Abstract
On the basis of phylogenetic analysis of nucleotide sequences, multiple genotypes and subtypes of hepatitis C virus (HCV) have been identified. Characterization of these genetic groups is likely to facilitate and contribute to the development of an effective vaccine against infection with HCV. Differences among HCV genotypes in geographic distributions have provided investigators with an epidemiologic marker that can be used to trace the source of HCV infection in a given population. HCV genotype 1 may represent a more aggressive strain and one that is less likely to respond to interferon treatment than HCV genotype 2 or 3. However, these observations require confirmation before HCV genotyping can be used in clinical settings.
Hepatitis C virus (HCV) infection has reached epidemic proportions. Worldwide, more than one million new cases of infection are reported annually, and HCV is believed to be more prevalent than hepatitis B virus infection (HBV) (26). In the United States alone, nearly four million persons are infected and 30,000 acute new infections are estimated to occur annually (89). Currently, HCV is responsible for an estimated 8,000 to 10,000 deaths annually in the United States, and without effective intervention, that number is predicted to triple in the next 10 to 20 years (89). Furthermore, HCV is the leading reason for liver transplantation in the United States and this has major implications in the present era of organ shortage. The ultimate goal is a universally effective vaccine to prevent new cases, especially in underdeveloped countries, where HCV infection is more prevalent and treatment is financially out of reach for most patients. The development of such a vaccine has been hampered, at least partly, by the great heterogeneity of the HCV genome, which is the focus of this review.
HCV was the first virus discovered by molecular cloning without the direct use of biologic or biophysical methods. This was accomplished by extracting, copying into cDNA, and cloning all the nucleic acid from the plasma of a chimpanzee infected with non-A, non-B hepatitis by contaminated factor XIII concentrate (24). The HCV genome is a positive-sense, single-stranded RNA genome approximately 10 kb long. It has marked similarities to those of members of the genera Pestivirus and Flavivirus. Different HCV isolates from around the world show substantial nucleotide sequence variability throughout the viral genome (25). Based on the identification of these genomic differences, HCV has been classified into multiple strains. It is thought that genetic heterogeneity of HCV may account for some of the differences in disease outcome and response to treatment observed in HCV-infected persons.
Before proceeding with the discussion, it is important to consider the shortcomings of studies related to the clinical importance of HCV genotypes. Although several studies have specifically evaluated the role of HCV genotypes and the clinical utility of genotyping, many questions have not been answered. Investigators have used several classification systems, especially before 1995, and have adopted different methods of genotyping. Furthermore, there has been no consistency among studies in the definition of study end points to allow for comparison and “collective experience.” This was most obvious in studies that addressed the role of genotypes in liver disease progression or response to interferon therapy. The severity of liver disease was based on histologic activity in some studies and on the development of cirrhosis or hepatocellular carcinoma in others. Similarly, in most trials before 1995, the response to interferon treatment was defined as normalization of transaminases at the end of therapy (biochemical response), but this was replaced by the virologic response, defined as the disappearance of PCR-detectable HCV RNA in plasma. Also, the clinical significance of HCV genotypes that are not common in the United States, Europe, or Japan has received minimal attention because most scientific investigations are being conducted in these countries. These genotypes, which include HCV types 4 through 9, have been found mostly in less industrialized countries (India and countries in Southeast Asia and the Middle East).
GENOMIC ORGANIZATION OF HCV
The original isolate (HCV-1) was a positive-sense RNA virus with approximately 9,400 ribonucleotides, containing a poly(A) tail at the 3′ end (Fig. 1). The sequence contained a 5′ untranslated region (5′ UTR) of 341 bases, a long open reading frame coding for a polyprotein of 3,011 amino acids, and a 3′ untranslated region (3′ UTR) of about 27 bases. This RNA structure is most similar to that of the family Flaviviridae, which encompasses numerous arthropod-borne viruses. Consistent with the known functions of most flavivirus proteins, the three N-terminal HCV proteins are probably structural (C, E1, and E2/NS2) and the four C-terminal proteins (NS2, NS3, NS4, and NS5) are believed to function in viral replication.
FIG. 1.
Genomic organization of HCV. First generation, second generation, and third generation refer to serologic assays for detection of HCV antibodies.
The open reading frame length of each genotype is characteristically different. Whereas the open reading frame in type 1 isolates is approximately 9,400 ribonucleotides, that of type 2 isolates is typically 9,099 nucleotides and that of type 3 isolates is typically 9,063 nucleotides (12). These differences may potentially account for some of the phenotypic differences among genotypes discussed below.
The 5′ UTR of HCV RNA is the most highly conserved portion of the genome and thus has been used in most laboratories to develop sensitive detection assays for HCV RNA (52). It is also thought to be important in the translation of the HCV open reading frame. The E1 and E2 regions of the HCV genome demonstrate the highest mutation rate at the nucleotide level as well as at the predicted amino acid level (49). The finding of a rapidly evolving region within one of the envelope proteins of HCV suggests that this region is under selective pressure by the host immune system. An extraordinarily high rate of nucleotide change that frequently resulted in codon changes was found in hypervariable region 1 of the E2 protein of the HCV genome and consists of 27 amino acids (93). The variability of this region resembles that of the V3 loop of human immunodeficiency virus (HIV), a domain that elicits anti-HIV type-specific neutralizing antibodies. Studies in chimpanzees and in patients with acute and chronic hepatitis C have demonstrated that these infected hosts mount a humoral immune response to epitopes of hypervariable region 1 of the HCV genome (110, 149). The presence of this rapidly changing region may permit a mechanism by which HCV evades host immune surveillance and establishes and maintains persistent infection.
Multiple protein antigens encoded by the viral RNA seem to produce serologic responses in the host. Serologic responses to four of these protein antigens are used in diagnostic laboratories for the detection of HCV infection (Fig. 1). The first two proteins, termed 5-1-1 and c100-3, are derived from nonstructural regions of the HCV genome, specifically the NS3 and NS4 regions, and together they form the basis of the first-generation antibody assays (enzyme-linked immunosorbent assay 1 [ELISA-1] and strip immunoassay 1 [SIA-1]). In addition to 5-1-1 and c100-3, proteins c33 and c22 are included in the second-generation antibody assays (ELISA-2 and SIA-2). Protein c33c is derived from the NS3 region, which is a nonconserved region of the viral genome. However, protein c22 is derived from the highly conserved nucleocapsid (C) region (Fig. 1). A recombinant NS5 antigen has been added to the above four antigens to improve the sensitivity of serologic assays for the detection of HCV antibodies (third-generation antibody assays).
GENOMIC HETEROGENEITY AND CLASSIFICATION SYSTEMS
After the complete HCV genome was determined by Choo et al. in 1991 (25), several HCV isolates from different parts of the world were obtained and sequenced (22, 31, 38, 60, 68, 69). Comparison of the published sequences of HCV has led to the identification of several distinct types that may differ from each other by as much as 33% over the whole viral genome (96). Sequence variability is distributed equally throughout the viral genome, apart from the highly conserved 5′ UTR and core regions and the hypervariable envelope (E) region (59, 95–97, 109, 126).
As different investigators developed and used their own classification system for HCV strains, a confusing literature developed (Table 1). However, at the 2nd International Conference of HCV and Related Viruses, a consensus nomenclature system was proposed that is to be used in future studies of HCV genotypes and subtypes (P. Simmonds, A. Alberti, H. J. Alter, F. Bonino, D. W. Bradley, C. Brechot, J. T. Brouwer, S. W. Chan, K. Chayama, D. S. Chen, Q.-L. Choo, M. Colombo, H. T. M. Cuypers, T. Date, G. M. Dusheiko, J. I. Esteban, O. Fay, S. J. Hadziyannis, J. Han, A. Hatzakis, E. C. Holmes, H. Hotta, M. Houghton, B. Irvine, M. Kohara, J. A. Kolberg, G. Kuo, J. Y. N. Lau, P. N. Lelie, G. Maertens, F. McOmish, T. Miyamura, M. Mizokami, A. Nomoto, A. M. Prince, H. W. Reesink, C. Rice, M. Roggendorf, S. W. Schalm, T. Shikata, K. Shimotohno, L. Stuyver, C. Trépo, A. Weiner, P. L. Yap, and M. S. Urdea, Letter, Hepatology 19:1321–1324, 1994). According to this system, HCV is classified on the basis of the similarity of nucleotide sequence into major genetic groups designated genotypes. HCV genotypes are numbered (arabic numerals) in the order of their discovery (Tables 1 and 2). The more closely related HCV strains within some types are designated subtypes, which are assigned lowercase letters (in alphabetic order) in the order of their discovery (Tables 1 and 2). The complex of genetic variants found within an individual isolate is termed the quasispecies (Table 2). The quasispecies composition of HCV results from the accumulation of mutations during viral replication in the host (135).
TABLE 1.
Classification systems for HCV genotypes
| Okamoto et al. (96) | Enomoto et al. (38) | Simmonds et al. (115) | Cha et al. (17) | Consensusa (Simmonds et al., Letter) | Example isolates |
|---|---|---|---|---|---|
| I | PT | 1a | I | 1a | HCV-1, HCV-H |
| II | K1 | 1b | II | 1b | HCV-J, HCV-JT, HCV-BK |
| 1c | HC-G9, YS-117 | ||||
| III | K2a | 2a | III | 2a | HC-J6, HC-J5, HCV-K2a |
| IV | K2b | 2b | III | 2b | HC-J8, HC-J7, HCV-K2b |
| III | 2c | S-83, T-983 | |||
| V | 3 | IV | 3a | HCV-K3a, T-1, T-7 | |
| VI | IV | 3b | HCV-TR, T-9, T-10 | ||
| 4 | 4a | Z4, Z8, Z5, Syr1, Syr2, N5, Cam600, Z1, N1, N2, DK13 | |||
| V | 5a | SA-1, SA-7 | |||
| 6a | HK-2 |
It has been recommended that this classification be used in all future publications.
TABLE 2.
Terminology commonly used in studies related to HCV genomic heterogeneity
| Terminology | Definition | % Nucleotide similaritya |
|---|---|---|
| Genotype | Genetic heterogeneity among different HCV isolates | 65.7–68.9 |
| Subtype | Closely related isolates within each of the major genotypes | 76.9–80.1 |
| Quasispecies | Complex of genetic variants within individual isolates | 90.8–99 |
% Nucleotide similarity refers to the nucleotide sequence identities of the full-length sequences of the HCV genome.
The genomic sequences of different HCV isolates vary by as much as 35% (96). The degrees of difference in nucleotide sequences among isolates vary from one genomic region to another. Sequence similarities between members of the different genotypes of a 222-bp segment of the NS5 region that we used in our laboratory range between 55 and 72%, whereas identities of subtypes range from 75 to 86% (Table 3) (Simmonds et al., Letter, Hepatology 19:1321–1324, 1994).
TABLE 3.
