Abstract
DNA fingerprinting methods have evolved as major tools in fungal epidemiology. However, no single method has emerged as the method of choice, and some methods perform better than others at different levels of resolution. In this review, requirements for an effective DNA fingerprinting method are proposed and procedures are described for testing the efficacy of a method. In light of the proposed requirements, the most common methods now being used to DNA fingerprint the infectious fungi are described and assessed. These methods include restriction fragment length polymorphisms (RFLP), RFLP with hybridization probes, randomly amplified polymorphic DNA and other PCR-based methods, electrophoretic karyotyping, and sequencing-based methods. Procedures for computing similarity coefficients, generating phylogenetic trees, and testing the stability of clusters are then described. To facilitate the analysis of DNA fingerprinting data, computer-assisted methods are described. Finally, the problems inherent in the collection of test and control isolates are considered, and DNA fingerprinting studies of strain maintenance during persistent or recurrent infections, microevolution in infecting strains, and the origin of nosocomial infections are assessed in light of the preceding discussion of the ins and outs of DNA fingerprinting. The intent of this review is to generate an awareness of the need to verify the efficacy of each DNA fingerprinting method for the level of genetic relatedness necessary to answer the epidemiological question posed, to use quantitative methods to analyze DNA fingerprint data, to use computer-assisted DNA fingerprint analysis systems to analyze data, and to file data in a form that can be used in the future for retrospective and comparative studies.
Interest in assessing the genetic relatedness of isolates of the same species has grown rapidly as we have delved deeper into the epidemiology of a variety of fungal diseases. Indeed, as molecular genetic approaches have evolved for unraveling the basic biology of particular fungal pathogens, so have methods for fingerprinting them at the genetic level. In 1985, there were 3 nonforensic publications that had “DNA fingerprinting” in the title or abstract, and in 1996, 11 years later, there were 318, and these did not include papers that used DNA fingerprinting techniques but did not reference them specifically as such. Although the recent availability of DNA fingerprinting techniques provides investigators and clinicians with tools for tracking strains and identifying the sources of particular infections, the variety of methods and the low level of sophistication applied in most cases to the analysis of data have led to problems with interpretation. Not all DNA fingerprinting methods are equally effective (see, e.g., references 151, 152, 208, and 288), and some can lead to misinformation. In the case of the infectious fungi, no single DNA fingerprinting technique has evolved as a dominant method, and in fact, each method has its own set of assets and limitations. In some cases, a method resolves differences between isolates, but because the method has not been adequately characterized, it is not clear how the differences can be interpreted in terms of genetic distance. In other words, it is not clear if resolved differences between isolates reflect minor changes representing the microevolution of a single strain over a short period or major differences between highly unrelated strains. When a potentially effective fingerprinting method is used, the user may not know how to interpret the results adequately. Even more worrisome is the continuous stream of published studies in which data that could have been quantitatively analyzed and then stored are dealt with by the authors in a superficial, qualitative, one-time manner. Indeed, the most wasteful aspect of DNA fingerprinting studies to date is the underutilization of data. With the advent of computer-assisted DNA fingerprint analysis systems, DNA fingerprint data can now not only be quantitatively compared but can also be normalized to a universal standard and then stored in a database so that every newly analyzed isolate can be compared retrospectively and quantitatively with every previously analyzed isolate of that species. Indeed, if a method is highly reproducible between laboratories, the data from different laboratories can be compared and pooled in a general data bank.
DNA fingerprinting of the infectious fungi has become an important subdiscipline of medical mycology. As DNA fingerprinting is more frequently applied to a variety of epidemiological problems, it becomes increasingly evident that there are “ins and outs” to the methods. Criteria can be used to assess the resolution of a particular fingerprinting method, and strategies have evolved to verify the efficacy of a fingerprinting method. Researchers can now assess beforehand whether a particular method will provide data that will answer the questions posed. In addition, criteria have evolved to assess whether a fingerprinting method is amenable to computer-assisted methods. Therefore, it seems timely that the various techniques now being used to fingerprint fungi infectious to humans be described, compared, and evaluated. In addition, the ground rules for selecting a method should be discussed and the major results of recent DNA fingerprinting studies should be reviewed within this context. These are the general objectives of this review. The discussion that follows will rely disproportionately upon examples from the fingerprinting literature of Candida albicans and related species, since I have worked primarily on these systems and can therefore draw more easily upon that literature in arguing particular points. However, the points that are made will be applicable to DNA fingerprinting of the infectious fungi in general.
QUESTIONS THAT REQUIRE DNA FINGERPRINTING
To understand the dynamics of an infectious organism in a human population, decipher the complex relationship between commensalism and infection, identify the origin of an infection, or monitor the emergence of drug-resistant strains, one must have a method for assessing genetic relatedness. Species typing is a necessary first step in all epidemiological studies, but one must have a way of assessing the relatedness of isolates within a species if one wants to understand many of the epidemiological questions that are posed. In the discussion that follows in this review, an isolate is defined as a clone collected independently of other isolates. Two independently collected isolates may be completely unrelated or genetically indistinguishable. In contrast, a strain refers to a collection of isolates of the same species that are highly related or genetically indistinguishable. To classify two isolates as belonging to the same strain or as members of different strains, one must have a DNA fingerprinting method that has been appropriately characterized to perform these functions. The need for sensitive DNA fingerprinting methods is especially important when diseases emanate from commensal organisms, when strains become specialized for particular body locations or compromising conditions, when strains undergo microevolution for rapid adaptation, or when strains are communicated between individuals. If a transplant recipient enters a hospital, is immunosuppressed, and then presents with a nosocomial bloodstream infection that proves to be due to a fungus, how is the origin of the infecting fungus elucidated? How does one distinguish an organism originating in the hospital from an organism that was a commensal in an anatomical location other than the bloodstream (e.g., the gastrointestinal tract) when the patient first entered the hospital? To discriminate between these alternatives, simply identifying the infecting strain for species is insufficient. If one identifies the strain as C. albicans, and over 50% of patients and hospital staff carry C. albicans as a commensal in one or more body locations, species typing tells us little about the origin of the infection. To identify the origin of the infecting organism, one must collect isolates from hospital workers who have interacted with the patient, the patient's immediate physical environment, visitors, and different body locations of the patient prior to and during hospitalization. One must then have a DNA fingerprinting system that is effective in discriminating between unrelated isolates, recognizing the same strain in different isolates, and recognizing highly related but nonidentical isolates (i.e., isolates that differ due to microevolution). The fingerprinting system must be verified for its effectiveness at these different levels of discrimination and must provide a database for estimating the probability that a particular strain identified by the particular DNA fingerprinting method will be isolated twice by chance in a particular geographical locale.
Consider another example that requires sophisticated fingerprinting. If a drug-resistant strain colonizes a patient and it becomes necessary to estimate the potential threat this strain poses to the general population, isolates from hundreds of individuals must be analyzed for both drug resistance and genetic relatedness. The DNA fingerprinting method must have sufficient resolving power to discriminate this particular strain from the majority of other strains in the geographical locale, and it must be amenable to computer-assisted analysis, especially if surveillance is to be performed over time with a significant number of isolates. Because comparisons of genetic relatedness between isolates fingerprinted at different times must be performed, the fingerprinting method must be reproducible and quantitative. There are many more examples of epidemiological problems and questions, each with its own set of demands on the technology of DNA fingerprinting and analysis. Fortunately, DNA fingerprinting methods that fulfill the majority of these demands and are within the technical capabilities of most medical mycologists have been developed for several pathogenic fungi. Criteria have been formulated that, if followed, will lead to the development or selection of fingerprinting methods that can be effectively used to obtain very good and sometimes definitive answers to many of the epidemiological questions medical mycologists might pose.
BIOTYPING IS INADEQUATE AS A DNA FINGERPRINTING METHOD
Before the development of DNA fingerprinting methods, researchers in medical mycology realized that not all fungal isolates of a given species exhibited the same general phenotype. Therefore, they reasoned that differences in phenotype reflected genetic differences between strains. Without methods for measuring differences or similarities at the level of DNA, they resorted to biotyping methods that reflected genotype. In 1950, Evans used agglutination tests to distinguish three serotypes, A, B, and C, of Cryptococcus neoformans (109). In 1968, Wilson and coworkers identified a fourth serotype, D (428). It was subsequently demonstrated that the most prevalent serotype in the United States was A (25) and that the dominant serotype in Thailand changed with the emergence of AIDS (378). The usefulness of serotyping C. neoformans has been confirmed by the large number of studies employing the method—more than 150 studies were identified in a MedLine search of the bibliography database in 1998, and that represented an underestimate. Serotyping alone, however, falls short as a strain-typing method. For instance, if the majority of isolates in a geographical locale are primarily one serotype, this method does not provide adequate strain resolution for most of the epidemiological questions posed.
One of the first biotyping strategies used to discriminate among C. albicans strains was also based on serotyping. Hasenclever and coworkers separated C. albicans strains into the two serotypes A and B (133, 134, 379), and this method has been used to type strains for the past 38 years (see, e.g., references 45, 103, 246, 258, 371, and 415). However, just as for Cryptococcus neoformans, separation of an entire species into a few groups does not provide meaningful resolution for the majority of epidemiological questions posed. In addition, it was demonstrated that different serotyping methods did not always group strains in the same manner. Three serotyping methods that had been established by 1990 for C. albicans included Hasenclever's original antisera HSN1 and HSN2 (133, 134, 379), the Iatron Candida Check factor 6 typing antiserum (IF6) (283), and agglutination with the monoclonal antibody H9 (45). Brawner (44) compared these serotype methods and demonstrated strong correlations between HSN1 and HSN2 and between these two antisera and H9 but not between IF6 and either HSN1, HSN2, or H9. Even more worrisome was the discovery that antigen expression could be affected by the phase of growth and, more importantly, that serotype B cells could produce serotype A antigen (283), which placed in question the entire methodology. For the A and B serotypes in C. albicans, the problem with variability probably lies in the fact that antigenicity is based on the polysaccharide moieties of the phosphomanno-protein complexes (227, 341, 394).
Realizing the shortcomings of serotyping, Odds and Abbott in the early 1980s developed the first complex biotyping protocol for distinguishing among Candida species and among strains of a species (240, 241). The rationale behind the Odds and Abbott method made sense. If one selects a large enough list of diverse phenotypic characteristics, the combined phenotype should reflect the genotype, even if a few of the selected characteristics proved to be sensitive to the environment and hence unreliable. The original method of Odds and Abbott (240) for species typing included nine assays that were quite easy to perform, involving the growth of cells of a cloned isolate on test agars with different compositions. These assays tested for growth at pH 1.4; production of secreted acid proteinase; resistance to flucytosine, boric acid, and safranin; assimilation of urea, sorbose, and citrate; and sensitivity to high salt. This list of assessed characteristics was effective in discriminating among species, and four additional tests, i.e., resistance to tetrazolium salts, sodium periodate, and cetrimide and growth on MacConkey agar, were used to supplement the preceding tests for discrimination among strains within a Candida species. Alterations in the method have also been developed to make it more amenable to general use (75). In the original application of this method to oral and vaginal isolates, 45 different types were identified (240), but the system had the potential for discriminating 512 types. The use of this biotyping strategy proved effective in a number of epidemiological studies (see, e.g., references 166, 189, 235, 244, 245, 247, 248, 276, 342, and 434), and if care was taken in standardizing methods, intralaboratory reproducibility could be achieved. Unfortunately, the Odds and Abbott biotyping system was found to have poor interlaboratory reproducibility among five laboratories (242). The conclusion of the authors of this latter study was that although this biotyping method was effective for research applications, it probably would not prove effective for clinical use. In addition to serotyping and the Odds and Abbott biotyping method, a number of other biotyping methods have been used to discriminate C. albicans strains, including morphotyping (145, 270, 293), resistotyping (144, 215), killer yeast typing (278, 279), enzyme typing (66, 424, 425), sugar assimilation typing (50, 105, 113, 120) and drug susceptibility typing (294). Isoenzyme biotyping has also been successfully applied to Candida species (52, 180, 288). However, because the different isoenzyme patterns in a method such as multilocus enzyme electrophoresis (MLEE) directly reflect allelic differences at defined loci and because MLEE outperforms several popular DNA fingerprinting methods in assessing genetic relatedness, it is dealt with separately in a following section.
Although biotyping methods became less popular with the emergence of DNA fingerprinting, the use of sugar assimilation profiles has remained the most popular method for rapid species identification. Commercial kits have been available for several years that assess the assimilation of more than 19 carbohydrates as the only source of carbon (50, 105, 113, 120), and their reliability in determining species is quite high (greater than 90%). In addition, an indicator agar referred to commercially as CHROMagar is now available that discriminates colorimetrically between species and is based upon the reduction of unnamed compounds (243, 263, 324). The CHROMagar method is probably not as discriminatory as the sugar assimilation kits, but the convenience of plating cells on a single indicator agar is unparalleled for preliminary identification of species and as an indicator of population homogeneity.
Although phenotype reflects genotype by definition, biotyping methods have fundamental problems that render them in some instances inadequate for discriminating among strains within a species. First, many of the assays in the Odds and Abbott method and in sugar assimilation profiles may be too sensitive to growth conditions; this is probably the reason why unacceptable levels of interlaboratory variability have been reported (242). As noted, even the expression of serotypes can be affected by growth (45, 283). Second, C. albicans and related species undergo spontaneous high-frequency switching among a limited number of general phenotypes (48, 239, 280, 305, 344, 345, 351, 359) that affects a variety of phenotypic traits, including antigenicity (2), sensitivity to antifungal agents (343, 356), uptake of dyes (4), secretion of acid proteinase (233) and assimilation of carbohydrates (350). These differences exist among the switch phenotypes of the same strain grown under the same growth conditions. For instance, C. albicans strain WO-1 switches reversibly and spontaneously at frequencies of approximately 10−3 between hemispheric white and flatter grey (opaque) colony phenotypes (345). The white-opaque transition results in dramatic changes in a variety of cellular traits (4, 351, 356) and the activation-deactivation of several white and opaque phase-specific genes (143, 232, 233, 248, 353, 354, 367, 418). White-phase cells will assimilate ribitol, xylitol, methyl-d-glucoside, and trehalose, but opaque-phase cells will not (350), and this pattern changes in a perfectly reversible fashion with each spontaneous switch. In addition, while white-phase cells secrete negligible amounts of aspartyl proteinase in either Lee's medium or serum-containing medium, opaque-phase cells secrete copious amounts (233); again, this change reverses with each spontaneous switch. Although most do not switch at high frequency, all isolates of C. albicans are capable of switching. In the minority of strains in which switching occurs at high frequency, the use of biotyping for strain discrimination could lead to erroneous results. Switching has also been demonstrated in Candida tropicalis (360), Candida glabrata (173), and Cryptococcus neoformans (127).
Recently, de Bernardis et al. (85) presented evidence that differences in pH in different host niches can dramatically affect the expression of several genes in C. albicans. As C. albicans multiplies in culture, it causes a continuous decrease in the pH of the medium. Cells may continually change their pattern of gene expression as the cell density increases and as the pH decreases. Therefore, if the growth conditions and time of harvesting are not precisely duplicated among experiments and laboratories, intralaboratory and interlaboratory reproducibility of biotyping methods will be compromised. In contrast, the basic DNA sequence of an organism can be presumed to be insensitive to short-term environmental change and thus should provide a more stable alternative for strain discrimination.
REQUIREMENTS FOR AN EFFECTIVE DNA FINGERPRINTING SYSTEM
To assess the relatedness of strains within a fungal species, a method must be used that measures relatedness at the genotypic level. Assessment at this level in most cases involves direct DNA measurements. Methods that directly measure DNA differences among strains within a species are henceforth referred to as DNA fingerprinting methods. For a DNA fingerprinting method to be effective for epidemiological purposes, it must fulfill certain general requirements. It should be kept in mind, however, that the stringency of these requirements will vary in relation to the defined objectives of each epidemiological study.
Resistance to Environmental Perturbations and High-Frequency Genomic Reorganization
Since the data obtained by any DNA fingerprinting method reflects the genetic relatedness among isolates, the genetic differences a particular method reveals must be due to evolutionary change. However, the rate of change of some DNA sequences may be inordinately affected by the environment or the physiological state of the cell. For instance, maintenance of a minichromosome or nonintegrated plasmid may be affected by growth conditions or cellular phenotype. A DNA fingerprinting method that targets such nonchromosomal DNA might therefore be unreliable. A DNA fingerprinting method must also be resistant to DNA reorganizational events that are affected by phenotype. For instance, C. albicans strain 3153A can undergo high-frequency phenotypic switching among seven general phenotypes (344, 351). In the dominant o-smooth phenotype, the organism switches to the other variant phenotypes at frequencies of less than 10−4, but when expressing a variant phenotype such as “star” or “irregular wrinkle,” the organism switches to other phenotypes at frequencies of approximately 10−2 (295, 344). When cells of strain 3153A are in the dominant and more stable o-smooth phenotype, their electrophoretic karyotype is relatively stable (295). However, when they express a less stable variant phenotype, their electrophoretic karyotype also becomes less stable, changing at extraordinarily high frequencies, primarily as a result of the nonreciprocal reorganization of rDNA cistrons in the two rDNA-containing chromosomes (295). Therefore, electrophoretic karyotypes of cells in a high-frequency mode of switching can rapidly diverge and converge in switching lineages (295). The convergence of karyotypic patterns leads to homoplasy, defined as common characteristics that do not have a common ancestry. Homoplasy is a characteristic that is inconsistent with the goals of a fingerprinting method. Recently, B. B. Magee and P. T. Magee (personal communication) discovered that the electrophoretic karyotypes of C. dubliniensis was hypervariable and that the reorganization of non-rDNA-containing chromosomes was also involved. Although high-frequency changes such as those noted for C. albicans and C. dubliniensis karyotypes may interfere with the capacity to distinguish between moderately related and unrelated strains, they can be used effectively to assess microevolution within an infecting strain (192). However, the demonstration of high-frequency reorganization and homoplasy reduces the effectiveness of karyotyping as a general DNA fingerprinting method for C. albicans.
If an organism undergoes reversible high-frequency DNA reorganization at specific loci in association with phenotypic switching, as occurs in Salmonella enterica serovar Typhimurium (125), Escherichia coli (125), and Borrelia hermsii (16), or if an organism undergoes irreversible DNA reorganization at specific loci in association with antigenic switching, as occurs in Trypanosoma brucei (99) and Neisseria gonorrhoeae (383), the sequences at these loci should not be employed in a general DNA fingerprinting method. In the setting of reversible reorganization, the system will inherently lead to homoplasy.
Data Should Reflect Genetic Distance
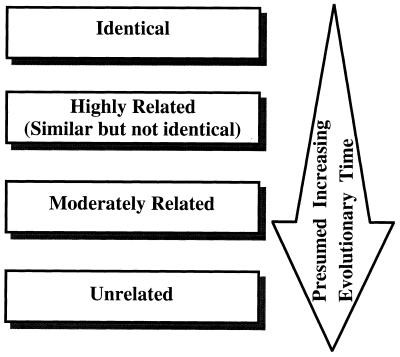
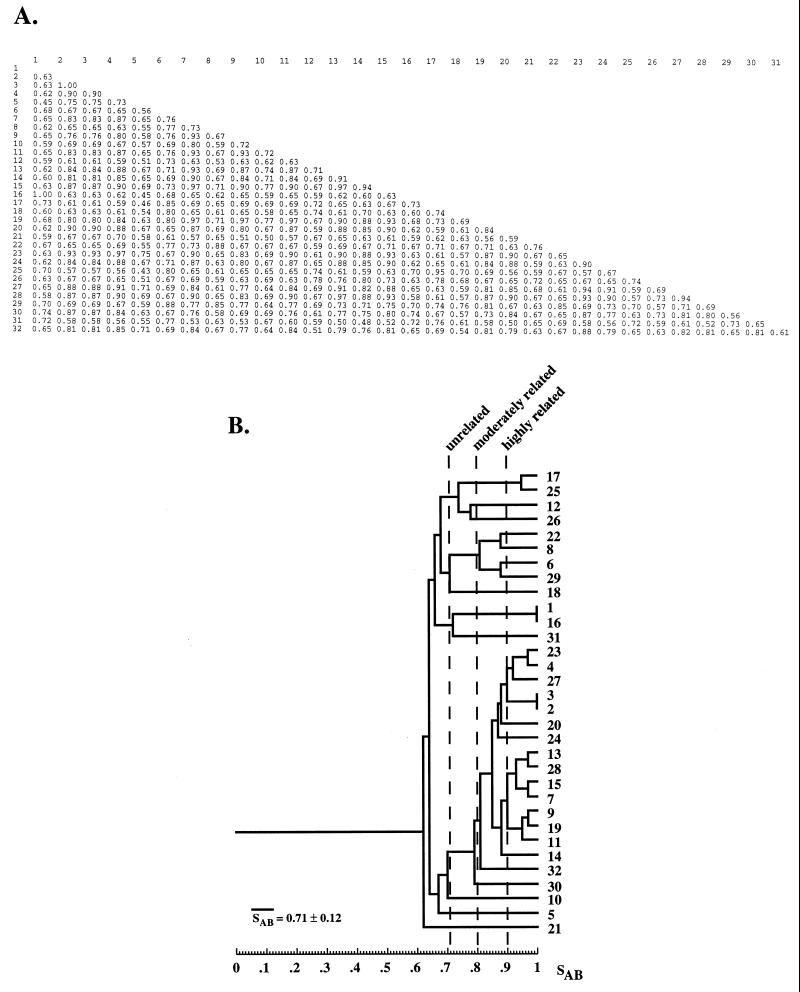
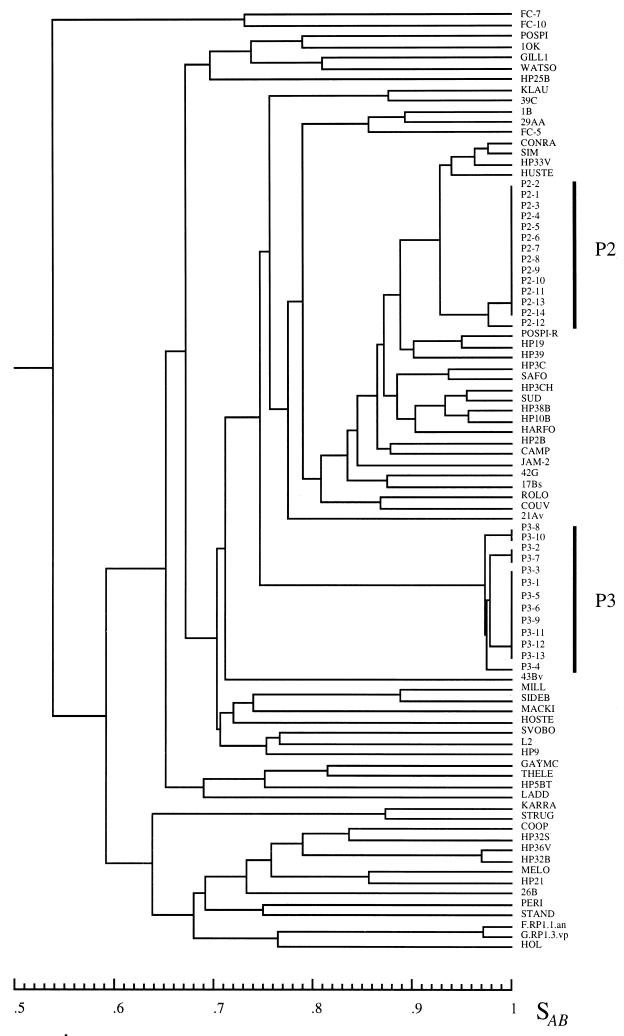
The requirement that the data should reflect genetic distance is, perhaps, the trickiest and most important requirement. Many methods elucidate genotypic traits that are stable within a strain but vary among strains and therefore are reasonably effective in identifying the same strain and distinguishing among unrelated strains in independent isolates. Problems arise, however, when the researcher wishes to group isolates into moderately related clusters or identify microevolution within a strain. An effective DNA fingerprinting system should (i) identify the same strain in independent isolates, (ii) identify microevolutionary changes in a strain, (iii) cluster moderately related isolates and (iv) identify completely unrelated isolates. These levels of resolution are diagrammed in relation to evolutionary time in Fig. 1. Although several DNA fingerprinting methods fulfill requirements i and iv and some fulfill i, ii, and iv, several of the popular DNA fingerprinting do not fulfill requirement iii. The capacity to measure genetic distance in moderately related isolates may, in fact, be the most difficult requirement to meet. Unfortunately, no DNA fingerprinting method provides a definitive measure of genetic distance between two isolates, although some come far closer than others. As Tibayrenc has so poignantly articulated, “there is actually no means to fully ascertain the identity of two microbial genotypes but to sequence their entire genome” (391). Because different molecular markers may have different molecular clocks (i.e., the evolutionary speed at which they change), a good DNA fingerprinting method should be based on a number of molecular markers. The method should also be resistant to homoplasy. A DNA fingerprinting method should provide quantitative data that reflect genetic distance. One way to assess whether a DNA fingerprinting method fulfills this requirement is to compare the data generated by one method with those generated by a completely unrelated DNA fingerprinting method for the same set of test isolates (392, 395). The set of test isolates should include ones which are identical, ones which are highly related but nonidentical, and ones which are independent with no known relationships, including moderately and completely unrelated isolates (288). If the two methods identify the identical isolates and nonidentical but highly related isolates as such, generally cluster less closely related isolates in a similar fashion, and distinguish among completely unrelated isolates, then, in essence, the two methods have cross-verified each other for all of the levels of resolution described in Fig. 1. However, such verification can be achieved only for species with a predominantly clonal population structure (see the next section).
FIG. 1.
Levels of relatedness that must be resolved by DNA fingerprinting methods. The order of relatedness is presented, with relatedness in each category defined methodologically. “Identical” means that the DNA fingerprints of isolates are indistinguishable by most methods. “Highly Related” means that the DNA fingerprints are highly similar but nonidentical. Usually, differences between highly related isolates are due to hypervariable changes reflecting microevolution. “Moderately Related” means that isolates group in a dendrogram in a cluster defined by a similarity coefficient threshold well above the average similarity coefficient for a set of presumed unrelated isolates. “Unrelated” means that the SAB of isolates is near or below the average similarity coefficient for a set of presumed unrelated isolates.
Stability over Time for Some but Not All Epidemiological Questions
The data generated for a strain by a DNA fingerprinting method (e.g., a gel pattern) must be relatively stable over many generations. This requires that there be little recombination between the sequences selected for analysis and the populations within the species under analysis undergo primarily clonal reproduction. If panmixia (random gene exchange) occurs at high frequencies due to sexual mating and recombination, the results of epidemiological studies employing standard DNA fingerprinting methods are more difficult to interpret. Fortunately, many infectious fungi undergo recombination or gene exchange at extremely low frequencies (386). It has been concluded that C. albicans reproduction is primarily clonal (130, 291) and that standard DNA fingerprinting methods are applicable.
In addition to the stability resulting from clonal reproduction, a DNA fingerprinting method should assess primarily sequences that are not highly reorganizational (i.e., are reasonably stable over time). For instance, a complex DNA fingerprinting probe such as the 11-kb Ca3 probe of C. albicans (288, 315, 359) contains both repetitive sequences dispersed throughout the genome (e.g., the RPS repetitive element) and unique sequences represented at only one locus (152, 289). Because sequences in the genome containing clusters of full-length RPS units (73, 150) undergo frequent reorganization through duplication and deletion of full-length units (289), frequent changes occur in bands in the Ca3 hybridization pattern containing full-length RPS sequences. Bands containing full-length RPS elements represent, on average, one-fifth of the pattern generated by Ca3. Because the remaining four-fifths of the Ca3 hybridization pattern represent less variable sequences (3), these latter bands tend to stabilize the pattern. In contrast, a probe consisting entirely of RPS elements will generate a far less stable fingerprint pattern than will the complex Ca3 probe (289), and therefore the former will be far less effective in accurately clustering moderately related isolates. Therefore, both the full-length Ca3 probe and a restricted RPS probe can be used in studies assessing rapid change due to microevolution, but only the full-length Ca3 probe can be used in studies in which moderately related isolates must be analyzed.
Amenability to Automated Computer-Assisted Analysis
Sophisticated computer-assisted systems have evolved for the automatic analysis and storage of DNA fingerprinting data in the form of banding patterns. These systems, which are discussed in more detail in a later section, automatically identify the lanes and bands in Southern blot patterns, density-scan the bands in a pattern, normalize patterns to universal standards, compute similarity coefficients for every pair of isolates, and generate phylogenetic trees (dendrograms). Computer programs have also evolved to store and analyze sequence data. The storage and rapid accessibility of data has emerged as a requirement for any fingerprinting method, since it provides a mechanism for retrospective analyses and for comparisons among studies.
THE MOST COMMON METHODS TO DNA FINGERPRINT THE INFECTIOUS FUNGI
Multilocus Enzyme Electrophoresis
Before considering the major DNA-based methods, we must consider MLEE and comparable methods that assess isozymes or allozymes, since they outperform many of the DNA-based methods at most levels of resolution. The power of MLEE is that if one is careful in selecting enzymes, one can discriminate among the gene products of different alleles for a number of loci. Thus, the method can assess codominant markers in diploids for each locus, a requirement for evolutionary biologists that is not achieved by a few of the popular DNA fingerprinting methods.
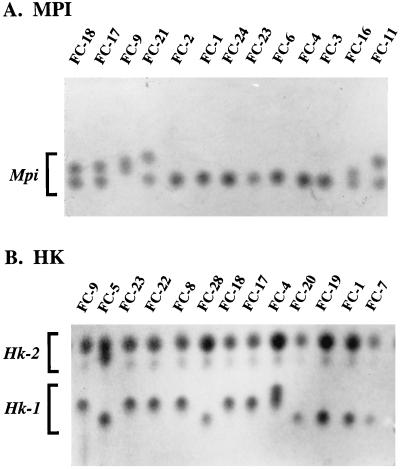
The MLEE method is straightforward. Cell extracts are separated by starch gel electrophoresis, polyacrylamide gel electrophoresis, or isoelectric focusing under native conditions, and the enzymes are visualized in the gels by specific enzyme-staining procedures. For haploids, one band is obtained, and for diploids, one or two are obtained. In some cases, the enzyme is active in multimeric forms, but in most of these cases the enzyme phenotypes can be assessed. To obtain complex data for computing a similarity coefficient between two isolates, several enzymes must be assessed. For instance, in a recent analysis by Pujol et al. (288) of C. albicans using MLEE, 21 enzymes were tested on 29 isolates. Thirteen exhibited variability and were therefore used in the analysis. In Fig. 2, an example is presented of starch gel electrophoresis patterns for mannose-6-phosphate isomerase and the two loci expressing hexokinase activity.
FIG. 2.
Examples of starch gel electrophoresis patterns of two enzymes, mannose-6-phosphate isomerase (MPI) (A) and hexokinase (HK) (B), used in an MLEE analysis of 13 C. albicans isolates. While the mannose-6-phosphate isomerase activity was expressed by a single locus, the hexokinase activity was expressed by two loci, Hk-1 and HK-2. Reproduced from reference 288 with permission of the publisher.
MLEE has been used to fingerprint C. albicans (8, 9, 17, 34, 35, 67, 91, 179, 180, 182, 234, 288, 291, 292, 301, 321), C. tropicalis (95, 180), Candida lusitaniae (220), Candida haemulonii (183), Candida parapsilosis (188), Candida guillermondii (325), Cryptococcus neoformans (28, 40, 41, 42, 43, 316), and Aspergillus fumigatus (307). This method was recently verified for C. albicans by a cluster analysis of a set of test isolates in which MLEE, randomly amplified polymorphic DNA (RAPD) and Southern blot hybridization with fingerprinting probes were compared and parity was demonstrated (288). It was further demonstrated that if enough markers are used, MLEE will reveal microevolution within strains (288). There is little question that MLEE is an excellent method for DNA fingerprinting infectious fungi, since information is provided at all of the levels of resolution outlined in Fig. 1. The only drawback to this method is that it is relatively time-consuming, because one must combine the data from at least 10 enzymes that provide variability among isolates.
Restriction Fragment Length Polymorphism without Hybridization
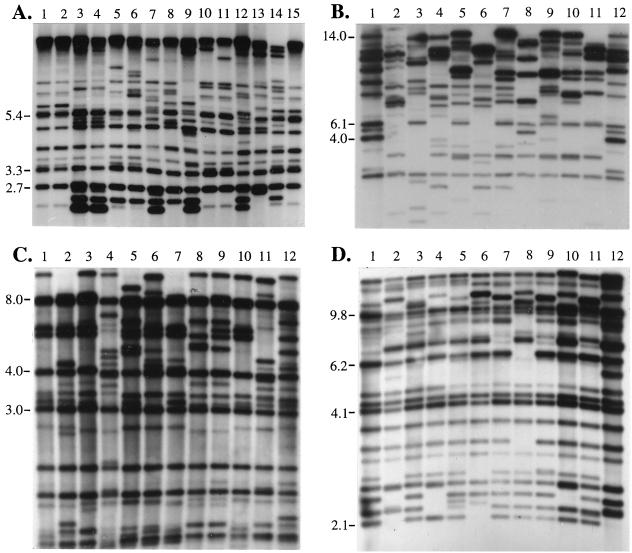
One of the first DNA fingerprinting methods used to assess strain relatedness in the infectious fungi is restriction fragment analysis, or restriction fragment length polymorphism (RFLP) comparisons, without probe hybridization. RFLP has been applied to a variety of infectious fungi, including C. albicans (21, 63, 77, 78, 146, 161, 208, 262, 326, 346, 382, 384, 399, 410), C. parapsilosis (39, 267), Candida rugosa (92), Candida utilis (1), C. tropicalis (94), C. lusitaniae (163, 266), Cryptococcus neoformans (82), Histoplasma capsulatum (405) and A. fumigatus (55, 186). The method is straightforward. DNA is extracted from spheroplasts, digested with one or more endonucleases, and separated by electrophoresis in an agarose gel. The banding pattern of digested DNA is then visualized, usually by staining with ethidium bromide. Separation depends upon the percentage of agarose in the gel, the electrophoresis time, the voltage, and the particular endonuclease(s) employed. All of the experimental conditions must be determined empirically. The pattern is based on different fragment lengths determined by the restriction sites identified by the particular endonuclease(s) employed. Variations among strains can occur as a result of changes in restriction site sequences, secondary modification of restriction sites, deletion of recognition sites, or deletions and insertions in the sequences between recognition sites.
