Abstract
Fungal pathogens deploy a barrage of secreted effectors to subvert host immunity, often by evading, disrupting, or altering key components of transcription, defense signaling, and metabolic pathways. However, the underlying mechanisms of effectors and their host targets are largely unexplored in necrotrophic fungal pathogens. Here, we describe the effector protein Ascochyta rabiei PEXEL-like Effector Candidate 25 (ArPEC25), which is secreted by the necrotroph A. rabiei, the causal agent of Ascochyta blight disease in chickpea (Cicer arietinum), and is indispensable for virulence. After entering host cells, ArPEC25 localizes to the nucleus and targets the host LIM transcription factor CaβLIM1a. CaβLIM1a is a transcriptional regulator of CaPAL1, which encodes phenylalanine ammonia lyase (PAL), the regulatory, gatekeeping enzyme of the phenylpropanoid pathway. ArPEC25 inhibits the transactivation of CaβLIM1a by interfering with its DNA-binding ability, resulting in negative regulation of the phenylpropanoid pathway and decreased levels of intermediates of lignin biosynthesis, thereby suppressing lignin production. Our findings illustrate the role of fungal effectors in enhancing virulence by targeting a key defense pathway that leads to the biosynthesis of various secondary metabolites and antifungal compounds. This study provides a template for the study of less explored necrotrophic effectors and their host target functions.
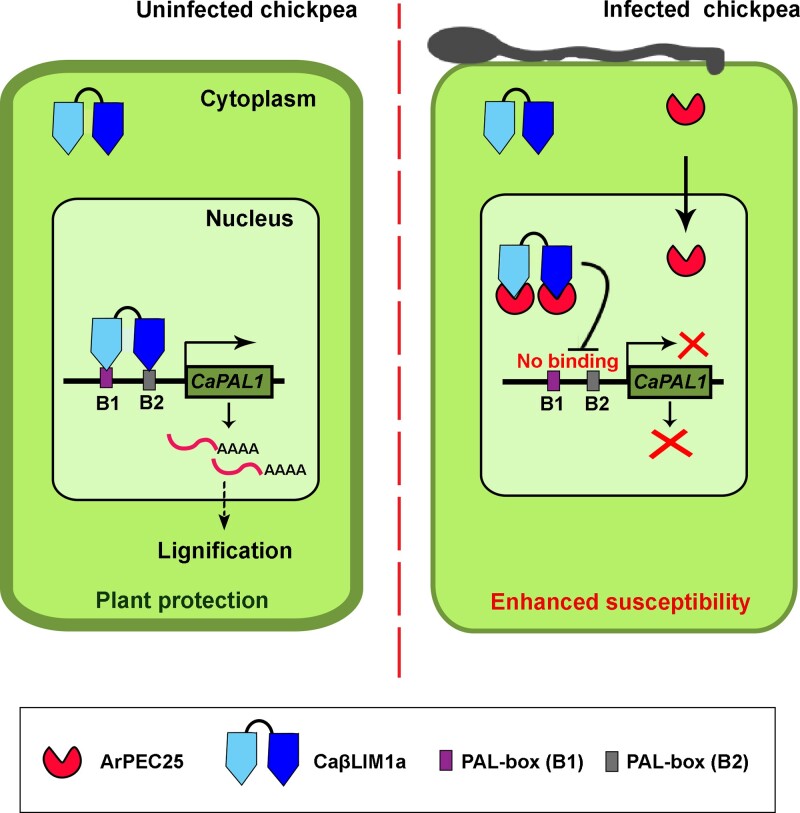
The Ascochyta rabiei effector ArPEC25 enters the host nucleus and targets the transcription factor CaβLIM1a to suppress the expression of CaPAL1, resulting in modulation of lignin biosynthesis.
IN A NUTSHELL.
Background: Adapted pathogens exploit plants to get nutrition and shelter. Thus, a pathogen must overcome preformed structural barriers and suppress the immune system of the host by evolving tactics to invade and survive inside the host environment. Pathogen-secreted molecules (termed effectors) manipulate the signaling or metabolic machinery of the host to benefit the pathogen. These secreted effectors can act inside (intracellular) or outside (extracellular) the host cells. The legume crop chickpea (Cicer arietinum) is infected by a devastating fungus Ascochyta rabiei, resulting in Ascochyta blight disease. Although genome sequencing and in planta expression studies have revealed various A. rabiei effectors, their targets inside chickpea remain unclear.
Question: Which effectors are secreted at the early stage of A. rabiei infection? What are the targets of these effectors inside the chickpea host? How do A. rabiei effectors manipulate chickpea cellular signaling or metabolism to the pathogen's advantage?
Findings: The early expressed effector A. rabiei PEXEL-like Effector Candidate 25 (ArPEC25) is essential for fungal virulence on chickpea. ArPEC25 is secreted by fungi and moves to the chickpea nucleus where it physically interacts with LIM transcription factors. The chickpea nuclear localization of ArPEC25 is essential for its virulence activity, since it disrupts the DNA-binding activity of a CaβLIM1a factor, resulting in reduced expression of a phenylalanine ammonia-lyase (PAL) gene. The PAL enzyme is an important protein of the phenylpropanoid pathway that produces various molecules including lignin to provide structural strength to the plant cell. Thus, one mechanism by which ArPEC25 manipulates the host is by suppressing lignin levels in chickpea.
Next steps: The next challenge is to determine whether manipulating chickpea targets of ArPEC25 by genome editing can result in tolerant chickpea plants. Also, we will try to explore the mechanism of ArPEC25 uptake in chickpea cells.
Introduction
Agricultural crops are continually exposed to biotic factors (pathogens) that can cause severe economic losses. Plant fungal pathogens have broadly evolved into two groups, as defined by their infection cycles: biotrophs (with a predominantly biotrophic phase) and necrotrophs (with a predominantly necrotrophic phase) (Seybold et al., 2020). During their pathogenesis and proliferation, biotrophs maintain a tightly regulated interaction with their hosts that keeps them alive, whereas necrotrophic fungal pathogens promote necrosis and the death of their hosts to feed on the released nutrients (Mengiste, 2012; Ökmen and Doehlemann, 2014). Nevertheless, results gathered from the necrotrophs Botrytis cinerea and Sclerotinia sclerotiorum have suggested that necrotrophs have a short biotrophic phase during the early stages of infection (Shlezinger et al., 2011; Williams et al., 2011; Seifbarghi et al., 2017; Rajarammohan, 2021). Instead of arbitrarily killing their hosts, necrotrophs, like biotrophs, elegantly manipulate crucial biological processes in their hosts to delay cell and tissue necrosis. Only at later stages does the infection enter the necrotrophic phase, which results in the onset of cell death to nourish the pathogen (Veloso et al., 2018).
Invading plant pathogenic fungi and oomycetes secrete an arsenal of specialized molecules called effectors that facilitate successful infection. The current view of plant–pathogen interactions suggests that the secreted effectors from biotrophs and necrotrophs circumvent host immunity by hijacking their diverse normal physiological functions such as cell wall composition and intracellular signaling. A compatible interaction between a host-specific receptor and its cognate effector induces the onset of effector-triggered immunity in biotrophic interactions. By contrast, the direct or indirect interaction of host-specific proteins with necrotrophic effectors (NEs) often triggers cell death, which culminates in host susceptibility (effector-triggered susceptibility) (Oliver and Solomon, 2010; Sung et al., 2021).
The victorin–LOCUS ORCHESTRATING VICTORIN EFFECTS 1 (LOV1) interaction that occurs during infection of Arabidopsis (Arabidopsis thaliana) by the necrotroph Cochliobolus victoriae is one of the few known examples of effector–target interaction leading to susceptibility (Winterberg et al., 2014). Similarly, the secreted effector SnTox3 from Parastagonospora nodorum facilitates disease progression in wheat (Triticum aestivum) upon interaction with its receptor Pathogenesis-related 1 (TaPR1) (Breen et al., 2016; Sung et al., 2021). Eight additional examples of interactions between NEs and their cognate host targets studied in the same pathosystem are SnToxA-Tsn1 (Liu et al., 2006), SnTox1-Snn1 (Liu et al., 2004), SnTox2-Snn2 (Friesen et al., 2007), SnTox4-Snn4 (Abeysekara et al., 2009), SnTox5-Snn5 (Friesen et al., 2012), SnTox6-Snn6 (Gao et al., 2015), and SnTox7-Snn7 (Shi et al. 2015). Likewise, interactions between NEs and their host targets that have been characterized for cell death phenotype upon pathogen attack in the Pyrenophora tritici-repentis-wheat pathosystem are PtrToxA-Tsn1, PtrToxB-Tsc2, and PtrToxC-Tsc1 (Corsi et al., 2020).
The current consensus about these pathosystems is that the pathogen secretes effector proteins to circumvent the host innate immunity pathway. Although the effectors secreted by pathogenic microbes are extremely diverse, they sometimes carry a conserved N-terminal amino acid sequence that plays a role in effector secretion and translocation. The few examples of characterized motifs include the RxLR, LFLAK-HVLVxxP, Crinkler (CRN), Y/F/WxC, CFEM, LysM, DELD, EAR, and RGD motifs (Boddey et al., 2016; Snelders et al., 2020). The RxLR motifs are well characterized for their role in the virulence of various oomycete phytopathogens (Liu et al., 2019). Additionally, the recent availability of genome sequences for various phytopathogenic fungi has revealed the presence of a fifth conserved amino acid residue in the RxLR motif that suggested its high similarity to the Plasmodium Export Element (PEXEL). PEXEL motifs from the malaria parasite (Plasmodium spp.) share the conserved sequence RxLxE/D/Q (with x being any amino acid), which is positioned close to the N-terminal secretory signal sequence (Hiller et al., 2004). This conserved sequence is often cleaved by the endoplasmic reticulum (ER) resident protease Plasmepsin V (PMV) (Boddey et al., 2009, 2010, 2016). This cleavage step is crucial for effector secretion from the parasite into host erythrocytes. The in silico comparative genome and secretome analyses of many phytopathogens suggested that fungal genomes encode more PEXEL-containing effectors than Plasmodium spp. (Choi et al., 2010).
A preformed physical barrier such as a thickened plant cell wall constitutes the first line of protection against most pathogens. To invade and colonize the host, attacking necrotrophs must overcome these physical barriers by penetrating directly through a natural opening or indirectly by using a penetration peg and by secreting cell wall-degrading enzymes (Majd, 2007; Łaźniewska et al., 2009). Plants respond to necrotrophic attack by inducing the reinforcement of their cell wall through lignification. The phenylpropanoid biosynthetic pathway produces monolignols (G-, H-, and S-lignin), which are the building blocks of polymerized lignin. Genes participating in the phenylpropanoid pathway are strongly transcriptionally induced upon pathogen invasion and belong to multigene families, with phenylalanine ammonia lyase (PAL) acting as a gateway enzyme (Bhuiyan et al., 2009; Zhang and Liu, 2015). Two-dimensional proteomic studies in the rapeseed (Brassica napus)-Alternaria brassicae pathosystem showed that cinnamyl alcohol dehydrogenase (CAD), which catalyzes the final step in the phenylpropanoid pathway specific to lignin formation, accumulates 48 h after pathogen infection (Sharma et al., 2007). The phytohormone salicylic acid (SA) provides resistance to a range of pathogens and is itself a product of the phenylpropanoid pathway, but its levels have been shown to be manipulated by secreted effectors from bacteria, oomycetes, and fungi (Liu et al., 2014; Shine et al., 2016; Yuan et al., 2019a; Bauters et al., 2021). Whether host susceptibility can result from NEs specifically targeting the monolignol biogenesis branch of the phenylpropanoid pathway remains unclear. However, a few studies showed that the B. cinerea elicitor BcGs1 and the P. nodorum NE SnTox3 caused upregulation of the phenylpropanoid pathway and increased lignin deposition (Winterberg et al., 2014; Yang et al., 2018). The Tin2 effector protein of corn smut (Ustilago maydis) indirectly modulates the phenylpropanoid pathway by rewiring metabolite flow into the anthocyanin pathway, which results in suppression of lignin accumulation (Brefort et al., 2014). Additionally, the Sta1 effector from the same pathogen suppresses the phenylpropanoid pathway, indicating modulation of lignin content (Tanaka et al., 2020). However, the molecular mechanism behind the modulation of lignin biogenesis by these NEs has not been completely deciphered.
The necrotrophic fungus Ascochyta rabiei causes Ascochyta blight (AB) disease and is a major constraint to chickpea (Cicer arietinum) production worldwide (Singh et al., 2022). Fungal conidia form germ tube-like structures that subsequently develop into an appressorium to penetrate host tissues (Fondevilla et al., 2015). Fungal hyphae grow sub-epidermally and produce necrotic lesions on chickpea leaves (Fondevilla et al., 2015). Several independent studies have examined the devastating effects of the pathogen on chickpea production (Kaiser et al., 2000; Galdames and Mera, 2003; Viotti et al., 2012). While breeding of genetically resistant chickpea cultivars has been attempted using quantitative trait loci for resistance to AB disease (Kumar et al., 2018), the fungal effectors involved in AB diseases have remained largely unexplored. Analyses of the A. rabiei genome, transcriptome, and secretome have revealed a variety of A. rabiei effectors with possible roles in pathogen virulence (Singh et al., 2012; Fondevilla et al., 2015; Verma et al., 2016; Maurya et al., 2020; Mohd Shah et al., 2020). Earlier studies on A. rabiei pathogenesis suggested a role for the fungal toxins solanapyrones A, B, and C as well as cytochalasin D in virulence (Alam et al., 1989; Hamid and Strange, 2000). However, the deletion of the solanopyrone biosynthesis gene cluster demonstrated that these phytotoxins are not required for pathogenicity (Kim et al., 2015). Additionally, a recent study also proposed the involvement of endocytosis in A. rabiei virulence and a role for the membrane curvature sensing protein ArF-BAR in effector secretion (Sinha et al., 2021).
One of the most important mechanisms to suppress host defense employed by pathogens entails the targeting of fungal effectors to the host nucleus to reprogram the host transcriptional network. Several fungal effectors such as See1 (U. maydis), PstGSRE1 (Puccinia striiformis f.sp. tritici), and CgEP1 (Colletotrichum graminicola) translocate to and function inside the host cell nucleus (Redkar et al., 2015; Vargas et al., 2016; Qi et al., 2019; Kim et al., 2020). Moreover, how nucleus-targeted effectors lead to host susceptibility is also poorly understood in fungi, especially in the case of necrotrophs. In this study, we report the identification and characterization of the PEXEL-like motif-containing nuclear effector ArPEC25 from A. rabiei, which is indispensable for virulence. Notably, ArPEC25 interferes with the activity of its host transcription factor target, CaβLIM1a, leading to susceptibility in chickpea. These findings pave the way to a better understanding of the mechanisms of effector action and provide new insights into necrotrophic fungal virulence.
Results
The candidate effector ArPEC25 is essential for A. rabiei virulence
In previous work on the genome sequencing of the necrotrophic fungal pathogen A. rabiei (ArD2), we revealed the presence of several putative secretory proteins with unknown function according to Gene Ontology software (Verma et al., 2016). Motif search analysis showed that [Y/F/W]xC is the most frequent among all characterized motifs. The PEXEL motif initially identified in the malaria parasite Plasmodium spp. was also highly abundant, being present in 122 A. rabiei secretory proteins, in contrast to the RxLR motif (present in 38 proteins) that is commonly found among effectors of oomycete pathogens (Supplemental Table 1). In silico predictions for effector localization determined that 17 out of 122 candidate PEXEL motif-containing effectors harbor either a monopartite or a bipartite nuclear localization signal (NLS; RKRRRRR), suggesting their nuclear localization in the host nucleus (Verma et al., 2016). We then analyzed transcript levels for the encoding fungal genes using data from our earlier differential transcript expression study using a suppression subtractive hybridization (SSH) library of A. rabiei upon oxidative stress: One gene, encoding the putative secretory protein ST47_g1734, was highly expressed. The corresponding ArPEC25 protein, which possessed a typical PEXEL motif, selected for further characterization (Singh et al., 2012).
ArPEC25 encodes a small cysteine-rich protein of 134 amino acids. Analysis of the amino acid sequence using SignalP 4.1 server predicted a signal peptide (SP) at the N-terminal end of ArPEC25, with a cleavage site between residues 28 and 29 (Figure 1A). ArPEC25 appears to lack any known functional or structural domains other than the PEXEL-like motif (RTLND), which was located 11 amino acid downstream of the SP cleavage site and an arginine-rich patch (RKRRRRR) (Figure 1A). Using ArPEC25 as a query, we identified its homologs in 29 fungal species representing diverse groups such as saprophytes, symbionts, animal pathogens, and plant pathogens with biotrophic, hemi-biotrophic, and necrotrophic lifestyles. Multiple alignments showed a high degree of conservation for PEXEL-like motif and arginine-rich patch in various fungal species (Supplemental Figure 1A). These homologs also have two highly conserved cysteine residues that are characteristic features of many known effector proteins. The sequence alignment further showed the presence of a conserved stretch of 10 residues (RECPIPRPGG) at the C terminus. To analyze the relationships among these identified ArPEC25 homologs, we reconstructed a phylogenetic tree, which showed a close association between ArPEC25 and a homolog from the pathogen Ascochyta lentis (Supplemental Figure 1B).
