Abstract
Establishing causal links between adaptive mutations and ecologically relevant phenotypes is key to understanding the process of adaptation, which is a central goal in evolutionary biology with applications for conservation, medicine, and agriculture. Yet despite recent progress, the number of identified causal adaptive mutations remains limited. Linking genetic variation to fitness-related effects is complicated by gene-by-gene and gene-by-environment interactions, among other processes. Transposable elements, which are often ignored in the quest for the genetic basis of adaptive evolution, are a genome-wide source of regulatory elements across organisms that can potentially result in adaptive phenotypes. In this work, we combine gene expression, in vivo reporter assays, CRISPR/Cas9 genome editing, and survival experiments to characterize in detail the molecular and phenotypic consequences of a natural Drosophila melanogaster transposable element insertion: the roo solo-LTR FBti0019985. This transposable element provides an alternative promoter to the transcription factor Lime, involved in cold- and immune-stress responses. We found that the effect of FBti0019985 on Lime expression depends on the interplay between the developmental stage and environmental condition. We further establish a causal link between the presence of FBti0019985 and increased survival to cold- and immune-stress. Our results exemplify how several developmental stages and environmental conditions need to be considered to characterize the molecular and functional effects of a genetic variant, and add to the growing body of evidence that transposable elements can induce complex mutations with ecologically relevant effects.
Keywords: cold-stress, immune-stress, drosophila, transposable elements
Introduction
Establishing causal links between mutations and their relevant fitness-related phenotypes is crucial in biology, with implications for evolution, development, and disease (Otwinowski and Nemenman 2013; Mackay and Huang 2018; Nelson et al. 2019). However, causal genotype-phenotype links are difficult to establish since the effect of a mutation can depend on the genetic background (epistasis) as well as on other contexts such as the environment and the developmental stage (Reddy et al. 2009; Kammenga 2017; Young et al. 2019). Because context dependance contributes to diverse traits in diverse organisms, a shift from identifying the impact of a mutation in a particular context to pinpointing the spectrum of effects of a mutation is needed to provide a more realistic picture of the genotype-phenotype map (Eguchi et al. 2019).
Identifying adaptive mutations and analyzing how context dependance influences their effects is even more relevant in the current scenario of rapid environmental change (Catullo et al. 2019; Nelson et al. 2019). To date, most studies that aim at characterizing adaptive mutations have focused on single-nucleotide polymorphism (SNP) variants that are easier to detect by the commonly used short-read sequencing techniques. However, other types of mutations such as transposable elements (TEs), which are known to be a source of adaptive mutations across organisms, have been understudied so far (Hoban et al. 2016). The increased availability of whole genome sequences and advances in sequencing technologies, such as the improvements in long-read sequencing techniques, are fostering the discovery of candidate adaptive TEs (e.g.Rech et al. 2022). Besides identifying adaptive mutations at the DNA level, and their fitness-related trait in a relevant ecological context, pinpointing the molecular mechanism by which the mutation influences the phenotype is key to conclude that a mutation has an adaptive effect. TEs can affect gene structure and expression through many different molecular mechanisms (Casacuberta and González 2013; Chuong et al. 2017), and some of these changes have been associated with adaptive phenotypes (Chuong et al. 2016; Ding et al. 2016; Van’t Hof et al. 2016; Huang et al. 2018; Esnault et al. 2019; Ullastres et al. 2021; Brosh et al. 2022; Green et al. 2022). In Drosophila, a dual role as enhancer and promoter of an adaptive insertion conferring tolerance to bacterial infection has been recently reported (Ullastres et al. 2021). There are also several examples of TE-induced mutations affecting more than one phenotype, such as the disruption of the Chkov1 gene leading to resistance to pesticides and to viral infection (Aminetzach et al. 2005; Magwire et al. 2011). Indeed, D. melanogaster is an unrivaled model organism to investigate genotype-phenotype links because it has one of the best functionally annotated genomes and powerful tools to genetically modify the organism in vivo, which allows not only to identify the molecular mechanism behind the adaptive effect of a mutation but also to demonstrate its causality (Anholt and Mackay 2018).
The roo solo-LTR element known as FBti0019985, has been previously identified as a candidate adaptive TE insertion: the patterns of nucleotide diversity in its flanking regions suggest that this insertion has increased in frequency due to positive selection (González et al. 2008; Merenciano et al. 2016). FBti0019985 is inserted in the promoter region of the Lime gene, which is located in the first intron of the cbx gene (fig. 1). Lime is a C2H2-type zinc finger transcription factor associated with chill-coma resistance and immune response (Telonis-Scott et al. 2009; Mihajlovic et al. 2019), and cbx is a ubiquitin-conjugating enzyme that has also been associated with immune-stress (Ayres et al. 2008; Ullastres et al. 2021). There is previous partial evidence for the association of FBti0019985 with changes in the expression of its nearby genes, for the molecular mechanisms that might underlie these expression changes, and for FBti0019985 potential phenotypic effects. Briefly, FBti0019985 is associated with changes in the expression of its two nearby genes: up-regulation of Lime in embryos under nonstress conditions, and down-regulation of cbx in adults in nonstress and immune-stress conditions (Merenciano et al. 2016; Ullastres et al. 2021). However, these results were based on the analysis of a limited number of genetic backgrounds, and whether FBti0019985 is indeed the causal mutation, and if that is the case, whether it also affects Lime expression under stress conditions, are still open questions.
Fig. 1.
Schematic representation of the genomic region where FBti0019985 is inserted. Lime gene is located in the first intron of cbx gene. Light boxes represent untranslated regions (UTRs) and darker boxes represent coding exons. FBti0019985 insertion overlaps with Lime gene. Transcription start sites, including the one that FBti0019985 adds, are represented as arrows. Transgenic insertion sites are represented with triangles.
The molecular mechanisms underpinning FBti0019985 effects on gene expression are also not clear yet. There is recent evidence for FBti0019985 inducing a depletion of the H3K9me3 repressive histone mark in its flanking regions in nonstress conditions (Ullastres et al. 2021). Moreover, while there is evidence for FBti0019985 providing an alternative transcription start site (TSS) to Lime in nonstress conditions (Batut et al. 2013; Merenciano et al. 2016), whether this new transcript is also present in stress conditions and whether it significantly contributes to the increased expression of Lime is unknown. Similarly, while FBti0019985 has been reported to act as an enhancer in response to immune-stress in adults (Villanueva-Cañas et al. 2019), whether the TE also acts as an enhancer in other environmental conditions is also unknown. Finally, previous evidence for the role of FBti0019985 in egg-to-adult viability is based on association analysis (Merenciano et al. 2016), thus whether FBti0019985 is the causal mutation and whether it also affects immune-stress survival remain open questions.
In this work, we characterized in detail the molecular and phenotypic effects of FBti0019985 under cold- and immune-stress conditions, both of them relevant to the survival of D. melanogaster in natural environments. We integrated gene expression analysis, in vivo reporter assays, in vivo Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR/Cas9) genome editing in outbred strains, and survival experiments, to establish a causal link between the presence of the insertion and its ecologically relevant phenotypic effects.
Results
Lime Expression Changes Affect Immune- and Cold-stress Survival and cbx Expression Changes Affect Immune-stress Survival
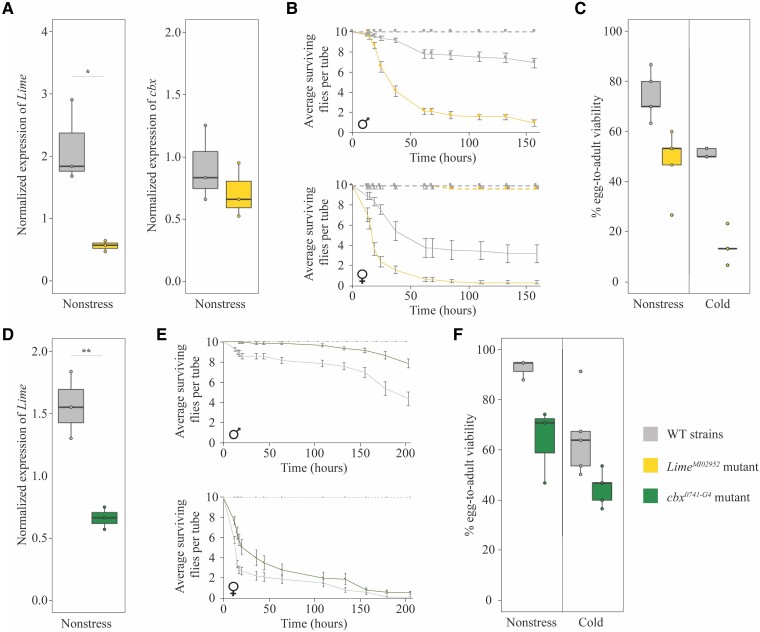
To provide further evidence for the role of Lime in cold- and immune-stress and for the role of cbx in immune-stress, we performed survival experiments in these two stress conditions using available mutant strains (see Material and Methods). Previous evidence for the role of Lime in immune-stress was obtained in larvae after wasp infection, where Lime mutants failed to induce systemic metabolic changes needed to develop the immune response (Mihajlovic et al. 2019). To test whether changes in Lime expression are also associated with differences in survival to bacterial infection in adult flies, we exposed Lime mutant flies to P. entomophila. P. entomophila is a gram-negative bacterium that orally infects and kills Drosophila in the wild (Vodovar et al. 2005). Using quantitative reverse transcription polymerase chain reaction (qRT-PCR), we first confirmed that LimeMI02952 mutant, which contains a MiMIC insertion in the first coding exon of Lime (fig. 1), down-regulates Lime expression (fig. 2A, supplementary tables S1A and S1B, Supplementary Material online). Because mutations in this genomic region have previously been reported to affect the expression of both genes (Merenciano et al. 2016; Ullastres et al. 2021), we also tested whether LimeMI02952 mutant affects the expression of cbx, but found that this was not the case (fig. 2A, supplementary table S1B, Supplementary Material online). We then performed survival experiments, and consistent with the previous evidence for the role of Lime in immune response (Mihajlovic et al. 2019), we found that Lime mutant flies, both male and females, were more sensitive to P. entomophila infection compared with wild-type (WT) flies (Log-rank test P-values < 0.001) (fig. 2B, supplementary table S1C, Supplementary Material online).
