A gain-of-function epigenetic switch that disrupts differentiation is triggered by inactivating KDM6A mutations in bladder cancer and can serve as a potential target for novel therapies.
Abstract
Disruption of KDM6A, a histone lysine demethylase, is one of the most common somatic alternations in bladder cancer. Insights into how KDM6A mutations affect the epigenetic landscape to promote carcinogenesis could help reveal potential new treatment approaches. Here, we demonstrated that KDM6A loss triggers an epigenetic switch that disrupts urothelial differentiation and induces a neoplastic state characterized by increased cell proliferation. In bladder cancer cells with intact KDM6A, FOXA1 interacted with KDM6A to activate genes instructing urothelial differentiation. KDM6A-deficient cells displayed simultaneous loss of FOXA1 target binding and genome-wide redistribution of the bZIP transcription factor ATF3, which in turn repressed FOXA1-target genes and activated cell-cycle progression genes. Importantly, ATF3 depletion reversed the cell proliferation phenotype induced by KDM6A deficiency. These data establish that KDM6A loss engenders an epigenetic state that drives tumor growth in an ATF3-dependent manner, creating a potentially targetable molecular vulnerability.
Significance:
A gain-of-function epigenetic switch that disrupts differentiation is triggered by inactivating KDM6A mutations in bladder cancer and can serve as a potential target for novel therapies.
Introduction
Bladder cancer is the sixth most common malignancy in the United States. Molecular characterization studies across bladder cancer cohorts have demonstrated recurrent inactivating alterations in chromatin modifier genes in nonmuscle-invasive and muscle-invasive disease, which implies that these mutations occur early in carcinogenesis and are maintained during progression (1–5). Moreover, inactivating mutations affect subunits of unique complexes (COMPASS-like and SWI/SNF) and their activities (methyltransferases, acetyltransferases, target binding), which may produce distinct alterations in epigenetic states that lead to the development of bladder cancer (6–8).
KDM6A is a histone H3 lysine 27 demethylase that plays a central role in regulating enhancer activity as part of the COMPASS-like complex, and its function is critical for embryonic stem cell differentiation and tissue development. Inactivating KDM6A mutations have been identified in multiple malignancies, including leukemias, lymphomas, and cancers of the esophagus, stomach, endometrium, cervix, head and neck, lung, pancreas, colon, and rectum (9, 10). Nonetheless, the highest frequency of KDM6A mutation is found in urothelial cancer, with 26% and 38% of cases affected in muscle-invasive bladder and upper tract urothelial carcinoma cohorts, respectively, and alteration rates in early-stage bladder cancer are as high as 52% (2, 4, 11).
The high frequency of KDM6A-inactivating mutations has led to its designation as a tumor suppressor in bladder cancer; however, the molecular mechanisms by which KDM6A limits tumor initiation and promotion are incompletely elucidated. KDM6A was shown to activate gene expression in a catalytic-independent manner; and in muscle-invasive bladder cancer, its loss cooperated with FGFR3 activation to repress a luminal gene-expression program (12). However, another study demonstrated that KDM6A loss leads to EZH2-mediated cell proliferation, which can be reversed through inhibition of EZH2 methyltransferase activity (13). More recently, the tumor suppressor activity of KDM6A in leukemia and pancreatic cancer models was found to be dependent on its ability to phase separate and form condensates that contain other members of the COMPASS-like complex (14). Here, we investigate the epigenetic alterations caused by KDM6A loss and discover decreased chromatin accessibility at motifs associated with transcription factors governing urothelial differentiation such as FOXA1, GATA3, and ELF3. In KDM6A-deficient bladder cancer cells, we identified concomitant, widespread changes in the chromatin binding of ATF3, a transcription factor associated with cellular stress response genes. Additionally, we found that ATF3 plays a dual role in tumor promotion by repressing a FOXA1-driven urothelial differentiation program and activating cell-cycle genes. ATF3 depletion decreased cell proliferation in KDM6A-deficient but not parental bladder cancer cells. Our findings suggest that targeting the gain-of-function epigenetic alterations that result from KDM6A loss may provide a novel therapeutic strategy for KDM6A-mutant bladder cancers.
Materials and Methods
Bladder cancer patient cohort
Written informed consent was obtained from bladder cancer patients for specimen collection under an Institutional Review Board-approved protocol adhering to U.S. Common Rule guidelines at Cleveland Clinic. The intraluminal aspect of tumors identified on cystoscopy was sampled using a cold cup biopsy forceps, and pathologic evaluation confirmed greater than 80% tumor content. The tissue was then snap frozen immediately in liquid nitrogen for downstream applications. Clinicopathologic characteristics of the cohort are described in Supplementary Table S1. Subjects did not undergo chemotherapy or radiotherapy prior to tissue collection.
Cell lines
RT4 (RRID:CVCL_0036), SW780 (RRID:CVCL_1728), and SCaBER (RRID:CVCL_3599) cell lines were purchased from ATCC. Cells were cultured in McCoy's 5A medium or DMEM supplemented with 10% fetal bovine serum as recommended by ATCC. All cell lines underwent short tandem repeat profiling by Labcorp to verify identity prior to downstream experiments and also prior to cryopreservation. Mycoplasma testing was performed regularly using the MycoAlert Mycoplasma Detection Kit (Lonza) to ensure that no contaminated cells were used for experimentation or cryopreservation. Cells were used for downstream experiments within six passages after thawing.
Mouse urothelial organoid culture and xenografts
All mouse studies were performed under an IACUC-approved animal use protocol. Mouse urothelial organoids were generated as previously described (15). Briefly, urothelium was dissected from male C57BL6/J mice bladders bearing wild-type, Kdm6aF, Trp53F/F, or Kdm6aF;Trp53F/F alleles, dissociated into single-cell suspension, and organoid cultures were maintained in growth factor–reduced Matrigel (Corning) tabs suspended in organoid medium with the following changes: final EGF concentration = 5 ng/μL, final A83-01 concentration = 20 nmol/L, and FGF10, FGF2, dihydrotestosterone, Y-27632, SB202190, and primocin were omitted. Cre recombinase was introduced into these organoids by infection with Ad5CMVCre-eGFP adenovirus (University of Iowa Viral Vector Core) in vitro, and recombination of floxed alleles was verified using PCR. Differentiation of mouse urothelial organoids was performed by treatment with 100 nmol/L all-trans retinoic acid in organoid medium for 2 weeks.
For xenograft studies, 50 μL of organoids in 50% Matrigel/50% organoid medium containing 1×106 cells were injected into a subcutaneous pocket in the flanks of C57BL6/J mice. Growth measurements of three independent organoid lines of each genotype were performed in triplicate. Xenografts from Trp53F/F and Kdm6aF;Trp53F/F organoids were harvested and processed for 4% paraformaldehyde fixation, paraffin embedding, and hematoxylin–eosin staining using standard protocols.
Orthotopic xenografts
NOD-scid IL2Rgammanull (NSG) mice (The Jackson Laboratory) received isoflurane (2%–3%) general anesthesia during xenograft implantation. For pain control, buprenorphine (0.05 mg/kg s.c.) was given preoperatively and postoperatively. Hair was removed by depilatory cream. Skin was prepared with betadine followed by 70% ethanol wipe. Local anesthesia was provided by subcutaneous injection of bupivacaine at the surgical site. A 6-mm midline incision was made above the pubis, subcutaneous tissues were dissected sharply, and the bladder was identified. 2×105 bladder cancer cells in 10 μL sterile saline was injected into the ventral wall of the bladder. The fascia and subcutaneous tissue were closed in one layer using 5–0 absorbable suture. Skin was closed with stainless steel clips, which were removed on day 10.
Bioluminescent imaging was performed using the Cleveland Clinic Small Animal Imaging Core. Mice were anesthetized using isoflurane and received an intraperitoneal injection of 6 mg luciferin prior to imaging with IVIS Spectrum (Perkin-Elmer).
Lentivirus infection
Lentiviral shRNA targeting ATF3, FOXA1, and control shNT were purchased from Sigma (#TRCN0000013572, #TRCN0000329689, #TRCN0000014878, #TRCN0000014881, #TRCN0000014879, and #SHC016). Luciferase, KDM6A, and ATF3 overexpression plasmids were purchased from Addgene (#17477) and VectorBuilder (#VB180814-1301, #VB181109-1119, and #VB180323-1141). Lentiviral packaging plasmids psPAX2 (#12260) and pMD2.G (#12259) vectors were obtained from Addgene. For KDM6A knockout experiments, sgRNAs (GGTATGCAGATAATGCTGAA, ACAGTTTACAGTCTGACTAC) (IDT) were cloned into lentiCRISPR v2 nickase vector (16). All plasmid sequences were verified using Sanger sequencing before use. Lentiviral plasmids were cotransfected with psPAX2 and pMD2.G into HEK293T cells to generate lentiviral particles, which were subsequently collected from the culture medium, concentrated, and used for target cell transduction. Transduced cells bearing shRNA and overexpression constructs were cultured in selective growth medium containing either puromycin or hygromycin and collected for experiments within 3 to 5 days. Transduced cells bearing CRISPR vectors were cultured in a selective growth medium containing puromycin for 48 hours and then maintained in a basal growth medium. Single clones were screened for KDM6A expression by immunoblot, and clones containing no detectable KDM6A protein levels were used for further experiments.
Cell viability
Cell proliferation experiments were performed by plating 3,000 cells per well in a 96-well plate. Three replicates per cell type were used at each time point. CellTiter-Glo (Promega) was used to measure cell viability according to the manufacturer's instructions. Each experiment was performed four times to ensure reproducibility, and data were normalized to day 0 counts and plotted as mean ± SEM.