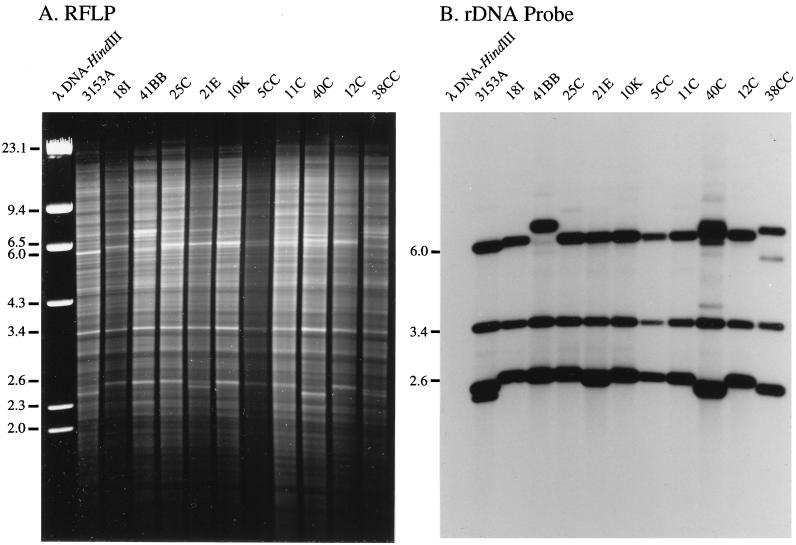
In bacteria, which have lower genomic complexity and contain less repetitive DNA than eukaryotes, the complex ethidium bromide-stained RFLP banding pattern can be more concise and the intensity of bands can be relatively equivalent. In spite of this, complex RFLPs have been used in only a limited number of DNA fingerprinting studies of bacteria (see, e.g., references 100, 132, 272, 273, and 380). In infectious fungi, the increased complexity of the genome increases the number of bands obtained with conventional endonucleases. This decreases the resolution of bands representing unique sequences. While the Escherichia coli genome contains approximately 3.0 × 107 bp (31), the C. albicans genome contains approximately 2 × 108 bp (304, 416), a 6.5-fold difference in size. The E. coli genome contains approximately 4,300 genes, while the estimate for C. albicans is at least 50% greater. Like all eukaryotic genomes, fungal genomes contain repetitive rRNA genes with relatively homologous sequences and intergenic regions. Eukaryotic ribosomal cistrons are usually clustered on one or two chromosomes. The C. albicans genome contains approximately 50 to 130 rDNA cistrons per diploid genome (149). Fungi also contain multiple copies of the mitochondrial genome (see, e.g., reference 373). rDNA sequences (326) and to a lesser extent mitochondrial DNA sequences (251, 427) represent the majority of intense bands in an RFLP pattern. A particularly good ethidium bromide-stained gel containing RFLP patterns of a number of C. albicans isolates is presented in Fig. 3A. The same gel after Southern blotting and hybridization with a radiolabeled probe of the C. albicans large-subunit, small-subunit, and 5S rDNAs is presented in Fig. 3B. Each lane of the ethidium bromide-stained gel (Fig. 3A) contains a complex background pattern of EcoRI fragments of various sizes and several intensely stained bands. While a subset of intensely stained bands was shown to represent mitochondrial DNA by hybridization (427), the majority of these bands were ribosomal, as is evident in a comparison of Fig. 3A and B. Variation is evident between isolates (Fig. 3A), but this variation is most readily resolved in the intensely hybridized rDNA bands (Fig. 3B). In contrast to the well-resolved low-intensity bands in the example in Fig. 3A, the low-intensity bands in most published RFLP patterns are poorly resolved. Therefore, comparisons of RFLP patterns rely in many cases only on differences between the most intense bands (i.e., the rDNA fragments). As will become clear in the next section, these fragments do not provide enough information to assess the relatedness of moderately related isolates. Even so, there is abundant evidence from the many RFLP studies of C. albicans (see, e.g., references 78, 262, 268, 326, and 346) that the method has been successful in identifying the same strain in independent isolates and in distinguishing among unrelated isolates. In a limited number of studies, the most intense bands have been manually digitized into databases for computer-assisted analysis (see, e.g., reference 78), but the general patterns are usually too dense and unresolved for automatic computer-assisted analyses. RFLP without a hybridization probe therefore represents a legitimate method for answering select epidemiological questions related to the infectious fungi, but it does not lend itself to studies in which cluster analyses of moderately related isolates is necessary. Therefore, RFLP is not well suited for large epidemiological studies. More importantly, the method has not been critically validated by comparison with another method for the levels of resolution listed in Fig. 1. It has not been critically tested by cluster analysis for how well it performs in grouping moderately related isolates, as have MLEE, RAPD and Southern blot hybridization with complex probes (288). It is faster, however, than all the alternative methods, including Southern blot hybridization, PCR-based methods, and sequencing methods.
FIG. 3.
(A) RFLP patterns for 11 C. albicans isolates. In each case, whole-cell DNA was digested with EcoRI, electrophoresed in an agarose gel, and stained with ethidium bromide. The resolution of bands in this gel is unusually good. (B) Southern blot hybridization of the gel in panel A with a radioactive ribosomal probe containing 28S, 18S, and 5S rDNA. Note that the most intense bands in the ethidium bromide-stained pattern are ribosomal.
Restriction Fragment Length Polymorphisms with Hybridization Probes
A general RFLP pattern of eukaryotic cell DNA visualized by ethidium bromide staining is poorly resolved primarily because all restriction fragments are stained. Fragments within the pattern are in fact highly resolved, and a method for selectively visualizing a limited number of fragments will provide a more highly resolved fingerprint pattern for analysis. To visualize particular fragments in the pattern, one can probe a Southern blot of the RFLP gel with a radiolabeled or biotinylated DNA sequence that recognizes one or more fragments as a result of sequence homology, as in Fig. 3B. The stringency of hybridization can be controlled by varying the salt concentration and/or temperature (318). If the probe identifies a unique sequence (e.g., a single gene), the resulting pattern will be simple. If the unique sequence is relatively intact within a single restriction fragment in the gel, the probe will hybridize to only that fragment. In a haploid organism, the pattern will include only one band. In a diploid organism, such as C. albicans, the pattern will include one or two bands. If a site for the restriction enzyme employed is contained within the unique sequence, more than two bands are possible. Single-gene probes can discriminate among some isolates based upon allelic polymorphisms. However, because single-gene probes generate patterns with only one or two bands, they do not alone provide the level of data complexity necessary for measuring genetic distance. For instance, the URA5 gene was used to probe HaeII-digested DNA of 17 clinical isolates of C. neoformans (82). Isolates were separated into four types based on the size of URA5-containing fragments, but the resolution (i.e., the number of isolates distinguished genetically) was far below that obtained in the same study with the repetitive CNRE-1 DNA probe (82).
rDNA and mDNA probes.
The low resolution of a single-copy probe was realized quite early by researchers attempting to DNA fingerprint fungi. It was apparent that by using a number of single-copy probes, one could obtain data similar to those of MLEE, but the process would be labor-intensive. Therefore, probes that hybridized to repeat sequences dispersed throughout the genome were sought, since it was believed that either the repeat sequences or bordering sequences would vary among strains and that the resulting data from one Southern blot hybridization pattern would be complex enough to reflect genetic distance. Labeled C. albicans rDNA was used to probe Southern blots of C. albicans total-cell DNA, and although the method provided some resolution of unrelated strains (206, 217), the complexity of the hybridization pattern was not that much greater than that of patterns obtained with single-copy probes. Using rDNA as a probe, Stein et al. (370) were able to distinguish five patterns in 18 isolates of C. albicans. The rDNA hybridization patterns with EcoRI-digested DNA had a maximum of three bands (370, 415) and were comparable to those in Fig. 3B. One, two, or all three rDNA bands were common to apparently unrelated isolates at a relatively high frequency, suggesting homoplasy. rDNA and spacer regions of rDNA have also been tested as DNA fingerprinting probes in a number of infectious fungi other than C. albicans (see, e.g., references 62, 111, 119, 363, and 366), but in no case were complex enough patterns generated to be considered effective fingerprinting probes.
In bacteria, rDNA probes do generate relatively complex fingerprint patterns (49, 88, 138, 286) and have been the basis of relatively elegant automated ribotyping systems, such as the Riboprinter Microbial Characterization System marketed by Qualicon (Wilmington, Del.). Because ribosomal cistrons are dispersed throughout the single-chromosome genome of bacteria (237), endonuclease digests of bacterial DNA will contain multiple fragments of different sizes containing rDNA sequences. Thus, variability in fragment size will depend on differences in sequences bordering the rDNA cistrons as well as rDNA polymorphisms. This has resulted in high levels of resolution for discriminating among bacterial species and strains. In contrast, eukaryotic ribosomal cistrons are clustered (126, 135, 339, 406). Endonuclease digestion of these tandem repeats, which are separated by spacer sequences, generates fragments of similar relative sizes, resulting in a far simpler Southern blot hybridization pattern (i.e., a pattern with fewer bands) (Fig. 3B) and therefore low in resolution for strain discrimination (see, e.g., reference 320). Therefore, although effective in DNA fingerprinting bacteria, rDNA probes have not been that effective in DNA fingerprinting fungi.
The pattern obtained by probing a Southern blot of EcoRI-digested whole-cell DNA of C. albicans with mitochondrial DNA appears to be more complex than that obtained with a rDNA probe. Wills et al. (427) obtained five distinct bands, and the pattern varied among isolates sufficiently to suggest that Southern blot hybridization with a mitochondrial DNA probe would be effective in identifying the same strain in independent isolates and in distinguishing among unrelated strains of C. albicans (251). In an analysis of type I and type II Candida stellatoidea, a close relative of C. albicans, the hybridization patterns of type I isolates were identical but those of type II isolates varied (170). The patterns of some type II C. stellatoidea isolates were indistinguishable from those of some C. albicans isolates, demonstrating a lack of species specificity or low resolving power, if one accepts C. stellatoidea as an independent species (170). It is more likely that C. stellatoidea type II represents a subgroup of C. albicans, as has been suggested (172, 290). rDNA and mitochondrial DNA probes, therefore, have not been generally used in broad epidemiological studies of the infectious fungi, and neither method has been validated for the different levels of genetic resolution.
Repetitive and complex DNA probes for C. albicans.
To date, the most successful and popular hybridization probes for the fungi have been cloned fragments containing repetitive genomic sequences. Two such probes for C. albicans, 27A (327) and Ca3 (315, 359), were cloned at approximately the same time in the late 1980s and subsequently were found to be related (Fig. 4) (289). Because both contain sequences of the C. albicans repetitive element RPS and common non-RPS upstream sequences (Fig. 4) (73, 150), these probes hybridize to a majority of the same bands in a Southern blot. However, the two probes are not identical. 27A contains sequences downstream of the RPS cluster that hybridize to unique bands, while Ca3 contains sequences upstream of the RPS cluster that hybridize to unique bands (Fig. 4) (289). By comparison, Ca3 is a more complex probe than 27A, containing an additional repetitive sequence, the B sequence (Fig. 4). Ca3 generates a pattern that is, on average, more complex than that generated by 27A (C. Pujol, S. Joly, S. Lockhart, and D. R. Soll, unpublished observations). Furthermore, DNA fingerprinting with Ca3 fulfills the general requirements set forth in a previous section of this review for effective fingerprinting methods (288). Ca3 and 27A have been used in a number of epidemiological studies of C. albicans and the highly related species C. dubliniensis (see, e.g., references 6, 80, 137, 164, 168, 192, 196, 216, 218, 227, 246, 265, 271, 297, 310, 315, 327, 328, 329, 330, 332, 334–336, 352, 357, 358, 359, 374, 377, and 419).
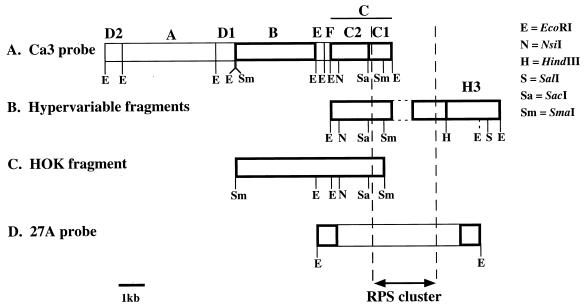
FIG. 4.
Physical maps of the complex DNA fingerprinting probe Ca3 (3, 192, 290), the HOK fragment (76), hypervariable fragments of isolates that hybridize to Ca3 (290), and an RPS cluster (150) of C. albicans. Known DNA sequences are represented by boxes with thick walls. Relative positions are assigned by DNA homology. RPS regions contain different numbers of full-length RPS units and are demarcated by dashed borders. The vertically dashed EcoRI site located to the right of the RPS region of the hypervariable fragments was observed in only one fragment (290). Reproduced from reference 76 with permission of the publisher.
The logic and methods used to clone and characterize 27A and Ca3 have also been used to develop similar complex probes for other infectious fungi, including A. fumigatus (122), C. tropicalis (152), C. glabrata (194), C. dubliniensis (151), C. parapsilosis (L. Enger and D. R. Soll, unpublished data), and C. lusitaniae (S. R. Lockhart, A. Hill, M. Pfaller, and D. R. Soll, unpublished data). The logic behind using a complex probe is relatively straightforward. In a Southern blot of endonuclease-digested genomic DNA, such a probe will hybridize to repetitive sequences dispersed throughout the genome, thus identifying variability among isolates at a variety of dispersed loci. It will also hybridize to additional sequences that are less variable, including sequences that vary as a result of allelic polymorphisms. Finally, it will hybridize to some hypervariable sequences, revealing microevolutionary changes within a strain. The virtue of the complex probe is that all this information is provided by a single Southern blot hybridization pattern. A complex probe should generate a pattern complex enough to provide an accurate and sensitive measure that reflects the relatedness of isolates. The major portion of the pattern it generates must be relatively stable over time for a particular strain. In addition, the probe should contain one or more sequences that hybridize to monomorphic fragments (i.e., fragments that exhibit the same size in all or most strains within a species). Monomorphic bands will facilitate normalization to a universal standard for computer-assisted storage for subsequent retrospective and comparative studies (see below).
Cloning complex DNA probes.
To clone complex DNA fingerprinting probes, genomic DNA is first prepared from spheroplasts, being careful not to shear the DNA unnecessarily. Contaminating RNA is digested with RNaseA. The genomic DNA is then incubated with a selected endonuclease (e.g., Sau3AI), aliquots are removed at short time intervals, and the reaction is stopped. The DNA of each sample is electrophoresed, and the time at which the majority of DNA falls in the size range of 10 to 20 kb is noted. The reaction is then scaled up. Fractions in the 10- to 20-kb range are ligated to lambda arms, packaged, titrated, and amplified. This library is then screened with radiolabeled genomic DNA for fragments that harbor at least one repetitive sequence. This can be accomplished either by limiting the time of hybridization or by limiting the concentration of probe. One can simultaneously screen duplicate blots for fragments that contain ribosomal sequences. In the blot probed with genomic DNA, intensely hybridizing clones that appear as dark dots in the autoradiogram (Fig. 5A) represent putative repetitive DNA inserts. Those dark dots with no correlates in the duplicate blot probed with rDNA (Fig. 5B) represent putative complex probes.
FIG. 5.
Hybridization screen used to select genomic clones of A. fumigatus that contained repetitive sequences that were nonribosomal. A genomic library was blotted on duplicate filters and hybridized with either radiolabeled genomic DNA (A) or a radiolabeled ribosomal probe (B). R, clones containing ribosomal DNA sequences; 1, 2, 3, potential complex probes containing repetitive sequences but devoid of ribosomal sequences. Reproduced from reference 123 with permission of the publisher.
When this method was recently applied to C. glabrata, seven clones were isolated that generated both unique and common bands when used to probe Southern blots of EcoRI-digested DNA of a set of test isolates (194). In all cases, the patterns appeared complex enough to compute meaningful similarity coefficients between pairs of isolates (194). However, the seven probes generated patterns for the same strain of C. glabrata with different degrees of band similarity. They also cross-hybridized, suggesting that they contained common sequences. One of these probes, Cg6, was tested for its capacity to discriminate among unrelated isolates, identify the same strain in different isolates, and identify microevolution within an infecting population (194). The effectiveness of Cg6 at the different levels of genetic relatedness (Fig. 1) was assessed by comparing it to the RAPD method in a cluster analysis (194). The two unrelated methods proved similar in identifying the same strain in different samples, distinguishing among unrelated isolates and clustering moderately related isolates into similar groups. In applying the same type of screen to C. tropicalis (152), 10 clones were isolated that generated complex banding patterns that varied among unrelated isolates. In these 10 clones, two general patterns emerged, suggesting that the probes could be separated into two groups, each containing a distinct family of repetitive elements. This was supported by the absence of cross-hybridization between probes from the two groups (152). Again, the effectiveness of the C. tropicalis probes Ct14 and Ct3, representing the two respective groups, was demonstrated through cross-comparison of their capacity to discriminate among unrelated isolates, identify the same strain in different isolates and cluster moderately related isolates (152).
In Fig. 6A through D, examples are presented of the DNA fingerprint patterns generated by the species-specific complex DNA probes Ca3 for C. albicans (192, 315, 332, 359), Ct14 for C. tropicalis (152), Cg6 for C. glabrata (194), and Cd25-1 for C. dubliniensis (151), respectively. Each probe hybridized to fragments of different molecular sizes, and each hybridized to one or more monomorphic bands. The major bands are more distinct than ethidium bromide-stained patterns of similar EcoRI-digested and electrophoretically separated genomic DNA (Fig. 3A).
FIG. 6.
Examples of Southern blot hybridization patterns generated with the complex probes Ca3 of C. albicans (A), Ct14 of C. tropicalis (B), Cg6 of C. glabrata (C), and Cd25-1 of C. dubliniensis (D).
Characterizing complex DNA probes.
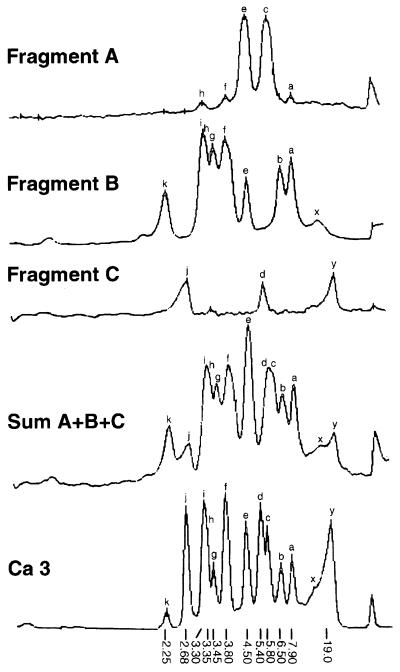
Once a complex probe is cloned, it should be physically mapped, analyzed for genomic distribution, and characterized for resolution. Since the 27A and Ca3 probes of C. albicans represent the most carefully characterized complex probes used to DNA fingerprint the infectious fungi, their characterization will be reviewed in some detail. Scherer and Stevens (327) first demonstrated that the 6.7-kb probe 27A, which was cloned from C. albicans strain 616, a fresh clinical isolate, contained a repeat sequence dispersed throughout the genome. They analyzed two clones from a partial Sau3A digest of genomic DNA that hybridized to the 27A probe and demonstrated that 27A and the two clones contained a common sequence (327), subsequently identified by Iwagushi et al. (150) to be the RPS repetitive element (Fig. 4). The 11-kb probe Ca3, first referred to as JH3 in an article on phenotypic switching in vaginitis isolates (359), was subsequently renamed Ca3 (360). Ca3 was cloned from laboratory strain 3153A and was shown to be dispersed on seven of the eight chromosomes of C. albicans (315). Anderson et al. (3) subsequently digested the Ca3 probe with EcoRI and obtained seven fragments, which, in descending order of size, were labeled A, B, C, D1, D2, E, and F (Fig. 4). The fragments were subsequently mapped in the 5′-to-3′ order (289) (Fig. 4). When EcoRI-digested DNA of C. albicans strain 3153A was probed with fragments A (∼4.2 kb), B (∼3.0 kb), and C (∼2.9 kb), three distinct patterns were obtained which, when combined, accounted for all of the bands obtained when the entire Ca3 probe was employed (Fig. 7). Fragment A generated three different hybridization patterns when used to probe Southern blots of EcoRI-digested DNA of a set of C. albicans isolates, the first consisting of one band at ∼5.8 kb, the second consisting of one band at ∼4.5 kb, and the third consisting of two bands at ∼5.8 and ∼4.5 kb. Thus, fragment A identified a single unique-copy gene, and these three patterns represented allelic variations of a single gene locus. Fragment B generated a pattern which included over half of the bands in the complete Ca3 pattern. The majority of B-pattern bands were polymorphic and represented most of the moderately variable bands necessary for cluster analyses used to demonstrate parity with MLEE and RAPD analysis (288). Fragment C generated patterns that included the highly variable high-molecular-weight bands that have proven valuable in assessing microevolution within an infecting strain (see, e.g., references 192, 196, 289, 330, and 336).
FIG. 7.
Pixel density scans of the Southern blot hybridization patterns generated with EcoRI fragments A, B, and C (Fig. 4) of the Ca3 probe and EcoRI-digested DNA of C. albicans strain 3153A. Note that the summed patterns of fragments A, B, and C (A+B+C) identify all of the major bands in the Ca3 pattern. Reproduced from reference 3 with permission of the publisher.
To determine the genomic distribution of sequences hybridizing to the three major fragments A, B, and C, Southern blots of electrophoretically separated C. albicans chromosomes were probed with the three fragments (3). Seven distinct chromosomal bands of C. albicans strain 3153A were separated by transverse alternating-field electrophoresis (TAFE) and numbered in descending order of size. Since C. albicans contains eight chromosomes (420), there was overlap in at least one position. The entire Ca3 probe hybridized strongly to bands 1, 3, 5, 6, and 7. Subfragment A hybridized strongly to band 7 and weakly to bands 1 and 3. Subfragment B hybridized strongly to bands 5 and 7, and weakly to band 6. Subfragment C hybridized strongly to bands 1, 3, and 6 and weakly to bands 5 and 7. These results demonstrated that both subfragments B and C contained sequences that were dispersed on more than one chromosome and that subfragment C, which contained RPS sequences, was more highly dispersed than was subfragment B.
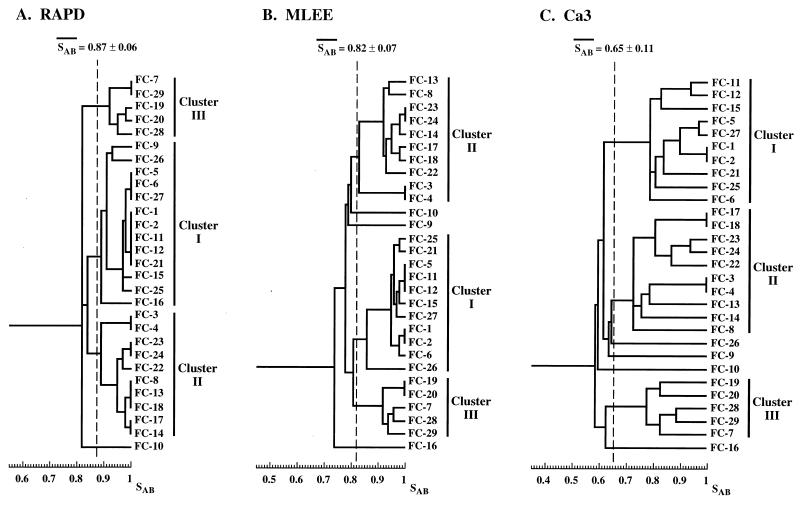
The physical relationships between the Ca3 probe (315), the large EcoRI genomic fragments to which the C fragment of Ca3 hybridizes (3, 289), the 27A probe (327), the recently characterized related HOK fragment (76), and the RPS element (73, 150) were recently determined (74, 76, 287, 289). A comparison of the physical maps is presented in Fig. 4. The three probes Ca3, 27A, and HOK all contain sequences of the C. albicans RPS element and the upstream C2 border of the RPS cluster. Therefore, all three identify a set of common bands in a Southern blot of EcoRI-digested genomic DNA. If the RPS element alone is used as a fingerprinting probe, the pattern is similar to that obtained with the C fragment of the Ca3 probe (150, 289). Although such a pattern is useful in assessing microevolution based on the hypervariability of full-length RPS sequences located in tandem at particular sites in the genome (289), its usefulness as a DNA fingerprinting probe falls short in clustering isolates that are not highly related (289). The problem of using a probe that contains primarily a repetitive dispersed genomic element was recently demonstrated in C. albicans. Lasker et al. (175) cloned the species-specific repetitive element CARE-2 from C. albicans genomic DNA. CARE-2 is a 1.06-kb sequence represented 10 to 14 times in the haploid C. albicans genome (175). It is dispersed on all chromosomes, and Southern blot hybridization demonstrated different copy numbers on different chromosomes (175). When Southern blots of EcoRI-digested DNA of a variety of isolates were probed with labeled CARE-2, complex hybridization patterns were generated that contained approximately the same number of bands as Southern blots probed with Ca3, but in contrast to Ca3, every band in the CARE-2 pattern was variable (288). CARE-2 distinguished unrelated isolates and identified the same strain in independent isolates. However, while the MLEE, RAPD, and Ca3 fingerprinting methods clustered the collection of isolates into three groups in a highly similar fashion (Fig. 8), two of the three groups fragmented into unrelated, smaller groups in the CARE-2 dendrogram (288). Therefore, because CARE-2 identifies only hypervariable fragments, it is far less effective in clustering moderately related isolates.
FIG. 8.
Dendrograms for a set of 29 isolates of C. albicans DNA fingerprinted by RAPD (A), MLEE (B), and Ca3 (C). Reproduced from reference 288 with permission of the publisher.
The results with CARE-2 (288) point to a common misconception regarding a useful DNA fingerprinting probe. There is a tendency to want to reduce a complex DNA fingerprinting probe to a single repeat element, as in the case of reducing Ca3 to the RPS element, based upon the misinformed notion that the more variable a pattern is, the better it will serve in DNA fingerprinting strains. When the bands in the Ca3 pattern were individually analyzed for variability, subsets identified by the subfragments exhibited hypervariability, moderate variability, low variability, and no variability. This combination strengthens the probe, such that the several levels of resolution we have proposed for an effective DNA fingerprinting method are attained (Fig. 1). Reducing Ca3 to RPS results in a probe useful primarily in studies of strain microevolution (289; Pujol et al., unpublished). Therefore, it is a misconception to strive for a probe composed exclusively of a repetitive element, and it is a misnomer to refer to a complex probe as a repetitive probe.
Repetitive and complex probes for fungi other than C. albicans.
As noted, complex species-specific probes have been cloned using the above general method for a variety of infectious fungi (122, 151, 152, 193, 194, 315, 327, 359; L. Enger, S. Joly, C. Pujol, and D. R. Soll, unpublished data). Both the patterns generated by these probes and the variability these probes identify among independent isolates (Fig. 6) suggest that future characterization of the non-C. albicans probes will reveal combinations of unique and repetitive elements similar to those contained in the complex C. albicans probe Ca3 (3, 289). A variety of other probes have been developed for C. albicans over the past several years that might be useful for DNA fingerprinting studies. Wilkinson et al. (422) first used poly(G-T) as a Southern blot hybridization probe and demonstrated variation among C. albicans isolates and differences between C. albicans and C. glabrata. Noting in 1993 that there were no good, accessible probes for Candida species other than for C. albicans, Sullivan et al. tested five oligonucleotides, (GGAT)4, (GGTG)5, (GATA)4, (GACA)4 and (GT)8, on a variety of Candida species and selected (CT)8 as the most effective for strain discrimination based on pattern complexity and variability (374). These sequences, which identify microsatellite regions, must still be characterized to determine whether they will serve as effective fingerprinting systems. Thrash-Bingham and Gorman (390) cloned two repetitive sequences, the 0.2-kb Rel-1 and the 2.8-kb Rel-2 sequences. Rel-2 belongs to the same repeat family as CARE-2. One subcloned fragment of Rel-2 that included bp 1478 to 2787 generated a complex pattern, but neither Rel-1 nor Rel-2 was characterized further for their potential use in DNA fingerprinting. Carlotti et al. (62) cloned the probe CkF1,2, which included the 3′ end of the large-subunit rRNA gene and the repeat sequence CKRS-1, from Candida krusei. Banding patterns appeared laddered, suggesting tandem deletion and duplication of the 165-bp kre element. The fingerprint pattern of CkF1,2, however, was not complex enough to be used to assess the relatedness of moderately related isolates in cluster analyses, and the probe has not been validated for fingerprinting. Such a probe, however, should be effective in assessing microevolution in infecting populations of C. krusei.
Probes containing repeat sequences have also been used to discriminate among strains of Cryptococcus neoformans. Spitzer and Spitzer (364) cloned CNRE-1, which hybridized to seven C. neoformans chromosomes and to 12 bands of a Southern blot of SstI-digested DNA of strain ATCC 6352. However, CNRE-1 generated far fewer bands and with far less intensity when hybridized to SstI-digested DNA of strain ATCC 28958 (171). This difference in hybridization appears to reflect genetic differences between serotypes A and D of C. neoformans. In addition, CNRE-1 exhibited only weak hybridization to serotype C strains. CNRE-1 fingerprinting was evaluated by comparing its discriminatory capacity to that of URA5 gene sequences (117). Groupings of URA5 alleles by parsimony analysis coincided with CNRE-1 groupings. These results suggested little genetic exchange between groups within a single geographical locale. A second DNA probe, the linear autonomous plasmid UT-4p, was used by Varma and Kwon-Chung to fingerprint C. neoformans (397). UT-4p generated hybridization patterns specific to the different serotypes of C. neoformans and showed minor variation among isolates within each group. The UT-4p pattern was stable in strains serially subcultured and identified the same strain in different isolates (397). As with CNRE-1 fingerprinting, UT-4p fingerprinting has also been used in a limited number of epidemiological studies (102). However, neither CNRE-1 nor UT-4p has been sufficiently characterized for the levels of resolution set forth in Fig. 1.
Several fingerprinting probes have been developed for Aspergillus flavus. pAF28 represents a 6.2-kb partial EcoRI digestion fragment that is species specific and generates a complex Southern blot hybridization pattern (214). This probe effectively discriminated among unrelated isolates. Repetitive sequences derived from Aspergillus nidulans and Neurospora crassa have also been used as probes for A. flavus, A. parasiticus, and A. nomius (230). Using the strategy employed to clone Ca3, several fingerprinting probes, including 3.11, 3.19, and 3.9, were developed for A. fumigatus (122). By limited cluster analysis, the three probes grouped the majority of moderately related A. fumigatus isolates in a similar fashion. These probes were subsequently demonstrated to be quite effective in the analysis of nosocomial infections (see, e.g., reference 124). Other probes have also been developed for Histoplasma capsulatum. rDNA and mitochondrial DNA probes have been used (363), but, again, they have not evolved for general use because of problems of resolution. One probe, yps-3, which hybridizes to yeast-phase-specific mRNAs of H. capsulatum (159), was subsequently used as a fingerprinting probe (158). yps-3 identified polymorphic differences between strains but generated only a very limited pattern of one or two bands. Therefore, yps-3 does not provide sufficiently complex data to be considered an effective fingerprinting probe.
In summary, complex DNA probes can be the basis for extremely effective DNA fingerprinting methods. Probes that generate patterns of only a few bands, however, do not provide enough complexity, and probes that contain exclusively a dispersed repetitive element may identify sequences too hypervariable for clustering moderately related isolates. The latter probes may, however, be very effective in studies of microevolution or in studies in which strains must be identified simply as related or unrelated. In some cases, while one probe may be more effective in clustering moderately related isolates, another may be more effective in identifying microevolutionary changes. In such cases, both probes can be used and the data can be combined, as has been suggested for the C. tropicalis probes Ct14 and Ct3 (152).