Comparative sequence analysis among HCV subtypes of a 222-nucleotide segment derived from the viral NS5 regiona
| Subtype | % Similarity to:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1a | 1b | 1c | 2a | 2b | 2c | 3a | 3b | 4a | 5a | 6a | |
| 1a | 100 | 81 | 85 | 65 | 66 | 63 | 67 | 66 | 68 | 69 | 64 |
| 1b | 100 | 77 | 64 | 67 | 64 | 67 | 71 | 64 | 70 | 65 | |
| 1c | 100 | 68 | 70 | 67 | 65 | 70 | 64 | 61 | 61 | ||
| 2a | 100 | 82 | 77 | 67 | 67 | 66 | 66 | 68 | |||
| 2b | 100 | 81 | 64 | 69 | 65 | 67 | 66 | ||||
| 2c | 100 | 64 | 65 | 65 | 66 | 65 | |||||
| 3a | 100 | 79 | 65 | 67 | 64 | ||||||
| 3b | 100 | 66 | 68 | 61 | |||||||
| 4a | 100 | 66 | 66 | ||||||||
| 5a | 100 | 68 | |||||||||
| 6a | 100 | ||||||||||
Nucleotide positions 7975 to 8196 of the prototype virus.
ROLE OF GENOMIC HETEROGENEITY IN HCV PERSISTENCE AND VACCINE DEVELOPMENT
With the rarity of severe acute or fulminant HCV infections, the significance of this infection in humans is its tendency to become persistent and to induce chronic liver disease. The mechanisms of HCV persistence are not known. However, in most human viral infections, the interaction of several arms of the immune system is important in limiting viral replication and preventing persistence. These arms of the immune system include humoral and cellular immunity.
Antibody responses are often directed against several viral proteins, although it is the antibodies directed against the viral envelope proteins that usually serve as neutralizing antibodies (90). Neutralizing antibodies are often specific for a particular serologic type of the virus, an issue that is particularly relevant in discussions of strategies for vaccination against highly variable viruses such as HIV or HCV.
Whether infection with HCV elicits protective immunity in the host remains unclear. Farci et al. (41) attempted to neutralize HCV in vitro with plasma obtained from a chronically infected patient. The source of HCV was the same patient during the acute phase of posttransfusion non-A, non-B hepatitis. The residual infectivity was evaluated by inoculation of seronegative chimpanzees. The authors showed that neutralization was achieved with plasma obtained 2 years after the initial exposure but not with plasma obtained 11 years later. Analysis of viral isolates for the same patient showed significant genetic divergence of HCV over time. These data support the quasispecies nature of HCV and the selection of strains to avoid immune pressure. This experiment also emphasized the possible role of genetic heterogeneity of HCV in escaping the immune system. It has been suggested that these antibodies are likely to be directed against epitopes of hypervariable region 1 located in the E2 region (41).
Similarly, cellular immune responses, particularly those mediated by cytotoxic T lymphocytes (CTLs), are important components of protective immunity against many viral infections, including hepatitis B. In HCV infection, the role of CTLs in protecting against viral persistence is unknown. HCV-specific, HLA class I-restricted CTLs were demonstrated within the liver (63, 64). Possible targets for HCV-specific CTL recognition within the conserved core protein and additional epitopes in the more highly variable region E2 protein were also identified (63, 64). HCV heterogeneity may also be important in escaping CTL-induced immunity. In a chronically infected chimpanzee, CTLs obtained from the liver were initially able to recognize an epitope in the NS3 protein. Over a period of several years, a new strain of the virus emerged with a mutation in the CTL epitope that was no longer recognized by the CTLs isolated earlier. Although direct evidence for the presence of CTL escape mutants in human HCV infection is lacking, it has been shown that single-amino-acid changes in CTL epitopes result in failure of recognition by HCV-specific CTLs (62). These single-amino-acid changes are found in natural isolates of HCV, hence the need to address the problem of type specificity of immune responses.
Irrespective of the specific type of immune response (humoral or cellular) that is associated with protection and clearance of HCV after an acute exposure, the response appears to be type specific. This conclusion could be extracted from a natural experiment in children with thalassemia who had undergone numerous transfusions (66). Lai et al. (66) reported multiple episodes of acute hepatitis C in these children. The second episode of acute hepatitis C appeared to result from infection with a different strain of HCV, suggesting that immune responses to the initial strain did not protect against an infection with another strain of HCV.
Analogously, the genetic heterogeneity of HCV is likely to make the development of an HCV vaccine difficult. A vaccine that consisted of recombinant E1 and E2 proteins of HCV-1 (genotype 1a) was tested in chimpanzees (23). Of seven chimpanzees that were challenged with HCV-1 after vaccination, five were protected against reinfection, compared with none of four control unvaccinated chimpanzees. The finding that these proteins, derived from hypervariable regions of the HCV genome, can elicit protective immunity poses a major challenge for the development of a broadly effective vaccine for the prevention of HCV.
METHODS FOR HCV GENOTYPING
Molecular Genotyping
Because differences in geographical distribution, disease outcome, and response to therapy among HCV genotypes have been suggested, reliable methods for determining the HCV genotype may become an important clinical test. The reference standard and most definitive method for HCV genotyping is sequencing of a specific PCR-amplified portion of the HCV genome obtained from the patient, followed by phylogenetic analysis. Investigators of HCV genotyping have used sequence analysis of HCV NS5, core, E1, and 5′ UTRs. However, direct sequencing is impractical on a large scale because of the complexity of the procedure. Even with the introduction of automated sequencing methods that do not require radioactive isotopes, only a few laboratories are equipped to perform these procedures on a regular basis. Finally, sequencing of amplified DNA does not usually identify mixed infections with two different HCV genotypes.
Other methods that have been reported depend mainly on the amplification of HCV RNA from clinical specimens, followed by either reamplification with type-specific primers or hybridization with type-specific probes (70, 98, 107, 134) or by digestion of PCR products with restriction endonucleases that recognize genotype-specific cleavage site (81; D. Murphy, B. Willems, and G. Delage, Letter, J. Infect. Dis. 169:473–475, 1994). HCV genotyping by using type-specific primers was first introduced by Okamoto et al. (98) and used primers specific for the core region. This method lacked sensitivity and specificity (135). Without modification, this method was able to detect subtypes 1a, 1b, 1c, 2a, 2b, and 3a. However, modifications have been introduced to improve the sensitivity and specificity of this method (94, 134), but more studies are required before the efficiency of this genotyping method can be compared with that of other methods. Several DNA hybridization assays for HCV genotyping have been described. A commercial kit (InnoLipa) for HCV genotyping has been introduced in Europe by Innogenetics (Zwijndre, Belgium) and is based on hybridization of 5′ UTR amplification products with genotype-specific probes (123). Although the initial version of InnoLipa had lower sensitivity, the newer version is capable of discriminating among HCV subtypes 1a, 1b, 2a to 2c, 3a to 3c, 4a to 4h, 5a, and 6a (75). It has been shown that genotyping methods using 5′ UTR, including InnoLipa, may not distinguish subtype 1a from 1b in 5 to 10% of cases and also may not distinguish between subtypes 2a and 2c (120).
Others have used restriction enzymes to determine a restriction fragment length polymorphism. In this method, a PCR-amplified DNA fragment is digested into fragments with different lengths by enzymes (restriction endonucleases) that recognize cleavage sites specific for each genotype (135). Investigators have used different regions of the HCV genome for restriction fragment length polymorphism, including NS5 and the 5′ UTR (14, 19, 87).
Although all these methods are able to identify correctly the major genotypic groups, only direct nucleotide sequencing is efficient in discriminating among subtypes (12, 113). Moreover, all of these PCR-based methods have the shortcomings and advantages of PCR. They are expensive and time-consuming and require specialized facilities to ensure accurate results and prevent contamination. Their reliability may further be compromised if viral RNA is lost in the serum or plasma through storage or improper laboratory handling or if it is absent from the circulation during sample collection. The advantages of PCR-based methods include reliability if performed accurately and the ability to obtain information relevant to the molecular pathogenesis of HCV.
Serologic Genotyping
More recently, investigators identified genotype-specific antibodies that could be used as indirect markers for the HCV genotype (serotyping or serologic genotyping) (74, 85, 119, 130). Serologic genotyping has several advantages that make it suitable for large epidemiologic studies. These advantages include the low risk for contamination and the simplicity of the assay. However, serologic typing seems to lack specificity and sensitivity, which limits its usefulness.
Two commercially available serologic genotyping assays have been introduced over the past 3 to 4 years. The RIBA SIA was introduced by Chiron Corp. and contained five different serotype-specific peptide sequences taken from the NS4 region and two serotype-specific peptide sequences taken from the core region of the HCV genomes for genotypes 1, 2, and 3 (34). The second serologic genotyping assay is the Murex HCV serotyping enzyme immune assay (Murex Diagnostics Ltd.), which is based on the detection of genotype-specific antibodies directed to epitopes encoded by the NS4 region of the genomes for genotypes 1 through 6. These two assays have been compared and showed a concordance rate of more than 96% for genotypes 1, 2, and 3 (44).
A recent study by Beld et al. (4) showed high reliability of HCV serotyping by the RIBA SIA (Chiron Corp., Emeryville, Calif.) in immunocompetent individuals infected with genotype 1a. However, the assay had low sensitivity in samples containing genotype 3a or in samples from patients coinfected with HIV. These findings suggest that the use of this assay may be limited at this time, particularly in geographic regions where genotype 1a is not prevalent. Similarly, Songsivilai et al. (122) showed that serotyping had poor sensitivity for samples from patients infected with HCV genotype 6. Unlike the two previous studies, a study conducted in the United States reported high concordance between serologic genotyping and molecular genotyping assays (44). These findings suggest variation in the reliability of these assays based on the distribution of HCV genotypes in a specific geographic area.
The choice of typing method for HCV should be based on the expertise in a specific laboratory or institution and the goal of typing. To identify all subtypes and to identify novel sequences if present, PCR amplification followed by sequencing should be the method of choice. However, the goal in treatment trials is frequently to separate patients infected with genotype 1 from those infected with other genotypes—a task that could be done adequately by any of the methods mentioned.
GEOGRAPHIC DISTRIBUTION OF HCV GENOTYPES
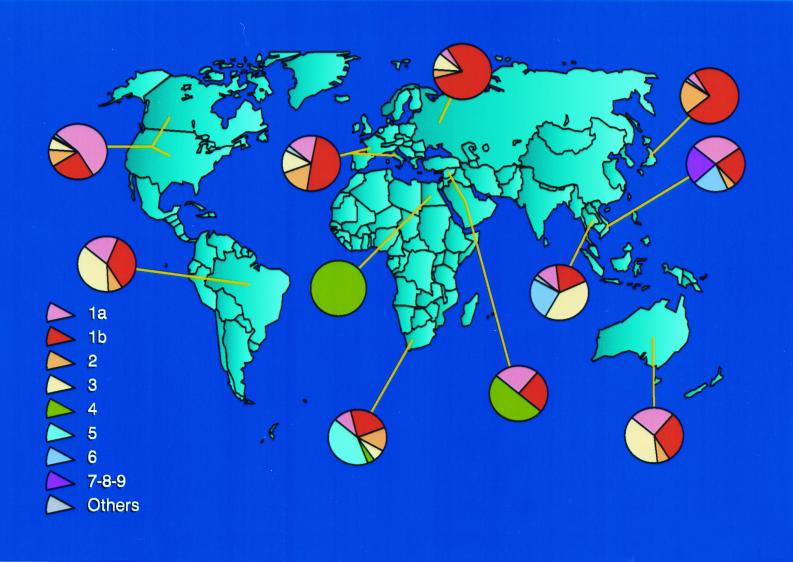
At least six major genotypes of HCV, each comprising multiple subtypes, have been identified worldwide (142). Substantial regional differences appear to exist in the distribution of HCV genotypes (Fig. 2). Although HCV genotypes 1, 2, and 3 appear to have a worldwide distribution, their relative prevalence varies from one geographic area to another (Fig. 2).