Whole-exome sequencing
DNA was isolated from bladder cancer tissue using the QIAamp Fast DNA Tissue Kit (Qiagen). Exome capture was performed using the SureSelect Human All Exon V5 kit (Agilent) prior to library generation using Illumina adapters. The samples were then sequenced on the NovaSeq 6000 System using an S4 flow cell in paired-end mode. Raw sequencing data were aligned to the GRCh38/hg38 genome build using the Burrows–Wheeler Aligner (BWA; ref. 17). Further indel realignment, base-quality score recalibration, and duplicate-read removal were performed using the Genome Analysis Toolkit (GATK) following raw reads alignments guidelines. SNV and indels were called by MuTect2 (GATK v2.2) according to the Genomic Data Commons best practices (https://docs.gdc.cancer.gov/Data/Bioinformatics_Pipelines/DNA_Seq_Variant_Calling_Pipeline/#tumor-only-variant-calling-workflow) and annotated with Ensembl Variant Effect Predictor. Variants with total coverage greater than 10 and alternative allele frequency greater than 4% were included. Common SNPs and variants were identified and excluded using dbSNP v142 and gnomAD v2.1.1, respectively. Indels in low mappability and blacklisted regions were filtered (18). Variants were then annotated using OncoKB and COSMIC, and the annotated Variant Call Format file was converted to Mutation Annotation Format using the vcf2maf tool (https://github.com/mskcc/vcf2maf).
ATAC-seq
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) was performed using the Omni-ATAC protocol with modifications (19). For cell lines, 50,000 cells were lysed in lysis buffer (10 mmol/L Tris-HCl, pH 7.4, 10 mmol/L NaCl, 3 mmol/L MgCl2, 0.1% NP-40, 0.1% Tween 20, 0.01% Digitonin). For bladder cancer surgical specimens, snap-frozen tissues were pulverized with a cold mortar and pestle, resuspended in lysis buffer, and nuclei were filtered using a 30-μm filter. 50,000 nuclei were then used for transposition. Transposition and fragmentation were performed by adding Tn5 transposase from the Nextera DNA Library Prep Kit (Illumina) and incubating at 37°C for 30 minutes. The tagmented chromatin was purified with the MinElute PCR purification kit (#28004, Qiagen). Purified DNA was then ligated with adapters, amplified by PCR, and size selected before sequencing on a NovaSeq 6000 System using an S4 flow cell in a paired-end mode.
Raw reads were subjected to adapter trimming and quality filtering using fastp (20). Filtered reads were then aligned to GRCh38/hg38 using Bowtie 2 (21). Duplicates were removed using Picard MarkDuplicates (https://broadinstitute.github.io/picard/) and aligned reads corresponding to mitochondrial and ENCODE blacklist regions were filtered using SAMtools (22). Peak calling was performed using MACS2 (23) using –shift -75 –nomodel –extsize 150 –call-summits. For human bladder cancer specimens, reads were assigned to peaks using featureCounts (24), and differential peaks between KDM6A-wt and KDM6A-mut samples were identified by DESeq2 (absolute log2FC > 0.58, P < 0.05; ref. 25). Differential peaks for cell lines were identified using MAnorm (26) by comparing reads from each parental cell line and its KDM6A knockout derivative. The top 5,000 statistically significant peaks (P < 0.05) by M value for each condition (parental vs. KDM6A knockout) were considered enriched. MAnorm was also used to identify private and common peaks between each parental cell line and its KDM6A knockout derivative using MACS2 peak calls as input, and the final peak sets were constructed by extending the peak summits in each direction for a final width of 500 bp. Overlap between cell line and patient peaks was calculated using Intervene (27). Transcription factor motif enrichment analysis was performed on differential peaks using the HOMER known motif analysis (28).
Footprinting analysis of cell line ATAC-seq data to predict transcription factor occupancy at regions of accessible chromatin was performed using the TOBIAS package (29). ATACorrect was used to correct Tn5 insertion bias, and ScoreBigwig was used to calculate footprinting scores. BINDetect was then used to predict transcription factor occupancy at differential peaks identified by MAnorm with motifs from JASPAR CORE collection.
Comparison of ATAC-seq data between Cleveland Clinic bladder cancer and The Cancer Genome Atlas (TCGA) cohorts (Supplementary Data S1; https://gdc.cancer.gov/about-data/publications/ATACseq-AWG) was performed as follows. Peaks in distal elements from Cleveland Clinic bladder cancer patients were identified first by extending peak summits to 250 bp in each direction for a final width of 501 bp and then filtering peaks that resided at transcription start sites. Significant peaks were identified by normalizing MACS2 peak scores (−log10(P)) according to Corces and colleagues so that samples with varying read depth and quality could be compared (30). These peaks were merged with TCGA BLCA, KIRC, KIRP, PRAD, and TGCT called peaks to generate a universe of peaks. Reads from Cleveland Clinic bladder cancer and TCGA cohorts were assigned to peaks using deepTools multiBigwigSummary, and the raw count table was normalized using DESeq2. Dimension reduction was performed using UMAP (https://github.com/lmcinnes/umap), and the coordinates were plotted for visualization.
Functional enrichment analysis for ATAC-seq peaks enriched in KDM6A-wt bladder cancer patient samples was performed by first assigning peaks to genes using GREAT (31). Assigned genes were then submitted to The Gene Ontology Resource (http://geneontology.org/). Gene ontology (GO) terms were manually curated into subnetworks, and the top terms from each subnetwork were displayed.
Principal component analysis of cell lines was performed as follows. Differential peaks in RT4 parental and K2 knockout cells were identified by MAnorm and combined into a universe of peaks bed file. The average ATAC-seq scores at these peaks were computed using deepTools multiBigwigSummary and output to a compressed numpy array, which was plotted onto two dimensions using deepTools plotPCA –transpose.
RNA-seq
Total RNA from cell lines and organoids was isolated with the RNeasy Mini Kit (#74104, Qiagen) according to the manufacturer's protocol. RNA-seq libraries were generated with the TruSeq Stranded mRNA Sample Prep Kit (#20020594, Illumina) and sequenced on the Illumina NovaSeq 6000 System using an S4 flow cell in paired-end mode. Adapters were trimmed and low-quality reads were filtered using fastp, which were then aligned to GRCh37/hg19 or GRCm38/mm10 using HISAT2 (32). Reads were assigned to exons using featureCounts and the GENCODE GRCh37 v34 or GRCm38 M25 comprehensive gene annotation file (https://www.gencodegenes.org/). DESeq2 was used to identify differentially expressed genes between each parental cell line and its KDM6A knockout derivative (i.e., RT4 vs. K2) with absolute log2FC > 0.58 and FDR < 0.05. For functional enrichment analysis, differentially expressed genes were submitted to g:Profiler with GO biological process as the data source, no electronic GO annotations, and FDR < 0.05 for significance threshold (33). Enrichment maps highlighting common GO terms and their relationships were generated using Cytoscape as described by Reimand and colleagues (34). Integration of transcriptome and transcription factor binding site data [chromatin immunoprecipitation sequencing (ChIP-seq) and CUT&RUN] was performed using BETA basic (35) with differential peaks identified using MAnorm (absolute M > 1) and differentially expressed genes in limma format as inputs.
ChIP-seq and CUT&RUN
H3K27ac, H3K4me1, H3K4me3, and FOXA1 ChIP-seq were performed by cross-linking cells with 1% formaldehyde for 15 minutes at room temperature. The reaction was quenched with glycine at a final concentration of 0.125M for 5 minutes. Cells were washed twice with ice cold 0.5% Igepal CA-630 with 1 mmol/L PMSF in PBS and snap frozen on dry ice. Chromatin was sheared by sonication, and an aliquot was reserved as input DNA. 30 μg chromatin was used for immunoprecipitation with the following antibodies: anti-H3K27ac (#39133, Active Motif, RRID:AB_2561016), anti-H3K4me1 (#39297, Active Motif, RRID:AB_2615075), anti-H3K4me3 (#39159, Active Motif, RRID:AB_2615077), anti-FOXA1 (#39837, Active Motif, RRID:AB_2793362). Crosslinks were reversed by heat, DNA was purified, and enrichment was confirmed by quantitative PCR. Sequencing libraries were generated and then sequenced on an Illumina HiSeq platform using single-end mode.
KDM6A- and ATF3-binding sites were identified using the CUT&RUN assay kit (#86652, Cell Signaling Technology). Thirty microliters of activated Concanavalin A magnetic beads and 1.5 μg anti-KDM6A (#33510, Cell Signaling Technology, RRID:AB_2721244) or anti-ATF3 antibody (#33593, Cell Signaling Technology, RRID:AB_2799039) were incubated with 3×105 cells overnight at 4°C. The enriched chromatin was digested by 150 μL pAG-MNase enzyme and followed by DNA purification (MinElute PCR Purification Kit, #28004, Qiagen). Sufficient input DNA for CUT&RUN experiments was generated by fragmented genomic DNA by sonication to the appropriate size before DNA purification. Sequencing libraries were generated with NEBNext Ultra II DNA Library Prep Kit for Illumina (#E7645, New England Biolabs) and sequenced on an NovaSeq 6000 System using an S4 flow cell in paired-end mode.
Using fastp, raw reads underwent adapter trimming and quality filtering. Filtered reads were then aligned to GRCh38/hg38 using Bowtie 2. Duplicates were removed using Picard MarkDuplicates and aligned reads corresponding to mitochondrial and ENCODE blacklist regions were filtered using SAMtools. Peak calling was performed using MACS2. Common and private peaks between parental and knockout cell lines were identified using MAnorm with MACS2 called peaks as input. Differential peaks were identified with MAnorm using absolute M > 1 as a cutoff. Transcription factor motif enrichment analysis was performed on differential peaks using HOMER known motif analysis. For UCSC genome browser visualization, bigwig files were generated using deepTools bamCoverage. Genomic contexts corresponding to called peaks were annotated using ChIPseeker and the GENCODE GRCh38 release 38 comprehensive gene annotation file (https://www.gencodegenes.org/human/release_38.html). Superenhancers were identified by stitching enhancers marked by H3K27ac using ROSE and plotting H3K27ac signal versus superenhancer rank (36, 37). Stitched enhancers with a position above a line with slope = 1 that is tangent to the plotted curve were labeled as superenhancers.
