeds.
Contributor Information
Yanmin Qian, State Key Laboratory of Plant Physiology and Biochemistry, Department of Plant Sciences, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Xiaohong Wang, State Key Laboratory of Plant Physiology and Biochemistry, Department of Plant Sciences, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Yimin Liu, State Key Laboratory of Plant Physiology and Biochemistry, Department of Plant Sciences, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Xiangfeng Wang, State Key Laboratory of Plant Physiology and Biochemistry, Department of Plant Sciences, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Tonglin Mao, State Key Laboratory of Plant Physiology and Biochemistry, Department of Plant Sciences, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Funding
The authors appreciate the funding support received from the National Science Fund for Distinguished Young Scholars (31625005 to T.M.) and the Natural Science Foundation of China (31872821 and 32170682 to T.M. and 31872644 to X.W.).
References
- Adamowski M, Li L, Friml J (2019) Reorientation of cortical microtubule arrays in the hypocotyl of Arabidopsis thaliana is induced by the cell growth process and independent of auxin signaling. Int J Mol Sci 20(13): 3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Ruan Y, Gardiner J, Tamblyn LM, Catching A, Kirik V, Marc J, Overall R, Wasteneys GO (2013) CLASP Interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev Cell 24(6): 649–659 [DOI] [PubMed] [Google Scholar]
- Baskin TI (2015) Auxin inhibits expansion rate independently of cortical microtubules. Trends Plant Sci 20(8): 471–472 [DOI] [PubMed] [Google Scholar]
- Bellini C, Pacurar DI, Perrone I (2014) Adventitious roots and lateral roots: similarities and differences. Annu Rev Plant Biol 65(1): 639–666 [DOI] [PubMed] [Google Scholar]
- Benkova E, Bielach A (2010) Lateral root organogenesis—from cell to organ. Curr Opin Plant Biol 13(6): 677–683 [DOI] [PubMed] [Google Scholar]
- Boruc J, Deng X, Mylle E, Besbrugge N, Van Durme M, Demidov D, Tomaštíková ED, Tan TC, Vandorpe M, Eeckhout D, et al. (2019) TPX2-LIKE PROTEIN3 is the primary activator of α-Aurora kinases and is essential for embryogenesis. Plant Physiol 180(3): 1389–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudaoud A, Burian A, Borowska-Wykret D, Uyttewaal M, Wrzalik R, Kwiatkowska D, Hamant O (2014) Fibriltool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat Protoc 9(2): 457–463 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Beeckman T, Graham N, Bhalerao R, Zhang H, Casero P, Sandberg G, Bennett MJ (2003) Dissecting Arabidopsis lateral root development. Trends Plant Sci 8(4): 165–171 [DOI] [PubMed] [Google Scholar]
- Castoldi M, Popov AV (2003) Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr Purif 32(1): 83–88 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S, Ang LH, Puente P, Deng XW, Wei N (1998) Arabidopsis bZIP protein HY5 directly interacts with light-responsive promoters in mediating light control of gene expression. Plant Cell 10(5): 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Grandont L, Li H, Hauschild R, Paque S, Abuzeineh A, Rakusova H, Benkova E, Perrot-Rechenmann C, Friml J (2014) Inhibition of cell expansion by rapid ABP1-mediated auxin effect on microtubules. Nature 516(7529): 90–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Peng Y, Tian J, Wang X, Kong Z, Mao T, Yuan M, Li Y (2016) TCS1, A microtubule-binding protein, interacts with KCBP/ZWICHEL to regulate trichome cell shape in Arabidopsis thaliana. PLoS Genet 12(10): e1006266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS (2004) The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38(2): 332–347 [DOI] [PubMed] [Google Scholar]
- Deng J, Wang X, Liu Z, Mao T (2021) The microtubule-associated protein WDL4 modulates auxin distribution to promote apical hook opening in Arabidopsis. Plant Cell 33(6): 1927–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditengou FA, Teale WD, Kochersperger P, Flittner KA, Kneuper I, van der Graaff E, Nziengui H, Pinosa F, Li X, Nitschke R, et al. (2008) Mechanical induction of lateral root initiation in Arabidopsis thaliana. Proc Natl Acad Sci USA 105(48): 18818–18823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou L, He K, Peng J, Wang X, Mao T (2021) The E3 ligase MREL57 modulates microtubule stability and stomatal closure in response to ABA. Nat Commun 12(1): 2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Scheres B (2018) Lateral root formation and the multiple roles of auxin. J Exp Bot 69(2): 155–167 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Rost TL, Colo´n-Carmona A, Doerner P (2001) Early primordium morphogenesis during lateral root initiation in Arabidopsis thaliana. Planta 214(1): 30–36 [DOI] [PubMed] [Google Scholar]
