Abstract
High citrus consumption may increase melanoma risk; however, little is known about the biological mechanisms of this association, or whether it is modified by genetic variants. We conducted a genome-wide analysis of gene-citrus consumption interactions on melanoma risk among 1,563 melanoma cases and 193,296 controls from the UK Biobank. Both the 2-degrees-of-freedom (df) joint test of genetic main effect and gene-environment (G-E) interaction and the standard 1-df G-E interaction test were performed. Three index SNPs (lowest p-value SNP among highly correlated variants [r2 >0.6]) were identified from among the 365 genome-wide significant 2-df test results (rs183783391 on chromosome 3 [MITF], rs869329 on chromosome 9 [MTAP], rs11446223 on chromosome 16 [DEF8]). Although all three were statistically significant for the 2-df test (4.25e-08, 1.98e-10, and 4.93e-13, respectively), none showed evidence of interaction according to the 1-df test (p=0.73, 0.24, 0.12, respectively). Eight non-index, 2-df test significant SNPs on chromosome 16 were significant (p<.05) according to the 1-df test, providing evidence of citrus-gene interaction. Seven of these SNPs were mapped to AFG3L1P (rs199600347, rs111822773, rs113178244, rs3803683, rs73283867, rs78800020, rs73283871), and one SNP was mapped to GAS8 (rs74583214). We identified several genetic loci that may elucidate the association between citrus consumption and melanoma risk. Further studies are needed to confirm these findings.
Keywords: citrus, melanoma, GWAS, gene-environment interaction
Introduction
Incidence of melanoma, the most deadly form of skin cancer, is increasing faster than any other cancer.1,2 The rapid observed growth in melanoma incidence is not artifactual,3 and with global rates increasing by 3–7% per year,2 incidence is doubling every 10–20 years.4 Melanoma is also associated with considerable mortality. In the US, there are projected to be 7,180 melanoma deaths in 2021,5 and, in the UK, melanoma mortality has increased by 144% since the early 1970s.6 Globally, melanoma is the fastest growing cause of cancer death save for non-Hodgkin’s lymphoma, lung cancer in women, and testicular cancer.1
The etiology of melanoma is multifactorial, involving environmental risk factors, genetic variants, and the interactions between them. High citrus consumption is an environmental factor that has received increased attention in recent years due to citrus products’ natural abundance of psoralen, a type of furocoumarin known to be photosensitizing and photocarcinogenic7 in mice8,9 and in humans.10,11 Research to this end has yielded inconsistent findings, as results from the Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS)12 have demonstrated an overall positive association between citrus consumption and melanoma risk, and findings from the Women’s Health Initiative (WHI)13 and the European Prospective Investigation into Cancer and Nutrition (EPIC)14 have suggested positive associations between citrus consumption and melanoma risk among those who spent the most time outdoors in summer and those with the highest consumption of citrus fruit, respectively. Our own previous research in the UK Biobank (UKBB) has also suggested a positive association, finding a 63% increased risk of melanoma among participants in the highest category of total citrus intake.15 The inconsistency of these findings is possibly due to heterogeneity of study populations and/or differences in methods of quantifying citrus consumption. None of these studies, however, have investigated the role of possible nutrient-gene interaction. It has also been established that an individual’s genotype may influence melanoma risk. Previous candidate gene studies and genome-wide association studies (GWAS) have identified several genes that are associated with melanoma risk,16,17 but there is currently no evidence as to whether genetic variation modulates the association between melanoma risk and citrus consumption.
As it is critical to identify nutrient-gene interactions that may influence the etiology and pathophysiology of melanoma, the purpose of the current study was to conduct, to our knowledge, the first genome-wide analysis of citrus-gene interactions on risk of melanoma. We believe the results of the current study will increase knowledge of melanoma pathology, identify genetic variants and gene-environment (G-E) interactions underlying photocarcinogenesis in melanoma, and, upon further validation, could serve as an empirical basis for the development of a skin cancer prediction model, eventually leading to improved precision prevention.
Methods
Data Collection, Genotyping, Imputation, and Quality Control
A more detailed description of UKBB genomic methodology is described elsewhere.18 The UKBB is a large, prospective cohort with extensive genomic data collected for all approximately 500,000 participants. Participants were all from the UK and aged 40–69 years at recruitment between 2006–2010. Each participant was assessed at one of 22 assessment centers throughout the UK and provided blood samples from which DNA was extracted and sent to Affymetrix Research Services Laboratory for genotyping. Upon receipt at Affymetrix, samples were then processed on the GeneTitan Multi-Channel Instrument in 96-well plates containing 94 UKBB samples and two control samples from the 1000 Genomes Project. Genotyping was carried out in 106 batches of approximately 4,700 samples. The first 50,000 participants were genotyped on the UK BiLEVE Axiom Array, and the following 450,000 participants were genotyped using the UK Biobank Axiom Array. Although different arrays were used, they share 95% of the same marker content, and only markers present on both arrays were used. Autosome phasing was performed using SHAPEIT3, with the 1000 Genomes phase 3 dataset as the reference panel. The UKBB was also imputed using merged UK10K and 1000 Genomes phase 3 reference panels. Principal components (PCs) were computed via an algorithm in which 407,219 high quality, unrelated samples and 147,604 high quality markers were used and pruned to minimize linkage disequilibrium.
Poor quality markers were identified using statistical tests to check for consistency of genotype calling. These included tests for batch effects, plate effects, departures from Hardy-Weinberg equilibrium, sex effects, array effects, and discordance across control replicates. Markers failing at least one test in a batch had the genotype calls in that batch set as missing, and if a marker was not reliable across all batches, it was excluded altogether. Markers were also removed if they had at least a 5% overall missing rate or if they had a minor allele frequency <0.0001.
Poor quality samples were identified using missing rate and heterozygosity that were computed using 605,876 high quality autosomal markers typed on both arrays. Samples that were outliers for heterozygosity or missing rate were removed, as were a small number of samples identified as duplicates and approximately 10 samples that were mishandled in the laboratory. Overall, these filters and exclusions resulted in a dataset with 93,095,623 autosomal SNPs, short indels and large structural variants in 487,442 samples.