Immunoblot and coimmunoprecipitation
Cells were lysed in RIPA buffer (#89900, Thermo Scientific) or Nuclear Extraction buffer (50 mmol/L Tris-HCl, pH 8, 200 mmol/L NaCl, 1.5% NP-40) with protease inhibitors (#A32955, Thermos Scientific). Protein (25–50 μg) was separated on 4% to 20% MOPS gel (GenScript) and blotted onto the PVDF membrane (Millipore). Coimmunoprecipitation was performed as follows: 500 to 1,000 μg of freshly extracted protein was incubated with 2 to 10 μg of antibody or rabbit IgG (#12-370, Millipore) overnight at 4°C with rotation. Protein A/G magnetic beads (#88802, Thermos Scientific) were added to this mixture and incubated for an additional 4 hours. The bead-protein complexes were washed three times with PBST wash buffer. The precipitated proteins were eluted from the beads with 2× Laemmli sample buffer (#1610737, Bio-Rad) and boiling for 10 minutes followed by western blot analyses or Mass Spec analysis. The following primary antibodies were used: anti-KDM6A (#33510, Cell Signaling Technology, RRID:AB_2721244), anti-ELF3 (#PA5-89261, Thermo Scientific, RRID:AB_2805451), anti-FOXA1 (#58613, Cell Signaling Technology, RRID:AB_2799548), anti-FOXA1 (#ab5089, Abcam, RRID:AB_304744), and anti-ATF3 (#33593, Cell Signaling Technology, RRID:AB_2799039). Anti-GAPDH (#2118, Cell Signaling Technology, RRID:AB_561053) was used as a loading control. The secondary antibodies used were as follows: HRP-linked donkey anti-rabbit (GE Healthcare, NA934V), HRP-linked mouse anti-rabbit (#18-8816-31, Rockland), and HRP-linked donkey anti-goat (#ab97110, Abcam).
AP-1 transcription factor assay
AP-1 activity was measured by colorimetric assay according to the manufacturer's directions (#44296, Active Motif). Briefly, nuclear extracts were prepared from luminal subtype parental (RT4 and SW780) and KDM6A knockout (K2 and S5) cells. Nuclear extracts were then incubated at room temperature for 1 hour with an oligonucleotide containing a 12-O-tetradecanoylphorbol 13-acetate (TPA) response element in a 96-well plate to capture activated AP1 transcription factor elements. After washing the wells 3 times, primary AP-1 antibodies supplied in the kit or anti-ATF3 antibody (#33593, Cell Signaling Technology, RRID:AB_2799039) were added. The wells were then washed 3 times, incubated with secondary anti-rabbit HRP-conjugated antibody (1:1,000 dilution), washed 4 times, developed with colorimetric reaction solution, and read by spectrophotometer at 450 nm wavelength (reference wavelength = 655 nm).
Statistical analysis
Statistical significance testing for cell proliferation assays was performed using unpaired two-tailed Student t tests. Statistical parameters are expressed as mean ± SEM with P values reported in the figure legends. Genes were differentially expressed if absolute log2FC was > 0.58 (fold change > 1.5) and FDR < 0.05.
Data and materials availability
All cell lines, plasmids, and other stable reagents generated in this study are available from the corresponding author with a completed Materials Transfer Agreement. The data generated in this study are publicly available at Sequence Read Archive with Accession: PRJNA822231.
Results
KDM6A mutation disrupts urothelial differentiation transcription factor circuitry in human bladder cancers
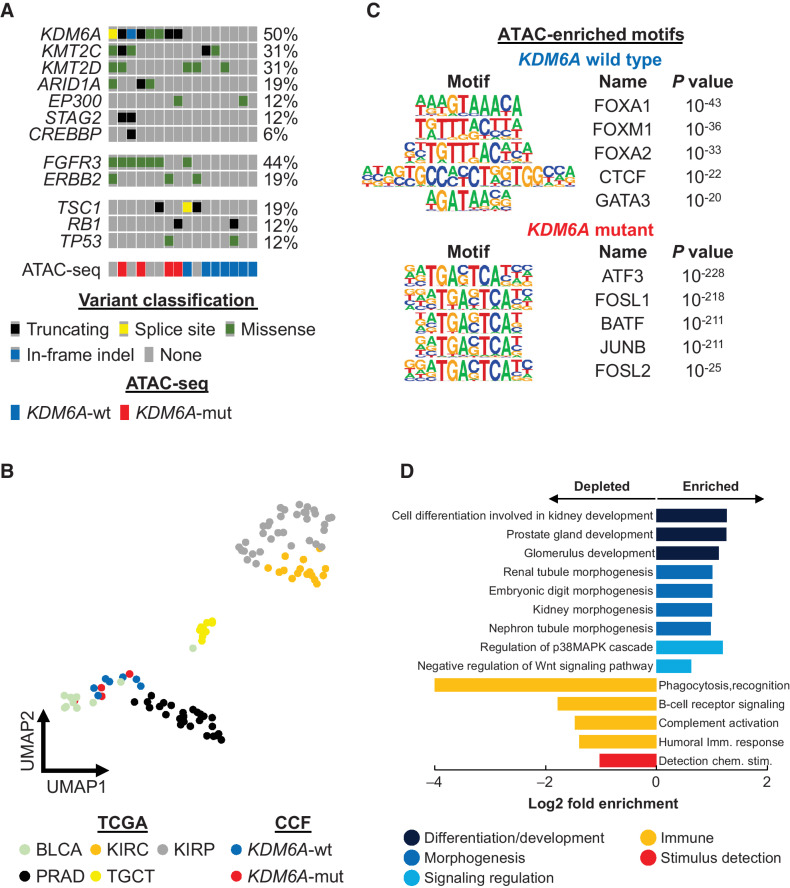
To identify changes in chromatin accessibility due to KDM6A mutation, we first performed whole-exome sequencing on 16 bladder cancer surgical specimens to determine their mutational landscape and classify cases as KDM6A mutant or wild-type (Fig. 1A; Supplementary. Table S1). These cancers showed mutational profiles similar to those seen in other muscle-invasive and nonmuscle-invasive bladder cancer cohorts, with recurrent chromatin modifier gene and FGFR3 mutations. We identified four cases with (KDM6A-mut) and seven cases without (KDM6A-wt) an inactivating KDM6A mutation. ATAC-seq was performed on these cases to ascertain genome-wide chromatin accessibility profiles. KDM6A mutation in bladder cancer did not result in the global alteration of chromatin accessibility (Fig. 1B). KDM6A-wt and KDM6A-mut cases were clustered with TCGA bladder cancer cases, and not with other genitourinary malignancies such as renal cell carcinoma, prostate adenocarcinoma, and testicular germcell tumor. Furthermore, KDM6A mutation was accompanied by distinct chromatin accessibility changes in regulatory elements associated with developmental and transcriptional regulation pathways (Fig. 1C). Called peaks from the Cleveland Clinic cohort were combined into a set of 499,265 total peaks, of which, 8,510 were differentially enriched in KDM6A-wt cases, and 6,292 were differentially enriched in KDM6A-mut cases (absolute log2FC > 0.58, P < 0.05). Peaks specific for KDM6A-wt cases were enriched for motifs of transcription factors that control urothelial identity including forkhead box (FOXA1/A2) and GATA Binding Protein 3 (GATA3). Genes linked to KDM6A-wt specific peaks were enriched for biological processes involved in the regulation of differentiation, development, morphogenesis, and signal transduction and depleted for immune terms (Fig. 1D). On the other hand, peaks specific for KDM6A-mut cases were enriched for basic-region leucine zipper (bZIP) transcription factors (ATF3, FOSL1/2, and JUNB; Fig. 1C). These data suggest that KDM6A inactivation is associated with the dysregulation of transcription factor circuitry that instructs urothelial cell identity and cell fate.
Figure 1.
KDM6A mutation alters chromatin accessibility in human bladder cancers. A, Whole-exome sequencing of bladder cancer surgical specimens (n = 16). Seven KDM6A-wt (blue) and four KDM6A-mut (red) cases were chosen for ATAC-seq analysis. B, UMAP analysis of Cleveland Clinic bladder cancer specimens (KDM6A-wt, blue; KDM6A-mut, red) and TCGA genitourinary cancer specimens based on ATAC-seq data. C, Enriched motifs in accessible chromatin peaks specific for KDM6A-wt (blue) and KDM6A-mut (red) cancers. D, Functional enrichment analysis of accessible chromatin peaks enriched in KDM6A-wt cancers.