Fig. 2.
Lime expression changes affect immune- and cold-stress survival and cbx expression changes affect immune-stress survival. (A) Normalized expression of Lime and cbx with Act5C in the whole-body of LimeMI02952 mutant females compared with wild-type (WT) females, which have the same genetic background (supplementary table S1A, Supplementary Material online). Boxplots show the median (horizontal line), first and third quartiles (lower and upper bounds, respectively), and minimum and maximum values (lower and upper whiskers, respectively). For each strain, each dot represents an independent biological replication of a pool of 25 female flies. One-tailed t-test P-value = 0.015 and 0.400, for Lime and cbx genes, respectively. (B) Survival curves of adult LimeMI02952 mutant males and females compared with the WT strain. Survival curves in noninfected conditions are depicted as dotted lines while survival curves after P. entomophila infection are depicted as continuous lines. For each strain and sex, each dot represents the average number of surviving flies per tube, for ten replicates of ten flies each (infected conditions), and for three replicates of ten flies each (noninfected conditions). Error bars represent SEM. No mortality was observed in nonstress conditions. Log-rank test was performed to analyze differences between survival curves in infected conditions (log-rank P-values < 0.05 both in males and females). (C) Egg-to-adult viability in nonstress and cold-stress conditions of LimeMI02952 mutants compared with the WT strain. For each strain, each dot represents an independent biological replication of a pool of 30 embryos. Two-way ANOVA: genotype (G: WT/mutant) P-value <0.001; experimental condition (EC: nonstress vs. cold-stress) P-value <0.001; genotype by experimental condition interaction (GxEC) P-value = 0.105. (D) Normalized expression of Lime with Act5C in the whole-body of cbx0741-G4 mutant females compared with WT females. For each strain, each dot represents an independent biological replication of a pool of 25 female flies. One-tailed t-test P-value = 0.005. (E) Survival curves of adult cbx0741-G4 mutant males and females compared with the WT strain. Survival curves in noninfected conditions are depicted as dotted lines while survival curves after P. entomophila infection are depicted as continuous lines. For each strain and sex, each dot represents the average number of survival flies per tube, for ten replicates of ten flies each (infected conditions), and for three replicates of ten flies each (noninfected conditions). Error bars represent SEM. No mortality was observed in nonstress conditions. Log-rank test was performed to analyze differences between survival curves in infected conditions (log-rank P-value < 0.001 and P-value = 0.002 for males and females, respectively. (F) Egg-to-adult viability in nonstress and cold-stress conditions of cbx0741-G4 mutants compared with the WT strain. For each strain, each dot represents an independent biological replication of a pool of 30 embryos. Two-way ANOVA: G P-value = 0.002; EC P-value = 0.003; GxEC P-value = 0.528.
Previous works also showed that Lime is up-regulated in strains selected for cold-tolerance, and that up-regulation of Lime in embryos is associated with increased egg-to-adult viability in nonstress and cold-stress conditions (Telonis-Scott et al. 2009; Merenciano et al. 2016). Consistent with these results, we found that LimeMI02952 mutant flies, in which Lime expression is down-regulated, showed reduced egg-to-adult viability in nonstress and in cold-stress conditions (fig. 2C, supplementary table S1D, Supplementary Material online).
Evidence for a role of cbx in immune response is based on the analysis of two mutant stocks: cbxc00428 and cbx0741-G4, that have a PiggyBac insertion in the first and third intron of cbx, respectively (fig. 1). However, although Ayres et al. (2008) found that cbxc00428 mutant flies were associated with increased sensitivity to the gram-positive bacteria S. aureus, we found that this mutant does not affect cbx expression (supplementary table S1B, Supplementary Material online) (Ullastres et al. 2021). On the other hand, we found that cbx0741-G4, a null mutant for cbx (Ullastres et al. 2021), also down-regulates Lime expression (fig. 2D, supplementary table S1B, Supplementary Material online). Moreover, we confirmed that cbx0741-G4 is associated with increased tolerance to P. entomophila infection, as previously suggested by Ullastres et al. (2021) (Log-rank test P-value > 0.001 and 0.002 for males and females, respectively) (fig. 2E, supplementary table S1C, Supplementary Material online). These results suggest that cbx indeed plays a role in immune response as Lime down-regulation alone is associated with increased sensitivity to P. entomophila infection (fig. 2B, supplementary table S1C, Supplementary Material online), while Lime down-regulation and knockout of cbx expression in the same genetic background was associated with increased tolerance to infection (fig. 2E, supplementary table S1C, Supplementary Material online).
Since we found that cbx0741-G4 mutant also down-regulates Lime expression, we also checked whether this mutant affects egg-to-adult viability in nonstress and cold-stress conditions, as we have shown that down-regulation of Lime is associated with decreased egg-to-adult viability in nonstress and cold-stress conditions (fig. 2C). Indeed, we found that cbx0741-G4 mutant showed reduced egg-to-adult viability in nonstress and cold-stress conditions (fig. 2F, supplementary table S1D, Supplementary Material online).
Overall, our results provide further evidence suggesting that changes in Lime expression affect cold- and immune-stress responses and changes in cbx expression affect immune-stress response (Ayres et al. 2008; Telonis-Scott et al. 2009; Mihajlovic et al. 2019; Ullastres et al. 2021).
FBti0019985 is Not Associated With Changes of Expression of cbx in Response to Immune-Stress
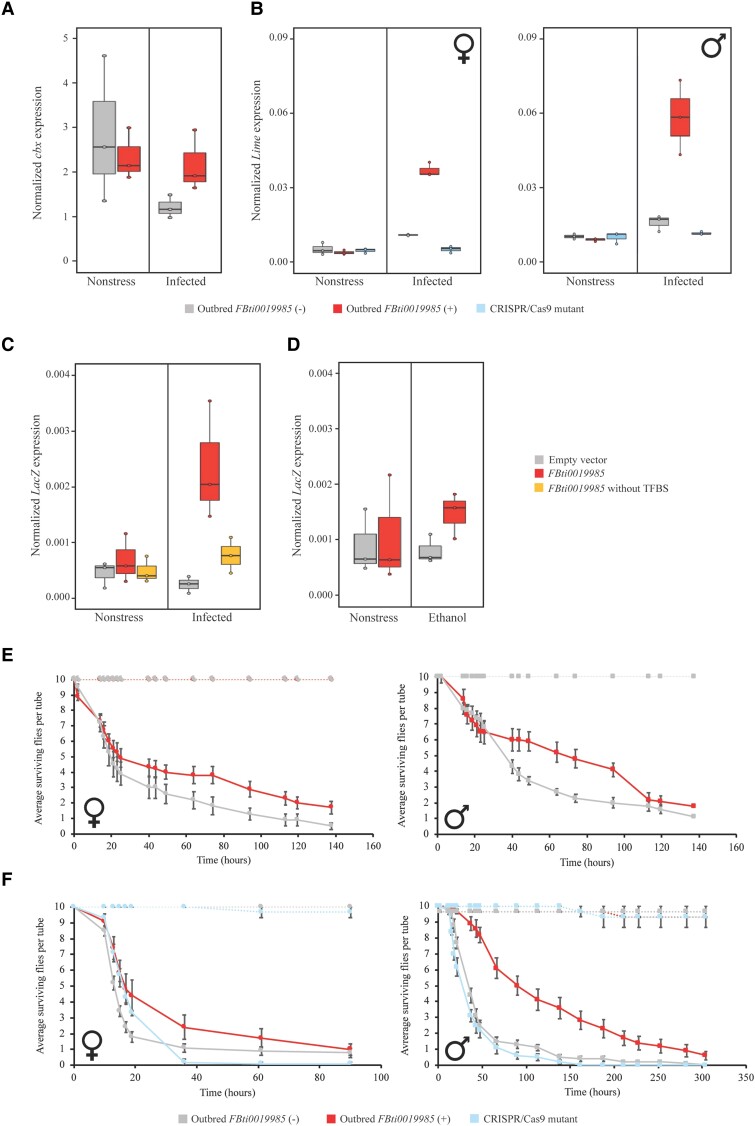
To test whether FBti0019985 is associated with changes of expression of cbx in response to immune-stress, we used two outbred populations: one homozygous for the presence and one homozygous for the absence of the FBti0019985 insertion in a heterogenous unlinked background (Behrman et al. 2018; Merenciano et al. 2019). Note that previous evidence for an association between FBti0019985 and cbx expression was based on allele specific expression analysis performed in adult guts of the offspring of crosses between an inbred strain homozygous for the presence of the insertion and an inbred strain homozygous for its absence, and concluded that the effect of the insertion was background dependent (Ullastres et al. 2021). We thus performed a qRT-PCR to compare the total expression of cbx in adult guts from the outbred populations with and without FBti0019985 that were orally infected with P. entomophila. Note that the gut epithelium is the first barrier that bacteria encounter in the organism and it is known that P. entomophila induces strong perturbations in this tissue (Vodovar et al. 2005). We found that, in outbred populations, cbx expression was not significantly affected by the insertion genotype (G: presence/absence of FBti0019985), the experimental condition (EC: nonstress vs. immune-stress), or the interaction between the genotype and the experimental condition (GxEC) (Analysis of variance (ANOVA) P-values > 0.05; fig. 3A and supplementary table S2, Supplementary Material online), suggesting that the TE does not affect cbx expression. We thus focused on the effect of FBti0019985 on Lime for the rest of this work.