Citrus Intake, Melanoma Ascertainment, and Measurement of Covariates
Citrus consumption in the UKBB were collected via five ‘rounds’ of 24-hour recall questionnaires. The five ‘rounds’ took place between April 2009-September 2010, February-April 2011, June-September 2011, October-December 2011, and April-June 2012, respectively, with the first round taking place in the assessment center and subsequent rounds administered electronically. This electronic 24-hour recall yielded a mean Spearman’s correlation coefficient of 0.6 (range 0.5–0.9) compared with an interviewer-assisted 24-hr recall.19 Although collection of citrus consumption data was more recent, we are confident using it as a proxy for past citrus consumption in the UK due to data from other wealthy nations suggesting that average fruit intake is staying consistent over time,20,21 and due to 82% of UKBB cases reporting the same or adjacent category of fresh fruit consumption between the first and last dietary assessments.22 A total of n=210,126 participants completed at least one 24-hour dietary recall to provide complete citrus consumption data. Consumption of orange, grapefruit, satsuma, orange juice, and grapefruit juice were categorized into four intake groups: 0, half, >half-1, and >1 serving. A cumulative average of this citrus consumption over the 5 ‘rounds’ of nutritional data collection was used to assess total citrus consumption. For this cumulative average, total citrus consumption was categorized into the following groups: ‘none’, ‘>0-half a serving’, ‘half-1 serving’, >1–2 servings’, ‘>2 servings’. Participants were not required to complete all five ‘rounds’ of data collection for inclusion in the analysis, and any untaken ‘rounds’ were treated as missing and therefore had no impact on participants’ cumulative average for citrus consumption. ICD codes (C43.0–9) were used to identify melanoma outcome data. Cases were acquired via these codes, which were linked with national registries that obtain cancer diagnosis data from various sources (such as hospitals, nursing homes, death certificates, general practices, etc.) and retrospectively dated back to the early 1970s. The current study focused on melanomas of the skin and case ascertainment was not limited to primary melanoma cases. Age of the participants was derived based on date of birth provided at the assessment center. It refers to participants’ age when they first visited the center, truncated to whole year. A participant’s sex was acquired from the National Health Service at recruitment, and, in some cases, updated by the participant. Due to its noted strength as a melanoma risk factor,23 tanning ability was also included as a covariate. Tanning ability was measured by asking “What would happen to your skin if it was repeatedly exposed to bright sunlight without any protection?”, with the following as our coded responses for this variable: “get very tanned”, “get moderately tanned”, “get mildly or occasionally tanned”, and “never tan, only burn”. There were no missing data for participants’ age or sex, and missing data for tanning ability was not substantial (1.6%), therefore it was simply excluded from the analysis.
Statistical Analysis
Of the n=210,126 with complete citrus data, n=11,162 non-Caucasian participants (5.3%) were excluded as done in previous analyses12,13 due to low melanoma incidence in ethnic minorities.24 Genetic data for another n=4,105 (2.0%) were also excluded due to quality control procedures, leaving n=194,859 Caucasian UKBB participants available for this analysis. From this sample, we included SNPs with a minor allele frequency >0.01 and an imputation quality score >0.3. After applying these filters, a total of 9,981,017 SNPs were analyzed in this study.
We created a quantile-quantile (QQ) plot and calculated lambda (λ) to assess genomic inflation. Interactions between citrus consumption and genetic variants were evaluated by performing the joint 2-degrees-of-freedom (df) test of genetic main effect and G-E interaction. The 2-df joint test performed a test of SNP marginal effects and their interaction with citrus consumption by conducting likelihood ratio tests between the full logistic regression model (Logit(Pr(melanoma=1)) = b0 + b1*total citrus consumption + b2*SNP + b3*total citrus consumption*SNP + b4*age + b5*gender + b6*tanning ability +b7*PC1+ … +b21*PC15) and the reduced model (excluding SNP and total citrus consumption*SNP). This approach has been found to be much more powerful than the standard test and better suited for discovering new genetic markers and investigating new potential G-E interactions, making it a valuable tool for large discovery scans in which true gene-environment associations are unknown.25,26 Of the significant results (p<5e-08), one index SNP was selected (SNP with the lowest p-value) from among highly correlated variants (r2 >0.6).
Next, to gauge whether the joint 2-df test results were being primarily driven by interaction or genetic main effect, we also performed the 1-df likelihood ratio test comparing the above full model vs. the model without the multiplicative interaction term. This test was performed on all significant SNPs from the 2-df test. SAS version 9.4 (SAS Institute, Cary, NC), PLINK version 2.0 (www.cog-genomics.org/plink/2.0/), and R version 3.6.0 (The R Foundation for Statistical Computing, Vienna, Austria) were used for the described analyses. All data storage and programming were performed in Karst, a high-performance computing cluster at Indiana University.
Results
Our analyses were conducted on a total of 194,859 Caucasian UKBB participants, including 1,563 cases and 193,296 controls. As illustrated in Figure 1, the genomic variants in our analysis have a fairly normal chi-squared distribution with little evidence of genomic inflation (λ=1.031). After adjusting for age, gender, tanning ability, and the first 15 PCs, the 2-df joint test revealed a total of 365 SNPs that were significant at a p-value <5e-08 (Table S1), including one on chromosome three, 270 on chromosome nine, and 94 on chromosome 16. Of these, 3 index SNPs were identified: rs183783391 (p=4.25e-08) on chromosome 3 (3p13), rs869329 (p=1.98e-10) on chromosome 9 (9p21.3), and rs11446223 (p=4.93e-13) on chromosome 16 (16q24.3) (Table 1). These three SNPs are mapped to MITF, MTAP, and DEF8, respectively. Although p-values for these index SNPs reached statistical significance for the joint test, neither rs183783391 (p=0.73), rs869329 (p=0.24), or rs11446223 (p=0.12) showed evidence of interaction with citrus consumption according to the conventional 1-df test. Evidence of interaction (p<0.05) was observed for 8 of the 365 SNPs found to be significant (p<5e-08) by the 2-df joint test (Table 2). These SNPs, all on chromosome 16, included rs199600347, rs111822773, rs113178244, rs3803683, rs73283867, rs78800020, and rs73283871 mapped to AFG3L1P and rs74583214 mapped to GAS8.