KDM6A inactivation induces a phenotypic switch from urothelial differentiation to cell proliferation
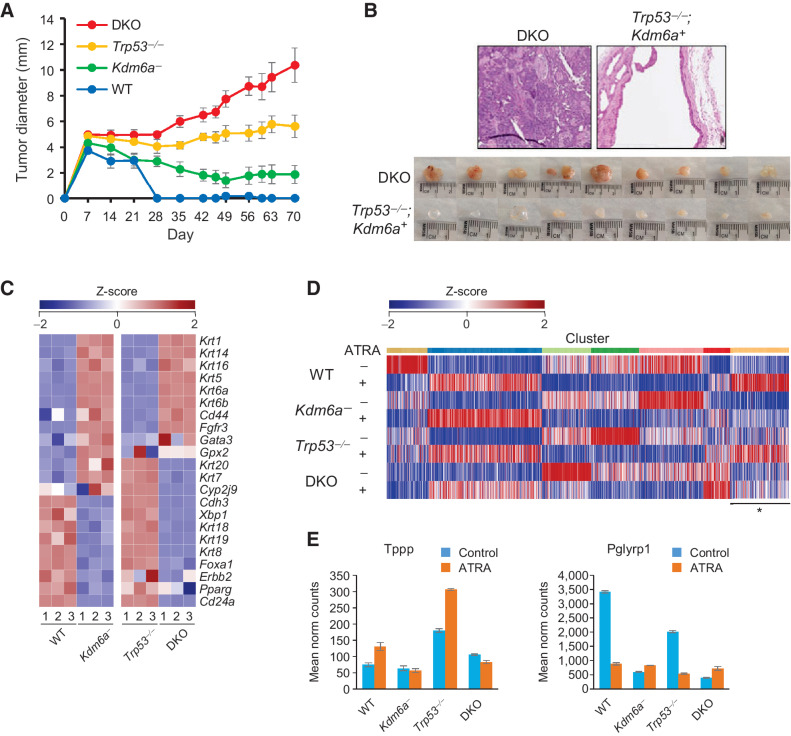
We previously generated mouse urothelial organoids from conditional knockout mice of the following genotypes: (i) Kdm6aF, (ii), Trp53F/F, and (iii) Trp53F/F; Kdm6aF and transduced them with Adeno-cre to induce gene deletion (15). Urothelial organoids from wild-type mice that underwent Adeno-cre treatment were used as a control, and data were collected from three independent organoid lines from each genotype. Organoids were implanted subcutaneously in the flanks of C57BL/6J mice to assess their tumorigenicity (Fig. 2A). Trp53-/-;Kdm6a− (DKO) organoids generated cancers with squamous differentiation; however, neither Kdm6a− nor Trp53−/− organoids formed tumors (Fig. 2B), which suggests that tumorigenicity due to Trp53 knockout requires a change in the epigenetic state caused by Kdm6a deletion in this model system.
Figure 2.
Kdm6a loss alters differentiation and cooperates with Trp53 deletion to confer tumorigenicity in mouse urothelial organoids. A, Effect of Kdm6a and Trp53 deletion on organoid growth as subcutaneous xenografts. DKO denotes combined Kdm6a and Trp53 knockout. B, Hematoxylin and eosin (top) and gross (bottom) images of Trp53−/− and DKO organoid xenografts. Trp53−/− organoids formed cystic masses, and DKO organoids formed cancers with squamous differentiation. C, Heat map of luminal and basal marker expression according to MDACC classification. D, K-means clustering of retinoic acid–responsive genes in mouse urothelial organoids. Cluster denoted by asterisk shows genes with altered response to 100 nmol/L ATRA in Kdm6a-deficient (Kdm6a and DKO) organoids. E, Examples of two retinoic acid target genes that exhibited an abnormal response to ATRA treatment in Kdm6a-deficient organoids.
We then used RNA-seq to examine the effects of Kdm6a deletion on organoid gene expression. Compared with wild-type organoids, both Kdm6a− and DKO organoids showed decreased expression of luminal genes and increased expression of basal genes with the exception of Fgfr3, which has been implicated in driving a luminal-to-basal shift (Fig. 2C; ref. 12). This luminal-to-basal switch led us to hypothesize that Kdm6a loss also causes a block in urothelial differentiation in these organoids. Retinoic acid signaling has been shown to induce embryonic stem cells to differentiate into urothelial cells in vitro and regulates the specification of urothelial progenitors during injury/regeneration. Additionally, KDM6A has been shown to facilitate the recruitment of MLL4 to retinoic acid–inducible genes (38). When treated with all-trans retinoic acid (ATRA), wild-type organoids increased expression of the terminal differentiation markers Upk1a, Krt13, and Krt20 (Supplementary. Fig. S1A). Conversely, Kdm6a− organoids treated with ATRA showed a decreased expression of these markers, which suggests that Kdm6a loss interferes with differentiation induced by retinoic acid signaling. Genes in Kdm6a− organoids that did not respond to ATRA treatment in the expected direction showed enrichment for biological processes governing development, apoptotic signaling, immune regulation, and stimulus response (Supplementary Fig. S1B). Globally, most retinoic acid–responsive genes in the wild-type organoids exhibited similar behavior in the Kdm6a− organoids upon ATRA treatment, and this behavior was preserved in a Trp53−/− background (Fig. 2D). Nonetheless, unsupervised clustering of RNA-seq data demonstrated a set of genes that showed an abnormal response to ATRA in Kdm6a− and DKO organoids when compared with WT organoids (Fig. 2D and E). Overall, these data demonstrate that Kdm6a deletion in mouse urothelial organoids partially disrupts a retinoic acid–dependent differentiation program and induces a permissive state for tumorigenesis that can be triggered by loss of tumor suppressor gene function.
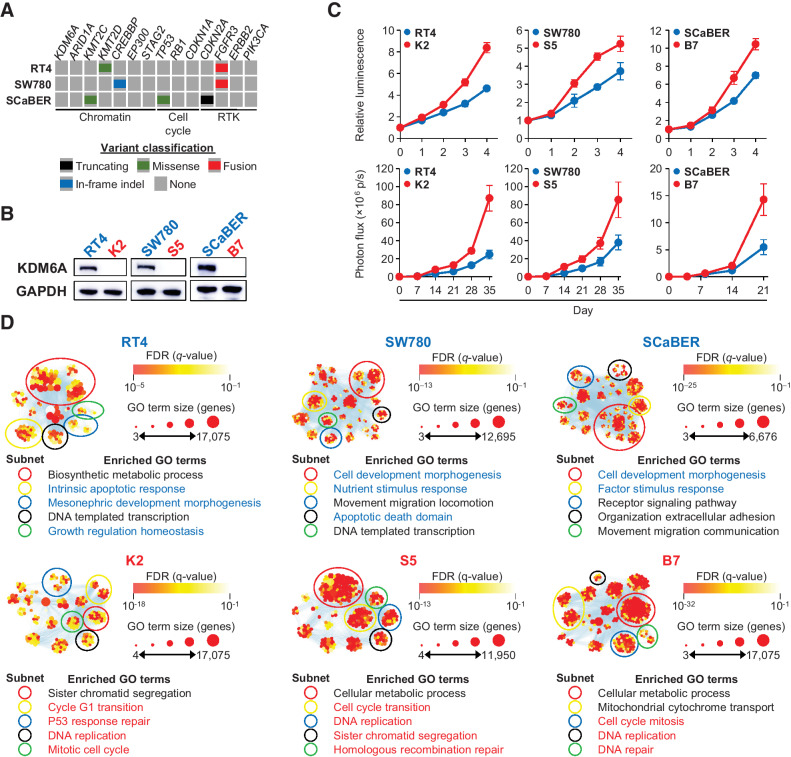
To examine the functional effects of KDM6A inactivation in human bladder cancer, we deleted KDM6A in three bladder cancer cell lines: RT4, SW780, and SCaBER (Fig. 3A and B). These cell lines were chosen because they capture the phenotypic diversity of bladder cancer and did not harbor inactivating chromatin modifier gene mutations. RT4 and SW780 were derived from low-grade papillary noninvasive cancers in a male and female, respectively; whereas SCaBER was derived from a high-grade muscle-invasive bladder cancer. Additionally, these cell lines represent both luminal (RT4 and SW780) and basal (SCaBER) bladder cancer molecular subtypes. Lastly, both RT4 and SW780 harbor FGFR3 fusions to TACC3 and BAIAP2L1, respectively; whereas SCaBER does not harbor any FGFR3 alterations. Compared with parental cells, KDM6A knockout cells showed increased proliferation in vitro and in vivo as orthotopic xenografts in immunodeficient mice (Fig. 3C). RNA-seq was performed to identify differentially expressed genes between each parental cell line and its KDM6A knockout derivative. Functional analyses of differentially expressed genes revealed that KDM6A deletion leads to downregulation of gene regulatory subnetworks governing development, morphogenesis, homeostasis, stimulus response, and apoptosis (Fig. 3D). In contrast, KDM6A deletion enriches subnetworks that control processes associated with mitosis such as DNA replication, spindle checkpoint, and DNA repair. Although KDM6A deletion upregulated the basal markers KRT5, KRT6A, CD44, TP63, ITGA6, and EGFR in the luminal subtype cell line RT4, it had the opposite effect on the basal subtype cell line SCaBER (Supplementary Fig. S1C). These data highlight the dysregulation in subtype identity caused by KDM6A loss in bladder cancer cells, resulting in an increased expression of basal markers in luminal subtype cells and vice versa. Nonetheless, a transcriptional state that supports increased cell proliferation was found consistently across the three cell lines.
Figure 3.
KDM6A deletion increases cell proliferation in luminal and basal subtype bladder cancer cells. A, Genomic alterations of the three bladder cancer cell lines used in this study. B, Immunoblot demonstrating loss of KDM6A protein after CRISPR-mediated gene deletion. C, Cell proliferation of parental (blue) and KDM6A knockout (red) bladder cancer cells in vitro (top) and in vivo (bottom) as orthotopic xenografts. D, Functional enrichment analysis of differentially expressed genes identified from RNA-seq (absolute log2FC > 0.58, FDR < 0.05) in parental (top) and KDM6A knockout (bottom) bladder cancer cells. Enriched GO terms list the most common GO terms in each subnetwork. Blue GO terms highlight subnetworks shared in parental cells, and red GO terms highlight subnetworks shared in KDM6A knockout cells.