Fig. 3.
FBti0019985 induces Lime up-regulation in guts under immune-stress conditions and increases tolerance to P. entomophila infection. (A) Normalized expression of cbx with Act5C in adult female guts in nonstress and after P. entomophila infection. Boxplots show the median (horizontal line), first and third quartiles (lower and upper bounds, respectively), and minimum and maximum values (lower and upper whiskers, respectively). For each strain, each dot represents an independent biological replicate of a pool of 25–35 guts. Two-way ANOVA: genotype (G: presence/absence of FBti0019985) P-value = 0.687; experimental condition (EC: nonstress vs. immune-stress) P-value = 0.138; genotype by experimental condition interaction (GxEC) P-value = 0.219. (B) Normalized expression of Lime with Act5C in nonstress and immune-stress conditions of adult female and male guts from outbred populations with and without FBti0019985, and for CRISPR/Cas9-mutant flies. For each strain, each dot represents an independent biological replicate of a pool of 25–35 guts. Two-way ANOVA outbred FBti0019985 (+) and outbred FBti0019985 (−) females: G P-value < 0.001, EC P-value < 0.001, GxEC P-value < 0.001. Two-way ANOVA outbred FBti0019985 (+) and CRISPR/Cas9 mutant females: G P-value < 0.001, EC P-value < 0.001, GxEC P-value < 0.001. Two-way ANOVA outbred FBti0019985 (+) and outbred FBti0019985 (−) males: G P-value = 0.002, EC P-value < 0.001, and GxEC P-value = 0.001. Two-way ANOVA outbred FBti0019985 (+) and CRISPR/Cas9 mutant males: G P-value = 0.001, EC P-value < 0.001, and GxEC P-value = 0.001. (C) Normalized expression of the reporter gene lacZ with Act5C in adult female guts under nonstress and immune-stress conditions from transgenic flies. Each dot represents an independent biological replicate of a pool of 25–35 guts. Two-way ANOVA empty vector and FBti0019985 complete sequence: G P-value = 0.009, EC P-value = 0.065, and GxEC P-value = 0.026. Two-way ANOVA FBti0019985 complete sequence and FBti0019985 without TFBSs: G P-value = 0.036, EC P-value < 0.024, and GxEC P-value = 0.084. (D) Normalized expression of the reporter gene lacZ with Act5C in transgenic adult female whole-body under nonstress conditions and after ethanol exposure. Each dot represents and independent biological replication of a pool of 25 flies. Two-way ANOVA: G P-value = 0.272, EC P-value = 0.669, and GxEC P-value = 0.494. (E) Survival curves of adult outbred female and male flies with and without FBti0019985 and (F) survival curves of adult outbred female and male flies with and without FBti0019985 and CRISPR/Cas9-mutant flies after P. entomophila infection. Survival curves in noninfected conditions are depicted as dotted lines while survival curves after P. entomophila infection are depicted as continuous lines. For each strain and sex, each dot represents the average number of survival flies per tube, for ten replicates of ten flies each (infected conditions), and for three replicates of ten flies each (noninfected conditions). Error bars represent SEM. No mortality was observed in nonstress conditions. Log-rank test was performed to analyze differences between survival curves in infected conditions (log-rank P-values < 0.05).
FBti0019985 is the Causal Mutation Inducing Lime Up-Regulation in Adult Flies Under Immune-Stress
To test for an association between FBti0019985 and Lime expression in nonstress and immune-stress conditions, we used the same two outbred populations described above. We quantified Lime expression in adult guts of these two populations as there is previous evidence showing that FBti0019985 acts as an enhancer in adults in response to immune-stress (Villanueva-Cañas et al. 2019). While no differences in Lime expression levels between outbred flies with and without the insertion were found in nonstress conditions, the outbred population with FBti0019985 had increased Lime expression under immune-stress conditions, both in male and female guts (fig. 3B, and supplementary table S2, Supplementary Material online). To confirm that the FBti0019985 is the mutation causing increased Lime expression, we generated a precise deletion of the insertion in the outbred population using the CRISPR/Cas9 technique (see Material and Methods). While no differences in expression levels of Lime were found in nonstress conditions, under immune-stress, the CRISPR/Cas9 mutant (in which the insertion was deleted) showed Lime down-regulation compared with the outbred strain with FBti0019985, as expected if this insertion is the causal mutation (fig. 3B and supplementary table S2, Supplementary Material online).
Overall, we found that FBti0019985 was associated with Lime up-regulation under immune-stress conditions, and we confirmed using CRISPR/Cas9 to precisely delete the insertion, that FBti0019985 was the causal mutation (fig. 3B).
FBti0019985 Harbors Functional Transcription Factor Binding Sites Related With Immune Response
To identify the molecular mechanism by which FBti0019985 up-regulates the expression of Lime in immune-stress conditions, we first performed in vivo enhancer assays. Previous studies have identified several transcription factor binding sites (TFBSs) in the FBti0019985 sequence (Merenciano et al. 2016; Villanueva-Cañas et al. 2019). Three of these predicted binding sites are for transcription factors DEAF-1, tin, and Dorsal, respectively, which are related to the immune response (Merenciano et al. 2016; Villanueva-Cañas et al. 2019). At least one of these transcription factors, DEAF-1, is expressed in the digestive system, which is the first barrier to face oral infections (Larkin et al. 2021). We thus tested whether these immune-related TFBSs could be responsible for Lime up-regulation in response to bacterial oral infection, by using directed mutagenesis to delete them from the FBti0019985 sequence (see Material and Methods), and cloning this modified FBti0019985 sequence in front of a reporter gene. As expected, we found that the complete sequence of FBti0019985 drives the expression of the reporter gene in guts in infected conditions (fig. 3C, supplementary table S3, Supplementary Material online) (Villanueva-Cañas et al. 2019). When we deleted the three immune-related TFBSs, the expression of the reporter gene was reduced, suggesting that the deleted binding sites were responsible for the enhancer activity of FBti0019985 in infected conditions (fig. 3C, supplementary table S3, Supplementary Material online).
To test whether the enhancer effect of FBti0019985 is stress-specific, we measured the expression of the reporter gene in adult flies exposed to ethanol-stress. We found no differences in expression under nonstress and ethanol-stress conditions (fig. 3D, supplementary table S3, Supplementary Material online). These results confirmed that FBti0019985 is not acting as an enhancer in nonstress conditions in adult flies, and suggested that the enhancer effect of FBti0019985 is stress-specific.
Besides acting as an enhancer, FBti0019985 could also be affecting Lime expression by adding a new TSS. TEs of the roo family, including FBti0019985, have previously been shown to add alternative TSS to nearby genes in embryonic stages (Batut et al. 2013). However, whether the TSS of FBti0019985 is also used in guts and whether this TSS could contribute to the differential expression of Lime in response to immune-stress is unknown. We found that the alternative transcript starting in FBti0019985 was also present in the gut of adult flies both in nonstress and in immune-stress conditions (supplementary table S2, Supplementary Material online). A transcript-specific qRT-PCR showed that the expression level of the transcript that starts in the TE does not increase significantly under infected conditions (one-tailed t-test P-value = 0.352). Moreover, if we compared the expression level of the transcript that starts in the TE with the total Lime expression, we found that its contribution, both in nonstress and immune-stress conditions, is low: <1% and 4%, respectively (supplementary table S2, Supplementary Material online).
Overall, we demonstrated that FBti0019985 harbors functional TFBSs related to immune response that is responsible for its enhancer activity in infected conditions and that this enhancer activity is stress-specific. Although FBti0019985 is adding a TSS in the gut, the transcript that starts in the TE does not significantly contribute to the increased expression of Lime in infected conditions.
FBti0019985 Increases Tolerance to P. entomophila Infection
To test whether the up-regulation of Lime in outbred flies with FBti0019985 under immune-stress conditions affects fly survival, we performed infection tolerance assays with P. entomophila. We found that both female and male flies from outbred populations with FBti0019985 were more tolerant to infection than flies without the insertion (Log-rank test P-value = 0.010 and 0.026, respectively) (fig. 3E, supplementary table S4, Supplementary Material online). We repeated the infection tolerance assay including this time the CRISPR/Cas9-mutant strain, which has a precise deletion of the FBti0019985 insertion. As before, we observed that outbred flies with FBti0019985 were more tolerant to infection compared with outbred flies without the insertion in both females and males (Log-rank test P-value = 0.002 and >0.001, respectively) (fig. 3F, supplementary table S4, Supplementary Material online). And, as expected if FBti0019985 was the causal mutation, the CRISPR/Cas9-mutant strain was more sensitive to infection compared to flies with FBti0019985 both in females and males (Log-rank test P-value = 0.004 and >0.001, respectively) (fig. 3F, supplementary table S4, Supplementary Material online).