Random Amplified Polymorphic DNA and Related PCR-Based Fingerprinting Methods
Although a variety of PCR-based strategies have been developed for DNA fingerprinting purposes (59), RAPD (59, 413, 423) has evolved as the most popular method for DNA fingerprinting the infectious fungi. Using random primers of approximately 10 bases, amplicons throughout the genome are targeted and amplified. Amplified products are separated on an agarose gel and stained with ethidium bromide. When a single random oligonucleotide primer is used in a reaction, it hybridizes to homologous sequences in the genome. If the primer hybridizes to sequences on alternative DNA strands within roughly 3 kb, the DNA region between the two hybridization sites will be amplified using Taq polymerase. In the first reaction on each strand, a sequence is replicated by the DNA polymerase beginning at the site of hybridization and extending beyond the point at which there is a cognate sequence on the opposite strand. However, in the second reaction, the primer finds the homologous site within the first amplified strand and the replication reaction extends the second strand to the terminus of the sequence, which is the homologous sequence on the opposite strand. This second reaction produces the first amplified sequences equal in length to the targeted amplicon. In the amplification reactions that follow, primers continue to promote exclusively the synthesis of fragments of the amplicon sequence.
In the development of a RAPD DNA fingerprinting system for a particular species, a number of oligonucleotide primers must be tested, and those that provide the best variability among independent isolates are selected. Although a single primer can generate a relatively complex pattern that varies among isolates, in most cases a single primer provides one to three intense bands that may differ among isolates. Therefore, one must usually select a number of primers, run each independently for each test isolate, and combine the information. This strategy is illustrated by the work of Pujol et al. (288), who tested 40 random primers (each 10 bases in length) on a limited number of C. albicans test isolates; 8 were selected that provided maximum variability. Only reproducible, intense bands in each pattern were used in the analysis. Each primer generated one to six intense bands. Examples of RAPD patterns with the primers OPE-3 and OPE-18 are presented in Fig. 9. The combined number of intense bands generated by eight primers selected by Pujol et al. for variability among isolates was 31, but only 16 were polymorphic for the 29 test isolates. When dendrograms were generated based on similarity coefficients computed from the combined data obtained with the eight probes, clustering of the 29 test isolates was similar to that obtained using either MLEE or Southern blot hybridization with the Ca3 probe. Parity has also been demonstrated between the RAPD method and Southern blot hybridization with complex probes of C. glabrata (194). The results of Pujol et al. (288) suggest that the RAPD method of fingerprinting attains the same data complexity and resolving power as complex DNA fingerprinting probes and MLEE. These findings are of no surprise. Complex probes such as Ca3 identify a number of homologous sequences throughout the genome homologous to different portions of the probe, and MLEE accumulates allelic data for a number of genes. A combination of random oligonucleotide primers provides similar data. Pujol et al. (288) demonstrated that RAPD, MLEE, and Ca3 fingerprinting of C. albicans not only clustered moderately related isolates in a similar fashion but also afforded similar levels of resolution of microevolution within a clonal population. However, it should be noted that the microevolutionary changes identified by the three methods were independent and that the methods therefore did not identify changes in the same highly related isolates. Rather, the methods measured similar frequencies of variants within the same strain. Recently, the RAPD method was used to analyze the level of homology among fragments amplified by the same primers in closely related species of the sunflower (303). A total of 91% of comigrating fragments exhibited homology. This represents a high level of homology, given that the test was interspecific. Thormann et al. (388) further demonstrated that RAPD fragments of cruciferous plant species with similar molecular weights were identical, but they were not always identical between species. Together, these results demonstrate the efficacy of the RAPD method in assessing genetic relatedness. Similar tests have not yet been reported for the fungi, although one can assume that the outcomes will be similar.
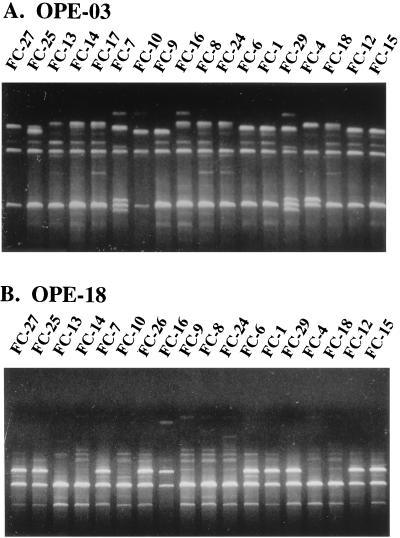
FIG. 9.
Examples of RAPD patterns with the primers OPE-3 (A) and OPE-18 (B) for different C. albicans isolates. Reproduced from reference 288 with permission of the publisher.
The RAPD method of DNA fingerprinting has become quite popular for all infectious fungi and has been successfully applied to C. albicans (see, e.g., references 22, 36, 37, 77, 83, 86, 131, 140, 141, 181, 223, 288, 306, 309, 369, and 393), C. dubliniensis (79), C. parapsilosis (199), C. lusitaniae (163, 181), C. tropicalis (181, 187), C. glabrata (195, 181, 337, 387), C. krusei (437), C. famata (437), C. rugosa, (437), A. fumigatus (5, 11, 178, 187, 200, 202, 228, 296), A. flavus (51), Cryptococcus neoformans (32, 33, 41, 68, 71, 72, 117, 197, 282, 311, 361, 362, 375), H. capsulatum (160, 431), and Blastomyces dermatitidis (435). However, several caveats of the procedure must be kept in mind. First, there is the problem of reproducibility not only among laboratories but within a laboratory over time, and this single problem, although not insurmountable, makes the development of a common database difficult. Virtually every methodological aspect of PCR can affect reproducibility. Artifactual variation can occur as a result of small differences in the primer-to-template-concentration ratio, the temperatures during the amplification reaction, and the concentration of magnesium in the reaction mixture (104). Changes in these parameters affect most notably the presence of low-intensity bands but can also affect the position and intensity of high-intensity bands. This may explain the general result that even in the same laboratory, using the same thermocycler and reagents, variation occurs in the low-intensity bands (see, e.g., reference 215). Even more disturbing are reports of variation due to the Taq enzyme source. Louden et al., who have used the RAPD methodology to fingerprint A. fumigatus isolates in a number of studies (202), reported that different lots of Taq polymerase resulted in great enough variability to generate what they termed “pseudoclusters” (201). Evidence was presented that one enzyme lot could not discriminate among isolates while the other could. Meunier and Grimont (222) reported artifactual variation due to the use of Taq DNA polymerase from different manufacturers. They also added the manufacturer of the thermocycler to the list of variables that could affect the RAPD pattern. The artifacts observed were primarily, but not exclusively, in low-intensity bands (222). It is therefore imperative that such factors be considered when comparing RAPD patterns from different laboratories. Such factors place constraints on the use of this technology in generating a global database.
One promising modification of RAPD is amplified fragment length polymorphism (408). This method selectively amplifies restriction fragments in a genomic DNA digest. Amplification is achieved by using the adapter and restriction sites for the annealing of primers, and fragment selection is achieved by adding selective bases to the 3′ end of the primers. By using stringent reaction conditions for primer annealing, the reliability of the method has been reported to be superior to that of the RAPD method, and careful selection of the restriction enzymes and selective nucleotides at the primer 3′ ends results in a complex fingerprint pattern in a sequencing gel with yeast DNA as template (408). This method may evolve into a superior DNA fingerprinting method, but at the time of this review there has been no meaningful development for the infectious fungi.
The PCR method can also be used to target specific sequences. Customized primers have been developed for the spacer regions between ribosomal cistrons, the spacer regions between tRNAs, and microsatellite sequences. Theoretically, the complexity of the information obtained with primers to these repetitive sequences should be similar to that using hybridization probes of the same sequences. Therefore, primers customized for ribosomal spacer sequences will most probably not provide great enough complexity. For microsatellites, primers can be designed outside of the regions (221), or primers like (GTG)5 can be used. In the former case, differences in size are due to insertions and deletions of the short tandem repeats of the microsatellite regions. For a given locus, these differences can be interpreted as alleles, but these regions may undergo recombination at extremely high frequencies and may therefore have their greatest value in the analysis of microevolution. In the case of primers like (GTG)5, the primers will amplify genomic fragments flanked by inversely oriented targeted motifs. This method, referred to as interrepeat PCR, should result in a higher level of reproducibility than RAPD because the annealing temperature is higher (19, 224).
Thanos et al. (387) compared the arbitrary 10-mer primer AP3, the primer T3B designed on the basis of the intergenic tRNA spacer region, and the primer (GTG)5 designed on the basis of a microsatellite sequence for their capacity to discriminate not only among Candida species but also among strains within a species. The discriminatory capacity of the arbitrary 10-mer primer was greater than that of the intergenic tRNA spacer primer (387). Although Thanos et al. (387) demonstrated that intergenic tRNA spacer sequences vary among species, variability among strains within a species did not appear to be great enough for clustering moderately related isolates. Microsatellite and minisatellite primers should be more effective, since these sequences are usually dispersed throughout the genome. However, as with repeat sequence probes, variability due to a high frequency of change in satellite DNA sequences may decrease the effectiveness of the method in clustering moderately related isolates. For C. albicans, repeat sequences targeted for amplification have included minisatellite and microsatellite sequences (90, 177, 333, 375), the CARE-2 element and the COM-21 element, a repeat element within the RPS1 repetitive element (298), repeat sequences found in prokaryotes and eukaryotes (395, 396), and rDNA (15, 376).
Methods that combine amplification of an rRNA cistron and RFLP have been used to fingerprint bacteria and, recently, yeast. Primers for a conserved region of the ribosomal cistron are used to amplify DNA of an isolate by PCR. The reaction products are digested with one or more endonucleases and separated by electrophoresis, and the digestion pattern is analyzed. Baleiras Couto et al. (15) developed primers for a number of food-borne yeasts based on a conserved region between bp 34 and 1268 of the small-subunit rRNA sequence of C. albicans. The primers used for C. albicans and which represented the model for primers of related species were between bp 34 and 53 and between bp 1250 and 1268. The species analyzed included C. albicans, C. guillermondii, C. kefyr, C. krusei, C. glabrata, C. lusitaniae, C. parapsilosis, C. tropicalis, Hansenula polymorpha, Kluyveromyces lactis, Saccharomyces cerevisiae, Zygosaccharomyces rouxii, Pichia membranafaciens, and Schizosaccharomyces pombe (15). RFLP patterns were visualized by ethidium bromide staining. Unfortunately, the resolving power for strains within a species was limited, and only one of the seven tested endonucleases, MseI, discriminated among the five species Z. bailii, Z. rouxii, S. cerevisiae, C. valida, and C. lipolytica. The patterns were not complex, and differences even among species included only a few bands maximally. Differences among strains within a species usually included one or two bands. Therefore, this method will probably not be effective for DNA fingerprinting the infectious fungi using the ribosomal cistrons. It may, however, prove more effective with other more polymorphic genomic sequences.
A PCR-based strategy based on a method developed by Karl and Avise (155) to measure genetic relatedness using anonymous genetic markers has recently been used to fingerprint the infectious fungi (57, 58, 64, 130, 432, 433). In this strategy, arbitrary primers are used to amplify random DNA sequences. RAPD bands are excised and partially sequenced, and customized pairs of primers are generated for each band. Either single-strand conformational polymorphism analysis (252) or RFLP is then used to analyze polymorphisms among isolates. The advantage of this method is that there are usually only two identifiable alleles and so alleles from different individuals can be unambiguously identified (386). However, the method may be problematic when applied to diploid organisms such as C. albicans.
Recently, Geiser et al. (122) developed a similar approach that targeted noncoding regions (e.g., 5′ upstream regions and introns) of protein-coding loci to analyze the population structure of 31 A. flavus isolates collected in Australia. In this study, noncoding sequences of 11 identified genes were amplified using primer pairs customized from the known sequences of the selected genes, and examined for polymorphisms by single-strand conformational polymorphism. Representative alleles were sequenced to identify polymorphic restriction sites. The 11 genes were then amplified in each test isolate, and the PCR product was digested with the restriction enzymes that generated polymorphisms. Each gene in an isolate was then coded 1 or 0, representing the presence or absence, respectively, of the restriction site, and multilocus genotypes were generated and compared. The added strength of this approach is that it targets identified genes as well as noncoding regions, which, it is argued, may not have the same restrictive evolutionary constraints of coding regions.
Electrophoretic Karyotyping
With the invention of pulsed-field gel electrophoresis (338), orthogonal-field-alternation gel electrophoresis (OFAGE), and variations upon these basic electrophoretic karyotyping systems (e.g., contour-clamped homogeneous electric field gel electrophoresis [CHEF] and TAFE), chromosome-sized fragments of the yeast genome were readily separated in a gel. These inventions seemed ideal for fungal epidemiology. The general technology is straightforward. Cells are mixed with enzymes to remove the cell wall and then embedded in an agarose plug. Protease and detergent are added, and the cells are incubated to remove membranes and digest protein. The agarose gel protects the large DNA molecules from shearing forces. The agarose plug is placed in a well at the top of an agarose slab gel, and electrophoresis is carried out according to the specifications of the particular separating system. Yeast chromosome-sized DNA fragments are separated according to size and can be visualized by ethidium bromide staining. Specific chromosomes can be identified using Southern blot hybridization with chromosome-specific DNA probes (e.g., rDNA). In the earliest application of this general method, which we refer to as electrophoretic karyotyping, it was demonstrated that patterns varied among unrelated isolates of C. albicans (see, e.g., references 171, 174, 198, 207, 219, 349, 381, and 407) and therefore provided a potential method for fingerprinting. Using Southern blot hybridization with cloned genes as probes, Thrash-Bingham and Gorman (389) subsequently demonstrated that in spite of karyotypic variability among strains of C. albicans, the general genomic organization was maintained and that translocations contributed to karyotypic variability. More importantly, Sangeorzan et al. (323) demonstrated that patterns generated by electrophoretic karyotyping were highly reproducible among experiments, relatively insensitive to preparation methods within the same laboratory, and unaffected by high-frequency phenotypic switching. However, Holmberg and Feroze (139) showed technical variability for CHEF due to reagents, sample preparation, and running conditions. Electrophoretic karyotyping has been used extensively to fingerprint C. albicans (10, 18, 21, 22, 23, 26, 38, 96, 97, 98, 107, 148, 174, 191, 198, 204, 207, 208, 209, 219, 226, 229, 266, 269, 274, 322, 349, 381, 382, 384, 399, 403, 407, 410, 426), C. stellatoidea (171), C. glabrata (81, 107, 337, 398), C. lusitaniae (163, 220, 398), C. tropicalis (95, 107, 399), C. parapsilosis (39, 63, 107, 267, 398, 412), C. kefyr (398), C. krusei (107, 108), C. shehatae (255), C. rugosa (92), C. dubliniensis (231), Pneumonocystis carinii (372), A. nidulans (46), Cryptococcus neoformans (2, 20, 32, 117, 118, 121, 147, 169, 256, 257, 277), H. capsulatum (368), and Coccidiodes immitis (253). In several studies, electrophoretic karyotyping was compared to other DNA fingerprinting methods for its ability to discriminate among different fungal isolates (37, 208, 399). In three studies (37, 208, 399), electrophoretic karyotyping was found to be superior to RFLP without a probe in discriminating among independent isolates. However, there has been no careful verification of the efficacy of electrophoretic karyotyping in clustering moderately related strains. In fact, there are indications, as noted above, that electrophoretic karyotyping may not fulfill the necessary requirements for DNA fingerprinting C. albicans since cells of this species can be present in low- and high-frequency modes of chromosomal reorganization, leading to low rates of karyotypic change in the former case and high rates in the latter case (295). Cells of C. albicans strain 3153A expressing a variant-switch phenotype undergo extremely frequent changes in the size of the two chromosomes harboring rDNA cistrons (295). This phenomenon is reminiscent of the increased rate of recombination of ribosomal cistrons in strains of S. cerevisiae with mutations in the silencing gene SIR2 (128). The capacity of a cell to express two frequencies of chromosomal reorganization and hence two frequencies of karyotypic change as a result of reversible differentiation challenges the use of electrophoretic karyotyping as an effective fingerprinting method for cluster analyses of moderately related isolates. Indeed, in the study demonstrating high- and low-frequency modes of karyotypic change, it was also demonstrated in switching lineages that karyotypic patterns could diverge and then converge (295), a form of homoplasy. If divergence is followed by convergence of a DNA fingerprinting pattern, then two similar or identical electrophoretic karyotypes may be interpreted as reflecting the same strain when, in fact, the compared isolates may be unrelated. High frequencies of karyotypic change have been demonstrated in C. albicans strain 3153A (312–314) and during Cryptococcus neoformans infection in a mouse model (121). However, even with this caveat, a modified version of electrophoretic karyotyping may eventually prove to be an effective DNA fingerprinting method. The resolution of electrophoretic karyotyping as a fingerprinting method has been increased dramatically by digesting chromosome-length DNA with endonucleases prior to pulsed-field gel electrophoresis (81, 93, 281). By increasing the complexity of the pattern in this way, electrophoretic karyotyping could conceivably fulfill the requirements laid down in this article for an effective fingerprinting system. However, even without modification, the high frequency of change that occurs in the electrophoretic karyotype of an infecting strain can be used to monitor microevolution, as demonstrated for C. neoformans (118).
Sequencing as a Basis for DNA Fingerprinting
There is little controversy among evolutionary biologists that theoretically the preferred method for measuring phylogenetic relationships is through comparison of the DNA sequences of a variety of protein-coding regions. The underlying assumption is that the rates at which mutations accumulate in coding regions reflect evolutionary clocks. Because particular coding regions may be highly resistant to change and therefore may have very slow evolutionary clocks while other coding regions, such as those of genes involved in pathogenesis, may be far less resistant to change and therefore may have very fast evolutionary clocks, more than one protein-coding region should be used in any comparison. In many strategies, noncoding regions are selected based on the assumption that they are less prone to biases resulting from selective pressure (155). Until recently, cloning and sequencing genes was a slow, technically demanding, and expensive undertaking that seemed beyond the technical capacity of most medical mycologists interested in DNA fingerprinting large collections of fungal isolates. However, the emergence of PCR, automated DNA-sequencing technologies, and gene data banks may together provide us with sequence-based fingerprinting strategies that will be amenable and affordable for broad epidemiological studies in the near future.
With the emergence of computer data banks providing the sequences of both coding and noncoding regions of a variety of genes of the infectious fungi, PCR-sequencing strategies can be targeted to identified genes (see, e.g., reference 122). There have been sporadic studies in which the sequence of one or more genes of a limited number of fungal isolates have been compared. For instance, Stringer et al. (372) compared the small-subunit rRNA gene and the α-tubulin gene of human- and rat-derived Pneumocystis carinii isolates and found dramatic differences suggesting that they represented different species. Coleman et al. (79) used a 500-bp sequence of the V3 variable region of the large-subunit rRNA gene to demonstrate that C. dubliniensis formed an independent cluster from a number of other Candida species, including C. albicans. Several additional studies have used single-gene sequences to separate collections of fungal isolates into groups or simply to explore allelic variability (see, e.g., references 65, 70, 79, 117, and 254). Recently, Kasuga et al. (156) used partial sequences of four genes to analyze the genetic relatedness of 46 H. capsulatum isolates and Koufopanou et al. (167) used partial sequences of five genes to assess the population structure of Coccidioides immitis. However, as yet there is no sequencing strategy in which regions of coding loci are selected for different levels of variability and, hence, their combined capacity to provide a highly accurate genotype. In addition, sequencing strategies have not evolved that accommodate diploids with clonal population structures.
DATA ANALYSIS AND THE ROLE OF COMPUTERS
When infectious fungi were first fingerprinted by RFLP without and then with probes in the late 1980s, the collections of isolates were restricted in size, the epidemiological questions were restricted in scope, and qualitative interpretations of data were the norm. In general, the patterns of two isolates were considered either identical or different, with little thought being given to what the differences really reflected in terms of genetic relatedness. Study results were used to answer very limited questions in a one-time fashion, with data from a new study rarely being compared to data from a preceding study. In the last 10 years, although several new DNA fingerprinting methods have emerged, the level of sophistication in interpreting data has not evolved in step. As we have become more sophisticated in our approach to DNA fingerprinting the infectious fungi, as the collections of isolates have become larger, and as the questions asked have become more complex, our expectations should have increased accordingly. First, we should expect interpretations to be based on the quantitative analysis of data. Second, we should expect the data to tell us more about genetic relatedness than simply whether the DNA fingerprints are the same or different. Third, we should expect the data from one study to be in a form comparable to that of the data from another study. These expectations lead to a number of requirements not only for the fingerprinting method but also for computer-assisted analysis.
Data Analysis
Because we have considered a number of DNA fingerprint methods, the form of the data obtained is not uniform. For RFLPs without probes, the data are in the form of a complex banding pattern in a single gel image, containing several intense bands against a backdrop of a large number of less intense bands (Fig. 3A). With respect to RFLPs with probes (Fig. 6), electrophoretic karyotypes, and RAPDs (Fig. 8), the data are in the form of a limited number of discrete bands in single gel images. The bands can vary in intensity in RFLPs with complex or repetitive probes (Fig. 6). When several different primers or primer pairs are used in the RAPD method, several gels must be independently analyzed (Fig. 9). Each of these latter gels will contain a limited number of bands, and only the most intense will usually be analyzed. The data from the multiple gels must then be combined for interpretation. For MLEEs, the data will be in the form of a number of starch gel electrophoresis images in which each lane usually contains one or two spots. Again, the data from the different gels must be combined for interpretation. Finally, for fingerprinting methods based on sequencing, the data will be in the form of nucleotide sequences. Each form of data, therefore, has its own peculiarities that must be accommodated for computer-assisted analysis, storage, and interpretation. In all cases, the final interpretation must involve a measurement of similarity of the data collected for every possible pair of analyzed isolates. These measurements are then used to generate phylogenetic trees (dendrograms) for cluster analyses. Other methods used to compare isolates not involving a direct measure of pattern similarity include the use of cladistic methods to generate phylogenetic trees and pictorial correspondence for analyzing correlations between genetic information and other phenotypic or collection characteristics. Since the last two methods have rarely been used in fingerprinting the infectious fungi, the discussion that follows will focus on the use of similarity coefficients to generate dendrograms.
Analysis of Complex Banding Patterns
As noted above, the complexity of low-intensity bands in ethidium bromide-stained RFLP patterns hampers automated computer-assisted analysis of the general pattern. Therefore, these patterns usually have to be manually digitized for subsequent computer-assisted analysis. In contrast, the patterns generated by Southern blots hybridized with fingerprinting probes, RAPD analysis, or electrophoretic karyotyping contain discrete bands that in most cases can be automatically identified and digitized by computer-assisted systems. Since the banding pattern generated by the C. albicans DNA fingerprinting probe Ca3 (Fig. 6A) has received intense scrutiny using computer-assisted systems (137, 164, 168, 192, 196, 268, 329, 330, 332, 336, 358), it will be used as a model to describe how complex banding patterns can be analyzed.
The Ca3 fingerprint pattern of a C. albicans isolate contains approximately 15 to 20 bands of different intensities (Fig. 6A) (332). The object of pattern comparison is to obtain a measure of commonness or difference between the gel patterns of two isolates. Many different measures of genetic distance, or similarity coefficients, exist. However, because the majority of fingerprinting methods applied to the infectious fungi generate banding patterns without defined loci, the formulas that are best suited for these methods take into consideration the presence or absence of bands and band intensity. These formulas can also be used to score the data obtained by methods that provide allelic information and thus represent the best formulas for comparing the results of different methods (288). For a pattern such as that generated by Ca3 (Fig. 6A), a similarity coefficient based simply on band positions is first calculated. The data for two banding patterns (lanes A and B) can be synopsized by the binary values 0 and 1, where 0 indicates no band at a position and 1 indicates a band at that position (348). The value nAB is the number of bands common in lanes A and B (coded 1,1), a is the total number of bands in lane A not present in lane B (coded 1,0), and b is the total number of bands in lane B not present in lane A (coded 0,1). The sample size (total number of band positions) in this case is nAB + a + b. The total number of mismatches is a + b. Perhaps the most widely used computation based on band position is the coefficient of Jaccard (Sj) (347, 348). SJ is computed by the formula Sj = nAB/(nAB + a + b). It varies between 0 and 1.00. A measure of 0 reflects no common bands, while a measure of 1.00 reflects all common bands. Measures of 0.01 to 0.99 represent increasing degrees of commonness. It should be realized that all bands are treated equally in the computation regardless of the speed of their evolutionary clocks, that different patterns need not contain the same number of bands, and that an increasing pattern complexity should provide increasing resolution, on average. This last point is best demonstrated by considering the simplest example, in which pattern A contains one band and pattern B contains one band. For patterns A and B, the possible SJ values in this case are either 1.00 (bands are common) or 0 (bands are not common). Next consider two bands in pattern A and two bands in pattern B. The possible SJ values in this case are 1.00 (both bands are common), 0.33 (one band is common), or 0 (no bands are common). Finally, consider three bands for both pattern A and pattern B. The possible values in this case are 1.00 (all three bands are common), 0.50 (two bands are common), 0.20 (one band is common), and 0 (no bands are common). Therefore, the possible number of different SJ values increases as the patterns increase in complexity.
The coefficient of Dice (SD) (348) is similar to the coefficient of Jaccard's in that it is based on band position alone but gives more relative weight to matches than to mismatches by the formula SD = 2nAB/(a + b + 2nAB). The possible SD values for patterns A and B both containing two bands are 1.00 (both bands are common), 0.50 (one band is common), and 0 (no bands are common), and the possible SD values for patterns A and B both containing three bands are 1.00 (both bands are common), 0.66 (two bands are common), 0.33 (one band is common), and 0 (no bands are common). Notice that weighting common bands in this case increases the SD values for partially common patterns compared to the coefficient of Jaccard. Other coefficients are discussed in detail by Sneath and Sokal in their excellent book on numerical taxonomy (348).
For some hybridization patterns, band intensities as well as band positions must be compared. Differences in intensity are readily identified in patterns generated by complex probes (Fig. 6). The Pearson product-moment correlation coefficient is one of the most frequently employed for this purpose and is discussed in detail by Sneath and Sokal (348). The Pearson product-moment correlation coefficient (348) computes intensity in relation to the average intensity of bands in a pattern. This coefficient is computed between pattern A and pattern B according to the formula
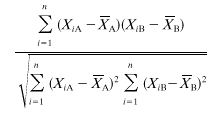 |
where XiA and XiB represent the intensities of band in patterns A and B, respectively, X---A and X---B represent the means of all intensities in patterns A and B, respectively, and n is the total number of band positions. The coefficients, which will range between −1 and +1, must then be converted to a range of 0 to 1 for generating dendrograms.
A simpler coefficient that compares absolute intensities (332) is computed by the formula
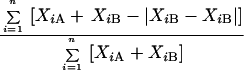 |
where XiA and XiB are the intensities of band in patterns A and B, respectively, and n is the number of bands (332). As in the case of coefficients for band position alone, a coefficient of 0 reflects no common bands, regardless of intensities, and a coefficient of 1.00 means not only that all bands are common but also that all matched bands have similar intensities. Coefficients ranging from 0.01 to 0.99 reflect increasing similarity that encompasses both an increasing proportion of common bands and similar intensities. It should be apparent that as in the coefficient of Dice, it is possible to develop formulas in which bands with common positions and intensities can be given more weight and in which band intensities can be normalized in a number of ways to account for small differences in loading, a problem not encountered when band position alone is the basis of the coefficient. In selecting or developing a particular similarity coefficient, one must keep in mind the characteristics of the patterns being compared. It should be obvious that each coefficient is imperfect and is based on one or more assumptions. For instance, it is assumed that bands in the same position in RFLP patterns of two strains represent the same genomic fragments. It should be obvious that if one relies on patterns of low complexity, this risk is high, but with increased complexity, the impact of a mistake on the final similarity coefficient decreases. Such concerns, however, should not deter the use of these coefficients in epidemiological studies, since whatever the failings of a comparison measurement, it is usually far superior to the alternative of qualitative and, in many cases, subjective interpretation. In the discussion that follows, a similarity coefficient, regardless of the formula used to compute it, will be referred to generically as SAB.
Once SAB values have been computed between all pairs of a collection of isolates, a matrix of similarity coefficients is generated (Fig. 10A). Based on this matrix of similarity coefficients, a dendrogram (Fig. 10B) can then be generated by the unweighted pair-group method using arithmetic averages (UPGMA), first used by Rohlf (308) and discussed in detail by Sneath and Sokal (348), or a comparable method. The UPGMA, which has been used frequently to generate dendrograms based on matrices of a variety of similarity coefficients, is relatively straightforward. The SAB matrix is scanned for the most similar isolates. If there is more than one group at the highest SAB, the first group is arbitrarily taken. These isolates are then joined at the appropriate position along an SAB axis. The initial cluster in the matrix is then scanned for the most similar group, and this is again repeated. A new connection can be made between an isolate and a cluster established in the preceding step. This connection is computed as the arithmetic mean between the new isolate and the isolates in the cluster. A number of alternative algorithms can be used to generate dendrograms from a matrix of similarity coefficients that result in different branch lengths for isolates emanating from a common isolate or group (112, 114, 185, 317) and may not be based on the assumption of a uniform evolutionary rate (12). However, it should be realized that all of these algorithms were developed to generate phylogenetic trees at the species level, not at the subspecies level or level of microevolution. Even so, such distance algorithms provide interpretable trees. The researcher can test whether the method used to generate a dendrogram is accurate by assessing the goodness of fit, which is a measure of how well the distances in the dendrogram match the distances in the matrix (12). Algorithms that adjust branch length will obviously perform better than the UGPMA (27), but there are trade-offs, which are discussed in detail elsewhere (12). For instance, the UPGMA automatically produces a rooted tree but does not accurately weight the branch lengths (12).
FIG. 10.
Example of a computer-generated matrix of similarity coefficients (SAB) computed between 32 isolates (A) and the dendrogram based on those SAB values (B).
Regardless of the algorithms that are used to generate a dendrogram, the accuracy of a fingerprinting method must be evaluated. This can best be achieved by a comparative cluster analysis in which the same distance algorithm is used to generate trees for the same set of test isolates from data obtained by the method of interest and a completely unrelated DNA fingerprinting method. If the general characteristics of the trees are similar, the methods in effect cross-verify each other. This form of cross-verification can be achieved only with a clonal population structure. Indeed, achieving such parity between completely unrelated fingerprinting methods is, in turn, strong evidence for a clonal population structure. This method of verification was performed by applying the UPGMA to data obtained by MLEE, RAPD, Southern blot hybridization with the complex probe Ca3, and Southern blot hybridization with the repetitive sequence CARE-2, in each case using the same common set of C. albicans isolates (288). The first three methods generated data sets that formed very similar dendrograms (Fig. 8). The three fingerprinting methods not only identified the same isolates as identical and the same isolates as completely unrelated but also clustered moderately related isolates in the same fashion, thus cross-verifying the efficacy of the three methods and verifying the distance algorithm employed. Unfortunately, this represents one of only a few examples in the literature of studies performed to evaluate the efficacy of fingerprinting methods and algorithm performance at the subspecies level for the infectious fungi. Hopefully, awareness of the need for such evaluation will lead to additional tests by others performing fingerprinting studies on infectious fungi.
Once a dendrogram is generated, the researcher must carefully consider how to interpret it. This interpretation will depend heavily on understanding the actual fingerprinting method and on empirical tests of relatedness. As noted above, the following four types of relatedness can be tested empirically: (i) identicalness, which requires a set of known identical isolates; (ii) high relatedness but nonidenticalness, which requires a set of isolates from a single strain that have diverged by microevolution; (iii) moderate relatedness, which requires a broad collection of isolates; and (iv) unrelatedness, which also requires a broad collection of isolates. By analyzing isolates in the above four categories, one can establish SAB thresholds for relatedness such as the ones noted in the dendrogram in Fig. 10B of a collection of C. albicans isolates fingerprinted with the complex probe Ca3 and compared with an SAB based on band position alone. The threshold at an SAB of 0.71 represents the average for a large collection of presumably unrelated isolates. The threshold at an SAB of 0.90 demarcates the lower limit of SAB values of several sets of isolates from individual strains that had undergone microevolution (192, 196, 265, 288, 330, 358). The threshold at an SAB of 0.80 is the most arbitrary one. It is approximately midway between the measure of unrelatedness (0.69) and the threshold for high relatedness (0.90). After analyzing a number of dendrograms based on the SAB values of isolates from both restricted and broad epidemiologic studies, an SAB of 0.80 was found to be a reasonable threshold for defining clusters of moderately related isolates (see, e.g., reference 195). Thresholds are specific for (i) a particular organism, (ii) a particular method of DNA fingerprinting, (iii) a particular coefficient of similarity, and (iv) a particular method of generating a phylogenetic tree. Thresholds, therefore, are not transferable among studies when any one of these components is changed.
Once established, thresholds can be used and adjusted for subsequent epidemiological studies. In dendrograms generated from SAB values computed from the patterns generated by complex probes like Ca3, the order of nodes in a tree should approximate the hierarchy of genetic relatedness. The distances between nodes or branch lengths, however, should not be considered direct reflections of genetic distance. The reason for this is the nature of a complex probe. By definition, a complex probe measures differences based on a variety of probed sequences (hypervariable, moderately variable, rarely variable, and invariable). All differences between two isolates, however, are weighted equally. The bands generated by the hypervariable C1 sequence of Ca3 (192) are weighted equally with bands far less variable in the computation of SAB. Therefore, dendrograms based on SAB values derived from Ca3 patterns should roughly reflect the evolutionary order. It is obvious that more information on the mechanism and frequency of reorganizational events identified by the different portions of a complex probe like Ca3 (Fig. 4) will provide a means of weighting band variation in a more complex SAB computation. Recently, Pujol et al. (289) measured the frequency of change of hypervariable bands in the Ca3 pattern of C. albicans in vitro to be 1 per 1,000 cell divisions. Although the frequency of change of less variable bands cannot be assessed in the same manner, their variability can be estimated by other means, and these estimates can be used to weight those bands in formulating a more accurate SAB.