FIG. 2.
Worldwide geographic distribution of HCV genotypes and subtypes. “Others” indicate unclassified sequences.
HCV subtypes 1a and 1b are the most common genotypes in the United States (Fig. 3) (145). These subtypes also are predominant in Europe (35, 82, 91). In Japan, subtype 1b is responsible for up to 73% of cases of HCV infection (124). Although HCV subtypes 2a and 2b are relatively common in North America, Europe, and Japan, subtype 2c is found commonly in northern Italy. HCV genotype 3a is particularly prevalent in intravenous drug abusers in Europe and the United States (101). HCV genotype 4 appears to be prevalent in North Africa and the Middle East (1, 18), and genotypes 5 and 6 seem to be confined to South Africa and Hong Kong, respectively (17, 116). HCV genotypes 7, 8, and 9 have been identified only in Vietnamese patients (129), and genotypes 10 and 11 were identified in patients from Indonesia (127). There has been disagreement about the number of genotypes into which HCV isolates should be classified. Investigators have proposed that genotypes 7 through 11 should be regarded as variants of the same group and classified as a single genotype, type 6 (30, 83, 118, 128).
FIG. 3.
HCV genotype distribution in patients with positive results in SIA-2 and in those with indeterminate results in SIA-2.
The geographic distribution and diversity of HCV genotypes may provide clues about the historical origin of HCV (121). The presence of numerous subtypes of each HCV genotype in some regions of the world, such as Africa and Southeast Asia, may suggest that HCV has been endemic for a long time. Conversely, the limited diversity of subtypes observed in the United States and Europe could be related to the recent introduction of these viruses from areas of endemic infection.
CLINICAL RELEVANCE OF HCV GENOTYPES
Although the impact of HCV heterogeneity and genotypes on the day-to-day clinical management of chronic HCV infection has not been established, its role as an epidemiologic marker has been clearly shown. Furthermore, the sensitivity and specificity of serologic and virologic assays for the detection of HCV may be influenced by the heterogeneity of HCV. However, the exact role of genotypes in the progression of liver disease, the outcome of HCV infection, and the response to interferon therapy are much less well understood than their role as an epidemiologic marker. The study of these issues has been hampered by the long natural history of HCV infection and the lack of information about the exact time of exposure to the infection. The following subsections in this section specifically address these issues on the basis of the information available.
Genotypes and HCV Genetic Heterogeneity as Epidemiologic Markers
Because of geographic clustering of distinct HCV genotypes, genotyping may be a useful tool for tracing the source of an HCV outbreak in a given population. Examples include tracing the source of HCV infection in a group of Irish women to contaminated anti-D immunoglobulins (104). All of these women were infected with HCV genotype 1b, a genotype identical to the isolate obtained from the implicated batch of anti-D immunoglobulin. Hohne et al. used genotyping to trace the sources of outbreaks in Germany (54). More recently, genotyping and molecular characterization of HCV isolates provided evidence for a patient-to-patient transmission of HCV during colonoscopy (11). The index case as well as the two other infected patients had HCV genotype 1b. Nucleotide sequencing of the NS3 region showed that the three patients had the same isolate (100% homology), strongly suggesting a common source of infection.
Suspected nonconventional routes of HCV transmission could also be investigated by molecular analysis of HCV strains from different persons. These include the vertical and sexual routes. Weiner et al. (133) showed that a single predominant HCV variant was transmitted to an infant born to a mother infected with multiple variants. Gish et al. reported similar findings (R. G. Gish, K. L. Cox, M. Mizokami, T. Ohno, and J. Y. Lau, Letter, J. Pediatr. Gastroenterol. Nutr. 22:118–119, 1996). A specific 12-nucleotide insertion in the E2 hypervariable region of the HCV genome was noted in the vertically transmitted sequence of an infant born to a mother infected with two different genotypes, each composed of multiple heterogeneous sequences (2). These data may suggest a potential role of HCV heterogeneity and genotypes in mother-to-infant transmission of HCV (138).
Reports on the sexual transmission of HCV infection are conflicting. The detection of anti-HCV positivity ranged from 0% in partners of transfusion-associated hepatitis patients (40) to 8% in male homosexuals (39) and 5% in household contacts (G. Ideo, G. Bellati, E. Pedraglio, R. Bottelli, T. Donzelli, and G. Putignano, Letter, Lancet 335:353, 1990). A possible explanation is that sexual transmission occurs only in association with specific HCV genotypes or in the presence of specific mutations along the HCV genome. As with vertical transmission, samples from patients with suspected sexual transmission of HCV have undergone nucleotide sequence analysis to confirm the similarity of sequences obtained from sexual partners and thus the common origin of these HCV strains (21, 51, 61). Although the data are suggestive of a role of HCV heterogeneity in sexual transmission, this speculation needs to be confirmed.
Although Zein et al. (145) found no association between HCV genotypes and the mode of HCV acquisition in their population, others have provided evidence for such an association (7, 101, 131). It has been suggested that genotypes 3a and 1a are closely associated with intravenous drug use and that genotype 1b is seen more often in patients who acquired HCV through blood transfusion. This information may be useful in tracing sources of HCV epidemics.
HCV Diagnostic Assays
Cloning of the HCV genome led to the identification of the 5-1-1 protein that was reactive with sera obtained from patients with non-A, non-B hepatitis. Using 5-1-1 as a hybridization probe, the recombinant antigen c100 was expressed in yeast and eventually was used to develop the first screening assay (65). The first-generation HCV antibody test approved by the Food and Drug Administration became commercially available in 1990 and was widely used. This was an ELISA that incorporated the c100 epitope from the NS4 region. Because of the high level of false positivity, SIAs were introduced as a supplemental test in patients with positive ELISA results. The ELISAs were affected by the level of circulating globulins in serum, particularly in patients with autoimmune chronic active hepatitis (79).
As more reactive recombinant antigens were identified from conserved regions of the HCV genome, newer serologic assays (second and third generation) were introduced. With improved sensitivity and specificity, the newer assays quickly replaced the first-generation assays. These second-generation assays included the ELISA-2 and SIA-2, which were approved by the Food and Drug Administration in 1992 and 1993, respectively. In addition to the 5-1-1 and c100-3 antigens, these assays incorporated the c22-3 antigen derived from the core region of the HCV genome and the c33c antigen derived from the nonstructural region NS3. Second-generation SIAs are widely used for the validation of second-generation ELISAs in the serologic diagnosis of HCV infection. Current criteria for a positive SIA require reactivity to at least two HCV antigens encoded by different parts of the HCV genome. A common problem is that of indeterminate results, defined as reactivity to a single antigen band or reactivity to two bands derived from the same coding region (i.e., 5-1-1 and c100-3). A significant proportion of samples (5 to 10%) that repeatedly tested reactive in ELISA-2 yielded indeterminate results when evaluated by SIA-2. Some patients with indeterminate SIA results may have detectable HCV RNA on PCR, confirming the presence of HCV viremia (46; E. A. Follett, B. C. Dow, F. McOmish, P. L. Yap, W. Hughes, R. Mitchell, and P. Simmonds, Letter, Lancet 338:1024, 1991; P. Halfon, S. Rousseau, C. Tamalet, M. Antoni, V. Gerolami, M. Levy, M. Bourliere, R. Planells, and G. Cartouzou, Letter, J. Infect. Dis. 166:449, 1992).
Recently, Zein et al. (140) reported on their experience with indeterminate second-generation SIAs. They found that of 720 ELISA-2-positive samples tested in the diagnostic virology laboratory between January 1994 and January 1995, 96 (13%) had indeterminate results. Of these indeterminate samples, 30 (31%) were HCV RNA positive on PCR. Next, they determined the HCV genotype distribution of SIA-2-indeterminate samples and found that it was significantly different from that of SIA-2-positive samples, suggesting that the HCV genotype affects the interpretation of these assays. Less common HCV genotypes in the United States, such as 2a, 2c, 3a, 4a, and 5a, were more prevalent in the samples with SIA-2-indeterminate results (Fig. 3); this would make the serologic detection of these genotypes less efficient. These findings are consistent with an earlier report that suggested the presence of differences in serologic reactivities to HCV antigens among HCV genotypes (146). Reactivities to protein 5-1-1 were significantly lower for patients infected with genotype 2b or 3a than for those infected with genotypes 1a or 1b. Antibody reactivity to the c100-3 protein was also reduced in patients infected with HCV types 2b and 3a. These genotype-dependent differences may have major implications for the current use of HCV diagnostic tests, especially in geographic areas with a high prevalence of HCV genotypes that are phylogenetically distant from genotype 1a, the prototype sequence used in the development of these assays. Studies related to the efficiency of these assays should be done locally in blood banks in different regions of the world before the assay is relied on for blood screening and prevention of new infections.
Third-generation assays (ELISA-3 and SIA-3) were introduced in Europe more than 3 years ago but are still not available commercially in the United States. In these assays, a recombinant NS5 antigen has been added to the four antigens used in the second-generation assays. These third-generation assays have higher sensitivities and specificities than second-generation assays and are much less strongly influenced by the infecting genotype (57, 76, 77).
The detection of HCV RNA by reverse transcriptase PCR has become essential for the diagnosis of HCV infection and for the selection of patients before therapy. The main advantages of reverse transcriptase PCR include early diagnosis after acute infection and detection of viremia in selected patients (those with indeterminate antibody results and immunosuppressed patients). The sensitivity of PCR for HCV RNA detection may vary according to the choice of primers and the handling of preextraction samples (13, 15). Most laboratories use primers specific for the 5′ UTR of the HCV genome because it represents the most highly conserved region among HCV genotypes. Choosing primers from less highly conserved regions of the HCV genome is likely to compromise the sensitivity of PCR in a population in which multiple genotypes are represented.