FBti0019985 Drives Lime Up-Regulation in Embryos Under Nonstress Conditions
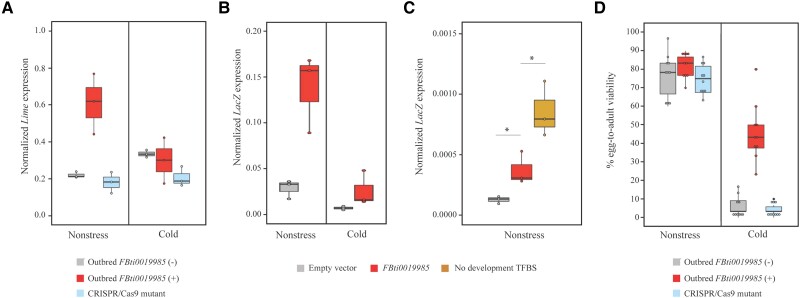
There is previous evidence associating the presence of FBti0019985 insertion with increased expression of Lime in nonstress conditions in embryos (Merenciano et al. 2016). However, this association was found to be background dependent, and whether FBti0019985 also affects Lime expression in cold-stress conditions has not been tested before (Merenciano et al. 2016). Thus, we checked whether FBti0019985 affects Lime expression in embryos in nonstress and cold-stress conditions. We confirmed that embryos from the outbred population with FBti0019985 showed Lime up-regulation in nonstress conditions compared to embryos from the outbred population without the insertion (fig. 4A, supplementary table S2, Supplementary Material online). However, we found no differences in Lime expression in cold-stress conditions associated with the presence of the TE (fig. 4A, supplementary table S2, Supplementary Material online), further suggesting that the effect of the TE is stress-specific. As expected if FBti0019985 is the causal mutation, CRISPR/Cas9-mutant embryos showed reduced expression levels of Lime in nonstress conditions (fig. 4A, supplementary table S2, Supplementary Material online).
Fig. 4.
FBti0019985 induces Lime up-regulation in embryos and increases viability in cold-stress conditions. (A) Normalized expression of Lime with Act5C in nonstress and cold-stress conditions in embryos from outbred populations, and from the CRISPR/Cas9-mutant strain. Boxplots show the median (horizontal line), first and third quartiles (lower and upper bounds, respectively), and minimum and maximum values (lower and upper whiskers, respectively). For each strain, each dot represents an independent biological replication of a pool of approximately 50 embryos. Two-way ANOVA outbred FBti0019985 (+) and outbred FBti0019985 (−): genotype (G: presence/absence of FBti0019985) P-value = 0.018, experimental condition (EC; nonstress vs. cold-stress) P-value P = 0.142, and genotype by experimental condition interaction (GxEC) P-value = 0.007. Two-way ANOVA outbred FBti0019985 (+) and CRISPR/Cas9 mutant: G P-value = 0.003, EC P-value = 0.056, and GxEC P-value = 0.029. (B) Normalized expression of the reporter gene lacZ with Act5C in embryos under nonstress and cold-stress conditions from transgenic flies. For each strain, each dot represents an independent biological replication of a pool of approximately 50 embryos. Two-way ANOVA: G P-value = 0.002, EC P-value = 0.001, and GxEC P-value = 0.012. (C) Normalized expression of the reporter gene lacZ with Act5C in embryos under nonstress conditions from transgenic flies. For each strain, each dot represents an independent biological replication of a pool of approximately 50 embryos. Two-sided t-test P-values = 0.038 and 0.035 for comparisons between transgenic embryos with the FBti0019985 complete sequence and transgenic embryos without the TE sequence or FBti0019985 without development-related TFBSs, respectively. (D) Egg-to-adult viability in nonstress and in cold-stress conditions of outbred populations and CRISPR/Cas9 mutant. For each strain, each dot represents an independent biological replication of a pool of 30 embryos. Two-way ANOVA outbred FBti0019985 (+) and outbred FBti0019985 (−): G P-value < 0.001, EC P-value < 0.001, and GxEC P-value < 0.001. Two-way ANOVA outbred FBti0019985 (+) and CRISPR/Cas9 mutant: G P-value < 0.001, EC P-value < 0.001, and GxEC P-value < 0.001.
Lime Up-Regulation in Embryos is Likely Not Due to Functional Binding Sites in FBti0019985 Sequence
Consistent with the effect of FBti0019985 on Lime expression in outbred populations, we found that transgenic embryos containing the FBti0019985 sequence showed significantly increased reporter gene expression only in nonstress conditions (fig. 4B, supplementary table S3, Supplementary Material online). In-silico predictions found TFBSs related to developmental processes in the FBti0019985 sequence that could be responsible for this enhancer activity of the TE in embryos (Merenciano et al. 2016). We thus performed an in vivo reporter assay deleting the seven development-related predicted binding sites from the TE sequence (Dorsal, Nub, ara/mirr (x2), Bap, Vnd, and Btd) (see Material and Methods). Contrary to our expectations, we did not find reduced expression of the reporter gene when the binding sites were removed from the FBti0019985 sequence (fig. 4C, supplementary table S3, Supplementary Material online). Indeed, we found an increased expression of the reporter gene suggesting that these binding sites might be functional but they were repressing the expression of the reporter gene (fig. 4C, supplementary table S3, Supplementary Material online).
Finally, the level of expression of the transcript that starts in the FBti0019985 insertion was not significantly different in nonstress and cold-stress conditions (one-tailed t-test P-value = 0.174). In embryos, the contribution to the total Lime expression of the transcript that starts in the TE was higher than the one observed in adults (13.6% and 10.2% in nonstress and stress, respectively vs. <1% and 4%; supplementary table S2, Supplementary Material online), as expected since roo elements are known to provide embryonic promoters (Batut et al. 2013).
FBti0019985 Increases Egg-to-Adult Viability Under Cold-Stress Conditions
We checked the egg-to-adult viability of outbred strains with and without FBti0019985 in nonstress and in cold-stress conditions. We found that the insertion genotype (presence/absence of FBti0019985), the experimental condition (nonstress vs. cold-stress), and the interaction between the genotype and the experimental condition were significant (fig. 4D, supplementary table S5, Supplementary Material online). As expected if FBti0019985 is the causal mutation, CRISPR/Cas9-mutants showed reduced viability compared with the outbred strain with the element in cold-stress conditions (fig. 4D, supplementary table S5, Supplementary Material online).
roo Insertions Are Not Enriched Nearby Immune-Stress Genes
Our results show that the FBti0019985 roo insertion increases the expression of Lime in response to immune-stress by adding functional immune-related TFBS, leading to increased survival to bacterial infection (fig. 3). We thus checked whether other roo insertions in the genome were enriched nearby immune-stress response genes. We focused on the 157 roo insertions present in the last release of the D. melanogaster reference genome (r.6.48) and identified those that are inserted nearby (<1 kb) candidate genes involved in immune-stress (see Material and Methods). Overall, we found that 10 of the 157 roo insertions were inserted nearby immune-related genes (supplementary table S7, Supplementary Material online). However, there was no significant enrichment of roo insertions nearby immune-related genes (one-side Fisher's exact test P = 0.094) (supplementary table S7, Supplementary Material online).
Discussion
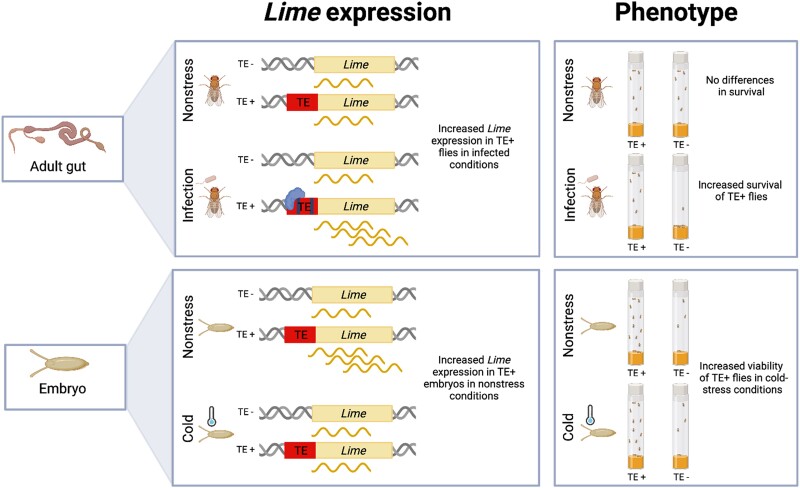
In this work, we characterized a naturally occurring TE-induced mutation and found that the effect of the mutation depends on the interplay between the environmental condition and the developmental stage (fig. 5). While in nonstress conditions the FBti0019985 insertion only affects Lime expression in embryos, in stress conditions FBti0019985 affects Lime expression in adults, and specifically in response to immune-stress (figs. 3–5). CRISPR/Cas9-mediated deletion of the insertion in an outbred population confirmed that the TE is the causal mutation leading to Lime up-regulation in nonstress in embryos and immune-stress in adults. Furthermore, we showed that FBti0019985 is the causal mutation leading to increased bacterial infection tolerance and increased viability in cold-stress conditions (figs. 3–5). Previous evidence for the complex effects of TE-induced mutations was mostly associated with coding mutations (Wang et al. 2018; Frank et al. 2022). For instance, Syncytin-1, a human-endogenous retrovirus gene with a clear role in human placental formation (Mi et al. 2000; Senft and Macfarlan 2021), has also been associated with the development of neuropsychological disorders and multiple sclerosis under certain environmental stress conditions such as infection, or drug application (Gröger and Cynis 2018; Wang et al. 2018). Similarly, Supressyn, another gene from a retroviral origin also involved in placental development, has been recently linked with resistance to viral infection, demonstrating that its effect is context-dependent (Frank et al. 2022). Our results thus showed that besides TE-induced coding mutations, the effect of TE-induced regulatory mutations is also complex, further suggesting that TEs could be more likely to have complex effects compared to SNPs (e.g., a single mutation can add both promoters and TFBS) (Schrader and Schmitz 2019; Baduel and Quadrana 2021).