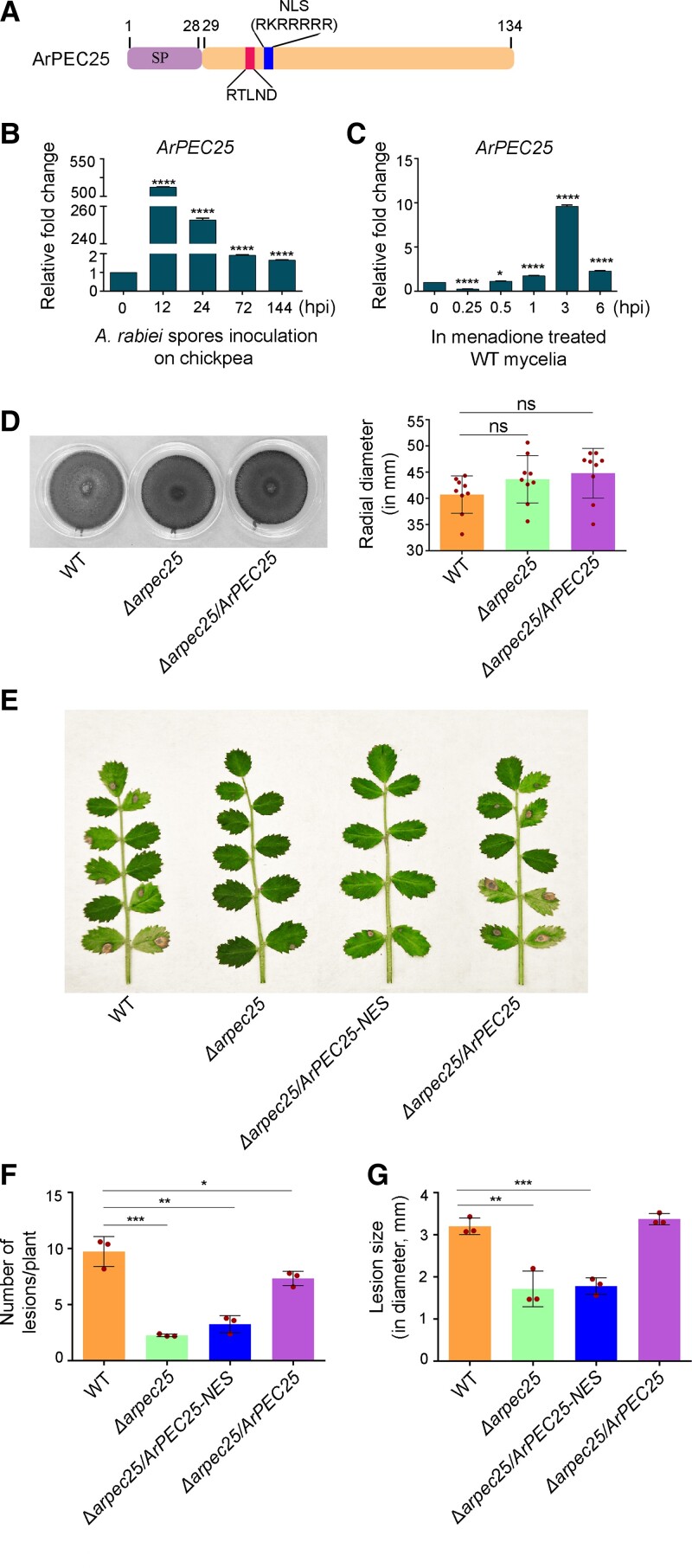
Figure 1.
The Ascochyta rabiei effector ArPEC25 is required for fungal virulence on chickpea plants. A, Schematic diagram of ArPEC25. The 134 amino acid protein has an N-terminal SP of 28 amino acids for secretion of the mature effector through the ER secretory pathway, a PEXEL-like motif sequence (RTLND), and a putative NLS (RKRRRRR). B, In planta expression of ArPEC25 in susceptible chickpea seedlings infected with wild-type A. rabiei. Two-week-old “Pusa 362” seedlings were spray inoculated with a conidial suspension (2 × 106 conidia mL−1) and the aerial tissue was harvested post-inoculation at the indicated time points for RT-qPCR analysis, with A. rabiei elongation factor 1α (ArEF1α, ST47_g4052) used for normalization. Relative expression was calculated using the 2−ΔΔCt method. Data are means ± Sd from three independent biologicals replicates (n = 3). Statistically significant differences were determined by unpaired two-tailed t-test; **** P ≤ 0.0001. C, ArPEC25 expression in menadione-treated wild-type A. rabiei. The broth culture of wild-type A. rabiei mycelia was treated with 250 µM menadione (an oxidative stress inducer) and tissue was harvested at the indicated time points. Ethanol-treated fungal mycelia were taken as control. Transcript levels were checked as in C. Data are means ± Sd of three independent experiments with three technical replicates each (n = 3). Statistically significant differences were determined by unpaired two-tailed t-test; **** P ≤ 0.0001 and * P ≤ 0.0160. D, Radial growth of A. rabiei (wild type), Δarpec25 (knockout) and Δarpec25/ArPEC25 (complementation) strains on PDA plates. The indicated fungal strains were grown for 7 days at 22°C. Each red dot represents data from a single plate. Ns, non-significant. E, AB disease symptoms on chickpea plants inoculated with different A. rabiei strains: wild-type A. rabiei, Δarpec25 (ArPEC25 KO mutant), Δarpec25/ArPEC25-NES (encoding ArPEC25 tagged at its C terminus with a nuclear export signal) and Δarpec25/ArPEC25 (ArPEC25 KO mutant complemented with ArPEC25). Conidial suspension (2 × 106 conidia mL−1) of each strain was spray inoculated on two-week-old “Pusa362” seedlings and photographs were taken at 7 days post-inoculation (dpi). F and G, Mean lesion size on chickpea seedlings spray inoculated with conidial suspension of the strains in (E). The red dots in bar graphs represent the average number of necrotic lesions per plant (F) or the average of lesion diameter per plant (G) in a biological replicate at 7 dpi. This bioassay data was obtained from 3 biological replicates with each replicate having 15 seedlings for each strain type. Data are means ± Sd of three independent experiments. For (F), statistically significant differences were determined by two-tailed t-test; ***P ≤ 0.0006, **P ≤ 0.0019 and *P ≤ 0.0483. Similarly, for (G) ***P ≤ 0.0009, **P ≤ 0.0052.
Further, we determined the virulence function of ArPEC25 during A. rabiei infection. ArPEC25 expression was highly induced by approximately 500-fold at 12 h post-inoculation (hpi) and 250-fold by 24 hpi relative to the 0 h time point during in planta expression analysis on susceptible chickpea, “Pusa 362” (Figure 1B). Moreover, ArPEC25 transcript levels also rose 10-fold in A. rabiei mycelia 3 h after menadione treatment, which is a chemical agent use to mimic host-induced oxidative stress and was also used in the SSH library earlier (Figure 1C).
We generated ArPEC25 gene deletion mutants in the wild-type A. rabiei strain (Δarpec25) using polyethylene glycol (PEG)-mediated homologous recombination (Supplemental Figure 2A). We confirmed the deletion of ArPEC25 from the wild-type A. rabiei strain by Southern blot hybridization (Supplemental Figure 2B) and reverse transcription quantitative PCR (RT-qPCR) (Supplemental Figure 2C). The vegetative growth rate and colony morphology of Δarpec25 mutant strain were similar to those of wild type as shown by radial growth assay (Figure 1D) and calcofluor white staining (Supplemental Figure 2D). However, the Δarpec25 mutant exhibited markedly reduced virulence on a susceptible chickpea variety compared to the wild type (Figure 1, E–G and Supplemental Figure 2E). Furthermore, to validate the loss of virulence in Δarpec25, we generated a complementation construct, where ArPEC25 was expressed under its native promoter. The complemented strain Δarpec25/ArPEC25 rescued the virulence of the Δarpec25 mutant and produced the characteristic AB symptoms on chickpea plants at a rate similar to that of the wild-type strain (Figure 1, E–G and Supplemental Figure 2E).
Bioinformatics analysis suggested that the probable location of ArPEC25 is the host cell nucleus. Therefore, to investigate the role in virulence played by ArPEC25 nuclear localization, we performed a complementation test of Δarpec25 with a construct encoding a mislocalized version of ArPEC25 i.e. ArPEC25 translationally fused at its C terminus with a nuclear export signal (Δarpec25/ArPEC25-NES). The resulting complementation strain was compromised in its virulence against chickpea to the same extent as the Δarpec25 mutant (Figure 1, E–G and Supplemental Figure 2E). Moreover, we mutated all conserved arginine (R) and lysine (K) residues in the predicted NLS sequence of ArPEC25 to alanine (A) residues, hereafter designated as ArPEC25mNLS. We then introduced the resulting complementation construct encoding the NLS mutant, ArPEC25mNLS, into the Δarpec25 mutant using the Agrobacterium (Agrobacterium tumefaciens)-mediated transformation (ATMT) method (Sinha et al., 2021). In contrast to the wild type, the Δarpec25/ArPEC25mNLS strain showed significantly reduced disease symptoms (Supplemental Figure 3, A and B), indicating that ArPEC25mNLS cannot restore virulence to the same extent as the wild type. Together, these results corroborate the notion that ArPEC25 is important for the full virulence of A. rabiei, possibly by functioning as an effector whose nuclear localization is a prerequisite for its virulence.
Instances of functional conservation among the effectors of class Dothideomycetes are very rare. Therefore, to further explore the functional conservation between ArPEC25 and its ortholog A. lentis PEC25 (AlPEC25) (Supplemental Figure 1B), we synthesized the AlPEC25 sequence (KAF9694412) and generated the complementation strain, Δarpec25/AlPEC25. Here, AlPEC25 was expressed under the ArPEC25 promoter. We verified the expression of AlPEC25 in the complementation strain by end-point RT-PCR (Supplemental Figure 4A) AlPEC25 failed to rescue the virulence of the Δarpec25 mutant (Supplemental Figure 4B), suggesting that these two effectors are not functionally conserved. Possibly, ArPEC25 and AlPEC25 may have evolved to target specific proteins in their host legumes.
The A. rabiei effector ArPEC25 is secreted and translocate to the host cell nucleus
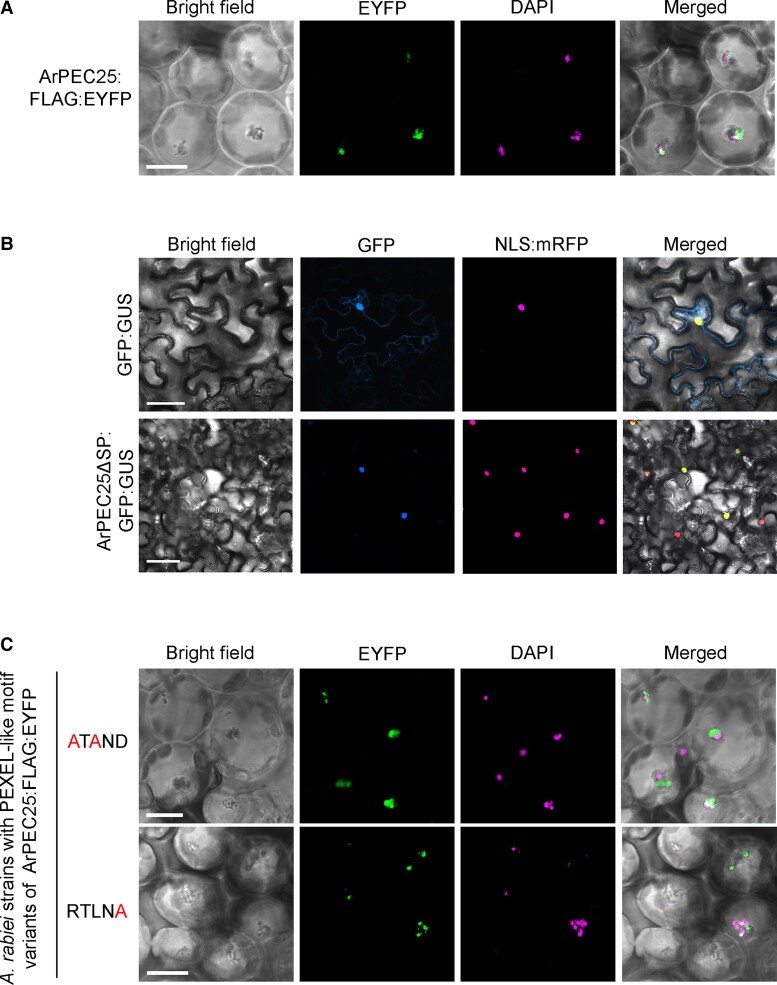
To investigate the subcellular localization of ArPEC25 during chickpea infection, we expressed a construct encoding the chimeric protein ArPEC25-FLAG-EYFP (a fusion between ArPEC25 with a FLAG tag and enhanced yellow fluorescent protein [EYFP]) in A. rabiei under the control of the Glyceraldehyde-3-phosphate dehydrogenase (GPDA) promoter. Chickpea leaves inoculated with the A. rabiei transformants exhibited nuclear localization of ArPEC25 (Figure 2A and Supplemental Figure 5). The chickpea nuclei were marked by staining with 4′,6-diamidino-2-phenylindole (DAPI). The nuclear effectors MoHTR1 and MoHTR2 from rice blast fungus (Magnaporthe oryzae) were previously shown to move to the nuclei of uninvaded rice cells (Kim et al., 2020). Similarly, we also detected EYFP-tagged ArPEC25 in the nuclei of uninvaded chickpea cells, suggesting cell-to-cell movement of the secreted effector (Supplemental Figure 5). Furthermore, we generated another construct encoding ArPEC25 without the SP (ArPEC25ΔSP) translationally fused to GFP and β-glucuronidase (GUS), ArPEC25ΔSP-GFP-GUS. We investigated the subcellular localization of ArPEC25ΔSP-GFP-GUS in Nicotiana benthamiana cells by confocal microscopy. We observed that ArPEC25ΔSP-GFP-GUS accumulates in the nucleus, as evidenced by co-localization with the nuclear marker NLS-mRFP consisting of red fluorescent protein (RFP) fused to a NLS (Figure 2B and Supplemental Figure 6A). Notably, ArPEC25 contains an NLS (Figure 1A). Therefore, to determine the function of this NLS in ArPEC25, we generated a construct encoding the variant protein ArPEC25ΔSPmNLS-GFP-GUS: we observed that ArPEC25ΔSPmNLS predominantly localizes in the cytoplasm by confocal microscopy (Supplemental Figure 6A). The subcellular localization pattern of ArPEC25ΔSPmNLS-GFP-GUS in N. benthamiana cells was similar to the chimeric protein EYFP-ArPEC25ΔSP-NES (Supplemental Figure 6B). Together, these results indicate that the NLS mediates the translocation of the ArPEC25 effector into the host cell nucleus.
Figure 2.
The A. rabiei effector ArPEC25 localizes to the plant cell nucleus. A, Confocal images of chickpea leaf cells showing the co-localization of ArPEC25-FLAG-EYFP with DAPI, marking nuclei. Wild-type A. rabiei expressing ArPEC25-FLAG-EYFP was inoculated on chickpea leaves and EYFP fluorescence signal was detected 48 h post-inoculation (hpi). Scale bar, 10 µm. B, Confocal images of Nicotiana benthamiana leaf cells co-infiltrated with Agrobacterium strain GV3101 harboring ArPEC25ΔSP-GFP-GUS or free GFP-GUS construct and NLS-mRFP (a nuclear marker). Images were taken at 48 hpi. Scale bar, 50 µm. C, ArPEC25 PEXEL-like motif variants (ArPEC25ATAND-FLAG-EYFP and ArPEC25RTLNA-FLAG-EYFP) expressed in A. rabiei localize to the cell nucleus in chickpea. DAPI staining was used to mark nuclei. Scale bar, 10 µm.
Effector secretion at the site of infection is a prerequisite for host susceptibility, as it alters normal host physiological functions. Effectors are secreted through the conventional or non-conventional pathway, the former requiring the cleavage of the functional N-terminal SP in the ER (Liu et al., 2014). To investigate ArPEC25 secretion, we employed a yeast secretion trap (YST) assay (Lee et al., 2006). Accordingly, we generated the constructs ArPEC25-Suc2 and ArPEC25ΔSP-Suc2 by cloning the coding sequence for full-length and SP-truncated ArPEC25 into the pYST1 vector, followed by transformation into the yeast suc2 mutant strain lacking the secreted invertase Suc2. The resulting ArPEC25-Suc2 transformants grew on medium supplemented with sucrose, while the pYST1 (EV) or the ArPEC25ΔSP construct failed to grow on sucrose-supplemented medium (Supplemental Figure 7A). Furthermore, we tested the secretion of the Suc2 invertase in yeast cells by adding 2,3,5-triphenyltetrazolium chloride to the growth medium: we only detected a strong red color from the insoluble product triphenylformazan from ArPEC25-Suc2, but not ArPEC25ΔSP-Suc2, yeast transformants, substantiating the idea that the SP of ArPEC25 is functional (Supplemental Figure 7B). Additionally, we performed an immunoblot assay with an anti-FLAG antibody to detect the secreted ArPEC25 in culture filtrate (CF, without fungal tissues) of wild-type A. rabiei and A. rabiei overexpression (OE) transformants expressing a C-terminally FLAG-tagged ArPEC25 under the control of the constitutive GPDA promoter. Indeed, we obtained a strong immunoblot signal from the CF and A. rabiei mycelial lysate of the OE transformants using an anti-FLAG antibody (Supplemental Figure 7, C and D).
The translocation of several Plasmodium effector proteins, such as histidine-rich protein II (HRPII), knob-associated histidine-rich protein (KAHRP), and glycophorin-binding protein 130 (GBP130) from the parasite to host erythrocytes is tightly regulated by the conserved PEXEL motif RxLxE/D/Q. This sequence appears to work as an internal signal for the specialized effector secretion pathway (Boddey et al., 2009). The secretion of these PEXEL motif-containing proteins of Plasmodium requires; (1) the recognition of the conserved 1st, 3rd, and 5th residues of this motif and (2) cleavage of this motif by ER resident PMV proteases at the conserved leucine (L) residue (Boddey et al., 2016). Therefore, to investigate whether the A. rabiei PEXEL-like sequence (RTLND) might perform a similar function during ArPEC25 effector secretion from the fungus, we generated two variants of RTLND by amino acid substitutions to investigate the role of PEXEL-like motif in ArPEC25 (Supplemental Figure 8A). To this end, we translationally fused ArPEC25 and its RTLND variants with FLAG-EYFP, hereafter referred to as ArPEC25RTLND-FLAG-EYFP, ArPEC25ATAND-FLAG-EYFP, and ArPEC25RTLNA-FLAG-EYFP, respectively (Supplemental Figure 8A). We confirmed the abundance of the chimeric proteins in A. rabiei by confocal microscopy and immunoblotting (Supplemental Figure 8, B and C). Next, we looked for the probable role of RTLND in ArPEC25 secretion from A. rabiei. Surprisingly, we detected bands of the expected size for both the intact PEXEL-like (ArPEC25RTLND-FLAG-EYFP) and the mutant PEXEL-like (ArPEC25ATAND-FLAG-EYFP and ArPEC25RTLNA-FLAG-EYFP) proteins in the CF of axenically grown fungal transformants. We used an antibody against Histone as a control for the potential contamination of purified CF with cytosolic proteins, which we did not observe (Supplemental Figure 8D).