Assays for the quantification of HCV viremia levels in serum (HCV RNA titer) have been developed and shown to be valuable in the selection of patients for interferon treatment and in the assessment of response during therapy (78, 99). Two different methods have commonly been used for the quantitation of HCV RNA and have become commercially available. The first was developed by Roche Diagnostics Systems (Branchburg, N.J.) and is based on a competitive PCR assay for HCV RNA quantitation (Roche Monitor assay). The second method is based on the coamplification of synthetically mutated target RNA (branched DNA [bDNA] assay; Quantiplex [Chiron Corp.]). The quantitative results of HCV RNA detected by both methods are reliable and reproducible (73).
Earlier studies have suggested a difference in the efficiency of the bDNA assay for quantification of HCV RNA of patients infected with different HCV genotypes (10). However, a second-generation bDNA assay was developed (Quantiplex HCV RNA 2.0) that incorporated a set of oligonucleotide probes to enhance the efficiency of binding to genotypic variants of HCV (32). The new assay was highly sensitive and virtually unaffected by HCV genotypes. A similar variation in the efficiency of detection of different HCV genotypes was recently observed for the Roche Monitor assay (50). These genotype-dependent differences in the efficiency of assays to accurately quantify HCV RNA may have to be considered if clinical decisions are based on the results obtained.
Outcome of Acute HCV Infection
After initial exposure to HCV, the infection fails to resolve in the majority of patients (80%) who become chronically infected. The ability to evolve into chronic disease associated with liver damage is by far the most striking feature of HCV. The spontaneous clearance of HCV following acute infection in a small proportion of patients has been the focus of intense investigations. It has been proposed that differences in the host cellular (84) or humoral (N. N. Zein, H. Li, and D. H. Persing, Letter, Gastroenterology 117:510, 1999) immune responses to HCV are important in spontaneous clearance, but these proposals remain unproved.
Amoroso et al. (3) specifically investigated the role of HCV genotypes in persistence of HCV infection following an acute exposure. The rate of evolution to chronicity after acute exposure to HCV was 92% in patients exposed to HCV genotype 1b infection, compared with 33% to 50% in patients exposed to other genotypes. These data provided evidence that viral factors, including the HCV genotype, may potentially play an important role in the development of chronic infection following acute exposure to HCV.
Progression of Liver Disease
The role of HCV genotypes in the progression of liver disease is one of the most controversial areas of HCV research. There appears to be significant biologic variation in HCV disease expression in the host over the length of the infection (typically the life of the patient). This variation among infected persons became apparent in studies on the natural history of HCV infection, for example, in a retrospective analysis of patients with chronic HCV infection whose time of HCV acquisition was known (N. N. Zein, A. S. Abdulkarim, D. Brandhagen, T. Therneau, and D. H. Persing, Abstract, Hepatology 24:150A, 1996). The mean times from exposure to HCV to the diagnosis of chronic active hepatitis, to compensated liver cirrhosis, to decompensated cirrhosis, and to hepatocellular carcinoma were 11, 18, 23, and 29 years, respectively (Fig. 4). What is striking is that severe complications such as cirrhosis and hepatocellular carcinoma can occur over a short period in some persons whereas others have no complication despite a much longer period of infection (Fig. 4). Therefore, it is likely that viral or host factors, including the infecting HCV genotype, contribute to these variations in the natural history among infected patients.
FIG. 4.
Mean time (years) between exposure to HCV and diagnosis of HCV-related complications in patients with known time of HCV acquisition.
Currently, investigators are divided into those who strongly believe in differences in pathogenicity among genotypes and those who do not. Conclusions have been derived from indirect evidence, because conducting accurate investigations to answer these questions has been difficult. Frequently, the role of genotypes as an independent factor in the progression of liver disease cannot be separated from the roles of other cofactors such as viral load, alcohol intake, and length of time of HCV infection. Patients may not provide accurate information about drug use or the amount of alcohol intake; therefore, the time of HCV acquisition often is not known. Because of the overall slow progression of liver disease in HCV-infected patients, prospective studies frequently are not possible.
For the purpose of this discussion, the data related to HCV genotypes and progression of liver disease in patients with chronic HCV infection were examined separately from those for liver transplant recipients.
In patients with chronic HCV, infection with genotype 1b is reportedly associated with a more severe liver disease and a more aggressive course than is infection with other HCV genotypes (91; G. Pozzato, M. Moretti, F. Franzin, L. S. Croce, C. Tiribelli, T. Masayu, S. Kaneko, M. Unoura, and K. Kobayashi, Letter, Lancet 338:509, 1991). Similar to others (102, 105, 112), Zein et al. (145, 147) found that HCV genotype 1b was significantly more prevalent among patients with liver cirrhosis and those with decompensated liver disease requiring liver transplantation than among those with chronic active hepatitis C. Although this is indirect evidence, it suggests an association between HCV genotype 1b and the development of these complications. Furthermore, a possible link to hepatocellular carcinoma has been proposed for HCV genotype 1b. There is compelling evidence that hepatocellular carcinoma occurs more frequently or emerges earlier among HCV-infected Japanese patients (125, 137) than among HCV carriers in western countries (33, 55). Because HCV genotype 1b is more common in Japan than in Europe or the United States, the hypothesis relating to genotype is attractive and appears to explain these differences. Furthermore, HCV genotype 1b was present in most of the patients with HCV-associated hepatocellular carcinoma studied by Zein et al. (143). Similarly, Reid et al. determined the HCV genotypes in 28 patients with hepatocellular carcinoma and found that 19 (68%) were infected with HCV genotype 1b and the rest were infected with a mixture of HCV genotypes that always included genotype 1b (A. E. Reid, L. J. Jeffers, K. R. Reddy, I. Aiza, E. R. Schiff, J. L. Dienstag, and T. J. Liang, Abstract, Hepatology 20:250A, 1994).
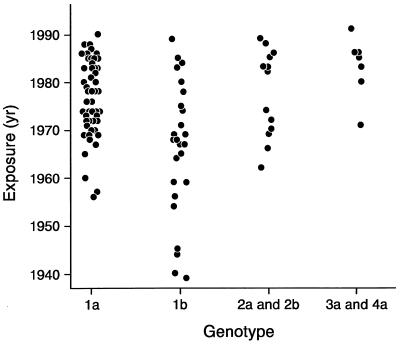
Some reports refute the associations mentioned above (6, 9, 48, 67, 88, 117, 136). A possible and simple explanation may reconcile these reported discrepancies. Zein et al. (145) found that patients infected with HCV genotype 1b were older than those infected with other genotypes and that genotype 1b may have been present before the other genotypes. Thus, patients infected with genotype 1b may have been infected for a longer time (Zein et al., Abstract, Hepatology 24:150A, 1996). As shown in Fig. 5, all of the patients who acquired HCV before 1955 were infected with genotype 1b. HCV genotype 1a was introduced into the United States in the late 1950s and then became the most prevalent genotype in the United States. It was not until the 1960s and 1970s that genotype 2 and genotypes 3 and 4, respectively, were introduced in the United States. After accounting for differences in the duration of HCV infection, HCV genotypes were distributed equally among patients with mild or advanced liver disease. Similar observations have been made in France and Spain (72, 103). According to this explanation, HCV genotype 1b is a marker for more severe HCV-associated liver disease, because it reflects a longer time of infection rather than a more aggressive form of hepatitis C. Future studies are still needed to rule out other host, viral, or environmental factors that may contribute to these differences.
FIG. 5.
Time of HCV acquisition in patients with different HCV genotypes.
Zein et al. (147) and Gordon et al. (45) reported that in liver transplant recipients, HCV genotype 1b is associated with earlier recurrence and more severe hepatitis than are other genotypes. Although others have reported similar findings (5, 100, 111), some authors have suggested that there is no association between genotype and HCV recurrence after transplantation (8, 20, 27, 148). The difference in the duration of infection that may have been a factor in non-transplant-associated HCV patients is not likely to explain the discrepancies in the literature about posttransplantation HCV. More studies are needed.
Response to Interferon Therapy
Since the discovery of HCV, considerable effort has been devoted to defining the factors that may be important in predicting the long-term response to interferon therapy (144). The interferon dose (16), duration of treatment (58), viral RNA level (132), and liver histology (71) all seem to play a role in predicting response. It has been suggested that patients infected with HCV genotypes 1b and, to a lesser degree, 1a are less likely to have a favorable response to interferon treatment than are those infected with genotype 2 or 3. Zein et al. (145) reported a complete biochemical response at the end of 6 months of treatment with interferon in 60 to 70% of patients infected with HCV genotype 2 and in 10 to 15% of those infected with genotype 1. This difference was also present for sustained response and was independent of liver histologic features or the pretreatment HCV RNA levels. This may partly explain the higher rates of long-term response to interferon treatment that have been reported in Europe, where HCV genotype 2 is more prevalent than in the United States or Japan. However, a meta-analysis of the most relevant studies was performed recently (29), and although there was a difference, the predictive value of the non-1 HCV genotype for response to treatment was low (58% for response at the end of treatment and 55% for sustained response). It has been suggested that the introduction of more effective therapies such as the combination of interferon and ribavirin may make the value of predictive factors for response to therapy less important and the differential response of HCV genotypes less obvious (29). However, more recent treatment trials using interferon plus ribavirin in interferon-naive patients with chronic HCV (80) or in patients in whom previous interferon treatment failed (28) showed higher rates of sustained response to therapy in patients with HCV genotypes other than 1. Among patients with HCV genotype 1, 48 weeks of treatment was required to achieve a response similar to that of patients infected with other genotypes treated for 24 weeks (80).
An “interferon-sensitive” region in the nonstructural portion of the HCV genome has been identified in Japanese patients infected with genotype 1b (37). However, studies from the United States and Europe failed to confirm these findings (36, 53, 92). The significance of these findings is not known, and the clinical application of such an expensive and labor-intensive procedure to predict the response to treatment is impractical.
CONCLUSIONS
Multiple genotypes of HCV have been isolated throughout the world. The identification and characterization of HCV types and subtypes have major implications for HCV vaccine development. It is clear that HCV genotypes are important epidemiologic markers and may alter the sensitivity and specificity of diagnostic assays for the detection of HCV. Differences among genotypes in pathogenicity are not clear and have not been proved. Although not efficient by itself, HCV genotyping in combination with other markers, such as quantitative evaluation of HCV RNA, may be beneficial in the management of chronic hepatitis C and in the selection of candidates for interferon treatment. At present, patients should not be excluded from treatment on the basis of the infecting genotype. However, genotype determination could potentially be used to decide the length of treatment. Patients infected with HCV genotype 1 are likely to achieve the best rate of sustained remission following a 48-week course of treatment with interferon and ribavirin. A 24-week course of therapy appears to be sufficient to achieve the maximal rate of responsiveness. More studies are needed before guidelines can be established for the routine use of genotyping outside clinical trials and research laboratories.