Figure 1.
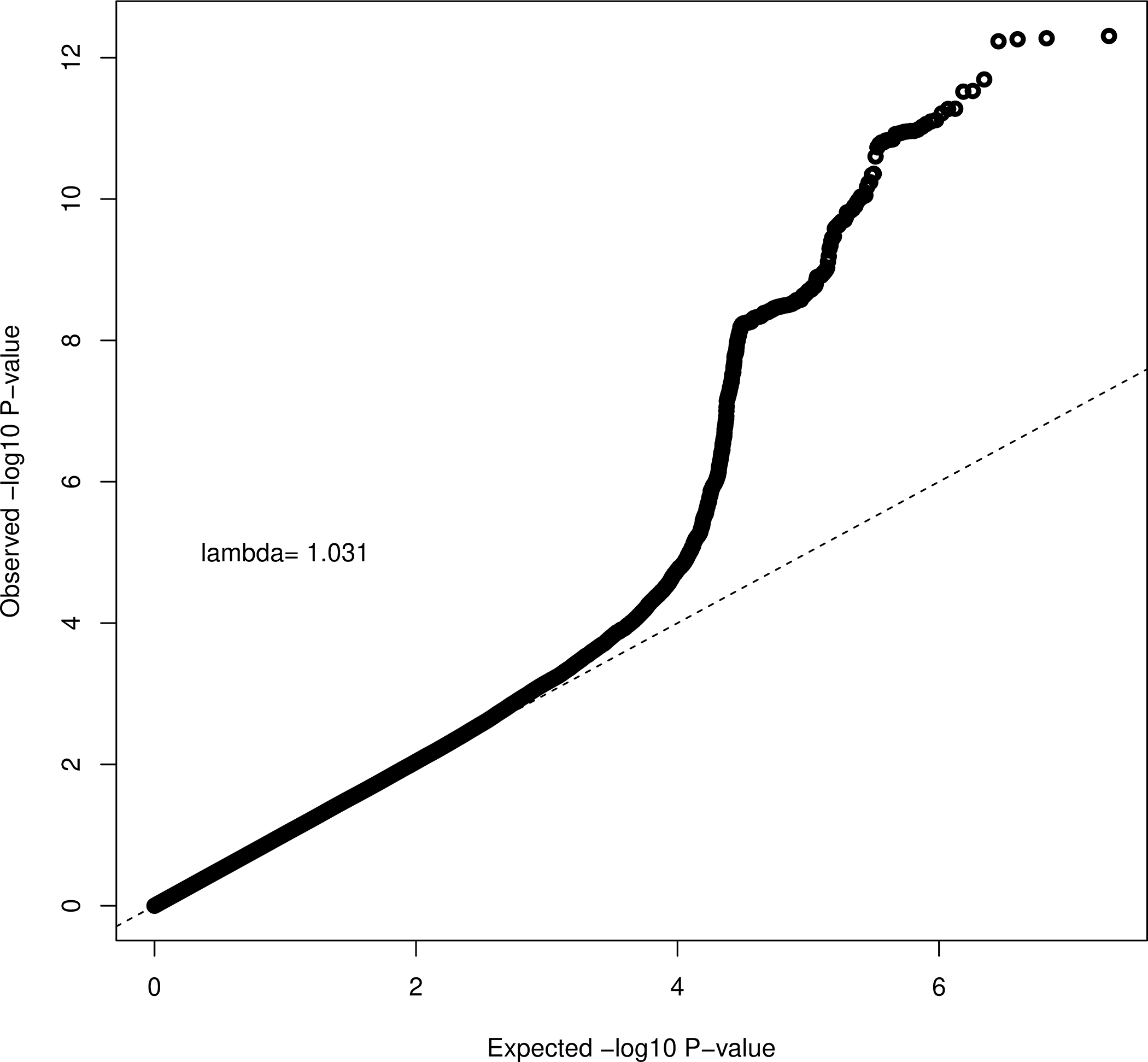
QQplot of Joint 2DF Test across 22 Chromosomes
Table 1.
Index single nucleotide polymorphisms (SNPs) resulting from a genome-wide analysis of gene-citrus consumption interaction on melanoma risk.
| SNP ID | Chr | Position | Region | Associated gene | Minor allele | MAF | Info score | p-2DF | p-1DF |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| rs183783391 | 3 | 69950451 | 3p13 | MITF | T | 0.013 | 0.95 | 4.25e-08 | 0.73 |
| rs869329 | 9 | 21804693 | 9p21.3 | MTAP | A | 0.48 | 0.99 | 1.98e-10 | 0.24 |
| rs11446223 | 16 | 90022484 | 16q24.3 | DEF8 | GA | 0.17 | 0.99 | 4.93e-13 | 0.12 |
Abbreviations: Chr, chromosome; Ref allele, reference allele; MAF, minor allele frequency; Info score, imputation quality score; p-1DF, p-value for the conventional 1 degree-of-freedom test; p-2DF, p-value for the 2 degree-of-freedom joint test
Table 2.
Non-index SNPs significant for the joint 2 degrees-of-freedom test and with evidence of citrus-gene interaction.