Fig. 5.
Schematic representation of the effect of FBti0019985 in nonstress, infected, and cold-stress conditions. Flies with the FBti0019985 insertion show increased Lime expression in guts in infected conditions due to the presence of functional immune-related TFBSs in the TE sequence. These differences in Lime expression are responsible for the increased survival from infection in flies with the insertion. Embryos with FBti0019985 show an increased Lime expression in nonstress conditions, and increased viability in cold-stress conditions. The molecular mechanism of increased expression in embryos in nonstress conditions remains to be uncovered. Big boxes represent Lime gene, while small boxes represent the FBti0019985 TE insertion. Wavy lines represent Lime transcripts. Functional TFBSs in the FBti0019985 sequence are depicted as small rectangles within FBti0019985.
Besides adding an alternative TSS to Lime, FBti0019985 also acts as an enhancer (Batut et al. 2013; Merenciano et al. 2016). There is increasing evidence across organisms that regulatory regions can have a dual function as both promoters and enhancers (Dao and Spicuglia 2018; Andersson and Sandelin 2020). Indeed, in Drosophila, 4.5% of enhancer sequences overlap with TSS (Arnold et al. 2014; Zabidi et al. 2015). TE insertions have been documented to contribute to gene regulation by functioning as enhancers (Barco et al. 2019; Sundaram and Wysocka 2020) and promoters (Faulkner and Carninci 2009; Batut et al. 2013), and recently a dual role as both enhancer and promoter in immune-stress conditions has been described for FBti0019386, a natural TE insertion in D. melanogaster (Ullastres et al. 2021). However, unlike FBti0019386, the alternative transcript starting in FBti0019985 does not seem to have a significant contribution to the changes in gene expression in the developmental stages and conditions analyzed in this work. Thus, characterization of other TEs with the potential to increase transcript diversity and to act as enhancers is needed to evaluate the contribution of this type of mutations to complex regulatory regions.
Our results also suggest that the molecular mechanism by which a TE increases the expression of its nearby gene could be different in nonstress versus stress conditions. While we found that the insertion harbors functional binding sites for immune-related transcription factors (fig. 3C), a mechanism other than adding binding sites appears to be responsible for the enhancer role in embryonic stages (fig. 4C). Although we cannot discard that TFBS not identified in this work are responsible for the enhancer activity of FBti0019985 in embryos, other mechanisms such as inducing epigenetic changes could also be responsible for the observed changes in expression (Rey et al. 2016; Guio et al. 2018; Ullastres et al. 2021).
Finally, we speculate that the effect of Lime on sugar metabolism might underlay the increased immune- and cold-stress survival observed (figs. 3 and 4) (Mihajlovic et al. 2019). Lime affects the levels of glucose and trehalose, which has been shown to affect immune cell proliferation and activation (Mihajlovic et al. 2019). Moreover, changes in glucose and trehalose have been reported in cold-shock and rapid cold-hardening treatments in D. melanogaster (Overgaard et al. 2007; Koštál et al. 2011). Thus, increased basal levels of Lime expression in embryos might be beneficial when flies are exposed to cold-stress. Indeed, distinct basal transcriptional states have been associated with different phenotypic responses to enteric infection and also to cold-stress in D. melanogaster (Telonis-Scott et al. 2009; Bou Sleiman et al. 2015). However, further experiments are needed to link the effect of FBti0019985 on Lime expression with changes in sugar metabolism and its subsequent effects on cold and immune tolerance.
Overall, this work provides evidence for FBti0019985 being the causal mutation responsible for Lime up-regulation as a result of the interplay between the developmental stage and environment. We further demonstrate that FBti0019985 increases viability in cold-stress and immune-stress. These results open up the question of how often tissue-specific and environment-dependent gene expression could be due to the presence of TEs and call for their inclusion in studies aimed at understanding the context-dependent effect of mutations. Furthermore, our results provide support for the need to expand the concept of the genotype-phenotype map from identifying the impact of a mutation in a particular context to exploring the spectrum of effects in different contexts, including different genetic backgrounds, environments, and developmental stages.
Materials and Methods
Fly Stocks
Fly stocks were reared on standard fly food medium containing glucose, fresh yeast, wheat flour, agar, propionic acid, and nipagin in a 12:12 h light/dark cycle at 25 °C.
Laboratory Mutant and RNAi Knock-Down Strains
We used three laboratory mutant strains and one RNAi knock-down strain that were likely to affect Lime and/or cbx genes. We also used two wild-type (WT) strains with the same genetic backgrounds as the mutant strains, and a strain with a GAL4 promoter to activate the RNAi (supplementary table S1A, Supplementary Material online). Briefly, we used LimeMI02952 mutants (Bloomington Drosophila Stock Center stock number #36170) that contain a MiMIC insertion in the first exon of Lime gene. As a WT strain, we used y[1] w[67c23] flies (stock number #6599) with the same genetic background (supplementary table S1A, Supplementary Material online). We used a RNAi knock-down strain for the same gene (stock number #33735) (supplementary table S1A, Supplementary Material online). To activate the RNAi, that strain was crossed to a strain containing a GAL4 promoter (w; act GAL4/TM6 + tb) (supplementary table S1A, Supplementary Material online). We also used cbx0741-G4 mutants (stock number #63767) and cbxc00428 mutants (stock number #10067) with a PiggyBac insertion in the first and third intron of cbx, respectively. For these two mutants, we used as a WT the strain w1118, which has the same genetic background as the two mutant strains (supplementary table S1A, Supplementary Material online).
Outbred Strains
We used the two outbred populations, one with and one without FBti0019985 TE insertion, generated by Merenciano et al. (2019). Briefly, they were generated by a round-robin cross-design of inbred lines from the Drosophila genetic reference panel (Mackay et al. 2012) and isofemale lines from different European populations (Merenciano et al. 2019). Outbred populations were maintained by random mating with a large population size for over five generations before starting the first experiments.
CRISPR/Cas9 Mutant Strains
To generate a CRISPR/Cas9 strain with a precise deletion of FBti0019985 in the natural outbred population we followed a two-step approach (Merenciano and González 2023). First, we substituted the insertion by the DsRed fluorescence marker and then, in a second step, we removed the visual marker to avoid any possible effect of the introduced DsRed sequence in the results (supplementary fig. S1, Supplementary Material online). For the substitution of FBti0019985 by the DsRed fluorescence marker, guide RNAs (gRNA1: 5′-tatctcaaataagtctagct-3′ and gRNA2: 5′-cagagaaacgtcgagctgcg-3′) were designed in the FBti0019985 flanking region using the flyCRISPR target finder (http://targetfinder.flycrispr.neuro.brown.edu/). Both gRNAs1 and gRNA2 were cloned into pCFD5 plasmid following the pCFD5 cloning protocol (www.crisprflydesign.com) (Port and Bullock 2016) using the primers 5′-gcggcccgggttcgattcccggccgatgcaagctagacttatttgagatagttttagagctagaaatagcaag-3′ and 5′-attttaacttgctatttctagctctaaaaccagagaaacgtcgagctgcgtgcaccagccgggaatcgaaccc-3′, that contain gRNA1 and gRNA2 sequences, respectively. Briefly, these primers are used on a pCFD5 template and the resulting PCR products are assembled with a previously linearized pCFD5 backbone (digested with the BbsI type II-S restriction enzyme [NEB]) in a single Gibson Assembly reaction. A donor DNA containing two homology arms flanking the DsRed sequence for homology repair was cloned into the pHD-ScarlessDsRed plasmid (https://flycrispr.org/scarless-gene-editing/) using the Q5® High-Fidelity DNA polymerase kit (New England Biolabs). Left homology arm contained the sequence in 2R:9870299–9871095 (FlyBase Release 6) (Larkin et al. 2021) while the right homology arm contained the sequence in 2R:9871529–9872365 (FlyBase Release 6) (Larkin et al. 2021) from the outbred population with FBti0019985. To avoid cleavage of the donor construct and mutagenesis after integration by CRISPR/Cas9, two single-nucleotide synonymous substitutions (C > G for sgRNA1; G > T for sgRNA2) were introduced into the two sgRNA target site protospacer adjacent motif (PAM) sequences, respectively. The pCFD5 plasmid containing the gRNAs, the donor pHD-ScarlessDsRed plasmid containing the homology arms, and a plasmid containing Cas9 endonuclease were co-injected as a unique mix into approximately 550 embryos from the outbred population with FBti0019985. All the injections were performed using the following mix concentrations: pCFD5 plasmid at 100 ng/µl, donor plasmid at 500 ng/µl, and Cas9 plasmid at 250 ng/µl. F0-emerged flies were backcrossed individually with the parental outbred population with FBti0019985, and F1 offspring was screened for eye fluorescence. These flies were backcrossed again individually with the parental line for a minimum of five generations to remove potential off-targets generated during the process (Bassett and Liu 2014; Port et al. 2020). Then, we performed crosses between flies expressing eye fluorescence, to establish an outbred population homozygous for the deletion of FBti0019985. The substitution of FBti0019985 by DsRed was checked by PCR with two combinations of primers in each cross: forward primer 5′-aacaatgcaagtccgtgctc-3′ and reverse primer 5′-gtggttcctccacccttgtg-3′ spanning the whole substituted region; and forward primer 5′-ggccgcgactctagatcataatc-3′ (inside DsRed sequence) and reverse primer 5′-gtggttcctccacccttgtg-3′ to check for the absence of substitution events. PCR bands were confirmed by Sanger sequencing.