REFERENCES
- 1.Abdulkarim A S, Zein N N, Germer J J, Kolbert C P, Kabbani L, Krajnik K L, Hola A, Agha M N, Tourogman M, Persing D H. Hepatitis C virus genotypes and hepatitis G virus in hemodialysis patients from Syria: identification of two novel hepatitis C virus subtypes. Am J Trop Med Hyg. 1998;59:571–576. doi: 10.4269/ajtmh.1998.59.571. [DOI] [PubMed] [Google Scholar]
- 2.Aizaki H, Saito A, Kusakawa I, Ashiwara Y, Nagamori S, Toda G, Suzuki T, Ishii K, Matsuura Y, Miyamura T. Mother-to-child transmission of a hepatitis C virus variant with an insertional mutation in its hypervariable region. J Hepatol. 1996;25:608–613. doi: 10.1016/s0168-8278(96)80227-3. [DOI] [PubMed] [Google Scholar]
- 3.Amoroso P, Rapicetta M, Tosti M E, Mele A, Spada E, Buonocore S, Lettieri G, Pierri P, Chionne P, Ciccaglione A R, Sagliocca L. Correlation between virus genotype and chronicity rate in acute hepatitis C. J Hepatol. 1998;28:939–944. doi: 10.1016/s0168-8278(98)80340-1. [DOI] [PubMed] [Google Scholar]
- 4.Beld M, Penning M, van Putten M, van den Hoek A, Lukashov V, McMorrow M, Goudsmit J. Hepatitis C virus serotype-specific core and NS4 antibodies in injecting drug users participating in the Amsterdam cohort studies. J Clin Microbiol. 1998;36:3002–3006. doi: 10.1128/jcm.36.10.3002-3006.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belli L S, Silini E, Alberti A, Bellati G, Vai C, Minola E, Rondinara G, de Carlis L, Asti M, Forti D, Ideo G. Hepatitis C virus genotypes, hepatitis, and hepatitis C virus recurrence after liver transplantation. Liver Transplant Surg. 1996;2:200–205. doi: 10.1002/lt.500020305. [DOI] [PubMed] [Google Scholar]
- 6.Benvegnu L, Pontisso P, Cavalletto D, Noventa F, Chemello L, Alberti A. Lack of correlation between hepatitis C virus genotypes and clinical course of hepatitis C virus-related cirrhosis. Hepatology. 1997;25:211–215. doi: 10.1053/jhep.1997.v25.pm0008985292. [DOI] [PubMed] [Google Scholar]
- 7.Berg T, Hopf U, Stark K, Baumgarten R, Lobeck H, Schreier E. Distribution of hepatitis C virus genotypes in German patients with chronic hepatitis C: correlation with clinical and virological parameters. J Hepatol. 1997;26:484–491. doi: 10.1016/s0168-8278(97)80411-4. [DOI] [PubMed] [Google Scholar]
- 8.Boker K H, Dalley G, Bahr M J, Maschek H, Tillmann H L, Trautwein C, Oldhaver K, Bode U, Pichlmayr R, Manns M P. Long-term outcome of hepatitis C virus infection after liver transplantation. Hepatology. 1997;25:203–210. doi: 10.1002/hep.510250137. [DOI] [PubMed] [Google Scholar]
- 9.Brechot C. Hepatitis C virus 1b, cirrhosis, and hepatocellular carcinoma. Hepatology. 1997;25:772–774. doi: 10.1002/hep.510250347. [DOI] [PubMed] [Google Scholar]
- 10.Bresters D, Cuypers H T, Reesink H W, Mauser-Bunschoten E P, van den Berg H M, Schaasberg W P, Wilber J C, Urdea M S, Neuwald P, Lelie P N. Comparison of quantitative cDNA-PCR with the branched DNA hybridization assay for monitoring plasma hepatitis C virus RNA levels in haemophilia patients participating in a controlled interferon trial. J Med Virol. 1994;43:262–268. doi: 10.1002/jmv.1890430313. [DOI] [PubMed] [Google Scholar]
- 11.Bronowicki J P, Venard V, Botte C, Monhoven N, Gastin I, Chone L, Hudziak H, Rhin B, Delanoe C, LeFaou A, Bigard M A, Gaucher P. Patient-to-patient transmission of hepatitis C virus during colonoscopy. N Engl J Med. 1997;337:237–240. doi: 10.1056/NEJM199707243370404. [DOI] [PubMed] [Google Scholar]
- 12.Bukh J, Miller R H, Purcell R H. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin Liver Dis. 1995;15:41–63. doi: 10.1055/s-2007-1007262. [DOI] [PubMed] [Google Scholar]
- 13.Bukh J, Purcell R H, Miller R H. Importance of primer selection for the detection of hepatitis C virus RNA with the polymerase chain reaction assay. Proc Natl Acad Sci USA. 1992;89:187–191. doi: 10.1073/pnas.89.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bukh J, Purcell R H, Miller R H. Sequence analysis of the 5′ noncoding region of hepatitis C virus. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Busch M P, Wilber J C, Johnson P, Tobler L, Evans C S. Impact of specimen handling and storage on detection of hepatitis C virus RNA. Transfusion. 1992;32:420–425. doi: 10.1046/j.1537-2995.1992.32592327714.x. [DOI] [PubMed] [Google Scholar]
- 16.Causse X, Godinot H, Chevallier M, Chossegros P, Zoulim F, Ouzan D, Heyraud J-P, Fontanges T, Albrecht J, Meschievitz C, Trepo C. Comparison of 1 or 3 MU of interferon alfa-2b and placebo in patients with chronic non-A, non-B hepatitis. Gastroenterology. 1991;101:497–502. doi: 10.1016/0016-5085(91)90030-o. [DOI] [PubMed] [Google Scholar]
- 17.Cha T A, Beall E, Irvine B, Kolberg J, Chien D, Kuo G, Urdea M S. At least five related, but distinct, hepatitis C viral genotypes exist. Proc Natl Acad Sci USA. 1992;89:7144–7148. doi: 10.1073/pnas.89.15.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chamberlain R W, Adams N, Saeed A A, Simmonds P, Elliott R M. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J Gen Virol. 1997;78:1341–1347. doi: 10.1099/0022-1317-78-6-1341. [DOI] [PubMed] [Google Scholar]
- 19.Chan S W, McOmish F, Holmes E C, Dow B, Peutherer J F, Follett E, Yap P L, Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992;73:1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- 20.Charlton M, Seaberg E, Wiesner R, Everhart J, Zetterman R, Lake J, Detre K, Hoofnagle J. Predictors of patient and graft survival following liver transplantation for hepatitis C. Hepatology. 1998;28:823–830. doi: 10.1002/hep.510280333. [DOI] [PubMed] [Google Scholar]
- 21.Chayama K, Kobayashi M, Tsubota A, Koida I, Arase Y, Saitoh S, Ikeda K, Kumada H. Molecular analysis of intraspousal transmission of hepatitis C virus. J Hepatol. 1995;22:431–439. doi: 10.1016/0168-8278(95)80106-5. [DOI] [PubMed] [Google Scholar]
- 22.Chen P J, Lin M H, Tai K F, Liu P C, Lin C J, Chen D S. The Taiwanese hepatitis C virus genome: sequence determination and mapping the 5′ termini of viral genomic and antigenomic RNA. Virology. 1992;188:102–113. doi: 10.1016/0042-6822(92)90739-c. [DOI] [PubMed] [Google Scholar]
- 23.Choo Q L, Kuo G, Ralston R, Weiner A, Chien D, Van Nest G, Han J, Berger K, Thudium K, Kuo C, Kansopon J, McFarland J, Tabrizi A, Ching K, Moss B, Cummins L B, Houghton M, Muchmore E. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc Natl Acad Sci USA. 1994;91:1294–1298. doi: 10.1073/pnas.91.4.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 25.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby R, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooreman M P, Schoondermark-Van de Ven E M. Hepatitis C virus: biological and clinical consequences of genetic heterogeneity. Scand J Gastroenterol Suppl. 1996;218:106–115. doi: 10.3109/00365529609094740. [DOI] [PubMed] [Google Scholar]
- 27.Crespo J, Carte B, Lozano J L, Casafont F, Rivero M, de la Cruz F, Pons-Romero F. Hepatitis C virus recurrence after liver transplantation: relationship to anti-HCV core IgM, genotype, and level of viremia. Am J Gastroenterol. 1997;92:1458–1462. [PubMed] [Google Scholar]
- 28.Davis G L, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J for the International Hepatitis Interventional Therapy Group. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 29.Davis G L, Lau J Y. Factors predictive of a beneficial response to therapy of hepatitis C. Hepatology. 1997;26(Suppl. 1):122S–127S. doi: 10.1002/hep.510260721. [DOI] [PubMed] [Google Scholar]
- 30.de Lamballerie X, Charrel R N, Attoui H, De Micco P. Classification of hepatitis C virus variants in six major types based on analysis of the envelope 1 and nonstructural 5B genome regions and complete polyprotein sequences. J Gen Virol. 1997;78:45–51. doi: 10.1099/0022-1317-78-1-45. [DOI] [PubMed] [Google Scholar]
- 31.Delisse A M, Descurieux M, Rutgers T, D'Hondt E, De Wilde M, Arima T, Barrera-Sala J M, Ercilla M G, Ruelle J L, Cabezon T. Sequence analysis of the putative structural genes of hepatitis C virus from Japanese and European origin. J Hepatol. 1991;13(Suppl. 4):S20–S23. doi: 10.1016/0168-8278(91)90017-6. [DOI] [PubMed] [Google Scholar]
- 32.Detmer J, Lagier R, Flynn J, Zayati C, Kolberg J, Collins M, Urdea M, Sanchez-Pescador R. Accurate quantification of hepatitis C virus (HCV) RNA from all HCV genotypes by using branched-DNA technology. J Clin Microbiol. 1996;34:901–907. doi: 10.1128/jcm.34.4.901-907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Bisceglie A M, Goodman Z D, Ishak K G, Hoofnagle J H, Melpolder J J, Alter H J. Long-term clinical and histopathological follow-up of chronic posttransfusion hepatitis. Hepatology. 1991;14:969–974. doi: 10.1016/0270-9139(91)90113-a. [DOI] [PubMed] [Google Scholar]
- 34.Dixit V, Quan S, Martin P, Larson D, Brezina M, DiNello R, Sra K, Lau J Y, Chien D, Kolberg J, Tagger A, Davis G, Polito A, Gitnick G. Evaluation of a novel serotyping system for hepatitis C virus: strong correlation with standard genotyping methodologies. J Clin Microbiol. 1995;33:2978–2983. doi: 10.1128/jcm.33.11.2978-2983.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dusheiko G, Schmilovitz-Weiss H, Brown D, McOmish F, Yap P L, Sherlock S, McIntyre N, Simmonds P. Hepatitis C virus genotypes: an investigation of type-specific differences in geographic origin and disease. Hepatology. 1994;19:13–18. [PubMed] [Google Scholar]
- 36.Duverlie G, Khorsi H, Castelain S, Jaillon O, Izopet J, Lunel F, Eb F, Penin F, Wychowski C. Sequence analysis of the NS5A protein of European hepatitis C virus 1b isolates and relation to interferon sensitivity. J Gen Virol. 1998;79:1373–1381. doi: 10.1099/0022-1317-79-6-1373. [DOI] [PubMed] [Google Scholar]
- 37.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Enomoto N, Takada A, Nakao T, Date T. There are two major types of hepatitis C virus in Japan. Biochem Biophys Res Commun. 1990;170:1021–1025. doi: 10.1016/0006-291x(90)90494-8. [DOI] [PubMed] [Google Scholar]
- 39.Esteban J I, Esteban R, Viladomiu L, López-Talavera J C, González A, Hernández J M, Roget M, Vargas V, Genescà J, Buti M, Guardia J, Houghton M, Choo Q-L, Kuo G. Hepatitis C virus antibodies among risk groups in Spain. Lancet. 1989;ii:294–297. doi: 10.1016/s0140-6736(89)90485-6. [DOI] [PubMed] [Google Scholar]
- 40.Everhart J E, Di Bisceglie A M, Murray L M, Alter H J, Melpolder J J, Kuo G, Hoofnagle J H. Risk for non-A, non-B (type C) hepatitis through sexual or household contact with chronic carriers. Ann Intern Med. 1990;112:544–545. doi: 10.7326/0003-4819-112-7-544. [DOI] [PubMed] [Google Scholar]
- 41.Farci P, Alter H J, Wong D C, Miller R H, Govindarajan S, Engle R, Shapiro M, Purcell R H. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reference deleted.