We next investigated the possibility of RTLND sequence cleavage by fungal PMV-like proteases at the conserved leucine residue. We thus purified ArPEC25RTLND-FLAG-EYFP secreted from the CF of axenically grown A. rabiei OE transformants using an anti-FLAG antibody for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (Supplemental Figure 9, A and B). In contrast to Plasmodium effectors where protein secretion requires cleavage of the conserved PEXEL-like motif at the leucine (L) residue, the LC-MS/MS spectra of ArPEC25 indicated that the secreted effector protein retains the RTLND sequence (Supplemental Figure 9C). These results suggest that the cleavage of the PEXEL-like motif is not required for the secretion of ArPEC25 in A. rabiei. Interestingly, some reports suggest that in oomycetes N-terminal RxLR motifs are directly involved in host cell entry (Wawra et al., 2017). Therefore, we also explored the possible role of the ArPEC25 RTLND sequence in host cell uptake. We used A. rabiei transformants expressing one of the two RTLND variant constructs (ArPEC25ATAND-FLAG-EYFP and ArPEC25RTLNA-FLAG-EYFP) to infect chickpea leaves before acquiring confocal images at 48 hpi. To our surprise, we observed EYFP fluorescence in chickpea nuclei (Figure 2C and Supplemental Figure 10), suggesting that the RTLND sequence is not involved in host cell uptake. Together, these results indicate that ArPEC25 is secreted and that unlike Plasmodium effectors, the PEXEL-like sequence of ArPEC25 is not cleaved during effector secretion from the A. rabiei fungus.
ArPEC25 interacts with the chickpea transcription factor CaβLIM1a
Pathogen-delivered effector molecules physically associate with an array of host proteins and interfere with their normal functions of signaling, transcription, and other physiological processes to render the host plant susceptible to infection. Identification of these targets is a promising approach to elucidate effector function during plant infection (Pogorelko et al., 2016). Accordingly, we used a split ubiquitin-based yeast two-hybrid (Y2H) assay using ArPEC25ΔSP as bait and screened a cDNA library prepared from total RNA extracted from chickpea tissues infected with A. rabiei. We obtained eight putative ArPEC25-interacting targets belonging to the LIM (Lin 11, Isl-1, and Mec-3) and TCP (TEOSINTE BRANCHED 1, CYCLOIDEA, and PROLIFERATING CELL NUCLEAR ANTIGEN FACTOR1/2) families of transcription factors (Supplemental Table 2). The chickpea genome harbors nine genes encoding proteins with similarity to the eukaryotic lineage-specific subfamily of two LIM domain (2LIM) proteins (Srivastava and Verma, 2015). Among them, CaβLIM1a exhibited a steep rise in its transcript levels immediately upon A. rabiei infection that remained high at 12 hpi (Srivastava and Verma, 2015).
Several reports have indicated that LIM transcription factors function as transcriptional activators or repressors in many plant species (Kim and Hwang, 2014; Li et al., 2018). Therefore, we used the yeast system to assess the transcriptional function of CaβLIM1a and other 2LIM proteins from chickpea (Ca2LIMs) identified as putative ArPEC25 targets through the Y2H assay above (Supplemental Table 2). In the transcriptional activator assay, we fused the full-length coding sequences of Ca2LIMs to the sequence encoding the yeast GAL4 DNA-binding domain (DB) in the pGBKT7 vector. Among the six Ca2LIM proteins tested in the assay (CaWLIM1a, CaWLIM1b, CaβLIM1a, CaβLIM1b, CaWLIM2, and CaδLIM2), only CaβLIM1a activated the transcription of reporter genes driven by GAL4 regulatory sequences, as evidenced by the growth of yeast colonies on selective growth medium (Supplemental Figure 11A), suggesting that CaβLIM1a is a transcriptional activator. We also performed a repressor assay in the yeast system. However, none of the Ca2LIMs showed transcriptional repressor activity when fused to a strong transactivator domain VP16 from herpes simplex virus (Supplemental Figure 11B). Therefore, we selected CaβLIM1a as the putative target of ArPEC25 for further molecular characterization.
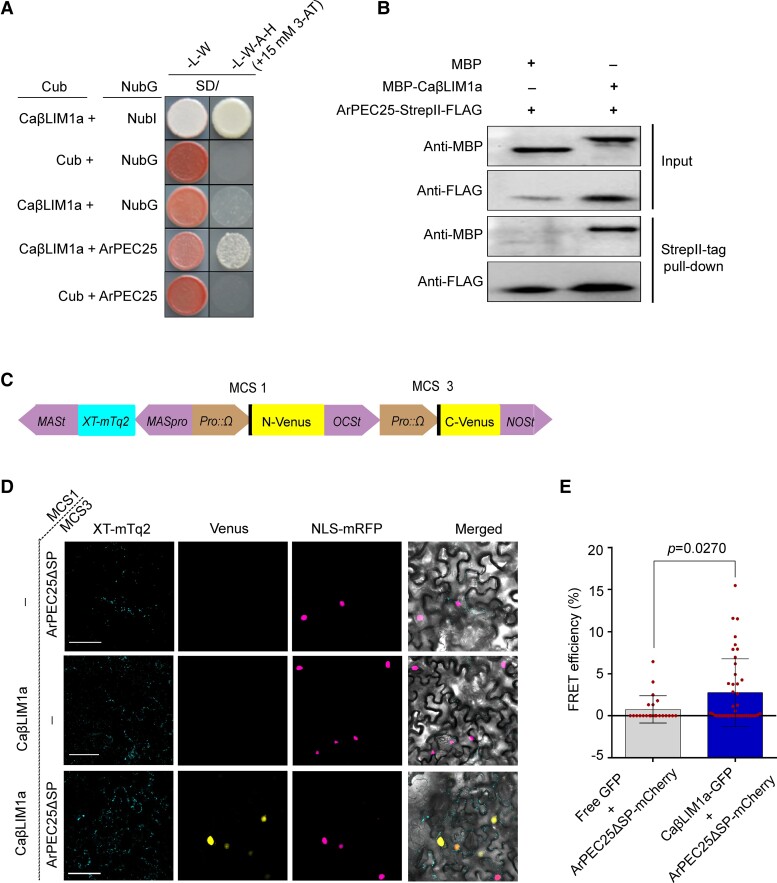
We generated a prey construct consisting of the full-length CaβLIM1a-coding sequence to confirm its interaction with ArPEC25 using a split ubiquitin-based Y2H assay (Figure 3A). Surprisingly, we found that ArPEC25 interacts with all the tested LIM proteins in the Y2H assay (Supplemental Figure 12). However, we focused on CaβLIM1a, as the expression of its encoding gene was induced during A. rabiei infection and the protein showed transactivation in a yeast one-hybrid (Y1H) assay. We also tested the interaction between ArPEC25 and CaβLIM1a by in vitro pull-down assay using recombinant purified maltose-binding protein (MBP)-CaβLIM1a and ArPEC25-StrepII-FLAG. The immunoblot analysis of proteins pulled-down by StrepII-tag showed that recombinant MBP-CaβLIM1a interacts with ArPEC25-StrepII-FLAG (Figure 3B). We independently validated their interaction in plant cells by single-vector bimolecular fluorescence complementation (BiFC) assay using the pDOE-05 vector (Gookin and Assmann, 2014). In this assay, we fused CaβLIM1a to the N-terminal half of Venus and ArPEC25 to the C-terminal half of Venus, resulting in the constructs CaβLIM1a-NVenus and ArPEC25-CVenus (Figure 3C). We then transiently expressed these constructs by Agrobacterium-mediated co-infiltration with the nuclear marker NLS-mRFP into N. benthamiana leaves. We observed the reconstitution of Venus fluorescence in the nucleus of leaf epidermal cells, but not in the cytoplasm (Figure 3D), indicating that ArPEC25 and CaβLIM1a interact in the plant cell nucleus.
Figure 3.
ArPEC25 physically associates with CaβLIM1a, a LIM family transcription factor. A, Split ubiquitin-based yeast two-hybrid assay for protein–protein interactions. NMY51 yeast cells co-transformed with pDHB1 (expressing “X” protein as Ost4p-X-Cub-LexA-VP16) and pPR3-N (expressing “Y” protein as Y-NubG-HA) clones were resuspended in 0.9% (w/v) NaCl and spotted on synthetic defined (SD) medium –W–L and SD–W–L–A–H + 15 mM 3-AT. The interaction between chimeric Cub and NubI served as a positive control while CaβLIM1a + NubG served as a negative control. B, In vitro pull-down assay for protein–protein interaction. MBP, MBP-tagged CaβLIM1a (MBP-CaβLIM1a), and StrepII-FLAG-tagged ArPEC25 (ArPEC25-StrepII-FLAG) recombinant proteins were used for the in vitro pull-down assay. In the input immunoblots, purified MBP or MBP-CaβLIM1a was detected with anti-MBP antibody and ArPEC25-StrepII-FLAG protein was detected with anti-FLAG antibody. MBP alone was taken as a control. C, Schematic diagram of the pDOE-05 vector used for single-vector BiFC, showing the cloning positions for CaβLIM1a and ArPEC25. CaβLIM1a was cloned in MCS1 to generate the translational fusion CaβLIM1a-NmVenus210. ArPEC25 was cloned in MCS3 to generate the translational fusion ArPEC25-CVenus210. XT-mTurquoise2 (XT-mTq2) served as Golgi marker and internal reference for transient expression. D, BiFC assay testing the in planta interaction between ArPEC25 and CaβLIM1a. Agrobacterium strain GV3101 harboring the pDOE-05 single plasmid BiFC clone was co-infiltrated with Agrobacterium cells harboring NLS-mRFP (nuclear marker) in N. benthamiana leaves. Scale bar, 5 µm. E, Analysis of ArPEC25 and CaβLIM1a interaction by forster resonance energy transfer (FRET) assay. Free GFP and CaβLIM1a-GFP served as donor for FRET, while ArPEC25-mCherry served as acceptor. All constructs were expressed from the CaMV 35S promoter. Agrobacterium colonies harboring these constructs individually were co-infiltrated in N. benthamiana leaves. FRET signal between the two partners (CaβLIM1a and ArPEC25) was analyzed 48 hpi by confocal microscopy. Data are means ± Sd from three independent biologicals replicates (n = 3). The acceptor only was used as a control representing the co-expression of ArPEC25-mCherry and free GFP. Significant differences were determined using unpaired two-tailed t-test.
We further explored their physical interaction in vivo using forster resonance energy transfer-acceptor photobleaching (FRET-APB) by constructing translational fusion constructs between CaβLIM1a and the FRET donor GFP while ArPEC25 was fused to the FRET acceptor mCherry. The resulting constructs were then transiently co-infiltrated in N. benthamiana leaves (Supplemental Figure 13). We measured strong FRET efficiency (mean value of 2.745 ± 0.62) compared to the control (mean value of 0.76 ± 0.33), which was in agreement with the interaction between CaβLIM1a and ArPEC25 (Figure 3E). Thus, Y2H, in vitro pull-down, BiFC, and FRET assays strongly indicate that ArPEC25 and CaβLIM1a physically interact in the chickpea nucleus.
To determine the ArPEC25 region(s) involved in its physical interaction with CaβLIM1a, we generated a series of ArPEC25 deletion variants, designated as T1 (ArPEC25ΔSP29–82), T2 (ArPEC25ΔSP65–134), T3 (ArPEC25ΔSP65–108), T4 (ArPEC25ΔSP83–134), and T5 (ArPEC25ΔSP29–108) and performed a Y2H assay against full-length CaβLIM1a. In contrast to ArPEC25ΔSP, the strength of association decreased between CaβLIM1a and deletion variants (Supplemental Figure 14A). We indirectly quantified the strength of the physical interaction by β-galactosidase assay. The interaction of ArPEC25ΔSP with CaβLIM1a was strongest, followed by the T4 and T2 deletion variants (Supplemental Figure 14B). Thus, the overall structure of ArPEC25 is required for its strong interaction with CaβLIM1a.
We then observed the localization of CaβLIM1a and ArPEC25 in plant cells. The CaβLIM1a-GFP fusion localized to the nucleus and cytoplasm of N. benthamiana cells (Supplemental Figure 15A). However, the transient co-expression of CaβLIM1a-GFP and ArPEC25-mCherry showed the nucleus as the site of co-localization (Supplemental Figure 15B). Moreover, we also investigated the effect of ArPEC25 on CaβLIM1a localization by checking the GFP fluorescence intensity in the nucleus. We determined that the nuclear fluorescence intensity of CaβLIM1a-GFP alone or in the presence of ArPEC25-mCherry is 113.1 ± 6.733 and 123.3 ± 9.3 A.U. (arbitrary units), respectively (Supplemental Figure 15C), indicating that the effector has no significant effect on the stability or nuclear localization of CaβLIM1a. Taken together, these results strongly suggest that the mature effector (ArPEC25ΔSP) targets CaβLIM1a inside the plant cell nucleus.
CaβLIM1a binds to the CaPAL1 promoter sequence
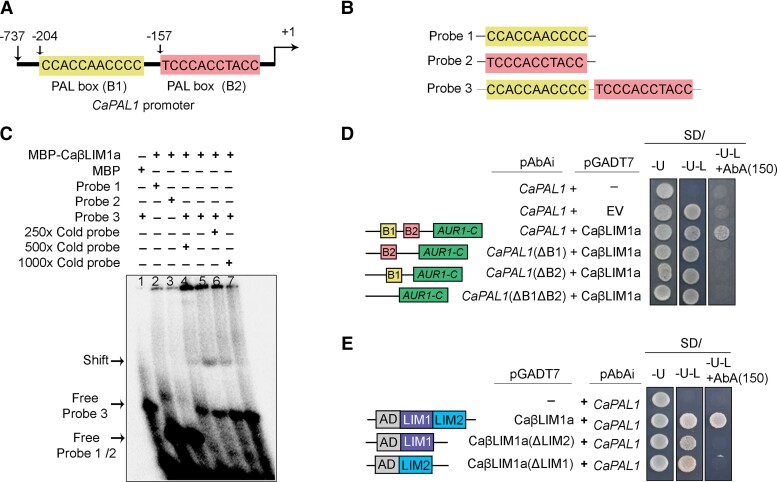
Plant LIM transcription factors have been shown to positively regulate the phenylpropanoid biosynthetic pathway by binding to the PAL-box element with the consensus sequence CCA(C/A)C(A/T)A(C/A)C(C/T)CC (Kawaoka et al., 2000; Kawaoka and Ebinuma, 2001). The PAL gene shows differential regulation during biotic stress such as fungal attacks (Zhang et al., 2017). The chickpea genome contains four PAL genes: CaPAL1 (LOC101507594), CaPAL2 (LOC101509831), CaPAL3 (LOC101496077), and CaPAL4 (LOC101493062). We investigated the expression of CaPAL genes in A. rabiei-infected chickpea plants. We only observed a strong biphasic induction for CaPAL1, as seen by RT-qPCR (Supplemental Figure 16). Additionally, we scanned the promoter sequence of all four CaPAL genes and identified two PAL-box elements [PAL-box (B1) and PAL-box (B2)] in the CaPAL1 promoter, but not in the other CaPAL promoters (Figure 4A).
Figure 4.
CaβLIM1a binds to the PAL-boxes of the CaPAL1 promoter. A, Schematic representation of the CaPAL1 promoter. The CaPAL1 promoter has two conserved PAL-boxes, PAL-box (B1) and PAL-box (B2) 204 and 157 bp upstream from the translation start site (+1), respectively. B, Oligonucleotide probes used for EMSA. Three different oligonucleotide probes, the 40-bp probe 1 (P1, with PAL-box B1), the 41-bp probe 2 (P2, with PAL-box B2), and the 70-bp probe 3 (P3, with both the PAL-box B1 and B2) were synthesized and used for EMSA in (C). C, EMSA for CaβLIM1a-DNA interaction analysis. MBP alone or MBP-CaβLIM1a recombinant proteins were incubated with different oligonucleotide probes shown in Figure 4B. Black arrows indicate the retarded protein-DNA complex and free labeled probes. Unlabeled DNA probes (cold probe) with 250, 500 and 1,000× concentration were used as competitors. Purified MBP served as a control. D, Yeast one-hybrid assay to check the binding of CaβLIM1a to PAL-boxes of the CaPAL1 promoter. The CaPAL1 promoter and its PAL-box deletion variants CaPAL1(ΔB1), CaPAL1(ΔB2) and CaPAL1(ΔB1 ΔB2) were cloned upstream of the reporter gene AUR1-C, which confers AbA resistance. CaβLIM1a was fused to the GAL4-AD domain in the vector pGADT7. The pAbAi constructs were integrated into the genome of yeast strain Y1H Gold. These individual yeast strains were transformed with pGADT7 vector (EV) or pGADT7-CaβLIM1a. E, Yeast one-hybrid assay testing the interaction between CaβLIM1a and its LIM domain deletion mutants to the CaPAL1 promoter. The sequence encoding CaβLIM1a and its LIM domain deletion variants (ΔLIM1 and ΔLIM2) were cloned in pGADT7.
To test whether CaβLIM1a can directly bind to the CaPAL1 promoter region, we performed an electrophoretic mobility shift assay (EMSA) using recombinant CaβLIM1a purified from Escherichia coli (Supplemental Figure 17). We synthesized three DNA probes: P1 (42 bp), P2 (41 bp), and P3 (70 bp) from the CaPAL1 promoter (Figure 4B) and radiolabeled each probe with γ-phosphate. By EMSA, we observed a notable shift in the presence of P3 and CaβLIM1a (Figure 4C). Binding of CaβLIM1a to radiolabeled P3 diminished gradually upon co-incubation with an increasing amount of unlabeled P3 as competitor (Figure 4C). However, we failed to observe shifted radiolabeled bands when we incubated CaβLIM1a with P1 or P2 (Figure 4C). This result suggests that efficient DNA binding of CaβLIM1a to the CaPAL1 promoter requires the presence both PAL-boxes.
We next performed an Y1H assay to assess the binding of CaβLIM1a to the CaPAL1 promoter. To this end, we cloned the CaPAL1 promoter fragment having PAL-boxes B1 and B2 and placed it upstream of the AUR1-C reporter gene in the yeast genome using the integrating vector pAbAi, which confers resistance to the antibiotic Aureobasidin A (AbA). In parallel, we cloned the full-length CaβLIM1a-coding sequence in-frame and downstream of the sequence of the GAL4 activation domain (AD) in the pGADT7 vector. We transformed the pAbAi-CaPAL1 linear plasmid into Y1H Gold strain to integrate the CaPAL1pro:AUR1-C cassette into the yeast genome. Yeast clones harboring both the CaPAL1 promoter and the CaβLIM1a construct showed activation of the AUR1-C reporter, as demonstrated by the growth of yeast on selection medium-containing AbA (150 ng mL−1). The two other chickpea LIM proteins, CaWLIM2 and CaWLIM1a, also bound to the same promoter fragment (Supplemental Figure 18A). We verified the accumulation of LIM proteins in yeast by immunoblot (Supplemental Figure 18B). We further tested the functional relevance of each PAL-box in the CaPAL1 promoter by cloning a promoter fragment lacking one or both of PAL-box elements: CaPAL1ΔBox1, CaPAL1ΔBox2, and CaPAL1ΔBox1ΔBox2. The Y1H result showed that both PAL-boxes are crucial for binding by CaβLIM1a (Figure 4D). Again, we confirmed that CaβLIM1a accumulates in yeast to rule out the possibility of negative results (Supplemental Figure 18C).