In the second step, to remove the DsRed sequence, we applied again the CRISPR/Cas9 technique designing two gRNAs (gRNA3: 5′-ttgaacactaatgacaattt-3′and gRNA4: 5′-gagctgcgcagttgtgagct-3) in the flanking regions of the DsRed sequence using the flyCRISPR target finder as before. Both gRNA3 and gRNA4 were cloned into the pCFD5 plasmid following the pCFD5 cloning protocol as mentioned above using the primers 5′-gcggcccgggttcgattcccggccgatgcttgaacactaatgacaatttgttttagagctagaaatagcaag-3′ and 5′-attttaacttgctatttctagctctaaaacagctcacaactgcgcagctctgcaccagccgggaatcgaaccc-3′, that contain the gRNA3 and gRNA4 sequences, respectively. A donor DNA containing only two homology arms for homology-directed repair was cloned into the pHD-ScarlessDsRed plasmid removing the DsRed sequence. Left homology arm contained the sequence in 2R:9870299–9871095 (FlyBase Release 6) (Larkin et al. 2021) while the right homology arm contained the sequence in 2R:9871529–9872365 (FlyBase Release 6) (Larkin et al. 2021) from the outbred population with FBti0019985 adding two single-nucleotide synonymous substitutions (G > A for sgRNA3 site; C > T for sgRNA4 site). The pCFD5 plasmid containing the gRNAs 3 and 4, the donor pHD-Scarless plasmid containing the homology arms, and a plasmid containing Cas9 endonuclease were co-injected as explained before into embryos from the previously generated CRISPR/Cas9 mutant containing the DsRed fluorescent marker and the FBti0019985 deletion. F0-emerged flies were backcrossed individually with the CRISPR/Cas9 mutant without FBti0019985, and F1 offspring was screened this time for the absence of eye fluorescence. These flies were backcrossed again individually for a minimum of five generations to remove potential off-targets generated during the process. Then, performing crosses between flies not expressing eye fluorescence, an outbred population containing a homozygous deletion of DsRed was established. The DsRed deletion was checked by PCR in each cross using the primer pair 5′-aacaatgcaagtccgtgctc-3′ and 5′-cgtaggatcagtgggtgaaaatg-3′, and finally, PCR bands were confirmed by Sanger sequencing. No polymorphism were found in the sequenced region except for the two single-nucleotide synonymous substitutions introduced into the two sgRNA target site PAM sequences.
Transgenic Strains
For the in vivo reporter assays, we used the transgenic flies with the FBti0019985 sequence cloned in front of the lacZ reporter gene generated in Ullastres et al. (2021), and we compared them with transgenic strains with the placZ.attB empty vector to control for possible lacZ expression driven by the vector sequence itself. To generate the transgenic flies with deleted TFBSs, we performed directed mutagenesis for each TFBS using the placZ.attB vector containing the FBti0019985 sequence as a template (Ullastres et al. 2021), with the Q5 site-directed mutagenesis kit (New England Biolabs). Since it is not possible to delete several TFBSs at the same time, we performed consecutive deletions until all the binding sites were deleted. Primers used for deleting each TFBSs can be found in supplementary table S6, Supplementary Material online. Then, the vector with the FBti0019985 sequence with the immune-related TFBSs deleted (DEAF-1, tin, and Dorsal) and the vector with the developmental TFBSs deleted (Dorsal, Nub, ara/mirr (x2), Bap, Vnd, and Btd) were microinjected separately at 350–500 ng/µl into a D. melanogaster strain with a stable docking site in the chromosome 2 (Bloomington Stock number: #24749). Offspring was screened for red eyes and the insertion of the construct was verified by PCR and Sanger sequencing with the primer pair 5′-ggtgggcataatagtgttgtttat-3′ and 5′-cgacgtgttcactttgcttgt-′3. Three independent homozygous stocks were generated and used as biological replicates for the qRT-PCR experiments.
Sample Collection for Expression Analysis
Laboratory Mutant and RNAi Knock-Down Strains
5–7 day-old females from each strain were harvested and placed in vials with fresh food in groups of 25. We allowed the flies to recover from CO2 anesthesia for 24 h at 25 °C. Then, the pool of 25 females of every strain was placed in three empty Eppendorf tubes and considered as independent replicates. Finally, samples were flash-frozen in liquid nitrogen and stored at −80 °C until sample processing. Lime and cbx expression changes were measured in the LimeMI02952 mutants compared with a strain with the same genetic background (y[1] w[67c23]) (supplementary table S1A and B, Supplementary Material online). We also measured Lime and cbx expression in the offspring of the cross between the Lime-RNAi strain and a strain expressing the GAL4 promoter for the activation of the RNAi (w; act GAL4/TM6 + tb). We compared the expression of the offspring with the maternal strain containing the GAL4 promoter (supplementary table S1A and B, Supplementary Material online). Lime expression was measured in cbx0741-G4 and cbxc00428 mutants, comparing both of them to a strain with the same genetic background (w1118) (supplementary table S1A and B, Supplementary Material online). Only LimeMI02952 mutants that affected Lime expression and cbx0741-G4 mutants that affected cbx and Lime expression were used in the phenotypic assays (supplementary table S1B, Supplementary Material online).
P. entomophila Infection in Outbred Populations and CRISPR/Cas9 Mutants
5–7 day-old flies from each strain tested were separated by sex and placed in vials with fresh food in groups of 25–35. We allowed flies to recover from CO2 anesthesia for 24 h at 25° C. To expose the flies to the gram-negative bacteria P. entomophila infection, we followed the protocol described in Neyen et al. (2014). Briefly, after two hours of starvation, we transfer 75–105 flies in groups of 25–35 into three vials with fresh food and a filter paper soaked with 120 µl of a solution containing 1.25% sucrose and bacterial preparation adjusted to a final OD600 = 50 for females and OD600 = 150 for males. These bacterial loads were chosen so that the mortality observed after the 12 h of treatment was <70%. Flies were kept at 29 °C, the optimal temperature condition for P. entomophila infection. Simultaneously, a total of 75–105 flies were also transferred in groups of 25–35 into three vials with fresh food and a filter paper soaked with 120 ul of a solution containing sterile lysogeny broth (LB) medium with 1.25% sucrose and kept at 29 °C as a control. Guts from males and from females of every one of the three vials were dissected after 12 h and considered as independent replicates. Samples were then flash-frozen in liquid nitrogen and stored at −80 °C until sample processing.
Cold-stress Treatment in Outbred Populations and CRISPR/cas9 Mutants
5–7 day-old flies from each strain tested were allowed to lay eggs at 25 °C in a fly cage with egg-laying medium (2% agar with apple juice and a piece of fresh yeast) for four hours. After these four hours, adults were removed and the plate containing embryos was kept at 1 °C for four additional hours. Simultaneously, another plate with embryos was kept at 25 °C for four additional hours as a control. 4–8 h-old embryos were then collected from both plates using the method described in Schou (2013). Briefly, eggs laid on the surface of the egg-laying medium were gently washed with a sucrose solution (29 g/100 mL sucrose/water) with a small brush and poured into an Erlenmeyer flask with a mesh to keep the embryos. Then, embryos were dechorionated for 10 min with 50% bleach. A pool of approximately 50 dechorionated embryos of every strain and treatment was placed in three different empty eppendorf tubes and considered as independent replicates. Finally, samples were flash-frozen in liquid nitrogen and stored at −80 °C until sample processing.
Ethanol Exposure in Transgenic Strains
5–7 days-old flies from each strain tested were separated by sex and placed in vials with fresh food in groups of 25. We allowed flies to recover from CO2 anesthesia for 24 h at 25 °C. To expose the flies to ethanol we followed the method described in Maples and Rothenfluh (2011). Briefly, 150 flies in groups of 25 were transferred into six empty vials. A coated cotton ball with 0.5 ml ethanol was inserted into three vials (exposure vials) for nine minutes ensuring that the alcohol was facing into the vial and not toward the wall. Simultaneously, coated cotton balls with 0.5 ml H2O were inserted into the remaining three vials (control vials). After that, flies of each vial were transferred to empty eppendorf tubes and considered as independent replicates. Finally, samples were flash-frozen in liquid nitrogen and stored at −80 °C until processing.