Chromatin accessibility alterations due to KDM6A loss affect distinct transcriptional circuitry and enhancer usage
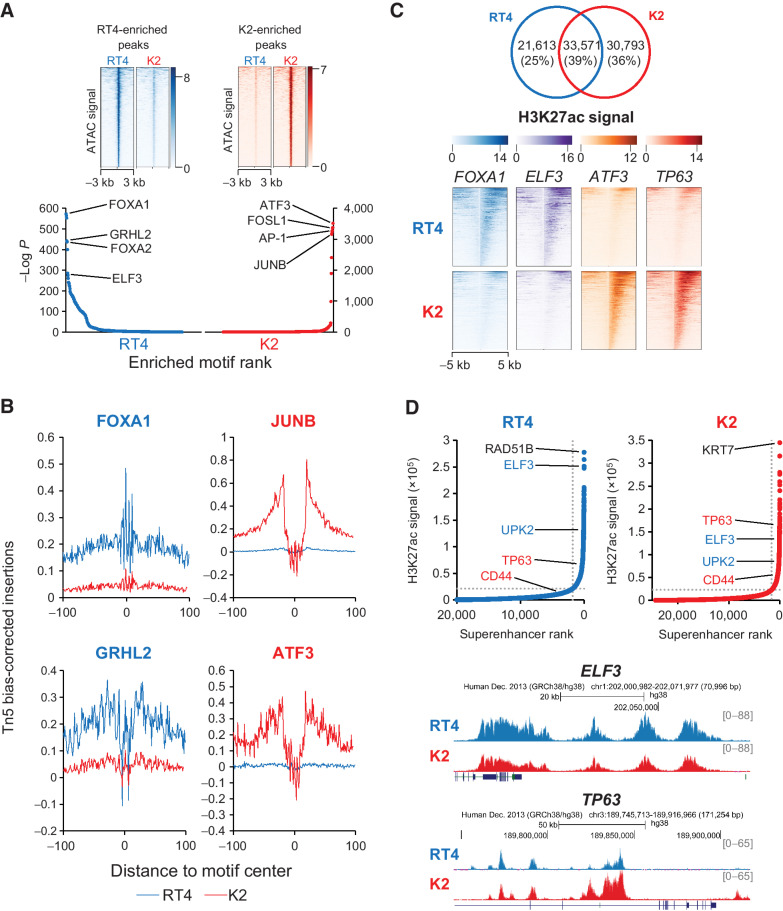
We ascertained the chromatin accessibility landscapes of parental and KDM6A knockout cells to identify the core transcriptional networks altered by KDM6A deletion. 82,135 ATAC-seq peaks were identified among the three parental cell lines, of which, 22,186 (27%) were shared (Supplementary. Fig. S2A). Globally, parental and knockout cells showed significant overlap in chromatin accessibility. For example, when RT4 was compared with its KDM6A knockout subclone K2, they shared 51% of combined called ATAC-seq peaks (data not shown). Similar to the chromatin accessibility data from primary bladder cancer specimens, wild-type–specific peaks were enriched for motifs of transcription factors that play a role in the formation and differentiation of the urothelium, notably FOXA1, GRHL2, ELF3, TP63, and KLF5, and peaks specific for KDM6A knockout subclones showed enrichment for bZIP transcription factors (Fig. 4A; Supplementary Fig. S2B and S2C). Sites bearing these motifs showed footprinting changes between parental and knockout cells, which suggests that a change in motif enrichment is accompanied by differential transcription factor binding (Fig. 4B; Supplementary Fig. S2D and S2E). These findings indicate that the chromatin accessibility changes arising from KDM6A deletion in bladder cancer cell lines affect similar transcription factor circuitry as primary cancer specimens, making these KDM6A knockout subclones suitable models for functional studies. Additional KDM6A knockout subclones generated from all three cell lines showed similar changes in proliferation, chromatin accessibility profiles, and motif enrichment (Supplementary Fig. S3). To further refine cell line selection, we examined ATAC-seq peak intersection between parental cells and KDM6A-wt primary cancer specimens as well as between knockout cells and KDM6A-mut primary cancer specimens (Supplementary Fig. S4A). RT4 and K2 showed the highest overlap with KDM6A-wt and KDM6A-mut cases, respectively. We next determined ATAC-seq signal enrichment of parental and KDM6A knockout subclones in peaks from KDM6A-wt and KDM6A-mut cases. RT4 showed the highest signal enrichment at KDM6A-wt patient peaks, whereas all three KDM6A knockout subclones showed similar enrichment at KDM6A-mut patient peaks (Supplementary Fig. S4B). Additionally, motif enrichment of ATAC-seq peaks in RT4 and K2 most closely resembled the motif enrichment of ATAC-seq peaks seen in KDM6A-wt and KDM6A-mut primary cancer specimens, respectively (Figs. 1C and 4A). Thus, we selected RT4 and K2 for further functional studies.
Figure 4.
KDM6A deletion alters chromatin accessibility and enhancer usage in bladder cancer cells. A, ATAC signal heat map (top) and motif analysis (bottom) of accessible chromatin peaks enriched in RT4 parental (blue) and K2 knockout (red) cells. B, Footprinting analysis at accessible chromatin peaks containing transcription factor motifs enriched in RT4 parental (blue) and K2 knockout (red) cells. Blue and red lines indicate Tn5 bias-corrected insertions in parental and knockout cells, respectively. C, Venn diagram (top) showing common and private H3K27ac peaks between RT4 and K2, and heat map (bottom) showing H3K27ac signal intensity at differential peaks containing transcription factor motifs for FOXA1, ELF3, ATF3, and TP63. D, Top, inflection plot of superenhancers identified in parental and knockout cells showing relative changes in luminal (blue) and basal (red) superenhancer rankings. The top-ranked superenhancers are labeled in black. Bottom, genome browser views of the ELF3 and TP63 superenhancers.
Next, we examined H3K27ac and H3K4me3 signal enrichment at ATAC-seq peaks by performing ChIP-seq for these marks in RT4 and K2 cells, and we found that 69.1% (RT4) and 71.4% (K2) of accessible chromatin peaks were associated with either H3K27ac or H3K4me3, which mark active enhancers and transcription, respectively (Supplementary. Fig. S5A). Sixty-one percent of H3K27ac peaks were unique to RT4 or K2 cells, indicating an alteration in enhancer usage after KDM6A loss (Fig. 4C). Motif analysis of RT4-specific H3K27ac peaks confirmed enrichment for transcription factors directing urothelial formation and differentiation, whereas K2-specific H3K27ac peaks showed enrichment for the bZIP transcription factors and the basal subtype transcription factor TP63 (Fig. 4C; Supplementary Fig. S5B). H3K27ac signals were stitched and ranked to identify superenhancers, which have been reported to control key aspects of cell identity and cancer biology (39). Superenhancers associated with luminal subtype such as ELF3 and UPK2 were highly ranked in RT4 and decreased after KDM6A deletion (Fig. 4D). Conversely, KDM6A deletion increased the ranking of superenhancers associated with basal subtype such as TP63 and CD44. These data demonstrate that KDM6A loss alters enhancer and superenhancer usage to shift luminal subtype cancers to a basal cell state, and this plasticity in cellular state may permit usage of alternate signaling pathways for cell growth.
KDM6A loss disrupts the FOXA1 transcription circuit in luminal subtype bladder cancer cells
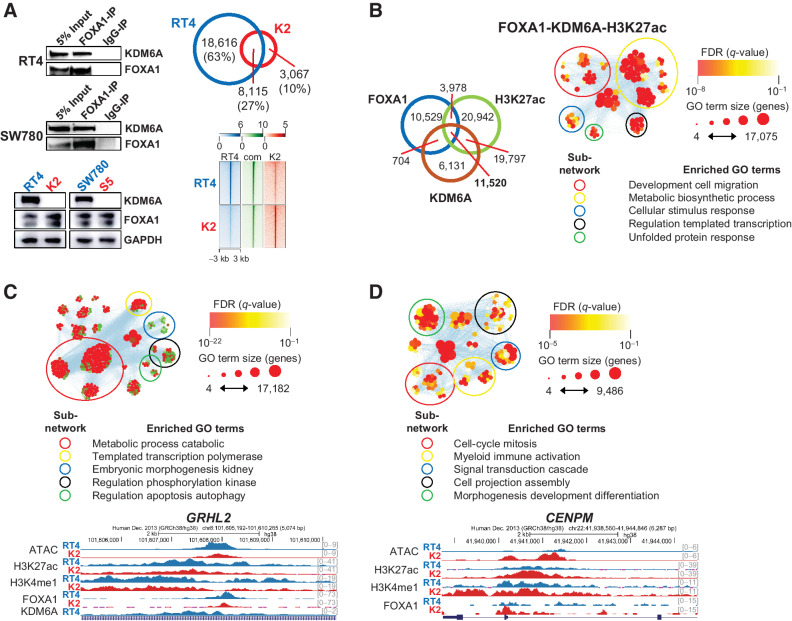
We then proposed that KDM6A interacts directly with luminal subtype transcription factors such as FOXA1 and ELF3, and KDM6A loss disrupts target binding. To explore this hypothesis, we performed coimmunoprecipitation using these transcription factors as bait. When ELF3 was used as bait, KDM6A was not detected (data not shown). However, KDM6A was detected in the two luminal subtype cell lines, RT4 and SW780, when FOXA1 was used as bait (Fig. 5A). Importantly, KDM6A deletion did not affect FOXA1 expression in these cell lines. FOXA1 ChIP-seq in RT4 and K2 cells demonstrated that KDM6A deletion results in a loss of 63% of peaks. Nonetheless, not all FOXA1 target binding was disrupted by KDM6A loss with K2 retaining 8,115 shared FOXA1 peaks and gaining 3,067 private peaks. To identify genomic locations bound by KDM6A, we performed KDM6A CUT&RUN in RT4 cells and found that 90% of peaks overlap with enhancers marked by either H3K27ac or H3K4me1. This finding is consistent with another study that showed the majority of KDM6A binding sites in UMUC1 bladder cancer cells fell within enhancers (12). Forty-six percent of FOXA1 binding sites showed the evidence of KDM6A binding, and 94% of sites bound by both FOXA1 and KDM6A colocalized with enhancers marked by H3K27ac. Enhancers bound by FOXA1 and KDM6A were associated with biological processes that govern development, metabolism, transcription regulation, and stimulus response (Fig. 5B). Integration of FOXA1 ChIP-seq and gene-expression data revealed that FOXA1 binding is associated with the upregulation of genes in both RT4 and K2. RT4-specific FOXA1 peaks controlled biological processes that regulate differentiation, metabolism, and apoptosis, whereas K2-specific FOXA1 peaks were associated with cell-cycle processes, signal transduction cascade, and immune activation (Fig. 5C and D). ATAC and H3K27ac signals in RT4 and K2 agreed with FOXA1 ChIP-seq signals at these private peaks, confirming that FOXA1 controls distinct gene-expression programs in parental and KDM6A knockout cells (Supplementary Fig. S6A). We next assessed phenotypic changes that arise from FOXA1 depletion. Interestingly, FOXA1 depletion led to increased proliferation in parental cells but had the opposite effect in KDM6A knockout cells (Supplementary Fig. S6B). Combined, these data support that KDM6A deletion leads to dysregulation of the FOXA1 transcription circuit by disrupting gene-expression programs associated with development and homeostasis, whereas programs associated with cell cycle, signal transduction, and immune activation remain intact.