Previous studies have shown that the LIM1 and LIM2 domains of the tobacco (Nicotiana tabacum) transcription factor NtLIM1 bind to promoter sequences independently (Kawaoka et al., 2000). We thus asked whether CaβLIM1a might behave similarly by generating truncated versions of CaβLIM1a lacking either the LIM1 domain (CaβLIM1aΔLIM1) or the LIM2 domain (CaβLIM1aΔLIM2) and testing the resulting truncated proteins by Y1H assay against the CaPAL1 promoter. In contrast to NtLIM1, CaβLIM1a appeared to require both the LIM1 and LIM2 domains to bind to the CaPAL1 promoter, as neither CaβLIM1aΔLIM1 nor CaβLIM1aΔLIM2 sustained yeast growth on medium-containing AbA (Figure 4E). This inability to activate the AUR1-C reporter gene was not due to protein instability, as an immunoblot assay on yeast cell extracts with an anti-HA antibody detected proteins of the expected molecular weights (Supplemental Figure 18D). Together, these results indicate that the LIM1 and LIM2 domains in CaβLIM1a bind directly to both PAL-box elements (B1 and B2) in CaPAL1 promoter.
CaβLIM1a regulates the transcription of the phenylpropanoid biosynthetic pathway gene CaPAL1
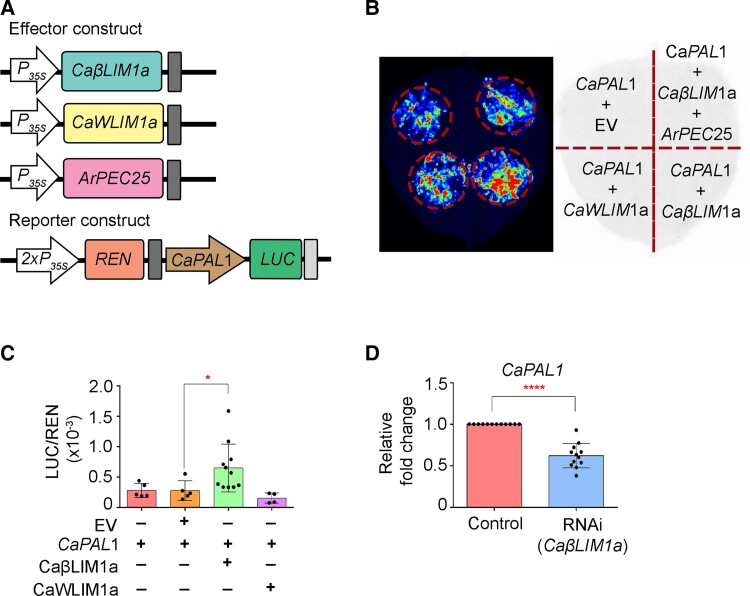
We further explored the relationship between chickpea CaβLIM1a and CaPAL1 through in planta reporter gene activation assays. To execute this experiment, we generated effector constructs of CaβLIM1a, CaWLIM1a, and ArPEC25 by mobilizing Gateway Entry clones into pGWB series destination vectors and verified the accumulation of the resulting encoded proteins by immunoblotting (Supplemental Figure 19). In the reporter construct, we cloned the CaPAL1 promoter upstream of the firefly luciferase reporter gene LUC (Figure 5A) and carried out a dual-luciferase reporter assay by co-expressing the reporter and effector constructs in N. benthamiana leaves. The initial qualitative assay showed activation of the LUC reporter gene in the presence of CaβLIM1a (Figure 5B). To verify this result in a highly sensitive quantitative assay, we measured relative luminescence (LUC/REN, with REN derived from Renilla LUC driven by the cauliflower mosaic virus [CaMV] 35S promoter) in the presence of different effector constructs. The luminescence ratio increased significantly in the presence of CaβLIM1a, whereas CaWLIM1a failed to activate the reporter gene as compared to controls (Figure 5C). CaWLIM1a lacked transactivation in Y1H and failed to activate the reporter gene in dual-luciferase assay, suggesting that it may not be involved in the regulation of CaPAL1. Therefore, CaβLIM1a is a strong candidate for the transcriptional regulation of CaPAL1 during pathogen infection.
Figure 5.
CaβLIM1a regulates CaPAL1 expression in luciferase reporter assays and chickpea. A, Schematic representation of reporter and effector constructs for dual-luciferase assay. In the reporter construct, the CaPAL1 promoter was cloned upstream of the firefly luciferase (LUC) reporter; in the effector constructs, CaβLIM1a, CaWLIM1a, and ArPEC25 were cloned downstream of the CaMV 35S promoter. Renilla LUC (REN) activity acts as internal reference. B, Qualitative analysis of luciferase reporter activity from the CaPAL1 promoter in the presence of effector constructs empty vector (EV), CaβLIM1a, CaWLIM1a, and ArPEC25. Different reporter and effector plasmid constructs shown in (A) were transiently expressed in N. benthamiana leaves by Agrobacterium-mediated co-infiltration. Images were collected after 48 hpi. C, In planta dual-luciferase assay to check the binding and regulation of the CaPAL1 promoter by CaβLIM1a and CaWLIM1a. Agrobacterium cells harboring reporter and effector constructs were co-infiltrated in the abaxial side of leaves. The combination of reporter and effector constructs (CaPAL1 alone and CaPAL1 + EV) served as experimental controls. Data are means ± Sd of up to 11 biological replicates. Significant differences were determined by unpaired one tailed t-test; *P ≤ 0.0324. Black dots represent individual experiments. D, RT-qPCR analysis of CaPAL1 expression in CaβLIM1a-RNAi hairy roots. Total RNA was extracted from the CaβLIM1a-RNAi hairy roots of chickpea plants. Relative CaPAL1 transcript levels were normalized to chickpea β-tubulin; relative expression was obtained using the 2−ΔΔCt method. Data are means ± Sd of three biological replicates along with three technical replicates in each set. Statistically significant differences were determined by unpaired two-tailed t-test; ****P ≤ 0.0001. The control represents the expression of CaPAL1 gene in untransformed hairy roots. Black dots represent technical replicates from independent experiments.
To further determine the relationship between the CaβLIM1a and CaPAL1, we knocked down the transcript levels of CaβLIM1a in AB-susceptible chickpea hairy roots. Chickpea roots were transformed with an RNA interference (RNAi) vector (pK7GWIWG2(II)-Red Root) carrying a specific 341-bp fragment of CaβLIM1a. RT-qPCR analysis of the transformed chickpea roots (RNAi) showed significant downregulation in CaβLIM1a transcript levels compared to non-transformed roots (control), suggesting effective knockdown in the accumulation of the transcript (Supplemental Figure 20). Next, we assessed the RNAi root samples for the expression of CaPAL1 by RT-qPCR. We observed that CaPAL1 transcript levels are significantly reduced in the CaβLIM1a-RNAi roots as compared to the vector control roots (Figure 5D), suggesting that CaβLIM1a functions directly upstream of CaPAL1 for its positive regulation.
ArPEC25 facilitates virulence in chickpea by inhibiting CaβLIM1a function
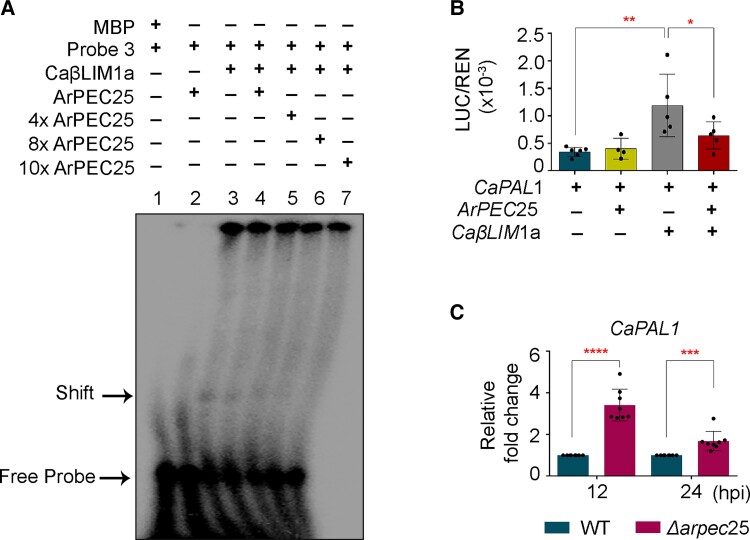
Secreted effectors modulate host immunity by direct binding or by enzymatically altering the function of host molecules such as NAC-type transcription factors (Yuan et al., 2019b). We speculated that ArPEC25 may have a similar mode of action during the establishment of AB disease in chickpea. To test this hypothesis, we first investigated whether ArPEC25 affect the DNA-binding ability of CaβLIM1a to the CaPAL1 promoter. We checked this possibility through EMSA using purified recombinant ArPEC25 (Supplemental Figure 21) and other components of the previously described EMSA (Figure 4C). In EMSA, we observed that the shifted band corresponding to P3 and the CaβLIM1a complex gradually disappears with increasing concentrations of ArPEC25 (1, 4, 8, and 10×) (Figure 6A), suggesting that the effector interferes with the DNA-binding capacity of CaβLIM1a. We validated this observation using the dual-luciferase assay by co-infiltrating N. benthamiana leaves with the reporter construct (ProCaPAL1:FUC) and effector constructs (ArPEC25 and CaβLIM1a) (Figure 6B). The co-expression of ArPEC25 and CaβLIM1a resulted in lower relative luminescence (LUC/REN) compared to CaβLIM1a alone, demonstrating that the presence of ArPEC25 interferes with the normal activities of transcriptional regulators like CaβLIM1a (Figure 6B). Furthermore, we assessed the ArPEC25-mediated interruption of CaPAL1 activation in the native chickpea system. To this end, we challenged chickpea plants with wild-type A. rabiei or the Δarpec25 mutant and analyzed CaPAL1 expression by RT-qPCR. We established that CaPAL1 transcript levels are significantly upregulated at 12 and 24 hpi in chickpea plants infected with Δarpec25 compared to those infected with wild-type A. rabiei (Figure 6C), suggesting ArPEC25-mediated inhibition of gene expression. Together, our data demonstrate that the effector ArPEC25 inhibits the transactivation function of CaβLIM1a to promote pathogenesis in chickpea.
Figure 6.
ArPEC25 inhibits the binding of CaβLIM1a to the CaPAL1 promoter and negatively regulates CaPAL1 expression in chickpea. A, ArPEC25 inhibits the binding of CaβLIM1a to the CaPAL1 promoter in EMSA. Purified MBP alone protein served as a negative control. B, Dual-luciferase activity assay to check the effect of ArPEC25 on CaβLIM1a-mediated regulation of CaPAL1 transcriptional activity. N. benthamiana leaves were co-infiltrated with different combinations of reporter and effector constructs. The indicated combination of reporter and effector constructs (CaPAL1 and CaPAL1 + ArPEC25) served as an experimental control. Data are means ± Sd of six independent experiments. Statistically significant differences were determined by unpaired two-tailed t-test; *P ≤ 0.0211, **P ≤ 0.0055. C, Relative CaPAL1 expression in chickpea seedlings inoculated with conidia suspension of wild-type A. rabiei and Δarpec25 KO mutant strain. Aerial tissue was harvested at the indicated time points post-inoculation for RT-qPCR analysis. Relative CaPAL1 expression was normalized to that of Caβ-tubulin and expression levels were obtained using the 2−ΔΔCt method. Data are means ± Sd from at least three independent biologicals replicates each having technical replicates. Statistically significant differences were determined by unpaired two-tailed Student’s t test; ****P ≤ 0.0001, ***P ≤ 0.0010.
The lignin biosynthetic pathway is severely compromised during A. rabiei infection
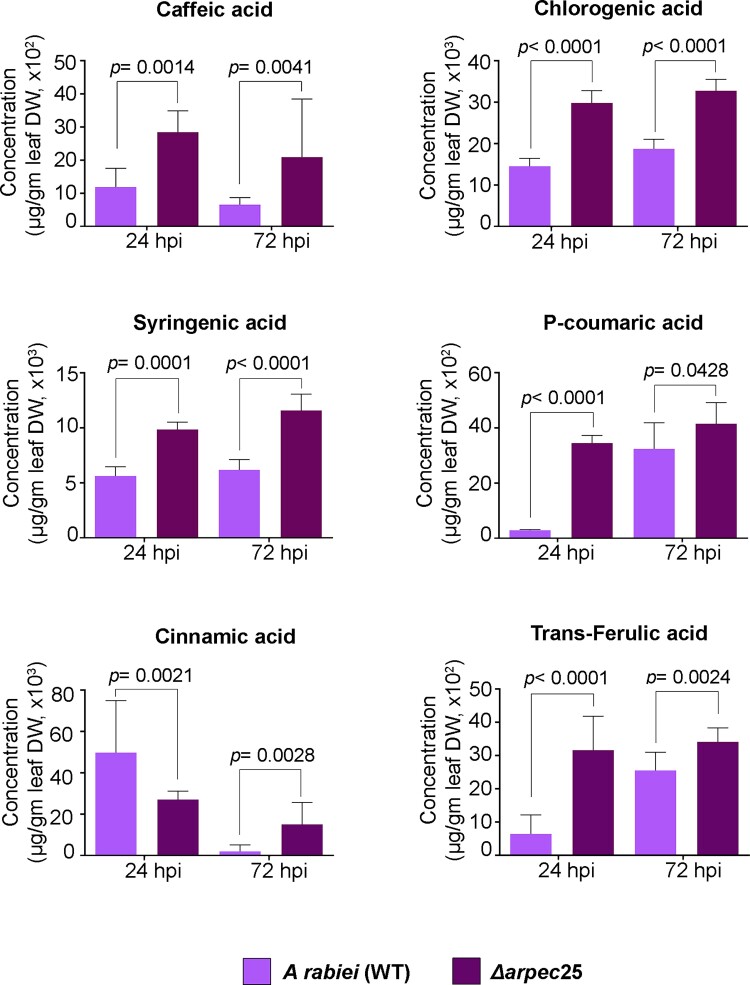
PAL is the regulatory enzyme that controls flux through the phenylpropanoid biosynthetic pathway and has been extensively studied in multiple plant systems in response to biotic stress such as the pathogenic fungus S. sclerotiorum (Ranjan et al., 2019). Reports show that the expression of several genes in the phenylpropanoid biosynthetic pathway is induced in chickpea upon A. rabiei infection (Kavousi et al., 2009), but the mechanism remains elusive. Our gene expression data also showed the induced expression of CaPAL1 upon A. rabiei infection, suggesting a direct connection between lignin biosynthesis and pathogenesis (Supplemental Figure 16). To further explore a direct connection between these two processes, we looked at the levels of different metabolites associated with the phenylpropanoid biosynthetic pathway in mock-inoculated AB-susceptible chickpea plants and plants inoculated with A. rabiei conidia, with a particular focus on the flavonoid and lignin biosynthetic branches. We also characterized amino acid biosynthesis of infected and mock-inoculated chickpea plants through ultra-high-performance liquid chromatography (UHPLC). We observed a significant reduction in the contents of lignin biosynthetic pathway intermediates; cinnamic acid, caffeic acid, syringic acid, and chlorogenic acid at 24 and 72 hpi in A. rabiei-infected plants relative to mock-inoculated plants (Supplemental Figures 22 and 23). However, most of the key intermediates in the flavonoid and amino acid biosynthetic pathways remained unaffected (Supplemental Figures 22, 24 and 25). To further establish the specific role of ArPEC25 in suppression of lignin biosynthesis, we assayed the accumulation of key lignin biosynthesis intermediates that were shown to be modulated in the above observations by using the Δarpec25 mutant strain. We challenged chickpea plants with wild-type A. rabiei or hte Δarpec25 mutant strain and assayed the accumulation of metabolites at 24 and 72 hpi. We observed that in contrast infection with wild-type A. rabiei, metabolites related to lignin biosynthesis intermediates such as cinnamic acid, caffeic acid, p-coumaric acid, chlorogenic acid, and ferulic acid are upregulated during infection by the Δarpec25 mutant (Figure 7). However, the metabolite cinnamic acid showed reduced accumulation at 24 hpi. Thus, the lower levels of monolignol precursors in wild-type fungus-infected chickpea compared to Δarpec25 mutant strain demonstrates the direct role of ArPEC25 in actively subverting host immunity by preventing new lignin biosynthesis, which would be expected to severely compromise the structural integrity of the host cell wall.
Figure 7.
The fungal effector ArPEC25 modulates metabolite levels in chickpea. Two-week old chickpea seedlings were spray inoculated with wild-type A. rabiei or Δarpec25 strains conidial suspensions (2 × 106 conidia mL−1). Aerial tissues were harvested at 24 and 72 hpi. Accumulation of the different metabolites caffeic acid, chlorogenic acid, cinnamic acid, syringic acid, p-coumaric acid, and trans-ferulic acid were checked from at least three independent biological replicates using UHPLC analysis of aqueous methanolic extracts from dried chickpea tissues. Accumulated metabolite values are plotted as fold-changes. Data are means ± Sd. Statistically significant differences were determined by unpaired two-tailed t-test.
Discussion
The constant need to evade host detection or to suppress its immune response has resulted in an array of highly diversified and functionally specific pathogen effectors. These secreted effectors suppress immunity by interacting and impairing the normal function of key host molecules. Surprisingly, for the majority of secreted effectors of fungal and oomycetes origin, the host targets and the mechanism of susceptibility remain unclear. Several studies have shown that these targets can have diverse functions, ranging from signaling components, transcriptional regulators, metabolic enzymes, or simply products of host R genes (Gimenez-Ibanez et al., 2014; Liu et al., 2014; Qin et al., 2018). This study on the nucleus-localized effector protein ArPEC25 demonstrated that its interaction with the host transcription factor CaβLIM1a is crucial to dampen immunity (Figure 1, F and G and Supplemental Figure 2E). As plant immunity also relies on transcription factors (both activators and repressors) to modulate the expression of many defense genes, it is not surprising that transcription factors are the target of secreted effectors. In fact, about 50% of all host proteins targeted by effectors participate in transcriptional regulation and in signaling (He et al., 2020). For example, the JASMONATE-ZIM-DOMAIN PROTEIN (JAZ) transcription factor is a negative modulator of jasmonic acid signaling that is targeted by the MiSSP7 effector from Laccaria bicolor (Plett et al., 2014). Likewise, the bacterial effector XopD from Xanthomonas campestris subverts Arabidopsis plant immunity by repressing the function of the transcription factor MYB30, a positive modulator of defense genes (Canonne et al., 2011). The P. infestans RxLR effector Pi03192 interacts with NAC transcription factors of potato (Solanum tuberosum) and promotes virulence by preventing their accumulation in the nucleus (McLellan et al., 2013).