- 43.Reference deleted.
- 44.Gish R G, Qian K P, Quan S, Xu Y L, Pike I, Polito A, DiNello R, Lau J Y. Concordance between hepatitis C virus serotyping assays. J Viral Hepatol. 1997;4:421–422. doi: 10.1046/j.1365-2893.1997.00069.x. [DOI] [PubMed] [Google Scholar]
- 45.Gordon F D, Poterucha J J, Germer J, Zein N N, Batts K P, Gross J B, Jr, Wiesner R, Persing D. Relationship between hepatitis C genotype and severity of recurrent hepatitis C after liver transplantation. Transplantation. 1997;63:1419–1423. doi: 10.1097/00007890-199705270-00009. [DOI] [PubMed] [Google Scholar]
- 46.Gretch D, Lee W, Corey L. Use of aminotransferase, hepatitis C antibody, and hepatitis C polymerase chain reaction RNA assays to establish the diagnosis of hepatitis C virus infection in a diagnostic virology laboratory. J Clin Microbiol. 1992;30:2145–2149. doi: 10.1128/jcm.30.8.2145-2149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reference deleted.
- 48.Han C J, Lee H S, Kim H S, Choe J H, Kim C Y. Hepatitis C virus genotypes in Korea and their relationship to clinical outcome in type C chronic liver diseases. Korean J Intern Med. 1997;12:21–27. doi: 10.3904/kjim.1997.12.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q L, Houghton M. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hawkins A, Davidson F, Simmonds P. Comparison of plasma virus loads among individuals infected with hepatitis C virus (HCV) genotypes 1, 2, and 3 by quantiplex HCV RNA assay versions 1 and 2, Roche Monitor assay, and an in-house limiting dilution method. J Clin Microbiol. 1997;35:187–192. doi: 10.1128/jcm.35.1.187-192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Healey C J, Smith D B, Walker J L, Holmes E C, Fleming K A, Chapman R W, Simmonds P. Acute hepatitis C infection after sexual exposure. Gut. 1995;36:148–150. doi: 10.1136/gut.36.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Ohkoshi S, Shimotohno K. Hypervariable regions in the putative glycoprotein of hepatitis C virus. Biochem Biophys Res Commun. 1991;175:220–228. doi: 10.1016/s0006-291x(05)81223-9. [DOI] [PubMed] [Google Scholar]
- 53.Hofgartner W T, Polyak S J, Sullivan D G, Carithers R L, Jr, Gretch D R. Mutations in the NS5A gene of hepatitis C virus in North American patients infected with HCV genotype 1a or 1b. J Med Virol. 1997;53:118–126. doi: 10.1002/(sici)1096-9071(199710)53:2<118::aid-jmv3>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 54.Hohne M, Schreier E, Roggendorf M. Sequence variability in the env-coding region of hepatitis C virus isolated from patients infected during a single source outbreak. Arch Virol. 1994;137:25–34. doi: 10.1007/BF01311170. [DOI] [PubMed] [Google Scholar]
- 55.Hopf U, Moller B, Kuther D, Stemerowicz R, Lobeck H, Ludtke-Handjery A, Walter E, Blum H E, Roggendorf M, Deinhardt F. Long-term follow-up of posttransfusion and sporadic chronic hepatitis non-A, non-B and frequency of circulating antibodies to hepatitis C virus (HCV) J Hepatol. 1990;10:69–76. doi: 10.1016/0168-8278(90)90075-3. [DOI] [PubMed] [Google Scholar]
- 56.Reference deleted.
- 57.Kao J H, Lai M Y, Hwang Y T, Yang P M, Chen P J, Sheu J C, Wang T H, Hsu H C, Chen D S. Chronic hepatitis C without anti-hepatitis C antibodies by second-generation assay. A clinicopathologic study and demonstration of the usefulness of a third-generation assay. Dig Dis Sci. 1996;41:161–165. doi: 10.1007/BF02208599. [DOI] [PubMed] [Google Scholar]
- 58.Karino Y, Matsushima T, Saga A, Tsuyuguchi M, Miyazaki T, Toyota J. Treatment of chronic non-A, non-B hepatitis with interferon. Gastroenterol Jpn. 1991;26(Suppl. 3):234–238. doi: 10.1007/BF02779308. [DOI] [PubMed] [Google Scholar]
- 59.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kato N, Ootsuyama Y, Ohkoshi S, Nakazawa T, Mori S, Hujikata M, Shimotohno K. Distribution of plural HCV types in Japan. Biochem Biophys Res Commun. 1991;181:279–285. doi: 10.1016/s0006-291x(05)81414-7. [DOI] [PubMed] [Google Scholar]
- 61.Koda T, Yonaha M, Hayashi A, Ishikawa K. Hepatitis C transmission between spouses. J Gastroenterol Hepatol. 1996;11:1001–1005. doi: 10.1111/j.1440-1746.1996.tb00021.x. [DOI] [PubMed] [Google Scholar]
- 62.Koziel M J. Immunology of viral hepatitis. Am J Med. 1996;100:98–109. doi: 10.1016/s0002-9343(96)90018-2. [DOI] [PubMed] [Google Scholar]
- 63.Koziel M J, Dudley D, Afdhal N, Choo Q L, Houghton M, Ralston R, Walker B D. Hepatitis C virus (HCV)-specific cytotoxic T lymphocytes recognize epitopes in the core and envelope proteins of HCV. J Virol. 1993;67:7522–7532. doi: 10.1128/jvi.67.12.7522-7532.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koziel M J, Dudley D, Wong J T, Dienstag J, Houghton M, Ralston R, Walker B D. Intrahepatic cytotoxic T lymphocytes specific for hepatitis C virus in persons with chronic hepatitis. J Immunol. 1992;149:3339–3344. [PubMed] [Google Scholar]
- 65.Kuo G, Choo Q-L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, Tegtmeier G E, Bonino F, Colombo M, Lee W-S, Kuo C, Berger K, Shuster J R, Overby L R, Bradley D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 66.Lai M E, Mazzoleni A P, Argiolu F, De Virgilis S, Balestrieri A, Purcell R H, Cao A, Farci P. Hepatitis C virus in multiple episodes of acute hepatitis in polytransfused thalassaemic children. Lancet. 1994;343:388–390. doi: 10.1016/s0140-6736(94)91224-6. [DOI] [PubMed] [Google Scholar]
- 67.Lau J Y, Davis G L, Prescott L E, Maertens G, Lindsay K L, Qian K, Mizokami M, Simmonds P the Hepatitis Interventional Therapy Group. Distribution of hepatitis C virus genotypes determined by line probe assay in patients with chronic hepatitis C seen at tertiary referral centers in the United States. Ann Intern Med. 1996;124:868–876. doi: 10.7326/0003-4819-124-10-199605150-00002. [DOI] [PubMed] [Google Scholar]
- 68.Li J S, Tong S P, Vitvitski L, Lepot D, Trepo C. Evidence of two major genotypes of hepatitis C virus in France and close relatedness of the predominant one with the prototype virus. J Hepatol. 1991;13(Suppl. 4):S33–S37. doi: 10.1016/0168-8278(91)90019-8. [DOI] [PubMed] [Google Scholar]
- 69.Li J S, Tong S P, Vitvitski L, Lepot D, Trepo C. Two French genotypes of hepatitis C virus: homology of the predominant genotype with the prototype American strain. Gene. 1991;105:167–172. doi: 10.1016/0378-1119(91)90147-4. [DOI] [PubMed] [Google Scholar]
- 70.Li J S, Vitvitski L, Tong S P, Trepo C. Identification of the third major genotype of hepatitis C virus in France. Biochem Biophys Res Commun. 1994;199:1474–1481. doi: 10.1006/bbrc.1994.1397. [DOI] [PubMed] [Google Scholar]
- 71.Lin R, Schoeman M N, Craig P I, Bilous M, Grierson J, McDonald J A, Batey R G, Farrell G C. Can the response to interferon treatment be predicted in patients with chronic active hepatitis C? Aust N Z J Med. 1991;21:387–392. doi: 10.1111/j.1445-5994.1991.tb04715.x. [DOI] [PubMed] [Google Scholar]
- 72.Lopez-Labrador F X, Ampurdanes S, Forns X, Castells A, Saiz J C, Costa J, Bruix J, Sanchez Tapias J M, Jimenez de Anta M T, Rodes J. Hepatitis C virus (HCV) genotypes in Spanish patients with HCV infection: relationship between HCV genotype 1b, cirrhosis and hepatocellular carcinoma. J Hepatol. 1997;27:959–965. doi: 10.1016/s0168-8278(97)80137-7. [DOI] [PubMed] [Google Scholar]
- 73.Lu R H, Hwang S J, Chan C Y, Chang F Y, Lee S D. Quantitative measurement of serum HCV RNA in patients with chronic hepatitis C: comparison between Amplicor HCV monitor system and branched DNA signal amplification assay. J Clin Lab Anal. 1998;12:121–125. doi: 10.1002/(SICI)1098-2825(1998)12:2<121::AID-JCLA8>3.0.CO;2-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Machida A, Ohnuma H, Tsuda F, Munekata E, Tanaka T, Akahane Y, Okamoto H, Mishiro S. Two distinct subtypes of hepatitis C virus defined by antibodies directed to the putative core protein. Hepatology. 1992;16:886–891. doi: 10.1002/hep.1840160406. [DOI] [PubMed] [Google Scholar]
- 75.Maertens G, Stuyver L. Genotypes and genetic variation of hepatitis C virus. In: Harrison T J, Zuckerman A J, editors. The molecular medicine of viral hepatitis. Chichester, England: John Wiley & Sons, Ltd.; 1997. pp. 182–233. [Google Scholar]
- 76.Maggi F, Vatteroni M L, Pistello M, Avio C M, Cecconi N, Panicucci F, Bendinelli M. Serological reactivity and viral genotypes in hepatitis C virus infection. J Clin Microbiol. 1995;33:209–211. doi: 10.1128/jcm.33.1.209-211.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Makris K, Kouvelis V, Drakopoulos I, Oikonomou E, Maniatis A. Frequency and characteristics of post-transfusion hepatitis in Greece with emphasis on hepatitis C: comparing second- and third-generation assays. Transfus Med. 1995;5:213–224. doi: 10.1111/j.1365-3148.1995.tb00231.x. [DOI] [PubMed] [Google Scholar]
- 78.Martinot-Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Le Breton V, Milotova V, Benhamou J P, Erlinger S. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22:1050–1056. [PubMed] [Google Scholar]
- 79.McFarlane I G, Smith H M, Johnson P J, Bray G P, Vergani D, Williams R. Hepatitis C virus antibodies in chronic active hepatitis: pathogenetic factor or false-positive result? Lancet. 1990;335:754–757. doi: 10.1016/0140-6736(90)90870-b. [DOI] [PubMed] [Google Scholar]
- 80.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K for the Hepatitis Interventional Therapy Group. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 81.McOmish F, Chan S W, Dow B C, Gillon J, Frame W D, Crawford R J, Yap P L, Follett E A, Simmonds P. Detection of three types of hepatitis C virus in blood donors: investigation of type-specific differences in serologic reactivity and rate of alanine aminotransferase abnormalities. Transfusion. 1993;33:7–13. doi: 10.1046/j.1537-2995.1993.33193142314.x. [DOI] [PubMed] [Google Scholar]
- 82.McOmish F, Yap P L, Dow B C, Follett E A, Seed C, Keller A J, Cobain T J, Krusius T, Kolho E, Naukkarinen R, Lin C, Lai C, Leong S, Medgyesi G A, Héjjas M, Kiyokawa H, Fukada K, Cuypers T, Saeed A A, Al-Rasheed A M, Lin M, Simmonds P. Geographical distribution of hepatitis C virus genotypes in blood donors: an international collaborative survey. J Clin Microbiol. 1994;32:884–892. doi: 10.1128/jcm.32.4.884-892.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mellor J, Walsh E A, Prescott L E, Jarvis L M, Davidson F, Yap P L, Simmonds P. Survey of type 6 group variants of hepatitis C virus in Southeast Asia by using a core-based genotyping assay. J Clin Microbiol. 1996;34:417–423. doi: 10.1128/jcm.34.2.417-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Missale G, Bertoni R, Lamonaca V, Valli A, Massari M, Mori C, Rumi M G, Houghton M, Fiaccadori F, Ferrari C. Different clinical behaviors of acute hepatitis C virus infection are associated with different vigor of the anti-viral cell-mediated immune response. J Clin Investig. 1996;98:706–714. doi: 10.1172/JCI118842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mondelli M U, Cerino A, Bono F, Cividini A, Maccabruni A, Aricò M, Malfitano A, Barbarini G, Piazza V, Minoli L, Silini E. Hepatitis C virus (HCV) core serotypes in chronic HCV infection. J Clin Microbiol. 1994;32:2523–2527. doi: 10.1128/jcm.32.10.2523-2527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reference deleted.