- Dvorak Tomaštíková E, Rutten T, Dvorak P, Tugai A, Ptoskova K, Petrovska B, van Damme D, Houben A, Dolezel J, Demidov D (2020) Functional divergence of microtubule-associated TPX2 family members in Arabidopsis thaliana. Int J Mol Sci 21(6):2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster R, Izawa T, Chua NH (1994) Plant bZIP proteins gather at ACGT elements. FASEB J 8(2): 192–200 [DOI] [PubMed] [Google Scholar]
- Fu Y, Li H, Yang Z (2002) The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14(4): 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF (2016) The multifaceted roles of HY5 in plant growth and development. Mol Plant 9(10): 1353–1365 [DOI] [PubMed] [Google Scholar]
- Goh T, Kasahara H, Mimura T, Kamiya Y, Fukaki H (2012) Multiple AUX/IAA-ARF modules regulate lateral root formation: the role of Arabidopsis SHY2/IAA3-mediated auxin signalling. Philos Trans R Soc Lond B Biol Sci 367(1595): 1461–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger CL, Cyr RJ (2001) Spatiotemporal relationships between growth and microtubule orientation as revealed in living root cells of Arabidopsis thaliana transformed with green-fluorescent-protein gene construct GFP-MBD. Protoplasma 216(3–4): 201–214 [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Vernos I (2004) The mechanism of spindle assembly: functions of Ran and its target TPX2. J Cell Biol 166(7): 949–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada T (2014) Microtubule organization and microtubule-associated proteins in plant cells. Int Rev Cel Mol Bio 312: 1–52 [DOI] [PubMed] [Google Scholar]
- Higaki T, Kutsuna N, Sano T, Kondo N, Hasezawa S (2010) Quantification and cluster analysis of actin cytoskeletal structures in plant cells: role of actin bundling in stomatal movement during diurnal cycles in Arabidopsis guard cells. Plant J 61(1): 156–165 [DOI] [PubMed] [Google Scholar]
- Landrein B, Hamant O (2013) How mechanical stress controls microtubule behavior and morphogenesis in plants: history, experiments and revisited theories. Plant J 75(2): 324–338 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW (2007) Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19(3): 731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HW, Kim NY, Lee DJ, Kim J (2009) LBD18/ASL20 Regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol 151(3): 1377–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Lei L, Somerville CR, Gu Y (2012) Cellulose synthase interactive protein 1 (CSI1) links microtubules and cellulose synthase complexes. Proc Natl Acad Sci USA 109(1): 185–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Li G, Gao S, Martinez C, He G, Zhou Z, Huang X, Lee JH, Zhang H, Shen Y, et al. (2010) Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell 22(11): 3634–3649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian N, Liu X, Wang X, Zhou Y, Li H, Li J, Mao T (2017) COP1 Mediates dark-specific degradation of microtubule-associated protein WDL3 in regulating Arabidopsis hypocotyl elongation. Proc Natl Acad Sci USA 114(46): 12321–12326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Qin T, Ma Q, Sun J, Liu Z, Yuan M, Mao T (2013) Light-regulated hypocotyl elongation involves proteasome-dependent degradation of the microtubule regulatory protein WDL3 in Arabidopsis. Plant Cell 25(5): 1740–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, Vob U, Swarup K, De Smet I, Van Damme D, Lawrence T, Peret B, Moscardi E, et al. (2013) Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proc Natl Acad Sci USA 110(13): 5229–5234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J (1995) Root architecture and plant productivity. Plant Physiol 109(1): 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malamy JE, Benfey PN (1997) Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124(1): 33–44 [DOI] [PubMed] [Google Scholar]
- Moreno-Risueno MA, Van Norman JM, Moreno A, Zhang J, Ahnert SE, Benfey PN (2010) Oscillating gene expression determines competence for periodic Arabidopsis root branching. Science 329(5997): 1306–1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motte H, Vanneste S, Beeckman T (2019) Molecular and environmental regulation of root development. Annu Rev Plant Biol 70(1): 465–488 [DOI] [PubMed] [Google Scholar]
- Osmont KS, Sibout R, Hardtke CS (2007) Hidden branches: developments in root system architecture. Annu Rev Plant Biol 58(1): 93–113 [DOI] [PubMed] [Google Scholar]
- Ovečka M, Luptovčiak I, Komis G, Šamajová O, Samakovli D, Šamaj J (2020) Spatiotemporal pattern of ectopic cell divisions contribute to Mis-Shaped phenotype of primary and lateral roots of katanin1 mutant. Front Plant Sci 11: 734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K (1997) The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev 11(22): 2983–2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Péret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett MJ (2009) Arabidopsis lateral root development: an emerging story. Trends Plant Sci 14(7): 399–408 [DOI] [PubMed] [Google Scholar]