| SNP ID | Chr | Position | Region | Closest gene | Minor allele | MAF | Info score | p-2DF | p-1DF |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| rs199600347 | 16 | 90052245 | 16q24.3 | AFG3L1P | TTA | 0.10 | 0.91 | 3.53e-10 | 0.023 |
| rs111822773 | 16 | 90054089 | 16q24.3 | AFG3L1P | C | 0.11 | 0.98 | 3.62e-09 | 0.043 |
| rs113178244 | 16 | 90056195 | 16q24.3 | AFG3L1P | G | 0.11 | 0.99 | 5.00e-09 | 0.049 |
| rs3803683 | 16 | 90060281 | 16q24.3 | AFG3L1P | C | 0.17 | 0.99 | 3.55e-10 | 0.047 |
| rs73283867 | 16 | 90066260 | 16q24.3 | AFG3L1P | G | 0.096 | 1.00 | 9.54e-12 | 0.050 |
| rs78800020 | 16 | 90067136 | 16q24.3 | AFG3L1P | C | 0.096 | 1.00 | 1.12e-11 | 0.045 |
| rs73283871 | 16 | 90067202 | 16q24.3 | AFG3L1P | C | 0.096 | 1.00 | 1.09e-11 | 0.045 |
| rs74583214 | 16 | 90110798 | 16q24.3 | GAS8 | T | 0.098 | 0.96 | 5.57e-09 | 0.043 |
Abbreviations: Chr, chromosome; Ref allele, reference allele; MAF, minor allele frequency; Info score, imputation quality score; p-1DF, p-value for the conventional 1 degree-of-freedom test; p-2DF, p-value for the 2 degree-of-freedom joint test
Note: all SNPs are in strong linkage disequilibrium with each other (R2 0.60–1)
Discussion
Although noted for challenges and some inconsistent/inconclusive results,27,28 genome-wide studies to identify G-E interactions influential to common traits and cancer are of critical importance and some have provided evidence of environment-associated genetic effects.29–31 Although previous epidemiological evidence has suggested that high citrus consumption may increase melanoma risk,12–15 little is known regarding the genetics of citrus metabolism, and the potential role of genetic variants involved in citrus-associated melanoma risk had been previously unexplored. In the current analysis, we tested the hypothesis that the increased risk of melanoma associated with high citrus consumption is an effect of G-E interaction. We identified three index SNPs that were highly significant for the joint 2-df test: rs183783391 (MITF), rs869329 (MTAP), and rs11446223 (DEF8).
Mapped to MITF on chromosome 3 (position 69950451), rs183783391 is an intronic variant located between the first and second exon. This SNP is not within a conserved motif or associated with any MITF regulatory elements. Other melanoma susceptibility variants have also been identified on chromosome 3, including rs3950296 mapped to TERC and rs149617956 mapped to the p.E318K functional variant on MITF;32,33 however, these variants are not in linkage disequilibrium with rs183783391 (r2 ≤ 0.2). Located at 3p13, MITF (microphthalmia-associated transcription factor) plays a major role in the development, function, and survival of pigment-producing melanocytes.34,35 As melanoma tumors are derived from melanocytes, MITF has been recognized for its role in driving melanoma progression and has been shown to regulate senescence, differentiation, proliferation, apoptosis, and migration of melanoma cells.35,36 Additionally, due to its role in the transformation of immortalized melanocytes and its expression in conjunction with BRAFV600E, MITF demonstrates oncogenic properties.37,38 Genetic epidemiology studies have also linked MITF with melanoma risk. Previous research has elucidated the role of MITF in human pigmentation,32,39 a known melanoma risk factor.23 Additional evidence has directly linked MITF with human melanoma risk, highlighting its role in familial melanoma.40 Therefore, although rare, we hypothesize that rs183783391 may play a role in melanoma etiology via its association with one of these mechanisms.
Mapped to MTAP on chromosome 9, rs869329 is an intronic variant at position 21804693 between the first and second exon. This SNP is not within a conserved motif of the genome or within any regulatory elements. Many melanoma susceptibility variants have been previously identified on chromosome 9, with particularly strong associations mapped to CDKN2A,32,41,42 a gene linked to an estimated 40% of familial melanoma cases.43 As MTAP is in close physical proximity to CDKN2A and rs869329 is in high linkage disequilibrium with previously identified CDKN2A SNPs (r2=0.76–1.00), it is possible that rs869329 serves as a CDKN2A tag. However, MTAP and CDK2NA are frequently co-deleted, causing MTAP loss to often be attributed to CDKN2A,44 and evidence has suggested that MTAP may have tumor suppressor function independent of CDK2NA.45,46 Previous genome-wide research has also found melanoma risk to be linked with MTAP, whether directly,47 or through its association with cutaneous nevi,44,48,49 another known melanoma risk factor.50 Additionally, a study by McMeniman et al. specifically found that single primary melanoma patients with melanoma at a site of visible UV-damage were significantly more likely to carry rs869329 relative to controls (odds ratio = 1.4 [CI=1.1–1.7]).51 MTAP (methylioadenosin phosphorylase), located at 9p21.3, is critical to polyamine metabolism and the salvage of adenine and methionine. It also acts in catalyzing the phosphorylation of methylthioadenosine, which plays a role in the inhibition of methyltransferases and polyamine aminopropyltransferase.46 Although typically expressed in cells and tissues, malignant cells tend to have decreased MTAP and have been shown to secrete methylthioadenosine rather than metabolize it.45,46 Because of this, we hypothesize that rs869329 influences melanoma risk independent of CDKN2A.
Our final index SNP, rs11446223, is an intronic variant at position 90022484. This SNP is not within any regulatory element and can be mapped to DEF8 on chromosome 16. Current knowledge is limited regarding the precise role and function of DEF8 (differentially expressed in FDCP 8 homolog) located at 16q24.3. DEF8 encodes an activator of intracellular signal transduction,52 however, as it is located just downstream of rs1805007 on MC1R (linkage disequilibrium r2 = 0.41), a melanoma-susceptibility gene known to play a role in skin pigmentation and sun sensitivity,53 it is unclear whether this signal is due to MC1R proximity or whether DEF8 has a more prominent independent role.54 Although likely this SNP tags for MC1R, a recent publication described a significant association between DEF8 and melanoma risk,55 demonstrating the possibility that DEF8 may have independent melanoma-susceptibility properties.
Although, these genes have been previously found to be associated with melanoma risk, none of the index SNPs identified by the 2-df joint test were significant for the standard 1-df test, indicating that these results are being primarily driven by the genetic main effects rather than citrus consumption or citrus-gene interaction. However, testing the remaining significant 2-df test significant SNPs revealed a possible G-E interaction between citrus consumption melanoma risk for eight SNPs on chromosome 16, including seven SNPs on the pseudogene gene AFG3L1P and one on GAS8. Of the seven AFG3L1P SNPs, rs199600347, rs111822773, rs113178244 are intronic variants, rs3803683 is a non-coding transcript variant, rs73283871 is a downstream transcript variant, and rs73283867 and rs78800020 are each split with two variants downstream of the gene and two non-coding transcripts. Most rs74583214 (GAS8) transcripts indicated 3 prime UTR variants, and the SNP is located within MYC and POLR2A elements.