RNA Extraction and cDNA Synthesis
RNA was extracted using the GeneEluteTM Mammalian Total RNA Miniprep Kit following manufacturer's instructions (Sigma). RNA was then treated with DNase I (Thermo 1 U) for 1 h at 37 °C. cDNA was then synthesized from a total of 250–1,000 ng of RNA using the NZY First-Strand cDNA synthesis kit (NZYTech).
qRT-PCR Analysis
Lime expression was measured using the forward primer 5′ -gagcagttggaatcgggttttac −3′ and the reverse primer 5′-gtatgaatcgcagtccagccata-3′ spanning 99 bp cDNA in the exon 1/exon 2 junction. cbx expression was measured with the forward primer 5′-gggaaaacgatctgggagca −3′ and the reverse primer 5′- gtcggagaagttgagtggga −3′ spanning 233 bp cDNA in the exon 2/exon 3 junction. lacZ reporter gene expression was measured using the forward primer 5′-cctgctgatgaagcagaacaact-3′ and the reverse primer 5′-gctacggcctgtatgtggtg-3′. Gene expression was normalized with Act5C (5′ -gcgcccttactctttcacca-3′ and 5′ -atgtcacggacgatttcacg-3′ primers). We performed the qRT-PCR analysis with SYBR green (BioRad) or with the qPCRBIO SyGreen Mix Lo-Rox (PCRBiosystems) on iQ5 and CFX384 Thermal cyclers, respectively. Results were analyzed using the difference between cycle threshold (dCT) method (Pfaffl 2001).
Transcript Start Site Detection
We performed RT-PCRs to detect whether FBti0019985 is adding an alternative TSS to the Lime gene in outbred flies carrying FBti0019985 after different stress conditions. We used the forward primer 5′- aaaactcaacgagtaaagtcttc −3′ and the reverse primer 5′- tataaagttccaacgcccagc −3′ to detect the Lime transcript starting in the TE. The forward primer 5′- cgcagagaaacgtcgagctg −3′ and the reverse primer 5′-cacgttaaattcactagggtggc −3′ were used to detect Lime total transcript. The outbred population without FBti0019985 was used as a control sample.
Phenotypic Assays
P. entomophila Infection
Ten tubes of ten 5–7 day-old male flies and ten tubes of ten 5–7 day-old females from each one of the strains tested were infected with the gram-negative bacteria P. entomophila as described before (total n = 100 flies per sex). Simultaneously, a total of three tubes of ten male and three tubes of ten female flies were tested as controls (total n = 30 flies per sex). We counted the number of dead flies in every vial at different time points for at least six days. Log-rank tests (Mantel-Cox) were performed to analyze pairwise differences between survival curves with SPSS v21 software.
Egg-to-Adult Viability Under Cold-Stress
5–7 day-old flies from each strain tested were allowed to lay eggs for 4 h at 25 °C in a fly cage with egg-laying medium (2% agar with apple juice and a piece of fresh yeast) for four hours. Then, adults were removed from the cage and plates were kept for four additional hours at 25 °C. After that, 4–8 h-old embryos were collected using the method described in Schou (2013) and placed in vials with fresh food in groups of 30. Briefly, eggs laid on the surface of the egg-laying medium were gently washed with a sucrose solution (29 g/100 mL sucrose/water) with a small brush and poured into an Erlenmeyer flask with a mesh to keep the embryos. Then, embryos were transferred with a small brush in the surface of a filter paper moistened with the sucrose solution, counted under a magnifying glass, and finally placed into tubes with fresh food. Then, a pool of 30 embryos of every strain was transferred to 20 empty vials with fresh food (ten vials for the experiment with mutant flies) and considered as independent replicates. Ten vials (five for the experiment with mutant flies) were kept at 1 °C for 15 h and then maintained at 25 °C until adult emergence. Simultaneously, ten control vials (five for the experiment with mutant flies) were kept at 25 °C and never exposed to cold. Percentage egg-to-adult viability was calculated based on the number of emerged flies to the total number of embryos placed in each vial. Statistical significance was calculated by performing ANOVA using SPSS v.21 combining all the data into a full model: experimental condition (stress and nonstress), insertion genotype (presence/absence of the insertion), and interaction between these two factors.
roo Enrichment Nearby Immune-Stress Candidate Genes
We used the last available TE annotation of the reference genome (http://ftp.flybase.net/genomes/Drosophila_melanogaster/dmel_r6.48_FB2022_05/fasta/dmel-all-transposon-r6.48.fasta.gz) to build a bed file with the coordinates and names of all the 157 roo insertions using the commands egrep and awk. To know how many of these copies were inserted near immune-stress candidate genes (<1 kb), we used previously available information that summarizes evidence from genome-wide association studies (GWAS), quantitative trait loci (QTL), gene expression, and protein–protein interactions, and candidate gene studies to identify genes involved in immune-stress (Rech et al. 2019). We thus generated a file with a list of 971 candidate genes (supplementary table S7A, Supplementary Material online). We then created a bed file with the coordinates of these genes using the gtf file of the last release of the reference genome (http://ftp.flybase.net/genomes/Drosophila_melanogaster/dmel_r6.48_FB2022_05/gtf/dmel-all-r6.48.gtf.gz) and the commands egrep and awk. Then, we used bedtools windows with default parameters to obtain the roo insertions present in <1 kb distance of our candidate immune-stress related genes (supplementary table S7B, Supplementary Material online). Statistical significance was calculated by performing Fisher's exact test.
Supplementary Material
Acknowledgments
We thank Fillip Port from the Division of Signaling and Functional Genomic led by Prof. Dr. Michael Boutros for his advice in the generation of the CRISPR/Cas9 mutants. This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (H2020-ERC-2014-CoG-647900).
Contributor Information
Miriam Merenciano, Institute of Evolutionary Biology (CSIC-Universitat Pompeu Fabra), Barcelona, Spain.
Josefa González, Institute of Evolutionary Biology (CSIC-Universitat Pompeu Fabra), Barcelona, Spain.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
M.M. and J.G. designed research; M.M. performed research; M.M. and J.G. analyzed data; and M.M. and J.G. wrote the paper.
Data Availability
The data is available within the Article and Supplementary Information.
Conflict of interest statement : The authors declare no competing interest.
References
- Aminetzach YT, Macpherson JM, Petrov DA. 2005. Pesticide resistance via transposition-mediated adaptive gene truncation in Drosophila. Science 309(5735):764–767. [DOI] [PubMed] [Google Scholar]
- Andersson R, Sandelin A. 2020. Determinants of enhancer and promoter activities of regulatory elements. Nat Rev Genet. 21(2):71–87. [DOI] [PubMed] [Google Scholar]
- Anholt RRH, Mackay TFC. 2018. The road less traveled: from genotype to phenotype in flies and humans. Mamm Genome. 29(1–2):5–23. [DOI] [PubMed] [Google Scholar]
- Arnold CD, Gerlach D, Spies D, Matts JA, Sytnikova YA, Pagani M, Lau NC, Stark A.. 2014. Quantitative genome-wide enhancer activity maps for five Drosophila species show functional enhancer conservation and turnover during cis-regulatory evolution. Nat Genet. 46(7):685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayres JS, Freitag N, Schneider DS. 2008. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics 178(3):1807–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baduel P, Quadrana L. 2021. Jumpstarting evolution: how transposition can facilitate adaptation to rapid environmental changes. Curr Opin Plant Biol. 61:102043. [DOI] [PubMed] [Google Scholar]
- Barco B, Kim Y, Clay NK. 2019. Expansion of a core regulon by transposable elements promotes Arabidopsis chemical diversity and pathogen defense. Nat Commun. 10(1):3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett A, Liu JL. 2014. CRISPR/Cas9 mediated genome engineering in Drosophila. Methods 69(2):128–136. [DOI] [PubMed] [Google Scholar]
- Batut P, Dobin A, Plessy C, Carninci P, Gingeras TR. 2013. High-fidelity promoter profiling reveals widespread alternative promoter usage and transposon-driven developmental gene expression. Genome Res. 23(1):169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrman EL, Howick VM, Kapun M, Staubach F, Bergland AO, Petrov DA, Lazzaro BP, Schmidt PS.. 2018. Rapid seasonal evolution in innate immunity of wild. Proc Biol Sci. 285(1870):20172599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bou Sleiman MS, Osman D, Massouras A, Hoffmann AA, Lemaitre B, Deplancke B. 2015. Genetic, molecular and physiological basis of variation in Drosophila gut immunocompetence. Nat Commun. 6:7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosh O, Fabian DK, Cogni R, Tolosana I, Day JP, Olivieri F, Merckx M, Akilli N, Szkuta P, Jiggins FM.. 2022. A novel transposable element-mediated mechanism causes antiviral resistance in Drosophila through truncating the Veneno protein. Proc Natl Acad Sci U S A. 119(29):e2122026119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, González J. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol. 22(6):1503–1517. [DOI] [PubMed] [Google Scholar]
- Catullo RA, Llewelyn J, Phillips BL, Moritz CC. 2019. The potential for rapid evolution under anthropogenic climate change. Curr Biol. 29(19):R996–R1007. [DOI] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2016. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 351(6277):1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 18(2):71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dao LTM, Spicuglia S. 2018. Transcriptional regulation by promoters with enhancer function. Transcription 9(5):307–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Berrocal A, Morita T, Longden KD, Stern DL. 2016. Natural courtship song variation caused by an intronic retroelement in an ion channel gene. Nature 536(7616):329–332. [DOI] [PubMed] [Google Scholar]