- Pierik R, Fankhauser C, Strader LC, Sinha N (2021) Architecture and plasticity: optimizing plant performance in dynamic environments. Plant Physiol 187(3): 1029–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos Teixeira JA, Ten Tusscher KH (2019) The systems biology of lateral root formation: connecting the dots. Mol Plant 12(6): 784–803 [DOI] [PubMed] [Google Scholar]
- Schutz LM, Louveaux M, Vilches Barro A, Bouziri S, Cerrone L, Wolny A, Kreshuk A, Hamprecht FA, Maizel A (2021) Integration of cell growth and asymmetric division during lateral root initiation in Arabidopsis thaliana. Plant Cell Physiol 62(8): 1269–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Park E, Choi G (2007) PIF3 Regulates anthocyanin biosynthesis in an HY5-dependent manner with both factors directly binding anthocyanin biosynthetic gene promoters in Arabidopsis. Plant J 49(6): 981–994 [DOI] [PubMed] [Google Scholar]
- Smertenko A, Clare SJ, Effertz K, Parish A, Ross A, Schmidt S (2021) A guide to plant TPX2-like and WAVE-DAMPENED2-like proteins. J Exp Bot 72(4): 1034–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockle D, Reyes-Hernandez BJ, Barro AV, Nenadic M, Winter Z, Marc-Martin S, Bald L, Ursache R, Fujita S, Maizel A, et al. (2022) Microtubule-based perception of mechanical conflicts controls plant organ morphogenesis. Sci Adv 8(6): eabm4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Ma Q, Mao T (2015) Ethylene regulates the Arabidopsis microtubule-associated protein WAVE-DAMPENED2-LIKE5 in etiolated hypocotyl elongation. Plant Physiol 169(1): 325–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JH, Shaw SL (2020) Exogenous auxin induces transverse microtubule arrays through TRANSPORT INHIBITOR RESPONSE1/AUXIN SIGNALING F-BOX receptors. Plant Physiol 182(2): 892–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D, De Rybel B, Gudesblat G, Demidov D, Grunewald W, De Smet I, Houben A, Beeckman T, Russinova E (2011) Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell 23(11): 4013–4024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Li P, Pierik R (2021) Regulation of lateral root development by shoot-sensed far-red light via HY5 is nitrate-dependent and involves the NRT2.1 nitrate transporter. Front Plant Sci 12: 660870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Paalman R, Keuskamp D, Hayes S, Pierik R (2018a) Far-red light detection in the shoot regulates lateral root development through the HY5 transcription factor. Plant Cell 30(1): 101–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Pierik R (2018b) Light signaling, root development, and plasticity. Plant Physiol 176(2): 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilches Barro A, Stockle D, Thellmann M, Ruiz-Duarte P, Bald L, Louveaux M, von Born P, Denninger P, Goh T, Fukaki H, et al. (2019) Cytoskeleton dynamics are necessary for early events of lateral root initiation in Arabidopsis. Curr Biol 29(15): 2443–2454 [DOI] [PubMed] [Google Scholar]
- Wang X, Mao T (2019) Understanding the functions and mechanisms of plant cytoskeleton in response to environmental signals. Curr Opin Plant Biol 52: 86–96 [DOI] [PubMed] [Google Scholar]
- Wang C, Shen L, Fu Y, Yan C, Wang K (2015) A simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genomics 42(12): 703–706 [DOI] [PubMed] [Google Scholar]
- Wang X, Zhang J, Yuan M, Ehrhardt DW, Wang Z, Mao T (2012) Arabidopsis microtubule destabilizing protein40 is involved in brassinosteroid regulation of hypocotyl elongation. Plant Cell 24(10): 4012–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhu L, Liu B, Wang C, Jin L, Zhao Q, Yuan M (2007) Arabidopsis MICROTUBULE-ASSOCIATED PROTEIN18 functions in directional cell growth by destabilizing cortical microtubules. Plant Cell 19(3): 877–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xun Q, Wu Y, Li H, Chang J, Ou Y, He K, Gou X, Tax FE, Li J (2020) Two receptor-like protein kinases, MUSTACHES and MUSTACHES-LIKE, regulate lateral root development in Arabidopsis thaliana. New Phytol 227(4): 1157–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang J, Shi H, Gu J, Dong J, Deng XW, Huang R (2016) Salt stress and ethylene antagonistically regulate nucleocytoplasmic partitioning of COP1 to control seed germination. Plant Physiol 170(4): 2340–2350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li C, Zhang J, Wang J, Yang J, Lv Y, Yang N, Liu J, Wang X, Palfalvi G, et al. (2017) Dissection of HY5/HYH expression in Arabidopsis reveals a root-autonomous HY5-mediated photomorphogenic pathway. PloS one 12(7): e0180449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang C, Xu H, Shi X, Zhen W, Hu Z, Huang J, Zheng Y, Huang P, Zhang KX, et al. (2019) HY5 Contributes to light-regulated root system architecture under a root-covered culture system. Front Plant Sci 10: 1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Zhang ML, Ma TL, Wang Y (2016) Phosphorylation of ARF2 relieves its repression of transcription of the K+ transporter gene HAK5 in response to low potassium stress. Plant Cell 28(12): 3005–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.