A couple of possibilities exist that could explain this evidence of interaction with AFG3L1P. Firstly, although these signals were strong for AFG3L1P, it is possible that this gene may not represent the true susceptibility variants, but rather tag MC1R within the same locus for which there is a biologically plausible association with melanoma56 (e.g, with the exception of rs3803683 [r2 = 0.39], all our AFG3L1P SNPs are in high linkage disequilibrium with rs1805007, an MC1R locus strongly associated with melanoma risk32 [r2 = 0.61–0.95]). Furocoumarins have been demonstrated to reach peak concentration in the skin within four hours of oral intake and remain detectable in cutaneous tissue for at least seven hours after intake.57 These furocoumarins, when exposed to UV radiation, can cause the formation of reactive oxygen species (ROS) and thereby induce photo-oxidative damage by accessing epidermal, dermal, and endothelial cells.57,58 UV exposure to furocoumarin-sensitized skin can also modify proteins, inactivate enzymes, and produce mutagenic and carcinogenic effects.59,60 Therefore, it is biologically plausible to observe an interaction between furocoumarins obtained through citrus intake and MC1R, which influences pigment metabolism and the ability to protect against UV radiation61 and also plays a role in the growth and development of melanoma cells via its influence on keratinocyte and melanocyte proliferation and differentiation.62,63
Secondly, it is also possible for AFG3L1P to play an independent role in this association. Although pseudogenes are typically superfluous and nonfunctional, evidence suggests that some of these genes are translated, and therefore may play a meaningful, functional role in human biology.64 Evidence from Bánfai et al. suggests that AFG3L1P is one of these rare exceptions, indicating it is transcribed and appears to be translated.65 Furthermore, Bánfai et al. conclusively mapped a consistently detected peptide to a novel exon downstream of the pseudogene transcriptional unit that is both beyond the parental gene similarity region and absent in the parental gene locus.65 This research provides meaningful support of AFG3L1P having a novel, protein-coding function distinct from the parental gene,65 and possibly having an independent function in the association between citrus consumption and melanoma risk. Although further validation studies are needed, AFG3L1P may possibly play a previously unknown role in citrus metabolism, photocarcinogenesis, or psoralen/furocoumarin absorption.
The interaction we found with rs74583214 on GAS8 may influence melanoma risk via similar mechanisms. GAS8 is a tumor suppressor gene that has been linked to several types of cancer,66–69 including melanoma.70,71 As GAS8 has been implicated in melanoma risk via its influence on pigmentation,71 furocoumarin exposure may exacerbate UV sensitivity due to GAS8 expression. Also, a rs7458314 is in strong linkage disequilibrium with rs1805007 on MC1R (r2=0.71), it is also possible that rs74583214 serves as a tag for MC1R, which also plays a significant role in pigmentation.61
Recent work by Landi et al. identified 86 genetic variants implicated in melanoma susceptibility.32 We further evaluated if the effect of these genetic variants may be modified by citrus consumption. Eight variants were found with potential gene-citrus interaction in our analysis (Table S2). Details for this additional analysis as well as variants found significant according to the 2-df test, and their associated 1-df test results, can be found in the supplementary material.
As with all studies, our study is subject to limitations. As with all environmental variables, possible population-to-population or country-to-country variance in citrus consumption may limit our results to this specific British cohort. We are limited by our use of recent citrus consumption as a proxy for past citrus consumption. We were also limited by our inability to control for personal or family melanoma history. Additionally, it is possible that our sample size (1,563 melanoma cases included in the analysis) may not be large enough to detect modest effects. Lastly, our analyses and estimates are based on common SNPs, and it is therefore plausible that rare variants could have contributed meaningful data to this analysis.
In conclusion, in this genome-wide analysis, we found that rs183783391 (MITF), rs869329 (MTAP), and rs11446223 (DEF8) are significantly associated with melanoma according to the joint 2-df test; however, 1-df tests found no evidence of citrus-gene interaction, demonstrating that these findings are primarily driven by gene main effects, and not by citrus consumption. Further analyses revealed that seven SNPs on AFG3L1P and one SNP on GAS8, which could possibly serve as tags for MC1R or have independent citrus metabolism or psoralen absorption effects, were significant on both the joint and standard tests, giving evidence of possible citrus-gene interaction.
Supplementary Material
Novelty and Impact:
Previous research has suggested an association between high citrus consumption and melanoma risk. Although inconsistent, findings from these large cohorts were based on the biologically plausible hypothesis that high consumption of citrus increases risk of melanoma due to photocarcinogenic psoralen and furocoumarins that are naturally abundant in citrus products. None of this research has investigated possible genetic bases for this association or identified genetic variants that may modify it. Here, authors conduct a novel genome-wide gene-environment interaction analysis, identifying several genetic loci that provide evidence of citrus-gene interaction for melanoma risk. These findings suggest a genetic basis for photosensitivity and photocarcinogenicity and could advance melanoma precision prevention.
Acknowledgements
This research was supported in part by Lilly Endowment, Inc., through its support for the Indiana University Pervasive Technology Institute, and in part by the Indiana METACyt Initiative. The Indiana METACyt Initiative at IU was also supported in part by Lilly Endowment, Inc.
Funding
This work was supported by the National Heart, Lung and Blood Institute under Grant K01HL14033 [to M.L.]; the Eunice Kennedy Shriver National Institute of Child Health and Human Development under Grant R03HD092854 [to M.L]; and the National Cancer Institute [T32CA117865-11 to V.C.].
Abbreviations:
- df
degrees-of-freedom
- EPIC
European Prospective Investigation into Cancer and Nutrition
- G-E
gene-environment
- GWAS
genome-wide association studies
- HPFS
Health Professionals Follow-up Study
- NHS
Nurses’ Health Study
- PC
principal components
quantile-quantile
- UKBB
UK Biobank
- WHI
Women’s Health Initiative
Footnotes
Conflict of Interest
The authors declare no competing interests.