Figure 5.
FOXA1 requires KDM6A in luminal subtype cells for the regulation of urothelial identity and homeostasis. A, Top left, coimmunoprecipitation of KDM6A in using FOXA1 as a bait. Bottom left, FOXA1 expression in parental (blue) and KDM6A knockout (red) cells. Top right, Venn diagram showing shared and private FOXA1 peaks in RT4 parental and K2 knockout cells. Bottom right, heat map showing FOXA1 ChIP-seq signal at shared and private peaks. B, Left, Venn diagram showing KDM6A CUT&RUN, FOXA1 ChIP-seq, and H3K27ac ChIP-seq shared and private peaks in RT4 cells. Right, functional enrichment analysis of enhancer peaks bound by both FOXA1 and KDM6A. C, Functional enrichment analysis of FOXA1-bound genes that are upregulated in RT4 cells and genome browser view of the GRHL2 locus. D, Functional enrichment analysis of FOXA1-bound genes that are upregulated in K2 cells and genome browser view of the CENPM locus.
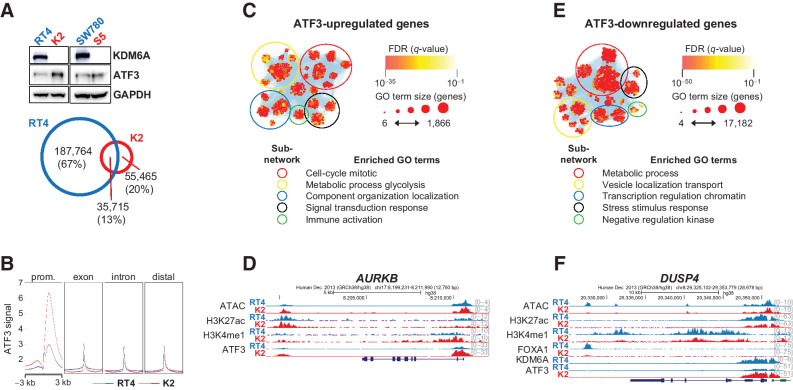
KDM6A loss activates the ATF3 transcription circuit in luminal subtype bladder cancer cells
Next, we focused on examining the changes in bZIP transcription factor binding after KDM6A deletion and how these changes affect gene expression. bZIP transcription factors comprise a large family of related members that can bind as homodimers or heterodimers. Combinatorial binding allows fine-tuning of site specificity and control of gene expression. Because bZIP transcription factors recognize similar motifs, we performed an in vitro transcription factor activity assay and found increased ATF3 activity after KDM6A deletion in both luminal subtype cell lines (Supplementary Fig. S7). ATF3 protein levels remained stable (S5) or increased (K2) after KDM6A deletion, and global analysis of ATF3 binding sites revealed that there were over 3-fold more RT4-specific peaks than K2-specific peaks (Fig. 6A). However, examination of ATF3 binding among different genomic contexts show that it binds weakly at peaks distributed throughout the genome in RT4 (Fig. 6B). After KDM6A deletion, there is a global shift in ATF3 binding away from exons, introns, and distal regions toward proximal promoter sequences, where it binds with high intensity. Whereas ATF3 peaks private to RT4 correlated with areas of inaccessible chromatin (data not shown); overall, there is an increase in chromatin accessibility and H3K27ac ChIP-seq signal at ATF3 peaks private to K2 (Supplementary. Fig. S8A). Nonetheless, ATF3 can act as both a transcriptional activator and repressor, depending on whether it homo- or heterodimerizes (40), and we observed this dual role in K2 cells. Upregulated ATF3 target genes in K2 belonged to regulatory subnetworks controlling biological processes such as cell cycle, signal transduction, and immune activation (Fig. 6C and D). Downregulated ATF3 target genes in K2 cells were involved in processes such as response to negative regulation of receptor tyrosine kinase signaling, cellular stress and stimulus response, and metabolic processes (Fig. 6E and F), and show decreased chromatin accessibility and H3K27ac ChIP-seq signal compared with upregulated ATF3 target genes (Fig. 6F). Interestingly, FOXA1-upregulated targets in RT4 and ATF3-downregulated targets in K2 shared enriched subnetwork terms, and we identified 1,170 ATF3 target genes downregulated in K2 that also harbored FOXA1 peaks in RT4 (Supplementary Data S2). Thus, genes repressed by ATF3 in K2 occur in three contexts (Supplementary Fig. S8B and 8D): contexts 1 and 2 are activated by FOXA1 in RT4, whereas context 3 is not. In the absence of KDM6A, context 1 genes show a complete loss of FOXA1 target binding, whereas context 2 genes show a partial loss of FOXA1 target binding. Compared with 1,170 genes in contexts 1 and 2, there were 787 genes in context 3. These data show that in the absence of KDM6A, ATF3-mediated repression of the FOXA1 regulatory network plays a prominent role in establishing this epigenetic switch. In summary, although FOXA1-dependent urothelial differentiation and homeostasis were disrupted by KDM6A loss, these data suggest that the gain-of-function ATF3 transcription circuit coordinates a gene-expression program that drives cell proliferation.
Figure 6.
ATF3 transcription circuit activation after KDM6A loss. A, Top, ATF3 expression in parental (blue) and KDM6A knockout (red) cells. Bottom, Venn diagram showing shared and private ATF3 peaks in RT4 parental and K2 knockout cells. B, ATF3 CUT&RUN signal at annotated genomic contexts. C, Functional enrichment analysis of ATF3-bound genes that are upregulated in knockout cells. D, Genome browser view of AURKB, a cell-cycle gene activated by ATF3 in knockout cells. E, Functional enrichment analysis of ATF3-bound genes that are downregulated in knockout cells. F, Genome browser view of DUSP4, a MAPK pathway regulator that is repressed in knockout cells.
Functional loss of either KDM6A or FOXA1 creates an epigenetic state that activates the gain-of-function ATF3 transcription circuit
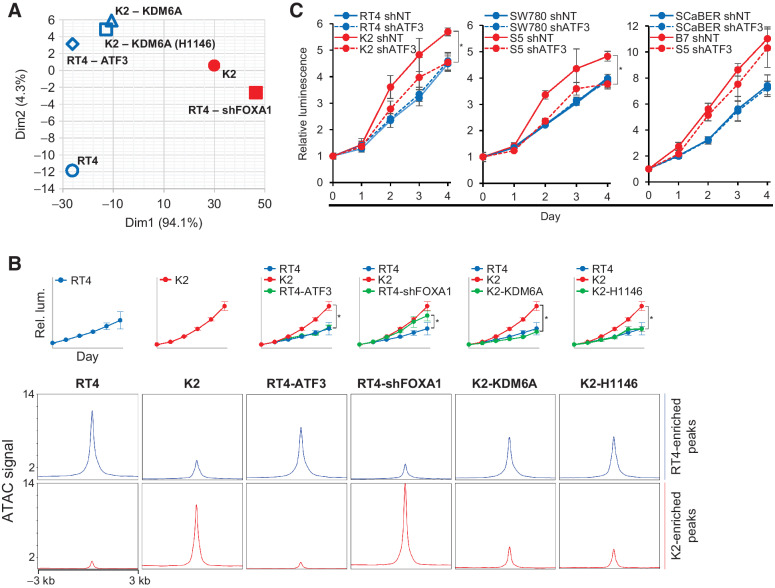
The most salient effect of KDM6A deletion on bladder cancer cells is increased proliferation, leading us to examine how a cell's chromatin state is related to this phenotype. Thus, we ascertained the contribution of each of the critical gene regulatory factors that we identified in determining the cell's proliferation and chromatin state. Although we observed increased ATF3 protein after KDM6A loss (Fig. 6A), principal component analysis of chromatin accessibility data (Fig. 7A) revealed that overexpression of ATF3 in RT4 cells did not significantly alter its chromatin state, which shows that the ATF3 transcription circuit cannot be activated by increasing protein levels alone. On the other hand, the depletion of FOXA1 in RT4 cells results in a chromatin state that is similar to the KDM6A knockout cell line K2, which shows that functional loss of either KDM6A or FOXA1 leads to a similar chromatin state. This chromatin state can be rescued by reintroducing KDM6A or a catalytically inactive mutant KDM6A (H1146A) back into K2 cells. Importantly, these chromatin state alterations were associated with expected changes in cell proliferation (Fig. 7B). In RT4 cells, proliferation was increased by depletion of FOXA1 but not overexpression of ATF3. In K2 cells, overexpression of either KDM6A or catalytically inactive KDM6A-H1146A decreased proliferation to the rate of RT4 cells. Because the gain-of-function ATF3 transcription circuit is active only in KDM6A knockout cells, we next examined whether disrupting this transcription circuit will affect the phenotype of KDM6A knockout cells specifically. Although parental cells showed no detectable changes in growth, ATF3 decreased cell proliferation in KDM6A-knockout luminal subtype cells (K2 and S5) but not basal subtype B7 cells (Fig. 7C). These data demonstrate that the gain-of-function ATF3 transcription circuit is luminal subtype-specific and can be activated only upon loss of KDM6A. Moreover, ATF3 disruption can reverse phenotypic changes induced by KDM6A deletion and does not affect cells with intact KDM6A.