CaβLIM1a belongs to the LIM family of transcription factors. In general, LIM family members are characterized by two LIM domains separated by a long spacer of around 40–50 amino acids, and they have been reported in various plants species to have different functions like cytoskeleton organization and transcriptional regulation (Weiskirchen and Günther, 2003; Han et al., 2013; Srivastava and Verma, 2015, 2017). LIM members localize to the cytoplasm, the nucleus, or both (Hoffmann et al., 2017; Sala and Ampe, 2018). Cytosol-localized LIMs function as actin bundlers (Han et al., 2013), while nucleus-localized LIMs are generally involved in the transcriptional regulation of genes with PAL-boxes such as PAL, CAD, and 4-COUMARATE:COA LIGASE (4CL) (Kawaoka et al., 2000). Here, we found that CaβLIM1a shows a dual localization in N. benthamiana leaves (Supplemental Figure 15A). A similar dual localization was also reported for WLIM1a in cotton (Gossypium hirsutum), whereby WLIM1a, which shuttles between the nucleus and the cytosol following H2O2 treatment, functions as an actin bundler and a transcriptional activator (Han et al., 2013). WLIM1a modulates the expression of genes involved in the biosynthesis of lignin and lignin-like phenolic compounds by binding to the PAL-boxes present in their promoter sequences (Han et al., 2013). However, we did not explore the role of CaβLIM1a in actin bundling in this study. Did showed that the secreted effector ArPEC25 enters the nucleus of chickpea cells to suppress immune gene expression by interacting and interfering with CaβLIM1a. Thus, we explored a strategy by which A. rabiei dampens chickpea immunity through a plant nucleus-localized effector. Based on these results and our previous finding on the upregulation of the CaLIM transcription factor genes upon A. rabiei infection (Srivastava and Verma, 2015), we speculate that CaβLIM1a plays a complex role in chickpea immunity.
Pathogens mediate effector entry into host cells via stomatal opening or by disrupting the physical integrity of the host. The various metabolic products of the phenylpropanoid biosynthetic pathway like lignin play crucial roles in plant immunity by maintaining the rigidity and integrity of the cell wall (Vance et al., 1980). Several reports also suggest the notion that besides its role in normal plant growth and development, lignin is also implicated in disease resistance (Malinovsky et al., 2014; Cesarino, 2019). The increased susceptibility of maize (Zea mays) bm (brown midrib) mutant plants to U. maydis support the idea that invading fungus find it easier to penetrate plant tissues with a less lignified cell wall (Vermerris et al., 2010; Tanaka et al., 2014). Furthermore, impaired lignin biosynthesis may also result in producing less effective defense-related lignin that could restrict or slow down the migration of invading pathogens (Tanaka et al., 2014; Lee et al., 2019). Another report in the S. sclerotiorum-B. napus pathosystem showed that the induced expression of BnaC.CCR2.b, an important gene involved in lignin biosynthesis, exhibits enhanced resistance to B. napus against the pathogen. The same report also indicated that the resistant line of B. napus (J964) exhibited higher lignin content in the stem, thereby confining the invasion and spread of the pathogen during early infection (Liu et al., 2021). Similarly, the GhDIR1 (Dirigent1) OE line in cotton restricts the spread of Verticillium dahlia by exhibiting increased accumulation of lignin (Shi et al., 2012). The expression of various genes in the phenylpropanoid biosynthetic pathway leading to the production of several metabolic products is modulated in response to biotic factors (Zhang et al., 2017). For instance, the invasion of camelina (Camelina sativa) by S. sclerotiorum strongly induces the expression of CsCCR2, which in turn increases lignin biosynthesis and resistance against the pathogen (Eynck et al., 2012). Evidence also suggests that secreted effectors target regulatory components of the phenylpropanoid pathway to subvert host responses. For example, the type-III effectors RipE1 and RipAY secreted by the necrotroph Ralstonia solanacearum promote infection in tobacco by enhancing the biosynthesis of SA, one of the products of the phenylpropanoid pathway (Sang et al., 2020). The Tin2 effector from U. maydis negatively modulates lignin deposition by stabilizing the Zea maize Tin2-targeting kinase1 (ZmTTK1) and refluxing the precursor towards the production of anthocyanin (Tanaka et al., 2014). Furthermore, to support colonization and infection, the NE ScQDO from the pathogen S. sclerotiorum selectively hydrolyzes flavonolaglycone, a product of the phenylpropanoid pathway that is normally toxic to the pathogen, to the nontoxic phloroglucinol carboxylic and phenolic acids (Chen et al., 2019). A recent study showed that in response to flg22 treatment, the SG2-type R2R3 MYB transcription factor MYB15 binds to ACC-rich promoter sequences from gene required for the biosynthesis of G-lignin and enhance lignification (Chezem et al., 2017). Likewise, LIM transcription factors have been reported to modulate the expression of phenylpropanoid biosynthetic pathway genes by binding to PAL-box (Kawaoka et al., 2000).
Among the four PAL genes identified in the chickpea genome, only CaPAL1 has PAL-box elements in its promoter. Our data demonstrate that CaβLIM1a appears to bind simultaneously to the two PAL-box elements in the CaPAL1 promoter via its LIM1 and LIM2 domains (Figure 4E) in contrast to NtLIM1, for which either LIM domain is sufficient to bind to the single PAL-box of the horseradish (Armoracia rusticana) peroxidase C2 (prxC2) promoter (Kawaoka et al., 2000). These possible differences between chickpea and tobacco LIMs may be due to the evolutionary distance separating the two plant families. Additionally, the reduced expression of CaPAL1 in CaβLIM1a knockdown hairy roots of chickpea suggests a direct positive relation between the two genes. The induced expression of lignin biosynthetic intermediates at 24 and 72 hpi in A. rabiei-infected chickpea compared to mock-treated plants is also in agreement with our hypothesis that CaPAL1 expression is positively modulated by CaβLIM1a.
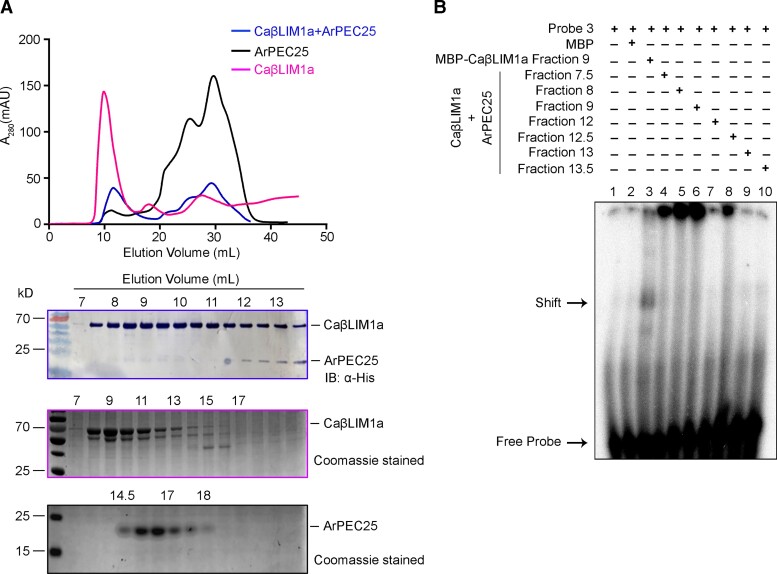
Here, we demonstrated that the physical interaction between the effector ArPEC25 and chickpea TF, CaβLIM1a negatively modulates CaPAL1 promoter activity (Figures 5B and 6B). We hypothesize two possible reasons for this downregulation. First, the interaction of ArPEC25 with CaβLIM1a may disrupt oligomerization of the transcription factor, turning it into a nonfunctional protein, as transcription factors typically work as oligomers (Sayou et al., 2016). Second, ArPEC25 may interfere with the DNA-binding ability of CaβLIM1a. However, gel filtration chromatography data suggested that the interaction between the two proteins has no influence on the oligomeric state of CaβLIM1a (Figure 8). Rather, the inability of CaβLIM1a to bind to the PAL-box elements of the CaPAL1 promoter in EMSA with increasing ArPEC25 concentrations suggests that ArPEC25 prevents the DNA binding and/or transactivation function of CaβLIM1a (Figure 6, A and B). Additionally, the increased expression of CaPAL1 in chickpea plants infected with the Δarpec25 mutant (Figure 6C) establishes that the transcriptional function of CaβLIM1a is negatively affected by ArPEC25. Since both LIM1 and LIM2 domains are required for DNA binding by CaβLIM1a, ArPEC25 may bind to one or both domains and thus interfere with CaβLIM1a function. However, additional data are required to make a conclusive statement.
Figure 8.
ArPEC25 binds CaβLIM1a and negatively regulates its DNA-binding activity to the CaPAL1 promoter. A, Gel filtration assay of recombinant CaβLIM1a and ArPEC25. Recombinant MBP-tagged CaβLIM1a and 6xHis-tagged ArPEC25 were incubated in an equal molar ratio and separated over a Superdex 200 Increase 10/300 GL column. The graph shows the absorption peak at 280 nm in milli absorbance unit (mAU) of purified CaβLIM1a, ArPEC25, and the mix of CaβLIM1a and ArPEC25 in pink, black and blue, respectively. The x-axis represents the time of elution for each protein in mL. Proteins present in different elution fractions were subjected to immunoblot analysis with an anti-His antibody and to Coomassie staining (lower panel). B, EMSA using oligonucleotide CaPAL1 probe P3 and different elution fractions of the gel filtration assay. Purified MBP served as negative control.
As CaβLIM1a induced CaPAL1 transcription by binding to the PAL-boxes in its promoter, we looked for any changes in metabolites derived from the phenylpropanoid biosynthetic pathway. In contrast to mock treatment, we detected lower levels of lignin biosynthesis intermediates at 24 and 72 hpi in A. rabiei-infected chickpea. However, the flavonoid and amino acid biosynthetic pathways remained unaffected. We speculate that the impaired lignin accumulation in A. rabiei-infected chickpea may be because of the inhibitory action of ArPEC25 on CaβLIM1a. To further test this hypothesis, we used the Δarpec25 mutant in a metabolite accumulation study. We observed increased accumulation of the lignin intermediates caffeic acid, chlorogenic acid, cinnamic acid, syringenic acid, P-coumaric acid, and trans-ferulic acid in Δarpec25-infected chickpea plants compared to plants infected with wild-type A. rabiei. Cinnamic acid is an intermediate compound in the pathway whose levels decreased at 24 hpi. As the lignin biosynthetic pathway is complex and its metabolites are interconnected, the reduced accumulation of cinnamic acid may be related to its conversion into other products in the pathway. Earlier reports on the chickpea-A. rabiei pathosystem also suggested a significant role for lignin and its derivative in protecting plants against fungal invasions. In an earlier report, it was shown through histological and cytopathological analysis that in contrast to less lignified tissues of the susceptible chickpea cultivar (Canitez-87), the highly lignified tissues in the leaflet and stem of resistant cultivar (ILC-195) were less prone to damage by the invading pathogen (Ilarslan and Dolar, 2002). Similarly, chickpea copper amine oxidase-mediated production of H2O2 has been attributed to its possible role in defense against A. rabiei probably by increasing the mechanical resistance of tissues through lignin and suberin polymerization (Rea et al., 2002). Thus, the differential modulation in the accumulation of these component in chickpea plants infected with wild type or the Δarpec25 mutant strain strongly support the speculation that the effector ArPEC25 is directly involved in lignin biosynthetic pathway alteration. We conclude that the biosynthesis of major lignin subunits is directly hampered during pathogenesis. The chemical composition and integrity of the lignin polymer largely depend upon the type and ratio of its constituent subunits. Lignin in conifers is predominantly composed of G subunits with a small fraction of H subunits, whereas woody dicots use mainly G and S subunits with an S/G ratio of 2 (Wang et al., 2014). It would be interesting to determine the relative biosynthesis rate of each type of lignin subunit and whether the ratios between subunit types change during AB disease progression.
Dothideomycetes presents an incredibly diverse group of fungi that includes many plant pathogens with broad host range. Many genes of Dothideomycetes encode effector proteins that are found often in close proximity to transposable elements and repetitive sequences, thereby potentially being exposed to a high rate of mutation (Ohm et al., 2012). This exposure could possibly speed up their independent rate of evolution, functional conservation and offers advantages in an arm race against the host. Also, the genome analysis of Blumeria graminis suggests that most of the effectors have undergone species-specific adaptation (Spanu et al., 2010). We have also shown that complementation of the Δarpec25 mutant strain with an ortholog, AlPEC25 from A. lentis failed to restore the virulence of fungi, suggesting that ArPEC25 has undergone host-specific adaptive evolution (Supplemental Figure 4B). Based on our Y2H analysis between CaβLIM1a and ArPEC25 truncated versions, we did not identify the specific regions of this fungal effector that targets the LIM family transcription factors. However, narrowed region swapping between ArPEC25 and AlPEC25 could possibly delineate the amino acids that are making AlPEC25 nonfunctional in A. rabiei. Additionally, ArPEC25 harbors two cysteine residues (C-35 and C-88) and homodimerizes (Supplemental Figure 26). The cysteine residues in effectors are known to facilitate various functions during host invasion. First, they form intermolecular disulfide bonds for homodimerization. Second, cysteine residues also assist in the interaction between the effector and its host targets, as was reported for the SsSSVP1 effector of S. sclerotiorum (Lyu et al., 2016). Third, these inter- and intra-molecular disulfide bonds prevent the degradation of effectors in the harsh chemical environment of the host (Liu et al., 2012). It will be interesting to explore the precise function of the two cysteine residues in ArPEC25. Although ArPEC25 contains the conserved PEXEL-like motif that was initially characterized in Plasmodium spp. for its role in effector secretion and translocation from the parasite to host erythrocytes (Boddey et al., 2009, 2010, 2016), we could not attribute any such role for this motif in ArPEC25 effector secretion from A. rabiei (Supplemental Figure 8D). The effector translocation data in host cells (Figure 2A and Supplemental Figure 10) also ruled out the possibility of a role for this motif in host entry. Many fungal genomes, including that of A. rabiei, encode an array of effectors with this conserved PEXEL-like motif (Hiller et al., 2004; Choi et al., 2010; Verma et al., 2016). Whether the PEXEL-like motif has some unexplored function is therefore a matter of conjecture.
In conclusion, we propose a mechanism whereby CaβLIM1a plays a role in host immunity by fortifying the physical barrier of the cell wall via lignin deposition in normal chickpea plants. The necrotrophic pathogen A. rabiei facilitates infection through its secreted virulence factor ArPEC25, which translocates to the host cell nucleus and modulates the DNA-binding activity of CaβLIM1a at target promoters by direct physical interaction (Figure 9). The impaired activity of the transcription factor results in reduced production of lignin subunits and a weakened cell wall to support successful penetration and virulence.
Figure 9.
Proposed working model for the role of ArPEC25 in chickpea susceptibility. The Ascochyta rabiei-secreted effector ArPEC25 interacts with CaβLIM1a in the nucleus of chickpea cells and inhibits its DNA-binding activity to the CaPAL1 promoter. In normal chickpea plants, CaβLIM1a binds directly to the PAL-boxes (B1 and B2) present upstream from CaPAL1. The binding results in the activation of CaPAL1 expression and accumulation of CaPAL1, thus producing metabolites that lead to the biosynthesis of lignin components. A. rabiei-secreted effector ArPEC25 translocates to the chickpea cell nucleus and interacts with the LIM domain-containing transcription factor CaβLIM1a and negatively regulates CaPAL1 expression by inhibiting the binding of CaβLIM1a to the CaPAL1 promoter, leading to lower CaPAL1 expression in infected chickpea, ultimately resulting in lower biosynthesis of lignin and hence compromised host resistance against the pathogen.
Materials and methods
Culture conditions, DNA isolation
Chickpea plants were grown under controlled conditions (D/N temperature: 24°C/18°C; D/N light duration: 14/10; Light intensity: 250 µE m−2 s−1 for day; Relative Humidity: 100%) in a plant growth chamber. The wild-type virulent A. rabiei isolate ArD2 (Indian Type Culture Collection No. 4638) was obtained from the Division of Plant Pathology, Indian Agricultural Research Institute (New Delhi, India). A. rabiei was routinely maintained on potato dextrose agar (PDA; Difco Laboratories, pH 5.2–5.5) at 22°C for 15–20 days. The conidial suspension of A. rabiei was prepared by extracting conidia/spores in sterile distilled water from a full-grown fungal culture PDA plate. To harvest fungal mycelia, the conidia were inoculated in potato dextrose broth (PDB; Difco Laboratories, pH 5.2–5.5) and allowed to grow at 22°C, 120 rpm for 5–7 days. Total genomic DNA from mycelial cells was isolated using a Quick-DN Fungal/Bacterial Miniprep Kit (Zymo Research, USA) as per the manufacturer's instructions.
For the cloning of the A. lentis AlPEC25 gene, the closest ortholog of ArPEC25, the DNA sequence of AlPEC25 (Accession, KAF9694412) was retrieved from NCBI (http://www.ncbi.nlm.nih.gov) and the complete 417-bp coding region was synthesized (Bio Basic Inc., Canada) and cloned into the pUC57 vector for further use.
RNA isolation and gene expression analysis
Two-week-old chickpea seedlings were spray inoculated with conidial suspension diluted to 2 × 106 conidia mL−1. Aerial tissues (stem and leaves) were detached from the inoculated chickpea plants at indicated time points using sterilized scalpel blade and immediately stored in liquid nitrogen for further use. Total RNA from stored infected chickpea tissues and PDB-grown fungal mycelia was isolated using TRIzol reagent (Invitrogen, USA). Briefly, tissues were crushed to a fine powder in liquid nitrogen and immediately resuspended in 1 mL of TRIzol reagent. For phase separation, 0.2 mL chloroform was added per mL of TRIzol reagent, mixed by shaking vigorously, and allowed to stand for 15 min at room temperature (RT). The resulting mixture was centrifuged at 12,000g for 15 min at 4°C. The colorless upper aqueous phase containing the RNA was collected in a fresh tube, and 0.5 mL of isopropanol was added per mL of TRIzol reagent. The samples were allowed to stand for 5–10 min at RT and centrifuged at 12,000g for 15 min at 4°C. The supernatant was discarded and the RNA pellet was washed twice by adding minimum of 1 mL of 75% (v/v) ethanol per 1 mL of TRIzol reagent. The RNA pellet was then air dried for 5–10 min and dissolved in DEPC-treated sterile water.