- 87.Nakao T, Enomoto N, Takada N, Takada A, Date T. Typing of hepatitis C virus genomes by restriction fragment length polymorphism. J Gen Virol. 1991;72:2105–2112. doi: 10.1099/0022-1317-72-9-2105. [DOI] [PubMed] [Google Scholar]
- 88.Naoumov N V, Chokshi S, Metivier E, Maertens G, Johnson P J, Williams R. Hepatitis C virus infection in the development of hepatocellular carcinoma in cirrhosis. J Hepatol. 1997;27:331–336. doi: 10.1016/s0168-8278(97)80179-1. [DOI] [PubMed] [Google Scholar]
- 89.National Institutes of Health Consensus Development Conference Panel. National Institutes of Health Consensus Development Conference Panel statement: management of hepatitis C. Hepatology. 1997;26(Suppl. 1):2S–10S. doi: 10.1002/hep.510260701. [DOI] [PubMed] [Google Scholar]
- 90.Nelson D R, Lau J Y N. Pathogenesis of chronic hepatitis C infection. In: Schinazi R F, Sommadossi J P, Thomas H C, editors. Therapies for viral hepatitis. London, United Kingdom: International Medical Press; 1998. pp. 65–76. [Google Scholar]
- 91.Nousbaum J B, Pol S, Nalpas B, Landais P, Berthelot P, Bréchot C the Collaborative Study Group. Hepatitis C virus type 1b (II) infection in France and Italy. Ann Intern Med. 1995;122:161–168. doi: 10.7326/0003-4819-122-3-199502010-00001. [DOI] [PubMed] [Google Scholar]
- 92.Odeberg J, Yun Z, Sonnerborg A, Weiland O, Lundeberg J. Variation in the hepatitis C virus NS5a region in relation to hypervariable region 1 heterogeneity during interferon treatment. J Med Virol. 1998;56:33–38. doi: 10.1002/(sici)1096-9071(199809)56:1<33::aid-jmv6>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 93.Ogata N, Alter H J, Miller R H, Purcell R H. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:3392–3396. doi: 10.1073/pnas.88.8.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Okamoto H, Kobata S, Tokita H, Inoue T, Woodfield G D, Holland P V, Al-Knawy B A, Uzunalimoglu O, Miyakawa Y, Mayumi M. A second-generation method of genotyping hepatitis C virus by the polymerase chain reaction with sense and antisense primers deduced from the core gene. J Virol Methods. 1996;57:31–45. doi: 10.1016/0166-0934(95)01960-x. [DOI] [PubMed] [Google Scholar]
- 95.Okamoto H, Kojima M, Sakamoto M, Iizuka H, Hadiwandowo S, Suwignyo S, Miyakawa Y, Mayumi M. The entire nucleotide sequence and classification of a hepatitis C virus isolate of a novel genotype from an Indonesian patient with chronic liver disease. J Gen Virol. 1994;75:629–635. doi: 10.1099/0022-1317-75-3-629. [DOI] [PubMed] [Google Scholar]
- 96.Okamoto H, Kurai K, Okada S, Yamamoto K, Lizuka H, Tanaka T, Fukuda S, Tsuda F, Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992;188:331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- 97.Okamoto H, Okada S, Sugiyama Y, Kurai K, Iizuka H, Machida A, Miyakawa Y, Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991;72:2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- 98.Okamoto H, Sugiyama Y, Okada S, Kurai K, Akahane Y, Sugai Y, Tanaka T, Sato K, Tsuda F, Miyakawa Y, Mayumi M. Typing hepatitis C virus by polymerase chain reaction with type-specific primers: application to clinical surveys and tracing infectious sources. J Gen Virol. 1992;73:673–679. doi: 10.1099/0022-1317-73-3-673. [DOI] [PubMed] [Google Scholar]
- 99.Orito E, Mizokami M, Suzuki K, Ohba K, Ohno T, Mori M, Hayashi K, Kato K, Iino S, Lau J Y. Loss of serum HCV RNA at week 4 of interferon-alpha therapy is associated with more favorable long-term response in patients with chronic hepatitis C. J Med Virol. 1995;46:109–115. doi: 10.1002/jmv.1890460205. [DOI] [PubMed] [Google Scholar]
- 100.Pageaux G P, Ducos J, Mondain A M, Costes V, Picot M C, Perrigault P F, Domergue J, Larrey D, Michel H. Hepatitis C virus genotypes and quantitation of serum hepatitis C virus RNA in liver transplant recipients: relationship with severity of histological recurrence and implications in the pathogenesis of HCV infection. Liver Transplant Surg. 1997;3:501–505. doi: 10.1002/lt.500030504. [DOI] [PubMed] [Google Scholar]
- 101.Pawlotsky J M, Taskiris L, Roudot-Thoraval F, Pellet C, Stuyver L, Duval J, Dhumeaux D. Relationship between hepatitis C virus genotypes and sources of infection in patients with chronic hepatitis C. J Infect Dis. 1995;171:1607–1610. doi: 10.1093/infdis/171.6.1607. [DOI] [PubMed] [Google Scholar]
- 102.Pistello M, Maggi F, Vatteroni L, Cecconi N, Panicucci F, Bresci G P, Gambardella L, Taddei M, Bionda A, Tuoni M, Bendinelli M. Prevalence of hepatitis C virus genotypes in Italy. J Clin Microbiol. 1994;32:232–234. doi: 10.1128/jcm.32.1.232-234.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pol S, Thiers V, Nousbaum J B, Legendre C, Berthelot P, Kreis H, Brechot C. The changing relative prevalence of hepatitis C virus genotypes: evidence in hemodialyzed patients and kidney recipients. Gastroenterology. 1995;108:581–583. doi: 10.1016/0016-5085(95)90088-8. [DOI] [PubMed] [Google Scholar]
- 104.Power J P, Lawlor E, Davidson F, Holmes E C, Yap P L, Simmonds P. Molecular epidemiology of an outbreak of infection with hepatitis C virus in recipients of anti-D immunoglobulin. Lancet. 1995;345:1211–1213. doi: 10.1016/s0140-6736(95)91993-7. [DOI] [PubMed] [Google Scholar]
- 105.Pozzato G, Moretti M, Croce L S, Sasso F, Kaneko S, Unoura M, Kobayashi K, Crovatto M, Santini G, Tiribelli C. Interferon therapy in chronic hepatitis C virus: evidence of different outcome with respect to different viral strains. J Med Virol. 1995;45:445–450. doi: 10.1002/jmv.1890450416. [DOI] [PubMed] [Google Scholar]
- 106.Reference deleted.
- 107.Qu D, Li J S, Vitvitski L, Mechai S, Berby F, Tong S P, Bailly F, Wang Q S, Martin J L, Trepo C. Hepatitis C virus genotypes in France: comparison of clinical features of patients infected with HCV type I and type II. J Hepatol. 1994;21:70–75. doi: 10.1016/s0168-8278(94)80139-8. [DOI] [PubMed] [Google Scholar]
- 108.Reference deleted.
- 109.Sakamoto M, Akahane Y, Tsuda F, Tanaka T, Woodfield D G, Okamoto H. Entire nucleotide sequence and characterization of a hepatitis C virus of genotype V/3a. J Gen Virol. 1994;75:1761–1768. doi: 10.1099/0022-1317-75-7-1761. [DOI] [PubMed] [Google Scholar]
- 110.Shimizu Y K, Hijikata M, Iwamoto A, Alter H J, Purcell R H, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shuhart M C, Bronner M P, Gretch D R, Thomassen L V, Wartelle C F, Tateyama H, Emerson S S, Perkins J D, Carithers R L., Jr Histological and clinical outcome after liver transplantation for hepatitis C. Hepatology. 1997;26:1646–1652. doi: 10.1002/hep.510260638. [DOI] [PubMed] [Google Scholar]
- 112.Silini E, Bono F, Cividini A, Cerino A, Bruno S, Rossi S, Belloni G, Brugnetti B, Civardi E, Salvaneschi L, Mondelli M U. Differential distribution of hepatitis C virus genotypes in patients with and without liver function abnormalities. Hepatology. 1995;21:285–290. [PubMed] [Google Scholar]
- 113.Simmonds P. Variability of hepatitis C virus. Hepatology. 1995;21:570–583. doi: 10.1002/hep.1840210243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Reference deleted.