Ethics statement
The current study is exempt ethical/institutional approval as it is a secondary analysis of previously collected, deidentified data and authors made no attempts to re-identify or contact study participants.
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process which may lead to differences between this version and the Version of Record. Please cite this article as doi:10.1002/ijc.33862
Data Availability Statement
This work has been conducted using the UK Biobank Resource under Application Number 49419. The UK Biobank is an open access resource and bona fide researchers can apply to use the UK Biobank dataset by registering and applying at http://ukbiobank.ac.uk/register-apply/. Further information is available from the corresponding author upon request.
References
- 1.Lens M, Dawes M. Global perspectives of contemporary epidemiological trends of cutaneous malignant melanoma. Br J Dermatol 2004;150: 179–85 [DOI] [PubMed] [Google Scholar]
- 2.Diepgen TL, Mahler V. The epidemiology of skin cancer. Br J Dermatol 2002;146: 1–6 [DOI] [PubMed] [Google Scholar]
- 3.Shaikh WR, Dusza SW, Weinstock MA, Oliveria SA, Geller AC, Halpern AC. Melanoma thickness and survival trends in the United States, 1989–2009. JNCI 2016;108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garbe C, McLeod GRC, Buettner PG. Time trends of cutaneous melanoma in Queensland, Australia and Central Europe. Cancer 2000;89: 1269–78 [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin 2021;71: 7–33 [DOI] [PubMed] [Google Scholar]
- 6.Cancer Research UK. Melanoma skin cancer mortality statistics.2018. Retrieved from https://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/melanoma-skin-cancer/mortality#heading-Zero. Accessed 30 April 2021
- 7.Ashwood-Smith M, Poulton G, Barker M, Mildenberger M. 5-Methoxypsoralen, an ingredient in several suntan preparations, has lethal, mutagenic and clastogenic properties. Nature 1980;285: 407. [DOI] [PubMed] [Google Scholar]
- 8.Mullen MP, Pathak MA, West JD, Harrist TJ, Dall’Acqua F. Carcinogenic effects of monofunctional and bifunctional furocoumarins. Natl Cancer Inst Monogr 1984;66: 205–10 [PubMed] [Google Scholar]
- 9.Griffin A, Hakim R, Knox J. The wave length effect upon erythemal and carcinogenic response in psoralen treated mice. J Invest Dermatol 1958;31: 289–95 [PubMed] [Google Scholar]
- 10.Stern RS. Photocarcinogenicity of drugs. Toxicol Lett 1998;102: 389–92 [DOI] [PubMed] [Google Scholar]
- 11.O’gorman SM, Murphy GM. Photosensitizing medications and photocarcinogenesis. Photodermatol Photoimmunol Photomed 2014;30: 8–14 [DOI] [PubMed] [Google Scholar]
- 12.Wu S, Han J, Feskanich D, Cho E, Stampfer MJ, Willett WC, Qureshi AA. Citrus consumption and risk of cutaneous malignant melanoma. J Clin Oncol 2015;33: 2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melough MM, Wu S, Li W-Q, Eaton C, Nan H, Snetselaar L, Wallace R, Qureshi AA, Chun OK, Cho E. Citrus Consumption and Risk of Cutaneous Malignant Melanoma in the Women’s Health Initiative. Nutr Cancer 2020;72: 568–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahamat-Saleh Y, Cervenka I, Al-Rahmoun M, Mancini FR, Severi G, Ghiasvand R, Veierod MB, Caini S, Palli D, Botteri E. Citrus intake and risk of skin cancer in the European Prospective Investigation into Cancer and Nutrition cohort (EPIC). Eur J Epidemiol 2020: 1–11 [DOI] [PubMed] [Google Scholar]
- 15.Marley A, Li M, Champion V, Song Y, Han J, Li X. The Association between Citrus Consumption and Melanoma Risk in the UK Biobank. Br J Dermatol 2021;185: 353–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monzon J, Liu L, Brill H, Goldstein AM, Tucker MA, From L, McLaughlin J, Hogg D, Lassam NJ. CDKN2A mutations in multiple primary melanomas. N Engl J Med 1998;338: 879–87 [DOI] [PubMed] [Google Scholar]
- 17.Njauw C-NJ, Kim I, Piris A, Gabree M, Taylor M, Lane AM, DeAngelis MM,Gragoudas E, Duncan LM, Tsao H. Germline BAP1 inactivation is preferentially associated with metastatic ocular melanoma and cutaneous-ocular melanoma families. PLoS One 2012;7: e35295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018;562: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Young H, Crowe FL, Benson VS, Spencer EA, Key TJ, Appleby PN, Beral V. Development and evaluation of the Oxford WebQ, a low-cost, web-based method for assessment of previous 24 h dietary intakes in large-scale prospective studies. Public Health Nutr 2011;14: 1998–2005 [DOI] [PubMed] [Google Scholar]
- 20.Dinnissen CS, Ocké MC, Buurma-Rethans EJ, van Rossum C. Dietary Changes among Adults in The Netherlands in the Period 2007–2010 and 2012–2016. Results from Two Cross-Sectional National Food Consumption Surveys. Nutrients 2021;13: 1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rehm CD, Peñalvo JL, Afshin A, Mozaffarian D. Dietary intake among US adults, 1999–2012. JAMA 2016;315: 2542–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradbury KE, Young HJ, Guo W, Key TJ. Dietary assessment in UK Biobank: an evaluation of the performance of the touchscreen dietary questionnaire. J Nutr Sci 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elwood J, Gallagher R, Hill G, Spinelli J, Pearson J, Threlfall W. Pigmentation and skin reaction to sun as risk factors for cutaneous melanoma: Western Canada Melanoma Study. Br Med J (Clin Res Ed) 1984;288: 99–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gloster HM Jr, Neal K. Skin cancer in skin of color. J Am Acad Dermatol 2006;55: 741–60 [DOI] [PubMed] [Google Scholar]
- 25.Kraft P, Yen Y-C, Stram DO, Morrison J, Gauderman WJ. Exploiting gene-environment interaction to detect genetic associations. Hum Hered 2007;63: 111–9 [DOI] [PubMed] [Google Scholar]