First-strand cDNA was synthesized using Superscript IV first-strand synthesis system (Thermo Fisher Scientific, USA). Briefly, the reaction mixture containing oligo d(T), dNTP mix, and purified total RNA was heated at 65°C for 5 min and then incubated on ice for at least 1 min. The 5xSSIV buffer, DTT, ribonuclease inhibitor, and Superscript IV reverse transcriptase enzyme were added to the reaction mixture and incubated at 50°C for 10 min. The reaction was inactivated by incubation at 80°C for 10 min. Targeted gene expression was determined by qPCR on an ABI7900 system (Applied Biosystem, USA) using gene-specific primers (Supplemental Data Set 1) and Brilliant III Ultra-Fast SYBR Green QPCR Master Mix (Agilent Technologies, USA). Relative gene expression levels were calculated using 2−ΔΔCt method, and the values were derived from independent samples with technical replicates. Caβ-tubulin (LOC101495306) and elongation factor 1-alpha (ArEF1α; ST47_g4052) were used as an internal controls for chickpea and A. rabiei, respectively.
PEG-mediated fungal genome editing and complementation in A. rabiei
To generate the ArPEC25 knockout construct (Δarpec25), a 773-bp fragment of 5′ flanking region of ArPEC25 was PCR amplified from A. rabiei genomic DNA using gene-specific primer pairs (Supplemental Data Set 1). The amplified PCR product was cloned into the XhoI and PstI restriction sites of the pGKO2-hph vector. The resulting construct, pGKO2-5′ArPEC25-hph, was confirmed through restriction digestion and Sanger sequencing. Similarly, a 1,023-bp fragment of the 3′ flanking sequence of ArPEC25 was PCR amplified from A. rabiei genomic DNA using gene-specific primers (Supplemental Data Set 1) and cloned into the BamHI and EcoRI sites in the pGKO2-5′ArPEC25-hph construct. The resulting construct, pGKO2-5′ArPEC25-hph-3′ArPEC25 was confirmed through restriction digestion and sequencing. The cloned ArPEC25 gene replacement construct of ∼3.5 kb, including 5′ and 3′ flanking sequences and sequence of the hygromycin resistance gene (hph), was amplified using the appropriate primer pair (Supplemental Data Set 1) and transformed into A. rabiei protoplasts through PEG-mediated transformation as described earlier (Sinha et al., 2021).
For the complementation tests of the Δarpec25 mutant (Δarpec25/ArPEC25) with a single copy of native ArPEC25, the full-length ArPEC25 gene including its native promoter was amplified using specific primers (Supplemental Data Set 1). The amplified fragment was cloned at the EcoRI and XbaI restriction sites in the pBIF2 vector. Similarly, the deletion mutant encoding a version of ArPEC25 with a nuclear export signal (Δarpec25/ArPEC25-NES) was prepared using specific primers (Supplemental Data Set 1) and cloned at the EcoRI and XbaI sites in the pBIF2 vector. For the complementation test of the Δarpec25 mutant with a single copy of AlPEC25 (Δarpec25/AlPEC25), the full-length AlPEC25 gene was synthesized from the A. lentis genome sequence and cloned at the SacI and XbaI sites in the pBIF2 vector. The expression of AlPEC25 in the complementation strain is driven by the ArPEC25 promoter. All generated constructs were confirmed through restriction enzyme digestion and Sanger sequencing. The complementation constructs were transformed into Agrobacterium (A. tumefaciens) strain GV3101. These Agrobacterium clones were used to transform the Δarpec25 mutant through Agrobacterium-mediated transformation (ATMT), as described earlier (Nizam et al., 2010). Knockout and complementation constructs were confirmed using Southern blot hybridization and PCR analysis as described earlier (Sinha et al., 2021).
Plant infection assay using the mini-dome technique
The mini-dome technique (Cho et al., 2004) was used to measure virulence of wild-type A. rabiei and its derived mutant Δarpec25, complementation transformants Δarpec25/ArPEC25, and Δarpec25/ArPEC25-NES on chickpea plants. Seedlings were grown for 2 weeks under control conditions (day/night [D/N] temperature: 24°C/18°C; D/N light duration: 14 h/10 h; light intensity: 250 µE m−2 s−1; relative humidity: 100%) in the plant growth chamber. Conidial suspension of wild-type A. rabiei and different mutant strains (Δarpec25, Δarpec25/ArPEC25, Δarpec25/ArPEC25-NES, and Δarpec25/AlPEC25) were harvested from 20-day-old full-grown cultures on PDA medium. Two-week-old chickpea seedlings were spray inoculated with conidial suspension diluted to 2 × 106 conidia mL−1. Inoculated seedlings were immediately covered with transparent plastic cups (referred to as mini-domes) to provide the uniform high level of relative humidity required for successful infection. The mini-domes were removed after 48 hpi. Disease severity was evaluated after 7 days post-inoculation (dpi).
For in vitro oxidative stress treatment, 250 µM menadione (Sigma-Aldrich, USA) was added to one-week-old broth culture of A. rabiei (inoculated with 100 µL of 1 × 104 conidia mL−1 suspension). Mycelial balls were harvested from PDB medium using three layers of Miracloth (EMD, Millipore Corp, Germany) at different time points (0.25, 0.5, 1, 3, and 6 h) post treatment. For control, ethanol-treated fungal mycelial balls were harvested. For ArPEC25 gene expression during in planta infection, A. rabiei conidial suspension (2 × 106 conidia mL−1) was harvested and prepared as mentioned above. The conidial suspension was spray inoculated on two-week-old Pusa 362 chickpea seedlings. Aerial tissues of inoculated seedlings were harvested at different time points (12, 24, 72, and 144 hpi).
Yeast two-hybrid (Y2H) assay
The interaction between the chickpea transcription factors CaβLIM1a, CaβLIM1b, CaδLIM2, CaWLIM1a, CaWLIM1b, CaWLIM2, and the A. rabiei effector ArPEC25 was investigated using a split ubiquitin-based DUALhunter system in yeast (Dualsystems Biotech, Switzerland). Full-length coding sequences were cloned into the bait (pGDHB1) and prey (pGPR3-N) vectors at the NotI and AscI restriction sites. The clones along with their controls were co-transformed into yeast strain NMY51, using an EZ-Yeast transformation kit (MP biomedicals, USA). Transformed yeast cells were selected on synthetic defined (SD) medium lacking Leu and Trp (SD/–L–W) for 3 days at 30°C. Yeast colonies were resuspended in 0.9% (q/v) NaCl and spotted onto SD/–L–W medium and SD/–L–W–H–A containing 15 mM 3-amino-1,2,4-triazole (3-AT) as selection medium for protein–protein interaction. The interaction between CaβLIM1a-Cub and NubI served as a positive control for bait expression while the co-expression of CaβLIM1a-Cub and NubG served as negative control for autoactivation. Primers used for cloning are listed in Supplemental Data Set 1.
Yeast one-hybrid assays
Binding of CaβLIM1a to PAL-box elements of the CaPAL1 promoter (LOC101507594) was confirmed by Y1H assay. The native promoter and its PAL-box deletion constructs were cloned upstream to the AUR1-C reporter gene into the yeast genome-integrating vector pAbAi. The full-length CaβLIM1a-coding sequence and its deletion variants were cloned in-frame and downstream of the sequence of the GAL4-AD in the pGADT7 vector (Clontech). Yeast strain Y1H gold was first transformed with linearized pAbAi and pAbAi:ProCaPAL1 DNA to integrate the DNA at specific genomic locations and support yeast growth on SD/–U medium. Subsequently, the pGADT7 clones were transformed and yeast transformants were selected on SD/–U–L medium. The interaction between DNA and protein was tested on SD/–U–L containing 150 ng mL−1 AbA. The optimal AbA concentration was checked with yeast cells transformed with pAbAi:ProCaPAL1 to block background growth. Primers used for cloning are listed in Supplemental Data Set 1.
Dual-luciferase reporter assay
A 737-bp promoter fragment of CaPAL1 was amplified from chickpea genomic DNA, cloned in entry vector (pENTR), and mobilized into the destination vector p635nRRF (Kumar et al., 2018) using Gateway LR Clonase II enzyme mix to generate the ProCaPAL1:LUC reporter construct. The coding sequence of CaβLIM1a and CaWLIM1a were cloned in pGWB420 and the coding sequence of ArPEC25 was cloned in pGWB411 vectors, respectively, to generate MYC- and FLAG-tagged effector constructs (CaβLIM1a-MYC, CaWLIM1a-MYC, and ArPEC25-FLAG). Agrobacterium cells at the same cell density based on absorbance at 600 nm with individual or combinations of reporter and effector constructs were co-infiltrated in N. benthamiana leaves. Infiltrated leaf discs were collected after 48 hpi. The infiltrated leaf discs were ground to a fine powder in liquid nitrogen and mixed with 1× passive lysis buffer (PLB) provided in the Dual-Luciferase reporter assay system (Promega, USA). Firefly luciferase and Renilla luciferase activity were measured following the manufacturer's instruction (Promega, USA) using a POLARstar Omega multimode plate reader (BMG Labtech, Germany). Relative LUC activity was calculated by normalizing LUC activity to REN activity. Immunoblot using total cell lysates of the same infiltrated leaf disc was used to confirm the accumulation of the effector constructs with respective antibodies. Primers used in cloning are listed in Supplemental Data Set 1.
Yeast transactivation assay
The cDNA of CaβLIM1a was mobilized from the entry clone to the destination vector pGBKT7g using Gateway LR Clonase II enzyme mix. The construct pGBKT7g-CaβLIM1a was transformed into the Y2H Gold yeast strain with an EZ-Yeast transformation kit (MP Biomedicals, USA). The positive clones selected on SD/–W medium were spotted onto SD/–W–H or SD/–W–H–A medium to observe transactivation of auxotrophic reporter genes. CaWRKY50 served as a positive control for yeast transactivation assay (Kumar et al., 2016). Empty pGBKT7 vector served as a negative control in the transactivation assay.
Subcellular localization
For subcellular localization study of CaβLIM1a and ArPEC25, fusion constructs were transiently expressed into N. benthamiana leaves. The coding sequence of ArPEC25 without SP (ArPEC25ΔSP) and CaβLIM1a were first cloned at NotI and AscI restriction enzyme sites of the entry vector pENTR. The entry clones were then mobilized into the destination vector pGWB405 using Gateway LR Clonase II enzyme mix to produce CaβLIM1a-GFP and ArPEC25-GFP fusion proteins. NLS-mRFP served as a nuclear marker. For co-localization of CaβLIM1a and ArPEC25, the ArPEC25ΔSP sequence was cloned into the binary vector pCAMBIA1301-mCherry. The plasmids were transformed into Agrobacterium strain GV3101. The desired constructs were transiently co-expressed in N. benthamiana leaves epidermal cells via Agrobacterium-mediated infiltration. Infiltrated plants were transferred to growth chamber at 22°C with a 16-h light/8-h dark photoperiod. The fluorescence signal was detected 48 hpi using a TCS SP8 confocal laser scanning microscope (Leica Microsystems, Germany). Primers used in cloning are listed in Supplemental Data Set 1.
Recombinant protein purification from E. coli
To construct the plasmid for recombinant protein production, the ArPEC25ΔSP sequence was cloned in-frame with that of StrepII by overlapping oligonucleotide-based amplification and cloned in the vector pET28a (+)to create pET-ArPEC25-StrepII-6xHis. The CaβLIM1a sequence was cloned into the pMAL2x-MBP vector to generate MBP-CaβLIM1a. The constructs were transformed into E. coli BL21-CodonPlus (DE3)-RIPL competent cells for protein production. Recombinant ArPEC25-StrepII-6xHis protein was produced in E. coli cells by adding 1 mM isopropyl β-D-thiogalactoside (IPTG) for 6 h at 28°C. The desired protein was affinity-purified with Strep-Tactin Sepharose beads (iba, Germany) following the manufacturer's instructions. For the purification of MBP-CaβLIM1a or MBP alone, E. coli cells were treated with 1 mM IPTG for 4 h at 28°C and the induced protein was affinity-purified with Amylose Resin (New England Biolabs). Primers used in cloning are listed in Supplemental Data Set 1.
EMSA
Complementary oligonucleotides 41, 42, and 70 bp in length with the PAL-box(s) of the CaPAL1 promoter were synthesized and labeled with [γ-32P] ATP as per the protocol of the gel shift assay system (Promega, USA). Briefly, the reaction mixture containing the synthesized oligonucleotides, T4 polynucleotide kinase buffer, [γ-32P] ATP, and T4 polynucleotide kinase enzyme was incubated at 37°C for 1 h. The reaction was stopped by adding 1 µL of 0.5 M EDTA. The labeled probes were mixed with recombinant purified MBP or MBP-CaβLIM1 and 5× gel shift binding buffer (from the gel shift assay system kit); the reaction was incubated for 20 min at RT. The reaction mixtures were separated on 6% (w/v) PAGE gels in 0.5× Tris-buffered EDTA buffer (pH 8.3) at 250–350 V. The gel was analyzed using phosphorimaging screen and Typhoon (GE Healthcare, USA).
YST assay
The ArPEC25 coding sequence with and without SP was independently cloned into the pYST1 vector (Lee et al., 2006) to generate pYST1-ArPEC25 and pYST1-ArPEC25ΔSP constructs, respectively. Empty pYST1 vector was used as negative control. The resulting constructs were transformed into the suc2 yeast mutant strain using a Yeastmaker Yeast transformation system 2 kit (Clontech). Positive transformants were selected on SD/–L medium for 3–5 days. The positive colonies from the primary selection plates were then resuspended in liquid SD/–L medium and grown to stationary phase overnight at 30°C and 250 rpm. Aliquots of these cultures were diluted in sterile distilled water to a cell density of OD600 = 1. Spotting for secretion analysis was performed on SD/–L medium supplemented with 2% (w/v) glucose alone or together with 2 μg mL−1 Antimycin A.
BiFC assay
The cDNAs of CaβLIM1a and ArPEC25 were cloned in-frame with the sequence encoding the N-terminal half and C-terminus half of Venus, respectively, in the pDOE-05 vector (Gookin and Assmann, 2014). NLS-RFP was used as nuclear marker. Agrobacterium colonies from strain GV3101 harboring the respective plasmids were grown in LB medium; for Agrobacterium-mediated infiltration, the bacterial cultures were incubated in infiltration medium (10 mM MES, 10 mM MgCl2, and 150–200 μM acetosyringone) at 28°C for ≥4 h. An equal volume of cultures resuspended at an OD600 = 0.4 was co-infiltrated in N. benthamiana leaf epidermal cells. Venus signal was detected using a TCS SP8 confocal laser scanning microscope (Leica Microsystems, Germany) after 48 hpi. The presence of mTurquoise2 signal indicates successful transient expression of the constructs in leaf cells. Primers used in cloning are listed in Supplemental Data Set 1.
Protein extraction and immunoblot analysis
Total protein was extracted from 0.2 g of infiltrated fresh N. benthamiana leaves by grinding to a fine powder in liquid nitrogen. Powder was then resuspended in 1× PLB provided in the Dual-Luciferase Reporter Assay System (Promega, USA). Extracts were vortexed and then centrifuged at 16,000g for 1 min at 4°C. The supernatant containing the total lysate was collected and separated by SDS-PAGE and analyzed through immunoblots.
For total protein extraction from yeast, 2 mL liquid culture was inoculated with fresh yeast cells and incubated at 30°C, with shaking at 200 rpm overnight. Freshly grown cells were harvested through centrifugation and resuspended in 2 M lithium acetate (LiAc). Cells were further harvested by centrifugation and then resuspended in 0.4 M NaOH for 5 min on ice. Harvested cells were finally resuspended in protein-loading dye and boiled for 5 min. The supernatant containing total proteins was collected and separated by SDS-PAGE and analyzed by immunoblots.
For total protein extraction of fungal mycelia, the conidial suspension (as mentioned earlier) was inoculated in PDB medium and incubated for 5–7 days at 22°C, with shaking at 120 rpm. Grown mycelia were treated with 250 µM menadione (Sigma-Aldrich, USA) for 3 h, and the tissues were harvested by passing through three layers of sterile Miracloth (EMD, Millipore Corp, Germany). Fungal tissue was ground to a fine powder in liquid nitrogen and homogenized in Tris-glycine buffer pH 8.3 (3 g Trizma and 14.4 g Glycine in 1 L of sterile distilled water). The lysate was centrifuged at 16,000g for 40 min at 4°C. The supernatant containing total fungal protein was collected and separated by SDS-PAGE and analyzed by immunoblots.
For extraction of total secreted proteins from CF of the fungus, 7-days-old fungal mycelia tissues grown in PDB medium were treated with 250 µM menadione (Sigma-Aldrich, USA) for 3 h. Axenic CF was separated from fungal mycelia by passing it through three layers of Miracloth as above. Further, CF was sequentially filtered with 0.45- and 0.22-µm filter discs (Durapore PVDF, Sigma-Aldrich, USA). Filtered CF was then concentrated using 3-kDa Amicon Ultra-15 Centrifugal Filter Units (Merck, USA). The concentrate was then snap-frozen and stored at −80°C for future use.
Proteins were separated on 12% (w/v) SDS-PAGE gels and transferred to polyvinylidene fluoride (PVDF) Transfer Membrane (MDI, India), at 4°C, 100 V for 1 h. Following transfer, the membrane was washed briefly in 1× phosphate-buffered saline 0.1% (v/v) Tween 20 (PBST, Sigma-Aldrich, USA). The membrane was blocked with 2% (w/v) nonfat powdered milk (Bio Basic, Canada Inc.) in 1× PBST overnight at 4°C. The membrane was then washed twice in 1× PBST and then the membrane was incubated for 1 h at RT in primary antibodies; antiMyc (Catalog No. CPA9004), anti-HA (Catalog No. CPA 9002), and anti-FLAG (Catalog No. CPA9001) from Cohesion Biosciences. Anti-Histone and anti-His antibodies were diluted to 1:2,000 in 1× PBST. Following incubation, membrane was washed five times in 1× PBST and incubated for 1 h at RT with goat anti-rabbit IgG (H + L) secondary antibody, HRP conjugated (Catalog No. 65-6120; Invitrogen, USA) at a dilution of 1:5,000 in 1× PBST. Following incubation, the membrane was washed five times in 1× PBST and the detection was performed with Clarity Western ECL substrate (BIO-RAD, USA).