- 115.Simmonds P, Holmes E C, Cha T A, Chan S W, McOmish F, Irvine B, Beall E, Yap P L, Kolberg J, Urdea M S. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J Gen Virol. 1993;74:2391–2399. doi: 10.1099/0022-1317-74-11-2391. [DOI] [PubMed] [Google Scholar]
- 116.Simmonds P, McOmish F, Yap P L, Chan S W, Lin C K, Dusheiko G, Saeed A A, Holmes E C. Sequence variability in the 5′ non-coding region of hepatitis C virus: identification of a new virus type and restrictions on sequence diversity. J Gen Virol. 1993;74:661–668. doi: 10.1099/0022-1317-74-4-661. [DOI] [PubMed] [Google Scholar]
- 117.Simmonds P, Mellor J, Craxi A, Sanchez-Tapias J M, Alberti A, Prieto J, Colombo M, Rumi M G, Lo Iacano O, Ampurdances-Mingall S, Forns-Bernhardt X, Chemello L, Civeira M P, Frost C, Dusheiko G. Epidemiological, clinical and therapeutic associations of hepatitis C types in western European patients. J Hepatol. 1996;24:517–524. doi: 10.1016/s0168-8278(96)80135-8. [DOI] [PubMed] [Google Scholar]
- 118.Simmonds P, Mellor J, Sakuldamrongpanich T, Nuchaprayoon C, Tanprasert S, Holmes E C, Smith D B. Evolutionary analysis of variants of hepatitis C virus found in South-East Asia: comparison with classifications based upon sequence similarity. J Gen Virol. 1996;77:3013–3024. doi: 10.1099/0022-1317-77-12-3013. [DOI] [PubMed] [Google Scholar]
- 119.Simmonds P, Rose K A, Graham S, Chan S W, McOmish F, Dow B C, Follett E A, Yap P L, Marsden H. Mapping of serotype-specific, immunodominant epitopes in the NS-4 region of hepatitis C virus (HCV): use of type-specific peptides to serologically differentiate infections with HCV types 1, 2, and 3. J Clin Microbiol. 1993;31:1493–1503. doi: 10.1128/jcm.31.6.1493-1503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Smith D B, Mellor J, Jarvis L M, Davidson F, Kolberg J, Urdea M, Yap P L, Simmonds P The International HCV Collaborative Study Group. Variation of the hepatitis C virus 5′ non-coding region: implications for secondary structure, virus detection and typing. J Gen Virol. 1995;76:1749–1761. doi: 10.1099/0022-1317-76-7-1749. [DOI] [PubMed] [Google Scholar]
- 121.Smith D B, Simmonds P. Molecular epidemiology of hepatitis C virus. J Gastroenterol Hepatol. 1997;12:522–527. doi: 10.1111/j.1440-1746.1997.tb00477.x. [DOI] [PubMed] [Google Scholar]
- 122.Songsivilai S, Kanistanon D, Dharakul T. A serotyping assay for hepatitis C virus in Southeast Asia. Clin Diagn Lab Immunol. 1998;5:737–739. doi: 10.1128/cdli.5.5.737-739.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stuyver L, Rossau R, Wyseur A, Duhamel M, Vanderborght B, Van Heuverswyn H, Maertens G. Typing of hepatitis C virus isolates and characterization of new subtypes using a line probe assay. J Gen Virol. 1993;74:1093–1102. doi: 10.1099/0022-1317-74-6-1093. [DOI] [PubMed] [Google Scholar]
- 124.Takada N, Takase S, Takada A, Date T. Differences in the hepatitis C virus genotypes in different countries. J Hepatol. 1993;17:277–283. doi: 10.1016/s0168-8278(05)80205-3. [DOI] [PubMed] [Google Scholar]
- 125.Takahashi M, Yamada G, Miyamoto R, Doi T, Endo H, Tsuji T. Natural course of chronic hepatitis C. Am J Gastroenterol. 1993;88:240–243. [PubMed] [Google Scholar]
- 126.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tokita H, Okamoto H, Iizuka H, Kishimoto J, Tsuda F, Lesmana L A, Miyakawa Y, Mayumi M. Hepatitis C virus variants from Jakarta, Indonesia classifiable into novel genotypes in the second (2e and 2f), tenth (10a) and eleventh (11a) genetic groups. J Gen Virol. 1996;77:293–301. doi: 10.1099/0022-1317-77-2-293. [DOI] [PubMed] [Google Scholar]
- 128.Tokita H, Okamoto H, Iizuka H, Kishimoto J, Tsuda F, Miyakawa Y, Mayumi M. The entire nucleotide sequences of three hepatitis C virus isolates in genetic groups 7-9 and comparison with those in the other eight genetic groups. J Gen Virol. 1998;79:1847–1857. doi: 10.1099/0022-1317-79-8-1847. [DOI] [PubMed] [Google Scholar]
- 129.Tokita H, Okamoto H, Tsuda F, Song P, Nakata S, Chosa T, Iizuka H, Mishiro S, Miyakawa Y, Mayumi M. Hepatitis C virus variants from Vietnam are classifiable into the seventh, eighth, and ninth major genetic groups. Proc Natl Acad Sci USA. 1994;91:11022–11026. doi: 10.1073/pnas.91.23.11022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tsukiyama-Kohara K, Yamaguchi K, Maki N, Ohta Y, Miki K, Mizokami M, Ohba K-I, Tanaka S, Hattori N, Nomoto A, Kohara M. Antigenicities of Group I and II hepatitis C virus polypeptides—molecular basis of diagnosis. Virology. 1993;192:430–437. doi: 10.1006/viro.1993.1058. [DOI] [PubMed] [Google Scholar]
- 131.Watson J P, Brind A M, Chapman C E, Bates C L, Gould F K, Johnson S J, Burt A D, Ferguson J, Simmonds P, Bassendine M F. Hepatitis C virus: epidemiology and genotypes in the north east of England. Gut. 1996;38:269–276. doi: 10.1136/gut.38.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Weiland O, Zhang Y Y, Widell A. Serum HCV RNA levels in patients with chronic hepatitis C given a second course of interferon alpha-2b treatment after relapse following initial treatment. Scand J Infect Dis. 1993;25:25–30. [PubMed] [Google Scholar]
- 133.Weiner A J, Thaler M M, Crawford K, Ching K, Kansopon J, Chien D Y, Hall J E, Hu F, Houghton M. A unique, predominant hepatitis C virus variant found in an infant born to a mother with multiple variants. J Virol. 1993;67:4365–4368. doi: 10.1128/jvi.67.7.4365-4368.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Widell A, Shev S, Mansson S, Zhang Y Y, Foberg U, Norkrans G, Fryden A, Weiland O, Kurkus J, Nordenfelt E. Genotyping of hepatitis C virus isolates by a modified polymerase chain reaction assay using type specific primers: epidemiological applications. J Med Virol. 1994;44:272–279. doi: 10.1002/jmv.1890440311. [DOI] [PubMed] [Google Scholar]
- 135.Xavier F, Bukh J. Methods for determining the hepatitis C genotype. Viral Hepatitis Rev. 1998;4:1–19. [Google Scholar]
- 136.Yamada M, Kakumu S, Yoshioka K, Higashi Y, Tanaka K, Ishikawa T, Takayanagi M. Hepatitis C virus genotypes are not responsible for development of serious liver disease. Dig Dis Sci. 1994;39:234–239. doi: 10.1007/BF02090191. [DOI] [PubMed] [Google Scholar]
- 137.Yano M, Yatsuhashi H, Inoue O, Inokuchi K, Koga M. Epidemiology and long term prognosis of hepatitis C virus infection in Japan. Gut. 1993;34(Suppl. 2):S13–S16. doi: 10.1136/gut.34.2_suppl.s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zein N N. Vertical transmission of hepatitis C: to screen or not to screen. J Pediatr. 1997;130:859–861. . (Editorial.) [PubMed] [Google Scholar]
- 139.Reference deleted.
- 140.Zein N N, Germer J J, Wendt N K, Schimek C M, Thorvilson J N, Mitchell P S, Persing D H. Indeterminate results of the second-generation hepatitis C virus (HCV) recombinant immunoblot assay: significance of high-level c22-3 reactivity and influence of HCV genotypes. J Clin Microbiol. 1997;35:311–312. doi: 10.1128/jcm.35.1.311-312.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reference deleted.
- 142.Zein N N, Persing D H. Hepatitis C genotypes: current trends and future implications. Mayo Clin Proc. 1996;71:458–462. doi: 10.4065/71.5.458. [DOI] [PubMed] [Google Scholar]
- 143.Zein N N, Poterucha J J, Gross J B, Jr, Wiesner R H, Therneau T M, Gossard A A, Wendt N K, Mitchell P S, Germer J J, Persing D H. Increased risk of hepatocellular carcinoma in patients infected with hepatitis C genotype 1b. Am J Gastroenterol. 1996;91:2560–2562. [PubMed] [Google Scholar]
- 144.Zein N N, Rakela J. Interferon therapy in hepatitis C. Semin Gastrointest Dis. 1995;6:46–53. [PubMed] [Google Scholar]
- 145.Zein N N, Rakela J, Krawitt E L, Reddy K R, Tominaga T, Persing D H the Collaborative Study Group. Hepatitis C virus genotypes in the United States: epidemiology, pathogenicity, and response to interferon therapy. Ann Intern Med. 1996;125:634–639. doi: 10.7326/0003-4819-125-8-199610150-00002. [DOI] [PubMed] [Google Scholar]
- 146.Zein N N, Rakela J, Persing D H. Genotype-dependent serologic reactivities in patients infected with hepatitis C virus in the United States. Mayo Clin Proc. 1995;70:449–452. doi: 10.4065/70.5.449. [DOI] [PubMed] [Google Scholar]
- 147.Zein N N, Rakela J, Poterucha J J, Steers J L, Wiesner R H, Persing D H. Hepatitis C genotypes in liver transplant recipients: distribution and 1-year follow-up. Liver Transplant Surg. 1995;1:354–357. doi: 10.1002/lt.500010603. [DOI] [PubMed] [Google Scholar]
- 148.Zhou S, Terrault N A, Ferrell L, Hahn J A, Lau J Y, Simmonds P, Roberts J P, Lake J R, Ascher N L, Wright T L. Severity of liver disease in liver transplantation recipients with hepatitis C virus infection: relationship to genotype and level of viremia. Hepatology. 1996;24:1041–1046. doi: 10.1002/hep.510240510. [DOI] [PubMed] [Google Scholar]
- 149.Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]