There is obvious difficulty in defining thresholds that identify clusters of moderately related isolates. However, it is not the definitions of the thresholds that really matter in these studies but, rather, the integrity of the interpreted clusters. As noted, one way to test the integrity of a cluster is to compare dendrograms generated by two or more unrelated fingerprinting methods with the same collection of isolates (288). A second way is to test the stability of one or more clusters. This can be accomplished by randomizing the order of isolates chosen in the genesis of a dendrogram (265), an important control for the UPGMA method, which can make mistakes in higher-order clusters (13). Second, different levels of random noise can be introduced (265). When these types of tests were applied to clusters above a threshold of 0.80 in a recent analysis of 21 C. albicans isolates from patients with bloodstream infections in hospitals in the northeastern United States, an unexpectedly high stability was obtained for the major clusters (265). Testing the stability of clusters should become a more common tool in DNA fingerprinting studies that involve the genesis of dendrograms.
Use of Computer Systems To Analyze DNA Fingerprint Patterns
Although a number of software programs are available for generating dendrograms from matrices automatically or manually entered, two programs, GelCompar, developed by Applied Maths BVBA in Belgium, and DENDRON, developed in our laboratory at the University of Iowa, combine the capabilities of image processing, gel image analysis, computation of similarity coefficients, genesis of dendrograms, and storage of the data for future retrospective analyses. In both cases, the program was developed to analyze complex multiband patterns such as those generated by Southern blot hybridization with repetitive elements or complex probes, RAPD and other PCR-based methods, and electrophoretic karyotyping. In both programs, data such as those obtained from MLEE and multiple RAPD gels can also be entered for subsequent analysis, thus bypassing image processing and automatic pattern analysis. Since the capabilities of the two programs have been compared and assessed as similar by an impartial third party (340), the following discussion considers a generic program that incorporates the capabilities of the two, as well as other programs developed for gel analysis. This generic version is referred to simply as the Program. The Program encompasses all the virtues that a pattern analysis program should possess, including advanced image processing, image analysis, computations, and display capabilities.
Digitizing and Unwarping the Gel Image
The first step in automated pattern analysis is to scan the gel image into the Program database. The scanner should transfer the image with at least 256 grey scales, especially if band intensities as well as band positions are used in the computation of similarity coefficients. In some gels a distortion may exist in the general pattern, sometimes in the form of a smile, frown, or droop (Fig. 11A). To store data for future use, patterns must be first compared to a universal standard. To do this, distortions must be removed. The Program allows the user to unwarp and straighten all lanes (Fig. 11B) and then reproduce the edited, or image-processed, version of the gel (Fig. 11C). In the unwarping process, known molecular weight markers or DNA from a reference strain of the same species are run in the same gel for purposes of unwarping and straightening. These standards can be run either in the outside lanes or in the outside and middle lanes of the gel. If they can be visualized by a method different from that used to visualize the test DNA bands, they can be run in every lane and the gel can be processed separately for the test DNA patterns and standards. The presence of monomorphic bands in the patterns (i.e., bands in the pattern with the same molecular weight in all or most strains) greatly facilitates unwarping and straightening, especially if there is at least one monomorphic band among the low-molecular-weight bands and one among the high-molecular-weight bands. This is one of the reasons why the presence of monomorphic bands should be included in the list of desirable characteristics for selecting an effective DNA fingerprinting probe.
FIG. 11.
Image-processing and analysis capabilities of a computer-assisted DNA fingerprint analysis Program. (A) Original digitized image with distortion. (B) The user aligns common bands by drawing horizontal connecting lines and traces interlane junctions. (C) The Program then straightens the gel according to the user-drawn lines. (D) The Program automatically identifies lanes and bands and uses the pixel density to assign an intensity class (1 to 10). (E) The Program then correlates bands with a universal standard. (F) The Program generates a model. (G) The Program aligns processed gel images with isolates in the dendrogram. (H) The Program can also align models with isolates in the dendrogram.
In addition to straightening and unwarping, the image-processing software of the Program can slide an entire windowed lane up or down in the vertical axis, stretch or compress the pattern of a lane, intensify the banding pattern of an underloaded lane, reduce the intensity of an overloaded lane, subtract background to accentuate bands, and erase a lane. The capacity to manipulate a digitized image and re-create what would appear to be an original gel, but which is in fact an edited gel, can be worrisome. The basic fear is that a manipulated gel can be fraudulently represented as original data. However, the same can be said of data placed in a bar graph, a line graph, or a table. In all cases, the integrity of the researcher must be assumed in presenting data. However, as a safeguard, the Program should have policing software that tags an edited gel image with the history of manipulations.
Automatic Detection, Scanning, and Band Classification
After the Program has corrected the gel image for distortions and processed it so that it is amenable to automatic analysis, the Program then identifies each lane, which can be labeled by the user. Lanes can be separated so that succeeding steps for aligning bands and normalizing the pattern to a global standard are made easier. The Program then scans each lane along a single or multiple pixel axis and through a variety of possible algorithms and thresholds, and it then identifies and scores the intensity of each band. Bands can also be identified through a set of size/threshold algorithms, and intensity can be summed for the encapsulated band area. The Program displays the pixel density scans so that decisions can be made to add or subtract bands from the pattern or to separate bands not discriminated automatically. The Program must be sufficiently interactive that computer-interpreted bands that are artifacts can be subtracted from the pattern, intensities misinterpreted because of inconsistencies in the electrophoretic path can be rectified, and closely associated bands misinterpreted by the computer as a single band can be individualized.
Once bands are identified and scanned, the pixel density of each can be converted into classes reflecting intensity. In the example in Fig. 11D, intensity classes range from 0 to 10, with 0 reflecting no band and 1 to 10 reflecting progressively increasing intensities. The range of intensities will depend upon the requirements of the study. For analyses based upon band position alone, 0 and 1 can represent the absence and presence, respectively, of a band.
Calibration to a Global Standard and Genesis of a Data Text Map
To compare patterns from different gels, the Program must first calibrate each pattern by comparing the local standard (in the gel being analyzed) to the global standard. The use of a reference strain or a defined set of molecular markers to generate global and local standards facilitates normalization and the assignment of molecular weights. Bands to be compared in the different patterns are then connected, as in Fig. 11E. The molecular weights and intensities of bands are then logged in a text map, or densitometry map, which in some programs is used for comparison of patterns. The information in a text map can also be used to generate a model of a gel, such as the one in Fig. 11F.
Once the banding pattern of an isolate is normalized to a global standard, the patterns of two or more isolates analyzed in separate gels can be windowed, normalized, and neighbored in the same image (Fig. 12). The capacity to compare gel images in this manner is valuable in visually demonstrating microevolution, the presence of common bands, or the general similarity or dissimilarity of patterns of isolates fingerprinted in different gels or experiments.
FIG. 12.
The Program can take DNA fingerprints of different isolates in different gels, normalize them to a universal standard, and then “neighbor” the processed gel images for visual comparison. Note that gels were selected in this example that ran very differently.
Genesis of Phylogenetic Trees or Dendrograms
The data in a gel text map are used by the Program to generate a dendrogram by computing a similarity coefficient between every possible pair of isolates, as discussed above. Because all patterns are normalized to a global standard, this can be done between patterns (isolates) on different gels years apart, as long as the gel information is stored in a comparable fashion in the data files. Similarity coefficients are then used to generate a matrix (Fig. 10A) from which a phylogenetic tree or dendrogram is generated (Fig. 10B).
Once a dendrogram is generated, the Program will provide the average similarity coefficient for the analyzed collection and the average similarity coefficient for each cluster defined by a particular SAB threshold. The Program will also label isolates and denote isolates in the dendrogram by a particular common characteristic (e.g., common hospital origin, geographical locale, or patient characteristic) according to user directions. Finally, the Program will test the stability of clusters according to a variety of methods, some of which have already been described in this review. Because the fingerprint pattern of each isolate is digitized and normalized, it can be moved at will, as described in the neighboring process (Fig. 12). This allows the Program to generate, next to a dendrogram, horizontal representations (Fig. 11G) or models (Fig. 11H) of the fingerprint pattern of each isolate. Text maps of each gel can be used for a number of searches and cross-comparisons. For instance, they can be screened for a particular pattern or for a particular molecular weight band. The program will provide a selection dialog box that can be filled out for such a search.
Patient and Strain Characteristics for Searches
Each isolate in an epidemiological study has a history, both for the host and for the pathogen. In many studies, this information is collected but rarely used to the fullest extent. Every strain fingerprinted and logged in the data file of the Program can be described according to host parameters (e.g., age, sex, weight, medical characteristics, predisposing conditions, nature of disease, prosthetic wear, geographical locale, socioeconomic factors, and association with other individuals with analyzed strains) and pathogen characteristics (e.g., sugar assimilation pattern, antigenicity, secretion of proteinase, drug susceptibility pattern, hypha formation, switch phenotype or switching repertoire, and antigenicity). One can then select strains for comparison from the data file that conform to one or several of these characteristics. For instance, one can select and generate a dendrogram for analyzed isolates in the database collected from the vaginal canals of all females who are human immunodeficiency virus (HIV) positive or all individuals in a specific surgical intensive care unit over a 10-year period. Alternatively, one can select and generate a dendrogram for all isolates that are resistant to a particular antifungal drug, that grow at temperatures above 40°C, or that undergo a particular type of phenotypic switching (e.g., the white-opaque transition). It is also possible to select all isolates with a particular fingerprint pattern and then compare host or strain characteristics for common traits.
Computer-Assisted Analyses Pave the Way for a Worldwide Database
Several times in this review, it has been lamented that in most epidemiological fingerprinting studies of the infectious fungi, the data are analyzed once usually without using quantitative methods and then discarded. For fingerprinting studies with only a few samples and limited objectives, this is quite acceptable. However, for large collections, this represents a waste of data. Computer-assisted studies set the stage not only for comparative and retrospective studies within a single laboratory but also among laboratories worldwide. For fingerprinting methods that involve banding patterns, the requirements for developing a worldwide database are that experiments be performed by the same method, patterns be normalized to the same universal standard, and participants use the same or compatible software. All users must also conform to a set of standards for image processing such that the text maps, which will be the foundation of a worldwide data base, are comparable among laboratories. The use of the same computer program will also ensure that host and pathogen characteristics are tagged in a common fashion. In developing a worldwide database for sequence-based fingerprinting studies, the requirements are similar. In this case, the same set of sequences must be used and the same program will be required for comparable analyses. However, sequence data are more absolute than banding patterns, and in their simplest form they require manageable methods for downloading or transferring sequences for comparison. The extraordinary growth in the area of genomics has provided a variety of programs for storage and sequence comparison (89).
BRIEF REVIEW OF APPLICATIONS
The development of DNA fingerprinting methods for the infectious fungi has in part been driven by the need to answer a discrete set of epidemiological questions. As noted above, the majority of studies to date have, unfortunately, been performed by fingerprinting methods that were not verified for the particular application and did not include quantitative methods for comparison. In spite of this, legitimate answers have been obtained for several basic epidemiological questions and several epidemiological trends have been identified. Because of the variety of questions that have been asked, the large number of studies that have been performed, and the size constraints of this article, the following review first considers the problems of collection as they impact DNA fingerprinting studies and then considers only a few epidemiological questions and the DNA fingerprinting studies that have been performed to answer them.
Problems of Collection
The ability to compare the genotypes of isolates within a species allows one to raise a number of epidemiological questions that impact on treatment. It has become evident that collection procedures and the design of the collection are as important as the validation of the particular DNA fingerprinting methods employed. Perhaps the most immediate and widespread problem of collection involves the manner in which primary samples are cultured. Most samples are not initially plated in a clonal fashion (i.e., single cells producing independent colonies). Rather, the original sample on a cotton swab or other collection tool is usually streaked on nutrient agar and grown to confluency. This expedient method is quite common and is based on the assumption that fungal colonization or infection involves a single strain. We now know that in many cases and for many pathogenic conditions, this assumption is not always correct. In oral and esophageal thrush, many of the infecting populations are composed of two or more unrelated strains or multiple species (26, 81, 87, 269, 322, 403, 404). Multiple-species carriage also increases in the elderly (195). The mean number of species carried in the oral cavity of individuals 60 to 69 years of age in the Iowa City, Iowa, area was recently determined to be 1.0 ± 0.0 (n = 8), but those of individuals 70 to 79 years and ≥80 years of age are 1.4 ± 0.7 (n = 25) and 1.5 ± 0.7 (n = 29), respectively (195). Growing original samples of mixed strains or species to confluency, therefore, may result in overgrowth of a strain or species with the shortest generation time under the conditions of culture and may lead to a misinterpretation of the prevalent strain or species at the original site of colonization or infection. It is therefore essential that cells in an original test sample be “clonally plated” (i.e., at a density low enough that single cells produce independent colonies). It is also important to realize that analyzing only one colony from the sample may be just as misleading. As noted, there is ample evidence not only of multiple-species and multiple-strain carriage but also of the fact that colonizing strains undergo microevolution (118, 121, 192, 196, 265, 327, 329, 330, 358, 375, 419). Therefore, analysis of a single clonal colony will not provide information on the genetic variability within an infecting strain resulting from microevolution, or the proportions of substrains and associated phenotypes. Finally, even if cells are clonally plated, the agar must be of the correct composition to discriminate among species, strains or switch phenotypes. As previously noted, CHROMagar (243, 263, 324) and similar indicator agars provide a first approximation of heterogeneity in a single plating. Most importantly, multiple clones from each sample should be isolated and analyzed whenever possible to assess the degree of genetic heterogeneity.
In addition to multiple-species, multiple-strain, and multiple-substrain carriage at a single collection site (anatomical location), there is evidence, especially for healthy individuals carrying commensal strains, that different species, strains, or substrains may be more highly adapted to different body locations (83, 358). For instance, in one study of 52 healthy women (358), 11 carried a Candida sp. in both the oral cavity and vaginal canal. In two cases (18%), different species colonized the two locales, and in five cases (45%), unrelated strains of C. albicans, distinguished by Ca3 fingerprinting, colonized the two locales. In the remaining four cases (36%), variants of the same strain, distinguished by differences in the hypervariable bands of the Ca3 hybridization patterns, inhabited the two locales. In a study of mucosal candidiasis in 32 HIV-positive women (83), isolates from the vagina and oral cavities differed genetically in 9 of the women. In this latter study, strains were discriminated by Southern blot hybridization with repeat probes and RAPD analysis (83). Species and strain differences in different anatomical locations can directly affect the interpretation of epidemiological studies. For instance, if samples only from the oral cavity were collected in a surveillance study on the origin of fungal bloodstream infections (BSI), one might find that the BSI isolate differed from the oral isolate and conclude that the BSI was due to a hospital strain. However, if the patient carried that strain in the vagina, it would have been missed in surveillance and the conclusion that the BSI was due to a hospital strain would be wrong. The solution to this potential problem is to sample as many body sites as is possible in a surveillance study (see, e.g., reference 358).
Another problem associated with collection is geographical specificity. In some studies, isolates of a particular species causing a particular disease (e.g., vulvovaginitis) or exhibiting a particular phenotype (e.g., drug resistance) have been obtained from different geographical locales. The possibility exists that the specificity of strains for a particular geographical locale may outweigh the specificity of strains for a particular disease. Indeed, evidence of geographical specificity has been documented for Cryptococcus neoformans serotypes (see, e.g., reference 25) and suggested for C. albicans (see, e.g., references 77 and 330). For this reason, the isolates may have to be collected within a single geographical locale when strain-specificity is being tested. In some broad epidemiological studies, isolates obtained from single geographical locales may still not be representative of the diversity within those geographical locales. For example, isolates from one hospital may include a strain endemic to the hospital and not representative of genetic diversity within the geographical locale.
Finally, the time of collection may affect outcomes. Indeed, a change in the dominant serotype of C. neoformans accompanied the emergence of AIDS in Thailand (378). In addition, there is evidence of rapidly emerging pathogens such as C. glabrata (136) and C. dubliniensis (79, 376) and changes in the proportions of dominant strains of C. albicans within a geographical locale over time (D. R. Soll, unpublished observations). Changes in strains within a geographical locale over a short period (e.g., 5 years) may, in some cases, be as great as the differences between strains in two geographical locales or between endemic strains in two different hospitals in a single geographical locale. Isolates that are compared in a study should therefore be collected, if possible, within a short time window.
Finally, in many epidemiological studies the DNA fingerprint data of a collection of test isolates must be compared to that of a parallel collection of control isolates. The control collection must be as similar to the test collection as is experimentally possible for all of the reasons just outlined. Because of geographical specificity, the control collection should originate from the same geographical locale as the test collection, and because of the possibility that dominating strains may change over short periods within a geographical locale, control and test isolates should be collected in the same time window. Finally, because there is mounting evidence for species and strain specificity related to age and gender (see, e.g., reference 195), these characteristics should be matched as closely as possible in control and test collections. Surprisingly, few if any of these precautions have been taken in the majority of DNA fingerprinting studies. In fact, most studies rarely include collections of control isolates for comparison.
Strain Maintenance during Persistent or Recurrent Infection
In spite of the absence of proper controls, the lack of precautions that must be taken in collecting samples, and the absence of verification experiments for the majority of DNA fingerprinting methods employed, several consistent answers to a number of basic epidemiological questions have been obtained. Perhaps the best example of this is the question of strain maintenance during persistent colonization or recurrent infections. Virtually every standard method of DNA fingerprinting has been used to examine this question for a variety of infectious fungi. The results of the majority of such studies have been surprisingly consistent. Three scenarios have emerged; single-strain maintenance, strain replacement, and mixed-strain colonization. The dominant scenario, however, has proven to be single-strain maintenance. This scenario was first suggested in biotyping studies (166, 235, 245, 247, 262, 342) and soon thereafter verified by a variety of genetic fingerprinting methods. In 1988, Soll et al. monitored a single patient with a C. tropicalis BSI over a 2-month period using Southern blot hybridization with a C. tropicalis-specific complex probe and demonstrated that two strains persistently infected the bloodstream (360). Similar results were obtained in a number of subsequent studies in which isolates collected from the same healthy individuals or candidemia patients were compared over time. For HIV-infected individuals, a number of studies using a variety of DNA fingerprinting methods have demonstrated that in a majority of cases one strain and in a minority of cases two or more strains of Candida spp. were maintained over time in the oral-esophageal cavity (18, 19, 35, 101, 269, 322, 419). Similar results have been obtained for patients with other compromising conditions (10, 18, 19, 67, 98, 101, 116, 196, 205, 218, 265, 274, 275, 288, 289, 333, 336, 365, 403, 404, 409, 419) and for patients infected with fungi other than C. albicans (see, e.g., references 43, 81, 118, 121, 178, 337, 365, 375, and 412). For instance, in 1989, Fox et al. used Southern blot hybridization of EcoRI-digested DNA with probe 27A to demonstrate that serial isolates from each of eight immunosuppressed patients collected over a 2- to 18-month period were genetically identical (116). Since 27A patterns are highly sensitive to microevolution (289), identical patterns in this case do indeed reflect the same strain. Rarer examples of strain replacement have been described. For instance, Caugant and Sandren, using MLEE, demonstrated that three of seven bone marrow transplant patients were colonized with more than one strain and that in one patient flucytosine-resistant isolates appeared that had a very different MLEE profile, suggesting strain replacement (67). In an analysis of isolates by Southern blot hybridization with probe Ca3, Schmid et al. (330) demonstrated strain replacement in one patient with recurrent vaginitis and strain maintenance in another, but in a larger study that included nine patients with recurrent vaginitis, Lockhart et al. (196) demonstrated that strain maintenance accompanied by microevolution was, in most cases, the common scenario.
In almost all of the studies in which sequential isolates from the same individual have been DNA fingerprinted, the methods were usually good enough to identify the same strain or discriminate between unrelated isolates. However, in the majority of studies, cluster analyses were not performed and microevolution was not assessed. In a few cases, although differences in DNA fingerprints were interpreted as reflecting unrelated strains, it was not clear if the differences actually reflected microevolution within a single strain. In most of the published studies on strain maintenance and replacement, the DNA fingerprinting methods were not characterized for resolution, and for this reason, many additional questions related to the basic one of strain maintenance could not be answered. In addition, in the majority of studies the DNA fingerprints were not performed or stored in a manner that would have facilitated comparison with fingerprints obtained in other studies or in retrospective studies. The lament, therefore, is not that the basic questions posed were not answered but that the design of the experiments and the methods employed resulted in less sophisticated questions and answers, underutilization of the data, and a one-time experimental attitude.
Microevolution in Infecting Populations
Although a variety of studies have demonstrated that the major scenario in prolonged fungal colonization or recurrent infection is strain maintenance, methods that have been characterized for resolution and demonstrated to resolve microevolutionary changes within infecting strains over time have been applied in only a few studies. With no knowledge of resolving power, researchers have in some cases considered any pattern difference to represent unrelated strains. Therefore, a single band difference in the electrophoretic karyotype patterns of two isolates has been interpreted as reflecting distinct unrelated strains, when the change most probably reflected a high-frequency microevolutionary change within a strain at the site of infection. In several studies, however, researchers with a greater understanding of their fingerprinting data have correctly interpreted differences in terms of microevolution. Scherer and Stevens (327), using Southern blot hybridization with probe 27A, observed minor differences in C. albicans isolates from different body locations of several patients. They correctly interpreted these differences as microevolutionary changes within an infecting strain. These observations were expanded in a study in which C. albicans isolates from 17 different body locations in 52 healthy women were compared by Southern blot hybridization with the Ca3 probe (358). Of 11 women carrying commensals in both the oral cavity and vaginal canal, four possessed strains at the alternative sites that were highly similar but nonidentical, suggesting that through microevolution, a progenitor strain had adapted to two different body niches and that the divergent populations could be distinguished genetically. Recently, Southern blot hybridization with the Ca3 probe demonstrated microevolution in strains cocolonizing vaginitis patients and their male partners (336).
In perhaps the most detailed analysis of microevolution within infecting C. albicans strains, 9 to 14 isolates were collected directly from each site of commensalism or colonization of nine test subjects (192). In only three of the collections did all of the isolates exhibit identical Ca3 fingerprint pattern. In the six remaining collections, the isolates were closely related but exhibited microevolution. The proportion of isolates exhibiting a minor variant genotype in each of the six latter collections ranged between 7 and 42%. In Fig. 13, a dendrogram is presented in which multiple commensal isolates collected simultaneously from the vulvas of two healthy women, P2 and P3, were compared to 60 unrelated C. albicans isolates obtained from a data bank at the University of Iowa (196). The dendrogram was generated from the SAB values computed between all pairs of isolates based on Ca3 Southern blot hybridization patterns. The P2 collection had only 1 variant genotype (P2-12) out of 14 isolates. The remaining 13 isolates had identical genotypes. The P3 collection contained one predominant phenotype that included eight isolates and three minor variant phenotypes of two, two, and one isolates, respectively. The SAB threshold defining the P3 cluster in the dendrogram was 0.98 (Fig. 13). Note that even though both collections contained one or more variants, none of the 60 unrelated isolates penetrated either the P2 or P3 cluster.
FIG. 13.
Dendrogram comparing the genetic relatedness of multiple commensal isolates from the vulvas of two healthy women, P2 and P3, and 60 previously analyzed unrelated strains drawn from a data bank at Iowa (192). The dendrogram is based on the SAB values computed between the Ca3 patterns of all listed strains. Reproduced from reference 192 with permission of the publisher.
In an extension of this study, Lockhart et al. monitored infecting C. albicans strains over time in patients with recurrent vaginitis, again using the complex probe Ca3 as a fingerprinting method (196). They found that isolates shuffled between a set of genotypes over time and concluded that the colonizing strains had undergone microevolution and therefore contained a variety of subgenotypes, any one of which may dominate at a particular time. They named this phenomenon “substrain shuffling” to denote that the colonizing strain contained minor genotypes due to microevolution that could be sampled at any particular time. White et al. (419) used Ca3 fingerprinting to analyze a clonal strain colonizing an HIV-positive individual over time and observed microevolution that was associated with the acquisition of fluconazole resistance. These researchers suggested that the acquisition of resistance may not only be accompanied by small genetic changes but may also in fact be due to such high-frequency changes. This interpretation is predicated on the assumption that minor changes are occurring randomly throughout the genome or that the high-frequency changes that are identified by the Ca3 probe occur at loci that directly affect drug resistance. Recent results suggest that the microevolutionary changes identified by the Ca3 probe are in fact not random, and involve the insertion or deletion of full-length RPS sequences at specific sites in the genome (289). The possibility, however, cannot be excluded that cells undergoing changes at RPS-containing loci at unusually high frequency may also be undergoing parallel high frequency changes at other genetic loci directly involved in drug susceptibility.
Because the Ca3 and 27A fingerprinting probes of C. albicans contain sequences of the repeat element RPS (289), they are highly effective in discriminating microevolution within strains. C. dubliniensis, a very close relative of C. albicans, also contains sequences homologous to RPS dispersed throughout their genome (151). C. dubliniensis contains an additional C. dubliniensis-specific repetitive element that can effectively detect microevolution (151). The recently cloned complex probe Cd25, which contains this latter repetitive element, is capable of identifying microevolutionary changes that occur in vitro and in vivo (151). Complex DNA fingerprinting probes have also been cloned from C. tropicalis (152) and C. glabrata (194) that effectively detect microevolutionary changes in the respective species. In addition to the analysis of hypervariable regions with repeat sequence probes, other methods can be equally effective in assessing microevolution. Pujol et al. (288) demonstrated that both MLEE and RAPD can effectively identify microevolutionary changes in infecting strains of C. albicans. In addition, changes in electrophoretic karyotypes occur quite frequently and can be used effectively in monitoring microevolution and distinguishing substrains within infecting populations (see, e.g., references 10 and 274). Microevolutionary changes have been demonstrated in Cryptococcus neoformans by using electrophoretic karyotyping (118, 121), RAPD (375), and Southern blot hybridization with microsatellite-containing oligonucleotide probes (375). Franzot et al. (118) found that in vitro passage of C. neoformans strain ATCC 24067 (strain 52D) resulted in concomitant changes in karyotype and attenuation of virulence. Isolates of the same strain maintained in six different laboratories had undergone significant phenotypic and genetic changes, and the authors warned that microevolution in vitro could affect both intralaboratory and interlaboratory comparisons (118).
There seems to be little doubt that colonizing strains of each infectious fungus undergo microevolutionary change that can be distinguished by the correct fingerprinting method. There is also reason to believe that some substrains in an infecting population may possess altered phenotypes that include changes in virulence traits (see, e.g., reference 419). If one can measure in vitro the rate of microevolutionary change for a particular sequence or set of sequences in the genome, one can extrapolate rates to the in vivo conditions and estimate, in some cases, the average number of generations that have occurred in vivo based on microevolution. As noted, by analyzing the reorganization of RPS sequences over 3,000 generations in each of four unrelated C. albicans strains, Pujol et al. (289) have estimated that the average rate of RPS insertion/deletion in a C. albicans strain is 1 reorganization per 1,000 generations. It has therefore become just as critical to possess DNA fingerprinting methods that identify microevolutionary changes as to possess methods that assess moderate levels of relatedness. As noted throughout this review, fingerprinting methods ineffective at achieving the latter may be highly effective at achieving the former.
Determining the Origin of Nosocomial Infections
One of the most important challenges of DNA fingerprinting in recent years has been to elucidate the origins of nosocomial infections (54, 260, 261, 268). The origins of such infections are not always obvious. Infectious fungi such as Aspergillus spp. live in the environment, thrive in damp areas, and represent the majority of airborne spores (176). In addition, many Candida spp. are carried as commensals into hospital settings by patients, health care workers, and visitors. Particular groups of patients in intensive care units are at risk, especially bone marrow transplant recipients and other groups of immunosuppressed patients (24, 47, 154, 165, 302, 402, 414, 421). Surveillance studies indicate that the frequency of BSI and other forms of invasive candidiasis acquired in hospital settings continues to increase (24, 264). Therefore, DNA fingerprinting techniques have become critical tools in elucidating the sources of nosocomial infections, and although the large number of studies on nosocomial infections cannot be adequately reviewed here, the general application of DNA fingerprinting methods is considered.
Outbreaks of systemic fungemias in neonatal and surgical intensive care units and in transplant units represent the most visible cases of nosocomial infections, but the problem is far greater when one considers single dispersed episodes and superficial infections. In the majority of DNA fingerprinting studies of nosocomial infections, the approaches have been quite similar. Isolates collected from a limited number of nosocomial infections in a single care unit of a hospital during a limited period are compared, and interpretations are made solely upon genetic identity. In 1985, Burnie et al. (56) used a complex biotyping method to compare 13 cases of systemic candidemias and 25 cases of superficial candidiasis that occurred in a 9-month period in a single intensive care unit. One apparent strain with a common set of phenotypic characteristics, including serotype A, morphotype A1, and biotype O/(1)5 5/7, was identified in 100% of the systemic isolates and 44% of the superficial isolates. This strain was also found on the hands and in the mouths of some nurses from the particular intensive care unit. The singular biotype was interpreted to represent a common endemic strain. Subsequent studies of nosocomial Candida infections using a variety of DNA fingerprinting methods, however, have provided a variety of scenarios. In the majority of these studies, it was demonstrated that no single Candida strain was responsible for the majority of candidemias in a particular hospital or intensive care unit (7, 29, 38, 161, 184, 400, 401, 410, 438). In a minority of these studies, however, single strains were found to be responsible for selective outbreaks (22, 84, 93, 94, 162, 187, 266, 297, 306, 319, 321, 412, 436). Similar results have been obtained in a more limited set of studies of nosocomial Aspergillus infection. In several of these studies, nosocomial aspergillosis cases in the same care units were caused primarily by unrelated strains (69, 124, 178), while in other studies, the same strain was cultured from the majority of patients in an intensive care unit (202, 385). In two similar studies of Cryptococcus neoformans, no indications were obtained of single strains causing an outbreak (147) or of single strains causing infections in a particular hospital or geographical locale (256).
The most common result of the majority of nosocomial studies using DNA fingerprinting, therefore, is that most fungal infections that arise in an intensive care unit over a period of less than 1 year are caused by unrelated strains. In addition to this general observation, several very interesting and potentially important observations have been made on the basis of DNA fingerprinting. In several studies of candidemias, the DNA fingerprints of isolates from some health care workers matched those causing the candidemias (8, 22, 29, 94, 410). For example, Doebbeling et al. (94) found, using RFLP patterns, that C. tropicalis isolates from eight sternal wound infections were similar to each other and to an isolate from the scrub nurse. In another study using a combination of DNA fingerprinting methods, Doi et al. (96) found that hospitalized leukemia patients shared strains with visiting parents. However, in most of these studies, it was usually not clear if the patient was infected by the health care worker or visitor, or vice versa. One study of the strains carried on the hands of health care workers suggested that no single genotype was endemic in a care unit (142).
Several DNA fingerprinting studies have examined the role of catheters (184, 310, 410), peritoneal dialysis (412), prosthetic valves (51), glycerin suppositories (412), hospital construction (51), and the general hospital environment (38, 124, 178, 203, 401). In most of these studies, there was proof or suggestion from the DNA fingerprinting data that either treatment or environment played a role in the transmission of nosocomial infections. DNA fingerprinting has also been used to investigate the origin of Candida spp. colonizing newborns or causing candidemias in infants in neonatal intensive care units. It must be assumed that at birth a neonate does not possess an established strain of Candida spp. Indeed, Reef et al. (299) found that all neonates were yeast free at birth. Therefore, yeast colonization must originate from the mother or the hospital environment. Reef et al. (299), using both RFLP and hybridization with the midrepeat sequence probe CARE-2, found that of six neonates who acquired C. albicans in a neonatal intensive care unit, four had C. albicans-positive mothers. However, none of the four were colonized by the strain carried by their respective mothers, suggesting that the origin of colonization in these four cases was the hospital environment. For healthy children 3 days after birth, Willinger et al. (426) demonstrated that in six cases in which mother and child provided Candida spp., the electrophoretic karyotypes were identical. This was not, however, the case for hospitalized infants with candidemias. Waggoner-Fountain et al. (411), using RFLP patterns, found that of seven infants with BSIs and mothers carrying Candida spp., three of the infants were colonized with the same strain as their mother while four had different strains, presumably of hospital origin. In a number of studies using a variety of DNA fingerprinting methods (e.g., electrophoretic karyotyping, RFLP, and RFLP with probes) results have been obtained suggesting that candidemia outbreaks in neonatal intensive care units have a high probability of single-strain origin (110, 297, 310, 400). However, Khatib et al. (161) found infrequent cases of neonatal intensive care unit clusters due to single strains.