Protein immunoprecipitation and analysis by mass spectrometry
For the immunoprecipitation of ArPEC25-FLAG secreted by the A. rabiei OE strain, the transformant was grown in PDB medium and the CF was collected and processed as mentioned above. The processed CF was incubated with anti-FLAG antibody-coated Dynabeads Protein G, (ThermoFisher, USA) for 1 h at 4°C in a rotating shaker. The mixture was then used for specific purification of ArPEC25-FLAG following the manufacturer's instructions. Briefly, antibody-bound ArPEC25-FLAG was separated from the solution using a magnetic stand. Further, nonspecifically attached proteins were removed from the beads through three rounds of washing buffer. Magnetic bead-attached ArPEC25-FLAG was then dissolved in G-buffer.
Aliquots of 25 µL were taken and reduced with 5 mM TCEP and further alkylated with 50 mM iodoacetamide and then digested with trypsin (1:50, trypsin/lysate ratio, w/v) for 16 h at 37°C. Digests were cleaned using a C18 silica cartridge and dried using a speed vac. The dried pellet was resuspended in buffer A (5% [v/v] acetonitrile, 0.1% [v/v] formic acid).
Processed samples were identified by mass spectrometry analysis using an EASY-nLC 1000 system (ThermoFisher Scientific, USA) coupled to a QExactive mass spectrometer (ThermoFisher Scientific, USA) equipped with a nano electrospray ion source. About 1.2 µg of the peptide mixture was resolved using a 15-cm PicoFrit column (360 µm outer diameter, 75 µm inner diameter, 10-µm tip) filled with 1.9 µm of C18-resin (Dr Maeisch, Germany). The peptides were loaded with buffer A and eluted with a 0%–40% gradient of buffer B (95% [v/v] acetonitrile, 0.1% [v/v] formic acid) at a flow rate of 300 nL min−1 for 45 min. MS data were acquired using a data-dependent top10 method dynamically choosing the most abundant precursor ions from the survey scan. For data processing, three reactions were processed and three RAW files generated were analyzed with Proteome Discoverer against the ArPEC25 sequence. For Sequence search, the precursor and fragment mass tolerances were set to 10 ppm and 0.5 Da, respectively. The protease used to generate peptides, i.e. enzyme specificity was set for trypsin/P (cleavage at the C terminus of “K/R”: unless followed by “P”) along with maximum missed cleavages value of two. Carbamidomethyl on cysteine was set as fixed modification and oxidation of methionine and N-terminal acetylation was considered as variable modifications for database search. Both peptide spectrum match and protein false discovery rate were set to 0.01.
Gel filtration assay
Different recombinant proteins (MBP-CaβLIM1a and ArPEC25-6xHis) were purified as mentioned above and dialyzed in PBS (Sigma-Aldrich, USA). For ArPEC25-mediated disruption of the CaβLIM1a oligomeric complex, equal amounts of both proteins were mixed and incubated for 30 min at RT. Proteins in mixture and or alone were subjected to gel filtration analysis using Superdex 200 Increase 10/300 GL column (GE Healthcare Bio-Sciences AB) with a flow rate of 0.2 mL min−1 and an injection volume of 2 mL and a fraction size of 0.5 mL. The eluted fractions were analyzed by immunoblot with anti-His antibody.
RNA interference
For the knockdown of CaβLIM1a in hairy roots of chickpea “Pusa 362,” a specific 341-bp fragment of CaβLIM1a was cloned in the entry vector pENTR into the NotI and AscI sites. The construct was recombined with the pK7GWIWG2(II)-RedRoot vector using Gateway cloning technology to construct the RNAi transformation vector. The resulting vector, harboring a 35S:DsRed cassette, was introduced into Agrobacterium rhizogenes strain ARqua1 and used for transient transformation of chickpea hairy roots as described previously (Singh and Verma 2022; Singh et al., 2022). Putative chickpea hairy roots were first screened for RFP fluorescence by microscopy. The downregulation of CaβLIM1a in roots was then confirmed by RT-qPCR analysis.
Metabolite analysis
Chickpea plants susceptible to AB (cultivar Pusa 362) were spray inoculated with conidial suspension (2 × 106 conidia mL−1) of wild-type A. rabiei or the Δarpec25 mutant strain as described earlier. Chickpea leaf and stem samples were harvested at 24 and 72 hpi and stored in liquid nitrogen for downstream processing. The samples were completely desiccated in a lyophilizer. The lyophilized samples were ground to a fine powder in a tissue lyser and ∼100 mg dry weight sample was used for metabolite extraction. For the total extraction of metabolites, 80% (v/v) methanol (HPLC grade) was added to the powder, followed by heating at 65°C for 15 min. The samples were incubated at 28°C overnight for complete extraction of metabolites, followed by centrifugation at 10,000g for 5 min at 4°C to remove cell debris. The supernatant was collected and evaporated in a speed vac at RT. The resulting pellet was dissolved in 1 mL 80% (v/v) methanol and filtered through a 0.45-µm filter disc (Durapore PVDF, Sigma-Aldrich, USA). UHPLC was performed using 50 µL of this filtered sample and the data were analyzed.
Phylogenetic analysis
The MUSCLE algorithm of MEGA X software was used for the alignment of different protein sequences used in the phylogenetic tree. The aligned sequences were used for the construction of phylogeny through MEGA X software using neighbor-joining method, with bootstrap value of 1,000. The alignment used for phylogenetic analysis is provided in Supplemental File 1 and the tree in Newick format is provided in Supplemental File 2.
Quantitative measurement and statistical analysis
Lesion size and diameter of AB infection were measured using ImageJ/Fiji software. FRET efficiency was calculated in Leica software. Statistical significance of means or difference among groups was obtained from the average of three biological replicates, having at least three technical replicates in each group. To calculate statistical significance, Student's t test and ANOVA followed by Tukey test between multiple groups were performed using GraphPad Prism 6. The P < 0.05 was accepted as significant. Statistical data are provided in Supplemental Data Set 2.
In silico bioinformatics analysis
All gene and protein sequences in this study were obtained from NCBI (http://www.ncbi.nlm.nih.gov). Multiple sequence alignment was performed by MUSCLE algorithm of MEGA X software using default parameters. Molecular evolution and phylogenetic studies of ArPEC25 were performed in MEGA X software using the neighbor-joining method, with a bootstrap value of 1,000. To determine the putative transcription factor binding sites in the promoter sequence of CaPAL1, PlantCARE online tools (Lescot et al., 2002) was used. The theoretical pI was obtained from the Expasy compute pI/Mw tool.
Accession numbers
Sequence data used in this study can be found from NCBI server (https://www.ncbi.nlm.nih.gov/) under the following accession numbers: ArPEC25, KZM27126; CaWLIM1a, XM_004498934; CaWLIM1b, XM_004501459; CaβLIM1a, XM_027335175; CaWLIM2, XM_004503166; CaδLIM2, XM_004503703; CaβLIM1b, XM_004508783; AlPEC25, KAF9694412; CaPAL1, XM_004497459; CaPAL2, XM_027333119; CaPAL3, XM_004493920; CaPAL4, XM_004491570.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Conservation and phylogenetic relationship between the A. rabiei effector ArPEC25 and orthologs from other organisms.
Supplemental Figure S2. The Ascochyta rabiei effector ArPEC25 is indispensable for virulence activity.
Supplemental Figure S3. ArPEC25 requires a functional nuclear localization signal (NLS; RKRRRRR) for its virulence activity on chickpea plants.
Supplemental Figure S4. The ArPEC25 ortholog from Ascochyta lentis, AlPEC25, fails to complement virulence activity of the A. rabiei KO mutant strain (Δarpec25).
Supplemental Figure S5. The ArPEC25-FLAG-EYFP effector translocates from A. rabiei fungus to the host cell nucleus.
Supplemental Figure S6. ArPEC25 has a functional NLS (RKRRRRR) and a nuclear export signal added at its C terminus directs it outside the nucleus.
Supplemental Figure S7. The ArPEC25 signal peptide (SP) directs the effector to the ER secretory pathway in yeast and A. rabiei cells.
Supplemental Figure S8. ArPEC25-FLAG-EYFP and two variants of its PEXEL-like motif are secreted by A. rabiei.
Supplemental Figure S9. LC-MS/MS analysis reveals that the PEXEL-like motif (RTLND) of A. rabiei effector ArPEC25 is not cleaved during fungal effector secretion.
Supplemental Figure S10. The ArPEC25 effector does not require PEXEL-like motif residues (RTLND) for translocation from A. rabiei to host cell.
Supplemental Figure S11. CaβLIM1a is activator.
Supplemental Figure S12. ArPEC25 interacts with CaLIMs.
Supplemental Figure S13. Confocal image of donor and acceptor proteins used for FRET experiment.
Supplemental Figure S14. Mature ArPEC25 interacts with CaβLIM1a strongly through its overall structure and the ArPEC2583–134 region contributes to this interaction.
Supplemental Figure S15. CaβLIM1a localizes to the host cell nucleus.
Supplemental Figure S16. The chickpea CaPAL genes CaPAL1, CaPAL2, CaPAL3, and CaPAL4 are induced upon A. rabiei infection.
Supplemental Figure S17. Purification of recombinant protein for EMSA.
Supplemental Figure S18. CaβLIM1a bind to CaPAL1 promoter.
Supplemental Figure S19. Immunoblot analysis of different effector constructs used in dual-luciferase assay.
Supplemental Figure S20. RNAi-mediated knockdown of CaβLIM1a in chickpea hairy roots.
Supplemental Figure S21. Purification of recombinant protein for EMSA.
Supplemental Figure S22. Schematic representation of various metabolite levels by heatmaps in A. rabiei (wild type) inoculated and mock-inoculated chickpea plants.
Supplemental Figure S23. Lignin components in chickpea plant infected with wild-type A. rabiei and mock treatment.
Supplemental Figure S24. Concentration of key phenylpropanoid pathway metabolites synthesizing flavonoids and anthocyanin in mock- and A. rabiei-inoculated chickpea seedlings.
Supplemental Figure S25. Modulation of the accumulation of different amino acids in chickpea treated with mock or A. rabiei.
Supplemental Figure S26. ArPEC25 forms dimers in Y2H assay.
Supplemental Table S1. Summary of candidate A. rabiei effector proteins containing conserved sequence motifs.
Supplemental Table S2. List of putative ArPEC25-interacting transcription factors found in chickpea library screening.
Supplemental Data Set S1. List of primers used in this study.
Supplemental Data Set S2. Summary of statistical analyses.
Supplemental File S1. Protein sequence alignment used for the phylogenetic tree shown in Supplemental Figure 1B.
Supplemental File S2. Newick file format of the phylogenetic tree shown in Supplemental Figure 1B.
Supplementary Material
Acknowledgments
We gratefully acknowledge a research grant from the Department of Biotechnology, Government of India (File No: BT/PR10605/PBD/16/791/2008 and BT/AGR/CG-Phase II/01/2014) and a core grant from the National Institute of Plant Genome Research (NIPGR), New Delhi, India for funding this work. Authors thank the Central Instrumental Facility of NIPGR for utilizing various instruments.
Contributor Information
Shreenivas Kumar Singh, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India; Plant Immunity Laboratory, School of Life Sciences, Jawaharlal Nehru University, New Delhi 110067, India.
Ankita Shree, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Sandhya Verma, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Kunal Singh, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Kamal Kumar, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Vikas Srivastava, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Ritu Singh, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Samiksha Saxena, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Agam Prasad Singh, National Institute of Immunology, Aruna Asaf Ali Marg, New Delhi 110067, India.
Ashutosh Pandey, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India.
Praveen Kumar Verma, Plant Immunity Laboratory, National Institute of Plant Genome Research, Aruna Asaf Ali Marg, New Delhi 110067, India; Plant Immunity Laboratory, School of Life Sciences, Jawaharlal Nehru University, New Delhi 110067, India.
References
- Abeysekara NS, Friesen TL, Keller B, Faris JD (2009) Identification and characterization of a novel host–toxin interaction in the wheat–Stagonospora nodorum pathosystem. Theor Appl Genet 120(1): 117–126 [DOI] [PubMed] [Google Scholar]
- Alam SS, Bilton JN, Slawin AMZ, Williams DJ, Sheppard RN, Strange RN (1989) Chickpea blight: production of the phytotoxins solanapyrones A and C by Ascochyta rabiei. Phytochemistry 28(10): 2627–2630 [Google Scholar]
- Bauters L, Stojilković B, Gheysen G (2021) Pathogens pulling the strings: effectors manipulating salicylic acid and phenylpropanoid biosynthesis in plants. Mol Plant Pathol 22(11): 1436–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuiyan NH, Selvaraj G, Wei Y, King J (2009) Gene expression profiling and silencing reveal that monolignol biosynthesis plays a critical role in penetration defence in wheat against powdery mildew invasion. J Exp Bot 60(2): 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddey JA, Hodder AN, Günther S, Gilson PR, Patsiouras H, Kapp EA, Pearce JA, De Koning-Ward TF, Simpson RJ, Crabb BS, et al. (2010) An aspartyl protease directs malaria effector proteins to the host cell. Nature 463(7281): 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddey JA, Moritz RL, Simpson RJ, Cowman AF (2009) Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic 10(3): 285–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddey JA, O’Neill MT, Lopaticki S, Carvalho TG, Hodder AN, Nebl T, Wawra S, Van West P, Ebrahimzadeh Z, Richard D, et al. (2016) Export of malaria proteins requires co-translational processing of the PEXEL motif independent of phosphatidylinositol-3-phosphate binding. Nat Commun 7(1): 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen S, Williams SJ, Winterberg B, Kobe B, Solomon PS (2016) Wheat PR-1 proteins are targeted by necrotrophic pathogen effector proteins. Plant J 88(1): 13–25 [DOI] [PubMed] [Google Scholar]
- Brefort T, Tanaka S, Neidig N, Doehlemann G, Vincon V, Kahmann R (2014) Characterization of the largest effector gene cluster of Ustilago maydis. PLoS Pathog 10(7): e1003866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canonne J, Marino D, Jauneau A, Pouzet C, Brière C, Roby D, Rivas S (2011) The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell 23(9): 3498–3511 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Cesarino I (2019) Structural features and regulation of lignin deposited upon biotic and abiotic stresses. Curr Opin Biotechnol 56: 209–214 [DOI] [PubMed] [Google Scholar]
- Chen J, Ullah C, Reichelt M, Gershenzon J, Hammerbacher A (2019) Sclerotinia sclerotiorum circumvents flavonoid defenses by catabolizing flavonol glycosides and aglycones. Plant Physiol 180(4): 1975–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chezem WR, Memon A, Li FS, Weng JK, Clay NK (2017) SG2-type R2R3-MYB transcription factor MYB15 controls defense-induced lignification and basal immunity in Arabidopsis. Plant Cell 29(8): 1907–1926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S, Chen W, Muehlbauer FJ (2004) Pathotype-specific genetic factors in chickpea (Cicer arietinum L.) for quantitative resistance to Ascochyta blight. Theor Appl Genet 109(4): 733–739 [DOI] [PubMed] [Google Scholar]
- Choi J, Park J, Kim D, Jung K, Kang S, Lee YH (2010) Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics 11(1): 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi B, Percival-Alwyn L, Downie RC, Venturini L, Iagallo EM, Campos Mantello C, McCormick-Barnes C, See PT, Oliver RP, Moffat CS, et al. (2020) Genetic analysis of wheat sensitivity to the ToxB fungal effector from Pyrenophora tritici-repentis, the causal agent of tan spot. Theor Appl Genet 133(3): 935–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eynck C, Séguin-Swartz G, Clarke WE, Parkin IAP (2012) Monolignol biosynthesis is associated with resistance to Sclerotinia sclerotiorum in Camelina sativa. Mol Plant Pathol 13(8): 887–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondevilla S, Krezdorn N, Rotter B, Kahl G, Winter P (2015) In planta identification of putative pathogenicity factors from the chickpea pathogen Ascochyta rabiei by de novo transcriptome sequencing using RNA-Seq and massive analysis of cDNA ends. Front Microbiol 6: 1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen Tl, Chu C, Xu SS, Faris JD (2012) Sntox5–Snn5: a novel Stagonospora nodorum effector–wheat gene interaction and its relationship with the SnToxA–Tsn1 and SnTox3–Snn3–B1 interactions. Mol Plant Pathol 13(9): 1101–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen TL, Meinhardt SW, Faris JD (2007) The Stagonospora nodorum-wheat pathosystem involves multiple proteinaceous host-selective toxins and corresponding host sensitivity genes that interact in an inverse gene-for-gene manner. Plant J 51(4): 681–692 [DOI] [PubMed] [Google Scholar]
- Galdames R, Mera M (2003) First report of Ascochyta Blight of Chickpea caused by Ascochyta rabiei in Chile. Plant Dis 87(5): 603. [DOI] [PubMed] [Google Scholar]
- Gao Y, Faris JD, Liu Z, Kim YM, Syme RA, Oliver RP, Xu SS, Friesen TL (2015) Identification and characterization of the SnTox6-Snn6 interaction in the Parastagonospora nodorum–wheat pathosystem. Mol Plant-Microbe Interact 28(5): 615–625 [DOI] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Boter M, Fernández-Barbero G, Chini A, Rathjen JP, Solano R (2014) The bacterial effector HopX1 targets JAZ transcriptional repressors to activate jasmonate signaling and promote infection in Arabidopsis. PLoS Biol 12(2): 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gookin TE, Assmann SM (2014) Significant reduction of BiFC non-specific assembly facilitates in planta assessment of heterotrimeric G-protein interactors. Plant J 80(3): 553–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamid K, Strange RN (2000) Phytotoxicity of solanapyrones A and B produced by the chickpea pathogen Ascochyta rabiei (Pass.) Labr. and the apparent metabolism of solanapyrone A by chickpea tissues. Physiol Mol Plant Pathol 56(6): 235–244 [Google Scholar]