- Eguchi Y, Bilolikar G, Geiler-Samerotte K. 2019. Why and how to study genetic changes with context-dependent effects. Curr Opin Genet Dev. 58–59:95–102. [DOI] [PubMed] [Google Scholar]
- Esnault C, Lee M, Ham C, Levin HL. 2019. Transposable element insertions in fission yeast drive adaptation to environmental stress. Genome Res. 29(1):85–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ, Carninci P. 2009. Altruistic functions for selfish DNA. Cell Cycle 8(18):2895–2900. [DOI] [PubMed] [Google Scholar]
- Frank JA, Singh M, Cullen HB, Kirou RA, Benkaddour-Boumzaouad M, Cortes JL, Garcia Pérez J, Coyne CB, Feschotte C. 2022. Evolution and antiviral activity of a human protein of retroviral origin. Science 378(6618):422–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA. 2008. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 6(10):e251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green L, Coronado-Zamora M, Radío S, Rech GE, Salces-Ortiz J, González J. 2022. The genomic basis of copper tolerance in Drosophila is shaped by a complex interplay of regulatory and environmental factors. BMC Biol. 20(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gröger V, Cynis H. 2018. Human endogenous retroviruses and their putative role in the development of autoimmune disorders such as multiple sclerosis. Front Microbiol. 9:265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guio L, Vieira C, González J. 2018. Stress affects the epigenetic marks added by natural transposable element insertions in Drosophila melanogaster. Sci Rep. 8(1):12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoban S, Kelley JL, Lotterhos KE, Antolin MF, Bradburd G, Lowry DB, Poss ML, Reed LK, Storfer A, Whitlock MC.. 2016. Finding the genomic basis of local adaptation: pitfalls, practical solutions, and future directions. Am Nat. 188(4):379–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Sun H, Xu D, Chen Q, Liang Y, Wang X, Xu G, Tian J, Wang C, Li D, et al. 2018. ZmCCT9 enhances maize adaptation to higher latitudes. Proc Natl Acad Sci U S A. 115(2):E334–E341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kammenga JE. 2017. The background puzzle: how identical mutations in the same gene lead to different disease symptoms. FEBS J. 284(20):3362–3373. [DOI] [PubMed] [Google Scholar]
- Koštál V, Korbelová J, Rozsypal J, Zahradníčková H, Cimlová J, Tomčala A, Šimek P.. 2011. Long-term cold acclimation extends survival time at 0° C and modifies the metabolomic profiles of the larvae of the fruit fly Drosophila melanogaster. PLoS One. 6(9):e25025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A, Marygold SJ, Antonazzo G, Attrill H, Dos Santos G, Garapati PV, Goodman JL, Gramates LS, Millburn G, Strelets VB, et al. 2021. Flybase: updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 49(D1):D899–D907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Huang W. 2018. Charting the genotype-phenotype map: lessons from the Drosophila melanogaster genetic reference panel. Wiley Interdiscip Rev Dev Biol. 7(1). doi: 10.1002/wdev.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482(7384):173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwire MM, Bayer F, Webster CL, Cao C, Jiggins FM. 2011. Successive increases in the resistance of Drosophila to viral infection through a transposon insertion followed by a duplication. PLoS Genet. 7(10):e1002337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maples T, Rothenfluh A. 2011. A simple way to measure ethanol sensitivity in flies. J Vis Exp 19(48):2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenciano M, González J. 2023. Two-step CRISPR-Cas9 protocol for transposable element deletion in D. melanogaster natural populations. EcoEvoRxiv. doi: 10.32942/X2P88M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenciano M, Iacometti C, González J. 2019. A unique cluster of roo insertions in the promoter region of a stress response gene in Drosophila Melanogaster. Mob DNA. 10:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merenciano M, Ullastres A, de Cara MA, Barrón MG, González J. 2016. Multiple independent retroelement insertions in the promoter of a stress response gene have variable molecular and functional effects in Drosophila. PLoS Genet. 12(8):e1006249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403(6771):785–789. [DOI] [PubMed] [Google Scholar]
- Mihajlovic Z, Tanasic D, Bajgar A, Perez-Gomez R, Steffal P, Krejci A. 2019. Lime is a new protein linking immunity and metabolism in Drosophila. Dev Biol. 452(2):83–94. [DOI] [PubMed] [Google Scholar]
- Nelson TC, Jones MR, Velotta JP, Dhawanjewar AS, Schweizer RM. 2019. UNVEILing connections between genotype, phenotype, and fitness in natural populations. Mol Ecol. 28(8):1866–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyen C, Bretscher AJ, Binggeli O, Lemaitre B. 2014. Methods to study Drosophila immunity. Methods. 68(1):116–128. [DOI] [PubMed] [Google Scholar]
- Otwinowski J, Nemenman I. 2013. Genotype to phenotype mapping and the fitness landscape of the E. coli lac promoter. PLoS One 8(5):e61570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overgaard J, Malmendal A, Sørensen JG, Bundy JG, Loeschcke V, Nielsen NC, Holmstrup M.. 2007. Metabolomic profiling of rapid cold hardening and cold shock in Drosophila melanogaster. J Insect Physiol. 53(12):1218–1232. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Bullock SL. 2016. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat Methods 13(10):852–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Port F, Strein C, Stricker M, Rauscher B, Heigwer F, Zhou J, Beyersdörffer C, Frei J, Hess A, Kern K, et al. 2020. A large-scale resource for tissue-specific CRISPR mutagenesis in. Elife 13(9):e53865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech GE, Bogaerts-Márquez M, Barrón MG, Merenciano M, Villanueva-Cañas JL, Horváth V, Fiston-Lavier AS, Luyten I, Venkataram S, Quesneville S, et al. 2019. Stress response, behavior, and development are shaped by transposable element-induced mutations in Drosophila. PLoS Genet. 15(2):e1007900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech GE, Radío S, Guirao-Rico S, Aguilera L, Horvath V, Green L, Lindstadt H, Jamilloux V, Quesneville H, González J.. 2022. Population-scale long-read sequencing uncovers transposable elements associated with gene expression variation and adaptive signatures in Drosophila. Nat Commun. 13(1):1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy KC, Andersen EC, Kruglyak L, Kim DH. 2009. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science 323(5912):382–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey O, Danchin E, Mirouze M, Loot C, Blanchet S. 2016. Adaptation to global change: a transposable element-epigenetics perspective. Trends Ecol Evol. 31(7):514–526. [DOI] [PubMed] [Google Scholar]
- Schou MF. 2013. Fast egg collection method greatly improves randomness of egg sampling in Drosophila melanogaster. Fly (Austin). 7(1):44–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Schmitz J. 2019. The impact of transposable elements in adaptive evolution. Mol Ecol. 28(6):1537–1549. [DOI] [PubMed] [Google Scholar]
- Senft AD, Macfarlan TS. 2021. Transposable elements shape the evolution of mammalian development. Nat Rev Genet. 22(11):691–711. [DOI] [PubMed] [Google Scholar]
- Sundaram V, Wysocka J. 2020. Transposable elements as a potent source of diverse cis-regulatory sequences in mammalian genomes. Philos Trans R Soc Lond B Biol Sci. 375(1795):20190347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telonis-Scott M, Hallas R, McKechnie SW, Wee CW, Hoffmann AA. 2009. Selection for cold resistance alters gene transcript levels in Drosophila melanogaster. J Insect Physiol. 55(6):549–555. [DOI] [PubMed] [Google Scholar]
- Ullastres A, Merenciano M, González J. 2021. Regulatory regions in natural transposable element insertions drive interindividual differences in response to immune challenges in Drosophila. Genome Biol. 22(1):265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van't Hof AE, Campagne P, Rigden DJ, Yung CJ, Lingley J, Quail MA, Hall N, Darby AC, Saccheri IJ. 2016. The industrial melanism mutation in British peppered moths is a transposable element. Nature 534(7605):102–105. [DOI] [PubMed] [Google Scholar]
- Villanueva-Cañas JL, Horvath V, Aguilera L, González J. 2019. Diverse families of transposable elements affect the transcriptional regulation of stress-response genes in Drosophila melanogaster. Nucleic Acids Res. 47(13):6842–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodovar N, Vinals M, Liehl P, Basset A, Degrouard J, Spellman P, Boccard F, Lemaitre B.. 2005. Drosophila host defense after oral infection by an entomopathogenic Pseudomonas species. Proc Natl Acad Sci U S A. 102(32):11414–11419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Huang J, Zhu F. 2018. Human endogenous retroviral envelope protein syncytin-1 and inflammatory abnormalities in neuropsychological diseases. Front Psychiatry. 9:422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young AI, Benonisdottir S, Przeworski M, Kong A. 2019. Deconstructing the sources of genotype-phenotype associations in humans. Science. 365(6460):1396–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabidi MA, Arnold CD, Schernhuber K, Pagani M, Rath M, Frank O, Stark A.. 2015. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature. 518(7540):556–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data is available within the Article and Supplementary Information.
Conflict of interest statement : The authors declare no competing interest.