- 26.Cornelis MC, Tchetgen Tchetgen EJ, Liang L, Qi L, Chatterjee N, Hu FB, Kraft P.Gene-environment interactions in genome-wide association studies: a comparative study of tests applied to empirical studies of type 2 diabetes. Am J Epidemiol 2011;175: 191–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraft P, Hunter D. Integrating epidemiology and genetic association: the challenge of gene–environment interaction. Philos Trans R Soc B Biol Sci 2005;360: 1609–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAllister K, Mechanic LE, Amos C, Aschard H, Blair IA, Chatterjee N, Conti D,Gauderman WJ, Hsu L, Hutter CM. Current challenges and new opportunities for gene-environment interaction studies of complex diseases. Am J Epidemiol 2017;186: 753–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Figueroa JD, Han SS, Garcia-Closas M, Baris D, Jacobs EJ, Kogevinas M, Schwenn M, Malats N, Johnson A, Purdue MP. Genome-wide interaction study of smoking and bladder cancer risk. Carcinogenesis 2014;35: 1737–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu C, Kraft P, Zhai K, Chang J, Wang Z, Li Y, Hu Z, He Z, Jia W, Abnet CC.Genome-wide association analyses of esophageal squamous cell carcinoma in Chinese identify multiple susceptibility loci and gene-environment interactions. Nat Genet 2012;44: 1090–7 [DOI] [PubMed] [Google Scholar]
- 31.Siegert S, Hampe J, Schafmayer C, Von Schönfels W, Egberts J-H, Försti A, Chen B, Lascorz J, Hemminki K, Franke A. Genome-wide investigation of gene–environment interactions in colorectal cancer. Hum Genet 2013;132: 219–31 [DOI] [PubMed] [Google Scholar]
- 32.Landi MT, Bishop DT, MacGregor S, Machiela MJ, Stratigos AJ, Ghiorzo P, Brossard M, Calista D, Choi J, Fargnoli MC. Genome-wide association meta-analyses combining multiple risk phenotypes provide insights into the genetic architecture of cutaneous melanoma susceptibility. Nat Genet 2020;52: 494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potrony M, Puig-Butille JA, Aguilera P, Badenas C, Tell-Marti G, Carrera C, Del Pozo LJ, Conejo-Mir J, Malvehy J, Puig S. Prevalence of MITF p. E318K in patients with melanoma independent of the presence of CDKN2A causative mutations. JAMA Dermatol 2016;152: 405–12 [DOI] [PubMed] [Google Scholar]
- 34.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med 2006;12: 406–14 [DOI] [PubMed] [Google Scholar]
- 35.Hartman ML, Czyz M. MITF in melanoma: mechanisms behind its expression and activity. Cell Mol Life Sci 2015;72: 1249–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yajima I, Kumasaka MY, Thang ND, Goto Y, Takeda K, Iida M, Ohgami N, Tamura H, Yamanoshita O, Kawamoto Y. Molecular network associated with MITF in skin melanoma development and progression. J Skin Cancer 2011;2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giuliano S, Cheli Y, Ohanna M, Bonet C, Beuret L, Bille K, Loubat A, Hofman V, Hofman P, Ponzio G. Microphthalmia-associated transcription factor controls the DNA damage response and a lineage-specific senescence program in melanomas. Cancer Res 2010;70: 3813–22 [DOI] [PubMed] [Google Scholar]
- 38.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 2005;436: 117–22 [DOI] [PubMed] [Google Scholar]
- 39.Morgan MD, Pairo-Castineira E, Rawlik K, Canela-Xandri O, Rees J, Sims D, Tenesa A, Jackson IJ. Genome-wide study of hair colour in UK Biobank explains most of the SNP heritability. Nat Commun 2018;9: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bertolotto C, Lesueur F, Giuliano S, Strub T, De Lichy M, Bille K, Dessen P,d’Hayer B, Mohamdi H, Remenieras A. A SUMOylation-defective MITF germline mutation predisposes to melanoma and renal carcinoma. Nature 2011;480: 94–8 [DOI] [PubMed] [Google Scholar]
- 41.Amos CI, Wang L-E, Lee JE, Gershenwald JE, Chen WV, Fang S, Kosoy R, Zhang M, Qureshi AA, Vattathil S. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet 2011;20: 5012–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrett JH, Iles MM, Harland M, Taylor JC, Aitken JF, Andresen PA, Akslen LA, Armstrong BK, Avril M-F, Azizi E. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet 2011;43: 1108–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, Azizi E, Bergman W, Bianchi-Scarra G, Bruno W. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007;44: 99–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mario F, Veronique B, Nicholas HK, David DL, Julia NBA, Tomi P, Alessandra C, Zhen ZZ, Panos D, Nicole S. Loci at 9p21 and 22q13 harbour alleles for development of cutaneous nevi and melanoma. Nat Genet 2009;41: 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Christopher SA, Diegelman P, Porter CW, Kruger WD. Methylthioadenosine phosphorylase, a gene frequently codeleted with p16cdkN2a/ARF, acts as a tumor suppressor in a breast cancer cell line. Cancer Res 2002;62: 6639–44 [PubMed] [Google Scholar]
- 46.Behrmann I, Wallner S, Komyod W, Heinrich PC, Schuierer M, Buettner R, Bosserhoff A-K. Characterization of methylthioadenosin phosphorylase (MTAP) expression in malignant melanoma. Am J Pathol 2003;163: 683–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bishop DT, Demenais F, Iles MM, Harland M, Taylor JC, Corda E, Randerson-Moor J, Aitken JF, Avril M-F, Azizi E. Genome-wide association study identifies three loci associated with melanoma risk. Nat Genet 2009;41: 920–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Newton-Bishop JA, Chang Y-M, Iles MM, Taylor JC, Bakker B, Chan M, Leake S, Karpavicius B, Haynes S, Fitzgibbon E. Melanocytic nevi, nevus genes, and melanoma risk in a large case-control study in the United Kingdom. Cancer Epidemiol Biomarkers Prev 2010;19: 2043–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang XR, Liang X, Pfeiffer RM, Wheeler W, Maeder D, Burdette L, Yeager M, Chanock S, Tucker MA, Goldstein AM. Associations of 9p21 variants with cutaneous malignant melanoma, nevi, and pigmentation phenotypes in melanoma-prone families with and without CDKN2A mutations. Fam Cancer 2010;9: 625–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tucker MA, Halpern A, Holly EA, Hartge P, Elder DE, Sagebiel RW, Guerry D, Clark WH. Clinically recognized dysplastic nevi: a central risk factor for cutaneous melanoma. JAMA 1997;277: 1439–44 [PubMed] [Google Scholar]