- Han LB, Li YB, Wang HY, Wu XM, Li CL, Luo M, Wu SJ, Kong ZS, Pei Y, Jiao GL, et al. (2013) The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 25(11): 4421–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Mclellan H, Boevink PC, Birch PRJ (2020) All roads lead to susceptibility: the many modes of action of fungal and oomycete intracellular effectors. Plant Commun 1(4): 100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller NL, Bhattacharjee S, van Ooij C, Liolios K, Harrison T, Lopez-Estraño C, Haldar K (2004) A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306(5703): 1934–1937 [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Brown-Clay J, Thomas C (2017) Subcellular localization and function of 2LIM proteins in plants and humans. Planta 246(6): 1243–1245 [DOI] [PubMed] [Google Scholar]
- Ilarslan H, Dolar FS (2002) Histological and ultrastructural changes in leaves and stems of resistant and susceptible chickpea cultivars to Ascochyta rabiei. J Phytopathol 150(6): 340–348 [Google Scholar]
- Kaiser WJ, Coca WF, Vega OS (2000) First report of Ascochyta blight of chickpea in Latin America. Plant Dis 84(1): 102. [DOI] [PubMed] [Google Scholar]
- Kavousi HR, Marashi H, Mozafari J, Bagheri AR (2009) Expression of phenylpropanoid pathway genes in chickpea defense against race 3 of Ascochyta rabiei. Plant Pathol J 8(3): 127–132 [Google Scholar]
- Kawaoka A, Ebinuma H (2001) Transcriptional control of lignin biosynthesis by tobacco LIM protein. Phytochemistry 57(7): 1149–1157 [DOI] [PubMed] [Google Scholar]
- Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H (2000) Functional analysis of tobacco LIM protein Ntlim1 involved in lignin biosynthesis. Plant J 22(4): 289–301 [DOI] [PubMed] [Google Scholar]
- Kim DS, Hwang BK (2014) An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J Exp Bot 65(9): 2295–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kim CY, Park SY, Kim KT, Jeon J, Chung H, Choi G, Kwon S, Choi J, Jeon J, et al. (2020) Two nuclear effectors of the rice blast fungus modulate host immunity via transcriptional reprogramming. Nat Commun 11(1): 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Park CM, Park JJ, Akamatsu HO, Peever TL, Xian M, Gang DR, Vandemark G, Chen W (2015) Functional analyses of the Diels-Alderase gene sol5 of Ascochyta rabiei and Alternaria solani indicate that the solanapyrone phytotoxins are not required for pathogenicity. Mol Plant-Microbe Interact 28(4): 482–496 [DOI] [PubMed] [Google Scholar]
- Kumar K, Purayannur S, Kaladhar VC, Parida SK, Verma PK (2018) mQTL-seq and classical mapping implicates the role of an AT-HOOK MOTIF CONTAINING NUCLEAR LOCALIZED (AHL) family gene in Ascochyta blight resistance of chickpea. Plant Cell Environ 41(9): 2128–2140 [DOI] [PubMed] [Google Scholar]
- Kumar K, Srivastava V, Purayannur S, Kaladhar VC, Cheruvu PJ, Verma PK (2016) WRKY domain-encoding genes of a crop legume chickpea (Cicer arietinum): comparative analysis with Medicago truncatula WRKY family and characterization of group-III gene (s). DNA Res 23(3): 225–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łaźniewska J, Ae Ł, Macioszek VK, Christopher AE, Ae BL, Kononowicz AK (2009) Fight to the death: Arabidopsis thaliana defense response to fungal necrotrophic pathogens. Acta Physiol Plant 32(1): 1–10 [Google Scholar]
- Lee MH, Jeon HS, Kim SH, Chung JH, Roppolo D, Lee HJ, Cho HJ, Tobimatsu Y, Ralph J, Park OK (2019) Lignin-based barrier restricts pathogens to the infection site and confers resistance in plants. EMBO J 38(23): e101948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Kim BD, Rose JK (2006) Identification of eukaryotic secreted and cell surface proteins using the yeast secretion trap screen. Nat Protoc 1(5): 2439–2447 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1): 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang NN, Wang Y, Liu D, Gao Y, Li L, Li XB (2018) The cotton XLIM protein (GhXLIM6) is required for fiber development via maintaining dynamic F-actin cytoskeleton and modulating cellulose biosynthesis. Plant J 96(6): 1269–1282 [DOI] [PubMed] [Google Scholar]
- Liu ZH, Faris JD, Meinhardt SW, Ali S, Rasmussen JB, Friesen TL (2004) Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by Stagonospora nodorum. Phytopathology 94(10): 1056–1060 [DOI] [PubMed] [Google Scholar]
- Liu Z, Friesen TL, Ling H, Meinhardt SW, Oliver RP, Rasmussen JB, Faris JD (2006) The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome 49(10): 1265–1273 [DOI] [PubMed] [Google Scholar]
- Liu T, Song T, Zhang X, Yuan H, Su L, Li W, Xu J, Liu S, Chen L, Chen T, et al. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun 5(1): 4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Wu J, Lin L, Li P, Li S, Wang Y, Li J, Sun Q, Liang J, Wang Y (2021) Overexpression of cinnamoyl-CoA reductase 2 in Brassica napus increases resistance to Sclerotinia sclerotiorum by affecting lignin biosynthesis. Front Plant Sci 12: 732733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Xu L, Jia Q, Pan R, Oelmüller R, Zhang W, Wu C (2019) Arms race: diverse effector proteins with conserved motifs. Plant Signal Behav 14(2): 1557008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang Z, Faris JD, Oliver RP, Syme R, McDonald MC, McDonald BA, Solomon PS, Lu S, Shelver WL, et al. (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog 8(1): e1002467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu X, Shen C, Fu Y, Xie J, Jiang D, Li G, Cheng J (2016) A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PLoS Pathog 12(2): 1–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majd A (2007) Light and scanning electron microscopy studies on the penetration and infection processes of Alternaria Alternata, causing brown spot on Minneola tangelo in the West Mazandaran - Iran. World Appl Sci J 2(1): 68–72 [Google Scholar]
- Malinovsky FG, Fangel JU, Willats WGT (2014) The role of the cell wall in plant immunity. Front Plant Sci 5: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurya R, Singh Y, Sinha M, Singh K, Mishra P, Singh SK, Verma S, Prabha K, Kumar K, Verma PK (2020) Transcript profiling reveals potential regulators for oxidative stress response of a necrotrophic chickpea pathogen Ascochyta rabiei. 3 Biotech 10(3): 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan H, Boevink PC, Armstrong MR, Pritchard L, Gomez S, Morales J, Whisson SC, Beynon JL, Birch PRJ (2013) An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog 9(10): e1003670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengiste T (2012) Plant immunity to necrotrophs. Annu Rev Phytopathol 50(1): 267–294 [DOI] [PubMed] [Google Scholar]
- Mohd Shah R, Williams AH, Hane JK, Lawrence JA, Farfan-Caceres LM, Debler JW, Oliver RP, Lee RC (2020) Reference genome assembly for Australian Ascochyta rabiei isolate ArME14. G3 (Bethesda) 10(7): 2131–2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizam S, Singh K, Verma PK (2010) Expression of the fluorescent proteins DsRed and EGFP to visualize early events of colonization of the chickpea blight fungus Ascochyta rabiei. Curr Genet 56(4): 391–399 [DOI] [PubMed] [Google Scholar]
- Ohm RA, Feau N, Henrissat B, Schoch CL, Horwitz BA, Barry KW, Condon BJ, Copeland AC, Dhillon B, Glaser F, et al. (2012) Diverse lifestyles and strategies of plant pathogenesis encoded in the genomes of eighteen dothideomycetes fungi. PLoS Pathog 8(12): 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ökmen B, Doehlemann G (2014) Inside plant: biotrophic strategies to modulate host immunity and metabolism. Curr Opin Plant Biol 20: 19–25 [DOI] [PubMed] [Google Scholar]
- Oliver RP, Solomon PS (2010) New developments in pathogenicity and virulence of necrotrophs. Curr Opin Plant Biol 10.1016/j.pbi.2010.05.003 [DOI] [PubMed] [Google Scholar]
- Plett JM, Daguerre Y, Wittulsky S, Vayssières A, Deveau A, Melton SJ, Kohler A, Morrell-Falvey JL, Brun A, Veneault-Fourrey C, et al. (2014) Effector MiSSP7 of the mutualistic fungus Laccaria bicolor stabilizes the Populus JAZ6 protein and represses jasmonic acid (JA) responsive genes. Proc Natl Acad Sci U S A 111(22): 8299–8304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogorelko G, Juvale PS, Rutter WB, Hewezi T, Hussey R, Davis EL, Mitchum MG, Baum TJ (2016) A cyst nematode effector binds to diverse plant proteins, increases nematode susceptibility and affects root morphology. Mol Plant Pathol 17(6): 832–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Guo J, Liu P, He F, Wan C, Islam MA, Tyler BM, Kang Z, Guo J (2019) Stripe rust effector PstGSRE1 disrupts nuclear localization of ROS-promoting transcription factor TaLOL2 to defeat ROS-induced defense in wheat. Mol Plant 12(12): 1624–1638 [DOI] [PubMed] [Google Scholar]
- Qin J, Wang K, Sun L, Xing H, Wang S, Li L, Chen S, Guo HS, Zhang J (2018) The plant-specific transcription factors CBP60G and SARD1 are targeted by a Verticillium secretory protein VDSCP41 to modulate immunity. eLife 7: 1–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarammohan S (2021) Redefining plant-necrotroph interactions: the thin line between hemibiotrophs and necrotrophs. Front Microbiol 12: 944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjan A, Westrick NM, Jain S, Piotrowski JS, Ranjan M, Kessens R, Stiegman L, Grau CR, Conley SP, Smith DL, et al. (2019) Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol J 17(8): 1567–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea G, Metoui O, Infantino A, Federico R, Angelini R (2002) Copper amine oxidase expression in defense responses to wounding and Ascochyta rabiei invasion. Plant Physiol 128(3): 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redkar A, Hoser R, Schilling L, Zechmann B, Krzymowska M, Walbot V, Doehlemann G (2015) A secreted effector protein of Ustilago maydis guides maize leaf cells to form tumors. Plant Cell 27(4): 1332–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala S, Ampe C (2018) An emerging link between LIM domain proteins and nuclear receptors. Cell Mol Life Sci 75(11): 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sang Y, Yu W, Zhuang H, Wei Y, Derevnina L, Yu G, Luo J, Macho AP (2020) Intra-strain elicitation and suppression of plant immunity by Ralstonia solanacearum Type-III effectors in Nicotiana benthamiana. Plant Commun 1(4): 100025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayou C, Nanao MH, Jamin M, Posé D, Thévenon E, Grégoire L, Tichtinsky G, Denay G, Ott F, Peirats LM, et al. (2016) A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat Commun 7(1): 11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifbarghi S, Borhan MH, Wei Y, Coutu C, Robinson SJ, Hegedus DD (2017) Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genomics 18(1): 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybold H, Demetrowitsch TJ, Hassani MA, Szymczak S, Reim E, Haueisen J, Lübbers L, Rühlemann M, Franke A, Schwarz K, et al. (2020) A fungal pathogen induces systemic susceptibility and systemic shifts in wheat metabolome and microbiome composition. Nat Commun 11(1): 1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N, Rahman MH, Strelkov S, Thiagarajah M, Bansal VK, Kav NNV (2007) Proteome-level changes in two Brassica napus lines exhibiting differential responses to the fungal pathogen Alternaria brassicae. Plant Sci 172(1): 95–110 [Google Scholar]
- Shi G, Friesen T L, Saini J, Xu S S, Rasmussen J B, Faris J D (2015) The Wheat Snn7 Gene Confers Susceptibility on Recognition of the Parastagonospora nodorum Necrotrophic Effector SnTox7. The Plant Genome 8(2). eplantgenome2015.02.0007 [DOI] [PubMed] [Google Scholar]
- Shi H, Liu Z, Zhu L, Zhang C, Chen Y, Zhou Y, Li F, Li X (2012) Overexpression of cotton (Gossypium hirsutum) dirigent1 gene enhances lignification that blocks the spread of Verticillium dahliae. Acta Biochim Biophys Sin 44(7): 555–564 [DOI] [PubMed] [Google Scholar]
- Shine MB, Yang JW, El-Habbak M, Nagyabhyru P, Fu DQ, Navarre D, Ghabrial S, Kachroo P, Kachroo A (2016) Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol 212(3): 627–636 [DOI] [PubMed] [Google Scholar]
- Shlezinger N, Minz A, Gur Y, Hatam I, Dagdas YF (2011) Anti-apoptotic machinery protects the necrotrophic fungus Botrytis cinerea from host-induced apoptotic-like cell death during plant infection. PLoS Pathog 7(8): 1002185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh J, Verma P Kumar (2022) Genome-wide identification, expression, and characterization of CaLysM-RLKs in chickpea root nodule symbiosis. Environ Exp Bot 202: 104999. [Google Scholar]
- Singh R, Kumar K, Purayannur S, Chen W, Verma PK (2022) Ascochyta rabiei: a threat to global chickpea production. Mol Plant Pathol 23(9): 1241–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, Nizam S, Sinha M, Verma PK (2012) Comparative transcriptome analysis of the necrotrophic fungus Ascochyta rabiei during oxidative stress: insight for fungal survival in the host plant. PLoS One 7(3): e33128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Shree A, Singh K, Kumar K, Singh SK, Kumar V, Verma PK (2021) Modulation of fungal virulence through CRZ1 regulated F-BAR-dependent actin remodeling and endocytosis in chickpea infecting phytopathogen Ascochyta rabiei. PLoS Genet 17(5): e1009137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelders NC, Rovenich H, Petti GC, Rocafort M, van den Berg GCM, Vorholt JA, Mesters JR, Seidl MF, Nijland R, Thomma BPHJ (2020) Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat Plants 6(11): 1365–1374 [DOI] [PubMed] [Google Scholar]
- Spanu PD, Abbott JC, Amselem J, Burgis TA, Soanes DM, Stüber K, Loren van Themaat EV, Brown JKM, Butcher SA, Gurr SJ, et al. (2010) Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330(6010): 1543–1546 [DOI] [PubMed] [Google Scholar]
- Srivastava V, Verma PK (2015) Genome wide identification of LIM genes in Cicer arietinum and response of Ca-2LIMs in development, hormone and pathogenic stress. PLoS One 10(9): e0138719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava V, Verma PK (2017) The plant LIM proteins: unlocking the hidden attractions. Planta 246(3): 365–375 [DOI] [PubMed] [Google Scholar]
- Sung Y, Outram MA, Breen S, Wang C, Dagvadorj B, Winterberg B, Kobe B, Williams SJ, Solomon PS (2021) PR1-mediated defence via C-terminal peptide release is targeted by a fungal pathogen effector. New Phytol 229(6): 3467–3480 [DOI] [PubMed] [Google Scholar]
- Tanaka S, Brefort T, Neidig N, Djamei A, Kahnt J, Vermerris W, Koenig S, Feussner K, Feussner I, Kahmann R (2014) A secreted Ustilago maydis effector promotes virulence by targeting anthocyanin biosynthesis in maize. Elife 3: e01355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Gollin I, Rössel N, Kahmann R (2020) The functionally conserved effector Sta1 is a fungal cell wall protein required for virulence in Ustilago maydis. New Phytol 227(1): 185–199 [DOI] [PubMed] [Google Scholar]
- Vance CP, Kirk TK, Sherwood RT (1980) Lignification as a mechanism of disease resistance. Annu Rev Phytopathol 18(1): 259–288 [Google Scholar]
- Vargas WA, Sanz-Martín JM, Rech GE, Armijos-Jaramillo VD, Rivera LP, Echeverria MM, Díaz-Mínguez JM, Thon MR, Sukno SA (2016) A fungal effector with host nuclear localization and DNA-binding properties is required for maize anthracnose development. Mol Plant Microbe Interact 29(2): 83–95 [DOI] [PubMed] [Google Scholar]
- Veloso J, Van Kan JAL, Nl JV, Van Kan JAL (2018) Many shades of grey in Botrytis-host hlant interactions. Trends Plant Sci 23(7): 613–622 [DOI] [PubMed] [Google Scholar]
- Verma S, Gazara RK, Nizam S, Parween S, Chattopadhyay D, Verma PK (2016) Draft genome sequencing and secretome analysis of fungal phytopathogen Ascochyta rabiei provides insight into the necrotrophic effector repertoire. Sci Rep 6(1): 24638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermerris W, Sherman DM, McIntyre LM (2010) Phenotypic plasticity in cell walls of maize brown midrib mutants is limited by lignin composition. J Exp Bot 61(9): 2479–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viotti G, Carmona MA, Scandiani M, Formento AN, Luque A (2012) First report of Ascochyta rabiei causing Ascochyta blight of chickpea in Argentina. Plant Dis 96(9): 1375. [DOI] [PubMed] [Google Scholar]
- Wang JP, Naik PP, Chen HC, Shi R, Lin CY, Liu J, Shuford CM, Li Q, Sun YH, Tunlaya-Anukit S, et al. (2014) Complete proteomic-based enzyme reaction and inhibition kinetics reveal how monolignol biosynthetic enzyme families affect metabolic flux and lignin in Populus trichocarpa. Plant Cell 26(3): 894–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawra S, Trusch F, Matena A, Apostolakis K, Linne U, Zhukov I, Stanek J, Koźmiński W, Davidson I, Secombes CJ, et al. (2017) The RxLR motif of the host targeting effector AVR3a of Phytophthora infestans is cleaved before secretion. Plant Cell 29(6): 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiskirchen R, Günther K (2003) The CRP/MLP/TLP family of LIM domain proteins: acting by connecting. BioEssays 25(2): 152–162 [DOI] [PubMed] [Google Scholar]
- Williams B, Kabbage M, Kim HJ, Britt R, Dickman MB (2011) Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog 7(6): e1002107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winterberg B, Du Fall LA, Song X, Pascovici D, Care N, Molloy M, Ohms S, Solomon PS (2014) The necrotrophic effector protein SnTox3 re-programs metabolism and elicits a strong defence response in susceptible wheat leaves. BMC Plant Biol 14(1): 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Liang Y, Qiu D, Zeng H, Yuan J, Yang X (2018) Lignin metabolism involves Botrytis cinerea BcGs1- induced defense response in tomato. BMC Plant Biol 18(1): 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Jiang T, Du K, Chen H, Cao Y, Xie J, Li M, Carr JP, Wu B, Fan Z, et al. (2019a) Maize phenylalanine ammonia-lyases contribute to resistance to Sugarcane mosaic virus infection, most likely through positive regulation of salicylic acid accumulation. Mol Plant Pathol 20(10): 1365–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan X, Wang H, Cai J, Li D, Song F (2019b) NAC Transcription factors in plant immunity. Phytopathol Res 1(1): 1–3 [Google Scholar]
- Zhang X, Liu CJ (2015) Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Mol Plant 8(1): 17–27 [DOI] [PubMed] [Google Scholar]
- Zhang C, Wang X, Zhang F, Dong L, Wu J, Cheng Q, Qi D, Yan X, Jiang L, Fan S, et al. (2017) Phenylalanine ammonia-lyase 2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci Rep 7(1): 7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.