In the large majority of studies of the relatedness of nosocomial strains collected in the same intensive care units, the basis of genetic discrimination has simply been identical versus nonidentical, or similar versus dissimilar, and in most cases the DNA fingerprinting method seems to have been adequate at this level of resolution. In almost none of these studies, however, has the DNA fingerprinting method been verified by the procedures described in this review, rarely have these studies employed computer-assisted methods, rarely have the researchers provided quantitative measures of relatedness, and rarely have controls been performed that would give some idea of the probability of collecting isolates with similar versus dissimilar patterns in a nonhospitalized control population. Even with these reservations, the conclusions drawn from the majority of studies are probably correct. However, there are lessons that can be learned from the literature reviewed on nosocomial infections that will, hopefully, influence future studies. One example of the confusion emanating from a lack of information on the fingerprinting methods employed can be found in a study reported by Faix et al. (110). These researchers used both RFLP and Southern blot hybridization with the probe 27A to analyze isolates from candidemias arising in five low-weight neonates in a neonatal intensive care unit. The RFLP patterns were interpreted to demonstrate that the isolates were similar, but the 27A hybridization patterns were interpreted to suggest that the isolates were dissimilar. If the authors had known that the 27A probe hybridizes primarily to hypervariable fragments containing the repeat sequence RPS (150, 289) and that the differences they observed may have been the result of microevolution, they may have concluded that the RFLP patterns represented a better indicator of genetic relatedness and that these patterns suggested that the isolates represented the same endemic strain. They might have also concluded that the 27A patterns demonstrated microevolution within that endemic strain.
By using methods that simply distinguish similar from dissimilar, the major epidemiological question that can be posed is whether a single strain or multiple strains are responsible for infections in the same intensive care unit or hospital and whether there is horizontal transfer. As we have seen, without having characterized a fingerprinting system for the levels of resolution outlined in Fig. 1, interpretations can be flawed. By characterizing a method, the interpretations become more valid and the answers become far more sophisticated. For instance, because no single strain is responsible for nosocomial infections in an intensive care unit, does that mean that the responsible isolates are the commensal strains carried to the hospital by the patients? If the same strain infects several patients in an intensive care unit over several months, is it undergoing microevolution? Can several strains be endemic in a single intensive care unit? Are particular strains in a general geographical locale responsible for nosocomial infections in a particular intensive care unit? Two recent studies have examined nosocomial infections using fingerprinting methods that were characterized for resolution. Schmid et al. (331) fingerprinted C. albicans isolates obtained from 32 patients hospitalized for 3 days or more in New Zealand by using Southern blot hybridization with the Ca3 probe. The similarity coefficient that they employed to generate dendrograms was based on both band position and intensity (332). By analyzing the frequency of isolates from different wards in clusters defined by an SAB threshold of 0.80, they demonstrated that collections of isolates from specific wards were more highly related than were general isolates, suggesting a decrease in diversity. Although it appeared that the isolates analyzed had been collected in surveillance procedures and not from candidemias, these results still suggested that several strains may have been transmitted to patients in the hospital setting. Pfaller et al. (265), using a similar fingerprinting method, examined the relatedness of C. albicans isolates from BSI collected throughout the continental United States. In this study, the relatedness of fingerprint patterns was computed by an SAB based on band position alone. Using cluster analyses, they demonstrated that particular strains were more highly concentrated in particular geographical locales and that in some hospitals established strains were endemic. They also presented evidence suggesting that the putative endemic strains underwent microevolution in the hospital settings. The average similarity coefficients for isolates from particular hospitals reflected the selective increases in relatedness or decreases in diversity. While the average SAB of 22 unrelated isolates was 0.65 ± 0.11, the average SAB values for 14, 13, and 6 BSI isolates from three hospitals in the northeastern United States were 0.83 ± 0.10, 0.76 ± 0.08, and 0.81 ± 0.07, respectively, suggesting decreases in diversity. In contrast, the average SAB values for six, five, and five isolates from three hospitals in the midwestern United States were 0.70 ± 0.08, 0.68 ± 0.09, and 0.67 ± 0.08, respectively, suggesting diversity similar to that in the control collection of unrelated isolates. Since the Schmid et al. study (331) and the Pfaller et al. study (265) both used the same fingerprinting protocols and filed their data in compatible text files, they could readily share data even though they used different formulas for the computation of SAB. However, in almost all of the other fingerprinting studies of nosocomial isolates, the diversity of methods and protocols, the absence of computer analysis, and the absence of quantitative methods of comparison either exclude the possibility or make it extremely difficult to compare data. In addition, the Schmid et al. (331) and Pfaller et al. (265) studies have taken advantage of quantitative methods to generate dendrograms and perform cluster analyses. In a recent extension of these studies, Marco et al. (211) examined in detail the cluster characteristics of dendrograms generated from BSI isolates from the intensive care units of four hospitals and mixed dendrograms of BSI isolates and isolates from health care workers in the same units. Although the average SAB values of the BSIs of isolates from the intensive care units reflected diversity in each case close to that of a collection of control isolates, the cluster characteristics of BSI isolates suggested hospital origins for some infecting strains. High levels of relatedness were observed between BSI and health care worker isolates from the same intensive care units. The unique suggestion from this study (211) is that endemic strains in a hospital may be diverse and that simple comparisons of average SAB values may therefore not be sufficient indicators of the origin of nosocomial infections. More importantly, this study and the Schmid et al. (331) study and the Pfaller et al. (265) study demonstrate the power of computer-assisted analyses of DNA fingerprinting data in examining such complex epidemiological problems as the origin of nosocomial infections.
Other Epidemiological Questions Amenable to DNA Fingerprinting
A variety of additional questions related to virulence and the epidemiology of fungal infections can be raised and in some cases answered by DNA fingerprinting methods. The relationship of virulence and genotype represents an important question that, although frequently considered, has rarely been approached in any systematic fashion. In the past, strains causing infection have been compared to strains carried as commensals, but the fingerprinting methods employed have rarely been sophisticated enough and the experimental strategies for collection have not been considered carefully enough to obtain meaningful answers. In many cases, isolates from particular disease states obtained from diverse geographical locales were analyzed without considering the impact of geographical specificity. In addition, most studies were performed in the past without quantitative measurements of genetic relatedness and without the capacity to resolve microevolution or perform cluster analyses. Finally, virulence has been ascribed to strains based on the disease state of the host rather than the virulence potential measured quantitatively in animal or other models. The necessary methods are now available for fingerprinting the major infectious fungal species and performing the necessary cluster analyses, for testing virulence in relevant animal models, for assessing the developmental repertoires (i.e., the bud-hypha transition and high-frequency switching), and for testing the expression of putative virulence genes. A marriage of all of these techniques can now be performed to investigate the relationship of virulence and genotype.
The same arguments can be made for correlating drug resistance and genotype. Data continue to mount that drug-resistant strains and species are emerging (236, 259, 285, 300, 312), especially in the setting of continued drug therapy, as in cases of recurrent oral-esophageal thrush in AIDS patients (21, 190, 226, 269, 419). At least two species, C. krusei and C. glabrata, have increased in frequency in immunodeficient patients, and at least one reason for their recent success may be their natural or acquired resistance to fluconazole (30, 115, 212, 238, 284, 429, 430). However, there have also been studies demonstrating both strain replacement (21, 190) and strain evolution (226, 269). It is clear from these latter studies that resistant substrains can evolve from susceptible strains and that a resistant strain or species can replace a susceptible strain. However, that is not where the story should end. It is quite possible that only specific strains of, for example, C. albicans with identifiable genotypes within a geographical locale are capable of becoming resistant after drug therapy. This may be the result of the particular alleles of drug resistance genes a strain carries (417), the capacity of a strain to undergo phenotypic switching, which has been demonstrated to regulate at least one drug resistance gene, CDR3 (14), or an associated phenotypic characteristic, such as adhesion, that indirectly affects the evolution of drug resistance. To resolve these issues, studies should be performed in which both the genotypic and phenotypic characteristics of strains infecting HIV-positive individuals before and after the initiation of drug therapy are characterized. Such studies should be performed by a characterized and verified DNA fingerprinting method, within a single geographical locale and within a defined time window. Such studies should include an equal number of control isolates from healthy individuals who are not undergoing fungal drug therapy, that are collected in the same geographical locale and in the same time window.
Several additional epidemiological questions related to fungal pathogenesis, some of which have been superficially touched upon in the above discussion, deserve far more detailed treatment in the future. First, the relationship between commensal and infecting strains has not been adequately dealt with. Although we have fairly good evidence that commensals can become pathogens, we still do not know if all commensals are capable of becoming pathogens, if commensals undergo genotypic and/or phenotypic changes in the transition to pathogenesis, and if commensals with particular genotypes and therefore potentially specific phenotypes are specific to particular pathogenic states. Second, the specificity of particular strains to specific host characteristics must be ascertained. Is there strain specificity for commensalism in the oral cavity, gastrointestinal tract, vaginal canal, and vulva, as has been suggested (358)? Is there strain specificity or strain enrichment for the oral cavity of individuals in different age groups, as has been suggested (195)? Is there strain specificity for individuals with particular long-term illnesses or conditions, such as HIV infection, diabetes, denture use, smoking, particular diets, poor hygiene, drug therapies for other conditions, pregnancy, gender, and geographical locale? Third, is there strain specificity in mixed fungal infections, which increase as a result of particular disease states and old age (195)? Fourth, how fast do strains enter and dominate a particular geographical locale? With the proper computer database and a common method of DNA fingerprinting, the composition of colonizing strains in a geographical locale can be monitored over time. Fifth, do some strains undergo microevolution faster than others, does the rate of microevolution vary with the frequency of switching, as has been demonstrated for electrophoretic karyotypes (295), and is an increased rate of microevolution associated with increased pathogenesis, selective colonization, or the capacity to rapidly replace commensal strains after the establishment of a predisposing condition? Sixth, how is the first commensal strain established in a newborn? Is the origin the mother or the hospital environment? Do different conditions, such as nursing or neonatal procedures specific to a hospital, affect the origin of the first commensal? Does the first commensal remain, or is it replaced when a baby leaves the hospital, stops nursing, changes diet, etc.? Does the method of infant delivery affect initial colonization? Seventh, are there endemic strains in particular intensive care units? If they indeed exist, how diverse are they? How heterogeneous are substrains resulting from microevolution? Do these strains differ phenotypically from the average commensal carried by an individual prior to hospitalization? Finally, a worldwide description of strains, substrains, relationships and the temporal dynamics of change must be attempted. All of the above questions are now amenable to investigation using high-resolution, validated fingerprinting methods that provide quantitative measures reflecting genetic distance, that are amenable to computer-assisted analysis and storage, that are highly reproducible among laboratories, that are rapid and affordable, and that are technically within the capabilities of the majority of medical mycologists.
CONCLUSIONS
In this review, the major methods for DNA fingerprinting the infectious fungi have been considered in terms of their efficacy for assessing strain relatedness at the four levels of resolution outlined in Fig. 1. It has been stressed that one cannot simply dismiss a method because it does not conform to a set of rigid requirements set forth by evolutionary biologists that, in essence, would eliminate the majority of DNA fingerprinting methods now being used to analyze the relatedness of the infectious fungi. To ascertain the levels of resolution provided by a DNA fingerprinting method, a strategy of validation has been outlined that compares two or more unrelated methods by cluster analysis. For C. albicans, this strategy has validated the use of MLEE, RAPD, and complex probes for assessing all levels of relatedness and has placed restrictions on the use of probes composed primarily of repetitive elements. It has been stressed that the effectiveness of a DNA fingerprinting method depends in large part on the requirements of the epidemiological questions posed. For instance, although a repetitive probe may be ineffective in grouping moderately related strains, it may be highly effective in assessing recent microevolutionary changes generating substrains within an infecting population. Conversely, some probes that are highly effective in grouping moderately related isolates may be completely ineffective in revealing microevolutionary changes. The message is the following: characterize your DNA fingerprinting method and know its capabilities, consider the questions you are asking, and define the level(s) of resolution necessary to answer them.
Computer-assisted methods have been introduced that are essential for data analysis and storage. It has been lamented that DNA fingerprinting data in the past have rarely been analyzed in a quantitative fashion, that cluster analyses are rarely performed, and that data are used only to answer the question posed. Data are rarely stored for future comparisons and retrospective studies. A generic Program was described that possesses all of the capabilities one would utilize in the analysis of DNA fingerprinting data. Such programs are commercially available and, for the most part, user friendly. Quantitative methods were described for comparing the fingerprinting data obtained for independent isolates, and the use of thresholds in cluster analyses has been discussed. The methods for computing similarity coefficients, for generating dendrograms (phylogenetic trees), and for analyzing these dendrograms are well within the means of medical mycologists, and there is every expectation that they will be used in future DNA fingerprinting studies of the infectious fungi. It has been stressed that in most of the studies that have been performed to date, the data have been underutilized, related questions to the basic ones posed could have been answered but were not, and the data have been lost or abandoned because of the absence of computer-assisted methods of storage.
In this review, the problems inherent in collecting isolates for DNA fingerprinting have been considered. It has been implied that one can apply the most sophisticated DNA fingerprinting methods, but if the collection is flawed, the final results and interpretations will also be flawed. Because of the caveats resulting from the carriage of different strains or species in different body locations, change in species and strains due to aging, mixtures of species and strains in the same body location, and microevolution in colonizing populations, the isolation of a single clone from a single body location at a single time point may sometimes be misrepresentative. The simple act of streaking rather than clonally plating the original sample can lead to the overgrowth of a major member of the mycoflora that happens to grow faster in vitro. Most importantly, DNA fingerprinting studies rarely include a comparable set of control isolates. It was stressed that the most effective set of control isolates is one that is collected from the same geographical location within the same time window and is matched for age and even sex of the patients.
Finally, the application of DNA fingerprinting to three epidemiological questions was reviewed. It was noted in these reviews that even though the majority of studies involved DNA fingerprinting methods that were not sufficiently characterized or validated for the different levels of resolution, relied on qualitative rather than quantitative methods of analysis, did not consider the problems of collection, and did not include valid controls, the results obtained in the majority of cases were astonishingly consistent. In most cases, results were obtained that were interpretable and answered, to some degree, the very basic questions posed. However, it is pointed out that if more refined fingerprinting methods had been used, if computer-assisted methods of analysis had been applied, if precautions had been taken in collection, and if the proper controls had been performed, the studies and the conclusions would have been that much better.
It should be evident from the discussions in this review that DNA fingerprinting has evolved from an art to a science. Methods now exist for obtaining representative collections of test and control isolates, for fingerprinting them by carefully characterized and validated methods, for analyzing the data quantitatively by computer-assisted methods, and for interpreting the data in a highly sophisticated fashion. This review should serve as a guide for medical mycologists interested in attaining these goals.
ACKNOWLEDGMENTS
I am indebted to David Morice and Jen Swails-Wenger for help in assembling the manuscript, to Claude Pujol for detailed comments, to Shawn Lockhart for reviewing the manuscript, and to Sophie Joly for help in assembling figures.
The current research reviewed from the Soll laboratory was supported by Public Health Service grants AI39735 and DE10758.
REFERENCES
- 1.Alsina A, Mason M, Uphoff R A, Riggsby W S, Becker J M, Murphy D. Catheter-associated Candida utilis fungemia in a patient with acquired immunodeficiency syndrome: species verification with a molecular probe. J Clin Microbiol. 1988;26:621–624. doi: 10.1128/jcm.26.4.621-624.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson J M, Mihalik R, Soll D R. Ultrastructure and antigenicity of the unique cell wall “pimple” of the Candida opaque phenotype. J Bacteriol. 1990;172:224–235. doi: 10.1128/jb.172.1.224-235.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson J M, Srikantha T, Morrow B, Miyasaki S H, White T C, Agabian N, Schmid J, Soll D R. Characterization and partial nucleotide sequence of the DNA fingerprinting probe Ca3 of Candida albicans. J Clin Microbiol. 1993;31:1472–1480. doi: 10.1128/jcm.31.6.1472-1480.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson J M, Soll D R. Unique phenotype of opaque cells in the “white-opaque transition” of Candida albicans. J Bacteriol. 1987;169:5579–5588. doi: 10.1128/jb.169.12.5579-5588.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson M J, Gull K, Denning D W. Molecular typing by random amplification of polymorphic DNA and M13 southern hybridization of related paired isolates of Aspergillus fumigatus. J Clin Microbiol. 1996;34:87–93. doi: 10.1128/jcm.34.1.87-93.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anthony R M, Midgley J, Sweet S P, Howell S A. Multiple strains of Candida albicans in the oral cavity of HIV positive and HIV negative patients. Microbiol Ecol Health Dis. 1995;8:23–30. [Google Scholar]
- 7.Arif S, Barkham T, Power E G, Howell S A. Techniques for investigation of an apparent outbreak of infections with Candida glabrata. J Clin Microbiol. 1996;34:2205–2209. doi: 10.1128/jcm.34.9.2205-2209.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnavielhe S, Blancard A, Mallie M, Quilici M, Bastide J M. Multilocus enzyme electrophoresis analysis of Candida albicans isolates from three intensive care units. An epidemiological study. Mycoses. 1997;40:159–167. doi: 10.1111/j.1439-0507.1997.tb00208.x. [DOI] [PubMed] [Google Scholar]
- 9.Arnavielhe S, Blancard A, Mallie M, Gouin F, Ottomani A, Manelli J C, Bastide J M. Mycological monitoring of Candida albicans infections in various hospital care units. Molecular typing of isolated strains and epidemiological survey. Pathol Biol. 1996;44:447–451. . (In French.) [PubMed] [Google Scholar]
- 10.Asakura K, Iwaguchi S, Homma M, Sukai T, Higashide K, Tanaka K. Electrophoretic karyotypes of clinically isolated yeasts of Candida albicans and C. glabrata. J Gen Microbiol. 1991;137:2531–2538. doi: 10.1099/00221287-137-11-2531. [DOI] [PubMed] [Google Scholar]
- 11.Aufauvre-Brown A, Cohen J, Holden D W. Use of randomly amplified polymorphic DNA markers to distinguish isolates of Aspergillus fumigatus. J Clin Microbiol. 1992;30:2991–2993. doi: 10.1128/jcm.30.11.2991-2993.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avise J C. Molecular markers, natural history and evolution. New York, N.Y: Chapman & Hall; 1994. [Google Scholar]
- 13.Backeljau T, De Bruyn L, De Wolf H, Jordaens S, Van Dongen S, Winnepenninckx B. Multiple UPGMA and neighbor-joining trees and the performance of some computer packages. Mol Biol Evol. 1996;13:309–313. [Google Scholar]
- 14.Balan I, Alarco A M, Raymond M. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baleiras Couto M M, Vogels J T, Hofstra H, Huis in't Veld J H, van der Vossen J M. Random amplified polymorphic DNA and restriction enzyme analysis of PCR amplified rDNA in taxonomy: two identification techniques for food-borne yeasts. J Appl Bacteriol. 1995;79:525–535. doi: 10.1111/j.1365-2672.1995.tb03173.x. [DOI] [PubMed] [Google Scholar]
- 16.Barbour A. Antigenic variation in relapsing fever Borrelia species: genetic aspects. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 783–790. [Google Scholar]
- 17.Barchiesi F, Arzeni D, Del Prete M S, Sinicco A, Falconi Di Francesco L, Pasticci M B, Lamura L, Nuzzo M M, Burzacchini F, Coppola S, Chiodo F, Scalise G. Fluconazole susceptibility and strain variation of Candida albicans isolates from HIV-infected patients with oropharyngeal candidosis. J Antimicrob Chemother. 1998;41:541–548. doi: 10.1093/jac/41.5.541. [DOI] [PubMed] [Google Scholar]
- 18.Barchiesi F, Hollis R J, Del Poeta M, McGough D A, Scalise G, Rinaldi M G, Pfaller M A. Transmission of fluconazole-resistant Candida albicans between patients with AIDS and oropharyngeal candidiasis documented by pulsed-field gel electrophoresis. Clin Infect Dis. 1995;21:561–564. doi: 10.1093/clinids/21.3.561. [DOI] [PubMed] [Google Scholar]
- 19.Barchiesi F, Di Francesco L F, Compagnucci P, Arzeni D, Cirioni O, Scalise G. Genotypic identification of sequential Candida albicans isolates from AIDS patients by polymerase chain reaction techniques. Eur J Clin Microbiol Infect Dis. 1997;16:601–605. doi: 10.1007/BF02447925. [DOI] [PubMed] [Google Scholar]
- 20.Barchiesi F, Hollis R J, Messer S A, Scalise G, Rinaldi M G, Pfaller M A. Electrophoretic karyotype and in vitro antifungal susceptibility of Cryptococcus neoformans isolates from AIDS patients. Diagn Microbiol Infect Dis. 1995;23:99–103. doi: 10.1016/0732-8893(95)00169-7. [DOI] [PubMed] [Google Scholar]
- 21.Bart-Delabesse E, Boiron P, Carlotti A, Dupont B. Candida albicans genotyping in studies with patients with AIDS developing resistance to fluconazole. J Clin Microbiol. 1993;31:2933–2937. doi: 10.1128/jcm.31.11.2933-2937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bart-Delabesse E, van Deventer H, Goessens W, Poirot J L, Lioret N, van Belkum A, Dromer F. Contribution of molecular typing methods and antifungal susceptibility testing to the study of a candidemia cluster in a burn care unit. J Clin Microbiol. 1995;33:3278–3283. doi: 10.1128/jcm.33.12.3278-3283.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton R C, van Belkum A, Scherer S. Stability of karyotype in serial isolates of Candida albicans from neutropenic patients. J Clin Microbiol. 1995;33:794–796. doi: 10.1128/jcm.33.4.794-796.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck-Sague C M, Jarvis W R. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 25.Bennett J E, Kwon-Chung K J, Howard D H. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol. 1977;105:582–586. doi: 10.1093/oxfordjournals.aje.a112423. [DOI] [PubMed] [Google Scholar]
- 26.Berenguer J, Diaz-Guerra T M, Ruiz-Diez B, Bernaldo de Quiros J C, Rodriguez-Tudela J L, Martinez-Suarez J V. Genetic dissimilarity of two fluconazole-resistant Candida albicans strains causing meningitis and oral candidiasis in the same AIDS patient. J Clin Microbiol. 1996;34:1542–1545. doi: 10.1128/jcm.34.6.1542-1545.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berlocher S H. A comparison of molecular and morphological data, and phenetic and cladistic methods, in the estimation of phylogeny in Rhagoletis (Diptera: Tephritidae) In: Stock W W, editor. Applications of genetics and cytology in insect systematics and evolution. Moscow: University of Idaho Press; 1981. pp. 1–31. [Google Scholar]
- 28.Bertout S, Renaud F, Swinne D, Mallie M, Bastide J M. Genetic multilocus studies of different strains of Cryptococcus neoformans: taxonomy and genetic structure. J Clin Microbiol. 1999;37:715–720. doi: 10.1128/jcm.37.3.715-720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Betremieux P, Chevrier S, Quindos G, Sullivan D, Polonelli L, Guiguen C. Use of DNA fingerprinting and biotyping methods to study a Candida albicans outbreak in a neonatal intensive care unit. Pediatr Infect Dis J. 1994;13:899–905. doi: 10.1097/00006454-199410000-00011. [DOI] [PubMed] [Google Scholar]
- 30.Blaschke-Hellmessen R. Fluconazole and intraconazole susceptibility testing with clinical yeast isolates and algae of the genus Prototheca by means of the E test. Mycoses. 1996;39:39–43. doi: 10.1111/j.1439-0507.1996.tb00526.x. [DOI] [PubMed] [Google Scholar]
- 31.Blattner F R, Plunkett G, Block C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 32.Boekhout T, van Belkum A. Variability of karyotypes and RAPD types in genetically related strains of Cryptococcus neoformans. Curr Genet. 1997;32:203–208. doi: 10.1007/s002940050267. [DOI] [PubMed] [Google Scholar]
- 33.Boekhout T, van Belkum A, Leenders A C, Verbrugh H A, Mukamurangwa P, Swinne D, Scheffers W A. Molecular typing of Cryptococcus neoformans: taxonomic and epidemiological aspects. Int J Syst Bacteriol. 1997;47:432–442. doi: 10.1099/00207713-47-2-432. [DOI] [PubMed] [Google Scholar]
- 34.Boerlin P, Boerlin-Petzold F, Durussel C, Addo M, Pagani J L, Chave J P, Bille J. Cluster of oral atypical Candida albicans isolates in a group of human immunodeficiency virus-positive drug users. J Clin Microbiol. 1995;33:1129–1135. doi: 10.1128/jcm.33.5.1129-1135.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boerlin P, Boerlin-Petzold F, Goudet J, Durussel C, Pagani J L, Chave J P, Bille J. Typing Candida albicans oral isolates from human immunodeficiency virus-infected patients by multilocus enzyme electrophoresis and DNA fingerprinting. J Clin Microbiol. 1996;34:1235–1248. doi: 10.1128/jcm.34.5.1235-1248.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonacorsi S P, Munck A, Gerardin M, Doit C, Brahimi N, Navarro J, Bingen E. In situ management and molecular analysis of candidaemia related to a totally implantable vascular access in a cystic fibrosis patient. J Infect. 1996;33:49–51. doi: 10.1016/s0163-4453(96)92795-4. [DOI] [PubMed] [Google Scholar]
- 37.Bostock A, Khattak M N, Matthews R, Burnie J. Comparison of PCR fingerprinting, by random amplification of polymorphic DNA, with other molecular typing methods for Candida albicans. J Gen Microbiol. 1993;139:2179–2184. doi: 10.1099/00221287-139-9-2179. [DOI] [PubMed] [Google Scholar]
- 38.Branchini M L, Geiger D C, Fischman O, Pignatari A C. Molecular typing of Candida albicans strains isolated from nosocomial candidemia. Rev Inst Med Trop Sao Paulo. 1995;37:483–487. doi: 10.1590/s0036-46651995000600002. [DOI] [PubMed] [Google Scholar]
- 39.Branchini M L, Pfaller M A, Rhine-Chalberg J, Frempong T, Isenberg H D. Genotypic variation and slime production among blood and catheter isolates of Candida parapsilosis. J Clin Microbiol. 1994;32:452–456. doi: 10.1128/jcm.32.2.452-456.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brandt M E, Bragg S L, Pinner R W. Multilocus enzyme typing of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2819–2823. doi: 10.1128/jcm.31.10.2819-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brandt M E, Hutwagner L C, Klug L A, Baughman W S, Rimland D, Graviss E A, Hamill R J, Thomas C, Pappas P G, Reingold A L, Pinner R W. Molecular subtype distribution of Cryptococcus neoformans in four areas of the United States. Cryptococcal Disease Active Surveillance Group. J Clin Microbiol. 1996;34:912–917. doi: 10.1128/jcm.34.4.912-917.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brandt M E, Hutwagner L C, Kuykendall R J, Pinner R W. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. Cryptococcal Disease Active Surveillance Group. J Clin Microbiol. 1995;33:1890–1895. doi: 10.1128/jcm.33.7.1890-1895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandt M E, Pfaller M A, Hajjeh R A, Graviss E A, Rees J, Spitzer E D, Pinner R W, Mayer L W. Molecular subtypes and antifungal susceptibilities of serial Cryptococcus neoformans isolates in human immunodeficiency virus-associated Cryptococcosis. Cryptococcal Disease Active Surveillance Group. J Infect Dis. 1996;174:812–820. doi: 10.1093/infdis/174.4.812. [DOI] [PubMed] [Google Scholar]
- 44.Brawner D L. Comparison between methods for serotyping of Candida albicans produces discrepencies in results. J Clin Microbiol. 1991;29:1020–1025. doi: 10.1128/jcm.29.5.1020-1025.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brawner D L, Cutler J E. Oral Candida albicans isolates from nonhospitalized normal carriers, immunocompetent hospitalized patients, and immunocompromised patients with or without acquired immunodeficiency syndrome. J Clin Microbiol. 1989;27:1335–1341. doi: 10.1128/jcm.27.6.1335-1341.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brody H, Carbon J. Electrophoretic karyotype of Aspergillus nidulans. Proc Natl Acad Sci USA. 1989;86:6260–6263. doi: 10.1073/pnas.86.16.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bross J, Talbot G H, Maislin G, Hurwitz S, Strom B L. Risk factors for nosocomial candidemia: a case-control study in adults without leukemia. Am J Med. 1989;87:614–620. doi: 10.1016/s0002-9343(89)80392-4. [DOI] [PubMed] [Google Scholar]
- 48.Brown-Thomsen J. Variability in Candida albicans (Robin) Berkhous. Hereditas. 1968;60:335–398. [Google Scholar]
- 49.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buesching W J, Kurek K, Roberts G D. Evaluation of the modified API 20C system for identification of clinically important yeasts. J Clin Microbiol. 1979;9:565–569. doi: 10.1128/jcm.9.5.565-569.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buffington J, Reporter R, Lasker B A, McNeil M M, Lanson J M, Ross L A, Mascola L, Jarvis W R. Investigation of an epidemic of invasive aspergillosis: utility of molecular typing with the use of random amplified polymorphic DNA probes. Pediatr Infect Dis J. 1994;13:386–393. [PubMed] [Google Scholar]
- 52.Bunetel L, Deunff J. Isoenzymes et mycologie médicale. J Mycol Med. 1993;3:45–48. [Google Scholar]
- 53.Burgener-Kairuz P, Zuber J P, Jaunin P, Buchman T G, Bille J, Rossier M. Rapid detection and identification of Candida albicans and Torulopsis (Candida) glabrata in clinical specimens by species-specific nested PCR amplification of a cytochrome P-450 lanosterol-alpha-demethylase (L1A1) gene fragment. J Clin Microbiol. 1994;32:1902–1907. doi: 10.1128/jcm.32.8.1902-1907.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burnie J P. Candida and hands. J Hosp Infect. 1986;8:1–4. doi: 10.1016/0195-6701(86)90099-x. [DOI] [PubMed] [Google Scholar]