Figure 7.
Activation of the ATF3 transcription circuit can be triggered by KDM6A or FOXA1 loss in luminal bladder cancer cells. A, Principal component analysis of ATAC-seq peaks demonstrating relationship among parental and knockout cells that have undergone the following manipulations: RT4-ATF3, RT4 with ATF3 overexpression; RT4-shFOXA1, RT4 with shRNA targeting FOXA1; K2-KDM6A, K2 with KDM6A overexpression; K2-KDM6A (H1146A), K2 with catalytically inactive KDM6A overexpression. B, Cell proliferation (top) and ATAC signal (bottom) at RT4-enriched (blue) and K2-enriched (red) accessible chromatin peaks. Green curve in cell proliferation plots indicates the growth of modified cell lines with RT4 (blue) and K2 (red) curves displayed for comparison. C, ATF3 depletion decreases cell proliferation in luminal subtype KDM6A knockout cells but not in parental cells. Solid lines, transduction with nontargeting construct; dashed lines, transduction with ATF3-targeting construct. *, P < 0.05.
Discussion
Chromatin modifier gene inactivating mutations occur frequently in bladder cancer, with most series reporting mutation rates between 70% and 80%. Many of these mutations arise in members of the COMPASS-like complex, with up to 60% of cases affected. Mutations in COMPASS-like complex genes occur early in bladder carcinogenesis. In an analysis of 140 stage Ta urothelial carcinomas, KDM6A mutations were present in 52% of cases (4). Although almost all papillomas, a benign urothelial proliferation, contained oncogenic HRAS or KRAS mutations, COMPASS-like complex member mutations in these lesions were rare (41). These data suggest that chromatin modifier gene inactivation constitutes a major pathway by which the urothelium undergoes malignant transformation.
The role of KDM6A inactivation in bladder carcinogenesis remains incompletely elucidated. In this study, we focused on investigating how KDM6A loss alters the epigenetic landscape and transcription circuitry usage of the urothelium to enable cellular transformation. Examination of human bladder cancer chromatin accessibility demonstrated that KDM6A mutation did not lead to global changes in the epigenome; but instead, targeted transcription factor circuitry was disrupted. Specifically, KDM6A-mut cases showed decreased chromatin accessibility in regions containing motifs of transcription factors that confer urothelial identity and increased accessibility in regions containing bZIP transcription factor motifs. Deletion of Kdm6a in organoids derived from mouse urothelium partially disrupted retinoic acid–induced differentiation and increased expression of basal markers. However, Kdm6a deletion alone did not confer tumorigenicity, which required both Kdm6a and Trp53 loss in this model. Similar findings were reported in attempts to delete ARID1A, another chromatin modifier gene commonly mutated in human cancers, in wild-type human gastric corpus organoids (42). In this model system, the deletion of TP53 was required for ARID1A-deficient organoids to undergo malignant transformation. These data support the model that Kdm6a loss results in an altered cellular state, which enables tumor formation in conjunction with either oncogene activation or tumor suppressor loss. One study found that KDM6A antagonizes FGFR3 signaling and suggests that other chromatin modifier gene mutations commonly seen in bladder cancer may also set the stage for tumor progression (12).
Bladder cancers can be broadly categorized as luminal and basal subtypes that express distinct markers and transcription factors. Although KDM6A mutations appear to be enriched in luminal subtype muscle-invasive disease, basal subtype cancers also harbor this alteration. Unlike pancreatic cancer where KDM6A mutations lead to activation of superenhancers regulated by transcription factors dictating squamous differentiation, we found distinct effects on differentiation markers in luminal (RT4 and SW780) versus basal (SCaBER) subtype bladder cancer cell lines. Basal markers showed increased expression in the luminal subtype cells but decreased expression in the basal subtype cells after KDM6A deletion, which suggests dysregulation of subtype identity. This observation is supported by the chromatin accessibility changes, which showed loss of peaks associated with differentiation-associated transcription factors after KDM6A deletion in both luminal and basal subtype cells. Some of these transcription factors have been reported to direct aggressive behavior, but it is unclear how the function of these specific circuits may be altered when KDM6A is lost. Nonetheless, our findings show that KDM6A deletion increases proliferation, regardless of bladder cancer molecular subtype.
FOXA1 has been shown to play a critical role in maintaining the luminal subtype, and loss of FOXA1 expression has been observed in cancers with squamous differentiation and in late-stage disease (43–45). FOXA1 can bind nucleosomal DNA and recruit coactivators, corepressors, and chromatin modifiers to initiate a transcriptional program in silent chromatin during development; however, we found that the FOXA1 regulatory network that drives luminal differentiation in bladder cancer requires KDM6A function. In K2 luminal subtype knockout cells, 63% of FOXA1 peaks were lost compared with parental cells despite no change in FOXA1 expression. This observation was not surprising, given that FOXA1 interacts with KDM6A in luminal subtype cancer cells as demonstrated by coimmunoprecipitation. These data are consistent with another study in breast cancer cells showing an interaction between FOXA1 and KMT2C, another key component of the COMPASS-like complex (46). FOXA1 peaks lost in K2 cells corresponded to active enhancers associated with urothelial identity, and KDM6A loss has been reported to disrupt the maintenance of active enhancers in embryonic stem cells (38). Our chromatin accessibility and gene-expression data also support FOXA1 shifting to a transcription program that mediates cell proliferation in the absence of KDM6A. Consistent with these findings, depletion of FOXA1 in K2 cells reduced cell proliferation. FOXA1 appears to play opposite roles depending on whether KDM6A is present; nonetheless, FOXA1 has been identified as a potential oncogene in other contexts. For example, Foxa1 is required for the maintenance of urothelial hyperplasia in transgenic mice that overexpress HRAS in the urothelium (47). In human prostate tissue, FOXA1 can reprogram the androgen receptor cistrome to promote malignant transformation, and FOXA1 mutations can further contribute to lineage plasticity during disease progression (48, 49). In bladder carcinogenesis, FOXA1 function appears to depend on the epigenetic context and may also contribute to transcriptional plasticity.
bZIP transcription factors have long been implicated in mediating a number of cellular processes related to carcinogenesis such as cell growth, survival, differentiation, stress response, and apoptosis since their discovery as viral oncoproteins. More recently, bZIP transcription factors have also been shown to participate in a gene regulatory network modulating cancer inflammation (50). Our data demonstrate that in luminal subtype bladder cancer cells, the bZIP transcription factor ATF3 directs an oncogenic regulatory circuit that drives cell proliferation and immune activation, inhibits urothelial differentiation, and alters cellular metabolism through its activator and repressor activities. Importantly, the combination of FOXA1 and KDM6A restrains this gain-of-function circuit in luminal subtype bladder cancer cells. FOXA1 depletion or KDM6A loss triggers an epigenetic switch that allows the ATF3 regulatory circuit to function. In the presence of FOXA1 and KDM6A, forced overexpression of ATF3 does not increase cell proliferation or alter accessible chromatin peaks. Disruption of either FOXA1 or KDM6A function induces a similar epigenetic state that is characterized by ATF3 repositioning to gene promoters. In KDM6A knockout cells, 60% of the ATF3 repressed targets were FOXA1-activated targets in wild-type cells, highlighting the switch from urothelial differentiation and identity to proliferation. Depletion of ATF3 reversed the cell proliferation phenotype in KDM6A knockout cells but had no discernible effect on cell proliferation in parental cells. Importantly, we did not observe the activation of the ATF3 regulatory circuit in KDM6A-deficient basal subtype cancer cells. Chromatin accessibility data for B7 cells showed enrichment for a number of bZIP transcription factors but not ATF3. Additionally, ATF3 depletion in B7 did not reduce cell proliferation. Although KDM6A deletion leads to cell proliferation in luminal and basal subtype bladder cancer cells, the regulatory circuits affected appear to be distinct.
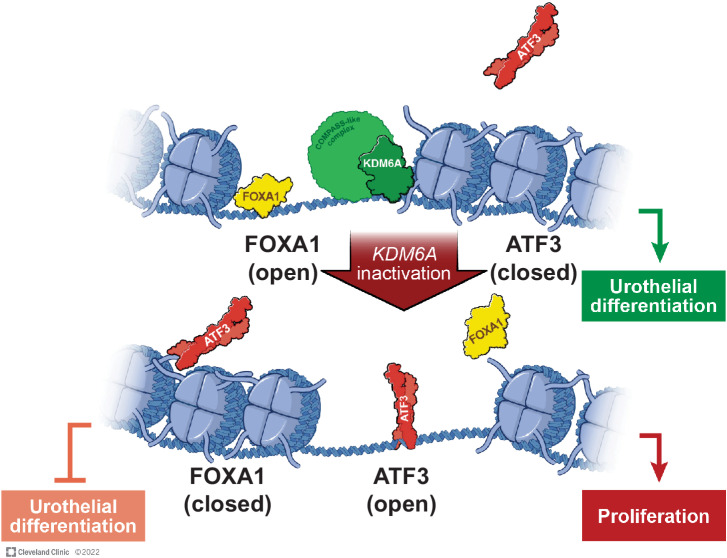
In summary, we show that KDM6A disruption triggers an epigenetic switch that perturbs urothelial identity and induces cell proliferation during bladder carcinogenesis (Fig. 8). In luminal subtype bladder cancer cells, disruption of the FOXA1 regulatory circuit through either depletion of FOXA1 or its binding partner, KDM6A, activates the ATF3 protumorigenic transcription program. ATF3 depletion reduced cell proliferation in KDM6A knockout cells but not wild-type cells, which suggests that this gain-of-function transcription circuit can be targeted directly rather than attempting to reconstitute KDM6A function in mutant cancers. KDM6A-mutant bladder cancers may hijack transcription circuits other than ATF3 to drive cell proliferation in different molecular subtypes. Identifying these alternate gain-of-function circuits will enable the discovery of subtype-specific epigenetic therapies.