- 51.McMeniman E, Duffy D, Jagirdar K, Lee K, Peach E, McInerney-Leo A, De’Ambrosis B, Rayner J, Smithers B, Soyer H. The interplay of sun damage and genetic risk in Australian multiple and single primary melanoma cases and controls. Br J Dermatol 2019 [DOI] [PubMed] [Google Scholar]
- 52.Asgari MM, Wang W, Ioannidis NM, Itnyre J, Hoffmann T, Jorgenson E, Whittemore AS. Identification of susceptibility loci for cutaneous squamous cell carcinoma. J Invest Dermatol 2016;136: 930–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu F, Visser M, Duffy DL, Hysi PG, Jacobs LC, Lao O, Zhong K, Walsh S, Chaitanya L, Wollstein A. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum Genet 2015;134: 823–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu JZ, Mcrae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG. A versatile gene-based test for genome-wide association studies. Am J Hum Genet 2010;87: 139–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fang S, Lu J, Zhou X, Wang Y, Ross MI, Gershenwald JE, Cormier JN, Wargo J, Sui D, Amos CI. Functional annotation of melanoma risk loci identifies novel susceptibility genes. Carcinogenesis 2020;41: 452–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Antonopoulou K, Stefanaki I, Lill CM, Chatzinasiou F, Kypreou KP, Karagianni F, Athanasiadis E, Spyrou GM, Ioannidis JP, Bertram L. Updated field synopsis and systematic meta-analyses of genetic association studies in cutaneous melanoma: the MelGene database. J Invest Dermatol 2015;135: 1074–9 [DOI] [PubMed] [Google Scholar]
- 57.Melough MM, Chun OK. Dietary furocoumarins and skin cancer: A review of current biological evidence. Food Chem Toxicol 2018;122: 163–71 [DOI] [PubMed] [Google Scholar]
- 58.Pathak MA, Joshi PC. Production of active oxygen species (1O2 and O2⨪) by psoralens and ultraviolet radiation (320–400 nm). Biochim Biophys Acta Gen Subj 1984;798: 115–26 [DOI] [PubMed] [Google Scholar]
- 59.Schiavon O, Veronese F. Extensive crosslinking between subunits of oligomeric proteins induced by furocoumarins plus UV-A irradiation. Photochem Photobiol 1986;43: 243–6 [DOI] [PubMed] [Google Scholar]
- 60.Melough MM, Cho E, Chun OK. Furocoumarins: A review of biochemical activities, dietary sources and intake, and potential health risks. Food Chem Toxicol 2018;113: 99–107 [DOI] [PubMed] [Google Scholar]
- 61.Healy E, Flannagan N, Ray A, Todd C, Jackson IJ, Matthews JN, Birch-Machin MA, Rees JL. Melanocortin-1-receptor gene and sun sensitivity in individuals without red hair. The Lancet 2000;355: 1072–3 [DOI] [PubMed] [Google Scholar]
- 62.De Luca M, Siegrist W, Bondanza S, Mathor M, Cancedda R, Eberle AN. Alpha melanocyte stimulating hormone (alpha MSH) stimulates normal human melanocyte growth by binding to high-affinity receptors. J Cell Sci 1993;105: 1079–84 [DOI] [PubMed] [Google Scholar]
- 63.Suzuki I, Cone R, Im S, Nordlund J, Abdel-Malek Z. Binding of melanotropic hormones to the melanocortin receptor MC1R on human melanocytes stimulates proliferation and melanogenesis. Endocrinology 1996;137: 1627–33 [DOI] [PubMed] [Google Scholar]
- 64.Ji Z, Song R, Regev A, Struhl K. Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. elife 2015;4: e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bánfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE, Kundaje A, Gunawardena HP, Yu Y, Xie L. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 2012;22: 1646–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pan W, Zhang N, Liu W, Liu J, Zhou L, Liu Y, Yang M. The long noncoding RNA GAS8-AS1 suppresses hepatocarcinogenesis by epigenetically activating the tumor suppressor GAS8. J Biol Chem 2018;293: 17154–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rashkin SR, Graff RE, Kachuri L, Thai KK, Alexeeff SE, Blatchins MA, Cavazos TB, Corley DA, Emami NC, Hoffman JD. Pan-cancer study detects genetic risk variants and shared genetic basis in two large cohorts. Nat Commun 2020;11: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Esfandi F, Rezaei FM, Taheri M, Gol MN, Oskooei VK, Namvar A, Ghafouri-Fard S. GAS8 and GAS8-AS1 expression in gastric cancer. Gastroenterol Hepatol Bed Bench 2019;12: 322. [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Y, Chu Y, Sun J, Song R, Li Y, Xu F. LncRNA GAS8-AS inhibits colorectal cancer (CRC) cell proliferation by downregulating lncRNA AFAP1-AS1. Gene 2019;710: 140–4 [DOI] [PubMed] [Google Scholar]
- 70.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, Turajlic S, Piperno-Neumann S, de la Grange P, Roman-Roman S. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer Discov 2013;3: 1122–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerstenblith MR, Shi J, Landi MT. Genome-wide association studies of pigmentation and skin cancer: a review and meta-analysis. Pigment Cell Melanoma Res 2010;23: 587–606 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This work has been conducted using the UK Biobank Resource under Application Number 49419. The UK Biobank is an open access resource and bona fide researchers can apply to use the UK Biobank dataset by registering and applying at http://ukbiobank.ac.uk/register-apply/. Further information is available from the corresponding author upon request.