- 55.Burnie J P, Coke A, Matthews R C. Restriction endonuclease analysis of Aspergillus fumigatus DNA. J Clin Pathol. 1992;45:324–327. doi: 10.1136/jcp.45.4.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burnie J P, Odds F C, Lee W, Webster C, Williams J D. Outbreak of systemic Candida albicans in intensive care unit caused by cross infection. Br Med J Clin Res Ed. 1985;290:746–748. doi: 10.1136/bmj.290.6470.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Burt A, Carter D A, Koenig G L, White T J, Taylor J W. Molecular markers reveal cryptic sex in the human pathogen Coccidioides immitis. Proc Natl Acad Sci USA. 1996;93:770–773. doi: 10.1073/pnas.93.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burt A, Dechairo B M, Koenig G L, Carter D A, White T J, Taylor J W. Molecular markers reveal differentiation among isolates of Coccidioides immitis from California, Arizona and Texas. Mol Ecol. 1997;6:781–786. doi: 10.1046/j.1365-294x.1997.00245.x. [DOI] [PubMed] [Google Scholar]
- 59.Caetano-Anollés G. Amplifying DNA with arbitrary oligonucleotide primers. Genome Res. 1993;3:85–94. doi: 10.1101/gr.3.2.85. [DOI] [PubMed] [Google Scholar]
- 60.Carle G F, Olson M V. Separation of chromosomal DNA molecules from yeast by orthogonal-field-alternation gel electrophoresis. Nucleic Acids Res. 1984;12:5647–5664. doi: 10.1093/nar/12.14.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carle G F, Olson M V. An electrophoretic karyotype of yeast. Proc Natl Acad Sci USA. 1985;82:3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Carlotti A, Grillot R, Couble A, Villard J. Typing of Candida krusei clinical isolates by restriction endonuclease analysis and hybridization with the CkF1,2 DNA probe. J Clin Microbiol. 1994;32:1691–1699. doi: 10.1128/jcm.32.7.1691-1699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carruba G, Pontieri E, De Bernardis F, Martino P, Cassone A. DNA fingerprinting and electrophoretic karyotype of environmental and clinical isolates of Candida parapsilosis. J Clin Microbiol. 1991;29:916–922. doi: 10.1128/jcm.29.5.916-922.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carter D A, Burt A, Taylor J W, Koenig G L, White T J. Clinical isolates of Histoplasma capsulatum from Indianapolis have a recombining population structure. J Clin Microbiol. 1996;34:2577–2584. doi: 10.1128/jcm.34.10.2577-2584.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Casadevall A, Freundlich L F, Marsh L, Scharff M D. Extensive allelic variation in Cryptococcus neoformans. J Clin Microbiol. 1992;30:1080–1084. doi: 10.1128/jcm.30.5.1080-1084.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Casal M, Linares M J. Preliminary investigations of Candida albicans biovars. J Clin Microbiol. 1983;18:430–431. doi: 10.1128/jcm.18.2.430-431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Caugant D A, Sandven P. Epidemiological analysis of Candida albicans strains by multilocus enzyme electrophoresis. J Clin Microbiol. 1993;31:215–220. doi: 10.1128/jcm.31.2.215-220.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaturvedi V, Flynn T, Niehaus W G, Wong B. Stress tolerance and pathogenic potential of a mannitol mutant of Cryptococcus neoformans. Microbiology. 1996;142:937–943. doi: 10.1099/00221287-142-4-937. [DOI] [PubMed] [Google Scholar]
- 69.Chazalet V, Debeaupuis J P, Sarfati J, Lortholary J, Ribaud P, Shah P, Cornet M, Vu Thien H, Gluckman E, Brucker G, Latge J P. Molecular typing of environmental and patient isolates of Aspergillus fumigatus from various hospital settings. J Clin Microbiol. 1998;36:1494–1500. doi: 10.1128/jcm.36.6.1494-1500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen F, Currie B P, Chen L C, Spitzer S G, Spitzer E D, Casadevall A. Genetic relatedness of Cryptococcus neoformans clinical isolates grouped with the repetitive DNA probe CNRE-1. J Clin Microbiol. 1995;33:2818–2822. doi: 10.1128/jcm.33.11.2818-2822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen S C, Brownlee A G, Sorrell T C, Ruma P, Nimmo G. Identification by random amplification of polymorphic DNA of a common molecular type of Cryptococcus neoformans var. neoformans in patients with AIDS or other immunosuppressive conditions. J Infect Dis. 1996;173:754–758. doi: 10.1093/infdis/173.3.754. [DOI] [PubMed] [Google Scholar]
- 72.Chen S C, Currie B J, Campbell H M, Fisher D A, Pfeiffer T J, Ellis D H, Sorrell T C. Cryptococcus neoformans var. gattii infection in northern Australia: existence of an environmental source other than known host eucalypts. Trans R Soc Trop Med Hyg. 1997;91:547–550. doi: 10.1016/s0035-9203(97)90021-3. [DOI] [PubMed] [Google Scholar]
- 73.Chibana H, Iwaguchi S-I, Homma M, Chindamporn A, Nakagawa Y, Tanaka K. Diversity of tandemly repetitive sequences due to short periodic repetitions in the chromosomes of Candida albicans. J Bacteriol. 1994;176:3851–3858. doi: 10.1128/jb.176.13.3851-3858.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chibana H, Magee B B, Grindle S, Ran Y, Scherer S, Magee P T. A physical map of chromosome 7 of Candida albicans. Genetics. 1998;149:1739–1752. doi: 10.1093/genetics/149.4.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Childress C M, Holder I A, Neely A N. Modifications of a Candida albicans biotyping system. J Clin Microbiol. 1989;27:1392–1394. doi: 10.1128/jcm.27.6.1392-1394.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chindamporn A, Nakagawa Y, Mizuguchi I, Chibana H, Doi M, Tanaka K. Repetitive sequences in (RPSs) in the chromosomes of Candida albicans are sandwiched between two novel stretches, HOK and RB2, common to each chromosome. Microbiology. 1998;144:849–857. doi: 10.1099/00221287-144-4-849. [DOI] [PubMed] [Google Scholar]
- 77.Clemons K V, Feroze F, Holmberg K, Stevens D A. Comparative analysis of genetic variability among Candida albicans isolates from different geographic locales by three genotypic methods. J Clin Microbiol. 1997;35:1332–1336. doi: 10.1128/jcm.35.6.1332-1336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clemons K V, Shankland G S, Richardson M D, Stevens D A. Epidemiologic study by DNA typing of a Candida albicans outbreak in heroin addicts. J Clin Microbiol. 1991;29:205–207. doi: 10.1128/jcm.29.1.205-207.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Coleman D, Sullivan D, Harrington B, Haynes K, Henman M, Shanley D, Bennett D, Moran G, McCreary C, O'Neill L. Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 1997;3:S96–S101. doi: 10.1111/j.1601-0825.1997.tb00384.x. [DOI] [PubMed] [Google Scholar]
- 80.Coleman D C, Sullivan D J, Bennett D E, Moran G P, Barry H J, Shanley D B. Candidiasis: the emergence of a novel species, Candida dubliniensis. AIDS. 1997;11:557–567. doi: 10.1097/00002030-199705000-00002. [DOI] [PubMed] [Google Scholar]
- 81.Cormican M G, Hollis R J, Pfaller M A. DNA macrorestriction profiles and antifungal susceptibility of Candida (Torulopsis) glabrata. Diagn Microbiol and Infect Dis. 1996;25:83–87. doi: 10.1016/s0732-8893(96)00083-1. [DOI] [PubMed] [Google Scholar]
- 82.Currie B P, Freundlich L F, Casadevall A. Restriction fragment length polymorphism analysis of Cryptococcus neoformans isolates from environmental (pigeon excreta) and clinical sources in New York City. J Clin Microbiol. 1994;32:1188–1192. doi: 10.1128/jcm.32.5.1188-1192.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dahl K M, Keath E J, Fraser V J, Powderly W G. Molecular epidemiology of mucosal candidiasis in HIV-positive women. 1997. AIDS Res Hum Retroviruses. 1997;13:485–491. doi: 10.1089/aid.1997.13.485. [DOI] [PubMed] [Google Scholar]
- 84.D'Antonio D, Violante B, Mazzoni A, Bonfini T, Capuani M A, D'Aloia F, Iacone A, Schioppa F, Romano F. A nosocomial cluster of Candida inspicua infections in patients with hematological malignancies. J Clin Microbiol. 1998;36:792–795. doi: 10.1128/jcm.36.3.792-795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.De Bernardis F, Mühlschlegel F A, Cassone A, Fonzi W A. The pH of the host niche controls gene expression in and virulence of Candida albicans. Infect Immun. 1998;66:3317–3325. doi: 10.1128/iai.66.7.3317-3325.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Del Castillo L, Bikandi J, Nieto A, Quindos G, Sentandreu R, Ponton J. Comparison of morphotypic and genotypic methods for strain delineation in Candida. Mycoses. 1997;40:445–450. doi: 10.1111/j.1439-0507.1997.tb00182.x. [DOI] [PubMed] [Google Scholar]
- 87.Denning D W, Shankland G S, Stevens D A. DNA fingerprinting of Aspergillus fumigatus isolates from patients with aspergilloma. J Med Vet Mycol. 1991;29:339–342. [PubMed] [Google Scholar]
- 88.Desjardins P, Picard B, Kaltenbock B, Elion J, Denamur E. Sex in Escherichia coli does not disrupt the clonal structure of the population: evidence from random amplified polymorphic DNA and restriction-fragment-length polymorphism. J Mol Evol. 1995;41:440–448. doi: 10.1007/BF00160315. [DOI] [PubMed] [Google Scholar]
- 89.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Diaz-Guerra T M, Martinez-Suarez J V, Laguna F, Rodriguez-Tudela J L. Comparison of four molecular typing methods for evaluating genetic diversity among Candida albicans isolates from human immunodeficiency virus-positive patients with oral candidiasis. J Clin Microbiol. 1997;35:856–861. doi: 10.1128/jcm.35.4.856-861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Diaz-Guerra T M, Martinez-Suarez J V, Laguna F, Valencia E, Rodriguez-Tudela J L. Change in fluconazole susceptibility patterns and genetic relationship among oral Candida albicans isolates. AIDS. 1998;12:1601–1610. doi: 10.1097/00002030-199813000-00006. [DOI] [PubMed] [Google Scholar]
- 92.Dib J C, Dube M, Kelly C, Rinaldi M G, Patterson J E. Evaluation of pulsed-field gel electrophoresis as a typing system for Candida rugosa: comparison of karyotype and restriction fragment length polymorphisms. J Clin Microbiol. 1996;34:1494–1496. doi: 10.1128/jcm.34.6.1494-1496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diekema D J, Messer S A, Hollis R J, Wenzel R P, Pfaller M A. An outbreak of Candida parapsilosis prosthetic valve endocarditis. Diagn Microbiol Infect Dis. 1997;29:147–153. doi: 10.1016/s0732-8893(97)81804-4. [DOI] [PubMed] [Google Scholar]
- 94.Doebbeling B N, Hollis R J, Isenberg H D, Wenzel R P, Pfaller M A. Restriction fragment analysis of a Candida tropicalis outbreak of sternal wound infections. J Clin Microbiol. 1991;29:1268–1270. doi: 10.1128/jcm.29.6.1268-1270.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Doebbeling B N, Lehmann P F, Hollis R J, Wu L C, Widmer A F, Voss A, Pfaller M A. Comparison of pulsed-field gel electrophoresis with isoenzyme profiles as a typing system for Candida tropicalis. Clin Infect Dis. 1993;16:377–383. doi: 10.1093/clind/16.3.377. [DOI] [PubMed] [Google Scholar]
- 96.Doi M, Homma M, Iwaguchi S, Horibe K, Tanaka K. Strain relatedness of Candida albicans strains isolated from children with leukemia and their bedside parents. J Clin Microbiol. 1994;32:2253–2259. doi: 10.1128/jcm.32.9.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Doi M, Mizuguchi I, Homma M, Tanaka K. Electrophoretic karyotypes of isolates of Candida albicans from hospitalized patients. J Med Vet Mycol. 1994;32:133–140. [PubMed] [Google Scholar]
- 98.Doi M, Mizuguchi I, Homma M, Tanaka K. Electrophoretic karyotypes of Candida yeasts recurrently isolated from single patients. Microbiol Immun. 1994;38:19–23. doi: 10.1111/j.1348-0421.1994.tb01739.x. [DOI] [PubMed] [Google Scholar]
- 99.Donelson J E. DNA rearrangements and antigenic variation in African trypanosomes. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 763–782. [Google Scholar]
- 100.Donovan W H, Hull R, Rossi C D. Analysis of gram negative recolonization of the neuropathic bladder among patients with spinal cord injuries. Spin Cord. 1996;34:587–591. doi: 10.1038/sc.1996.104. [DOI] [PubMed] [Google Scholar]
- 101.Dromer F, Improvisi L, Dupont B, Eliaszewicz M, Pialoux G, Fournier S, Feullie V. Oral transmission of Candida albicans between partners in HIV-infected couples could contribute to dissemination of fluconazole-resistant isolates. AIDS. 1997;11:1095–1101. doi: 10.1097/00002030-199709000-00003. [DOI] [PubMed] [Google Scholar]
- 102.Dromer F, Varma A, Ronin O, Mathoalin S, Dupont B. Molecular typing of Cryptococcus neoformans serotype D clinical isolates. J Clin Microbiol. 1994;32:2364–2371. doi: 10.1128/jcm.32.10.2364-2371.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Drouhet E, Mercier-Soucy L, Montplaisir S. Sensibilite et resistance des levures pathogenes aux 5-fluoropyrimidines. 1. Relation entre les phenotypes de resistance a la 5-fluorocytosine, le serotype de Candida albicans et l'ecologie de differentes especes de Candida d'origine humaine. Ann Microbiol (Paris) 1975;126:25–39. [PubMed] [Google Scholar]
- 104.Ellsworth D L, Rittenhouse K D, Honeycutt R L. Artifactual variation in randomly amplified polymorphic DNA banding patterns. BioTechniques. 1993;14:214–217. [PubMed] [Google Scholar]
- 105.El-Zaatari M, Pasarell L, McGinnis M R, Buckner J, Land G A, Salkin I F. Evaluation of the updated Vitek yeast identification data base. J Clin Microbiol. 1990;28:1938–1941. doi: 10.1128/jcm.28.9.1938-1941.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Reference deleted.
- 107.Espinel-Ingroff A, Quart A, Steele-Moore L, Metcheva I, Buck G A, Bruzzese V L, Reich D. Molecular karyotyping of multiple yeast species isolated from nine patients with AIDS during prolonged fluconazole therapy. J Med Vet Mycol. 1996;34:111–116. doi: 10.1080/02681219680000171. [DOI] [PubMed] [Google Scholar]
- 108.Essayag S M, Baily G G, Denning D W, Burnie J P. Karyotyping of fluconazole-resistant yeasts with phenotype reported as Candida krusei or Candida inconspicua. Int J Syst Bacteriol. 1996;46:35–40. doi: 10.1099/00207713-46-1-35. [DOI] [PubMed] [Google Scholar]
- 109.Evans E E. The antigenic composition of Cryptococcus neoformans. I. A serologic classification by means of the capsular and agglutination reactions. J Immunol. 1950;64:423–430. [PubMed] [Google Scholar]
- 110.Faix R G, Finkel D J, Andersen R D, Hostetter M K. Genotypic analysis of a cluster of systemic Candida albicans infections in a neonatal intensive care unit. Pediatr Infect Dis J. 1995;14:1063–1068. doi: 10.1097/00006454-199512000-00007. [DOI] [PubMed] [Google Scholar]
- 111.Fan M, Currie B P, Gutell R R, Ragan M A, Casadevall A. The 16S-like, 5.8S and 28S-like rRNAs of the two varieties of Cryptococcus neoformans: sequence, secondary structure, phylogenetic analysis and restriction fragment polymorphism. J Med Vet Mycol. 1994;32:163–180. doi: 10.1080/02681219480000231. [DOI] [PubMed] [Google Scholar]
- 112.Farris J S. Estimating phylogenetic trees from distance matrices. Am Nat. 1972;106:645–668. [Google Scholar]
- 113.Fenn J P, Segal H, Barland B, Denton D, Whisenant J, Chun H, Christofferson K, Hamilton L, Carroll K. Comparison of updated Vitek Yeast Biochemical Card and API 20C yeast identification systems. J Clin Microbiol. 1994;32:1184–1187. doi: 10.1128/jcm.32.5.1184-1187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fitch W M, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 115.Fortun J, Lopez-San Roman A, Velasco J J, Sanchez-Sousa A, de Vicente E, Quereda C, Barcena R, Monge G, Candela A, Honrubia A, Guerrero A. Selection of Candida glabrata strains with reduced susceptibility to azoles in four liver transplant patients with invasive candidiasis. Eur J Clin Microbiol Infect Dis. 1997;16:314–318. doi: 10.1007/BF01695638. [DOI] [PubMed] [Google Scholar]
- 116.Fox B C, Mobley H L, Wade J C. The use of a DNA probe for epidemiological studies of candidiasis in immunocompromised hosts. J Infect Dis. 1989;159:488–494. doi: 10.1093/infdis/159.3.488. [DOI] [PubMed] [Google Scholar]
- 117.Franzot S P, Hamdan J S, Currie B P, Casadevall A. Molecular epidemiology of Cryptococcus neoformans in Brazil and the United States: evidence for local genetic differences and a global clonal population structure. J Clin Microbiol. 1997;35:2243–2251. doi: 10.1128/jcm.35.9.2243-2251.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Franzot S P, Mukherjee J, Cherniak R, Chen L C, Hamdan J S, Casadevall A. Microevolution of a standard strain of Cryptococcus neoformans resulting in differences in virulence and other phenotypes. Infect Immun. 1988;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fraser V J, Keath E J, Powderly W G. Two cases of blastomycosis from a common source: use of DNA restriction analysis to identify strains. J Infect Dis. 1991;163:1378–1381. doi: 10.1093/infdis/163.6.1378. [DOI] [PubMed] [Google Scholar]
- 120.Fricker-Hidalgo H, Vandapel O, Duchesne M-A, Mazoyer M A, Monget D, Lardy B, Lebeau B, Freney J, Ambroise-Thomas P, Grillot R. Comparison of the new API Candida system to the ID 32C system for identification of clinically important yeast species. J Clin Microbiol. 1996;34:1846–1848. doi: 10.1128/jcm.34.7.1846-1848.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fries B C, Chen F, Currie B P, Casadevall A. Karyotype instability in Cryptococcus neoformans infection. J Clin Microbiol. 1996;34:1531–1534. doi: 10.1128/jcm.34.6.1531-1534.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Geiser D M, Pitt J L, Taylor J W. Cryptic speciation and recombination in the aflatoxin producing fungi Aspergillus flavus. Proc Natl Acad Sci USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Girardin H, Latge J-P, Srikantha T, Morrow B, Soll D R. Development of DNA probes for fingerprinting Aspergillus fumigatus. J Clin Microbiol. 1993;31:1547–1554. doi: 10.1128/jcm.31.6.1547-1554.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Girardin H, Sarfati J, Traore F, Camet J D, Derouin F, Latge J P. Molecular epidemiology of nosocomial invasive aspergillosis. J Clin Microbiol. 1994;32:684–690. doi: 10.1128/jcm.32.3.684-690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Glasgow A C, Hughes K T, Simon M I. Bacterial DNA inversion systems. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 637–660. [Google Scholar]
- 126.Glover D M. Genes for ribosomal DNA. In: Maclean M, Gregory S P, Flavell R A, editors. Eukaryotic genes: their structure and regulation. Cambridge, United Kingdom: Butterworth & Co.; 1983. pp. 207–224. [Google Scholar]
- 127.Goldman D, Fries B, Franzot S, Montella L, Casadevall A. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc Natl Acad Sci USA. 1998;95:14967–14972. doi: 10.1073/pnas.95.25.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gottfredsson M, Cox G M, Perfect J R. Molecular methods for epidemiological and diagnostic studies of fungal infections. Pathology. 1998;30:405–418. doi: 10.1080/00313029800169726. [DOI] [PubMed] [Google Scholar]
- 129.Gottlieb S, Esposito S. A new role for a yeast transcriptional silencer gene, SIR 2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- 130.Gräser Y, Volovsek M, Arrington J, Schönian G, Presber W, Mitchell T G, Vilgalys R. Molecular markers reveal that population structure of the human pathogen Candida albicans exhibits both clonality and recombination. Proc Natl Acad Sci USA. 1996;93:12473–12477. doi: 10.1073/pnas.93.22.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Gyanchandani A, Khan Z K, Farooqui N, Goswami M, Ranade S A. RAPD analysis of Candida albicans strains recovered from immunocompromised patients (ICP) reveals an apparently non-random infectivity of the strains. Biochem Mol Biol Int. 1998;44:19–27. doi: 10.1080/15216549800201022. [DOI] [PubMed] [Google Scholar]
- 132.Hall L M. Are point mutations or DNA rearrangements responsible for the restriction fragment length polymorphisms that are used to type bacteria? Microbiology. 1994;140:197–204. doi: 10.1099/13500872-140-1-197. [DOI] [PubMed] [Google Scholar]
- 133.Hasenclever H F, Mitchell W O. Antigenic studies of Candida. I. Observations of two antigenic groups in Candida albicans. J Bacteriol. 1961;82:570–573. doi: 10.1128/jb.82.4.570-573.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hasenclever H F, Mitchell W O. Antigenic studies of Candida. III. Comparative pathogenicity of Candida albicans group A, group B, and Candida stellatoidea. J Bacteriol. 1961;82:578–581. doi: 10.1128/jb.82.4.578-581.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Hatlan L E, Attardi G. Preparation of the HeLa cell genome complementary to tRNA and 5SRNA. J Mol Biol. 1971;56:535–554. doi: 10.1016/0022-2836(71)90400-1. [DOI] [PubMed] [Google Scholar]
- 136.Hazen K C. New and emerging yeast pathogens. Clin Microbiol Rev. 1995;8:462–478. doi: 10.1128/cmr.8.4.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hellstein J, Vawter-Hugart H, Fotos P, Schmid J, Soll D R. Genetic similarity and phenotypic diversity of commensal and pathogenic strains of Candida albicans isolated from the oral cavity. J Clin Microbiol. 1993;31:3190–3199. doi: 10.1128/jcm.31.12.3190-3199.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hesselbarth J, Schwarz S. Comparative ribotyping of Staphylococcus intermedius from dogs, pigeons, horses and mink. Vet Microbiol. 1995;45:11–17. doi: 10.1016/0378-1135(94)00125-g. [DOI] [PubMed] [Google Scholar]
- 139.Holmberg K, Feroze F. Comparative study of the GenePath group 4 reagent system and other CHEF systems for karyotype analysis of Candida spp. J Clin Lab Anal. 1995;9:184–192. doi: 10.1002/jcla.1860090307. [DOI] [PubMed] [Google Scholar]
- 140.Holmberg K, Feroze F. Evaluation of an optimized system for random amplified polymorphic DNA (RAPD)-analysis for genotypic mapping of Candida albicans strains. J Clin Lab Anal. 1996;10:59–69. doi: 10.1002/(SICI)1098-2825(1996)10:2<59::AID-JCLA1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 141.Howell S A, Anthony R M, Power E. Application of RAPD and restriction enzyme analysis to the study of oral carriage of Candida albicans. Lett Appl Microbiol. 1996;22:125–128. doi: 10.1111/j.1472-765x.1996.tb01124.x. [DOI] [PubMed] [Google Scholar]
- 142.Huang Y C, Lin T Y, Leu H S, Wu J L, Wu J H. Yeast carriage on hands of hospital personnel working in intensive care units. J Hosp Infect. 1998;39:47–51. doi: 10.1016/s0195-6701(98)90242-0. [DOI] [PubMed] [Google Scholar]
- 143.Hube B, Monod M, Schofield D A, Brown A J, Gow N A. Expression of seven members of the gene family encoding secretory aspartyl proteinases in Candida albicans. Mol Microbiol. 1994;14:87–99. doi: 10.1111/j.1365-2958.1994.tb01269.x. [DOI] [PubMed] [Google Scholar]
- 144.Hunter P R, Fraser C A. Use of modified resistogram to type Candida albicans isolated from cases of vaginitis and from feces in the same geographical area. J Clin Pathol. 1987;40:1159–1161. doi: 10.1136/jcp.40.10.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Hunter P R, Fraser C A, Mackenzie D W. Morphotype markers of virulence in human candidal infections. J Med Microbiol. 1989;28:85–91. doi: 10.1099/00222615-28-2-85. [DOI] [PubMed] [Google Scholar]
- 146.Hunter P R. A critical review of typing methods for Candida albicans and their applications. Crit Rev Microbiol. 1991;17:417–434. doi: 10.3109/10408419109115206. [DOI] [PubMed] [Google Scholar]
- 147.Ingram C W, Haywood H B, Morris V M, Allen R L, Perfect J R. Cryptococcal ventricular-peritoneal shunt infection: clinical and epidemiological evaluation of two closely associated cases. Infect Control Hosp Epidemiol. 1993;14:719–722. doi: 10.1086/646675. [DOI] [PubMed] [Google Scholar]
- 148.Iwaguchi S, Homma M, Tanaka K. Variation in the electrophoretic karyotype analysed by the assignment of DNA probes in Candida albicans. J Gen Microbiol. 1990;136:2433–2442. doi: 10.1099/00221287-136-12-2433. [DOI] [PubMed] [Google Scholar]
- 149.Iwaguchi S, Homma M, Tanaka K. Clonal variation of chromosome size derived from the rDNA cluster region in Candida albicans. J Gen Microbiol. 1992;138:1177–1184. doi: 10.1099/00221287-138-6-1177. [DOI] [PubMed] [Google Scholar]
- 150.Iwaguchi S-I, Homma M, Chibana H, Tanaka K. Isolation and characterization of a repeated sequence (RPS1) of Candida albicans. J Gen Microbiol. 1992;138:1893–1900. doi: 10.1099/00221287-138-9-1893. [DOI] [PubMed] [Google Scholar]
- 151.Joly S, Pujol C, Rysz M, Vargas K, Soll D R. Development and characterization of complex DNA fingerprinting probes for the infectious agent Candida dubliniensis. J Clin Microbiol. 1999;37:1035–1044. doi: 10.1128/jcm.37.4.1035-1044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Joly S, Pujol C, Schröppel K, Soll D R. Development and verification of two species fingerprinting probes for Candida tropicalis amenable to computer analysis. J Clin Microbiol. 1996;34:3063–3071. doi: 10.1128/jcm.34.12.3063-3071.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jordan J A. PCR identification of four medically important Candida species by using a single primer pair. J Clin Microbiol. 1994;32:2962–2967. doi: 10.1128/jcm.32.12.2962-2967.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Karabinis A, Hill C, Leclercq B, Tancrede C, Baume D, Andremont A. Risk factors for candidemia in cancer patients: a case-control study. J Clin Microbiol. 1988;26:429–432. doi: 10.1128/jcm.26.3.429-432.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Karl S A, Avise J C. PCR-based assays of Mendelian polymorphisms from anonymous single-copy nuclear DNA-techniques and applications for population genetics. Mol Biol Evol. 1993;10:342–361. doi: 10.1093/oxfordjournals.molbev.a040002. [DOI] [PubMed] [Google Scholar]
- 156.Kasuga T, Taylor J W, White T J. Phylogenetic relationships of varieties and geographical groups of the human pathogenic fungus, Histoplasma capsulatum Darling. J Clin Microbiol. 1998;37:2165–2168. doi: 10.1128/jcm.37.3.653-663.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kaufmann C S, Merz W G. Electrophoretic karyotypes of Torulopsis glabrata. J Clin Microbiol. 1989;27:2165–2168. doi: 10.1128/jcm.27.10.2165-2168.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Keath E J, Kobayashi G S, Medoff G. Typing of Histoplasma capsulatum by restriction fragment length polymorphisms in a nuclear gene. J Clin Microbiol. 1992;30:2104–2107. doi: 10.1128/jcm.30.8.2104-2107.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Keath E J, Painter A A, Kobayashi G S, Medoff G. Variable expression of a yeast-phase-specific gene in Histoplasma capsulatum strains differing in thermotolerance and virulence. Infect Immun. 1989;57:1384–1390. doi: 10.1128/iai.57.5.1384-1390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kersulyte D, Woods J P, Keath E J, Goldman W E, Berg D E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J Bacteriol. 1992;174:7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Khatib R, Thirumoorthi M C, Riederer K M, Sturm L, Oney L A, Baran J., Jr Clustering of Candida infections in the neonatal intensive care unit: concurrent emergence of multiple strains simulating intermittent outbreaks. Pediatr Infect Dis J. 1998;17:130–134. doi: 10.1097/00006454-199802000-00010. [DOI] [PubMed] [Google Scholar]
- 162.Khattak M N, Burnie J P, Matthews R C, Oppenheim B A. Clamped homogeneous electric field gel electrophoresis typing of Torulopsis glabrata isolates causing nosocomial infections. J Clin Microbiol. 1992;30:2211–2215. doi: 10.1128/jcm.30.8.2211-2215.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.King D, Rhine-Chalberg J, Pfaller M A, Moser S A, Merz W G. Comparison of four DNA-based methods for strain delineation of Candida lusitaniae. J Clin Microbiol. 1995;33:1467–1470. doi: 10.1128/jcm.33.6.1467-1470.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Kleinegger C, Lockhart S T, Vargas K, Soll D R. Frequency, intensity, species and strains of oral yeast vary as a function of host age. J Clin Microbiol. 1996;34:2246–2254. doi: 10.1128/jcm.34.9.2246-2254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Komshian S V, Uwaydah A K, Sobel J D, Crane L R. Fungemia caused by Candida species and Torulopsis glabrata in the hospitalized patient: frequency, characteristics, and evaluation of factors influencing outcome. Rev Infect Dis. 1989;11:379–390. doi: 10.1093/clinids/11.3.379. [DOI] [PubMed] [Google Scholar]
- 166.Korting H C, Ollert M, Georgii A, Fröschl M. In vitro susceptibilities and biotypes of Candida albicans isolates from the oral cavities of patients infected with human immunodeficiency virus. J Clin Microbiol. 1988;26:2626–2631. doi: 10.1128/jcm.26.12.2626-2631.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Koufopanou V, Burt A, Taylor J W. Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc Natl Acad Sci USA. 1997;94:5478–5482. doi: 10.1073/pnas.94.10.5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kuehnert M J, Clark E, Lockhart S R, Soll D R, Chia J, Jarvis W R. Candida albicans endocarditis traced to a contaminated aoertic valve allograft: implications for regulation of allograft processing. Clin Infect Dis. 1998;27:688–691. doi: 10.1086/514944. [DOI] [PubMed] [Google Scholar]
- 169.Kwon-Chung K J, Wickes B L, Stockman L, Roberts G D, Ellis D, Howard D H. Virulence, serotype, and molecular characteristics of environmental strains of Cryptococcus neoformans var. gattii. Infect Immun. 1992;60:1869–1874. doi: 10.1128/iai.60.5.1869-1874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Kwon-Chung K J, Riggsby W S, Uphoff R A, Hicks J B, Whelan W L, Reiss E, Magee B B, Wickes B L. Genetic differences between type I and type II Candida stellatoidea. Infect Immun. 1989;57:527–532. doi: 10.1128/iai.57.2.527-532.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Kwon-Chung K J, Wickes B L, Merz W G. Association of electrophoretic karyotype of Candida stellatoidea with virulence for mice. Infect Immun. 1988;56:1814–1819. doi: 10.1128/iai.56.7.1814-1819.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lacher D A, Lehmann P F. Application of multidimensional scaling in numerical taxonomy: analysis of isoenzyme types of Candida species. Ann Clin Lab Sci. 1991;21:94–103. [PubMed] [Google Scholar]
- 173.Lachke S A, Srikantha T, Tsai L K, Daniels K, Soll D R. Phenotypic switching in Candida glabrata involves phase-specific regulation of the metallothionein gene MT-II and the newly discovered hemolysin gene HLP. Infect Immun. 2000;68:884–895. doi: 10.1128/iai.68.2.884-895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lasker B A, Carle G F, Kobayashi G S, Medoff G. Comparison of the separation of Candida albicans chromosome-sized DNA by pulsed-field gel electrophoresis techniques. Nucleic Acids Res. 1989;17:3783–3793. doi: 10.1093/nar/17.10.3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lasker B A, Page L S, Lott T J, Kobayashi G S. Isolation, characterization, and sequencing of Candida albicans repetitive element 2. Gene. 1992;116:51–57. doi: 10.1016/0378-1119(92)90628-3. [DOI] [PubMed] [Google Scholar]
- 176.Latgé J-P. Aspergillus fumigatus and aspergillosis. Clin Microbiol Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Latouche G N, Daniel H-M, Lee O C, Mitchell T G, Sorrell T C, Meyer W. Comparison of use of phenotypic and genotypic characteristics for identification of species of the anamorph genus Candida and related teleomorph yeast species. J Clin Microbiol. 1997;35:3171–3180. doi: 10.1128/jcm.35.12.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Leenders A, van Belkum A, Janssen S, de Marie S, Kluytmans J, Wielenga J, Lowenberg B, Verbrugh H. Molecular epidemiology of apparent outbreak of invasive aspergillosis in a hematology ward. J Clin Microbiol. 1996;34:345–351. doi: 10.1128/jcm.34.2.345-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Le Guennec R, Reynes J, Mallie M, Pujol C, Janbon F, Bastide J M. Fluconazole- and Itraconazole-resistant Candida albicans strains from AIDS patients: multilocus enzyme electrophoresis analysis and antifungal susceptibilities. J Clin Microbiol. 1995;33:2732–2737. doi: 10.1128/jcm.33.10.2732-2737.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lehmann P F, Kemker B J, Hsiao C B, Dev S. Isoenzyme biotypes of Candida species. J Clin Microbiol. 1989;27:2514–2521. doi: 10.1128/jcm.27.11.2514-2521.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lehmann P F, Lin D, Lasker B A. Genotypic identification and characterization of species and strains within the genus Candida by using random amplified polymorphic DNA. J Clin Microbiol. 1992;30:3249–3254. doi: 10.1128/jcm.30.12.3249-3254.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lehmann P F, Wu L C, Mackenzie D W. Isoenzyme changes in Candida albicans during domestication. J Clin Microbiol. 1991;29:2623–2625. doi: 10.1128/jcm.29.11.2623-2625.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lehmann P F, Wu L C, Pruitt W R, Meyer S A, Ahearn D G. Unrelatedness of groups of yeasts within the Candida haemulonii complex. J Clin Microbiol. 1993;31:1683–1687. doi: 10.1128/jcm.31.7.1683-1687.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Levin A S, Costa S F, Mussi N S, Basso M, Sinto S I, Machado C, Geiger D C, Villares M C, Schreiber A Z, Barone A A, Branchini M L. Candida parapsilosis fungemia associated with implantable and semi-implantable central venous catheters and the hands of healthcare workers. Diagn Microbiol Infect Dis. 1998;30:243–249. doi: 10.1016/s0732-8893(98)00006-6. [DOI] [PubMed] [Google Scholar]
- 185.Li W-H, Grauer D. Fundamentals of molecular evolution. Sunderland, Mass: Sinauer Associates; 1991. [Google Scholar]
- 186.Lin D, Lehmann P F, Hamory B H, Padhye A A, Durry E, Pinner R W, Lasker B A. Comparison of three typing methods for clinical and environmental isolates of Aspergillus fumigatus. J Clin Microbiol. 1995;33:1596–1601. doi: 10.1128/jcm.33.6.1596-1601.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lin D, Lehmann P F. Random amplified polymorphic DNA for strain delineation within Candida tropicalis. J Med Vet Mycol. 1995;33:241–246. doi: 10.1080/02681219580000491. [DOI] [PubMed] [Google Scholar]
- 188.Lin D, Wu L C, Rinaldi M G, Lehmann P F. Three distinct genotypes within Candida parapsilosis from clinical sources. J Clin Microbiol. 1995;33:1815–1821. doi: 10.1128/jcm.33.7.1815-1821.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lipperheide V, Quindós G, Jiménez Y, Pontón J, Bagán-Sebastián J V, Auirre J M. Candida biotypes in patients with oral leukoplakia and lichen planus. Mycopathologia. 1996;134:75–82. doi: 10.1007/BF00436868. [DOI] [PubMed] [Google Scholar]
- 190.Lischewski A, Harmsen D, Hacker J, Morschhauser J. Standardized molecular typing of Candida albicans strains. Mycoses. 1997;40:369–372. doi: 10.1111/j.1439-0507.1997.tb00252.x. [DOI] [PubMed] [Google Scholar]
- 191.Lischewski A, Ruhnke M, Tennagen I, Schonian G, Morschhauser J, Hacker J. Molecular epidemiology of Candida isolates from AIDS patients showing different fluconazole resistance profiles. J Clin Microbiol. 1995;33:769–771. doi: 10.1128/jcm.33.3.769-771.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Lockhart S, Fritch J J, Meier A S, Schröppel K, Srikantha T, Galask R, Soll D R. Colonizing populations of Candida albicans are clonal in origin but undergo microevolution through C1 fragment reorganization as demonstrated by DNA fingerprinting and C1 sequencing. J Clin Microbiol. 1995;33:1501–1509. doi: 10.1128/jcm.33.6.1501-1509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Reference deleted.