Figure 8.
Proposed model of the epigenetic switch that is triggered upon KDM6A loss in luminal subtype bladder cancers. Reprinted with permission, Cleveland Clinic Foundation ©2022. All Rights Reserved.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Figure Legends
Supplementary Table S1
List of TCGA cases used in ATAC-seq analysis
ATF3 target genes downregulated in K2 that harbor FOXA1 peaks in RT4
Acknowledgments
This study was supported by NIH grant K08CA237842 (B.H. Lee), Sidney Kimmel Foundation Kimmel Scholars Award (B.H. Lee), Bladder Cancer Advocacy Network Young Investigator Award (B.H. Lee), and VeloSano Bike to Cure Pilot Award (B.H. Lee).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
V. Makarov reports a patent for EP3090066A2 issued. J.K. Bolzenius reports personal fees from Pfizer outside the submitted work. G.V. Iyer reports grants and personal fees from Bayer, Janssen, Mirati Therapeutics, Flare Therapeutics, personal fees from Basilea Pharmaceutica, Gilead Sciences, LOXO Oncology at Lilly, and Lynx Group, and grants from Debiopharm Group, Seattle Genetics, and Aadi Biosciences outside the submitted work. B. Ren reports other support from Arima Genomics, Inc. and Epigenome Technologies, Inc. outside the submitted work. T.A. Chan reports other support from Gritstone Bio, grants and personal fees from BMS, Illumina, and grants from Pfizer, AstraZeneca, Varian, and Gilead outside the submitted work. V.K. Arora reports grants from Damon Runyon Cancer Research Foundation during the conduct of the study and personal fees from Bristol Myers Squibb outside the submitted work. D.B. Solit reports personal fees from Pfizer, Loxo/Lilly Oncology, Vividion Therapeutics, Scorpion Therapeutics, Fore Therapeutics, BridgeBio, Fog Pharma, and Rain Therapeutics outside the submitted work. B.H. Lee reports grants from NIH, Sidney Kimmel Foundation, Bladder Cancer Advocacy Network, and VeloSano Bike to Cure during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
H. Qiu: Resources, formal analysis, methodology, writing–original draft. V. Makarov: Software, formal analysis, validation, investigation, methodology. J.K. Bolzenius: Resources, validation, investigation, methodology. A. Halstead: Resources, formal analysis, validation, investigation. Y. Parker: Formal analysis, validation, investigation, methodology. A. Wang: Software, methodology. G.V. Iyer: Resources. H. Wise: Validation, investigation. D. Kim: Validation, investigation. V. Thayaparan: Validation, investigation. D.J. Lindner: Methodology. G. Haber: Resources. A.H. Ting: Conceptualization, methodology. B. Ren: Conceptualization, software, methodology. T.A. Chan: Software. V. Arora: Conceptualization, methodology. D.B. Solit: Conceptualization, methodology. B.H. Lee: Conceptualization, software, formal analysis, supervision, funding acquisition, methodology, writing–original draft, writing–review and editing.
References
- 1. Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet 2011;43:875–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell 2017;171:540–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hedegaard J, Lamy P, Nordentoft I, Algaba F, Hoyer S, Ulhoi BP, et al. Comprehensive transcriptional analysis of early-stage urothelial carcinoma. Cancer Cell 2016;30:27–42. [DOI] [PubMed] [Google Scholar]
- 4. Hurst CD, Alder O, Platt FM, Droop A, Stead LF, Burns JE, et al. Genomic subtypes of non-invasive bladder cancer with distinct metabolic profile and female gender bias in KDM6A mutation frequency. Cancer Cell 2017;32:701–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pietzak EJ, Bagrodia A, Cha EK, Drill EN, Iyer G, Isharwal S, et al. Next-generation sequencing of nonmuscle invasive bladder cancer reveals potential biomarkers and rational therapeutic targets. Eur Urol 2017;72:952–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cenik BK, Shilatifard A. COMPASS and SWI/SNF complexes in development and disease. Nat Rev Genet 2021;22:38–58. [DOI] [PubMed] [Google Scholar]
- 7. Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17:487–500. [DOI] [PubMed] [Google Scholar]
- 8. Valencia AM, Kadoch C. Chromatin regulatory mechanisms and therapeutic opportunities in cancer. Nat Cell Biol 2019;21:152–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 2009;41:521–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive characterization of cancer driver genes and mutations. Cell 2018;173:371–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim K, Hu W, Audenet F, Almassi N, Hanrahan AJ, Murray K, et al. Modeling biological and genetic diversity in upper tract urothelial carcinoma with patient derived xenografts. Nat Commun 2020;11:1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barrows D, Feng L, Carroll TS, Allis CD. Loss of UTX/KDM6A and the activation of FGFR3 converge to regulate differentiation gene-expression programs in bladder cancer. Proc Nat Acad Sci U S A 2020;117:25732–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ler LD, Ghosh S, Chai X, Thike AA, Heng HL, Siew EY, et al. Loss of tumor suppressor KDM6A amplifies PRC2-regulated transcriptional repression in bladder cancer and can be targeted through inhibition of EZH2. Sci Transl Med 2017;9:eaai8312. [DOI] [PubMed] [Google Scholar]
- 14. Shi B, Li W, Song Y, Wang Z, Ju R, Ulman A, et al. UTX condensation underlies its tumour-suppressive activity. Nature 2021;597:726–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Halstead AM, Kapadia CD, Bolzenius J, Chu CE, Schriefer A, Wartman LD, et al. Bladder-cancer-associated mutations in RXRA activate peroxisome proliferator-activated receptors to drive urothelial proliferation. Elife 2017;6:e30862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al-Ahmadie HA, Iyer G, Lee BH, Scott SN, Mehra R, Bagrodia A, et al. Frequent somatic CDH1 loss-of-function mutations in plasmacytoid variant bladder cancer. Nat Genet 2016;48:356–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Amemiya HM, Kundaje A, Boyle AP. The ENCODE blacklist: Identification of problematic regions of the genome. Sci Rep 2019;9:9354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 2017;14:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018;34:i884–i90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012;9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, et al. Twelve years of SAMtools and BCFtools. Gigascience 2021;10:giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 25. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shao Z, Zhang Y, Yuan GC, Orkin SH, Waxman DJ. MAnorm: a robust model for quantitative comparison of ChIP-Seq data sets. Genome Biol 2012;13:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Khan A, Mathelier A. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinf 2017;18:287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010;38:576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bentsen M, Goymann P, Schultheis H, Klee K, Petrova A, Wiegandt R, et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun 2020;11:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Corces MR, Granja JM, Shams S, Louie BH, Seoane JA, Zhou W, et al. The chromatin accessibility landscape of primary human cancers. Science 2018;362:eaav1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 2010;28:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol 2019;37:907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res 2007;35:W193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, et al. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 2019;14:482–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc 2013;8:2502–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013;153:320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang SP, Tang Z, Chen CW, Shimada M, Koche RP, Wang LH, et al. A UTX-MLL4-p300 transcriptional regulatory network coordinately shapes active enhancer landscapes for eliciting transcription. Mol Cell 2017;67:308–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hai T, Wolfgang CD, Marsee DK, Allen AE, Sivaprasad U. ATF3 and stress responses. Gene Expr 1999;7:321–35. [PMC free article] [PubMed] [Google Scholar]
- 41. Isharwal S, Hu W, Sarungbam J, Chen YB, Gopalan A, Fine SW, et al. Genomic landscape of inverted urothelial papilloma and urothelial papilloma of the bladder. J Pathol 2019;248:260–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lo YH, Kolahi KS, Du Y, Chang CY, Krokhotin A, Nair A, et al. A CRISPR/Cas9-Engineered ARID1A-Deficient human gastric cancer organoid model reveals essential and nonessential modes of oncogenic transformation. Cancer Discov 2021;11:1562–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Warrick JI, Walter V, Yamashita H, Chung E, Shuman L, Amponsa VO, et al. FOXA1, GATA3 and PPAR cooperate to drive luminal subtype in bladder cancer: A molecular analysis of established Human Cell Lines. Sci Rep 2016;6:38531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. DeGraff DJ, Clark PE, Cates JM, Yamashita H, Robinson VL, Yu X, et al. Loss of the urothelial differentiation marker FOXA1 is associated with high grade, late stage bladder cancer and increased tumor proliferation. PLoS One 2012;7:e36669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Osei-Amponsa V, Buckwalter JM, Shuman L, Zheng Z, Yamashita H, Walter V, et al. Hypermethylation of FOXA1 and allelic loss of PTEN drive squamous differentiation and promote heterogeneity in bladder cancer. Oncogene 2020;39:1302–17. [DOI] [PubMed] [Google Scholar]
- 46. Jozwik KM, Chernukhin I, Serandour AA, Nagarajan S, Carroll JS. FOXA1 directs H3K4 monomethylation at enhancers via recruitment of the methyltransferase MLL3. Cell Rep 2016;17:2715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yee CH, Zheng Z, Shuman L, Yamashita H, Warrick JI, Wu XR, et al. Maintenance of the bladder cancer precursor urothelial hyperplasia requires FOXA1 and persistent expression of oncogenic HRAS. Sci Rep 2019;9:270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pomerantz MM, Li F, Takeda DY, Lenci R, Chonkar A, Chabot M, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet 2015;47:1346–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, Zhang Z, et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 2019;571:408–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ji Z, He L, Regev A, Struhl K. Inflammatory regulatory network mediated by the joint action of NF-kB, STAT3, and AP-1 factors is involved in many human cancers. Proc Natl Acad Sci U S A 2019;116:9453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Figure 8
Supplementary Figure Legends
Supplementary Table S1
List of TCGA cases used in ATAC-seq analysis
ATF3 target genes downregulated in K2 that harbor FOXA1 peaks in RT4