- 194.Lockhart S R, Joly S, Pujol C, Sobel J D, Pfaller M A, Soll D R. Development and verification of fingerprinting probes for Candida glabrata. Microbiology. 1997;143:3733–3746. doi: 10.1099/00221287-143-12-3733. [DOI] [PubMed] [Google Scholar]
- 195.Lockhart S, Joly S, Vargas K, Swails-Wenger J, Enger L, Soll D R. Natural defenses against Candida colonization break down in the oral cavity of the elderly. J Dent Res. 1998;78:857–868. doi: 10.1177/00220345990780040601. [DOI] [PubMed] [Google Scholar]
- 196.Lockhart S R, Reed B D, Pierson C L, Soll D R. Most frequent scenario for recurrent Candida vaginitis is strain maintenance with “substrain shuffling”: demonstration by sequential DNA fingerprinting with probes Ca3, C1, and CARE2. J Clin Microbiol. 1996;34:767–777. doi: 10.1128/jcm.34.4.767-777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Lo Passo C, Pernice I, Gallo M, Barbara C, Luck F T, Criseo G, Pernice A. Genetic relatedness and diversity of Cryptococcus neoformans strains in the Maltese Islands. J Clin Microbiol. 1997;35:751–755. doi: 10.1128/jcm.35.3.751-755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lott T J, Boiron P, Reiss E. An electrophoretic karyotype for Candida albicans reveals large chromosomes in multiples. Mol Gen Genet. 1987;209:170–174. doi: 10.1007/BF00329854. [DOI] [PubMed] [Google Scholar]
- 199.Lott T J, Keykendall R J, Welbel S F, Pramanik A, Lasker B A. Genomic heterogeneity in the yeast Candida parapsilosis. Curr Genet. 1993;23:463–467. doi: 10.1007/BF00312635. [DOI] [PubMed] [Google Scholar]
- 200.Loudon K W, Burnie J P, Coke A P, Matthews R C. Application of polymerase chain reaction to fingerprinting Aspergillus fumigatus by random amplification of polymorphic DNA. J Clin Microbiol. 1993;31:1117–1121. doi: 10.1128/jcm.31.5.1117-1121.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Loudon K W, Coke A P, Burnie J P. “Pseudoclusters” and typing by random amplification of polymorphic DNA of Aspergillus fumigatus. J Clin Pathol. 1995;48:183–184. doi: 10.1136/jcp.48.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Loudon K W, Coke A P, Burnie J P, Lucas G S, Liu Yin J A. Invasive aspergillosis: clusters and sources? J Med Vet Mycol. 1994;32:217–224. doi: 10.1080/02681219480000281. [DOI] [PubMed] [Google Scholar]
- 203.Loudon K W, Coke A P, Burnie J P, Shaw A J, Oppenheim B A, Morris C Q. Kitchens as a source of Aspergillus niger infection. J Hosp Infect. 1996;32:191–198. doi: 10.1016/s0195-6701(96)90145-0. [DOI] [PubMed] [Google Scholar]
- 204.Lupetti A, Guzzi G, Paladini A, Swart K, Campa M, Senesi S. Molecular typing of Candida albicans in oral candidiasis: karyotype epidemiology with human immunodeficiency virus-seropositive patients in comparison with that with healthy carriers. J Clin Microbiol. 1995;33:1238–1342. doi: 10.1128/jcm.33.5.1238-1242.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Maffei C M, Paula C R, Mazzocato T S, Franceschini S. Phenotype and genotype of Candida albicans strains isolated from pregnant women with recurrent vaginitis. Mycopathologia. 1997;137:87–94. doi: 10.1023/a:1006878908668. [DOI] [PubMed] [Google Scholar]
- 206.Magee B B, D'Souza T M, Magee P T. Strain and species identification by restriction length polymorphisms in the ribosomal DNA repeat of Candida species. J Bacteriol. 1987;169:1639–1643. doi: 10.1128/jb.169.4.1639-1643.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Magee B B, Magee P T. Electrophoretic karyotypes and chromosome numbers in Candida species. J Gen Microbiol. 1987;133:425–430. doi: 10.1099/00221287-133-2-425. [DOI] [PubMed] [Google Scholar]
- 208.Magee P T, Bowdin L, Staudinger J. Comparison of molecular typing methods for Candida albicans. J Clin Microbiol. 1992;30:2674–2679. doi: 10.1128/jcm.30.10.2674-2679.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Mahrous M, Lott T J, Meyer S A, Sawant A D, Ahearn D G. Electrophoretic karyotyping of typical and atypical Candida albicans. J Clin Microbiol. 1990;28:876–881. doi: 10.1128/jcm.28.5.876-881.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Maiwald M, Kappe R, Sonntag H G. Rapid presumptive identification of medically relevant yeasts to the species level by polymerase chain reaction and restriction enzyme analysis. J Med Vet Mycol. 1994;32:115–122. doi: 10.1080/02681219480000161. [DOI] [PubMed] [Google Scholar]
- 211.Marco F, Lockhart S, Pfaller M, Soll D R. Elucidating the origins of nosocomial infections of C. albicans by DNA fingerprinting with the probe Ca3. J Clin Microbiol. 1998;37:2817–2828. doi: 10.1128/jcm.37.9.2817-2828.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Marichal P, Vanden B H, Odds F C, Nobels G, Warnock D W, Timmerman V, et al. Molecular biological characterization of an azole-resistant Candida glabrata isolate. Antimicrob Agents Chemother. 1997;41:2229–2237. doi: 10.1128/aac.41.10.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Mason M M, Lasker B A, Riggsby W S. Molecular probe for identification of medically important Candida species and Torulopsis glabrata. J Clin Microbiol. 1987;25:563–566. doi: 10.1128/jcm.25.3.563-566.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.McAlpin C E, Mannarelli B. Construction and characterization of a DNA probe for distinguishing strains of Aspergillus flavus. Appl Environ Microbiol. 1995;61:1068–1072. doi: 10.1128/aem.61.3.1068-1072.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.McCreight M C, Warnock D W, Martin M V. Resistogram typing of Candida albicans isolates from oral and cutaneous sites in irradiated patients. Sabouraudia. 1985;23:403–406. [PubMed] [Google Scholar]
- 216.McCullough M J, Ross B C, Dwyer B D, Reade P C. Genotype and phenotype of oral Candida albicans from patients infected with the human immunodeficiency virus. Microbiology. 1994;140:1195–1202. doi: 10.1099/13500872-140-5-1195. [DOI] [PubMed] [Google Scholar]
- 217.Mercure S, Montplaisir S, Lemay G. Correlation between the presence of a self-splicing intron in the 25S rDNA of C. albicans and strain susceptibility to 5-fluorocytosine. Nucleic Acids Res. 1993;21:6020–6027. doi: 10.1093/nar/21.25.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Mercure S, Poirier S, Lemay G, Auger P, Montplaisir S, de Repentigny L. Application of biotyping and DNA typing of Candida albicans to the epidemiology of recurrent vulvovaginal candidasis. J Infect Dis. 1993;168:502–507. doi: 10.1093/infdis/168.2.502. [DOI] [PubMed] [Google Scholar]
- 219.Merz W G, Connelly C, Hieter P. Variation of electrophoretic karyotypes among clinical isolates of Candida albicans. J Clin Microbiol. 1988;26:842–845. doi: 10.1128/jcm.26.5.842-845.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Merz W G, Khazan U, Jabra-Rizk M A, Wu L C, Osterhout G J, Lehmann P F. Strain delineation and epidemiology of Candida (Clavispora) lusitaniae. J Clin Microbiol. 1992;30:449–454. doi: 10.1128/jcm.30.2.449-454.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Metzgar D, van Belkum A, Field D, Haubrich R, Wills C. Random amplification of polymorphic DNA and microsatellite genotyping of pre- and posttreatment isolates of Candida spp. from human immunodeficiency virus-infected patients on different fluconazole regimens. J Clin Microbiol. 1998;36:2308–2313. doi: 10.1128/jcm.36.8.2308-2313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Meunier J-R, Grimont P A. Factors affecting reproducibility of random amplified polymorphic DNA fingerprinting. Res Microbiol. 1993;144:373–379. doi: 10.1016/0923-2508(93)90194-7. [DOI] [PubMed] [Google Scholar]
- 223.Meyer W, Lieckfeldt E, Kuhls K, Freedman E Z, Borner T, Mitchell T G. DNA- and PCR-fingerprinting in fungi. Exs. 1993;67:311–320. doi: 10.1007/978-3-0348-8583-6_28. [DOI] [PubMed] [Google Scholar]
- 224.Meyer W, Mitchell T. Polymerase chain reaction fingerprinting in fungi using single primers specific to minisatellites and simple repetitive DNA sequences: strain variation in Cryptococcus neoformans. Electrophoresis. 1995;16:1648–1656. doi: 10.1002/elps.11501601273. [DOI] [PubMed] [Google Scholar]
- 225.Meyer W, Mitchell T G, Freedman E Z, Vilgalys R. Hybridization probes for conventional DNA fingerprinting used as single primers in the polymerase chain reaction to distinguish strains of Cryptococcus neoformans. J Clin Microbiol. 1993;31:2274–2280. doi: 10.1128/jcm.31.9.2274-2280.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Millon L, Manteaux A, Reboux G, Drobacheff C, Monod M, Barale T, Michel-Briand Y. Fluconazole-resistant recurrent oral candidiasis in human immunodeficiency virus-positive patients: persistence of Candida albicans strains with the same genotype. J Clin Microbiol. 1994;32:1115–1118. doi: 10.1128/jcm.32.4.1115-1118.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Miyakawa Y, Kagaya K, Kuribayashi T, Suzuki M, Fukazawa Y. Isolation and chemical and biological characterization of antigenic mutants of Candida albicans serotype A. Yeast. 1989;5:S225–S229. [PubMed] [Google Scholar]
- 228.Miyasaki S H, Hicks J B, Greenspan D, Polachek I, MacPhail L A, White T C, Agabian N, Greenspan J S. The identification and tracking of Candida albicans isolates from oral lesions in HIV-seropositive individuals. J Acquired Immune Defic Syndr. 1992;5:1039–1046. [PubMed] [Google Scholar]
- 229.Mondon P, Thelu J, Lebeau B, Ambroise-Thomas P, Grillot R. Virulence of Aspergillus fumigatus strains investigated by random amplified polymorphic DNA analysis. J Med Microbiol. 1995;42:299–303. doi: 10.1099/00222615-42-4-299. [DOI] [PubMed] [Google Scholar]
- 230.Monod M, Porchet S, Baudraz-Rosselet F, Frenk E. The identification of pathogenic yeast strains by electrophoretic analysis of their chromosomes. J Med Microbiol. 1990;32:123–129. doi: 10.1099/00222615-32-2-123. [DOI] [PubMed] [Google Scholar]
- 231.Moody S F, Tyler B M. Use of nuclear DNA restriction fragment length polymorphisms to analyze the diversity of the Aspergillus flavus group: A. flavus, A. parasiticus and A. nomius. Appl Environ Microbiol. 1990;56:2453–2461. doi: 10.1128/aem.56.8.2453-2461.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Moran G P, Sullivan D J, Henman M C, McCreary C E, Harrington B J, Shanley D B, Coleman D C. Antifungal drug susceptibilities of oral Candida dubliniensis isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected subjects and generation of stable fluconazole-resistant derivatives in vitro. Antimicrob Agents Chemother. 1997;41:617–623. doi: 10.1128/aac.41.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Morrow B, Srikantha T, Anderson J, Soll D R. Coordinate regulation of two opaque-phase-specific genes during white-opaque switching in Candida albicans. Infect Immun. 1993;61:1823–1828. doi: 10.1128/iai.61.5.1823-1828.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Morrow B, Srikantha T, Soll D R. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol Cell Biol. 1992;12:2997–3005. doi: 10.1128/mcb.12.7.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Nebavi F, Arnavielhe S, Le Guennec R, Menan E, Kacou A, Combe P, Aoussi E, Mallie M, Kone M, Bastide J M. Oropharyngeal candidiasis in AIDS patients from Abidjan (Ivory Coast): antifungal susceptibilities and multilocus enzyme electrophoresis analysis of Candida albicans isolates. Pathol Biol. 1998;46:307–314. [PubMed] [Google Scholar]
- 236.Neely A N, Odds F C, Basatia B K, Holder I A. Characterization of Candida isolates from pediatric burn patients. J Clin Microbiol. 1988;26:1645–1649. doi: 10.1128/jcm.26.9.1645-1649.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Nguyen M, Peacock J, Morris A, Tanner D, Nguyen M, Snydman D R, Wagener M, Rinaldi M, Yu V. The changing face of candidemia: emergence of non-Candida albicans species and antifungal resistance. Am J Med. 1996;100:617–623. doi: 10.1016/s0002-9343(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 238.Nomura M, Morgan E A. Genetics of bacterial ribosomes. Annu Rev Genet. 1977;11:297–347. doi: 10.1146/annurev.ge.11.120177.001501. [DOI] [PubMed] [Google Scholar]
- 239.Odds F C. Resistance of yeasts to azole-derivative antifungals. J Antimicrob Chemother. 1993;31:463–471. doi: 10.1093/jac/31.4.463. [DOI] [PubMed] [Google Scholar]
- 240.Odds F C. Switch of phenotype as an escape mechanism of the intruder. Mycoses. 1997;2:9–12. doi: 10.1111/j.1439-0507.1997.tb00556.x. [DOI] [PubMed] [Google Scholar]
- 241.Odds F C, Abbott A B. A simple system for the presumptive identification of Candida albicans and differentiation of strains within the species. Sabouraudia. 1980;18:301–317. [PubMed] [Google Scholar]
- 242.Odds F C, Abbott A B. Modification and extension of tests for differentiation of Candida species and strains. Sabouraudia. 1983;21:79–81. doi: 10.1080/00362178385380111. [DOI] [PubMed] [Google Scholar]
- 243.Odds F C, Auger P, Krogh P, Neely A N, Segal E. Biotyping of Candida albicans: results of an international collaborative survey. J Clin Microbiol. 1989;27:1506–1509. doi: 10.1128/jcm.27.7.1506-1509.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Odds F C, Bernaerts R. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J Clin Microbiol. 1994;32:1923–1929. doi: 10.1128/jcm.32.8.1923-1929.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Odds F C, Palacio-Hernanz A, Cuadra J, Sanchéz J. Disseminated Candida infection syndrome in heroin addicts—dominance of a single Candida albicans biotype. J Med Microbiol. 1987;23:275–277. doi: 10.1099/00222615-23-3-275. [DOI] [PubMed] [Google Scholar]
- 246.Odds F C, Kibbler C C, Walker E, Bhamra A, Prentice H G, Noone P. Carriage of Candida species and C. albicans biotypes in patients undergoing chemotherapy or bone marrow transplantation for haematological disease. J Clin Pathol. 1989;42:1259–1266. doi: 10.1136/jcp.42.12.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Odds F C, Schmid J, Soll D R. Proceedings of the Third Symposium of Topics in Mycology on Mycoses in AIDS Patients. New York, N.Y: Plenum Press; 1990. Epidemiology of Candida infections in AIDS; pp. 67–74. [Google Scholar]
- 248.Odds F C, Webster C E, Fisk P G, Riley V C, Mayuranathan P, Simmons P D. Candida species and C. albicans biotypes in women attending clinics in genitourinary medicine. J Med Microbiol. 1989;29:51–54. doi: 10.1099/00222615-29-1-51. [DOI] [PubMed] [Google Scholar]
- 249.Odds F C, Webster C E, Riley V C, Fisk P G. Epidemiology of vaginal Candida infection: significance of numbers of vaginal yeast and their biotypes. Eur J Obst Gynecol Reprod Biol. 1987;25:53–66. doi: 10.1016/0028-2243(87)90092-x. [DOI] [PubMed] [Google Scholar]
- 250.Olive D M, Bean P. Principles and applications of methods for DNA-based typing of microbial organisms. J Clin Microbiol. 1999;37:1661–1669. doi: 10.1128/jcm.37.6.1661-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Olivo P D, McManus E J, Riggsby W S, Jones J M. Mitochondrial DNA polymorphism in Candida albicans. J Infect Dis. 1987;156:214–215. doi: 10.1093/infdis/156.1.214. [DOI] [PubMed] [Google Scholar]
- 252.Orita M, Suzuki Y, Sekiya T, Hayashi K. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics. 1989;5:874–879. doi: 10.1016/0888-7543(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 253.Pan S, Cole G T. Electrophoretic karyotypes of clinical isolates of Coccidioides immitis. Infect Immun. 1992;60:4872–4880. doi: 10.1128/iai.60.11.4872-4880.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Pan S, Sigler L, Cole G T. Evidence for a phylogenetic connection between Coccidioidies immitis and Uncinocarpus reesii (Onygenaceae) Microbiology. 1994;140:1481–1494. doi: 10.1099/00221287-140-6-1481. [DOI] [PubMed] [Google Scholar]
- 255.Passoth V, Hansen M, Klinner U, Emeis C C. The electrophoretic banding pattern of the chromosomes of Pichia stipitis and Candida shehatae. Curr Genet. 1992;22:429–431. doi: 10.1007/BF00352445. [DOI] [PubMed] [Google Scholar]
- 256.Perfect J R, Ketabchi N, Cox G M, Ingram C W, Beiser C L. Karyotyping of Cryptococcus neoformans as an epidemiological tool. J Clin Microbiol. 1993;31:3305–3309. doi: 10.1128/jcm.31.12.3305-3309.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Perfect J R, Magee B B, Magee P T. Separation of chromosomes of Cryptococcus neoformans by pulsed field gel electrophoresis. Infect Immun. 1989;57:2624–2627. doi: 10.1128/iai.57.9.2624-2627.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Perrot M O, Grilloot R, Ambroise-Thomas P. Repartition dans l'organisme et sensibilite a la 5-fluorocytosine des deux serotypes de Candida albicans etude de 680 souches. Bull Soc Fr Mycol Med. 1977;6:305–309. [Google Scholar]
- 259.Pfaller M, Rex J, Rinaldi M. Antifungal susceptibility testing: technical advances and potential clinical applications. Clin Infect Dis. 1997;24:776–784. doi: 10.1093/clinids/24.5.776. [DOI] [PubMed] [Google Scholar]
- 260.Pfaller M A. Epidemiology of candiasis. Comp Ther. 1995;21:653–657. [PubMed] [Google Scholar]
- 261.Pfaller M A. Nosocomial candidiasis: emerging species, reservoirs, and modes of transmission. Clin Infect Dis. 1996;22:S89–S94. doi: 10.1093/clinids/22.supplement_2.s89. [DOI] [PubMed] [Google Scholar]
- 262.Pfaller M A, Cabezudo I, Hollis R, Huston B, Wenzel R P. The use of biotyping and DNA fingerprinting in typing Candida albicans from hospitalized patients. Diag Microbiol Infect Dis. 1990;13:481–489. doi: 10.1016/0732-8893(90)90080-f. [DOI] [PubMed] [Google Scholar]
- 263.Pfaller M A, Houston A, Coffmann S. Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, and Candida (Torulopsis) glabrata. J Clin Microbiol. 1996;34:58–61. doi: 10.1128/jcm.34.1.58-61.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Pfaller M A, Jones R N, Doern G V, Sader H S, Hollis R J, Messer S A. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and antifungal susceptibility of isolates collected in 1997 in the United States, Canada and South America for the SENTRY Program. J Clin Microbiol. 1998;36:1886–1889. doi: 10.1128/jcm.36.7.1886-1889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Pfaller M A, Lockhart S R, Pujol C, Swails-Wenger J A, Messer S A, Edmund M B, Jones R N, Wenzel R P, Soll D R. Hospital specificity, region specificity and fluconazole-resistance of Candida albicans blood stream isolates. J Clin Microbiol. 1998;36:1518–1529. doi: 10.1128/jcm.36.6.1518-1529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Pfaller M A, Messer S A, Hollis R J. Strain delineation and antifungal susceptibilities of epidemiologically related and unrelated isolates of Candida lusitaniae. Diagn Microbiol Infect Dis. 1994;20:127–133. doi: 10.1016/0732-8893(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 267.Pfaller M A, Messer S A, Hollis R J. Variations in DNA subtype, antifungal susceptibility, and slime production among clinical isolates of Candida parapsilosis. Diagn Microbiol Infect Dis. 1995;21:9–14. doi: 10.1016/0732-8893(94)00114-c. [DOI] [PubMed] [Google Scholar]
- 268.Pfaller M A, Messer S A, Houston A, Rangel-Frausto M S, Wiblin T, Blumberg H M, Edwards J E, Jarvis W, Martin M A, Neu H C, Saiman L, Patterson J E, Dibb J C, Roldan C M, Rinaldi M G, Wenzel R P. National epidemiology of mycoses survey: a multicenter study of strain variation and antifungal susceptibility among isolates of Candida species. Diagn Microbiol Infect Dis. 1998;31:289–296. doi: 10.1016/s0732-8893(97)00245-9. [DOI] [PubMed] [Google Scholar]
- 269.Pfaller M A, Rhine-Chalberg J, Redding S W, Smith J, Farinacci G, Fothergill A W, Rinaldi M G. Variations in fluconazole susceptibility and electrophoretic karyotype among oral isolates of Candida albicans from patients with AIDS and oral candidiasis. J Clin Microbiol. 1994;32:59–64. doi: 10.1128/jcm.32.1.59-64.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Phongpaichit S, Mackenzie D W, Fraser C. Strain differentiation of Candida albicans by morphotyping. Epidemiol Infect. 1987;99:421–428. doi: 10.1017/s0950268800067911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Pierson D L, Mehta S K, Magee B B, Mishra S K. Person-to-person transfer of Candida albicans in the spacecraft environment. J Med Vet Mycol. 1995;33:145–150. [PubMed] [Google Scholar]
- 272.Pineda-Garcia L, Ferrera A, Galvez C A, Hoffner S E. Drug-resistant Mycobacterium tuberculosis and atypical mycobacteria isolated from patients with suspected pulmonary tuberculosis in Honduras. Chest. 1997;111:148–153. doi: 10.1378/chest.111.1.148. [DOI] [PubMed] [Google Scholar]
- 273.Pineda-Garcia L, Ferrera A, Hoffner S E. DNA fingerprinting of Mycobacterium tuberculosis strains from patients with pulmonary tuberculosis in Honduras. J Clin Microbiol. 1997;35:2393–2397. doi: 10.1128/jcm.35.9.2393-2397.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Pittet D, Monod M, Filthuth I, Frenk E, Suter P M, Aukenthaler R. Contour-clamped homogeneous electric field gel electrophoresis as a powerful epidemiological tool in yeast infections. Am J Med. 1991;91:256S–263S. doi: 10.1016/0002-9343(91)90378-b. [DOI] [PubMed] [Google Scholar]
- 275.Pittet D, Monod M, Suter P M, Frenk E, Auckenthaler R. Candida colonization and subsequent infections in critically ill surgical patients. Ann Surg. 1994;220:751–758. doi: 10.1097/00000658-199412000-00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Poirier S, Auger P, Joly J, Steben M. Interest of biotyping Candida albicans in chronic vulvovaginitis. Mycoses. 1990;33:24–28. doi: 10.1111/myc.1990.33.1.24. [DOI] [PubMed] [Google Scholar]
- 277.Polacheck I, Lebens G A. Electrophoretic karyotype of the pathogenic yeast Cryptococcus neoformans. J Gen Microbiol. 1989;135:65–71. doi: 10.1099/00221287-135-1-65. [DOI] [PubMed] [Google Scholar]
- 278.Polonelli L, Archibusacci C, Sestito M, Morace G. Killer system: a simple method for differentiating Candida albicans strains. J Clin Microbiol. 1983;17:774–780. doi: 10.1128/jcm.17.5.774-780.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Polonelli L, Castoanola C, Rossetti D V, Morace G. Use of killer toxins for computer-aided differentiation of Candida albicans strains. Mycopathologia. 1985;91:175–179. doi: 10.1007/BF00446297. [DOI] [PubMed] [Google Scholar]
- 280.Pomes R, Gil C, Nombela C. Genetic analysis of Candida albicans morphological mutants. J Gen Microbiol. 1985;131:2107–2113. doi: 10.1099/00221287-131-8-2107. [DOI] [PubMed] [Google Scholar]
- 281.Pontieri E, Gregori L, Gennarelli M, Ceddia T, Novelli G, Dallapiccola B, De Bernardis F, Carruba G. Correlation of Sfil macrorestriction endonuclease fingerprint analysis of Candida parapsilosis isolates with source of isolation. J Med Microbiol. 1996;45:173–178. doi: 10.1099/00222615-45-3-173. [DOI] [PubMed] [Google Scholar]
- 282.Poonwan N, Mikami Y, Poosuwan S, Boon L J, Mekha N, Kusum M, Yazawa K, Tanaka R, Nishimura K, Konyama K. Serotyping of Cryptococcus neoformans strains isolated from clinical specimens in Thailand and their susceptibility to various antifungal agents. Eur J Epidemiol. 1997;13:335–340. doi: 10.1023/a:1007376917836. [DOI] [PubMed] [Google Scholar]
- 283.Poulain D, Hopwood V, Vernes A. Antigenic variability of Candida albicans. Crit Rev Microbiol. 1985;12:223–270. doi: 10.3109/10408418509104430. [DOI] [PubMed] [Google Scholar]
- 284.Powderly W G. Mucosal candidiasis caused by non-albicans species of Candida in HIV-positive patients. AIDS. 1992;6:604–605. [PubMed] [Google Scholar]
- 285.Price M F, LaRocco M T, Gentry L O. Fluconazole susceptibilities of Candida species and distribution of species recovered from blood cultures over a 5-year period. Antimicrob Agents Chemother. 1994;38:1422–1427. doi: 10.1128/aac.38.6.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Priest F G, Kaji D A, Rosato Y B, Canhos V P. Characterization of Bacillus thuringiensis and related bacteria by ribosomal RNA gene restriction fragment length polymorphisms. Microbiology. 1994;140:1015–1022. doi: 10.1099/13500872-140-5-1015. [DOI] [PubMed] [Google Scholar]
- 287.Reference deleted.
- 288.Pujol C, Joly S, Lockhart S, Noel S, Tibayrenc M, Soll D R. Parity of MLEE, RAPD and Ca3 hybridization as fingerprinting methods for Candida albicans. J Clin Microbiol. 1997;35:2348–2358. doi: 10.1128/jcm.35.9.2348-2358.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Pujol C, Joly S, Nolan B, Srikantha T, Lockhart S, Soll D R. Microevolutionary changes in Candida albicans identified by the complex Ca3 probe involve insertions and deletions of RPS units at specific genomic sites. Microbiology. 1999;145:2635–2646. doi: 10.1099/00221287-145-10-2635. [DOI] [PubMed] [Google Scholar]
- 290.Pujol C, Renaud F, Mallie M, de Meeus T, Bastide J M. Atypical strains of Candida albicans recovered from AIDS patients. J Med Vet Mycol. 1997;35:115–121. [PubMed] [Google Scholar]
- 291.Pujol C, Reynes J, Renaud F, Mallie M, Bastide J M. Genetic analysis of Candida albicans strains studied by isoenzyme electrophoresis. J Mycol Med. 1993;3:14–19. [Google Scholar]
- 292.Pujol C, Reynes J, Renaud F, Raymond M, Tibayrenc M, Ayala F J, Jambon F, Mallie M, Bastide J-M. The yeast Candida albicans has a clonal mode of reproduction in a population of infected human immunodeficiency virus-positive patients. Proc Natl Acad Sci USA. 1993;90:9456–9459. doi: 10.1073/pnas.90.20.9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Quindós G, Fernández-Rodríguez M, Burgos A, Tellaetxe M, Cisterna R, Ponton J. Colony morphotype on Sabouraud-triphenyltetrazolium agar: a simple and inexpensive method for Candida subspecies discrimination. J Clin Microbiol. 1992;30:2748–2752. doi: 10.1128/jcm.30.10.2748-2752.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Quindós G, Lipperheide V, Barturen B, Alonso R, Bikandi J, San Millán S, Tellaetxe M, Ribacoba L, Pontón J. A new method of antibiotyping yeasts for subspecies discrimination and distribution in human clinical specimens. Eur J Epidemiol. 1996;12:55–62. doi: 10.1007/BF00144429. [DOI] [PubMed] [Google Scholar]
- 295.Ramsey H, Morrow B, Soll D R. An increase in switching frequency correlates with an increase in recombination of the ribosomal chromosomes of Candida albicans strain 3153A. Microbiology. 1994;140:1525–1531. doi: 10.1099/13500872-140-7-1525. [DOI] [PubMed] [Google Scholar]
- 296.Rath P M, Marggraf G, Dermoumi H, Ansorg R. Use of phenotypic and genotypic fingerprinting methods in the strain identification of Aspergillus fumigatus. Mycoses. 1995;38:429–434. doi: 10.1111/j.1439-0507.1995.tb00015.x. [DOI] [PubMed] [Google Scholar]
- 297.Reagan D R, Pfaller M A, Hollis R J, Wenzel R P. Evidence of nosocomial spread of Candida albicans causing bloodstream infection in a neonatal intensive care unit. Diagn Microbiol Infect Dis. 1995;21:191–194. doi: 10.1016/0732-8893(95)00048-f. [DOI] [PubMed] [Google Scholar]
- 298.Redkar R J, Dubé M P, McCluskey F K, Rinaldi M G, Del Vecchio V G. DNA fingerprinting of Candida rugosa via repetitive sequence-based PCR. J Clin Microbiol. 1996;34:1677–1681. doi: 10.1128/jcm.34.7.1677-1681.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Reef S E, Lasker B A, Butcher D S, McNeil M M, Pruitt R, Keyserling H, Jarvis W R. Nonperinatal nosocomial transmission of Candida albicans in a neonatal intensive care unit: prospective study. J Clin Microbiol. 1998;36:1255–1259. doi: 10.1128/jcm.36.5.1255-1259.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Rex J, Rinaldi M, Pfaller M. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Reynes J, Pujol C, Moreau C, Mallie M, Renaud F, Janbon F, Bastide J M. Simultaneous carriage of Candida albicans strains from HIV-infected patients with oral candidiasis: multilocus enzyme electrophoresis analysis. FEMS Microbiol Lett. 1996;137:269–273. doi: 10.1111/j.1574-6968.1996.tb08117.x. [DOI] [PubMed] [Google Scholar]
- 302.Richet H M, Andremont A, Tancrede C, Pico J L, Jarvis W R. Risk factors for candidemia in patients with acute lymphocytic leukemia. Rev Infect Dis. 1991;13:211–215. doi: 10.1093/clinids/13.2.211. [DOI] [PubMed] [Google Scholar]
- 303.Rieseberg L H. Homology among RAPD fragments in interspecific comparisons. Mol Ecol. 1996;5:99–105. [Google Scholar]
- 304.Riggsby W S, Torres-Bauzá L J, Wills J W, Tomes T M. DNA content, kinetic complexity and the ploidy question in Candida albicans. Mol Cell Biol. 1982;2:853–862. doi: 10.1128/mcb.2.7.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Rikkerink E H, Magee B B, Magee P T. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J Bacteriol. 1988;170:895–899. doi: 10.1128/jb.170.2.895-899.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Robert F, Lebreton F, Bougnoux M E, Paugam A, Wassermann D, Schlotterer M, Tourte-Schaefer C, Dupouy-Camet J. Use of random amplified polymorphic DNA as a typing method for Candida albicans in epidemiological surveillance of a burn unit. J Clin Microbiol. 1995;33:2366–2371. doi: 10.1128/jcm.33.9.2366-2371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Rodriguez E, De Meeus T, Mallie M, Renaud F, Symoens F, Mondon P, Piens M A, Lebeau B, Viviani M A, Grillot R, Nolard N, Chapuis F, Tortorano A M, Bastide J M. Multicentric epidemiological study of Aspergillus fumigatus isolated by multilocus enzyme electrophoresis. J Clin Microbiol. 1996;34:2559–2568. doi: 10.1128/jcm.34.10.2559-2568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Rohlf F J. Classification of Aedes by numerical taxonomic methods (Diptera: Culcidae) Ann Entomol Soc Am. 1963;56:798–804. [Google Scholar]
- 309.Ruhnke M, Grosch-Worner I, Steinmuller A, Neubauer A. Molecular epidemiology of Candida infections in HIV-infected mothers and their offspring. Wien Med Wochenschr. 1997;147:446–449. . (In German.) [PubMed] [Google Scholar]
- 310.Ruiz-Diez B, Martinez V, Alvarez M, Rodriguez-Tudela J L, Martinez-Saurez J V. Molecular tracking of Candida albicans in a neonatal intensive care unit: long term colonizations versus catheter-related infections. J Clin Microbiol. 1997;35:3032–3036. doi: 10.1128/jcm.35.12.3032-3036.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Ruma P, Chen S C, Sorrell T C, Brownlee A G. Characterization of Cryptococcus neoformans by random DNA amplification. Lett Appl Microbiol. 1996;23:312–316. doi: 10.1111/j.1472-765x.1996.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 312.Rustchenko-Bulgac E P. Variations of Candida albicans electrophoretic karyotypes. J Bacteriol. 1991;173:6586–6596. doi: 10.1128/jb.173.20.6586-6596.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Rustchenko-Bulgac E P, Howard D H. Multiple chromosomal and phenotypic changes in spontaneous mutants of Candida albicans. J Gen Microbiol. 1993;139:1195–1207. doi: 10.1099/00221287-139-6-1195. [DOI] [PubMed] [Google Scholar]
- 314.Rustchenko-Bulgac E P, Sherman F, Hicks J B. Chromosomal rearrangements associated with morphological mutants provide a means for genetic variation in Candida albicans. J Bacteriol. 1990;172:1276–1283. doi: 10.1128/jb.172.3.1276-1283.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Sadhu C, McEachern M J, Rustchenko-Bulgac E P, Schmid J, Soll D R, Hicks J B. Telomeric and dispersed repeat sequences in Candida yeasts and their use in strain identification. J Bacteriol. 1991;173:842–850. doi: 10.1128/jb.173.2.842-850.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Safrin R E, Lancaster L A, Davis C E, Braude A I. Differentiation of Cryptococcus neoformans serotypes by isoenzyme electrophoresis. Am J Clin Pathol. 1986;86:204–208. doi: 10.1093/ajcp/86.2.204. [DOI] [PubMed] [Google Scholar]
- 317.Saitou N, Nei M. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 318.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 319.Sanchez V, Vazquez J A, Barth-Jones D, Dembry L, Sobel J D, Zervos M J. Epidemiology of nosocomial acquisition of Candida lusitaniae. J Clin Microbiol. 1992;30:3005–3008. doi: 10.1128/jcm.30.11.3005-3008.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Sandhu G S, Kline B C, Stockman L, Roberts G D. Molecular probes for diagnosis of fungal infections. J Clin Microbiol. 1995;33:2913–2919. doi: 10.1128/jcm.33.11.2913-2919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Sandven P, Bjorneklett A, Maeland A. Susceptibilities of Norwegian Candida albicans strains to fluconazole: emergence of resistance. The Norwegian Yeast Study Group. Antimicrob Agents Chemother. 1993;37:2443–2448. doi: 10.1128/aac.37.11.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Sangeorzan J A, Bradley S F, He X, Zarins L T, Ridenour G L, Tiballi R N, Kauffman C A. Epidemiology of oral candidiasis in HIV-infected patients: colonization, infection, treatment, and emergence of fluconazole resistance. Am J Med. 1994;97:339–346. doi: 10.1016/0002-9343(94)90300-x. [DOI] [PubMed] [Google Scholar]
- 323.Sangeorzan J A, Zervos M J, Donabedian S, Kauffman C A. Validity of contour-clamped homogeneous electric field electrophoresis as a typing system for Candida albicans. Mycoses. 1995;38:29–36. doi: 10.1111/j.1439-0507.1995.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 324.San-Millan R, Ribacoba L, Ponton J, Quindos G. Evaluation of a commercial medium for identification of Candida species. J Clin Microbiol Infect Dis. 1996;15:153–158. doi: 10.1007/BF01591489. [DOI] [PubMed] [Google Scholar]
- 325.San Millan R M, Wu L C, Salkin I F, Lehmann P F. Clinical isolates of Candida guilliermondii include Candida fermentati. Int J Syst Bacteriol. 1997;47:385–393. doi: 10.1099/00207713-47-2-385. [DOI] [PubMed] [Google Scholar]
- 326.Scherer S, Stevens D A. Application of DNA typing methods to epidemiology and taxonomy of Candida species. J Clin Microbiol. 1987;25:675–679. doi: 10.1128/jcm.25.4.675-679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Scherer S, Stevens D A. A Candida albicans dispersed, repeated gene family and its epidemiological applications. Proc Natl Acad Sci USA. 1988;85:1452–1456. doi: 10.1073/pnas.85.5.1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Schmid J, Hunter P R, White G C, Nand A K, Cannon R D. Physiological traits associated with success of Candida albicans strains as commensal colonizers and pathogens. J Clin Microbiol. 1995;33:2920–2926. doi: 10.1128/jcm.33.11.2920-2926.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Schmid J, Odds F C, Wiselka M J, Nicholson K G, Soll D R. Genetic similarity and maintenance of Candida albicans strains in a group of AIDS patients demonstrated by DNA fingerprinting. J Clin Microbiol. 1992;30:935–941. doi: 10.1128/jcm.30.4.935-941.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Schmid J, Rotman M, Reed B, Pierson C L, Soll D R. Genetic similarity of Candida albicans strains from vaginitis patients and their partners. J Clin Microbiol. 1993;31:39–46. doi: 10.1128/jcm.31.1.39-46.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Schmid J, Tay Y P, Wan L, Carr M, Parr D, McKinney W. Evidence for nosocomial transmission of Candida albicans obtained by Ca3 fingerprinting. J Clin Microbiol. 1995;33:1223–1230. doi: 10.1128/jcm.33.5.1223-1230.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Schmid J, Voss E, Soll D R. Computer-assisted methods for assessing strain relatedness in Candida albicans by fingerprinting with the moderately repetitive sequence Ca3. J Clin Microbiol. 1990;28:1236–1243. doi: 10.1128/jcm.28.6.1236-1243.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Schonian G, Meusel O, Tietz H J, Meyer W, Graser Y, Tausch I, Presber W, Mitchell T G. Identification of clinical strains of Candida albicans by DNA fingerprinting with the polymerase chain reaction. Mycoses. 1993;36:171–179. doi: 10.1111/j.1439-0507.1993.tb00746.x. [DOI] [PubMed] [Google Scholar]
- 334.Schoofs A, Odds F C, Colebunders R, Ieven M, Goossens H. Use of specialized isolation media for recognition and identification of Candida dubliniensis isolates from HIV-infected patients. Eur J Clin Microbiol Infect Dis. 1997;16:296–300. doi: 10.1007/BF01695634. [DOI] [PubMed] [Google Scholar]
- 335.Schoofs A, Odds F C, Colebunders R, Ieven M, Wouters L, Goossens H. Isolation of Candida species on media with and without added fluconazole reveals high variability in relative growth susceptibility phenotypes. Antimicrob Agents Chemother. 1997;41:1625–1635. doi: 10.1128/aac.41.8.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Schröppel K, Rotman M, Galask R, Mac K, Soll D R. Evolution and replacement of C. albicans strains during recurrent vaginitis demonstrated by DNA fingerprinting. J Clin Microbiol. 1994;32:2646–2654. doi: 10.1128/jcm.32.11.2646-2654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Schwab U, Chernomas F, Larcom L, Weems J. Molecular typing and fluconazole susceptibility of urinary Candida glabrata isolates from hospitalized patients. Diagn Microbiol Infect Dis. 1997;29:11–17. doi: 10.1016/s0732-8893(97)00076-x. [DOI] [PubMed] [Google Scholar]
- 338.Schwartz D C, Cantor C R. Separation of yeast chromosome-sized DNAs by pulsed field gradient gel electrophoresis. Cell. 1984;37:67–75. doi: 10.1016/0092-8674(84)90301-5. [DOI] [PubMed] [Google Scholar]
- 339.Schweizer E, MacKechnie C, Halvorson H D. The redundancy of ribosomal and transfer RNA genes in Saccharomyces cerevisiae. J Mol Biol. 1969;40:261–278. doi: 10.1016/0022-2836(69)90474-4. [DOI] [PubMed] [Google Scholar]
- 340.Seward R J, Ehrenstein B, Grundmann H J, Towner K J. Direct comparison of two commercially available computer programs for analysing DNA fingerprinting gels. J Med Microbiol. 1997;46:314–320. doi: 10.1099/00222615-46-4-314. [DOI] [PubMed] [Google Scholar]
- 341.Shibata N, Arai M, Haga E, Kikuchi T, Najima M, Satoh T, Kobayashi H, Suzuki S. Structural identification of an epitope of antigenic factor 5 in mannans of Candida albicans NIH B-792 (serotype B) and J-1012 (serotype A) as beta-1,2-linked oligomannosyl residues. Infect Immun. 1992;60:4100–4110. doi: 10.1128/iai.60.10.4100-4110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Skorepová M, Hauck H. Differentiation of Candida albicans biotypes by the method of Odds and Abbott. Mykosen. 1985;28:323–331. doi: 10.1111/j.1439-0507.1985.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 343.Slutsky B. A characterization of two high frequency switching systems in the dimorphic yeast Candida albicans. Ph.D. thesis. Iowa City: University of Iowa; 1986. [Google Scholar]
- 344.Slutsky B, Buffo J, Soll D R. High frequency “switching” of colony morphology in Candida albicans. Science. 1985;230:666–669. doi: 10.1126/science.3901258. [DOI] [PubMed] [Google Scholar]
- 345.Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll D R. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197. doi: 10.1128/jb.169.1.189-197.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Smith R A, Hitchcock C A, Evans E G, Lacey C J, Adams D J. The identification of Candida albicans strains by restriction fragment length polymorphism analysis of DNA. J Med Vet Micol. 1989;27:431–434. [PubMed] [Google Scholar]
- 347.Sneath P H A. Some thoughts on bacterial classification. J Gen Microbiol. 1957;17:184–200. doi: 10.1099/00221287-17-1-184. [DOI] [PubMed] [Google Scholar]
- 348.Sneath P H A, Sokal R R. Numerical taxonomy. W. H. San Francisco, Calif: Freeman & Co.; 1973. [Google Scholar]
- 349.Snell R G, Wilkins R J. Separation of chromosomal DNA molecules from C. albicans by pulsed field gel electrophoresis. Nucleic Acids Res. 1986;14:4401–4406. doi: 10.1093/nar/14.11.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Soll D R. Dimorphism and high frequency switching in Candida albicans. In: Kirsch D R, Kelly R, Kurtz M B, editors. The genetics of Candida. Boca Raton, Fla: CRC Press, Inc.; 1990. pp. 147–176. [Google Scholar]
- 351.Soll D R. High-frequency switching in Candida albicans. Clin Microbiol Rev. 1992;5:183–203. doi: 10.1128/cmr.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Soll D R. Molecular biology of Candida pathogenesis. In: Wright D M M, Archard L C, editors. Molecular biology of sexually transmitted diseases. London, United Kingdom: Chapman & Hall; 1992. pp. 131–172. [DOI] [PubMed] [Google Scholar]
- 353.Soll D R. The emerging molecular biology of switching in Candida albicans. ASM News. 1997;62:415–420. [Google Scholar]
- 354.Soll D R. Gene regulation during high frequency switching in Candida albicans. Microbiology. 1997;143:279–288. doi: 10.1099/00221287-143-2-279. [DOI] [PubMed] [Google Scholar]
- 355.Reference deleted.
- 356.Soll D R, Anderson J, Bergen M. The developmental biology of the white-opaque transition in Candida albicans. In: Prasad R, editor. The molecular biology of Candida albicans. Berlin, Germany: Springer-Verlag KG; 1991. pp. 20–45. [Google Scholar]
- 357.Soll D R, Galask R, Isley S, Rao T V G, Stone D, Hicks J, Schmid J, Mac K, Hanna C. “Switching” of Candida albicans during successive episodes of recurrent vaginitis. J Clin Microbiol. 1989;27:681–690. doi: 10.1128/jcm.27.4.681-690.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Soll D R, Galask R, Schmid J, Hanna C, Mac K, Morrow B. Genetic dissimilarity of commensal strains of Candida spp. carried in different anatomical locations of the same healthy women. J Clin Microbiol. 1991;29:1702–1710. doi: 10.1128/jcm.29.8.1702-1710.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Soll D R, Langtimm C J, McDowell J, Hicks J, Galask R. High-frequency switching in Candida strains isolated from vaginitis patients. J Clin Microbiol. 1987;25:1611–1622. doi: 10.1128/jcm.25.9.1611-1622.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Soll D R, Staebell M, Langtimm C J, Pfaller M, Hicks J, Rao T V G. Multiple Candida strains in the course of a single systemic infection. J Clin Microbiol. 1988;26:1448–1459. doi: 10.1128/jcm.26.8.1448-1459.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Sorrell T C, Brownlee A G, Ruma P, Malik R, Pfeiffer T J, Ellis D H. Natural environmental sources of Cryptococcus neoformans var. gattii. J Clin Microbiol. 1996;34:1261–1263. doi: 10.1128/jcm.34.5.1261-1263.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Sorrell T C, Chen S C, Ruma P, Meyer W, Pfeiffer T J, Ellis D H, Brownlee A G. Concordance of clinical and environmental isolates of Cryptococcus neoformans var. gattii by random amplification of polymorphic DNA analysis and PCR fingerprinting. J Clin Microbiol. 1996;34:1253–1260. doi: 10.1128/jcm.34.5.1253-1260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Spitzer E D, Lasker B A, Travis S J, Kobayashi G S, Medoff G. Use of mitochondrial and ribosomal DNA polymorphisms to classify clinical and soil isolates of Histoplasma capsulatum. Infect Immun. 1989;57:1409–1412. doi: 10.1128/iai.57.5.1409-1412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Spitzer E D, Spitzer S G. Use of a dispersed repetitive DNA element to distinguish clinical isolates of Cryptococcus neoformans. J Clin Microbiol. 1992;30:1094–1097. doi: 10.1128/jcm.30.5.1094-1097.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Spitzer E D, Spitzer S G, Freundlich L F, Casadevall A. Persistence of initial infection in recurrent Cryptococcus neoformans meningitis. Lancet. 1993;341:595–596. doi: 10.1016/0140-6736(93)90354-j. [DOI] [PubMed] [Google Scholar]
- 366.Spreadbury C L, Bainbridge B W, Cohen J. Restriction fragment length polymorphisms in isolates of Aspergillus fumigatus probed with part of the intergenic spacer region from the ribosomal RNA gene complex of Aspergillus nidulans. J Gen Microbiol. 1990;136:1991–1994. doi: 10.1099/00221287-136-10-1991. [DOI] [PubMed] [Google Scholar]
- 367.Srikantha T, Soll D R. A white-specific gene in the white-opaque switching system of Candida albicans. Gene. 1993;131:53–60. doi: 10.1016/0378-1119(93)90668-s. [DOI] [PubMed] [Google Scholar]
- 368.Steele P E, Carle G F, Kobayashi G S, Medoff G. Electrophoretic analysis of Histoplasma capsulatum chromosomal DNA. Mol Cell Biol. 1989;9:983–987. doi: 10.1128/mcb.9.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 369.Steffan P, Vazquez J A, Boikov D, Xu C, Sobel J D, Akins R A. Identification of Candida species by randomly amplified polymorphic DNA fingerprinting of colony lysates. J Clin Microbiol. 1997;35:2031–2039. doi: 10.1128/jcm.35.8.2031-2039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Stein G E, Sheridan V L, Magee B B, Magee P T. Use of rDNA restriction fragment length polymorphisms to differentiate strains of Candida albicans in women with vulvovaginal candidiasis. Diagn Microbiol Infect Dis. 1991;14:459–464. doi: 10.1016/0732-8893(91)90001-v. [DOI] [PubMed] [Google Scholar]
- 371.Stiller R L, Bennett J E, Scholer H J, Wall M, Polak A, Stevens D A. Susceptibility to 5-fluorocytosine and prevalence of serotype in 402 Candida albicans isolates from the United States. Antimicrob Agents Chemother. 1982;22:482–487. doi: 10.1128/aac.22.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Stringer J R, Stringer S L, Zhang J, Baughman R, Smulian A G, Cushion M T. Molecular genetic distinction of Pneumocystic carinii from rats and humans. J Eukaryot Microbiol. 1993;40:733–741. doi: 10.1111/j.1550-7408.1993.tb04468.x. [DOI] [PubMed] [Google Scholar]
- 373.Su C-S, Meyer S A. Characterization of mitochondrial DNA in various Candida species: isolation, restriction endonuclease reaction, size and base composition. Int J Syst Bacteriol. 1991;41:6–14. doi: 10.1099/00207713-41-1-6. [DOI] [PubMed] [Google Scholar]
- 374.Sullivan D, Bennett D, Henman M, Harwood P, Flint S, Mulcahy F, Shanley D, Coleman D. Oligonucleotide fingerprinting of isolates of Candida species other than C. albicans and of atypical Candida species from human immunodeficiency virus-positive and AIDS patients. J Clin Microbiol. 1993;31:2124–2133. doi: 10.1128/jcm.31.8.2124-2133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Sullivan D, Haynes K, Moran G, Shanley D, Coleman D. Persistence, replacement, and microevolution of Cryptococcus neoformans strains in recurrent meningitis in AIDS patients. J Clin Microbiol. 1996;34:1739–1744. doi: 10.1128/jcm.34.7.1739-1744.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Sullivan D J, Henman M C, Moran G P, O'Neill L C, Bennett D E, Shanley D B, Coleman D C. Molecular genetic approaches to identification, epidemiology and taxonomy of non-albicans Candida species. J Med Microbiol. 1996;44:399–408. doi: 10.1099/00222615-44-6-399. [DOI] [PubMed] [Google Scholar]
- 377.Sullivan D J, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 378.Sukroongreung S, Nilakul C, Ruangsomboon O, Chuakul W, Eampokalap B. Serotypes of Cryptococcus neoformans isolated from patients prior to and during the AIDS era in Thailand. Mycopathologia. 1996;135:75–78. doi: 10.1007/BF00436454. [DOI] [PubMed] [Google Scholar]
- 379.Summers D F, Grollman A P, Hasenclever H F. Polysaccharide antigens of Candida cell wall. J Immunol. 1964;92:491–499. [PubMed] [Google Scholar]
- 380.Sutherland S S, Hart R A, Buller N B. Genetic differences between nitrate-negative and nitrate-positive C. pseudotuberculosis strains using restriction fragment length polymorphisms. Vet Microbiol. 1996;49:1–9. doi: 10.1016/0378-1135(95)00146-8. [DOI] [PubMed] [Google Scholar]
- 381.Suzuki T, Kobayashi I, Mizuguchi I, Banno I, Tanaka K. Electrophoretic karyotypes in medically important Candida species. J Gen Appl Microbiol. 1988;34:409–416. [Google Scholar]
- 382.Swanson H, Hughes P A, Messer S A, Lepow M L, Pfaller M A. Candida albicans arthritis one year after successful treatment of fungemia in a healthy infant. J Pediatr. 1996;129:688–694. doi: 10.1016/s0022-3476(96)70151-8. [DOI] [PubMed] [Google Scholar]
- 383.Swanson J, Koomey J M. Mechanisms for variation of pili and outer membrane protein II in Neisseria gonorrhoea. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 743–762. [Google Scholar]
- 384.Tanaka K. Strain-relatedness among different populations of the pathogenic yeast Candida albicans analyzed by DNA-based methods. Nagoya J Med Sci. 1997;60:1–14. [PubMed] [Google Scholar]
- 385.Tang C M, Cohen J, Rees A J, Holden D W. Molecular epidemiological study of invasive pulmonary aspergillosis in a renal transplantation unit. Eur J Clin Microbiol Infect Dis. 1994;13:318–321. doi: 10.1007/BF01974610. [DOI] [PubMed] [Google Scholar]
- 386.Taylor J W, Geiser D M, Burt A, Koufopanou V. The evolutionary biology and population genetics underlying fungal strain typing. Clin Microbiol Rev. 1999;12:126–146. doi: 10.1128/cmr.12.1.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Thanos M, Schönian G, Meyer W, Schweynoch C, Gräser Y, Mitchell T G, Presber W, Tietz H-J. Rapid identification of Candida species by DNA fingerprinting with PCR. J Clin Microbiol. 1996;34:615–621. doi: 10.1128/jcm.34.3.615-621.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Thormann C E, Ferreira M E, Camargo L E, Tivang J G, Osborn T C. Comparison of RFLP and RAPD markers to estimate genetic relationships within and among cruciferous species. Theor Appl Genet. 1994;88:973–980. doi: 10.1007/BF00220804. [DOI] [PubMed] [Google Scholar]
- 389.Thrash-Bingham C, Gorman J A. DNA translocations contribute to chromosome length polymorphisms in Candida albicans. Curr Genet. 1992;22:93–100. doi: 10.1007/BF00351467. [DOI] [PubMed] [Google Scholar]
- 390.Thrash-Bingham C, Gorman J A. Identification, characterization, and sequence of Candida albicans repetitive DNAs Rel-1 and Rel-2. Curr Genet. 1993;23:455–462. doi: 10.1007/BF00312634. [DOI] [PubMed] [Google Scholar]
- 391.Tibayrenc M. Genetic epidemiology of parasitic protozoa and other infectious agents: the need for an integrated approach. Int J Parasitol. 1998;28:85–104. doi: 10.1016/s0020-7519(97)00180-x. [DOI] [PubMed] [Google Scholar]
- 392.Tibayrenc M, Neubauer K, Barnabé C, Guerrini F, Skarecky D, Ayala F J. Genetic characterization of six parasitic protozoa: parity between random-primer DNA typing and multilocus enzyme electrophoresis. Proc Natl Acad Sci USA. 1993;90:1335–1339. doi: 10.1073/pnas.90.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Tietz H J, Kussner A, Thanos M, De Andrade M P, Presber W, Schonian G. Phenotypic and genotypic characterization of unusual vaginal isolates of Candida albicans from Africa. J Clin Microbiol. 1995;33:2462–2465. doi: 10.1128/jcm.33.9.2462-2465.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Tojo M, Shibata N, Ban Y, Suzuki S. The phospho-d-mannan of Candida albicans NIH B-792 strain. Carbohydr Res. 1990;199:215–226. doi: 10.1016/0008-6215(90)84263-t. [DOI] [PubMed] [Google Scholar]
- 395.van Belkum A, Melchers W, de Pauw B E, Scherer S, Quint W, Meis J F. Genotypic characterization of sequential Candida albicans isolates from fluconazole-treated neutropenic patients. J Infect Dis. 1994;169:1062–1070. doi: 10.1093/infdis/169.5.1062. [DOI] [PubMed] [Google Scholar]
- 396.van Belkum A, Mol W, van Saene R, Ball L M, van Velzen D, Quint W. PCR-mediated genotyping of Candida albicans strains from bone marrow transplant patients. Bone Marrow Transplant. 1994;13:811–815. [PubMed] [Google Scholar]
- 397.Varma A, Kwon-Chung K J. DNA probe for strain typing of Cryptococcus neoformans. J Clin Microbiol. 1992;30:2960–2967. doi: 10.1128/jcm.30.11.2960-2967.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Vazquez J A, Beckley A, Donabedian S, Sobel J D, Zervos M J. Comparison of restriction enzyme analysis and pulsed-field gradient gel electrophoresis as a typing system for Torulopsis glabrata and Candida species other than C. albicans. J Clin Microbiol. 1993;31:2021–2030. doi: 10.1128/jcm.31.8.2021-2030.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Vazquez J A, Beckley A, Sobel J D, Zervos M J. Comparison of restriction enzyme analysis and pulsed-field gradient gel electrophoresis as typing systems for Candida albicans. J Clin Microbiol. 1991;29:962–967. doi: 10.1128/jcm.29.5.962-967.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Vazquez J A, Boikov D, Boikov S G, Dajani A S. Use of electrophoretic katyotyping in the evaluation of Candida infections in a neonatal intensive-care unit. Infect Control Hosp Epidemiol. 1997;18:32–37. doi: 10.1086/647498. [DOI] [PubMed] [Google Scholar]
- 401.Vazquez J A, Dembry L M, Sanchez V, Vazquez M A, Sobel J D, Dmuchowski C, Zervos M J. Nosocomial Candida glabrata colonization: an epidemiologic study. J Clin Microbiol. 1998;36:421–426. doi: 10.1128/jcm.36.2.421-426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Vazquez J A, Sanchez V, Dmuchowski C, Dembry L M, Sobel J D, Zervos M J. Nosocomial acquisition of Candida albicans: an epidemiologic study. J Infect Dis. 1993;168:195–201. doi: 10.1093/infdis/168.1.195. [DOI] [PubMed] [Google Scholar]
- 403.Vazquez J A, Sobel J D, Demitriou R, Vaishampayan J, Lynch M, Zervos M J. Karyotyping of Candida albicans isolates obtained longitudinally in women with recurrent vulvovaginal candidiasis. J Infect Dis. 1994;170:1566–1569. doi: 10.1093/infdis/170.6.1566. [DOI] [PubMed] [Google Scholar]
- 404.Verweij P E, Meis J F, Sarfati J, Hoogkamp-Korstanje J A, Latgé J P, Melchers W J. Genotypic characterization of sequential Aspergillus fumigatus isolates from patients with cystic fibrosis. J Clin Microbiol. 1996;34:2595–2597. doi: 10.1128/jcm.34.10.2595-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Vincent R D, Geowart R, Goldman W E, Kobayashi G S, Lambowitz A M, Medoff G. Classification of Histoplasma capsulatum isolates by restriction fragment polymorphisms. J Bacteriol. 1986;165:813–818. doi: 10.1128/jb.165.3.813-818.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 406.Vlad M. Quantitative studies of rDNA in amphibians. J Cell Sci. 1977;24:109–118. doi: 10.1242/jcs.24.1.109. [DOI] [PubMed] [Google Scholar]
- 407.Vollrath D, Davis R W. Resolution of DNA molecules greater than 5 megabases by contour-clamped homogeneous electric fields. Nucleic Acids Res. 1987;15:7865–7876. doi: 10.1093/nar/15.19.7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Voss A, Hollis R J, Pfaller M A, Wenzel R P, Doebbeling B N. Investigation of the sequence of colonization and candidemia in nonneutropenic patients. J Clin Microbiol. 1994;32:975–980. doi: 10.1128/jcm.32.4.975-980.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Voss A, Pfaller M A, Hollis R J, Rhine-Chalberg J, Doebbeling B N. Investigation of Candida albicans transmission in a surgical intensive care unit cluster by using genomic DNA typing methods. J Clin Microbiol. 1995;33:576–580. doi: 10.1128/jcm.33.3.576-580.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Waggoner-Fountain L A, Walker M W, Hollis R J, Pfaller M A, Ferguson J E, Wenzel R P, Donowitz L G. Vertical and horizontal transmission of unique Candida species to premature newborns. Clin Infect Dis. 1996;22:803–808. doi: 10.1093/clinids/22.5.803. [DOI] [PubMed] [Google Scholar]
- 412.Welbel S F, McNeil M M, Kuykendall R J, Lott T J, Promanik A, Silberman R, Oberle A D, Bland L A, Aguero S, Arduino M, Crow S, Jarvis W R. Candida parapsilosis bloodstream infections in neonatal intensive care unit patients: epidemiologic and laboratory confirmation of a common source outbreak. Pediatr Infect Dis J. 1996;15:998–1002. doi: 10.1097/00006454-199611000-00013. [DOI] [PubMed] [Google Scholar]
- 413.Welsch J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7224. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Wey S B, Mori M, Pfaller M A, Woolson R F, Wenzel R P. Risk factors for hospital-acquired candidemia. Arch Intern Med. 1989;149:2349–2353. [PubMed] [Google Scholar]
- 415.Whelan W L, Kirsch D R, Kwon-Chung K J, Wahl S M, Smith P D. Candida albicans in patients with the acquired immunodeficiency syndrome: absence of a novel or hypervirulent strain. J Infect Dis. 1990;162:513–518. doi: 10.1093/infdis/162.2.513. [DOI] [PubMed] [Google Scholar]
- 416.Whelan W L, Magee P T. Natural heterozygosity in Candida albicans. J Bacteriol. 1981;145:896–903. doi: 10.1128/jb.145.2.896-903.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.White T C, Marr K A, Bowden R A. Clinical, cellular and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.White T C, Miysaki S H, Agabian N. Three distinct secreted aspartyl proteinases in Candida albicans. J Bacteriol. 1993;175:6126–6133. doi: 10.1128/jb.175.19.6126-6133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.White T C, Pfaller M A, Rinaldi M G, Smith J, Redding S W. Stable azole drug resistance associated with a substrain of Candida albicans from an HIV-infected patient. Oral Dis. 1997;3:S102–S109. doi: 10.1111/j.1601-0825.1997.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 420.Wickes B, Staudinger J, Magee B B, Kwon-Chung K-J, Magee P T, Scherer S. Physical and genetic map of Candida albicans: several genes previously assigned to chromosome 1 map to chromosome R, the rDNA-containing linkage group. Infect Immun. 1991;59:2480–2484. doi: 10.1128/iai.59.7.2480-2484.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Wiley J M, Smith N, Leventhal B G, Graham M L, Strauss L C, Hurwitz C A, Modlin J, Mellits D, Baumgardner R, Corden B J, et al. Invasive fungal disease in pediatric acute leukemia patients with fever and neutropenia during induction chemotherapy: a multivariate analysis of risk factors. J Clin Oncol. 1990;8:280–286. doi: 10.1200/JCO.1990.8.2.280. [DOI] [PubMed] [Google Scholar]
- 422.Wilkinson B M, Morris L, Adams D J, Evans E G, Lacey C J, Walmsley R M. A new, sensitive polynucleotide probe for distinguishing Candida albicans strains and its use with computer assisted archiving and pattern comparison system. J Med Vet Mycol. 1992;30:123–131. [PubMed] [Google Scholar]
- 423.Williams J G, Kubelik A R, Livak K J, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Williamson M I, Samaranayake L P, MacFarlane T W. Biotypes of oral Candida albicans and Candida tropicalis isolates. J Med Vet Mycol. 1986;24:81–84. [PubMed] [Google Scholar]
- 425.Williamson M I, Samaranayake L P, MacFarlane T W. A new simple method for biotyping Candida albicans. Microbios. 1987;51:159–167. [PubMed] [Google Scholar]
- 426.Willinger B, Berger A, Li L, Hirschl A M, Aspock C, Makristathis A, Pruckl P M, Rotter M L. Pulsed-field gel electrophoresis for the epidemiological analysis of yeast isolates. Mycoses. 1994;37:S57–S59. [PubMed] [Google Scholar]
- 427.Wills J W, Lasker B A, Sirotkin K, Riggsby W S. Repetitive DNA of Candida albicans: nuclear and mitochondrial components. J Bacteriol. 1984;157:918–924. doi: 10.1128/jb.157.3.918-924.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 428.Wilson D E, Bennett J E, Bailey J W. Serologic grouping of Cryptococcus neoformans. Proc Soc Exp Biol Med. 1968;127:820–823. doi: 10.3181/00379727-127-32812. [DOI] [PubMed] [Google Scholar]
- 429.Wingard J R, Merz W G, Rinaldi M G, Miller C B, Karp J E, Saral R. Association of Torulopsis glabrata infections with fluconazole prophylaxis in neutropenic bone marrow transplant patients. Antimicrob Agents Chemother. 1993;37:1847–1849. doi: 10.1128/aac.37.9.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Wingard J R, Merz W G, Rinaldi M G, Johnson T R, Karp J E, Saral R. Increase in Candida krusei infection among patients with bone marrow transplantation and neutropenia treated prophylactically with fluconazole. N Engl J Med. 1991;325:1274–1277. doi: 10.1056/NEJM199110313251803. [DOI] [PubMed] [Google Scholar]
- 431.Woods J P, Kersulyte D, Goldman W E, Berg D E. Fast DNA isolation from Histoplasma capsulatum: methodology for arbitrary primer polymerase chain reaction-based epidemiological and clinical studies. J Clin Microbiol. 1993;31:463–464. doi: 10.1128/jcm.31.2.463-464.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Xu J, Mitchell T G, Vilgalys R. PCR-RFLP analyses reveal extensive clonality and local genetic differentiation in Candida albicans. Ecology. 1999;8:59–74. doi: 10.1046/j.1365-294x.1999.00523.x. [DOI] [PubMed] [Google Scholar]
- 433.Xu J, Vilgalys R, Mitchell T G. Lack of genetic differentiation between two geographically diverse samples of Candida albicans isolated from patients infected with human immunodeficiency virus. J Bacteriol. 1999;181:1369–1373. doi: 10.1128/jb.181.4.1369-1373.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Xu Y Y, Samaranayake L P. Oral Candida albicans biotypes in Chinese patients with and without oral candiosis. Arch Oral Biol. 1995;40:577–579. doi: 10.1016/0003-9969(94)00196-i. [DOI] [PubMed] [Google Scholar]
- 435.Yates S K, Sander D M, Keath E J. Genetic diversity in clinical isolates of the dimorphic fungus Blastomyces dermatitidis detected by a PCR-based random amplified polymorphic DNA assay. J Clin Microbiol. 1995;33:2171–2175. doi: 10.1128/jcm.33.8.2171-2175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Yuan K Y, Seto W H, Ching T Y, Cheung W C, Kwok Y, Chu Y B. An outbreak of Candida tropicalis peritonitis in patients on intermittent peritoneal dialysis. J Hosp Infect. 1992;22:65–72. doi: 10.1016/0195-6701(92)90131-5. [DOI] [PubMed] [Google Scholar]
- 437.Zeng S, Wu L C, Lehmann P F. Random amplified polymorphic DNA analysis of culture collection strains of Candida species. J Med Vet Mycol. 1996;34:293–297. doi: 10.1080/02681219680000501. [DOI] [PubMed] [Google Scholar]
- 438.Zhang J, Hollis R J, Pfaller M A. Variations in DNA subtype and antifungal susceptibility among clinical isolates of Candida tropicalis. Diagn Microbiol Infect Dis. 1997;27:63–67. doi: 10.1016/s0732-8893(97)00002-3. [DOI] [PubMed] [Google Scholar]